Abstract
Improvements in mass spectrometry (MS)-based peptide sequencing provide a new opportunity to determine whether polymorphisms, mutations, and splice variants identified in cancer cells are translated. Herein, we apply a proteogenomic data integration tool (QUILTS) to illustrate protein variant discovery using whole genome, whole transcriptome, and global proteome datasets generated from a pair of luminal and basal-like breast-cancer-patient-derived xenografts (PDX). The sensitivity of proteogenomic analysis for singe nucleotide variant (SNV) expression and novel splice junction (NSJ) detection was probed using multiple MS/MS sample process replicates defined here as an independent tandem MS experiment using identical sample material. Despite analysis of over 30 sample process replicates, only about 10% of SNVs (somatic and germline) detected by both DNA and RNA sequencing were observed as peptides. An even smaller proportion of peptides corresponding to NSJ observed by RNA sequencing were detected (<0.1%). Peptides mapping to DNA-detected SNVs without a detectable mRNA transcript were also observed, suggesting that transcriptome coverage was incomplete (∼80%). In contrast to germline variants, somatic variants were less likely to be detected at the peptide level in the basal-like tumor than in the luminal tumor, raising the possibility of differential translation or protein degradation effects. In conclusion, this large-scale proteogenomic integration allowed us to determine the degree to which mutations are translated and identify gaps in sequence coverage, thereby benchmarking current technology and progress toward whole cancer proteome and transcriptome analysis.
Massively parallel sequencing (MPS)1 of cancer genomes has demonstrated enormous complexity, and it is often unclear which somatic mutations drive tumor biology and which are nonfunctional passenger mutations that passively accumulate. RNA sequencing is frequently used to determine which nucleotide variants are transcribed and therefore have the potential for biological function. However, many mutations detected at the DNA level are not observed at the mRNA level, and their observation is dependent upon expression of the stability of the mRNA (1). Mutation detection at the peptide level clearly increases the confidence that any given variant is a potential biological driver, and by assessing peptide levels across all of an individual's polymorphisms, an independent assessment of transcriptome coverage can be obtained.
Integrated proteogenomic methods that combine MPS analysis and proteomics are of particular importance for identifying novel peptides resulting from somatic mutations or inherited polymorphisms. The identification of peptide sequences by mass spectrometry (MS) relies heavily on the quality of the protein sequence database. Use of databases with missing peptide sequences will fail to identify the corresponding peptides within the proteomic data; however, addressing this by including large numbers of irrelevant sequences in the search will decrease sensitivity. Therefore, it is essential that data acquired through MPS are used to create tumor-specific databases, incorporating the possibility of variant proteins arising through somatic mutation, inherited polymorphisms, alternatively spliced isoforms, and novel expression. The goal of this study was to analyze the flow of information though the central dogma of biology in an unbiased and comprehensive way to profoundly understand the aberrant information flux that underlies all cancer biology (2–5).
Integrated proteogenomic methodologies have been successfully implemented in several model systems, including mouse and human cell lines (2, 4–7), and in other model systems for gene annotation (3, 8–10). Sample-specific DNA databases in MS identification pipelines are a developing tool, but we lack a clear understanding of the proteomic depth and sensitivity required to obtain a truly comprehensive mutant or germline variant peptide identification. To address these issues, we used two patient-derived xenograft (PDX) breast cancer models that provided enough material for extensive genomic, transcriptomic, and proteomic analyses to determine the capabilities and limitations of MS/MS for novel protein isoforms identification in cancer cells.
EXPERIMENTAL PROCEDURES
Patient-Derived Xenograft (PDX) Tumors
Patient-derived xenograft (PDX) tumors from established Basal (WHIM2) and Luminal-B (WHIM16) breast cancer intrinsic subtypes (11) (12) were obtained from the Washington University Human and Mouse-Linked Evaluation of Tumors Core. The tumors were raised subcutaneously in 8-week-old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice (Jackson Labs, Bar Harbor, ME) as previously described (13) (14) and in compliance with NIH regulations and institutional guidelines and approved by the institutional review board at Washington University. These tumors have significantly different gene expression and proteomic signatures (14) that are related to their intrinsic biology and endocrine signaling. Tumors were harvested and processed to a frozen powder with minimal ischemia time as previously described (15, 16). Tumors from each animal were harvested by surgical excision at ∼1.5 cm3 with minimal ischemia time by immediate immersion in a liquid nitrogen bath. The tumor tissues were then placed in precooled tubes on dry ice and stored at −80 °C. A tissue “pool” of cryopulverized tumors was prepared in order to generate sufficient material that could be reliably shared and analyzed between multiple laboratories. Briefly, tumor pieces were transferred into precooled Covaris Tissue-Tube 1 Extra (TT01xt) bags (Covaris #520007) and processed in a Covaris CP02 Cryoprep device using different impact settings according to the total tumor tissue weight: <250 mg = 3; 250–350 mg = 4; 350–440 mg = 5; 440–550 mg = 6. Tissue powder was transferred to an aluminum weighing dish (VWR #1131–436) on dry ice, and the tissue was thoroughly mixed with a metal spatula precooled in liquid nitrogen. The tissue powder was then partitioned (∼ 100 mg aliquots) into precooled cryovials (Corning #430487). (Note: Cryopulverized tissue will melt if transferred to a plastic weighing boat.) All procedures were carried out on dry ice to maintain tissue in a powdered, frozen state.
Whole Genome Analysis and RNA Sequencing
Methodologies and data repositories have been published (14). Variant calling using the Whole Genome Sequencing (WGS) data were performed by an in-house developed pipeline combining several variant calling algorithms (VarScan (17), Genome Analysis Toolkit (GATK) (18), and Pindel (19)) to achieve better accuracy and sensitivity. For the RNA-Seq analysis, raw FASTQ files were trimmed by 1bp from both ends. All trimmed RNA-Seq reads were aligned to the human reference genome version hg19 using software TopHat version 2.0.3 with -g 1, -bowtie1 (version 0.12.7.0), -M, -x 1, -n 2, -no-coverage-search, and -fusion-search settings to generate Binary Sequence Alignment/Map (BAM) files, junction files, and fusion files. Then software TopHat-Fusion (20) version 2.0.3 and ChimeraScan fusion detection (17, 18) (default parameters) were used to generate final potential fusion genes. BedTools and in-house developed software were then used to generate the base pair counts for each exon based on Ensembl annotation file version GRCh37.68 using mapped reads in BAM format.
Global Proteomics
Tumor pieces were cryopulverized in precooled Covaris Tissue-Tube 1 Extra (TT01xt) bags and processed in a Covaris CP02 Cryoprep device, with impact setting derived from the weight of the tumor (<250 mg = 3; 250–350 mg = 4; 350–440 mg = 5; 440–550 mg = 6). The resulting powder was then transferred to a weighing dish, mixed using a precooled metal spatula, and partitioned into ∼100 mg aliquots. All procedures were completed on dry ice to maintain sample freezing. Proteins were reduced, alkylated, and subjected to trypsin digestion. Peptides were separated using an off-line high pH (7.5 or 10) reversed-phase column and analyzed by Thermo Fisher Q-Exactive and Orbitrap Velos instruments. The iTRAQ analysis was completed by three separate sites, and the label-free analysis was performed at two separate sites. The data are freely available through the Clinical Proteomic Tumor Analysis Consortium (CPTAC) Data Portal (https://cptacdcc.georgetown.edu/cptac/study/showDetails?accNum=S010).
Tumor-Specific Database Construction
QUILTS is a publicly available software (quilts.fenyolab.org) that can take in up to four inputs for sample-specific database creation: (i) a Browser Extensible Data (BED) file containing RNA-Seq predicted junctions; Variant Call Format (VCF) files containing (ii) somatic variants and (iii) germline variants; and (iv) a fusion file containing all predicted fusion genes. The output of QUILTS is a protein sequence FASTA file, using either Ensembl or RefSeq as a reference for the hg19 proteome and genome. Example VCF, BED (http://genome.ucsc.edu/FAQ/FAQformat.html), and fusion input files are available alongside the QUILTS webserver for mock database creation and file formatting.
Step 1. Creation of Variant Peptide Database
QUILTS parses out details of variant location and nucleotide change from the variant VCF file for subsequent incorporation into genomic sequences. In cases where both somatic and germline variants are present, tumor-specific variants are obtained by filtering out all germline variants from the somatic variant calls. Based on intron/exon boundaries from Ensembl (version 70) or RefSeq (version 20130727), sequences of annotated coding regions are extracted, and variant changes were incorporated into these regions based on genomic location. The modified sequences are then translated to proteins in a single reading frame and stored as a FASTA file (Fig. 1A). Stop codon removal and insertion due to single amino acid changes were accounted for and highlighted within file output. In this study, even low-quality variants were included in the variant database to allow for validation by mass spectrometric analysis.
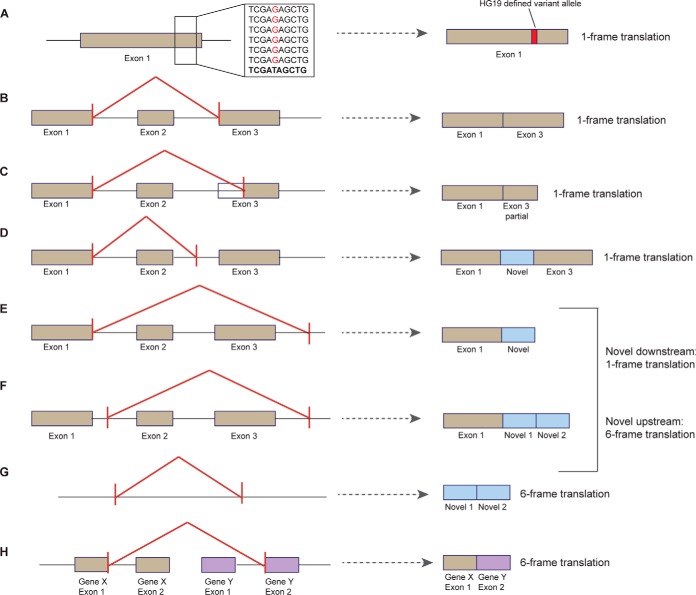
Fig. 1.
QUILTS processing of different variant scenarios. QUILTS treats each potential splicing situation differently in terms of how many frames are translated into protein. Single amino acid changes resulting from germline or somatic variants require only one reading frame (A). Junction-based changes include unannotated alternative splicing with conserved exon boundaries (B), truncation of an exon (C), and elongation of an exon within an intron (D), all with conserved reading frame. For elongation of an exon in the intergenic space (E) and regions of novel expression (F), frame translation is determined based on whether or not the novel exon boundary is up or downstream of the annotated exon. Junctions showing completely novel expression require a 6 reading frame translation (B). Fusion genes (H) are also translated in six frames. Situations with insertion or deletion of nucleotides are treated as junction changes as indicated above (B-D).
Step 2. Creation of Junction Protein Database
The RNA-Seq-derived junction BED file contains predicted boundaries of adjoining exons (chromosome, intron start, intron end) for each sample. QUILTS filters out splice junctions matching previously annotated boundaries (Ensembl or RefSeq), leaving only novel junction which are split into three categories, unannotated alterative splicing (two known exons, Fig. 1B), completely novel junctions (no known exons, Figs. 1F and 1G), and partially novel junctions (one known exon, Figs. 1C-1E). The required frame translation for in silico protein synthesis is indication in Fig. 2 for each scenario. Variant peptides are incorporated into the sequences of the alternatively spliced proteins as described in Step 1 (Fig. 1A). In this study, junctions with at least one supporting RNA read were included in database creation.
Fig. 2.
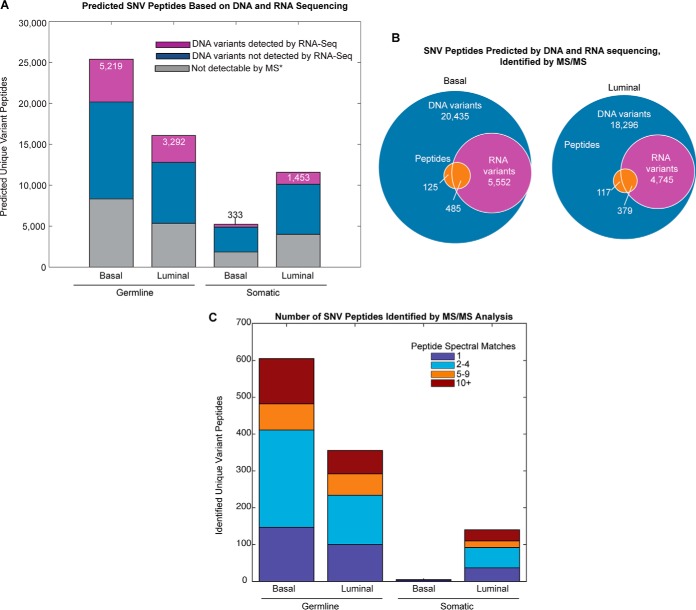
Single nucleotide variant (SNV) peptide expression in basal and luminal tumors. (A) Predicted proteomic change based on single nucleotide variants. Purple bars show the number of predicted unique peptides based on DNA variants that were also detected by RNA-Seq, blue bars indicate the number of predicted unique peptides based on DNA variants that were not detected by RNA-Seq, and gray bars indicate those peptides that are not detectable by mass spectrometry techniques due to size limitations (*peptides that have lengths <6 or >30 amino acids). (B) Proportion of DNA variants that were also identified by RNA sequencing and MS/MS proteomics for luminal and basal breast tumors. (C) Total basal and luminal variant peptides identified by MS/MS. Identification by iTRAQ, label-free MS/MS, or both is indicated by stacked bar color.
Step 3. Creation of Fusion Protein Database
QUILTS translates predicted fusion gene output by a six-frame translation (Fig. 1H). Protein coding regions with more than six consecutive amino acids are included in the fusion protein database.
Experimental Design and Statistical Rational
Proteowizard (release 2.2.3246 (2012–1-30)) was used to convert raw MS data files to mzML format for downstream data analysis. Peptide spectral matching was then done using the tumor-specific QUILTS databases with the database search engine X! Tandem CYCLONE (2013.02.01.1) using precursor tolerance of 20 ppm and fragment mass tolerances of 20 ppm for high-resolution data and 0.4 Da for low-resolution data, complete modifications of carbamidomethylated cysteines and iTRAQ 4-plex on peptide N termini and lysines, potential modifications of oxidation of methionine, and deamidation of aspartic and glutamic acid, allowing for one missed cleavage. The databases contained a total of 1,078,922 and 1,225,632 trypic peptides for basal and luminal, respectively. Both databases included a list of external contaminants from common Repository of Adventitious Proteins (cRAP) (maintained by the global proteome machine organization) in addition to Ensembl sequences for Mus musculus. The search was done against a concatenated database containing mouse and human Ensembl 37.70, cRAP, basal and luminal variant peptides, and NSJ peptides. Searches were completed independently for each center (A-E, Table I) using a simple False Discovery Rate (FDR) with separate target–decoy searches that were created through reversal of all protein sequences and searched in combination (21). The FDR was calculated as described by Käll, et al. (22) and was controlled for at a q-value of 0.01 (1%) at the peptide level. The FDR was calculated separately for variant, junction, and references peptide (23). Information detailing identified variant and junction peptides are included (Supplemental Table 4). The dataset was also searched against the same databases, allowing up to one amino acid substitution per tryptic peptide, and any spectrum that in this search matched better against a variant peptide not detected in the WGS data was removed from the subsequent analysis. Also, peptides with length shorter than eight amino acids were excluded. The results were further filtered on the percentage of the intensity in the fragment mass spectra that were explained by fragmentation of the assigned peptide sequence and only peptides with PSM where larger than 50% of the fragment ion intensity matched to the sequence (all peaks with intensity larger than 1% of the largest fragment peak) were included. Peptides were also required not to have continuous gaps of larger than two amino acids, i.e. peptides with three amino acids in a row without evidence of fragmentation between them were included, but peptides with four or more amino without fragment ions between them were excluded. In addition, if the total number of gaps throughout a peptide exceeded six, the peptide was excluded. The rationale for the filters based on gap size was the observation that, commonly when extensive fragmentation is observed for one end of a peptide but not for the other end, the identification is incorrect. In these cases, manual inspection often reveals for high-quality spectra that the search engine has assigned the incorrect sequence, and the correct sequence is missing from the database that was searched.
Table I. Mass spectrometry metrics and methodology by center.
| Resolution | iTRAQ | Label-Free |
|||
|---|---|---|---|---|---|
| High | Low | ||||
| Center | A | B | C | D | E |
| Instrument | Q-Exactive | Orbitrap Velos | Orbitrap Velos | Q-Exactive | Orbitrap Velos |
| Fractions | 25 | 25 | 24 | 24 | 15 |
| Replicates | 7 | 6 | 5 | 2 | 28 |
| Gradient (min) | 110 | 90 | 100 | 240 | 80 |
| Total peptides | 143,790 | 75,440 | 81,888 | 49,351 | 38,437 |
| SNV peptides | 642 | 267 | 274 | 79 | 72 |
Proteome analysis of basal and luminal PDX tumors was completed across five centers (A-E) using Thermo Fisher Q-Exactive or Orbitrap Velos instrumentation. The number of fractions, LC-MS/MS gradient length, MS/MS model, number of PDX sample process replicates, and the average number unique total, single nucleotide variant (SNV) and novel junction (NSJ) peptides are given for each center. All five centers used bRPLC for LC-MS/MS separation.
To assess the quality of the variant peptide identification, the distributions of the fraction of MS/MS intensity explained by the identified peptide, the peptide mass, the e-values, and total fragment ion intensity were compared with the distributions for all identified peptides in the reference protein sequence database (Supplemental Figs. 1A-1D, 1G-1J, and 1M-1P). Also, the distribution of the variant (Supplemental Figs. 1E, 1K, and 1Q) and reference (Supplemental Fig. 1F, 1L, and 1R) peptides in the four-dimensional space spanning MS/MS intensity explained by the identified peptide, peptide mass, e-value, and total MS/MS intensity were compared. Because no significant difference was observed, we conclude that the FDR for the variant peptides can be accurately estimated using the overall peptide FDR.
Also, for the NSJ peptides the quality was assessed by comparing the distributions of the fraction of MS/MS intensity explained by the identified peptide, the peptide mass, the e-values, and total fragment ion intensity to the distributions for all identified peptides in the reference protein sequence database (Supplemental Figs. 2A-2D, 2F-2I, 2K-2N). Also, the distribution of the NSJ (Supplemental Figs. 2E, 2J, and 2O) and reference (Supplemental Figs. 1F, 1L, and 1R) peptides in the four-dimensional space spanning MS/MS intensity explained by the identified peptide, peptide mass, e-value, and total MS/MS intensity were compared. Again no significant difference was observed between the NSJ peptides and the reference peptides, indicating that the overall quality of NSJ peptides is close to that of the reference peptides.
In addition, no significant difference was observed for germline and somatic variant peptides (Supplemental Figs. 3A-3R) or for variant peptides with or without mRNA evidence (Supplemental Figs. 4A-4R).
All the individual peptide spectrum matches are shown in Supplemental Fig. 5. The spectra are annotated with the assigned b- and y-ions and neutral losses. The evidence for fragmentation between pairs of amino acids is also shown by annotating the peptide sequence with bars with lengths proportional to the corresponding fragment ion intensity.
For variant and novel peptide identification, a total of 18 iTRAQ and 30 label-free sample process replicates (independent tandem MS experiments using identical sample material) were used to identify sample-specific peptides. Peptides with at least one peptide spectral match were considered as a positive identification. This is a reasonable assumption because of the stringent filter that includes the requirement of no gaps in the peptide sequence coverage larger than two consecutive amino acid pairs and a maximum of six gaps over the entire peptide. Following identification, peptides matching the reference human, mouse or cRAP proteomes were removed, leaving those peptides that map only to predicted variant or novel junction peptides. All variant and NSJ peptides were compared with the RefSeq Human + Mouse protein sequence database (downloaded December 1, 2011) to filter out any known peptides that were missed by the original filtering step. Additionally, all novel peptides were compared against the GenBank nonredundant translation database and the neXtProt protein database (24), and all matches were included in the associated tables (Supplemental Tables 1 and 2). Peptides with nonredundant matches were included in all analysis except in the creation of highly confident variant and novel junction peptide tables, for which they were removed. For each of the 48 process replicates, the number of unique variant or novel peptides identified was used for titration analysis. Robust regression was used to determine the correlation between total peptides identified and novel/variant peptides in each run. Variants were searched against the dbsnp database (build 142) and the Catalogue of Somatic Mutations in Cancer (COSMIC) to determine if the genomic variant had been previously identified (Supplemental Table 1C).
As per minimum guidelines for proteogenomic studies, suggested by Nesvizhskii (23), the custom protein sequence databases described here are available at http://openslice.fenyolab.org/data/compref/compref.zipand annotated spectrum for all novel splice junction, and single nucleotide variant peptides are provided (Supplemental Fig. 5).
RESULTS
PDX tumor lines were generated from primary breast tumors as previously described (25). Two PDX lines (one derived from a luminal tumor, WHIM16, and the other from a basal-like tumor, WHIM2) were included in this study, of which the whole genome sequences (WGS) and RNA-Seq analysis have already been published (13, 14). In the WGS analysis, 40x-50x coverage of 75 bp paired end reads (1.5 × 109-2 × 109) was obtained for germline, tumor, and xenograft. RNA-Seq data of 4 × 108, 75 bp paired end reads were obtained for each xenograft. Global proteome expression was acquired by 18 independent iTRAQ-labeled MS/MS sample process replicates (each analysis contained the two luminal and two basal samples, both of which were represented by two different reporter ions in the iTRAQ 4-plex) and 15 independent label-free MS/MS sample replicate pairs, containing each PDX sample. These experiments were completed across five centers, three using iTRAQ chemical labeling and two using label-free methods (Table I, Supplemental Fig. 6). The number of total peptides, variant peptides, and novel junction peptides identified by each center varied based on MS instrumentation, fraction number, and gradient length (Table I). A total of 184,182 unique human peptides and 9,597 proteins were identified across all datasets, representing the state-of-the-art proteomic coverage available with current techniques. Paired tumor-specific protein databases and corresponding deep proteomics tandem MS data were used to first identify novel protein isoforms involved in breast cancer progression and second to determine the depth of proteomic analysis required to capture comprehensive variant peptide identification.
Variant identification through whole genome sequencing permits the representation of nonsynonymous substitutions in our protein sequence database, increasing the number of tumor-specific peptides that can be identified by tandem MS (2, 5, 9). Most MS identification methods rely on reference databases, such as Ensembl, UniProt, or RefSeq, for peptide matching. Unfortunately, these databases do not allow novel peptide identification due to variations in the somatic or germline genome. In order to identify tumor-specific variant peptides, we used the proteogenomic integration tool Quantitative Integrated Library of Translated SNPs/Splicing (QUILTS), developed specifically for cancer proteome analysis (http://quilts.fenyolab.org). QUILTS uses protein coding variant calls (germline and somatic) and RNA-Seq-based junction predictions to build a customized, tumor-specific database containing peptide sequences that contain single nucleotide variants and bridging sequences from alternative splicing against the background of a reference human proteome database.
Variant Peptide Identification
Following its construction, each sample-specific database was used to identify tumor and sample-specific peptides attributable to both germline and somatic genomic variants. A total of 31,792 basal (26,421 germline and 5,371 somatic) and 28,635 luminal (16,662 germline and 11,973 somatic) variant peptides were predicted by WGS. Of these, we expect only a portion to be both verifiable by MS and also expressed at the mRNA level. The portion of variants identifiable by MS can be predicted based on peptide characteristics known to provide efficient MS identification, including low hydrophobicity and peptide length within 6–30 amino acids. Further, peptides that map to more than one proteomic location are difficult to attribute to a specific gene (26). Therefore, we considered only proteotypic tumor-specific changes with appropriate amino acid lengths as verifiable variant peptides. We found that ∼3.5% of theoretical variant peptides mapped to more than one gene for basal and luminal tumors, which were then removed from subsequent analysis. An additional 32% of all possible variant peptides were found to be outside of the peptide length limitations for both tumor types. The proportion of variants that are expressed at the mRNA level, however, cannot be easily predicted and relies on variant calling from whole transcriptome sequencing. Of the approximate 65% of peptides meeting the requirements for MS/MS, a minority had evidence for expression at the mRNA level (30.5% germline basal; 30.7% germline luminal; 10% somatic basal; 19% somatic luminal) (Figs. 2A and 2B).
A total of 772 unique variant peptides were identified across all 48 sample process replicates, 119 of which were found by both labeled and label-free analysis, representing a total of 667 genomic SNVs (Supplemental Table 1). Peptide spectral matching of iTRAQ and label-free global proteomics data using tumor-specific databases identified a total of 610 basal and 496 luminal variant peptides (including both somatic and germline variants). Each variant peptide contains at least one amino acid change resulting from a genomic SNV, with eight of the 772 containing two amino acid substitutions, two peptides with three substitutions, and one peptide with five substitutions (Supplemental Table 1). An overwhelming majority of variants identified were represented in dbsnp (96.8%) and neXtProt (76.8%) databases, leaving only 1.8% of identified protein variants being described by neither database. Interestingly, ∼20% of the variants identified by proteomics analysis lacked mRNA evidence based on RNA-Seq variant calling (Fig. 2B). This can be explained, at least partially, by the limitations associated with variant calling from RNA-Seq data due to the inherent complexity of the transcriptome (27).
Of the 610 variant peptides in the basal tumor, 605 were due to germline variants and only five were due to somatic variants (Fig. 2C). Two of the five basal somatic variants were identified by RNA-seq, one in the transcription factor GTF3C3 (ALGYMEGAAESYGK) and one in the nucleoprotein AHNAK2 (SFGVLAPGK) (Supplemental Table 1A). In the luminal tumor, 140 somatic and 356 germline variant peptides were identified, and more than 70% of the luminal somatic variants were found at the mRNA level. The difference in somatic variant expression between the basal and the luminal is considerable, implicating differential translation or increased protein degradation effects in the basal tumor. While ∼1/3 of the variant peptides were only identified by only one spectral match across all MS/MS runs, 20% had ten or more associated peptide spectral matches (PSMs) indicating consistently observable expression levels (Fig. 2C, Supplemental Table 1). Additionally, comparison of the proteomic variants with predicted SNVs in 105 of the breast The Cancer Genome Atlas (TCGA) tumors chosen for analysis by CPTAC found that 89% of the same genomic variants were present at the DNA level in at least one of the human breast tumors (Supplemental Table 1).
We identified a subset of highly confident somatic variant peptides that had more than ten peptide spectral matches identified in at least one process replicate, associated RNA expression, and evidence for expression in the associated protein (100+ PSM along the protein) (Table II). Fifteen variants were identified as tumor specific (somatic only) with high confidence, including variants in Keratin19 (ENSP00000408759: A60G), the mannose-6 phosphate receptor-binding protein Perilipin-3 (ENSP00000465596: V275A), and the interferon regulatory factor binding protein IRF2BP2 (ENSP00000355569: V275A) (Table II).
Table II. Highly confident SNV peptides.
Variant peptides identified by MS which were found to be somatic, with 10 or more peptide spectral matches (PSMs), variant expression at the RNA level, and evidence for protein expression in the associated gene (100+ PSMs across the gene). Includes associated gene name, chromosome, genomic start and end, genomic variant location, Ensembl protein identifier, original and variant amino acids, and the method it was identified in (iTRAQ, Label Free Low Resolution (LF-LR) or Label Free High Resolution (LF-HR)). All highly confident variants were found to be somatic in the luminal tumor only.
In order to determine the number of sample process replicates that are required to identify the maximum number of translated variants, we completed a titration analysis using 18 global iTRAQ and 28 label-free low-resolution MS/MS experiments. We demonstrated that additional MS analysis would likely result in further variant peptide identification (Figs. 3A and 3B). The number of total peptides identified in these replicates varied between 19,720 to 125,912 for iTRAQ labeled and 10,506 to 33,091 for label-free MS/MS, and the identification of variant peptides correlated to the total number of total peptides identified in each replicate (iTRAQ: r2 = 0.665; label free: r2 = 0.745). Despite this correlation, even process replicates with the highest peptide identifications identified less than 40% of the total unique variant peptides observed. (Supplemental Figs. 7A and 7B).
Fig. 3.
Titration analysis of SNV and novel junction peptide discovery. Titration to determine the number of tandem MS process replicates needed to identify all variant peptides for iTRAQ (A) and label free (B) and novel junction peptides for iTRAQ (C) and label free (D) MS/MS. iTRAQ center A corresponds to process replicates 2, 3, 8, 10, 11, 13, and 14; center B replicates 4, 6, 9, 15, 16 and 18; and center C replicates 1, 5, 7, 12, and 17. Only label-free analysis from center E was used in this analysis (B, D). Basal and luminal tumor peptide identifications were pooled for titration analysis. Total list of SNV and novel junction peptides are included in Supplemental Tables 1–2.
Novel Junction Peptide Identification
In addition to amino acid changes resulting from SNVs, alternative splicing and novel expression have been shown to effect tumor growth and disease progression (28). Intron/exon boundaries determined by RNA-Seq analysis were used to identify unannotated alternative splicing events (matching to two known exons Fig. 1B), partially novel splicing (matching to only one known exon Figs. 1C-1E), and completely novel splicing (matching to no known exons Figs. 1F and 1G) occurring in basal and luminal PDX tumors. The number of MS-verifiable peptides, being both the appropriate length and gene specific, was identified to determine the full proteomic potential due to novel splice junction (NSJ) peptides. Two percent of unannotated alternative splicing peptides, 1.3% of partially novel splicing peptides, and less than 0.01% of completely novel peptides were removed from the analysis due to their mapping to other known genes. An additional ∼24% of novel junction peptides were found to be outside of the MS peptide length limitations (Fig. 4A). Based on this analysis, we predicted a total of 22,187 and 20,442 unannotated alternative splicing peptides, 217,715 and 180,100 partially novel junction peptides, and 238,891 and 164,273 completely novel peptides to be verifiable by MS for luminal and basal, respectively (Fig. 4A). Using these custom databases for spectral matching, we were able to identify less than 0.05% of the novel junction peptides for both basal and luminal tumors (Fig. 4B).
Fig. 4.
Novel peptide expression in basal and luminal tumors. (A) Predicted proteomic change based on novel junction peptides. Gray bars indicates variant tryptic peptides with peptides with lengths <6 or >30 and orange/purple/green bars show the number of unique peptides which are verifiable by MS (B) Proportion of predicted junctions based on RNA sequencing (all junctions, purple; junctions 5+ reads, blue) that were identified by MS/MS proteomics for luminal and basal breast tumors (C) Total basal and luminal junction peptides identified. Identification by iTRAQ, label-free MS/MS, or both is indicated by stacked bar color.
We included all RNA-Seq junction predictions as input for protein database creation, without taking into account the number of reads supporting each junction. We used this liberal inclusion to create the most complete database with the available MPS data, containing all possible proteomic changes, with the idea that the proteomic data can be used to filter through transcriptional false positives. It is likely, however, that junctions confidently identified by proteomics analysis will have more supporting reads. We therefore considered the number of junction reads supporting these peptides postprocessing and found that novel junctions with MS identifications had significantly more RNA sequencing reads (median reads basal = 11, luminal = 6) compared with all junctions in the protein database (median reads basal = 2, p = 1.8e-4; luminal = 2, p = 2.898e-5). Upstream filtering of junction data containing five or more supporting reads drastically reduced our predicted novel junction peptide number to 76,053 basal and 91,609 luminal peptides, and ∼50% of novel junction peptides identified by MS/MS met this criteria (80 of the 129 basal and 68 of the 147 luminal peptides) (Fig. 4B). The ratio of peptides identified to peptides predicted was similar between each of the three junction classes (Supplemental Fig. 8).
A total of 86 unique novel splice junction (NSJ) peptides were identified in by least one sample process replicate. Of the 66 NSJ peptides identified in basal tumors, eight supported unannotated alternative splicing, 32 indicated splicing of a known exon with a novel coding region, and 26 demonstrated the splicing of two novel exons. Similarly, seven peptides supporting splicing of two known exons, 34 with one known exon, and 26 completely novel exon splicing events were identified in luminal tumors. Of these, ∼50% of the NSJ peptides were identified in both basal and luminal tumors (Supplemental Table 2). Approximately half of novel junction peptides were supported by only one peptide spectral match by only one process replicate (Fig. 4C).
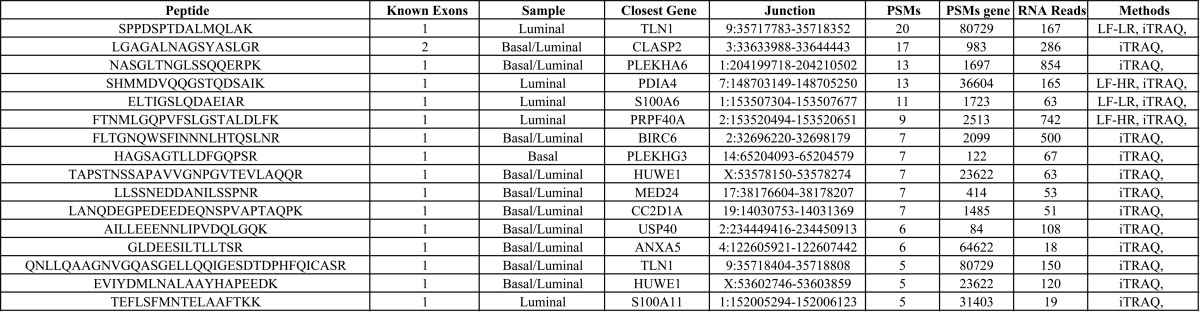
We identified 16 novel junction peptides we considered to be highly confident, having at least five peptide spectral matches, more than five supporting RNA read, and evidence for protein expression in the associated gene (>50 PSMs) (Table III). In a few cases, NSJ peptide appeared to be present in both the basal and the luminal tumor, i.e. iTRAQ reporter ions were observed for both samples and RNA-Seq evidence existed for both samples. In these cases, the sample is listed as both basal and luminal, but sometimes the identification from label-free analysis was able to determine the sample of origin. The peptide with the most evidence (highest PSM count) was found for a novel coding region near BHMT2 in luminal tumors (Supplemental Table 2).
Table III. Highly confident novel junction peptides.
Novel junction peptides identified by MS which were found to have more than 5 PSMs across iTRAQ and label free analysis, 5 or more RNA reads, and evidence for protein expression in the associated gene (50+ PSMs across the gene). Includes the number of known exons involved in splicing, peptide spectral matches, sample in which the peptide was identified, gene or the closest gene on the same strand, junction location, and the method it was identified in (iTRAQ, Label Free Low Resolution (LF-LR), or Label Free High Resolution (LF-HR)).
We completed similar titration analysis shown for the variant peptides to find the number of unique novel peptides cumulatively identified with each process replicate. As seen with the variant data, spectra from all replicates were required to attain the complete list of junction peptides in both iTRAQ and label-free analysis (Figs. 3C and 3D). The novel peptide identification was similarly correlated with total peptides identified than that seen with the variant peptide identification, with an R-squared value of 0.561 for iTRAQ and 0.385 for label-free tandem MS. Our results establish that the extent of novel and variant peptide identification is highly dependent on the depth of proteomic analysis, and advancements in MS sensitivity will result in more comprehensive variant peptide identification. To better understand this continued rise in SNV and NSJ peptides with increasing process replicate runs, we plotted a titration for reference peptide identification for iTRAQ and label free MS/MS (Supplemental Fig. 9). This analysis found a similar increase in reference peptide identification with each successive iTRAQ process replicate, though the label-free curve leveled out after ∼12 replicate MS/MS runs. This indicates that increasing the number of process replicates run, particularly using different platforms and methodologies, can identify a wider range of novel peptides.
Although splicing and variants were considered simultaneously in our tumor-specific databases, no peptides containing both a novel junction and SNV were identified from our analysis. We also used QUILTS to incorporate novel peptides from a six-frame translation of 38 gene fusion calls, 13 in basal and 25 in luminal. A total of 954 luminal fusion peptides and 483 basal fusion peptides were included in the protein sequence search database, but no positive identifications were found using our proteomics analysis. We expect this is, in part, a probabilistic issue, with relatively few potential fusion proteins represented in the protein sequence database. Although no fusion proteins were identified, of the 40 fusion gene junctions found within a known gene boundary, 26 had some evidence of protein expression (PSM>0 along the protein) (Supplemental Table 3).
DISCUSSION
The field of proteogenomics has seen considerable growth in the last five years, requiring a focus on the integration of genomics and proteomics through peptide mapping and the use of sequencing data to attain a comprehensive view of the proteome. Tools that utilize MPS data to better annotate and identify proteins are essential for broad protein identification. Ideally, integration of sequencing and proteomic analysis will vastly improve our understanding of complex biological systems, in that more comprehensive and diverse data from the same system can provide a clearer view of its biological landscape. Additionally, integrating multiple datasets can supplement the limitations associated with each method. For example, proteomics can be used to better annotate the protein coding regions of the genome (3, 8–10); DNA sequencing can predict protein coding variants in MS/MS (4); and RNA-Seq transcriptomics can supplement MS proteomic coverage (5, 6, 29).
This study demonstrates the current advantages and limitations of proteogenomic integration for interpreting cancer biology, specifically in the detection of novel protein isoforms. We determined the capacity of current MS proteomic methods for variant and novel peptide identification in a sample that has undergone both extensive proteomic (48 global MS/MS sample process replicates) and DNA/RNA MPS analysis. Though the ability to detect more than 700 peptides that confirm the expression of single nucleotide variations and 86 novel splicing events is notable, no one MS replicate was able to recover more than 40% of the full list of observable peptides, regardless of the instrumentation or methodology used. Further, titration curves for both variant and junction peptide analysis suggest that increased experimental process replicates would continue to increase the number of captured novel peptides (Fig. 3).
Although the sensitivity of MS-based proteomics has improved dramatically, the limited dynamic range of the analysis is obviously still an issue. For example, variant peptides of established breast cancer genes such as PIK3CA or TP53 were not detected in this study. Since peptide size and physiochemical properties have large effects on ionization and fragmentation (30), i.e. because some peptides are well suited for MS analysis while others are not, some issues with low protein coverage are currently unavoidable using tandem MS approaches. In addition, the overall dynamic range of the analysis is several orders of magnitude lower than the range of protein concentrations, even when extensive separation of peptides is performed. This is due to a low MS dynamic range, being only four orders of magnitude for a given measurement, and a known bottleneck in instrument speed (31). Although greater fractionation may partially resolve the issue, this approach is limited by sample quantity. Advancements in MS protein identification sensitivity must be made, either through improvements in sample preparation, instrumentation sensitivity and speed, or identification algorithms, if we are to ultimately achieve complete proteomic landscape.
The low recovery between predicted novel junction peptides and those identified in the proteomics data is particularly striking (Fig. 4B), and similar levels of discovery have been reported in other mammalian systems (5). This supports the notion that there is a substantial level of noise in the transcriptome, i.e. many transcripts never result in stable proteins. Thus, proteomics becomes an essential technology in validating novel protein isoforms and splice sites. This result also points to the high quality of the human genomic annotation, with minimal novel exon discoveries despite the great depth in sequencing and proteomics. Further, the overlap in novel junction peptides identified in the luminal and basal tumors (Supplemental Table 2) indicates that these peptides may correlate to either “normal” exon structure or, instead, may be attributed to a more cancer-specific splicing activity.
This study also demonstrates the current advantages and limitations in DNA/RNA MPS analysis. A portion of the peptides spanning novel junctions were only supported by a one or a few RNA-Seq but had strong MS peptide evidence. Similarly, many variants had only low quality score from WGS or did not have supporting RNA-Seq evidence but had strong MS evidence. This makes proteomics the method of choice for determining which variants will be incorporated into stable proteins or cause protein misfolding and degradation and in the search for drivers of cancer.
The paucity of translated somatic variants in the basal-like sample in comparison to the luminal tumor should be further investigated in larger datasets. Translational controls and degradation mechanisms may in play to restrict the repertoire of mutant genes that are translated, and these maybe another source of inter-tumor heterogeneity.
CONCLUSIONS
Proteomics is essential in validating genomic changes, including SNVs and novel splicing to assess the degree to which these genomic alterations are translated and therefore biologically active. Although peptide coverage is restricted by current technologies, with only 10% coverage of variant peptide sequence even with multiple fractions and process replicates, our ability to validate genomic variation in cancer using proteomics is still substantial. This low variant peptide coverage may occur for several reasons. Tryptic digestion is clearly one limitation, and the use of additional enzymes and fractionation approaches are under investigation. The low rates of peptide detection may not just be a sensitivity issue but reflect biological effects. For example, the very low detection rates for novel splicing events may reflect the fact that many are not efficiently translated or quickly degraded. Similarly, lack of peptide expression for some SNV may be due to somatic-mutation-driven translational effects or protein instability/degradation. The application of proteogenomic integration methods to larger datasets and improvements in peptide identification and MS/MS sensitivity will help clarify these issues in the future.
Supplementary Material
Acknowledgments
National Cancer Institute Clinical Proteomics Tumor Analysis Consortium (NCI CPTAC) investigators: Broad Institute of MIT and Harvard: Steven A. Carr, Michael A. Gillette, Karl R. Klauser, Eric Kuhn, D.R. Mani, Philipp Mertins; Enterprise Science and Computing, Inc.: Karen A. Ketchum; Fred Hutchinson Cancer Research Center: Amanda G. Paulovich, Jeffrey R. Whiteaker; Georgetown University: Nathan J. Edwards, Subha Madhavan, Peter B. McGarvey; Icahn School of Medicine at Mount Sinai: Pei Wang; Johns Hopkins University: Daniel Chan, Akhilesh Pandey, Ie-Ming Shih, Hui Zhang, Zhen Zhang, Heng Zhu; Leidos, Inc.: Gordon A. Whiteley; Massachusetts General Hospital and Harvard University: Steven J. Skates; Massachusetts Institute of Technology: Forest M. White; Memorial Sloan Kettering Cancer Center: Douglas A. Levine; National Cancer Institute: Emily S. Boja, Christopher R. Kinsinger, Tara Hiltke, Mehdi Mesri, Robert C. Rivers, Henry Rodriguez, Kenna M. Shaw; National Institute of Standards and Technology: Stephen E. Stein; New York University: David Fenyo; Pacific Northwest National Laboratory: Tao Liu, Jason E. McDermott, Samuel H. Payne, Karin D. Rodland, Richard D. Smith; Spectragen-Informatics: Paul Rudnick; Stanford University: Michael Snyder; University of Chicago: Yingming Zhao; University of North Carolina at Chapel Hill: Xian Chen, David F. Ransohoff; University of Washington: Andrew N. Hoofnagle; Vanderbilt University: Daniel C. Liebler, Melinda E. Sanders, Zhiao Shi, Robbert J.C. Slebos, David L. Tabb, Bing Zhang, Lisa J. Zimmerman; Virginia Tech: Yue Wang; Washington University in St. Louis: Sherri R. Davies, Li Ding, Matthew J. Ellis, R. Reid Townsend.
Footnotes
Author contributions: K.V.R., S.R.D., C.R.K., K.D.R., L.D., H.R., D.C., R.D.S., D.C.L., S.A.C., S.P., M.J.E., and D.F. designed the research; K.V.R., P.M., R.S., P.E., H.P.G., L.X., T.L., J.Z., S.S., C.M.P., X.C., S.R.D., H.Z., Z.Z., R.R.T., and D.F. performed the research; M.A. and K.R.C. contributed new reagents or analytic tools; K.V.R., Z.T., X.W., H.G., J.T., S.C., M.D.M., K.R.C., D.L.T., P.M., S.L., K.A.H., C.M.P., C.A.M., and D.F. analyzed data; and K.V.R., S.P., M.J.E., and D.F. wrote the paper.
* This work was supported by National Cancer Institute (NCI) CPTAC awards U24CA159988, U24CA160019, U24CA160034, U24CA160035, U24CA160036 and by CPTAC contract 13XS068 from Leidos Biomedical Research, Inc. This work has utilized computing resources at the High Performance Computing Facility of the Center for Health Informatics and Bioinformatics at the NYU Langone Medical Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental material Supplemental Tables 1–4 and Supplemental Figs. 1–9.
This article contains supplemental material Supplemental Tables 1–4 and Supplemental Figs. 1–9.
1 The abbreviations used are:
- iTRAQ
- Isobaric Tags for Relative and Absolute Quantitation
- LF
- label free
- MPS
- massively parallel sequencing
- NGS
- next generation sequencing
- NSJ
- novel splice junction
- PDX
- patient-derived xenograft
- PSM
- peptide spectral match
- QUILTS
- Quantitative Integrated Library of Translated SNPs/Splicing
- SNV
- single nucleotide variant.
REFERENCES
- 1. Cirulli E. T., Singh A., Shianna K. V., Ge D., Smith J. P., Maia J. M., Heinzen E. L., Goedert J. J., and Goldstein D. B. (2010) Screening the human exome: A comparison of whole genome and whole transcriptome sequencing. Genome Biol. 11, R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang X., Slebos R. J., Wang D., Halvey P. J., Tabb D. L., Liebler D. C., and Zhang B. (2012) Protein identification using customized protein sequence databases derived from RNA-Seq data. J. Proteome Res. 11, 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Castellana N. E., Shen Z., He Y., Walley J. W., Cassidy C. J., Briggs S. P., and Bafna V. (2013) An automated proteogenomic method utilizes mass spectrometry to reveal novel genes in Zea mays. Mol. Cell. Proteomics 13(1), 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li J., Su Z., Ma Z. Q., Slebos R. J., Halvey P., Tabb D. L., Liebler D. C., Pao W., and Zhang B. (2011) A bioinformatics workflow for variant peptide detection in shotgun proteomics. Mol. Cell. Proteomics 10, M110.006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sheynkman G. M., Shortreed M. R., Frey B. L., and Smith L. M. (2013) Discovery and mass spectrometric analysis of novel splice-junction peptides using RNA-Seq. Mol. Cell. Proteomics 12, 2341–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Omenn G. S., Yocum A. K., and Menon R. (2010) Alternative splice variants, a new class of protein cancer biomarker candidates: Findings in pancreatic cancer and breast cancer with systems biology implications. Disease Markers 28, 241–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Evans V. C., Barker G., Heesom K. J., Fan J., Bessant C., and Matthews D. A. (2012) De novo derivation of proteomes from transcriptomes for transcript and protein identification. Nat. Methods 9, 1207–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adamidi C., Wang Y., Gruen D., Mastrobuoni G., You X., Tolle D., Dodt M., Mackowiak S. D., Gogol-Doering A., Oenal P., Rybak A., Ross E., Sánchez Alvarado A., Kempa S., Dieterich C., Rajewsky N., and Chen W. (2011) De novo assembly and validation of planaria transcriptome by massive parallel sequencing and shotgun proteomics. Genome Res. 21, 1193–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Woo S., Cha S. W., Merrihew G., He Y., Castellana N., Guest C., Maccoss M., and Bafna V. (2013) Proteogenomic database construction driven from large scale RNA-seq data. J. Proteome Res. 13(1), 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xing X. B., Li Q. R., Sun H., Fu X., Zhan F., Huang X., Li J., Chen C. L., Shyr Y., Zeng R., Li Y. X., and Xie L. (2011) The discovery of novel protein-coding features in mouse genome based on mass spectrometry data. Genomics 98, 343–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parker J. S., Mullins M., Cheang M. C., Leung S., Voduc D., Vickery T., Davies S., Fauron C., He X., Hu Z., Quackenbush J. F., Stijleman I. J., Palazzo J., Marron J. S., Nobel A. B., Mardis E., Nielsen T. O., Ellis M. J., Perou C. M., and Bernard P. S. (2009) Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27, 1160–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perou C. M., Sørlie T., Eisen M. B., van de Rijn M., Jeffrey S. S., Rees C. A., Pollack J. R., Ross D. T., Johnsen H., Akslen L. A., Fluge O., Pergamenschikov A., Williams C., Zhu S. X., Lonning P. E., Borresen-Dale A. L., Brown P. O., and Botstein D. (2000) Molecular portraits of human breast tumours. Nature 406, 747–752 [DOI] [PubMed] [Google Scholar]
- 13. Ding L., Ellis M. J., Li S., Larson D. E., Chen K., Wallis J. W., Harris C. C., McLellan M. D., Fulton R. S., Fulton L. L., Abbott R. M., Hoog J., Dooling D. J., Koboldt D. C., Schmidt H., Kalicki J., Zhang Q., Chen L., Lin L., Wendl M. C., McMichael J. F., Magrini V. J., Cook L., McGrath S. D., Vickery T. L., Appelbaum E., Deschryver K., Davies S., Guintoli T., Crowder R., Tao Y., Snider J. E., Smith S. M., Dukes A. F., Sanderson G. E., Pohl C. S., Delehaunty K. D., Fronick C. C., Pape K. A., Reed J. S., Robinson J. S., Hodges J. S., Schierding W., Dees N. D., Shen D., Locke D. P., Wiechert M. E., Eldred J. M., Peck J. B., Oberkfell B. J., Lolofie J. T., Du F., Hawkins A. E., O'Laughlin M. D., Bernard K. E., Cunningham M., Elliott G., Mason M. D., Thompson D. M. Jr., Ivanovich J. L., Goodfellow P. J., Perou C. M., Weinstock G. M., Aft R., Watson M., Ley T. J., Wilson R. K., and Mardis E. R. (2010) Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li S., Shen D., Shao J., Crowder R., Liu W., Prat A., He X., Liu S., Hoog J., Lu C., Ding L., Griffith O. L., Miller C., Larson D., Fulton R. S., Harrison M., Mooney T., McMichael J. F., Luo J., Tao Y., Goncalves R., Schlosberg C., Hiken J. F., Saied L., Sanchez C., Giuntoli T., Bumb C., Cooper C., Kitchens R. T., Lin A., Phommaly C., Davies S. R., Zhang J., Kavuri M. S., McEachern D., Dong Y. Y., Ma C., Pluard T., Naughton M., Bose R., Suresh R., McDowell R., Michel L., Aft R., Gillanders W., DeSchryver K., Wilson R. K., Wang S., Mills G. B., Gonzalez-Angulo A., Edwards J. R., Maher C., Perou C. M., Mardis E. R., and Ellis M. J. (2013) Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell reports 4, 1116–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mertins P., Yang F., Liu T., Mani D. R., Petyuk V. A., Gillette M. A., Clauser K. R., Qiao J. W., Gritsenko M. A., Moore R. J., Levine D. A., Townsend R., Erdmann-Gilmore P., Snider J. E., Davies S. R., Ruggles K. V., Fenyo D., Kitchens R. T., Li S., Olvera N., Dao F., Rodriguez H., Chan D. W., Liebler D., White F., Rodland K. D., Mills G. B., Smith R. D., Paulovich A. G., Ellis M., and Carr S. A. (2014) Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol. Cell. Proteomics 13, 1690–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang B., Wang J., Wang X., Zhu J., Liu Q., Shi Z., Chambers M. C., Zimmerman L. J., Shaddox K. F., Kim S., Davies S. R., Wang S., Wang P., Kinsinger C. R., Rivers R. C., Rodriguez H., Townsend R. R., Ellis M. J., Carr S. A., Tabb D. L., Coffey R. J., Slebos R. J., and Liebler D. C. (2014) Proteogenomic characterization of human colon and rectal cancer. Nature 513(7518), 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koboldt D. C., Chen K., Wylie T., Larson D. E., McLellan M. D., Mardis E. R., Weinstock G. M., Wilson R. K., and Ding L. (2009) VarScan: Variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., and DePristo M. A. (2010) The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ye K., Schulz M. H., Long Q., Apweiler R., and Ning Z. (2009) Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25, 2865–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim D., and Salzberg S. L. (2011) TopHat-Fusion: An algorithm for discovery of novel fusion transcripts. Genome Biol. 12, R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Perkins D. N., Pappin D. J., Creasy D. M., and Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 22. Käll L., Storey J. D., MacCoss M. J., and Noble W. S. (2008) Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J. Proteome Res. 7, 29–34 [DOI] [PubMed] [Google Scholar]
- 23. Nesvizhskii A. I. (2014) Proteogenomics: Concepts, applications and computational strategies. Nat. Methods 11, 1114–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gaudet P., Michel P. A., Zahn-Zabal M., Cusin I., Duek P. D., Evalet O., Gateau A., Gleizes A., Pereira M., Teixeira D., Zhang Y., Lane L., and Bairoch A. (2015) The neXtProt knowledgebase on human proteins: Current status. Nucleic Acids Res. 43, D764–D770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kuperwasser C., Chavarria T., Wu M., Magrane G., Gray J. W., Carey L., Richardson A., and Weinberg R. A. (2004) Reconstruction of functionally normal and malignant human breast tissues in mice. Proc. Natl. Acad. Sci. U. S. A. 101, 4966–4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Craig R., Cortens J. P., and Beavis R. C. (2005) The use of proteotypic peptide libraries for protein identification. Rapid Commun. Mass Spectrom. 19, 1844–1850 [DOI] [PubMed] [Google Scholar]
- 27. Piskol R., Ramaswami G., and Li J. B. (2013) Reliable identification of genomic variants from RNA-seq data. Am. J. Human Genetics 93, 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Balmain A., Gray J., and Ponder B. (2003) The genetics and genomics of cancer. Nat. Genetics 33, 238–244 [DOI] [PubMed] [Google Scholar]
- 29. Ning K., and Nesvizhskii A. I. (2010) The utility of mass spectrometry-based proteomic data for validation of novel alternative splice forms reconstructed from RNA-Seq data: A preliminary assessment. BMC Bioinformatics 11, S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang Y., Triscari J. M., Tseng G. C., Pasa-Tolic L., Lipton M. S., Smith R. D., and Wysocki V. H. (2005) Statistical characterization of the charge state and residue dependence of low-energy CID peptide dissociation patterns. Anal. Chem. 77, 5800–5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eriksson J., and Fenyö D. (2007) Improving the success rate of proteome analysis by modeling protein-abundance distributions and experimental designs. Nature Biotechnol. 25, 651–655 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.