Highlights
-
•
G. lamblia has two nitroreductases with substrate specificities not only for nitro compounds, but also for quinones.
-
•
GlNR1 rather activates nitro drugs by forming toxic intermediates, GlNR2 rather inactivates them.
Keywords: Mode of action of nitro compounds, Functional assays, Nitroreduction
Graphical Abstract
Abstract
Giardia lamblia is a protozoan parasite that causes giardiasis, a diarrhoeal disease affecting humans and various animal species. Nitro drugs such as the nitroimidazole metronidazole and the nitrothiazolide nitazoxanide are used for treatment of giardiasis. Nitroreductases such as GlNR1 and GlNR2 may play a role in activation or inactivation of these drugs. The aim of this work is to characterise these two enyzmes using functional assays. For respective analyses recombinant analogues from GlNR1 and GlNR2 were produced in Escherichia coli. E. coli expressing GlNR1 and GlNR2 alone or together were grown in the presence of nitro compounds. Furthermore, pull-down assays were performed using HA-tagged GlNR1 and GlNR2 as baits. As expected, E. coli expressing GlNR1 were more susceptible to metronidazole under aerobic and semi-aerobic and to nitazoxanide under semi-aerobic growth conditions whereas E. coli expressing GlNR2 were susceptible to neither drug. Interestingly, expression of both nitroreductases gave the same results as expression of GlNR2 alone. In functional assays, both nitroreductases had their strongest activities on the quinone menadione (vitamin K3) and FAD, but reduction of nitro compounds including the nitro drugs metronidazole and nitazoxanide was clearly detected. Full reduction of 7-nitrocoumarin to 7-aminocoumarin was preferentially achieved with GlNR2. Pull-down assays revealed that GlNR1 and GlNR2 interacted in vivo forming a multienzyme complex. These findings suggest that both nitroreductases are multifunctional. Their main biological role may reside in the reduction of vitamin K analogues and FAD. Activation by GlNR1 or inactivation by GlNR2 of nitro drugs may be the consequence of a secondary enzymatic activity either yielding (GlNR1) or eliminating (GlNR2) toxic intermediates after reduction of these compounds.
1. Introduction
Giardia lamblia (syn. Giardia duodenalis; Giardia intestinalis), a flagellated protozoan, is the most common causative agent of persistent diarrhoea worldwide (Thompson, 2000, Robertson et al, 2010). Currently, anti-giardial chemotherapy is performed using a couple of effective drugs, namely, the nitroheterocyclic drugs tinidazole, metronidazole, furazolidone, quinacrine, the aminoglycoside paromomycin, and the benzimidazole albendazole (Wright et al, 2003, Lalle, 2010). Furthermore, the nitrothiazolide nitazoxanide has been introduced as an alternative option (Hemphill et al., 2006).
As frequently hypothesised, metronidazole and other nitro drugs are reduced to a nitro-radical. According to one of these hypotheses, electrons are provided by the enzyme pyruvate:flavodoxin/ferredoxin oxidoreductase (PFOR), representing a protein that is lacking in higher eukaryotic cells (Brown et al, 1998, Horner et al, 1999). Referring to this model, the electrons are transferred via PFOR from pyruvate to ferredoxin. The resulting reduced ferredoxin is then re-oxidised by ferredoxin:NAD-oxidoreductase transferring its electrons to NAD(P). The resulting NAD(P)H serves as a redox partner for subsequent reactions such as the reduction of O2 by NAD(P)H oxidase (Brown et al., 1998). Nitro drugs may interfere in this pathway and capture electrons directly from the reduced ferredoxin or from the NAD(P)H-oxidase. This process leads to the accumulation of toxic radicals that cause irreversible damage in the parasite. Further evidence for PFOR being a major target for nitro drugs in Giardia comes from metronidazole-resistant isolates with lower PFOR expression levels (Upcroft and Upcroft, 2001).
Since a few years, however, evidence is emerging that PFOR may not represent the exclusive target of nitro drugs in semi-aerobic or anaerobic pathogens. In the case of T. vaginalis, metronidazole and other nitroimidazoles were shown to covalently bind, and thus inactivate, proteins involved in the thioredoxin reductase pathway. Resistant cells compensate this blocking by re-regulating PFORs and other enzymes participating in oxidoreductive processes. Accordingly, down-regulation of PFOR seems not to be a prerequisite but rather a consequence of resistance formation (Leitsch et al., 2009). Moreover, our investigations on Giardia cell lines resistant to nitro drugs have demonstrated that resistance is not necessarily linked to down-regulation of PFOR (Müller et al, 2007a, Müller et al, 2008). Although some nitro drugs are supposed to interact with PFOR in a direct manner, direct reduction of the nitro group via ferredoxin is rather unlikely. Accordingly, catalysis of this reaction by nitroreductases is a more realistic scenario (Roldán et al., 2008).
Nitroreductases belong to the enzymatic repertoire of many archaebacteria and eubacteria (Nixon et al., 2002), where they contribute to the assimilation of nitro compounds as carbon sources (Johnson, Spain, 2003, Luque-Almagro et al, 2006). From a mechanistic point of view, nitroreductases are divided into two classes, namely oxygen-sensitive and oxygen-insensitive nitroreductases (Roldán et al., 2008). Oxygen-sensitive nitroreductases transfer electrons one by one from NAD(P)H to the nitro group. In presence of oxygen, the intermediate radicals are re-oxydised. Thus, there is NAD(P)H consumption without nitroreduction, and the reaction looks most like the one catalysed by a NAD(P)H oxydase. Oxygen-insensitive nitroreductases catalyse the full reduction of nitro compounds into the corresponding amines by two-electron transfers. Also this type of reaction produces toxic intermediates, namely, nitroso or hydroxylamine intermediates (Moreno and Docampo, 1985).
In anaerobic or microaerophilic pathogens, nitroreductases are also well documented as resistance factors. In Helicobacter pylori, resistance to metronidazole is associated with loss-of-function mutations of the gene rdxA encoding an oxygen-insensitive nitroreductase (Goodwin et al., 1998), which reduces metronidazole under anaerobic conditions (Olekhnovich et al., 2009). Other nitroreductases are found in enteric bacteria including Escherichia coli (Lee et al, 1994, Zenno et al, 1996a, Zenno et al, 1996b, Zenno et al, 1996c, Guillén et al, 2009, Tavares et al, 2009, Yanto et al, 2010).
Nitroreductases have also been identified in microaerophilic or anaerobic eukaryotic parasites such as Entamoeba histolytica and G. lamblia. These organisms may have acquired the respective genes from prokaryotes by lateral transfer (Nixon et al., 2002). G. lamblia (clone WB C6) harbours two genes encoding nitroreductases GlNR1 (accession N° EDO80257; Gl50803-22677, referred to as Fd-NR2 in the Giardia database) and GlNR2 (accession N° XM_764091.1; Gl50803-6175, referred to as Fd-NR1 in the Giardia database). The polypeptide sequence of GlNR2 is rather similar to that one of GlNR1. Both proteins contain a ferredoxin domain with four Fe-S-clusters at their N-terminus and a nitro-FMN-reductase domain at their C-terminus. Our previous results suggest that both enzymes have a different action on nitro drugs: GlNR1 behaving as an activator (Müller et al, 2007b, Nillius et al, 2011), GlNR2 more as an inactivator (Müller et al., 2013). The biological role of these enzymes is, however, completely unclear.
Here, we present results from functional assays showing that both nitroreductases are multifunctional with strong quinone reductase activities. Moreover, we show that both nitroreductases interact in vivo forming a multienzyme complex.
2. Materials and methods
2.1. Tissue culture media, biochemicals and drugs
If not otherwise stated, all biochemical reagents were from Sigma (St Louis, MO, USA). 7-Nitrocoumarin was purchased from Santa Cruz Biotechnology (Dallas, Texas, USA). Nitazoxanide was synthesised at the Department of Chemistry and Biochemistry, University of Berne (kindly provided by Ch. Leumann). The nitroimidazole C17 was kindly provided by J. A. Upcroft (Molecular Genetics Laboratory, Queensland Institute of Medical Research, Brisbane, Australia). CB1954 was purchased from Santa Cruz Biotechnology. The compounds were kept as 100 mM stock solutions in DMSO at −20 °C.
2.2. Overexpression of recombinant GlNR1 and 2 in E. coli and His-Tag-purification
Overexpression of recombinant GlNR1 and GlNR2 in E. coli BL21 (DE3) and their purification by His-Tag-affinity-chromatography was performed as previously described (Müller et al, 2007b, Müller et al, 2013).
2.3. Overexpression of recombinant HA-tagged GlNR1 and 2 in G. lamblia
Cloning of PCR-amplified GlNR1 and GlNR2 open reading frames into the XbaI and PacI sites from vector pPacV-Integ (Jiménez-García et al., 2008) was essentially done as previously described (Müller et al, 2007b, Müller et al, 2009). Briefly, GlNR1 and GlNR2-specific forward primer contained the XbaI site followed by the constitutive glutamate dehydrogenase (GDH) promoter sequence (Davis-Hayman and Nash, 2002) (Table S1). In the reverse primer, a sequence encoding three consecutive human influenza haemagglutinin (HA) tags was introduced 5′ of the PacI site (Table S1). PCRs for amplification of GlNR1 and GlNR2 open reading frames, insertion of amplification products into XbaI and PacI sites from pPacV thus yielding pPacV-GlNR1-3xHA or pPacV-GlNR2-3xHA, and transfection of G. lamblia WBC6 with Swa1-linearised recombinant plasmids were performed as previously described (Müller et al, 2007b, Müller et al, 2009).
2.4. Co-immunoprecipitation assay with HA-tagged nitroreductases
G. lamblia WBC6 GlNR1-3xHA and GlNR2-3xHA transgenic trophozoites were grown under anaerobic condition in triple flasks (Nunc, cat. 132867). The parasites were harvested by chilling in ice water for an hour followed by centrifugation (900 × g, 10 min, 4 °C), washed in 50 ml ice cold PBS, and counted in a Neubauer chamber. For co-immunoprecipitation assays, 109 parasites were then re-suspended in a 15-ml-Falcon-tube containing 5 ml of lysis buffer (RIPA) consisting of 50 mM Tris pH 7.4, 150 mM NaCl, 1% IGEPAL, 0.5% sodium deoxycholate, 0.1% SDS, 10 mM EDTA supplemented with 2 mM phenylmethylsulfonyl fluoride, PMSF and 1X Protease Inhibitor cocktail, PIC (cat. No. 539131, Calbiochem USA), and sonicated twice (60 pulses, 2 output control, 30% duty cycle and 60 pulses, 4 output control, 40% duty cycle). To solubilise the proteins, the Falcon tube was incubated on a rotating wheel (1 h, 4 °C). Cell lysate was transferred into 1.5 ml microtubes and the supernatant containing the solubilised protein was collected after high-speed centrifugation (14,000 × g, 10 min, 4 °C). The solubilised protein fraction was diluted 1:1 with detergent-free RIPA lysis buffer supplemented with 2% Triton-X-100. To this diluted protein lysate, 40 µl anti-HA agarose bead slurry from the Pierce HA Tag IP/Co-IP Kit (Thermo Fisher Scientific, Rockford, Il.) were added and incubated at 4 °C for 2 h on a rotating wheel in order to allow the HA-tagged proteins to bind to the agarose beads. Samples were pulse-centrifuged at 3500 g at 4 °C and 100 µl was stored as flow though control. Samples were washed 4 times with 3 ml of Tris-Buffered Saline (TBS) supplemented with 0.1% Triton-X-100 and once with 3 ml PBS. The agarose slurry was re-suspended in 350 µl PBS and transferred into the spin column provided in the kit and pulse-fuged at 14,000 × g for 10 s at 4 °C. The agarose beads (boiled beads) were then re-suspended in 30 µl PBS and transferred into a 1.5 ml microtubes and stored at −20 °C overnight for further analysis.
2.5. Protein analysis and sample preparation for mass spectrometry
For SDS-PAGE according to (Laemmli (1970), the samples collected as described above were suspended in one volume of SDS-PAGE sample buffer containing 100 mM dithiothreitol, and boiled for 5 min followed by high speed centrifugation (14,000 × g, 10 min, RT). For MS analysis, 25 µl of boiled bead sample were loaded on a 12% polyacrylamide gel under reducing conditions. The gel (see Fig. S1) was then stained with Instant blue (Expedeon, San Diego, CA), de-stained with sterile water, and subsequently sent to the Functional Genomics Center Zürich for mass spectrometry. For immunoblot analysis, approximately 107 trophozoites were processed and samples were collected as described above. Immunoblots were performed with rabbit anti-GlNR1 (Nillius et al., 2011) and mouse-anti-HA (Roche Diagnostics, Rotkreuz, Switzerland) antibodies as described before (Nillius et al., 2011).
2.6. Mass spectrometry
Gel lanes (see Fig. S1) were cut in 8 equal sections. Each section was further diced into smaller pieces and washed twice with 100 µl of 100 mM ammonium bicarbonate/50% acetonitrile for 15 min at 50 °C. The sections were dehydrated with 50 µl of acetonitrile. The supernatants of the washing and de-hydration steps were discarded. The gel pieces were re-hydrated with 20 µl trypsin solution (5 ng/ µl in 10 mM Tris/2 mM CaCl2 at pH 8.2) and 40 µl buffer (10 mM Tris/2 mM CaCl2 at pH 8.2). Microwave-assisted digestion was performed for 30 min at 60 °C with the microwave power set to 5 W (CEM Discover, CEM Corp., USA). Supernatants were collected in fresh tubes and the gel pieces were extracted with 150 µl of 0.1% trifluoroacetic acid/50% acetonitrile. Supernatants were combined, dried, and the samples were dissolved in 20 µl of 0.1% formic acid before being transferred to the autosampler vials for liquid chromatography-tandem mass spectrometry, 7 to 9 µl were injected. Samples were measured on a Q-exactive mass spectrometer (Thermo Fisher Scientific) equipped with a nanoAcquity UPLC (Waters Corporation, Milford, MA). Peptides were trapped on a trap column (Symmetry C18, 5 µm, 180 µm × 20 mm, Waters Corporation) before they were separated on a BEH300 C18, 1.7 µm, 75 µm × 150 mm column (Waters Corporation) by applying a gradient formed between solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile). On the mass spectrometer, a gradient starting at 1% solvent B and increasing to 40% within 60 min was established. Database searches were performed using the MASCOT search program against the Giardia database (http://giardiadb.org/giardiadb/) with a concatenated decoy database supplemented with commonly observed contaminants and the Swissprot database to increase size of the database. The identified hits were then loaded onto the Scaffold Viewer version 4 (Proteome Software, Portland, USA) and the hits were filtered based on high stringency parameters, namely, minimal mascot score of 95 for peptide probability, a protein probability of 95% and a minimum of 2 unique peptides per protein.
2.7. Quantification of nitroreductase activity
For enzymatic quantification, the nitroreductase activity was measured in 96-well microtiter plates containing 100 µl of a reaction mix containing buffer (50 mM Tris-Cl−, pH 7.0), 0.1 mM of the compounds to be tested, 0.5 mM thiazolyl blue tetrazolium (MTT), 0.5 mM NADH or NADPH and 0.1 to 0.2 µg of the recombinant enzymes. The plates were incubated at 37 °C under aerobic conditions or in an anaerobic growth chamber. Substrate and enzyme blanks were included. After different time points, the reaction was stopped by adding 100 µl of pure ethanol thus solubilising the product formed by the reduction of MTT, formazan. The absorbance at 590 nm was read on a 96-well plate spectrophotometer (Versamax; Molecular Devices, Sunnyvale, CA). After subtraction of substrate and enzyme blanks, nitroreductase activity was expressed as ΔA590/min/mg (Prochaska and Santamaria, 1988).
The reduction of 7-nitrocoumarin to 7-aminocoumarin was quantified using the reaction mix as described above, but without MTT, with the same volumes and under the same conditions of incubation. Enzyme and substrate blanks were included. The reaction was stopped by adding 100 µl of 50 mM HCl. The resulting solution had pH 2 resulting in full protonation of 7-aminocoumarin which was quantified by fluorimetry with excitation at 365 nm and emission at 455 nm (Wagner, 2009) using a 96-well-multimode plate reader (Enspire; Perkin-Elmer, Waltham, MA).
2.8. Determination of drug susceptibility in E. coli
Drug susceptibility of recombinant E. coli BL21 (DE3) lines (Invitrogen, Carlsbad, CA, USA) expressing either GlNR1 (recombinant plasmid pGlNR1), GlNR2 (pGlNR2), glucuronidase A (pGusA) alone (Nillius et al, 2011, Müller et al, 2013) or both nitroreductases (this study, see below) were tested as described (Müller et al., 2013). Single gene expression was achieved in vector system pET151 Directional TOPO® (Invitrogen) containing the ampicillin resistance marker for selection of transformants and allowing IPTG-inducible over-expression of recombinant proteins (see pET151 Directional TOPO® manual provided by the manufacturer) as described (Müller et al., 2013). In order to achieve double transfectants expressing both nitroreductases, the following cloning strategies were chosen: (i) the entire pET151 Directional TOPO® expression cassettes carrying GlNR1 and GusA open reading frames were amplified by PCR using T7 forward (5′-TAATACGACTCACTATAGGG-3′) and T7 reverse (5′-TAGTTATTGCTCAGCGGTGG-3′) primers (annealing to regions flanking the expression cassette of pET151 Directional TOPO®) and pGlNR1 and pGusA as DNA templates. (ii) PCR products were used for re-cloning of GlNR1 and GusA into pCR-Blunt II-TOPO® (Invitrogen) containing a kanamycin resistance marker. (iii) This re-cloning step provided plasmid constructs, pGlNR1-KanR and pGusA-KanR, suitable for subsequent transformation of ampicillin-resistant BL21 (DE3)/pGlNR2, and BL21 (DE3)/pGusA strains by selection for ampicillin (100 µg/ml)/kanamycin (50 µg/ml) double-resistant clones. E. coli BL21 (DE3) carrying pGlNR1, pGlNR2, pGusA, pNR1-KanR/pGlNR2 and pGusA-KanR/pGusA were tested under aerobic or microaerobic (5% O2, 10% CO2, 85% N2) conditions by a conventional disc diffusion agar procedure as described (Müller et al., 2013). Growth inhibition zone diameters were determined and the inhibition zone around the disc was calculated (in mm2).
2.9. Statistical methods
Statistical analysis of the results was done based on the tools from the open source software package R (R Core Team, 2012) Differences exhibiting p values < 0.01 were considered significant.
3. Results
3.1. E. coli expressing recombinant GlNR1 and GlNR2 have different susceptibilities to nitro drugs
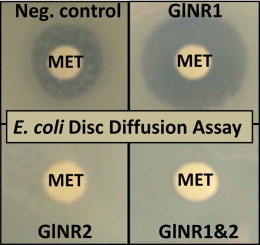
We generated recombinant E. coli lines producing GlNR1 or GlNR2 alone (Müller et al., 2013) or both (this study). As a control, we have created recombinant lines expressing glucuronidase A (Gus) alone (Müller et al., 2013) or double (Gus/Gus) (this study). In a pilot experiment, overproduction of proteins induced by IPTG strongly reduced growth of E. coli. Accordingly, non-induced, recombinant E. coli cultures were chosen for growth inhibition assays as described earlier (Nillius et al., 2011). With these five strains, disc diffusion assays were performed with metronidazole, nitazoxanide or tetracyclin as a positive control under aerobic or semi-aerobic conditions. Under aerobic as well as semi-aerobic growth conditions, metronidazole clearly inhibited growth of control strains transformed with Gus or Gus/Gus. In the presence of GlNR1 alone, this susceptibility was significantly enhanced. Bacteria transformed with GlNR2 alone or with both nitroreductases were completely resistant to metronidazole. Under aerobic conditions, nitazoxanide did not affect growth of bacteria. Under semi-aerobic conditions, however, bacteria expressing GlNR1 exhibited a significantly higher susceptibility to nitazoxanide than control bacteria. Other inserts than GlNR1 had no effects. There were no significant effects in nitroreductase-transformed strains vs. control strains in the presence of tetracycline (Fig. 1).
Fig. 1.
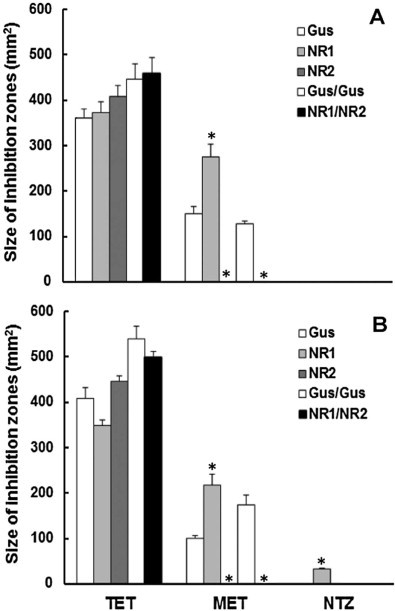
Susceptibility of E. coli BL21 (DE3) expressing GusA as a control (Gus), GlNR1 (NR1), and GlNR2 (NR2), two GusA (Gus/Gus), or both nitroreductases (NR1/NR2) to metronidazole (MET) and to nitazoxanide (NTZ). Tetracycline (TET) was included as a positive control. Plates with different cell lines exposed to discs containing the drugs were incubated under aerobic (A) or semi-aerobic (B) conditions. After 24 h, diameters of inhibition zones were determined. Mean values ± SE are given for 3 replicates. Values marked by asterisks are significantly different from the controls, i.e. to Gus for the single transformants and to Gus/Gus for the double transformant (paired t-test, two-sided, *p < 0.01).
3.2. Recombinant GlNR1 and GlNR2 are quinone reductases
The results obtained with metronidazole clearly suggested a role of both nitroreductases under aerobic conditions. Was their role thus restricted to the reduction of nitro compounds? To answer this question we implemented a functional assay based on the reduction of MTT to formazan by reduced nitrocompounds or quinones. In a first experiment, we incubated the assays in a 37 °C incubator under normal atmosphere or in an anaerobic culture chamber (100% N2). We offered NADH as electron donor, the quinone menadion and dinitrotoluene as electron acceptors or DMSO as a solvent control. Interestingly, both nitroreductases reduced menadione, and this even to much higher extents than dinitrotoluene. Both enzymes worked evenly well under anaerobic and under aerobic conditions (Fig. 2).
Fig. 2.
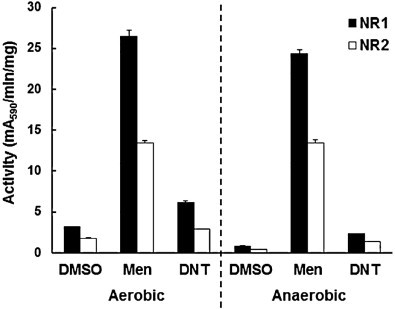
Activity of recombinant G. lamblia nitroreductases GlNR1 (NR1) and GlNR2 (NR2) with menadione (Men) or dinitrotoluene (DNT) as substrates (0.1 mM). DMSO was included as a solvent control, thus as a substrate blank. Functional assays were performed with NADH as an electron donor and MTT as chromogenic electron acceptor. The reaction was performed at 37 °C under normal atmosphere (aerobic) or in an anaerobic chamber (100% N2; anaerobic) and stopped after 2 h addition of one volume pure ethanol. Mean values (±SE) are given for three replicates.
3.3. Both nitroreductases are NADH dependent and have a preference for menadione
We tested the preference for NADH or NADPH as electron donors with menadione or dinitrotoluene as a substrate. Both nitroreductases had clearly a preference for NADH with both substrates (Table 1). In a next step, we determined the activities of both enzymes on a series of compounds including FAD, dicoumarol, quinacrine and various nitro compounds including the antigiardial drugs metronidazole and nitazoxanide as well as C17 and CB1954. Furthermore, we included 7-nitrocoumarin, a compound yielding the highly fluorescent 7-aminocoumarin upon complete reduction.
Table 1.
Activity of recombinant G. lamblia nitroreductases (GlNR1 and GlNR2) with various nitro- and non-nitro compounds (0.1 mM) as substrates. Functional assays were performed with MTT as chromogenic electron acceptor. Electron donor was NADH or NADPH if specified. The reaction was performed at 37 °C and stopped after various time points by addition of one volume pure ethanol. Mean values (±SE) are given for three replicates after subtraction of enzyme and substrate blanks.
| Substrate | GlNR1 | GlNR2 |
|---|---|---|
| (ΔA590 min−1 mg prot−1) | ||
| Menadione | 22.9 ± 0.4 | 16.2 ± 0.2 |
| Menadione NADPH | 1.8 ± 0.2 | 0.2 ± 0.1 |
| Dicoumarol | 3.1 ± 0.2 | 0.8 ± 0.1 |
| Menadione + dicoumarol | 27.2 ± 0.3 | 14.5 ± 0.1 |
| Ubiquinone (coenzyme Q10) | 2.8 ± 0.1 | 1.8 ± 0.2 |
| FAD | 11.8 ± 0.4 | 8.9 ± 0.9 |
| Dinitrotoluene | 5.2 ± 0.3 | 2.8 ± 0.2 |
| Dinitrotoluene NADPH | 0.9 ± 0.3 | 0.3 ± 0.1 |
| 7-Nitrocoumarine | 10.6 ± 0.3 | 6.5 ± 0.5 |
| Dinitrophenol | 5.9 ± 0.4 | 0.2 ± 0.1 |
| Nitrophenol | 2.9 ± 0.1 | 2.6 ± 0.1 |
| Metronidazole | 2.8 ± 0.2 | 1.7 ± 0.1 |
| Nitazoxanide | 2.7 ± 0.2 | 1.9 ± 0.1 |
| CB1954 | 3.0 ± 0.1 | 1.7 ± 0.1 |
| C17 | 2.1 ± 0.2 | 0.9 ± 0.1 |
The highest activities were observed for both enzymes with menadione as a substrate. Dicoumarol, a typical inhibitor of mammalian quinone reductases (Müller and Hemphill, 2011), had no effects. The second best substrate in our series was FAD. Ubiquinone (coenzyme Q10) was also reduced by both enzymes but with lower specific activities than meandione (Table 1).
With respect to nitro compounds as substrates, 7-nitrocoumarin was clearly the best. Interestingly, we could detect a reduction of all nitrocompounds with antigiardial activity, i.e. metronidazole, nitazoxanide, and C17. Dinitrophenol was only reduced by GlNR1. GlNR1 had a higher specific activity with all substrates (Table 1).
3.4. 7-nitrocoumarin is fully reduced preferentially by GlNR2
These results prompted us to investigate whether 7-aminocoumarin was fully reduced by both nitroreductases thus yielding the fluorescent 7-aminocoumarin. For this purpose, we performed the same assay as above without MTT, and quantified the fluorescent product. Although GlNR2 had a lower specific activity in the assay as described above, it was twice as active as GlNR1 in fully reducing 7-aminocoumarin. When added together, both enzymes were more active than the sum of the single activities (Fig. 3).
Fig. 3.
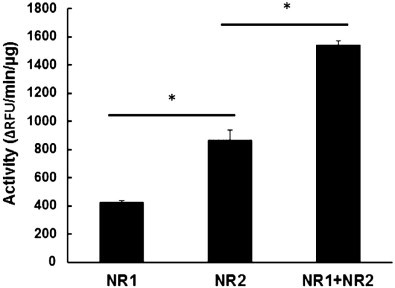
Activity of recombinant G. lamblia nitroreductases GlNR1 (NR1) and GlNR2 (NR2) with 7-nitrocoumarine as a substrate (0.1 mM) and with NADH as an electron donor. The reaction was performed at 37 °C with GlNR1, GlNR2 alone (125 ng each) or together (62.5 ng each) and stopped after various time points by addition of one volume HCl 0.05 M. Formation of 7-aminocoumarin was quantified by fluorimetry (excitation at 365 nm, emission at 455 nm). Mean values (±SE) are given for three replicates. Values marked by asterisks are significantly different from each other (paired t-test, two-sided, *p < 0.01).
3.5. GlNR1 and GlNR2 interact in vivo
The synergistic effect of the two nitroreductases (see Fig. 3) suggested a physical interaction of the two enzymes prompting us to perform pull-down assays using HA-tagged GlNR1 and GlNR2 as baits. These pull-down assays were used for the identification of proteins which specifically bind to these nitroreductases in G. lamblia. Crude extracts from G. lamblia WBC6 expressing the corresponding constructs were affinity-purified using anti-HA-antibodies immobilised on beads followed by mass spectrometry. Under high stringency conditions, HA-tagged nitroreductase GlNR1 co-purified with GlNR2 and with a couple of other proteins including fructose-bisphosphate aldolase (Table 2). Conversely, HA-tagged GlNR2 co-purified with GlNR1 and with two other proteins, namely, fructose-bisphosphate aldolase and ornithine carbamoyl-transferase (Table 3). Immunoblot analysis using an antibody specific for GlNR1 and an anti-HA antibody showed the presence of GlNR1 in the immunoprecipitate of GlNR2-3xHA. GlNR2 was not cross reactive with GlNR1. (Fig. 4).
Table 2.
Proteins interacting with HA-tagged GlNR1 in vivo. G. lamblia WBC6 was transformed with GlNR1-3xHA. Crude extracts were affinity-purified using anti-HA-antibodies immobilised on beads followed by mass spectrometry. A control experiment was performed with crude extract from WBC6 expressing no recombinant protein. Only hits with highest stringency, namely, minimal mascot score of 95 for peptide probability, a protein probability of 95% and a minimum of 2 unique peptides per protein, are shown.
| Name | Accession-N° | Molecular weight (kDa) | Unique peptides (N°) |
|---|---|---|---|
| Axoneme-associated protein GASP-180 | GL50803_137716 | 175 | 4 |
| Fructose-bisphosphate aldolase | GL50803_11043 | 35 | 3 |
| GlNR2 (Fd-NR1) | GL50803_6175 | 31 | 2 |
| Hypothetical protein | GL50803_9183 | 214 | 2 |
| TCP1-chaperon-subunit gamma | GL50803_17411 | 62 | 2 |
| Phosphoglycerate kinase | GL50803_90872 | 44 | 2 |
| Arginyl-tRNA-synthetase | GL50803_10521 | 70 | 2 |
| Vacuolar ATP-synthase catalytic subunit A | GL50803_7532 | 72 | 2 |
| Malate dehydrogenase | GL50803_3331 | 35 | 2 |
Table 3.
Proteins interacting with HA-tagged GlNR2 in vivo. G. lamblia WBC6 was transformed with GlNR2-3xHA. Crude extracts were affinity-purified using anti-HA-antibodies immobilised on beads followed by mass spectrometry. A control experiment was performed with crude extract from WBC6 expressing no recombinant protein. Only hits with highest stringency, namely, minimal mascot score of 95 for peptide probability, a protein probability of 95% and a minimum of 2 unique peptides per protein, are shown.
| Name | Accession-N° | Molecular weight (kDa) | unique peptides (N°) |
|---|---|---|---|
| GlNR1 (Fd-NR2) | GL50803_22677 | 29 | 15 |
| Fructose-bisphosphate aldolase | GL50803_11043 | 35 | 2 |
| Ornithine carbamoyl-transferase | GL50803_10311 | 36 | 2 |
Fig. 4.
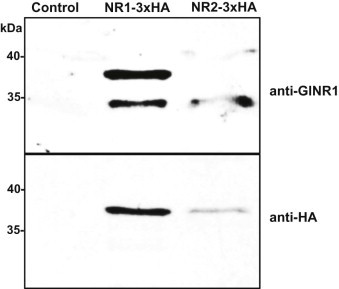
Immunoblot with anti-GlNR1 and anti HA-antibodies of immunoprecipitates from untransformed WBC6 (control), WBC6 transformed with GlNR1-3xHA as a bait, and WBC6 transformed with GlNR2-3xHA as a bait. Trophozoites were processed as described. The position of molecular weight markers is indicated in kDa.
4. Discussion
Like some other antiparasitic nitro drugs, metronidazole is considered as a prodrug that is activated by partial reduction. This reaction is supposed to form a toxic radical (Docampo and Moreno, 1984), or partially reduced nitroso- or hydroxylamine-intermediates (Moreno and Docampo, 1985), causing DNA damage (Sisson et al., 2000). Conversely, complete reduction results in detoxification of nitro compounds thus allowing various bacteria to use toxic compounds such as trinitrotoluene as carbon sources (Kutty and Bennett, 2005). Our results obtained with MTT as a final electron acceptor suggest that both nitroreductases are multifunctional and able to use a variety of nitro compounds as substrates. With 7-nitrocoumarin as a substrate, full reduction to the corresponding 7-aminocoumarin can be investigated (Wagner, 2009). In our hands, GlNR2 is twice as effective as GlNR1 in this reaction indicating that it performs the full reduction rather than a partial reduction. When mixed, both enzymes act synergistically.
In E. coli, expression of GlNR1 increases the susceptibility to nitazoxanide exclusively under semi-aerobic growth conditions whereas susceptibility to metronidazole is increased under semi-aerobic and aerobic conditions. In contrast, GlNR2 decreases the susceptibility to metronidazole under both growth conditions confirming previous results (Nillius et al, 2011, Müller et al, 2013). Interestingly, expression of both enzymes has the same effects as expression of GlNR2 alone suggesting that E. coli BL21(DE3) is able to express endogenous nitroreductases even under aerobic growth conditions what is fully in frame with previously published results (Zenno et al, 1996a, Zenno et al, 1996c, Valle et al, 2012). While these endogenous nitroreductases reduce metronidazole to toxic intermediates, GlNR2 acts antagonistically and eliminates metronidazole toxicity in E. coli. Susceptibility of E. coli to metronidazole has been reported earlier (Olekhnovich et al, 2009, Nillius et al, 2011). The results with nitroreductase-expressing E. coli and the results obtained with 7-nitrocoumarin in functional assays suggest that GlNR1 preferentially performs a partial reduction of nitro compounds yielding toxic intermediates. Conversely, GlNR2 is able to entirely reduce a nitro compound thus generating a non-toxic end product, e.g. the corresponding amine.
Our experiments show, however, that nitro compounds are not the best substrates for both enzymes having highest activities on menadione as substrate. This quinone reductase activity is not inhibited by dicoumarol as it is the case for typical mammalian nitroreductases such as the human quinone reductase NQO1 (Müller and Hemphill, 2011). Moreover, both enzymes reduce free FAD. Similar multifunctional nitroreductases have been identified in various bacteria including E. coli (Zenno et al, 1996a, Zenno et al, 1996b, Zenno et al, 1996c), Lactobacillus plantarum (Guillén et al., 2009), Salmonella typhimurium (Yanto et al., 2010) and in eukaryotes such as Trypanosoma cruzi (Hall et al., 2012), The genome of G. lamblia WBC6 contains three putative NADPH dependent quinone reductases, namely, Gl50803_15004 (18.6 kDa), Gl50803_17150 (18.5 kDa), and Gl50803_17151 (19.5 kDa). Only one, Gl50803_15004, has been previously characterised in detail. This enzyme reduces menadione using NADPH as preferred electron donor (Sanchez et al., 2001).
The biological function of GlNR1 and GlNR2 could be the reduction of quinones and other heterocyclic compounds in essential steps of intermediate metabolism. Menadione is not a quinone present in Giardia and is even toxic (Paget et al., 2004). This toxicity may be due to a partial reduction of menadione to the corresponding semiquinone. Other potential substrates such as FAD and ubiquinone (Ellis et al., 1994) are, however, present in Giardia and play an important role in intermediate metabolism.
This could explain why knock-down approaches have failed, so far in our hands, and why a nitroreductase is essential for Leishmania donovani (Voak et al., 2013). From an evolutionary point of view, the reduction of nitro compounds could be a side effect without negative selective pressure until a nitro compound yielding toxic intermediates after reduction comes into play. Then, the presence of an otherwise beneficial activity turns into a disaster for the cell. In the presence of sublethal concentrations of the nitro compound, resistance formation is then achieved not by a mere down-regulation of the nitroreductase responsible for the toxic intermediate formation, but rather by a complete re-organisation of cellular metabolism as exemplified by the resistance formation of WBC6 against nitazoxanide (Müller et al., 2008).
Pull-down assays with both nitroreductases as baits and immunoblot analysis show that both enzymes interact with each other. Moreover, they may also interact with fructose-bisphosphate aldolase, a key enzyme of glycolysis. The significance of this finding is unclear and will be studied in further immunoprecipitation experiments using fructose-bis-phosphate aldolase as a bait.
Novel promising techniques like conditional knock-outs (Wampfler et al., 2014) could open the way to understand the biological function of nitroreductases in G. lamblia.
Conflict of interest
The authors declared that there is no conflict of interest.
Acknowledgements
We would like to thank The Functional Genomics Center Zürich (FGCZ) for highly valuable technical support. We also wish to thank A. Hemphill (Institute of Parasitology, University of Berne, Berne, Switzerland) for proofreading of the manuscript and C. Huber as well as V. Balmer (Institute of Parasitology, University of Berne, Berne, Switzerland) for technical assistance. This study was supported by grants from the Swiss National Science Foundation (grants No. 31003A_138353 [NM, JM] and No. 31-140803/1 [AH]) and the Austrian Science Fund (project J3492 1 [DL].
Footnotes
Supplementary data to this article can be found online at doi:10.1016/j.ijpddr.2015.03.001.
Appendix. Supplementary material
The following is the supplementary data to this article:
SDS-PAGE-based fractionation of immunoprecipitated protein for mass spectrometry. Crude extracts from G. lamblia WBC6 transfected with GlNRl-3xHA (NRl-3xHA) or GlNR2-3xHA (NR2-3xHA) were affinity-purified using anti-HA-antibodies immobilised on beads followed by analysis of precipitated protein on Instant blue-stained SDS-polyacrylamide gels. A control experiment (control) was performed with crude extract from WBC6 expressing no recombinant protein. Gel lanes were cut in 8 equal sections, each section was diced into smaller pieces and further processed for mass spectrometry as described in ‘Materials and methods’. Molecular weight markers (M) are given in kDa. Interpretation of the visible bands: the upper and lower bands could correspond to IgG heavy and light chains. The two bands in the middle (visible only in the NRl-3xHA and NR2-3xHA-samples) could correspond to the tagged NRs and to a lower co-immunoprecipitation product (e.g. an untagged NR).
Overview of primers used in this study. Please note that the nitroreductases GlNR1 and GlNR2 are now annotated as nitroreductase Fd-NR2 (GL50803_22677) and nitroreductase family protein fused to ferredoxin domain Fd-NR1 (GL50803_6175) in the Giardia database (http://giardiadb.org/giardiadb/), respectively. We use GlNR1 and GlNR2 in order to be in line with our previous publications. The restriction sites are given in italics, GDH promoter regions and stop codons in bold, 3 × 27 nt HA-tag coding sequences in minuscule, and GlNR1 and GlNR2 coding sequences are underlined.
References
- Brown D.M., Upcroft J.A., Edwards M.R., Upcroft P. Anaerobic bacterial metabolism in the ancient eukaryote Giardia duodenalis. Int. J. Parasitol. 1998;28:149–164. doi: 10.1016/s0020-7519(97)00172-0. [DOI] [PubMed] [Google Scholar]
- Davis-Hayman S.R., Nash T.E. Genetic manipulation of Giardia lamblia. Mol. Biochem. Parasitol. 2002;122:1–7. doi: 10.1016/s0166-6851(02)00063-4. [DOI] [PubMed] [Google Scholar]
- Docampo R., Moreno S.N. Free radical metabolites in the mode of action of chemotherapeutic agents and phagocytic cells on Trypanosoma cruzi. Rev. Infect. Dis. 1984;6:223–238. doi: 10.1093/clinids/6.2.223. [DOI] [PubMed] [Google Scholar]
- Ellis J.E., Setchell K.D., Kaneshiro E.S. Detection of ubiquinone in parasitic and free-living protozoa, including species devoid of mitochondria. Mol. Biochem. Parasitol. 1994;65:213–224. doi: 10.1016/0166-6851(94)90073-6. [DOI] [PubMed] [Google Scholar]
- Goodwin A., Kersulyte D., Sisson G., Veldhuyzen van Zanten S.J., Berg D.E., Hoffman P.S. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 1998;28:383–393. doi: 10.1046/j.1365-2958.1998.00806.x. [DOI] [PubMed] [Google Scholar]
- Guillén H., Curiel J.A., Landete J.M., Muñoz R., Herraiz T. Characterization of a nitroreductase with selective nitroreduction properties in the food and intestinal lactic acid bacterium Lactobacillus plantarum WCFS1. J. Agric. Food Chem. 2009;57:10457–10465. doi: 10.1021/jf9024135. [DOI] [PubMed] [Google Scholar]
- Hall B.S., Meredith E.L., Wilkinson S.R. Targeting the substrate preference of a type I nitroreductase to develop antitrypanosomal quinone-based prodrugs. Antimicrob. Agents Chemother. 2012;56:5821–5830. doi: 10.1128/AAC.01227-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemphill A., Müller J., Esposito M. Nitazoxanide, a broad-spectrum thiazolide anti-infective agent for the treatment of gastrointestinal infections. Expert Opin. Pharmacother. 2006;7:953–964. doi: 10.1517/14656566.7.7.953. [DOI] [PubMed] [Google Scholar]
- Horner D.S., Hirt R.P., Embley T.M. A single eubacterial origin of eukaryotic pyruvate: ferredoxin oxidoreductase genes: implications for the evolution of anaerobic eukaryotes. Mol. Biol. Evol. 1999;16:1280–1291. doi: 10.1093/oxfordjournals.molbev.a026218. [DOI] [PubMed] [Google Scholar]
- Jiménez-García L.F., Zavala G., Chávez-Munguía B., Ramos-Godínez Mdel P., López-Velázquez G., Segura-Valdez Mde L. Identification of nucleoli in the early branching protist Giardia duodenalis. Int. J. Parasitol. 2008:1297–1304. doi: 10.1016/j.ijpara.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Johnson G.R., Spain J.C. Evolution of catabolic pathways for synthetic compounds: bacterial pathways for degradation of 2,4-dinitrotoluene and nitrobenzene. Appl. Microbiol. Biotechnol. 2003;62:110–123. doi: 10.1007/s00253-003-1341-4. [DOI] [PubMed] [Google Scholar]
- Kutty R., Bennett G.N. Biochemical characterization of trinitrotoluene transforming oxygen-insensitive nitroreductases from Clostridium acetobutylicum ATCC 824. Arch. Microbiol. 2005;184:158–167. doi: 10.1007/s00203-005-0036-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lalle M. Giardiasis in the post genomic era: treatment, drug resistance and novel therapeutic perspectives. Infect. Disord. Drug Targets. 2010;10:283–294. doi: 10.2174/187152610791591610. [DOI] [PubMed] [Google Scholar]
- Lee H., Cherng S.H., Liu T.Y. Bacterial mutagenicity, metabolism, and DNA adduct formation by binary mixtures of benzo[a]pyrene and 1-nitropyrene. Environ. Mol. Mutagen. 1994;24:229–234. doi: 10.1002/em.2850240312. [DOI] [PubMed] [Google Scholar]
- Leitsch D., Kolarich D., Binder M., Stadlmann J., Altmann F., Duchêne M. Trichomonas vaginalis: metronidazole and other nitroimidazole drugs are reduced by the flavin enzyme thioredoxin reductase and disrupt the cellular redox system. Implications for nitroimidazole toxicity and resistance. Mol. Microbiol. 2009;72:518–536. doi: 10.1111/j.1365-2958.2009.06675.x. [DOI] [PubMed] [Google Scholar]
- Luque-Almagro V.M., Blasco R., Paloma Sáez L., Roldán M.D., Moreno-Vivián C., Castello F. Interactions between nitrate assimilation and 2,4-dinitrophenol cometabolism in Rhodobacter capsulatus E1F1. Curr. Microbiol. 2006;53:37–42. doi: 10.1007/s00284-005-0185-9. [DOI] [PubMed] [Google Scholar]
- Moreno S.N., Docampo R. Mechanism of toxicity of nitro compounds used in the chemotherapy of trichomoniasis. Environ. Health Perspect. 1985;64:199–208. doi: 10.1289/ehp.8564199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Hemphill A. Identification of a host cell target for the thiazolide class of broad-spectrum anti-parasitic drugs. Exp. Parasitol. 2011;128:145–150. doi: 10.1016/j.exppara.2011.02.007. [DOI] [PubMed] [Google Scholar]
- Müller J., Sterk M., Hemphill A., Müller N. Characterization of Giardia lamblia WB C6 clones resistant to nitazoxanide and to metronidazole. J. Antimicrob. Chemother. 2007;60:280–287. doi: 10.1093/jac/dkm205. [DOI] [PubMed] [Google Scholar]
- Müller J., Wastling J., Sanderson S., Müller N., Hemphill A. A novel Giardia lamblia nitroreductase, GlNR1, interacts with nitazoxanide and other thiazolides. Antimicrob. Agents Chemother. 2007;51:1979–1986. doi: 10.1128/AAC.01548-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Ley S., Felger I., Hemphill A., Müller N. Identification of differentially expressed genes in a Giardia lamblia WB C6 clone resistant to nitazoxanide and metronidazole. J. Antimicrob. Chemother. 2008;62:72–82. doi: 10.1093/jac/dkn142. [DOI] [PubMed] [Google Scholar]
- Müller J., Nillius D., Hehl A., Hemphill A., Müller N. Stable expression of Escherichia coli β-glucuronidase A (GusA) in Giardia lamblia: application to high-throughput drug susceptibility testing. J. Antimicrob. Chemother. 2009;64:1187–1191. doi: 10.1093/jac/dkp363. [DOI] [PubMed] [Google Scholar]
- Müller J., Schildknecht P., Müller N. Metabolism of nitro drugs metronidazole and nitazoxanide in Giardia lamblia: characterization of a novel nitroreductase (GlNR2) J. Antimicrob. Chemother. 2013;68:1781–1789. doi: 10.1093/jac/dkt106. [DOI] [PubMed] [Google Scholar]
- Nillius D., Müller J., Müller N. Nitroreductase (GlNR1) increases susceptibility of Giardia lamblia and Escherichia coli to nitro drugs. J. Antimicrob. Chemother. 2011;66:1029–1035. doi: 10.1093/jac/dkr029. [DOI] [PubMed] [Google Scholar]
- Nixon J.E., Wang A., Field J., Morrison H., McArthur A.G., Sogin M.L. Evidence for lateral transfer of genes encoding ferredoxins, nitroreductases, NADH oxidase, and alcohol dehydrogenase 3 from anaerobic prokaryotes to Giardia lamblia and Entamoeba histolytica. Eukaryot. Cell. 2002;1:181–190. doi: 10.1128/EC.1.2.181-190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olekhnovich I.N., Goodwin A., Hoffman P.S. Characterization of the NAD(P)H oxidase and metronidazole reductase activities of the RdxA nitroreductase of Helicobacter pylori. FEBS J. 2009;276:3354–3364. doi: 10.1111/j.1742-4658.2009.07060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget T., Maroulis S., Mitchell A., Edwards M.R., Jarroll E.L., Lloyd D. Menadione kills trophozoites and cysts of Giardia intestinalis. Microbiology. 2004;150:1231–1236. doi: 10.1099/mic.0.26836-0. [DOI] [PubMed] [Google Scholar]
- Prochaska H.J., Santamaria A.B. Direct measurement of NAD(P)H:quinone reductase from cells cultured in microtiter wells: a screening assay for anticarcinogenic enzyme inducers. Anal. Biochem. 1988;169:328–336. doi: 10.1016/0003-2697(88)90292-8. [DOI] [PubMed] [Google Scholar]
- R Core Team . 2012. R: A language and environment for statistical computing. [Google Scholar]
- Robertson L.J., Hanevik K., Escobedo A.A., Mørch K., Langeland N. Giardiasis – why do the symptoms sometimes never stop? Trends Parasitol. 2010;26:75–82. doi: 10.1016/j.pt.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Roldán M.D., Pérez-Reinado E., Castillo F., Moreno-Vivián C. Reduction of polynitroaromatic compounds: the bacterial nitroreductases. FEMS Microbiol. Rev. 2008;32:474–500. doi: 10.1111/j.1574-6976.2008.00107.x. [DOI] [PubMed] [Google Scholar]
- Sanchez L.B., Elmendorf H., Nash T.E., Müller M. NAD(P)H:menadione oxidoreductase of the amitochondriate eukaryote Giardia lamblia: a simpler homologue of the vertebrate enzyme. Microbiology. 2001;147:561–570. doi: 10.1099/00221287-147-3-561. [DOI] [PubMed] [Google Scholar]
- Sisson G., Jeong J.Y., Goodwin A., Bryden L., Rossler N., Lim-Morrison S. Metronidazole activation is mutagenic and causes DNA fragmentation in Helicobacter pylori and in Escherichia coli containing a cloned H. pylori RdxA(+) (Nitroreductase) gene. J. Bacteriol. 2000;182:5091–5096. doi: 10.1128/jb.182.18.5091-5096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares A.F., Nobre L.S., Melo A.M., Saraiva L.M. A novel nitroreductase of Staphylococcus aureus with S-nitrosoglutathione reductase activity. J. Bacteriol. 2009;191:3403–3406. doi: 10.1128/JB.00022-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R.C.A. Giardiasis as a re-emerging infectious disease and its zoonotic potential. Int. J. Parasitol. 2000;30:1259–1267. doi: 10.1016/s0020-7519(00)00127-2. [DOI] [PubMed] [Google Scholar]
- Upcroft P., Upcroft J.A. Drug targets and mechanisms of resistance in the anaerobic protozoa. Clin. Microbiol. Rev. 2001;14:150–164. doi: 10.1128/CMR.14.1.150-164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle A., Le Borgne S., Bolívar J., Cabrera G., Cantero D. Study of the role played by NfsA, NfsB nitroreductase and NemA flavin reductase from Escherichia coli in the conversion of ethyl 2-(2′-nitrophenoxy)acetate to 4-hydroxy-(2H)-1,4-benzoxazin-3(4H)-one (D-DIBOA), a benzohydroxamic acid with interesting biological properties. Appl. Microbiol. Biotechnol. 2012;94:163–171. doi: 10.1007/s00253-011-3787-0. [DOI] [PubMed] [Google Scholar]
- Voak A.A., Gobalakrishnapillai V., Seifert K., Balczo E., Hu L., Hall B.S. An essential type I nitroreductase from Leishmania major can be used to activate leishmanicidal prodrugs. J. Biol. Chem. 2013;288:28466–28476. doi: 10.1074/jbc.M113.494781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner B.D. The use of coumarins as environmentally-sensitive fluorescent probes of heterogeneous inclusion systems. Molecules. 2009;14:210–237. doi: 10.3390/molecules14010210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wampfler P.B., Faso C., Hehl A.B. The Cre/loxP system in Giardia lamblia: genetic manipulations in a binucleate tetraploid protozoan. Int. J. Parasitol. 2014;44:497–506. doi: 10.1016/j.ijpara.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Wright J.M., Dunn L.A., Upcroft P., Upcroft J.A. Efficacy of antigiardial drugs. Expert Opin. Drug Saf. 2003;2:529–541. doi: 10.1517/14740338.2.6.529. [DOI] [PubMed] [Google Scholar]
- Yanto Y., Hall M., Bommarius A.S. Nitroreductase from Salmonella typhimurium: characterization and catalytic activity. Org. Biomol. Chem. 2010;8:1826–1832. doi: 10.1039/b926274a. [DOI] [PubMed] [Google Scholar]
- Zenno S., Koike H., Kumar A.N., Jayaraman R., Tanokura M., Saigo K. Biochemical characterization of NfsA, the Escherichia coli major nitroreductase exhibiting a high amino acid sequence homology to Frp, a Vibrio harveyi flavin oxidoreductase. J. Bacteriol. 1996;178:4508–4514. doi: 10.1128/jb.178.15.4508-4514.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenno S., Koike H., Tanokura M., Saigo K. Conversion of NfsB, a minor Escherichia coli nitroreductase, to a flavin reductase similar in biochemical properties to FRase I, the major flavin reductase in Vibrio fischeri, by a single amino acid substitution. J. Bacteriol. 1996;178:4731–4733. doi: 10.1128/jb.178.15.4731-4733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenno S., Koike H., Tanokura M., Saigo K. Gene cloning, purification, and characterization of NfsB, a minor oxygen-insensitive nitroreductase from Escherichia coli, similar in biochemical properties to FRase I, the major flavin reductase in Vibrio fischeri. J. Biochem. 1996;120:736–744. doi: 10.1093/oxfordjournals.jbchem.a021473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SDS-PAGE-based fractionation of immunoprecipitated protein for mass spectrometry. Crude extracts from G. lamblia WBC6 transfected with GlNRl-3xHA (NRl-3xHA) or GlNR2-3xHA (NR2-3xHA) were affinity-purified using anti-HA-antibodies immobilised on beads followed by analysis of precipitated protein on Instant blue-stained SDS-polyacrylamide gels. A control experiment (control) was performed with crude extract from WBC6 expressing no recombinant protein. Gel lanes were cut in 8 equal sections, each section was diced into smaller pieces and further processed for mass spectrometry as described in ‘Materials and methods’. Molecular weight markers (M) are given in kDa. Interpretation of the visible bands: the upper and lower bands could correspond to IgG heavy and light chains. The two bands in the middle (visible only in the NRl-3xHA and NR2-3xHA-samples) could correspond to the tagged NRs and to a lower co-immunoprecipitation product (e.g. an untagged NR).
Overview of primers used in this study. Please note that the nitroreductases GlNR1 and GlNR2 are now annotated as nitroreductase Fd-NR2 (GL50803_22677) and nitroreductase family protein fused to ferredoxin domain Fd-NR1 (GL50803_6175) in the Giardia database (http://giardiadb.org/giardiadb/), respectively. We use GlNR1 and GlNR2 in order to be in line with our previous publications. The restriction sites are given in italics, GDH promoter regions and stop codons in bold, 3 × 27 nt HA-tag coding sequences in minuscule, and GlNR1 and GlNR2 coding sequences are underlined.


