Abstract
Closely related sympatric species commonly develop different ecological strategies to avoid competition. Ctenomys minutus and C. flamarioni are subterranean rodents parapatrically distributed in the southern Brazilian coastal plain, showing a narrow sympatric zone. To gain understanding on food preferences and possible competition for food resources, we evaluated their diet composition performing DNA metabarcoding analyzes of 67 C. minutus and 100 C. flamarioni scat samples, collected along the species geographical ranges. Thirteen plant families, mainly represented by Poaceae, Araliaceae, Asteraceae and Fabaceae, were identified in the diet of C. minutus. For C. flamarioni, 10 families were recovered, with a predominance of Poaceae, Araliaceae and Asteraceae. A significant correlation between diet composition and geographical distance was detected in C. minutus, whereas the diet of C. flamarioni was quite homogeneous throughout its geographical distribution. No significant differences were observed between males and females of each species. However, differences in diet composition between species were evident according to multivariate analysis. Our results suggest some level of diet partitioning between C. flamarioni and C. minutus in the sympatric region. While the first species is more specialized on few plant items, the second showed a more varied and heterogeneous diet pattern among individuals. These differences might have been developed to avoid competition in the region of co-occurrence. Resource availability in the environment also seems to influence food choices. Our data indicate that C. minutus and C. flamarioni are generalist species, but that some preference for Poaceae, Asteraceae and Araliaceae families can be suggested for both rodents.
Introduction
According to the competitive exclusion principle, two complete competitor species cannot coexist in the same area at the same time under a limited resource (Gause, 1934). In real populations, complete ecological overlap is unexpected, as individuals within and between species can use the resources in their habitat differently. Ecological differences may vary depending on the population density, community composition, environment heterogeneity and throughout time and space. These ecological differences may allow the long-term coexistence of species in competitive communities, and hence add to community diversity. However, competition can be increased between sympatric congeners due to their similarities, particularly when resources become scarce. To reduce the ecological superposition and avoid competition, closely related species commonly use different habitats or microhabitats; can present differences in their diet composition; or can be active at different periods of time (Pianka, 2011).
The investigation of diet composition is one of the first steps to better understand the ecological mechanisms involved in the avoidance of competition between closely related sympatric species. However, to determine whether the diet composition of species with overlapping distribution is influenced by the presence of closely related competitors, it is important to assess feeding habits when species are distributed in allopatry. The burrowing rodents of the genus Ctenomys can provide a useful study system to explore questions related to ecological overlapping and competition for food resources.
Subterranean rodents of the genus Ctenomys commonly show allopatric or parapatric species distribution, and overlapping zones, when present, are very narrow (Lessa, 2000). The first reported case of sympatric species for the genus Ctenomys was that of C. australis and C. talarum (Reig et al., 1990). A comparative study of their diets revealed that these species have small microhabitat differences that influence their preferences for different plant species within their home ranges (Comparatore et al., 1995).
Little is known about the dietary composition of ctenomyid species. Some of the few existing studies were conducted using microhistological techniques to analyze fecal contents of C. australis, C. talarum, C. mendocinus and C. eremophilus (Comparatore et al., 1995; Valle et al., 2001; Rosi et al., 2009; Albanese et al., 2010). These subterranean rodents are herbivorous and usually collect their food above ground, around their burrows, cutting plants near the soil and feeding on them inside the burrows, but they can also forage within their tunnels for roots and subterranean stems (Comparatore et al., 1995; Valle et al., 2001; Martino et al., 2007). Ctenomyids are usually referred to as generalists, probably because of the high costs of foraging and digging burrows, and the predation pressure (Nevo, 1979; Reig et al., 1990; Comparatore et al., 1995; Busch et al., 2000). According to field observations, ctenomyids' predators are mainly represented by owls, foxes, southern crested caracara, hawks and snakes.
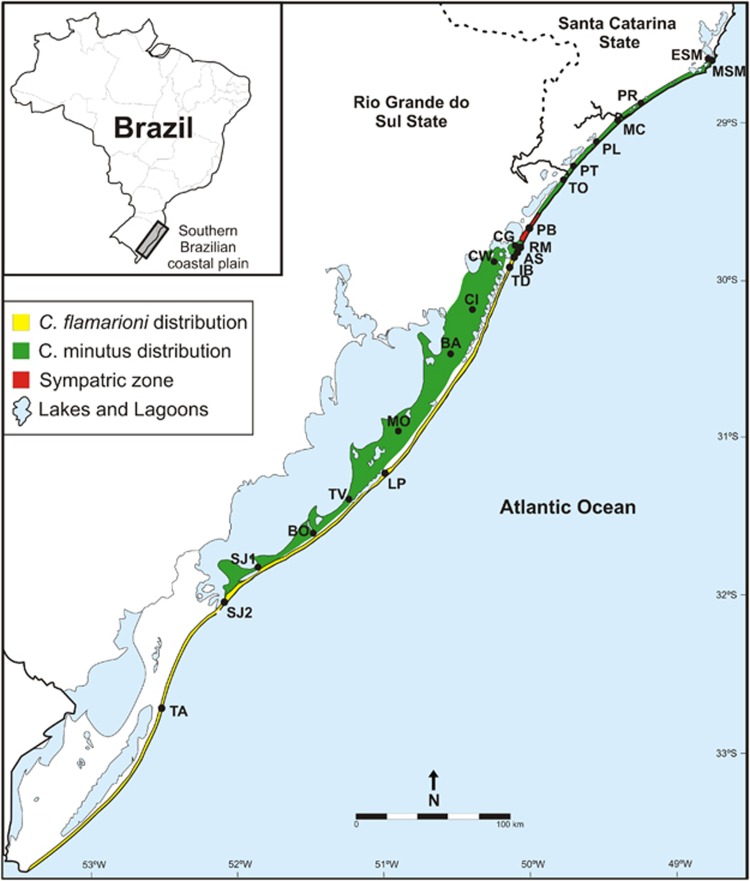
In this study, we focus on two parapatric ctenomyid species: C. flamarioni Travi, 1981 and C. minutus Nehring, 1887, which have a narrow sympatric zone recently described (Kubiak, 2013). C. flamarioni belongs to the mendocinus group, and C. minutus is recovered in the torquatus group within the genus (Parada et al., 2011). These two species inhabit the southern Brazilian coastal plain. C. flamarioni is endemic to the first line of sand dunes, the most recent depositional system in the coastal plain, and its range extends for 560 km (Fernández-Stolz et al., 2007; Figure 1). C. minutus occupies only the sand fields in the southern portion of its distribution, and in the north it preferentially inhabits the first-dune line, extending along 500 km (Lopes et al., 2013). The two species overlap at the northern range of C. flamarioni, in an area extending about 15 km in the sandy-dunes (Kubiak, 2013).
Figure 1.
Geographic distribution of C. minutus and C. flamarioni in the southern Brazilian coastal plain. Small circles represent the sampling sites. AS, Atlântida Sul; BA, Bacopari; BO, Bojuru; CG, Clube de Golfe; CI, Cidreira; CW, Campo dos Weber; ESM, Estrada Farol de Santa Marta; IB, Imbé LB, Lagoa do Peixe; LP, Lagoa do Peixe; MC, Morro dos Conventos; MO, Mostardas; MSM, Morro Farol de Santa Marta; PB, Praia do Barco; PL, Praia da Lagoinha; PR, Praia do Rincão; PT, Passo de Torres; RM, Rainha do Mar; SJ1, São José do Norte; TA, Taim; TD, Tramandaí TO, Torres; TV, avares.
Ctenomyids do not tend to be very selective about the food items they consume (Nevo, 1979; Reig et al., 1990; Comparatore et al., 1995; Busch et al., 2000). However, to avoid competition in the sympatric distribution, it would not be surprising that C. minutus and/or C. flamarioni would present at least some degree of food specialization, or microhabitat differences, unless there is enough food and space in the overlapping area. Moreover, rodent males and females can show distinct foraging strategies associated with differences in nutritional requirements during the growth and reproduction stages (Puig et al., 1999). To better understand how C. minutus and C. flamarioni can co-occur in the same habitat, and the level of possible competition for food resources, we evaluated the dietary composition of these two species, both when they are parapatrically and sympatrically distributed. In addition, we compared the diet composition of males and females within each species, to evaluate possible distinct dietary patterns associated with different diet requirements for each sex.
Materials and methods
Study area
The southern Brazilian coastal plain width ranges from 20 to 100 km and extends for about 800 km, from Cabo de Santa Marta, in the state of Santa Catarina, to the Chui River mouth, at the border between Brazil and Uruguay (Tomazelli et al., 2000; Dillenburg and Hesp, 2009). The region is a mosaic of lakes, lagoons, rivers, dunes and sand fields, and its geomorphology as well as the fauna and flora of the region are continuously influenced by fluctuations of the Atlantic Ocean sea level (Tomazelli et al., 2000).
This coastal plain is characterized by pioneer vegetation with marine influence in the sandy-dunes near to the coast, and with fluvial influence in more internalized sand fields (IBGE, 2012). There is a prevalence of herbaceous species, but shrub forms are also common, and trees become more frequent farther they are from the coast. Closer to the sea, the halophyte-psammophillous vegetation is predominant, whereas in sandy-dunes there is also occurrence of xerophyte species. Around constantly wet environments, it is possible to observe hygrophyte, geophyte and hemicryptophyte plants (IBGE, 1986). However, the natural landscape of the region has been progressively modified over many years, mainly by urbanization, cattle raising, rice cultivation and more recently by Pinus forestry (Fernandes et al., 2007; Boldrini et al., 2008).
Floristic inventories realized in the southern Brazilian coastal plain demonstrated that the families Poaceae, Asteraceae, Cyperaceae, Fabaceae and Solanaceae have the highest proportion of species among all families retrieved (Filho et al., 2013), and represent more than 50% of the species richness in the sand fields (Palma and Jarenkow, 2008; Boldrini, 2009).
Sample collection
Animals were trapped alive using the Oneida Victor no. 0 snap traps, and covered with rubber to avoid injuring the animals. They were anesthetized, their bionomic characteristics were recorded and feces samples were recovered once the animals defecated. Each individual was then released back within its burrow. Animals killed for use in other research projects had their digestive tracts dissected. The stool samples were preserved dried with silica gel, and were sent to the Laboratoire d'Ecologie Alpine (Université Joseph Fourier, Grenoble, France). We collected 67 stool samples for C. minutus and 100 samples for C. flamarioni, across the species entire distribution range (Figure 1 and Table 1).
Table 1. List of sampling sites (abbreviation within parentheses), habitat types, total number of samples, number of individuals of each sex and number of samples collected in different seasons, for C. minutus and C. flamarioni.
| Habitat | N°S | Sex | Season | |
|---|---|---|---|---|
| Ctenomys minutus | ||||
| Morro Farol de Santa Marta (MSM) | Dune | 6 | 3 F / 3 M | 6 Summer |
| Estrada Farol de Santa Marta (ESM) | Sand field | 7 | 4 F / 3 M | 7 Summer |
| Praia do Rincão (PR) | Dune | 2 | 2 F | 2 Summer |
| Morro dos Conventos (MC) | Dune | 2 | 2 F | 2 Summer |
| Praia da Lagoinha (PL) | Dune | 1 | 1 F | 1 Summer |
| Passo de Torres (PT) | Dune | 2 | 2 M | 2 Summer |
| Torres (TO) | Dune | 9 | 5 F / 4 M | 9 Autumn |
| Praia do Barco (PB) | Dune (sympatry) | 6 | 4 F / 2 M | 5 Summer / 1 Autumn |
| Clube de Golfe (CG) | Sand field | 2 | 2 M | 2 Summer |
| Campo dos Weber (CW) | Sand field | 1 | 1 M | 1 Spring |
| Cidreira (CI) | Sand field | 4 | 3 F / 1 M | 4 Summer |
| Bacopari (BA) | Sand field | 4 | 3 F / 1 M | 4 Summer |
| Mostardas (MO) | Sand field | 4 | 3 F / 1 M | 4 Summer |
| Tavares (TV) | Sand field | 2 | 2 F | 2 Summer |
| Bojuru (BO) | Sand field | 8 | 5 F / 3 M | 8 Autumn |
| São José do Norte (SJ1) | Sand field | 7 | 5 F / 2 M | 7 Autumn |
| Total | 67 | 42 F / 25 M | 41 Summer / 25 Autumn / 1 Spring | |
| Ctenomys flamarioni | ||||
| Praia do Barco (PB) | Dune (sympatry) | 13 | 6 F / 6 Ma | 5 Summer / 8 Autumn |
| Rainha do Mar (RM) | Dune | 8 | 4 F / 4 M | 8 Spring |
| Atlântida Sul (AS) | Dune | 11 | 8 F / 3 M | 11 Spring |
| Imbé (IB) | Dune | 21 | 13 F / 8 M | 21 Spring |
| Tramandaí (TD) | Dune | 10 | 7 F / 3 M | 10 Spring |
| Lagoa do Peixe (LP) | Dune | 5 | 3 F / 2 M | 5 Autumn |
| São José do Norte (SJ2) | Dune | 6 | 4 F / 2 M | 6 Summer |
| Taim (TA) | Dune | 26 | 17 F / 9 M | 18 Spring / 8 Summer |
| Total | 100 | 62 F / 37 M | 19 Summer 13 Autum / 68 Spring | |
Abbreviations: F, female; M, male.
There is one individual without sex identification.
Laboratory procedures
Total DNA was extracted from a portion of about 20 mg of stool sample, in a room dedicated to extractions of low-quantity DNA. Samples were initially incubated during 2 h at 55 °C in 1 ml of lysis buffer (Tris-HCl 0.1 m, EDTA 0.1 m, NaCl 0.01 m and N-lauroyl sarcosine 1%, pH 7.5–8). Extractions were then completed using the DNeasy Blood Tissue Kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's instructions. DNA extracts were recovered in a total volume of 300 μl. Blank extractions without samples were systematically performed to monitor possible contaminations.
The amplification of the P6 loop of the chloroplast trnL (UAA) intron was carried out using primers g and h (Taberlet et al., 2007; Table 2). Two other sets of primer pairs were used to amplify a portion of the first internal transcribed spacer (ITS1) of nuclear ribosomal DNA of the families Asteraceae (primers ITS1-F and ITS1Ast-R) and Poaceae (primers ITS1-F and ITS1Poa-R; Table 2; Baamrane et al., 2012). The primer pair 16SMAV-F and 16SMAV-R (Table 2; De Barba et al., 2014) was also applied to amplify a fragment of the 16S rDNA mitochondrial gene of mollusks, arthropods and vertebrates.
Table 2. Primer sequences used in this study and amplicon length of filtered diet sequences.
| Primer name | Primer sequence (5′–3′) | Reference | Sequence length (bp) |
|---|---|---|---|
| g | GGGCAATCCTGAGCCAA | Taberlet et al. (2007) | 31–60 |
| h | CCATTGAGTCTCTGCACCTATC | Taberlet et al. (2007) | |
| ITS1-F | GATATCCGTTGCCGAGAGTC | Baamrane et al. (2012) | |
| ITS1Poa-R | CCGAAGGCGTCAAGGAACAC | Baamrane et al. (2012) | 69–75 |
| ITS1Ast-R | CGGCACGGCATGTGCCAAGG | Baamrane et al. (2012) | 83–85 |
| 16SMAV-F | CCAACATCGAGGTCRYAA | De Barba et al. (2014) | 37–38 |
| 16SMAV-R | ARTTACYNTAGGGATAACAG | De Barba et al. (2014) | |
| Mam-MAVB1 | CCTAGGGATAACAGCGCAATCCTATT-C3 | De Barba et al. (2014) | |
| c | CGAAATCGGTAGACGCTACG | Taberlet et al. (1991) | |
| d | GGGGATAGAGGGACTTGAAC | Taberlet et al. (1991) |
PCR amplifications contained 1 U of AmpliTaq Gold DNA Polymerase (Life Technologies, Carlsbad, CA, USA), 10 mm Tris-HCl, 50 mm KCl, between 2 and 2.5 mm MgCl2, 0.2 mm each dNTP, between 0.2 and 0.3 μm of each primer and 0.005 mg bovine serum albumin (Roche Diagnostics, Basel, Switzerland). We also used 2 μm of Mam-MAVB1 mammalian blocking primers (De Barba et al., 2014) in the MAV PCR reactions to avoid amplifying the ctenomyids own DNA, which can reduce detection of mollusks and arthropods DNA. All DNA amplifications were carried out in a final volume of 30 μl, using 3 μl of DNA extract as template. The PCR mixtures were denatured during 10 min at 95 °C, followed by 50–55 cycles at 95 °C for 30 s and between 50 °C and 58 °C for 30 s. No elongation step at 72 °C was included as the DNA fragments are very short (Table 2).
Each DNA sample was amplified and sequenced two times independently, for each molecular marker described above. Moreover, two negative and two positive PCR controls were amplified and sequenced two times, to guide the selection of sequence filtering parameters and evaluate the filtering process in postsequencing bioinformatic analysis of the diet data sets (De Barba et al., 2014). The positive controls for the P6 loop marker were composed by six plant species (P161, P162, P172, P178, P184 and P185; see Supplementary Table S1) mixed in different known concentrations. The positive controls for ITS1 of Poaceae and Asteraceae consisted of two different plant species each (P161 and P162; and P172 and P178, respectively; see Supplementary Table S1), mixed in different known concentrations. All primer pairs used in a given PCR were modified by the addition of molecular identifier tags on the 5' end, to allow the assignment of sequence reads to their source samples (Valentini et al., 2009). PCR products were tagged differently on both ends. Tags were designed using the OligoTag program (Coissac, 2012) and were composed by eight nucleotides, containing at least five differences among them.
PCR products were titrated using capillary electrophoresis (QIAxel; Qiagen GmbH), purified using the MinElute PCR Purification Kit (Qiagen GmbH), and then mixed together in different concentrations, to obtain a proportion of 50% of the P6 loop, 25% of ITS1 of Poaceae, 10% of ITS1 of Asteraceae and 15% of 16S rDNA of mollusks and arthropods. The different proportions reflected the importance of the diet information provided by each marker (i.e. P6 loop overall herbivorous diet vs complementary information of ITS1 and 16S). The sequencing was carried out on HiSeq 2000 (Illumina Inc., San Diego, CA, USA), following the manufacturer's instructions, using a commercial service (http://www.fasteris.com).
DNA barcoding database
To improve the taxonomic identification of the plant species in the samples, we constructed a database of sequences from 195 specimens of plants collected in the surroundings of the sampling sites. The plant specimens were taxonomically identified, and deposited in the Herbarium and in the Departamento de Botânica of the Universidade Federal do Rio Grande do Sul (Porto Alegre, Brazil). DNA extractions were carried out using about 30 mg of leaves, following the protocol described by Roy et al. (1992) and using the DNeasy Plant Kit (Qiagen GmbH), according to the manufacturer's instructions. The trnL (UAA) intron was amplified using primers c and d for the plant samples P003 to P149 (Supplementary Table S1), the P6 loop of the chloroplast trnL (UAA) intron was sequenced using primers g and h for samples P160 to P269 (Supplementary Table S1) and the ITS1 rDNA of 37 Asteraceae and 63 Poaceae samples was amplified using the primers ITS1Ast-R, ITS1Poa-R and the ITS1-F (Table 2). The trnL (UAA) intron was amplified following Taberlet et al. (1991), and the PCR products were purified using the Exonuclease I and Shrimp Alkaline Phosphatase (Gibco-BRL Life Sciences/Invitrogen, Carlsbad, CA, USA), following the suppliers guidelines. Sequencing was conducted on an ABI 3730 (Applied Biosystems Inc., Foster City, CA, USA) automated sequencer, using both forward and reverse primers. Markers for the P6 loop and ITS1 rDNAs were analyzed following the same PCR, purification and sequencing conditions applied for the dietary samples described above.
Filtering and annotating sequences of the diet data set
Sequences obtained from the next-generation sequencer consisted of a mix of direct and reverse single-strand molecules of DNA, which were firstly filtered and annotated, using the programs available in the OBITools package (http://metabarcoding.org/obitools), following the steps described in Shehzad et al. (2012b). The forward and reverse strands corresponding to the same molecule were aligned and merged, using the illuminapairedend program. The ngsfilter program was applied to keep only sequences unambiguously identified by their tags (no error allowed), and showing a maximum of two errors on primers. Sequences were then sorted in four different files according to each of the four molecular markers analyzed, keeping the information of how many times each sequence was observed in each PCR product. Sequences were classified in the categories of ‘head' (the most common sequence within a group of sequences differing by a single indel/substitution), ‘internal' (sequences less frequent within the group of related sequences) or ‘singleton' (a sequence with no other variants differing by a single indel/substitution) for each PCR product, as described by Shehzad et al. (2012a) and De Barba et al. (2014), to detect possible amplification/sequencing errors. A sequence reference database was built for each molecular marker analyzed, applying the programs ecoPCR (Ficetola et al., 2010) and OBITools, using the sequences of the plant database generated for this study and recovering the relevant part of the molecular markers from the EMBL database (release 117). These reference databases were used to assign the taxon to the diet sequences using ecotag program (Pegard et al., 2009).
The software R 3.0.0 (http://www.R-project.org/) was used to continue the sequence filtering process, following the workflow detailed in De Barba et al. (2014), and to perform statistical analyses. To delete noisy sequence reads (i.e. incorrect sequences occurring at low frequencies), sequences with a frequency lower than 0.01 in a given PCR product were removed from that sample, based on the frequency of noisy sequences observed in the positive controls. To remove possible PCR and sequencing errors, only sequences classified as head in both replicates of at least one sample of each of the four sample groups (C. minutus, C. flamarioni, positive controls, negative controls) were kept for further analyses. We transformed the counts of the sequences into frequency data, and estimated the distance between the two replicates of each sample using the Renkonen similarity index (Renkonen, 1938). A threshold distance of 0.6 was used to exclude samples with highly dissimilar replicates from subsequent analyses, based on comparisons between replicate similarity of positive and negative controls, and dietary samples. A mean of the count of each sequence was calculated between the pair of sample replicates to obtain a sequence profile for all samples. We inspected the profile of negative controls and compared their read counts with that of dietary samples to exclude samples of possible low-quantity DNA (samples with <500 reads for P6 loop and the ITS1 of Poaceae family markers, and <250 for ITS1 of Asteraceae family and MAV markers). Finally, to avoid chimeric sequences and to increase the accuracy of taxonomic assignation, sequences were removed from the whole diet data set when the best identity of the query sequence did not match at least 95% of a sequence from the reference database, when the taxonomic classification of a given sequence did not reach at least the family level, or if the taxon assigned to the sequence did not occur in the region studied (only the Picea genus was excluded from the P6 loop data set).
We used the filtered sequence data generated with the P6 loop marker for characterizing diet composition and patterns of the study species. Sequences obtained from the other markers were merely used for genus/species level identification in Asteraceae and Poaceae (ITS1) and for verifying if the study species were strictly herbivorous (16S rDNA).
Data analyses
Multivariate analyses of the P6 loop diet data were performed using program R 3.0.0. Sample categories were determined based on: (i) the source species (C. flamarioni vs C. minutus); (ii) the habitat/region where the scats were collected (dunes, sand fields or sympatric zone); and (iii) the sex of the specimens (Table 1). The proportions of plant families in the diet composition of each category were plotted using the sum of sequence reads. The package ade4 1.4-17 (Dray and Dufour, 2007) was implemented to carry out correspondence analysis (CA) and between-class analysis (BCA), which performs a principal component analysis based on instrumental variables, to summarize the pattern of diet differentiation among groups of scat samples. Both the CA and BCA analyses were calculated based on the logarithm of the count of each sequence within each sample, to attenuate the effect of extreme values.
The Vegan 2.0-9 package (Oksanen et al., 2013) was implemented to perform an analysis of multivariate homogeneity of group dispersions (betadisper), to evaluate the pattern of dispersion of samples within each category (Anderson, 2006). The Renkonen index was used to calculate the distance between pair of samples, based on the frequency of the sequences in each sample. The average distance of the samples to the group centroid was pairwise compared, under a null hypothesis of no difference in dispersion between groups. We applied a classical t-test, under 999 random permutations, assuming α=0.05.
The relationship between the geographical distance and the diet composition was investigated using a simple Mantel test. The correlation between the Renkonen pairwise dissimilarity matrix and the matrix of pairwise geographical distances between samples was calculated using Vegan 2.0-9 package, applying the Pearson's correlation method, and assuming 999 permutations.
Results
The reference database
From the 195 specimens of plants sequenced to generate a reference database for the P6 loop of the chloroplast trnL (UAA) intron, 102 different sequences were recovered corresponding to 142 species distributed in 28 plant families (Supplementary Table S1) (some species had identical P6 loop sequence). For the ITS1 molecular marker of Poaceae and Asteraceae families, 28 and 16 different sequences were generated, belonging to 31 and 16 species, respectively (Supplementary Table S1) (some Poaceae had identical ITS1 sequence).
Diet composition of C. minutus and C. flamarioni
No sequence reads remained in the PCR-negative control samples of all final data sets, after the filtering process. Relevant sequences corresponding to the species mixed in the PCR-positive controls were found in all final data sets, with exception of the plant samples P172 and P184, which were lost during the filtering process for the P6 loop marker.
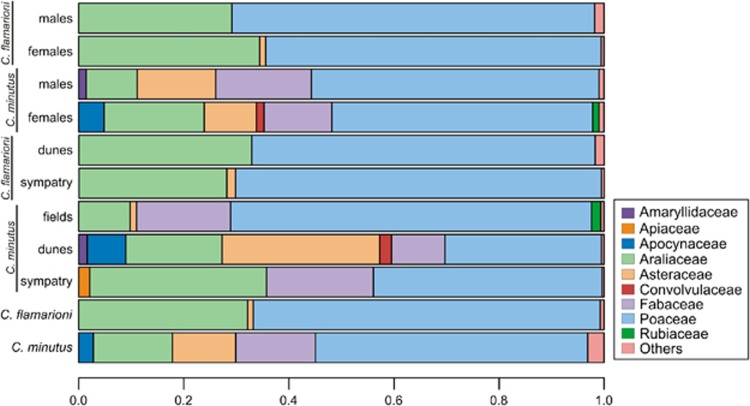
Next-generation sequencing of all molecular markers and samples (dietary samples, plant samples to reference database, PCR-negative and -positive controls) generated a total of about 19.5 million paired-end sequence reads. A total of 1 301 382 reads was retained for the whole diet data set of 67 specimens of C. minutus for the P6 loop marker after the sequence filtering process, corresponding to 41 different sequences distributed in 13 plant families (Table 3). Most of these reads were assigned to the Poaceae family (51.81%), followed by Fabaceae (15.18%), Araliaceae (15.06%), Asteraceae (12.06%) and Apocynaceae (2.78%). Each of the remaining families identified comprised <1% of the total C. minutus reads (Figure 2).
Table 3. Proportion of samples, in each habitat/region and species category, presenting in their diet composition at least one sequence identified in a given plant family for the P6 loop of the chloroplast trnL (UAA) intron.
| Sequence ID |
Taxonomic identification |
Best match |
C. minutus |
C. flamarioni |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family | Genus | Species | Fields (n=39) | Dunes (n=22) | Sympatry (n=6) | All samples (n=67) | Dunes (n=85) | Sympatry (n=13) | All samples (n=98) | ||
| gh0182 | Amaranthaceae | NA | NA | EF688748 | — | — | — | — | 1.18% | 7.69% | 2.04% |
| gh0059 | Amaryllidaceae | Nothoscordum | NA | AF508517 | — | 4.55% | — | 1.49% | — | — | — |
| gh0021 | Apiaceae | Centella | Centella asiatica | P147 | 2.56% | — | 16.67% | 2.99% | — | — | — |
| gh0145 | Apiaceae | Eryngium | Eryngium sanguisorba | P218 | |||||||
| gh0030 | Apocynaceae | NA | NA | AF214362 | — | 13.64% | — | 4.48% | — | — | — |
| gh0003 | Araliaceae | Hydrocotyle | Hydrocotyle bonariensis | P08_P39_P49_P118 | 35.90% | 72.73% | 83.33% | 52.24% | 64.71% | 53.85% | 63.27% |
| gh0011 | Asteraceae | NA | NA | GQ244965 | 20.51% | 22.73% | 16.67% | 20.90% | 10.59% | 15.38% | 11.22% |
| gh0018 | Asteraceae | NA | NA | AF098853 | |||||||
| gh0025 | Asteraceae | NA | NA | AB070918 | |||||||
| gh0036 | Asteraceae | NA | NA | AY952925 | |||||||
| gh0094 | Asteraceae | NA | NA | AF452505 | |||||||
| gh0263 | Asteraceae | Acanthospermum | Acanthospermum australe | P61 | |||||||
| gh0167 | Bignoniaceae | NA | NA | EF105090 | — | 4.55% | — | 1.49% | — | — | — |
| gh0068 | Caryophyllaceae | Cardionema | Cardionema ramosissimum | P88 | 5.13% | — | — | 2.99% | — | — | — |
| gh0073 | Convolvulaceae | NA | NA | AJ430942 | — | 9.09% | — | 2.99% | — | — | — |
| gh0188 | Ericaceae | Empetrum | NA | AY496911 | — | — | — | — | 1.18% | — | 1.02% |
| gh0005 | Fabaceae | Desmodium | Desmodium incanum | P27_P73_P124 | 25.64% | 9.09% | 33.33% | 20.90% | 2.35% | — | 2.04% |
| gh0064 | Fabaceae | Desmodium | NA | EU717290 | |||||||
| gh0143 | Fabaceae | Stylosanthes | NA | AJ230730 | |||||||
| gh0519 | Fabaceae | Vigna | NA | AB304049 | |||||||
| gh0200 | Iridaceae | Herbertia | Herbertia lahue | GQ214203 | — | — | — | — | 1.18% | — | 1.02% |
| gh0101 | Oxalidaceae | Oxalis | NA | AJ582295 | 2.56% | — | — | 1.49% | — | — | — |
| gh0001 | Poaceae | NA | NA | AM404244 | 100% | 95.45% | 100% | 98.51% | 98.82% | 100% | 98.98% |
| gh0006 | Poaceae | Axonopus | NA | AY769150 | |||||||
| gh0012 | Poaceae | NA | NA | EF156672 | |||||||
| gh0002 | Poaceae | NA | NA | AB223118 | |||||||
| gh0007 | Poaceae | NA | NA | AY116268 | |||||||
| gh0009 | Poaceae | NA | NA | AJ489466 | |||||||
| gh0004 | Poaceae | Paspalum | Paspalum notatum | AY769174 | |||||||
| gh0013 | Poaceae | NA | NA | AB571314 | |||||||
| gh0029 | Poaceae | Panicum | NA | AY142726 | |||||||
| gh0032 | Poaceae | NA | NA | AJ488528 | |||||||
| gh0034 | Poaceae | Leersia | Leersia hexandra | AY792527 | |||||||
| gh0014 | Poaceae | NA | NA | AY116257 | |||||||
| gh0066 | Poaceae | Garnotia | Garnotia tenella | GQ869934 | |||||||
| gh0069 | Poaceae | Cenchrus | NA | EU940008 | |||||||
| gh0072 | Poaceae | NA | NA | EF137560 | |||||||
| gh0082 | Poaceae | NA | NA | AM404244 | |||||||
| gh0092 | Poaceae | NA | NA | AB042240 | |||||||
| gh0046 | Poaceae | Luziola | NA | AY792536 | |||||||
| gh0151 | Poaceae | NA | NA | AY116268 | |||||||
| gh0223 | Poaceae | NA | NA | GQ870005 | |||||||
| gh0043 | Poaceae | NA | NA | AY769150 | |||||||
| gh0288 | Poaceae | NA | NA | AY116268 | |||||||
| gh0028 | Poaceae | NA | NA | AY177347 | |||||||
| gh0717 | Poaceae | NA | NA | AY792527 | |||||||
| gh0901 | Poaceae | NA | NA | AY142726 | |||||||
| gh1371 | Poaceae | NA | NA | GQ869934 | |||||||
| gh0061 | Polygonaceae | Rumex | NA | AB219660 | — | — | — | — | 1.18% | — | 1.02% |
| gh0120 | Polygonaceae | NA | NA | DQ860530 | |||||||
| gh0074 | Rubiaceae | Richardia | Richardia stellaris | P211 | 7.69% | — | — | 4.48% | — | — | — |
| gh0156 | Rubiaceae | Richardia | Richardia brasiliensis | P66 | |||||||
| gh0328 | Rubiaceae | Richardia | Richardia brasiliensis | P66 | |||||||
| gh0056 | Salicaceae | NA | NA | AF327597 | — | — | — | — | 1.18% | — | 1.02% |
| gh0505 | Solanaceae | NA | NA | AY266235 | 2.56% | — | — | 1.49% | — | — | — |
| gh0428 | Verbenaceae | NA | NA | AF231884 | — | — | — | — | 1.18% | — | 1.02% |
The taxonomic identification and the best match sequence identifier from GenBank are shown for each sequence. The number of samples for each category is within parentheses.
Figure 2.
Proportion of sequence reads amplified in each plant family with the P6 loop of the chloroplast trnL (UAA) intron. Samples were classified by species and by sex and habitat/region within each species. Families representing <1% of sequence reads were grouped as ‘others'.
Almost all specimens of C. minutus (66 individuals, ~98.51% of the samples) presented at least one sequence identified in the Poaceae family in their diet composition (Table 3). The Araliaceae family was identified in 35 samples (52.24%), and Asteraceae and Fabaceae families were identified in 14 samples (20.90%). The remaining families were less represented. The Amaryllidaceae, Apiaceae, Apocynaceae, Bignoniaceae, Caryophyllaceae, Convolvulaceae, Oxalidaceae, Rubiaceae and Solanaceae families were identified in samples of C. minutus but not in those of C. flamarioni (Table 3). On average, 3.60 different sequences were identified in samples of C. minutus for the P6 loop marker, ranging from 1 to 8 sequences per sample.
The whole diet data set of C. flamarioni for the P6 loop molecular marker was composed by 1 343 667 reads, corresponding to 29 different sequences, distributed in 10 plant families (Table 3). The mostly amplified sequences were attributed to Poaceae (66.04%), Araliaceae (32.15%) and Asteraceae (1.07%). Other families comprised <1% of sequences amplified (Figure 2).
We detected sequences identified in the Poaceae family in 97 of the 98 samples of C. flamarioni (98.98%) (Table 3). Sixty-two samples (~63.27%) showed the Araliaceae family in their diet composition, and the Asteraceae family was identified in 11 samples (11.22%). The families Amaranthaceae, Ericaceae, Iridaceae, Polygonaceae, Salicaceae and Verbenaceae were identified in samples of C. flamarioni only, but they were less represented. On average, 2.65 different sequences were identified in the samples for the P6 loop marker, ranging from 1 to 8 sequences per sample.
The taxonomic resolution of sequence identification for the P6 loop marker reached genus and species levels for 19.64% and 23.21% of the sequences, respectively. The single sequence amplified for the Apocynaceae family in this study was only classified to the level of tribe (Asclepiadeae). The single Araliaceae sequence amplified was identified without ambiguity as the species Hydrocotyle bonariensis. The Fabaceae family was mainly represented by the species Desmodium incanum, but the genera Desmodium, Stylosanthes and Vigna were also identified (Table 3). A greater taxonomic resolution for the Asteraceae family was achieved using the ITS1 molecular marker. Almost 21.00% of the sequences were identified to genus, and 37.50% were identified to species level. The genus Senecio was the taxon most frequently recovered in the scat samples of both C. minutus and C. flamarioni (Supplementary Table S2). The Poaceae family, according to the results of the ITS1 molecular marker, had 35.14% and 54.05% of the sequences identified to the genus and species level, respectively. Poaceae were mainly represented by the species Axonopus fissifolius and Stenotaphrum secundatum, and the genera Panicum and Paspalum (Supplementary Table S3).
The 16S rDNA mitochondrial gene of mollusks, arthropods and vertebrates data set consisted of eight samples (one C. flamarioni and seven C. minutus) and four sequences after the filtering process in R. Two sequences were classified in the Liposcelidae family of insects, and the remaining two sequences were classified in the Eriophyidae family of mites of plants (data not shown).
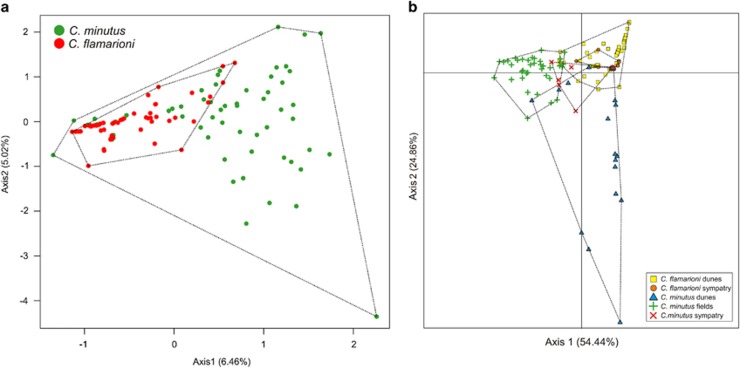
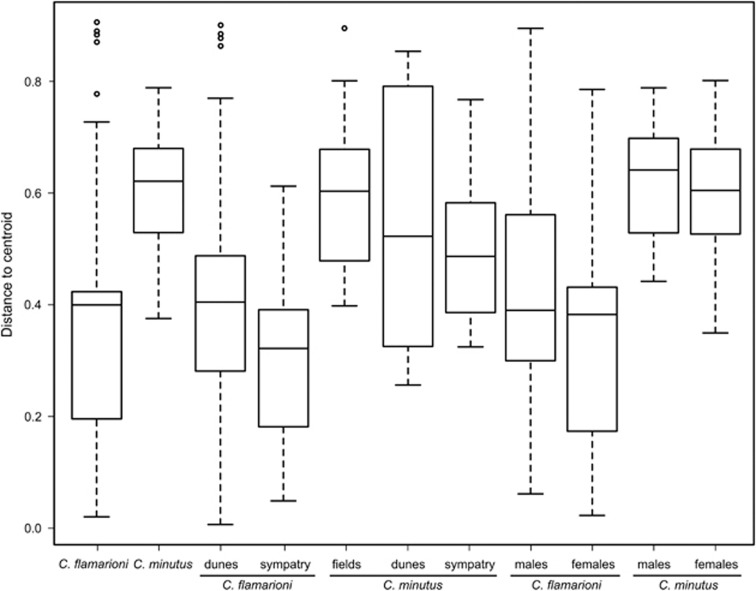
According to the CA plot, the distribution of C. minutus samples was more widespread in both axes 1 and 2 compared with C. flamarioni samples, which overlapped the distribution of C. minutus in the upper right position of the plot (Figure 3a). Moreover, the average distance of the samples to the group centroid (ADC) from the multivariate homogeneity test was significantly different between the two species, according to the permutation test results (ADC: C. flamarioni=0.38; C. minutus=0.61; P=0.001; Figure 4).
Figure 3.
Plots of CA (a) and BCA (b) for samples according to species (a) and habitat type (b).
Figure 4.
Boxplot of multivariate homogeneity of groups' dispersions (betadisper) of samples analyzed according to species, habitat type and sex of individuals.
In the BCA plot, most samples of C. minutus displayed a continuum distribution, except for samples collected at Morro dos Conventos (MC) and at the northern sites of Estrada Farol de Santa Marta (ESM) and Morro Farol de Santa Marta (MSM); this latter also had the most widespread plot (Supplementary Figure S1). For C. flamarioni, all samples grouped together, with exception of São José do Norte (SJ2) and Lagoa do Peixe (LP; Supplementary Figure S2).
The Mantel's test detected a positive and significant correlation between geographical distances and Renkonen distances between samples based on their diet composition, for the overall geographical range of C. minutus (r=0.2902, P=0.001), and for the samples separated by habitat type (dunes: r=0.597, P=0.001; fields: r=0.3537, P=0.001), even when the localities of MSM or ESM were excluded from the analyses (dunes: r=0.4782, P=0.009; fields: r=0.199, P=0.001). For C. flamarioni, there was a significant but weak correlation between geographical distance and sample composition for the whole diet data set (r=0.0783, P=0.013). However, when we excluded samples from SJ2 and LP, the correlation was not significant (r=0.0526, P=0.083).
Geographic distribution: habitats and sympatric zone
Most sequences amplified from C. minutus samples in the sympatric zone were identified in the Poaceae, Araliaceae and Fabaceae families (43.51%, 33.64% and 20.33%, respectively; Figure 2). All individuals showed Poaceae in their diet composition, and Araliaceae was also used by most part of the individuals (83.33% Table 3). A total of 11 different sequences were retrieved for the P6 loop molecular marker, with a mean of 3.5 sequences per sample (range 2–5). The count of sequence reads for samples from the sand fields were mainly represented by Poaceae (68.69%), Fabaceae (17.88%) and Araliaceae (9.77%) families. All individuals showed Poaceae in their diet composition (Table 3). Thirty different P6 loop sequences were retrieved, with a mean of four sequences amplified per sample (range 2–8). For C. minutus sampled in dunes, the Asteraceae family comprised 30.02% of the sequence reads, followed by Poaceae (29.80%) and Araliaceae (18.31%). Poaceae and Araliaceae were found in the diet composition of several individuals (95.45% and 72.73%, respectively) (Table 3). A total of 19 different sequences were identified, with an average of 2.91 sequences per sample (range 1–6).
Four families were identified in the diet of samples of C. flamarioni from the sympatric zone, and most of the sequences amplified were classified in the Poaceae (69.67%) and Araliaceae (28.14%) families (Figure 2). The families more frequently used by the individuals were Poaceae and Araliaceae (100% and 53.85% of individuals, respectively; Table 3). A total of five different sequences were retrieved for the P6 loop marker with a mean of two for each sample (range 1–4). Likewise, samples of C. flamarioni collected outside the sympatric zone showed a higher proportion of sequence reads identified in the Poaceae (65.37%) and Araliaceae (32.89%) families, and also most part of the individuals showed these two families in their diet composition (Table 3). Twenty nine different sequences were identified, averaging 2.75 sequences in each specimen (range 1–8). The number of distinct sequences shared between groups of specimens by habitat/region is given in Supplementary Table S5.
Samples of C. minutus from dunes were the most widespread in the BCA plot (Figure 3b). Samples of C. flamarioni from the sympatric zone showed a restricted distribution, overlapping the group of samples of C. flamarioni from dunes. C. minutus from the sympatric zone occupied a central position in the plot, slightly overlapping the distribution of other samples. However, there was no clear overlap between samples of C. minutus and C. flamarioni from the sympatric zone. The CA plots of axis 1 × axis 2 and axis 1 × axis 3 showed similar results for samples of the two species in the sympatric region (Supplementary Figure S3).
Average distances of the samples to the centroid of the groups were generally higher in C. minutus (ADC: fields=0.59; dunes=0.55; sympatry=0.51) than in C. flamarioni (ADC: dunes=0.39; sympatry=0.30). Five pairwise comparisons showed significant differences in the multivariate dispersion of the samples within categories: (i) C. minutus from dunes vs C. flamarioni from dunes (P=0.007); (ii) C. minutus from dunes vs C. flamarioni from sympatric zone (P=0.002); (iii) C. minutus from fields vs C. flamarioni from dunes (P=0.001); (iv) C. minutus from fields vs C. flamarioni from sympatric zones (P=0.001); and (v) C. minutus from sympatric zones vs C. flamarioni from sympatric zones (P=0.018).
Males vs females diet composition
The diet composition of males and females, within each species, showed similar variation according to the CA (data not shown) and BCA plots (Supplementary Figure S4), and the betadisper results (Figure 4). Moreover, there were no significant differences of samples dispersion between males and females of each species, based on the permutation tests for the betadisper results (Supplementary Table S4).
Discussion
Diet composition of C. minutus and C. flamarioni
Nineteen plant families were identified in this study, 13 in the samples of C. minutus and 10 in the samples of C. flamarioni. Despite differences in the families retrieved, Poaceae sequences comprised more than half of the sequence reads of both ctenomyid species, and were highly amplified in different habitats. The frequency of occurrence of different food items in the samples (i.e. the proportion of samples containing a given plant) was concordant with the information provided by the read counts, indicating that Poaceae sequences were identified in almost the totality of samples of both species. The Araliaceae and Asteraceae families were also commonly found in both species, together with Fabaceae for C. minutus. This may suggest that these subterranean rodents preferentially use these four plant families as food sources.
The final data set of the 16S rDNA mitochondrial gene of mollusks, arthropods and vertebrates consisted of a few samples and sequences. The four sequences retrieved were taxonomically identified in mite (Eriophyidae) and insect (Liposcelidae) families, which are commonly found on plants. It is likely that these organisms are not part of the diet composition of both ctenomyid species, but were only accidentally ingested while the individuals were feeding on plants. These results confirm that C. minutus and C. flamarioni are herbivorous species.
Some considerations apply to our inferences on the diet composition of the two species. The detection of DNA sequences of different food items and the sequence read counts observed can be subject to a number of methodological and biological bias (Deagle et al., 2013; Thomas et al., 2014). For example, biased amplification can occur when the primers match better sequences in certain species over others (Shehzad et al., 2012b), or because of preferential amplification of shorter sequences (Rayé et al., 2011). Biological factors, such as variation in the proportion of mitochondrial and chloroplast organelles among tissues and differential digestibility of foods, may also influence DNA quantity in the scats and therefore its detectability for diet analysis (Pegard et al., 2009; Deagle et al., 2010; Thomas et al., 2014). Even if these constraints preclude the use of sequence read counts for accurate absolute quantitative interpretations of the proportions of food ingested, various studies showed that consistent proportions of food DNA were estimated from fecal samples of animals fed a known diet (Deagle et al., 2010; Bowles et al., 2011), and that DNA sequence counts can be used for semiquantitative estimates of diet in comparative studies (Pompanon et al., 2012).
An additional biological aspect, important for diet analysis from feces of subterranean rodents, relates to their high digestive efficiencies, which allow enhancing the exploitation of even low-quality resources rich in fiber. This efficiency is associated with long retention times of food in the gut compared with other herbivorous, and, until the process of fermentation is not finished, the food is not eliminated (Buffenstein, 2000). In other words, the richer in fiber is a plant, the longer it will be retained in the gut, and thus it could be recovered in several feces samples, potentially affecting estimates of frequency of occurrence of plants in the samples and inferences of food preferences.
One limitation of our approach for identifying finer compositional differences among samples was the limited taxonomic resolution achieved, due to moderate variability of the P6 loop marker. In fact, despite several different sequences were retrieved in the diet composition of C. minutus and C. flamarioni, we were not able to identify all sequences to the species or genus level, and some species shared the same P6 loop sequence. However, this inconvenience is compensated by the fact that the primers g and h used to amplify the P6 loop region are extremely well conserved, from Gymnosperms to Angiosperms, which is particularly important when amplifying multiple species within the same PCR (Taberlet et al., 2007). Moreover, the P6 loop is short enough to allow the amplification of highly degraded DNA in fecal samples. We increased the taxonomic resolution for members of Poaceae and Asteraceae by using the more variable ITS1 markers within these two plant families.
Generalist vs specialist habits
The energetic cost of a subterranean life is high, mainly associated with the process of digging burrows for dispersion and foraging (Buffenstein, 2000). One way to decrease costs of foraging is to use food sources found inside and around the burrows, and be less selective about the food consumed (Altuna et al., 1998; Buffenstein, 2000; Martino et al., 2007). This is particularly true for subterranean rodents that carry out most of their activities belowground, and whose foraging patterns are usually associated to the probability of finding food along their path. In contrast, in subterranean rodents as ctenomyids, showing mainly an above-ground foraging behavior, some level of food specialization might be developed (Busch et al., 2000). Our data support this previous findings indicating that, whereas C. minutus and C. flamarioni are generalists, preference for particular plant species and plant families can also be suggested for both rodents.
Two of the plant families most frequently found in samples of C. minutus and C. flamarioni (Poaceae and Asteraceae) are abundant in the southern Brazilian coastal plain, figuring among the families with the highest species richness (Palma and Jarenkow, 2008; Boldrini, 2009; Filho et al., 2013). Moreover, the species Hydrocotyle bonariensis from the Araliaceae family is quite common in the coastal plain compared with other plant species (Palma and Jarenkow, 2008), and it is a food item frequently found in samples of C. minutus and C. flamarioni, even in distinct habitats. These results suggest that resource availability in the environment might influence food choices in these ctenomyid species. Curiously, even if the Cyperaceae family is quite common and represented by several species in the coastal plain (Boldrini, 2009), plants of this family were not identified as part of the diet of the subterranean rodents in this study. Unfortunately, at this point we are not able to confirm if this can be attributed to food choices or to detection bias for the Cyperaceae DNA.
Similar feeding behaviors were found among other ctenomyid species. C. mendocinus lives in Midwestern Argentina. Although this species has generalist habits, a preference for some food items was observed, mainly represented by above-ground parts of grasses, even in populations inhabiting environments with low food availability and harsh climatic conditions (Puig et al., 1999; Rosi et al., 2003). C. eremophilus has generalist behavior with a preference on aerial vegetative part of grasses, but in grazed paddocks their diet showed lower percentage of grasses and higher dietary diversities compared with ungrazed paddocks, which was associated with the competition for grasses with cattle (Rosi et al., 2009). C. talarum and C. australis are also generalists but prefer the aerial vegetative part of grasses over forbs throughout the year (Comparatore et al., 1995). Selective behaviors observed in ctenomyids have also been associated with specific nutritional requirements during the reproductive season, the time spent harvesting and handling the plants and the balance between the quality of the food to supply the energy, protein and fiber requirements vs the quantity of secondary components and plant defenses, which can influence their palatability and digestibility (Comparatore et al., 1995; Puig et al., 1999; Martino et al., 2007).
Differences in the diet composition between species, habitats and sex
The overall diet composition of C. minutus was more varied and heterogeneous among individuals than the overall diet of C. flamarioni, as inferred by the number of different sequences retrieved by sample and overall for each species, and results of multivariate analyses. C. minutus is distributed in the dunes near to the sea and in more internalized sand fields of the southern Brazilian coastal plain. These environments differ in their phytophysiognomy, with the first-dune line being characterized by more sparse vegetation cover, and plant species adapted to poor, salty and unstable soils. In contrast, the more internalized fields are a mosaic of natural landscape, anthropic disturbed areas and constantly wet regions of fluvial and lake influences (IBGE, 1986), and hence have higher species richness. Moreover, the diet among the individuals of C. minutus seems to diverge along the environment with increasing geographical distances, which can contribute to a more varied diet for this species. On the other hand, the diet of C. flamarioni was quite homogeneous all over its geographical distribution. Although Mantel test results for the whole diet data set of C. flamarioni were significant, the correlation between differences in diet composition and geographical distances seemed to be weak and affected by the distinct diet pattern of individuals sampled in the LP and SJ2 sites, both located in the southern species' distribution. The scats of C. flamarioni from LP were sampled in a sandy soil region constantly wet, as it is near to the margins of a lagoon (Lagoa do Peixe), whereas SJ2 is located in a sand field near to the first-dune line with vegetation cover similar to those fields inhabited by C. minutus in its southern range.
The specimens of C. minutus and C. flamarioni from the sympatric region diverged in all aspects considered in this study. Only five different sequences were retrieved in the fecal samples of 13 individuals of C. flamarioni from the sympatric zone, with a mean of two sequences amplified per individual for the P6 loop molecular marker, which was the lower value obtained among all groups of samples. This result could suggest that C. flamarioni inhabiting the sympatric region are more specialized on few plant items than C. minutus, which showed a higher mean of different sequences (3.5) per fecal sample, and a total of 11 sequences identified among the six individuals. The specimens of C. flamarioni showed a more homogeneous pattern of diet composition when compared with C. minutus, and no remarkable diet overlapping was detected between species, according to the low number of sequences shared (3 out of 13 sequences in the region of co-occurrence) and multivariate analyses. These results may indicate some level of dietary partitioning for C. minutus and C. flamarioni in the sympatric region, which can contribute to the coexistence of both rodents in the same area, by reducing major effects of competition between them. However, our small sample sizes and the limited number of sequences in the zone of sympatry limit definite conclusions on direct competition for and adaptation to food resources.
Distinct foraging strategies between males and females were described in rodents, and were mainly associated to differences in nutritional requirements during the growth and reproduction stages (Puig et al., 1999). The microhistological analysis of the diet of C. mendocinus demonstrated that males have a more varied diet in winter, attributed to changes in the behavior during the beginning of the reproductive season, while females showed a higher specialization on grasses in spring, probably associated to the high energetic costs of pregnancy and lactation occurring during this period of the year (Puig et al., 1999). Our results indicate no significant diet differences between males and females of both species, even in different habitats (Figures 2 and 4 and Supplementary Figure S4). Since we did not sample in different seasons in the same sampling sites, we could not perform seasonal analyses. Our results may be biased by the fact that males/females samples were analyzed all together, regardless of the season they were collected, and thus possible minor differences in diet composition between sexes could have not been detected.
Future research on diet analyses of these species based on DNA metabarcoding should focus on applying other molecular markers designed to amplify short and variable regions of specific plant families/genus of interest, which, combined with a more complete reference database, can increase the power of taxonomic resolution of this methodology. Moreover, sampling should be designed to assess the information of the diet composition in different seasons from the same sampling site, to allow inferences about differences related to seasonal environmental changes and in animal nutritional requirements during the growth and reproduction stages.
Data archiving
DNA sequences of trnL (UAA) intron for the plant samples P003 to P149: GenBank accession numbers KP057622–KP057680. NGS feces unfiltered data and sequences to reference databases: Dryad repository doi:10.5061/dryad.fp3fp.
Acknowledgments
This study was supported by Agence Nationale de la Recherche (ANR, France), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil) and Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS, Brazil). We thank Christian Miquel, Delphine Rioux and Carole Poillot for technical help in the laboratory, and Angelika Studeny, Marco Andrello, Lucie Zinger and Pierre de Villemereuil for help with statistics and R software.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
The authors note that LG and PT are two of the coinventors of patents related to the g/h primers and the use of the P6 loop of the chloroplast trnL (UAA) intron for plant identification using degraded template DNA. These patents only restrict commercial applications and have no impact on the use of this locus by academic researchers.
Supplementary Material
References
- Albanese S, Rodríguez D, Dacar MA, Ojeda RA. (2010). Use of resources by the subterranean rodent Ctenomys mendocinus (Rodentia, Ctenomyidae), in the lowland Monte desert, Argentina. J Arid Environ 74: 458–463. [Google Scholar]
- Altuna CA, Bacigalupe LD, Corte S. (1998). Food-handling and feces reingestion in Ctenomys pearsoni (Rodentia, Ctenomyidae). Acta Theriol 43: 433–437. [Google Scholar]
- Anderson MJ. (2006). Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62: 245–253. [DOI] [PubMed] [Google Scholar]
- Baamrane MAA, Shehzad W, Ouhammou A, Abbad A, Naimi M, Coissac E et al. (2012). Assessment of the food habits of the Moroccan dorcas gazelle in M'Sabih Talaa, west central Morocco, using the trnL approach. PLoS One 7: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldrini II. (2009). A flora dos campos do Rio Grande do Sul. In: Pillar VDP, Muller SC, Castilhos ZMS, Jacques AVA (eds). Campos Sulinos: Conservação e Uso Sustentável da Biodiversidade. Ministério do Meio Ambiente: Brazil, pp 63–77. [Google Scholar]
- Boldrini II, Trevisan R, Schneider AA. (2008). Estudo florístico e fitossociológico de uma área às margens da lagoa do Armazém, Osório, Rio Grande do Sul, Brasil. Rev Bras Biociênc 6: 355–367. [Google Scholar]
- Bowles E, Schulte PM, Tollit DJ, Deagle BE, Trites AW. (2011). Proportion of prey consumed can be determined from faecal DNA using real-time PCR. Mol Ecol Resour 11: 530–540. [DOI] [PubMed] [Google Scholar]
- Buffenstein R. (2000). Ecophysiological responses of subterranean rodents to underground habitats. In: Lacey EA, Patton JL, Cameron GN (eds). Life Underground: The Biology of Subterranean Rodents. The University of Chicago Press: Chicago. IL, USA, pp 62–110. [Google Scholar]
- Busch C, Antinuchi CD, del Valle JC, Kittlein MJ, Malizia AI, Vassallo AI et al. (2000). Population ecology of subterranean rodents. In: Lacey EA, Patton JL, Cameron GN (eds). Life Underground: The Biology of Subterranean Rodents. The University of Chicago Press: Chicago, IL, USA, pp 183–226. [Google Scholar]
- Coissac E. (2012). OligoTag: a program for designing sets of tags for next-generation sequencing of multiplexed samples. In: Pompanon F, Bonin A (eds). Data Production and Analysis in Population Genomics. Humana Press: New York, NY, USA, pp 13–31. [DOI] [PubMed] [Google Scholar]
- Comparatore VM, Cid MS, Busch C. (1995). Dietary preferences of two sympatric subterranean rodent populations in Argentina. Rev Chil Hist Nat 68: 197–206. [Google Scholar]
- De Barba M, Miquel C, Boyer F, Mercier C, Rioux D, Coissac E et al. (2014). DNA metabarcoding multiplexing and validation of data accuracy for diet assessment: application to omnivorous diet. Mol Ecol Resour 14: 306–323. [DOI] [PubMed] [Google Scholar]
- Deagle BE, Chiaradia A, McInnes J, Jarman SN. (2010). Pyrosequencing faecal DNA to determine diet of little penguins: is what goes in what comes out? Conserv Genet 11: 2039–2048. [Google Scholar]
- Deagle BE, Thomas AC, Shaffer AK, Trites AW, Jarman SN. (2013). Quantifying sequence proportions in a DNA-based diet study using Ion Torrent amplicon sequencing: which counts count? Mol Ecol Resour 13: 620–633. [DOI] [PubMed] [Google Scholar]
- Dillenburg SR, Hesp AP. (2009) Geology and Geomorphology of Holocene Coastal Barriers Of Brazil. Springer-Verlag: Berlin, Germany. [Google Scholar]
- Dray S, Dufour AB. (2007). The ade4 package: implementing the duality diagram for ecologists. J Stat Softw 22: 1–20. [Google Scholar]
- Fernandes FA, Fernández-Stolz GP, Lopes CM, Freitas TRO. (2007). The conservation status of the tuc-tucos, genus Ctenomys (Rodentia: Ctenomyidae), in southern Brazil. Braz J Biol 67: 839–847. [DOI] [PubMed] [Google Scholar]
- Fernández-Stolz G, Stolz JFB, de Freitas TRO. (2007). Bottlenecks and dispersal in the tuco-tuco das dunas, Ctenomys flamarioni (Rodentia: Ctenomyidae), in southern Brazil. J Mammal 88: 935–945. [Google Scholar]
- Ficetola GF, Coissac E, Zundel S, Riaz T, Shehzad W, Bessière J et al. (2010). An in silico approach for the evaluation of DNA barcodes. BMC Genom 11: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filho PJS, da S, Silva CC, da, Franco FP, Cavalli J, Bertholdo LM, Schmitt LA et al. (2013). Levantamento florístico de um fragmento de Floresta Ombrófila Densa no litoral norte do Rio Grande do Sul. Brasil Rev Bras Biociênc 11: 163–183. [Google Scholar]
- Gause GF. (1934) Struggle for Existence. Williams & Wilkins Co.: Baltimore, MD, USA. [Google Scholar]
- IBGE. (1986) Projeto RadamBrasil: Levantamento dos recursos naturais Vol 3, Instituto Brasileiro de Geografia e Estatística—IBGE: Rio de Janeiro, Brazil. [Google Scholar]
- IBGE. (2012) Manual técnico da vegetação brasileira,2nd edn. Instituto Brasileiro de Geografia e Estatística—IBGE: Rio de Janeiro, Brazil. [Google Scholar]
- Kubiak BB. (2013). Relação entre a distribuição espacial e características ambientais de duas espécies de tuco-tuco (Rodentia: Ctenomys em uma nova área de simpatria para o gênero na planície costeira do Sul do Brasil. MSc Thesis. Universidade Federal do Rio Grande do Sul. [Google Scholar]
- Lessa EP. (2000). The evolution of subterranean rodents. In: Lacey EA, Patoon JL, Cameron GN (eds). Life Underground: The Biology of Subterranean Rodents. The University of Chicago Press: Chicago, IL, USA, pp 389–420. [Google Scholar]
- Lopes CM, Ximenes SSF, Gava A, Freitas TRO. (2013). The role of chromosomal rearrangements and geographical barriers in the divergence of lineages in a South American subterranean rodent (Rodentia: Ctenomyidae: Ctenomys minutus). Heredity 111: 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino NS, Zenuto RR, Busch C. (2007). Nutritional responses to different diet quality in the subterranean rodent Ctenomys talarum (tuco-tucos). Comp Biochem Physiol Part A 147: 974–982. [DOI] [PubMed] [Google Scholar]
- Nevo E. (1979). Adaptive convergence and divergence of subterranean mammals. Annu. Rev Ecol Syst 10: 269–308. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB et al. (2013). Vegan: community ecology package. R package version 2.2-0, http://cran.r-project.org/web/packages/vegan/index.html.
- Palma CB, Jarenkow JA. (2008). Estrutura de uma formação herbácea de dunas frontais no litoral norte do Rio Grande do Sul, Brasil. Biociências 16: 114–124. [Google Scholar]
- Parada A, D'Elía G, Bidau CJ, Lessa EP. (2011). Species groups and the evolutionary diversification of tuco-tucos, genus Ctenomys (Rodentia: Ctenomyidae). J Mammal 92: 671–682. [Google Scholar]
- Pegard A, Miquel C, Valentini A, Coissac E, Bouvier F, François D et al. (2009). Universal DNA-based methods for assessing the diet of grazing livestock and wildlife from feces. J Agric Food Chem 57: 5700–5706. [DOI] [PubMed] [Google Scholar]
- Pianka ER. (2011). Competition. In: Pianka ER (ed.). Evolutionary Ecology - eBook.
- Pompanon F, Deagle BE, Symondson WOC, Brown DS, Jarman SN, Taberlet P. (2012). Who is eating what: diet assessment using next generation sequencing. Mol Ecol 21: 1931–1950. [DOI] [PubMed] [Google Scholar]
- Puig S, Rosi MI, Cona MI, Roig VG, Monge SA. (1999). Diet of Piedmont populations of Ctenomys mendocinus (Rodentia, Ctenomyidae): seasonal patterns and variations according sex and relative age. Acta Theriol 44: 15–27. [Google Scholar]
- Rayé G, Miquel C, Coissac E, Redjadj C, Loison A, Taberlet P. (2011). New insights on diet variability revealed by DNA barcoding and high-throughput pyrosequencing: chamois diet in autumn as a case study. Ecol Res 26: 265–276. [Google Scholar]
- Reig OA, Busch C, Ortells MO, Contreras JL. (1990). An overview of evolution, systematics, population biology and molecular biology in Ctenomys. In: Nevo E, Reig OA (eds). Evolution of Subterranean Mammals at the Organismal and Molecular Levels. Allan Liss: New York, NY, USA, pp 71–96. [PubMed] [Google Scholar]
- Renkonen O. (1938). Statisch-ökologische Untersuchungen über die terrestrische Käferwelt der finnischen Bruchmoore. Ann Zool Soc Zool Fenn Vanamo 6: 1–231. [Google Scholar]
- Rosi MI, Cona MI, Videla F, Puig S, Monge SA, Roig VG. (2003). Diet selection by the fossorial rodent Ctenomys mendocinus inhabiting an environment with low food availability (Mendoza, Argentina). Stud Neotrop Fauna Environ 38: 159–166. [Google Scholar]
- Rosi MI, Puig S, Cona MI, Videla F, Méndez E, Roig VG. (2009). Diet of a fossorial rodent (Octodontidae), above-ground food availability, and changes related to cattle grazing in the Central Monte (Argentina). J Arid Environ 73: 273–279. [Google Scholar]
- Roy A, Frascaria N, MacKay J, Bousquet J. (1992). Segregating random amplified polymorphic DNAs (RAPDs) in Betula alleghaniensis. Theor Appl Genet 85: 173–180. [DOI] [PubMed] [Google Scholar]
- Shehzad W, McCarthy TM, Pompanon F, Purevjav L, Coissac E, Riaz T et al. (2012. a). Prey preference of snow leopard (Panthera uncia in South Gobi, Mongolia. PLoS One 7: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehzad W, Riaz T, Nawaz MA, Miquel C, Poillot C, Shah SA et al. (2012. b). Carnivore diet analysis based on next-generation sequencing: application to the leopard cat (Prionailurus bengalensis in Pakistan. Mol Ecol 21: 1951–1965. [DOI] [PubMed] [Google Scholar]
- Taberlet P, Coissac E, Pompanon F, Gielly L, Miquel C, Valentini A et al. (2007). Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res 35: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17: 1105–1109. [DOI] [PubMed] [Google Scholar]
- Thomas AC, Jarman SN, Haman KH, Trites AW, Deagle BE. (2014). Improving accuracy of DNA diet estimates using food tissue control materials and an evaluation of proxies for digestion bias. Mol Ecol 23: 3706–3718. [DOI] [PubMed] [Google Scholar]
- Tomazelli LJ, Dillenburg SR, Villwock JA. (2000). Late quaternary geological history of Rio Grande do Sul coastal plain, southern Brazil. Rev. Bras Geociênc 30: 474–476. [Google Scholar]
- Valentini A, Miquel C, Nawaz MA, Bellemain E, Coissac E, Pompanon F et al. (2009). New perspectives in diet analysis based on DNA barcoding and parallel pyrosequencing: the trnL approach. Mol Ecol 9: 51–60. [DOI] [PubMed] [Google Scholar]
- Valle JC, Lohfelt MI, Comparatore VM, Cid MS, Busch C. (2001). Feeding selectivity and food preference of Ctenomys talarum (tuco-tuco). Mamm Biol 66: 165–173. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.