Abstract
Adaptation is defined as an evolutionary process allowing organisms to succeed in certain habitats or conditions. Chromosomal inversions have the potential to be key in the adaptation processes, since they can contribute to the maintenance of favoured combinations of adaptive alleles through reduced recombination between individuals carrying different inversions. We have analysed six genes (Pif1A, Abi, Sqd, Yrt, Atpα and Fmr1), located inside and outside three inversions of the O chromosome in European populations of Drosophila subobscura. Genetic differentiation was significant between inversions despite extensive recombination inside inverted regions, irrespective of gene distance to the inversion breakpoints. Surprisingly, the highest level of genetic differentiation between arrangements was found for the Atpα gene, which is located outside the O1 and O7 inversions. Two derived unrelated arrangements (O3+4+1 and O3+4+7) are nearly fixed for several amino acid substitutions at the Atpα gene that have been described to confer resistance in other species to the cardenolide ouabain, a plant toxin capable of blocking ATPases. Similarities in the Atpα variants, conferring ouabain resistance in both arrangements, may be the result of convergent substitution and be favoured in response to selective pressures presumably related to the presence of plants containing ouabain in the geographic locations where both inversions are present.
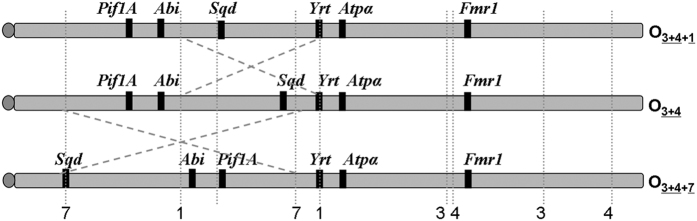
Adaptation is a major evolutionary mechanism that allows organisms to live in certain habitats or conditions1,2. Prime evidence for adaptation is the maintenance and reproducibility of latitudinal clines in the frequency of chromosomal inversions observed in continuous populations of several species of drosophilids across continents3,4,5,6. In particular, Drosophila subobscura has been studied extensively due to its abundant inversion polymorphism and its recent invasion of large areas of North and South America. The frequencies of most chromosomal inversions in D. subobscura are correlated with latitude in the Palearctic region. In addition, similar latitudinal clines were also detected in both colonized American regions, suggesting that the geographic distribution of its inversion polymorphism is adaptive and not a mere consequence of historic events3,7,8. The chromosomal arrangements of D. subobscura are formed by single and overlapped inversions that may differentially affect levels of nucleotide variability, since the overlapped inversions may effectively reduce recombination. Previous studies focused on chromosomal inversions located in the segment I of the O chromosome (Fig. 1) and the A chromosome9,10,11,12,13, but segment II of the O chromosome remains largely unexplored despite bearing some chromosomal inversions whose frequencies cycle seasonally and respond to acute environmental events14.
Figure 1.
The three chromosomal arrangements analysed in the present study (O3+4, O3+4+1 and O3+4+7) differ by the presence of single chromosomal inversions in the segment II of the O chromosome (Fig. 1). They can be considered medium size inversions since O1 includes 6.09 Mb and O7 inversion 11.76 Mb (estimated as in Pegueroles et al.15). The three arrangements are negatively correlated with latitude in the Palearctic region16, and one of them (O3+4+7) shows seasonal fluctuations17. These arrangements are found in sympatry in some regions around the Mediterranean Sea, although with different abundances8. The samples for the present research are from two well-studied localities, Barcelona and Mt. Parnes, where these arrangements coexist18.
Our aim is to test whether selection or drift are the evolutionary forces shaping genetic variability in single medium-size chromosomal inversions. We inferred population recombination and analysed patterns of DNA variation and linkage disequilibrium in six gene segments located within inverted and non-inverted regions, taking into account the age, the length of the inverted regions and the distance to the inversion breakpoint. We also evaluated whether variability patterns fit selectively neutral expectations using both evolutionary and protein structural approaches. We found significant genetic differentiation between arrangements despite extensive recombination being detected inside the inversions. Interestingly, we found nonsynonymous substitutions at the Atpα gene outside the inverted regions that appear to have been fixed by positive selection in association with both the O3+4+1 and O3+4+7 arrangements, and that occur at residues in the structure of the ATPase α subunit which are known to confer resistance to the plant toxin ouabain.
Results
Nucleotide variation and genetic differentiation
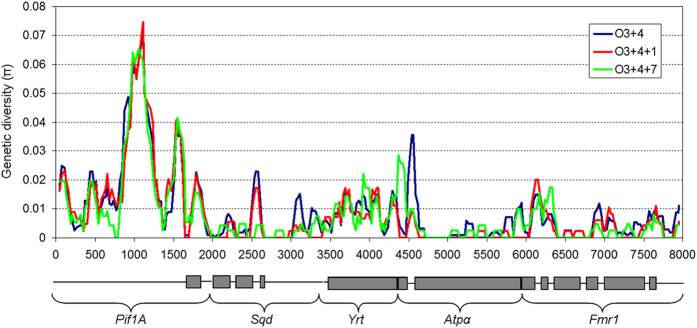
To characterize the genetic content of the O3+4+1 and the O3+4+7 arrangements, we first calculated nucleotide variation and divergence for each of the six gene fragments studied (Table 1). Sequences for O3+4 individuals are from Pegueroles et al.11 and diversity estimates are reported herein to facilitate comparison. The number of haplotypes was approximately the same as the number of analysed lines, except for Sqd and Atpα genes in O3+4+1 arrangement that had lower numbers of haplotypes (Table 1). Considering the variability of the O3+4 arrangement as a baseline, we observed a decrease in variability in the intronic regions of Sqd and Atpα genes at both inverted arrangements (Fig. 2). In contrast, we observed increased variability in the first exonic region of Atpα gene for the O3+4+7 arrangement. Diversity levels were highly variable between genes (Table 1, Fig. 2). The intronic regions of the Pif1A gene showed the highest diversity (π) for all three arrangements despite being located within the O7 inversion. The Yrt gene also showed high π levels despite most of the amplified fragment being exonic and located close to the O1 breakpoint and outside the inversion. Since the proportion of intronic-exonic regions amplified varied among genes, genetic variability was also estimated in silent positions exclusively (i.e., both synonymous and non-coding sites) to avoid biases in diversity estimates (Table 1). In agreement with π results, silent nucleotide variability (πsil) remained highly variable among genes, but quite similar when comparing the two inverted arrangements. No relationship was detected between π and distance to breakpoints, since Pearson correlation values were negative and non-significant for all arrangements.
Table 1. Nucleotide variation and divergence per chromosomal arrangement for the six genes studied.
| Gene | Pop | Arrangement | n | h | S | Singletons | π | πsil | θsil | Ksil |
|---|---|---|---|---|---|---|---|---|---|---|
| Pif1A | MP | O3+4+1 | 9 | 9 | 104 | 61 | 0.021 | 0.022 | 0.025 | 0.188 |
| O3+4 | 12 | 12 | 120 | 55 | 0.021 | 0.023 | 0.026 | 0.191 | ||
| BC | O3+4+7 | 9 | 9 | 89 | 54 | 0.017 | 0.018 | 0.021 | 0.187 | |
| O3+4 | 12 | 12 | 114 | 42 | 0.022 | 0.023 | 0.024 | 0.190 | ||
| Abi | MP | O3+4+1 | 3 | 3 | 8 | 8 | 0.003 | 0.008 | 0.008 | 0.174 |
| O3+4 | 12 | 12 | 32 | 20 | 0.005 | 0.012 | 0.017 | 0.173 | ||
| BC | O3+4+7 | 10 | 9 | 33 | 21 | 0.006 | 0.015 | 0.018 | 0.172 | |
| O3+4 | 7 | 7 | 24 | 15 | 0.006 | 0.014 | 0.015 | 0.173 | ||
| Sqd | MP | O3+4+1 | 10 | 5 | 12 | 8 | 0.002 | 0.005 | 0.008 | 0.115 |
| O3+4 | 12 | 12 | 25 | 16 | 0.005 | 0.006 | 0.008 | 0.116 | ||
| BC | O3+4+7 | 10 | 10 | 17 | 12 | 0.003 | 0.004 | 0.006 | 0.115 | |
| O3+4 | 10 | 10 | 21 | 14 | 0.005 | 0.006 | 0.007 | 0.115 | ||
| Yrt | MP | O3+4+1 | 11 | 11 | 28 | 14 | 0.009 | 0.031 | 0.035 | 0.327 |
| O3+4 | 12 | 12 | 33 | 18 | 0.010 | 0.034 | 0.042 | 0.327 | ||
| BC | O3+4+7 | 9 | 9 | 31 | 18 | 0.010 | 0.035 | 0.013 | 0.325 | |
| O3+4 | 12 | 12 | 35 | 21 | 0.010 | 0.035 | 0.041 | 0.326 | ||
| Atpα | MP | O3+4+1 | 9 | 4 | 6 | 6 | 0.001 | 0.003 | 0.005 | 0.204 |
| O3+4 | 12 | 12 | 22 | 9 | 0.005 | 0.015 | 0.013 | 0.208 | ||
| BC | O3+4+7 | 9 | 8 | 22 | 17 | 0.004 | 0.006 | 0.010 | 0.204 | |
| O3+4 | 12 | 11 | 27 | 16 | 0.005 | 0.016 | 0.019 | 0.209 | ||
| Fmr1 | MP | O3+4+1 | 10 | 10 | 30 | 24 | 0.004 | 0.007 | 0.010 | 0.150 |
| O3+4 | 11 | 11 | 38 | 25 | 0.005 | 0.009 | 0.012 | 0.151 | ||
| BC | O3+4+7 | 8 | 7 | 19 | 12 | 0.003 | 0.006 | 0.007 | 0.150 | |
| O3+4 | 10 | 10 | 28 | 18 | 0.004 | 0.008 | 0.010 | 0.151 |
Pop, population: Mt. Parnes (MP) and Barcelona (BC); n, sample size; h, number of haplotypes; S, number of polymorphic sites; π, nucleotide diversity in all sites; πsil, nucleotide diversity in synonymous sites and non-coding positions; θsil, heterozygosity in silent sites; Ksil, divergence per silent site between D. subobscura and D. pseudoobscura.
Figure 2. Genetic diversity (π) for the O3+4, O3+4+1 and O3+4+7 chromosomal arrangements using the concatenated genes data set.
Grey boxes and solid lines underneath mark exonic and intronic regions, respectively.
Most genes located inside the inverted regions showed significant genetic differentiation between arrangements (Table 2). However, the largest significant FST values were found for the Atpα gene, which is located close to but outside the studied inversions. Since recombination is expected to be constrained near inversion breakpoints, we plotted genetic differentiation with respect to the distance to nearest breakpoint. No relationship between genetic differentiation and distance to the nearest inversion breakpoint was found (Pearson correlation values were negative and non-significant) with most of the genes having similar FST values regardless of their location within or outside the inverted region (Suppl. Fig. 1).
Table 2. F ST values for each gene and the concatenated set and the statistical significance of Snn (ns, not significant; 0.01 <*P < 0.05; 0.001 <**P < 0.01; ***P < 0.001).
| O3+4 − O3+4+1 | O3+4 − O3+4+7 | O3+4+1 − O3+4+7 | |
|---|---|---|---|
| Pif1A | −0.028 ns | 0.146** | 0.076 ns |
| Abi | – | 0.065* | – |
| Sqd | 0.082* | 0.040* | 0.071** |
| Yrt | 0.017 ns | −0.001 ns | 0.024 ns |
| Atpα | 0.614*** | 0.525** | 0.367*** |
| Fmr1 | −0.033 ns | 0.004 ns | −0.005 ns |
| Concatenated | 0.099* | 0.182** | 0.092* |
Genes in grey are located inside inversions.
Linkage disequilibrium, gene flux and age of the inversions
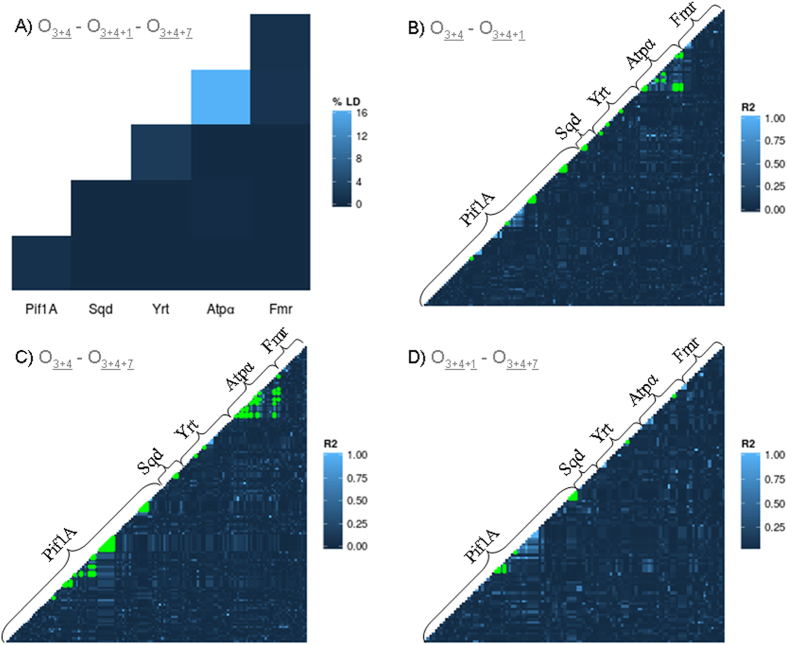
If chromosomal inversions are effectively reducing recombination levels, we would expect higher levels of LD within and between genes located inside them. However, levels of LD in these genes were very low and only a strong LD was observed within the Atpα gene (Fig. 3A). Significant associations after adjusting for multiple testing (in green) were obtained only within genes and never between genes regardless of their location in relation to the inversions (Fig. 3B–D). The highest ZnS values were obtained for the Atpα gene with 0.34 and 0.26 when analyzing together inversions O3+4–O3+4+1 and O3+4–O3+4+7, respectively (Table S1). Surprisingly, ZnS values were in general higher for O3+4+1 and O3+4+7 sequences alone, than compared to the O3+4 arrangement, suggesting the presence of recombination events between arrangements.
Figure 3.
(A) Percentage of significant LD among O3+4, O3+4+1 and O3+4+7arrangements within and between genes. (B–D) Pairwise LD measured as R2 for O3+4+7–O3+4, O3+4+1–O3+4 and O3+4+1–O3+4+7 comparisons. Green dots correspond to the significant associations after adjusting for multiple comparisons using Benjamini and Hochberg method (1995). Gene order in O3+4 and O3+4+1 is the same (Fig. 1) and has been used for homogeneity in all comparisons.
The low levels of linkage disequilibrium detected within inverted regions suggest that recombination between chromosomal arrangements may be frequent. Recombination was detected within and between arrangements for all genes (Rho, Table S1). Surprisingly, recombination estimates were higher when comparing different chromosomal arrangements than when comparing the same inversion. This result may simply be due to the higher number of informative sites when combining arrangements. Some gene conversion tracts (GCTs) were detected (Table S2) despite probabilities of a site to be informative for gene conversion events (ψ values) are low (10−3 to 10−4). A total of five and eight tracts were observed between O3+4–O3+4+1 and O3+4–O3+4+7, respectively. The lengths of the tracts were highly variable, ranging from 7 to 1573 bp, and the largest tracts were found in the Atpα gene. Since GCTs are expected to be small, the large tracts observed might be due to single or double crossover events, given that Atpα gene is located outside the inversion.
Sequence networks for all genes were highly reticulated, with the exception of the Atpα gene, suggesting high levels of recombination among individuals carrying different chromosomal arrangements for genes located inside and outside inversions (Suppl. Fig. 2). For the Sqd gene, despite being located within all three inversions and presenting significant Fst values between arrangements (Table 2), individual sequences of the same arrangement seldom clustered together suggesting high rates of exchange among chromosomal arrangements (Suppl. Fig. 2). For the Atpα gene, it was possible to distinguish three clades corresponding to each arrangement, although four recombinant individuals (FMP2, FBC49, FBC76 and MP36) could be identified matching those detected as GCT (Table S3). Two of them have the O3+4 arrangement (FMP2 and FBC49) and a GCT length larger than 1422 bp (Table S2), FBC76 has the O3+4+7 arrangement and also a large GCT (1573bp) and MP36 has the O3+4+1 arrangement and a small GCT (52 bp). In addition, for the Atpα gene, the number of recombination connections within O3+4+1 and O3+4+7 arrangements was lower than within the O3+4 arrangement (Suppl. Fig. 2), which could indicate a more recent origin of the former two arrangements.
Inversion ages may be overestimated from genes located in central positions of inverted regions, since they are more prone to be included in double crossovers consequently introducing additional variation from other arrangements. We estimated the age of the O3+4+7 arrangement using the Sqd gene, which is the closest to the proximal breakpoint of the O7 inversion (Fig. 1), to be 0.47 ± 0.12 Myr assuming that the divergence time between D. subobscura and D. pseudoobscura is 17.7 ± 4.4 Myr19. The age of the O3+4+1 arrangement was estimated to be 0.52 ± 0.13 Myr using the Sqd gene and the same divergence time. BEAST program could not be used to estimate the age of those two inversions due to the high recombination detected among the three arrangements (Suppl. Fig. 2).
Test of neutrality and adaptive evolution
To evaluate whether any of the six genes are under positive selection, we performed several statistical tests for departure from the expectations of an equilibrium neutral model of evolution. A majority (93%) of the Tajima’s D and Fu and Li’s D test statistics were negative (Table S4). These overall results suggest a general trend towards an excess of low frequency polymorphisms that could be due to population growth. It is worth noting that Tajima’s D and Fu and Li’s D test were only significant for the the Atpα gene in the O3+4+1 arrangement when using all positions (Table S4), although both tests failed to detect significant departures from neutrality when excluding the recombinant individual MP36 and/or using only silent sites (Table S5). In contrast, Tajima’s D test was statistically significant for Atpα in the O3+4+7 arrangement when excluding recombinant individuals but not when using silent sites only (Table S5), raising the possibility of negative selection on polymorphic amino acid replacements at this gene.
The McDonald and Kreitman test, which contrasts nonsynonymous and synonymous polymorphism and divergence, was only significant for the Atpα gene in the O3+4+7 arrangement (P = 0.0003). For this gene, the number of polymorphic sites was 9 (7 nonsynonymous and 2 synonymous), while the number of differences between species was 60 (9 nonsynonymous and 51 synonymous, Table S6, Supporting information). The Direction of Selection (DoS) statistic is −0.442 for the O3+4+7 arrangement of this gene (Table S6, Supporting information). If one assumes synonymous sites are neutral, then this pattern would indicate an excess of nonsynonymous polymorphism present at the Atpα gene within the O3+4+7 arrangement. Certain amino acid changes nearly fixed in this arrangement (99, 109, 111 and 122 probably implicated in resistance to plant toxins, see below) could be maintained by diversifying selection as polymorphisms, while the rest are low frequency variants that could be weakly deleterious and kept at low frequencies by negative selection.
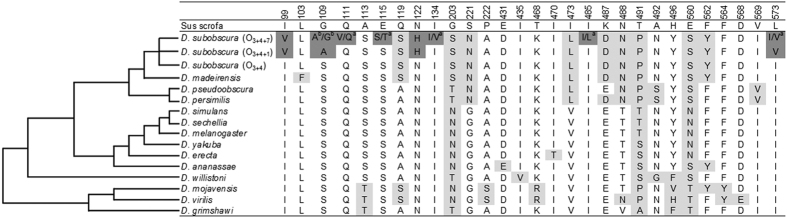
We tested for long-term positive selection at the Atpα gene using several site and branch-site tests implemented in CodeML of the PAML v4 package20, that were based on the consensus sequences of the arrangements (see methods). All positions in the consensus sequences correspond to nearly fixed substitutions between lineages except for amino acid position 109 in the O3+4+7 arrangement (Fig. 4) where the two equally likely substitutions (A/G) were evaluated separately. Site tests of the entire gene fragment (M1a vs M2a and M7 vs M8, see Materials and Methods section), which assume that the strength and direction of selection is uniform across all lineages, failed to detect positively selected sites in the Atpα gene (Table S7). However, the branch-site test 2, that allows to detect sites that evolved under positive selection in an specific lineage, inferred positive selection for several codons on the O3+4+7 arrangement regardless of which amino acid is present in position 109 (Table S7), but not on the O3+4 and O3+4+1 arrangements. Positions that showed departures from neutrality according to PAML in the O3+4+7 arrangement are 99, 109, 111 and 122, which are the positions highlighted in the structural protein model (Fig. 5). In fact, positions 111 and 122 are the ones affecting the ouabain–Atpα interaction (see below), and the nonsynonymous change at position 111 was only found in O3+4+7. Using BEAST, after removing recombinants, the time to most recent common ancestor (TMRCA) was estimated in 0.21 ± 0.009 Myr (mean ± SE) for O3+4+1, 0.46 ± 0.018 Myr for O3+4+7 and 1.69 ± 0.05 Myr for all O3+4. Using the average silent nucleotide diversity the time to most recent common ancestor (TMRCA) was estimated in 0.09 ± 0.02 Myr and 0.13 ± 0.03 Myr for O3+4+1 and O3+4+7 respectively considering that the divergence time between D. subobscura and D. pseudoobscura is 17.7 ± 4.4 Myr19.
Figure 4. Amino acid replacements detected in the amplified region of the Atpα gene in 14 Drosophila species.
alow frequency amino acid (<0.3); bequal frequency amino acid. Numbering corresponds to the mature pig enzyme (GenBank #: 1 × 03938). Shading is as follows: Pale grey: amino acid changes that occurred in the O3+4 arrangement of D. subobscura or in any other Drosophila species. Dark grey: changes found only in the O3+4+1 and/or O3+4+7 arrangements in comparison to the other Drosophila species.
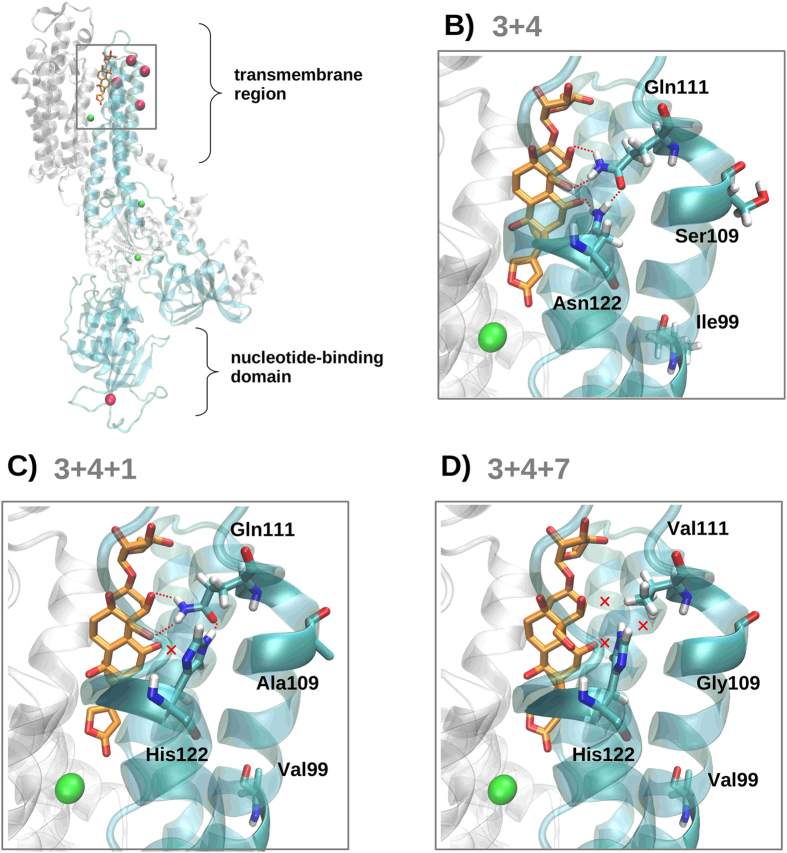
Figure 5.
(A) Structural model of the ATPase α-subunit. The gene region sequenced is depicted in cyan and ouabain in orange sticks. Pink spheres represent positions presenting variation between arrangements and green spheres show the ion positions derived from the crystal structure of the template. (B–D) Detailed view of mutations located in the transmembrane region of O3+4, O3+4+1 and O3+4+7 arrangements. Hydrogen bonding interactions are represented as red dotted lines, while lack of these interactions is signalled with a red cross.
We checked whether the variability in the Atpα gene, and specifically the nonsynonymous changes, were already present in other drosophilids or appeared de novo in D. subobscura. Amino acid replacements observed within the amplified region of the Atpα gene among 14 Drosophila species are summarized in Fig. 4. Most of the replacements detected can be assigned to specific lineages. For instance, a G to N replacement at position 221 and V to L replacement at position 473 were both detected for all species of the obscura group. Interestingly, some changes were only detected in O3+4+1 and O3+4+7 arrangements and not in any other of the thirteen drosophilids studied neither in the O3+4 arrangement of D. subobscura. These changes can be classified in three groups: shared mutations between the two arrangements (positions 99, 109 and 122), specific high frequency mutations in one arrangement (position 111 for O3+4+7 and 573 for O3+4+1), and specific low frequency mutations (115, 134, 485, 573 for the O3+4+7). The two equally frequent polymorphic amino acids in position 109 of the O3+4+7 arrangement (A and G) are small and non-polar, while the ancestral amino acid was polar (S) (Fig. 4). The O3+4 arrangement of D. subobscura is more similar to (closely) related species (i.e. D. madeirensis) than to O3+4+1 and O3+4+7 arrangements.
Three-dimensional structure of the ATPase α-subunit and putative functional consequences
At least two of the amino acid replacements observed in the ATPase α-subunit may impact the binding of the cardenolide ouabain, a plant toxin capable of blocking ATPases21,22. According to the crystallized ouabain–Na+, K+-ATPase complex23, ouabain would interact with a set of hydrophobic residues in helices αM4 and αM5 and would establish particular polar interactions with helices αM1, αM2 and αM6. Helices αM1, αM2 were sequenced in the present work and are depicted in cyan in Fig. 5. The positions contributing to variation between arrangements are located both in the transmembrane region of the protein as well as in its nucleotide-binding domain (depicted as magenta spheres in Fig. 5A). Of the variants located in the transmembrane region, we can expect different levels of impact regarding ouabain binding. Changes in positions 99 and 573 (I99 V and I573 V), which are located in the transmembrane and nucleotide-binding domain respectively, are similar in terms of hydrophobicity and shape and are not expected to have a big impact in terms of protein function. In the case of amino acid position 109 of the transmembrane segment, amino acid replacement results in a loss of a polar residue that could indirectly affect protein stability and insertion in the lipid bilayer (S109A in O3+4+1 and S109A, G in the O3+4+7 arrangement, Fig. 5B–D). Interestingly, we detected two further changes in the transmembrane region that could directly affect the binding of ouabain to the Atpα protein. In the O3+4 arrangement, ouabain establishes stabilizing hydrogen bonding interactions with residues Gln111 and Asn122 of the alpha subunit of the ATPase (Fig. 5B, red dashed lines). These interactions are further stabilized by a hydrogen bond between these two residues. In contrast, the replacement of Asn122 by His122 in both O3+4+1 and O3+4+7 arrangements destroys the interaction between this residue and ouabain (Fig. 5C, see red cross). In addition, mutation from Gln111 to Val111 in the O3+4+7 arrangement destroys the second stabilizing interaction as well as the intramolecular hydrogen bond formed by Atpα residues (Fig. 5D, see red crosses).
Discussion
Chromosomal inversions are known to strongly influence patterns of genetic diversity within their breakpoints. The degree of inversion variability and differentiation depends on the time since the formation of the inversion, on its size (large inversions are more likely to have double-crossovers within them), and on selection pressure4,24,25,26. Genetic differentiation for O3+4+1 and O3+4+7 arrangements was significant despite variability levels for most of the genes located within them seem to have recovered to the level observed in the O3+4 ancestral arrangement. We estimated the age of these derived inversions considering that the Sqd gene is roughly 0.50 Myr (assuming that the divergence time between D. subobscura and D. pseudoobscura is 17.7 ± 4.4 Myr)19. The age of the O3+4 arrangement was estimated to be 0.90 Myr using the same divergence time as mentioned above11. Thus, as expected, the O3+4+1 and O3+4+7 arrangements are younger than O3+4 from which they most likely derived (Fig. 1). However, the Sqd gene network showed recombination connections between different arrangements, further preventing estimating inversion ages using the coalescent process, which may result in an overestimation of their age. It is worth noting that the estimation of inversion ages may vary between markers9,11,27,28. Thus, our results should be interpreted with caution until more markers are available to confirm them.
The genetic differentiation that we found between O3+4+1–O3+4 and O3+4+7–O3+4 arrangements was smaller than between the older O3+4–OST arrangements9,11,27. The presence of two overlapped inversions (O3 and O4) in the later comparison may prevent crossovers formation more efficiently due to physical constraints. Overlapped inversions may be an important non-selective factor modulating nucleotide variability patterns and their absence may facilitate recombination. The small FST values obtained for individual genes located within O1 and O7 inverted regions suggest the presence of frequent genetic exchange with non-inverted arrangements for these regions, supporting recombination as the main contributor to variability recovery24.
We did not detect a significant relationship between genetic variability and distance to breakpoints, as observed in previous studies of D. subobscura9,10, D. buzzatti29, and Anopheles gambiae30. However, for D. melanogaster mixed results are obtained depending on the inversions and populations of origin evaluated, with peaks of high and low variability and differentiation interspersed31,32. We find that genetic differentiation close to inversion breakpoints can also be eroded through time at a gene specific rate, supporting previous experimental studies in D. subobscura15, and contrasting with those obtained in D. pseudoobscura33. As expected by the presence of high levels of recombination, linkage disequilibrium levels were low within inversions. Our results contrast with those obtained in D. pseudoobscura inversions, which generally show high levels of LD between genes associated with inversions that have been interpreted as an evidence for epistasis6,34. For D. melanogaster strong LD within the region spanned by In(3R)Payne has been detected although it is not uniformly distributed31. According to tests of neutrality based on frequency distributions (Tajima’s D and Fay and Wu’s H), there was a tendency towards an excess of low frequency polymorphisms for all genes consistent with a recent expansion of the species35, but only the Atpα gene departed from neutral expectations. Taken altogether, variability in all genes (with the only exception of the Atpα gene) in the O3+4+1 and O3+4+7 arrangements seem to be shaped mainly by extensive recombination rather than Darwinian (positive) selection.
Our results indicate that variability patterns of the Atpα gene seem to be strongly influenced by natural selection in the O3+4+1 and the O3+4+7 arrangements. Despite being located outside both studied inversions (but less than 2 Mb apart from the closest inversion breakpoint), we detected high levels of genetic differentiation when compared to the ancestral O3+4 arrangement due to fixed nonsynonymous differences. In D. melanogaster parallel geographic variation in regions inside and outside inversions have been observed across continents36. Besides that, SNPs varying in frequency seasonally throughout D. melanogaster genome–and not exclusively concentrated in inversions–have also been described37. Thus, spatial and temporal varying selection seems also to strongly influence regions outside inversions. Differences in the Atpα gene between O3+4+1 and O3+4+7 arrangements were significant at the nucleotide level although at the amino acid level, both arrangements are nearly identical. Interestingly, all specific changes of the D. subobscura lineage occurred in the O3+4+1 and/or O3+4+7 arrangements, since changes detected in the O3+4 arrangement were shared with D. madeirensis, which is consistent with their common ancestry. In addition, protein sequences of O3+4 and OST arrangements of D. subobscura were reported to be identical11.
The nature of natural selection acting on the Atpα gene is quite complex. The McDonald-Kreitman test was significant for Atpα gene and the Direction of Selection (DoS) statistic was negative in a direction consistent with strong selective constraint acting on most of the protein. These results are in agreement with the essential nature of this gene for individual survival. Nonetheless, PAML did reveal significant evidence for positive selection acting on several codons within the Atpα gene, including amino acid replacements at two codons that would confer resistance to a plant toxin. Positive selection acting on highly conserved genes has also been reported in other studies: according to Pupko & Galtier38, primate mitochondrial genomes evolved through episodes of positive selection at a few sites, enabling the fine-tuning of the three-dimensional protein structure to optimize the function of conserved genes. Similarly, Vasseur et al.39 found rare alleles with evidence of positive selection in some genes of the NLR family although this family is under strong purifying selection due to its vital role.
The case of the Atpα gene indicates that positive selection is able to act within a highly conserved gene to maintain adaptive mutations associated with certain chromosomal inversions. The structural analysis of the ouabain-ATPase α-subunit complex shows that two substitutions, both in the O3+4+1 and the O3+4+7 arrangements (111V and 122H), would reduce the affinity of the ATPase complex to bind the cardenolide ouabain due to the destruction of stabilizing hydrogen bonds. Remarkably, these observations are in line with mutagenesis studies showing a significantly increased survival of cells transfected with constructs having mutations 111V and 122H (from D. melanogaster) after ouabain treatment, and a 2.250-fold increased resistance to this toxin when bearing both mutations21,40. Previous studies demonstrated that adaptive mutations in Na,K-ATPase, such the ones in positions 111 and 122, were acquired in parallel in some cardenolide-feeding species21,22. Three hypotheses could explain the presence of convergent mutations in O3+4+1 and O3+4+7 arrangements in D. subobscura. (1) In a parallel scenario41, mutations may have occurred independently in the two new arrangements as the result of adaptation to similar environmental conditions. (2) In a collateral scenario41, variants from an ancestral polymorphism could have been independently captured during the formation of the two inversions and subsequently been maintained by selection. (3) Finally, amino acid substitutions that occurred in one of the two arrangements in response to selection could have been subsequently acquired by the other inversion through double recombination or gene conversion between arrangements, with those variants being subsequently driven to high frequency in both arrangements due to similar selective pressures. All three of these possible scenarios include natural selection and suggest that epistatic interactions between the ATPα gene and genes located inside both inversions (O1 and O7) are necessary to account for the maintenance of amino acid similarities despite ATPα gene being located outside both inversions. Furthermore the reduced number of recombinants with O3+4 can only be explained by selection if recombinant individuals are effectively purged from populations to maintain adaptive interactions. Currently available data does not allow us to discriminate between these three scenarios although the collateral hypothesis seems less likely since in D. madeirensis, O3+4 and OST share almost identical amino acid composition. Given the high chromosomal polymorphism in D. subobscura and the many inversion breakpoints in the neighbouring area of the Atpα gene8, future analysis of other chromosomal arrangements may help to reconstruct the process of acquisition of these adaptive substitutions, and to determine whether they were already present in a common ancestor (i.e. synapomorphy) or acquired by parallel evolution or through recombination.
Cardenolides have a huge diversity of chemical forms and are sporadically distributed across 12 families of angiosperms42. Cardenolide feeding species have been typically associated with plants of the family Apocynaceae, notably in the genera Asclepias and Apocynum21,22. Asclepias has a Neartic distribution and Apocynum a temperate Northern hemisphere distribution43, and cardenolides production seems to form latitudinal clines of different sign depending on the Asclepias species42,44. D. subobscura is a generalist saprophytic insect and its diet includes decaying plant material and fruits, fungi, yeast and microbials45, and it is known to be able to feed from decaying Digitalis purpurea46, a plant containing ouabain. We hypothesize that the appearance of mutations in the O3+4+7 and O3+4+1 arrangements conferring the ability to feed on cardenolide containing plants has changed the fitness of associated chromosomal inversions resulting in nonsynonymous polymorphism. Thus, in certain environments (i. e. in the presence of toxic plants) positive selection will favour the maintenance of adaptive variants. Future studies may help elucidate whether the observation of adaptive mutations in some arrangements of D. subobscura reflects geographical distribution of cardenolide-containing plants in the Mediterranean region and confirm whether these amino acid substitutions confer resistance to cardenolides in these insects.
Materials and Methods
Fly samples and DNA sequencing
A total of 45 isochromosomal lines for the O chromosome of D. subobscura derived in Araúz et al.18 were used: 11 O3+4+1 and 12 O3+4 lines from Mt. Parnes (Greece) and 10 O3+4+7 and 12 O3+4 lines from Barcelona (Spain). Genes were selected according to their chromosomal location within or nearby the studied inversions (Fig. 1). The six genes are Pif1A (PFTAIRE-interacting factor 1A), Abi (Abelson interacting protein), Sqd (Squid), Yrt (Yurt), Atpα (Na pump α subunit) and Fmr1 (Fragile X mental retardation). Genomic DNA extraction, DNA amplification and sequencing reactions for the O3+4+1 and O3+4+7 arrangements were carried out as reported in Pegueroles et al.11. Sequencing was done on a 3730 Analyzer (Applied Biosystems) at the Serveis Cientifico-Tècnics from Universitat de Barcelona. Sequences were assembled with SeqMan II (DNASTAR) and multiply aligned with Clustal W47 implemented in BioEdit v748. Sequences for the O3+4+7 and O3+4+1 arrangements are available at GenBank under the accession numbers KT318937- KT319043. Sequences for the O3+4 arrangement of D. subobscura and D. madeirensis were obtained from GenBank (accession # JN882382-JN882400, JN882406-JN882429, JN882441-JN882461, JN882472-JN882495, JN882508-JN882529, JN882541- JN882564 and JN882376-JN882381). Sequences from the other 12 Drosophila species with sequenced genome were downloaded from Flybase database (http://flybase.org).
Nucleotide polymorphism and genetic differentiation
Nucleotide polymorphism and genetic differentiation were estimated with DnaSP v549. We calculated the standard parameters of molecular diversity: number of haplotypes (h), number of polymorphic sites (S) and number of singletons, nucleotide diversity (π)50, nucleotide diversity in synonymous sites and non-coding positions (πsil)51, silent site heterozygosity (θsil)52 and divergence per silent site between D. subobscura and D. pseudoobscura (Ksil)51. Due to the presence of duplications in the Abi gene (data not shown), very few individuals could be sequenced for the O3+4+1 arrangement. Thus, the concatenated data set does not contain Abi gene sequences. Overall, 21 O3+4, 10 O3+4+1 and 8 O3+4+7 chromosomes were included in the concatenated data set and genes combined with Concatenator v153. Nucleotide diversity (π) across the concatenated data was calculated using a sliding window of 100 nucleotides with a step size of 25. Genetic distances were computed with FST54 and Snn55 and its significance estimated with 10,000 replicates. The distance of each gene to the nearest inversion breakpoint in bp was calculated assuming that all cytological bands contain the same genetic content and the length of the O chromosome of D. subobscura, but not its gene order56, is equivalent to that of the chromosome 2 of D. pseudoobscura as in Pegueroles et al.57.
Neutrality tests
Tajima’s D58 and Fu and Li’s D59 tests were carried out to assess whether the site frequency spectrum of variation within arrangements differ from their expectation under an equilibrium neutral model, using D. pseudoobscura as an outgroup. This species was used as outgroup instead of D. madeirensis since the level of divergence to D. subobscura for the latter is too low for these genes11. Furthermore, to test for footprints of selection we performed the McDonald and Kreitman60 test, the Direction of Selection (DoS) statistic61, and several site and branch-site tests implemented in CodeML of the PAML v4 package20. Site tests of the entire gene, allowing the ω ratio to vary among sites, were performed comparing two pairs of models, the nearly neutral model M1a (model = 0; NSsites = 1) with the alternative positive selection model M2a (model = 0; NSsites = 2), and the neutral model M7 (model = 0; NSsites = 7, ncatG = 10) with the alternative selection model M8 (model = 0; NSsites = 8, ncatG = 10). For the branch-site test 2, aiming to detect positive selection affecting a few sites, in the neutral model we used the parameters model = 2, NSsites = 2, fix_omega = 1 and omega = 1. and for the alternative selection model we used model = 2, NSsites = 2, fix_omega = 0 and omega = 1.5. All these tests were applied to the Atpα gene after excluding recombinant individuals and using the consensus sequences of the O3+4, O3+4+1 and O3+4+7 arrangements of D. subobscura with both D. madeirensis and D. pseudoobscura sequences as outgroups. Neutral and alternative models were compared using a likelihood ratio test and the P-value was assessed using a chi-squared test.
Linkage disequilibrium and recombination
For the concatenated data set we estimated the percentage of pairwise comparisons between informative sites presenting significant linkage disequilibrium (LD), and their statistical significance was analysed with Fisher’s exact test implemented in DnaSP v550. P-values were adjusted for multiple testing using the false discovery rate method of Benjamini & Hochberg62. LD between pairs of polymorphic sites was also measured with r2 parameter63 and ZnS64 as a global measure of LD obtained with DnaSP. LD plots were performed using ggplot2 package65. The population recombination rate (ρ = 4Ner, where Ne is the effective population size and r is the rate of recombination) was estimated using a composite likelihood method66 computed with LDhat v2.1 (http://www.stats.ox.ac.uk/~mcvean/LDhat/instructions.html). Recombination networks were constructed using SplitsTree4 program67. Gene conversion tracts (GCT) were identified using the method of Betrán et al.68 implemented in DnaSP. In order to avoid confounding effects due to the population of origin, FST, ρ, GCT and LD parameters were calculated between arrangements from the same population, despite the lack of genetic differentiation observed between O3+4 arrangements from different populations11.
Age of inversions
The ages of inversions were estimated for the Sqd gene, since it is located inside the inverted regions and close to the breakpoint (Fig. 1), using the average silent nucleotide diversity within inversions and excluding individuals carrying gene conversion tracts10,11,27. The number of substitutions per site per year was calculated using the divergence per silent site between D. subobscura and D. pseudoobscura, based on our sequences and using divergence time of 17.7 ± 4.4 Myr19. We dated the time to the most recent ancestor (TMRCA) for the O3+4+1 and O3+4+7 arrangements of the Atpα gene using the same method and also using BEAST 1.8.069. We used a lognormal relaxed clock model and considered the same divergence time and a mutation rate of 0.011 estimated for Drosophila species based on 176 nuclear genes19. The substitution model used was HKY + G + I, being the best substitution model for the Atpα gene inferred with jModelTest 2.070,71, with runs of 2 million steps, sampling a tree every 200 steps. Tracer v1.672 was used to check convergence of parameters and to obtain mean and standard errors (SE) of the time to the most common ancestor of all sequences for a given inversion. We discarded 10% of the steps as burn-in. In both methods we did not include recombinant individuals MP36 and FBC76.
Structural analysis of the Na+,K+-ATPase–ouabain complex
The crystal structure of a high-affinity Na+,K+-ATPase–ouabain complex (PDB ID 4HYT), which shows a 74% amino acid sequence identity with the predicted ATPα protein for D. subobscura, was selected for homology modelling. The model was built using the MOE package (http://www.chemcomp.com/software.htm). After sequence alignment (default settings), ten models were generated using the Amber12:EHT force field73. The best model for each arrangement was selected and superposed on the PDB ID 4HYT crystal structure in order to align the cardenolide ouabain, a plant toxin capable of blocking ATPases, to the newly obtained homology models. The resulting ouabain-receptor complexes were further refined by performing an energy minimization of ouabain and its binding pocket (defined as all residues at 4.5 Å of the compound) using the Amber12:EHT force field by applying gradient minimization until the RMS gradient was lower than 0.001 kcal mol−1 Å−1. Representations of ouabain-receptor complexes were created using VMD 1.9.174.
Additional Information
Accession codes: D. subobscura sequences data are available at GenBank under the accessions numbers KT318937- KT319043.
How to cite this article: Pegueroles, C. et al. Inversions and adaptation to the plant toxin ouabain shape DNA sequence variation within and between chromosomal inversions of Drosophila subobscura. Sci. Rep. 6, 23754; doi: 10.1038/srep23754 (2016).
Supplementary Material
Acknowledgments
We thank Ferran Palero, Celia Schunter, Casandra Riera, Iain Patten, Carolyn Newey and the entire Aquadro’s lab for their valuable comments on the manuscript. We also thank Pedro A. Araúz for the collection of the flies and his collaboration carrying out the genetic crosses, Joan Balanyà for the analyses of the polytene chromosomes and Gemma Calabria and Olga Dolgova for their collaboration in the in situ hybridizations. We are also grateful to two anonymous reviewers for helpful comments that improved the manuscript. This work was supported by a pre-doctoral fellowship to CP (2009FIC-00096) from the Generalitat de Catalunya (Spain). Research was funded by projects CGL2006-13423-C02-02 from the Ministerio de Ciencia y Tecnología (MCYT, Spain) and CTM2013-48163-C2-2-R from the Ministerio de Economía y Competitividad (MINECO, Spain). MP and FM are members of the research group 2014-SGR336 from the Generalitat de Catalunya (Spain).
Footnotes
Author Contributions C.P., C.F.A., M.P. and F.M. designed the research; C.P., A.F.C., M.M.S. and M.P. performed the experimental work and analyses; C.P. wrote the first draft of the manuscript, and subsequently, all authors contributed to the writing.
References
- Kawecki T. J. & Ebert D. Conceptual issues in local adaptation. Ecol. Lett. 7, 1225–1241 (2004). [Google Scholar]
- Savolainen O., Lascoux M. & Merilä J. Ecological genomics of local adaptation. Nat. Rev. Genet. 14, 807–820 (2013). [DOI] [PubMed] [Google Scholar]
- Prevosti A. et al. Colonization of America by Drosophila subobscura: Experiment in natural populations that supports the adaptive role of chromosomal inversion polymorphism. Proc. Natl. Acad. Sci. USA 85, 5597–5600 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A. A. & Rieseberg L. H. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu. Rev. Ecol. Evol. Syst. 39, 21–42 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapun M., van Schalkwyk H., McAllister B., Flatt T. & Schlötterer C. Inference of chromosomal inversion dynamics from Pool-Seq data in natural and laboratory populations of Drosophila melanogaster. Mol. Ecol. 23, 1813–1827 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer S. W. et al. Evolutionary genomics of inversions in Drosophila pseudoobscura: Evidence for epistasis. Proc. Natl. Acad. Sci. USA 100, 8319–8324 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balanyà J. et al. Evolutionary pace of chromosomal polymorphism in colonizing populations of Drosophila subobscura: an evolutionary time series. Evolution 57, 1837–1845 (2003). [DOI] [PubMed] [Google Scholar]
- Krimbas C. B. Drosophila subobscura: Biology, Genetics and Inversion polymorphism. Verlag Dr. Kovac, Hamburg, Germany (1993). [Google Scholar]
- Munté A., Rozas J., Aguadé M. & Segarra C. Chromosomal inversion polymorphism leads to extensive genetic structure: a multilocus survey in Drosophila subobscura. Genetics 169, 1573–1581 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nóbrega C., Khadem M., Aguadé M. & Segarra C. Genetic exchange versus genetic differentiation in a medium-sized inversion of Drosophila: the A2/AST arrangements of Drosophila subobscura. Mol. Biol. Evol. 25, 1534–1543 (2008). [DOI] [PubMed] [Google Scholar]
- Pegueroles C., Aquadro C. F., Mestres F. & Pascual M. Gene flow and gene flux shape evolutionary patterns of variation in Drosophila subobscura. Heredity 110, 520–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J. & Aguadé M. Gene conversion is involved in the transfer of genetic information between naturally occurring inversions of Drosophila. Proc. Natl. Acad. Sci. USA 91, 11517–11521 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Sabaté A., Aguadé M. & Segarra C. Excess of nonsynonymous polymorphism at Acph-1 in different gene arrangements of Drosophila subobscura. Mol. Biol. Evol. 20, 1833–1843 (2003). [DOI] [PubMed] [Google Scholar]
- Rodríguez-Trelles F., Tarrío R. & Santos M. Genome-wide evolutionary response to a heat wave in Drosophila. Biol. Lett. 9, 20130228 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegueroles C., Ordóñez V., Mestres F. & Pascual M Recombination and selection in the maintenance of the adaptive value of inversions. J. Evol. Biol. 23, 2709–2717 (2010). [DOI] [PubMed] [Google Scholar]
- Balanyà J., Solé E., Oller J. M., Sperlich D. & Serra L. Long-term changes in the chromosomal inversion polymorphism of Drosophila subobscura. II. European populations. J. Zool. Syst. Evol. Res. 42, 191–201 (2004). [DOI] [PubMed] [Google Scholar]
- Rodriguez-Trelles F., Alvarez G. & Zapata C. Time series analysis of seasonal changes of the O inversion polymorphism of Drosophila subobscura. Genetics 142, 179–187 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araúz P. A. et al. Tracking the origin of the American colonization by Drosophila subobscura: genetic comparison between Eastern and Western Mediterranean populations. J. Zool. Syst. Evol. Res. 47, 25–34 (2009). [Google Scholar]
- Tamura K., Subramanian S. & Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 21, 36–44 (2004). [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- Dobler S., Dalla S., Wagschal V. & Agrawal A. Community-wide convergent evolution in insect adaptation to toxic cardenolides by substitutions in the Na,K-ATPase. Proc. Natl. Acad. Sci. USA. 109, 13040–13045 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y., Aardema M. L., Medina E. M., Schumer M. & Andolfatto P. Parallel molecular evolution in an herbivore community. Science 337, 1634–1637 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen M., Yatime L., Nissen P. & Fedosova N. U. Crystal structure of the high-affinity Na + K +-ATPase-ouabain complex with Mg2 + bound in the cation binding site. Proc. Natl. Acad. Sci. USA. 110, 10958–10963 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A., Betran E., Barbadilla A. & Ruiz A. Recombination and gene flux caused by gene conversion and crossing over in inversion heterokaryotypes. Genetics 146, 695–709 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics of natural populations. XIX. Origin of heterosis through natural selection in populations of Drosophila pseudoobscura. Genetics 35, 288–302 (1950). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. R. Progress and prospects in Evolutionary Biology: The Drosophila model. Oxford University Press, New York. (1997). [Google Scholar]
- Rozas J., Segarra C., Ribó G. & Aguadé M. Molecular population genetics of the rp49 gene region in different chromosomal inversions of Drosophila subobscura. Genetics 151, 189–202 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Sabaté A., Aguadé M. & Segarra C. The relationship between allozyme and chromosomal polymorphism inferred from nucleotide variation at the Acph-1 gene region of Drosophila subobscura. Genetics 153, 871–889 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laayouni H., Hasson E., Santos M. & Fontdevila A. The evolutionary history of Drosophila buzzatii. XXXV. Inversion polymorphism and nucleotide variability in different regions of the second chromosome. Mol. Biol. Evol. 20, 931–944 (2003). [DOI] [PubMed] [Google Scholar]
- Cheng C. et al. Ecological genomics of Anopheles gambiae along a latitudinal cline: a population-resequencing approach. Genetics 190, 1417–1432 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane R. V, Rako L., Kapun M., Lee S. F. & Hoffmann A. A. Genomic evidence for role of inversion 3RP of Drosophila melanogaster in facilitating climate change adaptation. Mol. Ecol. 24, 2423–2432 (2015). [DOI] [PubMed] [Google Scholar]
- Daniel K. Fabian et al. Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol. Ecol. 21, 4748–4769 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer S. W. & Anderson W. W. Mechanisms of genetic exchange within the chromosomal inversions of Drosophila pseudoobscura. Genetics 171, 1729–1739 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace A. G., Detweiler D. & Schaeffer S. W. Molecular population genetics of inversion breakpoint regions in Drosophila pseudoobscura. G3 (Bethesda). 3, 1151–63 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratdesaba R., Segarra C. & Aguadé M. Inferring the demographic history of Drosophila subobscura from nucleotide variation at regions not affected by chromosomal inversions. Mol. Ecol. 24, 1729–1741 (2015). [DOI] [PubMed] [Google Scholar]
- Reinhardt J. A., Kolaczkowski B., Jones C. D., Begun D. J. & Kern A. D. Parallel geographic variation in Drosophila melanogaster. Genetics 197, 361–373 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland A. O., Behrman E. L., Schmidt P. S. & Petrov D. A. Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. Plos Genetics 10, e1004775 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pupko T. & Galtier N. A covarion-based method for detecting molecular adaptation: application to the evolution of primate mitochondrial genomes. Proc. Biol. Sci. 269, 1313–1316 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasseur E. et al. The evolutionary landscape of cytosolic microbial sensors in humans. Am. J. Hum. Genet. 91, 27–37 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla S., Swarts H. G. P., Koenderink J. B. & Dobler S. Amino acid substitutions of Na,K-ATPase conferring decreased sensitivity to cardenolides in insects compared to mammals. Insect Biochem. Mol. Biol. 43, 1109–1115 (2013). [DOI] [PubMed] [Google Scholar]
- Stern D. L. The genetic causes of convergent evolution. Nat. Rev. Genet. 14, 751–764 (2013). [DOI] [PubMed] [Google Scholar]
- Agrawal A. A, Petschenka G., Bingham R. A, Weber M. G. & Rasmann S. Toxic cardenolides: chemical ecology and coevolution of specialized plant-herbivore interactions. New Phytol. 194, 28–45 (2012). [DOI] [PubMed] [Google Scholar]
- Endress M. E. & Bruyn P. V. A revised classification of the Apocynaceae. Bot. Rev. 66, 1–56 (2000). [Google Scholar]
- Woods E. C., Hastings A. P., Turley N. E., Heard S. B. & Agrawal A. A. Adaptive geographical clines in the growth and defense of a native plant. Ecol. Monogr. 82, 149–168 (2012). [Google Scholar]
- Markow T. A. & O’Grady P. Reproductive ecology of Drosophila. Funct. Ecol. 22, 747–759 (2008). [Google Scholar]
- Shorrocks B., Ashburner M., Carson H. L. & Thompson J. N. Jr The breeding sites of temperate woodland Drosophila. In: The genetics and biology of Drosophila, pp. 385–428. Academic Press, London (1982). [Google Scholar]
- Thompson J. D., Higgins D. G. & Gibson T. J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 (1999). [Google Scholar]
- Librado P. & Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009). [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. (Columbia University Press, New York., 1987). [Google Scholar]
- Nei M. & Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426 (1986). [DOI] [PubMed] [Google Scholar]
- Watterson G. A. Number of segregating sites in genetic models without recombination. Theor. Popul. Biol. 7, 256–276 (1975). [DOI] [PubMed] [Google Scholar]
- Pina-Martins F. & Paulo O. S. CONCATENATOR: Sequence data matrices handling made easy. Mol. Ecol. Resour. 8, 1254–1255 (2008). [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Slatkin M. & Maddison W. P. Estimation of levels of gene flow from DNA-sequence data. Genetics 132, 583–589 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R. A new statistic for detecting genetic differentiation. Genetics 155, 2011–2014 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos J., Serra L., Solé E. & Pascual M. FISH mapping of microsatellite loci from Drosophila subobscura and its comparison to related species. Chrom. Res. 18, 213–226 (2010). [DOI] [PubMed] [Google Scholar]
- Pegueroles C., Araúz P. A., Pascual M. & Mestres F. A recombination survey using microsatellites: the O chromosome of Drosophila subobscura. Genetica 138, 795–804 (2010). [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical methods to test for nucleotide mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. X. & Li W. H. Statistical tests of neutrality of mutations. Genetics 133, 693–709 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J. H. & Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351, 652–654 (1991). [DOI] [PubMed] [Google Scholar]
- Stoletzki N. & Eyre-Walker A. Estimation of the neutrality index. Mol. Biol. Evol. 28, 63–70 (2011). [DOI] [PubMed] [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300 (1995). [Google Scholar]
- Hill W. G. & Robertson A. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38, 226–231 (1968). [DOI] [PubMed] [Google Scholar]
- Kelly J. K. A test of neutrality based on interlocus associations. Genetics 146, 1197–1206 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2: elegant graphics for data analysis. Springer: New York, (2009). [Google Scholar]
- Hudson R. R. Two-locus sampling distributions and their application. Genetics 159, 1805–1817 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D. H. & Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 (2006). [DOI] [PubMed] [Google Scholar]
- Betrán E., Rozas J., Navarro A. & Barbadilla A. The estimation of the number and the length distribution of gene conversion tracts from population DNA sequence data. Genetics 146, 89–99 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Suchard M. A., Xie D. & Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7 Mol. Biol. Evol. 29, 1969–1973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., Taboada G. L., Doallo R. & Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9, 772 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S. & Gascuel O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Systematic Biology 52, 696–704 (2003). [DOI] [PubMed] [Google Scholar]
- Rambaut A., Suchard M. A., Xie D. & Drummond A. J. Tracer v1.6, Available from http://beast.bio.ed.ac.uk/Tracer (2014). [Google Scholar]
- Gerber P. R. & Müller K. MAB, a generally applicable molecular force field for structure modelling in medicinal chemistry. J. Comput. Aided. Mol. Des. 9, 251–268 (1995). [DOI] [PubMed] [Google Scholar]
- Humphrey W., Dalke A. & Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.