Abstract
The integration of foreign DNA into algal and plant plastid genomes is a rare event, with only a few known examples of horizontal gene transfer (HGT). Plasmids, which are well-studied drivers of HGT in prokaryotes, have been reported previously in red algae (Rhodophyta). However, the distribution of these mobile DNA elements and their sites of integration into the plastid (ptDNA), mitochondrial (mtDNA), and nuclear genomes of Rhodophyta remain unknown. Here we reconstructed the complex evolutionary history of plasmid-derived DNAs in red algae. Comparative analysis of 21 rhodophyte ptDNAs, including new genome data for 5 species, turned up 22 plasmid-derived open reading frames (ORFs) that showed syntenic and copy number variation among species, but were conserved within different individuals in three lineages. Several plasmid-derived homologs were found not only in ptDNA but also in mtDNA and in the nuclear genome of green plants, stramenopiles, and rhizarians. Phylogenetic and plasmid-derived ORF analyses showed that the majority of plasmid DNAs originated within red algae, whereas others were derived from cyanobacteria, other bacteria, and viruses. Our results elucidate the evolution of plasmid DNAs in red algae and suggest that they spread as parasitic genetic elements. This hypothesis is consistent with their sporadic distribution within Rhodophyta.
Horizontal gene transfer (HGT) plays a significant role in the evolution of bacterial genomes, promoting environmental adaptation and speciation. Plasmids drive HGT by moving DNA from one genome to another, often between species, in the absence of sexual reproduction1,2,3. However, the mechanism of eukaryotic HGT is poorly understood, although it is known to occur from prokaryotes to eukaryotes4,5,6,7, between different eukaryotes8,9,10, and from eukaryotes to prokaryotes11,12. A special case of HGT, endosymbiotic gene transfer (EGT), is responsible for massive amounts of intracellular gene movement in eukaryotes. This is an outcome of organellogenesis, whereby 100 s to 1000 s of genes were transferred from the bacterium-derived organelle genomes (i.e., the mitochondrion and plastid) to the host nuclear chromosomes13,14,15,16.
Nuclear-encoded, plastid-derived genes have been studied in the glaucophyte alga Cyanophora paradoxa Korshikov (6–11%)17,18; the red alga Cyanidioschyzon merolae DeLuca, Taddei, & Varano (6–20%)19,20,21; the green algae Chlamydomonas reinhardtii P.C.A. Dangeard (6–14%) and Ostreococcus tauri C. Courties & M.-J. Chrétiennot-Dinet (11%)20,21,22; and in Arabidopsis thaliana (L.) Heynh. and other land plants (9–18%)20,21,23. EGT is essentially uni-directional. As a consequence, organelle (e.g., plastid) genomes have been reduced to 100 ~200 Kbp from their original size of several megabases in the cyanobacterial endosymbiont. It is not known, however, if plasmids may have facilitated EGT in algae and plants, thereby contributing significantly to their genome reduction.
HGTs have also been reported between organelle genomes of unrelated organisms. The plastid genome of the common milkweed Asclepias syriaca L. contains several mitochondrial genes24, whereas the mitochondrial genome of Amborella trichopoda Baill. contains mtDNAs from green algae (including the entire mitochondrial genome in three species), mosses, and other angiosperms25,26,27. The maize mitochondrial S-1 plasmid was found in the mitochondrial genome of the liverwort Marchantia polymorpha L.28,29. Interestingly, these sequences are similar to the mitochondrial dpo gene in the red alga Porphyra and the golden-brown alga Ochromonas danica E. Pringsheim30, suggesting the existence of HGT among different phyla. Plasmid-derived sequences were also reported from mtDNA in the brown alga Pylaiella littoralis (L.) Kjellman31 and two fungal species, Agaricus bisporus (J.E. Lange) Imbach and Gigaspora rosea T.H. Nicolson & N.C. Schenck32,33. The plastid genomes of photosynthetic haptophytes and cryptophytes contain bacterial-derived rpl36 genes7, and the cryptophyte Rhodomonas salina D.R.A. Hill & R. Wetherbee has a bacterial dnaX gene in its ptDNA34. These examples demonstrated significant HGT between organelles and between organelles and plasmids; however, no such data has been reported for the red algae (Rhodophyta).
In red algae, proteobacterial operons related to leucine biosynthesis (leuC and leuD subunits) are encoded in the plastid genome of Gracilaria tenuistipitata var. liui J. Zhang, & B. Xia35,36. This gene cluster (leuA/B/C/D) was traced to a plasmid from Buchnera, a genus of bacterial endosymbionts in aphids37,38. The ptDNA of the red alga Pyropia haitanensis (T. J. Chang & B.F. Zheng) N. Kikuchi & M. Miyata contains plasmid-derived sequences that were discovered in the plasmid of another red alga, Porphyra pulchra G.J. Hollenberg39,40. The mtDNA of other red algae, Gracilaria chilensis C.J. Bird, J.L. McLachlan, & E.C. de Oliveira, Gracilariopsis chorda (E.M. Holmes) Ohmi and Gracilariopsis lemaneiformis (Bory de Saint-Vincent) Dawson, Acleto, & Foldvik, contain partial plasmid sequences that have been reported in Gracilaria robusta Setchell41,42,43. However, less is known about the mechanisms of plasmid-derived HGT to the plastid genome.
Plasmids are extrachromosomal genetic materials that are generally referred to as autonomously replicating double-stranded, circular or linear DNA molecules44. About 25% of red algal genera contain more than two plasmids per species, and encode open reading frames (ORFs) that are transcriptionally active45. Eukaryotic plasmids are widely distributed throughout algae, land plants, fungi, yeast, and other eukaryotes. However, their origins are poorly understood and their functions, including pathogenicity, have been reported only in a few cases45,46,47,48. Of 35 red algal species assessed for plasmid sequences, 5 species contain 14 plasmid sequences: Porphyra pulchra (five plasmids); Pyropia tenera (Kjellman) N. Kikuchi, M. Miyata, M.S. Hwang & H.G. Choi (two plasmids); Gracilaria chilensis (three plasmids); G. robusta (two plasmids); and Gracilariopsis lemaneiformis (two plasmids)39,45,49,50,51,52. However, no comprehensive analysis has yet been done to investigate the evolutionary relationship between plasmid DNA and plastid genomes.
To this end, we sequenced five red algal ptDNAs, including two that are plasmid-rich from Gracilaria chilensis and Porphyra pulchra. We analyzed plasmid-derived sequences from a total of 21 available red algal plastid genomes35,36,40,53,54,55,56,57,58,59,60,61 to elucidate the impact of plasmids over the >1 billion year evolutionary history of red algae.
Results and discussion
Novel red algal plastid genomes
Five novel plastid genomes were completed using next-generation sequencing (NGS) data from Gelidium elegans (1,529 Mbp of total data), G. vagum (990 Mbp), Gracilaria chilensis (506 Mbp), Porphyra pulchra (263 Mbp) and Sporolithon durum (3,190 Mbp). The range of average genome coverage from the raw data was 52 ~445x (Supplementary Table S1). The plastid genome of P. pulchra (Supplementary Fig. S1) was the largest (194,175 bp) and had a higher GC-content (33.3%) than that of S. durum (191,465 bp, GC = 29.3%, Supplementary Fig. S2), G. elegans (174,748 bp, GC = 30.2%, Supplementary Fig. S3), G. vagum (179,853 bp, GC = 29.9%, Supplementary Fig. S4) and Gr. chilensis (185,637 bp, GC = 29.3%, Supplementary Fig. S5). Basic information about these plastid genomes is summarized in Supplementary Table S2. The plastid genome of P. pulchra, similar to those in other bangiophycean species, comprised 207 protein-coding genes, 37 tRNAs and 6 rRNAs; the rRNA operon (rrs, rrl and rrf) was duplicated. Among the florideophycean species, S. durum comprised 202 protein-coding genes, 30 tRNAs, 3 rRNAs, 3 rRNAs and 2 introns, as well as several pseudogenes (dnaB, syfB, ycf21 and ycf23). This genome lacked the syh gene and trnV tRNA, both of which are present in another member of the Corallinophycidae, Calliarthron tuberculosum. These two coralline algae have a unique group II intron in the chlB gene36 with intronic orfs. Gelidium vagum contained 201 protein-coding genes with pseudogenes of ycf34; G. elegans contained 202 protein-coding genes. These members of the order Gelidiales encode 30 tRNAs, 3 rRNAs and a group II intron in trnMe tRNA36. The plastid genome of Gracilaria chilensis (order Gracilariales) contained 203 protein-coding genes, 30 tRNAs, 3 rRNAs and a group II intron in trnMe tRNA, which had not been found previously in the plastid genomes of G. salicornia and G. tenuistipitata var. liui or in Grateloupia taiwanensis (Halymeniales).
The ML tree inferred from the concatenated dataset of 193 plastid protein-coding genes (Supplementary Table S3; Table S4) resolved phylogenetic relationships among red algae (Fig. 1A, Supplementary Fig. S6). The early diverging Cyanidiophyceae was chosen as the outgroup for this phylogeny62,63. The Bangiophyceae and the Florideophyceae grouped together with maximum ML bootstrap support value (MLB, 100%), and each class formed a strongly supported monophyletic clade, as previously reported62,63,64,65. Within the Bangiophyceae, Porphyra pulchra grouped within Pyropia clade (100% MLB) rather than Porphyra clade, suggesting a taxonomic revision of Porphyra pulchra as Pyropia pulchra. Relationships within the Florideophyceae were consistent with previous work64,65,66. For example, two Corallinophycidae species, Sporolithon durum (Sporolithales) and Calliarthron tuberculosum (Corallinales) grouped together (100% MLB) and were sister to the rest of florideophycean clades. Within the subclass Rhodymeniophycidae, Chondrus crispus (Gigartinales) diverged first, followed by Gelidium (Gelidiales), Grateloupia taiwanensis (Halymeniales) and Gracilaria (Gracilariales). Although internal relationships within the Rhodymeniophycidae were not resolved with the concatenated plastid dataset, we used this ML tree (Fig. 1A) as a reference for inferring the evolution of red algal plasmid DNAs.
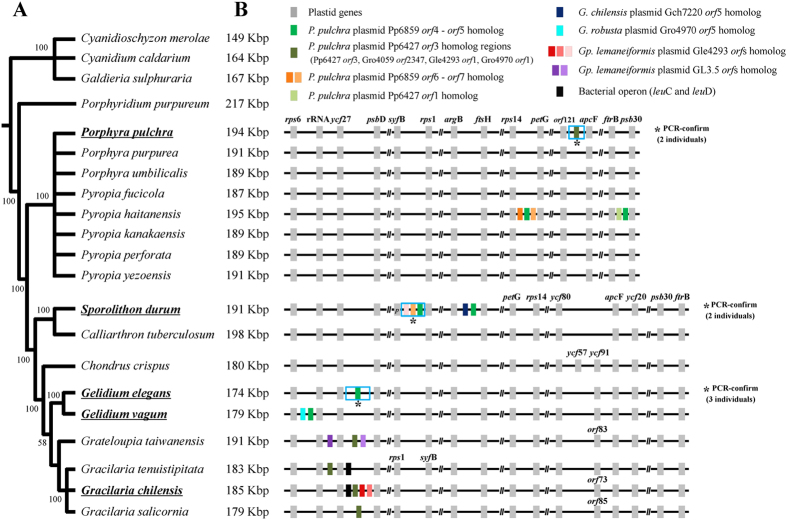
Figure 1. Phylogeny of red algae showing the distribution of plasmid-derived DNA.
(A) Simplified maximum likelihood (ML) tree topology for red algae based on concatenated 193 protein encoding genes of plastid genomes (see also Supplementary Fig. S6). (B) The plastid genome sizes (kilo base pair, Kbp) are shown beside the taxon names. Colored boxes indicate plastid genes (grey), plasmid-derived regions: Porphyra pulchra plasmid Pp6859 orf4 and orf5 homolog (green), P. pulchra plasmid Pp6427 orf3 homolog (dark green), P. pulchra plasmid Pp6859 orf6 and orf7 (orange and bright orange), P. pulchra plasmid Pp6427 orf1 homolog (bright green), Gracilaria chilensis plasmid Gch7220 orf5 homolog (blue), G. robusta plasmid Gro4970 orf5 homolog (cyan), Gracilariopsis lemaneiformis plasmid Gle4293 orfs homolog (red, pink and bright pink), Gp. lemaneiformis plasmid GL3.5 orfs homolog (violet and bright violet), and genes that encode the bacterial operon for leuC and leuD (black). Detailed information about plasmid-derived regions is given in Supplementary Fig. S7. The “p” in syfB gene of Sporolithon durum designates a pseudogenization of the gene. PCR-confirmed regions in different individuals are marked by asterisk. Syntenic diagrams for the Cyanidioschyzon, Cyanidium, Galdieria and Porphyridium are not shown because there was no plasmid-derived DNA.
Distribution of plasmid-derived genes in red algal ptDNA
We identified 22 plasmid-derived (PD) sequences in nine red algal species when 56 red algal plasmid-encoded proteins were used to query the available 21 red algal plastid genomes (using BLASTx, e-value ≤ 1.0e−05) (GI numbers of the 56 proteins are listed in Table 1). The putative origin, copy number, and distribution in the ptDNAs were different for each ORF (Fig. 1B; Supplementary Table S5). In addition to the previously reported bacterial operon leuC and leuD gene35,36 (two black blocks in Fig. 1B), out of the 22 PD orfs (including pseudogenized regions) identified here, six were homologous to orf4 and orf5 of the Porphyra pulchra plasmid Pp6859 (GI: 11466614; green region in Fig. 1B), five were homologous to the P. pulchra plasmid Pp6427 (GI: 11466608) orf3 (dark green region in Fig. 1B), and two were homologous to the P. pulchra plasmid Pp6859 orf6 (bright orange region in Fig. 1B). The rest of the PD orfs were unique to plasmids in their species of origin. Interestingly, six homologous PD sequences from Pp6859 orf4 and orf5 (green box in Fig. 1B) were found in four red algal plastid genomes but their copy number and position were not consistent with their phylogenetic relationships. For instance, two copies of the Pp6859 orf4-orf5 homolog were found in Pyropia haitanensis among eight Porphyra/Pyropia species, whereas a single copy was found in each Gelidium species, but at different locations. Sporolithon durum contained two homologous copies but one was pseudogenized. The sequences homologous to plasmid Pp6427 orf3 of P. pulchra39 were found in the plastid genomes of three Gracilaria species and Grateloupia taiwanensis (dark-green in Fig. 1B) in addition to that of P. pulchra, and were located near ribosomal RNAs and ycf27 genes. We note that half of the PD orfs were positioned near rRNA (rps6-rRNA-ycf27-psbD, see Fig. 1B), in particular in Gelidium, Grateloupia, and Gracilaria.
Table 1. Distribution of red algal plasmids and their homologous sequences (BLASTp results, cut-off = e−05).
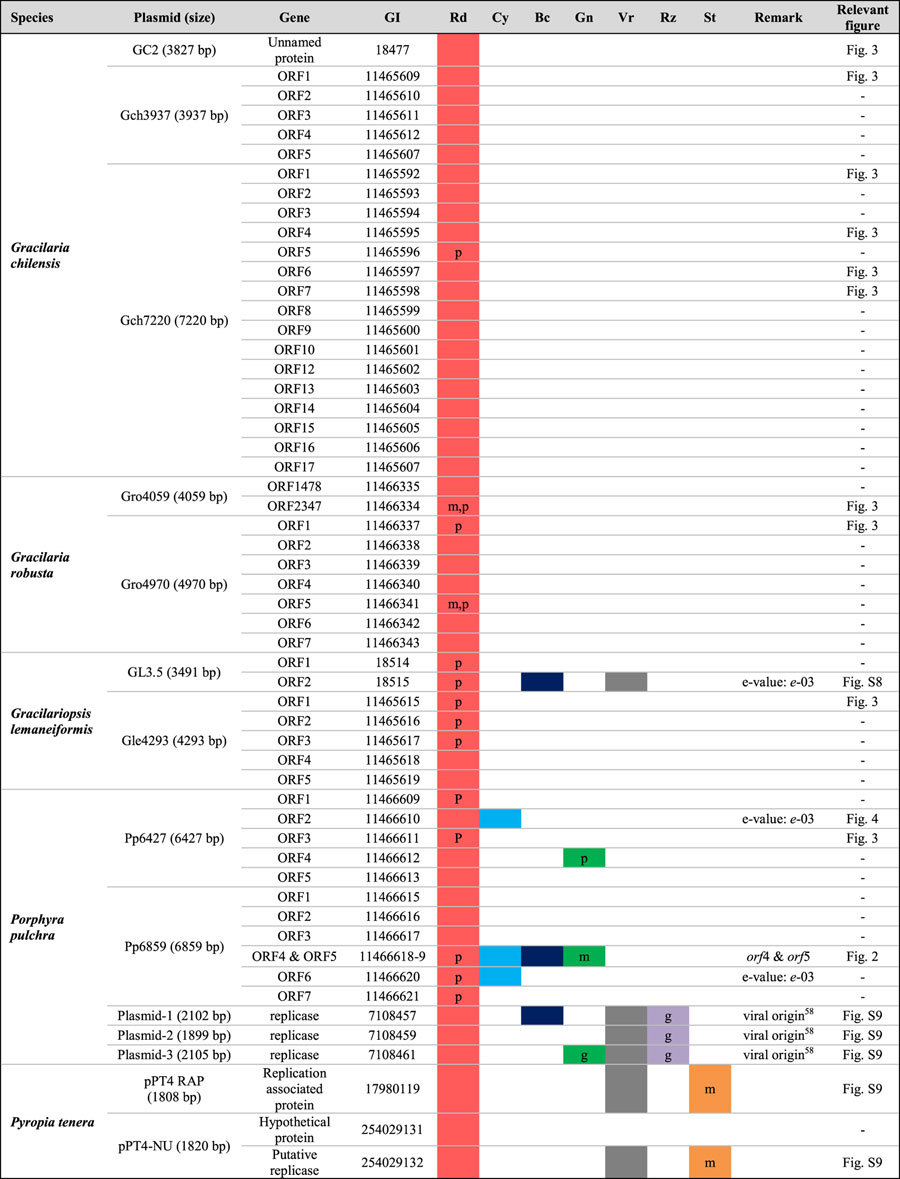
Rd = red algae, Cy = Cyanobacteria, Bc = Bacteria (excluding Cyanobacteria), Gn = green plant lineage (Viridiplantae), Vr = Virus, Rz = Rhizaria, St = Stramenopile. Letters beside the filled circles indicate origins: m for the mitochondrial homolog, p for the plastid homolog and g for the nuclear genome homolog. When the letters m, p, or g are absent, the origin of the gene is unknown.
We tested whether these PD orfs were conserved in populations within a species and in different individuals within a population. To this end, PCR was used to test three populations of G. elegans (SKKU18, SKKU22, SKKU28), two individuals of P. pulchra selected from a single population (UC1879714 and UC1454976), and three individuals of S. durum from a single population (SKKU_SD01, SKKU_SD02, and SKKU_SD03; Supplementary Table S6). All tested PD orfs were found in the same position with the same flanking region sequences. Therefore, these PD orfs are conserved across different individuals within one species.
For P. pulchra and G. chilensis, the PD orfs found in their ptDNA or their homologs were not detected in the draft genome data but rather only in the plasmid sequence. From draft genome data (NGS), five complete plasmid sequences (103 ~360× average coverage) were recovered from Porphyra pulchra (263 Mbp of reads) and three plasmids (401 ~590× average coverage) from Gracilaria chilensis (506 Mbp of reads). Thus, NGS data were useful for identifying plasmid sequences. However, we could not find red algal plasmid sequences in the published complete genome of Cyanidioschyzon merolae67,68, Galdieria sulphuraria69, Porphyridium purpureum70, Calliarthron tuberculosum71, Chondrus crispus56, and Gelidium vagum (Yoon et al. unpublished).
Origin of the plasmid-derived Pp6859 orf4-orf5 homologs in ptDNA
The origin of plasmid-derived orfs was difficult to determine because most plastid-encoded PD orfs matched only plasmid orf data, except for the following five cases (see, Figs 2, 3, 4, S8, S9). A BLAST search against the NCBI database using six homologous plastid genes of the P. pulchra plasmid Pp6859 orf4-orf5 resulted in 26 hypothetical proteins encoded in a bacterial genome, cyanobacterial genomes, cyanobacterial plasmids, and the mitochondrial genome of a liverwort. All homologous sequences of Pp6859 orf4-orf5 were used to reconstruct the ML phylogeny using RAxML (Fig. 2). In the best tree, red algal plastid PD orfs grouped together, including plasmid Pp6859 (98% MLB). It is interesting that plasmid genes of Pp6859 (P. pulchra) grouped with pseudogenized plastid genes from P. haitanensis (100% MLB), suggesting a possible ORF gene transfer mediated by a plasmid to a plastid genome (see discussion in previous study40).
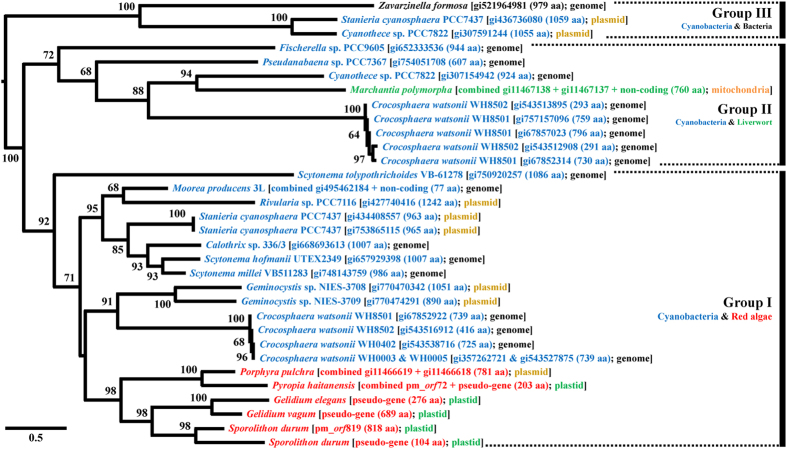
Figure 2. Maximum likelihood (ML) tree based on aligned amino acid sequences of homologous regions of Porphyra pulchra plasmid Pp6859 orf4 and orf5 with 2,000 ML bootstrap replications.
Species names are followed by GI, amino acid (aa) length, and location. Colored names indicate cyanobacteria (cyan), bacteria (black), liverwort (bright green) and red algae (red). Locations of the sequences are genome (black), plastid (green), mitochondria (orange) and plasmid (yellowish brown). Some orfs and pseudogenized or non-coding regions were combined and aligned with sampled taxon sequences (Supplementary Table S3; Table S4; Table S7). The clades of the ML tree are divided into three groups based on species composition. Group I includes cyanobacterial plasmids and genomes with red algal plasmid and plastid regions. Group II includes cyanobacterial genomes and mitochondrial regions of liverwort, Marchantia polymorpha). Group III includes cyanobacterial plasmids and a bacterial (Zavarzinella formosa) genome.
Figure 3. Maximum likelihood (ML) tree based on aligned amino acid sequences of homologous regions to Porphyra pulchra plasmid Pp6427 orf3 with 2,000 ML bootstrap replications.
Species names are followed by source, GI, amino acid (aa) length, and location. Some orfs and pseudogenized or non-coding regions were combined and aligned with sampled taxon sequences (Supplementary Table S7). Location of sequences is indicated by color: plasmid (underlined black), plastid (green) and mitochondria (orange). Synteny is shown with the schematic alignment on the right of the tree based on major regions of homology.
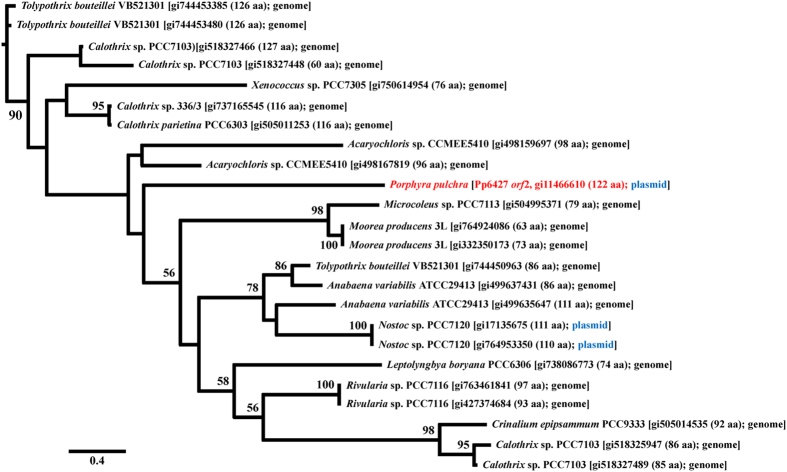
Figure 4. Maximum likelihood (ML) tree based on aligned amino acid sequences of homologous genes of Porphyra pulchra plasmid Pp6427 orf2 (red) with 2,000 ML bootstrap replications.
Blue indicates that the sequences found in the plasmid genome.
The red algal clade was positioned within cyanobacterial clade Group I (92% MLB) that included hypothetical proteins encoded in the cyanobacterial genome as well as cyanobacterial plasmids (Fig. 2). Group II (72% MLB) contained cyanobacterial species and mitochondrial sequences from the liverwort Marchantia polymorpha (combined with two fragmented genes with flanking region data). Moon and Goff39 reported the putative homologous relationship between Pp6859 and the liverwort mitochondrial region. Two cyanobacterial plasmid genes and a hypothetical gene from the Planctomycetes Zavarzinella formosa were grouped together (Group III, 100% MLB).
Because only 12 species (16 strains; Fig. 2) out of the 100 cyanobacterial genomes available in NCBI contain a homolog of Pp6859 orf4-orf5, it is unlikely to be a core cyanobacterial gene. If this orf was inherited from the primary endosymbiosis event, it should be retained in most red algal plastid genomes as well as those of other primary endosymbiotic lineages (i.e., green and glaucophyte algae). However, it is sporadically distributed in only a few species (e.g., Pyropia, Gelidium and Sporolithon) (Fig. 1). We postulate that this orf originated from an unknown cyanobacterial species, then spread independently to other cyanobacteria, to a bacterium (Z. formosa), to a liverwort (M. polymorpha), and to a few red algae.
The cyanobacterium Crocosphaera watsonii WH8501 contains three copies of this orf as a result of gene duplications72,73. However, it is likely that these red algal PD orfs originated independently, as a result of plasmid mobility. Alternatively, a red algal species inherited this orf from a cyanobacterial genome through the plasmid, after which it was transferred into the plastid genome in random genomic positions (e.g., see Fig. 1B), followed by pseudogenization or complete loss. This plasmid-mediated HGT may have occurred after speciation. For example, two Gelidium species both retain PD orfs, but they differ in size and genomic position. Similar cases were found in three Gracilaria species. If indeed the PD orfs were introduced during speciation, the presence and position of PD orfs could be used as species-specific markers.
Origin of the plasmid-derived Pp6427 orfs homologs in plastid genomes
Pp6427 orf3 homologs were found in five plastid regions (Fig. 1), nine plasmid orfs and a mitochondrial orf from seven red algal species. Unlike Pp6859, homologous sequences were not found in any other taxa; therefore, Pp6427 orf3 homologs are specific to red algae. In the ML tree using 16 homologs (Fig. 3), strong (>95% MLB) plasmid-plastid relationships were recovered, even though all plastid-encoded orfs were pseudogenized (see alignment in Fig. 3). For example, the plasmid Pp6427 orf3 (485 aa) grouped with a short pseudogenized gene (191 aa) in the plastid genome of P. pulchra (95% MLB), whereas the plasmid Gro4059 (GI: 11466333) orf2347 (190 aa) grouped with partial genes from G. taiwanensis (101 aa) within a clade of mitochondrial orf44 from Palmaria palmata (Linnaeus) F. Weber & Mohr (88% MLB) (Fig. 3). Gracilaria chilensis contained six orfs in three plasmids (Gch7220 [GI: 11465591], Gch3937 [GI: 11465608], and GC2 [GI: 18476]); however, those orfs did not group with the plastid-encoded homologs that were clustered (99% MLB) with the pseudogenized plastid gene of G. tenuistipitata (144 aa) and plasmid Gle4293 (GI: 11465614) orf1 of G. lemaneiformis.
Because the evolutionary trajectories of plasmid and plastid copies are very different (the former presumably functional and therefore subject to purifying selection, but the latter pseudogenized and under relaxed selective constraint), it is difficult to infer evolutionary relationships, since both rates and types of mutation (synonymous versus nonsynonymous) may be very different depending on the genetic background. We think it is likely that the plasmid orfs are ancestral because they contain complete orfs (405–485 aa), whereas plastids contain pseudogenized genes (up to 190 aa). On the other hand, plastid sequences occur in all the Gracilaria clades; the difference may be due to relaxed purifying selection on the shorter, non-functional (pseudogenized) plastid copies. Pp6427 orf3 homologs were found in the closely related genera Grateloupia, Gracilaria, and Gracilariopsis (multigene phylogeny using mitochondrial genes43), suggesting that an ancestral Pp6427 orf3 of P. pulchra was transferred into the ancestral plastid of these genera and the mitochondrial genome of Palmaria palmata. Some plasmid orfs were duplicated (e.g., Gch7220 orfs and Gch3937 orf1) and fragmented (orf1, orf6, and orf7) within a plasmid (Gch7220). Although the origin of the plasmid-derived sequences is unknown, they may have spread into red algal organelle genomes and subsequently undergone relaxed selective constraint.
Two other plasmid orfs of Pp6427, orf2 and orf4 showed exclusive homology to cyanobacteria and green plants species, respectively. Pp6427orf2 was homologous to a putative transcriptional regulator protein (GI: 495464247) from the cyanobacterium Moorea producens (e-value: 4e−08) and to other cyanobacterial genes. The red algal plasmid orf2 was likely transferred from cyanobacteria (Fig. 4; MLB 90% in basal clade). The combined region from Pp6427 orf4 (Supplementary Table S7, ML tree is not shown) and their flanking regions were homologous to a hypothetical protein (orf619) from the plastid genome of Ettlia pseudoalveolaris (T.R. Deason & H.C. Bold) J. Komárek (green alga; GI: 725650857; BLASTx result e-value: 5e−12) as well as orf436 of Mankyua chejuensis B.Y. Sun, M.H. Kim & C.H. Kim (fern; GI: 727397314; BLASTx result e-value: 7e−05). Therefore, orfs encoded in the plasmid Pp6427 originated from various sources, and some orfs were subsequently transferred to the red algal plastid and mitochondrial genomes. Both plasmid Pp6859 orf4-orf5 and Pp6427 orf2 were homologous to cyanobacterial orfs, including those from several common species, Calothrix sp. 336/3, Moorea producens 3L, and Rivularia sp. PCC7116. Thus, these two plasmids may have served as reservoirs for orfs from different sources that eventually were delivered to organelles.
Bacterial and viral origins of red algal plasmid ORFs
Bacterial or viral sequences were detected by a BLASTp search of the NCBI (nr) database using 22 PD red algal plastid orf queries (Table S5). The homologous sequence of Gracilariopsis lemaneiformis plasmid GL3.5 orf2 in the Grateloupia taiwanensis plastid genome showed a close phylogenetic relationship with bacterial and viral sequences (Supplementary Fig. S8). This red algal clade was positioned within the bacterial clades (100% MLB), suggesting the bacterial origin of the GL3.5 orf2 homologs. It was, however, unclear whether this plasmid-related sequence was transferred from bacteria directly or by a virus-mediated process, because the clade showed a sister relationship to the viral clade but with weak statistical support (48% MLB).
The ML tree based on red algal plasmid-encoded replicase genes (i.e., Pyropia tenera, GI: 17980119, 254029132; P. pulchra, GI: 7108457, 7108459, 7108461) and homologous sequences from a BLASTp search suggest a viral origin of these plasmid orfs (see Supplementary Fig. S9). Five red algal plasmid orfs, mitochondrial orf98 from Phytophthora sojae (stramenopile; GI: 145932354), and nuclear-encoded genes from Reticulomyxa filosa (rhizaria; GI: 569382219, 569382219) grouped together with diverse circular virus DNA (52% MLB), as well as with obligate parasitic bacteria (i.e., onion yellow phytoplasma) (78% MLB). A BLASTp search (cutoff e-value ≤ 1.0e−05, see Table 1) recognized eight sequences from the nuclear genome of Nicotiana tomentosiformis (green plant; GI: 697190580, 1587991, 697190578, 697159806, 697175541, 697190576, 697140845, 697149473) that were distantly related to the red algal plasmid clade. The replicase gene from the P. pulchra plasmid (GI: 7108457) was reported as a geminivirus-related sequence because it share five conserved motifs and phylogenetic affinities51,78.
Virus-derived plasmid genes (i.e., GL3.5 orf2, three replicase genes in P. pulchra plasmids, and two replicase genes in Py. tenera plasmids) were detected in both eukaryotic nuclear and organellar genomes. These were different from non-viral-derived red algal plasmid homolog sequences that were found only in organelle genomes (Table 1). It is likely that virus-derived plasmid genes could be transferred to the eukaryotic nuclear genome more easily than could non-viral plasmid genes.
Remnant DNA replication domain in plasmid-derived plastid genes
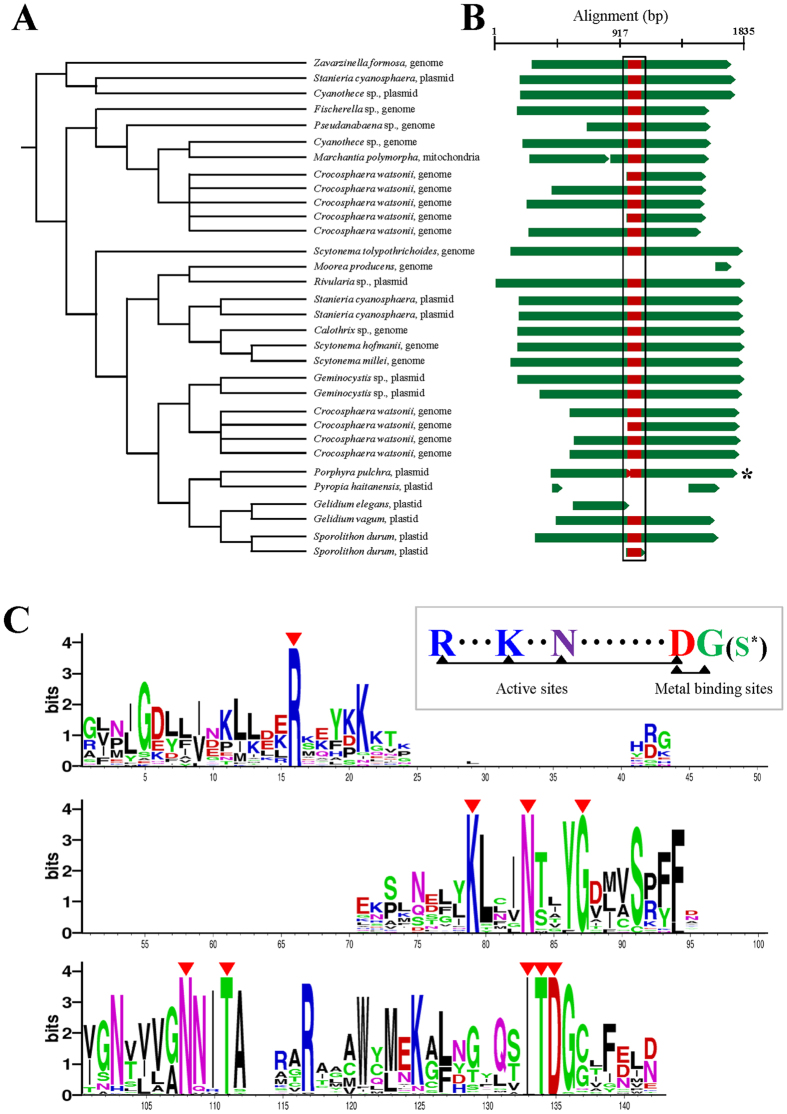
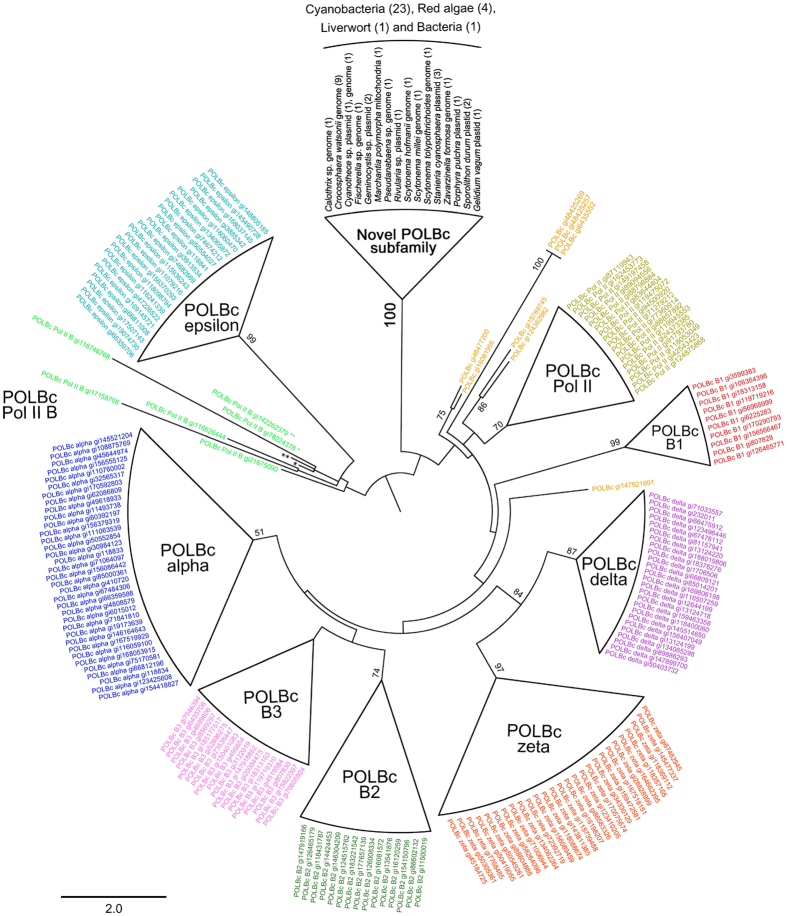
Plasmids are composed of three essential domains for replication, segregation and conjugation with additional accessory genes76,77. From the alignment of the Pp6859 orf4-orf5 homologs with the size range of 104 ~1,242 amino acid sequences, the functional domain was detected by a conserved domain database search79. One distinct domain is the DNA polymerase type-B family catalytic domain (POLBc) superfamily. Nine amino acid sequences were identical in this domain (aligned 142 aa), including highly conserved active sites (R-K-ND motif) and metal binding sites (DG motif) (see Fig. 5). The DNA polymerase type-B family consists of an editing active site and excision region for DNA replication (562 ~3,425 aa in size) that has been reported in a wide range of organisms, including Archaea, Bacteria, eukaryotes, bacteriophages and viruses80,81,82,83,84,85,86,87,88,89,90,91. Although the POLBc motif was generally conserved in nine major subfamilies85, we found differences in the catalytic domain of the Pp6859 orf4-orf5 homologs. These unique domains were represented in the ML tree that was reconstructed using homolog regions of the domain (aligned 222 aa) from the public POLBc superfamily database (Supplementary Table S8; Fig. 6). The ML tree showed that all POLBc domains in the Pp6859 orf4-orf5 homolog were grouped into a clade (100% MLB), but the clade did not belong to any other known POLBc subfamilies. This novel POLBc domain might contribute to the insertion of plasmid orfs into the red algal plastid genome.
Figure 5. Domain search of Porphyra pulchra plasmid Pp6859 orf4-orf5 homolog.
(A) Cladogram of the best maximum likelihood tree, Fig. 2. (B) Schematic of the DNA polymerase type-B family catalytic domain (POLBc) superfamily related regions (rectangular boundary) in the hypothetical homologous gene (green bars). Active and metal binding sites on the POLBc superfamily are shown below the alignment. In the domain of Porphyra pulchra (*), serine (S*) substitutes for glycine (G) in metal binding sites. (C) The composition of amino acids in alignment. Representative color of amino acid follows the Chemistry color scheme in WebLogo with probability-based size differences. The nine red arrowheads indicate conserved amino acids (100%) in the alignment. Blanks in the alignment indicate extremely low contribution by the hypothetical protein in Fischerella sp. (GI: 652333536) and in Scytonema tolypothrichoides (GI: 750920257).
Figure 6. Maximum likelihood (ML) tree based on aligned amino acid sequences of DNA polymerase type-B family catalytic domain (POLBc) superfamily with 2,000 ML bootstrap replications.
A public POLBc database was used from the conserved domain database (CDD) (Supplementary Table S8). Each cluster indicates a subfamily of POLBc superfamily. The novel POLBc subfamily clade comprises the POLBc-related partial domains of the hypothetical gene in this study. Eight public domain data are not identified to a specific subfamily in the POLBc superfamily (bright orange; GI: 16081956, 48477200, 15789745, 124362982, 48425269, 48425257, 6435552, 147921691). The DNA polymerase type-II B (Pol II B; bright green) subfamily is not monophyletic. Most subfamilies of POLBc are monophyletic; however, some inter-clade relationships differ slightly from the public POLBc superfamily database (CDD cl00145).
Conclusions
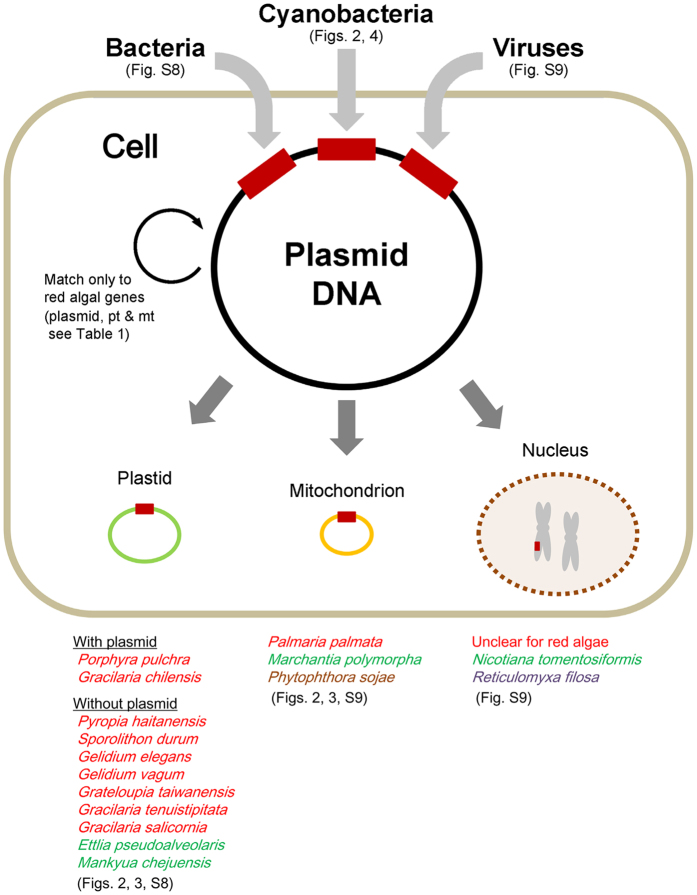
Plasmids have long been recognized as mobile elements but their origins in red algae remained unclear. Using a comprehensive database of 21 plastid genomes that included five novel red algal ptDNAs, we found evidence for the spread of plasmid DNA into plastid and mitochondrial genomes. There is currently insufficient nuclear genome data from species that contain plasmid-derived DNA to determine whether this compartment is also a major target for integration (Fig. 7). The distribution of plasmid-derived orfs showed a species-specific pattern, consistent with the evolution of a mobile genetic element. Because organelles are inherited maternally, foreign genetic DNA can be rapidly fixed in a population. Consistent with this idea, individual members of three lineages (i.e., Porphyra pulchra, Sporolithon durum, and Gelidium elegans) all showed plasmid DNA retention, although these orfs were absent or located in different genomic positions in closely related sister species (e.g., eight Porphyra/Pyropia species, Sporolithon-Calliarthron, two Gelidium species, see Fig. 1). It is known that the distribution of transposable elements can show variation within a single cyanobacterial species72,73,92. Therefore, plasmids may be regarded as analogous to transposable elements76,77,93,94,95,96, with mobility and loss contributing to variation in gain/loss among closely related genomes. For instance, Halary et al.97 demonstrated that plasmids are key vectors of genetic exchange between bacterial chromosomes on the basis of network analysis using sequences including phage, plasmid and environmental viral genomes.
Figure 7. The spread of plasmid DNA in eukaryote genomes.
The schematic cell includes the nucleus (dotted line circle), plastid (green circle), mitochondrion (orange circle) and plasmid (black circle) DNA. The plasmid-derived regions are indicated as red boxes in the genomes. The flow of plasmid DNA is indicated by the arrows. Plasmid-mediated HGT in plastid genomes are divided into two types: with plasmid and without plasmid (Figs 2, 3 and S8). Organisms with and without plasmid DNA are listed below as red algae (red taxa), green lineage (green taxa), stramenopiles (brown taxa), and rhizarians (violet taxa). Plastid genomes of Porphyra pulchra and Gracilaria chilensis include plasmid-derived homologs in both their plastid and plasmid genomes. The other red algae include plasmid-derived homologs only in the plastid genome. Mitochondrial HGT is found in red algae, the green lineage and stramenopiles (Figs 2, 3 and S9). Plasmid-mediated transfer to the nuclear genome is found only in Nicotiana tomentosiformis (plants) and Reticulomyxa filosa (rhizarian), with both regions related to viruses (Supplementary Fig. S9).
It should be noted that we were originally interested in testing the idea whether plasmids may have facilitated EGT in algae and thereby played a key role in their genome evolution. Analysis of the available data, however, suggests that plasmids are better thought of as parasitic elements (e.g., group II introns in red algal ptDNA98) that spread plasmid-derived DNA regions. As “mobile gene cassettes”75,76,77,78 it nonetheless remains possible that these selfish elements can mediate gene transfer between foreign DNA and organelles. As the databases of available organelle and nuclear genome data increase, plasmid involvement in recent instances of EGT may become apparent.
In summary, one of the major challenges in the field of microbial eukaryote genome evolution is to understand how genes move across the tree of life. Species such as Galdieria sulphuraria encode at least 5% foreign genes, many of which are clearly of adaptive value69. The halotolerant green alga Picochlorum SE3 has acquired at least 24 genes of bacterial provenance, putatively to deal with abiotic stress99. Plasmids, viruses, symbionts, and pathogens likely all play a role in the HGT process in protists. Therefore, the search for “smoking guns” of recent transfer will continue to fascinate biologists who seek to show that highways of gene sharing100, common in prokaryotes, are drivers of evolution in eukaryotic microbes.
Methods
Sample preparation, genome sequencing, and assembly
Thalli of the red algal species Gelidium elegans Kützing, G. vagum Okamura, Gracilaria chilensis C.J. Bird, J.L. McLachlan, & E.C. de Oliveira, and Sporolithon durum (Foslie) R. Townsend & W. Woelkerling were collected from nature and immediately dried in silica-gel. Tissue samples of Porphyra pulchra were taken from herbarium specimens collected in 1970 and housed at the University Herbarium, University of California at Berkeley (UC). Detailed information about the samples is shown in Table S6 in the Supplementary Information. Genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Next-Generation Sequencing (NGS) was carried out using Ion Torrent PGM platform (Life Technologies, San Francisco, California, USA). The Ion Xpress Plus gDNA Fragment Library Kit (Life Technologies) was used for 200 bp-sized or 400 bp-sized sequencing library preparation. Genome sequencing was done with the Ion PGM Template OT2 200 or 400 Kit and Ion PGM Sequencing 200 or 400 Kit (Life Technologies, San Francisco, California, USA).
The raw NGS reads were assembled using the CLC Genomics Workbench 5.5.1 (CLC bio, Aarhus, Denmark) and the MIRA assembler that was incorporated in the Ion Server. Contigs of plastid genes were sorted by customized Python scripts with local BLAST searches. Sorted contigs were re-assembled to construct consensus plastid genomes. A draft plastid genome was confirmed by the read-mapping method using CLC Genomics Workbench 5.5.1. Gaps were filled by PCR to generate intact genomes.
Gene annotation and plasmid-derived ORFs search
Putative ORFs in the five novel genomes were predicted using ORF Finder in Geneious 6.1.6101 and annotated based on BLASTx searches (e-value ≤ 1.0e−05) with codon table 11 (Bacterial, Archaeal and Plant Plastid Code). Ribosomal RNAs and transfer RNAs were predicted using the RNAmmer 1.2 Server102 and ARAGORN programs103. Group II intron and RNase P were searched using the program RNAweasel (http://megasun.bch.umontreal.ca/cgi-bin/RNAweasel/RNAweaselInterface.pl). Plasmid-derived sequences were searched by BLASTx (e-value ≤ 1.0e−05) using 56 proteins encoded in 14 red algal plasmids (Supplementary Table S9) derived from all available red algal ptDNAs. We also searched for plasmid-derived sequences in nuclear genome data. Here 56 plasmid-encoded genes were searched in the complete nuclear genomes of Cyanidioschyzon merolae67,68, Galdieria sulphuraria (Galdieri) Merola69, Porphyridium purpureum (Bory) K.M. Drew & R. Ross70, Calliarthron tuberculosum (Postels & Ruprecht) E.Y. Dawson71, Chondrus crispus Stackhouse56 and the 5 novel red algal draft genomes. Reported plasmid sequences from Gracilaria chilensis (GI: 11465591, 11465608 and 18476) and Porphyra pulchra (GI: 11466614, 11466608, 7108456, 7108458 and 7108460) were used as reference sequences for the read-mapping method for NGS data. To check consistency within individuals and populations, plasmid-derived sequences were determined from three individuals of Gelidium elegans from three different sites in Korea (SKKU18, SKKU22 and SKKU28), two individuals of Porphyra pulchra from Moss Beach, CA, USA (UC1454976 and UC1879714), and three individuals of Sporolithon durum from Army Bay, Whangaparaoa, NZ (SKKU_SD01, SKKU_SD02 and SKKU_SD03) using PCR with custom primer pairs (Supplementary Table S10).
Phylogenetic analysis of red algal plasmid-derived genes in plastid genome
Plastid-coding genes from 21 taxa (16 reference genomes and our five new genomes) were extracted and sorted by customized Python scripts with local BLAST searches. To identify the independent loss of plastid genes, each gene set was manually analyzed. A selection of 193 plastid-coding genes (e.g., homologous genes present in at least 16 different taxa) and plasmid-derived sequences were aligned using MAFFT 7.110104. All aligned plastid genes were concatenated for multigene phylogenetic analysis. Based on the alignment, fragmented plasmid-derived orfs were combined (Supplementary Table S7). To reconstruct the phylogenetic tree, an evolutionary model was selected using Modeltest implemented in MEGA 6.0105. Maximum likelihood (ML) tree search and ML bootstrap analysis were done using RaxML 8.0.0 with 2000 replications106 with the PROT + GAMMA + LG4MF model of evolution.
Domain prediction and phylogenetic analysis
Protein domain prediction was done using the conserved domain database CDD79. Predicted domain sequences were aligned and represented using Weblogo107. Aligned sequences of DNA polymerase type-B family catalytic domain (POLBc) subfamilies (POLBc_B1, POLBc_B2, POLBc_alpha, POLBc_delta, POLBc_zeta, POLBc_epsilon, POLBc_B3, POLBc_Pol_II, POLBc_Pol_II_B and unspecified POLBc domain (Supplementary Table S8) were used to find the inter-subfamily relationship based on the RAxML phylogeny.
Additional Information
How to cite this article: Lee, J.M. et al. Reconstructing the complex evolutionary history of mobile plasmids in red algal genomes. Sci. Rep. 6, 23744; doi: 10.1038/srep23744 (2016).
Supplementary Material
Acknowledgments
We thank the two anonymous reviewers for their constructive comments, which helped improve the manuscript. This study was supported by the Global Ph.D. Fellowship (GPF) Program from the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2013–034853) of Korea to JL, NRF (2013–0699 and MEST: 2014R1A2A2A01003588) and Marine Biotechnology Program (PJT200620) funded by Ministry of Oceans and Fisheries, Korea to ECY, SMB and HSY, Korean RDA Next-generation BioGreen21 (PJ011121) to HSY.
Footnotes
Author Contributions J.L., K.M.K., E.C.Y., K.A.M., S.M.B. and H.S.Y. collected samples from field and K.A.M. contributed tissue samples of Porphyra pulchra housed in the University Herbarium, University of California at Berkeley (UC). J.L., K.M.K. and E.C.Y. produced the genome sequencing. J.L. lead the plastid genome assembly, genome prediction, and wrote the manuscript. H.S.Y., S.M.B. and D.B. contributed to designing the work, interpreting the results, and editing the manuscript. All authors discussed the results and commented on the manuscript.
References
- Ochman H., Lawrence J. G. & Groisman E. A. Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304 (2000). [DOI] [PubMed] [Google Scholar]
- Lawrence J. G. Gene transfer in bacteria: speciation without species? Theor. Popul. Biol. 61, 449–460 (2002). [DOI] [PubMed] [Google Scholar]
- Wiedenbeck J. & Cohan F. M. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 35, 957–976 (2011). [DOI] [PubMed] [Google Scholar]
- Salzberg S. L., White O., Peterson J. & Eisen J. A. Microbial genes in the human genome: lateral transfer or gene loss? Science 292, 1903–1906 (2001). [DOI] [PubMed] [Google Scholar]
- Archibald J. M., Rogers M. B., Toop M., Ishida K. & Keeling P. J. Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid-containing alga Bigelowiella natans. Proc. Natl. Acad. Sci. USA 100, 7678–7683 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C., Brachat S. & Dietrich F. S. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot. Cell 4, 1102–1115 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice D. W. & Palmer J. D. An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol. 4, 31 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards T. A., Dacks J. B., Jenkinson J. M., Thornton C. R. & Talbot N. J. Evolution of filamentous plant pathogens: gene exchange across eukaryotic kingdoms. Curr. Biol. 16, 1857–1864 (2006). [DOI] [PubMed] [Google Scholar]
- Novo M. et al. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc. Natl. Acad. Sci. USA 106, 16333–16338 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards T. A. et al. Phylogenomic analysis demonstrates a pattern of rare and ancient horizontal gene transfer between plants and fungi. Plant Cell 21, 4897–1911 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guljamow A. et al. Horizontal gene transfer of two cytoskeletal elements from a eukaryote to a cyanobacterium. Curr. Biol. 17, 757–759 (2007). [DOI] [PubMed] [Google Scholar]
- Rogers M. B., Patron N. J. & Keeling P. J. Horizontal transfer of a eukaryotic plastid-targeted protein gene to cyanobacteria. BMC Biol. 5, 26 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D., Yoon H. S. & Hackett J. D. Photosynthetic eukaryotes unite: endosymbiosis connects the dots. BioEssays 26, 50- 60 (2004). [DOI] [PubMed] [Google Scholar]
- Timmis J. N., Ayliffe M. A., Huang C. Y. & Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Genetics 5, 123–135 (2004). [DOI] [PubMed] [Google Scholar]
- Richardson A. O. & Palmer J. D. Horizontal gene transfer in plants. J. Exp. Bot. 58, 1–9 (2007). [DOI] [PubMed] [Google Scholar]
- Keeling P. J. & Palmer J. D. Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 9, 605–618 (2008). [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A., Hackett J. D., Soares M. B., Bonaldo M. F. & Bhattacharya D. Cyanobacterial contribution to algal nuclear genomes is primarily limited to plastid functions. Curr. Biol. 16, 2320–2325 (2006). [DOI] [PubMed] [Google Scholar]
- Price D. C. et al. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science 335, 843–847 (2012). [DOI] [PubMed] [Google Scholar]
- Sato N., Ishikawa M., Fujiwara M. & Sonoike K. Mass identification of chloroplast proteins of endosymbiont origin by phylogenetic profiling based on organism-optimized homologous protein groups. Genome Inform. 16, 56–68 (2005). [PubMed] [Google Scholar]
- Deusch O. et al. Genes of cyanobacterial origin in plant nuclear genomes point to a heterocyst-forming plastid ancestor. Mol. Biol. Evol. 25, 748–461 (2008). [DOI] [PubMed] [Google Scholar]
- Dagan T. et al. Genomes of Stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol. Evol. 5, 31–44 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa A. & Bhattacharya D. PhyloSort: a user-friendly phylogenetic sorting tool and its application to estimating the cyanobacterial contribution to the nuclear genome of Chlamydomonas. BMC Evol. Biol. 8, 6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W. et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. USA 99, 12246–12251 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub S. C. K., Cronn R. C., Edwards C., Fishbein M. & Liston A. Horizontal transfer of DNA from the mitochondrial to the plastid genome and its subsequent evolution in milkweeds (Apocynaceae). Genome Biol. Evol. 5, 1872–1885 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulintchenko M., Konstantinov Y. & Dietrich A. Plant mitochondria actively import DNA via the permeability transition pore complex. EMBO J. 22, 1245–1254 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan M. B., McCurdy D. W. & Rose R. J. Mitochondria as a connected population: ensuring continuity of the mitochondrial genome during plant cell dedifferentiation through massive mitochondrial fusion. Plant J. 44, 744–755 (2005). [DOI] [PubMed] [Google Scholar]
- Rice D. W. et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 342, 1468–1473 (2013). [DOI] [PubMed] [Google Scholar]
- Paillard M., Sederoff R. R. & Levings C. S. III Nucleotide sequence of the S-1 mitochondrial DNA from the S cytoplasm of maize. EMBO J. 4, 1125–1128 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B., Börner T. & Weihe A. Remnants of a DNA polymerase gene in the mitochondrial DNA of Marchantia polymorpha. Curr. Genet. 27, 488–490 (1995). [DOI] [PubMed] [Google Scholar]
- Burger G., Saint-Louis D., Gray M. W. & Lang B. F. Complete sequence of the mitochondrial DNA of the red alga Porphyra purpurea: cyanobacterial introns and shared ancestry of red and green algae. Plant Cell 11, 1675–1694 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousvoal S., Oudot M. P., Fontaine J. M., Kloareg B. & Loiseaux-de Goër S. Witnessing the evolution of transcription in mitochondria: the mitochondrial genome of the primitive brown alga Pylaiella littoralis (L.) Kjellm. Encodes a T7-like RNA polymerase. J. Mol. Biol. 277, 1047–1057 (1998). [DOI] [PubMed] [Google Scholar]
- Férandon C., Xu J. & Barroso G. The 135kbp mitochondrial genome of Agaricus bisporus is the largest known eukaryotic reservoir of group I introns and plasmid-derived sequences. Fungal Genet. Biol. 55, 58–91 (2013). [DOI] [PubMed] [Google Scholar]
- Nadimi M., Beaudet D., Forget L., Hijri M. & Lang B. F. Group I intron–mediated trans-splicing in mitochondria of Gigaspora rosea and a robust phylogenetic affiliation of arbuscular mycorrhizal fungi with Mortierellales. Mol. Biol. Evol. 29, 2199–2210 (2012). [DOI] [PubMed] [Google Scholar]
- Khan H. et al. Plastid genome sequence of the cryptophyte alga Rhodomonas salina CCMP1319: lateral transfer of putative DNA replication machinery and a test of chromist plastid phylogeny. Mol. Biol. Evol. 24, 1832–1842 (2007). [DOI] [PubMed] [Google Scholar]
- Hagopian J. C., Reis M., Kitajima J. P., Bhattacharya D. & de Oliveira M. C. Comparative analysis of the complete plastid genome sequence of the red alga Gracilaria tenuistipitata var. liui provides insights into the evolution of rhodoplasts and their relationship to other plastid. J. Mol. Evol. 59, 464–447 (2004). [DOI] [PubMed] [Google Scholar]
- Janouškovec J. et al. Evolution of red algal plastid genome: ancient architectures, introns, horizontal gene transfer, and taxonomic utility of plastid markers. PLoS ONE 8, e59001 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann L., Baumann P., Moran N. A., Sandwtröm J. & Thao M. L. Genetic characterization of plasmids containing genes encoding enzymes of leucine biosynthesis in endosymbionts (Buchnera) of aphids. J. Mol. Evol. 48, 77–85 (1999). [DOI] [PubMed] [Google Scholar]
- Sabater-Muñoz B., van Ham R. C. H. J., Moya A., Silva F. J. & Latorre A. Evolution of the leucine gene cluster in Buchnera aphidicola: insights from chromosomal versions of the cluster. J. Bacteriol. 186, 2646–2654 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon D. A. & Goff L. J. Molecular characterization of two large DNA plasmids in the red alga Porphyra pulchra. Curr. Genet. 32, 132–138 (1997). [DOI] [PubMed] [Google Scholar]
- Wang L. et al. Complete sequence and analysis of plastid genomes of two economically important red algae: Pyropia haitanensis and Pyropia yezoensis. PLoS ONE 8, e65902 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. et al. Complete sequences of the mitochondrial DNA of the wild Gracilariopsis lemaneiformis and two mutagenic cultivated breeds (Gracilariaceae, Rhodophyta). PLoS ONE 7, e40241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E. C., Kim K. M., Kim S. Y. & Yoon H. S. Complete mitochondrial genome of agar-producing red alga Gracilariopsis chorda (Gracilariales). Mitochondrial DNA 25, 339–341 (2014). [DOI] [PubMed] [Google Scholar]
- Lee J.-M., Boo S. M., Mansilla A. & Yoon H. S. Unique repeat and plasmid sequences in the mitochondrial genome of Gracilaria chilensis (Gracilariales, Rhodophyta). Phycologia 54, 20–23 (2015). [Google Scholar]
- Brown G. G. & Finnegan P. M. RNA plasmids. Int. Rev. Cytol. 117, 1–56 (1989). [DOI] [PubMed] [Google Scholar]
- Goff L. J. & Coleman A. W. Red algal plasmids. Curr. Genet. 18, 557–565 (1990). [DOI] [PubMed] [Google Scholar]
- Esser K. et al. in Heidelberg Science Library, Plasmids of eukaryotes: Fundamentals and applications (eds Esser K et al.) 1–124 (Springer, 1986). [Google Scholar]
- Meinhardt F., Kempken F., Kämper J. & Esser K. Linear plasmids among eukaryotes: fundamentals and application. Curr. Genet. 17, 89–95 (1990). [DOI] [PubMed] [Google Scholar]
- Klassen R. & Meinhardt F. in Microbial Linear Plasmids, Vol. 7 (eds Meinhardt F. & Klassen R.) Linear protein-primed replicating plasmids in eukaryotic microbes, 188–226 (Springer, 2007). [Google Scholar]
- Villemur R. Circular plasmid DNAs from the red alga Gracilaria chilensis. Curr. Genet. 18, 251–257 (1990). [Google Scholar]
- Villemur R. The DNA sequence and structural organization of the GC2 plasmid from the red alga Gracilaria chilensis. Plant Mol. Biol. 15, 237–243 (1990). [DOI] [PubMed] [Google Scholar]
- Moon D. A. & Goff L. J. Small plasmids in Porphyra pulchra are related to flowering plant geminiviruses. J. Phycol. 32, 33–34 (1996). [Google Scholar]
- Choi H. S., Choi K. H., Kim T. H., Lee C. H. & Rhew T. H. Characterization of natural plasmid and construction of putative transformation vector using the plasmid in the Korean red alga, Porphyra tenera. Algae 16, 287–294 (2001). [Google Scholar]
- Reith M. & Munholland J. Complete nucleotide sequence of the Porphyra purpurea chloroplast genome. Plant Mol. Biol. Rep. 13, 333–335 (1995). [Google Scholar]
- Glöckner G., Rosenthal A. & Valentin K. The structure and gene repertoire of an ancient red algal plastid genome. J. Mol. Evol. 51, 382–390 (2000). [DOI] [PubMed] [Google Scholar]
- Ohta N. et al. Complete sequence and analysis of the plastid genome of the unicellular red alga Cyanidioschyzon merolae. DNA Res. 10, 67–77 (2003). [DOI] [PubMed] [Google Scholar]
- Collén J. et al. Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proc. Natl. Acad. Sci. USA 110, 5247–5252 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePriest M. S., Bhattacharya D. & López-Bautista J. M. The plastid genome of the red macroalga Grateloupia taiwanensis (Halymeniaceae). PLoS ONE 8, e68246 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M. A., Presting G., Bennett M. S. & Sherwood A. R. Highly conserved organellar genomes in the Gracilariales as inferred using new data from the Hawaiian invasive alga Gracilaria salicornia (Rhodophyta). Phycologia 53, 109–116 (2014). [Google Scholar]
- Hughey J. R. et al. Minimally destructive sampling of type specimens of Pyropia (Bangiales, Rhodophyta) recovers complete plastid and mitochondrial genomes. Sci. Rep. 4, 5113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain K. et al. Extreme features of the Galdieria sulphuraria organellar genomes: a consequence of polyextremophily? Genome Biol. Evol. 7, 367–380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima N. et al. Analysis of the complete plastid genome of the unicellular red alga Porphyridium purpureum. J. Plant Res. 127, 389–397 (2014). [DOI] [PubMed] [Google Scholar]
- Yoon H. S., Hackett J. D. & Bhattacharya D. A single origin of the peridinin- and fucoxanthin-containing plastids in dinoflagellates through tertiary endosymbiosis. Proc. Natl. Acad. Sci. USA 99, 11724–11729 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H. S., Hackett J. D., Ciniglia C., Pinto G. & Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 21, 809–818 (2004). [DOI] [PubMed] [Google Scholar]
- Le Gall L. & Saunders G. W. A nuclear phylogeny of the Florideophyceae (Rhodophyta) inferred from combined EF2, small subunit and large subunit ribosomal DNA: establishing the new red algal subclass Corallinophycidae. Mol. Phylogenet. Evol. 43, 1118–1130 (2007). [DOI] [PubMed] [Google Scholar]
- Verbruggen H. et al. Data mining approach identifies research priorities and data requirements for resolving the red algal tree of life. BMC Evol. Biol. 10, 16 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E. C. et al. Highly conserved mitochondrial genomes among multicellular red algae of the Florideophyceae. Genome Biol. Evol. 7, 2394–2406 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M. et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–657 (2004). [DOI] [PubMed] [Google Scholar]
- Nozaki H. et al. 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae. BMC Biol. 5, 28 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönknecht G. et al. Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 339, 1207–1210 (2013). [DOI] [PubMed] [Google Scholar]
- Bhattacharya D. et al. Genome of the red alga Porphyridium purpureum. Nat. Commun. 4, 1941 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. X. et al. Red and green algal monophyly and extensive gene sharing found in a rich repertoire of red algal genes. Curr. Biol. 21, 328–333 (2011). [DOI] [PubMed] [Google Scholar]
- Bench S. R., Ilikchyan I. N., Tripp H. J. & Zehr J. P. Two strains of Crocosphaera watsonii with highly conserved genomes are distinguished by strain-specific features. Front. Micorbiol. 2, 261 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bench S. R. et al. Whole genome comparison of six Crocosphaera watsonii strains with differing phenotypes. J. Phycol. 49, 786–801 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer K. M., Turner S. L. & Young J. P. W. Sequence diversity of the plasmid replication gene repC in the Rhizobiaceae. Plasmid 44, 209–219 (2000). [DOI] [PubMed] [Google Scholar]
- Sørensen S. J., Bailey M., Hansen L. H., Kroer N. & Wuertz S. Studying plasmid horizontal transfer in situ: a critical review. Nat. Rev. Microbiol. 3, 700–710 (2005). [DOI] [PubMed] [Google Scholar]
- Norman A., Hansen L. H. & Sørensen S. J. Conjugative plasmids: vessels of the communal gene pool. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 2275–2289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison E. & Brockhurst M. A. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 20, 262–267 (2012). [DOI] [PubMed] [Google Scholar]
- Vadivukarasi T., Girish K. R. & Usha R. Sequence and recombination analyses of the geminivirus replication initiator protein. J. Biosci. 32, 17–29 (2007). [DOI] [PubMed] [Google Scholar]
- Aron M. B. et al. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–229 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braithwaite D. K. & Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 21, 787–802 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner K. et al. Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius. Proc. Natl. Acad. Sci. USA 96, 3600–3605 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamoo Y. & Steitz T. A. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell 99, 155–166 (1999). [DOI] [PubMed] [Google Scholar]
- Iwai T., Kurosawa N., Itoh Y. H., Kimura N. & Horiuchi T. Sequence analysis of three family B DNA polymerases from the thermoacidophilic crenarchaeon Sulfurisphaera ohwakuensis. DNA Res. 7, 243–251 (2000). [DOI] [PubMed] [Google Scholar]
- Rodriguez A. C., Park H., Mao C. & Beese L. S. Crystal structure of a pol α family DNA polymerase from the hyperthermophilic archaeon Thermococcus sp. 9° N-7. J. Mol. Biol. 299, 447–462 (2000). [DOI] [PubMed] [Google Scholar]
- Albà M. M. 2001. Replicative DNA polymerases. Genome Biol. 2, 3002.1–3002.4 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers P. M. et al. Eukaryotic DNA polymerases: proposal for a revised nomenclature. J. Biol. Chem. 276, 43487–43490 (2001). [DOI] [PubMed] [Google Scholar]
- Hashimoto H. et al. Crystal structure of DNA polymerase from hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. J. Mol. Biol. 306, 469–477 (2001). [DOI] [PubMed] [Google Scholar]
- Filée J., Forterre P., Sen-Lin T. & Laurent J. Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J. Mol. Evol. 54, 763–773 (2002). [DOI] [PubMed] [Google Scholar]
- Freisinger E., Grollman A. P., Miller H. & Kisker C. Lesion (in) tolerance reveals insights into DNA replication fidelity. EMBO J. 23, 1494–1505 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V. Temporal order of evolution of DNA replication systems inferred by comparison of cellular and viral DNA polymerases. Biol. Direct 1, 39 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. et al. Crystal structure of the herpes simplex virus 1 DNA polymerase. J. Biol. Chem. 281, 18193–18200 (2006). [DOI] [PubMed] [Google Scholar]
- Barrick J. E. et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461, 1243–1247 (2009). [DOI] [PubMed] [Google Scholar]
- Thorsted P. B. et al. Complete sequence of the IncPb plasmid R751: implications for evolution and organisation of the IncP backbone. J. Mol. Biol. 282, 969–990 (1998). [DOI] [PubMed] [Google Scholar]
- Rawlings D. E. & Tietze E. Comparative biology of IncQ and IncQ-like plasmids. Microbiol. Mol. Biol. Rev. 65, 481–496 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines A. S., Jones K., Cheung M. & Thomas C. M. The IncP-6 plasmid Rms149 consists of a small mobilizable backbone with multiple large insertions. J. Bacteriol. 187, 4728–4738 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M. & Nielsen K. M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nature Rev. Microbiol. 3, 711–721 (2005). [DOI] [PubMed] [Google Scholar]
- Halary S., Leigh J. W., Cheaib B., Lopez P. & Bapteste E. Network analyses structure genetic diversity in independent genetic worlds. Proc. Natl. Acad. Sci. USA 107, 127–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrineau M. M., Price D. C., Mohr G. & Bhattacharya D. Recent mobility of plastid encoded group II introns and twintrons in five strains of the unicellular red alga Porphyridium. PeerJ 3, e1017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foflonker F. et al. Genome of the halotolerant green alga Picochlorum sp. reveals strategies for thriving under fluctuating environmental conditions. Environ. Microbiol. 17, 412–426 (2015). [DOI] [PubMed] [Google Scholar]
- Beiko R. G., Harlow T. J. & Ragan M. A. Highways of gene sharing in prokaryotes. Proc. Natl. Acad. Sci. USA 102, 14332–14337 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M. et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K. et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D. & Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32, 11–16 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K. & Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9, 286–298 (2008). [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G. E., Hon G., Chandonia J. & Brenner S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.