Abstract
Small molecules are extensively metabolized and cleared by the kidney. Changes in serum metabolite concentrations may result from impaired kidney function and can be used to estimate filtration (e.g., the established marker creatinine) or may precede and potentially contribute to CKD development. Here, we applied a nontargeted metabolomics approach using gas and liquid chromatography coupled to mass spectrometry to quantify 493 small molecules in human serum. The associations of these molecules with GFR estimated on the basis of creatinine (eGFRcr) and cystatin C levels were assessed in ≤1735 participants in the KORA F4 study, followed by replication in 1164 individuals in the TwinsUK registry. After correction for multiple testing, 54 replicated metabolites significantly associated with eGFRcr, and six of these showed pairwise correlation (r≥0.50) with established kidney function measures: C-mannosyltryptophan, pseudouridine, N-acetylalanine, erythronate, myo-inositol, and N-acetylcarnosine. Higher C-mannosyltryptophan, pseudouridine, and O-sulfo-L-tyrosine concentrations associated with incident CKD (eGFRcr <60 ml/min per 1.73 m2) in the KORA F4 study. In contrast with serum creatinine, C-mannosyltryptophan and pseudouridine concentrations showed little dependence on sex. Furthermore, correlation with measured GFR in 200 participants in the AASK study was 0.78 for both C-mannosyltryptophan and pseudouridine concentration, and highly significant associations of both metabolites with incident ESRD disappeared upon adjustment for measured GFR. Thus, these molecules may be alternative or complementary markers of kidney function. In conclusion, our study provides a comprehensive list of kidney function–associated metabolites and highlights potential novel filtration markers that may help to improve the estimation of GFR.
Keywords: CKD, metabolism, epidemiology, outcomes, GFR
Metabolites are small organic molecules involved in both systemic and organ-specific metabolic processes. The kidney receives about 20% of the cardiac output and is one of the most important excretory organs for a wide variety of metabolites, many of which are freely filtered as solutes of small molecular size. In addition, many metabolites are actively secreted into or reabsorbed from the tubular lumen. The kidney also has a role in the generation and metabolism of some metabolites such as amino acids. Changes in blood metabolite concentrations may therefore result from impaired kidney function via altered generation, filtration, secretion, reabsorption, or metabolism.
CKD is a major public health concern affecting approximately 10% of the population in Western countries1 and substantially increases the risk for cardiovascular morbidity and mortality.2 Despite the high prevalence and increasing incidence of CKD, the underlying pathophysiologic mechanisms are not fully understood.
Because blood metabolite concentrations are influenced by kidney function, some metabolites are currently used for its estimation—most importantly, serum creatinine is used to estimate the GFR. However, creatinine has important limitations, including a rise only after 50% of kidney function has been lost and a dependence on age, sex, and race, reflecting underlying differences in muscle mass.3 Estimates from creatinine-based equations can be inaccurate (especially in early stages of CKD) in elderly individuals and in those with extreme body mass index values. The identification of additional markers of kidney function is therefore clinically useful, as evidenced by the recent addition of a second marker to estimate GFR, cystatin C, which improves the ability to accurately estimate GFR4 and predict future risk of ESRD and death.5
Previous studies in the general population using targeted metabolomics approaches have found associations between lower eGFR and acylcarnitines.6 General population-based studies of eGFR decline and/or incident CKD have reported associations with spermidine and the kynurenine-to-tryptophan ratio7 as well as with kynurenic acid, choline, and citrulline, among others.8 Nontargeted metabolomics creates further opportunities for the discovery of CKD-associated metabolites because of its abilities to detect a wide spectrum of metabolites from different metabolic pathways, including metabolites of yet unknown identity. Earlier nontargeted metabolomics studies have either been conducted in small numbers of individuals with preexisting kidney disease9 or in a population-based study of African Americans without external replication.10
The goal of our study was to extend current knowledge by identifying and replicating novel and known metabolites that reproducibly associate with eGFR and incident CKD in European participants in large population-based studies. We used a nontargeted metabolomics approach to identify novel markers that are potentially useful to estimate kidney function and to gain additional insights into the pathophysiology of CKD and renal metabolite handling at a stage where preventive actions to slow kidney function decline may still be implemented.
Results
Table 1 shows the characteristics of the population samples studied. The distribution of most characteristics was similar in the two study samples used for discovery of metabolite associations. Participants in the replication study, the TwinsUK cohort, were on average younger and healthier than the Cooperative Health Research in the Region of Augsburg (KORA) participants, and the sample consisted only of women. Mean estimated creatinine-based GFR (eGFRcr) was higher and CKD prevalence was lower, with 3.1% in the TwinsUK cohort compared with 5%–7% in the KORA studies.
Table 1.
Demographic characteristics of the study populations
| Variable | KORA S4/F4 Sample at S4 Visit | KORA F4 Visit | TwinsUK Study |
|---|---|---|---|
| Sample size, n | 991 | 1735 | 1164 |
| Age, yr | 63.3 (5.4) | 60.8 (8.8) | 50.1 (11.0) |
| Men, % | 70.9 | 48.4 | 0.0 |
| Body mass index, kg/m2 | 28.2 (4.0) | 28.1 (4.8) | 26.3 (4.9) |
| Current smoking, % | 11.9 | 14.6 | 10 |
| Diabetes prevalence, % | 0.6 | 8.6 | 3.1 |
| Lipid-lowering medication use, % | 11.2 | 16.4 | N/A |
| Antihypertensive medication use, % | 32.3 | 37.1 | 3.8 |
| Systolic BP, mmHg | 134.3 (19.5) | 124.6 (18.6) | 126.8 (17.4) |
| eGFRcr, ml/min per 1.73 m2 | 81.0 (13.1) | 87 (15.8) | 93.8 (17.3) |
| eGFRcys, ml/min per 1.73 m2 | N/A | 101 (20.3) | N/A |
| CKD prevalence, eGFRcr <60 ml/min per 1.73 m2, % | 6.6 | 5.2 | 3.1 |
| eGFRcr change per year, ml/min per 1.73 m2 | 0.6 (1.6) | N/A | N/A |
Continuous measures are summarized as mean (SD) and categorical variables are given as percentages. Mean change in eGFRcr is reported for the subset of individuals with measures at both visits available; positive values correspond to a decrease in eGFRcr over time. The low prevalence of diabetes at the S4 visit reflects the fact that only fasting samples were selected for metabolomics analysis in S4. Such samples were available from KORA S4 participants who had been invited for an oral glucose tolerance test excluding participants with clinically diagnosed diabetes. N/A, not available.
Detailed information about the 493 metabolites examined in this study is given in Supplemental Table 1, including biochemical names, metabolic pathways, and quality control statistics. Of 321 identified metabolites with known chemical structure, there were 81 amino acids and related compounds, 14 carbohydrates, 15 cofactors and vitamins, 6 metabolites related to energy metabolism, 130 lipid metabolism derivatives, 14 purine and pyrimidine bases and related compounds, 27 peptides, and 34 xenobiotics.
Markers Cross-Sectionally Associated with eGFRcr
The analytical workflow of the cross-sectional association study between serum metabolites and eGFR in the KORA F4 study is shown in Supplemental Figure 1. The KORA F4 study was used to evaluate these associations because of the availability of a second, independent noncreatinine filtration marker for validation, serum cystatin C. Of 488 metabolites available in KORA F4 (see the Concise Methods), 112 were significantly associated with GFRcr and also with GFR estimated from cystatin C (eGFRcys) after correction for multiple testing (Bonferroni correction, P<1.0×10−4). Of these, 103 metabolites could be assessed in the smaller TwinsUK cohort. All but one showed effects in the same direction, and 54 of them were significantly associated (P<4.9×10−4, one-sided P value corrected for multiple testing) and thus considered replicated. This likely represents a conservative estimate, given the moderate size of the replication sample.
Table 2 highlights six of the replicated metabolites, which had a pairwise Pearson correlation coefficient of ≥0.50 with one or more of four established kidney function markers (eGFRcr, eGFRcys, creatinine measured by a standard test, and cystatin C): pseudouridine, C-mannosyltryptophan, N-acetylalanine, erythronate, myo-inositol, and N-acetylcarnosine. Associations with mass spectrometry (MS)–quantified creatinine are shown for comparison. C-mannosyltryptophan showed the highest pairwise correlations of −0.61 with eGFRcr and −0.71 with eGFRcys. The association of these six metabolites with CKD, as assessed by their association P values, was comparable in magnitude to that for MS-quantified serum creatinine (Table 2). In all instances, higher concentrations of the metabolites were associated with lower eGFRcr and higher CKD prevalence. Supplemental Table 2 provides complete association results, including both P values and false discovery rate Q values for all 488 metabolites with eGFRcr, eGFRcys, and CKD, as well as their pairwise correlations with established kidney function markers. Among them were previously reported metabolites such as kynurenine, erythritol,10 and glutaroylcarnitine,6 as well as several unidentified metabolites and novel metabolites (e.g., O-sulfo-L-tyrosine) that showed highly significant associations with kidney function but pairwise correlations with established kidney function markers of <0.5.
Table 2.
Highlighted metabolites significantly associated with eGFR in the KORA F4 study and replicated in the TwinsUK cohort
| Metabolite | KORA F4 Study | TwinsUK Cohort | Meta-Analysis | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Correlation | eGFRcr | eGFRcys | CKD | n | eGFRcr | eGFRcr | ||||||||||
| eGFRcr | eGFRcys | Creatinine | Cystatin C | Direction | P Value | Q Value | Direction | P Value | Direction | P Value | Direction | 1-Sided P Value | Direction | P Value | |||
| Creatinine (MS-quantified) | 1730 | −0.37 | −0.16 | 0.56 | 0.26 | − | 1.6E-100 | 3.7E-98 | − | 3.1E-31 | + | 1.3E-19 | 917 | − | 2.5E-29 | − | 4.5E-125 |
| Pseudouridine | 1726 | −0.56 | −0.59 | 0.50 | 0.64 | − | 9.1E-89 | 6.7E-87 | − | 7.6E-108 | + | 2.0E-21 | 915 | − | 2.6E-28 | − | 3.8E-113 |
| C-mannosyltryptophan | 1724 | −0.61 | −0.71 | 0.42 | 0.72 | − | 8.9E-89 | 6.7E-87 | − | 1.0E-163 | + | 1.1E-22 | 917 | − | 1.9E-26 | − | 7.5E-111 |
| N-acetylalanine | 1666 | −0.49 | −0.52 | 0.43 | 0.54 | − | 1.4E-65 | 5.1E-64 | − | 5.3E-81 | + | 3.9E-17 | 915 | − | 5.7E-25 | − | 1.2E-87 |
| Erythronate | 1703 | −0.55 | −0.55 | 0.49 | 0.60 | − | 3.6E-74 | 2.0E-72 | − | 2.3E-83 | + | 1.8E-21 | 900 | − | 4.0E-13 | − | 4.6E-80 |
| Myo-inositol | 1726 | −0.50 | −0.46 | 0.46 | 0.52 | − | 2.0E-68 | 8.8E-67 | − | 2.6E-68 | + | 8.8E-21 | 916 | − | 2.2E-15 | − | 2.0E-78 |
| N-acetylcarnosine | 1731 | −0.30 | −0.17 | 0.57 | 0.31 | − | 7.9E-65 | 2.5E-63 | − | 9.3E-35 | + | 4.3E-13 | 883 | − | 7.9E-16 | − | 3.7E-76 |
Directions of association refer to higher concentrations of the respective metabolites: Per unit increase in metabolite concentrations, negative association with eGFR denotes lower eGFR and positive association with CKD denotes higher odds of CKD.
Next, we assessed the cross-sectional association between pairwise metabolite ratios and eGFRcr and eGFRcys in the KORA F4 study. We previously showed that metabolite ratios can provide information above and beyond their individual components, as quantified by the P gain measure (the factor by which the P value decreases by taking the ratios compared with the respective single metabolites; see the Concise Methods).11,12 For example, these ratios can serve as a proxy for the activity of a metabolite conversion enzyme or a metabolite cotransporter or countertransporter. Of 115,476 pairwise metabolite ratios assessed, 60 showed significant associations after correction for multiple testing (P<4.3×10−7 or 0.05/115,476) with both eGFRcr and eGFRcys in the KORA F4 study, had a significant P gain (>10×115,476), were replicated in the TwinsUK study, and did not contain MS-quantified creatinine (Supplemental Table 3). Of these 60 ratios, 42 contained the creatinine precursor creatine, and many of the others were ratios of amino acids. The largest P gain was observed for the X-12094/creatine ratio (P gain=6×1024), and the largest P gain for a ratio that did not contain creatine was observed for 3-(4-hydroxyphenyl)lactate/tyrosine (P gain=2×1017). After the completion of our study, X-12094 was identified as N1-methyl-2-pyridone-5-carboxamide, which has been proposed as a uremic toxin.13 An example that illustrates the value of using ratios in this setting is the glycine/serine ratio (P gain =1.6×1016), which might reflect the activity of the enzyme serine hydroxymethyltransferase in the kidney, which converts glycine to serine. Several of the ratios with the largest P gains contained classic uremic solutes in addition to creatine, such as 3-indoxylsulfate and urea.
Sensitivity analyses excluding individuals with prevalent CKD at the KORA F4 visit did not change the number of significantly associated and replicated metabolites or metabolite ratios, but the magnitude of association was attenuated by approximately 25%.
Markers Associated with eGFRcr Decline over Time
To evaluate whether the eGFRcr-associated and/or additional metabolites also correlated with eGFRcr decline over time, we assessed their association with annual change in eGFRcr between the KORA S4 (baseline) and KORA F4 (follow-up after 7.1 years on average) study visits in a subsample of 991 individuals that attended both visits and for which measurements of 422 metabolites at the KORA S4 baseline survey were available. Three metabolites (C-mannosyltryptophan, pseudouridine, and O-sulfo-L-tyrosine) but no ratios were significantly associated with annual eGFRcr decline after adjusting for known correlates of kidney disease and baseline eGFRcr (Table 3). Higher serum concentrations of all three metabolites were associated with larger eGFRcr decline and incident CKD (P<0.05). Upon simultaneous inclusion followed by backward elimination, only C-mannosyltryptophan remained nominally associated with eGFRcr change (P=0.03). This was not unexpected, given the moderate to high correlation of the metabolites with each other (Table 3). The metabolites did not show significant interaction with baseline eGFRcr. The association between all tested metabolites and annual eGFRcr decline, as well as incident CKD, is shown in Supplemental Table 4.
Table 3.
Metabolites significantly associated with annual eGFRcr change and incident CKD in the KORA study between visits S4 and F4
| Metabolite | Annual eGFRcr Change | Incident CKD (n=95 cases) | ||||
|---|---|---|---|---|---|---|
| n | Direction | P Value | n | Direction | P Value | |
| C-mannosyltryptophan | 985 | + | 8.7E-06 | 920 | + | 1.1E-03 |
| Pseudouridine | 989 | + | 1.3E-05 | 924 | + | 3.7E-03 |
| O-sulfo-L-tyrosine | 987 | + | 6.8E-05 | 922 | + | 1.4E-02 |
A positive direction in eGFRcr change denotes a decline of kidney function over time per unit increase in metabolite concentration and a positive association with incident CKD with higher odds of CKD. For the analysis of incident CKD, patients with CKD at the S4 visit had been excluded (see the Concise Methods). Pairwise metabolite Pearson’s correlation coefficients in the eGFRcr change sample were 0.74 (C-mannosyl-tryptophan, pseudouridine), 0.41 (C-mannosyltryptophan, O-sulfo-L-tyrosine), and 0.46 (pseudouridine, O-sulfo-L-tyrosine). Mean eGFRcr among those who developed incident CKD was 73.4 (SD 9.5) at the S4 visit and 51.7 (SD 8.4) at the F4 follow-up visit.
Highlighted Markers Are Less Age and Sex Dependent than Serum Creatinine
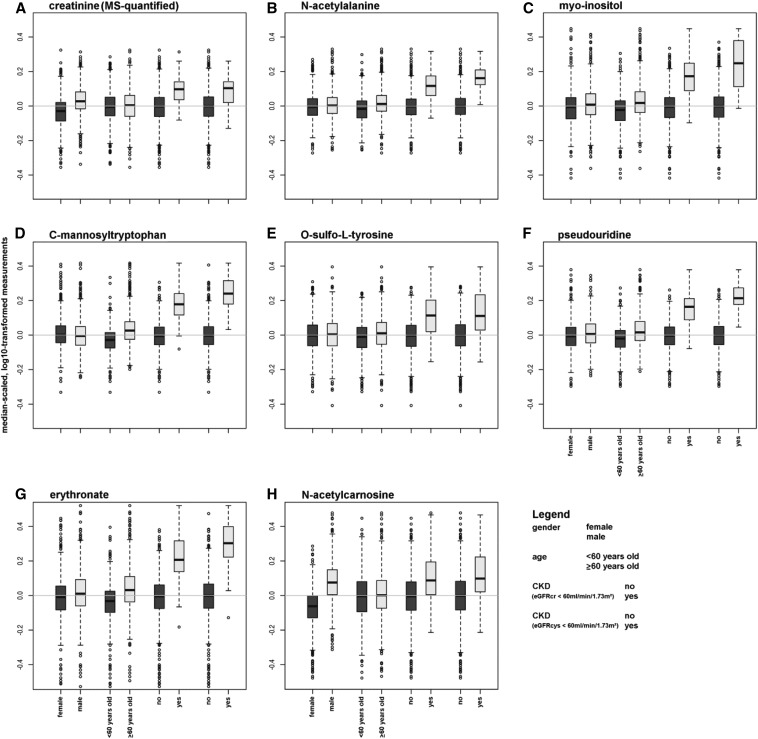
Because the novel eGFRcr-associated metabolites in Tables 2 and 3 represent potentially promising new kidney function markers, we evaluated their properties in more detail. Supplemental Table 5 contains detailed information on their distribution across strata of CKD defined by serum creatinine (quantified via standard assay) and by cystatin C, as well as important clinical CKD risk factors including male sex, older age, obesity, diabetes, hypertension, and smoking. CKD status had by far the most influence on differences in median metabolite concentrations. Figure 1 gives a graphical representation of the distribution of the metabolite concentration across strata of CKD, as well as of age and sex as established major determinants of serum creatinine concentrations. The difference in metabolite concentrations for individuals with and without CKD was most pronounced for erythronate, pseudouridine, and C-mannosyltryptophan, suggesting that the semiquantitative determination of metabolite abundance is sufficient to identify kidney function–related markers. Clear differences by sex were observed for concentrations of MS-quantified creatinine and N-acetylcarnosine, consistent with their high content in muscle tissue. C-mannosyltryptophan and erythronate showed higher median concentrations in old age compared with the other markers.
Figure 1.
Distribution of eGFR-associated markers across strata of CKD and its major risk factors. (A) Creatinine (MS-quantified). (B) N-acetylalanine. (C) Myo-inositol. (D) C-mannosyltryptophan. (E) O-sulfo-L-tyrosine. (F) Pseudouridine. (G) Erythronate. (H) N-acetylcarnosine. The two box plots on the right of each panel represent strata of cystatin-C based CKD and show that especially concentrations of erythronate, pseudouridine, and C-mannosyltryptophan differ more strongly by CKD status than does serum creatinine. Differences in the dependencies on sex and age can be observed for different metabolites.
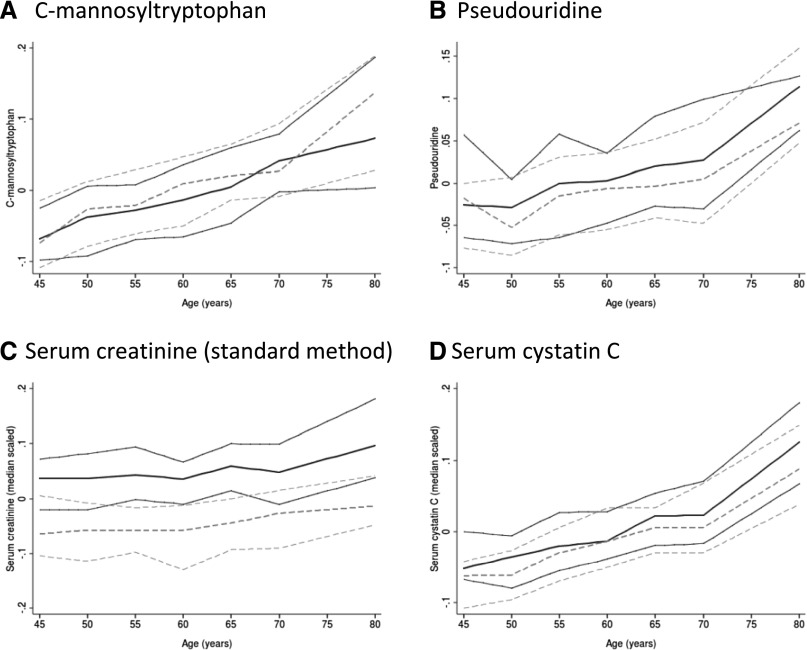
Because C-mannosyltryptophan and pseudouridine were associated with both eGFRcr and annual eGFRcr decline, we examined their distributions more closely. Figure 2 shows the concentrations of serum C-mannosyltryptophan, pseudouridine, serum creatinine (standard method), and cystatin C as a function of age and sex. As opposed to the other markers, serum creatinine concentrations increase less with advanced age and they are clearly higher in men than in women, suggesting that accounting for the effect of age but not sex would be important when considering whether to incorporate either C-mannosyltryptophan or pseudouridine into GFR-estimating equations.
Figure 2.
Comparison of serum C-mannosyltryptophan and pseudouridine with serum creatinine and cystatin C over age by sex. (A) C-mannosyltryptophan. (B) Pseudouridine. (C) Serum creatinine (standard method). (D) Serum cystatin C. Results are shown with solid lines for men and dashed lines for women. The three lines correspond to the median and 25th and 75th percentiles. Creatinine and cystatin C are transformed (median-scaled, log10 transformed) similarly to the MS-quantified metabolites to facilitate comparison. In contrast to other markers, serum creatinine concentrations are clearly higher in men than in women. They increase less with advanced age, when muscle mass typically declines.
C-Mannosyltryptophan and Pseudouridine Associate with Measured GFR
To further explore whether C-mannosyltryptophan or pseudouridine may represent potential novel filtration markers, we examined their concentrations in 200 participants in the African American Study of Kidney Disease and Hypertension (AASK) with three consecutive measures of GFR (mGFR; see the Concise Methods). The adjusted partial correlations of both markers with mGFR were −0.78, slightly lower than the corresponding correlations with eGFR (Table 4). Per doubling of metabolite concentration, both metabolites showed highly significant associations with incident ESRD, with hazard ratios of 11.4 (95% confidence interval [95% CI], 5.9 to 22.2) for C-mannosyltryptophan and 11.2 (95% CI, 5.3 to 23.7) for pseudouridine. Both associations were greatly attenuated when adjusting for eGFRcr at study baseline and were abolished completely when adjusting for mGFR (Table 4).
Table 4.
Correlation of C-mannosyltryptophan and pseudouridine with measured GFR and their association with ESRD in the AASK study
| C-Mannosyltryptophan | Pseudouridine | |||||||
|---|---|---|---|---|---|---|---|---|
| Correlation | P Value | Correlation | P Value | |||||
| Partial correlation with eGFR, r | -0.718 | 1.9E-31 | -0.714 | 7.0E-31 | ||||
| Partial correlation with mGFR, r | -0.783 | 1.2E-40 | -0.773 | 5.6E-39 | ||||
| N | HR | 95% CI | P Value | N | HR | 95% CI | P Value | |
|---|---|---|---|---|---|---|---|---|
| Association with incident ESRD | ||||||||
| Model 1: adjusted for age, sex, BP, treatment, and proteinuria | 199 (32) | 11.4 | (5.87 to 22.2) | 7.8E-13 | 199 (32) | 11.2 | (5.27 to 23.7) | 3.2E-10 |
| Model 1 plus adjustment for eGFRcr | 191 (31) | 3.39 | (1.25 to 9.19) | 0.02 | 191 (31) | 2.32 | (0.83 to 6.45) | 0.11 |
| Model 1 plus adjustment for mGFR | 199 (32) | 1.64 | (0.48 to 5.60) | 0.43 | 199 (32) | 0.77 | (0.21 to 2.78) | 0.69 |
Data are presented as n (number of events) and hazard ratios (95% CIs), unless otherwise indicated. Partial correlations were adjusted for age and sex. Sample sizes for individuals with complete covariate information and number of ESRD events are listed for each regression model. HR is per 2-fold increase in the metabolite. For comparison, corresponding correlations with mGFR were −0.784 for creatinine (Jaffe, P=7.4e-41) and −0.864 for cystatin C (P=8.1e-57).
Prediction of CKD
We also assessed whether a broad panel of metabolites could improve the prediction of incident CKD beyond that achieved with known clinical risk factors using unbiased variable selection (see the Concise Methods). For a model containing clinical variables only, age, systolic BP, antihypertensive medication use, smoking, and HDL were selected in addition to eGFRcr at the KORA S4 visit. For a model containing metabolites only, 19 metabolites were selected using a boosting approach (see the Concise Methods) and are listed in Supplemental Table 6. Finally, six metabolites were selected, including C-mannosyltryptophan, pseudouridine, and N-acetylornithine, for a model combining the selected clinical covariates as well as metabolites (Supplemental Table 6). The predictive ability for incident CKD, as measured by the area under the curve, was 0.82 (95% CI, 0.77 to 0.86) for the metabolites alone, 0.83 (95% CI, 0.79 to 0.87) for known clinical CKD risk factors alone, and 0.84 (95% CI, 0.80 to 0.88) for a model incorporating a combination of metabolites and clinical parameters (Supplemental Figure 2). The integrated discrimination improvement of 0.05 was not statistically significant (P=0.52). In addition, models with metabolites were less well calibrated, even after recalibration (Hosmer–Lemeshow test: P<0.05).
Genetic Associations for Highlighted Markers of eGFRcr
Finally, we queried the seven kidney function–associated metabolites in Tables 2 and 3 for their association with genome-wide genetic markers in an existing database to which the KORA study contributed,14 as well as in publicly available data on the association between the genetic markers and kidney function from the CKDGen Consortium,15,16 to gain further insights into factors influencing their serum concentrations beyond kidney function. Supplemental Table 7 displays all genetic variants that showed a significant association (P<7.1×10−9 or 5.0×10−8/7) with any of these metabolites. In several instances, the metabolite matched the function of the gene that contained metabolite-associated variants. For example, genetic variants in ISYNA1, encoding a protein important in myo-inositol synthesis, and in SLC5A11, encoding an inositol transporter, were significantly associated with myo-inositol concentrations. Thus, the concentrations of this biomarker are associated not only with kidney function but also with genetic variation related to its generation and handling. When evaluating the association between the genetic variants and eGFRcr, only rs6804368 in GADL1 showed a significant association with eGFRcr after correction for multiple testing (P<6.25×10−3 or 0.05/8). The associations of the same allele with higher concentrations of N-acetylcarnosine and lower eGFRcr complement the inverse association between N-acetylcarnosine and eGFRcr observed in our screen. The lack of association between the other genetic variants and eGFRcr is another piece of evidence that these metabolites may represent kidney function markers, rather than causing kidney disease.
Discussion
In this study of serum metabolites quantified using nontargeted metabolomics, we identified six metabolites that were reproducibly associated with eGFR and CKD in independent studies and showed high correlation with established kidney function markers. C-mannosyltryptophan and pseudouridine emerged as particularly interesting markers of kidney function that were highly correlated with measured GFR and showed less dependence on sex than serum creatinine, the marker currently most widely used to estimate GFR.
In Light of the Current Metabolomics Literature
We applied a nontargeted metabolomics approach, aimed at a broad coverage of the human metabolome. We previously investigated the association between kidney function and serum metabolites quantified using a lipid-focused targeted approach (Biocrates AbsoluteIDQ p150 and p180 assays) in the KORA and TwinsUK studies. We found that higher concentrations of multiple acylcarnitines were significantly associated with lower eGFR.6 Although targeted, absolute quantitative metabolomics techniques are generally more precise and accurate, they can only examine a predefined set of metabolites. The metabolite with the strongest association reported in our previous study, glutaroylcarnitine, was also identified in this project as strongly associated with eGFRcr and CKD, thus supporting the view that the nontargeted metabolomics approach used here has sufficient power in order to replicate previous findings from a targeted assay, while providing access to a much wider metabolite spectrum. The reported lack of a significant association between glutaroylcarnitine and kidney function decline7 could be confirmed in this study, indicating that acylcarnitines such as glutaroylcarnitine might be markers of kidney function rather than etiologic factors in CKD development or progression.
Other studies have identified kidney function– or disease–associated metabolites that were quantified using the same nontargeted metabolomics approach used here. Niewczas et al. conducted a small study of 80 individuals with diabetes and found 16 metabolites associated with incident ESRD, including myo-inositol, erythronate, pseudouridine, C-mannosyltryptophan, and glutaroylcarnitine.9 Our study extends these findings to show altered concentrations of these metabolites already at early stages of reduced kidney function in general population samples with a low prevalence of diabetes, supporting their potential use not only as progression markers of advanced kidney disease but also as early markers of reduced kidney function. Yu et al. conducted a population-based study among African-American participants in the Atherosclerosis Risk in Communities Study cohort.10 The authors reported some of the same markers to be associated with eGFR, including N-acetylalanine, which our study shows to be generalizable to European populations. They also highlighted but did not replicate associations between concentrations of 5-oxoproline and 1,5-anhydroglucitol with incident CKD among African Americans, neither of which was significantly associated with eGFR decline or incident CKD in our study. A potential explanation for this observation, other than or in addition to differences in statistical modeling, could be the presence of population-specific factors that influence these associations or the low prevalence of diabetes in our sample, because both markers have been linked to abnormal glucose metabolism.17
Biologic Mechanisms
A common theme among many of the associated metabolites identified here (N-acetylalanine, N-acetylcarnosine, C-mannosyltryptophan, erythronate, pseudouridine, and O-sulfo-L-tyrosine) is that they have all been linked to post-transcriptional or post-translational modifications. Although it is known that certain post-translational protein modifications such as carbamylation accumulate in the setting of renal failure and have been proposed to possess pathogenic properties,18 it is unclear whether the observed accumulation of metabolites that indicate specific post-translational modifications in the setting of a population-based study occurs as a consequence of an early decrease in kidney function. Altered renal handling of such metabolites in the setting of reduced GFR or a role in the development and progression of kidney disease, such as has been described for advanced glycation end products,19 are alternative explanations.
The C-glycosylation found in both C-mannosyltryptophan and pseudouridine is a glycosylation rarely observed in humans.20 While the sugar molecules are linked via oxygen or nitrogen to another molecule such as an amino acid or a nucleoside in most glycosylation products, C-glycosylic compounds connect the molecules in a C-C bond. C-glycosylation as a novel type of protein glycosylation has first been described for C-mannosylation of Trp-7 in human ribonuclease 2.21 C-2 mannosylation of tryptophan residues represents a novel enzymatic pathway in tryptophan metabolism in humans.22 C-mannosyltryptophan is found in human serum, urine, and cerebrospinal fluid as well as in various food products.22 It has been postulated as a marker of kidney function in individuals with kidney disease.23 The association between C-mannosyltryptophan and annual eGFR decline even after adjusting for baseline eGFRcr and clinical CKD risk factors may point toward an etiologic role in CKD progression. Alternatively, it may be a more sensitive kidney function marker compared with serum creatinine in early phases of CKD, when GFR is only mildly impaired and serum creatinine is still unchanged. In support of the latter hypothesis, Yonemura et al. documented a higher sensitivity of C-mannosyltryptophan compared with creatinine for the detection of normal renal function.24 Moreover, the high correlation of C-mannosyltryptophan with both mGFR and eGFRcr as well as the abolished association between C-mannosyltryptophan and incident ESRD upon inclusion of mGFR as a covariate in the AASK study further support C-mannosyltryptophan as a filtration marker.
Pseudouridine is the C-glycosidic derivative of uridine, a modified nucleoside found in RNA. Pseudouridine concentrations have long been recognized as elevated in uremia in studies of smaller sample sizes,25–28 but our study provides the first replicated association between pseudouridine concentrations and reduced kidney function and its progression in individuals from the general population. Similar to C-mannosyltryptophan, the high correlation of pseudouridine with mGFR and eGFRcr highlights its potential as an additional marker for the estimation of GFR.
Erythronic acid is an organic acid thought to be a breakdown product of glycated proteins or ascorbic acid, specifically in patients with diabetes.29 Erythronic acid is also a prominent urinary biomarker of transaldolase deficiency, a rare inherited inborn error of metabolism, which also features kidney disease.30,31
Two metabolites highlighted in our study, N-acetylalanine and N-acetylcarnosine, contain an N-acetylation as a post-translational modification.32 Higher concentrations of these metabolites were associated with lower eGFR. Previous studies have reported an association between genetic variants in NAT8 and both kidney disease15,33 and concentrations of N-acetylornithine.34,35 Likewise, we observed an association between a genetic variant in GADL1 and both higher concentrations of N-acetylcarnosine and lower eGFR. While not establishing causality, these combined findings justify the further study of the role of N-acetylation, an important mechanism for detoxification, in kidney disease.
Myo-inositol is a polyol produced from glucose-6-phosphate and serves as an important precursor of inositol phosphates, important intracellular second messengers, and as a component of phosphatidylinositols. The kidney is the most important organ in humans both for the synthesis as well as catabolism of myo-inositol.36 High blood concentrations of myo-inositol were found to associate with progression to ESRD among individuals with diabetes.9 In our study of population-based individuals, myo-inositol concentrations were associated with eGFR but not incident CKD, suggesting that its association with reduced kidney function is a reflection of the altered ability of the kidney to degrade or excrete myo-inositol rather than supporting it as a marker causally involved in the pathogenesis of CKD.
The only marker that showed a significant association with kidney function decline over time, but was not highly correlated with established kidney function markers cross-sectionally, was O-sulfo-L-tyrosine. Such a pattern of association may be expected from a marker that is related to the development and/or progression of kidney disease. Secreted and transmembrane proteins can contain O-sulfated tyrosine residues, where the transfer of sulfate to tyrosine residues is a post-translational modification catalyzed by tyrosylprotein sulfotransferases.37
Clinical Relevance and Future Work
Our results show that concentrations of several serum metabolites, most notably C-mannosyltryptophan and pseudouridine, discriminate individuals in early stages of cystatin C–based CKD at least as well as serum creatinine, with the advantage of being less dependent on sex. If the new markers described here, or a combination thereof, are more sensitive markers of early changes in kidney function compared with serum creatinine, they may be able to add information to already established kidney function markers in order to identify individuals prone to kidney function decline and CKD. Although the metabolites did not improve risk prediction for incident CKD above and beyond well known clinical risk factors in the general population, semiquantitative measurements of serum metabolites agnostic to the presence of clinical risk factors predicted risk equally well. A limitation of our study is that our approach does not provide absolute quantification of these markers; therefore, future studies utilizing targeted, absolute quantitative assays are needed to verify the reported associations and to establish reference ranges.
In conclusion, the metabolites C-mannosyltryptophan and pseudouridine are strongly and reproducibly associated with eGFR and CKD in population-based studies, are highly correlated with measured GFR in patients with CKD, and show favorable properties such as less dependence on sex compared with the established kidney function marker serum creatinine. Additional studies are required to establish absolute concentrations of the highlighted metabolites and to evaluate whether they represent kidney function markers whose serum concentrations increase at early stages of reduced kidney function.
Concise Methods
Study Design and Participants
Subjects in this study participated in the Fourth Cooperative Health Research in the Region of Augsburg Survey (KORA S4) and were between the ages of 55 and 74 years at their baseline visit (S4 visit, 1999–2001). A subsequent follow-up visit was conducted from 2006 to 2008 (F4 visit) 7.1 years after the baseline visit, on average.
Measurements of 488 serum metabolites that passed quality control were available for 1735 individuals at the F4 visit. Of those individuals that attended both the S4 and the F4 visits, 991 participants had available information on serum creatinine (standard method, see below) at both visits and on covariates and 422 serum metabolites at the S4 visit. Standardized examinations, interviews, and laboratory tests conducted in the KORA study were previously described in detail.38
The TwinsUK study is an adult twin registry: Unselected twins were recruited through national media campaigns in the United Kingdom. Participants were reported to be comparable to age-matched singleton members of the general population in disease-related and lifestyle characteristics.39 The 1164 participants for this study were selected from cross-sectional visits taking place between 1997 and 2008. Written informed consent was obtained from all participants. The studies were approved by their local ethics committees.
AASK was a two-by-three multicenter randomized clinical trial of BP control and specific agents (angiotensin-converting enzyme inhibitors versus metoprolol versus calcium channel blockers), as previously described. Participants were African-American men and women with hypertensive kidney disease and GFR between 20 and 65 ml/min per 1.73 m2.40,41 GFR was measured as urinary clearance of 125I-iothalamate.42 For our analysis, 200 AASK participants with consistent mGFR at the AASK 48-month follow-up visit (previous and next semiannual visits within 25% of the 48-month mGFR) were selected for metabolite quantification of samples collected at the 48-month follow-up visit. Characteristics of 188 individuals with data on the two candidate metabolites and serum creatinine measurements are provided in Supplemental Table 8.
Measurement of Metabolites
Serum metabolites from KORA and TwinsUK participants were quantified at Metabolon Inc. (Durham, NC) using a nontargeted metabolomics gas and liquid chromatography coupled to mass spectrometry (GC/MS and LC/MS, respectively) approach. Details of the methods applied for quantification and identification of metabolites were reported previously.43,44 Briefly, sample preparation was performed on a Hamilton MLStar (Hamilton Company, Salt Lake City, UT) robotics system: After thawing, 400 µl of extraction solvent (methanol containing recovery standards) was added to each 100 µl of serum samples in a 96-well plate format. Extraction was carried out by shaking for 2 minutes using a Geno/Grinder 2000 (Glen Mills Inc., Clifton, NJ). After centrifugation, the supernatant was split into four aliquots: two for LC/MS analysis (positive and negative electrospray ionization [ESI] mode), one for GC/MS analysis, and one reserve aliquot. Solvent was removed on a TurboVap (Zymark) and the samples were dried under vacuum overnight. Samples were reconstituted with 0.1% formic acid for LC/MS positive ion mode and 6.5 mM ammonium bicarbonate (pH 8.0) for negative ion mode. Both reconstitution solvents contained also internal standards. The GC/MS aliquots were derivatized for 1 hour at 60°C with N,O-bistrimethylsilyl-trifluoroacetamide in a solvent mixture of acetonitrile/dichloromethane/cyclohexane (5:4:1), containing 5% triethylamine and retention time markers.
LC/MS analysis was performed on an LTQ mass spectrometer (Thermo Fisher Scientific, Waltham, MA) equipped with a Waters Acquity UPLC system (Waters Corporation, Milford, MA). Two separate columns (2.1×100 mm, BEH C18 1.7 µm particle; Waters Corporation) were used for acidic (solvent A: 0.1% formic acid in H2O; solvent B: 0.1% formic acid in methanol) and basic (solvent A: 6.5 mM ammonium bicarbonate, pH 8.0; and solvent B: 6.5 mM ammonium bicarbonate in 98% methanol) mobile phase conditions, optimized for positive and negative ESI, respectively. After injection of the sample extracts, the columns were developed in a gradient of 100% A to 98% B in 11-minute runtime at a 350 µl/min flow rate. The eluent flow was directly connected to the ESI source of the LTQ mass spectrometer. Full-scan mass spectra (mass-to-charge ratio [m/z] of 99–1000) and data-dependent MS/MS scans with dynamic exclusion were recorded in turns.
GC/MS analysis was done on a Thermo-Finnigan Trace DSQ fast-scanning single-quadrupole mass spectrometer, equipped with a 20 m × 0.18 mm GC column with a 0.18-µm film phase consisting of 5% phenyldimethylsilicone. Electron ionization at 70 eV was used and the column temperature was ramped between 60°C and 340°C with helium as the carrier gas. Mass spectra in a scan range from 50 to 750 m/z were recorded.
Metabolites were identified from the LC/MS and GC/MS data by semiautomated multiparametric comparison with a proprietary library, containing retention times, m/z ratios, and related adduct/fragment spectra. To account for differences in the times of sample collection and because metabolite content for untargeted metabolomics experiments can vary somewhat over time for technical reasons, we applied stringent and consistent data cleaning procedures separately for the two data sets. In total, 493 metabolites measured at S4 (n=427) and/or F4 (n=488) could be matched to metabolites in the library, 321 with known and 172 with thus far unknown chemical identity. The majority of metabolites (86%) were available in both data sets.
For every metabolite, the raw area counts were normalized to the median value of the run day to correct for interday variation of the measurements. After log10 transformation, outlier values were removed for each metabolite by setting values of >4 SDs from the mean of the respective metabolite to missing. Metabolites that were measured in <300 individuals were excluded before analyses.
Untargeted, GC/MS-, and LC/MS‐based serum metabolite quantification in the AASK study was performed using the same approach as in the KORA and TwinsUK studies (global profiling v3; Metabolon Inc.).45
Outcome Definition
The primary outcomes of the discovery study were eGFR from serum creatinine measured by a standard method (eGFRcr) and eGFRcr decline defined by the annual change in eGFRcr between the S4 and F4 visits.
Serum creatinine was measured using an enzymatic method at the S4 visit and the Jaffe method at the F4 visit46 and in the TwinsUK study.47 To account for differences in measurement methods, Jaffe creatinine values in the KORA F4 and TwinsUK studies were first calibrated to representative estimates derived from the Third National Health and Nutrition Examination Survey, using age- and sex-stratified groups as described previously.48 Next, standard creatinine was calculated for the F4 measurements as follows: standard creatinine = 0.95 × calibrated serum creatinine.49 Enzymatic measurements carried out at the S4 visit did not require any standardization. The eGFRcr was then calculated from standard creatinine using the Chronic Kidney Disease Epidemiology Collaboration equation in all studies.50 Annual eGFRcr change in the KORA study, in which 7 years of follow-up were available for most participants, was defined as the difference in eGFRcr between the KORA S4 and F4 visits divided by the time between visits in years. Because serum creatinine constitutes an important component of the evaluated outcome, the creatinine measurements obtained by MS measurement were not followed up further but are presented for comparison in the result tables.
In the AASK study, serum creatinine was measured using the Jaffe method and cystatin C using the Siemens nephelometry method; both assays are traceable to international standards, as has been previously described.4 ESRD was defined as beginning maintenance dialysis therapy or receipt of a kidney transplant.51 Over a median follow-up time of 6.75 years, 32 ESRD events were observed among the 200 patients.
Definition of Covariates
Ten known clinical CKD risk factors were assessed at the KORA study visits and used as covariates in the statistical analyses: age (years), sex, body mass index (kilograms per square meter), systolic BP (millimeters of mercury), current smoking status, antihypertensive medication use, antihyperlipidemic medication use excluding herbal medications, serum triglycerides (milligrams per deciliter), serum HDL cholesterol (milligrams per deciliter), and fasting serum glucose (milligrams per deciliter). The natural logarithm of triglycerides and glucose was used in the statistical analyses. In the TwinsUK study, information for many of the covariables was not available from the same time at which the blood draw for the serum metabolite measurements occurred. Therefore, only age, sex, and cohort and twins status, the most important covariates, were used for adjustment in the TwinsUK study.
Statistical Analyses
The statistical analysis comprised two parts: the analysis of cross-sectional and of longitudinal data. The statistical analysis plan of the cross-sectional study in KORA F4 is outlined in Supplemental Figure 1. Cross-sectional associations between eGFR and serum metabolites were examined in the KORA F4 study, the only sample with available cystatin C measurements. We calculated multivariable-adjusted linear regression models of eGFRcr and eGFRcys on the single metabolites as well as the pairwise metabolite ratios (corresponding to the subtraction of the log-transformed metabolite quantities) among all 488 metabolites of 1735 individuals available at the F4 visit. This resulted in a total of 115,476 unique ratios after excluding ratios that could be obtained in <300 individuals and those that contained creatinine in either the numerator or denominator. Significantly associated metabolites were tested for association with the clinical entity CKD, defined as eGFRcr <60 ml/min per 1.73 m2.52 After the discovery screen of the metabolite ratios, we additionally computed the P gain for each ratio as follows: (minimum[P numerator, P denominator])/P ratio, reflecting the order of magnitude by which the P value for association decreased compared with the lower P value of its individual components. A significant P gain was defined as >1,154,760 (correction for 10 times the number of tested ratios) as described previously.11 Only significantly associated ratios with a significant P gain are presented throughout this article.
For the prospective analyses, linear regression models and logistic regression models were used to identify associations between annual eGFRcr decline and incident CKD, respectively, and the 422 metabolites and 87,762 metabolite ratios available at the S4 visit. Necessary data were available for 991 participants. Incident CKD was defined as eGFRcr <60 ml/min per 1.73 m2 at the F4 visit among 926 individuals without CKD at the S4 visit. Baseline covariates were used for multivariable adjustment, including baseline eGFRcr as a known important risk factor for eGFR decline.53 To assess whether identified metabolites associated with eGFRcr decline and incident CKD in the KORA study, a combined model including identified metabolites and adjusting factors was assessed, to which backward elimination using Akaike information criterion was applied.
Statistical significance was defined as a P value less than a Bonferroni-corrected threshold adjusting for the number of metabolites or metabolite ratios available at the applicable visit. For the cross-sectional analyses at the F4 visit, significance was defined as P<1.0×10−4 (0.05/488) for individual metabolites and P<4.3×10−7 (0.05/115,476) with a significant P gain for ratios. For the analyses of annual eGFRcr decline, statistical significance was defined accordingly as P<1.2×10−4 (0.05/422) for individual metabolites and P<5.7×10−7 (0.05/87,762), with a P gain >877,620. To provide information related to false discovery rate, Q values were additionally estimated from the P values obtained for the cross-sectional analyses using the R package QVALUE.54
Replication of significant metabolite associations obtained in the cross-sectional study in KORA F4 was attempted in the TwinsUK study. Successful replication was defined as a Bonferroni-corrected one-sided P value, namely P<4.9×10−4 (0.05/103). Subsequently, the estimates of replicated metabolites were meta-analyzed across the KORA and TwinsUK studies using a sample size–weighted fixed-effects model.
In addition, the predictive value of metabolites for incident CKD (KORA F4) was assessed using an imputed data set of patients without CKD at KORA S4. For imputation, metabolites with >20% missing values and samples with missing values for >10% of the remaining metabolites based on the complete S4 metabolomics data set were excluded. Missing values were imputed using the R package MICE with default settings (five imputations applying predictive mean matching), such that the data set used for this analysis comprised 914 patients and 363 metabolites. Three different prediction models were considered to evaluate the performance of metabolites either as alternative or as complementary information to known clinical risk factors: (1) a model based on clinical information only including known CKD risk factors (clinical model), (2) a model based on metabolites alone (metabolomic model), and (3) a model based on both sources of information (combined model). The derivation of the logistic regression models as well as the assessment of their predictive performance was done as previously described.7 Briefly, factors of the clinical model were unbiasedly selected from the set of available covariates using backward elimination while metabolites were selected out of 363 successfully imputed metabolites with a boosting approach.55 For each model, the area under the receiver operating curve was calculated together with a 95% CI and validated using a bootstrap approach (without replacement).56–58 In addition, calibration was checked using the Hosmer–Lemeshow test.58 To assess the added predictive ability in discrimination of the combined model compared with the clinical model, the integrated discrimination improvement was calculated.59 In addition to incident CKD (93 events), we also evaluated the more stringent outcome of incident CKD plus a >10 ml/min per 1.73 m2 decrease in eGFRcr between both visits (77 events) in a sensitivity analysis, with similar results (data not shown).
Statistical analyses were conducted using STATA (version 11.2, Special Edition; StataCorp LP, College Station, TX) and R (version 2.11.1, www.r-project.org; R Foundation for Statistical Computing) software.
In the AASK study, log-transformed mGFR defined as the average of the mGFR measurements at the 42-, 48-, and 54-month follow-up visits was correlated to log-transformed metabolite concentrations, adjusted for age and sex. Cox proportional hazards regression was used to relate log-transformed metabolites to time to ESRD, without and with adjustment for covariates including age, sex, BP, treatment and proteinuria (model 1). Model 1 was subsequently extended by either eGFRcr or mGFR.
Disclosures
A.S.L., L.A.I., and J.C. have applied for a patent for precise estimation of GFR using a panel of metabolomic filtration markers. R.P.M. is employed by Metabolon, Inc., and he contributed to the logistics and optimization of MS and to MS data interpretation. Metabolon Inc. was not involved in the design of the study, statistical analyses, or interpretation of the results.
Supplementary Material
Acknowledgments
We gratefully acknowledge the contribution of all members of field staffs who were involved in planning and conducting the KORA Augsburg studies. We thank all participants in the KORA S4 and F4 studies for their donation of blood and time. We also thank the participants in the TwinsUK study, and the staff involved in recruitment, laboratory work, and data management. Finally, we thank Harald Binder for support with the boosting analyses and Yingying Sang for analytical support with the AASK study data.
This study was supported in part by grants from the German Federal Ministry of Education and Research (BMBF) to the German Center Diabetes Research and from the European Commission Seventh Framework Programme (EurHEALTHAging Small-Scale Focused Research Collaborative Project 277849). The KORA research platform was initiated and financed by the Helmholtz Zentrum München, German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education, Science, Research, and Technology and by the state of Bavaria. The TwinsUK study was funded by the Wellcome Trust and the European Commission Seventh Framework Programme (FP7/2007-2013). The study also receives support from the National Institute for Health Research–funded BioResource Clinical Research Facility and Biomedical Research Centre based at Guy’s and St. Thomas’ National Health Service Foundation Trust, in partnership with King’s College London. Metabolomic analysis in the TwinsUK study was funded by Pfizer. The work of P.S., O.-N.G., and A.K. was funded by the Emmy Noether Program of the German Research Foundation (KO 3598/2-1 to A.K.) and the BMBF (Gerontosys II NephAge Project, 031 5896 A). C.B. is supported by a grant from the Spanish Society of Nephrology. W.R.-M. is supported by funds from the Helmholtz Association Cross Program Initiative in Individualized Medicine. J.K. is supported by a grant from the Helmholtz Postdoctoral Programme Initiative and Networking Fund. K.S. was supported by Biomedical Research Program funds at Weill Cornell Medical College in Qatar, a program funded by the Qatar Foundation. The work of J.C., L.A.I., and A.S.L. was supported by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases (R01-DK097020).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2014111099/-/DCSupplemental.
References
- 1.Eckardt KU, Coresh J, Devuyst O, Johnson RJ, Köttgen A, Levey AS, Levin A: Evolving importance of kidney disease: From subspecialty to global health burden. Lancet 382: 158–169, 2013 [DOI] [PubMed] [Google Scholar]
- 2.Gansevoort RT, Correa-Rotter R, Hemmelgarn BR, Jafar TH, Heerspink HJ, Mann JF, Matsushita K, Wen CP: Chronic kidney disease and cardiovascular risk: Epidemiology, mechanisms, and prevention. Lancet 382: 339–352, 2013 [DOI] [PubMed] [Google Scholar]
- 3.Stevens LA, Coresh J, Greene T, Levey AS: Assessing kidney function--measured and estimated glomerular filtration rate. N Engl J Med 354: 2473–2483, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Inker LA, Schmid CH, Tighiouart H, Eckfeldt JH, Feldman HI, Greene T, Kusek JW, Manzi J, Van Lente F, Zhang YL, Coresh J, Levey AS, CKD-EPI Investigators : Estimating glomerular filtration rate from serum creatinine and cystatin C. N Engl J Med 367: 20–29, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shlipak MG, Matsushita K, Ärnlöv J, Inker LA, Katz R, Polkinghorne KR, Rothenbacher D, Sarnak MJ, Astor BC, Coresh J, Levey AS, Gansevoort RT, CKD Prognosis Consortium : Cystatin C versus creatinine in determining risk based on kidney function. N Engl J Med 369: 932–943, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goek ON, Döring A, Gieger C, Heier M, Koenig W, Prehn C, Römisch-Margl W, Wang-Sattler R, Illig T, Suhre K, Sekula P, Zhai G, Adamski J, Köttgen A, Meisinger C: Serum metabolite concentrations and decreased GFR in the general population. Am J Kidney Dis 60: 197–206, 2012 [DOI] [PubMed] [Google Scholar]
- 7.Goek ON, Prehn C, Sekula P, Römisch-Margl W, Döring A, Gieger C, Heier M, Koenig W, Wang-Sattler R, Illig T, Suhre K, Adamski J, Köttgen A, Meisinger C: Metabolites associate with kidney function decline and incident chronic kidney disease in the general population. Nephrol Dial Transplant 28: 2131–2138, 2013 [DOI] [PubMed] [Google Scholar]
- 8.Rhee EP, Clish CB, Ghorbani A, Larson MG, Elmariah S, McCabe E, Yang Q, Cheng S, Pierce K, Deik A, Souza AL, Farrell L, Domos C, Yeh RW, Palacios I, Rosenfield K, Vasan RS, Florez JC, Wang TJ, Fox CS, Gerszten RE: A combined epidemiologic and metabolomic approach improves CKD prediction. J Am Soc Nephrol 24: 1330–1338, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niewczas MA, Sirich TL, Mathew AV, Skupien J, Mohney RP, Warram JH, Smiles A, Huang X, Walker W, Byun J, Karoly ED, Kensicki EM, Berry GT, Bonventre JV, Pennathur S, Meyer TW, Krolewski AS: Uremic solutes and risk of end-stage renal disease in type 2 diabetes: Metabolomic study. Kidney Int 85: 1214–1224, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu B, Zheng Y, Nettleton JA, Alexander D, Coresh J, Boerwinkle E: Serum metabolomic profiling and incident CKD among African Americans. Clin J Am Soc Nephrol 9: 1410–1417, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersen AK, Krumsiek J, Wägele B, Theis FJ, Wichmann HE, Gieger C, Suhre K: On the hypothesis-free testing of metabolite ratios in genome-wide and metabolome-wide association studies. BMC Bioinformatics 13: 120, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krumsiek J, Suhre K, Evans AM, Mitchell MW, Mohney RP, Milburn MV, Wägele B, Römisch-Margl W, Illig T, Adamski J, Gieger C, Theis FJ, Kastenmüller G: Mining the unknown: A systems approach to metabolite identification combining genetic and metabolic information. PLoS Genet 8: e1003005, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutkowski P, Słominska EM, Szołkiewicz M, Aleksandrowicz E, Smolenski RT, Wołyniec W, Renke M, Wisterowicz K, Swierczynski J, Rutkowski B: Relationship between uremic toxins and oxidative stress in patients with chronic renal failure. Scand J Urol Nephrol 41: 243–248, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang TP, Walter K, Menni C, Chen L, Vasquez L, Valdes AM, Hyde CL, Wang V, Ziemek D, Roberts P, Xi L, Grundberg E, Waldenberger M, Richards JB, Mohney RP, Milburn MV, John SL, Trimmer J, Theis FJ, Overington JP, Suhre K, Brosnan MJ, Gieger C, Kastenmüller G, Spector TD, Soranzo N, Multiple Tissue Human Expression Resource (MuTHER) Consortium : An atlas of genetic influences on human blood metabolites. Nat Genet 46: 543–550, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Köttgen A, Pattaro C, Böger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV, O’Connell JR, Li M, Schmidt H, Tanaka T, Isaacs A, Ketkar S, Hwang SJ, Johnson AD, Dehghan A, Teumer A, Paré G, Atkinson EJ, Zeller T, Lohman K, Cornelis MC, Probst-Hensch NM, Kronenberg F, Tönjes A, Hayward C, Aspelund T, Eiriksdottir G, Launer LJ, Harris TB, Rampersaud E, Mitchell BD, Arking DE, Boerwinkle E, Struchalin M, Cavalieri M, Singleton A, Giallauria F, Metter J, de Boer IH, Haritunians T, Lumley T, Siscovick D, Psaty BM, Zillikens MC, Oostra BA, Feitosa M, Province M, de Andrade M, Turner ST, Schillert A, Ziegler A, Wild PS, Schnabel RB, Wilde S, Munzel TF, Leak TS, Illig T, Klopp N, Meisinger C, Wichmann HE, Koenig W, Zgaga L, Zemunik T, Kolcic I, Minelli C, Hu FB, Johansson A, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Schreiber S, Aulchenko YS, Felix JF, Rivadeneira F, Uitterlinden AG, Hofman A, Imboden M, Nitsch D, Brandstätter A, Kollerits B, Kedenko L, Mägi R, Stumvoll M, Kovacs P, Boban M, Campbell S, Endlich K, Völzke H, Kroemer HK, Nauck M, Völker U, Polasek O, Vitart V, Badola S, Parker AN, Ridker PM, Kardia SL, Blankenberg S, Liu Y, Curhan GC, Franke A, Rochat T, Paulweber B, Prokopenko I, Wang W, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Shlipak MG, van Duijn CM, Borecki I, Krämer BK, Rudan I, Gyllensten U, Wilson JF, Witteman JC, Pramstaller PP, Rettig R, Hastie N, Chasman DI, Kao WH, Heid IM, Fox CS: New loci associated with kidney function and chronic kidney disease. Nat Genet 42: 376–384, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pattaro C, Köttgen A, Teumer A, Garnaas M, Böger CA, Fuchsberger C, Olden M, Chen MH, Tin A, Taliun D, Li M, Gao X, Gorski M, Yang Q, Hundertmark C, Foster MC, O’Seaghdha CM, Glazer N, Isaacs A, Liu CT, Smith AV, O’Connell JR, Struchalin M, Tanaka T, Li G, Johnson AD, Gierman HJ, Feitosa M, Hwang SJ, Atkinson EJ, Lohman K, Cornelis MC, Johansson Å, Tönjes A, Dehghan A, Chouraki V, Holliday EG, Sorice R, Kutalik Z, Lehtimäki T, Esko T, Deshmukh H, Ulivi S, Chu AY, Murgia F, Trompet S, Imboden M, Kollerits B, Pistis G, Harris TB, Launer LJ, Aspelund T, Eiriksdottir G, Mitchell BD, Boerwinkle E, Schmidt H, Cavalieri M, Rao M, Hu FB, Demirkan A, Oostra BA, de Andrade M, Turner ST, Ding J, Andrews JS, Freedman BI, Koenig W, Illig T, Döring A, Wichmann HE, Kolcic I, Zemunik T, Boban M, Minelli C, Wheeler HE, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Nöthlings U, Jacobs G, Biffar R, Endlich K, Ernst F, Homuth G, Kroemer HK, Nauck M, Stracke S, Völker U, Völzke H, Kovacs P, Stumvoll M, Mägi R, Hofman A, Uitterlinden AG, Rivadeneira F, Aulchenko YS, Polasek O, Hastie N, Vitart V, Helmer C, Wang JJ, Ruggiero D, Bergmann S, Kähönen M, Viikari J, Nikopensius T, Province M, Ketkar S, Colhoun H, Doney A, Robino A, Giulianini F, Krämer BK, Portas L, Ford I, Buckley BM, Adam M, Thun GA, Paulweber B, Haun M, Sala C, Metzger M, Mitchell P, Ciullo M, Kim SK, Vollenweider P, Raitakari O, Metspalu A, Palmer C, Gasparini P, Pirastu M, Jukema JW, Probst-Hensch NM, Kronenberg F, Toniolo D, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Siscovick DS, van Duijn CM, Borecki I, Kardia SL, Liu Y, Curhan GC, Rudan I, Gyllensten U, Wilson JF, Franke A, Pramstaller PP, Rettig R, Prokopenko I, Witteman JC, Hayward C, Ridker P, Parsa A, Bochud M, Heid IM, Goessling W, Chasman DI, Kao WH, Fox CS, CARDIoGRAM Consortium. ICBP Consortium. CARe Consortium. Wellcome Trust Case Control Consortium 2 (WTCCC2) : Genome-wide association and functional follow-up reveals new loci for kidney function. PLoS Genet 8: e1002584, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu J, Zhou J, Bao Y, Chen T, Zhang Y, Zhao A, Qiu Y, Xie G, Wang C, Jia W, Jia W: Serum metabolic signatures of fulminant type 1 diabetes. J Proteome Res 11: 4705–4711, 2012 [DOI] [PubMed] [Google Scholar]
- 18.Gillery P, Jaisson S: Post-translational modification derived products (PTMDPs): Toxins in chronic diseases? Clin Chem Lab Med 52: 33–38, 2014 [DOI] [PubMed] [Google Scholar]
- 19.D’Agati V, Schmidt AM: RAGE and the pathogenesis of chronic kidney disease. Nat Rev Nephrol 6: 352–360, 2010 [DOI] [PubMed] [Google Scholar]
- 20.Furmanek A, Hofsteenge J: Protein C-mannosylation: Facts and questions. Acta Biochim Pol 47: 781–789, 2000 [PubMed] [Google Scholar]
- 21.Doucey MA, Hess D, Cacan R, Hofsteenge J: Protein C-mannosylation is enzyme-catalysed and uses dolichyl-phosphate-mannose as a precursor. Mol Biol Cell 9: 291–300, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutsche B, Grun C, Scheutzow D, Herderich M: Tryptophan glycoconjugates in food and human urine. Biochem J 343: 11–19, 1999 [PMC free article] [PubMed] [Google Scholar]
- 23.Takahira R, Yonemura K, Yonekawa O, Iwahara K, Kanno T, Fujise Y, Hishida A: Tryptophan glycoconjugate as a novel marker of renal function. Am J Med 110: 192–197, 2001 [DOI] [PubMed] [Google Scholar]
- 24.Yonemura K, Takahira R, Yonekawa O, Wada N, Hishida A: The diagnostic value of serum concentrations of 2-(alpha-mannopyranosyl)-L-tryptophan for normal renal function. Kidney Int 65: 1395–1399, 2004 [DOI] [PubMed] [Google Scholar]
- 25.Asatoor AM: Retention of pseudouridine and 4-amino-5-imidazole carboxamide in uraemia. Clin Chim Acta 20: 407–411, 1968 [DOI] [PubMed] [Google Scholar]
- 26.Schoots AC, Gerlag PG, Mulder AW, Peeters JA, Cramers CA: Liquid-chromatographic profiling of solutes in serum of uremic patients undergoing hemodialysis and chronic ambulatory peritoneal dialysis (CAPD); high concentrations of pseudouridine in CAPD patients. Clin Chem 34: 91–97, 1988 [PubMed] [Google Scholar]
- 27.Niwa T, Takeda N, Yoshizumi H: RNA metabolism in uremic patients: Accumulation of modified ribonucleosides in uremic serum. Technical note. Kidney Int 53: 1801–1806, 1998 [DOI] [PubMed] [Google Scholar]
- 28.Dzúrik R, Lajdová I, Spustová V, Opatrný K, Jr: Pseudouridine excretion in healthy subjects and its accumulation in renal failure. Nephron 61: 64–67, 1992 [DOI] [PubMed] [Google Scholar]
- 29.Schleicher ED, Wagner E, Nerlich AG: Increased accumulation of the glycoxidation product N(epsilon)-(carboxymethyl)lysine in human tissues in diabetes and aging. J Clin Invest 99: 457–468, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engelke UF, Zijlstra FS, Mochel F, Valayannopoulos V, Rabier D, Kluijtmans LA, Perl A, Verhoeven-Duif NM, de Lonlay P, Wamelink MM, Jakobs C, Morava E, Wevers RA: Mitochondrial involvement and erythronic acid as a novel biomarker in transaldolase deficiency. Biochim Biophys Acta 1802: 1028–1035, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loeffen YG, Biebuyck N, Wamelink MM, Jakobs C, Mulder MF, Tylki-Szymańska A, Fung CW, Valayannopoulos V, Bökenkamp A: Nephrological abnormalities in patients with transaldolase deficiency. Nephrol Dial Transplant 27: 3224–3227, 2012 [DOI] [PubMed] [Google Scholar]
- 32.Karve TM, Cheema AK: Small changes huge impact: The role of protein posttranslational modifications in cellular homeostasis and disease. J Amino Acids 2011: 207691, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chambers JC, Zhang W, Lord GM, van der Harst P, Lawlor DA, Sehmi JS, Gale DP, Wass MN, Ahmadi KR, Bakker SJ, Beckmann J, Bilo HJ, Bochud M, Brown MJ, Caulfield MJ, Connell JM, Cook HT, Cotlarciuc I, Davey Smith G, de Silva R, Deng G, Devuyst O, Dikkeschei LD, Dimkovic N, Dockrell M, Dominiczak A, Ebrahim S, Eggermann T, Farrall M, Ferrucci L, Floege J, Forouhi NG, Gansevoort RT, Han X, Hedblad B, Homan van der Heide JJ, Hepkema BG, Hernandez-Fuentes M, Hypponen E, Johnson T, de Jong PE, Kleefstra N, Lagou V, Lapsley M, Li Y, Loos RJ, Luan J, Luttropp K, Maréchal C, Melander O, Munroe PB, Nordfors L, Parsa A, Peltonen L, Penninx BW, Perucha E, Pouta A, Prokopenko I, Roderick PJ, Ruokonen A, Samani NJ, Sanna S, Schalling M, Schlessinger D, Schlieper G, Seelen MA, Shuldiner AR, Sjögren M, Smit JH, Snieder H, Soranzo N, Spector TD, Stenvinkel P, Sternberg MJ, Swaminathan R, Tanaka T, Ubink-Veltmaat LJ, Uda M, Vollenweider P, Wallace C, Waterworth D, Zerres K, Waeber G, Wareham NJ, Maxwell PH, McCarthy MI, Jarvelin MR, Mooser V, Abecasis GR, Lightstone L, Scott J, Navis G, Elliott P, Kooner JS: Genetic loci influencing kidney function and chronic kidney disease. Nat Genet 42: 373–375, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suhre K, Wallaschofski H, Raffler J, Friedrich N, Haring R, Michael K, Wasner C, Krebs A, Kronenberg F, Chang D, Meisinger C, Wichmann HE, Hoffmann W, Völzke H, Völker U, Teumer A, Biffar R, Kocher T, Felix SB, Illig T, Kroemer HK, Gieger C, Römisch-Margl W, Nauck M: A genome-wide association study of metabolic traits in human urine. Nat Genet 43: 565–569, 2011 [DOI] [PubMed] [Google Scholar]
- 35.Yu B, Zheng Y, Alexander D, Morrison AC, Coresh J, Boerwinkle E: Genetic determinants influencing human serum metabolome among African Americans. PLoS Genet 10: e1004212, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Croze ML, Soulage CO: Potential role and therapeutic interests of myo-inositol in metabolic diseases. Biochimie 95: 1811–1827, 2013 [DOI] [PubMed] [Google Scholar]
- 37.Hoffhines AJ, Damoc E, Bridges KG, Leary JA, Moore KL: Detection and purification of tyrosine-sulfated proteins using a novel anti-sulfotyrosine monoclonal antibody. J Biol Chem 281: 37877–37887, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wichmann HE, Gieger C, Illig T, MONICA/KORA Study Group : KORA-gen--resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen 67[Suppl 1]: S26–S30, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Andrew T, Hart DJ, Snieder H, de Lange M, Spector TD, MacGregor AJ: Are twins and singletons comparable? A study of disease-related and lifestyle characteristics in adult women. Twin Res 4: 464–477, 2001 [DOI] [PubMed] [Google Scholar]
- 40.Beck GJ, Berg RL, Coggins CH, Gassman JJ, Hunsicker LG, Schluchter MD, Williams GW, The Modification of Diet in Renal Disease Study Group : Design and statistical issues of the Modification of Diet in Renal Disease Trial. Control Clin Trials 12: 566–586, 1991 [DOI] [PubMed] [Google Scholar]
- 41.Gassman JJ, Greene T, Wright JT, Jr, Agodoa L, Bakris G, Beck GJ, Douglas J, Jamerson K, Lewis J, Kutner M, Randall OS, Wang SR: Design and statistical aspects of the African American Study of Kidney Disease and Hypertension (AASK). J Am Soc Nephrol 14[Suppl 2]: S154–S165, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwong YT, Stevens LA, Selvin E, Zhang YL, Greene T, Van Lente F, Levey AS, Coresh J: Imprecision of urinary iothalamate clearance as a gold-standard measure of GFR decreases the diagnostic accuracy of kidney function estimating equations. Am J Kidney Dis 56: 39–49, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E: Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem 81: 6656–6667, 2009 [DOI] [PubMed] [Google Scholar]
- 44.Dehaven CD, Evans AM, Dai H, Lawton KA: Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform 2: 9, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coresh J, Inker LA, Levey AS: Precise estimation of glomerular filtration rate from multiple blood biomarkers [Abstract]. J Am Soc Nephrol 25: 52A, 2014 [Google Scholar]
- 46.Meisinger C, Stöckl D, Rückert IM, Döring A, Thorand B, Heier M, Huth C, Belcredi P, Kowall B, Rathmann W: Serum potassium is associated with prediabetes and newly diagnosed diabetes in hypertensive adults from the general population: The KORA F4-study. Diabetologia 56: 484–491, 2013 [DOI] [PubMed] [Google Scholar]
- 47.Junge W, Wilke B, Halabi A, Klein G: Determination of reference intervals for serum creatinine, creatinine excretion and creatinine clearance with an enzymatic and a modified Jaffé method. Clin Chim Acta 344: 137–148, 2004 [DOI] [PubMed] [Google Scholar]
- 48.Coresh J, Astor BC, McQuillan G, Kusek J, Greene T, Van Lente F, Levey AS: Calibration and random variation of the serum creatinine assay as critical elements of using equations to estimate glomerular filtration rate. Am J Kidney Dis 39: 920–929, 2002 [DOI] [PubMed] [Google Scholar]
- 49.Selvin E, Manzi J, Stevens LA, Van Lente F, Lacher DA, Levey AS, Coresh J: Calibration of serum creatinine in the National Health and Nutrition Examination Surveys (NHANES) 1988-1994, 1999-2004. Am J Kidney Dis 50: 918–926, 2007 [DOI] [PubMed] [Google Scholar]
- 50.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF, 3rd, Feldman HI, Kusek JW, Eggers P, Van Lente F, Greene T, Coresh J, CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) : A new equation to estimate glomerular filtration rate. Ann Intern Med 150: 604–612, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhavsar NA, Appel LJ, Kusek JW, Contreras G, Bakris G, Coresh J, Astor BC, AASK Study Group : Comparison of measured GFR, serum creatinine, cystatin C, and beta-trace protein to predict ESRD in African Americans with hypertensive CKD. Am J Kidney Dis 58: 886–893, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stevens PE, Levin A, Kidney Disease: Improving Global Outcomes Chronic Kidney Disease Guideline Development Work Group Members : Evaluation and management of chronic kidney disease: Synopsis of the kidney disease: improving global outcomes 2012 clinical practice guideline. Ann Intern Med 158: 825–830, 2013 [DOI] [PubMed] [Google Scholar]
- 53.Fox CS, Larson MG, Leip EP, Culleton B, Wilson PW, Levy D: Predictors of new-onset kidney disease in a community-based population. JAMA 291: 844–850, 2004 [DOI] [PubMed] [Google Scholar]
- 54.Storey JD, Tibshirani R: Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100: 9440–9445, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tutz G, Binder H: Boosting ridge regression. Comput Stat Data Anal 51: 6044–6059, 2007 [Google Scholar]
- 56.DeLong ER, DeLong DM, Clarke-Pearson DL: Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 44: 837–845, 1988 [PubMed] [Google Scholar]
- 57.Binder H, Schumacher M: Adapting prediction error estimates for biased complexity selection in high-dimensional bootstrap samples. Stat Appl Genet Mol Biol 7: Article 12, 2008 [DOI] [PubMed] [Google Scholar]
- 58.Steyerberg E: Clinical Prediction Models: A Practical Approach to Development, Validation, and Updating, New York, Springer-Verlag, 2009 [Google Scholar]
- 59.Pencina MJ, D’Agostino RB, Sr, D’Agostino RB, Jr, Vasan RS: Evaluating the added predictive ability of a new marker: From area under the ROC curve to reclassification and beyond. Stat Med 27: 157–172, discussion 207–212, 2008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.