Abstract
We introduce Pepper (Protein complex Expansion using Protein–Protein intERactions), a Cytoscape app designed to identify protein complexes as densely connected subnetworks from seed lists of proteins derived from proteomic studies. Pepper identifies connected subgraph by using multi-objective optimization involving two functions: (i) the coverage, a solution must contain as many proteins from the seed as possible, (ii) the density, the proteins of a solution must be as connected as possible, using only interactions from a proteome-wide interaction network. Comparisons based on gold standard yeast and human datasets showed Pepper’s integrative approach as superior to standard protein complex discovery methods. The visualization and interpretation of the results are facilitated by an automated post-processing pipeline based on topological analysis and data integration about the predicted complex proteins. Pepper is a user-friendly tool that can be used to analyse any list of proteins.
Availability: Pepper is available from the Cytoscape plug-in manager or online (http://apps.cytoscape.org/apps/pepper) and released under GNU General Public License version 3.
Contact: mohamed.elati@issb.genopole.fr
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Most cellular processes require a large number of proteins to assemble into functional complexes to perform their activity. Therefore, describing functional protein complexes taking part in given processes is critical to the underlying molecular mechanism understanding. Experimental protocols such as Affinity Purification followed by Mass-Spectrometry (AP-MS) have been devised to pull down a protein of interest (bait) together with all the interacting proteins within the same protein complex (preys). However, these sets of preys may contain both false positives, proteins detected despite not actually interacting with the bait, and omit false negatives (Gingras et al., 2007), proteins interacting in the cellular context studied but not detected. Effective control experiments and usage of contaminants repositories can remove some false positives. However, false negative interacting partners identification, thereby the definition of the entire protein complex, remains challenging. Protein–Protein Interaction (PPI) data represents abundant information that can be used for this purpose.
Protein complexes extraction from PPI networks is a very active area of research and many methodologies have been developed to tackle this problem. These computational methods generally model protein complexes as dense subnetworks within the complete set of PPIs and thus try to solve a graph clustering problem or to identify dense regions. Clustering approaches were shown to be efficient either on large PPI networks or with large-scale experimental settings in which big numbers of baits result in context-specific PPI networks (Bader and Hogue, 2003; Nepusz et al., 2012). However, these algorithms were not developed for use in small-scale AP-MS experiments (e.g. using only a single bait protein) and are unable to integrate experimental data with repositories of PPI.
We reasoned that although not all the protein partners may be detected in a given AP-MS experiment, these proteins may have been previously identified as interacting with either the bait or some of the preys of the experiment. Based on this hypothesis, we developed Pepper, which addresses the problem of finding protein complexes by combining the experimental results of a single AP-MS assay with the available information from protein interactions in a global PPI network. Pepper solves this non-trivial problem by using a multi-objective evolutionary algorithm (Elati et al., 2013), which was tested to demonstrate the relevance of our integrative approach. To do so, we used publicly available AP-MS datasets for yeast and human species and compared Pepper’s results with those of state-of-the-art protein complex discovery methods. Our findings highlight the relevance of integrating PPI repositories to the analysis of AP-MS experiments. We propose Pepper as a Cytoscape application to further refine protein complex predictions through functional and topological analyses.
2 METHODS AND IMPLEMENTATION
In the context of a single AP-MS experiment, Pepper aims to identify a dense subnetwork within the PPI network connecting as many of the proteins identified in this experiment as possible, referred to hereafter as the list of seed proteins. Pepper solves this problem by maximizing two objective functions: (i) coverage, a solution must contain as many proteins from the seed protein list as possible; (ii) density, a solution must contain as many interactions as possible. These objectives are often conflicting, and thus, no single solution can be considered to dominate over the others. Instead, the optimal solution is a Pareto optimal set with multiple solutions. SPEA2 (Zitzler et al., 2001), a popular Multi-Objective Evolutionary Algorithm, is used for the simultaneous optimization of the two objective functions and to identify solutions approximating the set of pareto-optimal solutions. These solutions are merged into a final predicted protein complex by maximizing the modularity with a greedy search (see SI algorithm section).
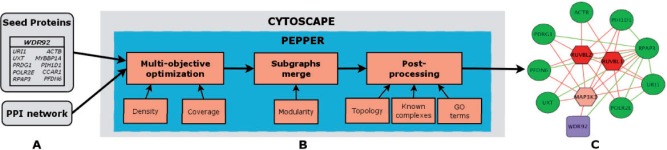
Pepper was developed as a Cytoscape application, which uses a seed list of proteins and a large-scale PPI network as inputs (Fig. 1A). In addition to the aforementioned subnetwork extraction procedure, Pepper includes a topological and function-based post-processing pipeline for ranking the added proteins (expansions) according to their relevance (Fig. 1B). The predicted complex and each of the proteins are annotated based on their cellular localization or function annotation specificity. Enrichment analysis is complemented by matching the solutions to a collection of reference protein complexes, and expansions are scored according to their co-occurrence with the seeds in these complexes. Topological scoring is based on the impact of the expansions on the overall connectivity of the subnetwork (see SI post-processing section). Pepper uses these scores to rank expansions and to facilitate results visualization and interpretation (Fig. 1C).
Fig. 1.
Schematic representation of the plug-in. (A) Example of input data, a large-scale PPI and the results of an AP-MS experiment with the bait and a list of prey proteins. (B) Context-specific protein complex extraction pipeline. (C) Output subnetwork representing a putative protein complex using only interactions from the input PPI network: example of WDR92. Purple squares and green circles correspond to bait and prey proteins, respectively. Hexagons indicate the expansions proposed by PEPPER and are shown in various shades of red, according to their post-processing score. Dark red indicates a high predicted relevance to the solution. The edges shown in the graph are exclusively those found in the input PPI network. Green edges are set between seed proteins. All edges involving an expansion protein are red
3 CASE STUDY
We assessed the performance of Pepper and two network clustering algorithms for protein complex discovery—MCODE (Bader and Hogue, 2003) and ClusterONE (Nepusz et al., 2012)—on a benchmark dataset of 135 yeast and 9 human single-bait AP-MS experiments and using a set of hand-curated protein complexes as gold standards. For network clustering methods, performance was assessed for each AP-MS experiment by selecting the predicted complex which best matched the seed (details in SI performance comparison section). For each experiment, the reference complex from the gold standard best matching the seed was used as the ground truth in a binary classification task. Compared with both of the clustering methods tested, the complexes predicted by Pepper scored higher in all of the performance measures for both organisms (details in SI performance comparison section) with notably an average increase of 16% of the geometric accuracy in human and 12% in yeast.
As an example, we describe here the results obtained for the human WDR92 protein. In the initial list of preys, WDR92 was identified as interacting with only one protein. Pepper expanded the seed with three new proteins (Fig. 1C) and greatly increased the overall density of the original solution (22 to 47%). The new expansion proteins were ordered on the basis of post-processing score. The first two proteins, RUVBL1 and RUVBL2, have both a high topological and Gene Ontology score. The lower scored protein, MAP3K3, still remains relevant according to its high topological score (connected to >90% of the predicted complex proteins). AP-MS experiments using RUVBL1 or RUVBL2 as baits both identified WDR92 as a prey protein (Choi et al., 2010). Moreover, in the raw WDR92 experimental data, the set of preys with lower processing scores (based on peptide counts) than the threshold contains RUVBL1 (see SI Case study section). Thus, the application of Pepper to this experiment led to the recovery of proteins that would not have been identified otherwise (potential false negatives).
Overall, these results demonstrate the feasibility of expanding the protein complexes identified in an AP-MS experiment through the use of PPI networks and the value of Pepper for this purpose.
Funding: This work was supported by the French National Cancer Institute (INCa_2960: PLBIO10) and the European Union/Framework Programme 7/2009 (“SYSCILIA” consortium, grant 241955). Funding for open access charge: SYSCILIA.
Conflict of interest: none declared.
Supplementary Material
REFERENCES
- Bader GD, Hogue CWV. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, et al. Saint: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods. 2010;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elati M, et al. Multi-objective optimization for relevant sub-graph extraction. In: Nicosia G, Pardalos P, editors. Learning and Intelligent Optimization (LION'7), LNCS. Vol. 7997. Springer, Berlin Heidelberg; 2013. pp. 104–109. [Google Scholar]
- Gingras A-C, et al. Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- Nepusz T, et al. Detecting overlapping protein complexes in protein-protein interaction networks. Nat. Methods. 2012;9:471–472. doi: 10.1038/nmeth.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitzler E, et al. SPEA2: Improving the strength Pareto evolutionary algorithm. Technical report, Athens, Greece. 2001 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.