Summary
Aim
KCNE2 encodes for the potassium voltage-gated channel, KCNE2. Mutations in KCNE2 have been associated with long-QT syndrome (LQTS). While KCNE2 has been extensively studied, the functions of its C-terminal domain remain inadequately described. Here, we aimed to elucidate the functions of this domain by identifying its protein interactors using yeast two-hybrid analysis.
Methods
The C-terminal domain of KCNE2 was used as bait to screen a human cardiac cDNA library for putative interacting proteins. Co-localisation and co-immunoprecipitation analyses were used for verification.
Results
Filamin C (FLNC) was identified as a putative interactor with KCNE2. FLNC and KCNE2 co-localised within the cell, however, a physical interaction was only observed under hypoxic conditions.
Conclusion
The identification of FLNC as a novel KCNE2 ligand not only enhances current understanding of ion channel function and regulation, but also provides valuable information about possible pathways likely to be involved in LQTS pathogenesis.
Keywords: LQTS, KCNE2, filamin C (FLNC), hypoxia, arrhythmia
Aim
Long-QT syndrome (LQTS) is a cardiac repolarisation disorder with an estimated global prevalence of 1:2 000 to 1:7 000.1,2 It is characterised by a prolonged QT interval on a surface electrocardiogram (ECG), with symptoms including syncope, cardiac arrest and sudden death.1,3,4 Occasionally, sudden cardiac death may be the first and only manifestation of LQTS.5,6
To date, different types of LQTS (LQT1–LQT13), classified according to the primary disease causal gene, have been identified, with more than 700 mutations leading to disease pathogenesis.7,8 Yet a large number of patients with clinically diagnosed LQTS have no mutations within any of the known LQTS causal genes,9-11 and numerous patients, despite carrying the same disease-causing mutation, display variable phenotypic expression and disease penetrance.12 To complicate matters further, LQTS can also be acquired through the use of certain prescribed medications, such as antipsychotics, antidepressants and antibiotics,13,14 adding to the growing challenge of clinical management and treatment of affected individuals.
The LQT type 6 (LQT6) causal gene, KCNE2 encoding for the potassium voltage-gated channel subfamily E member 2 (KCNE2) protein,15 has been implicated in the development of inherited, acquired and sporadic forms of LQTS.13,16-18 This protein consists of an extracellular N-terminal, a transmembrane and intracellular C-terminal domain. It comprises the beta-(β) subunits of ion channel complexes and co-assembles with many different alpha- (α) subunits, including the frequently studied human Ether-à-go-go-related (HERG) channel protein encoded for by the potassium voltage-gated channel, subfamily H (eag-related), member 2 (KCNH2) gene.15,17,19 In combination with KCNE2, properties of the different ion channel currents are modulated,20 assisting in cardiac pacemaker activity and repolarisation to ensure adequate myocardial recharging and the maintenance of a regular rhythm.15,21-23
A unique quality of many cardiac ion channels, including those containing KCNE2 and HERG, is their ability to adapt to hypoxic conditions. Hypoxia, defined as the decrease in available oxygen, causes changes in the electrical characteristics of ion channels and has been reported to predispose individuals to fatal arrhythmias.24-27 Additionally, hypoxic conditions affect the expression, folding, maturation and trafficking of various channels.28-30 In a recent study, it was noted that the expression of genes from the KCNE family (including KCNE2) could be affected by hypoxia in the heart.31 It has been observed that acute ischaemic hearts of rats after myocardial infarction show increased expression of KCNE proteins, attributable to hypoxia.31
The intricacy of processes causing and modifying cardiac arrhythmias highlights the importance of identifying the protein macromolecular complexes and pathways involved. Taking into consideration the relevance of KCNE2 in the context of ion channel regulation and LQTS, this study aimed to identify interactors with this β-subunit; specifically focusing on its cytoplasmic C-terminal domain, for which functional roles remain inadequately described.
Using yeast two-hybrid analysis, we identified filamin C (FLNC) as a KCNE2-interacting protein. FLNC and its paralogs, filamin A (FLNA) and filamin B (FLNB), act as scaffolding proteins and have been implicated in a number of cellular stress responses,32-38 including several hypoxia-related effects.35–38 For this reason, co-localisation and co-immunoprecipitation (Co-IP) analyses for verification of this interaction were conducted both under normoxic and hypoxic conditions.
Here, we show that, under normoxic and hypoxic conditions, FLNC and KCNE2c co-localised within the cell. However, FLNC and KCNE2 only co-immunoprecipitated under hypoxic conditions, suggesting that while these two proteins are located in close proximity to one another within the cell, it is only under conditions of cellular stress that a physical interaction between the two exists. The data presented here provide evidence to suggest that KCNE2 may play a role in hypoxia-induced arrhythmias.
Methods
KCNE2 construct
A fragment encoding the C-terminal of KCNE2 gene (amino acid 72-123) was amplified from human genomic DNA by means of polymerase chain reaction (PCR). The PCR reaction employed KCNE2 C-terminal-specific primers with two restriction enzyme sites (NdeI and EcoR1) (Table 1) for subsequent cloning into the CLONTECH yeast two-hybrid (Y2H) bait vector, pGBKT7 (pGBKT7-KCNE2), in-frame with the GAL4-DNA binding domain (GAL4BD). The integrity of the sequence and the conservation of the GAL4 domain reading frame of the resulting construct were verified via sequencing.
Table 1. Nucleotide sequences of primers used to amplify the C-terminal of KCNE2.
| Primer | Sequence (5’-3’) | Ta (°C) |
| KCNE2-forwardNde1 | 5’ - ACTGCAGAACATATGCTCAAATCCAAGAGACGG - 3’ | 50 |
| KCNE2-reverseEcoR1 | 5’ - ACTGCAGAAGAATTCCTATCAGGGGAACATTTTGAAC - 3’ | 51 |
°C: degrees Celsius; Ta: annealing temperature; KCNE2: potassium voltagegated ion-channel subfamily E member 2. The bold text represents a tag, which facilitates restriction enzyme digestion, while the underlined sequences correspond to the Nde1 and EcoR1 restriction enzyme sites, respectively. The short italic sequence (CTA) symbolises the stop codon, and the remaining text represents the sequence of the primer, which will anneal to the DNA in the PCR amplification reaction.
Yeast two-hybrid (Y2H) library screen
The Saccharomyces cerevisiae strain, AH109 (BD Biosciences, Clontech, USA), was transformed with the pGBKT7-KCNE2 construct and mated with the S cerevisiae strain, Y187, which was pre-transformed with a MATCHMAKER human cardiac cDNA library (BD Biosciences, Clontech, USA). Subsequently, the library screen was conducted according to manufacturer’s recommendations.
The prey plasmids, from colonies expressing the three essential reporter genes (HIS3, ADE2 and MEL1), were isolated from the diploid yeast cells and were retransformed into S cerevisiae strain Y187 to analyse their ability to activate the reporter genes when mated with heterologous baits (Table 2). Prey peptides showing specific interaction with the KCNE2 C-terminal domain were sequenced and the in-frame open reading frame (ORF) sequences were analysed using BLASTN and BLASTP against public databases (http://ncbi.nlm.nih.gov/blast).
Table 2. S cerevisiae bait strains.
| S cerevisiae bait strains | Plasmid type |
| AH109 pGBKT7-KCNE2 | Positive control plasmid |
| AH109 pGBKT7 | Non-recombinant plasmid |
| AH109 pGBKT-53* | Control bait plasmid |
| AH109 pGBKT7-WFS1 | Negative control plasmid |
*The pGBKT7 vector containing the human p53 gene. KCNE2: potassium voltage-gated ion-channel subfamily E member 2; WFS1: Wolfram syndrome 1.
Cell culture
The H9C2 rat-derived cardiac myoblasts (American Typer Culture Collection, USA) were grown in Dulbecco’s modified Eagle medium (DMEM, Lonza, CHE) containing 10% foetal bovine serum (FBS, Biochrom, GER) and 1% penicillin/streptomycin (Pen/Strep, Biochrom, GER) until they reached 80% confluency. For co-localisation, 10 000 cells were seeded onto glass cover slips in each well of a six-well plate (8-cm2 culture dishes) and incubated until 80% confluency was reached, while for Co-IP, cells were grown in 175-cm2 flasks until they reached 80% confluency. Differentiation medium (DMEM containing 1% horse serum and 1% Pen/Strep) was subsequently added to each well of the six-well plate and the 175-cm2 flasks. Cells were differentiated for 10–14 days.
For hypoxia induction, the differentiation medium was removed and replaced with Esumi buffer (138.6 mM NaCl, 12 mM KCl, 1 mM MgCl2, 1 mM CaCl2.H2O, and 4 mM Hepes, pH 6.2).39 Culture dishes and flasks were then placed in a chamber where a hypoxic environment was created by flushing the system with a 1% O2 gas mixture at a flow rate of 20 l/min, for approximately four minutes. The cells were then incubated in the hypoxic chamber at 37°C for two hours.
For Co-IP experiments, 5 ml of pre-warmed trypsin was used to detach the cells from the growth surface of the flasks. The cells were then centrifuged at 4ºC for three minutes at 2 500 rpm. The supernatant was discarded and the pellet resuspended in 1 ml of phosphate-buffered saline (PBS) and re-pelleted at 9 000 rpm for two minutes. The PBS was removed and the cells were then lysed with ice-cold lysis buffer (50 mM Hepes, 5 M NaCl, 0.5 M EDTA, 1% Triton X-100, 1 M Na3VO4) containing protease inhibitor cocktail tablets [one tablet EDTA-free protease inhibitor cocktail tablet per 20 ml lysis buffer and 1 mM phenylmethylsulfonylfluoride (PMSF) (Sigma-Aldrich, USA)].
Approximately 0.5 ml of ZROB05 Ceria zirconium oxide beads (0.5 mm diameter) (Next Advance Inc, USA) was added to the suspension and it was placed in a Bullet blender® (Gentaur, GBR) for one minute. The blending step was repeated three times at five-minute intervals. The cells were then pelleted by centrifugation at 9 000 rpm for two minutes, after which the supernatant was collected. A Bradford assay was used for protein concentration determination,40 to ensure equivalent amounts of protein per sample were subjected to sodium dodecyl sulphatepolyacrylamide gel electrophoresis (SDS-PAGE) analysis.
Co-localisation
For co-localisation experiments, the differentiation media and Esumi buffer was removed from the differentiated H9C2 rat-derived cardiomyocytes on the glass cover slips and the cells were rinsed with PBS. The cells were permeabilised with methanol for five minutes at –20°C and fixed with 4% paraformaldehyde for five minutes at room temperature. The cells were then washed three times with PBS for 10 minutes and blocked in 1% BSA for one hour at room temperature. Following the blocking step, the cells were again washed three times with PBS for 10 minutes and incubated at 4°C overnight with rabbit anti-KCNE2 (Abcam, Biocom Biotech, RSA, 1:50) and goat anti-FLNC (Santa Cruz Biotechnology Inc, USA, 1:50) primary antibodies diluted in 1% BSA.
The cells were then washed three times with PBS for 10 minutes and stained with Alexa 488 donkey anti-rabbit (Jackson ImmunoResearch Laboratories Inc, USA, 1:500) and Cy3 donkey anti-goat (Jackson ImmunoResearch Laboratories Inc, USA, 1:500) secondary antibodies in PBS for 90 minutes in the dark at room temperature. Afterwards, the cells were washed three times with PBS for 10 minutes, and Hoechst H-33342 was added for nuclear staining [Sigma-Aldrich (Pty) Ltd, RSA, 1:200; 10 mg/ml], followed by a 10-minute incubation at room temperature.
Subsequently, the cover slips with the stained cells were mounted onto glass slides using Mowiol (Jackson ImmunoResearch Laboratories Inc, USA) containing n-propylgallate as the anti-fade reagent and kept at 4°C in the dark until viewing. Samples were acquired using the Carl Zeiss Confocal LSM 780 Elyra S1, equipped with a LSM780 GaAsP detector, using a Plan Apochromat 63×/1.4 Oil DIC M27 or an alpha Plan- Apochromat 100×/1.46 oil DIC objective (Central analytical facility, Cell Imaging Unit, Stellenbosch University, RSA). The samples were excited with a 488-nm and 561-nm laser underutilisation of a MBS 488/561 beam splitter.
Images were acquired through z-stacking with an increment of 0.3-μm step width, and projected as maximum-intensity projections using ZEN software (black edition, 2011). Thresholds were determined using appropriate control images acquired for cells individually stained (single-stain) for KCNE2 and FLNC, respectively. The background was adjusted for all acquired images using images of cells only stained with secondary control antibodies.
Co-immunoprecipitation
Cells were harvested and the lysates were pre-cleared with protein G agarose beads (KPL Inc, USA) for 30 minutes at 4°C. The pre-cleared lysates (150 μg/total protein) were incubated with 1 μg of either rabbit polyclonal anti-KCNE2 (Santa Cruz Biotechnology Inc, USA) or goat polyclonal anti-FLNC (Santa Cruz Biotechnology Inc, USA) antibody rotating overnight at 4°C.
To capture the protein complexes, 60 μl of protein G agarose beads were added to the lysate and incubated for an additional hour rotating at 4°C. The complexes were then washed three times, each time removing the supernatant after centrifugation and adding fresh lysis buffer that contained protease inhibitors and PMSF. Proteins were eluted by addition of 1× SDS-PAGE sample buffer [95% Laemmli sample buffer (Bio-Rad Laboratories Inc, USA), 5% β-mercapto-ethanol], denatured for five minutes at 95°C and separated using 4–15% SDS-PAGE gels for Western blot analysis. Two negative controls, a non-relevant antibody control (HA-probe; Santa Cruz Biotechnology Inc, USA) and a protein G agarose control (without antibody) were included in all Co-IP experiments
Western blot analysis
Following co-IP, proteins were separated on 4–15% SDS-PAGE gels and transferred to a polyvinylidene difluoride (PVDF) membrane (Thermo Scientific, USA) by means of the iBlot® system (Invitrogen, USA). Membranes were blocked with 5% fat-free powdered milk, supplemented with Tris-buffered saline Tween-20 (TBST, 0.01% Tween-20), for one hour at room temperature. Membranes were then incubated at 4°C overnight with the appropriate primary antibodies (Santa Cruz Biotechnology Inc, USA, 1:200 anti-KCNE2; 1:1 000 anti-FLNC), diluted with 5% milk in TBST.
Subsequently, the membranes were washed with TBST and incubated for one hour at room temperature with the corresponding horseradish peroxidase (HRP) conjugated secondary antibodies (Santa Cruz Biotechnology Inc, USA, 1:2 000 donkey anti-rabbit; 1:2 000 donkey anti-goat), diluted with 5% milk in TBST. Following incubation with the secondary antibody, the membranes were washed for 30 minutes at room temperature.
The SuperSignal® West Pico chemiluminescence substrate kit (Thermo Scientific, USA) was then used according to the manufacturer’s instructions and the membranes were exposed for two minutes to CL-Xposure™ autoradiography film (Thermo Scientific, USA). The autoradiography film was developed using an Amersham hyperprocessor automatic autoradiography film processor (Amersham Pharmacia Biotech UK Ltd, UK) prior to final analyses.
Results
FLNC as a novel interactor with KCNE2 under hypoxic conditions
The Y2H screen identified FLNC (GenBank: NP_001449.3) as a KCNE2 (GenBank: NP_751951.1) interactor with the binding regions located between amino acids 2637–2725 of FLNC and amino acids 72–123 of KCNE2. These 88 amino acids of FLNC are positioned at the end of the C-terminal domain, shown to be involved in self-dimerisation.32
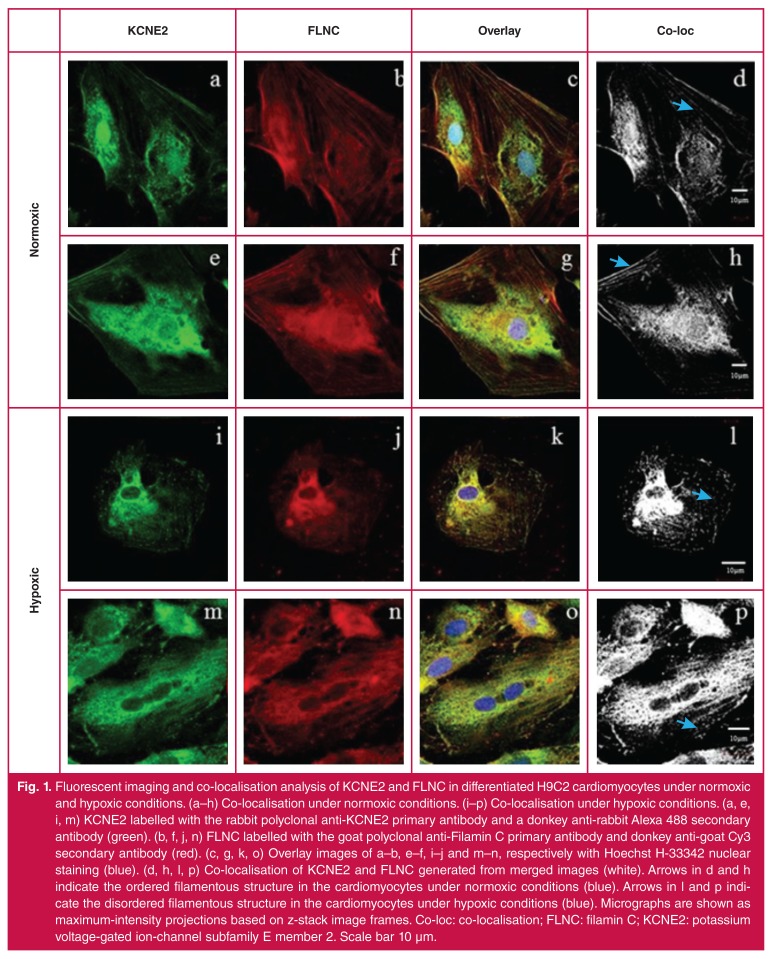
Imaging analysis revealed a strong co-localisation signal between KCNE2 and FLNC at the cell membrane, filamentous structures and the cytoplasm of differentiated H9C2 rat-derived cardiomyocytes under normoxic conditions (Fig. 1a-h), while the co-localisation of these two proteins was mainly restricted to the cytoplasm under conditions of hypoxia (Fig. 1i-p). When the cells were subjected to hypoxic stress, the pattern of co-localisation, relative to that during normoxia, changed considerably at the plasma membrane (Fig. 1i-p), where decreased co-localisation of these proteins was observed. Following hypoxic stress, the internal cellular structure became disrupted and the filaments and cytoskeleton showed clear signs of disarray (Fig. 1i-p). The less well-defined signal appearance of co-localisation seen in Fig. 1l and p may be attributable to this disarray.
Fig. 1.
Fluorescent imaging and co-localisation analysis of KCNE2 and FLNC in differentiated H9C2 cardiomyocytes under normoxic and hypoxic conditions. (a–h) Co-localisation under normoxic conditions. (i–p) Co-localisation under hypoxic conditions. (a, e, i, m) KCNE2 labelled with the rabbit polyclonal anti-KCNE2 primary antibody and a donkey anti-rabbit Alexa 488 secondary antibody (green). (b, f, j, n) FLNC labelled with the goat polyclonal anti-Filamin C primary antibody and donkey anti-goat Cy3 secondary antibody (red). (c, g, k, o) Overlay images of a–b, e–f, i–j and m–n, respectively with Hoechst H-33342 nuclear staining (blue). (d, h, l, p) Co-localisation of KCNE2 and FLNC generated from merged images (white). Arrows in d and h indicate the ordered filamentous structure in the cardiomyocytes under normoxic conditions (blue). Arrows in l and p indicate the disordered filamentous structure in the cardiomyocytes under hypoxic conditions (blue). Micrographs are shown as maximum-intensity projections based on z-stack image frames. Co-loc: co-localisation; FLNC: filamin C; KCNE2: potassium voltage-gated ion-channel subfamily E member 2. Scale bar 10 μm.
While co-localisation analysis provided convincing evidence that KNCE2 and FLNC are located in close proximity to one another within the cell, this does not necessarily mean that they physically interact. For this reason, Co-IP analysis was used to determine whether a physical interaction exists between the two.
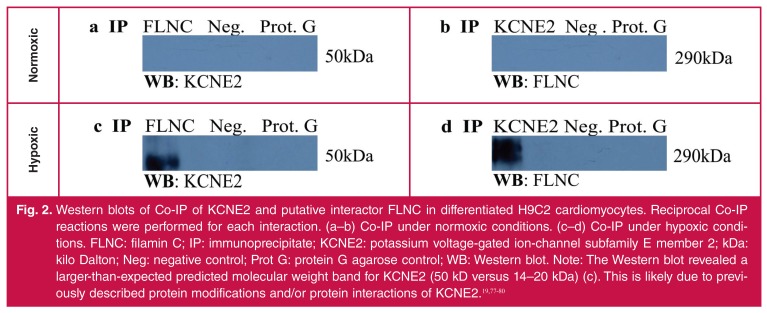
Under normoxic conditions, reciprocal Co-IP experiments failed to show any physical interaction between KCNE2 and FLNC (Fig. 2c, d). However, during hypoxia an interaction between KCNE2 and FLNC was observed (Fig. 2c, d). These findings suggest that the induction of stress is essential to the interaction and one could speculate that hypoxia-induced conformational changes of FLNC are necessary for the KCNE2–FLNC interaction.
Fig. 2.
Western blots of Co-IP of KCNE2 and putative interactor FLNC in differentiated H9C2 cardiomyocytes. Reciprocal Co-IP reactions were performed for each interaction. (a–b) Co-IP under normoxic conditions. (c–d) Co-IP under hypoxic conditions. FLNC: filamin C; IP: immunoprecipitate; KCNE2: potassium voltage-gated ion-channel subfamily E member 2; kDa: kilo Dalton; Neg: negative control; Prot G: protein G agarose control; WB: Western blot. Note: The Western blot revealed a larger-than-expected predicted molecular weight band for KCNE2 (50 kD versus 14–20 kDa) (c). This is likely due to previously described protein modifications and/or protein interactions of KCNE2.19,77-80
Discussion
This study identified a novel protein–protein interaction between the cytoplasmic C-terminal domain of KCNE2 and FLNC during conditions of acute hypoxia. To date, the intracellular C-terminal domain residues of KCNE2 have been implicated in modulating HERG current density,13,41 current deactivation rates,41 and phosphorylation-dependant channel degradation.19 However, studies elaborating on specific regulatory roles for this domain remain scarce, highlighting the importance of the current findings.
The interactor identified in this study, FLNC, is located in the cytoplasm at the Z-line of the sarcomere and functions in the cytoskeleton, where it is involved in crosslinking actin filaments into networks and anchoring membrane proteins.32,42 This filamin and its main paralogs, FLNA and FLNB, act as scaffolding proteins and have been implicated in a number of cellular stress responses,32-34,43-46 including several hypoxia-related effects.33,34,45,46 FLNC specifically, is predominantly expressed in muscle tissue and is associated with cardiac abnormalities such as desminopathy, characterised by muscle weakness, conduction blocks, arrhythmias and chronic heart failure, frequently resulting in sudden cardiac death.35,36
Filamins also play an important part in cell signalling by disrupting existing interactions or by the introduction of novel interactions.32,37,38,47 Interestingly, there are several reports detailing interactions of filamin family members with ion channel subunits.48-50 Particularly noteworthy is a previously descibed association in neuronal tissue between FLNC and the potassium voltage-gated channel subfamily D member 2 (KCND2),48 the α-subunit of the Kv4.2 channel. That study proposed that FLNC mediates the direct link between KCND2 and the actin cytoskeleton and showed that this interaction is essential for the generation of appropriate current densities.48
Both neuronal and cardiac tissue contain voltage-gated ion channels responsible for controlling the excitability of neurons and cardiomyocytes. These channels allow for communication between cells in these tissues.51-53 Furthermore, a KCNE2– KCND2 interaction has been described, implicating KCNE2 in the regulation of the rapidly inactivating KCND2 α-subunit.54,55
A common theme in the observation of the ion channel interactions with filamin is the ability of filamin to influence membrane localisation.48-50 For FLNC, this process has also been shown to involve other actin-binding and auxiliary ion channel proteins.56
In the present study, the C-terminal of FLNC, specifically amino acids 2637–2725 (GenBank: NP_001449.3), bound to the cytoplasmic C-terminal domain of KCNE2, exclusively during conditions of hypoxic stress. This finding is consistent with a number of other studies, indicating that the C-terminal region of filamins is involved in protein interactions.57 The FLNC amino acid residues defined to interact with KCNE2 in this investigation correspond to a domain that is responsible for protein dimer formation and is important for actin filament bundling and cross-linking activities.58,59
The introduction of hypoxic stress is known to have profound effects on the cell.60 These include the disruption of ionic homeostasis, mitochondrial dysfunction resulting in impaired ATP production, induction of cell death by apoptosis or necrosis, and the generation of reactive oxygen species (ROS).61 Excess ROS leads to cardiac cell damage and post-ischaemic contractile dysfunction by attacking virtually all cellular components.62 This results in the degradation of intracellular proteins, rupture of cellular membranes (including the sarcolemma), as well as intracellular calcium ion overload,63 however, it has been show that cells remain viable even after extended periods of hypoxic stress.64 In addition to this, the actin cytoskeleton is also severely compromised and may therefore be a driving force for novel interactions. Additionally, Kesner et al. indicated that stressinduced conformational changes in filamins could have a direct effect on existing interactions or may influence the presence of novel interactions.47
Co-localisation analysis revealed that KCNE2 and FLNC co-localise under both normoxic and hypoxic conditions. However, no physical interaction could be confirmed between these two proteins during normoxic conditions using Co-IP assays. Therefore, these findings suggest that the induction of stress is essential to the interaction. Given that hypoxic stress compromises the integrity of the cellular membranes and that FLNC has also been shown to interact with the actin cytoskeleton, it is tempting to speculate that during this time of cellular stress, the interaction between FLNC and KNCE2 may be an attempt by the cell to restore cellular membrane integrity, thereby promoting cellular survival during these conditions.
If this is the case, one could further speculate that mutations in the genes encoding for KCNE2 or FLNC, or both, which weaken or abrogate their interaction could result in the cell being unable to restore membrane integrity, which could lead to acute myocardial ischaemic arrhythmogenesis. Furthermore, it may be that hypoxia-induced conformational changes of FLNC are necessary for the novel KCNE2–FLNC interaction.
During hypoxia, the pattern of co-localisation changed and the differentiated H9C2 cardiomyocytes showed signs of internal structural disruption. Both KCNE2 and FLNC displayed reduced localisation at the surface membrane (Fig. 1i, j, m, n), while the intracellular co-localisation signal was intensified. Hypoxic conditions are known to initiate extensive variations in gene expression, alter protein sub-cellular localisation, and cause the attenuation of membrane protein translation.65-67
Furthermore, these findings are consistent with reports that the HERG α-subunit, known to bind KCNE2,16 showed reduced membrane localisation during hypoxia.30,68 The reason for the observed decrease of these proteins at the membrane requires further investigation; however, it is interesting to note that both KCNE2 and filamins have been implicated in processes involving the internalisation of membrane proteins.19,69,70 Additionally, given the evidence of filamins aiding channel localisation,69,70 and the increase in KCNE2 gene expression during hypoxia,31 it would be intriguing to investigate if this interaction serves a compensatory role to try to restore internally localised channels to the membrane.
The role of cytoskeletal components, including actin-binding proteins, in ion channel function and regulatory processes is a rapidly expanding field of study. Evidence supports their significant contribution towards channel trafficking and activity at the plasma membrane itself.71-73 Furthermore, there are numerous studies linking the dysfunction of cytoskeletal proteins with conduction defects and arrhythmias.72,73
This study is the first to identify the cytoskeletal protein, FLNC, as a constituent of an ion channel macromolecular complex, specifically forming part of the KCNE2 interactome. This observation was only valid during conditions of hypoxia, although it remains to be seen if other stimuli can elicit the same association. Together, FLNC and KCNE2 most likely modulate KCNE2-containing channels, especially pertaining to their surface expression.
Conclusion
It has long been known that all cells have the ability to adapt and respond to hypoxic conditions in order to prevent the harmful effects of oxygen deprivation.74 In cardiac cells, potassium channels play a central role in this adaptation. However, inadequate adaptive responses may lead to serious cellular damage and cardiac arrhythmias. This has previously been shown in the cardiovascular system where lack of oxygen contributes to cardiac arrhythmia.75,76
This study identified and validated FLNC as an interactor with KCNE2 under conditions of hypoxia. This finding points towards new insight and understanding into the mechanisms in which KCNE2 functions, and could contribute to our understanding of the interactome in cardiovascular conditions such as LQTS. Through identification of novel KCNE2 interacting proteins, new genes can be included in searches for causal and modifying effects of novel arrhythmia disorders. These findings ultimately advocate intriguing possibilities that might lead to new therapeutic avenues being discovered.
Acknowledgments
The KCNE2 Y2H screen was enabled by funding provided by the National Research Foundation grant FA2006040400017 (VAC). The authors thank Derick van Vuuren for his assistance with the hypoxic chamber (Division of Medical Physiology, Stellenbosch University).
Contributor Information
Annika Neethling, Email: aneethling@sun.ac.za, DST /NRF Centre of Excellence in Biomedical Tuberculosis Research, SA MRC Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Department of Biomedical Sciences, Faculty of Medicine and Health Sciences, Stellenbosch University, South Africa.
Jomien Mouton, DST /NRF Centre of Excellence in Biomedical Tuberculosis Research, SA MRC Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Department of Biomedical Sciences, Faculty of Medicine and Health Sciences, Stellenbosch University, South Africa.
Valerie Corfield, Email: , DST /NRF Centre of Excellence in Biomedical Tuberculosis Research, SA MRC Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Department of Biomedical Sciences, Faculty of Medicine and Health Sciences, Stellenbosch University, South Africa.
Carin de Villiers, DST /NRF Centre of Excellence in Biomedical Tuberculosis Research, SA MRC Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Department of Biomedical Sciences, Faculty of Medicine and Health Sciences, Stellenbosch University, South Africa.
Craig Kinnear, DST /NRF Centre of Excellence in Biomedical Tuberculosis Research, SA MRC Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Department of Biomedical Sciences, Faculty of Medicine and Health Sciences, Stellenbosch University, South Africa.
Ben Loos, Department of Physiological Sciences, Faculty of Science, Stellenbosch University, Stellenbosch, South Africa.
References
- 1.Schwartz PJ. Prevalence of the congenital long-QT syndrome. Circulation. 2009;120(18):1761–1767. doi: 10.1161/CIRCULATIONAHA.109.863209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schulze Bahr E. Long QT syndromes: genetic basis. Card Electrophysiol Clin. 2012;4(1):1–16. [Google Scholar]
- 3.Goldenberg I, Zareba W, Moss AJ. Long QT syndrome. Curr Probl Cardiol. 2008;33(11):629–694. doi: 10.1016/j.cpcardiol.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz PJ, Crotti L, Insolia R. Long-QT syndrome from genetics to management. Circ Arrhythm Electrophysiol. 2012;5(4):868–877. doi: 10.1161/CIRCEP.111.962019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zipes DP, Wellens HJJ. Sudden cardiac death. Circulation. 1998;98(21):2334–2351. doi: 10.1161/01.cir.98.21.2334. [DOI] [PubMed] [Google Scholar]
- 6.Bhuiyan MZA, Al-Shahrani S, Al-Aama J, Wilde A, Momenah TS. Congenital long QT syndrome: an update and present perspective in Saudi Arabia. Genet Disord. 2013;1:39. doi: 10.3389/fped.2013.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedley PL, Jørgensen P, Schlamowitz S. et al. The genetic basis of long QT and short QT syndromes: a mutation update. Hum Mutat. 2009;30(11):1486–1511. doi: 10.1002/humu.21106. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz PJ, Ackerman MJ, George AL Jr, Wilde AAM. Impact of genetics on the clinical management of channelopathies. J Am Coll Cardiol. 2013;62(3):169–180. doi: 10.1016/j.jacc.2013.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Napolitano C. Genetic testing in the long QT syndrome: Development and validation of an efficient approach to genotyping in clinical practice. J Am Med Assoc Chic Ill. 2005;294(23):2975–2980. doi: 10.1001/jama.294.23.2975. [DOI] [PubMed] [Google Scholar]
- 10.Medeiros-Domingo A, Iturralde-Torres P, Ackerman MJ. Clinical and genetic characteristics of long QT syndrome. Rev Esp Cardiol Engl Ed. 2007;60(7):739–752. [PubMed] [Google Scholar]
- 11.Newton-Cheh C, Eijgelsheim M, Rice KM. et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41(4):399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brink PA, Crotti L, Corfield V. et al. Phenotypic variability and unusual clinical severity of congenital long-QT syndrome in a founder population. Circulation. 2005;112(17):2602–2610. doi: 10.1161/CIRCULATIONAHA.105.572453. [DOI] [PubMed] [Google Scholar]
- 13.Sesti F, Abbott GW, Wei J. et al. A common polymorphism associated with antibiotic-induced cardiac arrhythmia. Proc Natl Acad Sci. 2000;97(19):10613–10618. doi: 10.1073/pnas.180223197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanguinetti MC, Mitcheson JS. Predicting drug–hERG channel interactions that cause acquired long QT syndrome. Trends Pharmacol Sci. 2005;26(3):119–124. doi: 10.1016/j.tips.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Abbott GW, Sesti F, Splawski I. et al. MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell. 1999;97(2):175–187. doi: 10.1016/s0092-8674(00)80728-x. [DOI] [PubMed] [Google Scholar]
- 16.Splawski I, Shen J, Timothy KW. et al. Spectrum of mutations in long-QT syndrome genes : KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 2000;102(10):1178–1185. doi: 10.1161/01.cir.102.10.1178. [DOI] [PubMed] [Google Scholar]
- 17.Larsen LA, Andersen PS, Kanters J. et al. Screening for mutations and polymorphisms in the genes KCNH2 and KCNE2 encoding the cardiac HERG/MiRP1 ion channel: implications for acquired and congenital long Q-T syndrome. Clin Chem. 2001;47(8):1390–1395. [PubMed] [Google Scholar]
- 18.Mazhari R, Greenstein JL, Winslow RL, Marbán E, Nuss HB. Molecular interactions between two long-QT syndrome gene products, HERG and KCNE2, rationalized by in vitro and in silico analysis. Circ Res. 2001;89(1):33–38. doi: 10.1161/hh1301.093633. [DOI] [PubMed] [Google Scholar]
- 19.Zhang M, Wang Y, Jiang M. et al. KCNE2 protein is more abundant in ventricles than in atria and can accelerate hERG protein degradation in a phosphorylation-dependent manner. Am J Physiol – Heart Circ Physiol. 2012;302(4):H910–922. doi: 10.1152/ajpheart.00691.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tinel N. KCNE2 confers background current characteristics to the cardiac KCNQ1 potassium channel. Eur Mole Biol Org J. 2000;19(23):6326–6330. doi: 10.1093/emboj/19.23.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Towbin JA, Vatta M. Molecular biology and the prolonged QT syndromes. Am J Med. 2001;110(5):385–398. doi: 10.1016/s0002-9343(00)00715-4. [DOI] [PubMed] [Google Scholar]
- 22.Chun KRJ, Koenen M, Katus HA, Zehelein J. Expression of the IKr components KCNH2 (rERG) and KCNE2 (rMiRP1) during late rat heart development. Exp Mol Med. 2004;36(4):367–371. doi: 10.1038/emm.2004.48. [DOI] [PubMed] [Google Scholar]
- 23.Kawakami K, Nagatomo T, Abe H. et al. Comparison of HERG channel blocking effects of various β-blockers – implication for clinical strategy. Br J Pharmacol. 2006;147(6):642–652. doi: 10.1038/sj.bjp.0706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark RE, Christlieb I, Sanmarco M, Diaz-Perez R, Dammann JF, Zipser ME. Relationship of hypoxia to arrhythmia and cardiac conduction hemorrhage: an experimental study. Circulation. 1963;27(4):742–747. [Google Scholar]
- 25.Ju YK, Saint DA, Gage PW. Hypoxia increases persistent sodium current in rat ventricular myocytes. J Physiol. 1996;497(Pt 2):337–347. doi: 10.1113/jphysiol.1996.sp021772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prandota J. Possible pathomechanisms of sudden infant death syndrome: key role of chronic hypoxia, infection/inflammation states, cytokine irregularities, and metabolic trauma in genetically predisposed infants. Am J Ther. 2004;11(6):517–546. doi: 10.1097/01.mjt.0000140648.30948.bd. [DOI] [PubMed] [Google Scholar]
- 27.Neary MT, Mohun TJ, Breckenridge RA. A mouse model to study the link between hypoxia, long QT interval and sudden infant death syndrome. Dis Model Mech. 2013;6(2):503–507. doi: 10.1242/dmm.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conforti L, Millhorn DE. Selective inhibition of a slow-inactivating voltage-dependent K+ channel in rat PC12 cells by hypoxia. J Physiol. 1997;502((Pt 2)):293–305. doi: 10.1111/j.1469-7793.1997.293bk.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Weigand L, Wang W, Sylvester JT, Shimoda LA. Chronic hypoxia inhibits Kv channel gene expression in rat distal pulmonary artery. Am J Physiol Lung Cell Mol Physiol. 2005;288(6):L1049–1058. doi: 10.1152/ajplung.00379.2004. [DOI] [PubMed] [Google Scholar]
- 30.Nanduri J, Bergson P, Wang N, Ficker E, Prabhakar NR. Hypoxia inhibits maturation and trafficking of HERG K+ channel protein: Role of Hsp90 and ROS. Biochem Biophys Res Commun. 2009;388(2):212–216. doi: 10.1016/j.bbrc.2009.07.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia S, Wang Y, Zhang Y. et al. Dynamic changes in HCN2, HCN4, KCNE1, and KCNE2 expression in ventricular cells from acute myocardial infarction rat hearts. Biochem Biophys Res Commun. 2010;395(3):330–335. doi: 10.1016/j.bbrc.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Van der Ven PF, Obermann WM, Lemke B, Gautel M, Weber K, Fürst DO. Characterization of muscle filamin isoforms suggests a possible role of gamma-filamin/ABP-L in sarcomeric Z-disc formation. Cell Motil Cytoskeleton. 2000;45(2):149–162. doi: 10.1002/(SICI)1097-0169(200002)45:2<149::AID-CM6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 33.Cho K-O, Lee K-E, Youn D-Y. et al. Decreased vulnerability of hippocampal neurons after neonatal hypoxia-ischemia in bis-deficient mice. Glia. 2012;60(12):1915–1929. doi: 10.1002/glia.22407. [DOI] [PubMed] [Google Scholar]
- 34.Nissou M-F, El Atifi M, Guttin A. et al. Hypoxia-induced expression of VE-cadherin and filamin B in glioma cell cultures and pseudopalisade structures. J Neurooncol. 2013;113(2):239–249. doi: 10.1007/s11060-013-1124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kley RA, Hellenbroich Y, van der Ven PFM. et al. Clinical and morphological phenotype of the filamin myopathy: a study of 31 German patients. Brain. 2007;130(12):3250–3264. doi: 10.1093/brain/awm271. [DOI] [PubMed] [Google Scholar]
- 36.Goldfarb LG, Olivé M, Vicart P, Goebel HH. Intermediate filament diseases: desminopathy. Adv Exp Med Biol. 2008;642:131–164. doi: 10.1007/978-0-387-84847-1_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004;6(11):1034–1038. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- 38.Ithychanda SS, Hsu D, Li H. et al. Identification and characterization of multiple similar ligand-binding repeats in filamin implication on filamin-mediated receptor clustering and cross-talk. J Biol Chem. 2009;284(50):35113–35121. doi: 10.1074/jbc.M109.060954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lecour S, Suleman N, Deuchar GA. et al. Pharmacological preconditioning with tumor necrosis factor-α activates signal transducer and activator of transcription-3 at reperfusion without involving classic prosurvival kinases (Akt and extracellular signal-regulated kinase). Circulation. 2005;112(25):3911–3918. doi: 10.1161/CIRCULATIONAHA.105.581058. [DOI] [PubMed] [Google Scholar]
- 40.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 41.Abbott GW. Disease-associated mutations in KCNE potassium channel subunits (MiRPs) reveal promiscuous disruption of multiple currents and conservation of mechanism. Fed Am Soc Exp Biol J. 2002;16(3):390–400. doi: 10.1096/fj.01-0520hyp. [DOI] [PubMed] [Google Scholar]
- 42.Fujita M, Mitsuhashi H, Isogai S. et al. Filamin C plays an essential role in the maintenance of the structural integrity of cardiac and skeletal muscles, revealed by the medaka mutant zacro. Dev Biol. 2012;361(1):79–89. doi: 10.1016/j.ydbio.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 43.Nakagawa K, Sugahara M, Yamasaki T. et al. Filamin associates with stress signalling kinases MKK7 and MKK4 and regulates JNK activation. Biochem J. 2010;427(2):237–245. doi: 10.1042/BJ20091011. [DOI] [PubMed] [Google Scholar]
- 44.Razinia Z, Mäkelä T, Ylänne J, Calderwood DA. Filamins in mechanosensing and signaling. A Rev Biophys. 2012;41(1):227–246. doi: 10.1146/annurev-biophys-050511-102252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boraldi F, Annovi G, Carraro F. et al. Hypoxia influences the cellular cross-talk of human dermal fibroblasts. A proteomic approach. Biochim Biophys Acta BBA – Proteins Proteomics. 2007;1774(11):1402–1413. doi: 10.1016/j.bbapap.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 46.Hastie LE, Patton WF, Hechtman HB, Shepro D. H2O2-induced filamin redistribution in endothelial cells is modulated by the cyclic AMP-dependent protein kinase pathway. J Cell Physiol. 1997;172(3):373–381. doi: 10.1002/(SICI)1097-4652(199709)172:3<373::AID-JCP11>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 47.Kesner BA, Ding F, Temple BR, Dokholyan NV. N-terminal strands of filamin Ig domains act as a conformational switch under biological forces. Proteins. 2010;78(1):12–24. doi: 10.1002/prot.22479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petrecca K, Miller DM, Shrier A. Localization and enhanced current density of the Kv4.2 potassium channel by interaction with the actinbinding protein filamin. J Neurosci Off J Soc Neurosci. 2000;20(23):8736–8744. doi: 10.1523/JNEUROSCI.20-23-08736.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sampson LJ, Leyland ML, Dart C. Direct interaction between the actin-binding protein filamin-A and the inwardly rectifying potassium channel, Kir2.1. J Biol Chem. 2003;278(43):41988–41997. doi: 10.1074/jbc.M307479200. [DOI] [PubMed] [Google Scholar]
- 50.Kim EY, Ridgway LD, Dryer SE. Interactions with filamin A stimulate surface expression of large-conductance Ca2+-activated K+ channels in the absence of direct actin binding. Mol Pharmacol. 2007;72(3):622–630. doi: 10.1124/mol.107.038026. [DOI] [PubMed] [Google Scholar]
- 51.An WF, Bowlby MR, Betty M. et al. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000;403(6769):553–556. doi: 10.1038/35000592. [DOI] [PubMed] [Google Scholar]
- 52.Zaika O, Zhang J, Shapiro MS. Functional role of M-type (KCNQ) K+ channels in adrenergic control of cardiomyocyte contraction rate by sympathetic neurons. J Physiol. 2011;589((Pt 10)):2559–2568. doi: 10.1113/jphysiol.2010.204768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaczmarek LK. Slack, slick, and sodium-activated potassium channels. Int Sch Res Not. 2013;2013:e354262. doi: 10.1155/2013/354262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang M, Jiang M, Tseng G-N. MinK-related peptide 1 associates with Kv4.2 and modulates its gating function: potential role as subunit of cardiac transient outward channel? Circ Res. 2001;88(10):1012–1019. doi: 10.1161/hh1001.090839. [DOI] [PubMed] [Google Scholar]
- 55.Abbott GW. KCNE2 and the K channel: the tail wagging the dog. Channels. 2012;6(1) doi: 10.4161/chan.19126. doi:10.4161/chan.6.1.19126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chan F-C, Cheng C-P, Wu K-H. et al. Intercalated disc-associated protein, mXin-alpha, influences surface expression of ITO currents in ventricular myocytes. Front Biosci Elite Ed. 2011;3:1425–1442. doi: 10.2741/e344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van der Flier A, Sonnenberg A. Structural and functional aspects of filamins. Biochim Biophys Acta BBA – Mol Cell Res. 2001;1538((2-3)):99–117. doi: 10.1016/s0167-4889(01)00072-6. [DOI] [PubMed] [Google Scholar]
- 58.Himmel M, Van der Ven PFM, Stöcklein W, Fürst DO. The limits of promiscuity: isoform-specific dimerization of filamins. Biochemistry (Mosc) 2003;42(2):430–439. doi: 10.1021/bi026501+. [DOI] [PubMed] [Google Scholar]
- 59.Vorgerd M, Van der Ven PFM. et al. A mutation in the dimerization domain of filamin c causes a novel type of autosomal dominant myofibrillar myopathy. Am J Hum Genet. 2005;77(2):297–304. doi: 10.1086/431959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zieseniss A. Hypoxia and the modulation of the actin cytoskeleton – emerging interrelations. Hypoxia. 2014;11 doi: 10.2147/HP.S53575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuznetsov AV, Javadov S, Sickinger S, Frotschnig S, Grimm M. H9c2 and HL-1 cells demonstrate distinct features of energy metabolism, mitochondrial function and sensitivity to hypoxia–reoxygenation. Biochim Biophys Acta. 2015;1853(2):276–284. doi: 10.1016/j.bbamcr.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dhalla NS, Elmoselhi AB, Hata T, Makino N. Status of myocardial antioxidants in ischemia–reperfusion injury. Cardiovasc Res. 2000;47(3):446–456. doi: 10.1016/s0008-6363(00)00078-x. [DOI] [PubMed] [Google Scholar]
- 63.Sanada S, Komuro I, Kitakaze M. Pathophysiology of myocardial reperfusion injury: preconditioning, postconditioning, and translational aspects of protective measures. Am J Physiol - Heart Circ Physiol. 2011;301(5):H1723–1741. doi: 10.1152/ajpheart.00553.2011. [DOI] [PubMed] [Google Scholar]
- 64.Bonavita F, Stefanelli C, Giordano E. et al. H9c2 cardiac myoblasts undergo apoptosis in a model of ischemia consisting of serum deprivation and hypoxia: inhibition by PMA. FEBS Lett. 2003;536((1-3)):85–91. doi: 10.1016/s0014-5793(03)00029-2. [DOI] [PubMed] [Google Scholar]
- 65.Nakamura M, Wang NP, Zhao ZQ. et al. Preconditioning decreases Bax expression, PMN accumulation and apoptosis in reperfused rat heart. Cardiovasc Res. 2000;45(3):661–670. doi: 10.1016/s0008-6363(99)00393-4. [DOI] [PubMed] [Google Scholar]
- 66.Hong Z, Weir EK, Nelson DP, Olschewski A. Subacute hypoxia decreases voltage-activated potassium channel expression and function in pulmonary artery myocytes. Am J Respir Cell Mol Biol. 2004;31(3):337–343. doi: 10.1165/rcmb.2003-0386OC. [DOI] [PubMed] [Google Scholar]
- 67.Henke RM, Dastidar RG, Shah A. et al. Hypoxia elicits broad and systematic changes in protein subcellular localization. Am J Physiol – Cell Physiol. 2011;301(4):C913–928. doi: 10.1152/ajpcell.00481.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nanduri J, Wang N, Bergson P, Yuan G, Ficker E, Prabhakar NR. Mitochondrial reactive oxygen species mediate hypoxic down-regulation of hERG channel protein. Biochem Biophys Res Commun. 2008;373(2):309–314. doi: 10.1016/j.bbrc.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 69.Minsaas L, Planagumà J, Madziva M. et al. Filamin A binds to CCR2B and regulates its internalization. PLoS ONE. 2010;5(8):e12212. doi: 10.1371/journal.pone.0012212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Noam Y, Ehrengruber MU, Koh A. et al. Filamin A promotes dynamindependent internalization of hyperpolarization-activated cyclic nucleotide- gated type 1 (HCN1) channels and restricts Ih in hippocampal neurons. J Biol Chem. 2014;289(9):5889–5903. doi: 10.1074/jbc.M113.522060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mazzochi C, Benos DJ, Smith PR. Interaction of epithelial ion channels with the actin-based cytoskeleton. Am J Physiol Renal Physiol. 2006;291(6):F1113–1122. doi: 10.1152/ajprenal.00195.2006. [DOI] [PubMed] [Google Scholar]
- 72.Vatta M, Faulkner G. Cytoskeletal basis of ion channel function in cardiac muscle. Future Cardiol. 2006;2(4):467–476. doi: 10.2217/14796678.2.4.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Steele DF, Fedida D. Cytoskeletal roles in cardiac ion channel expression. Biochim Biophys Acta. 2014;1838(2):665–673. doi: 10.1016/j.bbamem.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 74.López-López J, González C, Ureña J, López-Barneo J. Low pO2 selectively inhibits K channel activity in chemoreceptor cells of the mammalian carotid body. J Gen Physiol. 1989;93(5):1001–1015. doi: 10.1085/jgp.93.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Keating MT, Sanguinetti MC. Molecular and cellular mechanisms of cardiac arrhythmias. Cell. 2001;104(4):569–580. doi: 10.1016/s0092-8674(01)00243-4. [DOI] [PubMed] [Google Scholar]
- 76.Sanguinetti MC. When the KChIPs are down. Nat Med. 2002;8(1):18–19. doi: 10.1038/nm0102-18. [DOI] [PubMed] [Google Scholar]
- 77.Um SY, McDonald TV. Differential association between HERG and KCNE1 or KCNE2. PLoS ONE. 2007;2(9):e933. doi: 10.1371/journal.pone.0000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang M, Zhang M, Tang DG. et al. KCNE2 protein is expressed in ventricles of different species, and changes in its expression contribute to electrical remodeling in diseased hearts. Circulation. 2004;109(14):1783–1788. doi: 10.1161/01.CIR.0000124225.43852.50. [DOI] [PubMed] [Google Scholar]
- 79.Chandrasekhar KD, Bas T, Kobertz WR. KCNE1 subunits require co-assembly with K+ channels for efficient trafficking and cell surface expression. J Biol Chem. 2006;281(52):40015–40023. doi: 10.1074/jbc.M604398200. [DOI] [PubMed] [Google Scholar]
- 80.Chandrasekhar KD, Lvov A, Terrenoire C, Gao GY, Kass RS, Kobertz WR. O-glycosylation of the cardiac I(Ks) complex. J Physiol. 2011;589((Pt 15)):3721–3730. doi: 10.1113/jphysiol.2011.211284. [DOI] [PMC free article] [PubMed] [Google Scholar]