Abstract
Pig domestication began around 9000 YBP in the Fertile Crescent and Far East, involving marked morphological and genetic changes that occurred in a relatively short window of time. Identifying the alleles that drove the behavioural and physiological transformation of wild boars into pigs through artificial selection constitutes a formidable challenge that can only be faced from an interdisciplinary perspective. Indeed, although basic facts regarding the demography of pig domestication and dispersal have been uncovered, the biological substrate of these processes remains enigmatic. Considerable hope has been placed on new approaches, based on next-generation sequencing, which allow whole-genome variation to be analyzed at the population level. In this review, we provide an outline of the current knowledge on pig domestication by considering both archaeological and genetic data. Moreover, we discuss several potential scenarios of genome evolution under the complex mixture of demography and selection forces at play during domestication. Finally, we highlight several technical and methodological approaches that may represent significant advances in resolving the conundrum of livestock domestication.
Introduction
Domestication, which is difficult to define in a comprehensive and objective manner, may be understood as a mutualistic long-term relationship between humans and plant or animal species that implies a selective advantage for both parties (Zeder et al., 2006). This evolutionary process involves a series of behavioural (for example, decreased aggression, stress and watchfulness), morphological (for example, brain size and skull and teeth shape) and physiological (for example, increased growth and prolificacy) changes aimed to satisfy human needs. In addition, domestication can be defined as gradual, continuous and extremely complex with many potential trajectories and detours (Albarella et al., 2006; Zeder et al., 2006). Indeed, domestication comprises a wide spectrum of intermediate stages that exhibit diffuse borders and a dynamic and reversible nature. This is particularly true for the pig, which is a species that can rapidly retrace the steps that led to the development of its domestic form over millennia and become feral (White, 2011).
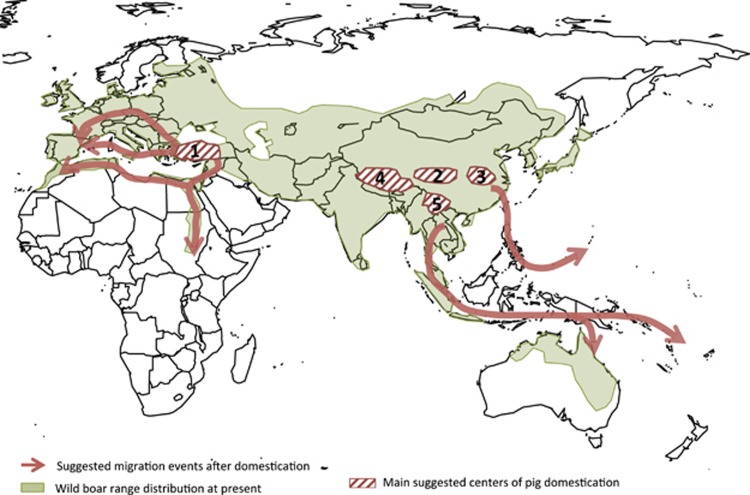
There is a general consensus that the Eurasian wild boar (Sus scrofa) and other sister species, such as Sus celebensis (Celebes warty pig), Sus verrucosus (Java warty pig), Sus cebifrons (Visayan warty pig), Sus philippensis (Philippine warty pig) and Sus barbatus (Bornean bearded pig), emerged in Southeast Asia in the early Pliocene (Figure 1), approximately 5.3–3.5 Myr ago (Larson et al., 2007a, 2011; Frantz et al., 2013). As we will discuss later, the spatial and temporal coexistence of these suid species involved frequent hybridization events (Frantz et al., 2013). This circumstance complicates the reconstruction of the evolutionary history of the genus Sus and calls into question whether species such as Sus celebensis, which currently live in the wild but may have been domesticated in the past, could have contributed to the gene pool of Asian pigs (Albarella et al., 2006; Larson et al., 2011; Frantz et al., 2013).
Figure 1.
Map depicting Southeast Asia, as the origin of suid species (Sus scrofa, Sus celebensis, Sus verrucosus, Sus barbatus, Sus philippensis and Sus cebifrons), and the current geographic distribution of wild boar. We also indicate (lined areas) pig domestication centres at the Near East (Anatolia, #1) and China (multiple putative centres have been reported at the upstream #2 and middle-downstream #3 regions of the Yangtze river, the Tibetan Plateau #4 and the Mekong region #5, but so far data are not conclusive). The potential dispersal routes from these domestication centres are also shown with brown arrows. Cryptic domestication centres, without zooarchaeological support, have been also proposed at Southeast Asia, India and other locations (Larson et al., 2011).
Wild boars are extremely adaptable, presenting a current geographic range that comprises territories from three continents (Figure 1). In strong contrast with other suids, Sus scrofa were very successful at expanding and colonizing new and highly differentiated ecological niches. From their place of origin, they migrated northwards, crossed the Kra isthmus and entered mainland Asia (Larson et al., 2007a, 2010). Taking advantage of land bridges caused by the reduction of sea levels, wild boars were able to colonize insular territories such as Taiwan, Lanyu, Japan and the Ryukyu chain (Larson et al., 2010). During the Pleistocene, wild boars spread westwards, reaching Europe approximately 0.8 Mya (Frantz et al., 2013). Indeed, wild boar remains corresponding to the late Early Pleistocene have been found at Atapuerca (Spain), as reported by Van der Made (2001). Genetic analyses showing a much higher level of diversity in Sus scrofa populations from Asia than in those from Europe (Larson et al., 2005; Ramírez et al., 2009) are consistent with the scenario outlined above. Migration to Europe was most likely followed by a long period of geographic isolation, linked to the colder and drier climate that characterizes the Calabrian stage, which led to the establishment of two highly differentiated eastern (Asia) and western (Europe, Near East and North Africa) Sus scrofa gene pools (Figure 2). In a pioneering study that was subsequently confirmed by many others, Giuffra et al. (2000) sequenced several mitochondrial and nuclear loci and concluded that European and Asian pigs diverged long before domestication, 500 000 YBP. This divergence estimate has recently been updated to the mid-Pleistocene (1.6–0.8 Myr) within the framework of the pig genome sequencing project (Groenen et al., 2012).
Figure 2.
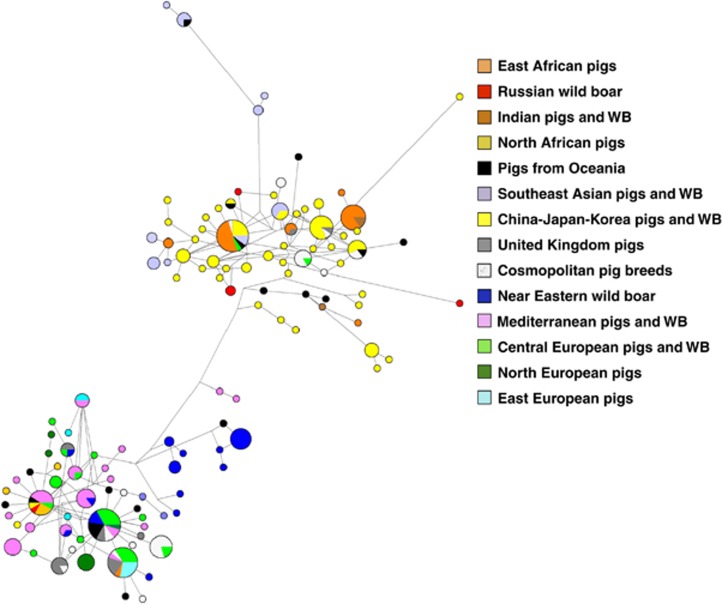
A median-joining network of 304 mitochondrial D-loop sequences of pigs and wild boars from Vietnam, Poland, Majorca, Nigeria, Tunisia, Morocco, Zimbabwe and Kenya (retrieved from Ramírez et al., 2009) as well as from specimens described in Supplementary file 1. The strong genetic divergence between Western and Far Eastern Sus scrofa can be observed. As a general trend, Near Eastern wild boars and European pigs form separate clusters, a feature attributable to the fact that pigs from the Fertile Crescent that entered Europe during the Neolithic were rapidly replaced by those domesticated locally. Some populations of European origin cluster with the Far Eastern ones, indicating the occurrence of bidirectional introgression events. Pigs from Eastern Africa group with both Far Eastern and Indian Sus scrofa evidencing a mixed ancestry.
Archaeologists and population geneticists infer past events through very different approaches: while the former use zoogeographic, biometric and sex-specific demographic profiling data, the latter fundamentally rely on the analysis of ancient and modern patterns of genetic variation (Zeder et al., 2006). Many constraints limit the resolution of these procedures, and their convergence is therefore essential to reliably identify primary domestication sites and the routes of dispersal of livestock populations. Two theories have been proposed to explain pig domestication (Larson et al., 2011), which basically differ in the number of putative domestication sites, either being limited to a few sites (largely in the Near East and China, Figure 1), or to multiple locations around the world, including Europe. Several of the proposed domestication sites, such as Southeast Asia and India, are cryptic because they have been inferred on the basis of mitochondrial analysis, but there is a lack of complementary zooarchaeological evidence (Larson et al., 2011). On the contrary, there is great consensus regarding the fundamental role of the Near East and China as major centres of pig domestication during the Neolithic (Larson et al., 2005, 2007a, 2007b, 2011).
The Fertile Crescent and China as the main centres of pig domestication
The domestication of pigs in the Near East and their spread into Europe
Interdependence between humans and wild boars may have developed very early, as much as 12 000 years ago, in the Fertile Crescent (Redding, 2005). How this relationship began is a mystery that will most likely never be solved, but there are reasons to believe that wild boar were attracted to human settlements because they fed on crops and waste (Zeder, 2012). The identification and biometric analyses of Sus scrofa remains found at numerous Neolithic sites in Eastern Anatolia (that is, Çayönü Tepesi, Hallan Çemi Tepesi, Hayaz Tepe, Tell Hallula and Gürcütepe) unequivocally suggest that this area was the earliest centre of pig domestication in the West (Larson et al., 2011). The investigation of a comprehensive chronological sequence of pig remains covering two millennia at Çayönü Tepesi has been particularly illuminating. This assemblage provides compelling testimony regarding the morphological changes (for example, reduction in the third molar length and snout shortening) that accompanied pig domestication in the Near East (Albarella et al., 2006).
In contrast, the archaeological evidence for the local domestication of pigs in Mesolithic Europe is quite weak (Rowley-Conwy, 2003). Genetic studies (Larson et al., 2005; Ramírez et al., 2009) have highlighted that Near Eastern wild boar harbour mitochondrial haplotypes that are not present in modern European pig breeds (Figure 2). High-throughput analysis of the autosomal genomes of a limited number of pig and wild boar populations also demonstrated this marked genetic divergence between Near East and European Sus scrofa (Manunza et al., 2013). Moreover, European pigs and wild boars share mitochondrial haplotypes (Larson et al., 2005), a feature that would suggest that Europe was a primary domestication centre for pigs. This situation is much more complex than it appears, as an analysis of ancient European pig remains from the Neolithic showed that they, in fact, carry Near Eastern mitochondrial haplotypes (Larson et al., 2007b). These findings proved beyond a doubt that domestic pigs from the Fertile Crescent were introduced into Europe, most likely through the Danubian and Mediterranean corridors, as early as 5500 BC (Larson et al., 2007b). Within a short window of time, possibly 500 years, Near Eastern pigs were completely replaced by their European counterparts, explaining why modern European swine breeds do not harbour Near Eastern mitochondrial haplotypes. In principle, these findings would confirm Europe as a secondary centre of pig domestication.
In an attempt to clarify the timeline of the population dynamics of Near Eastern pigs, Ottoni et al. (2013) used a powerful approach based on the analysis of ancient mitochondrial DNA and dental geometric and morphometric data obtained from 393 specimens from 48 archaeological sites. In doing so, they demonstrated that the European Neolithic pig remains carried NE2 haplotypes that are native to Anatolia, thus identifying this geographic location as the main centre of Near Eastern pig dispersal into Europe (Figure 1). Illustrating the complexity of the migratory movements that followed pig domestication, these authors also contributed evidence that, during the Bronze Age, European pigs, which were morphologically different from their Near Eastern counterparts, spread eastwards, reaching Anatolia.
The spread of pigs and other domestic animals into Europe involved complex social and cultural interactions between the incoming Neolithic farmers and Mesolithic hunter-gatherers. In this context, it is particularly contentious whether hunter-gatherer societies used domestic animals, either captured in the wild or acquired through trading with Neolithic farmers, as a source of food. In a recent study, Krause-Kyora et al. (2013) combined molecular and morphometric data from 63 Sus specimens from 17 Neolithic and Mesolithic (Ertebølle, Northern Germany) sites and provided proof of the presence of both European and Near Eastern pigs displaying a variety of colourings and sizes in the Ertebølle assemblage. Although the status of these pigs (that is, feral, domestic or intercrosses between wild and domestic pigs) has not been conclusively determined, it can be deduced that these pigs were acquired by Mesolithic hunter-gatherers. Importantly, domestic swine most likely had a role in the transition of hunter-gatherer nomadic communities to an agricultural and sedentary lifestyle.
China as a major centre of pig domestication in Asia
Similar to the Near East, archaeological and genetic evidences consistently depict China as a major pole of pig domestication. Some controversy about the timing of this process exists; diverse claims suggest an early (10 000–9000 YBP) domestication event at Zengpiyan (Nelson, 1998), but the current view (Jing and Flad, 2002) is that the oldest assemblage corresponds to the Cishan site (8000 YBP). Morphometric measurements of Sus scrofa remains and determination of their age at slaughter demonstrated the presence of domestic pigs at this site (Jing and Flad, 2002). Indeed, hundreds of earth-walled pits were found at Cishan, and many of them contained pig and dog skeletons covered by millet remains (Jing and Flad, 2002; Jing et al., 2008). It is likely that the successful cultivation of cereals promoted pig breeding in China because crop surpluses and byproducts could be used to feed livestock. In fact, carbon isotope analysis of pig bones from the Taoshi assemblage (4000 YBP) has shown that C4 plants, such as foxtail millet, formed part of the diet of swine (Jing and Flad, 2002).
It has been proposed that there were multiple pig domestication sites in China (Figure 1), distributed along the Yellow (Northern China) and Yangtze (Southern China) rivers (Jiang, 2004), although this has not been formally tested. A mitochondrial analysis of a wide array of Chinese pig breeds and wild boars revealed a geographic distribution of D haplogroups that is consistent with two independent domestication events in the Mekong region and in the middle and downstream regions of the Yangtze River (Wu et al., 2007). The highlands of the Tibetan plateau (Yang et al., 2011) and the upstream region of the Yangtze River (Jin et al., 2012) have also been claimed as putative domestication centres. In any case, the strong identity shared between mitochondrial haplotypes obtained from ancient and modern samples from Chinese pigs indicates that current Chinese porcine breeds descend directly from the local stocks of pigs domesticated during the Neolithic (Larson et al., 2011).
In the mid-Holocene, approximately 5000–4000 YBP, Austronesian-speaking rice agriculturalists from southern China migrated, either via Taiwan or Sundaland, to Southeast Asia and Oceania as well as westward to Madagascar. This large-scale demographic expansion also left a footprint in the genomes of domestic animals that can be observed today. Two potential routes of human-mediated pig dispersal to islands in Southeast Asia and Oceania have been identified (Larson et al., 2007a). One of these routes connects mainland Southeast Asia with Java, Sumatra, Wallacea and Oceania, and the other links East Asia with Western Micronesia, Taiwan and the Philippines (Figure 1). A third migratory route may have involved the translocation of Sus celebensis from Sulawesi to Flores and Timor (Larson et al., 2007a). Analysis of the genome sequences of wild pigs from insular and mainland Southeast Asia support the migration scenario depicted above (Frantz et al., 2013). Indeed, the long distance dispersal patterns (even across significant geographic barriers) observed for Sus scrofa and, more intriguingly, Sus celebensis can be understood only in the context of human-mediated translocations. As a whole, data obtained from pigs and other domestic species (Miao et al., 2013, Sacks et al., 2013) point to a Southeast Asian origin for the majority of domesticated animals found around the Pacific, thus illuminating the prehistoric past of this region and contributing to clarification of the migratory movements that shaped its demography.
Analysis of mitochondrial data and the existence of additional centres of pig domestication
Although the crucial role of the Near Eastern and Chinese Neolithic farmers in swine domestication is indisputable, the participation of additional human populations in this process currently lacks sufficient zooarchaeological support. Through the analysis of patterns of mitochondrial variation, cryptic domestication centres have been identified in Southeast Asia and India (Figures 1 and 2). The sharing of mitochondrial haplotypes between Southeast Asian wild boars and feral Australian swine may suggest Southeast Asia as a possible domestication site (Larson et al., 2011). However, the transfer of wild animals from Southeast Asia to Australia is also conceivable, so it is difficult to derive any firm conclusions from this finding. Another intriguing observation that may point to Southeast Asia as a domestication centre comes from the identification of a Pacific mitochondrial clade, which is widely represented on the islands of Southeast Asia, New Guinea and Remote Oceania and clusters with haplotypes from mainland Southeast Asia, but not with those found in Chinese pigs (Larson et al., 2007a). This would imply that pigs domesticated in Southeast Asia spread to islands in Southeast Asia and Oceania, possibly in the context of the Austronesian expansion discussed above (Larson et al., 2011). With regard to the Indian subcontinent, pigs from this region harbour haplotypes that are found in indigenous wild boar populations, but not in Sus scrofa from Europe, the Near East, Southeast Asia or Mainland Asia, providing preliminary evidence of a local domestication event (Larson et al., 2011).
Africa is the site of origin of several species from the Suidae family, none of which has ever been domesticated to our knowledge (although signs of semidomestication have been reported for bushpigs from Madagascar (Blench, 2007). The geographic distribution of wild boars is restricted to North Africa, but there is a general lack of zooarchaeological records and genetic information to reconstruct the history of swine breeding in Africa (Blench, 2000; Amills et al., 2013). The Nile Delta has been proposed as a potential centre of pig domestication (Gautier, 2002), although this hypothesis is highly controversial. Indeed, it is quite possible that Egyptian pigs descended from Near Eastern populations transported across the Sinai Peninsula or the Mediterranean Sea (Gautier, 2002). In sub-Saharan Africa, ancient pig production sites have been identified in Senegambia and in the West African and Angola extensions (Blench, 2000). Similar to Europe, no obvious Near Eastern signature has been found in the few African pig populations sampled so far (Ramírez et al., 2009), suggesting that the genetic background of Neolithic African domestic pigs was progressively replaced by that of swine conveyed by European and Asian colonizers and traders. For instance, Iberian pigs were introduced in large numbers in the south of Chad by missionaries coming from Cameroon (Logténé et al., 2006). Interestingly, Ramírez et al. (2009) reported a high frequency of Asian mitochondrial and Y-chromosome haplotypes in East Africa, which may suggest the existence of ancient contact between these regions, possibly in the context of the Austronesian expansion.
Moving towards an autosomal and paternal marker-based definition of pig domestication
The current molecular perspective regarding pig domestication has been fundamentally derived from the analysis of the mitochondrial genomes of pigs and wild boars. Mitochondrial markers are poor predictors of whole-genome variation because they simply reflect the matrilineal history of species. Given their reduced effective size, mitochondrial markers are strongly affected by genetic drift (Zhang and Hewitt, 2003). These limitations emphasize the need to perform complementary analyses based on autosomal and Y-chromosome markers. In this regard, Ramírez et al. (2009) provided data about Y-chromosome variation in a broad array of pig and wild boar populations and detected three main haplotypes. One of the haplotypes (HY3) was exclusively found in Asia, confirming that pigs were independently domesticated in the Far East. In European and Near Eastern wild boars, two HY1 and HY2 haplotypes were detected. However, these haplotypes segregated at similar frequencies, thereby making it impossible to discern the paternal ancestry of modern European swine breeds. The variation of pigs has been also analyzed with microsatellites (reviewed in Amills et al., 2010), but these analyses generally occur on a regional scale because the genotyping of these nuclear markers is time consuming and laborious. Such studies have detected a postglacial demographic expansion signature in European wild boars and have highlighted that, in contrast with previous mitochondrial analyses, the status of Italy as a primary domestication site is doubtful (Scandura et al., 2008). They have also confirmed the strong genetic divergence that exists between Western and Far Eastern Sus scrofa (Megens et al., 2008; Ramírez et al., 2009) and the high variation present in Chinese breeds (Li et al., 2004). However, a fine-grained nuclear perspective on pig domestication is still lacking. In the near future, the analysis of a large worldwide sample of domestic and wild pigs is expected to fill this gap (Megens et al., 2010).
Pig domestication and breeding from a genomic perspective
Domestication and selective breeding involve a series of phenotypic changes modulated by genetic factors that begin to be deciphered. Indeed, the extraordinary phenotypic diversity of livestock combined with the availability of resource populations and high-throughput molecular tools offers a unique opportunity to understand domestication from a molecular perspective (Andersson, 2009). Although behavioural traits were the most important component of the adaptation of pigs to a human-controlled environment (Mignon-Grasteau et al., 2005) and some of these traits exhibit moderate heritabilities (Johnson and McGlone, 2011), no genetic variant influencing swine temperament or docility has been found to date (Table 1, but see other associated domestication traits on Figure 2 in Trut, 1999). In contrast, mutations associated with pig breeding and selection (that is, post-domestication changes) have been identified through QTL and candidate gene approaches.
Table 1. Genomic regions identified by QTL and GWAS studies as associated with traits (directly or secondarily) related to events of domestication in pigs (sorted by year of publication).
| SSC/candidate region | Breeds involved | Trait | Genetic signal detected | Identification method | Reference |
|---|---|---|---|---|---|
| SSC1 | Wild boar × Pietrain Wild boar × Meishan | Teat number | QTL at 68.3 and at 171.2 cM | QTL mapping | Beeckmann et al., 2003 |
| SSC1: NR6A1 | Large white × Japanese wild boar, Jinhua × Duroc | Number of vertebrae | Mutation: C748T | QTL mapping, candidate gene | Mikawa et al., 2005, 2007 |
| SSC2, 3, 7, 8, 9 | White Duroc × Chinese Erhualian | Feeding behaviour | Multiple regions | QTL mapping | Zhang et al., 2009 |
| SSC5, 7, 8, 16 | Pietrain × Meishan | Behaviour under healthy and infection conditions | Multiple regions | QTL mapping | Reiner et al., 2009 |
| SSC2, 6, 14, 15, X | White Duroc × Chinese Erhualian | Maternal infanticide | Multiple regions | QTL mapping | Chen et al., 2009 |
| SSC7: PPARD | Domestic breeds; Chinese and European wild boars | Ear size | Mutation: missense G32A | QTL mapping | Ren et al., 2011 |
| SSC1 | Domestic breeds | Number of vertebrae | Between SJ7126 and SJ7099 markers (41 kb) on SSC1 | QTL mapping, candidate gene | Mikawa et al., 2011 |
| SSC13: MUC13 | Domestic breeds | ETEC susceptibility (immunity) | QTL at Chr 13 between 139.29—141.59 Mba | QTL mapping, GWAS | Ren et al., 2012 |
| SSC7: VRTN | Domestic breeds and wild boar | Number of thoracic vertebrae | Mutations: g.19034A>C g.20311_20312ins291 | QTL mapping, GWAS | Fan et al., 2013 |
Abbreviations: ETEC, enterotoxigenic Escherichia coli; GWAS, genome-wide association study; QTL, quantitative trait loci; SSC, Sus scrofa chromosome.
Genome positions using assembly 10.2 of the pig genome.
This notion is best exemplified by the enormous diversity of coat colours observed in pig breeds, a feature that contrasts strongly with that observed in wild boars, which display a brown pigmentation that helps to camouflage them from their predators. Genetic analyses of MC1R variation in pigs and wild boars indicate an excess of non-synonymous substitutions in the former that only can be explained in the context of positive selection for certain pigmentation patterns (Fang et al., 2009). Another pigmentation gene targeted by artificial selection is KIT. Variation in the number of KIT copies and one polymorphism associated with the skipping of exon 17 are the major determinants of the white coat that has been fixed in several cosmopolitan breeds, such as Large White and Landrace (Giuffra et al., 2002, Andersson, 2009). It is not known why certain pigmentation patterns were selected by ancient farmers, but it is possible that coat colours were used as markers to identify improved variants (Andersson, 2003). Religious and cultural preferences may also have driven this diversification process (Fang et al., 2009).
The application of next-generation sequencing techniques to high-throughput analyses of genome variation in pigs
The rapid advances in sequencing and genotyping technologies have allowed genomes to be scanned for footprints produced by candidate selective regions, founder effects, population admixture and other evolutionary processes. This strategy has also been used to identify candidate regions for domestication traits. The Swine Genome Sequencing Consortium (SGSC) began in 2003 (Schook et al., 2005) and published the assembly of the genome sequence of a domestic pig 9 years later (Groenen et al., 2012). Before the completion of the genome sequence, several methodologies for retrieving genome information were applied, such as the sequencing of reduced genomic libraries and the construction of dense genotyping panels (for example, Ramos et al., 2009). These approaches allowed rapid and inexpensive studies to be performed on the genotypes of a large number of individuals. High-density single-nucleotide polymorphism (SNP) panels are especially suitable to carry out association studies, replacing other marker panels with a lower resolution (for example, Archibald et al., 1995; Rohrer et al., 1996).
The analysis of variability based on a priori-established polymorphic markers (SNP genotyping panels) suffers from the bias introduced by the specific samples where the examined SNPs were discovered and thus ignores variants present in breeds or samples that were not included in the assay (Nielsen et al., 2004). This is the case for the 60K SNP panel (Ramos et al., 2009), which was built on the basis of sequence variation found in a pool of six populations (Duroc, Landrace, Pietrain and Large White breeds and a wild boar pool containing mainly European and a few Japanese samples). Ascertainment bias has been observed in the 60K SNP panel, especially in non-European pig populations (Ai et al., 2013; Burgos-Paz et al., 2013). Population structure analyses based on this panel are restricted to the detection of differences exclusively targeting ascertained SNPs. Consequently, these analyses are expected to capture the most relevant components of the population structure but not all the information contained in the genome.
To date, only a limited number of population genomic studies based on whole-genome resequencing are available, and these studies are based on a small numbers of individuals and populations. This contrasts strongly with the large number of studies performed with mitochondrial DNA. Results based on genome studies still need to be complemented with more data, thereby allowing us to disentangle the history and the effects of the variants on individuals and populations.
Nucleotide variability and signatures of demographic events inferred from genomic sequences suggest a major role for population admixture
Groenen et al. (2012) studied the demographic history of Sus scrofa inferred through a Hidden Markov model approach (Li and Durbin, 2011) and identified differences in population size over time between Asian and European wild boars, observing a demographic decline during the last 20 000 years (especially in Europe). Phylogenomic analyses estimated a split between Asian and European lineages at 0.8–1.6 MYA, a result supported by the considerable number of fixed differences between these groups and consistent with previous studies (for example, Megens et al., 2008). These differences in population sizes are reflected on the estimates of nucleotide diversity (π) calculated at the whole-genome level (Bosse et al., 2012), suggesting that wild boars from Asia are the most diverse specimens (π ∼0.0030 variants per base pair). In contrast, European wild boars (π ∼0.0010) would be the least diverse population. Asian pig breeds (π ∼0.0023) display lower diversity than Asian wild boars, while European local pigs (π=0.0017) that are more diverse than European wild boars are in between. The increased diversity in European pig breeds compared with wild boars suggests that admixture has had a relevant role in the history of European pigs.
Population differentiation analyses using genome sequences revealed that differentiation between European wild and domestic pigs is similar to or even lower than that observed among pig breeds (Amaral et al., 2011). Importantly, admixture analyses (Groenen et al., 2012) revealed significant gene flow from Chinese populations to Europe in the Pleistocene period and frequent recent admixture in domesticated pig breeds in Europe.
Population structures determined in wild boars and domestic pigs using the high-density genotyping panel (for example, Ai et al., 2013, Burgos-Paz et al., 2013, Wilkinson et al., 2013) confirm previous findings primarily based on mitochondrial (Figure 2) and microsatellite markers, evidencing a strong differentiation between Asian and European individuals. Moreover, the presence of admixture from Asian breeds into commercial European breeds and vice versa was also obvious. These introgression events likely reflect recent historical processes initiated in the eighteenth and nineteenth centuries that altered the way pigs were bred, especially in England; for example, mast feeding was almost entirely replaced by intensive production in enclosures and herd sizes increased substantially (Wealleans, 2013). British pig breeds were extensively introgressed with Chinese sows to improve fertility and fattening, and genomic footprints left by this admixture event have been detected recently with molecular tools (for example, Giuffra et al., 2000; Megens et al., 2008; Ai et al., 2013). Many of these breeds have become cosmopolitan given their excellent growth and reproductive abilities. Often, these breeds have replaced less productive local varieties, occasionally bringing local breeds to extinction (Amills et al., 2010). Without a doubt, this practice has reduced the gene pool of the porcine species to an important extent.
In summary, a complex demographic pattern becomes apparent from genomic data with recent mixtures of commercial breeds between geographically distant locations and recurrent introgression events of commercial breeds into wild populations (Goedbloed et al., 2013). The intricate genetic relationship among domesticated populations suggests that complex breeding practices, including genetic exchange with wild animals and/or multiple origins of domestication, have had an important role in delineating the genealogical relationships within this species.
Identifying signatures of artificial selection in the pig genome
It is generally assumed that artificial selection, which is implicit during the domestication process, has a significant effect on the genomes of the targeted populations. First, a bottleneck results from the selection of a small number of individuals, causing a reduction of variability across the entire genome in the domesticated population. Second, signals of selective sweeps are expected around strongly implicated loci associated with desired phenotypic traits (for example, Wright et al., 2005). Under this hypothetical framework, researchers have attempted to identify signals of selective sweeps in domesticated specimens that are not observed in wild populations (for example, Wright et al., 2005, Rubin et al., 2012, Wilkinson et al., 2013). A number of methods have been used to detect candidate regions, including analyses of population differentiation (for example, Beaumont and Balding, 2004; Foll and Gaggiotti, 2008; Green et al., 2010), excess homozygosity and differences in linkage disequilibrium (for example, Sabeti et al., 2002; Voight et al., 2006; Tang et al., 2007), and decreased variability and skewed spectrum frequency patterns (for example, Tajima, 1989; Braverman et al., 1995; Fay and Wu, 2000; Rubin et al., 2010).
The complex demographic history of the pig led researchers to use the empirically observed distributions of a number of statistics as a method for identifying candidate regions affected by selective sweeps (for example, Rubin et al., 2012; Li et al., 2013; Wilkinson et al., 2013). Nevertheless, this method does not rely on an underlying evolutionary model (see review in Ross-Ibarra et al., 2007), and candidate regions must therefore be validated in some way using an independent approach. The use of broadly reported QTL regions for a number of phenotypes affected by artificial selection has been the most commonly used (and perhaps the most robust) method to obtain such support. However, other approaches have also been used, such as Gene Ontology or pathway annotation, to detect enrichment in a given function (Amaral et al., 2011) or locate candidate genes associated with a trait of interest (a common method for selecting candidate genes in QTL studies). These other methods are more vulnerable to subjectivity and should be used carefully (Pavlidis et al., 2012).
Genome analyses have identified a number of regions and candidate loci that may be responsible, to some degree, for traits selected during and after domestication (Amaral et al., 2011; Mikawa et al., 2011; Ren et al., 2011; Rubin et al., 2012; Ai et al., 2013; Fan et al., 2013; Moutou et al., 2013; Wilkinson et al., 2013). In Table 2, genomic regions associated (directly or secondarily) with domestication traits are presented. These regions have been identified using either genome sequence or the 60K SNP panels in European or Asian specimens. These candidate loci are associated with coat colour, ear morphology, immunity, behaviour and other traits.
Table 2. Relevant genomic regions identified in genome diversity studies as associated with traits (directly or secondarily) related to events of domestication in pigs (sorted by year of publication).
| SSC/region/loci | Inference method and statistics | Traits associated | Support/validation | Genome data and references |
|---|---|---|---|---|
| SSC8, near KIT | Reduced levels of variability, using simulations of a neutral model | Neural functions | Previous description of the biological role of this locusa | RRL pool. Amaral et al. (2011)b |
| SSC12, near PPP1R1B | Gene implicated in motivated behaviour, working memory, and reward-related learningc | |||
| SSC1, near NR6A1, SSC4, near PLAG1 SSC8, near LCORL | Highly homozygous Regions, using the observed empirical distribution. | Body length | Within QTL intervals for traits of interest. Validation using individual genome sequences. | Whole-genome sequence of pools. Rubin et al. (2012)d |
| SSC8, KIT segmental duplications | Differential patterns in CNV, using the empirical distribution. | Body coat colour | Previous description of the biological role of this locusa Validation using individual genome sequences. | |
| SSC1 near to THBS2 and SMOC2 | High values of Fst, using the empirical distribution | Skull shape | Orthologous to a region of the canine genome associated with brachycephaly in dog breedse,f | 60 K SNP panel. Wilkinson et al. (2013)g |
| SSC5, close to LEMD3 | Ear morphology | Orthologous to a region associated with canine ear morphologyh,i | ||
| SSC7, near ADAMTSL3 | Ear morphology | Proximal to the PPARD gene where a missense mutation affecting ear size has been identified in pigsj | ||
| SSC7 (31.30–38.89 Mbk) | Immunity | Extreme patterns of differentiation associated with the major histocompatibility complex genesl | ||
| SSC8, KIT locus | Coat colour | Previous description of the biological role of this locusa | ||
| SSCX, near AR | Backfat thickness | QTL associated to backfat thicknessm | ||
| SSC12 (26.83–26.96 Mbk) | Teat number | Comparative analysis of teat number individual groups. | ||
| SSC11, upstream EDNRB | Bayesian approach using Fst components (Bayescan) | Belted versus non-belted pigs | Comparative analysis of belted vs non-belted. | 60 K SNP panel. Ai et al. (2013)n |
| SSC13, TRPC1 SSC14, KCNMA1 | Higher differentiation, lower diversity and skewed frequency, using combined empirical distributions | Genes related with saliva secretion | Combined analyses, using functional annotation tools. Enrichment of several functional categories. | Whole-genome sequence of individuals Li et al. (2013)o |
Abbreviations: CNV, copy number variants; RRL, reduced representation library; SSC, Sus scrofa chromosome.
Western pig pools plus one Asian-European mixed wild boar pool.
Western and Asian pigs pools plus one European wild boar pool.
Western and Asian pig populations plus one European wild boar populations.
Genome positions using assembly 10.2 of the pig genome.
See Moutou et al. (2013) and Amaral et al. (2011) for divergent results.
Chinese domestic and wild boars plus western pig populations.
Tibetan wild boars, Chinese and western pigs.
The genomic consequences of domestication were not evident from Reduced Representation Libraries. Although Amaral et al. (2011) observed a certain level of differentiation between wild and domesticated pigs, they did not detect an excess of candidate regions in any group. By mean of whole-genome resequencing, Rubin et al. (2012) observed a reduced number of candidate regions with fixed or high-frequency nonsense mutations, leading to the conclusion that gene inactivation did not had an important role in the pig domestication and breeding processes. A similar pattern was found in regions containing copy number variants (Rubin et al., 2012; see also Paudel et al., 2013). Nevertheless, Rubin et al. (2012) identified an excess of nonsynonymous-derived substitutions in domesticated pigs, a result that is expected if selective events predominate during and after domestication. Approximately half of these nonsynonymous differences were potentially harmful, suggesting that wild and domestic animals underwent a different evolutionary pattern as a consequence of selection from different environments. The authors argued that the phenotypic evolution of domestic pigs may be governed by a number of variants with large phenotypic effects.
Li et al. (2013) aimed to avoid the ascertainment bias associated with the Duroc pig reference sequence using a Tibetan wild boar genome sequenced de novo. Genetic comparison of Tibetan wild boars versus Chinese domestic pigs evidenced a close relationship between both populations and revealed that domestic pigs display reduced levels of synonymous variability but an increased ratio of nonsynonymous substitutions and more extreme Tajima's D values compared with Tibetan wild boars. This outcome suggests a higher efficiency of artificial versus natural selection in fixing functional variants.
Potential pathways of porcine genome evolution under domestication
The analyses performed on pigs and wild boars to date have not focused on the dynamics of genome evolution of populations undergoing domestication. Domestication implies a strong selective stage, and it is therefore of interest to understand whether and how this process modifies genome variation. A profound environmental change is associated with domestication, and the fitness of traits under selection is consequently and substantially modified. For example, neutral variants in wild animals may be deleterious or favourable under the new environment, or vice versa (that is, variants that were deleterious may become advantageous or neutral). This sudden change may have consequences on the patterns of variability across the genomes of the domesticated individuals, thus promoting genetic differentiation between wild and domesticated groups. For example, if domestication significantly affects a large number of variants influencing traits implicated in such process, then the patterns of genetic variation in domesticated populations may be greatly modified. In contrast, if domestication affects only a small subset of variants, genomic variation patterns in domesticates may resemble those of wild animals, except at a few loci.
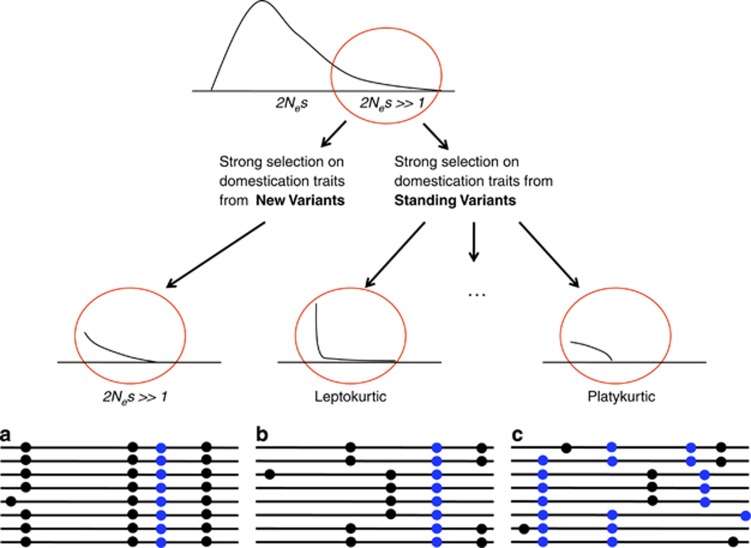
Although the process of domestication in pigs and other species is complex, we can hypothesize several scenarios for the evolution of the pig genome. These scenarios are based on three simple considerations: the number of loci directly affected by domestication (number), the strength of the domestication process (strength) and the initial frequency of the variants involved at the time of domestication (frequency). The strength and the number of loci involved in a trait under domestication are defined by the specific distribution of selective effects for that trait. An exponential distribution of selective effects may be expected over new selective variants (that is, an exponential decay of new variants with higher fitness) (Orr, 1998; Piganeau and Eyre-Walker, 2003). Furthermore, a leptokurtic distribution (a more L-shaped distribution, Caballero et al., 1991; Keightley, 1994; Loewe and Charlesworth, 2006; Boyko et al., 2008), where many loci have small effects and few loci have large effects, or a platykurtic distribution (a flatter distribution in relation to the expected exponential distribution), where many loci have moderate selective effects (for example, Chevin and Hospital, 2008; Yang et al., 2010), are also contemplated here.
Nevertheless, before considering the effects of domestication on the genome, it should be remembered that this process involves both selection and demographic effects. Thus, the effect of demographic processes on the patterns of variability across the genome must be considered. We outline several possible demographic and selection scenarios that may help to interpret the observations presented thus far.
Expected footprints of genome evolution under simple demographic scenarios
As a general principle, the effects of any demographic or selective processes in a population are expected to leave signatures on the genomes of the individuals in that population. The level of variation and other patterns, such as differences in the frequencies of mutations, linkage disequilibrium or population differentiation, may be affected. Here, we will consider the differential patterns that may be observed when two populations are compared under a few simple evolutionary models, one stationary scenario and the remaining ones based on population reduction, expansion or subdivision (Table 3).
Table 3. Variability patterns associated with diverse demographic scenarios versus a stationary model.
| Scenario | New mutations | Neutral variation | Site frequency spectrum | Linkage disequilibrium |
|---|---|---|---|---|
| Population contraction | Reduced | Increased ratio of neutral alleles versus non-neutral | Lack of low frequency variants | Increased |
| Population expansion | Increased | Reduced ratio of neutral alleles versus non-neutral | Excess of low frequency variants | Decreased |
| Subdivision and isolation | Equal at global population scale | Increased ratio of neutral alleles versus non-neutral | Excess of intermediate frequencies | Increase within populations and drastic increase at global population scale |
| Subdivision and posterior admixture | Equal at global population scale | Increased ratio of neutral alleles versus non-neutral | Excess of intermediate frequencies, also within populations | Drastic increase, also within populations |
Population reduction implies a change in the effective population size. At the genome level, the number of new variants appearing in the population is reduced, and the frequency spectrum skews to medium-high frequencies (Ramírez-Soriano et al., 2008). In addition, linkage disequilibrium will increase (Wright et al., 2005) through reduced Ner (where Ne is the effective population size and r is the recombination rate). It is expected that selective effects over the population gene pool would be attenuated because the value |Nes| (s is the coefficient of selection) would be reduced (see review at Lanfear et al., 2013). Therefore, a relatively large proportion of variants would be neutral (that is, |2Nes|⩽1), and a relatively large number of slightly deleterious mutations would thus be fixed through drift, possibly increasing the ratio of fixed nonsynonymous versus synonymous mutations (Eyre-Walker, 2002). Furthermore, although more speculative, the correlation patterns (if present) between neutral variability and recombination rates may be weaker if the elimination of deleterious mutations is less effective in regions of high recombination.
In contrast, population expansion would increase the efficiency of selection. The number of variants (Neμ) would be augmented, and the proportion of neutral variants (|2Nes|⩽1) would be reduced (Figure 3). A general pattern characterized by an excess of low-frequency-derived variants is expected. Linkage disequilibrium would be reduced, and the patterns of correlation between neutral variability and recombination rates may be accentuated.
Figure 3.
Distribution of 2Nes (Ne is the effective population size and s is the strength of selection). The central vertical grey line indicates 2Nes=0 and the left and right grey lines determine the limits of the distributions at which |2Nes|⩽1 (the range of neutral effects is shown with a horizontal green arrow). (a) Assuming an infinite and independent sites scenario, the distribution of s and 2Nes is equivalent. (b) In case Neincreases, the size of the distribution also increases, that is, there are more mutations and more extreme 2Nes values. In relation to the total mutations, the frequency of neutral mutations decreases. (c) In the case of relaxing the assumption of independent sites, different Ne may coexist across the genome if we assume interference among positions (for example, Santiago and Caballero, 1998). Interference is generally associated with recombination, gene density and other factors. Therefore, the distribution of 2Nes may modify its shape with respect to the fitness distribution by the effect of interference, here represented with blue arrows.
Subdivision and isolation cause the accumulation of fixed differences among sub-populations, significantly increasing linkage disequilibrium across the genome. The expected frequency spectrum is skewed to intermediate frequencies because the fixed variants in each sub-population will segregate in the global population. With regard to populations in complete isolation, the number of neutral variants in relation to detrimental variants is expected to increase because Ne is lower in each population (although these variants remain high in the whole species). If posterior admixtures between sub-populations were allowed, significant linkage disequilibrium would still be observed across extensive regions of the genome. In addition, a large number of derived variants at intermediate-high frequencies would be observed. The reduction of the effective population size in subdivided populations (Whitlock and Barton, 1997) can aid in fixing slightly deleterious mutations at a relative higher frequency. With regard to domestic pigs, the demographic scenario of subdivision and admixture appears to be the most realistic; however, several of its predictions, such as changes in the ratio of neutral fixations, are difficult to assess.
Expected footprints of genome evolution under simple scenarios of artificial selection
Domestication implies the artificial selection of a number of traits of interest in wild individuals. Here, we consider the possibility that selected phenotypes are due to new or standing variants with each trait being modulated by a number of loci that may display a different distribution of selective effects. We consider several scenarios for artificial selection and their possible consequences on the levels and patterns of variability and divergence. In addition, we consider constant fitness effects across time, which would result in different expectations than a framework with fitness associated to a changing environment (Lourenço et al., 2013).
The first scenario is domestication via the selection of new rare alleles in wild individuals (new variants at low frequencies, Figure 4a, Table 4). Here, we assume that the new variants are few and should have a strong effect for selection in the new environment; otherwise, selection through domestication will not occur in a short period of time. The domesticated population suffers a severe bottleneck(s) associated with a global reduction of variability in domesticated individuals plus a strong selective sweep around the selected loci (via hitchhiking, Maynard Smith and Haigh, 1974). Each new variant introduced in the domesticated population may still improve the selected trait to some degree. The probability of introducing new beneficial variants for a given trait in such a short time is quite small, unless the new variants disrupt or damage functions that are unnecessary in farming conditions but essential in the natural environment. Therefore, selected regions are more likely to result from a loss of function rather than from the acquisition of a new function. Under this assumption, considerable nucleotide differentiation of domestic pigs versus wild boars is expected, and this effect would be even more pronounced in the selected regions. Increased linkage disequilibrium in the domesticated population is anticipated. Association studies should detect causative regions that explain a high percentage of the variance in the trait.
Figure 4.
Diagram for hypothetical cases of selective events occurred via domestication under different scenarios. The top cartoon displays the distribution of 2Nes, the red circle shows the distribution of variants that are beneficial in the domesticated environment. The bottom drawings represent sequencing alignments for domesticated individuals. Blue spots represent beneficial variants in domesticated environment and black spots represent neutral variants. (a) Selection for domestication traits is mainly acting on new beneficial variants. These variants are selected starting from low frequencies and their frequencies increase rapidly. A drop in the levels of variability is expected. Given the short time since domestication, it is expected that most of these selective variants may be harmful in a wild environment (loss-of-function variants). (b, c) Standing variants segregating at wild populations mainly drive selection for domestication traits. Variants that are beneficial in a domestication context can segregate at any frequency. If only few variants contribute with major effect on the trait (b), we expect to find selective sweeps with a moderate reduction of variability. In the other extreme case, that is, if most variants have a modest effect on the trait (c) no strong selective sweeps are expected.
Table 4. Effects of artificial selection acting on new variants (appearing at domestic) and on standing variants (initially polymorphic in wild), of domestic individuals in relation to wild.
| Type of mutation involved in artificial directional selection | Patterns at whole-genome scale |
Patterns around causative regions |
|||
|---|---|---|---|---|---|
| Features | Neutral variability | Site frequency spectrum | Linkage disequilibrium | QTL detection | |
| New variants | No important effects | Reduced | Excess of high-intermediate frequency variants | Increased | Possibly detected |
| Standing variants (L-shaped distribution) | No important effects | Minor reduction | Possible excess of high-intermediate frequency variants | Increased. Possibly, few haplotypes highly differentiated | May be detected |
| Standing variants (Flatter distribution) | Depending on the number of causative variants, produce patterns similar to those observed around causative regions | Similar to that found in the wild ancestor | All causative regions may present some excess of high-intermediate frequency variants | Similar to wild animals | Undetected |
Abbreviation: QTL, quantitative trait loci.
In the case of posterior mixture with wild boars, the domesticated population may recover the initial levels of variability, with the exception of the regions affected by selective sweeps, in which loci would still be strongly selected. If the new variants were harmful in the wild environment (for example, loss-of-function mutations), we would find low differentiation between domestic pigs and wild boars, except in a few highly differentiated regions. Hard and soft selective sweeps (multiple adaptive alleles that sweep the population simultaneously, Hermisson and Pennings, 2005) may occur in these regions (Messer and Petrov, 2013). Association studies may be able to detect these few loci. If we consider multiple domestication events, selection for different alleles is expected at each location, possibly affecting related traits (for example, tameness and aggressiveness). Admixture would result in a combination of features from both populations. High linkage disequilibrium can be anticipated in a population that has incorporated individuals from a highly divergent group (for example, Asia versus Europe).
The second scenario is domestication through the selection of specific traits already segregating in wild individuals (standing variants at any frequency). In this scenario, a number of standing variants related to certain domestication traits are beneficial in the new environment. The distribution of beneficial variants is difficult to define under this assumption. However, we hypothesize that high-, medium- or low-frequency variants may become beneficial with different strengths. If only a few variants have strong effects (presenting a leptokurtic or L-shaped distribution, Figure 4b, Table 4), selection acting on standing variants would leave a more diffuse signal on average (for example, soft selective sweeps starting from intermediate frequencies). Nevertheless, this signal should be detectable given the strong effect on these few loci. Statistics based on population differentiation and haplotype signals of homozygosity are expected to be the more powerful methods applied to detect these signatures (Yi et al., 2010; Garud et al., 2013; Messer and Petrov, 2013).
Alternatively, a large number of loci may moderately contribute to the trait under selection (showing a platykurtic or flatter distribution, Figure 4c, Table 4), mimicking the infinitesimal effect characteristic of genetic quantitative models (Bulmer, 1971). The selective effect will be distributed across the genome, and strong selective sweeps therefore would not be observed. Nevertheless, an excess of functional substitutions may be observed in domesticated populations if the number of variants affected by selection in relation to the total gene pool is sufficient. This pattern would be similar to that observed in the case of population reduction (that is, increase of the ratio of fixed nonsynonymous versus synonymous mutants). Association studies may detect only a small percentage of the variance for a given trait. Strong differentiation between domestic and wild individuals caused by isolation and the bottleneck process in the domesticated population is expected.
Importantly, continuous admixture of domestic and wild individuals should reduce the differentiation between these two groups. The regions affected by selection may still be shared between these two groups but at different frequencies. If the fitness of the variants affecting a domestication trait exhibits an L-shaped distribution, soft selective sweeps may be observed. Otherwise, in case of a flatter distribution, no clear signal of polymorphism would be seen.
Finally, if we consider independent domestication events at different locations, standing variants for related traits would be selected at different domestication centres. These variants may be shared because they segregate in the wild population. If a trait subjected to domestication affects loci with an L-shaped fitness distribution, it is expected that different loci from the total gene pool involved in selection for domestication traits will be fixed at each centre of domestication. Posterior admixture among domestication centres would generate a soft sweep pattern. If the distribution of fitness effects impacting a domestication trait resembles a flatter distribution, the effects of domestication will be difficult to observe. Admixture between very isolated centres of domestication (for example, Asia and Europe) would lead to strong linkage disequilibrium in admixed individuals.
Post-domestication selective processes may have masked part of the genomic consequences of domestication, depending on the strength and the number of affected regions. Furthermore, recent artificial selection may have altered the shape of the distribution of fitness effects via the progressive modification of the environment towards favouring the selected trait.
How does (and did) the pig genome evolve under domestication?
We are interested in understanding the effect of domestication across the entire genome. Our main assumption is that domestication has driven important modifications of the phenotypic variation of domesticated populations with respect to wild populations through artificial selection. Thus, we are particularly interested in knowing whether domestication has focused on the selection of new or standing mutations that affect few or numerous loci.
Considering the expectations of each scenario and the related observations, we can potentially infer the general variability patterns that should be theoretically found in loci that had a role in the process of domestication. First, we should consider the background genome pattern produced by historical and demographic events. At the genome level, wild boars and domestic pigs from the same geographic location (Europe, Asia) exhibit low population differentiation but high differentiation among locations. Reduced variability in domestic pigs compared with wild boars has been observed in Asia (Groenen et al., 2012; Li et al., 2013), suggesting a genome-wide effect of the demographic population reduction produced by domestication. Nevertheless, this general reduction of variability is not observed in domesticated European populations. In fact, variability is higher in European domestic pigs (Bosse et al., 2012) mainly due to the intense admixture with Asian pigs and the selection of advantageous Asian mutations that has occurred in these lineages (Ojeda et al., 2011). In addition, historical admixture with wild boars may have also had a role until a few hundred years ago (White, 2011; Goedbloed et al., 2013; Manunza et al., 2013).
Amaral et al. (2011) observed a positive correlation between the level of variability and recombination (also observed in Badke et al., 2012; Bosse et al., 2012 and Esteve-Codina et al., 2013), suggesting that selection (positive or negative) has an active role in shaping the variability of Sus scrofa and that interference among genomic positions has an impact on variability. The significant presence of candidate mutations that are functionally harmful suggests that many of the variants that have been artificially selected were new variants with strong effects (as indicated in Rubin et al., 2012). The significant excess of substitutions observed in Chinese domestic pigs relative to wild boars may not only indicate a multiloci effect of domestication but also reflect the population size reduction experienced by domesticated populations. As argued by Rubin et al. (2012), few studies have been performed on early domestication traits using wild boars (Table 1), and few candidate regions have been identified. In this regard, it is worth highlighting studies on tameness in rats (Albert et al., 2009) and on the expression analysis of domestication traits in pigs, dogs and rabbits (Albert et al., 2012). These results suggest that these phenotypes correspond to complex quantitative traits that also segregate in wild populations.
Final remarks and future perspectives
Our understanding of the events involved in pig domestication and their genetic consequences has rapidly evolved, providing a picture that, although incomplete, probably recapitulates the most important facts. The majority of these studies are based on the analysis of a few selected markers (primarily mitochondrial DNA). The recent advent of sequencing technologies that allow for whole-genome characterization at an affordable cost has revolutionized the field of population genetics. These technologies provide unparalleled resolution for detecting the complex signatures that selection and demographic processes have left throughout the ages in plant and animal genomes. Even with such powerful tools at hand, the interpretation of genetic data to reconstruct the history of pigs is a challenging task plagued with many potential pitfalls, mirages and shortcomings. First, inference of the remote past through the analysis of modern samples can be very misleading. Thus, the development of techniques that allow for the high-throughput sequencing of ancient nuclear DNA would be crucial to circumvent this limitation. Currently, the number of ancient populations sampled is quite limited, and we have explored only the mitochondrial variation within these populations (Larson et al., 2007b; Ottoni et al., 2013). Third-generation sequencing platforms, which allow for the sequencing of single DNA molecules without an intermediary amplification step, may have a fundamental role in characterizing the autosomal genomes of ancient pig specimens (Rizzi et al., 2012). For instance, DNA from a Pleistocene horse bone was recently sequenced using the Helicos HeliScope and Illumina GAIIx platforms, and it was demonstrated that the former generated a higher proportion of data that aligned to the horse genome (Orlando et al., 2011). Regardless, accurate population genetic inferences regarding demographic and selective events will require a large number of samples and large genome fragments. These criteria are not readily achievable with ancient DNA, but the approach may be possible if extant data are also used.
For contemporary data, the forthcoming next-generation sequencing technologies (for sequencing long haplotypes) will facilitate the analysis of numerous de novo-assembled sequences, eliminating the ascertainment bias arising from the reference sequence. With the completed pig genome, accurate approaches for detecting differences in the effective population size across the genome and between closely related groups should aid in understanding the effect of selection on populations (for example, Gossmann et al., 2011). Differences in the frequencies of adaptive mutations between wild and domestic pigs and possible differences in the inferred distributions of fitness effects may provide answers regarding the general evolution of traits associated with domestication. Importantly, determining the functional implications of genome variation remains a challenge and will require a great deal of progress. Another challenge for future work is the improvement of methods for functional annotation, which are essential for interpreting variability across the genome. It is also worth emphasizing that to understand domestication from a molecular perspective, it is not sufficient to focus on changes at the sequence level. Epigenomic modifications may have also had a relevant role that still needs to be elucidated (for example, Gokhman et al., 2014).
Population genomics and divergence analyses can only partially answer questions about the process of domestication. Population genomics focuses on the variability of genomes but not on the effects this variability has on the phenotypes. Numerous statistical methods for quantitative genetics have been developed during the last several decades (see the review by Vinkhuyzen et al., 2013). Nevertheless, these methods may fail to map a large proportion of the genetic heritability of traits under study perhaps because models are inaccurate or the data are not sufficiently informative. Furthermore, the complexity of metabolic and interaction networks generally does not permit the precise localization of the causative loci for a given trait, but it does allow for highly confident predictions on the effect of a combination of variants in a genome on the phenotype of interest (Meuwissen et al., 2001). Accordingly, research on the consequences of domestication on genomic variability patterns could also be reoriented; in the near future, we may assume that we will not be able to detect most of the causative mutations involved in the domestication process, but we may be able to detect the global genomic effects of selection for a given trait involved in domestication. A challenge in forthcoming studies will be the combined use of quantitative methods to obtain probabilistic information for the entire genome and population genomic methods to adequately weight the data and determine the general effect of the trait of interest versus the overall genome pattern regardless of the loci involved.
Data archiving
There were no data to deposit.
Acknowledgments
We thank Humberto Quesada for critical comments and helpful discussion. WB-P is funded by COLCIENCIAS (Francisco José de Caldas fellowship 497/2009, Colombia). This work was funded by Grant CGL2009-09346 (MICINN, Spain) to SERO and a Consolider Grant from Spanish Ministry of Research (CSD2007-00036) from the Centre for Research in Agricultural Genomics. AM received a postdoctoral fellowship from the Center of Research in Agricultural Genomics.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Ai H, Huang L, Ren J. (2013). Genetic diversity, linkage disequilibrium and selection signatures in Chinese and Western pigs revealed by genome-wide SNP markers. PLoS One 8: e56001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albarella U, Dobney K, Rowley-Conwy P. (2006). The domestication of the pig (Sus scrofa): new challenges and approaches. In: Zeder MA, Decker-Walters D, Bradley D, Smith BD, (eds) Documenting Domestication: new Genetic and Archaeological Paradigms. University of California Press: Berkeley, CA, USA. pp 209–227. [Google Scholar]
- Albert FW, Carlborg O, Plyusnina I, Besnier F, Hedwig D, Lautenschläger S et al. (2009). Genetic architecture of tameness in a rat model of animal domestication. Genetics 182: 541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert FW, Somel M, Carneiro M, Aximu-Petri A, Halbwax M, Thalmann O et al. (2012). A comparison of brain gene expression levels in domesticated and wild animals. PLoS Genet 8: e1002962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral AJ, Ferretti L, Megens HJ, Crooijmans RP, Nie H, Ramos-Onsins SE et al. (2011). Genome-wide footprints of pig domestication and selection revealed through massive parallel sequencing of pooled DNA. PLoS One 6: e14782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amills M, Ramírez O, Galman-Omitogun O, Clop A. (2013). Domestic pigs in Africa. Afr Archaeol Rev 30: 73–82. [Google Scholar]
- Amills M, Clop A, Ramírez O, Pérez-Enciso M. (2010). Origin and genetic diversity of pig breeds. Encyclopedia Life Sci doi:10.1002/9780470015902.a0022884.
- Andersson L, Haley CS, Ellegren H, Knott SA, Johansson M, Andersson K et al. (1994). Genetic mapping of quantitative trait loci for growth and fatness in pigs. Science 263: 1771–1774. [DOI] [PubMed] [Google Scholar]
- Andersson L. (2003). Melanocortin receptor variants with phenotypic effects in horse, pig, and chicken. Ann N Y Acad Sci 994: 313–318. [DOI] [PubMed] [Google Scholar]
- Andersson L. (2009). Studying phenotypic evolution in domestic animals: a walk in the footsteps of Charles Darwin. Cold Spring Harb Symp Quant Biol 74: 319–325. [DOI] [PubMed] [Google Scholar]
- Andersson-Eklund L, Marklund L, Lundström K, Haley CS, Andersson K, Hansson I et al. (1998). Mapping quantitative trait loci for carcass and meat quality traits in a wild boar x Large White intercross. J Anim Sci 76: 694–700. [DOI] [PubMed] [Google Scholar]
- Archibald AL, Haley CS, Brown JF, Couperwhite S, McQueen HA, Nicholson D et al. (1995). The PiGMaP consortium linkage map of the pig (Sus scrofa). Mamm Genome 6: 157–175. [DOI] [PubMed] [Google Scholar]
- Badke YM, Bates RO, Ernst CW, Schwab C, Steibel JP. (2012). Estimation of linkage disequilibrium in four US pig breeds. BMC Genomics 13: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannasch D, Young A, Myers J, Truve K, Dickinson P et al. (2010). Localization of canine brachycephaly using an across breed mapping approach. PLoS One 5: e9632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont MA, Balding DJ. (2004). Identifying adaptive genetic divergence among populations from genome scans. Mol Ecol 13: 969–980. [DOI] [PubMed] [Google Scholar]
- Beeckmann P, Schröffel J, Moser G, Bartenschlager H, Reiner G, Geldermann H. (2003). Linkage and QTL mapping for Sus scrofa chromosome 1. J Anim Breed Genet 120: 1–10. [Google Scholar]
- Blench RM. (2000). A history of pigs in Africa. In: Blench RM, Mac Donald K, (eds) Origins and Development of African Livestock: Archaeology, Genetics, Linguistics and Ethnography. Routledge Books: Abingdon, UK. pp 355–367. [Google Scholar]
- Blench RM. (2007). New palaeozoogeographical evidence for the settlement of Madagascar. Azania: Archaeological Res Africa 42: 69–82. [Google Scholar]
- Bosse M, Megens HJ, Madsen O, Paudel Y, Frantz LA, Schook LB et al. (2012). Regions of homozygosity in the porcine genome: consequence of demography and the recombination landscape. PLoS Genet 8: e1003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko AR, Quignon P, Li L, Schoenebeck JJ, Degenhardt JD, Lohmueller KE et al. (2010). A simple genetic architecture underlies morphological variation in dogs. PLoS Biol 8: e1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko AR, Williamson SH, Indap AR, Degenhardt JD, Hernandez RD, Lohmueller KE et al. (2008). Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet 4: e1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman JM, Hudson RR, Kaplan NL, Langley CH, Stephan W. (1995). The hitchhiking effect on the site frequency spectrum of DNA polymorphisms. Genetics 140: 783–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer MG. (1971). The effect of selection on genetic variability. Am Nat 105: 201–211. [Google Scholar]
- Burgos-Paz W, Souza CA, Megens HJ, Ramayo-Caldas Y, Melo M, Lemus-Flores C et al. (2013). Porcine colonization of the Americas: a 60k SNP story. Heredity 110: 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero A, Toro MA, López-Fanjul C. (1991). The response to artificial selection from new mutations in Drosophila melanogaster. Genetics 128: 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Guo Y, Yang G, Yang Z, Zhang Z, Yang B et al. (2009). A genome wide detection of quantitative trait loci on pig maternal infanticide behavior in a large scale white Duroc × Erhualian resource population. Behav Genet 39: 213–219. [DOI] [PubMed] [Google Scholar]
- Chevin LM, Hospital F. (2008). Selective sweep at a quantitative trait locus in the presence of background genetic variation. Genetics 180: 1645–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Codina A, Paudel Y, Ferretti L, Raineri E, Megens HJ, Silió L et al. (2013). Dissecting structural and nucleotide genome-wide variation in inbred Iberian pigs. BMC Genomics 14: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A. (2002). Changing effective population size and the McDonald-Kreitman test. Genetics 162: 2017–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Xing Y, Zhang Z, Ai H, Ouyang Z, Ouyang J et al. (2013). A further look at porcine chromosome 7 reveals vrtn variants associated with vertebral number in Chinese and western pigs. PLoS One 8: e62534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M, Larson G, Ribeiro HS, Li N, Andersson L. (2009). Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet 5: e1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu CI. (2000). Hitchhiking under positive Darwinian selection. Genetics 155: 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll M, Gaggiotti O. (2008). A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180: 977–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz LA, Schraiber JG, Madsen O, Megens HJ, Bosse M, Paudel Y et al. (2013). Genome sequencing reveals fine scale diversification and reticulation history during speciation in Sus. Genome Biol 14: R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garud NR, Messer PW, Buzbas EO, Petrov DA. (2013). Soft selective sweeps are the primary mode of recent adaptation in Drosophila melanogaster. ArXiv 1303: 0906. [Google Scholar]
- Gautier A. (2002). The evidence for the earliest livestock in North Africa: or adventures with large bovids, ovicaprids, dogs and pigs. In: Hassan FA, (ed) Droughts, Food and Culture. Ecological Change and Food Security in Africa's Later Prehistory. Springer: Berlin, Germany. pp 195–207. [Google Scholar]
- Giuffra E, Kijas JM, Amarger V, Carlborg O, Jeon JT, Andersson L. (2000). The origin of the domestic pig: independent domestication and subsequent introgression. Genetics 154: 1785–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuffra E, Törnsten A, Marklund S, Bongcam-Rudloff E, Chardon P, Kijas JMH et al. (2002). A large duplication associated with Dominant White color in pigs originated by homologous re combination be tween LINE elements flanking KIT. Mamm Genome 13: 569–577. [DOI] [PubMed] [Google Scholar]
- Goedbloed DJ, Megens HJ, Van Hooft P, Herrero-Medrano JM, Lutz W, Alexandri P et al. (2013). Genome-wide single nucleotide polymorphism analysis reveals recent genetic introgression from domestic pigs into Northwest European wild boar populations. Mol Ecol 22: 856–866. [DOI] [PubMed] [Google Scholar]
- Gokhman D, Lavi E, Prüfer K, Fraga F, Riancho J, Kelso J et al. (2014). Reconstructing the DNA methylation maps of the Neandertal and the Denisovan. Science 344: 523–524. [DOI] [PubMed] [Google Scholar]
- Gossmann TI, Woolfit M, Eyre-Walker A. (2011). Quantifying the variation in the effective population size within a genome. Genetics 189: 1389–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M et al. (2010). A draft sequence of the Neandertal genome. Science 328: 710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen MA, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF et al. (2012). Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491: 393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlizius B, Rattink AP, de Koning DJ, Faivre M, Joosten RG, van Arendonk JA et al. (2000). The X Chromosome harbors quantitative trait loci for backfat thickness and intramuscular fat content in pigs. Mammalian Genome 11: 800–802. [DOI] [PubMed] [Google Scholar]
- Hermisson J, Pennings PS. (2005). Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics 169: 2335–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L. (2004) Kuahuqiao. Wenwu Press: Beijing. [Google Scholar]
- Jin L, Zhang M, Ma J, Zhang J, Zhou C, Liu Y et al. (2012). Mitochondrial DNA evidence indicates the local origin of domestic pigs in the upstream region of the Yangtze River. PLoS One 7: e51649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y, Flad R. (2002). Pig domestication in ancient China. Antiquity 76: 724–732. [Google Scholar]
- Jing Y, Jianlin H, Blench R. (2008). Livestock in ancient China: an archaeozoological perspective. In: Sanchez-Mazas A, Blench RM, Ross M, (eds) Human Migrations in Continental East Asia and Taiwan. Genetic, Linguistic and Archaeological Evidence. Routledge: Abingdon. [Google Scholar]
- Johansson Moller M, Chaudhary R, Hellmén E, Höyheim B, Chowdhary B, Andersson L. (1996). Pigs with the dominant white coat color phenotype carry a duplication of the KIT gene encoding the mast/stem cell growth factor receptor. Mamm Genome 7: 822–830. [DOI] [PubMed] [Google Scholar]
- Johnson AK, McGlone JJ. (2011). Behavior genetics of the domestic pig. In: Rothschild MF, Ruvinsky A, (eds) The Genetics of the Pig. CABI Publishing: Oxfordshire. pp 200–213. [Google Scholar]
- Keightley PD. (1994). The distribution of mutation effects on viability in Drosophila melanogaster. Genetics 138: 1315–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause-Kyora B, Makarewicz C, Evin A, Flink LG, Dobney K, Larson G et al. (2013). Use of domesticated pigs by Mesolithic hunter-gatherers in northwestern Europe. Nat Commun 4: 2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Kokko H, Eyre-Walker A. (2013). Population size and the rate of evolution. Trends Ecol Evol in press doi:10.1016/j.tree.2013.09.009. [DOI] [PubMed]
- Larson G, Albarella U, Dobney K, Rowley-Conwy P, Schibler J, Tresset A et al. (2007. b). Ancient DNA, pig domestication, and the spread of the Neolithic into Europe. Proc Natl Acad Sci USA 104: 15276–15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G, Cucchi T, Dobney K. (2011). Genetic aspects of pig domestication. In: Rothschild MF, Ruvinsky A, (eds) The Genetics of the Pig. CABI Publishing: Oxfordshire. pp 14–37. [Google Scholar]
- Larson G, Cucchi T, Fujita M, Matisoo-Smith E, Robins J, Anderson A et al. (2007. a). Phylogeny and ancient DNA of Sus provides insights into neolithic expansion in Island Southeast Asia and Oceania. Proc Natl Acad Sci USA 104: 4834–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G, Dobney K, Albarella U, Fang M, Matisoo-Smith E, Robins J et al. (2005). Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science 307: 1618–1621. [DOI] [PubMed] [Google Scholar]
- Larson G, Liu R, Zhao X, Yuan J, Fuller D, Barton L et al. (2010). Patterns of East Asian pig domestication, migration, and turnover revealed by modern and ancient DNA. Proc Natl Acad Sci USA 107: 7686–7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. (2011). Inference of human population history from individual whole-genome sequences. Nature 475: 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Tian S, Jin L, Zhou G, Li Y, Zhang Y et al. (2013). Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat Genet 45: 1431–1438. [DOI] [PubMed] [Google Scholar]
- Li SJ, Yang SL, Zhao SH, Fan B, Yu M, Wang HS et al. (2004). Genetic diversity analyses of 10 indigenous Chinese pig populations based on 20 microsatellites. J Anim Sci. 82: 368–374. [DOI] [PubMed] [Google Scholar]
- Loewe L, Charlesworth B. (2006). Inferring the distribution of mutational effects on fitness in Drosophila. Biol Lett 2: 426–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logténé YM, Koussou MO, Kaboré-Zoungrana CY. (2006). L'élevage porcin au Tchad: Bilan de l'introduction, de l'amélioration et de la diffusion des races exotiques. AGRI 38: 87–98. [Google Scholar]
- Lourenço JM, Glémin S, Galtier N. (2013). The rate of molecular adaptation in a changing environment. Mol Biol Evol 30: 1292–1301. [DOI] [PubMed] [Google Scholar]
- Manunza A, Zidi A, Yeghoyan S, Balteanu VA, Carsai TC, Scherbakov O et al. (2013). A high throughput genotyping approach reveals distinctive autosomal genetic signatures for European and Near Eastern wild boar. PLoS One 8: e55891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J, Haigh J. (1974). The hitch-hiking effect of a favourable gene. Genet Res 23: 23–35. [PubMed] [Google Scholar]
- Megens HJ, Crooijmans RP, San Cristobal M, Hui X, Li N, Groenen MA. (2008). Biodiversity of pig breeds from China and Europe estimated from pooled DNA samples: differences in microsatellite variation between two areas of domestication. Genet Sel Evol 40: 103–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megens HJ, Crooijmans RPMA, Larson G, Scandura M, Iacolina L, Apollonio M et al. (2010) The Porcine HapMap projects: genome-wide analysis of pig, wild boar and suiforme diversity. European Wild Boar 8th International Symposium on Wild Boars and Other Suids. York, United Kingdom.
- Messer PW, Petrov DA. (2013). Population genomics of rapid adaptation by soft selective sweeps. Trends Ecol Evol 28: 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen TH, Hayes BJ, Goddard ME. (2001). Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Straub RE, Lipska BK, Verchinski BA, Goldberg T, Callicott JH et al. (2007). Genetic evidence implicating DARPP-32 in human frontostriatal structure, function, and cognition. J Clin Invest. 117: 672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao YW, Peng MS, Wu GS, Ouyang YN, Yang ZY, Yu N et al. (2013). Chicken domestication: an updated perspective based on mitochondrial genomes. Heredity 110: 277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignon-Grasteau S, Boissy A, Bouix J, Faure J-M, Fisher AD, Hinch GN et al. (2005). Genetics of adaptation and domestication in livestock. Livest Prod Sci 93: 3–14. [Google Scholar]
- Mikawa S, Hayashi T, Nii M, Shimanuki S, Morozumi T, Awata T. (2005). Two quantitative trait loci on Sus scrofa chromosomes 1 and 7 affecting the number of vertebrae. J Anim Sci 83: 2247–2254. [DOI] [PubMed] [Google Scholar]
- Mikawa S, Morozumi T, Shimanuki S, Hayashi T, Uenishi H, Domukai M et al. (2007). Fine mapping of a swine quantitative trait locus for number of vertebrae and analysis of an orphan nuclear receptor, germ cell nuclear factor (NR6A1). Genome Res 17: 586–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa S, Sato S, Nii M, Morozumi T, Yoshioka G, Imaeda N et al. (2011). Identification of a second gene associated with variation in vertebral number in domestic pigs. BMC Genet 12: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutou K, Koutsogiannouli E, Stamatis C, Billinis C, Kalbe C, Scandura M et al. (2013). Domestication does not narrow MHC diversity in Sus scrofa. Immunogenetics 65: 195–209. [DOI] [PubMed] [Google Scholar]
- Nielsen SM (ed). (1998) Ancestors for the Pigs: Pigs in Prehistory – MASCA Research Papers in Science and Archaeology vol. 15. University of Pennsylvania Museum of Archaeology and Anthropology: Philadelphia. [Google Scholar]
- Nielsen R, Hubisz MJ, Clark AG. (2004). Reconstituting the frequency spectrum of ascertained single-nucleotide polymorphism data. Genetics 168: 2373–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojeda A, Ramos-Onsins SE, Marletta D, Huang LS, Folch JM, Pérez-Enciso M. (2011). Evolutionary study of a potential selection target region in the pig. Heredity 106: 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando L, Ginolhac A, Raghavan M, Vilstrup J, Rasmussen M, Magnussen K et al. (2011). True single-molecule DNA sequencing of a pleistocene horse bone. Genome Res 21: 1705–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. (1998). Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics 149: 2099–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottoni C, Flink LG, Evin A, Geörg C, De Cupere B, Van Neer W et al. (2013). Pig domestication and human-mediated dispersal in western Eurasia revealed through ancient DNA and geometric morphometrics. Mol Biol Evol 30: 824–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudel Y, Madsen O, Megens HJ, Frantz LA, Bosse M, Bastiaansen JW et al. (2013). Evolutionary dynamics of copy number variation in pig genomes in the context of adaptation and domestication. BMC Genomics 14: 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Jensen JD, Stephan W, Stamatakis A. (2012). A critical assessment of storytelling: gene ontology categories and the importance of validating genomic scans. Mol Biol Evol 29: 3237–3248. [DOI] [PubMed] [Google Scholar]
- Piganeau G, Eyre-Walker A. (2003). Estimating the distribution of fitness effects from DNA sequence data: implications for the molecular clock. Proc Natl Acad Sci USA 100: 10335–10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilez J, Short AD, Martinez V, Kennedy LJ, Ollier W et al. (2011). A selective sweep of.8 Mb on chromosome 26 in the Boxer genome. BMC Genomics 12: 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos AM, Crooijmans RP, Affara NA, Amaral AJ, Archibald AL, Beever JE et al. (2009). Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS One 4: e6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez O, Ojeda A, Tomàs A, Gallardo D, Huang LS, Folch JM et al. (2009). Integrating Y-chromosome, mitochondrial, and autosomal data to analyze the origin of pig breeds. Mol Biol Evol 26: 2061–2072. [DOI] [PubMed] [Google Scholar]
- Ramírez-Soriano A, Ramos-Onsins SE, Rozas J, Calafell F, Navarro A. (2008). Statistical power analysis of neutrality tests under demographic expansions, contractions and bottlenecks with recombination. Genetics 179: 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding RW. (2005). Breaking the mold, a consideration of variation in the evolution of animal domestication. In: Vigne JD, Peters J, Helmer D, (eds) The First Steps of Animal Domestication. Oxbow Books: Oxford. pp 41–48. [Google Scholar]
- Reiner G, Köhler F, Berge T, Fischer R, Hübner-Weitz K, Scholl J et al. (2009). Mapping of quantitative trait loci affecting behaviour in swine. Animal Genet 40: 366–376. [DOI] [PubMed] [Google Scholar]
- Ren J, Duan Y, Qiao R, Yao F, Zhang Z, Yang B et al. (2011). A missense mutation in PPARD causes a major QTL effect on ear size in pigs. PLoS Genet 7: e1002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Yan X, Ai H, Zhang Z, Huang X, Ouyang J et al. (2012). Susceptibility towards enterotoxigenic Escherichia coli F4ac diarrhea is governed by the MUC13 gene in pigs. PLoS One 7: e44573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzi E, Lari M, Gigli E, De Bellis G, Caramelli D. (2012). Ancient DNA studies: new perspectives on old samples. Genet Sel Evol 44: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer GA, Alexander LJ, Hu Z, Smith TP, Keele JW, Beattie CW. (1996). A comprehensive map of the porcine genome. Genome Res 6: 371–391. [DOI] [PubMed] [Google Scholar]
- Ross-Ibarra J, Morrell PL, Gaut BS. (2007). Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc Natl Acad Sci USA 104: 8641–8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley-Conwy P. (2003). Early domestic animals in Europe: imported or locally domesticated? In: Ammerman AJ, Biagi P, (eds) The Widening Harvest: The Neolithic Transition in Europe: Looking Back, Looking Forward. Archaeological Institute of America: Boston. pp 99–117. [Google Scholar]
- Rubin CJ, Megens HJ, Martinez Barrio A, Maqbool K, Sayyab S, Schwochow D et al. (2012). Strong signatures of selection in the domestic pig genome. Proc Natl Acad Sci USA 109: 19529–19536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin CJ, Zody MC, Eriksson J, Meadows JR, Sherwood E, Webster MT et al. (2010). Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 464: 587–591. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF et al. (2002). Detecting recent positive selection in the human genome from haplotype structure. Nature 419: 832–837. [DOI] [PubMed] [Google Scholar]
- Sacks BN, Brown SK, Stephens D, Pedersen NC, Wu JT, Berry O. (2013). Y chromosome analysis of dingoes and southeast asian village dogs suggests a neolithic continental expansion from Southeast Asia followed by multiple Austronesian dispersals. Mol Biol Evol 30: 1103–1118. [DOI] [PubMed] [Google Scholar]
- Santiago E, Caballero A. (1998). effective size and polymorphism of linked neutral loci in populations under directional selection. Genetics 149: 2105–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandura M, Iacolina L, Crestanello B, Pecchioli E, Di Benedetto MF, Russo V et al. (2008). Ancient vs. recent processes as factors shaping the genetic variation of the European wild boar: are the effects of the last glaciation still detectable? Mol Ecol 17: 1745–1762. [DOI] [PubMed] [Google Scholar]
- Schook LB, Beever JE, Rogers J, Humphray S, Archibald A, Chardon P et al. (2005). Swine genome sequencing consortium (SGSC): a strategic roadmap for sequencing the pig genome. Comp Funct Genom 6: 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K, Thornton KR, Stoneking M. (2007). A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol 5: e171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trut LN. (1999). Early canid domestication: the farm-fox experiment. Am Scit 87: 160–169. [Google Scholar]
- Van der Made J. (2001). The ungulates from Atapuerca: stratigraphy and biogeography. Anthropology 105: 95–113. [Google Scholar]
- Vaysse A, Ratnakumar A, Derrien T, Axelsson E, Rosengren Pielberg G, Sigurdsson S et al. (2011). Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLoS Genet 7: e1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkhuyzen AA, Wray NR, Yang J, Goddard ME, Visscher PM. (2013). Estimation and partition of heritability in human populations using whole-genome analysis methods. Annu Rev Genet 47: 75–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. (2006). A map of recent positive selection in the human genome. PLoS Biol 4: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wealleans AL. (2013). Such as pigs eat: the rise and fall of the pannage pig in the UK. J Sci Food Agric 93: 2076–2083. [DOI] [PubMed] [Google Scholar]
- White S. (2011). From globalized pig breeds to capitalist pigs: a study in animal cultures and evolutionary history. Environ Hist 16: 94–120. [Google Scholar]
- Whitlock MC, Barton NH. (1997). The effective size of a subdivided population. Genetics 146: 427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson S, Lu ZH, Megens HJ, Archibald AL, Haley C, Jackson IJ et al. (2013). Signatures of diversifying selection in European pig breeds. PLoS Genet 9: e1003453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Bi IV, Schroeder SG, Yamasaki M, Doebley JF, McMullen MD et al. (2005). The effects of artificial selection on the maize genome. Science 308: 1310–1314. [DOI] [PubMed] [Google Scholar]
- Wu GS, Yao YG, Qu KX, Ding ZL, Li H, Palanichamy MG et al. (2007). Population phylogenomic analysis of mitochondrial DNA in wild boars and domestic pigs revealed multiple domestication events in East Asia. Genome Biol 8: R245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR et al. (2010). Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Zhang H, Mao H, Yan D, Lu S, Lian L et al. (2011). The local origin of the Tibetan pig and additional insights into the origin of Asian pigs. PLoS One 6: e28215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE et al. (2010). Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329: 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeder MA, Emshwiller E, Smith BD, Bradley DG. (2006). Documenting domestication: the intersection of genetics and archaeology. Trends Genet 22: 139–155. [DOI] [PubMed] [Google Scholar]
- Zeder MA. (2012). Pathways to domestication. In: Gepts P, Famula RL, Bettinger RL, Brush SB, Damania AB, McGuire PE, Qualset CO, (eds) Biodiversity in Agriculture: Domestication, Evolution, and Sustainability. Cambridge University Press: Cambridge. pp 227–259. [Google Scholar]
- Zhang DX, Hewitt GM. (2003). Nuclear DNA analyses in genetic studies of populations: practice, problems and prospects. Mol Ecol. 12: 563–584. [DOI] [PubMed] [Google Scholar]
- Zhang ZY, Ren J, Ren DR, Ma JW, Guo YM, Huang LS. (2009). Mapping quantitative trait loci for feed consumption and feeding behaviors in a White Duroc × Chinese Erhualian resource population. J Animal Sci 87: 3458–3463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.