ABSTRACT
Following phagocytosis, microbes are exposed to an array of antimicrobial weapons that include reactive oxygen species (ROS) and cationic fluxes. This is significant as combinations of oxidative and cationic stresses are much more potent than the corresponding single stresses, triggering the synergistic killing of the fungal pathogen Candida albicans by “stress pathway interference.” Previously we demonstrated that combinatorial oxidative plus cationic stress triggers a dramatic increase in intracellular ROS levels compared to oxidative stress alone. Here we show that activation of Cap1, the major regulator of antioxidant gene expression in C. albicans, is significantly delayed in response to combinatorial stress treatments and to high levels of H2O2. Cap1 is normally oxidized in response to H2O2; this masks the nuclear export sequence, resulting in the rapid nuclear accumulation of Cap1 and the induction of Cap1-dependent genes. Here we demonstrate that following exposure of cells to combinatorial stress or to high levels of H2O2, Cap1 becomes trapped in a partially oxidized form, Cap1OX-1. Notably, Cap1-dependent gene expression is not induced when Cap1 is in this partially oxidized form. However, while Cap1OX-1 readily accumulates in the nucleus and binds to target genes following high-H2O2 stress, the nuclear accumulation of Cap1OX-1 following combinatorial H2O2 and NaCl stress is delayed due to a cationic stress-enhanced interaction with the Crm1 nuclear export factor. These findings define novel mechanisms that delay activation of the Cap1 transcription factor, thus preventing the rapid activation of the stress responses vital for the survival of C. albicans within the host.
IMPORTANCE
Combinatorial stress-mediated synergistic killing represents a new unchartered area in the field of stress signaling. This phenomenon contrasts starkly with “stress cross-protection,” where exposure to one stress protects against subsequent exposure to a different stress. Previously we demonstrated that the pathogen Candida albicans is acutely sensitive to combinations of cationic and oxidative stresses, because the induction of H2O2-responsive genes is blocked in the presence of cationic stress. We reveal that this is due to novel mechanisms that delay activation of the Cap1 AP-1-like transcription factor, the major regulator of the H2O2-induced regulon. Cap1 becomes trapped in a partially oxidized form following simultaneous exposure to oxidative and cationic stresses. In addition, cationic stress promotes the interaction of Cap1 with the Crm1 nuclear export factor, thus inhibiting its nuclear accumulation. These mechanisms probably explain the potency of neutrophils, which employ multiple stresses to kill fungal pathogens.
INTRODUCTION
Candida albicans is a major fungal pathogen of humans. Recent estimates indicate that invasive C. albicans infections are associated with disturbingly high mortality rates of between 46 and 75% and are responsible for over 400,000 life-threatening systemic infections each year (1). Immunocompromised patients are most at risk of systemic candidiasis, such as those receiving immunosuppressive treatments for cancer or transplant surgery. In contrast, in healthy hosts, robust immune protection mechanisms prevent such systemic infections, with innate immune cells such as macrophages and neutrophils providing the first line of defense.
A major antimicrobial defense mechanism mounted by innate immune cells is the production of superoxide anions (O2−) by the NADPH oxidase complex (2). The importance of this oxidative burst in fungal killing is exemplified by patients with chronic granulomatous disease (CGD). CGD is a genetic disorder in which patients have a defective phagocytic NADPH oxidase complex. These patients are significantly more susceptible to systemic Candida infections (3). The levels of O2− generated by neutrophils in the phagocytic vacuole are estimated to range between 1 (4) and 4 mol liter−1 (5). The steady-state levels of O2− are, however, likely to be much lower (4) due to its rapid dismutation to the more reactive hydrogen peroxide, H2O2 (6). Moreover, the resultant H2O2 can also generate hypochlorous acid (HOCl) by the action of myeloperoxidase. O2− can also react with the nitric oxide radical, generated by nitric oxide synthase, to form peroxynitrite (ONOO−). Thus, the production of superoxide leads to the generation of a range of reactive oxygen, nitrogen, and chloride species (reviewed in references 4 and 6).
The prevailing view that reactive oxygen species (ROS) are a major factor underlying the fungicidal action of phagocytes does, however, conflict with previous studies which have demonstrated that C. albicans is more resistant to multiple oxidative stress-inducing agents than other fungi (7, 8). Following exposure to ROS, the activation of several pathways allows C. albicans to detoxify the stress and repair the oxidative damage to cellular components (9). One major mechanism involves the rapid induction of genes with antioxidant properties (10–14), and this is largely regulated by the AP-1-like transcription factor Cap1 (15, 16). Similar to the homologous Saccharomyces cerevisiae Yap1 and Schizosaccharomyces pombe Pap1 transcription factors (17, 18), H2O2-mediated-Cap1 activation is triggered by the oxidation of redox-active cysteine residues located within two cysteine-rich domains (n-CRD and c-CRD). Based on studies in S. cerevisiae (19, 20), this is predicted to trigger a conformational change within Cap1 that masks the nuclear export sequence (NES) from the Crm1 nuclear export factor, thereby allowing the nuclear accumulation of this transcription factor. Once in the nucleus, Cap1 is phosphorylated, and the induction of Cap1-dependent genes ensues (21). As many key antioxidant genes, including CAT1 encoding catalase and TRX1 encoding thioredoxin, are direct Cap1 targets (22), C. albicans cells lacking Cap1 are exquisitely sensitive to ROS (11, 23, 24) and to phagocyte-mediated killing (25, 26).
As C. albicans mounts a robust response to oxidative stress in vitro and is more resistant to ROS than many other fungi, why is this pathogen unable to survive phagocytosis in the immunocompetent host? Recently, we demonstrated that the sensitivity of C. albicans to oxidative stress increases dramatically if cells are simultaneously exposed to cationic stress (27). This is relevant in the context of innate immune defenses, as following phagocytosis, there is an increased flux of the K+ cation into the phagosome to compensate for the anionic charge that accumulates due to the high levels of O2− generated (5). Thus, the potency of innate immune defenses against C. albicans can be attributed to exposure to both oxidative and cationic stresses within the phagosome. At the molecular level, this combinatorial oxidative and cationic stress-mediated synergistic killing of C. albicans is due to stress pathway interference (28). Specifically, Cap1 fails to accumulate in the nucleus following exposure of C. albicans to combinatorial oxidative and cationic stress, and thus Cap1-dependent antioxidant genes are not induced (28). Importantly, the cationic stress-mediated inhibition of oxidative stress responses appears to be of physiological relevance, as the high fungicidal activity of human neutrophils is impaired to similar extents when either the oxidative burst or the cationic flux is inhibited (28).
Here, we dissect the mechanisms underlying the combinatorial stress-mediated inhibition of Cap1 activation. We show that Cap1 becomes trapped in a partially oxidized form for sustained periods following combinatorial oxidative and cationic stress and also in response to high levels of oxidative stress. Significantly, Cap1-dependent gene expression does not occur when Cap1 is in this partially oxidized state. However, the failure of Cap1 to accumulate in the nucleus is specific to the combinatorial cationic and oxidative stress, due to the cation-mediated stabilization of the interaction between Cap1 and the Crm1 nuclear export factor. We propose that these previously uncharacterized mechanisms, which prevent the rapid activation of Cap1, underlie the exquisite sensitivity of C. albicans to combinatorial cationic and oxidative stress and hence the potency of innate immune defenses.
RESULTS
Differential oxidation of Cap1 following combinatorial stress.
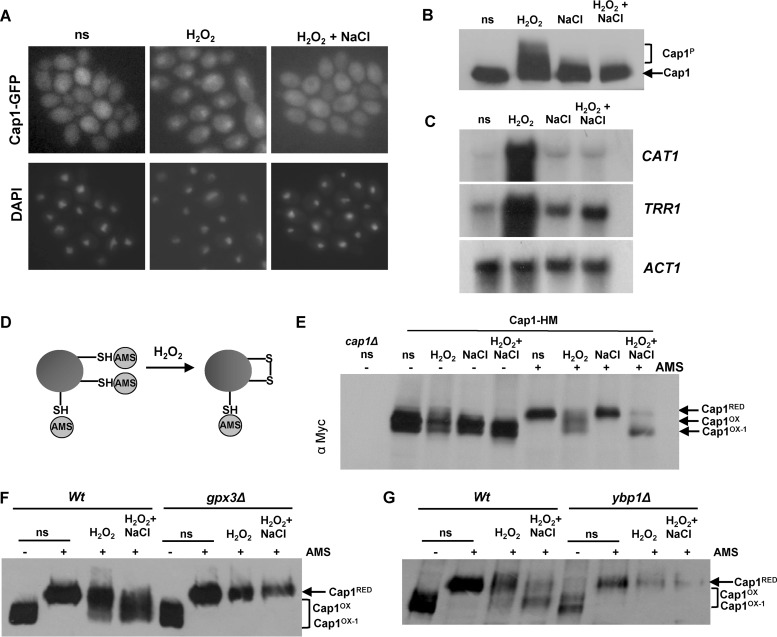
Previously we demonstrated that the normal transcriptional response to oxidative stress is not induced following the simultaneous exposure of C. albicans cells to cationic (1 M NaCl) and oxidative (5 mM H2O2) stress and that the major regulator of oxidative stress response gene expression, Cap1, fails to accumulate in the nucleus following such combinatorial stress treatments (28). However, the mechanisms underlying this inhibition of Cap1 function are unknown. As described above, the oxidative stress-induced nuclear accumulation of fungal AP-1-like transcription factors, such as Cap1, is triggered by the oxidation of redox-active cysteine residues. Therefore, we examined Cap1 oxidation alongside other readouts of Cap1 activation following combinatorial oxidative plus cationic stress treatments. Cells expressing Cap1 tagged with 2 copies of the Myc epitope were collected following a 10-min treatment with H2O2 or combinatorial H2O2 plus NaCl, and samples were simultaneously processed to examine Cap1 oxidation, phosphorylation, and Cap1-dependent gene expression. In addition, cells expressing Cap1-green fluorescent protein (GFP) were exposed to the same stress treatments. As expected, Cap1 failed to accumulate in the nucleus following a 10-min combinatorial H2O plus NaCl treatment but rapidly accumulated in the nucleus following H2O2 treatment alone (Fig. 1A). Furthermore, consistent with the effects of these stress conditions on nuclear accumulation, Cap1 was not phosphorylated following combinatorial stress treatment but was robustly phosphorylated following exposure to H2O2 alone (Fig. 1B). Analysis of the Cap1-dependent transcripts CAT1 and TRR1 reaffirmed our previous microarray data (28) that exposure of C. albicans to oxidative stress in the presence of cationic stress prevents the rapid induction of Cap1-regulated genes (Fig. 1C). As Cap1 oxidation is essential to drive the nuclear accumulation of Cap1 and Cap1-dependent gene expression, it was possible that cationic stress interferes with the H2O2-induced oxidation of Cap1. To examine this hypothesis, the redox status of Cap1 was determined using the alkylating agent 4-aceto-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) (Fig. 1D), which reacts specifically with the thiol groups of reduced cysteine residues, thereby increasing the molecular mass of thiol-modified proteins by 0.64 kDa/cysteine (21). The oxidation of cysteine residues prevents AMS binding, and consequently oxidized proteins have a lower molecular mass and faster mobility on nonreducing PAGE compared to the corresponding reduced proteins. We detected this mobility shift by Western blotting. Cells were subjected to acid lysis and reduced cysteine residues labeled with AMS. Strikingly, AMS-treated Cap1 exhibited a faster mobility following the combinatorial oxidative and cationic stress, compared to oxidative stress alone (Fig. 1E). Importantly the increased mobility of Cap1 following combinatorial stress was AMS dependent (Fig. 1E), indicating this was due to a greater number of cysteine residues being oxidized in Cap1 under these conditions, thus preventing AMS binding. We designated this differentially oxidized form “Cap1OX-1” and the form generated following H2O2 treatment alone “Cap1OX” (Fig. 1E). We also confirmed that the faster-mobility forms seen following oxidative and combinatorial stresses were not due to proteolysis. These forms were not seen in the absence of AMS following acid lysis (Fig. 1E) or following analysis of native extracts (see Fig. S1A in the supplemental material), and the cellular levels of Cap1 were not affected by any of the stress treatments employed above (see Fig. S1A and S1B).
FIG 1 .
The lack of antioxidant gene expression following combinatorial stress is accompanied by a differentially oxidized form of the Cap1 transcription factor. (A) Cap1 does not accumulate in the nucleus following combinatorial stress. Localization of Cap1 was detected by fluorescence microscopy of cells expressing Cap1-GFP (JC1060) under non-stress conditions (ns) and after exposure to 5 mM H2O2 or 5 mM H2O2 plus 1 M NaCl for 10 min. The position of the nuclei is shown by DAPI staining. (B) Cap1 is phosphorylated following H2O2 exposure but not combinatorial stress. Lysates from cells expressing 2Myc- and 6His-tagged Cap1 (Cap1-MH [JC948]), before (ns) and after the indicated stress treatments, were analyzed by Western blotting using an anti-Myc antibody. The positions of nonphosphorylated (Cap1) and phosphorylated (Cap1P) Cap1 are indicated. (C) Combinatorial stress inhibits H2O2-induced antioxidant gene expression. Northern blot analysis of RNA isolated from wild-type (Wt [JC747]) cells before (ns) and following a 10-min treatment with 5 mM H2O2, 1 M NaCl, or 5 mM H2O2 plus 1 M NaCl. Blots were analyzed with probes specific for the catalase (CAT1) and thioredoxin reductase (TRR1) genes. A probe against ACT1 was used as the loading control. (D) Diagram illustrating the detection of protein oxidation by AMS binding to reduced thiols. (E) Cap1 is differentially oxidized following combinatorial stress. Cap1 oxidation was analyzed by nonreducing SDS-PAGE and Western blotting of AMS-modified or untreated proteins prepared from cells expressing Cap1-HM before (ns) and following the stress treatments described above. Extracts from cap1Δ cells were included as a control. The positions of reduced (Cap1RED), oxidized (Cap1OX), and differentially oxidized (Cap1OX-1) Cap1 are indicated. (F) Differential oxidation of Cap1 is dependent on Gpx3. Cap1 oxidation was determined as described above in wild-type (Wt [JC948]) and gpx3Δ (JC1311) cells expressing Cap1-MH before (ns) and following the stress treatments described above. (G) Differential oxidation of Cap1 is dependent on Ypb1. Cap1 oxidation was determined as described above in wild-type (Wt [JC948]) and ybp1Δ (JC954) cells.
The oxidation of C. albicans Cap1 in response to H2O2 requires the thiol peroxidase Gpx3 (26). Hence, we next investigated whether the formation of Cap1OX-1 following combinatorial cationic and oxidative stress is also dependent on Gpx3. Wild-type cells and cells lacking GPX3 were exposed to both oxidative and combinatorial oxidative and cationic stresses, and Cap1 oxidation was examined. Interestingly, the formation of Cap1OX-1 following combinatorial stress, as well as Cap1OX following oxidative stress, was abolished in gpx3Δ cells (Fig. 1F). Gpx3-mediated Cap1 oxidation also requires an orthologue of the S. cerevisiae Yap1 binding protein, Ybp1, which additionally functions to prevent the degradation of these AP-1-like transcription factors (26). Similar to what was observed in gpx3Δ cells, no Cap1 oxidation was evident in cells lacking YBP1 following exposure to either oxidative or combinatorial stress (Fig. 1G). Furthermore, consistent with previous findings, Cap1 levels were significantly reduced in ybp1Δ cells (Fig. 1G). Based on these results, we conclude that differentially oxidized forms of Cap1 are generated following a 10-min exposure to oxidative and combinatorial stresses and that Gpx3 and Ybp1 are crucial for this differential Cap1 oxidation.
Cap1OX contains more disulfides than Cap1OX-1.
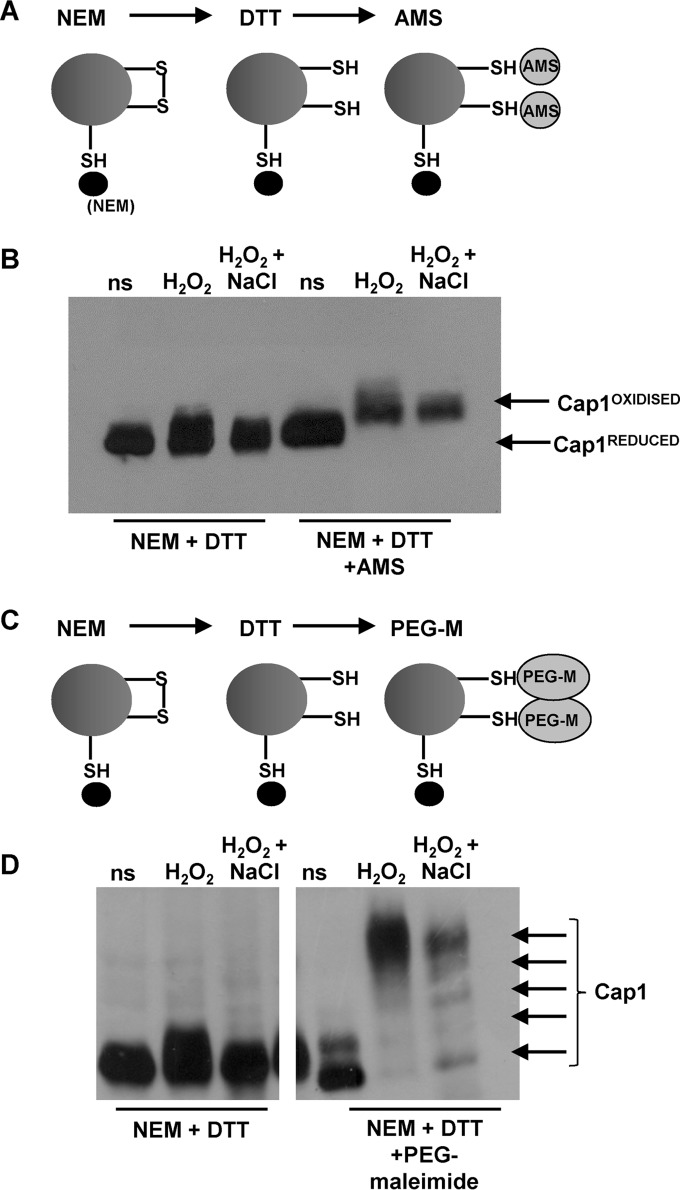
In S. cerevisiae, the stepwise oxidation of all six redox-active cysteines within Yap1, leading to three interdomain disulfides between the n-CRD and c-CRD cysteine-rich domains, is necessary for maximal activation (29). Hence, to investigate the nature of the Cap1OX-1 form, labeling experiments were performed to allow for the detection of H2O2-induced disulfide bond formation. Cells were subjected to acid lysis, and free reduced cysteines were blocked with the low-molecular-mass thiol-binding reagent N-ethylmaleimide (NEM). Any disulfides present were then reduced with dithiothreitol (DTT), and subsequent free thiols were labeled with AMS (Fig. 2A). Thus, in this experiment, the presence of oxidized intramolecular disulfides is indicated by the reduced mobility of Cap1 upon SDS-PAGE. Cap1 exhibited reduced mobility after the sequential NEM-DTT-AMS treatment in cells treated with either oxidative stress or combinatorial oxidative plus cationic stress (Fig. 2B; see Fig. S2A in the supplemental material). Thus, both Cap1OX and Cap1OX-1 forms contain oxidized intramolecular disulfides. Based on our previous experiment, in which CapOX-1 displayed a faster mobility than Cap1OX following AMS treatment alone (Fig. 1E), we predicted that this form of Cap1 may have more disulfides than Cap1OX. If this was the case, then following sequential NEM-DTT-AMS treatment, Cap1OX-1 would have a slower mobility than Cap1OX due to more AMS binding. However, this was not observed: Cap1 displayed a slightly slower mobility following oxidative stress than following combinatorial stress (Fig. 2B). This suggested that Cap1OX-1 has fewer disulfides than Cap1OX. It was possible, however, that this slower mobility of Cap1 following oxidative stress was due to residual phosphorylation of this transcription factor, even after phosphatase treatment, as phosphorylation occurs following H2O2, but not combinatorial, stress treatments (Fig. 1C). To avoid this complication, we repeated the experiment by monitoring mobility retardation mediated by another thiol-alkylation probe, polyethylene glycol (PEG)-linked maleimide, which has a higher molecular mass (2 kDa) than AMS (Fig. 2C). Using PEG-maleimide, clear differences were observed in the oxidized forms of Cap1 generated following oxidative and combinatorial stress treatment (Fig. 2D). Following H2O2 stress, Cap1 was present predominantly as a single species with significantly retarded mobility following the sequential NEM-DTT-PEG maleimide treatment. In contrast, following combinatorial stress, multiple differentially oxidized forms of Cap1 were observed, with only a fraction displaying the same retarded mobility as that seen for Cap1OX following oxidative stress alone (Fig. 2D). Thus, although the previous AMS binding experiment indicated that Cap1OX-1 was more oxidized than Cap1OX (Fig. 1E), these experiments indicate that Cap1OX has more H2O2-induced disulfides than Cap1OX-1, generated following combinatorial stress. This seemingly contradictory observation could be explained by the hyperoxidation of cysteine thiols to sulfinic or sulfonic acid derivatives in the Cap1OX-1 form.
FIG 2 .
Cap1 oxidation following oxidative and combinatorial stresses. (A) Diagram showing the sequential NEM-DTT-AMS treatments employed to allow for the detection of disulfide bonds. (B) The oxidized forms generated following both oxidative and combinatorial stresses contain disulfide bonds. Cap1 mobility was monitored by nonreducing SDS-PAGE and Western blotting of proteins prepared from cells expressing Cap1-HM, under non-stress conditions (ns [Cap1REDUCED]) or following treatment with the indicated compounds. Samples were phosphatase treated prior to loading. The presence of disulfide bonds is indicated by the retarded mobility of Cap1 due to AMS binding to DTT-resolved disulfides (Cap1OXIDISED). (C) Diagram showing the sequential NEM-DTT-PEG-maleimide (PEG-M) treatments that allow for the detection of disulfide bonds. (D) Comparison of Cap1 oxidation before (ns) and following oxidative and combinatorial stresses by PEG-maleimide binding to DTT-resolved thiols. As in panel B, samples were phosphatase treated prior to loading. This shows that different oxidized forms of Cap1 are present following combinatorial stress, whereas a single form containing multiple disulfides is prevalent following oxidative stress.
Differential oxidation and inactivation of Cap1 following combinatorial stress is transient.
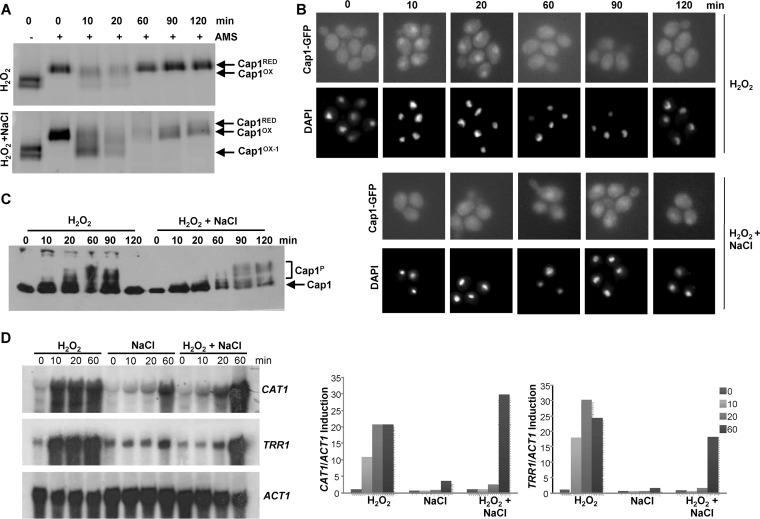
Next, we determined whether the differential oxidation and inactivation of Cap1 following combinatorial stress was short-lived or irreversible. Cells expressing either Myc-tagged Cap1 or Cap1-GFP were treated with H2O2 or H2O2 plus NaCl, and samples were collected over a 2-h period. First of all, we determined the redox status of Cap1 using the AMS alkylating agent as described in the legend to Fig. 1D. The double band of Cap1 seen in the non-AMS-treated time zero sample is likely due to oxidation during protein extraction as this is prevented by the addition of the low-molecular-mass thiol binding agent NEM during extraction (see Fig. S2B in the supplemental material). Consistent with previous findings, the Cap1OX form was quickly generated following H2O2 stress and then resolved back to the reduced form within 60 min. Similarly, the Cap1OX-1 form appeared rapidly after H2O2 plus NaCl stress (Fig. 3A). However, by 60 min, the combinatorial stress-induced Cap1OX-1 form was resolved to a form with mobility similar to that of Cap1OX (Fig. 3A). This indicates that the Cap1OX-1 form generated following combinatorial stress is transient.
FIG 3 .
Combinatorial stress-mediated inhibition of Cap1 activation is transient. (A) Differential oxidation of Cap1 following combinatorial stress is not sustained. Cap1 oxidation was measured as described in the legend to Fig. 1E following exposure of Cap1-MH cells to 5 mM H2O2 or 5 mM H2O2 plus 1 M NaCl for the indicated times. (B) Cap1 nuclear accumulation is delayed following combinatorial stress. Cap1 localization was detected as described in the legend to Fig. 1B following treatment of Cap1-GFP cells for the indicated times with the stress treatments described above. (C) Cap1 phosphorylation is delayed following combinatorial stress treatment. Cap1 phosphorylation was detected as described in the legend to Fig. 1C, following treatment of Cap1-MH cells with the stress treatments described above for the indicated times. (D) The inhibition of Cap-dependent gene expression following combinatorial stress is transient. Northern blots were performed as described in the legend to Fig. 1A, after wild-type cells were treated with the stress treatments described above for the indicated times (left panel). The levels of CAT1 and TRR1 mRNA were quantified relative to the ACT1 loading control (right panel).
To examine whether the inactivation of Cap1 following combinatorial stress was coincident with the presence of the Cap1OX-1 form, the kinetics of Cap1 nuclear accumulation, phosphorylation, and Cap1-dependent gene expression were determined. As illustrated in Fig. 3B, Cap1 did accumulate in the nucleus following combinatorial H2O2 plus NaCl stress, but with significantly delayed kinetics compared to H2O2 stress. Cap1 located to the nucleus 10 min following H2O2 stress, whereas nuclear accumulation of Cap1 post-combinatorial H2O2 and NaCl stress was not evident until 60 min. Notably the appearance of Cap1 in the nucleus coincided with Cap1 being resolved to the Cap1OX form (Fig. 3A and B). We next examined Cap1 phosphorylation, as this posttranslational modification is associated with the nuclear accumulation of fungal AP-1-like transcription factors (30). Cap1 phosphorylation was only seen 60 min after the combinatorial H2O2 plus NaCl stress, coincident with the point at which Cap1 accumulated in the nucleus (Fig. 3C). Significantly, the delayed nuclear accumulation and phosphorylation of Cap1 observed following combinatorial stress was mirrored by a delay in Cap1-dependent gene induction (Fig. 3D). Northern analysis revealed that the Cap1-dependent genes CAT1 and TRR1 are significantly induced following the combinatorial H2O2 plus NaCl stress, but not until 60 min after the combinatorial stress treatment. In contrast, these key antioxidant-encoding genes were induced within 10 min of H2O2 stress exposure (Fig. 3D). This is entirely consistent with our previous microarray data which failed to detect Cap1-dependent gene expression following a 10-min exposure to the combinatorial oxidative plus cationic stress (28). The significant delay in Cap1-dependent gene expression following simultaneous exposure to H2O2 plus NaCl likely underlies the inability of C. albicans to survive this combination of stresses.
Differential Cap1 oxidation triggered by high levels of H2O2.
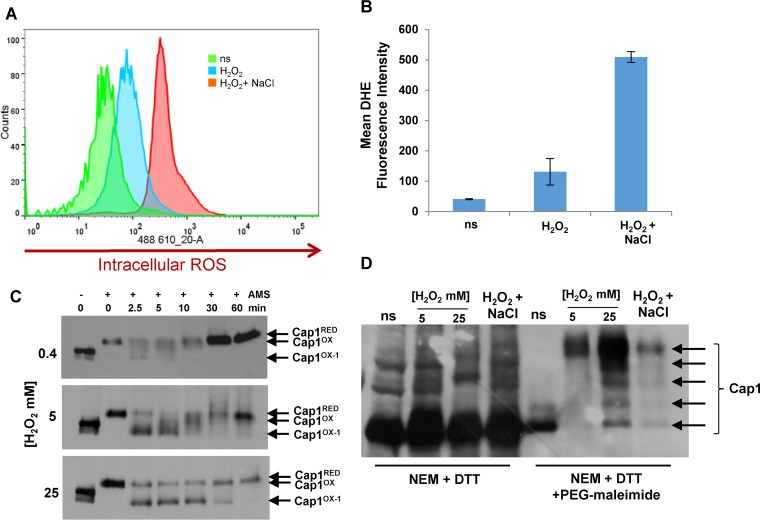
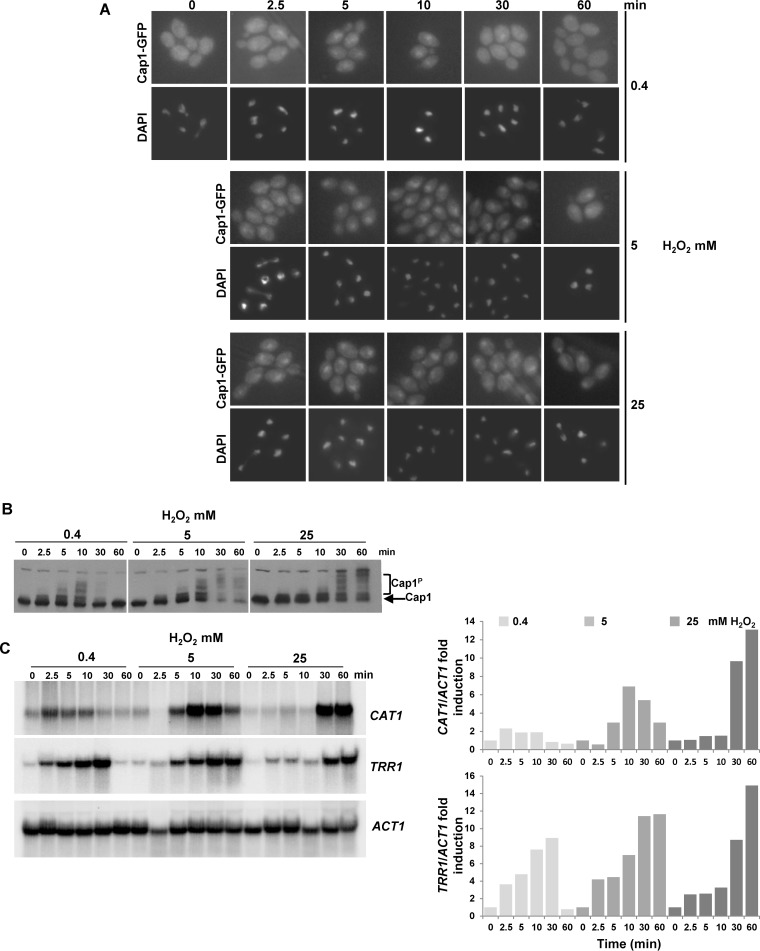
Why does Cap1 become differentially oxidized following combinatorial H2O2 and NaCl stress? We previously reported that there is a dramatic increase in intracellular ROS levels following exposure to combinatorial oxidative plus cationic stress, compared to oxidative stress alone (28). This rise in intracellular ROS could drive the differential oxidation and inactivation of Cap1. To explore this, we first quantified the increase in intracellular ROS following treatment of cells with H2O2 or combinations of H2O2 and NaCl. Approximately 5-fold-higher levels of intracellular ROS were observed following exposure of cells to 5 mM H2O2 in the presence of 1 M NaCl, compared to 5 mM H2O2 alone (Fig. 4A and B). Based on this observation, we hypothesized that Cap1 may also become differentially oxidized to the Cap1OX-1 form following exposure of cells to high levels of ROS. To investigate this, cells expressing Myc-tagged Cap1 were treated with low (0.4 mM), medium (5 mM), and high (25 mM) levels of H2O2, and Cap1 oxidation was monitored over a 60-min period. Strikingly, this kinetic analysis revealed that, at all levels of H2O2 tested, a faster-mobility Cap1OX-1 form is rapidly observed after stress treatment. However, following exposure to low or medium levels of H2O2, the presence of Cap1OX-1 is short-lived, whereas following exposure to high levels of H2O2, Cap1OX-1 persists for up to 30 min (Fig. 4C). Significantly, as a Cap1OX-1 form is seen at all H2O2 concentrations, albeit with different kinetics, this indicates that such a form may in fact be a normal intermediate in the formation of the Cap1OX form following H2O2 stress.
FIG 4 .
Combinatorial stress induces high levels of intracellular ROS, and high levels of H2O2 also result in sustained differential Cap1 oxidation. (A) Fluorescence-activated cell sorter (FACS) analysis of intracellular ROS levels in DHE-treated C. albicans cells before stress (ns) or following the treatment with 5 mM H2O2 or 5 mM H2O2 plus 1 M NaCl for 60 min. (B) Quantification of intracellular ROS production before (ns) and following treatment with the indicated stresses by calculating the mean DHE fluorescence intensity of the area under the curve. The mean ± standard deviation (SD) from three independent experiments is shown. (C) Cap1OX-1 is sustained following treatment with higher doses of H2O2. Cap1 oxidation was measured as described in the legend to Fig. 1D following exposure of Cap1-MH cells to 0.4, 5, or 25 mM H2O2 for the times indicated. (D) Comparison of Cap1 oxidation before (ns) and following the indicated stress treatments by PEG-maleimide binding to DTT-resolved thiols. This shows that different oxidized forms of Cap1 are similarly triggered following high-H2O2 and combinatorial stresses, whereas a single form containing multiple disulfides is prevalent following medium-H2O2 stress.
We explored whether the Cap1OX-1 form, generated for a sustained period following high levels of H2O2, was similar to the Cap1OX-1 form observed after combinatorial cationic and oxidative stress, using the sequential NEM-DTT-PEG maleimide treatments described above. As observed previously (Fig. 2D), exposure of cells to medium (5 mM) doses of H2O2 results in a single species of Cap1 predicted to contain multiple disulfide bonds, as evidenced by the slow, PEG-maleimide-dependent mobility on SDS-PAGE (Fig. 4D). In contrast, treatment of cells with either high (25 mM) doses of H2O2 or combinatorial stress results in the formation of multiple different oxidized forms of Cap1. For unknown reasons, we consistently recovered more Cap1 following high-H2O2 stress than other treatments (Fig. 4D). However, it is clear that the same differentially oxidized forms of Cap1 are observed following either high-H2O2 or combinatorial stress (Fig. 4D).
As Cap1-dependent gene expression is delayed following combinatorial oxidative and osmotic stress when the Cap1OX-1 form is prevalent (Fig. 3D), we hypothesized that the delayed conversion of Cap1OX-1 to Cap1OX following higher levels of H2O2 (Fig. 4C) may also result in an H2O2 concentration-dependent lag in Cap1-dependent gene expression. To investigate this, we examined the kinetics of Cap1 nuclear accumulation, phosphorylation, and Cap1-dependent gene expression following the exposure of cells to low (0.4 mM), medium (5 mM), and high (25 mM) doses of H2O2. The first observation made was that, in contrast to combinatorial H2O2 and NaCl stress, Cap1 rapidly accumulated in the nucleus irrespective of the level of H2O2 stress (Fig. 5A). Following exposure of cells to 0.4 mM H2O2, this nuclear accumulation is short-lived, consistent with the transient oxidation of Cap1 (Fig. 4C). However, Cap1 nuclear accumulation persists for 60 min following treatment with either 5 or 25 mM H2O2 (Fig. 5A). Thus, while the Cap1OX-1 form generated following combinatorial stress fails to accumulate in the nucleus, Cap1OX-1 formed in response to high levels of H2O2 rapidly accumulates in the nucleus (compare Fig. 5A with Fig. 3B).
FIG 5 .
Cap1 rapidly accumulates in the nucleus in response to high levels of H2O2, but Cap1 phosphorylation and Cap1-dependent gene expression are delayed. (A) Cap1 localization was detected as described in the legend to Fig. 1B following treatment of Cap1-GFP cells with 0.4, 5, or 25 mM H2O2 for the times indicated. (B) Cap1 phosphorylation was detected as described in the legend to Fig. 1C following treatment of wild-type cells with 0.4, 5, or 25 mM H2O2 for the times indicated. (C) Northern blots were performed as described in the legend to Fig. 1A, using the same cells that were processed for Cap1 phosphorylation analysis in panel B (left panel). The levels of CAT1 and TRR1 mRNA were quantified relative to the ACT1 loading control (right panel).
Strikingly, however, despite the fast nuclear accumulation following high levels of H2O2, Cap1 phosphorylation (Fig. 5B) and Cap1-dependent gene expression (Fig. 5C) were not observed until 30 min post-stress treatment. The delayed phosphorylation of Cap1 appeared to coincide with the resolution of the Cap1OX-1 form to the Cap1OX form (Fig. 4C), which in turn correlated with the timing of Cap1-dependent gene expression. Such observations are indicative of a connection between these Cap1 modifications and the activity of this transcription factor. In contrast, consistent with previous findings (31), following low and medium doses of H2O2, the kinetics of induction of Cap1-dependent genes (Fig. 5C) largely correlated with the oxidation, phosphorylation, and nuclear accumulation profiles of Cap1 (Fig. 4C and Fig. 5A and B).
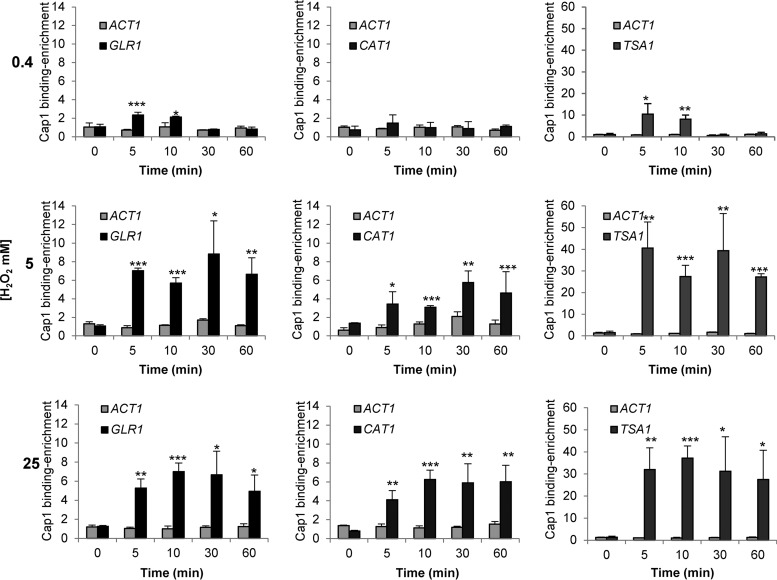
Cap1-dependent gene expression is delayed following high levels of H2O2, despite the clear nuclear accumulation of Cap1. Therefore, we explored whether Cap1 was bound to its target genes under such conditions. To examine Cap1 promoter binding, three targets were selected—CAT1, GLR1, and TSA1—based upon previous studies showing Cap1 enrichment at their promoters (22) and the Cap1-dependent induction of such genes in response to 5 mM H2O2 (11, 16, 32). C. albicans cells expressing Myc-tagged Cap1 together with an untagged control strain were treated with the low, medium, and high levels of H2O2 employed above, and Cap1 binding to the CAT1, GLR1, and TSA1 promoters was determined by chromatin immunoprecipitation (ChIP) and quantitative PCR analysis. As illustrated in Fig. 6, Cap1 was found to rapidly associate with CAT1, GLR1, and TSA1 promoters irrespective of the level of H2O2 (Fig. 6). Moreover, considerably higher levels of Cap1 were detected at these promoters following treatment with medium or high levels H2O2 compared to low doses (Fig. 6). Clearly therefore, the unphosphorylated Cap1OX-1 form generated following high levels of H2O2 is able to bind to the promoter region of its target genes. However, in contrast to the effect seen at low and medium levels of H2O2, this is not sufficient to drive Cap1-dependent antioxidant gene expression (Fig. 5C).
FIG 6 .
Quantification of Cap1 enrichment at the GLR1, CAT1, and TSA1 promoter targets. Cells expressing Cap1-MH (JC948) were treated with 0.4, 5, or 25 mM H2O2 for the indicated times and subjected to ChIP. The recovered DNA samples were analyzed by qPCR using primers specific for promoter regions within GLR1, CAT1, and TSA1. Relative Cap1-MH enrichment values (n-fold) are presented (mean ± SD; n = 3). The following P values were considered: *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001.
Taken together, these results highlight a number of significant findings regarding Cap1 regulation in C. albicans. First, it is apparent that Cap1-dependent gene expression is delayed following exposure of cells to high levels of H2O2. Importantly, this may underlie the delayed Cap1-dependent gene expression seen following the combinatorial H2O2 and NaCl stress, as this combination of stresses triggers high levels of intracellular ROS. Second, the nuclear Cap1OX-1 form, generated following high levels of oxidative stress, although bound to the promoters of Cap1-regulated genes, is not phosphorylated and fails to activate its target genes. These findings suggest that the resolution of the Cap1OX-1 form to Cap1OX and Cap1 phosphorylation are necessary prerequisites for the induction of the Cap1-mediated oxidative stress regulon.
Cationic stress promotes the interaction of Cap1 with the Crm1 nuclear export factor.
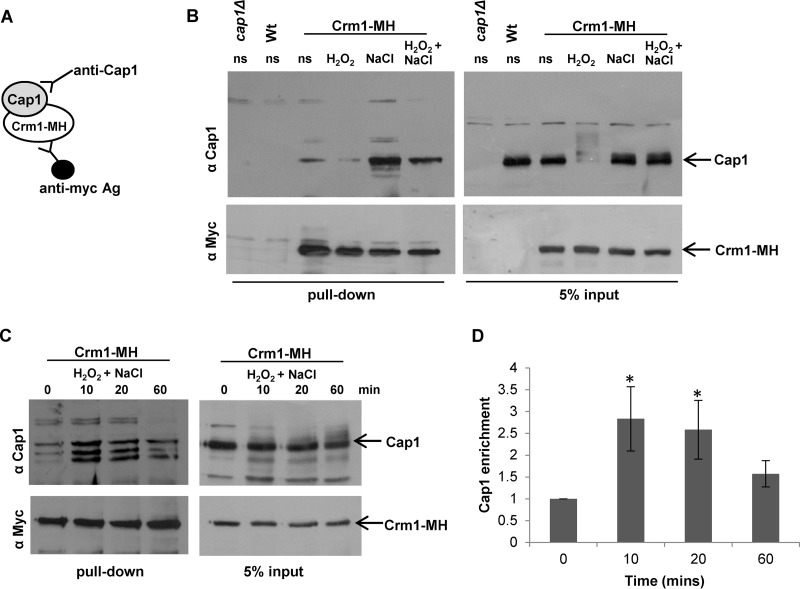
The results presented above illustrate that exposure of cells to either high levels of H2O2 (25 mM) or medium doses of H2O2 (5 mM) in the presence of NaCl results in the sustained formation of Cap1OX-1 and a delay in the activation of Cap1-dependent gene expression. However, while Cap1OX-1 rapidly accumulates in the nucleus in response to high levels of H2O2 (Fig. 5A), Cap1OX-1 does not accumulate in the nucleus until 1 h following combinatorial H2O2 and NaCl treatment (Fig. 3B). The H2O2-induced nuclear accumulation of fungal AP-1-like transcription factors is triggered by the oxidation of key cysteine residues, which results in a conformational change that masks the NES from the nuclear export factor Crm1 (20). Thus, the Cap1OX-1 forms generated following high-level H2O2 stress and combinatorial stress could conceivably be different, with the combinatorial stress Cap1OX-1 form adopting a structural conformation that still permits interaction with the Crm1 nuclear export factor. However, our data indicate that the oxidation profiles of Cap1OX-1 observed following high-H2O2 and combinatorial stresses are similar (Fig. 4D). Alternatively, it was possible that cationic stress modulates the interaction between Cap1 with the Crm1 nuclear export factor and thus affects the nuclear accumulation of Cap1OX-1. To test the latter hypothesis, we first created strains in which Crm1 tagged with 6His residues and 2Myc epitopes (Crm1-MH) was expressed from its native chromosomal locus. Crm1-MH was immunoprecipitated from extracts prepared from cells before and after exposure to H2O2, NaCl, and H2O2 plus NaCl. The interaction between Cap1 and Crm1 was then examined by Western blot analysis of these coimmunoprecipitates (Fig. 7A; see Fig. S3A in the supplemental material). Similar to previous studies of Yap1 in S. cerevisiae (33), Cap1 was found to interact with Crm1 in vivo, and moreover, this interaction was reduced in the presence of medium (5 mM) levels of H2O2 after 10 min (Fig. 7B; see Fig. S3A). Strikingly, the interaction of Cap1 with Crm1 was dramatically enhanced following the cationic stress (NaCl) and the combinatorial H2O2 and NaCl stress treatments (Fig. 7B; see Fig. S3A). Input controls confirmed that the cationic stress-induced interaction with Crm1 was not due to differences in Cap1 levels (Fig. 7B). The input controls, in contrast to the immunoprecipitated samples, were not phosphatase treated, which explains the slower mobility of Cap1 following oxidative stress seen in the input panel (Fig. 7B). The enhanced interaction between Cap1 and Crm1 following the combinatorial H2O2 and NaCl stress was transient, declining 1 h post-stress treatment (Fig. 7C). Quantification of the interaction of Cap1 with Crm1 from four biological replicates indicated a significant increase in Cap1 binding 10 and 20 min after combinatorial stress, but not after 60 min (Fig. 7D). This is consistent with the timing of Cap1 nuclear accumulation, which is observed 1 h following combinatorial H2O2 and NaCl stress (Fig. 3B). However, it is noteworthy that multiple species of Cap1 were often detected during the immunoprecipitation experiment (Fig. 7C; see Fig. S3A). These may reflect degradation products generated during the immunoprecipitation procedure. Taken together, these results support a model in which Cap1OX-1 fails to accumulate in the nucleus following exposure of cells to H2O2 in the presence of NaCl, due to the cation-enhanced interaction between Cap1 and the Crm1 nuclear export factor.
FIG 7 .
Cationic stress promotes Cap1 interaction with the Crm1 nuclear export factor. (A) Strategy used to examine Cap1 association with Crm1. (B) Stress effects on Cap1 interaction with Crm1. Extracts were prepared from wild-type cells (Wt [JC747]), cap1Δ cells (JC842), and wild-type cells expressing 2Myc- and 6His-tagged Crm1 (Crm1-MH [JC1925]) before (ns) and following exposure to 5 mM H2O2, 1 M NaCl, or combinations of the stresses for 10 min. Crm1-MH was immunoprecipitated using anti-Myc agarose, and precipitated proteins and 5% input were subjected to SDS-PAGE. Coprecipitation of Cap1 was assayed by Western blotting using an anti-Cap1 antibody (top panel), and precipitation of Crm1-MH was assayed using anti-Myc antibodies (bottom panel). (C) The increased interaction of Cap1 with Crm1 following combinatorial stress is transient. Cap1 interaction with Crm1 was analyzed as described for panel B before (ns) and following treatment of Crm1-MH cells with 5 mM H2O2 plus 1 M NaCl for the times indicated. (D) Quantification of the increased interaction of Cap1 with Crm1 following combinatorial stress. Quantitative densitometric analysis of Western blots from four biological replicates was conducted to determine the fold enrichment of Cap1 interaction with Crm1 relative to time zero. Mean values (±standard errors of the mean [SEM]) are shown, and ANOVA was used to determine statistically significant differences in Cap1 enrichment levels (*, P < 0.01).
DISCUSSION
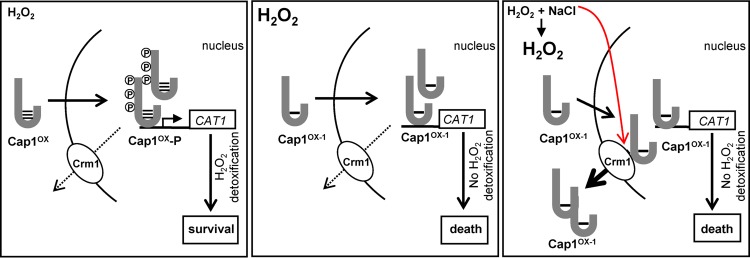
In this study, we have demonstrated that exposure of C. albicans cells to combinatorial H2O2 plus NaCl stress, which triggers high levels of intracellular ROS, results in the sustained formation of a differentially oxidized form of Cap1, Cap1OX-1. Furthermore, we show that Cap1OX-1 contains less oxidized disulfide bonds compared to those seen in the active Cap1OX form. This differentially oxidized Cap1OX-1 form is also observed following treatment of C. albicans cells with a range of H2O2 doses but is only sustained following high doses of H2O2 (Fig. 4). Notably, Cap1-dependent gene expression is not induced when Cap1 is in the Cap1OX-1 form. However, while Cap1OX-1 readily accumulates in the nucleus following high-H2O2 stress, the nuclear accumulation of Cap1OX-1 following combinatorial H2O2 and NaCl stress is delayed due to an enhanced interaction with the Crm1 nuclear export factor. Thus, at least two distinct mechanisms exist to inhibit Cap1 activation following combinatorial stress. This may explain why Cap1 inhibition is more sustained following combinatorial stress than high-H2O2 stress. A model comparing the mechanisms underlying the delayed activation of Cap1 following combinatorial H2O2 plus NaCl stress and high-H2O2 stress is shown in Fig. 8. We predict that the significant delay in Cap1-dependent gene expression following exposure of cells to H2O2 in the presence of NaCl underlies the exquisite sensitivity of C. albicans to this combinatorial stress. This is supported by the observation that ectopic expression of the Cap1-target gene, CAT1, can restore C. albicans resistance to combinatorial H2O2 and NaCl stress (28). In addition, our findings reported here that Cap1-dependent gene expression is also delayed following high-level H2O2 stress, albeit not to the same extent as following combinatorial stress, likely contributes to the increased sensitivity of C. albicans to high doses of oxidative stress. Moreover, the failure to respond rapidly to high intracellular ROS likely results in the activation of cell death programs (34).
FIG 8 .
Impact of combinatorial stress and high H2O2 on Cap1 activation. When C. albicans cells are exposed to medium (5 mM) levels of H2O2, a Cap1OX form containing multiple disulfides is swiftly generated, followed by the rapid accumulation of Cap1 in the nucleus, where it is phosphorylated. The ensuing efficient induction of Cap1-dependent antioxidant genes follows, leading to stress adaptation and survival (left panel). However, when C. albicans cells are exposed to high (25 mM) levels of H2O2, a Cap1OX-1 form containing less disulfides is prevalent, and although this rapidly accumulates in the nucleus and binds to target genes, Cap1 is not phosphorylated, and the induction of Cap1-dependent genes is significantly impaired, resulting in cell death (middle panel). Exposure of C. albicans to medium (5 mM) levels of H2O2 in the presence of NaCl triggers high intracellular ROS accumulation. This also results in the formation of Cap1OX-1, which is seemingly unable to induce Cap1-dependent genes. However, following combinatorial stress, an additional inhibitory mechanism is seen as Cap1 fails to accumulate in the nucleus due to a cationic stress-mediated stabilization of the interaction between Cap1 and the Crm1 export factor. Consequently, the induction of Cap1 target genes is prevented, leading to cell death (right panel).
How is Cap1 activated in C. albicans? Genetic and biochemical studies in S. cerevisiae support the stepwise oxidation of multiple redox-active cysteine residues in Yap1 following H2O2 exposure. This is initiated by the H2O2-induced oxidation of the catalytic cysteine (Cys36) of the thiol-peroxidase Gpx3, which subsequently reacts with Cys598 located within the c-CRD of Yap1 to form a mixed Yap1-Gpx3 disulfide intermediate. This is followed by thiol-disulfide exchange with Cys303 of Yap1 to generate the first interdomain disulfide bond between Cys303 and Cys598 (21). This disulfide is sufficient to mask the NES within the c-CRD of Yap1, prevent interaction with the Crm1 nuclear export factor, and thus stimulate nuclear accumulation (30). However, it is clear that disulfides in addition to Cys303 and Cys598 are required for optimal Yap1 function. Mass spectrometry analysis of oxidized Yap1 identified Cys303-Cys598 and Cys310-Cys629 disulfides (35), both of which are required to recruit the mediator component Rox3 to the TRX2 promoter (36). Furthermore, an in vitro analysis of Yap1 oxidation identified a third disulfide, Cys315-Cys620, which appears to sustain Yap1 activation following H2O2 exposure (29). Thus, the stepwise oxidation of all six redox-active cysteines is necessary for maximal Yap1 activation in S. cerevisiae. In this study, we present evidence for a similar stepwise formation of multiple disulfides in C. albicans Cap1. The Cap1OX-1 form would appear to be a normal intermediate in the formation of active Cap1OX, and our experiments, in which we examine the binding of PEG-maleimide to DTT-resolved disulfides (Fig. 4D), show that Cap1OX has more disulfides than Cap1OX-1. This result, however, seemingly conflicts with the oxidation profiles of Cap1 observed when we use the thiol binding agent AMS to detect differentially oxidized forms of this transcription factor. In such experiments, oxidation of cysteine residues precludes the binding of AMS, and thus oxidized proteins run with a faster mobility. Employing such an approach, we observe a faster-mobility form generated immediately following H2O2 stress (Cap1OX-1), which is then resolved into the Cap1OX slower-mobility state (Fig. 4C). How can these findings be reconciled with our observations that Cap1OX has more H2O2-induced disulfides than Cap1OX-1? Oxidation events, in addition to disulfide bond formation, such as the hyperoxidation of cysteine thiols to sulfinic or sulfonic acid derivatives, would preclude AMS binding. Alternatively, AMS binding to the CapOX-1 form may induce a conformation change that results in the observed faster mobility. Further investigations are required to distinguish between these hypotheses and identify the precise oxidation states of the Cap1OX and Cap1OX-1 forms in C. albicans.
The sustained formation of Cap1OX-1 following high-H2O2 and combinatorial stresses in C. albicans correlates with a lack of Cap1-dependent gene expression. Why is Cap1-dependent gene expression inhibited when Cap1 is in the Cap1OX-1 form? Following combinatorial oxidative plus cationic stress, Cap1OX-1 fails to accumulate in the nucleus, which would explain the lack of induction of Cap1-dependent genes. However, following high levels of H2O2, Cap1OX-1 rapidly accumulates in the nucleus, where it binds to the promoters of its target genes (Fig. 6), and yet still, Cap1-dependent gene induction is not observed (Fig. 5C). Here it is interesting to note that although Cap1OX-1 accumulates in the nucleus, it is not phosphorylated. Studies of S. cerevisiae revealed that the nuclear accumulation of Yap1 was necessary for this posttranslational modification (30). However, as both the kinase and target phosphorylation sites on Yap1 are unknown, it has not been possible to establish whether this posttranslational modification is important for the activation of this or other fungal AP-1-like transcription factors. Results from this study which show that there is a coordinated delay in Cap1 phosphorylation and Cap1-dependent gene expression following high-H2O2 stress, despite the nuclear accumulation of Cap1, are consistent with a model in which Cap1 phosphorylation is important for function. In this scenario, either the kinase responsible is inactivated following high-H2O2 stress, or it fails to recognize the Cap1OX-1 form. Indeed, Cap1 phosphorylation coincides with the appearance of the active Cap1OX form. However, other, phosphorylation-independent, mechanisms may also contribute to the delay in Cap1-induced gene expression. For example, in S. cerevisiae, the correct oxidation of Yap1 is necessary to recruit the polymerase II (Pol II) mediator subunit Rox3 (36). Thus, the Cap1OX-1 form may not be competent to recruit the general transcriptional machinery. Alternatively, high levels of intracellular ROS, generated following combinatorial and high-H2O2 stresses, may have a global inhibitory effect on transcription. Indeed, high levels of ROS have been previously demonstrated to result in a global inhibition of protein translation in S. cerevisiae (37). To explore whether a global inhibition of transcription occurs following combinatorial and high-H2O2 stresses, we examined the phosphorylation status of RNA Pol II. Phosphorylation of serine 2 and 5 within the repetitive YSPTSPS sequence, located in the carboxyl-terminal domain (CTD), is associated with transcriptionally active RNA Pol II (38). No obvious change in RNA Pol II phosphorylation status was seen following combinatorial and high-H2O2 stresses, suggesting that under such conditions, the polymerase is active (see Fig. S4 in the supplemental material). However, additional experiments are needed to further explore the potential global impacts of these stress treatments on gene expression.
The inhibition of Cap1 nuclear accumulation following combinatorial stress represents a clear difference in Cap1 regulation following high-H2O2 stress and combinatorial cationic and oxidative stress. Strikingly, in this study we find that cationic stress appears to enhance the interaction between Cap1 and Crm1 in C. albicans, and this interaction is maintained following combinatorial cationic and oxidative stress treatments (Fig. 7B; see Fig. S3A in the supplemental material). Structural analysis of Yap1 revealed that in the active oxidized form, the nuclear export signal (NES) in the c-CRD is masked by disulfide-bond-mediated interactions with a conserved amino-terminal α-helix. Upon reduction of the disulfide bonds, Yap1 undergoes a change to an unstructured conformation that exposes the NES and allows redistribution to the cytoplasm (20). As cationic stress alone can promote the interaction of Cap1 with Crm1, this indicates that this phenomenon occurs independently of changes in the oxidation status of Cap1. We reasoned that the production of osmolytes, triggered by cationic stress, may be responsible for the enhanced interaction between Cap1 and Crm1. However, the cation-induced interaction between Cap1 and Crm1 is maintained in cells lacking the Hog1 stress-activated protein kinase (SAPK) (see Fig. S3B), which fail to produce glycerol following osmotic stress (39). Furthermore, the enhanced interaction appears to be cation specific, as treatment of cells with the osmotic stress agent sorbitol fails to stimulate Cap1 interaction with Crm1 (see Fig. S3C). Although the molecular basis underlying the cation-stimulated Cap1-Crm1 association is unknown, this observation can account for the significant delay in Cap1 nuclear accumulation following combinatorial oxidative and osmotic stress. Interestingly, it is possible that the cationic stress-stimulated interaction with Crm1 may be Cap1 specific, rather than a global phenomenon. For example, we have previously shown that the Hog1 SAPK, whose cellular localization is predicted to regulated by Crm1 (40), rapidly accumulates in the nucleus following combinatorial stress (28). Further investigation into the mechanisms underlying the cationic-stress mediated association between Cap1 and Crm1 is clearly warranted. Although seemingly Hog1 dependent, the Rim101 pathway is worthy of consideration as this pathway is implicated in cationic but not osmotic stress. C. albicans cells lacking the Rim101 transcription factor are sensitive to cationic stress (41), and a role of Rim101 in regulating the Ena1 Na+-ATPase in the model yeast S. cerevisiae is well documented (42).
In conclusion, our results provide significant new insight into why combinatorial cationic plus oxidative stress treatments delay the activation of the Cap1 transcription factor, leading to stress pathway interference. Indeed, cationic stress appears to inhibit oxidative stress responses in two ways. First, cations inhibit catalase activity in C. albicans, thereby leading to significant increases in intracellular ROS levels observed following combinations of cationic and oxidative stresses (28). Here we show that such high levels of intracellular ROS trap Cap1 in an intermediate Cap1OX-1 form that fails to induce target antioxidant-encoding genes. Second, we find that cations stimulate the interaction of Cap1 with the nuclear export factor Crm1, thus delaying the oxidative stress-induced nuclear accumulation of this transcription factor. This combinatorial stress-mediated stress pathway interference that we have dissected in vitro is thought to contribute to the potency of host defenses in combating fungal infections in vivo. Indeed, pharmacological inhibition of ROS production and cationic fluxes, either separately or in combination, results in a similar, drastic, impaired ability of neutrophils to kill C. albicans (28). Thus, the efficacy of combinatorial stress-mediated killing of C. albicans may explain why this fungus only causes systemic infections in immunocompromised hosts. How does the exquisite sensitivity to combinatorial stress benefit C. albicans? It could be speculated that it is nonbeneficial for this pathogen to cause systemic infections as the likely outcome is killing of the host. It seems more likely that Candida albicans has evolved as a commensal organism in the human gut and urogenital tracts. In these environments, such combinations of oxidative and cationic stresses may not be encountered. Finally, it is important to note that C. albicans is exposed to many other combinations of stresses, in addition to oxidative and cationic stresses, following phagocytosis, such as reactive nitrogen and chloride species, antimicrobial peptides, pH fluctuations, and nutrient limitation (reviewed in reference 43). Therefore, future work will be directed at investigating the impact of other combinations of physiologically relevant stress conditions on killing C. albicans and other important fungal pathogens.
MATERIALS AND METHODS
Strains and media.
The strains used in this study are listed in Table S1 in the supplemental material. C. albicans cells were grown in Tris-buffered yeast extract-peptone-dextrose medium (YPDT [pH 7.4]) (27, 44). Oxidative stress was imposed by treating cells with a range of H2O2 (Sigma) concentrations as indicated, and cationic stress was imposed with 1 M NaCl. To stress cells with a combination of oxidative and cationic stresses, medium was supplemented with 5 mM H2O2 plus 1 M NaCl, as detailed previously (27, 28).
Tagging of Crm1.
To C-terminally tag Crm1, expressed from its native chromosomal locus, with 2 copies of the Myc epitope and 6 His residues, the 3′ region of CRM1 was amplified by PCR using the oligonucleotide primers Crm1MHPstF (AATGTCTGCAGCAATTATCTGGAGAAGC) and CrmMHPstR (AATGTCTGCAGTTCGTCATCCATTTCAGAAGG) and genomic DNA template. The resulting PCR product was digested with PstI and ligated into the PstI site of CIp-MH-PstI plasmid (31) to generate pCRM1-MH. pCRM1-MH was linearized by digestion with EcoRV to target integration at the CRM1 locus in SN148 wild-type cells (45) or hog1Δ cells (JC45) to generate strains JC1925 and JC1940, respectively. Correct insertion and tagging of Crm1 were verified by PCR and DNA sequencing.
Cap1 localization.
C. albicans cells expressing GFP-tagged Cap1 (JC1060) were prepared as described previously (11, 46). 4′,6-Diamidino-2-phenylindole (DAPI) and GFP fluorescence was captured using a Zeiss Axioscope with a 63× oil immersion objective and the AxioVision imaging system.
Cap1 phosphorylation and oxidation assays.
To monitor Cap1 phosphorylation, protein extracts were prepared as described previously (47) from exponentially growing C. albicans cells expressing 2Myc- and 6His-tagged Cap1 (JC948) before and after treatments with 5 mM H2O2, 1 M NaCl, or 5 mM H2O2 plus 1 M NaCl for the indicated times. Twenty-five micrograms of protein was subjected to SDS-PAGE, and Cap1-MH was detected using anti-Myc antibodies (9E10 [Sigma]) (31).
To monitor Cap1 oxidation, protein extracts were prepared under acid lysis conditions and treated with the thiol alkylating agent AMS, as described previously (31). To determine the nature of Cap1 oxidation following combinatorial stress and high ROS levels, protein extracts were obtained under acid lysis conditions and incubated with N-ethylmaleimide (NEM [Thermo Scientific, Paisley, United Kingdom]) at a final concentration of 10 mM at 30°C for 30 min in order to block free thiols, precipitated with 1 vol of 20% trichloroacetic acid (TCA) on ice for 30 min, and washed extensively three times with acetone. NEM-labeled samples were resuspended in sample buffer (200 mM Tris-HCl [pH 8], 1% SDS, 1 mM EDTA) supplemented with 20 mM DTT and incubated at 37°C for 60 min in order to reduce the existing disulfides. Acid precipitation and acetone washes were repeated, and protein pellets were resuspended in sample buffer containing methoxy PEG-maleimide (mPEG-Mal) (PEG-maleimide with a molecular weight of 2,000 [Nanocs, Inc., Boston, MA]) at a final concentration of 10 mM at 30°C for 30 min (29) in order to label free thiols that were involved in the formation of disulfides. Acid precipitation and acetone washes were repeated, and samples were treated with 5 U of alkaline phosphatase (New England Biolabs) at 37°C for 1 h to prevent phosphorylation from having an impact on AMS/PEG-maleimide-dependent mobility shifts (26). Samples were subjected to SDS-PAGE on 8% gels under nonreducing conditions, and Cap1-MH was detected as described above.
Northern blotting.
Northern blotting was performed as described previously (11). Gene-specific probes were amplified by PCR from genomic DNA using oligonucleotide primers specific for ACT1, CAT1, and TRR1 (11). RNA levels were quantified using the Typhoon phosphorimaging system (Amersham Biosciences) and Image Quant software (GE Healthcare Life Sciences).
Detection of intracellular ROS levels.
For intracellular ROS detection, exponentially growing C. albicans cells were treated with 20 µM of the fluorescent probe dihydroethidium (DHE [Sigma]) and incubated at 30°C for 45 min in the dark. Cells were washed with phosphate-buffered saline (PBS), sonicated for 15 s, and subjected to fluorescence-activated cell sorting using a FACSAria Fusion (BD Biosciences, Sydney, Australia) with an argon 488-nm laser emitting at 595 nm. Data were analyzed from three independent biological replicates using FlowJo software (TreeStar, Inc., Ashland, OR).
ChIP-qPCR.
Chromatin immunoprecipitation (ChIP) assays were performed as described by Liu et al. (48), with slight modifications. Briefly, three independent cultures of exponentially growing C. albicans JC465 (untagged control strain) and JC948 (Cap1-MH-tagged strain) cells were collected and chromatin fixed by the addition of formaldehyde (1% vol/vol) for 30 min at room temperature with agitation at 20 rpm. Formaldehyde was quenched by the addition of 125 mM glycine at room temperature at 20 rpm for 10 min. Samples were centrifuged, washed twice with ice-cold Tris-buffered saline (TBS) buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl), and snap-frozen in liquid nitrogen. The preparation of the total cell extracts and sonication was performed as described previously (48), followed by the overnight immunoprecipitation of the soluble chromatin extracts from both tagged and untagged strains with monoclonal mouse c-Myc antibody (9E10) (Santa Cruz Biotech) coupled to magnetic beads (Dynabeads [Invitrogen Life Technologies]) at 4°C. Beads were washed and reverse cross-linked, and DNA was recovered as described previously (48). The DNA concentrations were quantified using Quant-iT Picogreen double-stranded DNA assay kit (Molecular Probes/Invitrogen) as described previously (48). The DNA concentrations ranged from 0.03 ng/µl to 0.69 ng/µl for the tagged strains and 0.49 ng/µl to 0.72 ng/µl for the untagged strains. Quantitative PCRs (qPCRs) were performed on the Mastercycler ep realplex (Eppendorf) qPCR system using the Takyon ROX Probe MasterMix (Eurogentec) and the conditions described previously (49). Oligonucleotides for the CAT1, TSA1, and GLR1 promoter regions (targets) (see Table S2 in the supplemental material) were optimized using Primer3Web software (version 4.0.0.). ACT1 was used as a control for statistical analyses by t test, with TEF1 (orf19.1435) as a reference for normalization (50). All PCR products had melting curves indicating the presence of a single amplicon. qPCR data were analyzed using the threshold cycle (2−ΔΔCT) method (51), and statistical significance was determined using Welsh’s two-sample t test.
Crm1-Cap1 coimmunoprecipitation assay.
Exponentially growing wild-type (JC1925) or hog1Δ (JC1940) cells expressing 2Myc- and 6His-tagged Crm1 were harvested before and after stress treatments. Total protein extracts were prepared as described previously (47), and Crm1-MH was immunoprecipitated using anti-Myc (9E10) antibody-coupled agarose (Santa Cruz Biotechnologies), followed by incubation with 200 U of Lambda protein phosphatase (New England Biolabs) for 30 min at 30°C. Precipitated proteins and 5% of protein input were subjected to SDS-PAGE. Coprecipitation of Cap1 was monitored by Western blotting using anti-Cap1 polyclonal antibodies (kindly provided by Scott Moye-Rowley, University of Iowa). Membranes were subsequently probed with anti-Myc antibodies (9E10 [Sigma]) to determine the levels of precipitated Crm1 levels. Quantitative densitometric analysis of Western blots was conducted using ImageJ 1.44 to determine the fold enrichment of Cap1 interaction with Crm1 relative to time zero. Analysis of variance (ANOVA) was used to determine statistically significant differences in Cap1 enrichment levels.
Detection of RNA Pol II phosphorylation.
To monitor RNA Pol II phosphorylation, protein extracts were prepared as described previously (47) from exponentially growing C. albicans cells expressing 2Myc- and 6His-tagged Cap1 (JC948) before and after treatments with 5 mM H2O2, 25 mM H2O2, or 5 mM H2O2 plus 1 M NaCl for the indicated times. Fifteen micrograms of protein was subjected to SDS-PAGE, and Pol II phosphorylation was detected using anti-RNA Pol II phospho S2 (ab5095 [Abcam]) and anti-RNA Pol II phospho S5 (ab5408 [Abcam]) antibodies. Total levels of RNA Pol II were detected using an anti-RNA Pol II CTD repeat antibody (ab817 [Abcam]). Blots were stripped, and Cap1-MH was detected using anti-Myc antibodies (9E10 [Sigma]) (31).
SUPPLEMENTAL MATERIAL
Quantification of Cap1 levels pre- and post-stress treatment. (A) Cap1 protein levels were analyzed by SDS-PAGE and Western blotting of native extracts prepared from cells expressing Cap1-HM before (ns) and following exposure to 5 mM H2O2, 1 M NaCl, or combinations of these stresses for 10 min. Blots were stripped and reprobed with an anti-Hog1 antibody as a loading control. (B) Quantification of Cap1 levels. Quantitative densitometric analysis of Western blots from five biological replicates was conducted to determine the relative levels of Cap1 following the stress treatments described above, compared to nonstress (ns) levels. Mean values (±SEM) are shown, and ANOVA was used to determine statistically significant differences in Cap1 levels. No significant differences were observed (P > 0.05). Download
Cap1 is differentially oxidized following combinatorial stress. (A) Cap1 mobility was monitored by nonreducing SDS-PAGE and Western blotting of proteins prepared from cells expressing Cap1-HM exposed to 5 mM H2O2, 1 M NaCl, or combinations of these stresses for 10 min. Disulfide bonds are indicated by the retarded mobility of Cap1 due to AMS binding to DTT-resolved disulfides (compare NEM-DTT- with NEM-DTT-AMS-treated lanes). (B) Cap1 mobility was monitored by nonreducing SDS-PAGE and Western blotting of proteins prepared from unstressed cells (ns) expressing Cap1-HM. Protein extracts were obtained under acid lysis conditions and incubated or not with N-ethylmaleimide, which blocks free thiol groups, thus preventing oxidation. Download
The NaCl-enhanced interaction between Cap1 and Crm1 is Hog1 independent and cationic stress specific. (A) Biological replicate of the pulldown shown in Fig. 7B. (B) Hog1 is dispensable for the salt-induced Cap1-Crm1 interaction. Extracts were prepared from hog1Δ cells (JC45), and hog1Δ cells expressing 2Myc- and 6His tagged Crm1 (JC1940) before and following exposure to 5 mM H2O2, 1 M NaCl, or combinations of these stresses for 10 min. Coprecipitation of Cap1 was detected as described in the legend to Fig. 7B. The asterisk designates a nonspecific band as seen in cap1Δ cells. (C) Sorbitol does not stimulate Cap1 binding to Crm1. Extracts were prepared from wild-type cells expressing 2Myc- and 6His tagged Crm1 (Crm1-MH [JC1925]) before and following exposure to 1 M NaCl for 10 min or 2 M sorbitol for the indicated times. Coprecipitation of Cap1 was detected as described in the legend to Fig. 7B. Download
RNA Pol II phosphorylation is maintained following high-H2O2 exposure and combinatorial stress. Lysates from cells expressing 2Myc- and 6His-tagged Cap1 (Cap1-MH [JC948]), before (ns) and after the indicated stress treatments were analyzed by Western blotting using antibodies that detect RNA Pol II CTD phosphorylation at serine 2 or serine 5 of the repeat sequence YSPTSPS. Total levels of RNA Pol II were determined using an antibody that recognizes the CTD repeat sequence. As a control, a time zero sample was prepared that was treated with lambda phosphatase prior to loading (+pptase). The positions of nonphosphorylated (RNA Pol II) and phosphorylated (RNA-Pol II S2-P, RNA-Pol II S5-P) Pol II are indicated and are consistent with the predicted molecular mass of C. albicans Pol II (192 kDa). The RNA Pol II blot was stripped and reprobed with anti-Cap1 antibodies. This shows that although Cap1 phosphorylation (a marker of active Cap1) is significantly delayed following high-H2O2 stress and combinatorial stress, RNA Pol II phosphorylation is not reduced following such treatments. This indicates that the defect in Cap1-mediated gene expression is not due to a global inhibition of RNA Pol II activity. Download
Strains used in this study.
Oligonucleotides used for qPCR.
ACKNOWLEDGMENTS
We are grateful to Brian Morgan and Elizabeth Veal for insightful discussions, Mélanie Ikeh for experimental assistance, and Scott Moye-Rowley (University of Iowa) for the gift of the anti-Cap1 antibody.
This work was funded by the NIHR Newcastle Biomedical Research Centre (I.K.), a BBSRC DTG studentship (M.J.P.), the Wellcome Trust (grants 089930 and 097377 to J.Q. and 080088 and 097377 to A.J.P.B.), the BBSRC (grants BB/K016393/1 to J.Q. and BB/F00513X/1 and BB/K017365/1 to A.J.P.B.), the European Research Council (STRIFE Advanced grant ERC-2009-AdG-249793 to A.J.P.B.), the ANR (grant CANDIHUB, ANR-14-CE14-0018-01, to C.D.), and the French Government’s Investissement d’Avenir program (grant IBEID, ANR-10-LABX-62-IBEID, to C.D.).
Footnotes
Citation Kos I, Patterson MJ, Znaidi S, Kaloriti D, da Silva Dantas A, Herrero-de-Dios CM, d’Enfert C, Brown AJP, Quinn J. 2016. Mechanisms underlying the delayed activation of the Cap1 transcription factor in Candida albicans following combinatorial oxidative and cationic stress important for phagocytic potency. mBio 7(2):e00331-16. doi:10.1128/mBio.00331-16.
REFERENCES
- 1.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 2.Babior BM. 2004. NADPH oxidase. Curr Opin Immunol 16:42–47. doi: 10.1016/j.coi.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Cohen MS, Isturiz RE, Malech HL, Root RK, Wilfert CM, Gutman L, Buckley RH. 1981. Fungal infection in chronic granulomatous disease. The importance of the phagocyte in defense against fungi. Am J Med 71:59–66. doi: 10.1016/0002-9343(81)90259-X. [DOI] [PubMed] [Google Scholar]
- 4.Hampton MB, Kettle AJ, Winterbourn CC. 1998. Inside the neutrophil phagosome: oxidants, myeloperoxidase, and bacterial killing. Blood 92:3007–3017. [PubMed] [Google Scholar]
- 5.Reeves EP, Lu H, Jacobs HL, Messina CG, Bolsover S, Gabella G, Potma EO, Warley A, Roes J, Segal AW. 2002. Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature 416:291–297. doi: 10.1038/416291a. [DOI] [PubMed] [Google Scholar]
- 6.Segal AW. 2005. How neutrophils kill microbes. Annu Rev Immunol 23:197–223. doi: 10.1146/annurev.immunol.23.021704.115653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jamieson DJ, Stephen DW, Terrière EC. 1996. Analysis of the adaptive oxidative stress response of Candida albicans. FEMS Microbiol Lett 138:83–88. doi: 10.1111/j.1574-6968.1996.tb08139.x. [DOI] [PubMed] [Google Scholar]
- 8.Nikolaou E, Agrafioti I, Stumpf M, Quinn J, Stansfield I, Brown AJ. 2009. Phylogenetic diversity of stress signalling pathways in fungi. BMC Evol Biol 9:44. doi: 10.1186/1471-2148-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dantas AS, Day A, Ikeh M, Kos I, Achan B, Quinn J. 2015. Oxidative stress responses in the human fungal pathogen, Candida albicans. Biomolecules 5:142–165. doi: 10.3390/biom5010142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enjalbert B, MacCallum DM, Odds FC, Brown AJ. 2007. Niche-specific activation of the oxidative stress response by the pathogenic fungus Candida albicans. Infect Immun 75:2143–2151. doi: 10.1128/IAI.01680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enjalbert B, Smith DA, Cornell MJ, Alam I, Nicholls S, Brown AJ, Quinn J. 2006. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol Biol Cell 17:1018–1032. doi: 10.1091/mbc.E05-06-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fradin C, De Groot P, MacCallum D, Schaller M, Klis F, Odds FC, Hube B. 2005. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol Microbiol 56:397–415. doi: 10.1111/j.1365-2958.2005.04557.x. [DOI] [PubMed] [Google Scholar]
- 13.Lorenz MC, Bender JA, Fink GR. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell 3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miramón P, Dunker C, Windecker H, Bohovych IM, Brown AJ, Kurzai O, Hube B. 2012. Cellular responses of Candida albicans to phagocytosis and the extracellular activities of neutrophils are critical to counteract carbohydrate starvation, oxidative and nitrosative stress. PLoS One 7:e52850. doi: 10.1371/journal.pone.0052850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kusch H, Engelmann S, Albrecht D, Morschhäuser J, Hecker M. 2007. Proteomic analysis of the oxidative stress response in Candida albicans. Proteomics 7:686–697. doi: 10.1002/pmic.200600575. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Cao YY, Jia XM, Cao YB, Gao PH, Fu XP, Ying K, Chen WS, Jiang YY. 2006. Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans. Free Radic Biol Med 40:1201–1209. doi: 10.1016/j.freeradbiomed.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 17.Moye-Rowley WS. 2003. Regulation of the transcriptional response to oxidative stress in fungi: similarities and differences. Eukaryot Cell 2:381–389. doi: 10.1128/EC.2.3.381-389.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toone WM, Morgan BA, Jones N. 2001. Redox control of AP-1-like factors in yeast and beyond. Oncogene 20:2336–2346. doi: 10.1038/sj.onc.1204384. [DOI] [PubMed] [Google Scholar]
- 19.Kuge S, Arita M, Murayama A, Maeta K, Izawa S, Inoue Y, Nomoto A. 2001. Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol Cell Biol 21:6139–6150. doi: 10.1128/MCB.21.18.6139-6150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood MJ, Storz G, Tjandra N. 2004. Structural basis for redox regulation of Yap1 transcription factor localization. Nature 430:917–921. doi: 10.1038/nature02790. [DOI] [PubMed] [Google Scholar]
- 21.Delaunay A, Pflieger D, Barrault MB, Vinh J, Toledano MB. 2002. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111:471–481. doi: 10.1016/S0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- 22.Znaidi S, Barker KS, Weber S, Alarco AM, Liu TT, Boucher G, Rogers PD, Raymond M. 2009. Identification of the Candida albicans Cap1p regulon. Eukaryot Cell 8:806–820. doi: 10.1128/EC.00002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alarco AM, Raymond M. 1999. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J Bacteriol 181:700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, De Micheli M, Coleman ST, Sanglard D, Moye-Rowley WS. 2000. Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p. Mol Microbiol 36:618–629. doi: 10.1046/j.1365-2958.2000.01877.x. [DOI] [PubMed] [Google Scholar]
- 25.Jain C, Pastor K, Gonzalez AY, Lorenz MC, Rao RP. 2013. The role of Candida albicans AP-1 protein against host derived ROS in vivo models of infection. Virulence 4:67–76. doi: 10.4161/viru.22700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patterson MJ, McKenzie CG, Smith DA, da Silva Dantas A, Sherston S, Veal EA, Morgan BA, MacCallum DM, Erwig LP, Quinn J. 2013. Ybp1 and Gpx3 signaling in Candida albicans govern hydrogen peroxide-induced oxidation of the Cap1 transcription factor and macrophage escape. Antioxid Redox Signal 19:2244–2260. doi: 10.1089/ars.2013.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaloriti D, Tillmann A, Cook E, Jacobsen M, You T, Lenardon M, Ames L, Barahona M, Chandrasekaran K, Coghill G, Goodman D, Gow NA, Grebogi C, Ho HL, Ingram P, McDonagh A, de Moura AP, Pang W, Puttnam M, Radmaneshfar E, Romano MC, Silk D, Stark J, Stumpf M, Thiel M, Thorne T, Usher J, Yin Z, Haynes K, Brown AJ. 2012. Combinatorial stresses kill pathogenic Candida species. Med Mycol 50:699–709. doi: 10.3109/13693786.2012.672770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaloriti D, Jacobsen M, Yin Z, Patterson M, Tillmann A, Smith DA, Cook E, You T, Grimm MJ, Bohovych I, Grebogi C, Segal BH, Gow NA, Haynes K, Quinn J, Brown AJ. 2014. Mechanisms underlying the exquisite sensitivity of Candida albicans to combinatorial cationic and oxidative stress that enhances the potent fungicidal activity of phagocytes. mBio 5:e01334-14. doi: 10.1128/mBio.01334-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okazaki S, Tachibana T, Naganuma A, Mano N, Kuge S. 2007. Multistep disulfide bond formation in Yap1 is required for sensing and transduction of H2O2 stress signal. Mol Cell 27:675–688. doi: 10.1016/j.molcel.2007.06.035. [DOI] [PubMed] [Google Scholar]
- 30.Delaunay A, Isnard AD, Toledano MB. 2000. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J 19:5157–5166. doi: 10.1093/emboj/19.19.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.da Silva Dantas A, Patterson MJ, Smith DA, MacCallum DM, Erwig LP, Morgan BA, Quinn J. 2010. Thioredoxin regulates multiple hydrogen peroxide-induced signaling pathways in Candida albicans. Mol Cell Biol 30:4550–4563. doi: 10.1128/MCB.00313-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urban C, Xiong X, Sohn K, Schröppel K, Brunner H, Rupp S. 2005. The moonlighting protein Tsa1p is implicated in oxidative stress response and in cell wall biogenesis in Candida albicans. Mol Microbiol 57:1318–1341. doi: 10.1111/j.1365-2958.2005.04771.x. [DOI] [PubMed] [Google Scholar]
- 33.Kuge S, Toda T, Iizuka N, Nomoto A. 1998. Crm1 (XpoI) dependent nuclear export of the budding yeast transcription factor yAP-1 is sensitive to oxidative stress. Genes Cells 3:521–532. doi: 10.1046/j.1365-2443.1998.00209.x. [DOI] [PubMed] [Google Scholar]
- 34.Phillips AJ, Sudbery I, Ramsdale M. 2003. Apoptosis induced by environmental stresses and amphotericin B in Candida albicans. Proc Natl Acad Sci U S A 100:14327–14332. doi: 10.1073/pnas.2332326100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood MJ, Andrade EC, Storz G. 2003. The redox domain of the Yap1p transcription factor contains two disulfide bonds. Biochemistry 42:11982–11991. doi: 10.1021/bi035003d. [DOI] [PubMed] [Google Scholar]
- 36.Gulshan K, Rovinsky SA, Coleman ST, Moye-Rowley WS. 2005. Oxidant-specific folding of Yap1p regulates both transcriptional activation and nuclear localization. J Biol Chem 280:40524–40533. doi: 10.1074/jbc.M504716200. [DOI] [PubMed] [Google Scholar]
- 37.Shenton D, Smirnova JB, Selley JN, Carroll K, Hubbard SJ, Pavitt GD, Ashe MP, Grant CM. 2006. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J Biol Chem 281:29011–29021. doi: 10.1074/jbc.M601545200. [DOI] [PubMed] [Google Scholar]
- 38.Heidemann M, Hintermair C, Voß K, Eick D. 2013. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta 1829:55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 39.Alonso-Monge R, Navarro-García F, Molero G, Diez-Orejas R, Gustin M, Pla J, Sánchez M, Nombela C. 1999. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J Bacteriol 181:3058–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferrigno P, Posas F, Koepp D, Saito H, Silver PA. 1998. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J 17:5606–5614. doi: 10.1093/emboj/17.19.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Homann OR, Dea J, Noble SM, Johnson AD. 2009. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet 5:e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruiz A, Arino J. 2007. Function and regulation of the Saccharomyces cerevisiae ENA sodium ATPase system. Eukaryot Cell 6:2175–2183. doi: 10.1128/EC.00337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miramón P, Kasper L, Hube B. 2013. Thriving within the host: Candida spp. interactions with phagocytic cells. Med Microbiol Immunol 202:183–195. doi: 10.1007/s00430-013-0288-z. [DOI] [PubMed] [Google Scholar]
- 44.Sherman F. 1991. Getting started with yeast. Methods Enzymol 194:3–21. doi: 10.1016/S0076-6879(02)50954-X. [DOI] [PubMed] [Google Scholar]
- 45.Noble SM, Johnson AD. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell 4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barelle CJ, Manson CL, MacCallum DM, Odds FC, Gow NA, Brown AJ. 2004. GFP as a quantitative reporter of gene regulation in Candida albicans. Yeast 21:333–340. doi: 10.1002/yea.1099. [DOI] [PubMed] [Google Scholar]
- 47.Smith DA, Nicholls S, Morgan BA, Brown AJ, Quinn J. 2004. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol Biol Cell 15:4179–4190. doi: 10.1091/mbc.E04-03-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu TT, Znaidi S, Barker KS, Xu L, Homayouni R, Saidane S, Morschhäuser J, Nantel A, Raymond M, Rogers PD. 2007. Genome-wide expression and location analyses of the Candida albicans Tac1p regulon. Eukaryot Cell 6:2122–2138. doi: 10.1128/EC.00327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Znaidi S, Weber S, Al-Abdin OZ, Bomme P, Saidane S, Drouin S, Lemieux S, De Deken X, Robert F, Raymond M. 2008. Genomewide location analysis of Candida albicans Upc2p, a regulator of sterol metabolism and azole drug resistance. Eukaryot Cell 7:836–847. doi: 10.1128/EC.00070-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cabral V, Znaidi S, Walker LA, Martin-Yken H, Dague E, Legrand M, Lee K, Chauvel M, Firon A, Rossignol T, Richard ML, Munro CA, Bachellier-Bassi S, d’Enfert C. 2014. Targeted changes of the cell wall proteome influence Candida albicans ability to form single- and multi-strain biofilms. PLOS Pathog 10:e1004542. doi: 10.1371/journal.ppat.1004542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantification of Cap1 levels pre- and post-stress treatment. (A) Cap1 protein levels were analyzed by SDS-PAGE and Western blotting of native extracts prepared from cells expressing Cap1-HM before (ns) and following exposure to 5 mM H2O2, 1 M NaCl, or combinations of these stresses for 10 min. Blots were stripped and reprobed with an anti-Hog1 antibody as a loading control. (B) Quantification of Cap1 levels. Quantitative densitometric analysis of Western blots from five biological replicates was conducted to determine the relative levels of Cap1 following the stress treatments described above, compared to nonstress (ns) levels. Mean values (±SEM) are shown, and ANOVA was used to determine statistically significant differences in Cap1 levels. No significant differences were observed (P > 0.05). Download
Cap1 is differentially oxidized following combinatorial stress. (A) Cap1 mobility was monitored by nonreducing SDS-PAGE and Western blotting of proteins prepared from cells expressing Cap1-HM exposed to 5 mM H2O2, 1 M NaCl, or combinations of these stresses for 10 min. Disulfide bonds are indicated by the retarded mobility of Cap1 due to AMS binding to DTT-resolved disulfides (compare NEM-DTT- with NEM-DTT-AMS-treated lanes). (B) Cap1 mobility was monitored by nonreducing SDS-PAGE and Western blotting of proteins prepared from unstressed cells (ns) expressing Cap1-HM. Protein extracts were obtained under acid lysis conditions and incubated or not with N-ethylmaleimide, which blocks free thiol groups, thus preventing oxidation. Download
The NaCl-enhanced interaction between Cap1 and Crm1 is Hog1 independent and cationic stress specific. (A) Biological replicate of the pulldown shown in Fig. 7B. (B) Hog1 is dispensable for the salt-induced Cap1-Crm1 interaction. Extracts were prepared from hog1Δ cells (JC45), and hog1Δ cells expressing 2Myc- and 6His tagged Crm1 (JC1940) before and following exposure to 5 mM H2O2, 1 M NaCl, or combinations of these stresses for 10 min. Coprecipitation of Cap1 was detected as described in the legend to Fig. 7B. The asterisk designates a nonspecific band as seen in cap1Δ cells. (C) Sorbitol does not stimulate Cap1 binding to Crm1. Extracts were prepared from wild-type cells expressing 2Myc- and 6His tagged Crm1 (Crm1-MH [JC1925]) before and following exposure to 1 M NaCl for 10 min or 2 M sorbitol for the indicated times. Coprecipitation of Cap1 was detected as described in the legend to Fig. 7B. Download
RNA Pol II phosphorylation is maintained following high-H2O2 exposure and combinatorial stress. Lysates from cells expressing 2Myc- and 6His-tagged Cap1 (Cap1-MH [JC948]), before (ns) and after the indicated stress treatments were analyzed by Western blotting using antibodies that detect RNA Pol II CTD phosphorylation at serine 2 or serine 5 of the repeat sequence YSPTSPS. Total levels of RNA Pol II were determined using an antibody that recognizes the CTD repeat sequence. As a control, a time zero sample was prepared that was treated with lambda phosphatase prior to loading (+pptase). The positions of nonphosphorylated (RNA Pol II) and phosphorylated (RNA-Pol II S2-P, RNA-Pol II S5-P) Pol II are indicated and are consistent with the predicted molecular mass of C. albicans Pol II (192 kDa). The RNA Pol II blot was stripped and reprobed with anti-Cap1 antibodies. This shows that although Cap1 phosphorylation (a marker of active Cap1) is significantly delayed following high-H2O2 stress and combinatorial stress, RNA Pol II phosphorylation is not reduced following such treatments. This indicates that the defect in Cap1-mediated gene expression is not due to a global inhibition of RNA Pol II activity. Download
Strains used in this study.
Oligonucleotides used for qPCR.