Abstract
Background
HIPK2 (homeodomain-interacting protein kinase 2) has been identified as a nuclear serine/threonine kinase. A central function of HIPK2 is repressing transcription of homeodomain containing transcription factors.
Results and Conclusions
We show here that HIPK2 activates transcription mediated by tumor suppressor p53 responsive promoter elements. Overexpression of HIPK2 leads to an increase of p53 protein expression or stability, which becomes enhanced further in the presence of the DNA damaging drug doxorubicin. The effects of HIPK2 on p53 are not observed with kinase deficient HIPK2 mutants. However, HIPK2 is not sufficient for phosphorylation of three crucial serine residues of p53, suggesting that HIPK2-induced p53 activation does not involve phosphorylation of p53. Instead, HIPK2 leads to a downregulation of p53-induced Mdm2 protein and this may lead to stabilization of p53. Overexpression of HIPK2 does not lead to a change of Mdm2 mRNA expression. The data suggest that HIPK2 plays a critical role in p53 mediated cellular responses by removing the p53 inhibitor protein Mdm2 via modification of the protein itself or its intracellular movement.
Background
The tumor suppressor protein p53 contributes to the control of cell cycle checkpoints and apoptosis and is frequently lost or mutated in multiple types of human cancers [1]. DNA damaging agents induce p53 accumulation and induction of p53-mediated transcription [2,3]. Several proteins are known to play a crucial role in the stabilization and activation of p53 [4,5]. The association with murine double minute clone 2 (Mdm2) leads to a susceptibility of p53 for proteolysis [6,7] and thus, p53 protein levels are regulated post-transcriptionally [8].
Homeodomain-interacting protein kinase 2 (HIPK2) has been recently described as a member of a family of nuclear kinases that act as co-repressors for homeodomain transcription factors [9] and it is a potential interaction partner for interferon type I induced Mx GTPases with antiviral activity against several RNA viruses [10]. HIPK2 is regulated by the ubiquitin-like protein SUMO-1 and the covalent SUMO-1 modification correlates with its localization to nuclear speckles or nuclear dots [11]. Here we address the question whether HIPK2 modifies transcription regulated by transcription factors other than homeoproteins.
We found that HIPK2 activates transcription mediated by p53 specific promoter elements. HIPK2 also enhances expression levels of p53 protein. Both effects on p53 are not observed with a kinase defective mutant and large amino and carboxy-terminal deletion mutants of HIPK2. The DNA damaging anti-cancer drug doxorubicin enhances HIPK2-induced expression or stability of p53. HIPK2 overexpression downregulates Mdm2 protein levels. Therefore, the observed effects of HIPK2 on p53 seem to be mediated by Mdm2. We conclude that an important function of HIPK2 is the stabilization and activation of p53 by inducing degradation of Mdm2.
Results
HIPK2 activates p53-mediated transcription
Since HIPK2 has been described as a nuclear protein that interacts with homeodomain containing transcription factors [9,11], we looked for an influence of HIPK2 on transcription mediated by several transcription factors such as p53, NFκB and Elk1.
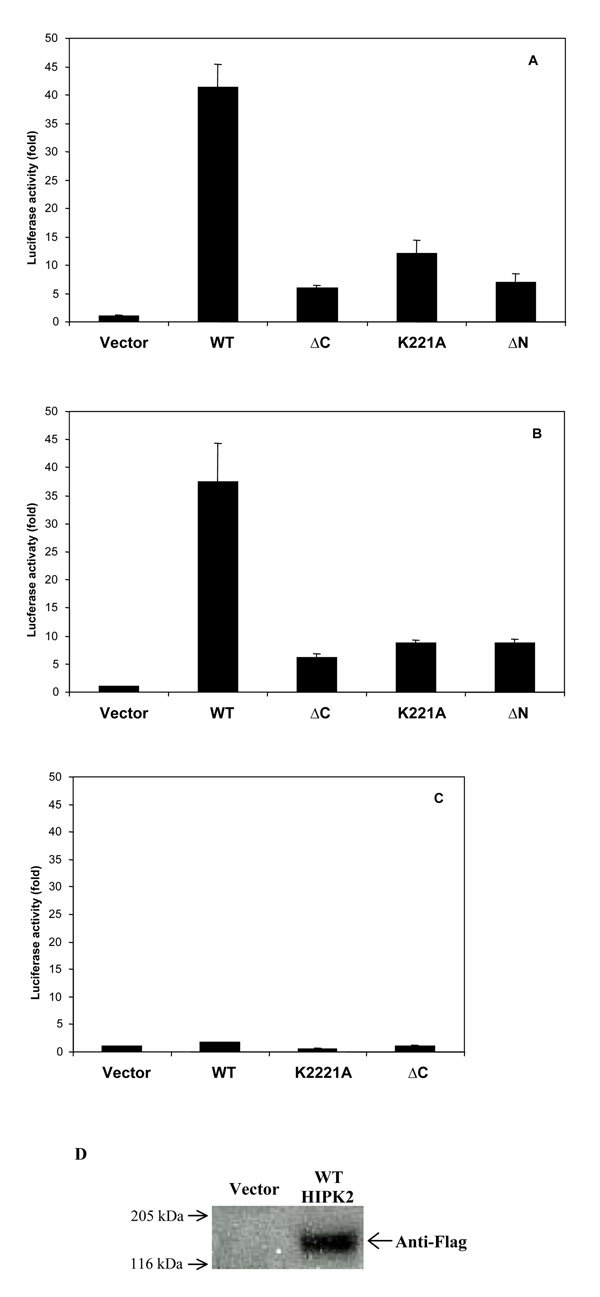
The tumorsuppressor p53 is a transcription factor that binds to DNA and is involved in the cell cycle and apoptosis. To test a possible transcriptional transactivation activity of HIPK2, expression vectors encoding wildtype or the mutated HIPK2 proteins (AC, AN and K221A) were transfected into 293 (Fig. 1A) and HeLa (Fig. 1B) cells together with p53-luc, a luciferase reporter gene under the control of the p53-specific enhancer element (TGCCTGGACTTGCCTGG)15. The p53-specific enhancer element is derived from the sequence comparison of promoters of p53-inducible genes [12,13]. HeLa cells have been found to contain endogenous transcriptionally active p53 protein by the intrinsic ability to transactivate p53-responsive promoter elements [14]. In line with this, endogenous p53 of the HeLa cells used in our experiments had transcriptional activity and wildtype sequence (data not shown). Activation of the luciferase activity by cotransfection of 50 ng of the reporter plasmid encoding p53 (pFC-p53) was 250 fold for 293 and 160 fold for HeLa cells, respectively (data not shown). The luciferase value of vector and p53-luc transfected cells were standardized for a value of 1 in each assay, to control for activation of the p53 specific enhancer element by endogenous p53. The intrinsic activity of endogenous p53 was approximately 20 fold above the blank controls (data not shown). Wildtype HIPK2 induced a 40 fold increase of p53-mediated transcription in 293 cells compared to the increase in vector transfected control cells (Fig. 1A). No such strong increase was observed with the kinase defective HIPK2 mutant K221A, a carboxy-terminal HIPK2 deletion mutant AC or an amino-terminal deletion mutant AN (Fig. 1A and 1B). But luciferase activity induced by the HIPK2 mutants was above the vector controls. Protein expression of recombinant HIPK2 in HeLa cells was shown in Western analysis after transient transfection of a Flag-tagged wildtype HIPK2 expression construct as described [26] (Fig. 1D). Expression of overexpressed and endogenous HIPK2 protein in HeLa cells was not detected by using a polyclonal anti-HIPK2 antiserum [26] (data not shown).
Figure 1.
Activation of p53-mediated transcription by HIPK2 overexpression. 293 (A), HeLa (B) and Saos2 (C) cells were cotransfected with a p53-luciferase reporter construct and the indicated HIPK2 constructs or vector. Results represent the mean of three independent experiments. Bars, SD. (D) Western analysis of transient overexpression of Flag-tagged wildtype HIPK2 in HeLa cells (right lane) compared to vector-transfected control cells (left lane) with an anti-Flag antibody.
To test whether HIPK2 activates transcription mediated by p53 responsive elements via p53, we transiently transfected human Saos2 cells, which lack p53 expression [15], with different HIPK2 expression constructs (Fig. 1C). Expression of wildtype HIPK2 did not lead to a significant increase in luciferase activity compared to its mutants, K221A and AC, and the vector control. However, transfection of 50 ng of pFC-p53 into Saos2 cells resulted in a 200 fold increase in luciferase activity and this increase was not further enhanced in the presence of HIPK2 (data not shown). Taken together, HIPK2 activates transcription mediated by p53 specific promoter elements and p53 is required for this effect. The enhancement of transcription is diminished by different mutations of HIPK2.
Wildtype HIPK2 leads to an increase of p53 expression or stability
Since HIPK2 activated p53-mediated transcription, we looked whether HIPK2 leads to an increase in the level of p53 protein after transient transfection of HeLa cells. Wildtype HIPK2 enhanced protein expression or stability of p53 compared to the vector transfected controls (Fig. 2A). The kinase-defective mutant K221A and the amino-terminal deletion AN which lacks the kinase domain led to a decrease of p53 protein. The carboxy-terminal HIPK2 deletion mutant AC which contains the kinase part did not influence p53 protein levels (Fig. 2A). This is a hint that the increase of p53 protein induced by wildtype HIPK2 could be responsible for the enhanced transcriptional activity mediated by p53 binding promoter elements (Fig. 1).
Figure 2.
Enhancement of p53 protein levels by wildtype HIPK2. HeLa cells were transiently transfected with the indicated HIPK2 expression constructs. Cellular extracts were analyzed for p53 expression by Western blotting. The signals corresponding to p53 or phosphorylated p53 (Phospho-p53) are indicated by an arrow. Equal loading was confirmed by reprobing the blots with an anti-β-actin antibody as shown below. (A) Cells were left untreated after transfection. (B) 24 h after transfection, doxorubicin was added to a final concentration of 0.1 μg/ml for 3 and 6 h as indicated. (C) 24 h after transfection doxorubicin was added to a final concentration of 0.1 μg/ml for 6 h and anti-phospho-p53 (Ser 15)-antibody was used for detection of phosphorylated serine 15 in p53. wt, wildtype; vec, vector.
Doxorubicin enhances the HIPK2-mediated increase of p53 expression or stability
Since p53 is stabilized in response to DNA damage, we tested whether HIPK2 has an effect on stabilization of p53 mediated by the DNA damaging drug doxorubicin in HeLa cells. Doxorubicin induced a significant increase of the p53 protein level after 6 h in the vector-transfected controls (Fig. 2B). When cells were transiently transfected with wildtype HIPK2, the increase of p53 protein was similar to that observed after 6 h doxorubicin treatment. The combination of overexpression of HIPK2 and treatment with doxorubicin for 6 h resulted in an increase in p53 protein levels compared to either treatment alone, i. e. the HIPK2-mediated increase of the p53 protein level was further enhanced by the addition of doxorubicin (Fig. 2B).
In summary, these results show that wildtype HIPK2 but not kinase deficient or severely truncated mutants led to an increase of p53 protein. Treatment with doxorubicin for at least 6 h further increased the amount of p53 protein.
HIPK2 is not sufficient for phosphorylation of serine 15, serine 20 and serine 392 of p53
Since HIPK2 contains a serine/threonine kinase domain and since multiple serine/threonine phosphorylation sites are present in p53 protein, we used antibodies that specifically detect single phosphorylated serine residues of p53 to look for a putative HIPK2-mediated phosphorylation of p53. Upon treatment with doxorubicin, serine 15 is phosphorylated in HeLa cells (Fig. 2C). However, no serine 15 phosphorylation was detected after transient transfection of HIPK2 in the absence of doxorubicin treatment. The amount of phosphorylated protein after doxorubicin treatment reflects the total amount of p53 protein (Fig. 2A, B and 2C), which suggests that almost all p53 protein present became phosphorylated on serine 15. Since the kinase defective HIPK2 mutant K221A probably acts in a dominant negative way, the amount of p53 which was induced by endogenous HIPK2 in the vector-transfected controls decreased. Similar to the decrease in the amount of p53 protein in the presence of the K221A mutant (Fig. 2A) there was a decrease in phosphorylated p53 after treatment with doxorubicin (Fig. 2C). As expected from these findings, overexpression of the HIPK2 mutants AN and AC yielded less phosphorylated p53 compared to overexpression of wildtype HIPK2 in the presence of doxorubicin (Fig. 2C). Similar results were obtained using an antibody against serine residue 20 (data not shown).
Using an antibody that specifically detects phosphorylation of serine 392 in p53 we could not detect phosphorylated p53 either in the presence of doxorubicin or during overexpression of HIPK2 (data not shown). Taken together, HIPK2 is not sufficient for phosphorylation of p53 at serine residues 15 and 20, where doxorubicin seems to be required, and for phosphorylation of serine 392.
HIPK2 leads to a decrease of p53-induced Mdm2 expression
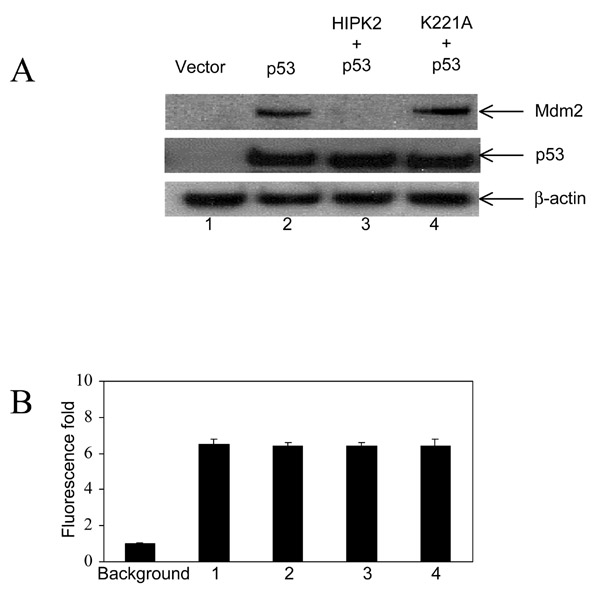
To further characterize the interaction of HIPK2 and p53, Saos2 cells were transiently cotransfected with constructs coding for p53, p53 and wildtype HIPK2 or p53 and the kinase defective HIPK2 mutant K221A together with pEGFP-Nl coding for a green fluorescence protein (Fig. 3A). Mdm2 and p53 proteins were not detectable in Saos2 cells (lane 1). In agreement with previous observations [16,17], wildtype p53 led to an induction of endogenous Mdm2 expression (lane 2). Interestingly, a p53-mediated induction of Mdm2 expression was not observed in the presence of wildtype HIPK2 (lane 3). Therefore, HIPK2 overexpression led to a decrease of p53-induced Mdm2 protein levels. This decrease in Mdm2 protein was not observed by coexpression of the kinase defective HIPK2 mutant K221A (lane 4), which suggests that the kinase domain is necessary for Mdm2 downregulation. Equal protein loading was shown by reprobing the filters with an antibody detecting β-actin. pAGFP-Nl was cotransfected in all cases to normalize overexpression. GFP fluorescence was not significantly different (Fig. 3B), indicating similar transfection efficiencies.
Figure 3.
Downregulation of p53-induced Mdm2 by overexpressed HIPK2. (A) Saos2 cells were transiently cotransfected with pEGFP-N and expression constructs coding for wildtype 53, wildtype HIPK2 or the kinase defective HIPK2 mutant K221A as indicated. After 24 h cells were lysed and analysed by Western blotting with the indicated antibodies. (B) GFP fluorescence was measured by quantitative fluorometry to show equal transfection efficiencies. Background fluorescence was set to 1 (lane 0). Fluorescence fold is increased above background (lanes 1 to 4). Bars, SD.
HIPK2 overexpression does not change Mdm2 mRNA expression
Next we asked the question whether HIPK2 downregulates Mdm2 by decreasing Mdm2 transcription. We transiently transfected Saos2 cells either with vector alone or with expression plasmids coding for p53 or HIPK2. In addition, the plasmid coding for p53 was transiently transfected in the presence of either wildtype HIPK2 or the kinase defective HIPK2 K221A mutant (Fig. 4). Relative Mdm2 mRNA levels were detected by quantitative TaqMan RT-PCR with sequence specific primers for human Mdm2 (GenBank accession AF201370). As expected, p53 led to an approximately 4 fold increase of Mdm2 mRNA levels [16,17]. In contrast, overexpression of HIPK2 did not lead to a change of Mdm2 mRNA compared to vector transfected controls (Fig. 4). Overexpression of p53 together with either wildtype HIPK2 or the kinase defective HIPK2 mutant K221A showed no significant decrease in p53-induced Mdm2 mRNA levels. Similar data were obtained with HeLa cells (data not shown). Therefore, HIPK2 does not downregulate Mdm2 at the transcriptional level. It seems that HIPK2 destabilizes Mdm2 by modifying the Mdm2 protein.
Figure 4.
Quantitative mRNA Expression levels of Mdm2. Saos2 cells were transiently transfected with the indicated expression constructs. Mdm2 mRNA expression levels were determined by TaqMan RT-PCR as described in Materials and Methods. As internal reference, mRNA expression of GAPDH mRNA was used. Relative Mdm2 mRNA expression is shown and Mdm2 mRNA expression of vector transfected cells was set to 1. Bars, SD.
HIPK2 induces Elk1- but not NFκB-mediated transcription
To test the expected influence of HIPK2 on other transcription factors we used a chimeric protein consisting of the GAL4 DNA binding domain fused to the activation domain of the transcription factor Elk1. The transcription activator Elk1 becomes phosphorylated and activated by the MAP (mitogen-activated protein) kinase. Direct or indirect phosphorylation of this transactivator by HIPK2 would activate transcription of the luciferase gene from a reporter plasmid with the GAL4-binding element. Cotransfection of the trans-activator plasmid pFA-Elk1, the luciferase reporter plasmid pFR-Luc and the positive control MEK1 encoding plasmid pFC-MEK1 resulted in an 115 fold increase in luciferase activity in 293 cells compared to pFA2-Elk1 and pFR-Luc transfected control cells (data not shown). In comparison, cotransfection of pFA2-Elk1 and the wildtype HIPK2 expression construct gave only a 6 fold increase (Fig. 5A). This increase was reduced to 3 fold when the kinase defective HIPK2 mutant K221A was used. However, using the carboxy-terminal deletion AC with an intact kinase domain, the increase of luciferase activity was about 17 fold. A possible direct effect of HIPK2 on the luciferase construct has been tested by cotransfection of wildtype HIPK2 and pFR-Luc into 293 cells. In this control, the luciferase activity was similar to the pFR-Luc and pFA2-Elk1 transfected control cells (data not shown). These results suggest that HIPK2 may weakly activate the transactivator Elk1-GAL4 and that the kinase function of HIPK2 is necessary for this activation. The carboxy-terminus of HIPK2 may contain inhibitory sequences for Elk1-phosphorylation. The conclusion is, that HIPK2 seems to activate components of the MAP kinase pathway. The exact role remains to be elucidated.
Figure 5.
Activation of Elk 1-mediated transcription but no activation of NFκB by HIPK2 overexpression. (A) 293 cells were cotransfected with a GAL4-luciferase reporter construct, an Elk1-GAL4 transactivator expression construct and the indicated HIPK2 constructs or vector. (B) 293 cells were cotransfected with an NFκB-luciferase reporter construct and the indicated HIPK2 constructs or vector. Results represent the mean of three independent experiments. Bars, SD.
In a similar assay, which we have used to study HIPK2-mediated activation of p53 specific promoter elements, we looked for a role of HIPK2 in activation of NFκB-mediated transcription. 293 cells were cotransfected with a luciferase reporter plasmid containing five tandem NFκB binding sites and different HIPK2 expression constructs (Fig. 5B). Only wildtype HIPK2 activated NFκB-mediated transcription slightly but not significantly. No effect was seen with the HIPK2 mutants (Fig. 5B). NFκB-mediated luciferase activation was 100 fold with a MEK kinase positive control plasmid (data not shown). The luciferase values of vector and pNF-κB-luc transfected cells were set as 1. Taken together, we conclude that HIPK2 does not activate NFκB.
Discussion
HIPK2 enhances p53 activity and p53 expression or stability
HIPK2 acts as a co-repressor for homeodomain containing transcription factors. We show here that HIPK2 has additional functions, namely the activation of transcription mediated by p53 specific promoter elements. Thereby, overexpression of HIPK2 leads to an increase of the p53 protein level. Neither the transcriptional activation nor the enhancement of p53 protein amounts is detected with overexpressed HIPK2 mutants.
The p53 responsive enhancer element becomes activated by HIPK2 in cell lines with p53 but not in a cell line devoid of p53 (Fig. 1). We conclude, that the activation of the enhancer element is indeed mediated by p53 and not by another transcription factor of the p53 family. The kinase defective mutant of HIPK2, K221A, leads to a decrease of p53 protein amounts (Fig 2A). However, this decrease does not correspond with a further decrease on p53-mediated transcription compared to HIPK2 mutants AC and AN (Fig. 1A and 1B).
HIPK2 is not sufficient for phosphorylation of crucial serine residues of p53
The nuclear phosphoprotein p53 is regulated and stabilized by different modifications [3,18], e. g. serine 15 phosphorylation has been detected as prerequisite for phosphorylation of tyrosine residue 18 [19]. HIPK2 could not activate or stabilize p53 either directly or indirectly by phosphorylation. DNA damaging agents such as doxorubicin induce increased p53 phosphorylation by as yet undefined mechanisms. In the presence of wildtype HIPK2 and doxorubicin, serine residue 15 and 20 of p53 become phosphorylated, which could be either due to transfected or endogenous HIPK2 of HeLa cells. In the presence of doxorubicin alone or together with mutants of HIPK2 serines 15 and 20 could be phosphorylated by endogenous HIPK2 (Fig. 2C). Thus, we conclude that HIPK2 is not sufficient for serine 15 and 20 phosphorylation, for which doxorubicin may be required. HIPK2 is also not sufficient for phosphorylation of residue 392, but doxorubicin requirement is excluded. Additional phosphorylation sites have not been tested since the absolute requirement of phosphorylation for DNA-damage induced stabilization of p53 remains controversial [20]. Other proteins, which regulate p53, like e. g. Mdm2, could be the targets for phosphorylation by HIPK2.
Doxorubicin treatment and HIPK2 overexpression show an additive effect on p53 protein expression or stability (Fig. 2B). The relative amounts of the increase of the p53 protein levels and the increase in doxorubicin induced phosphorylation of serine residue 15 are comparable and give rise to the assumption that the total amount of p53 protein present becomes phosphorylated under doxorubicin treatment. Since p53 has a high turnover rate [4], the increase of p53 protein levels seems to be due to stabilization of p53 rather than induction of enhancement of transcription of p53. An increase of p53 mRNA stability or translation efficiency could play additional roles.
HIPK2 downregulates Mdm2 protein
HIPK2 may exert all its effects on p53 via Mdm2, since it leads to a decrease of the p53-induced Mdm2 protein level (Fig. 3). HIPK2 has no influence on Mdm2 transcription (Fig. 4). Therefore, HIPK2 regulates Mdm2 stability at the protein level. The most likely mechanism is serine/threonine phosphorylation of Mdm2 either directly or indirectly by HIPK2. At present, we did not get convincing data showing a direct serine/threonine phosphorylation of Mdm2 by HIPK2.
Mdm2 shuttles between the nucleus and the cytoplasm and this nucleo-cytoplasmic shuttling of Mdm2 is essential for its ability to promote p53 degradation by the cytoplasmic proteasome [21]. Mdm2 seems to have a role in exporting p53 from the nucleus [22,23]. HIPK2 did not directly interact with either p53 or Mdm2 in yeast and 293T cells (data not shown). Whether HIPK2 can influence the shuttling of Mdm2 remains to be clarified.
Effect of HIPK2 on target genes of p53
We have addressed the issue to check expression of described downstream targets of p53 at the protein level in HeLa cells. The best characterized target gene that becomes transactivated by p53 is the cyclin/Cdk inhibitor p21 (Waf1/Cip1) [3]. But we did not detect a significant increase of expression of p21 (Waf1/Cip1) mediated by overexpression of HIPK2 or treatment with doxorubicin in Western blotting analysis. As a positive control 2 μg of the wildtype p53 expression construct pFC-p53 was transiently transfected and transient overexpression of wildtype p53 led to a weak increase of p21 (Waf1/Cip1) expression (data not shown). Western blotting does not seem to be sensitive enough to detect a significant difference in p21 expression between vector and wildtype HIPK2 transfected cells. In the same experimental setting an increase of expression of the pro-apoptotic Bcl2 family member Bax, whose gene has been described as a p53 target [3], was not detectable (data not shown). Therefore, induction of p53 target genes seems to be cell type specific. The recently described p53-dependent pro-apoptotic Bcl-2 family member Noxa [24] is a further candidate that could be regulated via HIPK2.
In this respect, a further difficulty is that HIPK2 downregulates the p53-induced Mdm2 protein level. This may also be the case for other p53-induced proteins.
Other functions of HIPK2
HIPK2 is also termed PKM (for Mx-interacting protein kinase) since it has been cloned in a yeast two-hybrid screen as an interaction partner for the interferon type I-induced protein MxA, that displays antiviral activity against a variety of RNA viruses [10]. The nuclear distribution of murine Mx1 is similar to HIPK2/PKM, but a direct interaction of the Mx protein with HIPK2/PKM and phosphorylation of Mx by HIPK2/PKM could not be demonstrated so far [10]. HIPK2/PKM phosphorylates several phosphoproteins expressed in E. coli. This reaction is inhibited by a kinase defective mutant K221W of HIPK2/PKM. Thereby it was also shown that HIPK2/PKM is a true serine/threonine kinase, rather then a dual specificity protein kinase which is able to phosphorylate both, serine/threonine and tyrosine residues [10].
HIPK2 interacts with the death factor receptor CD95 in the yeast two-hybrid system [25]. When overexpressed in mammalian cells it associates with TRADD [26]. TRADD has been described to bind to the tumor necrosis factor type I and thereby executing essential functions in signaling [27,28]. Whether HIPK2 plays an important role in death receptor mediated apoptosis is not clear at present. Recently, it has been described that HIPK3, a member of the HIPK family, binds to CD95 and thereby inhibits c-Jun N-terminal kinase activation [29].
An additional function of HIPK2 may be the activation of the mitogen-activated protein kinase pathways since we detected a weak activation of Elk1 mediated transcription. The carboxy-terminal deletion of HIPK2 ΔC augments the activation of Elk1-mediated transcription in comparison to wildtype HIPK2. The carboxy-terminal deletion of HIPK2 ΔC is equally distributed in the nucleus. In contrast, wildtype HIPK2 and the kinase defective K221A mutant localize to nuclear speckles [25]. This suggests that the equal nuclear distribution of the HIPK2 mutant ΔC is responsible for the increase in Elk1 mediated transcription. It has been shown that DNA damage in addition to stabilization of p53 leads to an activation of a MAP kinase homolog, the c-Jun N-terminal kinase or stress activated protein kinase [30].
Conclusions
HIPK2 is a nuclear kinase that activates transcription mediated by tumor suppressor p53 responsive promoter elements upon overexpression. Furthermore, it plays a role in the induction of expression or stability of p53 protein. Overexpression of HIPK2 does not lead to a change of Mdm2 mRNA levels, but it leads to a downregulation of p53-induced Mdm2 protein. Most likely, this HIPK2-induced decrease of Mdm2 is responsible for the stabilization of p53. The involvement of HIPK2 kinase activity remains to be identified.
Materials and methods
Cell lines and cell culture
HeLa cells were maintained in RPMI 1640 containing 10% heat-inactivated fetal bovine serum, L-glutamine, and penicillin/streptomycin (all from Gibco Life Technologies, Karlsruhe, Germany). The stock solution of doxorubicin (Amersham-Pharmacia, Freiburg, Germany) was 2 mg/ml in H20. Human osteosarcoma Saos2 cells and human embryonic kidney 293 cells were cultured in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum, L-glutamine, and penicillin/streptomycin (all from Gibco Life Technologies, Karlsruhe, Germany).
Plasmids, construction of expression plasmids, mutagenesis and transient transfection
The full length human HIPK2 cDNA [25] was subcloned into the EcoR1 and XhoI sites of pcDNA3.1-zeo(+) (Invitrogen, Groningen, The Netherlands). The DNA fragments for the deletion constructs were generated by PCR using Deep Vent polymerase (New England Biolabs, Frankfurt, Germany) and primers with appropriate restriction sites and epitope tags. Details about the used primers can be obtained upon request. The carboxy-terminal HIPK2 deletion AC contains amino acids until position 527, the amino-terminal deletion AN from amino acid position 559. The PCR fragments were subcloned in pBSIIsk(-) (Stratagene, Offenbach, Germany), sequenced using an automated sequencer (Applied Biosystem, Langen, Germany) and inserted into pcDNA3 (Invitrogen, Groningen, The Netherlands). The point mutant HIPK2 K221A was generated using the QuickChangeTM kit (Stratagene, Offenbach, Germany) according to the manufacturer's instructions. The plasmid pEGFP-Nl coding for a green fluorescent protein was obtained from Clontech (Heidelberg, Germany). The plasmid coding for wildtype p53 was a kind gift from M. Oren (The Weizmann Institute of Science, Israel).
HeLa and Saos2 cells were transiently transfected by using the GenePORTER TM Transfection Reagent (Gene Therapy Systems, San Diego, CA, USA) according to the protocol of the manufacturer. Transient transfections of 293 cells were performed by the calcium phosphate method. Prior to transfection approximately 5 × 105 cells were plated onto a 60 mm dishes to a confluency of 20%. Cells were harvested at 20–30 h following transfection and lysed in 500 μl of lysis buffer (20 mM HEPES, pH 7.4, 150 mM NaCl, 5 mM EDTA, 10 % glycerol, 0.5% Triton X-100, 20 μl/ml protease inhibitor cocktail (Sigma, Taufkirchen, Germany).
Elk1 activation assay
293 cells have been transiently cotransfected with 1 μg pFR-Luc (Stratagene, Offenbach, Germany) and 50 ng pFA2-Elk1 (PathDetect in vivo signal transduction pathway trans-reporting systems, Stratagene, Offenbach, Germany) with or without 1 μg of the indicated plasmids coding for HIPK2. 50 ng of pFC-MEK1 (Stratagene, Offenbach, Germany) served as positive control. After 24 h cells were starved for 8 h. Then lysophosphatidic acid (Sigma, Taufkirchen, Germany) was added to a final concentration of 10 μM for 30 min. Cells lysis and measurement of luciferase activity were done according to the instructions for the luciferase assay system (Promega, Mannheim, Germany).
p53 and NFκB activation assay
293 or HeLa cells have been transiently cotransfected with 1 μg pNF-κB-Luc (PathDetect in vivo signal transduction pathway cis-reporting systems, Stratagene, Offenbach, Germany) with or without 1 μg of the indicated plasmids coding for HIPK2. 50 ng of pFC-MEKK (Stratagene, Offenbach, Germany) served as positive control for activation of NFκB-mediated transcription. For measuring activation of p53, 1 μg pf p53-Luc (PathDetect in vivo signal transduction pathway cis-reporting systems, Stratagene, Offenbach, Germany) has been used for cotransfection instead of pNF-κB-Luc. The positive control for activation of p53-mediated transcription was 50 ng pFC-p53 (Stratagen, Offenbach, Germany), which contains the p53 cDNA under the control of a cytomegalovirus promoter. Cells were collected after 24 h. Cell lysis and measurement of luciferase activity were done according to the instructions for the luciferase assay system (Promega, Mannheim, Germany).
Western blotting
HeLa or Saos2 cells were transiently transfected with either the vector pcDNA3.1-zeo+ (Invitrogen, Groningen, The Netherlands), the p53 (pFC-p53, Stratagene, Offenbach, Germany) or the HIPK2 expression construct. Cells were harvested 20–30 h following transfection and lysed in 100 μl of lysis buffer (20 mM HEPES, pH 7.4, 150 mM NaCl, 5 mM EDTA, 10% glycerol, 0.5% Triton X-100, 20 μl/ml protease inhibitor cocktail (Sigma, Taufkirchen, Germany)). The protein concentration was determined by the BCA-Pierce-Protein Assay kit according to the protocol of the manufacturer. For Western analysis, 20–30 μg of protein was separated on 10–20% gradient gels (Bio-Rad, Hercules, CA, USA) and blotted onto nitrocellulose membranes (Amersham-Pharmacia, Freiburg, Germany) according to standard protocols. The monoclonal anti-p53 antibody DO-7 and the monoclonal anti-p21 (Waf1/Cip1) antibody SX118 were purchased from PharMingen International (Hamburg, Germany), the polyclonal anti-Bax antibody Ab-1 (150-165) from Calbiochem (Schwalbach, Germany), the polyclonal phospho-p53 (Ser 15) (Ab-3), phospho-p53 (Ser 392) (Ab-4) and the monoclonal Mdm2 (Ab-1) antibodies from OncogeneTM (Schwalbach, Germany) research products, the polyclonal phospho-p53 (Ser 20) from NewEngland Biolabs (Frankfurt, Germany), the monoclonal anti-Flag antibody M2 from Stratagene (Offenbach, Germany) and the monoclonal anti-β-actin antibody AC-15 from Sigma (Taufkirchen, Germany). An anti-mouse secondary IgG1 antibody coupled to horseraddish peroxidase (Santa Cruz Biotechnology, Heidelberg. Germany) was used at a dilution 1:5000. Antibody reactive bands were detected by using the enhanced chemiluminescence Western detection system (Amersham-Pharmacia, Freiburg, Germany). Fluorometric protein quantification was done by excitation at 485 nm and emission measurement at 510 nm of cellular extracts in a fluorometer (Victor, Wallac, Freiburg, Germany). For reprobing, membranes were treated with stripping buffer (62.5 mM Tris-HCl pH 6.7, 2% SDS, 100 mM β-mercaptoethanol) at 50°C for 30 min.
Quantitative TaqMan RT-PCR
Quantitative TaqMan RT-PCR (real time PCR) was carried out by Toplab GmbH (München, Germany) by using an ABI PRISM Sequence Detection System 7700. Sequence specific internal Primers were labelled with FAM/TAMRA. Endogenous reference mRNA was human GAPDH mRNA.
List of Abbreviations
HIPK2, homeodomain-interacting protein kinase 2; MAP, mitogen-activated protein; Mdm2, murine double minute clone 2.
Acknowledgments
Acknowledgments
We are very grateful to Moshe Oren for the wildtype p53 expression construct and Robert Zeillinger for the sequencing and functional assays of HeLa-p53. We thank Jörg Fellenberg for Saos2 cells. We are indebted to Irina Duttenhofer for technical assistance and Ina Menzl for critical reading of the manuscript. This work was supported by Deutsche Forschungsgemeinschaft (DFG) grant HU 674/2-1 to H.H.
Contributor Information
Ying Wang, Email: ying.wang@medizin.uni-ulm.de.
Klaus-Michael Debatin, Email: klaus-michael.debatin@medizin.uni-ulm.de.
Hubert Hug, Email: hubert.hug@medizin.uni-ulm.de.
References
- Brown JM, Wouters BG. Apoptosis, p53 and tumor cell sensitivity to anticancer agents. Cancer Res. 1999;59:1391–1399. [PubMed] [Google Scholar]
- Albrechtsen N, Domreiter I, Grosse F, Kim E, Wiesmüller L, Deppert W. Maintenance of genomic integrity by p53: complementary roles for activated and non-activated p53. Oncogene. 1999;18:7706–7717. doi: 10.1038/sj.onc.1202952. [DOI] [PubMed] [Google Scholar]
- Burns TF, El-Deiry WS. The p53 pathway and apoptosis. J Cell Physiol. 1999;181:231–239. doi: 10.1002/(SICI)1097-4652(199911)181:2<231::AID-JCP5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Oren M. Regulation of the p53 tumor suppressor protein. J Biol Chem. 1999;274:36031–36034. doi: 10.1074/jbc.274.51.36031. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Momand J, Wu HH, Dasgupta G. Mdm2-master regulator of the p53 tumor suppressor protein. Gene. 2000;242:15–29. doi: 10.1016/S0378-1119(99)00487-4. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Kim YH, Choi CY, Lee S-J, Conti MA, Kim Y. Homeodomain-interacting protein kinases, a novel family of co-repressors for homeodomain transcription factors. J Biol Chem. 1998;273:25875–25879. doi: 10.1074/jbc.273.40.25875. [DOI] [PubMed] [Google Scholar]
- Trost M, Kochs G, Haller O. Characterization of a novel serine/threonine kinase associated with nuclear bodies. J Biol Chem. 2000;275:7373–7377. doi: 10.1074/jbc.275.10.7373. [DOI] [PubMed] [Google Scholar]
- Kirn YH, Choi CY, Kim Y. Covalent modification of the homeodomain-interacting protein kinase 2 (HIPK2) by the ubiquitin-like protein SUMO-1. Proc Natl Acad Sci USA. 1999;96:12350–12355. doi: 10.1073/pnas.96.22.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- Tokino T, Thiagalingam S, el-Deiry WS, Waldman T, Kinzler KW, B Vogelstein. P53 tagged sites from human genomic DNA. Hum Mol Genet. 1994;3:1537–1542. doi: 10.1093/hmg/3.9.1537. [DOI] [PubMed] [Google Scholar]
- Hoppe-Seyler F, Butz K. Repression of endogenous p53 transactivation function in HeLa cervical carcinoma cells by human papillomavirus type 16 E6, human MDM-2, and mutant p53. J Virol. 1993;67:3111–3117. doi: 10.1128/jvi.67.6.3111-3117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcellus RC, Teodoro JG, Charbonneau R, Shore GC, Branton PE. Expression of p53 in Saos-2 osteosarcoma cells induces apoptosis which can be inhibited by Bcl-2 or the adenovirus E1B-55 kDa protein. Cell Growth Differ. 1996;7:1643–1650. [PubMed] [Google Scholar]
- Barak Y, Juven T, Haffher R, Oren M. Mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ. The p53-Mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18:7644–7655. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K, Saito S, Higashimoto Y, Roy S, Anderson CW, Appella E. Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase. Effect on Mdm2 binding. J Biol Chem. 2000;275:9278–9283. doi: 10.1074/jbc.275.13.9278. [DOI] [PubMed] [Google Scholar]
- Ashcroft M, Kubbutat MH, Vousden KH. Regulation of p53 function and stability by phosphorylation. Mol Cell Biol. 1999;19:1751–1758. doi: 10.1128/mcb.19.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W, Levine AJ. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc Natl Acad Sci USA. 1999;96:3077–3080. doi: 10.1073/pnas.96.6.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer RK, Yu ZK, Maki CG. The MDM2 RING-fmger domain is required to promote p53 nuclear export. Nat Cell Biol. 2000;2:569–573. doi: 10.1038/35023507. [DOI] [PubMed] [Google Scholar]
- Melchior F, Hengst L. Mdm2-SUMO1: is bigger better? Nat Cell Biol. 2000;2:E161–E163. doi: 10.1038/35023639. [DOI] [PubMed] [Google Scholar]
- Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, T Taniguchi, Tanaka N. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hofmann TG, Runkel L, Haaf T, Schaller H, K-Debatin M, Hug H. Isolation and characterization of cDNAs for the protein kinase HIPK2. Biochim Biophys Acta. 2001;1518:168–172. doi: 10.1016/S0167-4781(00)00308-0. [DOI] [PubMed] [Google Scholar]
- Li X, Wang Y, Debatin K-M, Hug H. The serine/threonine kinase HIPK2 interacts with TRADD, but not with CD95 or TNF-R1 in 293T cells. Biochem Biophys Res Commun. 2000;277:513–517. doi: 10.1006/bbrc.2000.3700. [DOI] [PubMed] [Google Scholar]
- Hsu H, Xiong J, Goeddel DV. The TNF receptor 1-associated protein TRADD signals cell death and nf-kb activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- V Rochat-Steiner, Becker K, Micheau O, Schneider P, Bums K, Tschopp J. FIST/HIPK3. A Fas/FADD-interacting serine/threonine kinase that induces FADD phosphorylation and inhibits Fas-mediated Jun NH(2)-terminal kinase activation. J Exp Med. 2000;192:1165–1174. doi: 10.1084/jem.192.8.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, Karin M, Davis RJ. JNK1: a protein kinase stimulated by UV light and Ha-ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]