Abstract
α-Actinin is an essential actin cross-linker involved in cytoskeletal organization and dynamics. The molecular conformation of α-actinin’s actin-binding domain (ABD) regulates its association with actin and thus mutations in this domain can lead to severe pathogenic conditions. A point mutation at lysine 255 in human α-actinin-4 to glutamate increases the binding affinity resulting in stiffer cytoskeletal structures. The role of different ABD conformations and the effect of K255E mutation on ABD conformations remain elusive. To evaluate the impact of K255E mutation on ABD binding to actin we use all-atom molecular dynamics and free energy calculation methods and study the molecular mechanism of actin association in both wild-type α-actinin and in the K225E mutant. Our models illustrate that the strength of actin association is indeed sensitive to the ABD conformation, predict the effect of K255E mutation—based on simulations with the K237E mutant chicken α-actinin—and evaluate the mechanism of α-actinin binding to actin. Furthermore, our simulations showed that the calmodulin domain binding to the linker region was important for regulating the distance between actin and ABD. Our results provide valuable insights into the molecular details of this critical cellular phenomenon and further contribute to an understanding of cytoskeletal dynamics in health and disease.
Introduction
The cytoskeleton mediates cell motility and adhesion that are critical for many cellular functions such as differentiation, proliferation, and growth (1, 2). Cytoskeletal dynamics is central to the force transmission machinery involved in converting mechanical stimuli into biochemical signals via the process of mechanotransduction (3, 4). α-Actinin accounts for the highly dynamic behavior of several actin assemblies (5, 6) and is crucial for cytoskeletal stabilization in both muscle and nonmuscle cells (7). Previous studies have shown that mutations in α-actinin lead to several diseases including muscular dystrophies and cardiomyopathy (8). Mutation at lysine 255 to glutamate in human α-actinin-4 results in a form of kidney failure, which occurs due to an increased affinity of α-actinin for actin that directly affects the cytoskeletal dynamics (9, 10).
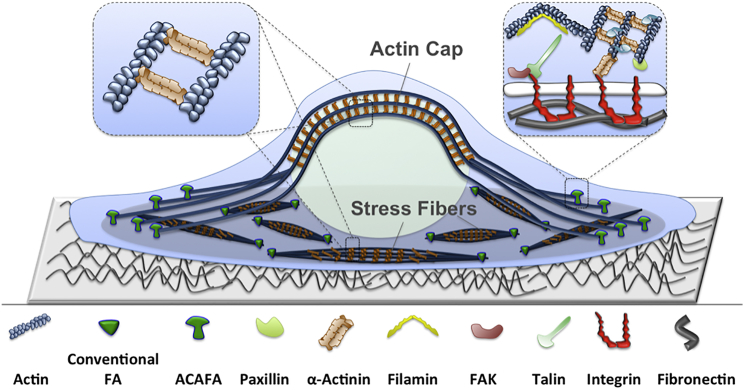
α-Actinin can cross-link both parallel and antiparallel actin filaments in various cytoskeletal structures and allow emergent actin bundles to exhibit different polarities (11). For instance, stress fibers are force-bearing actin bundles formed by highly organized α-actinin networks (12). Actin caps are also actin assemblies that directly connect the cell-substrate adhesions to the nucleus and are extended along the polarization axis of the cell as shown in Fig. 1 (6, 13). It has also been observed that α-actinin could link two points on a single actin filament, suggesting that the α-actinin function is tightly associated with its angular and linear elastic properties (11).
Figure 1.
α-Actinins are involved in formation and regulation of various cytoskeletal structures in adherent cells including stress fibers and actin caps. Stress fibers link two conventional focal adhesions and are made up of actin filaments cross-linked by α-actinins (shown in the left insert). The actin cytoskeleton can also be directly linked to focal adhesions through α-actinins (shown in the right insert). Actin caps also have a similar composition as stress fibers but are connected to ACAFA at their ends and are directly associated with the nucleus. To see this figure in color, go online.
One of the most widely studied pathways of cellular mechanotransduction is the integrin-mediated focal adhesions, which directly link the cytoskeleton to the extracellular matrix (14, 15). Focal adhesions are dynamic molecular complexes that can sense and transmit mechanical forces to the cytoskeleton (16, 17). α-Actinins also regulate stress fiber- and actin cap-associated focal adhesions, also referred to as conventional and ACAFA, respectively (Fig. 1) (6, 12, 18).
α-Actinin is a highly mechanosensitive molecule and has many important binding partners among focal adhesion players (5, 11, 12, 19, 20, 21). For instance, vinculin has a crucial role in force transmission and is recruited to the high stress regions in focal adhesion (22, 23, 24, 25). Its simultaneous interaction with actin and α-actinin reinforces the α-actinin-actin complex and possibly affect the local stiffness of the cytoskeleton (20). Vinculin’s binding site on α-actinin is within the fourth spectrin repeat (R4) and is likely to be mechanically regulated (26). α-Actinin can also directly link the actin cytoskeleton to the integrin receptors through its rod domain and contribute to both formation and maturation of focal adhesions (27, 28, 29).
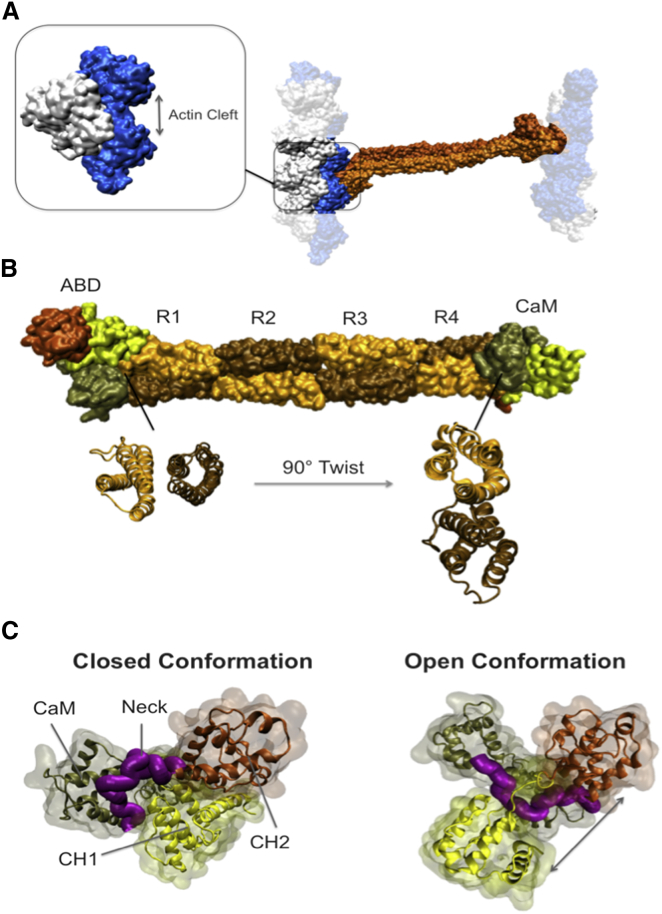
α-Actinin is an antiparallel homodimer as confirmed by its full-length structure resolved by Liu et al. in 2004 (30). Each α-actinin monomer has an actin-binding domain (ABD) at its N-terminus that is composed of two calponin homology (CH) domains. The rod domain is connected to the ABD domain via a flexible neck region and consists of four spectrin repeats (R1–R4) interacting strongly with the neighboring monomer (affinity ∼10 pM) as depicted in Fig. 2 (19, 31, 32). Of importance, there is an intrinsic 90° left-hand twist in the rod domain that puts ABDs of the two ends in different orientations relative to each other (Fig. 2, A and B).
Figure 2.
Important domains of the full-length α-actinin structure (PDB: 1SJJ). (A) The α-actinin (orange) molecule can cross-link two actin filaments each made up of two strands shown in white and blue. The F-actin model used in our simulations consisted of three actin monomers, which represents the simplest form of filamentous actin. Only two out of the three actin monomers can form a direct contact with α-actinin, which are referred to as the proximal actin monomers throughout this article. A cleft between the proximal actin monomers provided a space for α-actinin binding. (B) The actin binding domain of α-actinin is located at the N-terminus of each monomer consisting of two calponin homology domains, CH1 (yellow) and CH2 (dark orange), followed by four spectrin repeats forming the rod domain (sequentially colored in light orange and ochre) and the CaM domain at the C-terminal end (tan). A 90° twist of the rod domain is indicated by the cross-sectional views of the two ends. (C) The ABD domain takes two distinct conformations referred to as the closed (left) and open (right) conformations. In the closed conformation, CH1 (yellow) and CH2 (orange) are in contact and the neck region (purple) is positioned out of the CH1-CH2 interface, whereas in the open conformation CH domains are separated and the neck region sits in between them. The CaM domain (tan) also inserts itself between CH1 and CH2 in the open conformation. To see this figure in color, go online.
The C-terminal end of each α-actinin monomer contains a calmodulin-like domain (CaM) with two EF-hand motifs, which are next to the ABD domain of the other monomer. The CaM domain is involved in regulating the α-actinin function in a cell-dependent manner: in the nonmuscle α-actinin (isoforms 1,4), the actin binding affinity is controlled by the interaction between the Ca2+ ions and the EF-hand motifs (33), whereas muscle isoforms 2,3 are regulated by phosphatidylinositol 4,5-bisphosphate (PIP2) and have lost the capability of engaging with the Ca2+ ions throughout the evolution (33, 34, 35).
The CH domains are likely to be mechanically regulated and can either take a closed or an open conformation. It has been shown that the open conformation, in which the CH domains are dissociated, has a higher affinity for actin as shown in Fig. 2 C (10, 36). The CH1 domain contains the main actin binding sites that are mostly located on the protein surface except for ABS1, which is buried within the CH1-CH2 interface and only becomes accessible in the open conformation (36), thereby mutations that destabilize the interface, e.g., K255E may increase the actin binding affinity. It has also been shown that the CH1-CH2 fragment has a higher affinity for actin compared to the isolated CH1 suggesting a regulatory role for CH2 (37). However, the dynamic behavior of α-actinin during actin binding and the effect of ABD conformation on that are still elusive.
In this work, we study and characterize the binding between actin and two distinct ABD conformations of the full-length α-actinin using molecular dynamics (MD) simulations. We also investigate the effect of the K237E mutation in chicken α-actinin, which corresponds to the K255E mutation in humans, on the actin binding strength by exploring interactions of the mutant α-actinin during actin association and comparing the free energy profile of the mutant against the wild-type (WT). Our results are consistent with recent experimental studies and provide new, to our knowledge, insight on the molecular details that can further be employed to design modern therapeutics for impaired cellular functions associated with the cytoskeletal structure and dynamics.
Materials and Methods
All simulations were performed using the NAMD MD package (38) and CHARMM27 force field (39). The structure of the full-length α-actinin (Protein Data Bank (PDB): 1SJJ) was used (30) to model interactions with three actin monomers (PDB: 3LUE) as shown in Fig. 2 A (10). In the initial configuration of all binding simulations, the CH domains of the full-length α-actinin were positioned near two of the actin monomers isolated from the rest of the F-actin structure such that the ABD orientation relative to actin was similar to that in the structure of the actin-ABD complex (PDB: 3LUE). α-Actinin was then moved away from actin until a distance of 10 Å between the closest atoms of actin and those of α-actinin was reached. Constraints on the proteins were imposed during equilibration simulations, allowing water layers to form between them.
Due to the elongated structure of α-actinin, a triclinic box was used with the box vectors of 41.2 × 18.2 × 15.8 nm3, which satisfied the minimum image convention in all directions. A consistent version of the TIP3P water model with the CHARMM27 force field was used to solvate the simulation cell (40). The charge of the system was neutralized and the final ion concentration of the simulation cell was set to 150 mM of KCl to mimic the physiological condition inside the cytoplasm. The total number of atoms including protein, water, and ions reached 1,411,817.
The SHAKE algorithm was used to manage the bonds between hydrogens and heavy atoms. Electrostatic interactions were modeled using the particle mesh Ewald method and switching functions were applied to nonbonded interactions within the cut-off distance. All systems were initially minimized for 100,000 steps to remove all bad contacts and equilibrated for 1 ns using the NPT ensemble after which energy, pressure, and the root mean-square deviation of all simulations were monitored. The temperature was maintained at 310 using the Langevin dynamics and the pressure was kept at 1 bar with the Langevin piston. Periodic boundary conditions were applied in all three directions and a timestep of 2 fs was used. All production simulations ran for 60 ns using the NPT ensemble.
The umbrella sampling method (41) was used to calculate the potential of mean force (PMF) for both WT and mutated α-actinins. A range of different spring constants for the umbrella potential was examined to generate a series of smooth histograms with acceptable overlaps. The reaction coordinate was defined as the center of mass distance between the Cα atoms of CH1 (residues 26 to 146) and CH2 (residues 147 to 250) domains. To produce the initial configuration of each umbrella window, a steered MD simulation was performed in which the CH domains were pulled against each other with a constant rate of 0.005 nm/ps until the open conformation was reached. The step size along the reaction coordinate was set to 1 Å, which resulted in an optimized overlap between histograms. The total sampling time for each umbrella window was 1 ns as the CH1-CH2 distance was restrained using a spring constant of 3000 kJ Å−2 mol−2. It should be noted that for some umbrella windows a stiffer spring constant (3500–4500 kJ Å−2 mol−2) was necessary to avoid off-center displacement of the histograms. The trajectory was saved every 0.2 ps to gather sufficient sampling data. The final PMF and histograms were obtained using the g_wham analysis tool in GROMACS.
The visual MD software package (VMD) was used for visualizing and postprocessing the results (42). Furthermore, residue K237 was mutated to glutamate using the VMD plugin (42). The mutated α-actinin structure was carefully minimized and equilibrated before putting it in complex with actin to allow structural adjustments and remove bad contacts around the mutation site. The normal mode analysis and root mean-square fluctuations (RMSFs) were calculated using the Bio3D package in R (43). This work used resources of Extreme Science and Engineering Discovery Environment (44).
Results
Cell motility is highly dependent on the dynamic nature of the actin cytoskeleton at both cellular and molecular levels. Networks of actin filaments cross-linked by α-actinin form both two-dimensional and three-dimensional cytoskeletal structures with various sizes and shapes. It has been shown that the dynamics of actin assemblies are determined by the timescale of cross-linking (45, 46). Furthermore, it has been shown that the ABD conformation of α-actinin directly impacts actin association, thus mutations at the CH1-CH2 interface may interfere with the actin binding affinity of α-actinin. The K255E mutation in human α-actinin-4 is associated with a type of kidney disease and, as shown by the fluorescence microscopy, this mutation triples the affinity of α-actinin for F-actin and increases the average contractile stress by generating larger forces on the substrate. This mutation maps onto K237E in chicken α-actinin used in our simulations (Fig. S1 in the Supporting Material). We investigated the underlying mechanism of α-actinin binding to F-actin and the effect of K237E mutation on both the ABD conformation and the strength of actin binding using all-atomic MD simulations. Our results showed that the open conformation of ABD associated to actin with a higher strength and predicted a potential role for the CaM domain in regulating actin binding. Furthermore, our simulations indicated that the K237E mutation in chicken α-actinin decreased the level of forces required for opening the CH domains.
In the crystal structure of the smooth muscle α-actinin (PDB: 1SJJ), the ABD domain of each α-actinin monomer holds a different conformation (Fig. 2). We explored the binding between each of the ABD conformations with an actin trimer, however only two actin monomers were proximal to the ABD and the third integrated them together (Fig. 2 A). Also, in the initial configurations of all simulations, the distance between the surface atoms of F-actin and α-actinin was at least 10 Å while binding sites faced each other to reduce the diffusion time. It should be noted that the actin trimer, also referred to as the trimer nucleus, is a core from which actin polymerization initiates and thus can be considered as the simplest form of the filamentous actin. Moreover, initial trials showed that the actin trimer was likely to drift away before it could form a complex with α-actinin. Therefore, constraints were applied to the center of masses of actin monomers to shorten the time of dynamic complex formation. The high diffusivity of the actin trimer is not physiologically realistic, since actin monomers are usually confined within larger filaments. Therefore, we put soft harmonic restraints on Cα of three residues namely G13, G74, and G156 of each actin monomer to inhibit large translational and rotational motions throughout the simulations. Using the full-length α-actinin instead of the isolated ABD allowed us to study correlations between the structure and dynamics of different α-actinin domains upon actin binding. It should be noted that the actin trimer was placed only at one end of the α-actinin molecule in each simulation. To avoid confusion and repetitive terminology, we labeled our simulations and ABDs as summarized in Table 1.
Table 1.
Labels Used to Refer to Simulations and ABD Conformations
| Definition | Label |
|---|---|
| Simulations in which the open ABD of the wild-type α-actinin was placed near actin | OW |
| Simulations in which the closed ABD of the wild-type α-actinin was placed near actin | CW |
| Simulations in which the open ABD of the mutant α-actinin was placed near actin | OM |
| Simulations in which the closed ABD of the mutant α-actinin was placed near actin | CM |
| The open conformation of ABD | oABD |
| The closed conformation of ABD | cABD |
The effect of ABD conformation on the strength of actin binding and the cleft size
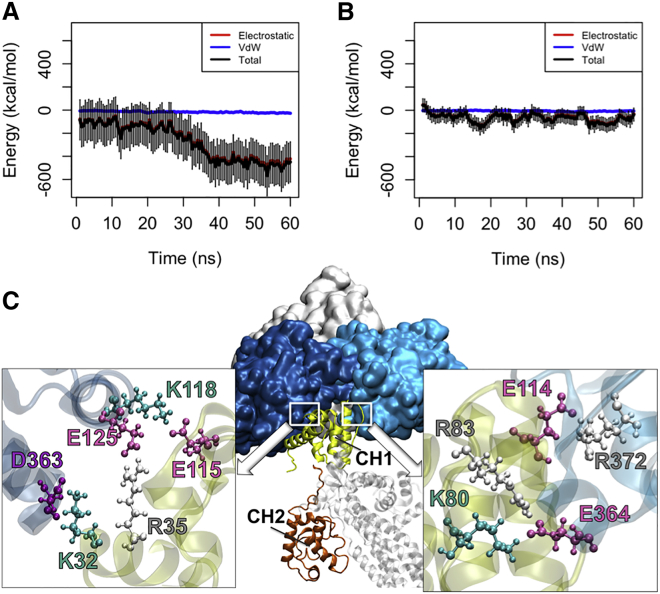
In the OW simulations (see Table 1), the CH1 domain formed a strong interaction (the average energy: −500 ± 100 kcal/mol) with actin after 30 ns that remained stable for the following 30 ns (Fig. 3 A). On the contrary, CH1 formed a much weaker interaction with actin in the CW simulations (Table 1) with the average interaction energy of −68.8 ± 37.7 kcal/mol (Fig. 3 B). Moreover, CH1 in the actin-associated open conformation of ABD (oABD) became engaged with both proximal actin monomers and formed three salt bridges with each as shown in Fig. 3 C, whereas part of the CH1 binding site in the closed conformation of ABD (cABD) was inhibited by CH2 and could only interact with a single actin monomer. Although the initial orientation of CH1 relative to actin was adjusted based on the cryo-electron microscopy structure, the CH1-actin interface was formed during the simulation and was not taken from the docked results reported by Galkin et al.
Figure 3.
The interaction between different ABD conformations and actin. (A) The CH1 domain of the oABD associated with actin after 30 ns with the final interaction energy of 550 kcal/mol averaged over three trials in the OW simulations. The interaction was stable for the last 20 ns of all trials. (B) On the contrary, the CH1 domain of the cABD did not strongly associate with actin within the 60 ns of simulation. (C) The interfaces between CH1 (yellow) of the oABD and two proximal actin monomers (blue) in the OW simulations are shown after 60 ns. Residues involved in all three salt bridges formed between CH1 and each actin monomer are highlighted (colors: arginine, white; lysine, turquoise; glutamic acid, pink; aspartic acid, purple). To see this figure in color, go online.
The solvent accessible surface area of the cABD decreased for the first 20 ns after which the average solvent accessible surface area did not notably change. This implied that the contact area of the cABD with actin remained intact during that time (Fig. S2). However, the detailed analysis of the binding trajectory revealed that the contact area of the cABD with actin dynamically changed in the last 35 ns and most interactions were transient.
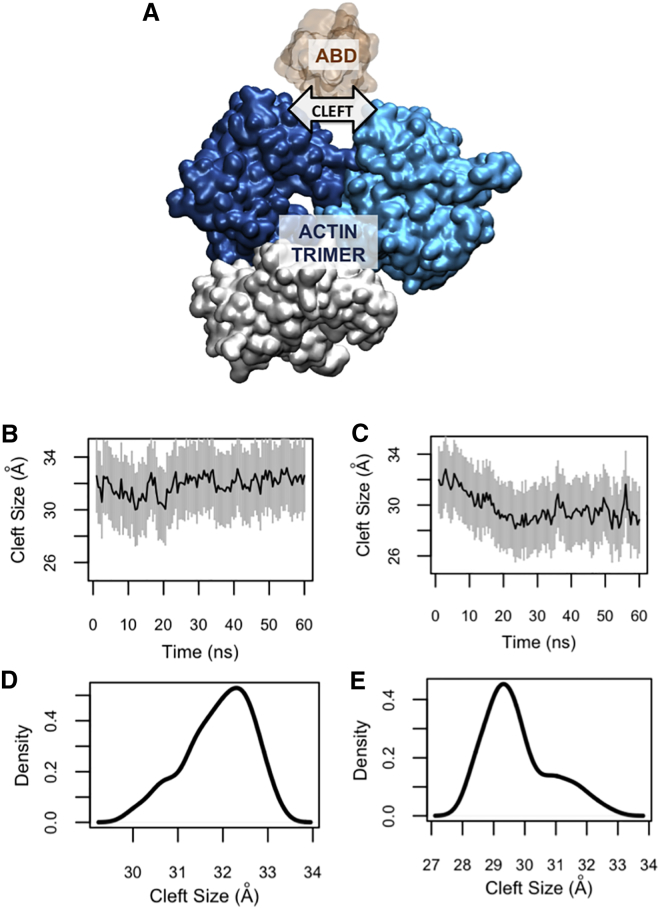
The actin cleft between the two proximal actin monomers served as the binding pocket for the ABD (Fig. 4 A). The size of the cleft was notably decreased by the oABD binding, while it only slightly increased upon the cABD association (Fig. 4, B and C). Comparing the density plots of the cleft size after the cABD and oABD binding showed a clear shift in the peak value of the cleft size indicating the sensitivity of the cleft to the ABD conformation (Fig. 4, D and E).
Figure 4.
The size of the actin cleft was affected by CH1 only after binding to the open conformation of ABD (oABD). (A) The structure of the actin trimer (blue and white) is shown in the presence of the ABD (orange). (B) The actin cleft size remained fairly constant upon binding to the closed conformation of ABD (cABD), whereas (C) it notably decreased after associating with the oABD. (D and E) The cleft size density plots are shown for a clear comparison. The peak value of the cleft after the oABD binding was 29 Å, while it was 32.5 Å for that after cABD binding. To see this figure in color, go online.
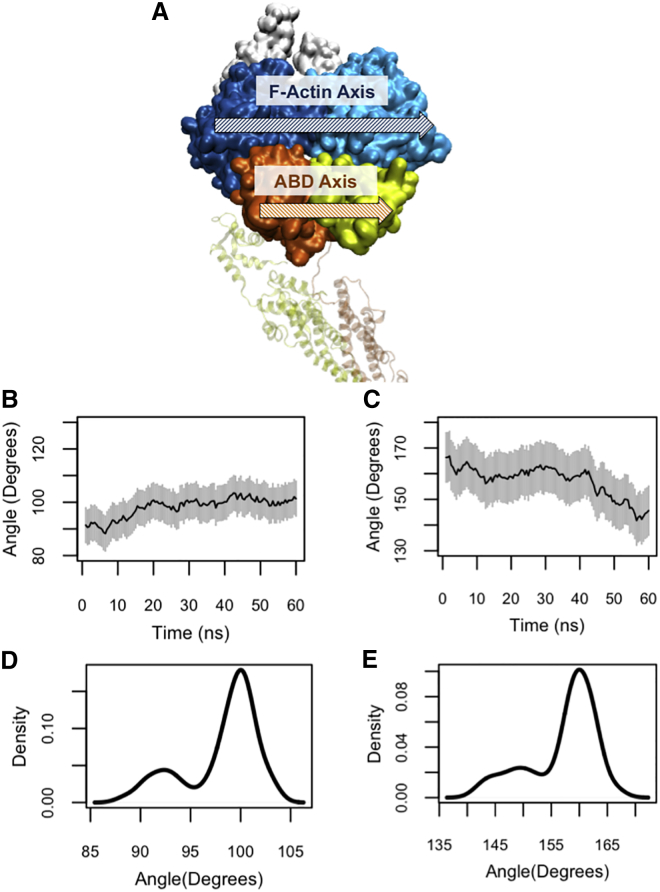
The ABD orientation relative to actin
The F-actin axis corresponds to the principal axis of the two proximal actin monomers in the actin trimer, therefore the relative orientation of the ABD along F-actin was easily quantified (Fig. 5 A). The oABD orientation in the OW simulations (Table 1) did not change drastically (Fig. 5 B), whereas the cABD rotated ∼30° relative to its initial orientation with respect to actin in the CW simulations (see Table 1 and Fig. 5 C). Of importance, the initial and final orientations of cABD and oABD were different from one another as shown by the position of peaks in the density plots shown in Fig. 5, D and E.
Figure 5.
(A) The principal axes of F-actin (blue) and the ABD domain (orange and yellow). (B and C) The angle between the principle axes of cABD and oABD relative to actin in the CW and OW simulations, respectively, showed a clear difference. (B) This angle did not change significantly after the oABD binding and ranged between 90° to 100°, whereas (C) it changed from 170° to 145° after the cABD binding. (D and E) Density plots show the peak value differences between the oABD (90°) and cABD (160°). Note that plots are presented in the same scale but different ranges. To see this figure in color, go online.
We also examined three other initial orientations of the oABD relative to actin to study other potentially favorable orientations for actin binding. The oABD was isolated from the rest of the structure and rotated around an axis orthogonal to F-actin for 90°, 180°, and 270° with respect to the original orientation used in all other binding simulations (Fig. S3, A and B). The oABD was equilibrated in complex with the actin trimer in four different simulations and energies were calculated and summarized after 5 ns as shown in Fig. S3 C. The results showed that the 180° rotation of the oABD resulted in a very favorable binding to actin in a short time. Furthermore, the 270°-rotated oABD also formed a strong interaction suggesting that various binding modes may occur between actin and α-actinin.
The role of the CaM domain in regulating actin binding
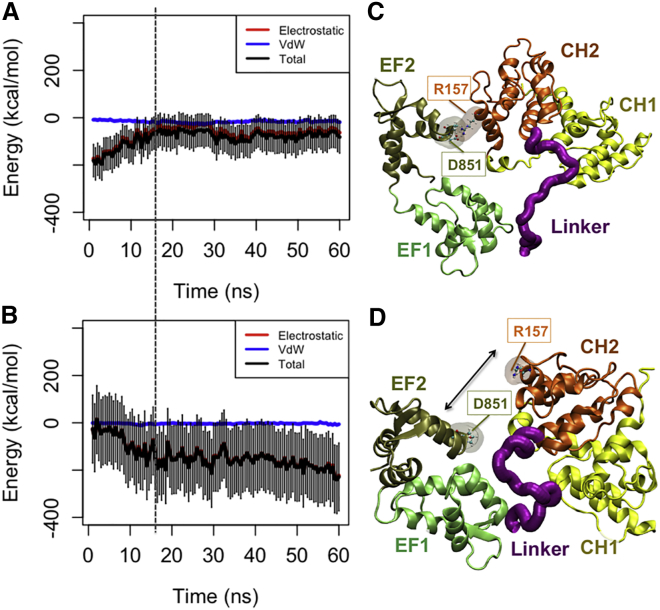
Since the CaM domain closely interacts with the ABD, it can directly influence both the orientation and distance of the ABD from actin. The CaM domain next to cABD was initially associated with the flexible neck region linking the cABD to the rod domain in the CW simulations (see Table 1) but was disassociated (Fig. 6 A) as CH2 started to interact with actin around 15 ns (Fig. 6 B). Specifically, an important electrostatic interaction between K265 in the neck region and D760 in the CaM domain was broken upon actin binding. In all simulations, the end-to-end distance of the neck region decreased pulling the cABD toward the rod domain. The EF1-hand motif engaged with the CH1 domain of the neighboring monomer after dissociating from the neck region.
Figure 6.
The CaM domain was released from the neck region as CH2 associated with actin in the CW simulations. (A) The interaction energy between CaM and the neck region was reduced around 18 ns (indicated by the dashed line), as (B) the CH2 domain got engaged with actin. (C and D) In one of the three CW trials, a salt bridge between D851 in CaM and R157 in CH2 was released allowing CH2 to form a more effective interaction with actin. (C) A salt bridge between R157 and D851 linked the CaM domain and the cABD in two out of three CW trials (see Table 1). CH1 is loosely associated with the second actin monomer blocking it from binding to CH2. Furthermore, the CaM domain itself associated with actin only in the presence of the salt bridge. (D) The disruption of the salt bridge between R157 and D851 linked the CaM domain and the cABD in one of the CW simulations released from CaM and resulted in a more effective binding between CH2 and actin. Only, in this trial, CH2 engaged with both proximal actin monomers, and the interaction with the second monomer followed the disruption of the salt bridge. To see this figure in color, go online.
A salt bridge between R157 and D851 linked the CaM domain and the cABD in two out of three CW trials but its disruption in the third CW simulation released the CaM domain and resulted in a more effective binding between CH2 and actin (Fig. 6, C and D). Only in this trial CH2 engaged with both proximal actin monomers and the interaction with the second monomer followed the disruption of the salt bridge. In two other trials, CH1 loosely associated with the second actin monomer blocking it from binding to CH2. Furthermore, the CaM domain itself associated with actin only in the presence of the salt bridge.
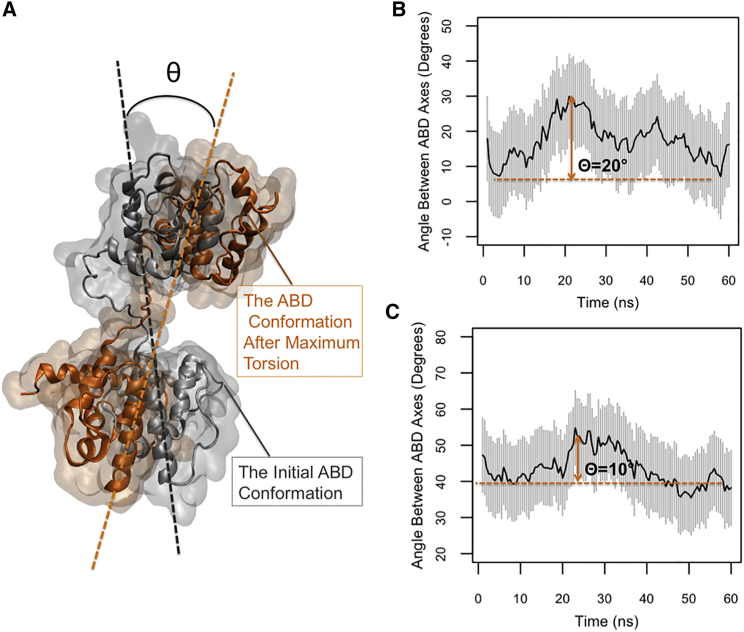
Torsion angle and allosteric effects
The torsional motion was illustrated by calculating the angle between the ABDs of the two ends of α-actinin in both CW and OW simulations (Fig. 7 A). The maximum change in the angle relative to the first frame of the CW simulations was 20° (Fig. 7 B), which was two times higher than that of the OW simulations (Fig. 7 C). This difference probably suggests that actin binding at one end can be transmitted as a mechanical signal to the other end by triggering the torsional mode of the molecule. Both plots in Fig. 7, B and C, indicate a single cycle of a periodic motion but the peaks occurred at different time points of the OW and CW simulations (Fig. 7, B and C).
Figure 7.
(A) The symbol θ represents the maximum change in the angle between the ABD axes relative to the initial frame (maximum torsion). (B) In the CW simulations, the value of θ averaged across three trials was 20° that was two times larger than that in (C) the OW simulations. To see this figure in color, go online.
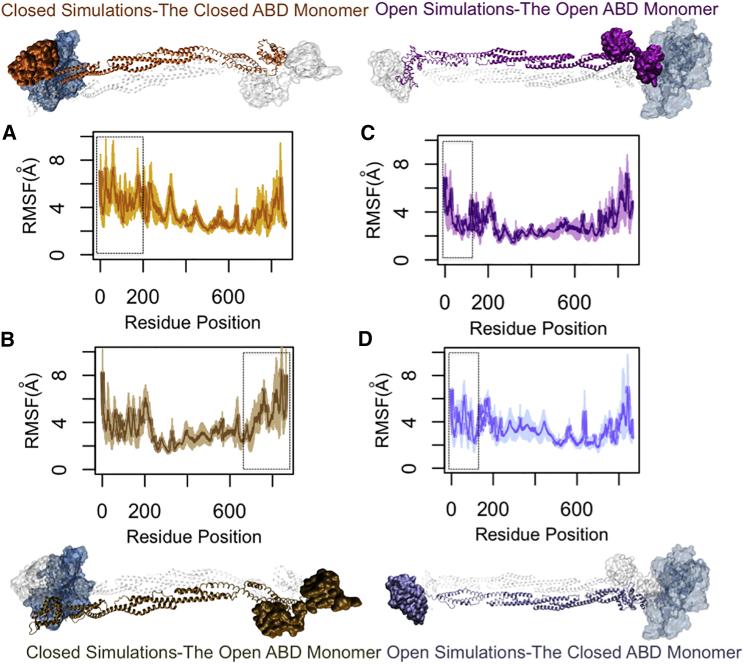
The RMSFs were calculated for both α-actinin monomers in the CW and OW simulations (Fig. 8). The rod domain consistently showed the lowest fluctuations in all simulations. The cABD of the actin-bound monomer and the CaM domain of the neighboring monomer seemed to be correlated in the CW simulations (dashed boxes in Fig. 8, A and B). The CH1 domain of the actin-bound oABD (dashed box in Fig. 8 C) showed lower fluctuations compared to that of the other monomer with the closed ABD (dashed box in Fig. 8 D).
Figure 8.
The RMSFs of α-actinin monomers both in bound and unbound states to actin. The rod domain had the lowest level of fluctuations in both CW and OW simulations. (A) The cABD domain (residues 25–250) in the CW simulations (the orange α-actinin monomer) underwent a slightly lower fluctuation compared to (B), the open ABD that was relative to actin (the tan α-actinin monomer). However, the CaM domain near actin (the C-terminal of the monomer with open ABD) showed a similar level of fluctuations to the cABD of its neighboring monomer in the CW simulations (dashed boxes). (C) In the OW simulations, the RMSF of the CH1 domain of the oABD was lowered upon actin binding (the purple α-actinin monomer) and was clearly different from that of the CH2 domain in the same monomer, whereas (D) the closed ABD that was far from actin did not show any clear difference between RMSFs of CH1 and CH2 (the ice blue α-actinin monomer). All plots are averaged over three trials. To see this figure in color, go online.
The role of the K237E mutation and the PMF
A mutant model of α-actinin was generated from the WT (PDB: 1SJJ) and aligned with the initial configuration of α-actinin in both CW and OW simulations to investigate the effect of the K237E mutation on actin binding. The mutant model was minimized and equilibrated in isolation before using it in the actin binding simulations to allow for possible structural adjustments. Our results showed that the CH1 domain of the mutant α-actinin in OM simulations associated with actin with an average interaction energy of −188.9 ± 74.2 kcal/mol that was slightly lower than that in the OW simulations (−284.6 ± 155.6 kcal/mol). Furthermore, our results indicated that the binding between the mutant α-actinin and actin was less abrupt and gradually strengthened over the course of 60 ns, whereas that binding with the WT α-actinin formed within the first 20 ns and remained stable for the following 20 ns (Fig. S4).
Our results suggested that the interface of the CH domains did not significantly change upon the mutation. We also examined all interactions of K237 at the interface that could potentially be altered after the mutation. It was previously suggested that W147 in the human CH1, which maps to W129 in the chicken CH1 (Fig. S1), forms a hinge-like connection with K255 in the human CH2 (K237 in the chicken CH2) maintaining the closed conformation (9). Our simulations also showed that the most significant interaction of K237 was with W129 (−6.1 ± 2.3 kcal/mol) that was lowered after the mutation (−2.7 ± 1.7 kcal/mol). Other nearby residues were W30, Q34, T37, and F38 but none of them showed a significant interaction. However, only Q34 formed a notable interaction with E237 in the mutant, which was not very stable (−7.3 ± 6.3 kcal/mol). Furthermore, the interaction energy between CH2 of the mutant oABD and R1 of the same monomer was lowered to −66.8 ± 26.2 kcal/mol from −173.7 ± 34 kcal/mol in the WT. Also, the distance between CH1 and CH2 was slightly smaller in the mutant than that of the WT, which may allow for more spontaneous regulatory interactions.
To estimate the free energy cost of changing the ABD conformation from closed to open, we used the steered MD simulations to separate the CH domains of the cABD and produce an artificial open conformation. The closed-to-open trajectory was then used to extract initial configurations of the umbrella windows from which the PMF was calculated. Because the structure of individual CH domains are independent of whether they are paired or not, the distance between them seemed to be a reasonable reaction coordinate for the PMF calculation.
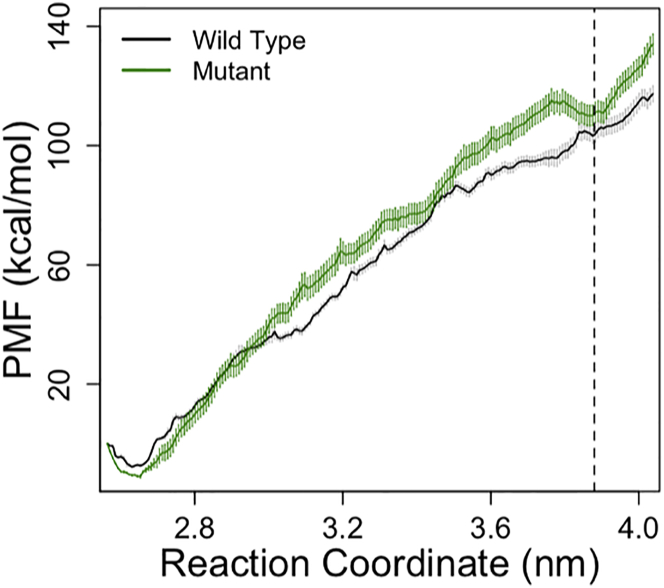
The PMF profile of both WT (black) and mutant (green) α-actinins are shown in Fig. 9 in which the horizontal axis is the separation distance between the centers of masses of the CH domains. Although, the energy landscape is quite rough, the minimum corresponding to the closed conformation (∼2.7 nm) was clearly similar between the WT and mutant. The PMF profiles followed each other closely up to 3 nm but deviated from that point on and a new minimum was formed for the mutant in the open state.
Figure 9.
Comparing the PMF profiles of the WT (black) and mutant (green) α-actinins shows that a new minimum (at 3.9 nm) was created upon the K237E mutation. The CH1-CH2 distance was taken as the reaction coordinate for the umbrella sampling calculations. Profiles are fairly similar up to 3 nm but deviate after that. The slope of the mutant PMF is slightly higher but reaches a short plateau around 3.3 nm. The error bars were calculated using the bootstrap analysis. To see this figure in color, go online.
Discussion
α-Actinin is a major cytoskeletal component involved in cross-linking actin filaments in stress fibers and linking them to focal adhesions as well as forming actin caps between the nucleus and adhesion sites. Therefore, any dysfunction of this protein can lead to serious pathogenic conditions mostly caused by the changes in its actin binding affinity. Although, it was shown that the ABD conformation is a deterministic factor in regulating the α-actinin function, current studies including the available crystal structures of ABD do not provide much information on the ABD dynamics neither in isolation nor during associating with actin. Certain point mutations in the ABD domain have been linked to human diseases (10), however, possible regulatory mechanisms targeted by those mutations are not yet fully understood. In this study, we investigated a mutation at residue K255 in the ABD of human α-actinin that results in a late onset form of kidney failure. To our knowledge, this is the first computational study on characterizing the effect of the K255E mutation in humans, which maps to the K237E mutation in chickens, on the binding between the full-length α-actinin and F-actin compared against the WT. Our simulations showed that the K237E mutation in chicken α-actinin reduced the forces required for opening the CH domains. Moreover, our results indicated a clear favorability of the open conformation of ABD for actin binding and also predicted a role for the CaM domain in regulating actin association.
Hampton et al. suggested that α-actinin had to be a flexible cross-linker as opposed to a rigid molecular component to allow the observed mechanical properties of cytoskeletal structures (11). Therefore, it is important to understand how structural features of α-actinin influence its molecular rigidity as well as its binding to actin in both health and disease. Furthermore, it was shown that α-actinin can also link two points on a single actin filament (11), thus the orientation of α-actinin relative to actin may alter based on it binding status.
There are various hypotheses on the mechanism of binding between different ABD conformations to actin. In this study, we explored the binding mechanisms of both ABD conformations to actin using MD simulations. However, it should be noted that as comparably high-resolution structural data that can uniquely resolve the atomic positions of our proteins is not available, both α-actinin and actin structures were extensively minimized and equilibrated until the system energy became stable. Furthermore, as MD simulations take atomic scale interactions into account, it can be used to predict protein-protein binding interfaces using reliable force fields such as CHARMM27 (47, 48). Protein structures will presumably reach an equilibrium state after a sufficiently long simulation time even if the simulations start off a low-resolution structure. It should be noted that atomic-level predictions have a lower confidence due to the resolution of the initial structures, however, both coarse and fine analyses can provide insight into the dynamics of the system and can further be validated in future experimental studies.
Since the CH1 domain containing the main actin binding sites is in contact with CH2 in the closed conformation, it has been suggested that the open conformation of ABD should have a higher affinity for actin due to a lower steric hindrance imposed by CH2 (10). Our simulations confirmed that the open conformation was indeed more favorable for actin binding as CH1 of the oABD formed six stable salt bridges with both proximal actin monomers, while the interaction between CH1 of the cABD and actin was much weaker. Furthermore, we observed that the actin cleft was narrowed down upon binding to the oABD, possibly generating a higher local stress. This suggests that the accumulation of cross-linkers may decrease the average length and affect the stress distribution along actin filaments.
Moreover, the final configuration of the CH1-actin complex in the OW simulations was similarly oriented compared to the structure of the CH1-decorated F-actin (PDB: 3LUE). Our results suggested that CH2 prevents CH1 from effective binding to actin by the following mechanisms: 1) blocking part of the actin binding site on CH1, i.e., residues K32 and R35, which are exposed in oABD (10); and 2) interacting with one of the actin monomers across the cleft and inhibiting it from engaging with CH1.
Borrego-Diaz et al. (36) proposed a mechanism for actin binding by which CH2 of the cABD first associates with actin filaments releasing CH1 for further interactions with actin. We also observed that CH2 in the CW simulations interacted relatively strongly with actin before CH1, but we did not capture full dissociation of CH1-CH2 in 60 ns most likely because either the timescale of such conformational change is longer or axial stress along actin is also necessary (36).
Because the CaM domain is in the close proximity and most often interacting with the cABD, it has an important regulatory role in the actin binding activity of α-actinin but the molecular mechanism is not yet fully understood. In the muscle isoform (α-actinin 2,3), PIP2 regulates the ABD function by disrupting the hydrophobic interaction between the CaM domain and the neck region (49), whereas in the nonmuscle α-actinin isoforms 1 and 4, the CaM domain regulates the actin binding activity of the neighboring ABD via calcium binding (33). A similar function has also been observed for some other members of the spectrin family (50).
We did not include the calcium ions within the EF-hand motifs in our simulations but still observed some correlations between the actin binding activity of ABD and interactions of the CaM domain (51). Specifically, the CaM domain associated with both CH2 and actin placing the cABD next to just one actin monomer in two trials of the CW simulations, whereas dissociated CaM allowed CH2 to engage simultaneously with both proximal actin monomers pushing CH1 away from actin in the third trial. It should be noted that this was the only result not consistent among all trials. Interaction of CH2 with two actin monomers may prevent effective transmission of forces to the cABD since, most likely, CH1 and CH2 need to be pulled against one another via axial tension along F-actin for complete opening of the cABD. Therefore, we suggest that the CaM domain is likely to be involved in regulating the ABD conformation by controlling the transmission of mechanical forces to the cABD. Furthermore, we observed that the association of the CaM domain to the same actin monomer as CH2 resulted in stronger anchorage to actin and thus may play a role in the early stages of the binding between the cABD and actin. However, in the OW simulations the relative positions of the CaM domain and the neck region with respect to the actin-bound oABD remained intact implying that no further conformational changes were required for regulating the actin binding activity of the oABD. Again, it should be noted that the dynamics of α-actinin binding to actin can also be highly influenced by Ca2+ and PIP2 and other regulatory factors.
We observed several distinct motions of α-actinin in the OW and CW simulations upon actin binding. Specifically, deviation of the cABD axis from its original orientation after actin binding was larger than that of the oABD in all trials (Fig. 3). This suggests that the ABD can reorient even in its bound state to actin in a conformation-dependent manner and is not confined to its initial alignment. Furthermore, we investigated the binding between different oABD orientations and actin for a shorter timescale to gain insight into the effect of the initial ABD angle on the strength of its actin binding. We observed that the 180°-rotated oABD that was the reversed orientation relative to the initial configuration of oABD used in all OW simulations (also referred to as the 0° rotation) and the 270°-rotated oABD formed the most favorable interactions in 5 ns (Fig. S3). Because α-actinin can cross-link both parallel and antiparallel actin filaments that easily rearrange and move in all possible directions, dynamic adaptation to the new arrangements is most likely a critical characteristic of α-actinin as an actin cross-linker also suggested by our simulations.
Interestingly, in all OW simulations the rod domain was curved in the opposite direction of that in the CW simulations (Fig. S5). This suggests that the bending mode of the α-actinin molecule can be triggered during actin binding but the phase of such motion may be dependent on the ABD conformation. Also, twisting is one of the top molecular modes of α-actinin that may be signified by the angle between the principle axes of ABDs at the two ends of the molecule. Our results showed that the oABD binding to actin caused a larger twist in the molecule compared to that of the cABD. This may imply that strong actin binding anchors the molecule at one end but still allows for large changes in the ABD orientation at the other end, i.e., α-actinin does not act like a rigid body after binding to actin. Furthermore, since α-actinin monomers are tied together via strong hydrophobic interactions within the coiled-coil structure of the rod domain, it is conceivable to expect some allosteric communication between the two ends of the molecule. We suggest that a mechanical signal is transferred upon actin binding between the two ends of the molecule via triggering the natural modes of the rod domain, i.e., twisting (Fig. 7 and Movie S1) and bending (Fig. S5 and Movie S2).
We further studied the effect of the K237E mutation on both the ABD conformation and actin binding. No appreciable conformational changes were observed in the cABD domain upon the mutation per se, however, the average interaction energy of the CH1-CH2 interface over time was changed from −428.50 ± 58.99 in the WT compared against that −319.67 ± 80.10 in the mutant (Fig. S6). This may imply that the mutation alone is not sufficient to initiate the closed-to-open conformational change, at least not in the nanosecond timescale, and mechanical tension may either be necessary to expedite or complete this process. We also observed that the interactions between the mutant α-actinin and actin formed more gradually compared to the WT (Fig. S6). This may suggest that the mutation created more transition states between the bound and unbound states with actin resulting in a smoother binding. Specifically, the probability of actin association may increase upon the mutation since introducing a higher number of transition states tends to expedite both physical and chemical reactions.
It has been shown that the CH1-CH2 fragment has a higher affinity than the isolated CH1 for actin suggesting an important regulatory role for CH2, however it is not yet clear how mutations in CH2 can influence the binding between CH1 and actin (37). We suggest that this could be due to the following: CH2 anchors the ABD to actin placing CH1 in the right orientation and contributes to regulating the size of the actin cleft, whereas CH1 alone is small and globular in shape and would probably have to rely on diffusion for adjusting to a favorable orientation relative to actin.
Our simulations showed that CH2 of the mutant α-actinin was released from the R1 spectrin repeat in the rod domain in all trials and thus experienced lower hindrance for spatial movements toward CH1, which may be important for regulating its orientation and binding. We would like to further emphasize that our results provide an insight into the dynamics of the system in the nanosecond timescale and thus different average behaviors may be expected in longer times, e.g., microseconds and milliseconds.
The constant rate pulling simulations indicated that the force required to separate CH1 and CH2 was markedly lower for the mutant compared to that for the WT (Fig. S7). Specifically, a notable difference in the pulling forces was observed as the CH1-CH2 interface was being disrupted, although before and after that time force curves followed each other closely (Fig. S7). Since there was no negatively charged residues in the vicinity of K237, we did not expect any direct influence of the charge on the CH1-CH2 interface. However, the pulling trajectories showed that the K237/E237 residue moved very close to D27 during opening and causing a strong attraction/repulsion between glutamate and aspartate. Furthermore, the pulling simulations showed that an interaction between residue 237 and R134 on the CH1-CH2 linker region occurred around the same time as the trend of the pulling forces between the CH domains of the mutant deviated from that of the WT (Fig. S7). The bond between CH2 and the linker region reduces the likelihood of structural reformation to the closed conformation. This is another potential reason why the K237E mutation stabilizes the open conformation as suggested by the minimum in the PMF plot (Fig. 9). This indicates that the mutation does not directly induce a conformational change in the ABD. However, as the CH1-CH2 interface is altered in response to mechanical stimuli or other physical cues, the interaction pattern may change upon the mutation.
Moreover, the PMF profile of α-actinin was calculated using the umbrella sampling method and the error bars were estimated by the bootstrap analysis to quantify the free energy cost of the closed-to-open conformational change in the ABD for both WT and mutant α-actinins. The free energy difference between the closed and open conformations of the CH domains were −112.6 ± 2.6 kcal/mol and −119.8 ± 4.9 kcal/mol for the WT and mutant, respectively. One obvious difference between the two PMF profiles is the appearance of a local minimum where the open conformation was reached for the mutant. This may suggest that the open conformation of the mutant is relatively more stable compared to the WT as the system gets trapped within a local minimum. It should be noted that actin monomers were not included in the umbrella sampling simulations, because they would introduce many extra degrees of freedom to the system making it increasingly more difficult to carry out the free energy calculation with a reasonable accuracy. However, the direction of tension and the presence of F-actin may affect the PMF profile, especially the first potion where CH1 and CH2 are still in contact. Our results suggested that the open conformation of the mutant α-actinin is more stable (Fig. 9) and also requires lower forces (Fig. S7).
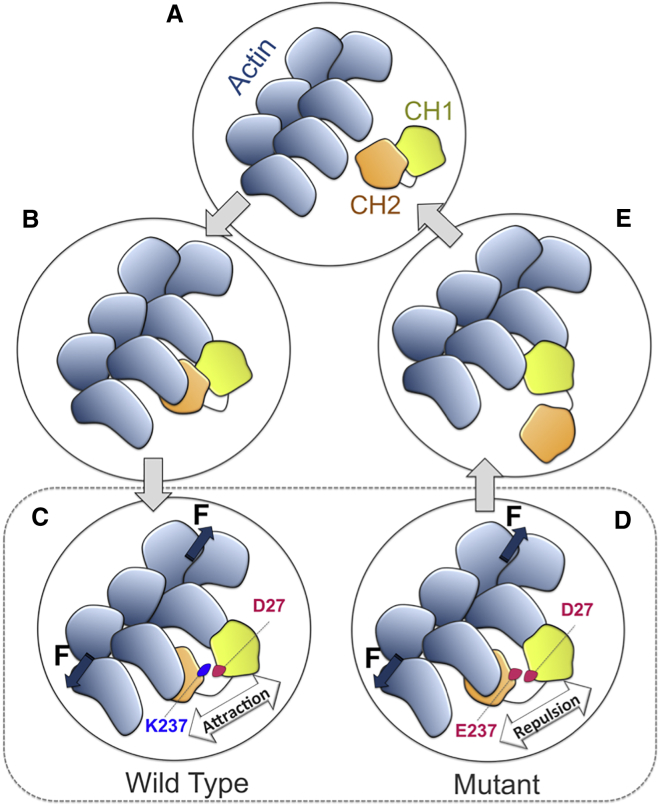
In summary, we proposed a molecular mechanism for the binding between α-actinin and actin based on a consensus among all our simulation trials as follows: 1) the α-actinin molecule diffuses toward the actin filament most likely with ABDs in the closed conformation and adjusts its orientation (Fig. 10 A). 2) The binding between CH2 and one actin monomer occurs first, which may be regulated by the CaM domain (Fig. 10 B, the CaM domain is not shown for clarity), and is followed by the CH1 interaction with the other monomer across the actin cleft. 3) Axial tension along the actin filament broadens the cleft causing the distance between CH1 and CH2 to change as they are bound to two different actin monomers. A transition state is reached where the CH1-CH2 interface is altered by the mechanical stimuli and D27 is pulled toward the interface. In the WT, D27 will face K237 creating a strong attraction that may prevent further opening of the ABD unless it is overcome by the axial tension along F-actin (Fig. 10 C). However, the negative charge of D27 faces E237 in the mutant creating a repulsive force that may expedite and reduce the amount of forces required for further ABD opening (Fig. 10 D). 4) As the CH2 domain is completely dissociated, CH1 moves further into the actin cleft and replaces CH2 due to its high affinity for actin (Fig. 10 E).
Figure 10.
The proposed mechanism of the ABD binding to actin. (A) The cABD diffuses toward the actin filament (B) allowing CH2 to engage with an actin monomer along F-actin that is followed by a weak interaction of CH1 with a neighboring actin monomer. (C) Axial tension along F-actin broadens the actin cleft and partially opens the cABD. Residue D27 on CH1 moves toward the interface with CH2 resulting in either a strong attraction toward K237 in the WT or a repulsion against E237 in the mutant and thus further opening of the ABD would require a different amount of forces depending on the type of residue 237. (D) As CH1 and CH2 are completely separated by axial tension and the steric hindrance imposed by CH2 is removed, (E) CH1 moves further into the actin cleft and replaces CH2 on the other monomer. Therefore, the open conformation can be maintained in the bound state to actin. To see this figure in color, go online.
Conclusions
The conformation and dynamics of the ABD domain of α-actinin play an important regulatory role in actin binding and here we proposed a mechanism for that. Our simulations predicted that the K237E mutation in the chicken α-actinin, which corresponds to K255E mutation in humans associated with a type of kidney failure, change the free energy landscape and most likely stabilize the open conformation of the ABD. Furthermore, the flexibility of the ABD orientation relative to actin suggests that α-actinin can cross-link F-actins with different angles leading to formation of various actin assemblies. Also, actin association at one end of the α-actinin molecule may be transmitted as a mechanical signal via the natural modes of the rod domain. Our simulations shed light on understanding controversies about the effect of the ABD conformation and mutations in that domain on the actin binding activity of α-actinin. Therefore, the insight from our simulations can be used to develop novel techniques and therapeutics to interfere with the α-actinin function and the kidney disease associated with the K255E mutation in humans.
Author Contributions
Conceived and designed the experiments: H.S., J.G., and M.R.K.M. Performed the experiments: H.S., J.G., and K.G. Analyzed the data: H.S., J.G., and M.R.K.M. Contributed reagents/materials/analysis tools: M.R.K.M. Wrote the article: H.S., J.G., and M.R.K.M.
Acknowledgments
We thank Amer Abdulla, Zeinab Jahed, and other members of the Molecular Cell Biomechanics Laboratory for fruitful discussions.
This work was supported by the National Science Foundation CAREER Award CBET-0955291 to M.R.K.M. In addition, this research used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant No. ACI-1053575.
Editor: Andrew McCulloch.
Footnotes
Seven figures and two movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)00212-5.
Supporting Material
In this mode, the rod domain of α-actinin twists such that the actin binding domains move in opposite directions.
In this mode, the rod domain of α-actinin bends in the middle such that the actin binding domains move in the same direction.
References
- 1.Mofrad M.R.K., Kamm R.D. Cambridge University Press; New York: 2011. Cytoskeletal Mechanics Models and Measurements in Cell Mechanics. [Google Scholar]
- 2.Anitei M., Hoflack B. Bridging membrane and cytoskeleton dynamics in the secretory and endocytic pathways. Nat. Cell Biol. 2012;14:11–19. doi: 10.1038/ncb2409. [DOI] [PubMed] [Google Scholar]
- 3.Jahed Z., Shams H., Mofrad M.R.K. Mechanotransduction pathways linking the extracellular matrix to the nucleus. Int. Rev. Cell Mol. Biol. 2014;310:171–220. doi: 10.1016/B978-0-12-800180-6.00005-0. [DOI] [PubMed] [Google Scholar]
- 4.DeMali K.A., Sun X., Bui G.A. Force transmission at cell-cell and cell-matrix adhesions. Biochemistry. 2014;53:7706–7717. doi: 10.1021/bi501181p. [DOI] [PubMed] [Google Scholar]
- 5.Edlund M., Lotano M.A., Otey C.A. Dynamics of alpha-actinin in focal adhesions and stress fibers visualized with alpha-actinin-green fluorescent protein. Cell Motil. Cytoskeleton. 2001;48:190–200. doi: 10.1002/1097-0169(200103)48:3<190::AID-CM1008>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 6.Kim D.-H., Khatau S.B., Wirtz D. Actin cap associated focal adhesions and their distinct role in cellular mechanosensing. Sci. Rep. 2012;2:555. doi: 10.1038/srep00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchard A., Ohanian V., Critchley D. The structure and function of alpha-actinin. J. Muscle Res. Cell Motil. 1989;10:280–289. doi: 10.1007/BF01758424. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi T., Arimura T., Kimura A. Tcap gene mutations in hypertrophic cardiomyopathy and dilated cardiomyopathy. J. Am. Coll. Cardiol. 2004;44:2192–2201. doi: 10.1016/j.jacc.2004.08.058. [DOI] [PubMed] [Google Scholar]
- 9.Weins A., Schlondorff J.S., Pollak M.R. Disease-associated mutant alpha-actinin-4 reveals a mechanism for regulating its F-actin-binding affinity. Proc. Natl. Acad. Sci. USA. 2007;104:16080–16085. doi: 10.1073/pnas.0702451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galkin V.E., Orlova A., Egelman E.H. Opening of tandem calponin homology domains regulates their affinity for F-actin. Nat. Struct. Mol. Biol. 2010;17:614–616. doi: 10.1038/nsmb.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hampton C.M., Taylor D.W., Taylor K.A. Novel structures for alpha-actinin:F-actin interactions and their implications for actin-membrane attachment and tension sensing in the cytoskeleton. J. Mol. Biol. 2007;368:92–104. doi: 10.1016/j.jmb.2007.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hotulainen P., Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J. Cell Biol. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chambliss A.B., Khatau S.B., Wirtz D. The LINC-anchored actin cap connects the extracellular milieu to the nucleus for ultrafast mechanotransduction. Sci. Rep. 2013;3:1087–1093. doi: 10.1038/srep01087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harburger D.S., Calderwood D.A. Integrin signalling at a glance. J. Cell Sci. 2009;122:159–163. doi: 10.1242/jcs.018093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puklin-Faucher E., Sheetz M.P. The mechanical integrin cycle. J. Cell Sci. 2009;122:179–186. doi: 10.1242/jcs.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galbraith C.G., Yamada K.M., Sheetz M.P. The relationship between force and focal complex development. J. Cell Biol. 2002;159:695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S.E., Kamm R.D., Mofrad M.R.K. Force-induced activation of talin and its possible role in focal adhesion mechanotransduction. J. Biomech. 2007;40:2096–2106. doi: 10.1016/j.jbiomech.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Khatau S.B., Hale C.M., Wirtz D. A perinuclear actin cap regulates nuclear shape. Proc. Natl. Acad. Sci. USA. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golji J., Collins R., Mofrad M.R. Molecular mechanics of the alpha-actinin rod domain: bending, torsional, and extensional behavior. PLoS Comput Biol. 2009;5:e1000389. doi: 10.1371/journal.pcbi.1000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bois P.R.J., Borgon R.A., Izard T. Structural dynamics of alpha-actinin-vinculin interactions. Mol. Cell. Biol. 2005;25:6112–6122. doi: 10.1128/MCB.25.14.6112-6122.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 21.Zaman M.H., Kaazempur-Mofrad M.R. How flexible is alpha-actinin’s rod domain. Mech Chem Biosyst. 2004;1:291–302. [PubMed] [Google Scholar]
- 22.Carisey A., Tsang R., Ballestrem C. Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr. Biol. 2013;23:271–281. doi: 10.1016/j.cub.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S.E., Chunsrivirot S., Mofrad M.R.K. Molecular dynamics study of talin-vinculin binding. Biophys. J. 2008;95:2027–2036. doi: 10.1529/biophysj.107.124487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carisey A., Ballestrem C. Vinculin, an adapter protein in control of cell adhesion signalling. Eur. J. Cell Biol. 2011;90:157–163. doi: 10.1016/j.ejcb.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holle A.W., Tang X., Engler A.J. In situ mechanotransduction via vinculin regulates stem cell differentiation. Stem Cells. 2013;31:2467–2477. doi: 10.1002/stem.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shams H., Golji J., Mofrad M.R.K. A molecular trajectory of α-actinin activation. Biophys. J. 2012;103:2050–2059. doi: 10.1016/j.bpj.2012.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roca-Cusachs P., del Rio A., Sheetz M.P. Integrin-dependent force transmission to the extracellular matrix by α-actinin triggers adhesion maturation. Proc. Natl. Acad. Sci. USA. 2013;110:E1361–E1370. doi: 10.1073/pnas.1220723110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otey C.A., Vasquez G.B., Erickson B.W. Mapping of the alpha-actinin binding site within the beta 1 integrin cytoplasmic domain. J. Biol. Chem. 1993;268:21193–21197. [PubMed] [Google Scholar]
- 29.Tadokoro S., Nakazawa T., Tomiyama Y. A potential role for α-actinin in inside-out αIIbβ3 signaling. Blood. 2011;117:250–258. doi: 10.1182/blood-2009-10-246751. [DOI] [PubMed] [Google Scholar]
- 30.Liu J., Taylor D.W., Taylor K.A. A 3-D reconstruction of smooth muscle alpha-actinin by CryoEm reveals two different conformations at the actin-binding region. J. Mol. Biol. 2004;338:115–125. doi: 10.1016/j.jmb.2004.02.034. [DOI] [PubMed] [Google Scholar]
- 31.Kahana E., Gratzer W.B. Properties of the spectrin-like structural element of smooth-muscle alpha-actinin. Cell Motil. Cytoskeleton. 1991;20:242–248. doi: 10.1002/cm.970200307. [DOI] [PubMed] [Google Scholar]
- 32.Mofrad M.R.K., Kamm R.D. Cambridge University Press; New York: 2014. Cellular Mechanotransduction: Diverse Perspectives from Molecules to Tissues. [Google Scholar]
- 33.Burridge K., Feramisco J.R. Non-muscle alpha actinins are calcium-sensitive actin-binding proteins. Nature. 1981;294:565–567. doi: 10.1038/294565a0. [DOI] [PubMed] [Google Scholar]
- 34.Fukami K., Endo T., Takenawa T. alpha-Actinin and vinculin are PIP2-binding proteins involved in signaling by tyrosine kinase. J. Biol. Chem. 1994;269:1518–1522. [PubMed] [Google Scholar]
- 35.Fraley T.S., Tran T.C., Greenwood J.A. Phosphoinositide binding inhibits alpha-actinin bundling activity. J. Biol. Chem. 2003;278:24039–24045. doi: 10.1074/jbc.M213288200. [DOI] [PubMed] [Google Scholar]
- 36.Borrego-Diaz E., Kerff F., Dominguez R. Crystal structure of the actin-binding domain of alpha-actinin 1: evaluating two competing actin-binding models. J. Struct. Biol. 2006;155:230–238. doi: 10.1016/j.jsb.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 37.Way M., Pope B., Weeds A.G. Evidence for functional homology in the F-actin binding domains of gelsolin and alpha-actinin: implications for the requirements of severing and capping. J. Cell Biol. 1992;119:835–842. doi: 10.1083/jcb.119.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacKerell A.D., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 40.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 41.Torrie G.M., Valleau J.P. Nonphysical sampling distributions in Monte Carlo free-energy estimation: umbrella sampling. J. Comput. Phys. 1977;23:187–199. [Google Scholar]
- 42.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 43.Grant B.J., Rodrigues A.P.C., Caves L.S.D. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 44.Towns J., Cockerill T., Wilkins-Diehr N. XSEDE: Accelerating Scientific Discovery. Comput. Sci. Eng. 2014;16:62–74. [Google Scholar]
- 45.Ehrlicher A.J., Krishnan R., Pollak M.R. Alpha-actinin binding kinetics modulate cellular dynamics and force generation. Proc. Natl. Acad. Sci. USA. 2015;112:6619–6624. doi: 10.1073/pnas.1505652112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tojkander S., Gateva G., Lappalainen P. Actin stress fibers--assembly, dynamics and biological roles. J. Cell Sci. 2012;125:1855–1864. doi: 10.1242/jcs.098087. [DOI] [PubMed] [Google Scholar]
- 47.Zhao C.L., Mahboobi S.H., Mofrad M.R.K. The interaction of CRM1 and the nuclear pore protein Tpr. PLoS One. 2014;9:e93709. doi: 10.1371/journal.pone.0093709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piana S., Lindorff-Larsen K., Shaw D.E. How robust are protein folding simulations with respect to force field parameterization? Biophys. J. 2011;100:L47–L49. doi: 10.1016/j.bpj.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sjöblom B., Salmazo A., Djinović-Carugo K. Alpha-actinin structure and regulation. Cell. Mol. Life Sci. 2008;65:2688–2701. doi: 10.1007/s00018-008-8080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Travé G., Lacombe P.J., Pastore A. Molecular mechanism of the calcium-induced conformational change in the spectrin EF-hands. EMBO J. 1995;14:4922–4931. doi: 10.1002/j.1460-2075.1995.tb00175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang J., Taylor D.W., Taylor K.A. The three-dimensional structure of alpha-actinin obtained by cryoelectron microscopy suggests a model for Ca(2+)-dependent actin binding. J. Mol. Biol. 2001;310:845–858. doi: 10.1006/jmbi.2001.4789. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In this mode, the rod domain of α-actinin twists such that the actin binding domains move in opposite directions.
In this mode, the rod domain of α-actinin bends in the middle such that the actin binding domains move in the same direction.