Summary
Histopathological evaluation of melanocytic tumours usually allows reliable distinction of benign melanocytic naevi from melanoma. More difficult is the histopathological classification of Spitz tumours, a heterogeneous group of tumours composed of large epithelioid or spindle-shaped melanocytes. Spitz tumours are biologically distinct from conventional melanocytic naevi and melanoma, as exemplified by their distinct patterns of genetic aberrations. Whereas conventional naevi and melanoma often harbour BRAF mutations, NRAS mutations, or inactivation of NF1, Spitz tumours show HRAS mutations, inactivation of BAP1 (often combined with BRAF mutations), or genomic rearrangements involving the kinases ALK, ROS1, NTRK1, BRAF, RET, and MET. In Spitz naevi, which lack significant histological atypia, all of these mitogenic driver aberrations trigger rapid cell proliferation, but after an initial growth phase, various tumour suppressive mechanisms stably block further growth. In some tumours, additional genomic aberrations may abrogate various tumour suppressive mechanisms, such as cell-cycle arrest, telomere shortening, or DNA damage response. The melanocytes then start to grow in a less organised fashion, may spread to regional lymph nodes, and are termed atypical Spitz tumours. Upon acquisition of even more aberrations, which often activate additional oncogenic pathways or reduce and alter cell differentiation, the neoplastic cells become entirely malignant and may colonise and take over distant organs (spitzoid melanoma). The sequential acquisition of genomic aberrations suggests that Spitz tumours represent a continuous biological spectrum, rather than a dichotomy of benign versus malignant, and that tumours with ambiguous histological features (atypical Spitz tumours) might be best classified as low-grade melanocytic tumours. The number of genetic aberrations usually correlates with the degree of histological atypia and explains why existing ancillary genetic techniques, such as array comparative genomic hybridisation (CGH) or fluorescence in situ hybridisation (FISH), are capable of accurately classifying histologically benign and malignant Spitz tumours, but are not very helpful in the diagnosis of ambiguous melanocytic lesions. Nevertheless, we expect that progress in our understanding of tumour genomics and progression will refine the classification of melanocytic tumours in the near future. By integrating clinical, pathological, and genetic criteria, distinct tumour subsets will be defined within the heterogeneous group of Spitz tumours, which will eventually lead to improvements in diagnosis, prognosis and therapy.
Keywords: Biomarkers, BAP1, BRAF, classification, diagnosis, genetics, genomics, melanocytic tumours, melanoma, molecular biology, pathology, precision oncology, RAS, Spitz tumours, spitzoid neoplasms, targeted therapy
INTRODUCTION
Melanocytic neoplasms include several tumour types that are characterised by distinct clinical features, histopathological appearances, genetic aberrations, and clinical behaviour.1 In most melanocytic tumours, accurate pathological distinction between benign (melanocytic naevus) and malignant (melanoma) is possible based on histological criteria. However, there are diagnostically challenging melanocytic neoplasms with conflicting morphological criteria, in which it is difficult to predict clinical behaviour with certainty. This difficulty leads to under-diagnosis as naevus, to over-diagnosis as melanoma, or to a diagnosis of ‘melanocytic tumour of uncertain malignant potential’ or ‘borderline melanocytic tumour’.
Spitzoid melanocytic neoplasms (hereafter designated ‘Spitz tumours’) represent an uncommon group of melanocytic skin lesions that constitute diagnostic problems for dermatopathologists on a regular basis.2 These tumours were first described by Sophie Spitz as ‘melanomas of childhood’, because they occurred predominantly in children and adolescents.3 The lesions were composed of large epithelioid and/or spindled-shaped melanocytes that contained large nuclei with vesicular chromatin and prominent nucleoli. After it became clear that these tumours may also arise later in life and behave in a benign fashion, they were re-named ‘Spitz naevi’ to indicate their benign nature. Melanocytic tumours with spitzoid features and marked histological atypia that show often an aggressive clinical course similar to conventional melanomas were termed ‘spitzoid melanomas’.
The term ‘atypical Spitz tumours’ (ASTs) refers to melanocytic tumours that exhibit morphological characteristics of Spitz naevi, as well as some features associated with malignancy, but to an insufficient degree as to classify them as ‘spitzoid melanomas’. ASTs have the ability to disseminate, but their spread is often limited to regional lymph nodes and has little impact on patient survival.4–6 Therefore, the histological features of ASTs mirror their clinical behaviour, which is intermediate between benign (no metastasis) and malignant (aggressive clinical behaviour with widespread metastasis) melanocytic tumours, and argues that these lesions might be best classified as low-grade melanocytic tumours.
Pathological classification of Spitz tumours as ‘Spitz naevus’, ‘AST’ or ‘spitzoid melanoma’ and predicting their clinical behaviour can be challenging, even for experts.2,7,8 This diagnostic uncertainty has stimulated numerous efforts to characterise the underlying genetic and epigenetic aberrations of Spitz tumours with the goal of finding new biomarkers and better explaining their biology. In this review, we summarise the current knowledge of the genetic landscape of Spitz tumours and describe how genetic aberrations influence their morphological appearance and their clinical behaviour. We also discuss the usefulness and the limitations of ancillary genetic methods in the diagnostic work-up of Spitz tumours.
INTEGRATED CLASSIFICATION OF MELANOCYTIC TUMOURS
The main goals of tumour classification systems are: (1) to distinguish tumours that are malignant and behave aggressively from those that are benign and pose no threat to patients; and (2) to establish relatively distinct disease entities or subsets that can be used for personalised therapeutic decisions. Since the development of drug therapies targeting mitogenic driver aberrations (such as BRAF mutations) has improved the treatment of patients with metastatic melanoma,9 the classification of melanocytic tumours has recently been integrated with genomic data. Thus, melanocytic tumours are currently classified according to their clinical and histological appearance, their biological behaviour, and their mitogenic driver aberrations.10
Several genetic aberrations are associated with specific clinical and histopathological subtypes of melanocytic tumours. For example, blue naevi and Spitz tumours show distinctive clinical and histological appearances, and have very different spectra of genetic aberrations to common acquired naevi and cutaneous melanoma.1,11 While morphological evaluation often provides clues about the probability of genetic aberrations in a given tumour, genomic or immunohistochemical methods are necessary for confirmation. No histological feature is entirely specific, because other factors, such as additional genetic aberrations or the tumour microenvironment can mask or distort the morphological features associated with specific genetic aberrations.
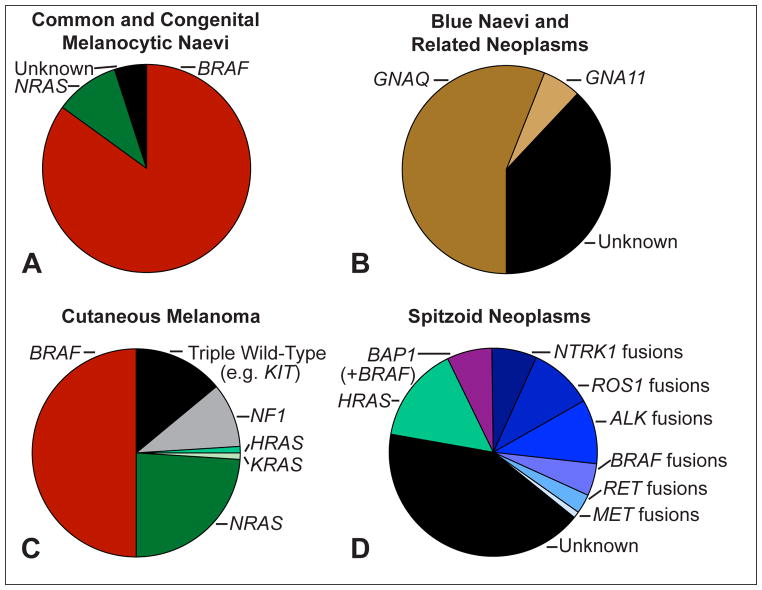
Common melanocytic naevi arise predominantly in the first three decades of life and normally appear as uniformly brown-pigmented maculae with a diameter of less than 6 mm on sun-exposed areas of the skin. The vast majority (>80%) show activating hotspot mutations leading to an amino acid exchange at codon 600 of BRAF (BRAFV600E/K).12 Congenital naevi arise in utero or post-natally, tend to be significantly larger than common acquired naevi, and harbour activating NRAS hotspot mutations (~75%), most commonly affecting codon 61 (Fig. 1A).13
Fig. 1.
Frequent genomic aberrations in cutaneous melanocytic tumours. (A) Common acquired naevi show BRAF hotspot mutations in the vast majority of cases. Congenital naevi frequently harbour NRAS hotspot mutations. (B) Blue naevi and related melanocytic neoplasms share activating mutations of GNAQ and GNA11. While GNAQ mutations dominate in cutaneous proliferations, GNA11 and GNAQ mutations account each for ~40% of uveal melanomas. (C) Cutaneous melanomas show BRAF hotspot mutations in ~50%, NRAS mutations in ~25% and rarely KRAS and HRAS mutations, and NF1 loss in ~10% of cases. The ‘triple wild-type melanoma’ subtype is a heterogeneous subgroup of melanoma with infrequent driver mutations such as KIT, CTNNB1, or genomic rearrangements. (D) Compared to cutaneous melanoma and to common and blue naevi, Spitz tumours show different chromosomal aberrations: translocations involving the kinases ALK, ROS1, NTRK1, BRAF, RET, MET are observed in up to 50% of cases, but are rarely observed in other subtypes of melanocytic tumours. HRAS mutations are often associated with a desmoplastic histological phenotype (desmoplastic Spitz naevus). Aberrations of BAP1 occur frequently with activating BRAF mutations and are associated with an epithelioid morphology.
Blue naevi and related skin neoplasms are heterogeneous melanocytic proliferations that vary in size from a few millimetres in acquired lesions to several centimetres in congenital lesions (e.g., Mongolian spot). Most tumours are histologically characterised by dendritic, spindled, or epithelioid melanocytes without significant epidermal involvement. Most tumours show activating mutations of GNAQ or GNA11, commonly affecting codon 209 (Fig. 1B).14–16
The Cancer Genome Atlas proposed that cutaneous melanoma be classified into four subgroups.10 The largest subgroup (~50%) of melanoma is characterised by BRAFV600E mutations (BRAF subtype).10 BRAF mutations are most frequent in melanoma arising on intermittently sun-exposed skin.17 Activating mutations in RAS genes account for approximately 25% of melanoma (RAS subtype), and subsume cases with NRAS mutations (~24%), as well as HRAS and KRAS hot-spot mutations (<1%).10 The NF1 subtype shows inactivation of NF1, which encodes a negative regulator of RAS.10 NF1 aberrations are found in approximately 10% of melanoma and more than half of the aberrations lead to complete loss-of-function due to nonsense, frame-shift, or splice-site mutations.18 NF1 mutations are associated with co-mutations affecting genes that mildly activate the MAPK/ERK pathway and occur more frequently in desmoplastic melanoma and in melanoma of chronically sun-exposed skin.19,20 Melanomas lacking BRAF, N/H/KRAS, or NF1 mutations comprise the heterogeneous group of ‘triple wild-type subtype’, which includes tumours with KIT mutations (more frequently found in acral and mucosal melanoma21), GNAQ/GNA11 mutations (well established drivers in uveal melanoma14,15), or genomic rearrangements involving BRAF or RAF1 (Fig. 1B).
GENETIC ABERRATIONS IN SPITZ TUMOURS
Spitz tumours are a heterogeneous group of melanocytic neoplasms that commonly arise under the age of 20, but may occur also in older individuals. Spitz tumours may be solitary or have an agminated or eruptive disseminated distribution,22 have an average diameter of ~10mm, and are usually well-circumscribed, dome-shaped papules with a homogenous colour ranging from reddish to dark brown. Spitz tumours show genomic aberrations that are rarely observed in the tumours described above, or in other melanocytic neoplasms (Fig. 1D). BAP1 inactivation occurs in ~5% of Spitz tumours and is associated with an epithelioid phenotype. Spitz tumours with BAP1 loss are associated with a hereditary tumour predisposition syndrome.23 HRAS mutations occur in ~15% of Spitz tumours and are often associated with desmoplastic histological phenotype.24 Genomic rearrangements (translocations, kinase fusions) of various receptor tyrosine kinases, including ALK, ROS1, NTRK1, RET, and MET, or the serine-threonine kinase BRAF are observed in up to 50% of Spitz tumours.25,26
BAP1 loss
BAP1 germline mutations
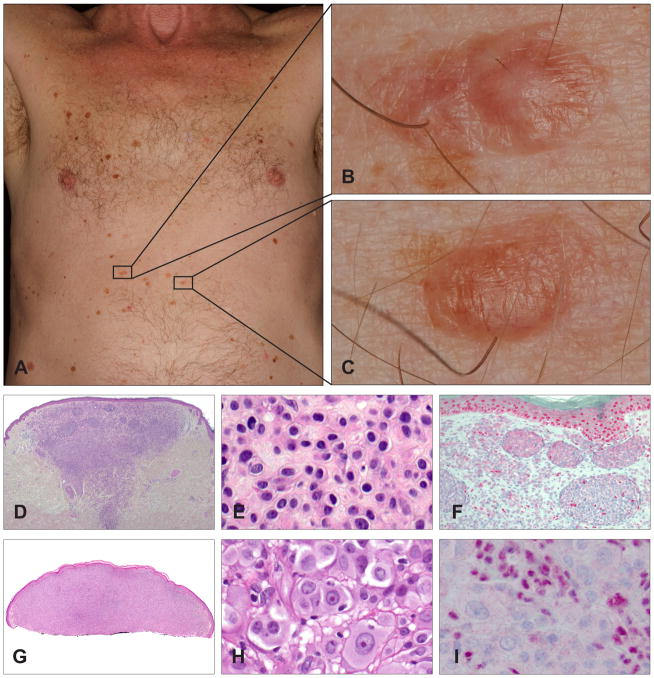
Germline mutations of the BAP1 gene were first described in two unrelated families in which several members developed multiple cutaneous epithelioid melanocytic tumours in an autosomal dominant pattern, and less commonly, cutaneous and uveal melanomas.23 The epithelioid melanocytic tumours are located predominantly on the sun-exposed skin and the number of lesions per affected individual ranges from a few to more than 50 (Fig. 2A). The characteristic skin lesions are skin-coloured to reddish-brown, 5–10 mm large, dome-shaped to pedunculated papules (Fig. 2B,C).23,27
Fig. 2.
Clinical and histological appearance of melanocytic tumours with BAP1 loss. (A) A 65-year-old man with a BAP1 germline mutation. Numerous flat and homogeneously brown-pigmented melanocytic skin lesions dominate the clinical picture. (B,C) Melanocytic tumours with BAP1 loss usually present as inconspicuous, small, skin-coloured to slightly pink plaques or dome-shaped papules. (D) Wedge-shaped, symmetrical tumour with melanocytes in nests and sheets. (E) The neoplastic cells present with well-defined cytoplasmic borders, moderate amounts of amphophilic cytoplasm, and slightly enlarged, round to oval nuclei with condensed chromatin without atypia. (F) BAP1 immunohistochemistry is negative in the naevoid melanocytes, but positive in nuclei of epidermal keratinocytes and scattered lymphocytes. (G) Shave biopsy of a predominantly dermal melanocytic tumour. (H) Large epithelioid and atypical melanocytes (same magnification as E; note the difference in size of the neoplastic cells). The neoplastic cells show well-defined cytoplasmic borders and abundant amphophilic cytoplasm. The large nuclei show vesicular chromatin with prominent nucleoli, and vary in size and shape. (I) The nuclei of the large, epithelioid melanocytes are negative for BAP1 immunohistochemistry, whereas the admixed lymphocytes are positive. These two cases illustrate the two extremes in the biological spectrum of melanocytic neoplasms with BAP1 inactivation, ranging from clearly benign lesions with only very slightly atypical naevoid cells (D–F) to borderline malignant tumours with significant atypia (G–I).
Histologically, the tumours are predominantly intradermal, with occasional involvement of the junctional epidermis. They are composed of cells with varying degrees of atypia ranging from naevus-like melanocytes with minimal atypia (Fig. 2D–F) to very large, epithelioid cells with large amounts of amphophilic cytoplasm and well-defined cytoplasmic borders, and pleomorphic vesicular nuclei with prominent nucleoli (Fig. 2G–I). Some tumours display marked atypical features (nuclear pleomorphism, high cellularity, increased mitotic activity), and cannot be confidently classified as benign or malignant on histological grounds. These findings suggest a histological spectrum ranging from clearly benign (Fig. 2D–F) to potentially malignant tumours (Fig. 2G–I).23 The cytological characteristics resemble those seen in epithelioid cells in Spitz tumours, but lack histological features of Spitz naevi, such as epidermal hyperplasia, hypergranulosis, clefting around junctional nests and Kamino bodies. Some tumours consist of an admixture of small naevus cells, which are seen in common acquired naevi, and larger epithelioid cells, and are thus classified as combined melanocytic tumours.28
The vast majority of familial epithelioid Spitz tumours show bi-allelic loss of BAP1. One BAP1 allele is inactivated through the BAP1 germline mutation and the other allele is somatically inactivated either by chromosomal deletion involving the wild-type BAP1 locus at 3p21, uniparental disomy of chromosome 3 with the mutated BAP1 gene, or an additional inactivating mutation in the wild-type BAP1. The biallelic BAP1 gene inactivation correlates with loss of nuclear BAP1 immunohistochemical expression (Fig. 2F,I). In addition to BAP1 loss, the epithelioid cells harbour BRAFV600E mutations in the vast majority of cases.23
These clinically and histologically distinctive melanocytic skin lesions are useful markers for a hereditary tumour syndrome (tumour predisposition syndrome; OMIM #614327), which predisposes to uveal and cutaneous melanoma,23,29 mesothelioma,27,30 renal cell cancer,31 and possibly also to other cancer types32 such as cholangiocarcinoma33 or basal cell carcinoma.34–36 The association of multiple epithelioid melanocytic skin lesions with BAP1 loss in a patient with a BAP1 germline mutation is comparable to the multiple sebaceous tumours associated with Muir–Torre syndrome, or to the multiple hyperpigmented macules on the lips and oral mucosa seen in Peutz–Jeghers syndrome. Consequently, patients with melanocytic tumours with a prominent epithelioid cell component as described above should be screened for BAP1 and BRAF status by immunohistochemistry or genotyping.37 If multiple tumours in one patient show loss of BAP1 expression, genetic counselling and testing for germline BAP1 mutations should be considered. Patients with BAP1 germline mutations should be screened for cancers on a regular basis.27,38
Sporadic tumours with BAP1 loss
A subset of sporadic epithelioid melanocytic tumours also shows loss of BAP1, along with BRAF mutations.39 The sporadic BAP1 negative tumours have the same histological appearance as those seen in patients with germline BAP1 mutations. The neoplastic melanocytes show loss of nuclear BAP1 expression by immunohistochemistry, which is a good surrogate for biallelic BAP1 inactivation. In combined melanocytic tumours, all melanocytes harbour BRAFV600E mutations, but only the epithelioid cells show a loss of BAP1 expression, demonstrating that these tumours represent a progression from common acquired naevi.23,40
Tumours in the setting of germline BAP1 mutations do not differ pathologically from those with somatic BAP1 mutations.41 However, the morphological features of BAP1 negative and BAP1 positive tumours overlap and without the aid of immunohistochemistry or ancillary genetic investigations, it is not possible to predict the underlying genetic basis. In addition, a recent study reported that BAP1 deficiency can also be detected in melanocytic tumours that have histological characteristics different from the majority of BAP1 inactivated epithelioid tumours.41,42
The nomenclature of this genetically distinct subset of melanocytic tumours remains in flux. One group designated them as ‘melanocytic BAP1-mutated atypical intradermal tumours’ (MBAITs).43 However, this term is inaccurate because the tumours are not exclusively intradermal and may exhibit junctional involvement,39,41 and it also suggests that these tumours are pathologically distinguishable from other spitzoid tumours, which is often not possible without BAP1 immunohistochemistry. Another group suggested the term ‘naevoid melanoma-like melanocytic proliferations’ (NEMMPs),44 which equates these tumours with naevoid melanomas and implies a high level of biological aggression. However, in our experience these lesions progress infrequently to melanoma27 and very rarely metastasise. The term ‘BAPoma’45 is also problematic because it implies that these lesions have activating BAP1 mutations, similar to lesions harbouring BRAF mutations (BRAFoma) or ALK fusions (ALKoma). As these lesions lost BAP1, the term ‘BAP1-inactivated spitzoid naevus’ might be more appropriate.46 Hopefully, the nomenclature will be clarified in the next edition of the World Health Organization Classification of Skin Tumours.
HRAS mutation and 11p gains
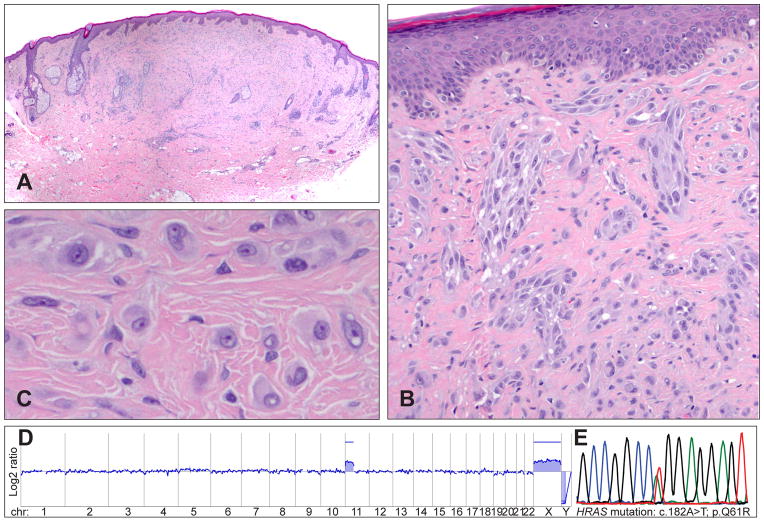
Increased copy numbers of 11p (which contains the HRAS locus) were first described in ~17% of Spitz naevi.48 The majority of tumours with 11p gains have activating HRAS mutations, whereas they are quite uncommon in tumours without 11p gains.24 Tumours with 11p gains show characteristic morphological features, including marked desmoplasia and stromal sclerosis (desmoplastic Spitz naevi, Fig. 3A–E). These tumours are predominantly intradermal and often show an infiltrating growth pattern and a low proliferation rate.24,49 Clinically, most desmoplastic Spitz naevi show a favourable prognosis, but some of them might also progress to melanoma.49,50
Fig. 3.
Desmoplastic Spitz tumours frequently show HRAS mutations and gains of the short arm of chromosome 11. (A) Quite symmetrical, intradermal tumour with low cellularity from the retro-auricular region of a 43-year-old man. (B) Desmoplasia with single cells and clusters of spindle-shaped and epithelioid melanocytes between collagen bundles. (C) Large, epithelioid melanocytes between collagen bundles with vesicular chromatin and prominent nucleoli. (D) The array CGH profile indicates a gain of the short arm of chromosome 11, which harbours the HRAS gene. (E) The electropherogram shows a mutation affecting codon 61 of the HRAS gene (HRASQ61R).
HRAS mutations usually occur in exon 3, causing replacement of the glutamine at amino acid position 61 with a lysine (HRASQ61K) or an arginine (HRASQ61R). Mutations affecting exon 2 (codon 12 and 13) are less frequent. The amino acid exchange leads to a constitutively active protein stimulating cell proliferation via the MAP/ERK and the PI3K/AKT/mTOR pathway.
HRAS belongs with KRAS and NRAS to the family of RAS genes. HRAS mutations are found in less than 1% of melanomas 10 and it is not known why HRAS mutations occur almost exclusively in Spitz tumours. One explanation could be that HRAS mutations activate the PI3K/AKT/mTOR pathway more vigorously than NRAS mutations.51 The PI3K/AKT/mTOR pathway is central to regulating mammalian cell size and cell survival.52 Stronger activation of the PI3K/AKT/mTOR pathway by HRAS mutations could explain why melanocytes are larger in Spitz tumours with HRAS mutations compared to the small, naevoid cells in common acquired naevi with BRAF mutations. The different activation of signalling pathways may also explain why many Spitz tumours appear amelanotic.53
Kinase fusions
Translocations of the receptor tyrosine kinases ALK, ROS1, NTRK1, RET, or MET, and the serine-threonine kinase BRAF are observed in up to 50% of Spitz tumours.25,26 Patients with fusion positive Spitz tumours are younger than patients whose tumours lack translocations,25 a feature also shared by patients with kinase fusion-driven lung cancers,54 thyroid cancers,55 and astrocytomas.56
The genomic rearrangements in Spitz tumours fuse the intact kinase domains to a wide range of partner genes, which lead to high expression of chimeric fusion proteins. Most of the fusion partners have coiled-coil domains, suggesting that these domains support ligand independent dimerisation and auto-phosphorylation of the kinase domain.25 The phosphorylated kinases activate multiple oncogenic signalling pathways, including the MAPK/ERK, PI3K/AKT/mTOR, and JAK-STAT pathways, which induce cell proliferation, improve cell survival, and increase cell size.25 While BRAF mutations predominantly activate the MAPK/ERK pathway, the major activation of the PI3K/AKT/mTOR pathway, which is a key regulator of cell size, could explain why the melanocytes are larger in Spitz tumours with kinase fusions than in common acquired naevi with BRAF mutations.
Kinase fusions occur in a mutually exclusive pattern and are not detected in tumours with HRAS mutations or BAP1 inactivation. Although the majority of Spitz tumours behave in an indolent fashion, some Spitz tumours (spitzoid melanomas) metastasise and require systemic therapy. Lung cancers, lymphomas, and sarcomas with ALK or ROS1 fusions57,58 and thyroid cancers with RET fusions can be successfully treated using kinase inhibitors such as crizotinib, cabozantinib, and vandetanib. Similarly, metastatic spitzoid melanomas harbouring kinase fusions might be potentially treatable with kinase inhibitors.
ALK fusions
ALK rearrangements occur in up to 10% of Spitz tumours and were reported in patients aged between 5 months and 64 years.25,59 ALK fusions have been described in other tumours, including anaplastic large cell lymphoma,60 lung cancer,61 inflammatory myofibroblastic tumours62 and acral melanoma.63 ALK positive Spitz tumours are usually solitary, dome-shaped lesions and occur slightly more frequently on the extremities.59,64 The majority of tumours are amelanotic, but they can occasionally be heavily pigmented. The lesions are often clinically suspected to be irritated melanocytic naevi, virus induced lesions (verruca, molluscum contagiosum), or vascular lesions (angioma, pyogenic granuloma).59,64
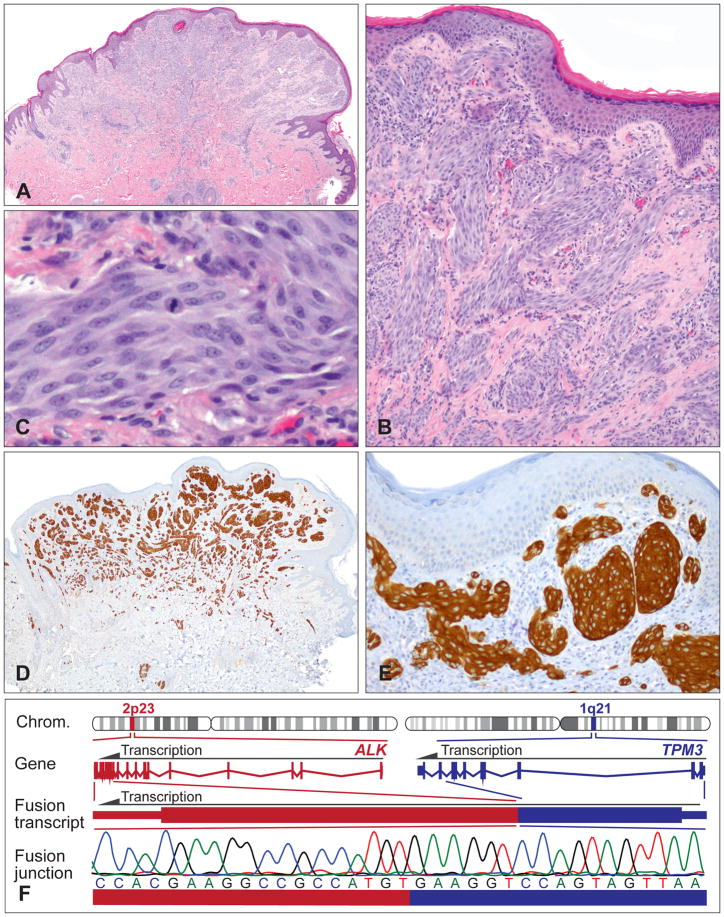
Histopathologically, the most characteristic features are plexiform, intersecting fascicles of predominantly fusiform melanocytes in the dermis (Fig. 4A).64 The cell nuclei usually show smooth contours and a slightly vesicular chromatin pattern without marked pleomorphism (Fig. 4B,C). However, some tumours are composed predominantly of large epithelioid cells with enlarged nuclei and nuclear pleomorphism, displaying a plexiform growth pattern only focally. In patients who undergo sentinel lymph node biopsy, nests of neoplastic melanocytes are occasionally found in the lymph node.64
Fig. 4.
Plexiform Spitz tumours frequently show ALK translocations. (A) Relatively symmetrical, exophytic, predominantly intradermal melanocytic tumour with focal epidermal hyperplasia from the buttock of a 14-year-old boy. (B) Plexiform growth pattern and intersecting fascicles of fusiform melanocytes. (C) Proliferation of cytologically fairly bland spindle and epithelioid melanocytes. Note mitosis in the centre. (D,E) The neoplastic melanocytes are positive for ALK immunohistochemistry, with staining of the cytoplasm. (F) Illustration of the TPM3-ALK kinase fusion. The ALK gene is located on chromosome 2p23; the TPM3 gene on chromosome 1q21. Because of the genomic rearrangement, the TPM3 exons 1–8 are fused with the ALK exons 20–29 containing the transforming tyrosine kinase domain. The in-frame fusion junction of the chimaeric transcript is confirmed by Sanger sequencing.
Spitz tumours with ALK translocations are strongly positive for ALK by immunohistochemistry (Fig. 4D,E). The plexiform morphology together with positive ALK immunohistochemistry enables the accurate identification of this morphologically and genetically distinct subset of Spitz tumours. ALK rearrangements can also be confirmed by fluorescence in situ hybridisation (FISH), reverse transcription polymerase chain reaction (RT-PCR), or next-generation sequencing (NGS; Fig. 4F). The recently described ALK isoform in melanoma, ALKATI,65 which originates from an alternative transcriptional initiation site within the ALK gene and consists primarily of the intracellular tyrosine kinase domain, has so far not been described in Spitz tumours.
The most prominent ALK fusion partners are TPM3 and DCTN1.25,59,64 ALK positive Spitz tumours analysed by FISH are negative for copy number changes of 6p, 6q, 9p, or 11q, and do not meet FISH criteria for melanoma.64,66 In line with these results, array comparative genomic hybridisation (CGH) often shows a balanced profile with no copy number aberrations, or only few chromosomal changes, including losses on chromosome 2 (usually between the ALK and the DCTN1 locus) or loss on chromosome 1p.59 Functional studies with murine melanocytes expressing ALK fusions showed activation of the MAPK/ERK and PI3K/AKT/MTOR pathways compared to controls.25 Crizotinib inhibited the ALK fusion induced activation of these oncogenic signalling pathways, which is in line with signalling data in lymphoma60 and lung cancer.61
ROS1 fusions
ROS1 fusions are seen in up to 10% of Spitz tumours, which occur predominantly on the extremities in patients aged between 1 and 59 years.25 ROS1 fusions have also been described in lung carcinoma,67 glioblastoma,68 and cholangiocarcinoma.69 The rearrangements fuse the intact tyrosine kinase coding sequence of ROS1 to a wide variety of fusion partners.
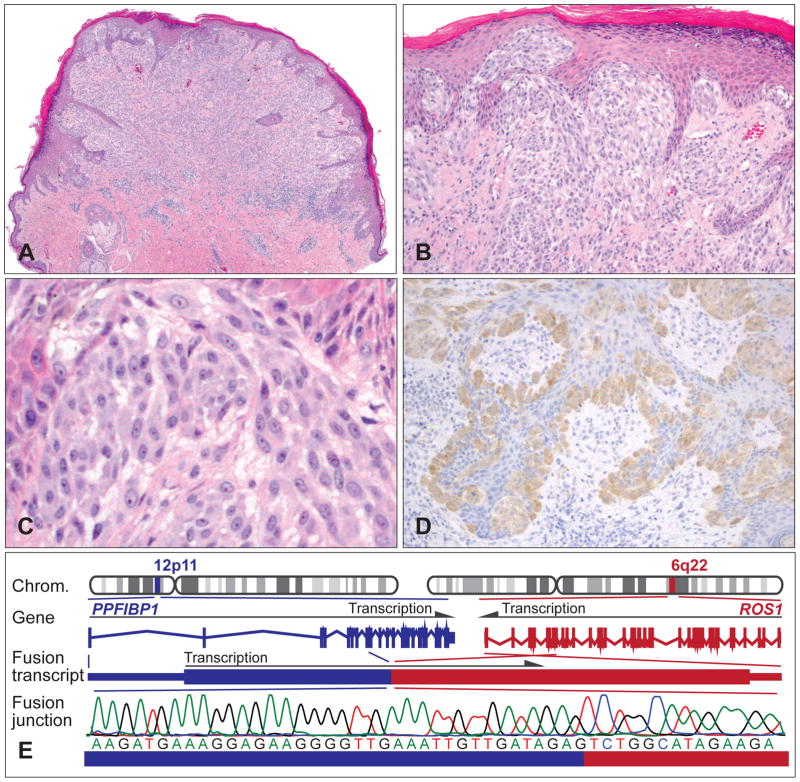
Spitz tumours with ROS1 fusions usually present as dome-shaped, well-circumscribed melanocytic compound proliferation (Fig. 5A,B). Irregular epidermal hyperplasia is frequently seen, along with occasional Kamino bodies. The lesions are composed of large spindled and epithelioid melanocytes with vesicular nuclei and variable atypia (Fig. 5C).25 However, as no specific cytological and histological features are associated with ROS1 fusions, the identification of tumours with ROS1 positive Spitz tumours is difficult. An antibody to detect the expression of the chimeric ROS1 protein by immunohistochemistry is available, but the staining is often quite weak (Fig. 5D). Positive ROS1 immunohistochemistry is seen exclusively in cases with ROS1 rearrangements (high specificity), but cases with genetically validated ROS1 kinase fusions can lack ROS1 immunoexpression (low sensitivity). Alternatively, ROS1 rearrangements can be detected by FISH, RT-PCR or NGS (Fig. 5E).
Fig. 5.
Spitz tumours with a ROS1 kinase fusion. (A) Exophytic, compound melanocytic tumour with irregular epidermal hyperplasia and permeative lymphocytic infiltrate from the lower left arm of a 19-year-old woman. (B) Epidermal and dermal nests of (C) epithelioid melanocytes with minor atypia. (D) The neoplastic melanocytes are positive for ROS1 immunohistochemistry. (E) Illustration of the PPFIBP1–ROS1 kinase fusion. The ROS1 gene is located on chromosome 6q22, and the PPFIBP1 gene on chromosome 12p11. Owing to genomic rearrangements, PWWP2A exons 1–8 are fused to ROS1 exons 35–43 containing the tyrosine kinase domain. Sanger sequencing over the junction confirms the chimaeric transcript.
Functional studies expressing ROS1 fusions in melanocytes revealed activation of the MAPK/ERK and PI3K/AKT/MTOR pathways,25 which is in line with data reported in various other cancer types with ROS1 fusions.58,70 Similar to ALK fusions, crizotinib inhibited the activation of the oncogenic signalling pathways, suggesting that inhibition of ROS1 might be a rational therapeutic option for metastatic tumours.25
NTRK1 fusions
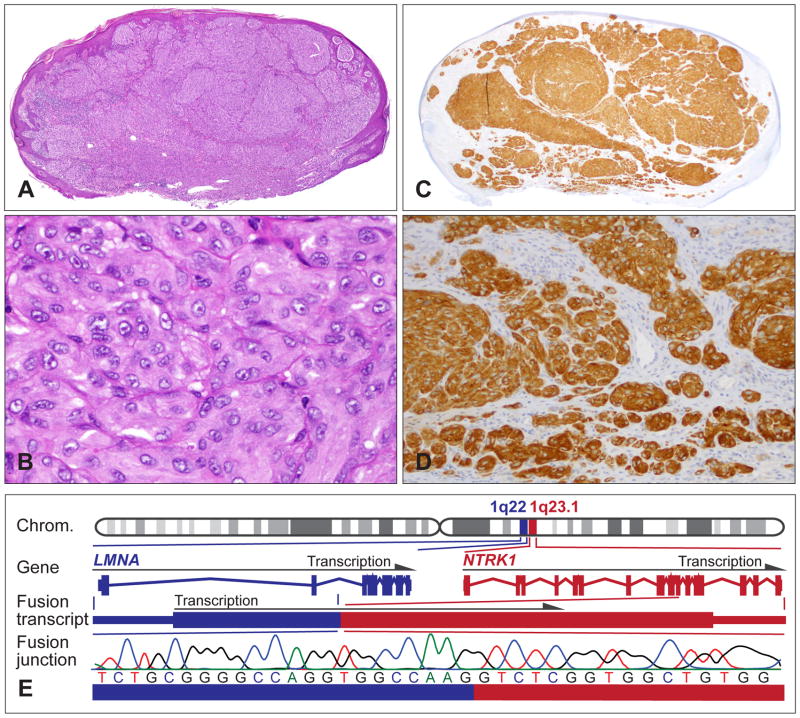
NTRK1 rearrangements occur in up to 10% of Spitz tumours, and are seen in patients aged between 2 and 73 years.25 NTRK1 fusions have been described at low frequency in lung carcinoma,71 papillary thyroid cancer,72 and paediatric glioma.73 Histopathologically, Spitz tumours with NTKR1 fusions show classical spitzoid features, but no specific morphological characteristics (Fig. 6A,B). Intersecting fusiform cellular growth, as seen in ALK positive tumours, is infrequent. Tumours with NTRK1 fusions show strong staining for NTRK1 by immunohistochemistry, which helps to identify cases with NTRK1 fusions (Fig. 6C,D). Although the antibody shows high specificity and sensitivity, some melanocytic lesions without NTRK1 fusions might show weak background staining due to low endogenous expression.
Fig. 6.
Spitz tumours with a NTRK1 kinase fusion. (A) Oval-shaped, compound melanocytic tumour with moderate epidermal hyperplasia from the upper arm of a 9-year-old girl. (B) Large epithelioid melanocytes with vesicular nuclei and prominent nucleoli, and moderate nuclear pleomorphism. (C,D) The neoplastic melanocytes are positive for NTRK1 immunohistochemistry and show cytoplasmic staining. (E) The LMNA–NTRK1 kinase fusion is caused by a 743 kb intrachromosomal deletion on chromosome 1q, joining the first two exons of LMNA with exon 11–17 of NTRK1. Sanger sequencing confirms the in-frame junction of the fusion transcript.
The LMNA-NTRK1 fusion is by far the most frequent NTRK1 fusion in Spitz tumours (Fig. 6E). Compared to the control cells, the expression of the LMNA-NTRK1 fusion in murine melanocytes showed increased phosphorylation of the chimaeric fusion protein and activation of the MAPK/ERK and PI3K/AKT/MTOR pathways.25 The oncogenic activity of NTRK1 is further supported by reports showing that autocrine neurotrophin signalling involving NTRK1 promotes proliferation and migration of melanocytic cell lines,74 and that constitutively active NTRK1 also activates oncogenic signalling pathways in other cell types.75 NTRK1 inhibitors suppress the oncogenic signalling.25
RET fusions
Genomic rearrangements of RET are seen in less than 5% of Spitz tumours and commonly involve the fusion partners KIF5B and GOLGA5.25 The KIF5B-RET fusion has been shown to drive lung cancer formation,54,76,77 and the GOLGA5-RET fusion was first described in papillary thyroid carcinomas occurring in children exposed to radioactive fallout from the Chernobyl nuclear accident.55 As RET positive Spitz tumours are quite rare and no RET antibody is reliable for immunohistochemistry, little data is available on their clinical and histopathological characteristics.
In mice, RET overexpression results in a generalised proliferation of melanocytes, naevi formation, and ultimately in melanomas.78 Murine melanocytes expressing RET fusions show activation of PLCγ-1 and of the MAPK/ERK and PI3K/AKT/MTOR pathways.25 RET inhibitors such as vandetanib or cabozantinib, which are both in clinical use for medullary thyroid cancer with RET fusions, suppress the oncogenic activity.25
MET fusions
Genomic rearrangements that fuse the MET kinase domain to various fusion partners were described in six Spitz tumours26 and are probably quite rare. Functional studies revealed that MET fusions constitutively activate the MAPK/ERK and PI3K/AKT/MTOR pathways. Kinase inhibitors such as cabozantinib block oncogenic MET signalling and may provide treatment options for patients with metastatic tumours.26
BRAF fusions and amplification
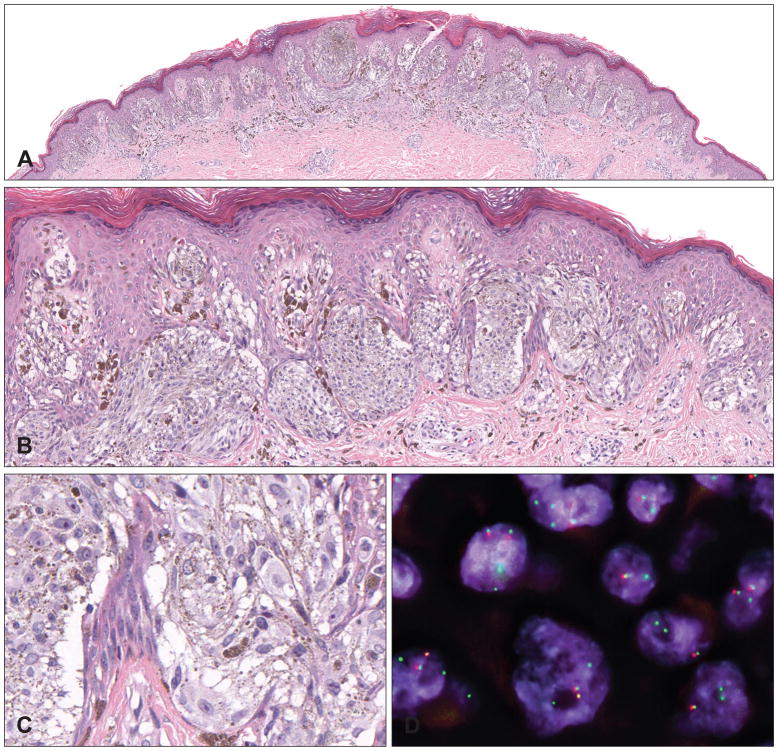
BRAF fusions are seen in approximately 5% of Spitz tumours and are not associated with specific morphological characteristics (Fig. 7A–C).25 BRAF rearrangements result in a loss of the auto-inhibitory, N-terminal RAS-binding domain and have been identified in pilocytic astrocytomas,56 papillary thyroid carcinoma,79 and rarely also in melanocytic tumours.10,80,81 As BRAF is constitutively expressed in melanocytic tumours, immunohistochemistry for BRAF fusions is not specific and the kinase fusions have to be detected with genomic methods such as FISH or NGS (Fig. 7D). In addition to BRAF fusions and BRAF mutations, amplification of wild-type BRAF has also been described in a small number of Spitz tumours.25
Fig. 7.
Pigmented spindle cell naevus with a BRAF fusion. (A) Symmetrical and well-circumscribed compound proliferation with slight epidermal hyperplasia from the lower right leg of a 17-year-old woman. (B) Elongated melanocytic nests with numerous melanophages in the papillary dermis. (C) Spindled and epithelioid pigmented melanocytes. (D) Interphase FISH with break-apart probes flanking the BRAF locus confirms the BRAF rearrangement (individual green and red signals). The paired red/green signals indicate the wild-type BRAF alleles.
MULTI-STEP TUMORIGENESIS AND TUMOUR PROGRESSION
Neoplastic cells evolve from normal cells through the sequential acquisition of randomly striking somatic genetic and epigenetic aberrations.82–84 Tumour evolution and progression are evolutionary processes that select for specific abilities and advantages. The selected genomic aberrations predominantly affect genes controlling cell proliferation, survival, differentiation, and additional traits associated with malignant cells. Multiple complex and interconnected barriers exist to prevent uncontrolled cell proliferations. These cell-autonomous defence mechanisms (such as apoptosis or senescence) efficiently hinder uncontrolled cell growth and cancer.85
In addition to cell-autonomous mechanisms, the immune system has the ability to monitor tissue homeostasis and to destroy neoplastic cells. Under normal conditions, the immune system is tamed by inhibitory immune checkpoints to maintain self-tolerance and to limit collateral tissue damage during viral and microbial immune responses. However, interrupting these inhibitory immune checkpoints with drugs, such as anti-CTLA-4, anti-PD-1, anti-PD-L1, unshackles the immune system, which may lead to durable cancer regression.86
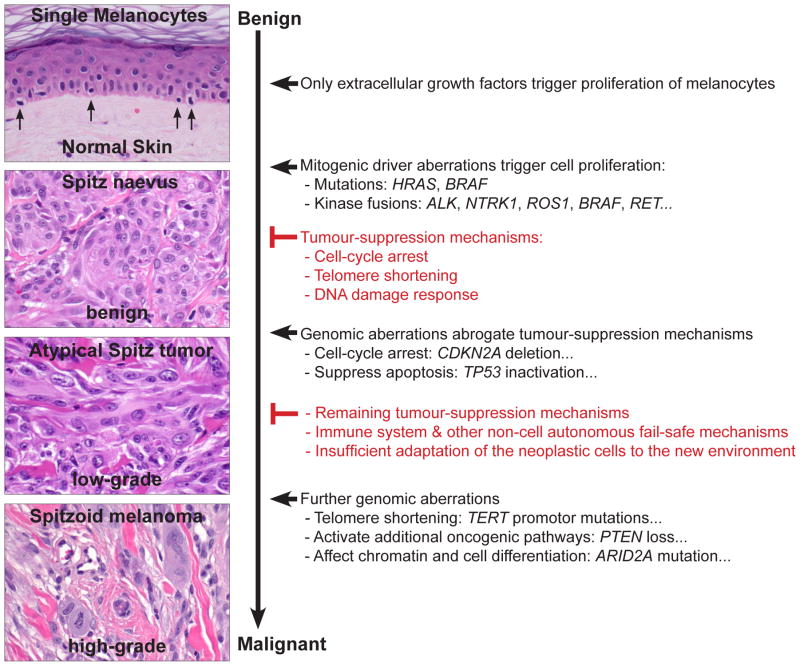
Mitogenic drivers cause Spitz naevi
Under physiological conditions, melanocytes are located in the basal layer of the epidermis and grow only when stimulated by extracellular growth factors, which are provided by the surrounding cells. These growth factors bind to receptor tyrosine kinases (or to other receptors) and activate various intracellular signalling cascades such as the MAP/ERK and the PI3K/AKT/mTOR pathway (Fig. 8). Early steps in tumour progression are often genomic aberrations, which activate the same signalling cascades that are stimulated by extracellular growth factors.87 These mitogenic driver aberrations stimulate cell proliferation by mimicking growth factors and thereby transform normal cells to clonal proliferations. In Spitz tumours, these mitogenic driver aberrations are HRAS mutations, BRAF mutations (in conjunction with BAP1 inactivation) and rearrangements of the kinases ALK, ROS1, NTRK1, RET, MET, and BRAF.
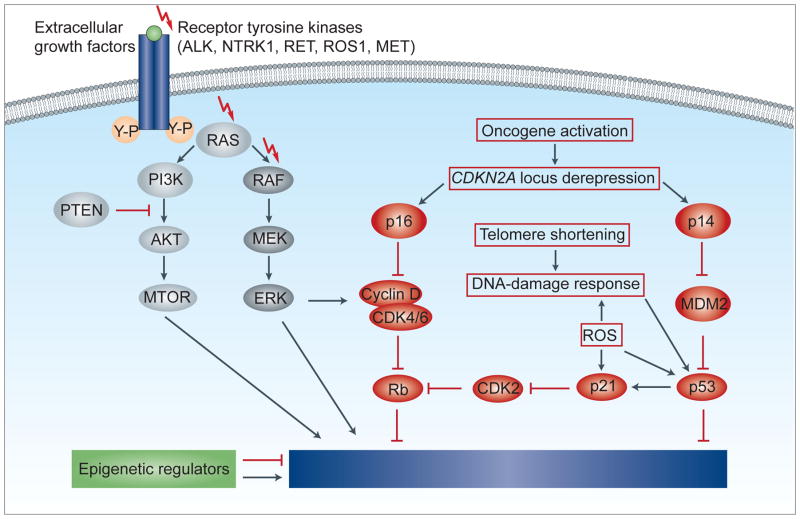
Fig. 8.
Molecular pathways of proliferation, survival and senescence. Extracellular growth factors trigger the dimerisation of receptor tyrosine kinases, leading to autophosphorylation and activation of intracellular signalling cascades such as the MAPK/ERK or the PI3K/AKT/mTOR pathway. Among a variety of other functions, these signalling pathways increase cell proliferation and survival. Many of the components, especially of the MAPK/ERK pathway, show genetic aberrations in spitzoid melanocytic tumours, which lead to a constitutive pathway activation. However, the activation of oncogenic pathways epigenetically de-represses the CDKN2A locus, encoding the proteins p16 and p14. Oncogene activation and cell proliferation may also lead to telomere shortening and increased concentrations of reactive oxygen species (ROS), which causes a DNA-damage response and activates p53 and p21. The signalling cascades of the p16 and p53 pathway converge on the tumour suppressor retinoblastoma (Rb) and on cell cycle inhibitors. After initial cell proliferation, these signalling pathways cause a durable proliferative arrest, termed senescence.
Mitogenic driver mutations generate small benign tumours such as Spitz naevi, but these become growth arrested through senescence. The mitogenic driver aberrations and the associated cellular stress usually leads to an initial phase of cell proliferation, followed by permanent growth arrest (senescence), which is characterised by failure to re-enter the cell cycle in response to mitogenic stimulation.88 Senescence is triggered by various mechanisms such as telomere erosion, DNA damage, increased concentration of reactive oxygen species (ROS), or epigenetic changes through oncogene activation such as de-repression of tumour suppressor genes.88 Mechanistically, the p16 and p53 pathways are key components in inducing senescence and suppressing tumour growth. When the p16 and p53 pathways are activated, cell cycle inhibitors counterbalance the oncogenic signalling from the MAPK/ERK and PI3K/AKT/mTOR pathways (Fig. 8). The senescent cells are then often removed by the immune system, partly by direct recognition of the senescent cells by T helper cells89 or by phagocytic cells, which are attracted by a pro-inflammatory response induced by senescent cells.90
Hence, mitogenic driver mutations are necessary, but not sufficient, for malignant transformation. This explains why mitogenic driver mutations are found across the entire biological spectrum of melanocytic neoplasms, from clearly benign to obviously malignant lesions, and illustrates that they are not useful in determining the biological behaviour of melanocytic tumours (Fig. 9). Nevertheless, they provide important therapeutic targets for therapy.57,58,91
Fig. 9.
Tumour progression model of Spitz tumours. Most tumours, including melanocytic neoplasms, develop through sequential acquisition of genomic aberrations. This tumour progression model suggests a continuous biological tumour spectrum rather than a clear dividing line between benign and malignant. The acquisition of genomic aberrations usually correlates with increased histological atypia and, consequently, tumours with conflicting histological criteria (atypical Spitz tumours) show more genomic aberrations than benign (Spitz naevi), but fewer than malignant (spitzoid melanoma) tumours. The order of genetic aberrations described here is likely to be a common sequence, because cell proliferation (induced by mitogenic genetic aberrations) is associated with a high probability to acquire additional genetic aberrations. However, the genetic changes may also occur in a different order and the sequence displayed here denotes that spitzoid melanomas have additional mutations, but not that all melanomas arise from naevi. Spitzoid melanoma arising without an obvious antecedent naevus may suggest that aberrations in the fail-safe mechanisms develop before the mitogenic driver. Spitz naevi usually only have a strong proliferation signal such as activating HRAS mutations or kinase fusions. These mitogenic genetic aberrations initiate tumour formation, but after initial cell proliferation, multiple fail-safe mechanisms stably block further growth (Spitz naevus). Atypical Spitz tumours abrogate some of these fail-safe mechanisms by gaining additional genomic aberrations so that the cells may continue to grow or to survive in distant organs, such as the lymph nodes. For example, aberrations of CDKN2A, CDK4, or CCND1 undermine the cell-cycle arrest, and TERT promoter mutations may prevent telomere shortening, and consequently senescence. The acquisition of these aberrations is reflected by an increase of cytological and histological atypia. Spitzoid melanomas acquire even more genetic and epigenetic aberrations, which may activate additional oncogenic pathways, affect the chromatin landscape, or reduce cell differentiation, so that the neoplastic cells may colonise and replace the infiltrated organs.
Atypical Spitz tumours escape senescence
Mitogenic drivers have been identified in naevi and melanoma. The key difference between benign and malignant melanocytic tumours is the ability of the latter to escape cell-autonomous (e.g., senescence or apoptosis) and non-cell autonomous (e.g., immune system) tumour suppressing mechanisms. The efficiency of transformation barriers in eliminating neoplastic cells is highlighted by multiple observations: naevi are much more common than melanomas; families with germline mutations in tumour suppressor genes, such as TP53, CDKN2A, or BAP1, have an increased risk for malignancies;92 and tumour suppressor genes are among the most commonly inactivated genes in cancer.83
The CDKN2A gene, for example, encodes the proteins p14 and p16. Loss of CDKN2A therefore affects two major pathways involved in cellular senescence. p14 stabilises p53 by preventing MDM2 mediated degradation of p53 (Fig. 8). p53 can then activate various tumour suppression pathways and thereby triggers apoptosis and senescence.87 p16 sequesters CDK4/6, which inhibits the phosphorylation of Rb and causes an arrest of the cell-cycle (Fig. 8). However, somatic disruption of CDKN2A alone, without mitogenic driver aberrations, has no biological effect, because normal melanocytes express p16 only as stress response;87 but if melanocytes with CDKN2A loss acquire additional mitogenic driver aberrations, atypical or malignant melanocytic tumours may develop directly without precursor lesions. This might explain why only a minority of melanomas arise from naevi.93
The accumulation of genetic aberrations abrogating transformation barriers induces selective survival benefits. The cells become tolerant to abnormal cellular functions, start to show cytological atypia, and may survive in foreign microenvironments such as lymph nodes. The neoplasms grow in a less organised fashion, deviate from the normal growth pattern of benign melanocytic lesions, and may become invasive. Because of these cytological and histological atypical, the lesions are usually classified as ASTs. However, additional transformation barriers are still intact and counteract unrestrained proliferation and metastasis, which explains why a few disseminated neoplastic melanocytes in the lymph node is not synonymous with cells that metastasise, grow and take over the infiltrated distant organs. One might speculate that the strong activation of PI3K/AKT/mTOR in Spitz tumours might lead to survival benefits so that the cells can survive in the lymph node, but that they have not acquired the abilities to grow efficiently and to spread to other organs. It also illustrates that the presence of isolated genetic aberrations in tumour-suppressive mechanisms, such as deletion of CDKN2A at 9p21,94 TERT promoter mutations,95 or isolated chromosomal aberrations,96 may increase the risk for dissemination (e.g., to regional lymph nodes), but are not sufficient for full malignant transformation and distant metastasis/colonisation (Fig. 9).
Spitzoid melanomas require additional aberrations
In contrast to benign proliferations, malignant tumours have the ability to grow indefinitely, to invade adjacent tissues, and to colonise distant tissues and organs. The aforementioned mitogenic driver aberrations provide proliferative signalling, and genetic aberrations abrogating tumour suppression pathways provide survival benefits. However, melanocytes have to acquire several additional abilities before they can widely spread, colonise and grow in different organs. The acquisition of these abilities occurs usually via genetic aberrations, epigenetic aberrations and/or adaptations to the new environment and is accompanied by increased cytological atypia.83,85
For example, to resist cell death and to adapt to unfamiliar and hostile microenvironments in diverse organs, melanocytes have to up-regulate additional pro-survival pathways such as the PI3K/AKT/mTOR pathway (e.g., through loss of PTEN or amplification of AKT), or they have to suppress pro-apoptotic pathways (e.g., by inactivating p53 or amplifying MDM2). Another example is that melanocytes enable replicative immortality by avoiding shortening of their telomeres and up-regulating the telomerase reverse transcriptase (TERT). TERT promoter mutations can up-regulate TERT, and thus, may confer to immortality of melanocytic tumours.97 TERT promoter mutations have not been reported in naevi, but occur in 33% of primary melanomas and in 85% of melanoma metastases, suggesting that these mutations also play an important role in tumour progression.97 Moreover, TERT promoter mutations have been associated with poorer prognosis in patients with melanoma98 and Spitz tumours.95 Additional complex characteristics that drive tumour progression, such as invasion and metastasis, induction of angiogenesis, reprogramming of energy metabolism, evading immune destruction, and genomic instability are often summarised under the term ‘hallmarks of cancer’. The characteristics are not specific to melanocytic tumours, but are acquired by all cancers and are reviewed in detail elsewhere.83,85
DIAGNOSIS OF SPITZ TUMOURS
Clinical and histopathological criteria
Histopathological examination remains the gold standard for the diagnosis of melanocytic neoplasms. Histological characteristics that are associated with malignancy in Spitz tumours are cytological atypia with marked nuclear pleomorphism, mitoses (especially near the base), pagetoid spread, confluence of nests, sheets of melanocytes, ulceration of the epidermis, necrosis, inflammation, poor circumscription, and asymmetry.8 An in-depth description of the clinicopathological criteria used for diagnosis of Spitz tumours is beyond the scope of this article, but these criteria are discussed in detail elsewhere.2,4,5,8,99
Ancillary diagnostic tools
Immunohistochemistry
Immunohistochemistry is widely accessible and plays an important role in the diagnosis of melanocytic tumours. Markers, such as S100, SOX10, Melan-A, MITF, Mart-1, HMB45, and tyrosinase are widely used to establish melanocytic differentiation. These markers also help to better assess the growth pattern when melanocytes are poorly visible. Proliferation markers, such as Ki-67 or pHH-3, are used to assess the percentage of proliferating cells.
Antibodies to detect specific genomic aberrations have gained relevance in recent years. The VE1 antibody recognises the mutant BRAFV600E protein.37,40 An antibody is also available for the protein product of NRASQ61R mutations.100 Both of these antibodies have high specificity and sensitivity, but recognise only the most prevalent mutations of BRAF and NRAS. BAP1 immunohistochemistry is commonly used for lesions with epithelioid cells to validate loss of BAP1 (Fig. 2F,I). Despite their sometimes worrisome histological picture, Spitz tumours with BAP1 loss often have a favourable prognosis. In addition, the presence of multiple melanocytic tumours with BAP1 loss is suspicious for BAP1 germline mutations (BAP1 hereditary predisposition syndrome) and therefore should be investigated further.38
Many of the translocated receptor kinases detected in Spitz tumours are normally not expressed in human tissue. Therefore, immunohistochemical expression of these kinases correlates well with the presence of genomic rearrangements. The ALK antibody has high sensitivity and specificity (Fig. 4D,E).64 The NTRK1 antibody may show some weak background staining due to low endogenous NTRK1 expression, but moderate to strong staining is usually very specific for NTRK1 fusions (Fig. 6C,D).25 ROS1 fusion can also be detected with immunohistochemistry, but in our experience, the antibody often shows weak reactivity, and is not 100% sensitive (Fig. 5D).25
Immunohistochemistry for p16 is controversial. As discussed above, p16 has been associated with aggressive clinical behaviour of ASTs. However, p16 expression is often heterogeneously expressed, and not even in common acquired melanocytic naevi do all melanocytes express p16.101 This may reflect the numerous mechanisms of tumour suppression, so that some cells become senescent by other mechanisms. The use of p21 immunohistochemistry in the diagnostic work-up of melanocytic tumours with borderline morphological features also remains debatable. At present, it appears that no single senescence marker can reliably distinguish naevi from melanomas.102
Array comparative genomic hybridisation
Array CGH is a genetic technique that analyses the entire genome for copy number alterations (CNA) by comparing tumour DNA to normal, reference DNA. In brief, the isolated tumour and reference DNA are each labelled with either a red or a green fluorescent-dye. The differentially fluorescent-labelled DNA samples are then mixed at a 1:1 ratio and co-hybridised to a microscope slide that is spotted with many thousand DNA fragments (DNA microarray). During co-hybridisation, the red- and green-labelled DNA probes compete for the binding site on each spot on the DNA microarray. Subsequently, the intensities of the red and green fluorescent-dye at each spot are individually measured and quantified. The resulting ratio of the fluorescence intensities is proportional to the ratio of the copy number of DNA sequences in the tumour and reference DNA; if the green and red fluorescence intensities are equal at one spot, that genome region is interpreted as having equal quantity of DNA in the tumour and reference samples, i.e., the tumour has no CNA at this region. An altered ratio of the red and green fluorescent dyes indicates DNA copy number loss or gain.
Array CGH works reasonably well on formalin fixed, paraffin embedded tissue, but in cases with suboptimal DNA quality (poor fixation, degraded material), it can be difficult to obtain reliable results.103 Array CGH screens the entire genome for CNAs, but false-negative results occur when the percentage of tumour cells in the specimens is below 50%, e.g., due to tumour-infiltrating lymphocytes or other admixed non-neoplastic (e.g., stromal) cells. Most importantly, array CGH only detects CNAs, but no other genomic aberrations such as small indels, point mutations, and or balanced translocations.
Naevi usually lack CNA, with a few exceptions such as gains of 11p in desmoplastic Spitz naevi24,104,105 and isolated loss of chromosome 3 in epithelioid melanocytic tumours with BAP1 loss.23 In contrast, most melanomas usually exhibit multiple CNAs of whole chromosomes and subchromosomal regions.104,105 Melanomas frequently have homozygous deletions of regions containing tumour suppressor genes (CDKN2A, 9p21; PTEN, 10q23) and hemizygous losses affecting entire chromosome parts, including 6q, 8p, 9p and 10. Regions containing oncogenes (BRAF, 7q34; MITF, 3p13) are frequently amplified and low copy number gains of the chromosome parts 6p, 7 and 8q are also often observed. In addition, specific subtypes of melanoma are associated with distinct patterns of CNAs.17
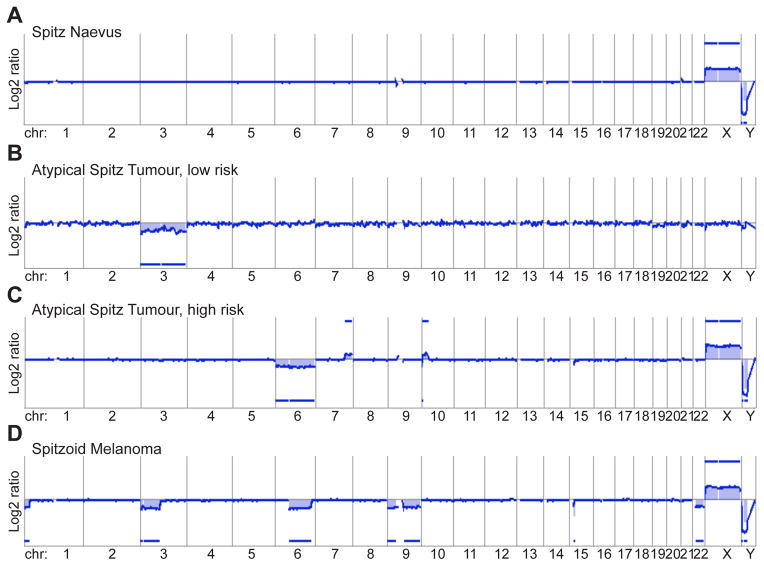
Since the vast majority of melanocytic tumours are histologically distinguishable as clearly benign or malignant, there is no need for CGH of these tumours in diagnostic settings. Few published data are available on ambiguous histological tumours,106 but from these data and our own experience it seems that the detected CNA reflect the histology quite well. The array CGH profiles of ASTs usually show more genomic aberrations than benign tumours, but fewer than malignant tumours. Therefore, array CGH may help to down-grade (low-risk) or up-grade (high-risk) the risk of malignant behaviour in ASTs, but usually does not give clear answers if a given lesion is benign or malignant (Fig. 10).
Fig. 10.
Array comparative genomic hybridisation (aCGH) as an ancillary diagnostic tool in evaluating Spitz tumours. The aCGH profile of atypical Spitz tumours usually shows more genomic aberrations than of Spitz naevi, but not as many as spitzoid melanoma. (A) The aCGH profile of benign Spitz naevi shows usually a flat line indicating no chromosomal gains or losses (here the profile of the X- and Y-chromosome indicates that the tumour is from a female patient: 2 X-chromosomes, no Y-chromosome). (B) Atypical Spitz tumours at the benign end of the biological spectrum show mild histological atypia, and usually no more than gains or losses of one or two chromosomes or chromosome arms (here loss of the entire chromosome 3). (C) Atypical Spitz tumours at the malignant end of the biological spectrum show a considerable amount of histological atypia, including anisonucleosis and pleomorphism. In aCGH, gains and losses of several chromosome parts can be observed (here loss of the entire chromosome 6, a gain on the long arm of chromosome 7, a small loss and a larger gain on chromosome 10). (D) Spitzoid melanoma shows severe histological atypia, pleomorphism, and numerous, atypical mitoses; aCGH shows numerous chromosomal aberrations (here loss of parts of chromosome 1, 3, 6, 15, and loss of the entire chromosome 9 and 22).
Fluorescence in situ hybridisation
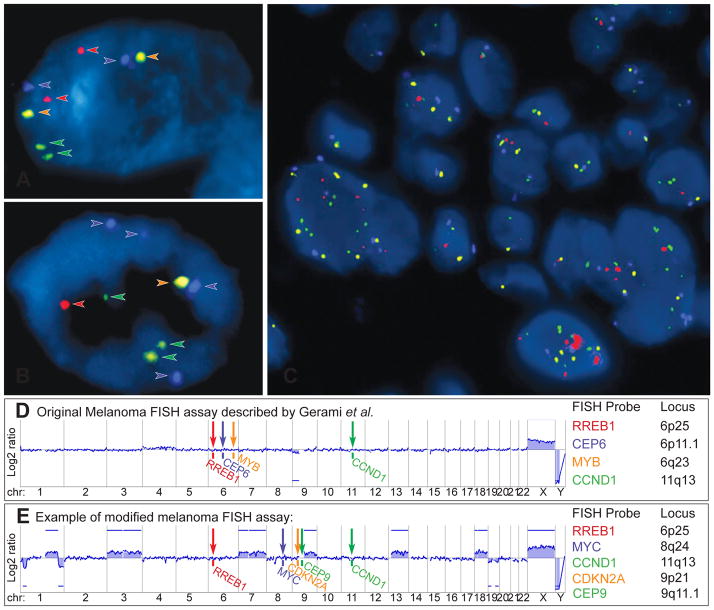
The identification of chromosomal gains and losses by array CGH led to the development of interphase FISH assays. While array CGH is mainly performed at academic centres, because it is labour-intensive and associated with significant costs, FISH is more widely available and requires only basic laboratory equipment and a fluorescence microscope.107 In FISH, fluorescent-labelled DNA probes are hybridised to formalin fixed, paraffin embedded histological sections on glass slides. The fluorescent-labelled FISH probes bind to their complementary DNA sequences on specific chromosome loci. The number of fluorescent signals in each cell nucleus then is evaluated under a fluorescence microscope, and is equal to the copy number of the chromosomal regions targeted by the FISH probes (Fig. 11A–C).
Fig. 11.
Fluorescence in situ hybridisation (FISH) as ancillary diagnostic tool for ambiguous melanocytic tumours. Melanoma FISH assays are often based on 4 probes, which are labelled with 4 different fluorescent dyes (red, green, orange, aqua) and bind to 4 specific chromosomal regions. (A) In normal cells without chromosomal gains and losses 2 signals of each probe are detected. (B) In cells with chromosomal aberrations, which affect the FISH probe-binding regions, losses or gains of the FISH signals can be observed (here 4 aqua, 3 green, and 1 red and orange signal). (C) FISH of a melanoma showing several cell nuclei with a varying number of FISH probes indicating chromosomal aberrations and chromosomal instability. (D) Binding sites of the original 4 FISH probes described by Gerami et al.: RREB1 (6p25), MYB (6q23), CCND1 (11q13) genes and of centromere 6. This aCGH profile of an AST shows loss of chromosome arm 9p, which would not be detected by FISH. (E) Modified and commercially used assay with FISH probes for RREB1 (6p25), MYC (8q24), CDKN2A (9p21), centromere 9 (CEP 9) and CCND1 (11q13). The corresponding aCGH profile detects several chromosomal aberrations involving chromosome 1, 3, 7, 9, 13, 18, 19, all of which would be missed by FISH. These examples illustrate that melanoma FISH has major blind spots. On the other hand, the detection of a chromosomal aberration by FISH does not prove that the tumour is malignant (see Fig. 9). Consequently, FISH is not very helpful in the diagnosis of ASTs.
The originally described melanoma FISH assay (Fig. 11D), which is still commercially available, uses four FISH probes to evaluate the CCND1 region (11q13, green) on chromosome 11 and the regions RREB1 (6p25, red), centromere 6 (blue), MYB (6q23, orange) on chromosome 6.66,108 This assay was reported to distinguish histologically unambiguous melanomas from naevi with a sensitivity of 87% and a specificity of 95%.108 Subsequent studies reported that this assay also distinguishes melanoma from adjacent naevi in 78% of cases,109 naevoid melanomas from mitotically active naevi in 100 % of cases,110 and epithelioid blue naevi from blue naevus-like cutaneous melanoma metastases with a sensitivity of 90%.111
More recently, homozygous deletions of the CDKN2A locus at 9p21 were found in spitzoid melanomas and associated with a more aggressive clinical course in ASTs.94,112,113 Other groups also reported fatal outcomes of spitzoid tumours with heterozygous CDKN2A deletions detected by FISH,114 suggesting that the other CDKN2A allele was inactivated by other genetic mechanisms such as mutations, genomic rearrangements, epigenetic mechanisms (e.g., promoter hypermethylation), or small deletions, which are often not detected by FISH.115 Nevertheless, FISH probes targeting 9p21 and centromere 9 (as a control) were added to the original FISH panel, resulting in a 6-probe assay (RREB1, CCND1, MYB, CEP6, CDKN2A, and CEP9).108 Other labs omitted two probes on chromosome 6 of the original FISH assay, and instead added probes for MYC at 8q24, CDKN2A at 9p21, and centromere 9, resulting in a 5-probe assay (RREB1, MYC, CDKN2A, CCND1 and CEP9). However, the sensitivity and specificity of all melanoma FISH assays in discriminating between melanoma and benign naevi seems to be quite similar to the originally described FISH panel (approximately 85% and 95%, respectively), and tends to be even lower in spitzoid melanomas and superficial spreading melanoma.116,117
Although a number of commercial labs and academic centres are promoting FISH as an ancillary diagnostic test for borderline melanocytic tumours such as ASTs, it has not been proven that FISH improves the diagnostic accuracy of difficult melanocytic lesions. FISH only evaluates 4–6 genomic loci for CNAs, which explains its limited sensitivity and specificity,118 with a false negative rate of 15% in unambiguous melanocytic tumours. CNAs in other chromosomal regions, and all other types of genomic aberrations, including mutations, small indels, and genomic rearrangements, cannot be detected with FISH. The original FISH assay does not discriminate ASTs from Spitz naevi.114 Many FISH negative melanomas meet criteria for melanoma when analysed with array CGH (Fig. 11E).119 Consequently, atypical histopathological tumours concerning for malignancy are usually diagnosed on the basis of pathological evaluation as melanoma, and negative FISH results are usually disregarded.
On the other hand, a single chromosomal aberration in FISH does not equate to the diagnosis of melanoma, although they are often incorrectly equated with malignancy.120 Specific examples are cases with loss of MYB on 6q2394,121,122 or BAP1 on 3p2123,39 which usually appear to show an indolent clinical behaviour. As we explained above, tumour progression is a complex multistep process, and although single genomic aberrations might increase the risk for malignant progression, a single aberration is not equivalent to malignancy. Lastly, the accuracy of FISH is highly operator dependent (e.g., due to factors such as biased selection of nuclei for evaluation and the presence of polyploidy),123,124 which is also exemplified by the fact that several different analysis algorithms are published.119,125 Overall, based on available data, FISH has a very limited role in the diagnostic evaluation of Spitz tumours.96
Next-generation sequencing
In recent years, NGS has revolutionised the molecular analysis of cancers. NGS assays can assess most relevant genomic aberrations, including CNAs, point mutations, small deletions and insertions, and genomic rearrangements across the entire genome (Fig. 12),126 which contrasts with the more limited range of aberrations evaluable with array CGH and FISH. NGS assays can be scaled to the required tasks, ranging from sequencing the entire genome (whole-genome sequencing), to sequencing only the protein-coding regions (whole-exome sequencing), to sequencing only a few selected genes (targeted sequencing).127
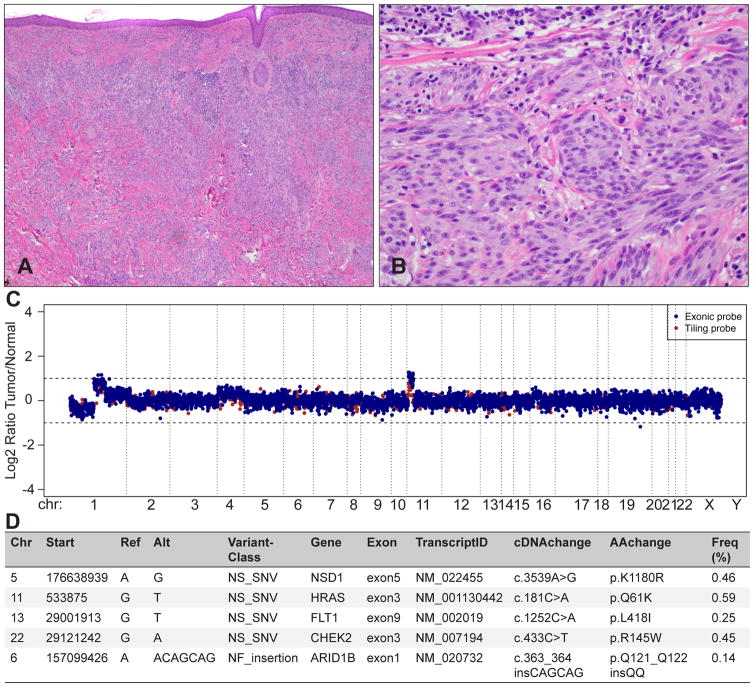
Fig. 12.
Integrated molecular diagnostics with targeted next-generation sequencing for ambiguous melanocytic tumours. The tumour metastasised to the regional lymph nodes. (A) Predominantly intradermal melanocytic tumour with large nests and prominent desmoplasia. (B) Large, epithelioid melanocytes with vesicular chromatin and prominent nucleoli between collagen bundles; mitosis. (C) A targeted next-generation sequencing assay, termed MSK-IMPACT, reveals a loss of chromosome 1p, and partial gains on chromosome 1q and 11p, which is characteristic for desmoplastic Spitz tumours. The log2 ratio was calculated across all targeted regions by comparing the coverage in tumour versus matched normal DNA. (D) Mutational profiling of 341 genes revealed 5 somatically acquired mutations, including an activating HRASQ61K mutation, which is in line with the 11p gain, and a non-frameshift insertion of 6 nucleotides in exon 1 of ARID1B. The 3 missense mutations exchange a single amino acid in the genes NSD1, FLT1, and CHEK2. These mutations are of uncertain functional significance and need further evaluation.
Challenges of NGS include: the need for infrastructure with considerable computing power and capacities for data storage; expertise in bioinformatics and data interpretation, especially because the data analysis pipelines are not standardised; and high costs, certainly in comparison to immunohistochemistry, CGH and FISH, although the costs of NGS assays are decreasing rapidly. Currently, only a limited number of commercial and academic laboratories offer NGS-based mutational testing, mainly for management of patients with advanced stage malignancies. As the costs of NGS assays continue to decrease, their availability and use as ancillary tests to help with diagnosis, prognostic assessment, and evaluation of therapeutic targets can be expected to increase significantly.
CONCLUSIONS
Advances in tumour genomics have helped us to better understand the complex biology and distinct morphological features of Spitz tumours. However, diagnosis still relies primarily on the clinical and histopathological features, because the morphological phenotype reflects an enormous number of pathobiological processes, including the concerted expression of all genes, the sum of all epigenetic effects (gene methylation, post-translational modifications, interactions of proteins and other gene products), the tumour microenvironment, and the immune response with specific infiltration patterns and composition of immune cells. Currently available genomic techniques are not able to capture the complexity of the mechanisms involved in malignant progression.
The histopathological diagnosis of ASTs remains challenging and ancillary genetic techniques have been evaluated in the diagnostic work-up of spitzoid melanocytic tumours.96,128 However, after initial enthusiasm, the results of ancillary genetic tests are quite sobering. Array CGH and FISH are quite proficient at separating tumours that are readily classifiable as benign or malignant by pathologists after microscopic evaluation, but they are usually not very helpful in determining the dignity of histologically ambiguous melanocytic tumours such as ASTs. These disappointing results can be partially explained by the tumour progression model characterised by the sequential acquisition of genomic aberrations, which suggests the existence of a continuous biological spectrum rather than a clear-cut dividing line between benign and malignant. ASTs with conflicting histological criteria usually show more genomic aberrations than Spitz naevi, but fewer than spitzoid melanomas, and are probably best classified as low-grade melanocytic tumours.
With the greater impact of genomic techniques in the diagnosis, prognosis and therapy of Spitz neoplasms, the concept of ‘Spitz tumours’ may change in the future, mainly because it is a genetically very heterogeneous group of diseases. Following an integrated clinical, histopathological and molecular classification system, Spitz tumours with HRAS, BAP1, and specific types of kinase fusions will probably be considered and reported as distinct disease entities. Histopathological evaluation can help to determine the likelihood of specific genetic aberrations within a given tumour, but because the specificity of morphological features is limited, genomic and immunohistochemical assays should be used to validate the underlying aberrations. Even more importantly, genomic aberrations should be correlated with the histopathology. For example, a plexiform Spitz tumour with an ALK kinase fusion can be expected to have a good prognosis, but a tumour with the same morphological characteristics with a BRAF mutation is quite concerning; BRAF mutations are usually found in common acquired melanocytic naevi with small naevoid cells, but not in tumours with large epithelioid or spindled cells in a plexiform pattern, and such discrepancies should raise the suspicion of malignancy.
In the near future, NGS can be expected to provide a more comprehensive picture of the genomic aberrations in melanocytic tumours in general, and Spitz tumours in particular. Integration of clinical, histopathological and molecular data will likely play an essential role in diagnosis, prognostic assessment and targeted treatment of melanocytic tumours (‘precision oncology’).
Acknowledgments
Sources of funding: Thomas Wiesner is funded by the Jubilaeumsfonds of the Oesterreichische Nationalbank OeNB, a Harry J. Lloyd Trust - Translational Research Grant, and a Klaus Wolff Fellowship. The work is also supported by the National Institutes of Health (P30 CA008748; MSK Cancer Center Support Grant).
Footnotes
Conflicts of interest
The authors state that there are no conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bastian BC. The molecular pathology of melanoma: an integrated taxonomy of melanocytic neoplasia. Annu Rev Pathol. 2014;9:239–71. doi: 10.1146/annurev-pathol-012513-104658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnhill RL. The Spitzoid lesion: rethinking Spitz tumors, atypical variants, ‘Spitzoid melanoma’ and risk assessment. Mod Pathol. 2006;19(Suppl 2):S21–33. doi: 10.1038/modpathol.3800519. [DOI] [PubMed] [Google Scholar]
- 3.Spitz S. Melanomas of childhood. Am J Pathol. 1948;24:591–609. [PMC free article] [PubMed] [Google Scholar]
- 4.Ludgate MW, Fullen DR, Lee J, et al. The atypical Spitz tumor of uncertain biologic potential: a series of 67 patients from a single institution. Cancer. 2009;115:631–41. doi: 10.1002/cncr.24047. [DOI] [PubMed] [Google Scholar]
- 5.Murali R, Sharma RN, Thompson JF, et al. Sentinel lymph node biopsy in histologically ambiguous melanocytic tumors with spitzoid features (so-called atypical spitzoid tumors) Ann Surg Oncol. 2008;15:302–9. doi: 10.1245/s10434-007-9577-3. [DOI] [PubMed] [Google Scholar]
- 6.Lallas A, Kyrgidis A, Ferrara G, et al. Atypical Spitz tumours and sentinel lymph node biopsy: a systematic review. Lancet Oncol. 2014;15:e178–83. doi: 10.1016/S1470-2045(13)70608-9. [DOI] [PubMed] [Google Scholar]
- 7.Barnhill RL, Argenyi ZB, From L, et al. Atypical Spitz nevi/tumors: lack of consensus for diagnosis, discrimination from melanoma, and prediction of outcome. Hum Pathol. 1999;30:513–20. doi: 10.1016/s0046-8177(99)90193-4. [DOI] [PubMed] [Google Scholar]
- 8.Cerroni L, Barnhill R, Elder D, et al. Melanocytic tumors of uncertain malignant potential: results of a tutorial held at the XXIX Symposium of the International Society of Dermatopathology in Graz, October 2008. Am J Surg Pathol. 2010;34:314–26. doi: 10.1097/PAS.0b013e3181cf7fa0. [DOI] [PubMed] [Google Scholar]
- 9.McArthur GA, Ribas A. Targeting oncogenic drivers and the immune system in melanoma. J Clin Oncol. 2013;31:499–506. doi: 10.1200/JCO.2012.45.5568. [DOI] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas N. Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–96. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiesner T, Kutzner H. Morphological and genetic aspects of Spitz tumors (German.) Pathologe. 2015;36:37–43. 45. doi: 10.1007/s00292-014-1984-1. [DOI] [PubMed] [Google Scholar]
- 12.Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 13.Bauer J, Curtin JA, Pinkel D, et al. Congenital melanocytic nevi frequently harbor NRAS mutations but no BRAF mutations. J Invest Dermatol. 2007;127:179–82. doi: 10.1038/sj.jid.5700490. [DOI] [PubMed] [Google Scholar]
- 14.Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–9. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Raamsdonk CD, Bezrookove V, Green G, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murali R, Wiesner T, Rosenblum MK, et al. GNAQ and GNA11 mutations in melanocytomas of the central nervous system. Acta Neuropathol. 2012;123:457–9. doi: 10.1007/s00401-012-0948-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 18.Shain AH, Garrido M, Botton T, et al. Exome sequencing of desmoplastic melanoma identifies recurrent NFKBIE promoter mutations and diverse activating mutations in the MAPK pathway. Nat Genet. 2015;47:1194–9. doi: 10.1038/ng.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krauthammer M, Kong Y, Bacchiocchi A, et al. Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat Genet. 2015;47:996–1002. doi: 10.1038/ng.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiesner T, Kiuru M, Scott SN, et al. NF1 mutations are common in desmoplastic melanoma. Am J Surg Pathol. 2015;39:1357–62. doi: 10.1097/PAS.0000000000000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curtin JA, Busam K, Pinkel D, et al. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24:4340–6. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 22.Gantner S, Wiesner T, Cerroni L, et al. Absence of BRAF and HRAS mutations in eruptive Spitz naevi. Br J Dermatol. 2011;164:873–7. doi: 10.1111/j.1365-2133.2011.10210.x. [DOI] [PubMed] [Google Scholar]
- 23.Wiesner T, Obenauf AC, Murali R, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43:1018–21. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bastian BC, LeBoit PE, Pinkel D. Mutations and copy number increase of HRAS in Spitz nevi with distinctive histopathological features. Am J Pathol. 2000;157:967–72. doi: 10.1016/S0002-9440(10)64609-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiesner T, He J, Yelensky R, et al. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat Commun. 2014;5:3116. doi: 10.1038/ncomms4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeh I, Botton T, Talevich E, et al. Activating MET kinase rearrangements in melanoma and Spitz tumours. Nat Commun. 2015;6:7174. doi: 10.1038/ncomms8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiesner T, Fried I, Ulz P, et al. Toward an improved definition of the tumor spectrum associated with BAP1 germline mutations. J Clin Oncol. 2012;30:e337–40. doi: 10.1200/JCO.2011.41.2965. [DOI] [PubMed] [Google Scholar]
- 28.Murali R, Wiesner T, Scolyer RA. Tumours associated with BAP1 mutations. Pathology. 2013;45:116–26. doi: 10.1097/PAT.0b013e32835d0efb. [DOI] [PubMed] [Google Scholar]
- 29.Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–3. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Testa JR, Cheung M, Pei J, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022–5. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popova T, Hebert L, Jacquemin V, et al. Germline BAP1 mutations predispose to renal cell carcinomas. Am J Hum Genet. 2013;92:974–80. doi: 10.1016/j.ajhg.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abdel-Rahman MH, Pilarski R, Cebulla CM, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet. 2011;48:856–9. doi: 10.1136/jmedgenet-2011-100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiao Y, Pawlik TM, Anders RA, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45:1470–3. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de la Fouchardiere A, Cabaret O, Savin L, et al. Germline BAP1 mutations predispose also to multiple basal cell carcinomas. Clin Genet. 2015;88:273–7. doi: 10.1111/cge.12472. [DOI] [PubMed] [Google Scholar]
- 35.Mochel MC, Piris A, Nose V, et al. Loss of BAP1 expression in basal cell carcinomas in patients with germline BAP1 mutations. Am J Clin Pathol. 2015;143:901–4. doi: 10.1309/AJCPG8LFJC0DHDQT. [DOI] [PubMed] [Google Scholar]
- 36.Wadt KA, Aoude LG, Johansson P, et al. A recurrent germline BAP1 mutation and extension of the BAP1 tumor predisposition spectrum to include basal cell carcinoma. Clin Genet. 2015;88:267–72. doi: 10.1111/cge.12501. [DOI] [PubMed] [Google Scholar]
- 37.Llamas-Velasco M, Perez-Gonzalez YC, Requena L, et al. Histopathologic clues for the diagnosis of Wiesner nevus. J Am Acad Dermatol. 2014;70:549–54. doi: 10.1016/j.jaad.2013.10.032. [DOI] [PubMed] [Google Scholar]
- 38.Busam KJ, Wanna M, Wiesner T. Multiple epithelioid Spitz nevi or tumors with loss of BAP1 expression: a clue to a hereditary tumor syndrome. JAMA Dermatol. 2013;149:335–9. doi: 10.1001/jamadermatol.2013.1529. [DOI] [PubMed] [Google Scholar]
- 39.Wiesner T, Murali R, Fried I, et al. A distinct subset of atypical Spitz tumors is characterized by BRAF mutation and loss of BAP1 expression. Am J Surg Pathol. 2012;36:818–30. doi: 10.1097/PAS.0b013e3182498be5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Busam KJ, Sung J, Wiesner T, et al. Combined BRAFV600E-positive melanocytic lesions with large epithelioid cells lacking BAP1 expression and conventional nevomelanocytes. Am J Surg Pathol. 2013;37:193–9. doi: 10.1097/PAS.0b013e318263648c. [DOI] [PubMed] [Google Scholar]
- 41.Yeh I, Mully TW, Wiesner T, et al. Ambiguous melanocytic tumors with loss of 3p21. Am J Surg Pathol. 2014;38:1088–95. doi: 10.1097/PAS.0000000000000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murali R, Wilmott JS, Jakrot V, et al. BAP1 expression in cutaneous melanoma: a pilot study. Pathology. 2013;45:606–9. doi: 10.1097/PAT.0b013e3283653818. [DOI] [PubMed] [Google Scholar]
- 43.Carbone M, Ferris LK, Baumann F, et al. BAP1 cancer syndrome: malignant mesothelioma, uveal and cutaneous melanoma, and MBAITs. J Transl Med. 2012;10:179. doi: 10.1186/1479-5876-10-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Njauw CN, Kim I, Piris A, et al. Germline BAP1 inactivation is preferentially associated with metastatic ocular melanoma and cutaneous-ocular melanoma families. PloS One. 2012;7:e35295. doi: 10.1371/journal.pone.0035295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Engen-van Grunsven AC, Kusters-Vandevelde H, Groenen PJ, et al. Update on molecular pathology of cutaneous melanocytic lesions: what is new in diagnosis and molecular testing for treatment? Front Med (Lausanne) 2014;1:39. doi: 10.3389/fmed.2014.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vilain RE, McCarthy SW, Thompson JF, et al. BAP1-inactivated spitzoid naevi. Am J Surg Pathol. 2015;39:722. doi: 10.1097/PAS.0000000000000407. [DOI] [PubMed] [Google Scholar]
- 47.Hafner C. Wiesner nevus: A new melanocytic tumor defined by molecular genetic analysis. Hautarzt. 2014;65:653–5. doi: 10.1007/s00105-014-2827-y. [DOI] [PubMed] [Google Scholar]
- 48.Bastian BC, Wesselmann U, Pinkel D, et al. Molecular cytogenetic analysis of Spitz nevi shows clear differences to melanoma. J Invest Dermatol. 1999;113:1065–9. doi: 10.1046/j.1523-1747.1999.00787.x. [DOI] [PubMed] [Google Scholar]
- 49.van Engen-van Grunsven AC, van Dijk MC, Ruiter DJ, et al. HRAS-mutated Spitz tumors: A subtype of Spitz tumors with distinct features. Am J Surg Pathol. 2010;34:1436–41. doi: 10.1097/PAS.0b013e3181f0a749. [DOI] [PubMed] [Google Scholar]
- 50.van Dijk MC, Bernsen MR, Ruiter DJ. Analysis of mutations in B-RAF, N-RAS, and H-RAS genes in the differential diagnosis of Spitz nevus and spitzoid melanoma. Am J Surg Pathol. 2005;29:1145–51. doi: 10.1097/01.pas.0000157749.18591.9e. [DOI] [PubMed] [Google Scholar]
- 51.Yan J, Roy S, Apolloni A, et al. Ras isoforms vary in their ability to activate Raf-1 and phosphoinositide 3-kinase. J Biol Chem. 1998;273:24052–6. doi: 10.1074/jbc.273.37.24052. [DOI] [PubMed] [Google Scholar]
- 52.Fingar DC, Salama S, Tsou C, et al. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–87. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ross AL, Sanchez MI, Grichnik JM. Molecular nevogenesis. Dermatol Res Pract. 2011;2011:463184. doi: 10.1155/2011/463184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18:378–81. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 55.Rabes HM, Demidchik EP, Sidorow JD, et al. Pattern of radiation-induced RET and NTRK1 rearrangements in 191 post-chernobyl papillary thyroid carcinomas: biological, phenotypic, and clinical implications. Clin Cancer Res. 2000;6:1093–103. [PubMed] [Google Scholar]
- 56.Jones DT, Kocialkowski S, Liu L, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673–7. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–70. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yeh I, de la Fouchardiere A, Pissaloux D, et al. Clinical, histopathologic, and genomic features of Spitz tumors with ALK fusions. Am J Surg Pathol. 2015;39:581–91. doi: 10.1097/PAS.0000000000000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lamant L, Dastugue N, Pulford K, et al. A new fusion gene TPM3-ALK in anaplastic large cell lymphoma created by a (1;2)(q25;p23) translocation. Blood. 1999;93:3088–95. [PubMed] [Google Scholar]
- 61.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 62.Lawrence B, Perez-Atayde A, Hibbard MK, et al. TPM3-ALK and TPM4-ALK oncogenes in inflammatory myofibroblastic tumors. Am J Pathol. 2000;157:377–84. doi: 10.1016/S0002-9440(10)64550-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Niu HT, Zhou QM, Wang F, et al. Identification of anaplastic lymphoma kinase break points and oncogenic mutation profiles in acral/mucosal melanomas. Pigment Cell Melanoma Res. 2013;26:646–53. doi: 10.1111/pcmr.12129. [DOI] [PubMed] [Google Scholar]
- 64.Busam KJ, Kutzner H, Cerroni L, et al. Clinical and pathologic findings of spitz nevi and atypical spitz tumors with ALK fusions. Am J Surg Pathol. 2014;38:925–33. doi: 10.1097/PAS.0000000000000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wiesner T, Lee W, Obenauf AC, et al. Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature. 2015;526:453–7. doi: 10.1038/nature15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morey AL, Murali R, McCarthy SW, et al. Diagnosis of cutaneous melanocytic tumours by four-colour fluorescence in situ hybridisation. Pathology. 2009;41:383–7. doi: 10.1080/00313020902915875. [DOI] [PubMed] [Google Scholar]
- 67.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 68.Birchmeier C, Sharma S, Wigler M. Expression and rearrangement of the ROS1 gene in human glioblastoma cells. Proc Natl Acad Sci USA. 1987;84:9270–4. doi: 10.1073/pnas.84.24.9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gu TL, Deng X, Huang F, et al. Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. PloS One. 2011;6:e15640. doi: 10.1371/journal.pone.0015640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jun HJ, Johnson H, Bronson RT, et al. The oncogenic lung cancer fusion kinase CD74-ROS activates a novel invasiveness pathway through E-Syt1 phosphorylation. Cancer Res. 2012;72:3764–74. doi: 10.1158/0008-5472.CAN-11-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vaishnavi A, Capelletti M, Le AT, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med. 2013;19:1469–72. doi: 10.1038/nm.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Greco A, Miranda C, Pierotti MA. Rearrangements of NTRK1 gene in papillary thyroid carcinoma. Mol Cell Endocrinol. 2010;321:44–9. doi: 10.1016/j.mce.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 73.Wu G, Diaz AK, Paugh BS, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46:444–50. doi: 10.1038/ng.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Truzzi F, Marconi A, Lotti R, et al. Neurotrophins and their receptors stimulate melanoma cell proliferation and migration. J Invest Dermatol. 2008;128:2031–40. doi: 10.1038/jid.2008.21. [DOI] [PubMed] [Google Scholar]
- 75.Reuther GW, Lambert QT, Caligiuri MA, et al. Identification and characterization of an activating TrkA deletion mutation in acute myeloid leukemia. Mol Cell Biol. 2000;20:8655–66. doi: 10.1128/mcb.20.23.8655-8666.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18:375–7. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Medicine. 2012;18:382–4. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kato M, Takahashi M, Akhand AA, et al. Transgenic mouse model for skin malignant melanoma. Oncogene. 1998;17:1885–8. doi: 10.1038/sj.onc.1202077. [DOI] [PubMed] [Google Scholar]
- 79.Ciampi R, Knauf JA, Kerler R, et al. Oncogenic AKAP9-BRAF fusion is a novel mechanism of MAPK pathway activation in thyroid cancer. J Clin Invest. 2005;115:94–101. doi: 10.1172/JCI23237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Palanisamy N, Ateeq B, Kalyana-Sundaram S, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16:793–8. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Botton T, Yeh I, Nelson T, et al. Recurrent BRAF kinase fusions in melanocytic tumors offer an opportunity for targeted therapy. Pigment Cell Melanoma Res. 2013;26:845–51. doi: 10.1111/pcmr.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349:1483–9. doi: 10.1126/science.aab4082. [DOI] [PubMed] [Google Scholar]
- 83.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 84.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 85.Weinberg RA. The Biology of Cancer. 2. New York: Garland Science/Taylor and Francis; 2014. [Google Scholar]
- 86.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–61. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bennett DC. Genetics of melanoma progression: the rise and fall of cell senescence. Pigment Cell Melanoma Res. 2015 Sep 19; doi: 10.1111/pcmr.12422. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 88.Munoz-Espin D, Serrano M. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol. 2014;15:482–96. doi: 10.1038/nrm3823. [DOI] [PubMed] [Google Scholar]
- 89.Kang TW, Yevsa T, Woller N, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479:547–51. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- 90.Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev Cancer. 2009;9:81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- 91.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aoude LG, Wadt KA, Pritchard AL, et al. Genetics of familial melanoma: 20 years after CDKN2A. Pigment Cell Melanoma Res. 2015;28:148–60. doi: 10.1111/pcmr.12333. [DOI] [PubMed] [Google Scholar]
- 93.Bevona C, Goggins W, Quinn T, et al. Cutaneous melanomas associated with nevi. Arch Dermatol. 2003;139:1620–4. doi: 10.1001/archderm.139.12.1620. [DOI] [PubMed] [Google Scholar]
- 94.Gerami P, Scolyer RA, Xu X, et al. Risk assessment for atypical spitzoid melanocytic neoplasms using FISH to identify chromosomal copy number aberrations. Am J Surg Pathol. 2013;37:676–84. doi: 10.1097/PAS.0b013e3182753de6. [DOI] [PubMed] [Google Scholar]
- 95.Lee S, Barnhill RL, Dummer R, et al. TERT promoter mutations are predictive of aggressive clinical behavior in patients with spitzoid melanocytic neoplasms. Sci Rep. 2015;5:11200. doi: 10.1038/srep11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gaiser T, Kutzner H, Palmedo G, et al. Classifying ambiguous melanocytic lesions with FISH and correlation with clinical long-term follow up. Mod Pathol. 2010;23:413–9. doi: 10.1038/modpathol.2009.177. [DOI] [PubMed] [Google Scholar]
- 97.Horn S, Figl A, Rachakonda PS, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–61. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 98.Griewank KG, Murali R, Puig-Butille JA, et al. TERT promoter mutation status as an independent prognostic factor in cutaneous melanoma. J Natl Cancer Inst. 2014;106:dju246. doi: 10.1093/jnci/dju246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Massi G, LeBoit PE. Histological Diagnosis of Nevi and Melanoma. 2. Berlin: Springer; 2014. [Google Scholar]
- 100.Massi D, Simi L, Sensi E, et al. Immunohistochemistry is highly sensitive and specific for the detection of NRASQ61R mutation in melanoma. Mod Pathol. 2015;28:487–97. doi: 10.1038/modpathol.2014.137. [DOI] [PubMed] [Google Scholar]
- 101.Gray-Schopfer VC, Cheong SC, Chong H, et al. Cellular senescence in naevi and immortalisation in melanoma: a role for p16? Br J Cancer. 2006;95:496–505. doi: 10.1038/sj.bjc.6603283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tran SL, Haferkamp S, Scurr LL, et al. Absence of distinguishing senescence traits in human melanocytic nevi. J Invest Dermatol. 2012;132:2226–34. doi: 10.1038/jid.2012.126. [DOI] [PubMed] [Google Scholar]
- 103.Fried I, Artl M, Cota C, et al. Clinicopathologic and molecular features in cutaneous extranodal natural killer-/T-cell lymphoma, nasal type, with aggressive and indolent course. J Am Acad Dermatol. 2014;70:716–23. doi: 10.1016/j.jaad.2013.11.028. [DOI] [PubMed] [Google Scholar]
- 104.Bastian BC, LeBoit PE, Hamm H, et al. Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res. 1998;58:2170–5. [PubMed] [Google Scholar]
- 105.Bastian BC, Olshen AB, LeBoit PE, et al. Classifying melanocytic tumors based on DNA copy number changes. Am J Pathol. 2003;163:1765–70. doi: 10.1016/S0002-9440(10)63536-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kutzner H, Metzler G, Argenyi Z, et al. Histological and genetic evidence for a variant of superficial spreading melanoma composed predominantly of large nests. Mod Pathol. 2012;25:838–45. doi: 10.1038/modpathol.2012.35. [DOI] [PubMed] [Google Scholar]
- 107.Wiesner T, Streubel B, Huber D, et al. Genetic aberrations in primary cutaneous large B-cell lymphoma: a fluorescence in situ hybridization study of 25 cases. Am J Surg Pathol. 2005;29:666–73. doi: 10.1097/01.pas.0000155163.40668.e7. [DOI] [PubMed] [Google Scholar]
- 108.Gerami P, Jewell SS, Morrison LE, et al. Fluorescence in situ hybridization (FISH) as an ancillary diagnostic tool in the diagnosis of melanoma. Am J Surg Pathol. 2009;33:1146–56. doi: 10.1097/PAS.0b013e3181a1ef36. [DOI] [PubMed] [Google Scholar]
- 109.Newman MD, Lertsburapa T, Mirzabeigi M, et al. Fluorescence in situ hybridization as a tool for microstaging in malignant melanoma. Mod Pathol. 2009;22:989–95. doi: 10.1038/modpathol.2009.72. [DOI] [PubMed] [Google Scholar]
- 110.Gerami P, Wass A, Mafee M, et al. Fluorescence in situ hybridization for distinguishing nevoid melanomas from mitotically active nevi. Am J Surg Pathol. 2009;33:1783–8. doi: 10.1097/PAS.0b013e3181ba6db6. [DOI] [PubMed] [Google Scholar]
- 111.Gammon B, Beilfuss B, Guitart J, et al. Fluorescence in situ hybridization for distinguishing cellular blue nevi from blue nevus-like melanoma. J Cutan Pathol. 2011;38:335–41. doi: 10.1111/j.1600-0560.2010.01667.x. [DOI] [PubMed] [Google Scholar]
- 112.Gammon B, Beilfuss B, Guitart J, et al. Enhanced detection of spitzoid melanomas using fluorescence in situ hybridization with 9p21 as an adjunctive probe. Am J Surg Pathol. 2012;36:81–8. doi: 10.1097/PAS.0b013e31822d5ff8. [DOI] [PubMed] [Google Scholar]
- 113.Gerami P, Busam K, Cochran A, et al. Histomorphologic assessment and interobserver diagnostic reproducibility of atypical spitzoid melanocytic neoplasms with long-term follow-up. Am J Surg Pathol. 2014;38:934–40. doi: 10.1097/PAS.0000000000000198. [DOI] [PubMed] [Google Scholar]
- 114.Massi D, Tomasini C, Senetta R, et al. Atypical Spitz tumors in patients younger than 18 years. J Am Acad Dermatol. 2015;72:37–46. doi: 10.1016/j.jaad.2014.09.049. [DOI] [PubMed] [Google Scholar]
- 115.Wiesner T, Obenauf AC, Geigl JB, et al. 9p21 deletion in primary cutaneous large B-cell lymphoma, leg type, may escape detection by standard FISH assays. J Invest Dermatol. 2009;129:238–40. doi: 10.1038/jid.2008.224. [DOI] [PubMed] [Google Scholar]
- 116.Gerami P, Li G, Pouryazdanparast P, et al. A highly specific and discriminatory FISH assay for distinguishing between benign and malignant melanocytic neoplasms. Am J Surg Pathol. 2012;36:808–17. doi: 10.1097/PAS.0b013e31824b1efd. [DOI] [PubMed] [Google Scholar]
- 117.Gerami P, Mafee M, Lurtsbarapa T, et al. Sensitivity of fluorescence in situ hybridization for melanoma diagnosis using RREB1, MYB, Cep6, and 11q13 probes in melanoma subtypes. Arch Dermatol. 2010;146:273–8. doi: 10.1001/archdermatol.2009.386. [DOI] [PubMed] [Google Scholar]
- 118.Busam KJ. Molecular pathology of melanocytic tumors. Semin Diagn Pathol. 2013;30:362–74. doi: 10.1053/j.semdp.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 119.Kerl K, Palmedo G, Wiesner T, et al. A proposal for improving multicolor FISH sensitivity in the diagnosis of malignant melanoma using new combined criteria. Am J Dermatopathol. 2012;34:580–5. doi: 10.1097/DAD.0b013e3182433f3a. [DOI] [PubMed] [Google Scholar]
- 120.North JP, Garrido MC, Kolaitis NA, et al. Fluorescence in situ hybridization as an ancillary tool in the diagnosis of ambiguous melanocytic neoplasms: a review of 804 cases. Am J Surg Pathol. 2014;38:824–31. doi: 10.1097/PAS.0000000000000189. [DOI] [PubMed] [Google Scholar]
- 121.Gerami P, Cooper C, Bajaj S, et al. Outcomes of atypical spitz tumors with chromosomal copy number aberrations and conventional melanomas in children. Am J Surg Pathol. 2013;37:1387–94. doi: 10.1097/PAS.0b013e31828fc283. [DOI] [PubMed] [Google Scholar]
- 122.Shen L, Cooper C, Bajaj S, et al. Atypical spitz tumors with 6q23 deletions: a clinical, histological, and molecular study. Am J Dermatopathol. 2013;35:804–12. doi: 10.1097/DAD.0b013e31828671bf. [DOI] [PubMed] [Google Scholar]
- 123.Gerami P, Barnhill RL, Beilfuss BA, et al. Superficial melanocytic neoplasms with pagetoid melanocytosis: a study of interobserver concordance and correlation with FISH. Am J Surg Pathol. 2010;34:816–21. doi: 10.1097/PAS.0b013e3181dd1e72. [DOI] [PubMed] [Google Scholar]
- 124.Isaac AK, Lertsburapa T, Pathria Mundi J, et al. Polyploidy in spitz nevi: a not uncommon karyotypic abnormality identifiable by fluorescence in situ hybridization. Am J Dermatopathol. 2010;32:144–8. doi: 10.1097/DAD.0b013e3181b72d6f. [DOI] [PubMed] [Google Scholar]
- 125.Kerl K, Leo S, Hesse R, et al. Two-dimensional visualization of multicolor FISH-generated data as a helpful tool for the analysis and understanding of cytogenetic and chromosomal alterations in melanocytic lesions. Am J Dermatopathol. 2013;35:151–8. doi: 10.1097/DAD.0b013e318265fd08. [DOI] [PubMed] [Google Scholar]
- 126.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–64. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Murali R, Chandramohan R, Moller I, et al. Targeted massively parallel sequencing of angiosarcomas reveals frequent activation of the mitogen activated protein kinase pathway. Oncotarget. 2015;6:36041–52. doi: 10.18632/oncotarget.5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wiesner T, Fried I, Cerroni L, et al. Molecular biology methods to improve diagnosis and prognosis of melanocytic tumors. J Dtsch Dermatol Ges. 2013;11(Suppl 4):19–24. doi: 10.1111/ddg.12083_supp. [DOI] [PubMed] [Google Scholar]