Abstract
Increasing data have supported the importance of divergence with gene flow (DGF) in the generation of biological diversity. In such cases, lineage divergence occurs on a shorter timescale than does the completion of reproductive isolation. Although it is critical to explore the mechanisms driving divergence and preventing homogenization by hybridization, it is equally important to document cases of DGF in nature. Here we synthesize data that have accumulated over the last dozen or so years on DGF in the chipmunk (Tamias) radiation with new data that quantify very high rates of mitochondrial DNA (mtDNA) introgression among para- and sympatric species in the T. quadrivittatus group in the central and southern Rocky Mountains. These new data (188 cytochrome b sequences) bring the total number of sequences up to 1871; roughly 16% (298) of the chipmunks we have sequenced exhibit introgressed mtDNA. This includes ongoing introgression between subspecies and between both closely related and distantly related taxa. In addition, we have identified several taxa that are apparently fixed for ancient introgressions and in which there is no evidence of ongoing introgression. A recurrent observation is that these introgressions occur between ecologically and morphologically diverged, sometimes non-sister taxa that engage in well-documented niche partitioning. Thus, the chipmunk radiation in western North America represents an excellent mammalian example of speciation in the face of recurrent gene flow among lineages and where biogeography, habitat differentiation and mating systems suggest important roles for both ecological and sexual selection.
Introduction
Increasing evidence has accumulated over the last two decades (for example, Rice and Hostert, 1993) that supports divergence with gene flow (DGF) as an important driver of biological diversity. This process differs from classical models of speciation, in which divergence is postulated to occur in allopatry or in the absence of gene flow between diverging lineages and in which hybridization on secondary contact causes homogenization of diverging lineages (for example, Taylor et al., 2006; Zitari et al., 2012). However, as pointed out by Jiggins (2008) and Smadja and Butlin (2010), allopatric versus sympatric models of speciation represent extremes of a continuum, with no possibility of gene flow during divergence in the former and no (geographic) barriers to gene flow during divergence in the latter. Because range fluctuations often occur over shorter temporal scales than that of the evolution of complete reproductive isolation, speciation may frequently involve ephemeral periods of allopatric differentiation that are punctuated by intermittent periods of contact during which introgression between and among diverging lineages may take place (for example, Fitzpatrick and Turelli, 2006). This may be especially critical in instances of rapid radiations simply due to the fact that multiple radiating lineages can provide many opportunities for range fluctuations to result in temporally variable range overlap. Therefore, important questions in speciation focus on the extent and nature of gene flow that is occurring or has occurred during the process of lineage divergence and on the genomic architecture and trait associations characterizing that differentiation (Smadja and Butlin, 2010; Wu, 2001); that is, how common is DGF (for example, Pinho and Hey, 2010) and how does it occur?
Here we present a synthesis of current data on DGF in a diverse group of ground squirrels, chipmunks of the genus Tamias, including previously unpublished genetic data and analyses that indicate that the many instances of mitochondrial introgression we have identified illustrate a gradual attenuation of gene flow with increasing depth of divergence (Reid et al., 2012), which is one of the easiest to test predictions of DGF. Thus, lineage differentiation is occurring at a more rapid rate in this radiation than is the evolution of complete reproductive isolation and this has resulted in multiple examples of gene flow between morphologically and ecologically differentiated taxa that are not sister species.
Study system
Interest in the biology of the 23 described species of western chipmunks (Tamias, subgenus Neotamias; but see Baker et al., 2003; Piaggio and Spicer, 2001) predates the modern synthesis (for example, Merriam, 1897). This group is one of the more speciose mammalian genera in North America and has the hallmarks of a recent, rapid radiation (for example, Reid et al., 2012; this study). Western chipmunks are widely distributed in a diverse array of habitats across western North America (Hall, 1981) and in some areas (for example, Sierra Nevada) up to four species may be co-distributed. One of the ecologically intriguing features of chipmunks is the strong niche partitioning observed among broadly co-distributed species, typically resulting in altitudinal zonation and extremely narrow zones of parapatry between some species (Heller, 1970, 1971; Chappell, 1978; Bergstrom, 1992). Early observations of this phenomenon (for example, Grinnell and Storer, 1924) in the eastern Sierra Nevada among least, yellow-pine, lodgepole and alpine chipmunks (T. minimus, T. amoenus, T. speciosus and T. alpinus, respectively) led to extensive studies that documented altitudinal zonation and examined the ecological and physiological mechanisms underlying niche partitioning (for example, Howell, 1929; Johnson, 1943; Heller, 1971; Heller and Gates, 1971; Chappell, 1978). Other instances of habitat partitioning between sympatric chipmunks are well documented, although physiological and ecological mechanisms have not been as thoroughly examined: Uinta and cliff chipmunks (T. umbrinus and T. dorsalis) in Nevada (Brown, 1971); dusky and Merriam's chipmunks (T. obscurus and T. merriami) in southern California (Callahan, 1977; Blankenship, 1985; Best and Granai, 1994a, 1994b); yellow-pine and red-tailed chipmunks (T. amoenus and T. ruficaudus) in the Inland Northwest (Gambs, 1965; Best, 1993); and least, Colorado and Uinta chipmunks (T. minimus, T. quadrivittatus and T. umbrinus) in the Front Range of Colorado (Bergstrom and Hoffmann, 1991; Bergstrom, 1992). These examples often include a narrowly distributed habitat specialist (for example, red-tailed chipmunks in the Inland Northwest) that excludes a more widely distributed habitat generalist (for example, yellow-pine chipmunks in the Inland Northwest) from the specialist's preferred habitat (for example, mesic forests).
Reproductive biology, sexual selection and genital morphology
Although details of the reproductive biology are not well known for every species of western chipmunk, all studied appear to be predominantly monestrous and each female mates only on a single day per year (Callahan, 1981). In preparation for mating, a female represses her territoriality and engages in visual (tail waving) and vocal (chip vocalizations) advertisement of approaching estrus (Callahan, 1981). Once in estrus, the female initiates a mating chase and subsequently copulates with one or more pursuing males (Callahan, 1981; Compton and Callahan, 1995). A very high incidence of multiple paternity has been reported in the yellow-pine chipmunk (T. amoenus), with more than 90% of litters sired by multiple males (Schulte-Hostedde et al., 2004). Such high levels of promiscuity could result in intense sexual selection through direct sperm competition and female choice (but see Schulte-Hostedde et al., 2004), and indeed eastern chipmunks (T. striatus) exhibit significant positive Bateman gradients for both genders (Bergeron et al., 2012). Consistent with this, patterns of variation for several reproductive characters within (Schulte-Hostedde and Millar, 2004) and between species (White, 1953) suggest a strong influence of sexual selection.
The most striking cases of rapid phenotypic evolution in chipmunks are found in patterns of genital evolution. In particular, the male penile bone (that is, the baculum or os penis) shows rapid differentiation in size and shape between closely related species (White, 1953; Patterson and Thaeler, 1982; Sutton and Patterson, 2000; Figure 3). Within Tamias, the baculum occupies an extremely distal position in the penis and bacular morphology is characterized by modest variation within species, but strong discontinuities among species (White, 1953; Patterson and Thaeler, 1982; Sutton and Patterson, 2000). Such discontinuities occur in parapatry and, in some instances, have been correlated with allozyme differentiation (for example, Sullivan and Petersen, 1988) and discontinuities in chip vocalizations (Gannon and Lawlor, 1989). These results, along with the lack of clear intermediates, have been interpreted as evidence that bacular discontinuities are reliable indicators, and perhaps even the primary mechanism, of reproductive isolation (for example, Patterson and Thaeler, 1982; Sutton, 1992; Sutton and Patterson, 2000). Consequently, bacular morphology has emerged as the defining morphological character for species delimitation within the genus.
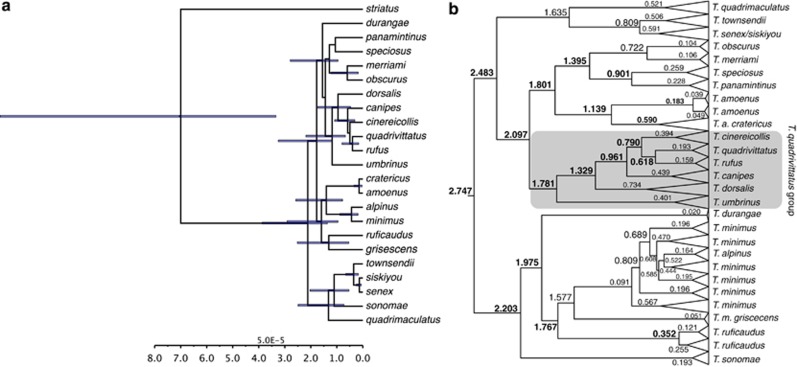
History of Tamias phylogenetics
Because of their conspicuous nature and importance in biogeography (for example, Lomolino et al., 2006) the 25 species of chipmunks have been the focus of numerous phylogenetic studies (for example, Ellis and Maxson, 1979; Levenson et al., 1985; Piaggio and Spicer, 2000, 2001; Reid et al., 2012). Earlier studies recognized three major clades within Tamias (classified as either subgenera or separate genera; for example, Jameson, 1999; Piaggio and Spicer, 2001), one in Asia, one in eastern North America and the large radiation (subgenus Neotamias) in western North America that may have originated ∼2.75 Myr (Figure 1, although with a large credibility interval). In addition, previous studies have recognized conflicting species groups within subgenus Neotamias, in part because congruence among data sets for these taxa has been elusive. Since the first application of mitochondrial DNA (mtDNA) sequences to Tamias phylogeny (Piaggio and Spicer, 2000, 2001), there have been indications of incongruence between the mtDNA gene tree, allozyme estimates and taxonomy (defined primarily by bacular morphology). Subsequent phylogeographic studies with denser geographic sampling have demonstrated that this incongruence is a recurring observation in the subgenus (for example, T. ruficaudus—Good and Sullivan, 2001; T. amoneus—Demboski and Sullivan, 2003, in the Inland Northwest). Furthermore, these studies have demonstrated that hybridization (between either species or subspecies) is the cause of incongruence (Good et al., 2003, 2008). The most comprehensive phylogenetic data set to date is that of Reid et al. (2012), which we have reanalyzed in this study using relaxed-clock approaches (Figure 1). This phylogeny will be used to structure the synthesis presented here; these data were generated from a broad geographic and taxonomic sample from four nuclear genes involved in reproduction.
Figure 1.
Chronograms for 20 of 23 species of western chipmunks in the subgenus Neotamias (missing are T. bulleri, T. palmeri and T. ochrogenys) based on reanalysis of four nuclear genes (acrosin, zonadhesin, zona pellucida proteins 2 and 3) published by Reid et al. (2012). (a) Species-tree estimate derived using *BEAST from four nuclear loci reported in Reid et al. (2012). These four phased nuclear loci from individuals assigned to species based on bacular morphology were used to estimate a posterior distribution of species trees using *BEAST (Heled and Drumond, 2010). The analysis assumed an uncorrelated lognormal clock prior and birth–death tree prior. The MCMC chain was run for 50 000 000 generations and sampled every 5000 generations. The posterior distribution of species trees was summarized into a point estimate of topology using TreeAnnotator v1.7.5 (available: www.beast2.org/wiki/index.php/TreeAnnotator) after removing 1000 samples (10%) as a burn-in, setting a posterior probability threshold of 0.5 and using median node heights. The tree was visualized using FigTree v1.4.0 (available: http://tree.bio.ed.ac.uk/software/figtree/), and the root has been rescaled to a depth of 7 Myr. (b) The concatenated tree was rooted with T. striatus and rescaled the root node to be 7 Myr based on Late Miocene date of Dahlquist et al. (1996). PartitionFinder 1.0.1 (Lanfear et al., 2012) was used to choose a partitioning scheme based on the Bayesian Information Criterion (BIC) that placed zonadhesin, zona pellucida protein 2 and zona pellucida protein 3 under a K80+I+Γ model of sequence evolution and acrosin under an HKY+Γ model of sequence evolution. A posterior distribution of topologies was generated using BEAST 1.7.4 (Drummond et al., 2012). We ran two independent runs of 50 000 000 generations and sampled every 5000 generations. Convergence was assessed by comparing traces, comparing posterior distributions of parameter estimates and ensuring the effective sampling size of each parameter was at least 200 or greater. The two runs were then combined using LogCombiner (available: https://github.com/SeleniumHQ/selenium/blob/.../LogCombiner.java) after removing 10% of the samples as a burn-in. A maximum clade credibility tree was generated using TreeAnnotator from the combined distribution of topologies with a posterior probability threshold of 0.5 and median node heights. Associated with nodes are divergence times (in Myr), and those in bold font are nodes that span introgression events. Shaded taxa represent the T. quadrivittatus group.
Our conceptualization of DGF
We view the concept of DGF in its broadest sense (for example, Smadja and Butlin, 2010). That is, DGF certainly includes instances of ecological speciation, say between host races (for example, Pecoud et al., 2009), but it can apply to any instance where divergence has occurred or at least persisted in the face of introgression. One of the most straightforward predictions of the hypothesis that DGF has occurred in the chipmunk radiation is that hybridization is expected to be more common between recently diverged taxa and should attenuate with time since divergence. Wu's (2001) verbal model of DGF discretizes this expected gradual attenuation of interspecific gene flow into four stages. However, because these stages are arbitrary and other aspects of this verbal model are controversial (for example, the importance of divergence hitchhiking; Feder and Nosil, 2010), we synthesize existing data on DGF in western chipmunks using relative divergence order provided by relaxed molecular clock analyses (Figure 1).
Detection of hybridization
Incongruence between mtDNA gene trees and bacular morphology is likely a good indicator of introgression in Tamias given that species limits defined by bacular morphology have proven to be ecologically and morphologically well circumscribed. However, in our studies described below, we have taken a more rigorous approach to support our conclusions that introgression is the cause of most of the phylogenetic incongruence. Specifically, we examine one or more of four lines of evidence; these are ordered by level of rigor, with the last being the most explicit rejection of coalescent stochasticity. The first is the demonstration that the putatively introgressed haplotypes are geographically structured, which should not be the case for incongruence due to incomplete lineage sorting (Good et al., 2003). Second, congruence between morphology and nuclear microsatellite data (that is, correct assignment of individuals to morphologically defined taxa in Bayesian clustering analyses) support introgression of mtDNA in cases where the mtDNA gene tree is incongruent with morphology (Good et al., 2008; Hird and Sullivan, 2009; Reid et al., 2010). Third, commonly sequenced nuclear genes (for example, Rag-1, c-myc, Acp) from individuals with putatively introgressed mtDNA are expected to sort more slowly due to their larger effective population size (Ne). That is, if nuclear genes have sorted (that is, their gene trees are congruent with morphology), incongruence between mtDNA and morphology is likely due to introgression (Good et al., 2008). Fourth, coalescent simulations on a species tree estimated from nuclear genes can rule out phylogenetic uncertainty and coalescent stochasticity as the cause of incongruence between the mtDNA gene tree and morphology (Reid et al., 2012), leaving introgression as the likely cause. In all the cases described below, one or (usually) more of these lines of evidence has implicated introgression of mtDNA.
Numerous differentially aged introgressions
In over a decade of field collection and sequencing, we have demonstrated that mtDNA introgression appears to be common between many different species of Tamias. These discoveries of introgression are summarized in Table 1. Over 1800 individuals (20 species) have been sequenced, and nearly 300 of them (16%), representing 9 species (40%), exhibit some type of introgressed mtDNA. Note that some of our sampling has targeted potential hybrid zones (for example, Good et al., 2003) and that this may bias these overall estimates of the extent of introgression. Nevertheless, examples of hybridization include intraspecific introgressions between recently diverged, but morphologically differentiated subspecies (for example, T. r. ruficaudus and T. r. simulans), interspecific introgression between and among several members of a closely related clade or species group (for example, T. quadrivittatus species group; T. umbrinus, T. dorsalis, T. cinereicollis and T. quadrivittatus) and introgression between more distantly related taxa that perhaps span the deepest nodes in the subgenus phylogeny and belong to different species group (for example, T. ruficaudus and T. amoenus; Figure 1b). Furthermore, there are also instances of apparently complete mtDNA capture in which all individuals of a species or subspecies appear to have introgressed mtDNA (for example, T. speciosus, T. panamintinus, T. amoenus canicaudus). We now discuss each of these case studies in turn.
Table 1. Summary of mtDNA introgression detected in all 23 species of western chipmunks (subgenus Neotamias).
| Taxon | N | # Int. (%) | Source | Div. time (Myr) | Maximum mtDNA % Div | Ref |
|---|---|---|---|---|---|---|
| T. r. ruficaudus | 131 | 0 (0) | NA | 1,2 | ||
| T. r. simulans | 175 | 63 (36) | T. r. ruficaudus | 0.35 | 4.7 | 1,2 |
| T. a. canicaudus | 46 | 46 (100) | T. r. simulans | 2.75 | 3.2a | 2,3 |
| T. a. luteiventris | 148 | 43 (29) | T. r. ruficaudus | 2.75 | 10.0 | 2,3 |
| Other T. amoenus | 122 | 0 (0) | NA | 4 | ||
| T. dorsalis | 54 | 35 (65) | T. cinereicollis, | 1.33 | 4.6 | 5 |
| T. umbrinus, | 1.78 | 4.7 | 5 | |||
| T. quadrivittatus | 1.33 | 4.7 | 5 | |||
| Not assignableb | 4.7 | 5 | ||||
| T. cinereicollis | 25 | 7 (28) | Not assignableb | 4.7 | 5 | |
| T. umbrinus | 64 | 9 (19) | Not assignableb | 4.7 | 5 | |
| T. canipes | 8 | 0 (0) | NA | 5 | ||
| T. rufus | 14 | 0 (0) | NA | 5 | ||
| T. quadrivittatus | 38 | 14 (37) | Not assignableb | 4.7 | 5 | |
| T. townsendii | 6 | 0 (0) | NA | 6 | ||
| T. ochrogenys | 0 | NA | NA | NA | ||
| T. sonomae | 4 | 0 (0) | NA | 6 | ||
| T. siskiyou | 5 | 0 (0) | NA | 6 | ||
| T. senex | 155 | 0 (0) | NA | 5,6 | ||
| T. quadrimaculatus | 221 | 0 (0) | NA | 5,6 | ||
| T. minimus | 305 | 0 (0) | NA | 5 | ||
| T. panamintinus | 15 | 15 (100) | T. minimus | 2.75 | 10.0a | 6,7 |
| T. alpinus | 34 | 0 (0) | NA | 6,7 | ||
| T. speciosus | 56 | 56 (100) | T. minimus | 2.75 | 10.0a | 6,7 |
| T. merriami | 155 | 0 (0) | NA | 7 | ||
| T. obscurus | 87 | 0 (0) | NA | 7 | ||
| T. bulleri | 1 | ? | NA | 6 | ||
| T. durangae | 1 | ? | NA | 6 | ||
| T. palmeri | 1 | ? | NA | 5 | ||
| Totals | 1871 | 298 |
Abbreviations: mtDNA, mitochondrial DNA; Div, divergence; Int, number of introgressed individuals; NA, not applicable.
5–This study.
7–Demboski et al. (in prep).
Only introgressed mtDNA has been identified.
The introgressed mtDNA is clade C in Figure 2 and cannot be assigned as native to any taxon.
Chipmunks of the Inland Northwest
The first documentation of hybridization in Tamias resulted from comparative phylogeographic studies of forest-dwelling rodents in the Inland Northwest (Good and Sullivan, 2001; Demboski and Sullivan, 2003). To date, four independent instances of introgressive hybridization have been documented in the region.
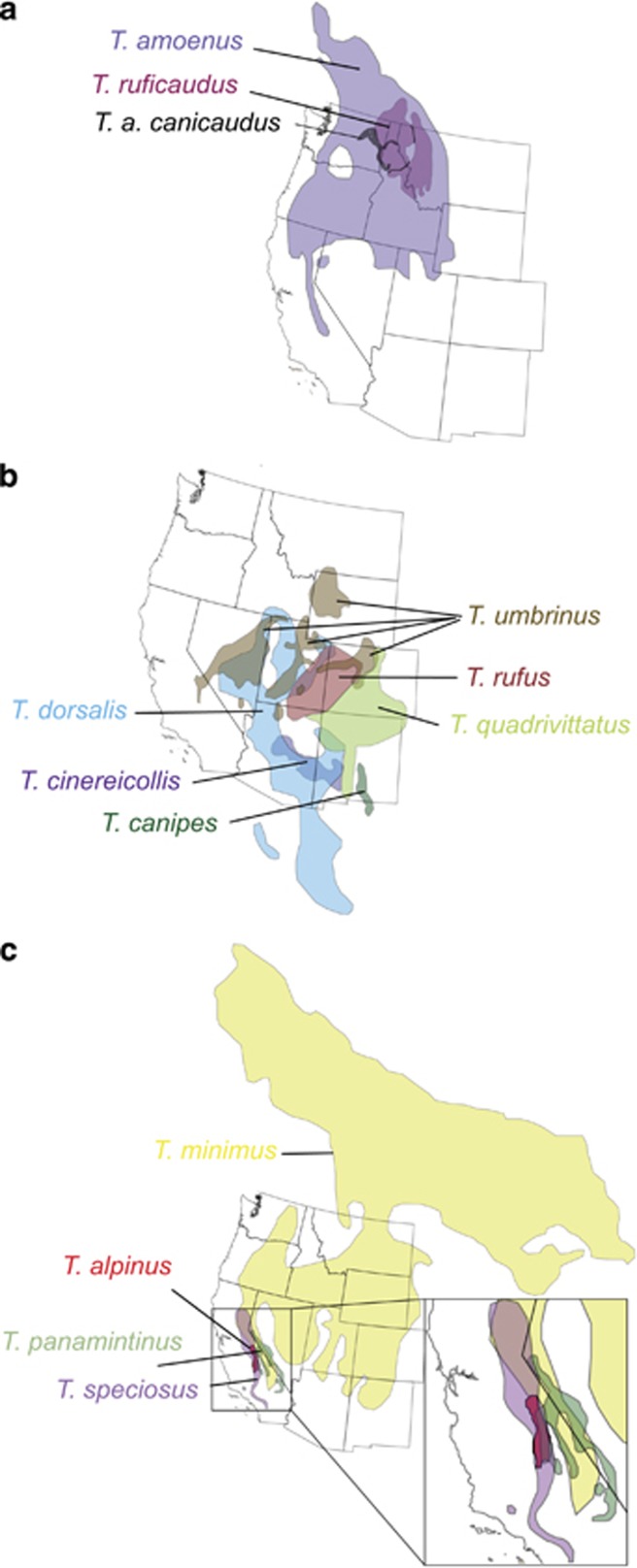
The first two introgressions occur between the two subspecies of red-tailed chipmunks (T. ruficaudus) that were suggested by Patterson and Heaney (1987) to be reproductively isolated species based on a large difference in bacular morphology. This species has a doughnut-shaped distribution in northern Idaho, NW Montana, NE Washington, southern British Columbia and Alberta (Figure 2a), and is endemic to mesic (that is, inland temperate rainforests) and subalpine forests (Gambs, 1965). Coalescent analyses of microsatellite data suggest that these subspecies diverged ∼325 000 years ago (Hird and Sullivan, 2009), and this date is supported independently by our relaxed-clock analyses of nuclear gene sequences (Figure 1b). The two subspecies (T. r. ruficaudus in the south and east and T. r. simulans in the west) contact each other at two distinct hybrid zones (Figure 2a). The southern zone occurs along the Lochsa River in the Clearwater Drainage, which remained free of ice during Pleistocene glacial maxima (Daubenmire, 1975). Populations along the north bank exhibit the T. r. simulans baculum and populations along the south bank exhibit the T. r. ruficaudus baculum. However, T. r. ruficaudus haplotypes have introgressed north of the Lochsa River and all populations between this river and the next drainage to the north (the North Fork of the Clearwater River) exhibit T. r. simulans bacula but are fixed for one or more T. r. ruficaudus haplotypes (Good and Sullivan, 2001; Good et al., 2003). Microsatellite data (Hird and Sullivan, 2009) indicate that there is virtually no gene flow across the Lochsa River, suggesting that it is an effective barrier to gene flow. However, T. r. rufucaudus and T. r. simulans are exchanging genes over the Bitterroot Divide (at Lolo Pass) east of the headwaters of the Lochsa River (Hird and Sullivan, 2009). The northern contact zone between these two subspecies occurs near Whitefish Montana, north of the southern extent of Pleistocene glaciation. Palynology from localities of similar latitude in the region suggests that forests had not been established until only ∼3000 years ago (Mack et al., 1978), establishing a lower bound for the age of the Whitefish contact zone. In this northern zone, all data (morphology, mtDNA and microsatellites; Good and Sullivan, 2001; Good et al., 2003; Hird et al., 2010) are congruent in indicating a narrow hybrid zone with very little introgression.
Figure 2.
Range maps for various introgressing species. (a) T. amoenus and T. ruficaudus in the Inland Northwest. T. r. ruficaudus occurs east of the black line and T. r. simulans occurs west of it. T. a. canicaudus, in gray shading, has experienced ancient capture of mtDNA that is sister to T. r. simulans mtDNA. (b) The T. quadrivittatus group of the central and southern Rockies. Although T. rufus has a partially overlapping range with T. umbrinus and T. dorsalis, there are no known localities where it is sympatric with either. (c) Ranges of T. minimus and taxa that are monophyletic for hapolgroups nested within widespread T. minimus. T. panamintinus and T. speciosus (in the inset) each appear to have become fixed on introgressions from T. minimus, whereas T. alpinus is likely a very recent derivative of T. minimus.
The other introgressions in the Inland Northwest both involve interspecific introgression between T. ruficaudus and T. amoenus (yellow-pine chipmunk), a broadly distributed (Figure 2a) ecological generalist (Sutton, 1992). In the Northern Rockies, a single mtDNA haplotype from T. r. ruficaudus has introgressed into populations of T. a. luteiventris along the spine of the range in southern British Columbia and Alberta (Good et al., 2003). This event has resulted in the T. r. ruficaudus haplotype introgressing outside the species range and apparently (based on somewhat limited sampling) replacing the native T. a. luteiventris mtDNA in a few populations. Interestingly, the introgressed haplotype is identical to that found in the northernmost populations of T. r. ruficaudus, indicating a relatively recent (relative to the mtDNA mutation rate) introgression event. Reid et al. (2010) examined 10 microsatellite loci and sequences from a single nuclear gene (acrosin) in this contact zone. The nuclear data convincingly support nearly complete reproductive isolation, except for a single T. a. luteiventris individual with appreciable T. r. ruficaudus coancestry. Thus, although the mtDNA introgression appears to have been recent based on shared identical haplotypes (Good et al., 2003), only a hint of introgression was detectable in the nuclear data (Reid et al., 2010).
The fourth introgression in the Inland Northwest is perhaps the most interesting as it involves an ancient but nonrecurrent mtDNA introgression event, resulting in complete fixation across a subspecies range (Good et al., 2003, 2008). The ash-tailed yellow-pine chipmunk, T. a. canicaudus, is geographically isolated from other forms of T. amoenus and occurs in forest patches east of the Columbia River in central Washington to the Palouse Formation in eastern Washington and into northern Idaho and western Montana. Its pelage is distinct (hence the common name) and its bacular morphology is indistinguishable from populations of T. a. luteiventris in the Columbia Mountains to the north and across the Columbia River (Good et al., 2003). All 46 T. a. canicaudus individuals we have sequenced (from across the subspecies range) have mtDNA haplotypes that form a reciprocally monophyletic clade with T. r. simulans, and these two sister phylogroups are sister to the T. r. ruficaudus haplotypes. Thus, this appears to be the result of fixation on an ancient hybridization event between T. a. canicaudus and T. r. simulans. Microsatellite data confirm that there is no significant contemporary gene flow between these two taxa, nor is there any between T. a. canicaudus and its geographically closest conspecific populations, T. r. luteiventris (Good et al., 2008). However, gene trees from four nuclear loci (acr, c-myc, rag-1 and acp-1) have largely sorted between T. amoenus and T. ruficaudus, supporting the hypothesis of an ancient mtDNA introgression from T. r. simulans to T. a. canicaudus (Good et al., 2008).
Thus, these four instances of introgression in the Inland Northwest are differentially aged. They include two instances of introgression between subspecies of T. ruficaudus (the Lochsa and Whitefish introgressions) that have very different bacular morphologies, presumably following secondary contact. The other two involve interspecific introgression from T. ruficuadus to T. amoenus; one of these is very recent, and possibly ongoing (the Northern Rockies introgression of T. r. ruficaudus into T. a. luteiventris), and the other represents fixation in T. a. canicaudus of an ancient introgression of T. r. simulans-like mtDNA.
Chipmunks of the southern Rockies/Great Basin—T. quadrivittatus group
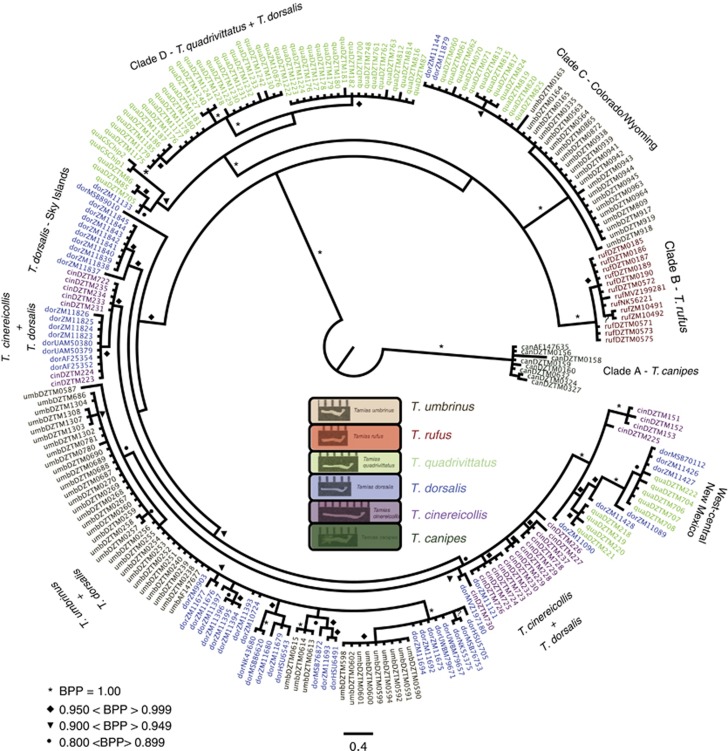
The T. quadrivittatus group has been circumscribed based on mtDNA to include T. bulleri and T. durangae, two species endemic to the highlands of northern Mexico, as well as T. umbrinus, T. palmeri, T. dorsalis, T. cinereicollis, T. rufus, T. canipes and T. quadrivittatus (Piaggio and Spicer, 2000, 2001). The latter seven species are all distributed across the Great Basin and central and southern Rocky Mountains (Figure 2b), and Reid et al. (2012) based on sequences from nuclear reproductive protein coding genes demonstrated that six of these species (T. palmeri was not included) form a pectinate tree (Figure 1), with T. umbrinus as sister to the rest, followed by T. dorsalis. These relationships are strongly supported in both the species tree (Figure 1a) estimated with *BEAST (Heled and Drummond, 2010) and the concatenated tree (Figure 1b). The rest of the quadrivittatus group tree is less well supported. Poorly supported nodes suggest that T. rufus is sister to T. quadrivittatus, followed by T. cinereicollis and then followed by T. canipes (which is allopatric with respect to the rest of the group). Critically, in the concatenated tree, where monophyly of species can be assessed, each of these species was monophyletic based on geographically dispersed sampling (Figure 1b). This is in stark contrast to the mtDNA gene tree (Figure 3). We report here analysis of 231 mitochondrial cytochrome b sequences from members of this group (including 43 sequences first published by Reid et al. (2012)): 8 T. canipes sequences (5 novel, not previously published); 13 T. rufus (10 novel); 64 T. umbrinus (53 novel); 54 T. dorsalis (42 novel); 67 T. quadrivittatus (61 novel); and 25 T. cinereicollis sequences (17 novel). Collection localities and Genbank accession numbers are listed in Supplementary Material. The only species that are monophyletic with respect to their mtDNA gene tree are T. canipes and T. rufus, two species with allopatric distributions or that lack documented contact zones (Clades A and B: Figure 3). The other four taxa have partially overlapping distributions (as well as documented contact zones) and are strongly polyphyletic with respect to the mtDNA gene tree. There is one haplogroup (Clade C in Figure 3) that is restricted to central Colorado and Wyoming. T. dorsalis, T. quadrivittatus and T. umbrinus variously overlap in that region, and this haplogroup is found in all three of these species; we cannot identify to which species this haplogroup is native, and it is possible that this haplogroup has been introgressed from an extinct taxon (that is, a ghost lineage). Throughout the rest of the gene tree, it is possible to identify haplogroups primarily affiliated with individual species, but each of these is also carried by individuals from other species (for example, Clade D; Figure 3). A remarkable example of this is T. dorsalis; wherever it comes into contact with one of the other species, individuals carry that species' mtDNA. Conversely, where T. dorsalis occurs in isolated populations (for example, southern Arizona sky islands), individuals carry unique, T. dorsalis-specific haplotypes (Figure 3). Thus, it appears that mtDNA introgression is widespread in the T. quadrivittatus group and coalescent simulations (Reid et al., 2012) exclude incomplete lineage sorting as the source of incongruence between the species tree and the mtDNA gene tree.
Figure 3.
The mtDNA gene tree for the T. quadrivittatus group species from the central and southern Rockies estimated from 231 individuals were collected largely from our own fieldwork. Sequences were trimmed and DT-ModSel (Minin et al., 2003), which selects among models of nucleotide sequence evolution using a decision-theoretic approach, indicated the HKY+Γ model. Phylogenetic inference was obtained using MrBayes 3.1.2 (Ronquist and Huelsenbeck, 2003). Two independent runs of seven million generations, sampling every 500, were performed. Convergence was assessed through visual inspection of plots of log-likelihood scores vs generation, high effective sample sizes, a low (<0.01) s.d. of split frequencies among runs and a potential scale reduction factor near unity for all parameter estimates. A majority-rule consensus tree was generated from the posterior distribution of topologies after removing 25% of samples as a burn-in.
A more direct comparison of intogression can be conducted by direct estimation of gene flow (that is, interspecific migration rates) for mtDNA using MIGRATE-N (Beerli, 2009). Initial, relatively short runs were conducted for all six species; however, both because of monophyly of and small samples for T. canipes and T. rufus, the likelihood surfaces for θ and M were flat for these two species and they were excluded from the longer runs. Further, because Strasburg and Rieseberg (2010) have shown that ignoring unsampled phylogenetic structure does not compromise estimates of gene flow, we estimated migration for all pairwise comparisons of the four overlapping species. For each analysis we ran five Metropolis-coupled chains for 10 000 000 generations, after a burn-in of 1 000 000 generations.
The migration rates estimated from mtDNA (Nm=θ × M) suggest substantial migration (that is, introgression) between T. umbrinus and T. dorsalis (Table 2). Similar high levels of interspecific mtDNA gene flow were obtained for analyses of other pairs of taxa in this group (Table 2). In fact, these analyses support the observational conclusion from the phylogeny that T. dorsalis has received introgressed mtDNA from three different species: T. umbrinus, T. quadrivittatus and T. cinereicollis. Similarly, T. umbrinus has apparently contributed its mtDNA to the other three species. Thus, it is very clear that there is extensive mtDNA introgression occurring in this group on a time scale that is faster than the mutation rate of mtDNA because identical tip haplotypes are shared between and among species.
Table 2. Direct estimates of mtDNA introgression, Nm (95% CI), in the T. quadrivittatus group.
| Source species |
Receiving species |
|||
|---|---|---|---|---|
| cinereicollis | dorsalis | quadrivittatus | umbrinus | |
| cinereicollis | — | 1.768 (0.77–3.14) | 2.3 × 10−9 (∼0–0.38) | 1.148 (0.55–2.05) |
| dorsalis | 0.326 (0.05–1.01) | — | 0.796 (0.25–1.96) | 0.383 (0.10–0.98) |
| quadrivittatus | 0.326 (0.05–1.01) | 1.010 (0.31–2.35) | — | 2.55 × 10−16 (∼0–0.02) |
| umbrinus | 1.304 (0.60–2.42) | 2.020 (0.92–3.76) | 1.393 (0.60–2.69) | — |
Abbreviations: CI, confidence interval; mtDNA, mitochondrial DNA.
T. minimus introgressions
The least chipmunk (T. minimus) is the most widely distributed of the western chipmunks (Figure 2c), and occurs in the widest variety of habitats in North America (Verts and Carraway, 2001). To date, we have sequenced several hundred individuals (Demboski et al., in prep), and it is clear that the species is not monphyletic with respect to its mtDNA. In addition to the possible presence of an unrecognized species (probably corresponding to the subspecies T. m. girsescens; Reid et al., 2012), T. panamintinus (Panamint chipmunk), T. speciosus (lodgepole chipmunk) and T. alpinus (alpine chipmunk) samples are entirely composed of individuals whose mtDNA sequences are nested within those of T. minimus sequences (Reid et al., 2012). The mtDNA sequences for the Panamint and lodgepole chipmunks each form monophyletic groups, suggesting the possibility of fixation on a single, ancient introgression akin to the T. a. canicaudus scenario discussed above. Furthermore, Reid et al. (2012) conducted coalescent simulations and were able to reject phylogenetic uncertainty and incomplete lineage sorting (that is, mutational and coalescent stochasticity) as explanations, at least for the nesting of lodgepole and Panamint chipmunk sequences within T. minimus. Panamint and lodgepole chipmunks are each monophyletic with respect to their nuclear genes (Reid et al., 2012), and they are sister taxa that are not particularly closely related to T. minimus in the species-tree estimates (Figure 1); rather, the speciosus–panamintinus clade is sister to a clade containing two geographically adjacent species, T. obscurus and T. merriami, which occur in southern California into Mexico (Reid et al., 2012). Panamint chipmunks are primarily restricted to pinyon-juniper habitats in the southwestern Great Basin and contact T. minimus at the transitions between pinyon and sagebrush (Best et al., 1994a). Lodgepole chipmunks typically occur (obviously) in lodgepole pine habitats at moderate to high elevations in western and south-central California (Best et al., 1994b). T. minimus and T. speciosus come into contact at several places along the Eastern Sierras (Figure 2c), but no recent/current mtDNA introgression has been documented (JL Patton, personal communication).
Interestingly, individuals of the narrow endemic alpine chipmunk (T. alpinus) have one of a number of mtDNA haplotypes that are nested within those found in nearby populations of T. minimus (roughly corresponding to T. m. scrutator). It is also the case that T. alpinus nuclear sequences are nested within those of T. minimus (Figure 1b; Reid et al., 2012). This suggests that T. alpinus is likely a very recent offshoot of T. minimus isolated in the upper elevations of the eastern Sierra Nevada. The two species have strongly diverged bacula, and we hypothesize that they actually provide an example of incomplete lineage sorting (rather than hybridization), a conclusion we are testing with genomic data.
Summary
Thus, the conclusion that has emerged from over a decade's worth of intensive field collecting, generation of mtDNA sequences and a small set of nuclear loci (sequences and microsatellites) is that ecological, morphological and behavioral differentiation have in many instances outpaced the evolution of complete reproductive isolation in the western North American chipmunks. Therefore, DGF has played a role in this radiation, and we can putatively allocate different introgression events to various stages along the continuum of speciation.
The introgression between T. r. ruficaudus and T. r. simulans fits the expectations of early speciation. This applies to both the older Lochsa Zone (Good and Sullivan, 2001; Hird and Sullivan, 2009), as well as the more recent Whitefish Zone (Hird et al., 2010), where gene flow is occurring at both mtDNA and microsatellite loci. Genome-wide data will permit more accurate estimates of gene flow and timing of divergence.
In intermediate stages of speciation, more of the genome may be resistant to introgression, but there still may be substantial introgression. The extensive mtDNA introgressions we have identified between (and among) the predominantly parapatric members of the T. quadrivittatus group (T. umbrinus, T. dorsalis, T. quadrivittatus and T. cinereicollis) in the central/southern Rockies and Great Basin (Figure 3, Table 2) fit the expectations for intermediate stages. To date, we only have data from four nuclear loci from which to make inferences regarding gene flow among these taxa, and likelihood surfaces are too flat to make direct inferences of gene flow from these four loci (results not shown). If genome-wide data fit the predictions of DGF, this group may provide an important and rarely examined stage in the speciation process, where ecological and morphological differentiation have been solidly (and perhaps irreversibly) established, but where neutral loci may be subject to moderate gene flow. This may be particularly important because the species-tree estimate produced by Reid et al. (2012), especially the *BEAST tree and the concatenated tree shown in Figure 1, provides a temporal scale to testing the predictions within the group.
Later in divergence, blocks of the genome that are resistant to introgression may have grown, such that any hybridization results only in very small amounts of introgression. This therefore may be a difficult phase to identify, but it appears that the northern Rockies introgression between T. r. ruficaudus and T. a. luteiventris exhibits this pattern. There is restricted introgression of mtDNA along the spine of the northern Rockies along the BC/Alberta border (Good et al., 2003), no detectable introgression at the nuclear locus acrosin and a mere suggestion of a late generation backcross at microsatellite loci (Reid et al., 2010). Again, detection of divergence at this stage may require data of the scale only possible via genomic approaches.
The final stage of DGF, where reproductive isolation is complete and there are no genomic regions introgressing, is common and easy enough to identify. Perhaps the most interesting cases are those that exhibit evidence of ancient post-divergence introgression between taxa that currently are reproductively isolated. The best-documented example of this in the Tamias radiation involves fixation on an ancient mtDNA introgression in T. a. canicaudus (the Palouse Introgression; Good et al., 2003, 2008). Here, T. a. canicaudus is fixed for a haplogroup that is sister to haplotypes found in T. r. simulans, whereas the microsatellite data and nuclear sequences indicate that there is no current gene flow between sympatric populations and that there is little chance that anomalous lineage sorting can explain the pattern (Good et al., 2008). Other possible examples are the apparent fixations on introgressed T. minimus mtDNA in T. panamintinus and T. speciosus (Table 2; Reid et al., 2012), although we currently lack extensive nuclear data to test these hypotheses further.
One of the more intriguing aspects of DGF in Tamias is the mosaic nature of some widespread taxa with respect to completion of reproductive isolation. T. a. canicaudus and T. r. simulans appear to have evolved complete reproductive isolation (Good et al., 2003, 2008), but T. a. luteiventris and T. r. ruficaudus appear to have experienced a recent introgression event and may be currently exchanging genes at a very low rate (Good et al., 2003; Reid et al., 2010). Although speculative, this contrast could be related to the length of time that each pair has been in contact with each other. In the case of T. a. canicaudus and T. r. simulans, conditions of intermittent parapatry have probably recurred throughout much of the Pleistocene. Good et al. (2008) estimated a mid-to-late Pleistocene date for the Palouse Introgression event and the two taxa partition habitats across a broad range of sympatry. In this case, therefore, the evolution of reproductive character displacement (or specifically reinforcement if there is selection against hybrids) has perhaps had sufficient time to effect the completion of reproductive isolation. Conversely, in the Northern Rockies Introgression, forest habitat for any species of chipmunk has only been established for ∼3000 years (after glacial retreat) and T. a. luteiventris and T. r. ruficaudus have probably not experienced a long history of parapatry; it is therefore possible that selection has had insufficient time to influence reproductive isolation between this pair of taxa. This mosaic nature of reproductive isolation is perhaps further enabled because each of the two species involved, T. amoenus and T. ruficaudus, exhibit populations that are highly structured across geography (Demboski and Sullivan, 2003; Good and Sullivan, 2001, respectively).
DGF also appears to be operating in other mammalian radiations. For example, the genus Lepus (hares) has been documented to exhibit similar introgressions to those described in Tamias. In particular, Melo-Ferriera et al. (2012) have demonstrated introgression between non-sister pairs of species, including species-wide fixation on introgressed mtDNA (for example, in L. castroviojoi and L. corsicanus). Thus, DGF may prove to be relatively more common during mammalian speciation than is often thought and additional examples will almost certainly be discovered. However, it is important to note that most evidence for introgression in mammals comes from the study of mtDNA and perhaps a small number of other genetic markers. The nonrecombinant maternal inheritance and rapid evolution of mtDNA make it a powerful population genetic marker (for example, Zink and Barrowclough, 2008), but mtDNA is also highly prone to asymmetric and extensive introgression (for example, Petit and Excoffier, 2009; Toews and Brelsford, 2012). As even rare hybridization events can yield extensive mtDNA gene flow (Currat et al., 2008), it is likely that mtDNA provides a highly biased view of overall genetic contribution of introgressive hybridization. Thus, an important question going forward is to determine both the frequency and overall genetic contribution of introgressive hybridization in chipmunks and other mammals.
Future directions
In addition to testing the hypotheses discussed above, a number of remaining questions arise. First, these hypotheses rest on phylogenies (Figure 1) estimated from reproductive protein coding genes (Reid et al., 2012). As these are based on just four loci, additional phylogenomic data and additional sampling of species are required to provide a better topological and temporal framework for testing DGF in chipmunks. Further, a better understanding of species population history and contact zones is also critical in moving forward. Second, what is the role of sexual selection (for example, female choice), and perhaps genital evolution and sperm competition in governing chipmunk speciation (Hosken and Stockley, 2004; Ramm, 2007)? Third, what genes or genomic regions are prone to introgression versus resistant to gene flow? Finally, what is the role of genomic islands of divergence and what are the dynamics of selection and linkage in shaping the genomic structure of introgression (for example, Noor and Bennett, 2009; Nosil and Feder, 2012)? Most of these issues will require some form of comparative genomic data to be addressed. Powerful next-generation sequencing techniques now enable the collection of large-scale genomic data in non-model species (Good, 2011), including the recent development of large-scale targeted resequencing tools for chipmunks (Bi et al., 2012, 2013). With these resources in hand, we anticipate that the next decade of research should resolve many of these questions.
Data archiving
GenBank accession numbers are given for each individual in Supplementary Table S1 of the Supplementary material. Nexus files available from the Dryad Digital Repository: doi:10.5061/dryad.52mp1.
Acknowledgments
We thank the following for assistance in the field over several years: W Bell, I Demboski, M Fraker, D Good, P Good, J Good, J Harper, A Hornsby, S Poler, A Runck, D Sullivan and the 1999-03 University of Idaho Mammalogy classes. We thank various museums and individuals for loans of tissue samples, including the Denver Museum of Nature & Science, Museum of Southwestern Biology (J Cook), Humboldt State University Vertebrate Museum, University of Alaska Museum (L Olson), Burke Museum (J Bradley), Museum of Vertebrate Zoology (J Patton) and D Burkett. Peter Beerli and two anonymous reviewers have provided advice and comments that improved the paper greatly. This research was conducted in compliance with University of Idaho Animal Care and Use Committee under protocol UIACUC-2005-40 and was supported by a seed grant from the University of Idaho Research Foundation, the NSF EPSCoR program (NSF cooperative agreement number EPS-9720634), the Institute for Bioinformatics and Evolutionary Studies (IBEST) at the University of Idaho (by NIH NCRR 1P20RR016454-01; NIH NCRR 1P20RR016448-01; NSF EPS-809935), NSF DEB-0717426, NSF DEB-0716200, NSF Cooperative Agreement No. DBI-0939454 and the Denver Museum of Nature & Science. JS and BS received funding through BEACON, an NSF-funded Center for the Study of Evolution in Action (DBI-0939454). Any opinions, findings and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Baker RJ, Bradley LC, Dragoo JW, Engstrom MD, Hoffman RS, Jones CA et al. (2003). Revised checklist of North American Mammals North of Mexico. Occas Pap Mus Texas Tech Univ 229: 1–24. [Google Scholar]
- Beerli P. (2009). How to use MIGRATE, or why are Markov chain Monte Carlo programs difficult to use? In: Bertoelle G, Bruford MW, Hauffe HC, Rizzoli A, Versesi C (eds). Population Genetics for Animal Conservation, vol. 17 of Conservation Biology. Cambridge University Press: Cambridge, UK, pp 42–79.
- Bergeron P, Montiglio P-O, Réale D, Humphries MM, Garant D. (2012). Bateman gradients in a promiscuous mating system. Behav Ecol Sociobiol 66: 1125–1130. [Google Scholar]
- Bergstrom BJ, Hoffmann RS. (1991). Distribution and diagnosis of three species of chipmunks (Tamias) in the Front Range of Colorado. Southwest Nat 36: 14–28. [Google Scholar]
- Bergstrom BJ. (1992). Parapatry and competition between chipmunk (Tamias) species and the hypothesized role of parasitism. Am Midl Nat 128: 168–179. [Google Scholar]
- Best TL. (1993). Tamias ruficaudus. Mamm Species 452: 1–7. [Google Scholar]
- Best TL, Granai NJ. (1994. a). Tamias obscurus. Mamm Species 472: 1–6. [Google Scholar]
- Best TL, Granai NJ. (1994. b). Tamias merriami. Mamm Species 476: 1–9. [Google Scholar]
- Best TL, Clawson RG, Clawson JA. (1994. a). Tamias panamintinus. Mamm Species 468: 1–7. [Google Scholar]
- Best TL, Clawson RG, Clawson JA. (1994. b). Tamias speciosus. Mamm Species 478: 1–9. [Google Scholar]
- Bi K, Linderoth T, Vanderpool D, Good JM, Nielsen R, Moritz C. (2013). Unlocking the vault: next-generation museum population genomics. Mol Ecol 22: 6018–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi K, Vanderpool D, Singhal S, Linderoth T, Moritz C, Good JM. (2012). Transcriptome-based exon capture enables highly cost-effective comparative genomic data collection at moderate evolutionary scales. BMC Genomics 13: 403–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship DJ. (1985) Reproductive isolating mechanisms of southern California chipmunks: a systematic evaluation of Tamias obscurus Allen, 1890 and T. merriami Allen, 1890 (Rodentia: Sciuridae). PhD dissert, Loma Linda University, Loma Linda, CA, USA, pp 137.
- Brown JH. (1971). Mechanisms of competitive exclusion between two species of chipmunks. Ecology 52: 305–311. [Google Scholar]
- Callahan JR. (1977). Diagnosis of Eutamias obscurus (Rodentia: Sciuridae). J Mammal 58: 188–201. [Google Scholar]
- Callahan JR. (1981). Vocal solicitation and parental investment in female Eutamias. Am Nat 118: 872–875. [Google Scholar]
- Chappell MA. (1978). Behavioral factors in the altitudinal zonation of chipmunks (Eutamias). Ecology 59: 565–579. [Google Scholar]
- Compton SB, Callahan JR. (1995). Reproductive behavior in Merriam's chipmunk (Tamias merriami). Great Basin Nat 55: 89–91. [Google Scholar]
- Currat M, Ruedi M, Petit RJ, Excoffier L. (2008). The hidden side of invasions: massive introgression by local genes. Evolution 62: 1908–1920. [DOI] [PubMed] [Google Scholar]
- Dalquest WW, Baskin JA, Schultz GE. (1996). Fossil mammals from a late Miocene (Clarendonian) site in Beaver County, Oklahoma. In: Genoways HH, Baker RJ (eds). Contribution in Mammalogy. A Memorial Volume Honoring Dr. J. Konx Jones, Jr. Museum of Texas Tech University: Lubbock, TX, USA, pp 107–137.
- Daubenmire R. (1975) Floristic Plant Geography of Eastern Washington and Northern Idaho. Brigham Young University Press: Provo, UT, USA. [Google Scholar]
- Demboski JR, Sullivan J. (2003). Extensive mtDNA variation within the yellow-pine chipmunk, Tamias amoenus (Rodentia: Sciuridae), and phylogeographic inferences for northwest North America. Mol Phylogenet Evol 26: 389–408. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambeaut A. (2012). Bayesian phylogenetic with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis LS, Maxson LR. (1979). Evolution of the chipmunk genera Eutmias and Tamias. J Mammal 60: 331–334. [Google Scholar]
- Fitzpatrick BM, Turelli M. (2006). The geography of mammalian speciation: mixed signals from phylogenies and range maps. Evolution 60: 601–615. [PubMed] [Google Scholar]
- Feder JL, Nosil P. (2010). The efficacy of divergence hitchhiking in generating genomic islands during ecological speciation. Evolution 64: 1729–1747. [DOI] [PubMed] [Google Scholar]
- Gambs RD. (1965) Zoogeography of Chipmunks in the Pacific Northwest. MS Thesis, University of Idaho, Moscow, ID, USA, p 109.
- Gannon WL, Lawlor TE. (1989). Variation of the chip vocalization of three species of Townsend chipmunks (genus Eutamias). J Mammal 70: 740–753. [Google Scholar]
- Good J. (2011). Reduced representation methods for subgenomic enrichment and next-generation sequencing. In: Orgogozo V, Rockman MV (eds). Methods in Molecular Biology, vol. 772. Humana Press: New York, NY, USA. pp 85–103. [DOI] [PubMed]
- Good JM, Sullivan J. (2001). Phylogeography of red-tailed chipmunks (Tamias ruficaudus), a northern Rocky Mountains endemic. Mol Ecol 10: 2683–2696. [DOI] [PubMed] [Google Scholar]
- Good JM, Demboski JR, Nagorsen DM, Sullivan J. (2003). Phylogeography and introgressive hybridization: chipmunks (Tamias) in the northern Rocky Mountains. Evolution 57: 1900–1916. [DOI] [PubMed] [Google Scholar]
- Good J, Hird S, Reid N, Demboski J, Steppan SJ, Sullivan J. (2008). Ancient introgression and mtDNA capture in non-sister species of chipmunks (Tamias). Mol Ecol 17: 1313–1327. [DOI] [PubMed] [Google Scholar]
- Grinnell J, Storer TI. (1924) Animal Life in the Yosemite. University of California Press: Berkeley, CA, USA. [Google Scholar]
- Hall ER. (1981) The Mammals of North America 2nd edn. John Wiley & Sons, Inc.: New York, NY, USA. [Google Scholar]
- Heled J, Drumond AJ. (2010). Bayesian inference of species trees from multilocus data. Mol Biol Evol 27: 570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller HC. (1970) Altitudinal zonation of chipmunks (Eutamias): interspecific aggression, water balance, and energy budgets. PhD Thesis, Yale University, New Haven, CT, USA.
- Heller HC. (1971). Altitudinal zonation of chipmunks (Eutamias): interspecific aggression. Ecology 52: 312–319. [Google Scholar]
- Heller HC, Gates DM. (1971). Altitudinal zonation of chipmunks (Eutamias): energy budgets. Ecology 52: 424–433. [Google Scholar]
- Hird S, Sullivan J. (2009). Assessment of gene flow across a hybrid zone in red-tailed chipmunks (Tamias ruficaudus). Mol Ecol 18: 3097–3109. [DOI] [PubMed] [Google Scholar]
- Hird S, Reid N, Demboski JR, Sullivan J. (2010). Introgression at differentially aged hybrid zones in red-tailed chipmunks. Genetica 138: 869–883. [DOI] [PubMed] [Google Scholar]
- Hosken DJ, Stockley P. (2004). Sexual selection and genital evolution. Trends Ecol Evol 19: 87–93. [DOI] [PubMed] [Google Scholar]
- Howell AH. (1929). Revision of the American chipmunks (genera Tamias and Eutamias). North Am. Fauna 52: 1–157. [Google Scholar]
- Jameson EW. (1999). Host-ectoparasite relationships among North American chipmunks. Acta Theriol 44: 225–231. [Google Scholar]
- Jiggins CD. (2008). Sympatric speciation: why the controversy? Curr Biol 16: R333–R334. [DOI] [PubMed] [Google Scholar]
- Johnson DH. (1943). Systematic review of the chipmunks (genus Eutamias) of California. Univ Calif Publ Zool 48: 63–148. [Google Scholar]
- Lanfear R, Calcott B, Ho SWY, Guindon S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol 29: 1695–1701. [DOI] [PubMed] [Google Scholar]
- Levenson H, Hoffmann RS, Nadler CF, Deutsch L, Freeman SD. (1985). Systematics of the Holarctic chipmunks (Tamias). J Mammal 66: 219–242. [Google Scholar]
- Lomolino MV, Riddle BR, Brown JH. (2006) Biogeography 3rd edn Sinauer Associates. Sunderland, MA, USA, p 752. [Google Scholar]
- Mack RN, Rutter NM, Bryant VM Jr, Valastro S. (1978). Reexamination of postglacial vegetation history in northern Idaho: Hager Pond, Bonner Co. Quat Res 10: 241–255. [Google Scholar]
- Melo-Ferriera J, Boursot P, Carneiro M, Esteves PJ, Farelo L, Alves PJ. (2012). Recurrent introgression of mitochondrial DNA among hares (Lepus spp.) revealed by species-tree inference and coalescent simulations. Syst Biol 61: 367–381. [DOI] [PubMed] [Google Scholar]
- Merriam CH. (1897). Notes on the chipmunks of the genus Eutamias occurring west of the east base of the Cascade-Sierra system, with descriptions of new forms. Proc Biol Soc Wash 11: 189–212. [Google Scholar]
- Minin V, Abdo Z, Joyce P, Sullivan J. (2003). Performance-based selection of likelihood models for phylogeny estimation. Syst Biol 52: 674–683. [DOI] [PubMed] [Google Scholar]
- Noor MAF, Bennett SA. (2009). Island of speciation or mirages in the desert? Examining the role of reduced recombination in maintaining species. Heredity 103: 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Feder JL. (2012). Genomic divergence during speciation: causes and consequences. Philos Trans R Soc Lond B Biol Sci 367: 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson BD, Heaney LR. (1987). Preliminary analysis of geographic variation in red-tailed chipmunks (Eutamias ruficaudus). J Mammal 68: 782–791. [Google Scholar]
- Patterson BD, Thaeler CS. (1982). The mammalian baculum: hypotheses on the nature of bacular variability. J Mammal 63: 1–15. [Google Scholar]
- Pecoud J, Ollivier A, Plantegenest M, Simon J-C. (2009). A continuum of genetic divergence from sympatric host races to species in the pea aphid complex. Proc Natl Acad Sci USA 106: 7495–7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit RJ, Excoffier L. (2009). Gene flow and species delimitation. Trends Ecol Evol 24: 386–393. [DOI] [PubMed] [Google Scholar]
- Piaggio AJ, Spicer GS. (2000). Molecular phylogeny of the chipmunk genus Tamias based on the mitochondrial cytochrome oxidase subunit II gene. J Mammal Evol 7: 147–166. [Google Scholar]
- Piaggio AJ, Spicer GS. (2001). Molecular phylogeny of the chipmunks inferred from mitochondrial cytochrome b and cytochrome oxidase II gene sequences. Mol Phylogenet Evol 20: 335–350. [DOI] [PubMed] [Google Scholar]
- Pinho C, Hey J. (2010). Divergence with gene flow: models and data. Annu Rev Ecol Evol Syst 41: 215–230. [Google Scholar]
- Ramm SA. (2007). Sexual selection and genital evolution in mammals: a phylogenetic analysis of baculum length. Am Nat 169: 360–369. [DOI] [PubMed] [Google Scholar]
- Reid N, Hird S, Schulte-Hostedde A, Sullivan J. (2010). Examination of nuclear loci across a zone of mitochondrial introgression between Tamias ruficaudus and T. amoenus. J Mammal 91: 1389–1400. [Google Scholar]
- Reid N, Demboski JR, Sullivan J. (2012). Phylogeny estimation of the radiation western American chipmunk (Tamias) in the face of introgression using reproductive protein genes. Syst Biol 61: 44–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR, Hostert EE. (1993). Laboratory studies on speciation: what have we learned in 40 years. Evolution 47: 1637–1653. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003). Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Schulte-Hostedde AI, Millar JS. (2004). Intraspecific variation of testis size and sperm length in the yellow-pine chipmunk (Tamias amoenus): implications for sperm competition and reproductive success. Behav Ecol Sociobiol 55: 272–277. [Google Scholar]
- Schulte-Hostedde AI, Millar JS, Gibbs HL. (2004). Sexual selection and mating patterns in a mammal with female-biased sexual size dimorphism. Behav Ecol 15: 351–356. [Google Scholar]
- Smadja CM, Butlin RK. (2010). A framework for comparing processes of speciation in the presence of gene flow. Mol Ecol 20: 5123–5140. [DOI] [PubMed] [Google Scholar]
- Strasburg JL, Rieseberg LH. (2010). How robust are “isolation with migration” analyses to violations of the IM model? A simulation study. Mol Biol Evol 27: 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan RM, Petersen KE. (1988). Systematics of southwestern populations of least chipmunks (Tamias minimus) reexamined: a synthetic approach. Occ Pap Mus Southwestern Biol 5: 1–27. [Google Scholar]
- Sutton DA. (1992). Tamias amoenus. Mamm Species 390: 1–8. [Google Scholar]
- Sutton DA, Patterson BD. (2000). Geographic variation of the western chipmunks Tamias senex and T. siskiyou, with two new subspecies from California. J Mammal 81: 316. [Google Scholar]
- Taylor EB, Boughman JW, Groenenboom M, Sniatynski M, Schluter D, Gow JL. (2006). Speciation in reverse: morphological and genetic evidence of the collapse of a three-spined stickleback (Gasterosteus aculeatus) species pair. Mol Ecol 15: 343–355. [DOI] [PubMed] [Google Scholar]
- Toews DPL, Brelsford A. (2012). The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol 21: 3907–3930. [DOI] [PubMed] [Google Scholar]
- Verts BJ, Carraway LN. (2001). Tamias minimus. Mamm Species 653: 1–10. [Google Scholar]
- White JA. (1953). The baculum in the chipmunks of western North America. Univ Kansas Pub Mus Nat Hist 5: 611–631. [Google Scholar]
- Wu CI. (2001). The genic view of the process of speciation. J Evol Biol 14: 851–865. [Google Scholar]
- Zink RMR, Barrowclough GFG. (2008). Mitochondrial DNA under siege in avian phylogeography. Mol Ecol 17: 2107–2121. [DOI] [PubMed] [Google Scholar]
- Zitari A, Scopece G, Helal AN, Widmer A, Cozzolino S. (2012). Is floral divergence sufficient to maintain species boundaries upon secondary contact in Mediterranean food-deceptive orchids? Heredity 108: 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.