Abstract
OBJECTIVES:
Bowel preparations (BPs) taken before colonoscopy may introduce a confounding effect on the results of gastrointestinal microbiota studies. This study aimed to determine the effect of bowel preparation on the mucosa-associated and luminal colonic microbiota in healthy subjects (HC) and inflammatory bowel disease (IBD) patients.
METHODS:
Biopsy samples (n=36) and fecal samples (n=30) were collected from 10 HC and 8 IBD subjects pre- and post-BP. 16S rRNA gene was pyrosequenced using 454 Titanium protocols. We compared the differences between the pre- and post-BP samples (i.e., comparisons-across-bowel-prep); we examined the effect of BP on the expected separation of the mucosal vs. the luminal compartments (i.e., comparisons-across-compartments). Last, we compared the baseline differences between the HC vs. IBD groups (a secondary analysis), and examined whether the differences between the HC vs. IBD changed after BP.
RESULTS:
In comparisons-across-bowel-prep, the Shannon's index (SI) decreased only in the biopsy samples of IBD subjects post-BP (P=0.025) and phylogenetic diversity-whole tree (PD-WT) metric decreased in biopsy samples of HC subjects post-BP (P=0.021). In secondary comparisons, the subtle differences between the fecal samples of the HC vs. IBD groups, in terms of evenness and the SI, were not apparent post-BP. In terms of β-diversity, in comparisons-across-bowel-prep, the proportion of shared operational taxonomic units (OTUs) in pre- and post-BP samples was low (~30%) and unweighted Unifrac distances between pre- and post-BP specimens ranged from 0.52 to 0.66. HC biopsies were affected more than IBD biopsies with BP (P=0.004). In comparisons-across-compartments, the proportion of shared OTUs between biopsy and fecal samples increased and Unifrac distances decreased post-BP in IBD subjects, reducing the differences between the mucosal and luminal compartments of the gut microbiota. Interindividual differences in Unifrac distances were preserved even with BP effects, although the effects were greater on weighted Unifrac distances. Bacteroidetes and its subtypes increased post-BP in both the luminal and mucosal compartments.
CONCLUSIONS:
Bowel preparations affect the composition and diversity of the fecal and luminal microbiota in the short term, introducing potential bias into experiments examining the gut microbiota. The magnitude of the effect of BP is not greater than that of interindividual variation. Both the luminal and mucosal compartments of the gut microbiota get affected, and samples from controls and IBD subjects may get affected differently. Studies of the colonic microbiota should take into account the direction and the magnitude of the change introduced by BP during the design stage of the experiments, and consider sample sizes so that potential bias is minimized.
INTRODUCTION
The human gastrointestinal tract harbors a large number of diverse microbial cells, some of which are associated with the feces, while others are associated with the mucosa. Most of these microbial cells are bacterial and form complex communities that constitute a unique and dense ecosystem,1, 2, 3 collectively referred to as the gastrointestinal microbiota. Until recently, little was known about the gastrointestinal microbiota because a significant majority of its members are not cultivable.4, 5, 6 Owing to the recent development of novel, culture-independent genomic techniques along with the advancements in bioinformatics tools, researchers have now been enabled to explore and unravel the hidden secrets of the complex gastrointestinal ecosystem in human health and disease. These recent developments have made the gastrointestinal microbiota one of the biggest new frontiers in research.
Recent studies have been conducted worldwide, in an effort to understand the composition, behavior and unique traits of the gastrointestinal microbiota and have noted the human colon as one of the most dense and diverse communities and one of the largest reservoirs of human body-associated microbes. Many of these studies including the Human Microbiome Project in the United States have involved obtaining stool samples to characterize the community composition of the gastrointestinal microbiota.7 Fewer studies have involved obtaining tissue samples from the intestinal tract during endoscopy or surgery. Surgical tissues are probably not ideal specimens for the study of the gastrointestinal microbiota in humans, as most if not all patients are expected to receive antibiotics of various kinds and varying intervals before surgical procedures. Antibiotics have been shown to affect the bacterial microbiota community significantly in the gastrointestinal tract.8 Alternatively, the study of the colonic microbiota using endoscopic tissue biopsies typically involves a colonoscopy, which requires a bowel preparation (BP) consisting of large doses of laxatives to evacuate most if not all of the stool from the colon. Typically, such a preparation is taken by the patient overnight before the procedure and results in 10–20 bowel movements, most of which are diarrheal stools. When the appearance of the stool is yellow or clear liquid without any solid particles within it, a patient is clinically considered to have taken an adequate BP. Hence, BPs can be considered a major disruptor of the colonic ecosystem, and it is unclear whether specimens obtained during colonoscopy are suitable for the study of the gastrointestinal tract microbiota and whether any bias is being introduced by the BP taken before the colonoscopy in colonic microbiota studies.
Prior studies suggest that BPs affect markers of proliferation in the intestinal epithelium, and thus could theoretically affect the mucosa-associated microbiota.9 It has also been found that polyethylene glycol-type BPs result in moderate-to-severe loss of superficial mucus in 96% of patients, which could also affect colonic microbiota composition.10 In fact, microbiota on the mucosal surface probably has more interactions with the host through immune and non-immune mechanisms, and thereby could affect host health to a greater extent than the microbiota found in the luminal compartment. As such, studies of the effects of BPs on the mucosal surface as well as the luminal/fecal compartment are needed.
The concern about the effect of BPs on the intestinal microbiota has been raised in the past by several researchers; however, the magnitude and nature of this potential effect has not been well characterized. It is logical to think that the effects of BP on the microbiota could differ between different compartments of the gastrointestinal tract and could be altered in biopsies that reflect the mucosa-associated compartment vs. fecal samples, which predominantly reflect the luminal microbiota. Additionally, the effects could be variable in a disease state vs. in health. To date, studies that have been carried out have primarily used fecal samples,11, 12, 13, 14, 15 and have used low-resolution techniques such as bacterial fingerprinting.11, 12, 13, 16 None of these studies have concurrently tried to characterize the changes in the mucosal and luminal compartments, and none have separated the effects seen in healthy control (HC) subjects vs. those with a disease.12, 15, 17
In our study, we aimed to concurrently characterize the effects of BP on the mucosa-associated microbiota, with colonic biopsies and the luminal microbiota with fecal samples obtained during colonoscopy, using high-throughput pyrosequencing. We also aimed to determine whether the magnitude or type change induced by BP would differ in a health vs. a disease state (healthy patients without colitis vs. inflammatory bowel disease (IBD) patients). Thus, we hypothesized that BPs cause changes in the composition and diversity of the colonic mucosa-associated and luminal microbiota and that these changes may differ between healthy subjects vs. those with IBD.
METHODS
Human subjects and research ethics and inclusion and exclusion criteria
The study proposal and its consent form were approved by the Institutional Review Board of Rush University Medical Center under the ORA numbers: L06082106 and L06082107. Patients who were scheduling for a routine surveillance colonoscopy for colorectal cancer were approached for recruitment. Subjects were recruited from the Rush University outpatient gastroenterology clinics. All subjects gave verbal and written informed consent. IBD subjects were identified and confirmed by chart review, through documented colonoscopy, pathology reports, lab tests, and medical history consistent with IBD. The inclusion criteria were as follows: HC patients who did not have any evidence of IBD were eligible for participation if they did not have any symptomatic organic gastrointestinal disorder other than a hiatus hernia, gastroesophageal reflux disease, or hemorrhoids and were having a colonoscopy for colon cancer screening. IBD patients were eligible for participation if the patient had: (1) documented ulcerative colitis (UC) or Crohn's disease (CD), based on classical medical history, endoscopic and/or surgical findings, and definite documented histopathology consistent with IBD at Rush University; (2) endoscopically inactive or mild disease at the time of the study for at least 2 weeks, as determined by a UC disease activity index of <4 (a modified Powell–Tuck index) or a Harvey–Bradshaw activity index <5 for CD patients. All patients were required to have stable dietary habits over the past 1 year before enrollment. The extensive exclusion criteria are found in the supportive Supplementary Table S1 online and include use of any antibiotics within the past 30 days and many comorbid conditions. While not part of the inclusion or exclusion criteria, review of patient records showed that our patients were not on any probiotics or antibiotics within the past 3 months before the study, and more than 1 year had passed since any of our IBD patients had a BP for an endoscopic procedure. Eight IBD subjects (n=5 for CD; n=3 for UC) and 10 non-IBD asymptomatic healthy volunteers (HC subjects) were enrolled in the study. All UC subjects had pancolitis, 1 CD subject had Crohn's colitis only, and the other 4 CD subjects had ileocolonic disease location. A majority (75%) of the IBD patients were on 5-aminosalicylates, and 5/8 (63%) were on immunosuppressive medications (with one patient on a stable dose of steroids, three patients on purine analogs, and one patient on a tumor necrosis factor inhibitor). Seven of the eight IBD patients were in remission and one subject with CD had mildly active disease. All patients except two had excellent or good BPs: One patient in the IBD group and one patient in the HC group had fair but adequate BPs. One UC subject had mild patchy disease in the descending colon on endoscopy and histopathology. Three CD patients had endoscopic involvement with disease at the time of colonoscopy: one of these patients had moderate ileocolonic anastomotic disease and few aphthae in the rectosigmoid. Of the other two CD subjects who had endoscopic involvement, one had terminal ileal involvement, few punctuate ulcerations in the cecal area, and a few left colon colocolonic anastomotic ulcerations. The other had gross involvement of the terminal ileum only on endoscopy with patchy distortion of vascular pattern and atrophy of the mucosa throughout the colon. Although subjects had mild endoscopic disease, biopsies for this study were obtained from endoscopically normal/non-ulcerated appearing areas of the colon. Additional subject characteristics are shown in Table 1. Clinical characteristics related to IBD subjects are shown in Table 2. Characteristics related to BP and the endoscopic procedures are shown in Table 3. For IBD patients, the histopathological involvement of tissue specimens obtained during colonoscopy is shown in Supplementary Table S2.
Table 1. Demographic characteristics of study subjects.
| Subject characteristics | HC (n=10) | IBD (n=8) | P-value |
|---|---|---|---|
| Age (years) | 55.4 (±8.27) | 49 (±14.45) | 0.3 |
| Gender (male/female) | 1/9 | 4/4 | 0.06 |
| Race | 0.2 | ||
| White (Caucasian) | 5 (50%) | 7 (87.5%) | — |
| African American | 4 (40%) | 1 (12.5%) | — |
| Asian | 1 (10%) | 0 (0%) | — |
| Ethnicity | 0.6 | ||
| Non-Hispanic/Latino | 8 (80%) | 7 (87.5%) | — |
| Hispanic/Latino | 2 (20%) | 1 (12.5%) | — |
| BMI | 32.2 (±8.2) | 22.15 (±5.34) | 0.2 |
| Smoking status | 0.2 | ||
| Current smoker | 0 (0%) | 2 (25%) | — |
| Non-smoker | 8 (80%) | 4 (50%) | — |
| Past-smoker | 2 (20%) | 2 (25%) | |
| Alcohol use | 0.06 | ||
| Non-user of alcohol | 5 (50%) | 1 (12.5%) | — |
| <1 drink per week | 5 (50%) | 2 (25%) | — |
| >1 drink per week | 0 (0%) | 4 (50%) | — |
| >1 drink per day | 0 (0%) | 1 (12.5%) | — |
BMI, body mass index; HC, healthy control; IBD, inflammatory bowel disease.
n (%) or mean±s.d.
Table 2. Special clinical characteristics related to IBD subjects.
| Clinical characteristics and medications | IBD subjects (n=8) |
|---|---|
| IBD type | |
| CD/UC | 5 (62.5%)/3 (37.5%) |
| Medicationsa | |
| Steroids | 1 (12.5%) |
| Oral 5-ASAs | 6 (75%) |
| Purine analogs | 3 (37.5%) |
| TNF inhibitors | 1 (12.5%) |
| Disease activity | |
| HBAI <5 (CD subjects) (remission) | 4 (80%) |
| HBAI <7 (CD subjects) (mild disease) | 1 (20%) |
| HBAI ≥8 (CD subjects) (moderate-to-severe disease) | 0 |
| UAI <3 (UC subjects) (remission) | 3 (100%) |
| UAI ≥4 (UC subjects) (active disease) | 0 |
| Time since diagnosis (years) | 18.3 (±7.2) |
| Time since last flare (weeks) | 5.87 (±2.4) |
| Number of flares since diagnosis | 6.7 (±6.3) |
| Symptoms | |
| Abdominal pain | 1 (12.5%) |
| Blood in stool | 1 (12.5%) |
| Extraintestinal manifestations | 1 (12.5%) |
| Disease behavior (CD subjects) | |
| Inflammatory (B1)/stricturing (B2)/fistulizing (B3)/perianal disease (p) | 2 (40%)/2 (40%)/1 (20%)/1 (20%) |
| Disease location (CD subjects) | |
| Ileal (L1)/colonic (L2)/iIeocolonic (L3)/isolated upper disease (L4) | 0/1 (20%)/4 (80%)/0 |
| Maximal disease extent (UC subjects) | |
| Ulcerative proctitis (E1)/left sided (E2)/pancolitis (E3) | 0/0/3 (100%) |
ASAs, aminosalicylates; CD, Crohn's disease; HBAI, Harvey–Bradshaw activity index; IBD, inflammatory bowel disease; TNF, tumor necrosis factor; UAI, ulcerative colitis disease activity index; UC, ulcerative colitis.
n (%) or mean±s.d.
Numbers may not add up to 100% since some patients were using more than one type of medication.
Table 3. Characteristics related to bowel prep and procedure.
| Characteristic | Healthy (n=10) | IBD (n=8) |
|---|---|---|
| Type of bowel prep | ||
| Polyethylene glycol-based bowel prep | 5 (50%) | 6 (75%) |
| Sodium phosphate-based bowel prep | 5 (50%) | 2 (25%) |
| Subject compliance with bowel prep | ||
| 100% bowel prep completed | 9 (90%) | 7 (87.5%) |
| 50% bowel prep completed | 1 (10%) | 1 (12.5%) |
| Results from bowel prep | ||
| Excellent prep | 3 (30%) | 1 (12.5%) |
| Good prep | 6 (60%) | 6 (75%) |
| Fair prep | 1 (10%) | 1 (12.5%) |
| Mean time between flexible sigmoidoscopy and colonoscopy | 4.4 days | 4.0 days |
IBD, inflammatory bowel disease.
Study design and sample collection
A repeated-measures design was used. Subjects underwent two endoscopic procedures: First, a limited unprepped flexible sigmoidoscopy was performed to 20–25 cm from the anal verge to obtain the pre-BP samples. Second, within a week (<7 days) after the first procedure, a prepped colonoscopy was performed to obtain the post-BP samples. The subjects were specifically asked not to alter their diet between the procedures. None of the subjects took any antibiotics in between or during the procedures. During both procedures, mucosal biopsies were taken from the distal sigmoid at 20 cm from the anal verge and fecal samples were also collected. During the flexible sigmoidoscopy, pre-BP biopsies were obtained at about 20 cm from the anal verge in an area of the colon that appeared pink and was grossly devoid of solid or mucoid stool, with a standard sterile 2.2 mm colonic biopsy forceps. Pre-BP fecal samples were collected during sigmoidoscopy with a sterile disposable Roth net. During colonoscopy, post-BP fecal samples were suctioned into a standard sterile Luken's trap (Allegiance Lukens Tube by Cardinal Health, Dublin, OH) via the scope. During colonoscopy, post-BP biopsies were obtained about 1–2 in away from the prior sampling area in normal looking mucosa, at about 20 cm from the anal verge, with a standard sterile 2.2 mm colonic biopsy forceps. We made sure to sample normal mucosa and did not sample any areas that are grossly inflamed or ulcerated because of the prior biopsies or because of IBD. Both endoscopic procedures were performed using Olympus video scopes (Olympus America, Center Valley, PA). All samples were snap frozen in liquid nitrogen at the time of collection within minutes in the endoscopy room and then stored in a −80 °C freezer until the time of analysis. Subjects were allowed to use their clinically prescribed BP. At the time of the colonoscopy, after BP, subject's medical and surgical and social and family history, medications (drugs taken within the past 24 h, the past week, the past month, and the past 3 months, along with the duration of use of these), type of BP consumed, and the amount of BP consumed were recorded. The quality of the BP was assessed and documented by the physician performing the procedure using the Aronchick scale.18 Subjects with fair or inadequate BPs were excluded from the study. Disease activity of IBD patients were characterized using the Harvey–Bradshaw index for the CD patients19 and a modified Powell-Tuck index for the UC patients.20, 21 Disease location and behavior was characterized using the Montreal classification for CD.22 Maximal disease extent was characterized using the Montreal classification for UC.23 Additionally, endoscopic findings were also recorded along with the endoscopic appearance of the site of sampling. For IBD patients, the endoscopic involvement of the site where the biopsies were obtained, was graded on a scale from 0–3 by the physician performing the endoscopic procedure: 0=no involvement with disease (i.e., normal appearing mucosa); 1=mild colitis (<5 aphthous ulcerations that have a diameter of 5 mm or less, or mild loss of vascular pattern with granularity); 2=moderate colitis (multiple small and large aphthous ulcerations with edema, erythema, loss of vascular pattern, and contact friability); and 3=severe colitis (majority of the mucosa ulcerated with small and large ulcerations, and spontaneous friability).
Sequencing
Total genomic DNA was extracted from tissue and fecal samples using the FastDNA Spin Kit for Soil (MP Biomedicals, Solon, OH), as per the manufacturer's instructions. The adequacy of the amount of extracted DNA from samples was verified with fluorometric quantitation (Qubit, Life Technologies, Grand Island, NY; 14072). 28F forward primer, 5′-GAGTTTGATCNTGGCTCAG-3′ and 519R reverse primer, 5′-GTNTTACNGCGGCKGCTG-3′ were used to pyrosequence the 16S rRNA gene on a 454 GS FLX platform (454 Life Sciences/Roche, Branford, CT), with barcoding, using Titanium Kits (454 Life Sciences/Roche).24, 25
Sequence processing and quality assessment was performed using custom C++ and python scripts as well as python scripts in the Quantitative Insights Into Microbial Ecology (QIIME) software pipelines (VirtualBox versions 1.5 and 1.6) (http://qiime.org).26, 27 Two sequence runs were performed with subjects of both groups in each run. The sequence outputs were filtered for low-quality sequences (defined as any sequences that are <200 or >1,000 bps, sequences with any nucleotide mismatches to either the barcode or primer, sequences with homopolymer runs >6, sequences with an average quality score of <25, and sequences with ambiguous bases) and were truncated at the reverse primer. Sequences were denoized using USEARCH,28 and chimera checked with UCHIME.29 Operational taxonomic units (OTUs) were picked using uclust28 at a 97% similarity level, and representative sequences were selected. Sequences were aligned with PyNAST30 and filtered alignments were generated in QIIME. Taxonomy assignment was performed in QIIME 1.6VB against the QIIME 1.6 version of Greengenes database31 using the RDP (Ribosomal Database Project) classifier32 at a 80% confidence threshold.
Sixty-six samples (36 tissues and 30 fecal samples) were analyzed. We obtained 368,283 raw sequences, and 104,645,122 raw bases with an average of 5,580 sequences per sample at an average length of 284 bps per sequence in two separate runs on a 454. After quality filtering (as described above), 187,275 total sequences and an average of 2,837 sequences per sample were available, which were denoised, greater than 250- bp-long, demultiplexed, reverse-primer-truncated and chimera-filtered for the rest of the analysis. The sequences were rarified to the minimum number of high-quality sequences (n=1,016) in all samples and normalized by total count for the α- and β-diversity analyses conducted below.
Statistical analyses
Richness
Richness was calculated at the OTU level. Richness reflects the number of unique taxa that are found within a given sample and tries to answer the question: “How many different bacteria are found in the sample?”. QIIME VB1.5 was used to generate α-rarefaction curves for richness.
Evenness
Evenness was calculated at the OTU level. Evenness reflects how uniformly the unique taxa found in a given sample are distributed and tries to answer the question: “Are there some bacteria that are dominating the sample or are the various different and unique bacteria present in relatively equal amounts within the sample?”. Evenness was calculated on rarified and log-transformed data using Multivariate Statistical Package version 3.13.33
Shannon index
Shannon index (SI) was calculated at the OTU level. The SI is a composite index that takes into account both richness and evenness of the bacterial composition within a sample. Shannon's diversity index was calculated on rarified and log-transformed data using Multivariate Statistical Package version 3.13.33
PD-WT
While richness, evenness, and the SI individually examine the counts of bacterial taxa detected in a sample, these indices cannot inform us about the relatedness of the bacterial taxa within a sample. For example, it is possible that a particular phylogenetic group of bacteria may be getting affected by BP more than any other phylogenetic group. In such a case, the diversity in a sample may be not changing in terms of counts but changing in terms of the phylogenetic relatedness of the bacteria. Phylogenetic diversity-whole tree (PD-WT) metric measures changes in diversity also in relation to the relatedness of bacteria within a sample. QIIME VB1.5 was used to generate α- rarefaction curves for PD-WT.
Proportion of shared OTUs
The proportion of shared OTUs between before and after samples was calculated using the following equation: proportion of shared OTUs=number of shared OTUs between S1 and S2/(number of OTUs in S1+number of OTUs in S2−number of shared OTUs between S1 and S2), with S1 representing pre-BP samples and S2 representing post-BP samples. Shared OTUs were calculated in QIIME VB1.5 and Microsoft Excel was used to calculate the proportion.
Unifrac analyses
Unifrac is a distance metric that uses phylogenetic information to measure dissimilarities in microbial community composition between samples.34, 35, 36 To calculate a Unifrac distance, a given sample's phylogenetic composition is mapped onto a phylogenetic tree and is compared with that of another sample. The Unifrac metric is a calculation of the branch length of the phylogenetic tree that is shared between two samples. If the value of Unifrac is 1, the two samples compared are as unrelated as they can be in terms of their bacterial composition. As Unifrac values approach zero, the two samples that are being compared get closer and closer in terms of their bacterial composition. Therefore, in general, the higher the Unifrac distance is, the higher the compositional differences between two samples. An unweighted Unifrac calculation gives equal importance to rare and common bacteria within a sample and is similar to a presence/absence type analysis. Unifrac can be weighted based on the percent abundance of bacteria within a sample. A weighted Unifrac calculation gives a higher importance to the most abundant bacteria in the sample.37 Unweighted and weighted Unifrac distance matrices were calculated in QIIME VB1.5.
Ordination analyses
Principal coordinate analysis (PCO) is an ordination technique that uses calculated distances between groups/or samples in terms of bacterial composition, and arranges them in an unsupervised manner into two or three visual axes, based on how the variability within the sample distances are explained. We used PCO to order samples based on their Unifrac distances and then presented this order in the form of three-dimensional graphs. On PCO graphs, each sample is represented by a dot. Differential coloring of the samples on the graph allows one to see if samples with a particular characteristic are clustering together or are lying far apart. Samples with very different bacterial taxonomic compositions are far apart on the graphs, and samples that contain similar bacteria are closer together. QIIME VB1.5 and KingViewer KiNG (Kinemage Next Generation) Display Software (Richardon Lab, Duke University, Durham, NC) were used to perform PCOs and generate graphs.
Indicator species analysis
Indicator species analysis (ISA)38 is a nonparametric procedure that is adjusted for multiple comparisons and is useful for distinguishing groups based on both relative frequency and abundance of particular bacterial taxa, reflecting the bacterial composition within a sample. Indicator species are species that, due to their niche preferences, can be used as ecological indicators of community or habitat types, environmental conditions, or environmental changes (such as BP perturbation in this study). The aim is to identify those species that show high fidelity to a particular group or environmental condition, and in our case, to the pre- or post-BP state, and as such serve as an indicator for the latter. ISA was performed at the phylum and genus levels in PC-ORD (MjM Software Design, Gleneden Beach, OR) using the Dufréne and Legendre method and the significance of each indicator value (observed maximum indicator value for each species) was tested by the Monte Carlo randomization test at 4,999 permutations. Secondary ISA analyses were also conducted at class, order, and family levels to verify the extent of changes along phylogenetic lineages for various bacterial taxa.
Additional graphs were made using Graph Pad Prism V6 (GraphPad Software, La Jolla, CA) and Microsoft Excel and Adobe Illustrator. SPSS v.19 (SPSS, Chicago, IL) was used to conduct independent and paired t-tests, and nonparametric tests as necessary.
RESULTS
Study subjects
Ten HC subjects and eight IBD subjects (three subjects with UC and five subjects with CD) provided samples for the study. The mean age of the subjects was 52.5 years; 5/18 (28%) were males; 12/18 (67%) were White; 5/18 (28%) were African American; and 3/18 (17%) were Hispanic. Subject characteristics have been further outlined in the Methods section and are also shown in Tables 1, 2, 3 and Supplementary Table S2.
Changes in microbiota composition within samples (i.e., α-diversity)
α-Diversity measures the diversity within a particular ecosystem or environment. In our case, it measures diversity in the bacterial composition within each of our samples or sample groupings. We analyzed α-diversity using OTU-based methods as well as a phylogenetic tree-based method: the α-diversity metrics examined were richness, evenness, and the Shannon diversity index, which are count-based, and PD-WT metric, which is tree-based. We performed three comparisons for each index. First of all, we compared the differences between the pre- vs. post-BP biopsy samples and pre- vs. post-BP fecal samples (which we termed comparisons-across-bowel-prep). Second, as previous data suggested that bacterial communities on the mucosal surface are different than those found in the luminal compartment,39 we examined the effect of BP on the expected separation of the mucosal vs. the luminal compartments (which we termed comparisons-across-compartments). Third, we compared the baseline differences between the HC vs. IBD groups (a secondary analysis), and examined whether the differences between the HC vs. IBD changed after BP, because the effect of BP could differ owing to the inherent differences between a healthy colon or a colon with colitis.
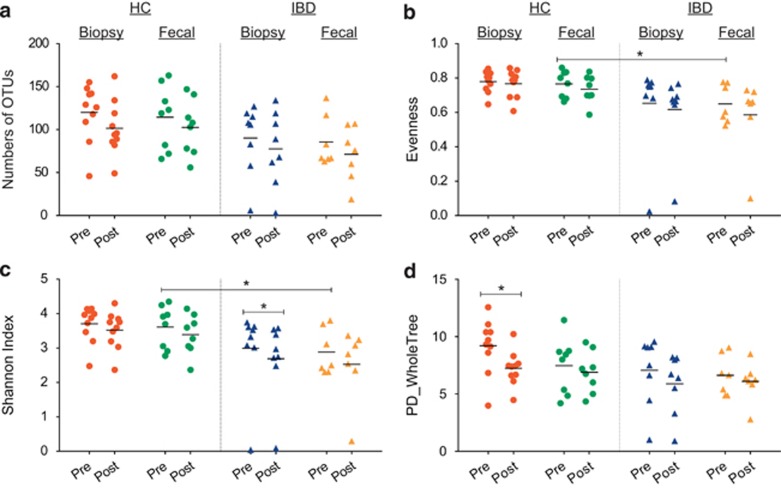
The differences in richness, i.e., mean number of OTUs are shown in Figure 1a; the differences in evenness are shown in Figure 1b; the differences in the SI are shown in Figure 1c; and the differences in PD-WT are shown in Figure 1d. Supplementary Figure S1 shows a comparison of the changes in each of the diversity indices of each individual subject pre- and post-BP. As can be seen in the graphs, the direction of change in PD-WT metric post-BP closely paralleled that of OTU richness in both the biopsy and fecal samples in both the IBD and HC subjects. The changes due to BP in the SI and evenness were more subtle in general, especially for the biopsy samples.
Figure 1.
Operational taxonomic unit (OTU)-based diversity indices of pre- and post-bowel preparation (BP) samples. (a) Richness of OTUs pre- and post-BP. Each marker represents a single sample. The lines represent the mean value for each sample group. The color red represents biopsy samples of healthy controls (HCs). The color green represents fecal samples of HCs. The color blue represents biopsy samples of inflammatory bowel disease (IBD) subjects. The color yellow represents the fecal samples of IBD. In comparisons-across-bowel-prep, a trend towards reduced richness in samples collected post-BP is seen in both HC and IBD groups, but the differences are not statistically significant (P>0.05). (b) The evenness pre- and post-BP. There was a difference in the evenness between pre-BP fecal samples of HC vs. IBD subjects (P=0.033), and this difference was not apparent after BP. (c) The Shannon's diversity index pre- and post-BP. The baseline (pre-BP) Shannon's index for the fecal samples of IBD was significantly lower compared with those for HCs (P=0.04). This difference was not apparent after BP. Although there was a trend towards reduced Shannon's indices in the post-BP samples (compared with pre-BP samples) in general, this reduction was only significant in the biopsy samples of the IBD subjects (P=0.025, paired t-test). (d) The phylogenetic diversity-whole tree (PD-WT) index pre- and post-BP. The effect of BP was more pronounced in the biopsy samples, especially in the HC group (P=0.021, related-samples sign test). The apparent lesser effect on fecal samples may be explained by the greater inconsistency in the direction of response to BP among the fecal samples.
In comparisons-across-bowel-prep, there was a trend towards reduced mean number of OTUs (i.e., richness) in the biopsy and fecal samples in both the HC and IBD subjects following BP (P>0.05 for all). Evenness did not change (P>0.05 for all). This may stem from the variable effects of BP on richness and evenness in individual patients and the inconsistency in the direction of change due to BP, in both HC and IBD groups and in both fecal and biopsy samples. In comparisons-across-bowel-prep, the SI decreased only in the biopsy samples of IBD subjects post-BP (P=0.025). There was also a significant decrease in PD-WT in biopsy samples overall (P=0.001, related-samples sign test), but not in the fecal samples. When IBD and HC biopsy samples were separately examined for the effect of BP, the PD-WT metric was significantly lower after BP in the biopsy samples of HC subjects (P=0.021, related-samples sign test), and there was a significant trend for the biopsy samples of IBD subjects (P=0.07, related-samples sign test). The changes in phylogenetic diversity due to BP were also not consistent in all patients: the reduction in phylogenetic diversity was seen in 9/10 biopsy samples in the HC subjects and 6/8 biopsy samples in the IBD subjects (Supplementary Figure S1). Furthermore, when comparing phylogenetic α-diversity changes in fecal samples and biopsy samples of each individual before and after BP, the direction of change in PD-WT of the biopsy sample was not necessarily congruent to that in the fecal sample of the same individual.
In comparisons-across-compartments, no differences were seen for any of the diversity indices examined (P>0.05 for all). However, after BP, in PD-WT, there was a significant correlation between the mucosal and the luminal samples (P=0.023), whereas there was none before BP, in the HC group (P=0.613). Similarly, in the IBD group after BP, there was a significant correlation between the mucosal and luminal samples (P=0.009), whereas there was none before BP (P=0.257).
In secondary comparisons, both fecal and biopsy samples obtained at pre- and post-BP time points from the HC subjects had a numerically higher richness (i.e., mean number of OTUs) compared with samples from IBD subjects (P>0.05 for all). In secondary comparisons, there was also a significant reduction in evenness in the IBD group's pre-BP fecal samples compared with those of HCs (P=0.033). This difference was not observed post-BP (P=0.097). When the biopsy samples were looked at, there was also a trend toward heightened differences in evenness in the HC and IBD samples post-BP (P=0.061). In secondary comparisons, at baseline before BP, the SI in the fecal IBD samples was significantly lower compared with that in the fecal HC samples (P=0.044), suggesting a lower overall diversity in IBD in the feces. This difference was not observed after BP (P=0.067). When the biopsy samples were looked at, the SI in the HC and IBD samples were not different before BP (P=0.118); however, there was a trend toward heightened differences after BP (P=0.056).
Changes in the microbiota composition between samples (i.e., β-diversity)
β-Diversity measures the diversity between environments, i.e., between sample groups. To look at group differences, we calculated shared OTUs between samples, which is based solely on the number count of shared organisms. We also used the Unifrac distance, a phylogenetic tree-based method, to assess β-diversity (to not only understand shared bacterial counts but also to look for changes in phylogenetic relatedness of the samples). Similar to our α-diversity analyses, we performed two calculations for each of the shared OTUs and the Unifrac distance: First, we calculated the two measures looking at paired biopsy or fecal samples before vs. after prep in each of our study groups (comparisons-across-bowel-prep). Then, we calculated the two measures looking at the mucosal and luminal compartments with pairing the biopsy vs. stool samples, collected pre-BP and post-BP (comparisons-across-compartments).
Shared OTUs between samples
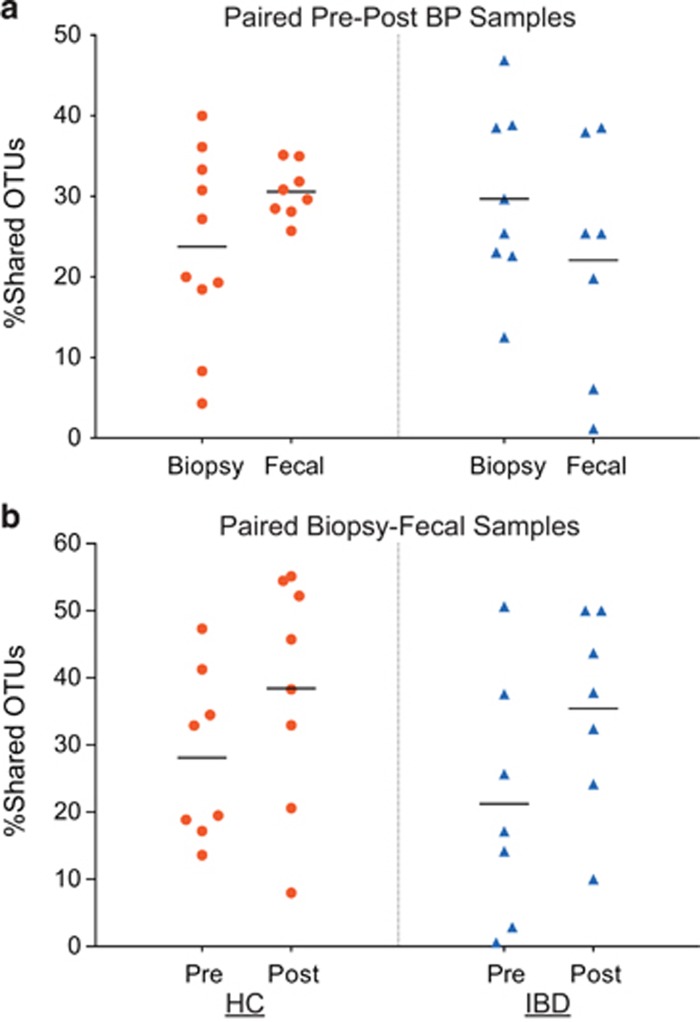
In comparisons-across-bowel-prep, the proportions of shared OTUs between paired samples pre- and post-BP are shown in Figure 2a. The proportion of shared OTUs between pre- and post-BP was around a mean of 30% (range=4.3–49.60% for biopsies; range=1.17–38.51% for fecal samples) in both the HC and IBD groups. This finding suggests a change of ~70% of OTUs post-BP. This effect was not specific to a particular sample type, or the presence of a disease state: There were no statistical differences between the HC and IBD subjects in the proportions of shared OTUs between pre- and post-BP for either biopsy or fecal samples (P>0.05, all).
Figure 2.
The proportion of shared operational taxonomic units (OTUs) between samples. (a) Shared OTUs between the pre- and post-bowel prep (BP) samples in the healthy control (HC) and inflammatory bowel disease (IBD) groups. Each marker plotted represents the proportion of shared OTUs between paired samples (before and after BP) of the same subject. Red markers represent shared OTUs in HCs. Blue markers represent shared OTUs in the IBD subjects. The lines represent the mean values for the proportion of shared OTUs. The proportion of shared OTUs between pre- and post-bowel prep samples in all sample sets was low (~20–30%). (b) Shared OTUs between the mucosal and luminal compartments (i.e., biopsy and fecal samples) collected at the same visit in the HC and IBD groups. Each marker plotted represents the proportion of shared OTUs between paired samples (fecal and biopsy samples in the same visit) of the same subject. Red markers represent shared OTUs in HCs. Blue markers represent shared OTUs in the IBD subjects. There was an increase in the proportion of shared OTUs between the biopsy and fecal samples after BP (P=0.016).
In comparisons-across-compartments, the proportions of shared OTUs between fecal and biopsy samples within the same visit are shown in Figure 2b. The average proportion of shared OTUs between biopsy and fecal samples collected after BP was greater than the average proportion of shared OTUs between biopsy and fecal samples collected before BP (mean proportion of shared OTUs=37.0% vs. 24.9%, respectively, P=0.016), suggesting that mucosal samples and fecal samples became more similar after BP for the entire study group. However, when IBD and HC samples were separately examined, there were no differences when the shared OTUs in biopsy and fecal samples were compared pre- and post-BP. Furthermore, the direction of change in the number of shared OTUs and the % of shared OTUs between sample types because of BP was not consistent in all of the subjects (Supplementary Table S3).
Comparison of bacterial composition using Unifrac distances
We used both unweighted and weighted Unifrac distance measures to obtain a better insight into the nature of community differences in our sample set.
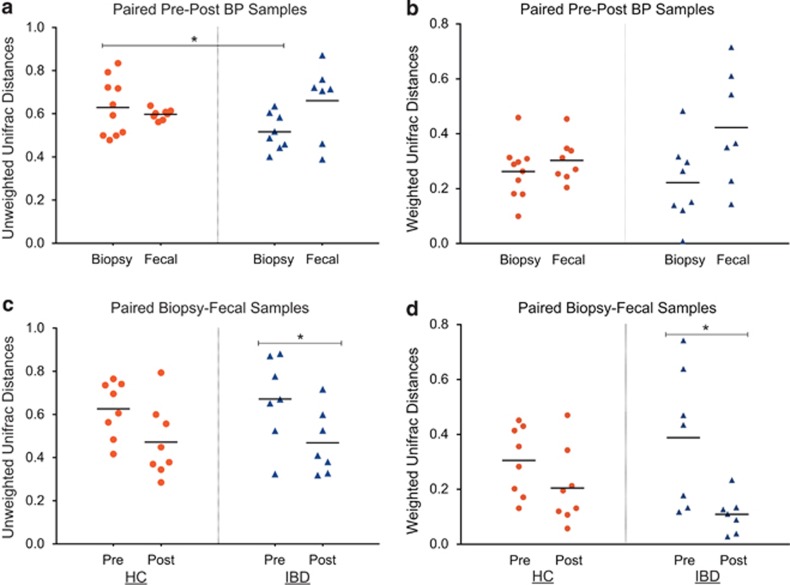
Unweighted Unifrac results: Unweighted Unifrac distances between pre- and post- BP samples are given in Figure 3a. The mean-unweighted Unifrac distances between paired pre- and post-BP samples ranged from 0.52 to 0.66 in the IBD and HC groups in their fecal and biopsy samples, suggesting an average of 50–60% dissimilarity of the samples overall. The fecal samples of the IBD group had the most dissimilarity, whereas the biopsy samples of the IBD group had the least dissimilarity pre- and post-BP. This was in contrast to the HC group in which the biopsy samples appeared to be affected more than the fecal samples by BP. When the distances between biopsy samples pre- and post-BP were statistically compared, the mean-unweighted Unifrac distance in biopsies was significantly greater in the HC group, compared with the IBD group (mean-unweighted Unifrac distance=0.629 vs. 0.5165, HC vs. IBD, respectively, P=0.04, t-test), suggesting a significant effect of BP on the mucosal surface in non-colitis patients over and above that is seen in IBD subjects. When the distances between fecal samples pre- and post-BP were compared, the mean-unweighted Unifrac distance in the fecal samples was not different in the IBD subjects, compared with the HCs (mean-unweighted Unifrac distance=0.597 vs. 0.66, HC vs. IBD, respectively, P=0.37, t-test).
Figure 3.
Unifrac distances between paired samples. (a) The unweighted Unifrac distances between the pre- and post-bowel prep (BP) samples in the healthy control (HC) and inflammatory bowel disease (IBD) groups. Each marker plotted represents the Unifrac distance between paired pre- and post-BP samples of the same type for the same subject. Red markers represent Unifrac distances in the HCs. Blue markers represent Unifrac distances in the IBD subjects. The lines represent the mean Unifrac distances. The unweighted Unifrac distances between the pre- and post-BP biopsy samples was significantly greater in the HC group, when compared with the IBD group (P=0.04). (b) The weighted Unifrac distances between the pre- and post-BP samples in the HC and IBD groups. (c) The unweighted Unifrac distances between the mucosal and luminal compartments (i.e., biopsy and fecal samples) collected at the same visit in the HC and IBD groups. Overall, the mean-unweighted Unifrac distance between the biopsy and fecal samples decreased after bowel prep (P=0.003). This reduction in unweighted Unifrac distances between sample types because of bowel prep was significant in the IBD group (P=0.03), but not in the HC group (P=0.068). (d) The weighted Unifrac distances between the mucosal and luminal compartments (i.e., biopsy and fecal samples) collected at the same visit in the HC and IBD groups. Although there was a trend towards reduced weighted Unifrac distances between the mucosal and luminal compartments after BP, this reduction was only significant in the IBD group (P=0.036).
When the effects on the differences between the mucosal and luminal compartments were compared pre- and post-BP, the mean-unweighted Unifrac distances between biopsy and fecal samples decreased overall after BP (mean-unweighted Unifrac distance=0.647 vs. 0.470, pre-BP mucosal-luminal distance vs. post-BP mucosal–luminal distance, respectively, P=0.003) compared with before BP, suggesting that the composition of the fecal samples approached that of the biopsies following a BP. When the HC and IBD groups were examined separately, the reduction of the unweighted Unifrac distances between biopsy-fecal samples was not significant in the HC group (mean-unweighted Unifrac distance=0.626 vs. 0.472, pre-BP mucosal–luminal distance vs. post-BP mucosal–luminal distance, respectively, P=0.068), whereas there was a significant reduction in the IBD group (mean-unweighted Unifrac distance=0.672 vs. 0.469, pre-BP mucosal–luminal distance vs. post-BP mucosal–luminal distance, respectively, P=0.03) (Figure 3c).
Weighted Unifrac results: Weighted Unifrac distances between samples obtained pre- and post-BP are given in Figure 3b. The range of mean-weighted Unifrac distances was 0.22 to 0.42 between paired biopsy and fecal samples either before or after BP in the HC and IBD groups, suggesting an average of 20–40% dissimilarity of the samples overall. This overall level of dissimilarity is less than what was observed in terms of unweighted Unifrac distances, suggesting that the rarer taxa are likely to be affected more by BP. The fecal samples of the IBD group had the most dissimilarity, whereas the biopsy samples of the IBD group had the least dissimilarity pre- and post-BP. This was in contrast to the HC group, where both biopsy and fecal samples appeared to be equally affected by BP. The weighted Unifrac distances between pre- and post-BP samples were not statistically significantly different between the HC and IBD groups for both biopsy and fecal samples (P>0.05, all), despite slightly higher numerical weighted Unifrac values in the fecal samples from the IBD group.
Mucosal surface vs. luminal compartment differences were compared by looking at weighted Unifrac distances between the biopsy and the fecal samples collected within the same visit (Figure 3d). In the HC group, the mean-weighted Unifrac mucosal–luminal distance pre-BP was numerically greater than that for the samples collected after BP (mean-weighted Unifrac distance=0.3 vs. 0.2, pre-BP mucosal–luminal distance vs. post-BP mucosal–luminal distance, respectively, P=0.162). In the IBD patients, there was a significant reduction in the weighted Unifrac mucosal–luminal distance after BP (mean-weighted Unifrac distance=0.388 vs. 0.1, pre-BP mucosal–luminal distance vs. post-BP mucosal–luminal distance, respectively, P=0.036). This finding is similar to the above observations with unweighted Unifrac, and suggests that both abundant taxa and rare taxa unique to each compartment are affected by BP in the IBD group.
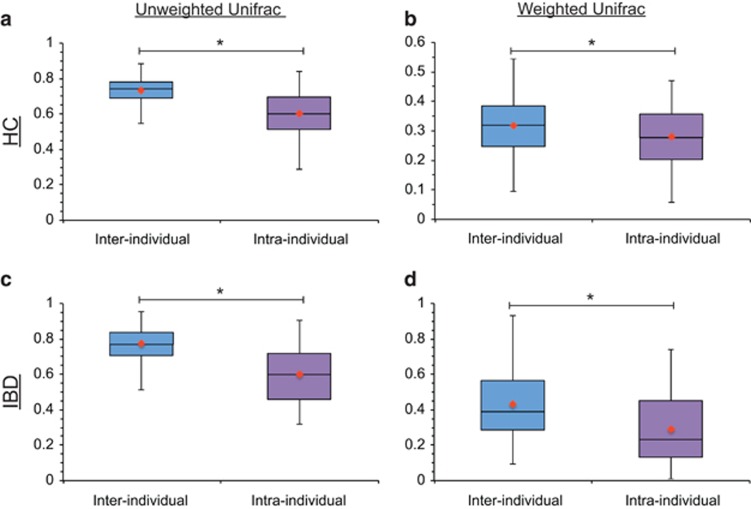
The magnitude of BP effect on Unifrac distances: To gain a deeper insight into the magnitude of the BP effect on β-diversity and to determine whether the effect of BP is greater than the differences between individual subjects, we compared intra- and inter-individual Unifrac distances (Figure 4). The differences between intra- and inter-individual unweighted Unifrac distances were preserved in both the HC and the IBD group (P<0.001, for both groups). The differences between intra- and inter-individual weighted Unifrac distances were also preserved in both the HC and the IBD group (P=0.005 and <0.001, respectively). These findings indicate that even though our results suggest that BP induces a lot of changes in the colonic microbiota, these changes were not sufficient to eliminate differences between individuals in our sample set.
Figure 4.
Unifrac distances between samples from the same subject (i.e., intra-individual differences) vs. those from other subjects (i.e., inter-individual differences). (a) The unweighted Unifrac distances between samples of the same subject (intra-individual distances) vs. the unweighted Unifrac distances between samples of different subjects (inter-individual distances) in the healthy control (HC) group (P⩽0.0001). (b) The weighted Unifrac distances between samples of the same subject (intra-individual distances) vs. the weighted Unifrac distances between samples of different subjects (inter-individual distances) in the HC group (P=0.0047). (c) The unweighted Unifrac distances between samples of the same subject (intra-individual distances) vs. the unweighted Unifrac distances between samples of different subjects (inter-individual distances) in the inflammatory bowel disease (IBD) group (P≤0.0001). (d) The weighted Unifrac distances between samples of the same subject (intra-individual distances) vs. the weighted Unifrac distances between samples of different subjects (inter-individual distances) in the IBD group (P≤0.0001). The inter-individual distances were significantly greater than the intra-individual distances for all comparisons. The line in each box represents the median and the orange marker represents the mean.
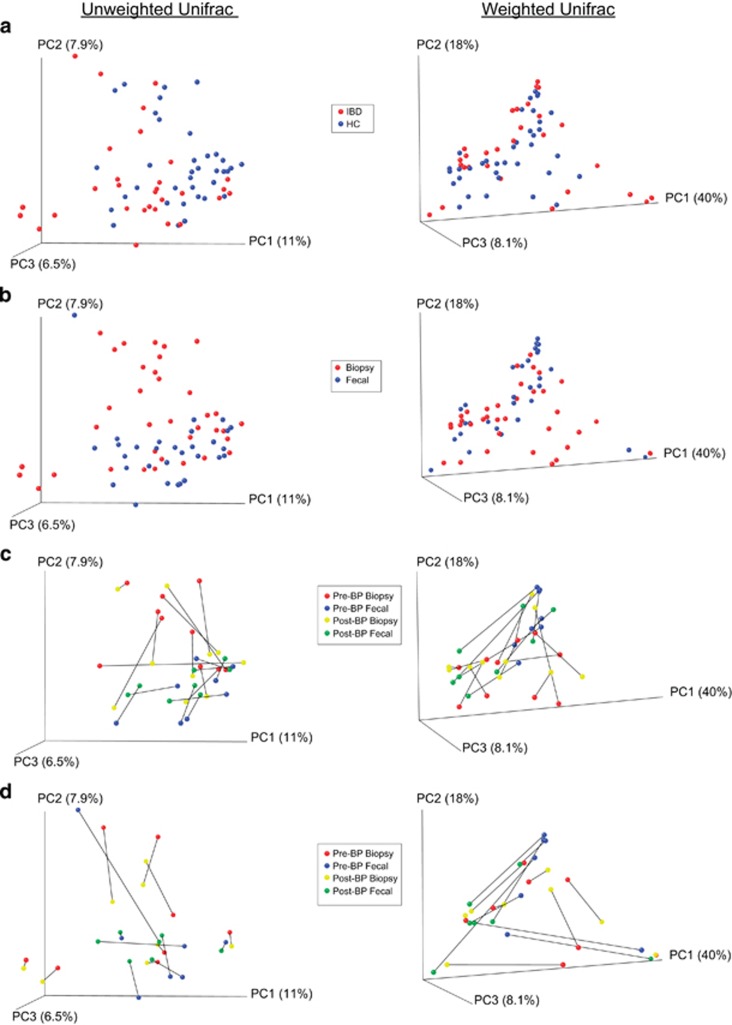
Ordination analyses: We performed PCO analyses using both unweighted and weighted Unifrac distances and examined whether samples cluster by demographics such as age; gender; race; ethnicity; body mass index; any history of gastrointestinal surgery; smoking history; alcohol consumption history; BP type (sodium phosphate-based prep vs. polyethylene glycol-based prep); BP quality rating at colonoscopy; sampling interval between the two study procedures (Supplementary Figures S2 and S3); and the presence of disease (HC vs. IBD) (Figure 5a). There were no visual differences noted in the graphs except for sample type (Figure 5b): biopsies were separated from the fecal samples when examined with unweighted Unifrac distances, suggesting that rare taxa between the two sample types differ. This graphical difference was also confirmed with ANOSIM (analysis of similarity; R-statistic=0.0666, P=0.003, permutations=1,000).
Figure 5.
Principal coordinate analysis (PCoA) plots of samples by Unifrac distances. (a) Plots of samples collected from both groups (healthy control (HC) and inflammatory bowel disease (IBD)) colored by disease state. Each marker denotes a single sample. PCoA plots for the unweighted Unifrac distances are shown on the left, whereas the PCoA plots for the weighted Unifrac distances are shown on the right. (b) Plots of all the samples colored by sample type (biopsy vs. fecal). (c) Plots of paired pre- and post-bowel prep samples of the HC group only. (d) Plots of paired pre- and post-bowel prep samples of IBD group only. The lines in (c and d) connect paired pre- and post-bowel prep samples of the same type for the same individual subject.
When we looked at the effect of BP on the samples, when all samples were examined, there was some clustering of the pre-BP biopsy samples. When broken down by the presence of disease, the difference lessened. Interindividual variability in patients was close to the variability induced by BP for the HC patients (Figure 5c), whereas in the IBD group, the interindividual variability visually was larger than the variability induced by BP (Figure 5d). As can be seen in the Figure 5c, the magnitude of the effect of BP was variable among the subjects (as indicated by the different lengths of lines between the corresponding pre- and post-BP samples) in both groups. ANOSIM using unweighted Unifrac distances showed a statistically significant difference between the pre- and post-BP biopsy samples in the HC group (R-statistic=0.1222, P=0.005, permutations=1,000), and between the pre- and post-BP fecal samples in the IBD group (R-statistic=0.1497, P=0.05, permutations=1,000). ANOSIM using weighted Unifrac distances showed that the fecal samples in both the HC and the IBD patients were affected by BP (R-statistic=0.3337, P=0.012, permutations=1,000 for comparing pre- and post-BP fecal samples of the HC group; R-statistic=0.2070, P=0.026, permutations=1,000 for comparing pre- and post-BP fecal samples of the IBD group), whereas the biopsy samples were not affected.
Examination of the taxonomic composition of samples
A total of 16 phyla were identified in our samples, out of which 6 phyla were present in 50% or more of our samples. Five of them were present in the samples at an average abundance of >1%. These five major phyla in order of abundance are: Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Tenericutes. As expected, Firmicutes and Bacteroidetes made up the majority of the microbial composition in the samples. The relative bacterial composition of the samples are shown in Supplementary Figures S4–S7.
Indicator species analysis
We performed an ISA38 to determine whether the observed differences after BP were in particular bacterial taxa or reflect global changes in many of them.
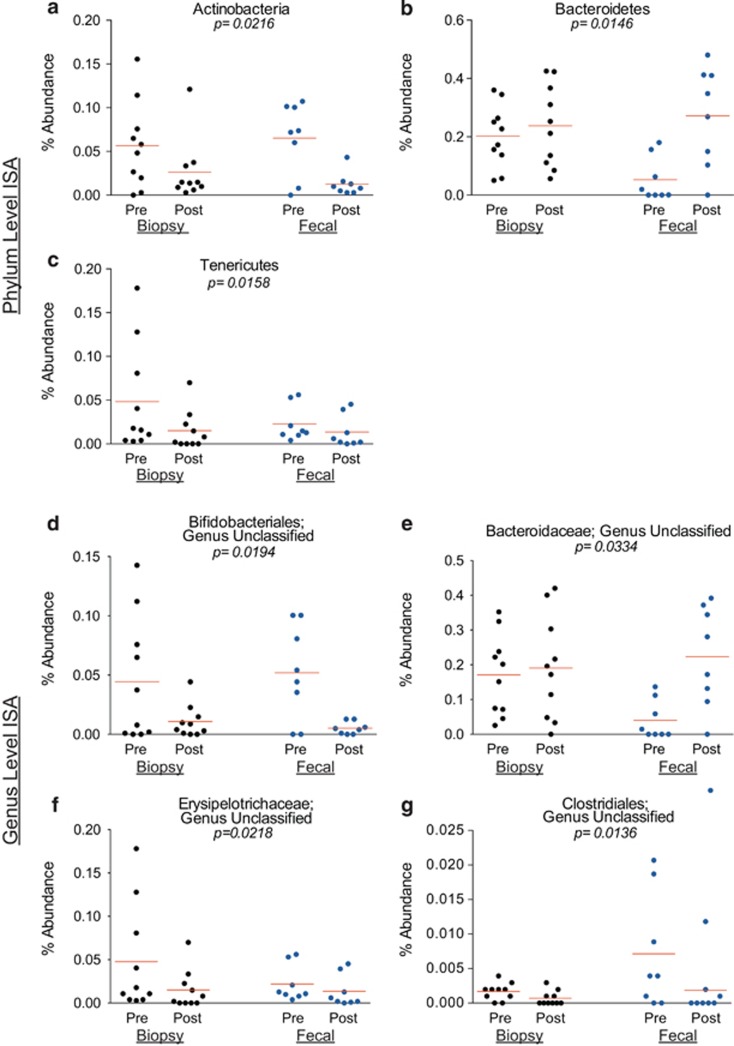
ISA of samples collected from HC subjects: In the HC group, after controlling for sample type, we detected 17 phylotypes as significant indicators of either the pre- or the post-BP state (Table 4). BP had a global effect on three major phyla (Figure 6): Actinobacteria and Tenericutes were indicators of the pre-BP state; and Bacteroidetes was detected as an indicator for the post-BP state, with a greater effect noted in the fecal samples. BP had a global effect on four genera (Figure 6). Furthermore, when differences in fecal vs. biopsy samples were examined, the class Epsilonproteobacteria was detected as an indicator for the pre-BP state in the biopsy samples only (P=0.0096, observed indicator value=40).
Table 4. Significant indicator species for sampling time (pre- vs. post-bowel prep) controlled for sample type in healthy subjects.
| Phylum | Class | Order | Family | Genus | Observed indicator value | P-value | Effect of bowel prep |
|---|---|---|---|---|---|---|---|
| Actinobacteriaa | 67.6 | 0.022 | Decreased | ||||
| Actinobacteria | 67.6 | 0.030 | Decreased | ||||
| Bifidobacteriales | 66.6 | 0.022 | Decreased | ||||
| Bifidobacteriales unclassified | 66.6 | 0.019 | Decreased | ||||
| Bifidobacteriales unclassified | 66.6 | 0.019 | Decreased | ||||
| Bacteroidetesa | 65.1 | 0.015 | Increased | ||||
| Bacteroidia | 64.7 | 0.018 | Increased | ||||
| Bacteroidales | 64.7 | 0.014 | Increased | ||||
| Bacteroidaceae | 61.1 | 0.033 | Increased | ||||
| Bacteroidaceae unclassified | 61.1 | 0.033 | Increased | ||||
| Tenericutesa | 69.5 | 0.016 | Decreased | ||||
| Erysipelotrichi | 68.9 | 0.018 | Decreased | ||||
| Erysipelotrichales | 68.9 | 0.016 | Decreased | ||||
| Erysipelotrichaceae | 68.9 | 0.021 | Decreased | ||||
| Erysipelotrichaceae unclassified | 68.9 | 0.022 | Decreased | ||||
| Clostridiales unclassifieda | 64.8 | 0.012 | Decreased | ||||
| Clostridiales unclassified | 58.4 | 0.014 | Decreased |
Denotes the highest level of taxonomy at which changes occur.
Figure 6.
Relative abundances of significant indicator phyla and genera in healthy subjects. Each marker represents the relative (%) abundance of the denoted phylum or genus in one sample. Each panel shows the relative (%) abundance in the pre- and post-bowel prep samples collected from healthy subjects. The lines represent the mean relative abundance. Black circles represent the biopsy samples and blue circles represent the fecal samples. (a) The relative abundance of the phylum Actinobacteria (an indicator of the pre-bowel prep (BP) state). (b) The relative abundance of the phylum Bacteroidetes (an indicator of the post-BP state). (c) The relative abundance of the phylum Tenericutes (an indicator of the pre-BP state). (d) The relative abundance of an unclassified genus within the Bifidobacteriales order (an indicator of the pre-BP state). (e) The relative abundance of an unclassified genus within the Bacteroidaceae family (an indicator of the post-BP state). (f) The relative abundance of an unclassified genus within the Erysipelotrichaceae family (an indicator of the pre-BP state). (g) The relative abundance of an unclassified genus within the Clostridiales order (an indicator of the pre-BP state). ISA, indicator species analysis.
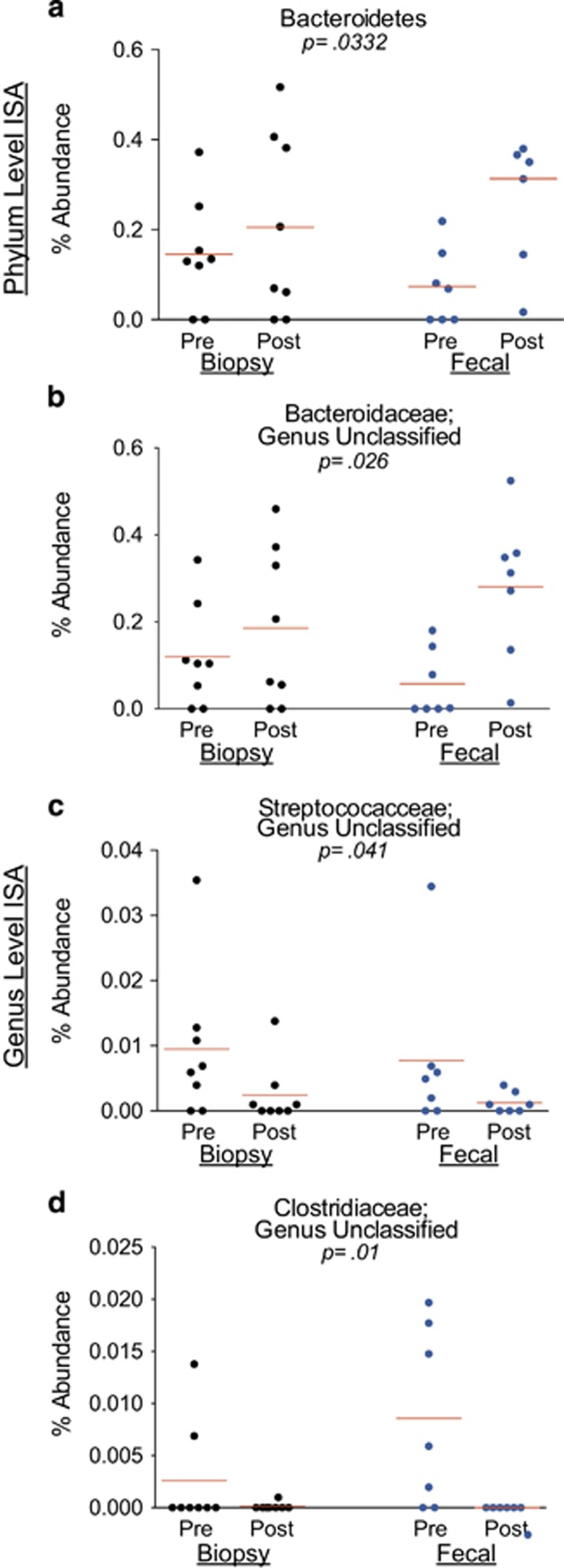
ISA of samples collected from IBD subjects: In the IBD group, after controlling for sample type, we detected 12 phylotypes as significant indicators of either the pre- or the post-BP state (Table 5). BP had a global effect on the phylum Bacteroidetes (Figure 7), which was an indicator of the post-BP state, with a greater effect in the fecal samples. BP had a global effect on three genera (Figure 7).
Table 5. Significant indicator species for sampling time (pre- vs. post-bowel prep) controlled for sample type in IBD subjects.
| Phylum | Class | Order | Family | Genus | Observed indicator value | P-value | Effect of bowel prep |
|---|---|---|---|---|---|---|---|
| Bacteroidetesa | 61.2 | 0.033 | Increased | ||||
| Bacteroidia | 62.4 | 0.032 | Increased | ||||
| Bacteroidales | 62.4 | 0.027 | Increased | ||||
| Bacteroidaceae | 63.1 | 0.022 | Increased | ||||
| Bacteroidaceae unclassified | 63.1 | 0.026 | Increased | ||||
| Bacillia | 65.1 | 0.028 | Decreased | ||||
| Lactobacillales | 68.5 | 0.016 | Decreased | ||||
| Streptococcaceae | 61.3 | 0.042 | Decreased | ||||
| Streptococcaceae unclassified | 61.3 | 0.041 | Decreased | ||||
| Clostridiaceaea | 45.6 | 0.010 | Decreased | ||||
| Clostridiaceae unclassified | 45.6 | 0.010 | Decreased |
Denotes the highest level of taxonomy at which changes occur.
Figure 7.
Relative abundances of significant indicator phyla and genera in inflammatory bowel disease (IBD) subjects. Each marker represents the relative (%) abundance of the denoted phylum or genus in one sample. Each panel shows the relative (%) abundance in the pre- and post-bowel prep samples collected from IBD subjects. The lines represent the mean relative abundance. Black circles represent the biopsy samples and blue circles represent the fecal samples. (a) The relative abundance of the phylum Bacteroidetes (the only significant indicator phylum in IBD subjects). (b) The relative abundance of an unclassified genus within the Bacteroidaceae family (an indicator of the post-bowel prep (BP) state). (c) The relative abundance of an unclassified genus within Streptococcaceae family (an indicator of the pre-BP state). (d) The relative abundance of an unclassified genus within the Clostridiaceae family (an indicator of the pre-BP state). ISA, indicator species analysis.
Effect of bowel preparation on the Bacteroidetes to Firmicutes ratio
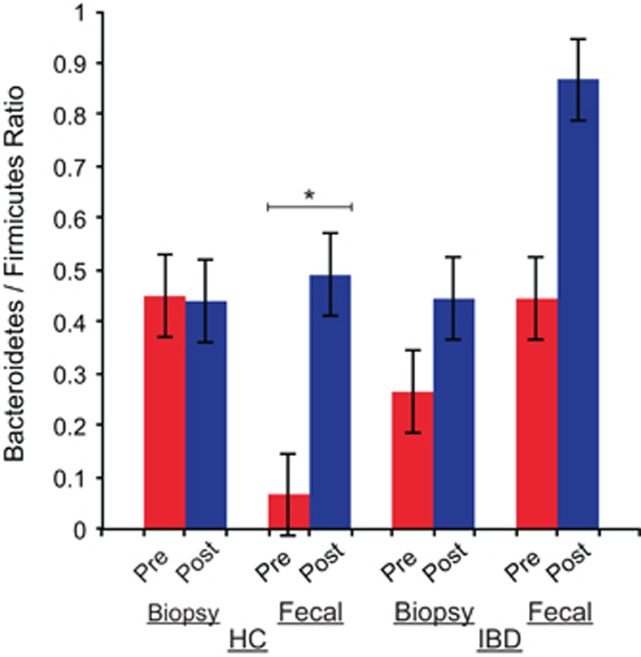
Because we have observed consistent increases in the Bacteroidetes in both our HC and IBD groups in our ISA, we also examined shifts in the ratio of Bacteroidetes/Firmicutes because of BP, considering that this ratio has been previously reported to be important to the study of a variety of disease states such as obesity and diabetes.40, 41, 42 The average Bacteroidetes/Firmicutes ratio increased after BP in the fecal samples of HC subjects (P=0.023, related-samples t-test) (Figure 8).
Figure 8.
Bacteroidetes/Firmicutes ratio in samples. The bar chart shows the ratio of Bacteroidetes to Firmicutes in each sample group and compares the pre-bowel prep samples to the post-bowel prep samples. The average Bacteroidetes/Firmicutes ratio increased after bowel prep in the fecal samples of healthy control (HC) subjects (P=0.023, related-samples t-test). IBD, inflammatory bowel disease.
DISCUSSION
The development of high-throughput sequencing technology and consequent massive reductions in time and financial costs of sequencing in the past 10 years has initiated a new era in microbial research focused on understanding the gut microbial ecosystem and how it is altered in health and in disease states. One important gut ecosystem perturbation, not only clinically but also in terms of obtaining undistorted samples for meaningful research, is that induced by BP. We found that BP causes qualitative and quantitative changes in the bacterial composition in both biopsies and fecal samples, i.e., in both the mucosa-associated and the luminal microbiota compartments of the colon. Such changes in both compartments highlight the need to pay careful attention to sample collection procedures in studies of the colonic microbiome.
There was a trend for reduction in α-diversity after BP (in comparisons-across-bowel-prep); however, the effects were variable in individual patients and did not reach statistical significance for most metrics. Furthermore, the changes in one sample type (i.e., biopsy vs. fecal samples) did not appear to mirror what was seen in the other sample type in individual subjects. When apparent, the reductions in α-diversity were seen in composite indices, mostly in the mucosa-associated compartment, and the type of metric that was altered post-BP varied in health and disease: a reduction in PD-WT was seen in the biopsies of HC subjects, and a reduction in the SI was seen in the biopsies of IBD subjects after BP. It appears that in health i.e., when a sample is inherently expected to be highly diverse compared with a disease state,43, 44, 45, 46 changes in α-diversity can be most noticeable with loss of rare phylotypes at high levels of taxonomy, resulting in reductions in the PD-WT metric. In a disease state similar to IBD, i.e., when a sample is expected to have reduced diversity inherent to the presence of disease,46, 47, 48, 49, 50, 51, 52, 53, 54, 55 BP-related perturbations appear to be more noticeable in count-based composite diversity indices such as the SI, and only trends are observed in the PD-WT metric. In fecal samples reflecting the luminal component, the direction of change in α-diversity with BP was highly variable and less consistent compared with the biopsy samples reflecting the mucosa-associated compartment. In fact, in secondary comparisons, subtle reductions in evenness in IBD (compared with HC) noticeable in the fecal samples before BP was not apparent after BP, suggesting that perturbations of the gastrointestinal tract bacteria with BP could adversely affect experiments studying differences in health vs. colitis with fecal samples obtained at the time of colonoscopy, and can potentially lead to false-negative results. Alternatively, one may require larger sample sets to discover differences between IBD and HCs with such samples.
An important consideration is the magnitude of the short-term effect of BP. Examination of the shared OTUs pre- and post-BP samples revealed that there was little overlap between the two time points. This suggests a massive change of 2/3 of the microbial OTUs within the colonic ecosystem after a seemingly temporary and nonselective perturbation of the ecosystem with BP. This change was not only apparent in the fecal samples as expected but also in the biopsy samples reflecting the mucosa-associated compartment. The latter finding suggests that the short-term effect of BP can be profound on the colon ecosystem at the OTU level. This change also appeared to affect more rare taxa, reflected by numerically higher unweighted Unifrac distances vs. weighted Unifrac distances. In HC, the differences post-BP was most apparent in the biopsy samples and in rare taxa. In IBD, the changes were mostly in the fecal compartment after BP, and appeared to affect both abundant and rare taxa, revealing reductions in both the unweighted and the weighted Unifrac distances.
Notably, the shared OTUs between the luminal and the mucosa-associated components increased after BP in general, making luminal samples similar to the mucosa-associated microbiota, leading to a loss of the expected separation of the bacteria on the mucosal surface vs. that in the lumen. In this regard, the effects of BP differed in health and disease: rare taxa (expected to carry unique functions) and abundant taxa were affected with BP, especially in the IBD group, revealing notable reductions in the unweighted and weighted Unifrac distances between biopsy and fecal samples after BP.
The effect of BP was consistently seen in particular phylotypes: specifically there were significant increases noted in Bacteroidetes and its lineages (across all the phylogenetic levels from the phylum level down to the genus level) after BP in both the healthy state and IBD. Additionally in the healthy state, there were notable reductions in families within the Bifidobacteriales order; in Erysipelotrichaceae; in unclassified families and genera from the Clostridiales; and in an unclassified genus from Campylobacteraceae. In IBD subjects, reductions were seen in the Clostridiaceae and in the Bacilli. Studies examining these particular phylotypes should take into account the effects of BPs. For example, our results suggest that studies examining Bifidobacteriales are less likely to find differences between healthy non-colitis patients and a disease state if conducted with samples obtained post-BP. Similarly, studies conducted with relatively rarer bacteria such as those belonging to unclassified taxa are less likely to find differences between cases and controls, if conducted using samples obtained post-BP. Therefore, the short-term effects with BPs mentioned above suggest that the effects and their magnitude in health and disease have to be taken into account when designing clinical translational studies of the lower gut microbiota.
Our findings also highlight the importance of taking a history of use of bowel cleansing practices, such as use of high-dose laxatives over the counter or colonics, and so on, which may have similar effects on the microbiota. Additionally, some of the changes that we have observed on the mucosal surface such as trends for reduction in diversity, and loss of rare taxa call into question what can be expected from the practice of colon cleansing to promote gut health. For example, in IBD, it is a well-known clinical fact that severe colitis cases can clinically worsen significantly after administration of BP.56 Our findings demonstrating a loss of separation between the mucosal and luminal compartments post-BP in IBD strongly suggest that a barrage of abundant and rare commensal bacteria that are normally kept in the luminal compartment can gain access to the mucosal surface to worsen the inflammatory reaction in this compartment.
Our results partially mirror those seen in prior studies, which have yielded conflicting data about the effects of BP. Prior studies with BP have a large number of limitations: first and foremost, most studies have examined only fecal samples and only one has concomitantly looked at mucosal biopsies and luminal samples.17 Some of these studies have not even sampled control and case subjects at similar intervals.15 It is also notable that most studies have been conducted using relatively insensitive techniques that have lower resolution of bacterial composition such as denaturing gradient gel electrophoresis (DGGE), temporal temperature gradient gel electrophoresis or terminal restriction fragment length polymorphism compared with high-throughput sequencing.11, 12, 13, 16
Furthermore, the subjects in most of these studies have not been well characterized, and the results obtained from samples of patients with gastrointestinal inflammation have been analyzed together with those samples obtained from individuals without colonic inflammation in all the studies to date. All of these differences could certainly account for the variability in the results seen in the published literature. Therefore, our study fills a major void in examining the effects of BP and is the first study that examined IBD and non-IBD cases separately across a consistent sampling interval, has had an adequate number of sequences obtained, and has used a high-resolution technique to study the effects of BP.
Work using low-resolution techniques in fecal samples has demonstrated the following: using culture and DNA hybridization for specific strains of Bacteroides, Morotomi et al.11 studied the effects of BP on the fecal microbiota in nine subjects: no alterations in the bacterial counts or in the percent distribution of 10 Bacteroides strains in the colonic aspirates collected after polyethylene glycol-based BP were seen compared with the fecal samples collected before prep. Similarly, Bibiloni et al.16 compared three specimen types (namely washed biopsy samples after BP, fecal samples obtained during colonoscopy, and fecal samples before BP) from 15 subjects using temporal temperature gradient gel electrophoresis: there were no differences in bacterial fingerprints after BP. Contrary to these, Mai et al.13 examined the fecal microbiota before, at 2–4 weeks, and at 6–8 weeks after colonoscopy with DGGE in five subjects with various endoscopic findings (including polyps and colonic inflammation) and showed that bacterial fingerprint profiles were similar only in 2/5 subjects pre- and post-colonoscopy. Furthermore, in the latter study, when DGGE, fluorescent in situ hybridization with various bacterial primers, and cloning and sequencing techniques were compared, inconsistent results were observed across examination techniques, highly supporting the notion that the inherent resolution of a particular technique significantly impacts the results obtained. Goossens et al.12 studied the effects of BP on the composition of the fecal samples obtained before and after BP with DGGE in 22 subjects and showed that bacterial profile similarities were high with a mean around 86%, and that overall banding patterns did not change significantly but some changes in individual bands occurred, mainly in the first week after BP, suggesting that BP could have an influence on specific bacterial species. When fecal samples obtained before and after colonoscopy were examined by analysis of DGGE profiles, O'Brien et al.15 found that the similarity coefficients between pre- and post-colonoscopy samples were within the range of normal temporal variation in 11/14 of the subjects, whereas the rest (constituting about 21% of the subjects) had short-term changes.
Three recent studies had data with more sensitive techniques in a limited number of subjects:15, 17, 57 O'Brien et al.15 compared 10 subjects who underwent colonoscopy to five subjects who did not, using pyrosequencing with fecal samples. In general, the normal temporal variation in fecal samples obtained from “HCs” 3–6 months apart appeared similar in magnitude to the changes induced by BP over 1–3 months. When the authors sought to determine whether there were any clinical factors that may explain the findings in the patients who had received BPs, three patients with UC showed short-term changes in their microbiota after BP, with two of them showing a lasting effect upto 3 months. O' Brien et al.15 also assessed the OTUs contributing to change in bacterial composition in their samples and did not find any one OTU showing a rise or fall after BP. When the distribution of the two major phyla, Firmicutes and Bacteroidetes were examined, there were large but inconsistent shifts (>32%) in the relative proportions of Firmicutes and Bacteroidetes, observed in the cases in which samples over time were dissimilar. Interestingly, 4/5 of the unprepped cases had rises in Firmicutes, which is compatible with our data demonstrating an increase in Bacteroidetes with BP. O'Brien et al.15 also did not note a difference in Jaccard distances (a metric that is based on the presence and absence of organisms) in fecal samples obtained pre- and post-BP. In this regard, their conclusions could be different than ours because of a variety of factors: they did not have a consistent sampling interval between controls who were not prepped and the cases who were prepped; they only examined fecal samples; their shortest sampling intervals in the prepped cases were as high as 2 weeks; their prepped cases were analyzed together and contained a mixture of IBD and non-colitis patients and the effects can vary in terms of disease as well as its activity at the time of sampling, which is not detailed; the Jaccard index used in their study differs from the various metrics that are more frequently used in microbiota studies that we have used and is a presence and absence analysis; and the study had half the number of subjects relative to ours, with three comparison points that could have led to a statistically negative result.
O'Brien et al.15 findings contradict that of Harrell et al.,57 who used terminal restriction fragment length polymorphism, a fingerprinting technique, as well as limited cloning and sequencing in seven subjects and studied mucosal biopsies as opposed to fecal samples. The latter study clearly demonstrated that terminal restriction fragment length polymorphism, the highest resolution technique among fingerprinting techniques, failed to show differences compared with cloning and sequencing. This finding confirms that the false-negative results obtained from low-resolution analyses are because of the limitations of the analysis technique and not because of the lack of differences from BP. In the limited cloning and sequencing part of the latter study, mucosal biopsy samples were obtained a week apart in three subjects who were given a BP and in four subjects who were controls (n=2 controls given a liquid diet and n=2 controls given a general diet); 288 colonies were analyzed per sample, thus the sequencing was limited compared with our study. There was a reduction in richness and the Shannon diversity index on the mucosal surface after BP. Although we did not observe a significant overall reduction in richness, there was also a trend toward reduced richness in our sample set. Harrell et al.57 also studied Unifrac distances in the three prepped samples and their distance calculations are similar to ours, suggesting a mean Unifrac distance around 0.6, which can be interpreted as a similarity of about 30–40% between the pre- and post-BP samples. While Harrell et al.57 did not find one particular phylotype to be consistently altered, their sample size and depth of sampling were not adequate to detect these types of changes.
Momozowa et al.17 characterized the bacterial composition in colonic biopsies from six different regions, fecal samples collected at colonoscopy and fecal samples collected (at an unspecified period) before colonoscopy in two healthy subjects and two subjects with IBD. Technical replicates of two separate nucleic acid extractions from the biopsy and fecal samples were pyrosequenced and Unifrac distances were examined using 400 sequences per sample. Results revealed that the different colonic regions were just as similar as technical replicates within the same colonic region. Contrary to this, technical replicates of fecal samples were found disparate enough to result in significant changes. In unadjusted comparisons, the biopsy samples also differed from fecal specimens, and these differences were most apparent when fecal samples before colonoscopy were compared with colonic biopsy samples after BP, when both rare and abundant taxa were analyzed using weighted and unweighted Unifrac distances. Owing to the limited number of sequences and subjects, this study did not examine any of the taxa that differed pre- and post-BP. Overall, their findings are similar to the results of our study, which is conducted at more than two times the sampling depth, in a much larger sample set of healthy patients and IBD patients, and we have also analyzed alterations in specific phylotypes.
Most recently, Jalanka et al.58 examined bacterial composition in fecal samples collected from 23 healthy volunteers before and immediately after BP, and in follow-up 14 and 28 days after BP, using microarrays. The results showed no significant change in richness, evenness and the SI in the fecal samples similar to our findings; however, variability in the α-diversity metrics were not reported to understand whether this occurs because of inconsistency in the response from one individual to another. There were notable decreases within orders of the Firmicutes phylum also reported. Our findings of reduced Clostridiales and Erysipelotrichaceae are congruent with the reduction of the Firmicutes.
The strength of our study stems from well-characterized subjects, a high-resolution technique to study the microbiome, concomitant study of the mucosal and luminal compartments, and evaluation of clinical parameters. Our study's limitations include its short-term evaluation as we had at most 7 days between two sampling times and that we had two sampling points. Therefore, the findings pertain to short-term changes in the microbiome. We also did not control for random drift over the few days of sampling interval in our study; therefore, given our study design, we cannot fully exclude the possibility that variations in the sampling site or random drift within a few days can result in profound differences in microbiome composition. This, however, is unlikely as random drift and sampling site variations and dietary changes were previously studied and the observed effects were significantly smaller compared with the effects of BP.57 Additionally, our findings are limited to sequencing depths in the few thousands. Last, our sample size was small and may have negatively affected our ability to detect differences in most of the α-diversity metrics.
In summary, our study demonstrates that short-term changes in microbiome composition occur with BP, and that the changes affect various microbiota-related diversity metrics in IBD and non-IBD samples and the mucosal and luminal compartments, differently. Microbiome studies using fecal and luminal samples should take these changes into account when designing studies as well as selecting metrics for data analysis. Microbiome studies that examine the differences between the mucosal and the luminal compartments are best suited to be carried out on unprepped specimens.
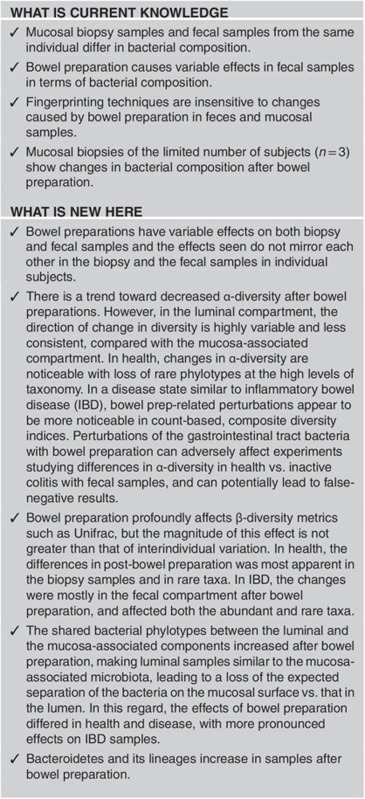
Study Highlights
Guarantors of the article: Rima M. Shobar, MD, Lars Koenig, PhD and Ece A. Mutlu, MD.
Specific author contributions: Rima M. Shobar recruited subjects, collected samples, analyzed data, and wrote the manuscript. Suresh Velineni, Ali Keshavarzian, Mark T. DeMeo, Joshua E. Melson, and John Losurdo recruited subjects and collected samples. Philip A. Engen coordinated sample inventories and extracted nucleic acids. Yan Sun sequenced samples. Lars Koenig performed bioinformatics and data analysis. Ece A. Mutlu generated hypothesis, collected samples, analyzed data, and wrote the manuscript. All authors participated in revising the manuscript.
Financial support: This study was partially supported with funds through NIH DK071838 as well as departmental funds at Rush University Medical Center. Rima M. Shobar was supported by the Canadian Bureau for International Education through the Libyan-North American Scholarship Program (SN3015). Ece A. Mutlu and Philip A. Engen received salary support from NIH grants AT001628 (not used to support this study directly) and DK071838, and from the DOD grant BC074932 (not used to support this study directly); Ece A. Mutlu has current salary support from NIH grant DK071838, CDC grant CK00161, and the Broad Foundation (none of which are used to support this study directly). Ali Keshavarzian received salary support from NIH grants AA019405, AA018729, AT003939, AA013745, AT001628 (none of which were used to support this study directly), and DK071838. Departmental funds were used to conduct the study. Yan Sun and Lars Koenig worked for Research and Testing Laboratory, LLC, Lubbock, Texas, United States of America at the time the study of the study, and Research and Testing Laboratory, LLC, Lubbock, Texas, United States of America was contracted and received payment to perform pyrosequencing and bioinformatics on the samples.
Potential competing interests: None.
Footnotes
Supplementary Information accompanies this paper on the Clinical and Translational Gastroenterology website (http://www.nature.com/ctg)
Supplementary Material
References
- Sears CL. A dynamic partnership: celebrating our gut flora. Anaerobe 2005; 11: 247–251. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN et al. Diversity of the human intestinal microbial flora. Science (New York, NY) 2005; 308: 1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Ley RE, Sonnenburg JL et al. Host–bacterial mutualism in the human intestine. Science (New York, NY) 2005; 307: 1915–1920. [DOI] [PubMed] [Google Scholar]
- Rajilic-Stojanovic M, Smidt H, de Vos WM. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol 2007; 9: 2125–2136. [DOI] [PubMed] [Google Scholar]
- Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol 2003; 57: 369–394. [DOI] [PubMed] [Google Scholar]
- Lewis K, Epstein S, D'Onofrio A et al. Uncultured microorganisms as a source of secondary metabolites. J Antibiot 2010; 63: 468–476. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M et al. The human microbiome project. Nature 2007; 449: 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA 2011; 108 (Suppl 1): 4554–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher LJ, Bury JP, Williams EA et al. Commonly used bowel preparations have significant and different effects upon cell proliferation in the colon: a pilot study. BMC Gastroenterol 2008; 8: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher P, Gervaz P, Egger JF et al. Morphologic alterations associated with mechanical bowel preparation before elective colorectal surgery: a randomized trial. Dis Colon Rectum 2006; 49: 109–112. [DOI] [PubMed] [Google Scholar]
- Morotomi M, Guillem JG, Pocsidio J et al. Effect of polyethylene glycol-electrolyte lavage solution on intestinal microflora. Appl Environ Microbiol 1989; 55: 1026–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens DAM, Jonkers DMAE, Russel GMVM et al. Bowel cleansing with subsequent intake of Lactobacillus plantarum 299v does not change the composition of the faecal flora. Microb Ecol Health Dis 2006; 18: 139–146. [Google Scholar]
- Mai V, Greenwald B, Morris JGJr et al. Effect of bowel preparation and colonoscopy on post-procedure intestinal microbiota composition. Gut 2006; 55: 1822–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Smukalla SM, Ni J et al. Variable recovery of the major phyla of the gut microbiome after colonoscopy bowel preparation. Gastroenterology 2011; 140: S-362. [Google Scholar]
- O'Brien CL, Allison GE, Grimpen F et al. Impact of colonoscopy bowel preparation on intestinal microbiota. PLoS One 2013; 8: e62815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibiloni R, Tandon P, Vargas-Voracka F et al. Differential clustering of bowel biopsy-associated bacterial profiles of specimens collected in Mexico and Canada: what do these profiles represent? J Med Microbiol 2008; 57: 111–117. [DOI] [PubMed] [Google Scholar]
- Momozawa Y, Deffontaine V, Louis E et al. Characterization of bacteria in biopsies of colon and stools by high throughput sequencing of the V2 region of bacterial 16S rRNA gene in human. PLoS One 2011; 6: e16952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronchick CA, Lipshutz WH, Wright SH et al. Validation of an instrument to assess colon cleansing. Am J Gastroenterol 1999;94:2667. [Google Scholar]
- Harvey RF, Bradshaw JM. A simple index of Crohn's-disease activity. Lancet 1980; 1: 514. [DOI] [PubMed] [Google Scholar]
- Powell-Tuck J, Bown RL, Lennard-Jones JE. A comparison of oral prednisolone given as single or multiple daily doses for active proctocolitis. Scand J Gastroenterol 1978; 13: 833–837. [DOI] [PubMed] [Google Scholar]
- Powell-Tuck J, Day DW, Buckell NA et al. Correlations between defined sigmoidoscopic appearances and other measures of disease activity in ulcerative colitis. Dig Dis Sci 1982; 27: 533–537. [DOI] [PubMed] [Google Scholar]
- Satsangi J, Silverberg MS, Vermeire S et al. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut 2006; 55: 749–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverberg MS, Satsangi J, Ahmad T et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Canad J Gastroenterol 2005;19 (Suppl A): 5A–36A. [DOI] [PubMed] [Google Scholar]
- Smith DM, Snow DE, Rees E et al. Evaluation of the bacterial diversity of pressure ulcers using bTEFAP pyrosequencing. BMC Med Genom 2010; 3: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu EA, Keshavarzian A, Losurdo J et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathogens 2014; 10: e1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczynski J, Stombaugh J, Walters WA et al. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Current Protocols in Bioinformatics 2011; 36:10.7: 10.7.1–10.7.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010; 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011; 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 2010; 26: 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007; 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach W. MVSP (Multivariate Statistical Package), Version 3.13m. Wales, UK: Kovach Computing Service, 2004. [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 2005; 71: 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinform 2006; 7: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Lladser ME, Knights D et al. UniFrac: an effective distance metric for microbial community comparison. ISME J 2011; 5: 169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Hamady M, Kelley ST et al. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 2007; 73: 1576–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrene M, Legendre P. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr 1997; 67: 345–366. [Google Scholar]
- Zoetendal EG, von Wright A, Vilpponen-Salmela T et al. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol 2002; 68: 3401–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 2005; 102: 11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S et al. Microbial ecology: human gut microbes associated with obesity. Nature 2006; 444: 1022–1023. [DOI] [PubMed] [Google Scholar]
- Larsen N, Vogensen FK, van den Berg FW et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 2010; 5: e9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haahtela T, Holgate S, Pawankar R et al. The biodiversity hypothesis and allergic disease: world allergy organization position statement. World Allergy Organ J 2013; 6: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S. Reduced bacterial biodiversity is associated with increased allergy. Environ Health Perspect 2012; 120: a304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher JU, Ubeda C, Artacho A et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol (Hoboken, NJ) 2015; 67: 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott SJ, Musfeldt M, Wenderoth DF et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 2004; 53: 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott SJ, Plamondon S, Hart A et al. Dynamics of the mucosa-associated flora in ulcerative colitis patients during remission and clinical relapse. J Clin Microbiol 2008; 46: 3510–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez C, Antolin M, Santos J et al. Unstable composition of the fecal microbiota in ulcerative colitis during clinical remission. Am J Gastroenterol 2008; 103: 643–648. [DOI] [PubMed] [Google Scholar]
- Dicksved J, Halfvarson J, Rosenquist M et al. Molecular analysis of the gut microbiota of identical twins with Crohn's disease. ISME J 2008; 2: 716–727. [DOI] [PubMed] [Google Scholar]
- Fujimoto T, Imaeda H, Takahashi K et al. Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn's disease. J Gastroenterol Hepatol 2013; 28: 613–619. [DOI] [PubMed] [Google Scholar]
- Walker AW, Sanderson JD, Churcher C et al. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol 2011; 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wang C, Tang C et al. Molecular–phylogenetic characterization of the microbiota in ulcerated and non-ulcerated regions in the patients with Crohn's disease. PLoS One 2012; 7: e34939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang W, Zhou R et al. Characteristics of fecal and mucosa-associated microbiota in Chinese patients with inflammatory bowel disease. Medicine (Baltimore, MD) 2014;93:e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedin C, van der Gast CJ, Rogers GB et al. Siblings of patients with Crohn's disease exhibit a biologically relevant dysbiosis in mucosal microbial metacommunities. Gut 2015. doi:10.1136/gutjnl-2014-308896. [DOI] [PubMed]
- Rehman A, Rausch P, Wang J et al. Geographical patterns of the standing and active human gut microbiome in health and IBD. Gut 2016; 65: 238–248. [DOI] [PubMed] [Google Scholar]
- Menees S, Higgins P, Korsnes S et al. Does colonoscopy cause increased ulcerative colitis symptoms? Inflamm Bowel Dis 2007; 13: 12–18. [DOI] [PubMed] [Google Scholar]
- Harrell L, Wang Y, Antonopoulos D et al. Standard colonic lavage alters the natural state of mucosal-associated microbiota in the human colon. PLoS One 2012;7:e32545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalanka J, Salonen A, Salojärvi J et al. Effects of bowel cleansing on the intestinal microbiota. Gut 2014; 64: 1562–1568. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.