Abstract
How metastases develop is not well understood and no genetic mutations have been reported as specific metastatic drivers. Here we have addressed the idea that epigenetic reprogramming by GLI-regulated pluripotent stemness factors promotes metastases. Using primary human colon cancer cells engrafted in mice, we find that transient expression of OCT4, SOX2, KLF4 +/− cMYC establishes an enhanced pro-metastatic state in the primary tumor that is stable through sequential engraftments and is transmitted through clonogenic cancer stem cells. Metastatic reprogramming alters NANOG methylation and stably boosts NANOG and NANOGP8 expression. Metastases and reprogrammed EMT-like phenotypes require endogenous NANOG, but enhanced NANOG is not sufficient to induce these phenotypes. Finally, reprogrammed tumors enhance GLI2, and we show that GLI2high and AXIN2low, which are markers of the metastatic transition of colon cancers, are prognostic of poor disease outcome in patients. We propose that metastases arise through epigenetic reprogramming of cancer stem cells within primary tumors.
Keywords: metastases, epigenetic reprogramming, NANOG, GLI, cancer stem cells
Introduction
Metastases represent the major cause of cancer-related deaths (Pantel and Brakenhoff, 2004) but what promotes their appearance is not clear. Cancers often progress through the acquisition of genetic mutations (Fearon and Vogelstein, 1990) but none are commonly associated with metastases (Jones et al., 2008). Analyses of primary tumors and derived metastases have also shown that the overwhelming majority of primary tumor and metastasis pairs show a similar mutational spectrum, with metastases being similar to at least one component of heterogeneous primary tumors (Goswami et al., 2015; Kogita et al., 2015).
Epigenetic reprogramming by nuclear transfer or by transcription factor function has been shown to alter normal cell fate revealing an unexpected degree of cell plasticity (e.g. Hochedlinger et al., 2004; Gurdon and Melton, 2008; Buganim et al., 2013). For instance, OCT4, SOX2, KLF4 (OSK), and cMYC (M) can, together, epigenetically reprogram adult normal cells towards embryonic-like states in vitro (Takahashi and Yamanaka, 2006; Wernig et al., 2007) and in vivo (Abad et al., 2013; Ohnishi et al., 2014). These factors can also trigger tumorigenesis from normal cells (Ohnishi et al., 2014) but have similarly been suggested to make colon cancer cells less tumorigenic, enhancing stem cell properties (Miyoshi et al., 2010; Wang et al., 2013; Zhang et al., 2013; Miyazaki et al., 2014; Oshima et al., 2014). Any possible role in metastases, however, remains unknown.
HEDGEHOG (HH)-GLI signaling can drive cellular epithelial-to-mesenchymal transition (EMT), is required for metastases and regulates endogenous OSKM gene expression in colon cancer cells (Varnat et al., 2009). This, together with the finding that HH-GLI, SOX2, and KLF4 levels are elevated in metastatic vs. non-metastatic colon cancers (Varnat et al., 2010), raised the possibility that epigenetic reprogramming by GLI-regulated pluripotent stemness factors, rather than specific genetic mutations, promotes metastases.
Results
Transient elevated OSKM activity in primary colon cancer cells in vitro drives EMT, invasive behavior, and enhanced numbers of clonogenic spheroids
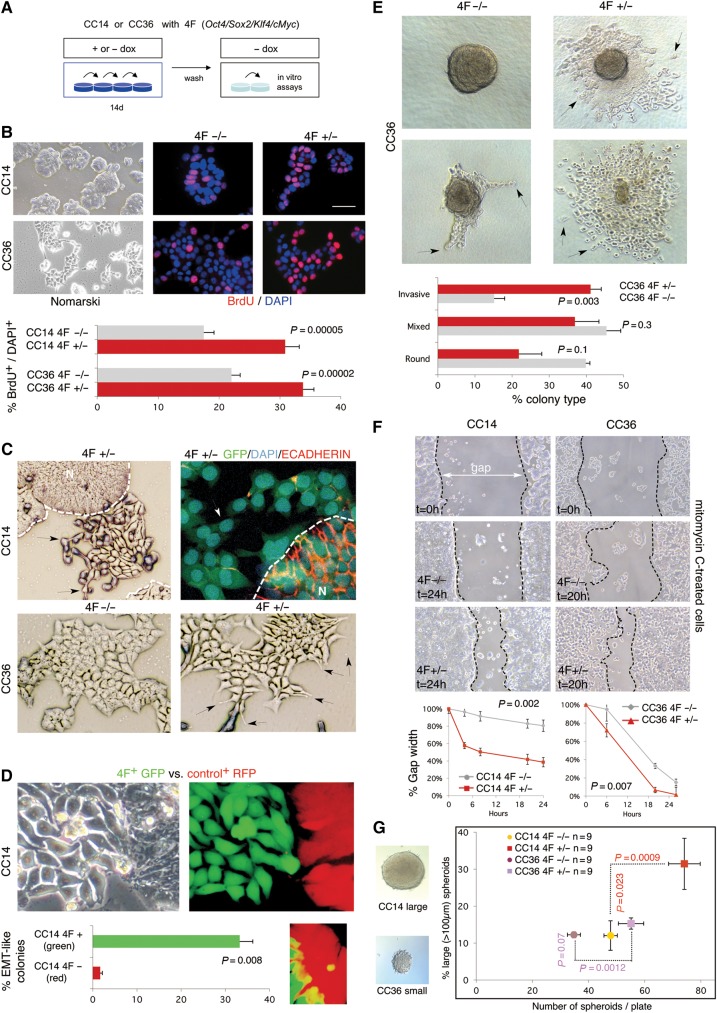
To begin to test a possible role of reprogramming in metastases, we used a doxycycline (dox)-inducible polycistronic lentivector encoding mouse OSKM (hereforth 4F) (Sommer et al., 2009) together with co-transduced rtTA-GFP in early passage primary colon adenocarcinoma CC14 (TNM4) and CC36 (TNM3) cells (Varnat et al., 2009) (Figure 1A and B). This construct allowed the distinction of endogenous from exogenous 4F expression. 4F+ cells exhibited increased BrdU incorporation (Figure 1B, right), and activated Caspase3+ apoptosis was reduced from 0.9% to 0.15% (P = 0.035) for CC14 and from 3.7% to 0.4% (P = 0.036) for CC36 on average. Cultures induced for 14 or 30 days (+dox) displayed EMT-like phenotypes with dispersing and elongated cells instead of the tight, compact islands of controls (−dox) (Figure 1C). This phenotype was first observed after 5−7 days and was not detected at similar expression levels in cells with any of these genes singly. Analyses of the archetypal epithelial marker ECADHERIN by immunolabeling showed that cells acquiring an EMT-like phenotype, dispersing and flattening, lost expression whereas those remained in the normal compact epithelial islands exhibited high membrane expression (Figure 1C, right).
Figure 1.
In vitro 4F-induced phenotypes in human primary colon cancer cells. (A and B) Human primary colon cancer cells CC14 and CC36 were transduced with the inducible STEMCCA lentivector expressing 4F (Oct4, Sox2, Klf4, cMyc) under doxycycline (dox) regulation (A). These cells, which are epithelial (B, top left), were tested for BrdU incorporation via immunocytochemistry (B, top right) and quantified (B, bottom). (C) EMT-like phenotypes observed in 2D culture of CC14 4F+/− as compared with wt islands (B, left), and in CC36 in 4F+/− vs. 4F−/−. Dispersing cells with protrusions are noted with arrows. Broken lines denote the extent of the normal area (N). Top right panel shows 4F+/− CC14 cells after immunolabeling with ECADHERIN antibodies. Note the loss of epithelial ECADHERIN expression (red) in reprogrammed 4F+/− cells with EMT-like phenotypes that disperse from the normal islands (broken line, N). All cells were GFP+ and nuclei were co-labeled with DAPI. (D) Red-Green tracing assay showing the dispersion of CC14 green (GFP+) cells, which normally form compact islands (RFP+), seen under Nomarski (top left) and fluorescent (top right) illumination. Quantification of EMT-like green and red colonies is shown in the bottom left panel. An example of the invasion of a green cell into the compact red area is depicted in the bottom right panel. (E) Invasion phenotypes of CC36 cells seen in colonies in collagen. Two examples are shown without (4F−/−, left column) or with (4F+/−, right column) transient dox induction (see A and text for +/− nomenclature) resulting in compact round (top left) or invasive (right panels) phenotypes. Mixed phenotypes (bottom left) were also observed. Arrows point to invasive cells leaving the clone and moving into the collagen. Quantification of the type of colonies formed is shown in the bottom panel. (F) Gap-closure assay following a scratch in 2D cultures in CC14 and CC36 with or without previous transient expression of 4F. The gap present at the start of the experiment is shown in the upper panels. The bottom panels show the differences between uninduced (−/−) and induced (+/−) samples at the times indicated. Quantification of the width of the gap is shown in the bottom panel. (G) Clonogenic spheroid assays with previously induced and control CC14 and CC36 4F+/− and 4F−/− cells. The graph (right) shows the total number of spheroids formed compared with the percentage of large spheroids. 4F expression induced more spheroids from both cell types, but only in CC14 were they larger. Images of large and small spheroids are shown in left panels. Scale bar, 150 µm (B left, E, G), 60 µm (B center and right, C top left and bottom), 15 µm (D, C top right), 250 µm (F).
To further analyze EMT-like phenotypes, CC14 4F cells (and thus GFP+) were mixed with control cells only carrying a lentivector expressing RFP. Induction of the mixed culture for 14 days revealed that 33% of GFP+ colonies, and <5% in RFP+ controls, showed EMT-like phenotype with single spreading cells (Figure 1D). Single GFP+ cells could be found invading RFP+ colonies (Figure 1D, inset). CC36, which are normally more dispersive than CC14, were instead plated at a frequency of 100 single cells/well inside a collagen cushion and allowed to form colonies. Cells that were not induced previously (−dox) formed compact groups that had limited protrusions and few if any cells moving in to the collagen after 10 days of culture without dox (4F−/− cells) (Figure 1E). By contrast, those cells that were previously induced (+dox) for 14d and placed in the collagen cushion without dox (4F+/− cells) yielded twice the number of colonies with spreading cells (Figure 1E) as compared with 4F−/− controls. CC14 aggregates showed much more limited collagen spreading and were not quantified. Scratch or gap closure assays with mitomycin-C to block cell proliferation showed that both CC14 and CC36 4F+/− cells moved faster into the gap than control 4F−/− cells (Figure 1F).
Both CC14 and CC36 4F+/− cells also produced a greater number of clonogenic 3D spheroids as compared with their respective 4F−/− controls (Figure 1G), suggesting a promotion of stem cell numbers, but only CC14 also produced larger (>100 µm) spheroids in comparison with 4F−/− controls (Figure 1G).
Enhanced metastases after injection into the circulation of in vitro reprogrammed colon cancer cells
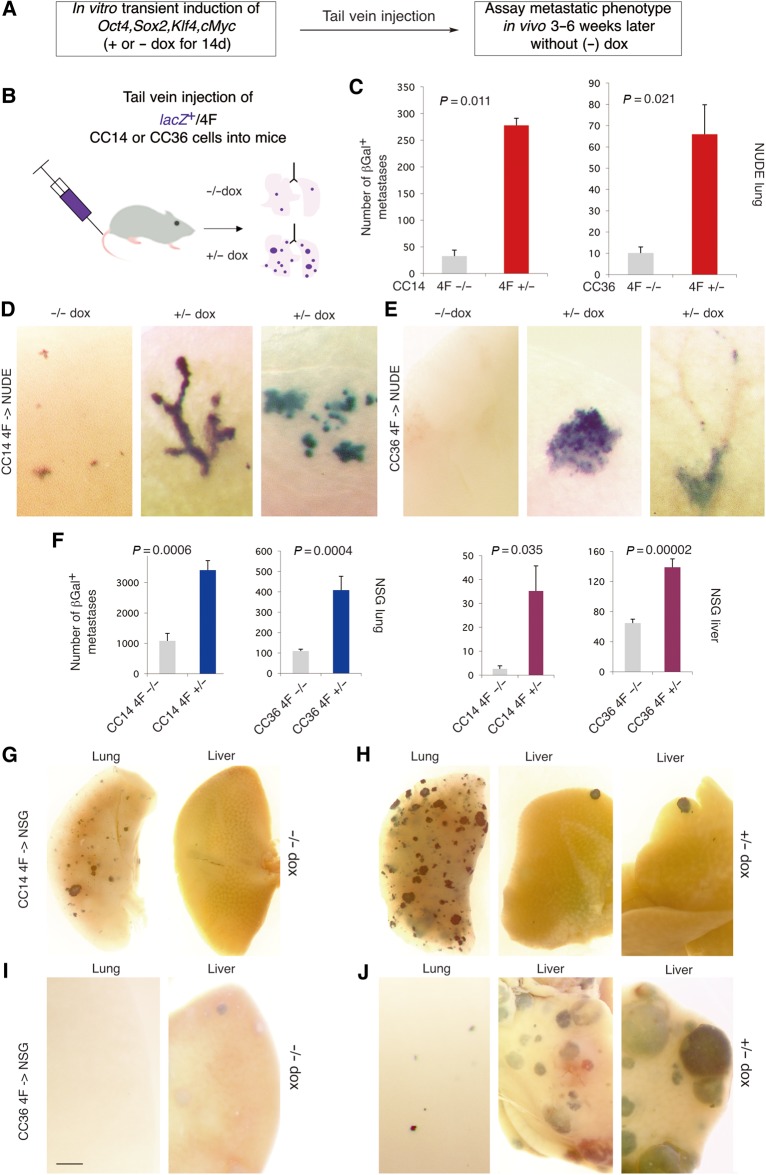
The ability of in vitro reprogrammed cells to induce in vivo metastases was first tested by directly seeding tumor cells in the lungs of recipient immunocompromised mice via tail vein injection (Figure 2A and B). Injected cells were genetically marked by insertion of lacZ-expressing lentivectors (Stecca et al., 2007). Both CC14 or CC36 4F+/− cells yielded many more βGal+ lung metastases in NUDE mice as compared with 4F−/− cells (Figure 2C and D, E left panels). Similar results were obtained with NSG mice, which are fully immunocompromised: injection of CC14 or CC36 yield a higher baseline of metastatic βGal+ colonies in the lungs and also in the liver as compared with NUDE mice (Figure 2F, G, and I). Using this model, CC14 4F+/− tumor-bearing mice had a larger increase in lung metastases whereas CC36 4F+/− cells produced a higher number of large metastases in the liver (Figure 2F−J). Analyses of the numbers of CC14 4F+/− vs. 4F−/− lung colonies per size showed a ∼3-fold increase in single cell micrometastases, small as well as large metastases with over 100 cells (Supplementary Figure S1).
Figure 2.
Transient in vitro expression of 4F increases metastases in mice after injection into the venous circulation. (A and B) Scheme (A) and diagram (B) of the experimental design in which dox is only given in vitro before grafting in mice. (C) Quantification of βGalactosidase+ (βGal+) colonies in the lungs of NUDE mice injected with control (4F−/−) or induced (4F+/−) cells as indicated. n = 4 for dox− and n = 4 for dox+, for both CC14 and CC36. (D and E) Examples of metastatic colonies in the lungs as indicated for CC14 (D) and CC36 (E) cells. Colonies are blue following the XGal reaction. (F) Quantification of βGal+ colonies in the lungs and livers of NSG mice injected with control (4F−/−) or induced (4F+/−) cells as indicated. n = 5 for dox− and n = 5 for dox+ for CC14, and n = 11 for dox− and n = 11 for dox+ CC36. (G−J) Representative images of metastatic colonies in the lungs and livers as indicated for CC14 (G, H) and CC36 (I, J). Scale bar, 150 µm (D, E), 1.5 mm (G, H), 600 µm (I, J).
In vivo OSKM reprogramming promotes distant metastases
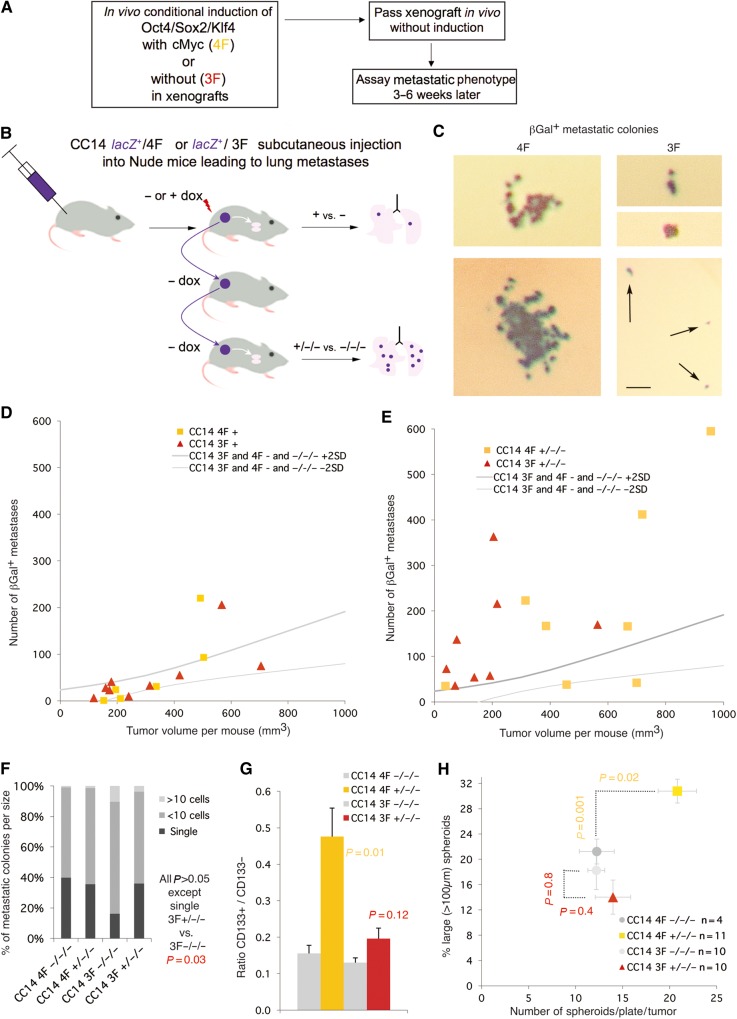
To test for in vivo reprogramming and to analyze full metastatic spread from a local tumor to a distant organ, we engrafted CC14 cells subcutaneously in NUDE mice and analyzed the appearance of distant metastases (Figure 3A). Moreover, to test for the establishment of a stable altered state by the transient action of the reprogramming factor cohort, we passed the induced tumors into secondary and tertiary hosts in the absence of dox (Figure 3A). Dox treatment in the first host was started at engraftment and continued for the duration of xenograft growth before reaching the local legal limit, for a total of ∼30 days. The lungs of these first, dox-induced mice (+ or −) were analyzed for the presence of βGal+ metastases. In parallel, the xenograft bulk was dissected and passed onto additional sequential hosts without dox. The lungs of third generation hosts (−/−/− or +/−/−) were then assayed for βGal+ metastases and the xenograft bulk analyzed (Figure 3B and C). To quantify and delineate the control conditions graphically we demarcated the area split by the trend line of all control values (− and −/−/− of all experiments) plus and minus two standard deviations for the number of βGal+ metastases and for tumor size (Supplementary Figure S2), with control samples outside of this zone being considered outliers. In vivo 4F induction of CC14 tumors generally resulted in a metastatic phenotype similar to controls (orange squares in Figure 3D), with an increased metastatic penetrance of 12%. This situation changed in +/−/− lungs (orange squares in Figure 3E), where there was a net increase in the number of metastases, correlated with an increase of tumor size, and a penetrance of 70%. Quantification of the distribution of metastases of different sizes showed a similar distribution between −/−/− and +/−/− samples (Figure 3F). Analyses of +/−/− vs. −/−/− xenograft tumor bulk revealed an increase in the percentage of CD133+ cells directly sorted (Figure 3G) and an increase in the number of clonogenic spheroids (Figure 3H). Consistent with an increase in the size of +/−/− vs. −/−/− tumors, the former yielded a higher proportion of large (>100 µm) spheroids as compared with the latter (Figure 3H).
Figure 3.
Restricted in vivo expression of 4F/3F in xenografts induces a stable-pro-metastatic state in subcutaneous xenografts. (A) Scheme of the experimental set-up for B−E. (B) Diagram of the flow chart of serial transfer in vivo, and analyses of induced vs. non-induced tumors (+ vs. −) and those after two generations without additional induction of the expression of 4F/3F (+/−/− vs. −/−/−). (C) Examples of βGal+ metastatic colonies detected in the lungs of host mice as indicated (see B). Scale bar, 100 µm (C). (D and E) Plots of the number of βGal+ lung metastases vs. total tumor load (size) of individual mice. All uninduced controls were pooled to create a robust control zone delineated by +/− 2SD borders as shown (see Supplementary Figure S2). P-values were determined by the unpaired two-tailed Student's t-test comparing all samples within a given class (e.g. 4F+) vs. the entire control pool (all uninduced samples) and were as follows: (D) Tumor volume 4F+ vs. controls P = 0.3, and 3F+ vs. controls P = 0.7. Number of metastases 4F+ vs. controls P = 0.4, and 3F+ vs. controls P = 0.05. (E) Tumor volume 4F+/−/− vs. controls P = 0.05, and 3F+/−/− vs. controls P = 0.2. Number of metastases 4F+/−/− vs. controls P = 0.04, and 3F+/−/− vs. controls P = 0.04. (F) Quantification of the size of metastases showing little variation between samples. (G) Histogram of the ratios of CD133+/CD133− cells obtained by MACS for the different conditions noted. P-values are experimental vs. control (e.g. 4F+/−/− vs. 4F−/−/−). (H) Plot of the number of spheroids vs. the number of large ones (diameter >100 µm) as noted. 4F but not 3F +/−/− cells produce more and larger clonal spheroids.
Transient function of OSK but not of MYC alone in the tumor bulk in vivo drives full metastases without increases in clonogenic spheroids
cMYC can be a powerful oncogene and might drive the action of 4F. We thus tested for the ability of the transient expression of cMYC in vivo from a dox-regulated lentivector to increase metastases from CC14 xenografts. cMYC+/−/− tumors (from two independent infections resulting in 3- or 30-fold expression of cMYC as compared with control cells) were similar to cMYC−/−/− in both size and number of lung metastases they produced; all cMYC tumors fell within the area defined by all −/−/− tumors (Supplementary Figure S3).
In contrast, in vivo induction of OSK (hereforth 3F) yielded more metastases in CC14 3F+/−/− as compared with the −/−/− controls but not in 3F+ as compared with 3F− tumor-bearing mice, much as with 4F (Figure 3D and E). However, 3F+/−/− xenografts were similar to controls in size, but 3-fold smaller (P = 0.014) at equivalent times than 4F+/−/− xenografts (when the first tumors in the cohort reached the local legal limit). Importantly, however, both 3F+/−/− and 4F+/−/− tumors yielded similar average numbers of metastases (105 vs. 130 metastasis per mouse, respectively, P = 0.39) above controls (37 metastases per animal, P = 0.04) for both 3F+/−/− and 4F+/−/− vs. the −/−/− control group. We observed also an increase in the number of single cell micrometastases in 3F+/−/− vs. 3F−/−/− (Figure 3F), suggesting slower growth of tumors and metastatic colonies, but similar spreading activity from the primary xenograft. A further difference between 3F+/−/− and 4F+/−/− was detected in the lack of an increase in the CD133+ population in 3F+/−/− tumors (Figure 3G) and the lack of increase of clonal spheroid numbers or tumor size (Figure 3H), both observed in 4F+/−/− tumors. These in vivo results are consistent with results from CC14 3F cells in vitro, which showed a very weak enhancement of BrdU incorporation (14% vs. 20%, P = 0.047), no increase in the number of clonogenic spheroids (39 vs. 37 per 96 well-plate on average, P = 0.62) but an increase in scratch assay gap closure with mitomycin-C treated cells (89% vs. 70% at 26 h, P = 0.02) in 3F+/− vs. 3F−/−.
Clonogenic cancer stem cells stably transmit OSK-induced pro-metastatic memory
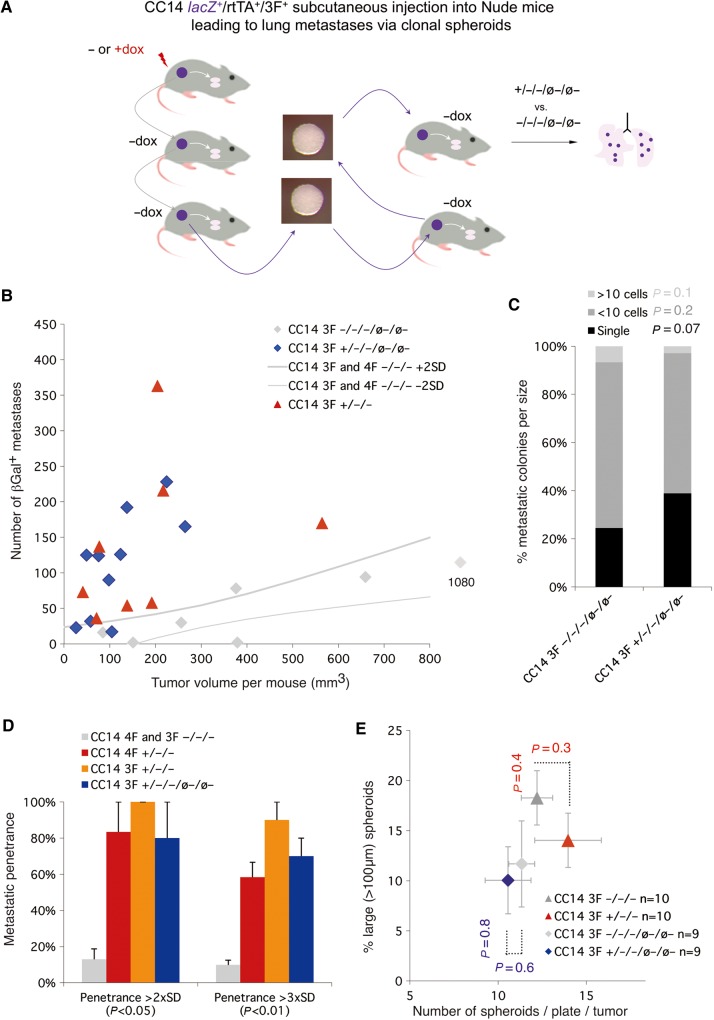
Since metastases may result from changes in the behavior of clonogenic cancer stem cells we asked if these could transmit 3F-imposed pro-metastatic memory. Floating clonogenic spheroids (ø) derived from 3F+/−/− and 3F−/−/− xenografts were grown in batches at clonal density (Figure 4A) and injected subcutaneously into primary and then secondary hosts in the absence of dox. 3F−/−/−/ø−/ø− mice behaved as all other −dox controls (Figure 4A and B). In contrast, 3F+/−/−/ø−/ø− mice (as well as 3F+/−/−/ø− mice) recapitulated the 3F+/−/− phenotype, with smaller tumor volumes as compared with 4F+/−/− (Figure 4B) and with high penetrance per animal (Figure 4D). The distribution of metastatic colonies per size was maintained (Figure 4C), without increases in clonogenic spheroid number or size (Figure 4E).
Figure 4.
Clonogenic tumor cells transmit stable pro-metastatic memory triggered by transient 3F function. (A) Diagram showing the strategy used. Tumors induced and passed as in Figure 3 (+/−/− vs. −/−/−) were dissociated and cloned to form stem cell cell-derived spheroids (ø) in the absence of additional dox induction. These were collected, dissociated, and injected into new host mice. The clonogenic passage and in vivo injection of the resulting spheroids were repeated and the lungs of last host mice (+/−/−/ø−/ø− vs. −/−/−/ø−/ø−) were stained to reveal βGal+ metastatic colonies. (B) Plot of individual mice for number of metastases vs. total tumor load (size). P-values were determined by the unpaired two-tailed Student's t-test comparing all samples within a given class vs. the entire control pool (all uninduced samples) and were as follows: Tumor volume 3F+/−/−/ø−/ø− vs. controls P = 0.05. Number of metastases 3F+/−/−/ø−/ø− vs. controls P = 0.03. (C) Quantification of the size of βGal+ metastases as noted. (D) Histogram of the per animal penetrance of metastases under the different conditions indicated, considering samples that fall outside the control areas of +/− 2SD (as in panel B) or +/− 3SD. (E) Plot of the number of total clonal spheroids vs. the number of large clonal spheroids (>100 µm) as indicated. Note the similar behavior of 3F+/−/− and 3F+/−/−/ø−/ø− cells in all cases.
Reprogramming-driven changes in stemness, WNT and HH pathway components
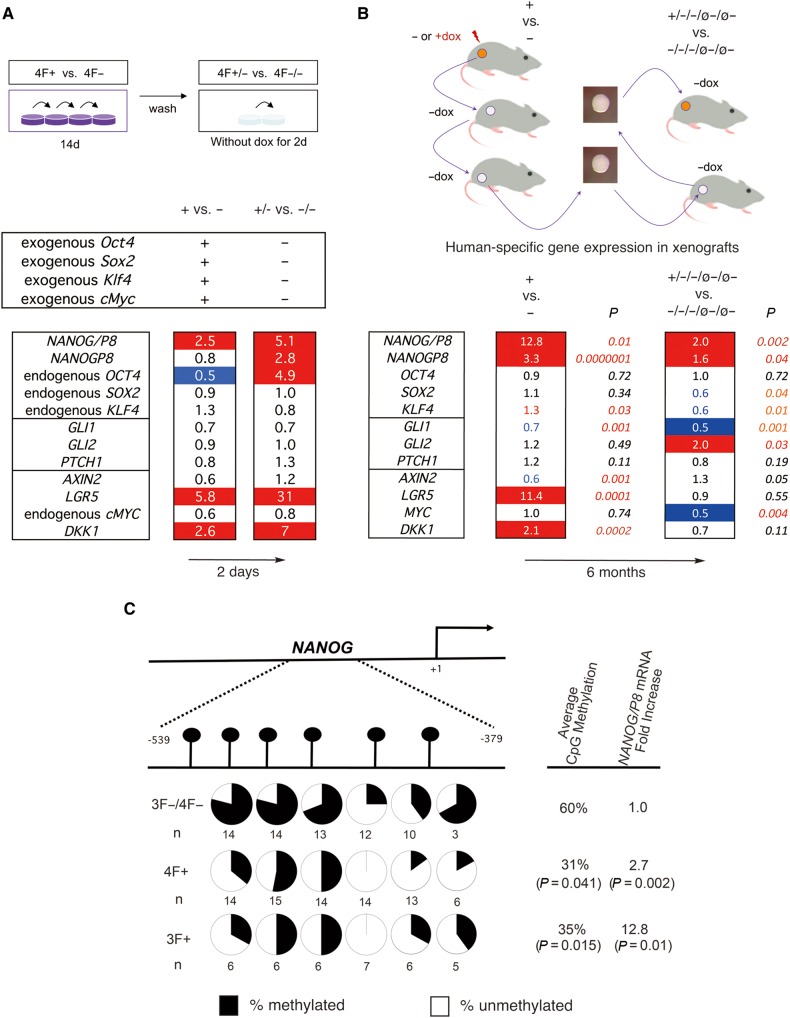
Analysis of gene expression focused on selected stemness, HH and WNT pathway components as changes in the expression of these define the metastatic transition of human colon cancers (Varnat et al., 2010). RT-qPCR of CC14 4F+ cells after 14 days of dox induction in vitro (+ vs. − cells; Figure 5A, top) used for tail vein injections (Figure 2) showed the selective exogenous enhanced expression of the mouse OSKM genes. 4F expression led to the induction of NANOG/P8 2.5-fold over control 4F− cells, but not to that of the second NANOG encoding gene NANOGP8, or of the endogenous OSKM genes (Figure 5A, bottom). Whereas the HH pathway components GLI1, GLI2, and PTCH1 were not affected, the WNT pathway targets LGR5 and DKK1 were upregulated 5.8- and 2.6-fold, respectively, although AXIN2 and endogenous cMYC were slightly repressed. Removal of dox for 2 days in vitro (+/− vs. −/− cells) further induced expression of endogenous NANOG/P8 (to 5.1-fold over control cells) and the levels of endogenous NANOGP8 and of endogenous OCT4 were enhanced 2.8- and 4.9-fold. HH pathway components were unchanged and the expression levels of LGR5, a colon stemness marker, and DKK1, encoding a WNT inhibitor, were also further enhanced (31- and 7-fold; Figure 5A).
Figure 5.
Selected gene expression changes induced by metastatic reprogramming and demethylation of NANOG. (A) In vitro gene expression changes. Top: Diagram of the strategy for in vitro reprogramming for 14 days followed by 2-day culture without dox. Bottom: Heat map of endogenous human gene expression changes after normalization shown as fold change (ratios over controls as indicated, e.g. + vs. −). Endogenous genes were detected with 5′UTR primers (as in B). Exogenous OSKM genes were tested with specific primers and found to be expressed only during dox treatment. (B) In vivo gene expression changes. Top: Diagram of the strategy used to derive the material used for gene expression analyses in vivo. Bottom: Heat map of fold change in experimental over control samples as indicated. All genes noted are endogenous genes of engrafted human tumor cells. Numbers are the geometrical mean of normalized individual fold changes. The number of individual tumors analyzed for each case were n = 10 for 3F+, n = 10 for 3F−, n = 10 for 3F+/−/−/ø−/ø−, and n = 8 for 3F−/−/−/ø−/ø−. P-values were derived from t-tests using the normalized Ct values without outliers outside the +/−2SD space. The heat maps of A and B show increased (1.5-fold or more) expression highlighted in red and decreased (0.5 or less) expression in blue. Numbers in red or blue in B bottom denote significative up (red) or down (blue) changes within the <0.5-to-<1.5-fold interval. (C) Reprogramming factors induce DNA methylation changes in NANOG. The upper diagram shows the position of analyzed CpG dinucleotides in the 5′ promoter region of NANOG and their methylation status (represented in pie charts in the lower part of the diagram) as determined by bisulfite sequencing. All uninduced controls were pooled as there were no differences between 4F− vs. 3F− samples. P-values are for experimental vs. control samples as indicated using t-tests. The overall CpG methylation of all the samples tested is noted, as is the average fold change in NANOG/P8 expression in the same samples. See Supplementary Figures S4−S6 for OCT4 data and MSPs.
In vivo analysis of gene expression (Figure 5B, top) focused on CC14 3F+ vs. 3F− NUDE mouse xenografts. These tumors showed a small but significant elevation of the levels of endogenous KLF4, but not of endogenous OCT4 or SOX2. The levels of NANOG/P8, and specifically of NANOGP8, were also enhanced as were those of LGR5 and DKK1 whereas AXIN2 was slightly repressed (Figure 5B, bottom), largely paralleling the in vitro 4F results (Figure 5A). Comparison of the profile of the expression in 3F+/−/−/ø−/ø− vs. 3F−/−/−/ø−/ø− tumors (Figure 5B, top), taken as the most extreme case for the presence of stable pro-metastatic memory (since reprogramming factor function and the tested samples are separated by 6 months and 5 mouse hosts) showed that NANOG/P8 and NANOGP8, but not LGR5 or DKK1, remained upregulated and that GLI2 levels were enhanced 2-fold (Figure 5B, bottom).
Reprogramming drives changes in NANOG and SFRP1 methylation
Together, these in vivo and in vitro data raised the possibility that NANOG might be a key player in metastatic epigenetic reprograming of cancer cells. We thus first tested for changes in its methylation in tumor cells through analyses of promoter region CpG dinucleotides (Figure 5C), given that it lacks CpG islands. Sequencing bisulfite-converted DNA revealed a decrease of NANOG DNA methylation in 4F+ and 3F+ vs. control (4F− and 3F−) tumors by about 50%, which was correlated with increased levels of expression (Figure 5C). Additional analysis by bisulfite sequencing did not reveal changes in OCT4 (Supplementary Figure S4), and human-specific methylation-specific PCR (MSP) of CpG islands of HH and WNT pathway genes in CC14 4F+/−/− vs. 4F−/−/− tumors also did not reveal clear changes for SHH, PTCH1, SMOH, GLI1, GLI2, DKK1, or LGR5 (Supplementary Figures S4 and S5). There was, however, a modest increase in unmethylated SFRP1 and in methylated AXIN2 (Supplementary Figure S4). In CC14 3F+/−/− tumors PTCH1 and AXIN2 showed small increases in their methylation status as compared to 3F−/−/− controls, but SFRP1, encoding a WNT inhibitor, displayed a strong increase in the unmethylated signal (Supplementary Figures S4 and S6).
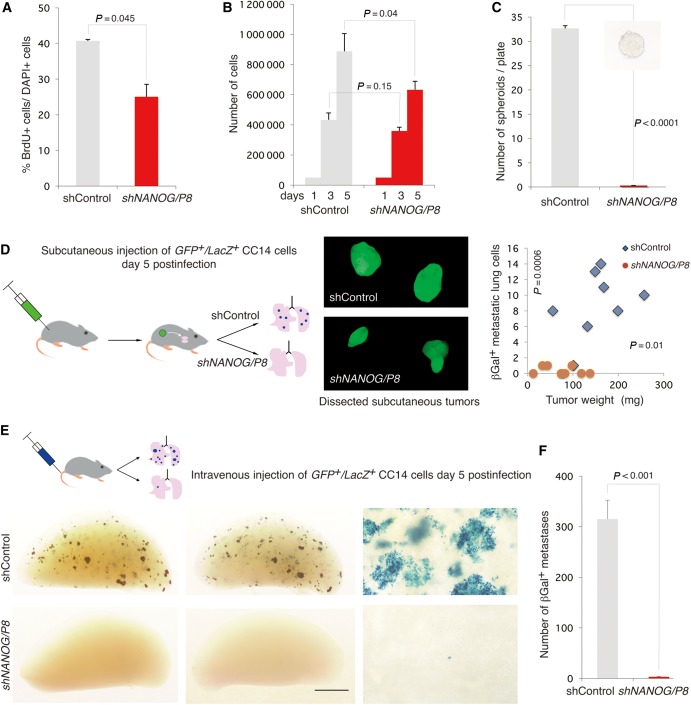
NANOG is required for metastases and for reprogramming-driven EMT-like phenotypes
NANOG was knocked-down with a fully validated GFP+ shRNA lentivector targeting both NANOG and NANOGP8 (NANOG/P8; Zbinden et al., 2010) with a targeting efficiency of >90%. This caused a modest decrease in CC14 cell proliferation on Day 5 as assessed by BrdU incorporation and total cell counts (Figure 6A and B). In contrast, clonogenic tests using spheroids revealed a total dependency of these primary colon cancer stem cells on NANOG (Figure 6C). Blocking NANOG in vivo (through subcutaneous grafting of viable CC14 LacZ cells expressing shNANOG/P8 in NUDE mice) showed that tumor growth ensued although shControl tumors were larger than those with repressed NANOG (Figure 6D; Jeter et al., 2009). However, whereas metastases were detected in each shControl animal, only one single βGAL+ cell was detected in the lungs of 3/9 shNANOG/P8 mice (Figure 6D, right), which could well be from rare LacZ+ cells not infected with shNANOG/P8 lentivectors. Metastases elsewhere were not detected.
Figure 6.
NANOG function is required for metastases. (A and B) Knock-down of NANOG (with a lentivector-encoded shRNA targeting both NANOG and NANOGP8) slightly reduces in vitro CC14 cell proliferation as measured by BrdU incorporation (A) and total cell counts (B). (C) Knock-down of NANOG completely inhibits CC14 cancer stem cell self-renewal, as cells with compromised NANOG function does not form spheroids, unlike control cells. The inset shows a representative spheroid from control conditions. (D) Left: Strategy used for the growth of subcutaneous GFP+/LacZ+ CC14 tumors (middle panel seen under GFP fluorescence) in NUDE mice carrying control or shNANOG-expressing lentivectors as indicated. Right: Quantification of tumor weights and the number of βGal+ lung metastases reveal a drastic decrease under shNANOG conditions. Images show representative tumors. (E and F) Diagram for the strategy to seed the lungs with control and shNANOG CC14 cells (E, top), resulting in a large number of metastases under control but nearly none under shNANOG conditions (E, bottom). A single blue cell is detected and shown in the bottom right panel for shNANOG. (F) Quantification of the number of βGal+ (blue) metastases per left lung lobe. Note the presence of many large metastases (with >100 cells) peppering the lungs of control NSG mice. n = 6 mice per condition. Error bars are SEM. Scale bar, 400 µm (C inset), 10 mm (D middle panels), 2.8 mm (E bottom left and middle panels), 350 µm (E right panel).
To extend these findings, we directly seeded shControl and shNANOG/P8 cells into the lungs of recipient NSG mice through injection into the venous circulation (Figure 6E). Control cells yielded hundreds of metastases in each of 6 mice, including single cells and micro- as well as macrometastases (Figure 6E). In contrast, blocking NANOG function abolished metastases, with only single βGal+ cells detected in the lungs (Figure 6E). Identical results were obtained analyzing GFP+ metastases (Supplementary Figure S7).
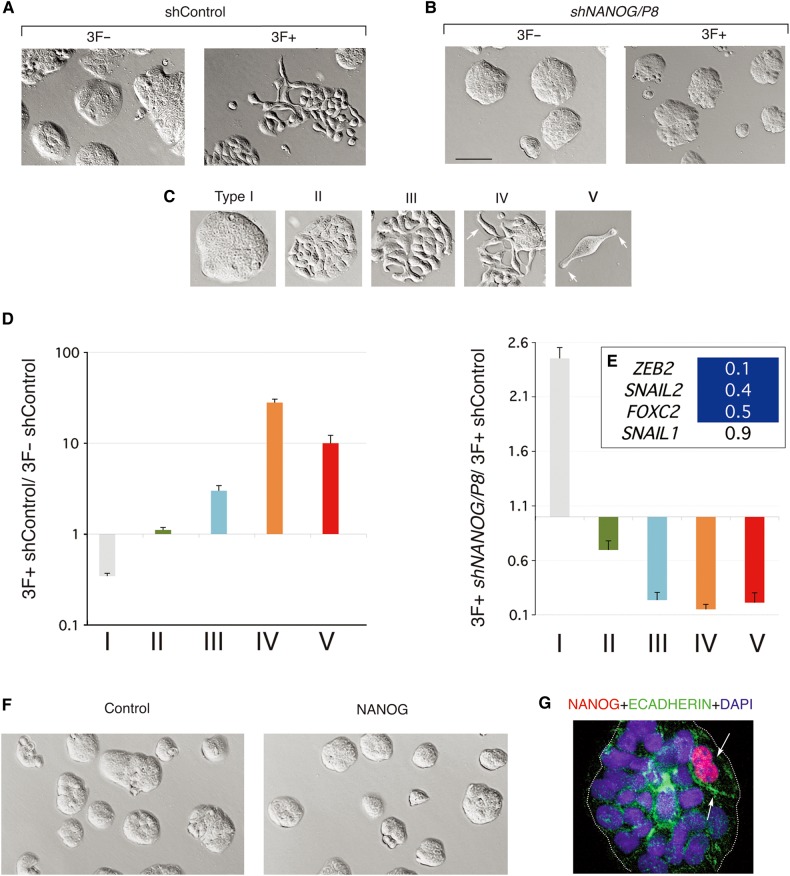
Given that NANOG function is essential for colon cancer metastases, we used a proxy assay to test for the requirement of NANOG in metastatic reprogramming. To this end we quantified reprogramming-induced EMT-like phenotypes at 14-day of dox treatment (Figure 1). Control CC14 cells (3F− and 3F−/shControl) grew largely in compact islands (Figure 7A), as did 3F− cells expressing shNANOG/P8 although the islands were smaller (Figure 7B) consistent with an effect on cell proliferation (see above). 3F+ cells displayed the expected reprogramming factor-induced EMT-like and cell-spreading phenotypes (Figures 1 and 7A) and their quantification revealed five types (Figure 7C and D): Type I: wild type, tight, highly adherent epithelial island morphology characteristic of CC14 cells; Type II: continuous but flat islands; Type III: discontinuous flat islands with internal gaps suggestive of diminished adhesion; Type IV: groups of dissociated or barely linked cells often with long protrusions; Type V: isolated single cells with fusiform morphology typical of migrating cells. Comparison of the occurrence of these various phenotypes in 3F−/shControl vs. 3F+/shControl cells revealed a diminution of the number of Type I compact islands and an increase of type II-V dispersing phenotypes (Figure 7D).
Figure 7.
NANOG function is required but not sufficient to induce reprogrammed EMT-like phenotypes. (A and B) Images of in vitro CC14 colon cancer cultures control and reprogrammed cells showing diverse phenotypes with normal (A) or compromised (B) NANOG function. Nomarksi images of 3F+ or 3F− cells with control or shNANOG lentivectors after 14 days of dox treatment show the typical growth of CC14 epithelial islands under control conditions without dox and the appearance of EMT-like cell phenotypes after dox treatment (shControl; 3F+). (C and D) Classification and quantification of resulting phenotypes after dox induction and reprogramming as described in the text. Arrows in C point to cell extensions (in type IV, V). The histograms in D show phenotypes that appear during reprogramming in control 3F cells after dox addition (3F+ control vs. 3F− control) but not in 3F+ shNANOG cells as compared with 3F+ control cells (3F+ shNANOG vs. 3F+ control). Error bars are SEM. (E) Heat map of the expression of selected genes as shown after normalization with housekeeping genes. Blue cells denote decreased values as compared with control as noted. (F) Images of control (cells transduced with a control lentivector) and NANOG-expressing cells (cells transduced with a lentivector expressing NANOG) show similar morphologies 18 days after infection. (G) Double immunofluorescence of an epithelial CC14 island, which contains two cells expressing high levels of exogenous NANOG (red) and endogenous ECADHERIN (green). Nuclei are counterstained with DAPI (blue). Scale bar, 140 µm (A, B, F), 70 µm (C types I−IV), 30 µm (C type V), 25 µm (G).
Previous work has shown that overexpression of NANOG can enhance the expression of SNAIL1/2 (Meng et al., 2010), but it is not known whether endogenous NANOG is required for its expression. The levels of key transcription factors involved in inducing EMT, including ZEB2, FOXC2, SNAIL1/2, and TWIST1 (Sánchez-Tilló et al., 2012), were then tested in reprogrammed cells with or without NANOG knock-down. 3F+/shControl cells showed increased normalized expression of ZEB2 and SNAIL2 by 6- and 3-fold, respectively, in comparison with 3F−/shControl cells. VIMENTIN expression was also induced by 1.5-fold (data not shown) but ECADHERIN was unaltered, likely due to the heterogeneity of the cultures following 3F induction, as much as with 4F (Figure 1C). Importantly, the expression of ZEB2, SNAIL2, and FOXC2, were repressed in cells with compromised NANOG (Figure 7E).
Enhanced NANOG expression is not sufficient to induce EMT-like phenotypes or metastases
Constitutively enhanced NANOG levels in colon or liver cancer cells have been suggested to promote metastases (Meng et al., 2010; Sun et al., 2013). We thus overexpressed NANOG in CC14 cells from a lentivector (Zbinden et al., 2010) by over 100 fold over endogenous levels and failed to detect enhanced EMT-like phenotypes (Figure 7F). This result strongly suggests that NANOG is not sufficient to mimic the in vitro effects of 3F-induced metastatic reprogramming. Moreover, immunolabeling with anti-NANOG antibodies to detect the overexpressed protein in cultures showing varying degrees of infection revealed that cells with high NANOG levels did not generally repress membrane expression of ECADHERIN and remained (92% of cases, >100 cells counted) within epithelial islands (Figure 7G). Consequently, injection of these CC14 cells overexpressing NANOG did not result in an increased number of metastases in the lungs from subcutaneous xenografts (mean metastatic index = 0.068 for control and 0.064 for NANOG overexpression, P = 0.88; Supplementary Figure S8).
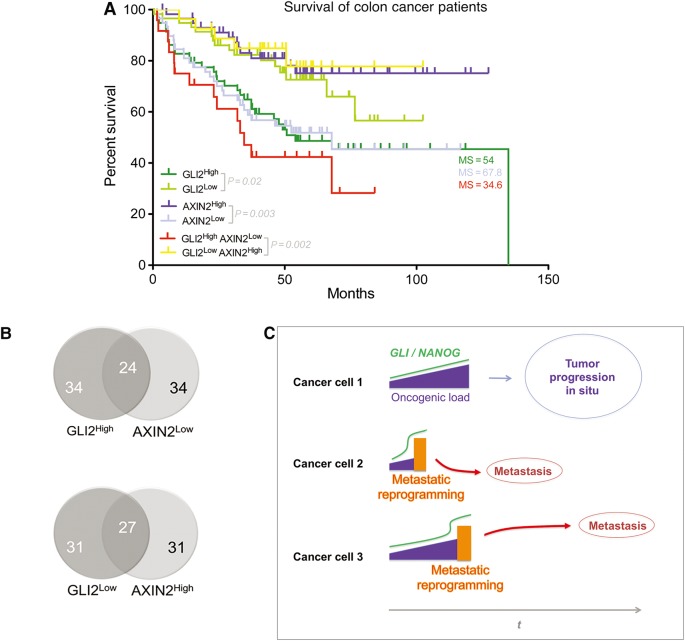
High GLI2 and low AXIN2 levels are prognostic of disease outcome in patients
If metastatic reprogramming in our experimental conditions recapitulates events of metastatic cancers in patients, reprogramming-induced alterations in gene expression might be evident in patient samples. Analyses of the expression levels of the genes showing variations in 3F+/−/−/ø−/ø− vs. 3F−/−/−/ø−/ø− (Figure 5A) in a published cohort of patients with known survival endpoints (GSE17537) focused our attention on GLI2. Comparison between high vs. low quartile expression groups revealed high GLI2 levels in colon cancers from patients with shorter survival times (Figure 8A). Of the others genes analyzed above by RT-qPCR, AXIN2 showed a significant correlation with survival, although this was inverted: High AXIN2 patient quartiles showed improved survival over low expressors (Figure 8A). Top and bottom GLI2 and AXIN2 quartiles partially overlapped (Figure 8B). NANOG high vs. low quartiles did not show significative differences, and NANOGP8 was not reported in the cohort. Interestingly, combined GLI2highAXIN2low vs. GLI2lowAXIN2high populations revealed an enhanced correlation with the shortest survival times (34.6 months) as compared with that of single gene cohorts (Figure 8A).
Figure 8.
High GLI2 and low AXIN2 predict colon cancer patient survival and a model of metastatic reprogramming in the tumor bulk. (A) Kaplan–Meier plots for different high and low expressor patient groups as indicated. Data were from the public colon cancer GSE17537 cohort, which comprises 232 patients with survival and gene expression data. P-values and median survival length in months are given for each case. GLI2 is reliably represented by probe 228537_at. AXIN2 is represented by probe 222695_s_at. A second probe for AXIN2, 222696_at, gives the same quartile results. (B) Venn diagrams of high and low expressor patient pools for the genes indicated showing their partial overlap. The source data were from the GSE17537 cohort of colon cancer patients. (C) A model for the action of metastatic reprogramming in cells within primary tumors. Three exemplary cells are shown. In most cells (1), the number of oncogenic events, the oncogenic load, increases during tumor progression (t) enhancing activating GLI levels (Stecca and Ruiz i Altaba, 2010; Aberger and Ruiz i Altaba, 2014). In a subset of cells, notably cancer stem cells, GLI-regulated OSK factors drive NANOG-dependent epigenetic reprogramming (orange boxes), EMT, migration, and metastases. Metastatic reprogramming results in further enhanced activating GLI and NANOG levels and can take place at different times during the life of a tumor.
Discussion
Here we have addressed the idea that epigenetic reprogramming by pluripotent stemness factors promotes metastases from primary tumors (Ruiz i Altaba, 2011) using the transient enhanced function of OSK/M to reprogram tumor cells. These factors are endogenously expressed and GLI-regulated in colon cancer cells (Varnat et al., 2010). We find that in vivo reprogramming of primary human colon xenografts in mice enhances a pro-metastatic state, which is stable, transmitted by cancer stem cells, and detected after sequential engraftments in multiple hosts, resulting in the promotion of distant metastases. Consistently, in vitro metastatic reprogramming enhances EMT-like phenotypes, cell dispersion, and lung and liver metastases after injection into the circulation. Our data also highlight NANOG since metastatic reprogramming stably enhances NANOG/P8 expression levels and demethylates NANOG. However, whereas we find that metastases and reprogramming-induced EMT phenotypes require endogenous NANOG function, enhanced NANOG is not sufficient on its own to induce these phenotypes.
Analyses of patient cancers obtained directly from the operating room indicate that colon cancers undergo a metastatic transition characterized by enhanced HH-GLI and repressed WNT-TCF target levels (Varnat et al., 2010). The gene expression changes we report here suggest that metastatic reprogramming partially mimics this low-to-high HH-GLI and high-to-low WNT-TCF switch. Moreover, we find that high GLI2 and low AXIN2 levels, markers of high HH-GLI and low WNT-TCF signaling, respectively, are predictive of poor colon cancer patient survival, which is mostly due to metastatic spread. Therefore, we suggest that epigenetic reprogramming underlies the metastatic transition and thus the development of metastases, overtaking the earlier role of genetic mutations in tumor progression.
Given that cell fate changes driven by pluripotent factors may be context-dependent, metastatic and iPSC fates could be two outcomes of OSK/M function with conserved features in radically different milieus, and a number of points can draw parallels between metastatic and iPSC reprogramming. (i) NANOG is a marker of consolidated iPSC reprogramming (Brambrink et al., 2008; Polo et al., 2012) and we find NANOG/P8 upregulated following the transient expression of exogenous OSK (3F) in cancer cells in vivo. (ii) NANOG function is important although not essential for iPSC formation (Silva et al., 2009; Schwarz et al., 2014), and we show that it is critical for metastases and EMT-like OSK-induced phenotypes, although not sufficient to induce these phenotypes. (iii) Reprogramming is epigenetically driven (reviewed in Apostolou and Hochedlinger, 2013) and we note that the pro-metastatic state driven by reprogramming factors is stable and involves methylation changes in NANOG early and SFRP1 late, although we have not detected demethylation of OCT4, which is typical of iPSC reprogramming. (iv) iPSC reprogramming converts differentiated cells into a stable embryonic-like (iPSC) stem cell state (Takahashi and Yamanaka, 2006) and we find that clonogenic cancer stem cells exhibit a stable pro-metastatic memory imposed by previous transient enhanced OSK expression. (v) The transient upregulation of LGR5 and DKK1 (but not of AXIN2) we report could reflect an involvement of aspects of WNT signaling in early metastatic reprogramming, much as in iPSC reprogramming (Lluis et al., 2008; Marson et al., 2008).
Nevertheless, it is difficult to directly compare metastatic and iPSC reprogramming since not only the contexts but also the protocols involved are distinct: OSKM reprogramming in normal mice leads to the appearance of iPSC-like cells after in vitro culture (Abad et al., 2013) whereas metastatic reprogramming does not select reprogrammed colonies in vitro and does not require ES media or feeder layers. Indeed, reprogramming of human colon cancer cell lines, following a full iPSC reprogramming protocol with ES media in vitro, has been reported to render them less or non-tumorigenic (Miyoshi et al., 2010; Miyazaki et al., 2014), a result radically different from our in vivo and in vitro data. Metastatic and iPSC reprogramming also diverge in that the first induces EMT-like phenotypes whereas the second involves the opposite (MET) transition to yield epithelial ES-like colonies.
The context but also the manner in which a cell is reprogrammed by pluripotent factors may be therefore crucial for the phenotype it acquires. OSK reprogrammed tumor cells in normal media or, critically, in vivo become pro-metastatic and this, we suggest, relates to the normal condition in colon cancers, in which SOX2 and KLF4 levels increase in metastatic vs. non-metastatic tumors whereas those of OCT4 are maintained (Varnat et al., 2010). We argue that metastatic reprogramming may be the consequence of high GLI function above a given threshold, driven by the oncogenic load (Stecca et al., 2007; Stecca and Ruiz i Ataba, 2010; Aberger and Ruiz i Altaba, 2014), that triggers repression of WNT-TCF signaling and enhanced pluripotent gene expression.
In contrast, under the experimental pressure of the in vitro protocol for iPSC formation, the same cells might instead respond to OSK function by becoming ES-like through the regulation of different pathways. As with colon cancer cells (Miyoshi et al., 2010; Miyazaki et al., 2014), recent data with leukemias suggest that tumoral cells can become normal through reprogramming in vitro (McClellan et al., 2015). Together, these studies highlight exciting aspects of cancer cell plasticity and have raised the possibility that an attractive anti-cancer therapy may be the in vivo reprogramming of cancer cells in patients. However, our data in vivo strongly suggests that such attempts will entail an unwanted enhancement of invasion and metastases resulting in worse prognosis.
Metastases include diseases with different clinical presentations. Full metastases can be rare events in patients since large primary tumors with millions of cells can yield single or a handful of metastatic lesions. However, small tumors can also sometimes spread quickly and in some cases the primary tumor is so small (or has regressed) that it cannot be found while its metastatic derivative is clinically problematic. It is therefore notable that whereas both OSKM (4F) and OSK (3F) drive the acquisition of pro-metastatic states, they do so in different manners, mimicking different disease presentations. Both yield large multicellular lesions as well as single cell micrometastases, but only OSKM drive an increase in tumor cell proliferation and an increase in the number of clonogenic cancer stem cells. OSKM function may therefore partly increase metastases by enhancing tumor growth although in colon cancer there does not appear to be a clinical correlation between tumor size and metastatic spread (Miller et al., 1985). In contrast, the ability of cancer stem cells to transmit the stable pro-metastatic phenotype of the primary tumor imposed by transient expression of OSK (without enhancing tumor growth or the number of clonogenic stem cells) supports the idea that reprogramming human cancers by OSK promotes metastases through a change in cancer stem cell identity or behavior. Recent findings indicating that different combination of reprogramming factors establish distinct embryonic-like fates (Tonge et al., 2014) may parallel our findings of different metastatic phenotypes.
Our present and previous data can suggest a novel model for the development of metastases. In colon cancer, positive GLI (GLI1/GLI2) activity is required for and can drive metastases as well as enhanced stemness and EMT-like phenotypes (Varnat et al., 2009, 2010). OSK and NANOG expression in colon cancer cells requires and is enhanced by positive HH-GLI activity (Varnat et al., 2010), and endogenous SOX2 and KLF4 levels are enhanced whereas those of OCT4 are maintained in patient cancers with metastatic spread as compared with non-metastatic tumors (Varnat et al., 2010). Together, these findings raise the possibility that endogenous GLI-regulated stemness factors, notably OSK and NANOG, may mediate pro-metastatic GLI function. Our model (Figure 8C) posits that enhancements in GLI activity induced by HH signaling and the oncogenic load (the sum of all tumor suppressor and oncogenic events in that cell) drive tumor progression up to advanced pre-metastatic stages. High GLI levels would then boost pluripotent, stemness factor function, leading to epigenetic reprogramming, EMT, invasion and metastasis.
Enhanced NANOG has been suggested to promote metastases in different cancers and its levels may be predictive for bad disease outcome (Meng et al., 2010; Lu et al., 2013; Sun et al., 2013). However, we find that its overexpression is not able to mimic the action of OSK in inducing EMT-like phenotypes, decreasing ECADHERIN levels, or enhancing distant metastases. Moreover, we did not find a correlation between NANOG levels and disease outcome in the data set we mined. NANOG may thus be required but not sufficient for metastases and metastatic reprogramming.
Our model may be applicable to other cancers as positive GLI activity is required for multiple human malignancies, HH-GLI regulates pluripotent stemness factors in various cancers (Clement et al., 2007; Ben-Porath et al., 2008; Varnat et al., 2010; Santini et al., 2014), and GLI and NANOG form a positive feed-forward regulatory loop in glioblastoma, a tumor that is rarely metastatic but is highly invasive (Zbinden et al., 2010). Preventing reprogramming and/or inhibiting GLI and NANOG function may provide considerable benefits to cancer patients.
The finding that local advanced colon carcinomas with linked metastatic spread (TNM3/4 tumors) and liver metastases have similar gene expression signatures (Varnat et al., 2009, 2010), may suggest that most if not all cells in the primary tumor have metastatic potential, much as in other cancers (Ramaswamy et al., 2003). However, not all cells in a tumor become metastatic. This plus the fact that most primary tumors are heterogeneous and metastases are often clonal (e.g. Fidler and Kripke, 1977; Yachida et al., 2010; Merlos-Suárez et al., 2011), poses the problem of the origin of metastases within the primary cancer. We suggest that the paradox of the derivation of rare metastases from cells within primary tumors with detectable (and thus likely largely homogeneous) metastatic signatures is resolved by very low-per-cell-penetrance metastatic reprogramming in cancer stem cells. There may thus be intrinsic barriers to metastatic behavior encoded by metastatic reprogramming suppressors, which might be infrequently or stochastically broken down. Defining suppressors of metastatic reprogramming and their variation in the human population may suggest new therapeutic approaches and reveal insights as to why the penetrance of metastases varies in patients with similar primary tumor diseases.
Materials and methods
Cell culture, BrdU incorporation, immunocytochemistry, and lentivectors
Primary colon cancer CC14 and CC36 were described in Varnat et al. (2010). Following a BrdU (3 µg/ml) pulse for 15 min, cells were fixed with fresh 4% paraformaldehyde, acid denatured, and immunostained with anti-BrdU antibodies (G3G4, University of Iowa Hybridoma Bank) at 1/5000 followed by fluorescent-secondary antibodies. Immunocytochemistry for ECADHERIN used Cell Signaling antibodies followed by fluorescent-labeled secondary antibodies and nuclear DAPI stain. The STEMCCA lentivector was used with vectors encoding rtTA to allow for dox induction (Sommer et al., 2009). Single factor (OCT4, SOX2, KLF4) lentivectors were from Addgene and the lacZ vector was described previously (Stecca et al., 2007). pLL3.7 parental (shControl), pLL3.7-shNANOG/P8 (GGGTTAAGCTGTAACATACT), and CMV-NANOG cDNA lentivectors were as described in Zbinden et al. (2010). Lentivector supernatants and concentrated particles were obtained by standard methods (Duquet et al., 2014). Transient expression of OSKM ranged from 2- to 12-fold and the knock-down afforded by shNANOG was 80% or greater in all experiments. At least four independent infections were performed for each vitro analysis and >2 for each vivo experiment.
3D. Culture in collagen, CD133 sorting, scratch assays and clonal spheroids
One hundred dissociated single cells were plated per well in 24-well plates with Type I Collagen (1 mg/ml). Cells proliferated for 10 days at which time colonies were counted and photographed. Indirect CD133 MACS (Miltenyi Biotec) was as in Varnat et al. (2009). Scratch assays were performed as described (Duquet et al., 2014): four observation areas were marked and the width of the scratched gap measured at different times. Clonal spheroids were obtained in batch after 14 days following the plating of 10000 dissociated cells in low adherent T75 flasks with DMEM/F12 plus B27 as in Varnat et al. (2009, 2010), or singly by plating one cell per well in low adherent 96 well plates.
Tail vein injections and detection of lacZ+ metastases
2D cultured cells were detached, dissociated, and resuspended in Ca- and Mg-free HBSS. 1 × 106 cells in 300 µl were injected in the lateral tail vein of host NUDE or NSG mice following approved protocols. Dissected organs were collected after 3 weeks for NSG or 7–8 weeks for NUDE mice, visualized for GFP fluorescence (see below) and stained with X-Gal (Stecca et al., 2007), photographed and the number of blue lacZ+ cells counted.
Subcutaneous xenografts and tumor processing
3 × 105–5 × 105 cells from 2D culture were detached and resuspended in 100 µl Ca- and Mg-free HBSS and injected in the flank of a NUDE mouse following approved protocols. The resulting tumors were allowed to grow and taken before reaching the local legal limit. The subcutaneous tumors were then dissociated and re-injected into a new host as above. Tumor samples were chopped and incubated in HBSS supplemented with 5 mM CaCl2, 1.5 mg/ml Type 2 Collagenase, and 0.1 mg/ml Hyaluronidase at 37°C for 30 min with brief intermittent vortexing. DMEM/F12 with 10% fetal bovine serum and penicillin/streptomycin was added and tissues mechanically dissociated by pipetting in the presence of 0.25 mg/ml DNAse I. Cells were harvested after washing in HBSS twice and passed through a 70 µm nylon cell strained prior to re-injection. Fluorescence of GFP+ xenografts were recorded using a color CCD camera (Lightools) and an appropriate laser.
RT-qPCR, methylation-specific PCR, and bisulfite sequencing
RNA and DNA extractions were performed with Trizol. cDNA preparation and PCR reactions using SYBRGreen were as in Duquet et al. (2014). Primers are described in Supplementary Table S1. The MethPrimer program (Li and Dahiya, 2002) was used to analyze presence of CpG islands, and to design MSP and bisulfite sequencing primers. MSP primer regions and primers are listed in Supplementary Figures S4 and S6. For MSP, PCR products were analyzed by agarose gel electrophoresis. Unmethylated and methylated DNAs from cells treated with 5 μM 5-azacytidine for 72 h and DNA methylated in vitro by M.SssI CpG methyltransferase were used as controls. Bisulfite DNA modification was performed with EZ DNA Methylation (Zymo Research). For NANOG and OCT4 sequencing PCR products were amplified with PfuturboCxHotstart DNA Polymerase (Agilent), TOPO TA (Invitrogen) cloned, and sequenced. To confirm that the primers for each target recognize only human bisulfite converted DNA, PCR reactions using bisulfite-converted mouse DNA as template were performed and shown to be negative.
Data mining and statistics
Human colon cancer cohort GSE17537 as well as the platform gene name list GPL570 were downloaded from the NCBI site. The expression level of genes of interest, as well as the corresponding survival times and the patient number were extracted. For each gene, the total cohort of patients was clustered in 25% high and 25% low expressors. The survival time and the death status were plotted using the Kaplan–Meier estimator. The Kaplan–Meier curves were tested for significant differences using a log-rank test. All experiments were done at least in triplicate and P-values were obtained by applying unpaired t-tests with two tails.
Supplementary material
Supplementary Material is available at Journal of Molecular Cell Biology online.
Funding
This work was funded by grants from the Swiss National Science Foundation, the Swiss Cancer League, the European Union grant HEALING, a James McDonnell 21st Century Brain Cancer Award, and funds from the Département d'Instruction Publique de la République et Canton de Genève, Switzerland, to A.R.A. C.B. was a recipient of postdoctoral Beca Chile and a EU-HEALING fellow. M.K. was a EU-HEALING fellow. I.S.-C. was a recipient of a long-term postdoctoral Human Frontier Science Program grant.
Conflict of interest: none declared.
Supplementary Material
Acknowledgements
We are grateful to all members of the Ruiz i Altaba lab for discussion and/or comments on the manuscript including Christophe Mas, Arnaud Duquet, Marie Zbinden, Chandan Seth, and Sonakshi Mishra. We thank G. Mostoslavsky (Massachusetts General Hospital Cancer Center, Harvard University, USA) for the STEMCCA vector.
References
- Abad M., Mosteiro L., Pantoja C. et al. (2013). Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature 502, 340–345. [DOI] [PubMed] [Google Scholar]
- Aberger F., Ruiz i Altaba A. (2014). Context-dependent signal integration by the GLI code: the oncogenic load, pathways, modifiers and implications for cancer therapy. Semin. Cell Dev. Biol. 33, 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou E., Hochedlinger K. (2013). Chromatin dynamics during cellular reprogramming. Nature 502, 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I., Thomson M.W., Carey V.J. et al. (2008). An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 40, 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambrink T., Foreman R., Welstead G.G. et al. (2008). Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell 2, 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y., Faddah D.A., Jaenisch R. (2013). Mechanisms and models of somatic cell reprogramming. Nat. Rev. Genet. 14, 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement V., Sanchez P., de Tribolet N. et al. (2007). HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr. Biol. 17, 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquet A., Melotti A., Mishra S. et al. (2014). A novel genome-wide in vivo screen for metastatic suppressors in human colon cancer identifies the positive WNT-TCF pathway modulators TMED3 and SOX12. EMBO Mol. Med. 6, 882–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon E.R., Vogelstein B. (1990). A genetic model for colorectal tumorigenesis. Cell 61, 759–767. [DOI] [PubMed] [Google Scholar]
- Fidler I.J., Kripke M.L. (1977). Metastasis results from preexisting variant cells within a malignant tumor. Science 197, 893–895. [DOI] [PubMed] [Google Scholar]
- Goswami R.S., Patel K.P., Singh R.R. et al. (2015). Hotspot mutation panel testing reveals clonal evolution in a study of 265 paired primary and metastatic tumors. Clin. Cancer Res. 21, 2644–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon J.B., Melton D.A. (2008). Nuclear reprogramming of cells. Science 322, 1811–1815. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K., Blelloch R., Brennan C. et al. (2004). Reprogramming of a melanoma genome by nuclear transplantation. Genes Dev. 18, 1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeter C.R., Badeaux M., Choy G. et al. (2009). Functional evidence that the self-renewal gene NANOG regulates human tumor development. Stem Cells 27, 993–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S., Chen W.D., Parmigiani G. et al. (2008). Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl Acad. Sci. USA 105, 4283–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogita A., Yoshioka Y., Sakai K. et al. (2015). Inter- and intra-tumor profiling of multiregional colon cancer and metastasis. Biochem. Biophys. Res. Commun. 458, 52–56. [DOI] [PubMed] [Google Scholar]
- Li L.C., Dahiya R. (2002). MethPrimer: designing primers for methylation PCRs. Bioinformatics 18, 1427–1431. [DOI] [PubMed] [Google Scholar]
- Lluis F., Pedone E., Pepe S. et al. (2008). Periodic activation of Wnt/beta-catenin signaling enhances somatic cell reprogramming mediated by cell fusion. Cell Stem Cell 3, 493–507. [DOI] [PubMed] [Google Scholar]
- Lu X., Mazur S.J., Lin T. et al. (2013). The pluripotency factor nanog promotes breast cancer tumorigenesis and metastasis. Oncogene 33, 2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A., Foreman R., Chevalier B. et al. (2008). Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell 3, 132–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan J.S., Dove C., Gentles A.J. et al. (2015). Reprogramming of primary human Philadelphia chromosome-positive B cell acute lymphoblastic leukemia cells into nonleukemic macrophages. Proc. Natl Acad. Sci. USA 112, 4074–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H.-M., Zheng P., Wang X.-Y. et al. (2010). Overexpression of nanog predicts tumor progression and poor prognosis in colorectal cancer. Cancer Biol. Ther. 9, 295–302. [DOI] [PubMed] [Google Scholar]
- Merlos-Suárez A., Barriga F.M., Jung P. et al. (2011). The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8, 511–524. [DOI] [PubMed] [Google Scholar]
- Miller W., Ota D., Giacco G. et al. (1985). Absence of a relationship of size of primary colon carcinoma with metastasis and survival. Clin. Exp. Metastasis 3, 189–196. [DOI] [PubMed] [Google Scholar]
- Miyazaki S., Yamamoto H., Miyoshi N. et al. (2014). A cancer reprogramming method using microRNAs as a novel therapeutic approach against colon cancer: research for reprogramming of cancer cells by microRNAs. Ann. Surg. Oncol. doi:10.1245/s10434-014-4217-1. [DOI] [PubMed] [Google Scholar]
- Miyoshi N., Ishii H., Nagai K. et al. (2010). Defined factors induce reprogramming of gastrointestinal cancer cells. Proc. Natl Acad. Sci. USA 107, 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K., Semi K., Yamamoto T. et al. (2014). Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell 156, 663–677. [DOI] [PubMed] [Google Scholar]
- Oshima N., Yamada Y., Nagayama S. et al. (2014). Induction of cancer stem cell properties in colon cancer cells by defined factors. PLoS One 9, e101735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantel K., Brakenhoff R.H. (2004). Dissecting the metastatic cascade. Nat. Rev. Cancer 4, 448–456. [DOI] [PubMed] [Google Scholar]
- Polo J.M., Anderssen E., Walsh R.M. et al. (2012). A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151, 1617–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S., Ross K.N., Lander E.S. et al. (2003). A molecular signature of metastasis in primary solid tumors. Nat. Genet. 33, 49–54. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A. (2011). Hedgehog signaling and the Gli code in stem cells, cancer and metastases. Sci. Signal. 4, pt9. [DOI] [PubMed] [Google Scholar]
- Sánchez-Tilló E., Liu Y., de Barrios O. et al. (2012). EMT-activating transcription factors in cancer: beyond EMT and tumor invasiveness. Cell. Mol. Life Sci. 69, 3429–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini R., Pietrobono S., Pandolfi S. et al. (2014). SOX2 regulates self-renewal and tumorigenicity of human melanoma-initiating cells. Oncogene 33, 4697–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz B.A., Bar-Nur O., Silva J.C. et al. (2014). Nanog is dispensable for the generation of induced pluripotent stem cells. Curr. Biol. 24, 347–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Nichols J., Theunissen T.W. et al. (2009). Nanog is the gateway to the pluripotent ground state. Cell 138, 722–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer C.A., Stadtfeld M., Murphy G.J. et al. (2009). Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells 27, 543–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecca B., Ruiz i Ataba A. (2010). Context regulation of the GLI code in cancer by HEDGEHOG and non-HEDGEHOG signals. J. Mol. Cell Biol. 2, 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecca B., Mas C., Clement V. et al. (2007). Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc. Natl Acad. Sci. USA 104, 5895–5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Sun L., Jiang K. et al. (2013). NANOG promotes liver cancer cell invasion by inducing epithelial-mesenchymal transition through NODAL/SMAD3 signaling pathway. Int. J. Biochem. Cell Biol. 45, 1099–1108. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Tonge P.D., Corso A.J., Monetti C. et al. (2014). Divergent reprogramming routes lead to alternative stem-cell states. Nature 516, 192–197. [DOI] [PubMed] [Google Scholar]
- Varnat F., Duquet A., Malerba M. et al. (2009). Human colon cancer epithelial cells harbour active HEDGEHOG-GLI signaling that is essential for tumour growth, recurrence, metastasis and stem cell survival and expansion. EMBO Mol. Med. 1, 338–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnat F., Siegl-Cachedenier I., Malerba M. et al. (2010). Loss of WNT-TCF addiction and enhancement of HH-GLI1 signalling define the metastatic transition of human colon carcinomas. EMBO Mol. Med. 2, 440–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.Q., Ng R.K., Ming X. et al. (2013). Epigenetic regulation of pluripotent genes mediates stem cell features in human hepatocellular carcinoma and cancer cell lines. PLoS One 8, e72435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M., Meissner A., Foreman R. et al. (2007). In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448, 318–324. [DOI] [PubMed] [Google Scholar]
- Yachida S., Jones S., Bozic I. et al. (2010). Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbinden M., Duquet A., Lorente-Trigos A. et al. (2010). NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. EMBO J. 29, 2659–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Espinoza L.A., Kinders R.J. et al. (2013). NANOG modulates stemness in human colorectal cancer. Oncogene 32, 4397–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.