Abstract
Background and Aims Studies on oaks (Quercus spp.) have often been hampered by taxonomic confusion, a situation further compounded by the occurrence of extensive interspecific hybridization. In the present study, a combination of genetic and morphological analyses was used to examine sympatric populations of Q. petraea and Q. robur at the north-western edge of their ranges in Northern Ireland, since it had previously been suggested that hybridization could facilitate the apparent rapid, long-distance dispersal of oaks following the glaciations.
Methods Samples were collected from 24 sites across Northern Ireland that had been previously designated as ancient or semi-natural woodland. Genotypes were obtained from a total of 950 trees using 12 nuclear microsatellite loci, and admixture coefficients were calculated based on a Bayesian clustering approach. Individuals were also classified as Q. petraea, Q. robur or hybrids based on two objective morphometric characters shown previously to delineate pure individuals effectively. Genetically ‘pure’ individuals of both species, as defined by the Bayesian clustering, were also genotyped for five chloroplast microsatellites.
Key Results Genetic and morphological analyses both indicated the presence of pure individuals of both species, as well as a continuum of intermediates. There was a good agreement between the molecular and morphological classification, with a generally clear separation between pure individuals.
Conclusions Despite millennia of hybridization and introgression, genetically and morphologically pure individuals of both Q. petraea and Q. robur can be found at the edge of their range, where both species occur sympatrically. The high proportion of individuals exhibiting introgression compared with previous studies may reflect the historical role of hybridization in facilitating dispersal following the glaciations. This is further supported by the significantly higher chloroplast diversity in Q. robur compared with Q. petraea.
Keywords: Hybridization, introgression, microsatellites, morphological analysis, oak, Quercus petraea, Quercus robur, species delineation
INTRODUCTION
The taxonomy of oaks (Quercus spp.) has intrigued and perplexed many eminent scientists since Linnaeus described 12 species in 1753 (Linnaeus, 1753). Darwin (1859) highlighted the difficulty in delimiting oak species across Europe in his On the origin of species, while more regional taxonomic difficulties within the British Isles were addressed by Babington (1862). These taxonomic struggles have resulted in the number of recognized oak species varying over time, culminating in Schwarz’s (1964) Flora Europaea entry, which distinguished 320 separate species. The challenge of separating various species complexes into taxonomic units across their geographical ranges remains an area of intensive research and debate, to the extent of proposals conflicting with the conventional Biological Species Concept sensu Mayr (1942; van Valen, 1976; Kremer and Petit, 1993; Manos et al., 1999).
One consequence of the sympatric occurrence of many closely related Quercus species, and one of the causes of taxonomic confusion, is extensive hybridization (Whittemore and Schaal, 1991; Rushton, 1993). Within the European white oak (Section Quercus) complex, Q. petraea (Matt.) Liebl. (sessile oak) and Q. robur L. (pedunculate oak) are the two most widespread and economically important species. They occur sympatrically across temperate Europe, with the range of Q. petraea being largely coincident with that of Q. robur as far as the eastern borders of Poland and Romania, beyond which Q. robur extends to the Urals. Hybrids between the two have been reported throughout their distribution (reviewed in Gardiner, 1970; Ortiz-Barrientos and Baack, 2014), despite the demonstration of both pre- and post-zygotic reproductive barriers (Steinhoff, 1993; Bacilieri et al., 1996; Streiff et al., 1999; Abadie et al., 2011). Quercus robur is an early-successional tree that is associated with base-rich, clay soils, often poorly drained sites and is tolerant of waterlogging, whilst Q. petraea prefers upland peaty soils and grows in mature forests. Under a model of density-dependent hybridization (‘Hubbs’ Principle’; Hubbs, 1955), patterns of hybridization between Q. petraea and Q. robur might be expected to reflect the species’ ecological preferences (Lagache et al., 2013).
Quercus petraea, Q. robur and their hybrid Q. × rosacea (Bechst.) are the only forms within the European white oak complex that occur in Ireland, which represents the extreme north-western limit of the species’ ranges. This species complex recolonized Ireland around 9500 BP from refugia located on the Iberian peninsula (Dumolin-Lapègue et al., 1997; Petit et al., 2002; Mitchell, 2003; Kelleher et al., 2004a; Muir et al., 2004; Lowe et al., 2005). Discriminating between the two species by pollen morphology is not possible, and thus it is not possible to determine which species arrived in Ireland first and the lag time, if any, until the arrival of the subsequent species. However, due to the recognized status of Q. robur as a colonizing species, it may be postulated that this species was the first to arrive. Oak woodlands subsequently dominated Ireland until the Neolithic, when use of the trees for building instigated a steep decline, and by the 1600s only 2–3 % of Ireland’s forest cover remained (Mitchell, 1995; Rackham, 1995), making Ireland the least wooded region in Europe, with the exception of Iceland.
The occurrence of oak at high latitudes soon after the retreat of the ice formed the basis of a long-standing debate in biogeography (‘Reid’s Paradox’), since the apparent speed of recolonization was believed vastly to exceed the species’ dispersal capacities (Reid, 1899; Provan and Bennett, 2008). It has been suggested, however, that hybridization between Q. petraea and Q. robur could facilitate rapid colonization through a process of differential dispersal and asymmetric pollen-mediated introgression (Petit et al., 2003). Ireland, being located on the north-westen edge of the species’ distribution, would have to have been recolonized particularly quickly due to the increase in sea levels following the glaciations. The aim of the present study was to determine the genetic composition of oaks in Northern Ireland. Here, we investigate genetic and morphological variation in Irish oak to examine how Q. petraea and Q. robur maintain genetic and phenotypic integrity while indulging in promiscuity during alternative periods of extreme environmental change and stasis. We sampled ‘blind’ (sensu Lepais et al., 2009), rather than a priori identifying individuals as either Q. petraea or Q. robur, in order to give a true representation of both species and the entire spectrum of genetic intermediates resulting from nearly 10 000 years of hybridization and backcrossing. We also analysed levels and patterns of nuclear and chloroplast genetic diversity in genetically ‘pure’ individuals of both species to determine if there was any bias in levels of chloroplast diversity. Under the scenario outlined by Petit et al. (2003), we would expect to see similar levels of nuclear diversity, but far higher levels of chloroplast diversity in Q. robur, since acorns from this species would be responsible for the majority of colonization events.
MATERIALS AND METHODS
Sampling and DNA extraction
Samples were collected from 24 sites across Northern Ireland that had been previously designated as ancient or semi-natural woodland based on data collected for the Woodland Trust Inventory of ancient and long-established woodland in Northern Ireland (www.backonthemap.org.uk; Fig. 1; Table 1). For genetic analyses, a single leaf was collected from up to 48 trees per site and stored in silica gel, and GPS co-ordinates were recorded for every tree. DNA was extracted using the cetyltrimethylammonium bromide (CTAB) method of Doyle and Doyle (1987). Nuclear genotypes were obtained for between 17 and 47 individuals per population (Table 1; total = 950; mean = 39·583). For morphometric analyses, three leaves were taken from the same 48 trees per population, with the exception of Banagher Glen (23) and Correl Glen (47; total = 1126; mean 46·875). Fully expanded, open canopy leaves were collected to minimize any effects of environmental factors such as exposure/shade.
Fig. 1.
Locations of sites sampled in this study. Numbers correspond to those in Table 1.
Table 1.
Details of populations studied
| No. | Name | Latititude (N) | Longitude (W) | n |
|---|---|---|---|---|
| 1 | Portaferry | 54·391 | 5·565 | 34 |
| 2 | Hollymount | 54·322 | 5·751 | 41 |
| 3 | Glenarm | 54·964 | 5·955 | 37 |
| 4 | Barnett’s Demesne | 54·552 | 5·960 | 46 |
| 5 | Hillsborough | 54·459 | 6·083 | 41 |
| 6 | Rostrevor | 54·093 | 6·190 | 44 |
| 7 | Rea’s Wood | 54·705 | 6·229 | 47 |
| 8 | Breen Wood | 55·138 | 6·238 | 32 |
| 9 | Portglenone | 54·863 | 6·472 | 41 |
| 10 | Gosford Park | 54·304 | 6·523 | 34 |
| 11 | Peatlands Park | 54·483 | 6·612 | 35 |
| 12 | Errigal Glen | 54·971 | 6·733 | 40 |
| 13 | Drum Manor | 54·639 | 6·815 | 43 |
| 14 | Roe Valley | 55·025 | 6·939 | 38 |
| 15 | Banagher Glen | 54·884 | 6·954 | 17 |
| 16 | Ness Wood | 54·947 | 7·181 | 42 |
| 17 | Gortin Glen | 54·667 | 7·233 | 41 |
| 18 | Fardross Forest | 54·374 | 7·268 | 45 |
| 19 | Crom | 54·170 | 7·451 | 43 |
| 20 | Belle Isle | 54·245 | 7·564 | 46 |
| 21 | Sloughan Glen | 54·622 | 7·564 | 34 |
| 22 | Castle Archdale | 54·484 | 7·722 | 46 |
| 23 | Marble Arch | 54·264 | 7·812 | 43 |
| 24 | Correl Glen | 54·439 | 7·885 | 40 |
n, number of individuals analysed.
Genotyping
All trees were genotyped for 12 nuclear microsatellite loci: MsQ13, QpZAG15, QpZAG110, QrZAG7, QrZAG20, QrZAG96, QrZAG112, PIE020, PIE102, PIE223, PIE239 and PIE271 (Guichoux et al., 2011). Forward primers included a 19 bp M13 tail (CACGACGTTGTAAAACGAC) and reverse primers included a 7 bp tail (GTGTCTT). PCR was carried out in a total volume of 10 μL containing 100 ng of genomic DNA, 5 pmol of 6-FAM-, HEX- or PET-labelled M13 primer, 0·5 pmol of each M13-tailed forward primer, 5 pmol of each reverse primer, 1× PCR buffer, 200 μm each dNTP, 2·5 mm MgCl2 and 0·25 U of GoTaq Flexi DNA polymerase (Promega, Sunnyvale, CA, USA). PCR was carried out on an MWG Primus thermal cycler (Ebersberg, Germany) using the following conditions: initial denaturation at 94 °C for 3 min followed by 40 cycles of denaturation at 94 °C for 30 s, annealing at 56 °C for 30 s, extension at 72 °C for 30 s and a final extension at 72 °C for 5 min. Genotyping was carried out on an AB3730×l capillary genotyping system (Applied Biosystems, Foster City, CA, USA). Allele sizes were scored using the GeneMapper software package (V4.1; Applied Biosystems) using LIZ-500 size standards, and were checked by comparison with previously sized control samples. Chromatograms were all inspected visually.
All trees were also genotyped for five chloroplast microsatellite loci: μdt1, μdt3, μdt4, μcd5 and μkk4 (Deguilloux et al., 2003). PCR and genotyping were carried out as described above, except that an annealing temperature of 44 °C was used over 30 cycles.
Genetic data analysis
GenePop (V3.4; Raymond and Rousset, 1995) was used to test for linkage disequilibrium between nuclear microsatellite loci. To assess the levels of admixture within individuals, Bayesian model-based clustering based on nuclear microsatellites was carried out using Structure (V2.2; Pritchard et al. 2000). The number of clusters was set to K = 2 to represent the two putative parental species, Q. petraea and Q. robur. The program was run using 50 000 burn-in iterations followed by 500 000 Markov Chain Monte Carlo iterations. The analysis was carried out ten times, and mean values of the admixture coefficient, Q, were calculated for each individual.
Levels and patterns of genetic diversity were calculated for populations containing at least ten ‘pure’ individuals of either species, based on the Q values (Q ≤ 0·1 for Q. petraea and Q ≥ 0·9 for Q. robur; Table 2). Levels of observed (HO) and expected (HE) heterozygosity and of allelic richness (AR) were calculated using the Fstat software package (V2.9.3.2; Goudet, 2001). Chloroplast microsatellite allele sizes were combined into haplotypes, and levels of genetic diversity (H) based on haplotype frequencies were calculated using the Arlequin software package (V3.5.1.2; Excoffier and Lischer, 2010). To account for differences in sample sizes, levels of haplotype richness (RH) were also calculated using Haplotype Analysis (V1.05; Eliades and Eliades, 2009).
Table 2.
Diversity statistics by population for ‘pure’ individuals (see text for details)
| Species | No. | Name | Nuclear |
Chloroplast |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | AR | HO | HE | n | H1 | H2 | H3 | H4 | H5 | H6 | H7 | H8 | Un | H | RH | |||
| Q. petraea | 2 | Hollymount | 14 | 5·256 | 0·714 | 0·693 | 14 | 14 | – | – | – | – | – | – | – | – | – | – |
| 3 | Glenarm | 13 | 4·914 | 0·642 | 0·711 | 13 | 2 | 11 | – | – | – | – | – | – | – | 0·282 | 0·923 | |
| 6 | Rostrevor | 23 | 5·472 | 0·680 | 0·725 | 23 | 23 | – | – | – | – | – | – | – | – | – | – | |
| 8 | Breen Wood | 17 | 5·045 | 0·641 | 0·731 | 17 | 17 | – | – | – | – | – | – | – | – | – | – | |
| 12 | Errigal Glen | 12 | 4·858 | 0·699 | 0·677 | 12 | 12 | – | – | – | – | – | – | – | – | – | ||
| 14 | Roe Valley | 10 | 5·600 | 0·697 | 0·766 | 9 | 9 | – | – | – | – | – | – | – | – | – | – | |
| 16 | Ness Wood | 24 | 5·197 | 0·557 | 0·721 | 24 | 24 | – | – | – | – | – | – | – | – | – | – | |
| 21 | Sloughan Glen | 14 | 4·995 | 0·634 | 0·706 | 14 | – | 14 | – | – | – | – | – | – | – | – | – | |
| 22 | Castle Archdale | 20 | 4·788 | 0·619 | 0·692 | 20 | – | 20 | – | – | – | – | – | – | – | – | – | |
| 24 | Correll Glen | 22 | 5·391 | 0·642 | 0·721 | 22 | 22 | – | – | – | – | – | – | – | – | – | – | |
| Q. robur | 1 | Portaferry | 12 | 5·007 | 0·588 | 0·740 | 12 | 9 | 23 | – | – | – | – | – | – | 1 | 0·204 | 1·705 |
| 4 | Barnett’s Demesne | 12 | 4·703 | 0·729 | 0·732 | 12 | 11 | – | – | – | – | – | – | – | 1 | 0·167 | 0·750 | |
| 9 | Portglenone | 18 | 4·728 | 0·705 | 0·720 | 18 | 16 | 2 | – | – | – | – | – | – | – | 0·209 | 0·765 | |
| 10 | Gosford Park | 16 | 4·556 | 0·575 | 0·679 | 15 | 2 | 10 | 1 | 1 | – | 1 | – | – | – | 0·562 | 2·657 | |
| 13 | Drum Manor | 13 | 4·727 | 0·668 | 0·713 | 11 | – | 6 | – | 4 | – | – | – | – | 1 | 0·618 | 1·818 | |
| 17 | Gortin Glen | 25 | 4·564 | 0·579 | 0·676 | 24 | – | 1 | 16 | – | 3 | – | 1 | 2 | 1 | 0·723 | 2·520 | |
| 18 | Fardross Forest | 31 | 5·163 | 0·680 | 0·741 | 31 | 9 | 13 | 1 | 6 | – | 1 | – | – | 1 | 0·551 | 2·743 | |
| 20 | Belle Isle | 18 | 5·011 | 0·781 | 0·760 | 16 | 5 | 8 | 1 | – | – | – | 1 | – | 1 | 0·683 | 2·683 | |
n, number of individuals analysed; AR, allelic richness; HO, observed heterozygosity; HE, expected heterozygosity; H1–H8, frequency of chloroplast haplotypes; Un, unique haplotype; H, gene diversity; RH, haplotype richness.
Morphometric analysis
Morphometric analysis employed two objective characters shown previously to delineate pure individuals of Q. petraea and Q. robur effectively, namely the presence (Q. petraea) or absence (Q. robur) of stellate hairs (Aas, 1995) and the ratio of petiole length to lamina length (Cousens, 1963; Kelleher et al., 2004b; Q. petraea ≥0·1, Q. robur <0·1). Individuals exhibiting both species-specific characters (petiole/lamina ratio averaged across the three leaves examined) were assigned to that species, with the remainder being classed as hybrids. Although our approach is not directly comparable with most previous morphological studies, which use multiple characters and multivariate analysis to classify individuals, it is categorical, which makes delineation simpler.
RESULTS
Genetic analysis
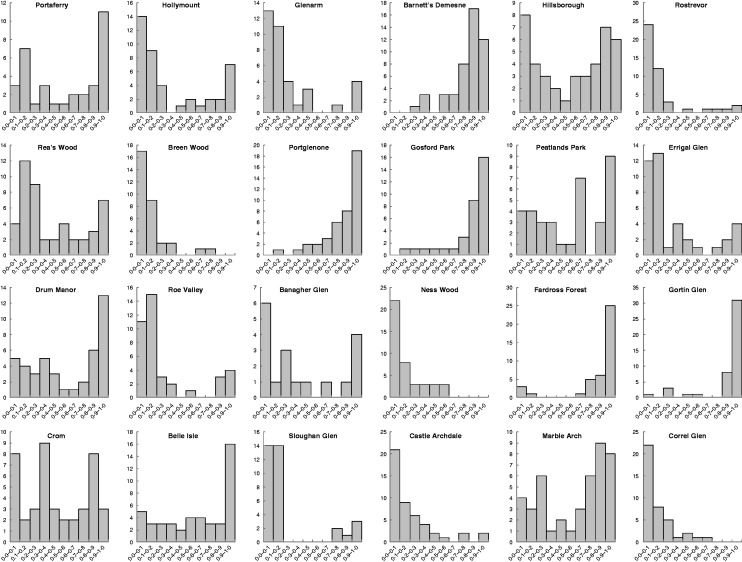
No significant evidence of consistent linkage disequilibrium (i.e. involving the same loci) was detected between any of the 12 nuclear microsatellites analysed (58 out of 1586 tests). The Structure analysis indicated a range of admixture (Q) values. Comparison with the morphometric analysis indicated that Q = 0·00 corresponded to Q. petraea and Q = 1·00 to Q. robur. The overall spectrum of Q values is shown in Fig. 2. Spectra for individual populations are shown in Fig. 3.
Fig. 2.
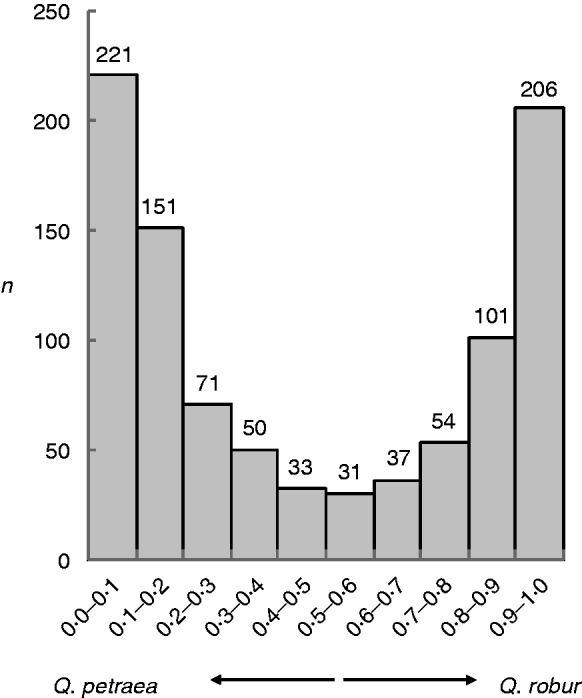
Distribution spectrum of admixture coefficient (Q) values across all samples (n = 955).
Fig. 3.
Distribution spectra of admixture coefficient (Q) values by population. Q value classes are given on the x-axis, while numbers of individuals are given on the y-axis. Note that values on the y-axis vary from site to site.
For genetically ‘pure’ individuals, levels of allelic richness (AR) ranged from 4·788 (Castle Archdale) to 5·600 (Roe Valley; mean = 5·152) in Q. petraea, and from 4·556 (Gosford Park) to 5·163 (Fardross Forest; mean = 4·807) in Q. robur (Table 2). Levels of observed (HO) and expected (HE) heterozygosity ranged from 0·619 (Castle Archdale) to 0·714 (Hollymount; mean = 0·653) and from 0·677 (Errigal Glen) to 0·766 (Roe Valley; mean = 0·714), respectively, in Q. petraea, and from 0·575 (Gosford Park) to 0·781 (Belle Isle; mean = 0·663) and from 0·676 (Gortin Glen) to 0·760 (Belle Isle; mean = 0·720), respectively, for Q. robur (Table 2).
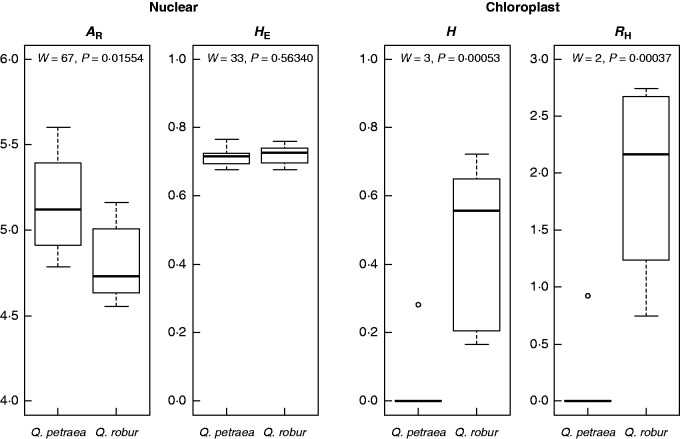
A total of 14 chloroplast haplotypes were detected, six of which were found in a single individual (Table 2). Only the two most common haplotypes (H1 and H2) were found in Q. petraea, and nine of the ten populations were fixed for a single haplotype, the exception being Glenarm, which exhibited a haplotype diversity (H) of 0·282 and a haplotype richness (RH) of 0·923. Quercus robur populations exhibited far higher chloroplast genetic diversity, with levels of H and RH ranging from 0·167 (Barnett’s Demesne) to 0·723 (Gortin Glen; mean = 0·464), and from 0·750 (Barnett’s Demesne) to 2·743 (Fardross Forest; mean = 1·955), respectively. Levels of AR were significantly lower in Q. robur than in Q. petraea (Wilcoxon test: W = 67; P = 0·01554), whereas there was no significant difference in HE (Wilcoxon test: W = 33; P = 0·56340; Fig. 4). Levels of both H and RH were significantly lower in Q. petraea than in Q. robur (Wilcoxon test: W = 3; P = 0·00053 and W = 2; P = 0·00037, respectively).
Fig. 4.
Boxplots showing comparisons of levels of nuclear (AR and HE) and chloroplast (H and RH) genetic diversity between ‘pure’ Q. petraea and Q. robur.
Morphometric analysis
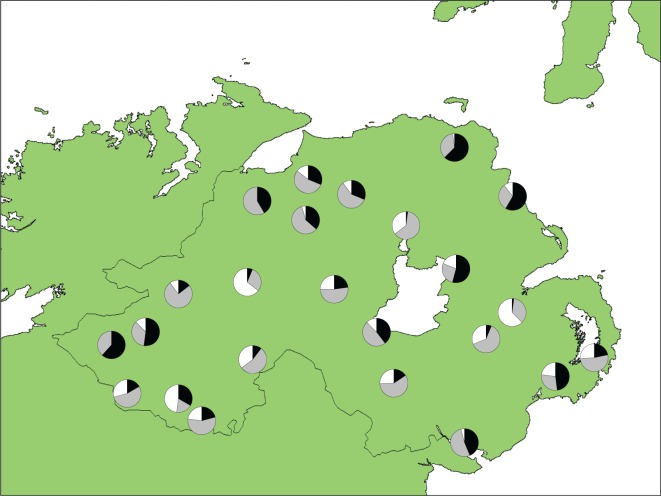
Morphometric analysis based on two discriminatory characters classified 345 individuals (30·67 % of individuals studied) as Q. petraea and 251 individuals (22·31 %) as Q. robur. The remaining 529 (47·02 %) were classified as hybrids. The geographical distribution of the three classes by population is shown in Fig. 5. Based on morphology, only two of the 24 populations studied (Ness Wood and Correl Glen) had pure individuals of only one species (Q. petraea) along with putative hybrid individuals. Q values for trees morphometrically assigned to Q. petraea (mean = 0·212; median = 0·125) were significantly different from those for trees assigned to Q. robur (mean = 0·821; median = 0·906; Fig. 6). Boxplots showing the distributions of Q values by population are given in Supplementary Data Figure S1. In 15 out of 18 populations where more than one individual of each species was analysed, Q values for Q. petraea and Q. robur were significantly different (Mann–Whitney U-test).
Fig. 5.
Geographical distribution of Quercus petraea (black), Q. robur (white) and hybrids (grey) by population based on morphological analyses.
Fig. 6.
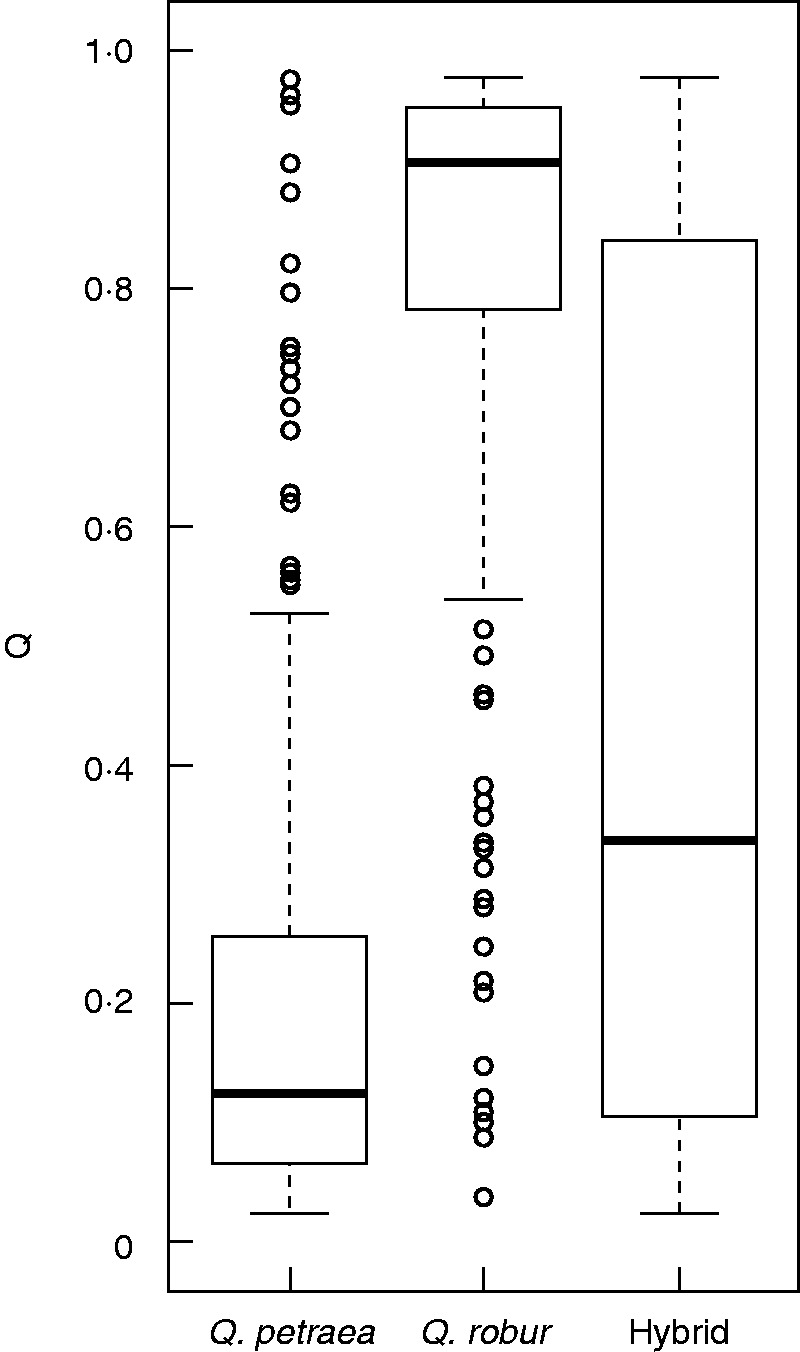
Boxplot showing admixture coefficient (Q) values for Quercus petraea, Q. robur and hybrids. The solid bar represents the median.
DISCUSSION
The results of the present study indicate that oak woodlands in Northern Ireland, which represents the north-western edge of their distribution range, comprise both a morphological and a genetic continuum of individuals that range from pure Q. petraea to pure Q. robur, with varying degrees of hybridization and introgression in intermediate individuals. Using the genetic criteria for ‘pure’ individuals as defined by Lepais et al. (2009; admixture coefficient Q < 0·1 for Q. petraea or Q > 0·9 for Q. robur), approx. 23 % of trees were classified as pure Q. petraea, while approx. 22 % were classified as pure Q. robur. There was good agreement between the molecular and morphological classification, with a generally clear separation between pure individuals. This was particularly apparent in the two populations from which no individuals were identified morphologically as Q. robur, Ness Wood and Correl Glen, with no Q scores of >0·6 in the former or >0·7 in the latter. Previous morphological studies in Ireland had suggested that the two form distinct species, but also that some hybrids occur (Carlisle and Brown, 1965; Rushton, 1983, 1993; Minihan and Rushton, 1984; Kelleher et al., 2004b). In their Flora of County Fermanagh, Forbes and Northridge (2012) stated that due to difficulty in species identification as a result of introgression, they had amalgamated their accounts of both oak species under Q. petraea. Their reasoning was that Q. petraea was ‘… generally regarded as a good, homogenous species …’, but a comparison of our morphological and genetic data indicate that both species are represented by genetically and morphologically distinct individuals. A previous study on 24 populations across Ireland using amplified fragment length polymorphisms (AFLPs) found little separation of the two species, despite morphological distinctiveness (Kelleher et al., 2005). This contrast with the findings of the present study could be due to the use of dominant markers and the low number of samples (five) analysed from each population, although an earlier AFLP study on Flemish populations using larger sample sizes (mean = 18·8 per population) showed clear molecular and morphological discrimination (Coart et al., 2002).
The use in the present study of 12 microsatellite markers that share many alleles between the two species means that it is not possible to assign individuals to various hybrid and backcross classes with any degree of certainty (Vaha and Primmer, 2006). Nevertheless, based on the distribution of admixture coefficients, it would seem that relatively few first-generation (F1) hybrids are represented in the samples, and that backcrossing to either putative parental species predominates in individuals exhibiting evidence of introgression. A similar scenario was described previously by Lepais et al. (2009), and thought to originate through a combination of pre-reproductive barriers such as density-dependent pollination and intrinsic pollen discrimination favouring intraspecific crosses and backcrosses over interspecific mating (Lepais and Gerber, 2011). Both the genetic and the morphological data indicate that the majority of the woodlands examined in the present study tend to be comprised of one or other of the two parental species, plus hybrids. Again, this is most probably a result of the relative abundance of parental species and pollen discrimination (Lepais et al., 2009; Lepais and Gerber, 2011; Legache et al., 2014), but it has also been shown that other environmental factors including fine-scale spatial organization and selection can control hybridization (Gugerli et al., 2007; Lagache et al., 2013). This complex interplay of factors may explain not only the regular occurrence of single species-/hybrid-dominated woodlands, but also the occasional sites where all three classes are found, such as Hillsborough.
Although 45 % of the individuals analysed genetically were assigned to pure Q. petraea or Q. robur, the remaining 55 % exhibiting evidence of introgression is far higher than the levels reported in a previous ‘blind’ study by Lepais et al. (2009), which also analysed two other white oak species, Q. pubescens and Q. pyrenaica. In it, the authors assigned 48·9 and 32·1 % of individuals to Q. robur and Q. petraea, respectively, from an exhaustively sampled stand containing only the two species, with 5·7 % of individuals classed as hybrids between the two, and the remaining 14·7 % exhibiting introgression from either Q. pubescens or Q. pyrenaica as a result of pollen immigration. The high values observed in the present study could reflect the historical importance of hybridization in facilitating the long-distance recolonization of Ireland from Iberia after the last glacial period, which ended approx. 11·5 kya. Although most palynological analyses do not discriminate between the two species, the pioneer nature of Q. robur, with its acorns more likely to be dispersed over long distances than those of Q. petraea (Jones, 1959; Petit et al., 2003), suggests that it colonized first. Subsequent pollen flow from Q. petraea, followed by hybridization, would facilitate the dispersal of the later successional species into the area colonized by the pioneer, and rapid unidirectional backcrossing could restore both species over time (Petit et al., 2003). The levels and patterns of genetic diversity observed at nuclear and chloroplast loci in genetically ‘pure’ individuals are completely consistent with this scenario. Previous studies have demonstrated that chloroplast haplotypes are shared between the two species (Dumolin-Lapègue et al., 1997; Cottrell et al., 2002; Petit et al., 2002) but, despite this, chloroplast genetic diversity is significantly higher in Q. robur, suggesting a great predominance of Q. robur acorns during the recolonization process. Generations of assortative mating, as a result of environmental and intrinsic barriers, would facilitate adaptation to prevailing climatic and edaphic conditions, and ultimately lead to the present-day observed distributions of the species and their hybrids at the limit of their distribution ranges.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of Figure S1: boxplots showing admixture coefficient (Q) values for Quercus petraea, Q. robur and hybrids by population.
ACKNOWLEDGEMENTS
This study was commissioned and funded by the Natural Heritage Research Partnership between the Northern Ireland Environment Agency (NIEA), Department of the Environment Northern Ireland and Quercus, Queen’s University Belfast. We are grateful to all the landowners who gave us permission to sample on their land, to all who helped with sampling, to Remy Petit for helpful discussions, and to John Farren, who acted as NIEA Client Officer.
LITERATURE CITED
- Aas G. 1995. Die Behaarung der Blätter von Traubeneiche und Stieleiche (Quercus petraea und Quercus robur): Variabilität und taxonomische Bedeutung. Mitteilungen aus der Forstlichen Versuchsanstalt Rheinland-Pflaz 34: 297–309. [Google Scholar]
- Abadie P, Roussell G, Dencausse B, et al. 2011. Strength, diversity and plasticity of postmating reproductive barriers between two hybridizing oak species (Quercus robur L. and Quercus petraea (Matt) Liebl.). Journal of Evolutionary Biology 25: 157–173. [DOI] [PubMed] [Google Scholar]
- Babington CC. 1862. Manual of British botany, 5th edn London: John van Voorst. [Google Scholar]
- Bacilieri R, Ducousso A, Petit RJ, Kremer A. 1996. Mating system and asymmetric hybridization in a mixed stand of European oaks. Evolution 50: 900–908. [DOI] [PubMed] [Google Scholar]
- Carlisle A, Brown AHF. 1965. The assessment of the taxonomic status of mixed oak (Quercus spp.) populations. Watsonia 6: 120–127. [Google Scholar]
- Coart E, Lamote V, de Loose M, van Bockstaele E, Lootens P, Roldán-Ruiz I. 2002. AFLP markers demonstrate local genetic differentiation between two indigenous oak species (Quercus robur L. and Quercus petraea (Matt) Liebl.) in Flemish populations. Theoretical and Applied Genetics 105: 431–439. [DOI] [PubMed] [Google Scholar]
- Cottrell JE, Munro RC, Tabbener HE, et al. 2002. Distribution of chloroplast DNA variation in British oaks (Quercus robur and Quercus petraea): the influence of postglacial colonisation and human management. Forest Ecology and Management 156: 181–195. [Google Scholar]
- Cousens JE. 1963. Variation of some diagnostic characters of the sessile and pedunculate oaks and their hybrids in Scotland. Watsonia 5: 273–286. [Google Scholar]
- Darwin CR. 1859. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London: John Murray. [PMC free article] [PubMed] [Google Scholar]
- Deguilloux MF, Pemonge MH, Bertel L, Kremer A, Petit RJ. 2003. Checking the geographical origin of oak wood: molecular and statistical tools. Molecular Ecology 12: 1629–1636. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- Dumolin-Lapègue S, Demesure B, Finseche S, Le Corre V, Petit RJ. 1997. Phylogoegraphic structure of white oaks throughout the European continent. Genetics 146: 1475–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliades N-G, Eliades DG. 2009. Haplotype Analysis: software for analysis of haplotype data.www.uni-goettingen.de/en/134935.html. [Google Scholar]
- Excoffier L, Lischer HEL. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10: 564–567. [DOI] [PubMed] [Google Scholar]
- Forbes RS, Northridge RH. 2012. The flora of County Fermanagh. National Museums; Northern Ireland. [Google Scholar]
- Gardiner AS. 1970. Pedunculate and sessile oak (Quercus robur L. and Quercus petraea (Mattuschka) Liebl.): a review of the hybrid controversy. Forestry 43: 151–160. [Google Scholar]
- Goudet J. 2001. Fstat, version 2.9.3, a program to estimate and test gene diversities and fixation indices. http://www2.unil.ch/popgen/softwares/fstat.htm. [Google Scholar]
- Gugerli F, Walser J-C, Dounavi K, Holderegger R, Finkeldey R. 2007. Coincidence of small-scale spatial discontinuities in leaf morphology and nuclear microsatellite variation of Quercus petraea and Q. robur in a mixed forest. Annals of Botany 99: 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichoux E, Lagache L, Wagner S, Léger P, Petit RJ. 2011. Two highly validated multiplexes (12-plex and 8-plex) for species delimitation and parentage analysis in oaks (Quercus spp.). Molecular Ecology Resources 11: 578–585. [DOI] [PubMed] [Google Scholar]
- Hubbs CL. 1955. Hybridization between fish in nature. Systematic Zoology 4: 1–20. [Google Scholar]
- Jones EW. 1959. Biological flora of the British Isles. Quercus L. Journal of Ecology 47: 169–222. [Google Scholar]
- Kelleher CT, Hodkinson TR, Kelly DL, Douglas GC. 2004a. Characterisation of chloroplast DNA haplotypes to reveal the provenance and genetic structure of oaks in Ireland. Forest Ecology and Management 189: 123–131. [Google Scholar]
- Kelleher CT, Kelly DL, Hodkinson TR. 2004b. Species status, hybridisation and geographic distribution of Irish populations of Quercus petraea (Matt.) Liebl. and Q. robur L. Watsonia 25: 83–97. [Google Scholar]
- Kelleher CT, Hodkinson TR, Douglas GD, Kelly DL. 2005. Species distinction in Irish populations of Quercus petraea and Q. robur: morphological versus molecular analyses. Annals of Botany 96: 1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer A, Petit RJ. 1993. Gene diversity in natural populations of oak species. Annales des Sciences Forestières 50: 186s–202s. [Google Scholar]
- Lagache L, Klein EK, Guichoux E, Petit RJ. 2013. Fine-scale environmental control of hybridization in oaks. Molecular Ecology 22: 423–436. [DOI] [PubMed] [Google Scholar]
- Lagache L, Klein EK, Ducousso A, Petit RJ. 2014. Distinct male reproductive strategies in two closely related oak species. Molecular Ecology 23: 4331–4343. [DOI] [PubMed] [Google Scholar]
- Lepais O, Petit RJ, Guichoux E, et al. 2009. Species relative abundance and direction of introgression in oaks. Molecular Ecology 18: 2228–2242. [DOI] [PubMed] [Google Scholar]
- Lepais O, Gerber S. 2011. Reproductive patterns shape introgression dynamics and species succession within the European white oak species complex. Evolution 65: 156–170. [DOI] [PubMed] [Google Scholar]
- Linnaeus C. 1753. Species Plantarum 2. Stockholm, Sweden. [Google Scholar]
- Lowe A, Unsworth C, Gerber S, et al. 2005. Route, speed and mode of oak postglacial colonisation across the British Isles: integrating molecular ecology, palaeoecology and modelling approaches. Botanical Journal of Scotland 57: 59–91. [Google Scholar]
- Manos PS, Doyle JJ, Nixon KC. 1999. Phylogeny, biogeography and processes of molecular differentiation in Quercus Subgenus Quercus (Fagaceae). Molecular Phylogenetics and Evolution 12: 333–349. [DOI] [PubMed] [Google Scholar]
- Mayr E. 1942. Systematics and the origin of species from the viewpoint of a zoologist. New York: Columbia University Press. [Google Scholar]
- Minihan VB, Rushton RS. 1984. The taxonomic status of oaks (Quercus spp.) in Breen Wood, Co. Antrim, Northern Ireland. Watsonia 15: 27–32. [Google Scholar]
- Mitchell FJG. 1995. The dynamics of Irish post-glacial forests. In: Mac an tSoir SS, ed. Wood, trees and forests in Ireland. Dublin: Royal Irish Academy, 13–22, [Google Scholar]
- Mitchell FJG. 2003. Natural invaders: the postglacial tree colonization of Ireland. In: Murray DA, ed. Biological invaders: the impact of exotic species. Dublin: Royal Irish Academy, 2–13. [Google Scholar]
- Muir G, Lowe AJ, Fleming CC, Vogl C. 2004. High nuclear genetic diversity, high levels of outcrossing, and low differentiation among remnant populations of Quercus petraea at the margin of its range in Ireland. Annals of Botany 93: 691–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Barrientos D, Baack EJ. 2014. Species integrity in trees. Molecular Ecology 23: 4188–4191. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Csaikl UM, Bordács S, et al. 2002. Chloroplast DNA variation in European white oaks: phylogeography and patterns of diversity based on data from over 2600 populations. Forest Ecology and Management 156: 5–26. [Google Scholar]
- Petit RJ, Bodénès C, Ducousso A, Roussel G, Kremer A. 2003. Hybridization as a mechanism of invasion in oaks. New Phytologist 161: 151–164. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics Society of America 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provan J, Bennett KD. 2008. Phylogeographic insights into cryptic glacial refugia. Trends in Ecology and Evolution 23: 564–571. [DOI] [PubMed] [Google Scholar]
- Rackham O. 1995. Looking for ancient woodland in Ireland. In: Mac an tSoir SS, ed. Wood, trees and forests in Ireland. Dublin: Royal Irish Academy, 1–12. [Google Scholar]
- Raymond M, Rousset F. 1995. GenePop (version 1.2): population genetic software for exact tests and ecumenicism. Journal of Heredity 86: 248–249. [Google Scholar]
- Reid C. 1899. The origin of the British flora. London: Dulau. [Google Scholar]
- Rushton BS. 1983. An analysis of variation of leaf characters in Quercus robur L. and Quercus petraea (Matt.) Liebl. population samples from Northern Ireland. Irish Forestry 40: 52–77. [Google Scholar]
- Rushton BS. 1993. Natural hybridization within the genus Quercus L. Annales des Sciences Forestières 50: 73s–90s. [Google Scholar]
- Schwarz O. 1964. Quercus L. In: Tutin TG, Heywood VH, Burges NA, Valentine DH, Walters SN, Webb DA, eds. Flora Europaea Vol. 1: Lycopodiaceae to Plantanaceae. Cambridge: Cambridge University Press, 61–64. [Google Scholar]
- Steinhoff S. 1993. Results of species hybridization with Quercus robur L. and Quercus petraea (Matt.) Liebl. Annals of Forest Science 50: 137s–143s. [Google Scholar]
- Streiff R, Ducousso A, Lexer C, Steinkellner H, Gloessl J, Kremer A. 1999. Pollen dispersal inferred from paternity analysis in a mixed oak stand of Quercus robur L. and Q. petraea (Matt.) Liebl. Molecular Ecology 8: 831–841. [Google Scholar]
- Vaha JP, Primmer CR. 2006. Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Molecular Ecology 15: 63–72. [DOI] [PubMed] [Google Scholar]
- van Valen L. 1976. Ecological species, multi-species and oaks. Taxon 25: 233–239. [Google Scholar]
- Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Whittemore AT, Schaal BA. 1991. Interspecific gene flow in sympatric oaks. Proceedings of the National Academy of Sciences, USA 88: 2540–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.