Abstract
The hydrothermal vent crab Shinkaia crosnieri is considered to obtain nutrition from the epibiotic bacteria found on the setae, but previous studies have not shown how nutrients can be transferred from the epibionts to the host. In this study, microscopic observations of S. crosnieri intestinal components detected autofluorescent setae fragments and pigmentation derived from the digestion of epibionts in a dye-stained epibiont tracer experiment. An in vitro digestion experiment with epibiotic populations using an intestinal extract demonstrated the degradation of epibiotic cells by digestive enzymes. A phylogenetic analysis showed that many of the bacterial 16S ribosomal RNA gene sequences obtained from the intestine were closely related to the sequences of the epibionts, thus they were probably derived from the epibionts. A stable isotope tracer experiment also indicated that 13C assimilated by the epibionts provided a carbon (nutrition) source for the host. Both activity measurements and isotope studies showed that chemosynthetic metabolism by the gut microbial components were inactive. Together with the feeding behaviour of living S. crosnieri, these results indicate that S. crosnieri ingests the epibionts using maxillipeds and assimilates them via its digestive organs as a nutrient source. The results of this study elucidate the mechanism of nutritional transfer in ectosymbiosis between chemosynthetic bacteria and deep-sea invertebrates.
Introduction
Since the first discovery of a dense, unique and diverse ecosystem in deep-sea hydrothermal vents in 1977 (Corliss et al., 1979), many studies have shown that deep-sea vent invertebrates such as tubeworms and Bathymodiolus mussels are sustained nutritionally by intracellular symbiotic bacteria (endosymbionts) as primary producers (Cavanaugh et al., 1981, 1987; Felbeck, 1981; Childress et al., 1986). In addition, several species of deep-sea vent invertebrates harbour bacteria (epibionts) that colonize the surfaces of specialized tissues such as the dorsal setae of the polychaete Alvinella pompejana, the gill chambers of the shrimp Rimicaris exoculata and the setae of the galatheid crabs Shinkaia crosnieri, Kiwa hirsuta and K. puravida (Polz and Cavanaugh, 1995; Cary et al., 1997; Goffredi et al., 2008; Watsuji et al., 2010; Thurber et al., 2011). The epibiotic microbial communities associated with these hosts are believed to have chemolithoautotrophic and methanotrophic products that support their hosts nutritionally, as well as the endosymbionts themselves (Goffredi et al., 2008; Grzymski et al., 2008; Watsuji et al., 2010; Thurber et al., 2011; Ponsard et al., 2012). Radioisotope-labelled tracer experiments suggest that R. exoculata epibionts achieve chemolithoautotrophic production by oxidizing reduced sulphur compounds and ferrous iron (Ponsard et al., 2012). A combination of 13C tracer experiments, microscopic fluorescence in situ hybridization and nano-scale secondary ion mass spectrometry also indicate that the predominant Sulfurovum-affiliated epibiotic population engages in thioautotrophic productivity in the epibiotic community associated with S. crosnieri (Watsuji et al., 2012). In addition, a combination of 13C tracer experiments and transcriptomic analysis showed that gammaproteobacterial type Ia and Ib methanotrophs in the S. crosnieri epibiotic community also support primary production using CH4 as energy and carbon sources (Watsuji et al., 2010, 2012).
Although the community composition, productivity and metabolic activity of the epibionts associated with deep-sea vent invertebrates have been relatively well characterized, the nutritional relationship between the epibionts and their hosts requires further elucidation. Radioisotope-labelled tracer experiments using living R. exoculata demonstrated that labelled bicarbonate and organic compounds (14C acetate and 3H lysine) were incorporated into R. exoculata tissues as well as the epibiotic community (Ponsard et al., 2012). In addition, these experiments demonstrated that the shrimps incorporated the labelled organic compounds across their integuments (Ponsard et al., 2012). These results suggest that the products assimilated by the epibionts were transported nutritionally to the host body of R. exoculata, possibly via non-gastric tissues, such as the gill chamber integument, although there is no evidence of the transport of organic compounds released from the epibionts (Ponsard et al., 2012). On the other hand, R. exoculata is known to possess a gut microbial community in its intestinal tract (Zbinden and Cambon-Bonavita, 2003; Durand et al., 2009) and appreciable incorporation of labelled bicarbonate into the intestine containing the gut microbial community has been confirmed (Polz et al., 1998). Thus, it has been suggested that primary production by the gut microbial community also contributes to nutrition in this shrimp.
13C-labelled tracer experiments using living S. crosnieri individuals showed that 13C-labelled bicarbonate and methane were incorporated not only into the epibiotic community but also into the muscle of S. crosnieri (Watsuji et al., 2010). In addition, it has been reported that living S. crosnieri individuals frequently exhibit feeding-like behaviour (Watsuji et al., 2010). For example, they comb the setae covered with dense epibionts using their third maxilliped and then bring the maxilliped to their mouth. Therefore, it has been predicted that nutrient acquisition by the host S. crosnieri is achieved by digesting chemosynthetic (thioautotrophic and methanotrophic) epibionts. However, no clear evidence has been obtained to support this hypothesis. Furthermore, the intestinal tract of S. crosnieri hosts a potential gut microbial community in addition to the epibiotic microbial community on its setae (Watsuji et al., 2010). Therefore, it is unclear whether nutritional transfer to the host S. crosnieri is predominantly mediated by feeding on and digesting the epibionts, or via the gut-endemic autotrophic and methanotrophic microbial populations (Watsuji et al., 2010).
Among the previously examined Okinawa Trough hydrothermal systems, the Iheya North field hosts dominant S. crosnieri populations with epibiotic microbial communities that exhibit dual thiotrophic and methanotrophic productivity patterns (Watsuji et al., 2010, 2012). In this study, we used S. crosnieri from the Iheya North field to perform microscopic observations and phylogenetic analysis of the gut microbial assemblages, as well as in vitro digestion experiments, thereby verifying whether the epibiotic bacterial population was ingested and digested by S. crosnieri. The autotrophic productivity and sulphide- and methane-oxidizing metabolisms of the dissected intestines were evaluated to determine whether sufficient chemolithotrophic activity and productivity could be achieved only by the potential gut microbial community. 13C-labelled tracer experiments using living S. crosnieri individuals were performed to determine whether S. crosnieri obtains nutrition from its epibionts.
Materials and methods
Collection of S. crosnieri from a deep-sea hydrothermal field
S. crosnieri individuals were collected from the Iheya North hydrothermal field in the Okinawa Trough, Japan, during dive #1335 on 19 March 2012 (27° 47.46′N, 126° 53.81′E, depth 986 m) and dive #1619 on 29 January 2014 (27° 47.45′N, 126° 53.80′E, depth 990 m) using the JAMSTEC remotely operated vehicle (ROV) ‘HyperDolphin'. Individuals were collected from the deep-sea vent habitats using a suction sampler and stored in a confined box filled with chilled seawater in the ROV. The S. crosnieri individuals obtained from dive #1619 were used in sulphide and methane consumption experiments, 16S ribosomal RNA (rRNA) gene sequence analysis of the gut microbial components and epibiotic community, in vitro digestion experiments with epibionts and intestinal extract activity measurements. The individuals from dive #1335 were used in the other experiments described below.
Fluorescence and light microscopic examination of the intestines and setae
The intestines and setae of S. crosnieri were observed by light and fluorescence microscopy (BX53; Olympus, Tokyo, Japan). The full-length intestines and setae fragments were dissected from 10 and several individuals, respectively, of S. crosnieri stored at −80 °C. The intestines and setae fragments were placed on glass slides (76 × 26 mm; Matsunami Glass, Osaka, Japan) and gently pressed with oblong cover glasses (24 × 45 mm; Matsunami Glass). The preparations of the intestinal components and setae were examined by optical and fluorescence light microscopy (BX53; Olympus). The fluorescence was monitored at 515–550 nm with excitation at 460–490 nm.
Dye-stained epibiont tracer experiment
A dye-stained epibiont tracer experiment was conducted in the onboard laboratory. The epibionts on the setae of two living S. crosnieri individuals were stained with crystal violet (Wako, Tokyo, Japan), as follows. First, moisture was removed from the wet setae with paper towels. Artificial seawater containing 0.4% (final concentration) crystal violet was infiltrated into the setae using a pipette. The artificial seawater was prepared as described previously (Watsuji et al., 2012). The individuals were laid on their backs on crushed ice for 5 min. Excess artificial seawater containing the dye was removed with paper towels before two dye-labelled and two non-labelled S. crosnieri individuals were maintained simultaneously for 24 h at 5 °C in a 54-l tank containing 40 l of artificial seawater. The seawater in the tank was filtered continuously using a canister filter (MEGA power 9012; GEX Co., Osaka, Japan) filled with 2 kg of activated charcoal (Wako) at a flow rate of 6.6 l min–1 to eliminate free dye from the dye-labelled S. crosnieri individuals. After incubation for 24 h, the whole intestines were removed from the dye-labelled and non-labelled S. crosnieri individuals, and then placed on glass slides (76 × 26 mm; Matsunami Glass) before gently pressing them with oblong cover glasses (24 × 45 mm; Matsunami Glass). The preparations were observed under an optical stereo zoom microscope (YDZ-3F; Yashima Optical Co., Tokyo, Japan) on board the ship.
Preparation of intestinal extracts
Full-length intestines were removed from two individuals (carapace lengths=57 and 62 mm), which had been preserved at −80 °C, and then homogenized in 500 μl of 40 mM Tris-HCl (pH 7.5) containing 160 mM KCl. The homogenates were centrifuged at 7000 × g for 10 min at 4 °C. The supernatants were used as crude extracts for the enzyme assays and in vitro digestion experiments with epibiotic cells. The protein contents were determined using a BCA protein assay kit (Thermo Scientific, Rockford, IL, USA).
In vitro digestion of epibionts using intestinal extracts
Portions of S. crosnieri setae with epibionts were removed from an individual (carapace length =62 mm), which had been stored at −80 °C, and incubated directly for 3 days at 5 °C in microtubes containing 100 μl of the crude intestinal extract (6.4 mg protein ml–1) with 0.05% (w/v) sodium azide, or 100 μl of buffer solution containing 40 mM Tris-HCl (pH 7.5), 160 mM KCl and 0.05% sodium azide. The morphological changes in the epibiotic microbial populations on the setae samples were observed before and after incubation using a light microscope (BX53; Olympus).
In addition, portions of setae of S. crosnieri with epibionts were removed from an individual (carapace length =62 mm), which had been stored at −80 °C, and brushed with a paintbrush and suspended in a buffer that comprised 40 mM Tris-HCl (pH 7.5), 160 mM KCl and 0.05% (w/v) sodium azide. This epibiont cell suspension was also used in the in vitro digestion experiments with the crude intestinal extract as the substrate. The reaction mixture contained 900 μl of the cell suspension and 100 μl of the crude extracts (6.0 mg protein ml–1). Following incubation for 3 days at 5 °C, the reaction mixture was centrifuged at 7000 × g for 10 min at 4 °C. Another sample was prepared in exactly the same manner, but it was centrifuged immediately without incubation. The pellets were used for DNA content quantification and the phylogenetic analysis. The DNA extraction procedure and the analysis of the bacterial 16S rRNA gene phylotype compositions of the pellets before and after incubation were performed as described in the Supplementary Information. The DNA concentrations were determined using a Quant-iT double-stranded DNA high-sensitivity assay kit and a Qubit Fluorometer (Invitrogen, Carlsbad, CA, USA).
Measurement of digestive enzyme activity levels in intestinal extracts
The activity levels of digestive enzymes in the intestinal extracts were measured as described in the Supplementary Information.
Measurement of methane and sulphide oxidation
The intestine and setae of one S. crosnieri individual (carapace length =43 mm) and the intestines of two other S. crosnieri individuals (carapace lengths =44 mm) were used to measure the methane-oxidizing activity in the onboard laboratory. The full-length intestines and whole setae were removed from living S. crosnieri individuals and homogenized using a Dounce-type homogenizer (glass–teflon) in 5 ml of ice-cold artificial seawater. Each of the intestinal homogenates from three individuals and from the whole setae of one individual was added to 95 ml of artificial seawater with dissolved methane in a 120-ml glass bottle, which was sealed with a butyl rubber stopper. The methane-containing artificial seawater was prepared by adding 1 ml of methane to the bottle and incubating at 5 °C for 2 days to dissolve the methane completely. The bottles with the methane-containing artificial seawater and the homogenate were sealed with butyl rubber stoppers and pressurized to 0.2 MPa with N2 after adding another 1 ml of methane to the headspace gas. Although the bottles were incubated at 5 °C, 0.5 ml of the gas phase was subsampled using a syringe at specific time intervals. These subsamples were injected into a 7-ml vial (V-5A; Nichiden-Rika Glass Co., Kobe, Japan) with a butyl rubber stopper and frozen at −20 °C. The methane concentration was assayed as described previously in the onshore laboratory (Watsuji et al., 2014).
Similarly, the intestine and setae from one S. crosnieri individual (carapace length =36 mm) and the intestines from two S. crosnieri individuals (carapace length =42 and 47 mm) were used to measure the sulphide-oxidizing activity. The full-length intestines and half of the setae assemblage were removed from the living S. crosnieri individuals and homogenized as described above. Sulphide oxidation was measured using the homogenates according to the previously described batch method (Watsuji et al., 2012).
Nucleic acid extraction and bacterial 16S rRNA gene clone analysis
The nucleic acid extraction procedure and the bacterial 16S rRNA gene clone analysis of the epibiotic microbial community and the cells in the in vitro digestion experiment and the gut microbial assemblage were conducted as described in the Supplementary Information.
13C-bicarbonate tracer experiments and stable carbon isotope analysis
In the onboard laboratory, a living S. crosnieri individual (carapace length =49 mm) and the full-length intestine removed from an individual (carapace length =49 mm) were used immediately in the 13C-bicarbonate tracer experiment. The living specimen and the intestine cut into six pieces were incubated in artificial seawater containing 1 mM of NaH13CO3 and 200 μM of sodium sulphide, as described previously (Watsuji et al., 2010), and stored at −80 °C after incubation. The S. crosnieri individual was dissected onshore to collect the whole tissue samples of the setae, intestine and muscle. The dry weights of the whole tissue samples were determined as 141 mg for the setae, 66 mg for the intestine and 1152 mg for the muscle. The dry weight of the cut intestine used in the 13C-bicarbonate tracer experiment was 61 mg. In addition, three S. crosnieri individuals, which were collected from the same colony as the individuals used in the tracer experiments and stored at −80 °C, were dissected onshore to analyse natural stable isotope abundances. The 13C composition and the total organic carbon contents of the samples were determined using a mass spectrometer (Delta Plus XP; Thermo Finnigan, Bremen, Germany), which was coupled online via a Finnigan ConFlo III interface with an elemental analyser (FlashEA 1112; Thermo-Quest, Milan, Italy). All of the samples were analysed in triplicates.
Results
Microscopic observations of setae and intestines
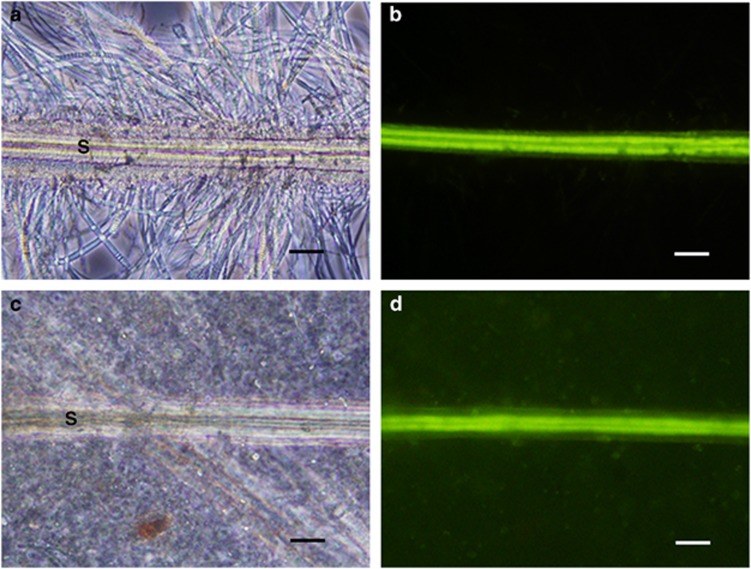
The fluorescence microscopy observations showed that S. crosnieri setae exhibited intrinsic fluorescence at excitation wavelengths of 460–490 nm, whereas the epibiotic microbial community associated with the setae exhibited no detectable autofluorescence (Figures 1a and b). Thus, the autofluorescence of S. crosnieri setae was used as a bio-tracer for setae in the intestines and their components. We detected setae fragments (1–5 fragments) in the components of each S. crosnieri intestine (8/10 individuals examined) based on optical and fluorescence microscopy observations (Figures 1c and d). In addition, the setae found in the S. crosnieri intestinal components lacked evident filamentous epibiotic assemblages (Figure 1c).
Figure 1.
Optical and fluorescence microscopic observation of S. crosnieri setae. Optical and fluorescence microscopy of setae cut from a living S. crosnieri and of setae found in an S. crosnieri intestine is shown in the top panels (a, b) and in the bottom panels (c, d), respectively. Fluorescence microscopy shows the intrinsic fluorescence of setae (b, d). Optical microscopy shows the dense filamentous epibiotic populations and the typical morphological appearance of setae in a living individual (a) but the absence of epibionts on setae in the intestine (c). Capital S indicates a seta. Scale bars=50 μm (a–d).
Dye-stained epibiont tracer experiment
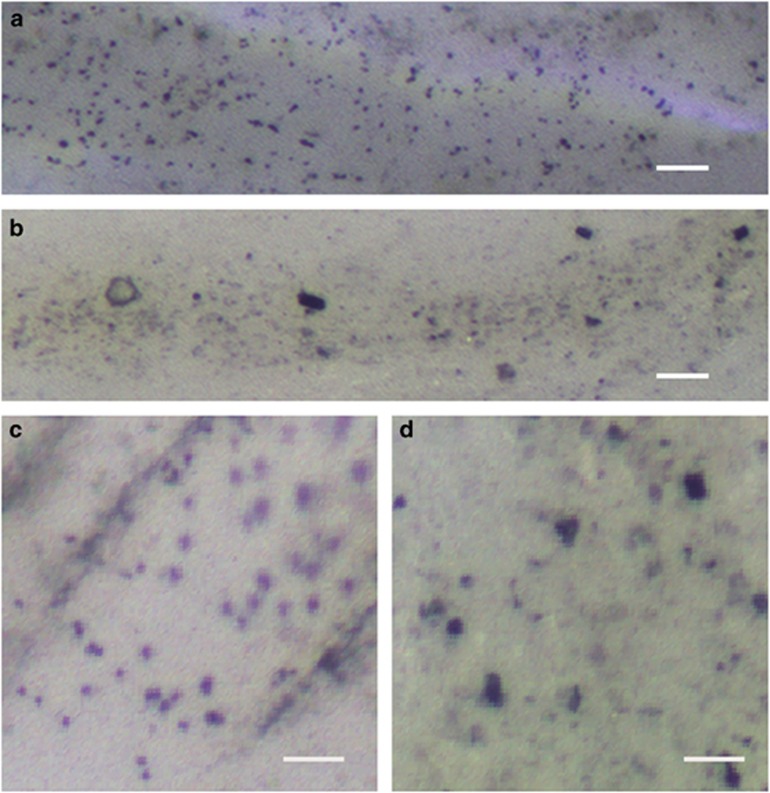
Living S. crosnieri individuals with dye-stained and unstained epibiotic communities were incubated together in artificial seawater, which was filtered through activated charcoal (Supplementary Figure S1). During incubation for 24 h, we found that no body parts of S. crosnieri were stained with crystal violet, except the dye-stained setae. After incubation, the S. crosnieri individuals were dissected and the dissected full-length intestines and their components were observed under an optical stereo zoom microscope. According to the microscopic observations of the S. crosnieri intestines and their components with the dye-stained epibiotic community, the dye pigmentation and many granules faintly coloured by crystal violet were observed (Figures 2a and c). However, no pigmentation was observed in the S. crosnieri intestines and their components with the unstained epibiotic community (Figures 2b and d).
Figure 2.
Microscopic observation of intestine ingredients in living S. crosnieri individuals with and without dye-labelled epibiont tracer experiment. Optical microscopy was performed for the intestines and their ingredients obtained from a S. crosnieri individual with crystal-violet-stained epibionts (a, c) and with unstained epibionts (b, d). Scale bars=50 μm (a, b) and 20 μm (c, d).
In vitro digestion experiment
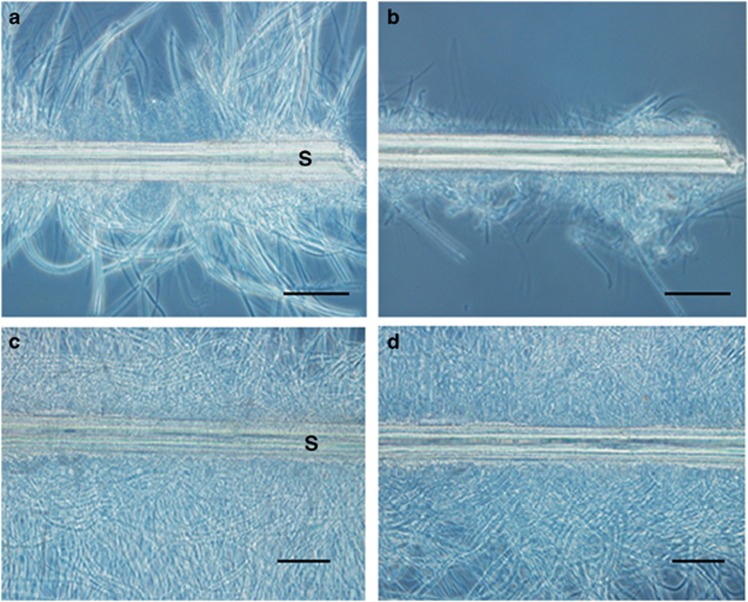
Portions of setae covered with epibiotic microbial populations were statically incubated in the presence or absence of the crude S. crosnieri intestinal extract. Microscopic observations before and after incubation showed that the abundance of epibiotic microbial populations decreased significantly in the setae incubated with the intestinal extract (Figures 3a and b), but they were apparently unchanged in the setae incubated without the intestinal extracts (Figures 3c and d).
Figure 3.
Microscopic observations of setae incubated with and without intestinal extract. Optical microscopy was performed to analyse setae dissected from a S. crosnieri individual before (a, c) and after (b, d) incubation with (a, b) and without (c, d) intestinal extract. Capital S indicates a seta. Scale bars=50 μm (a and b).
The epibiont cell suspensions were also incubated with the crude S. crosnieri intestinal extract. The DNA content of the cell pellet decreased drastically during incubation with the intestinal extract (that is, 13% of the DNA content in the cell pellet before incubation) (Supplementary Table S1). However, the bacterial 16S rRNA gene phylotype composition did not differ significantly between the cell pellets before and after incubation with the intestinal extract (that is, the clonal frequencies of the Sulfurovum, Thiotrichaceae and Methylococcales groups were 20.9%, 59.5% and 3.5% in the cell pellets before incubation and 29.9%, 47.1% and 3.4% in the cell pellets after digestion, respectively) (Supplementary Tables S2 and S3).
Determination of digestive enzyme activity levels in intestinal extracts
The crude intestinal extracts exhibited various digestive enzyme activities, such as α-amylase, lipase and protease (Supplementary Table S4), where protease had the highest specific activity level (Supplementary Table S4). In addition, the nuclease activity was determined on the basis of agarose gel electrophoresis of genomic DNA from Escherichia coli, which was incubated with the crude intestinal extract (Supplementary Figure S2).
Chemolithotrophic activity of epibiotic and gut microbial communities
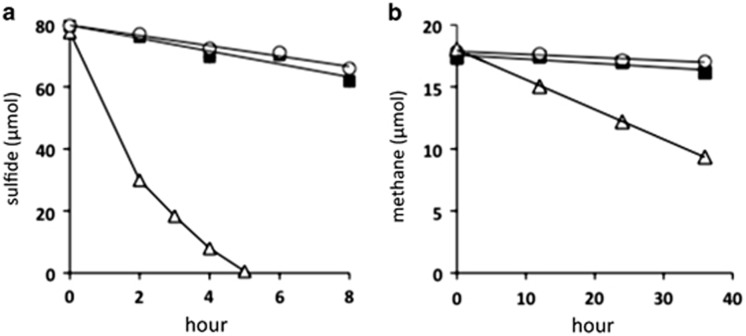
The rates of sulphide and methane consumption were measured using the homogenized setae and intestines (Supplementary Figure S1). The sulphide and methane concentrations drastically decreased during incubation of the homogenized setae (Figure 4). In contrast, the sulphide and methane concentrations slightly decreased during incubation in the presence of the homogenized intestines of three different individuals, although the changes in the concentrations did not differ significantly compared with those in the absence of homogenized intestines and setae (at each of the interval points, unpaired t-test, P>0.1) (Figure 4).
Figure 4.
Time course of consumption of sulphide and methane by homogenates of S. crosnieri intestine and setae with epibionts. The concentrations of sulphide (a) and methane (b) were examined at the indicated intervals in the absence of the homogenate sample (▪) and in the presence of homogenized setae with epibionts (Δ) and homogenized intestines (○).
Comparison of phylotype compositions and sequences in the gut and epibiotic microbial DNA assemblages
In total, 176 and 163 bacterial 16S rRNA gene clones were sequenced using the DNA extracts from the intestine and setae, respectively, of the same S. crosnieri individual (Supplementary Table S5). The clones that shared ⩾97% sequence identity were classified according to the same phylotype. Among the 38 gut bacterial phylotypes, eight phylotypes that represented 19% of the gut bacteria clones were very closely related to the S. crosnieri epibiont clones sequenced in previous studies and the present study (>99% identity) (Supplementary Table S5). The phylotypes that represented 54.5% of the gut bacterial clones and 88.9% of the epibiotic clones were affiliated with the Sulfurovum group within Epsilonproteobacteria, the Thiotrichaceae group and the Methylococcales group within Gammaproteobacteria (Supplementary Tables S5 and S6).
The number of phylotypes related to the Thiotrichaceae and Methylococcales groups in the epibiotic clone library was much higher than that in the gut bacterial clone library (Supplementary Table S5). Similarly, the Chao index showed that species richness levels of both the Thiotrichaceae and Methylococcales groups in the epibiotic 16S rRNA gene clone library (104 species) were much higher than those in the gut bacterial 16S rRNA gene clone library (29 species) (Supplementary Table S7). Most of the phylotypes (8/9 phylotypes) related to Thiotrichaceae and Methylococcales in the gut bacterial 16S rRNA gene clone library were found in the epibiotic and gut bacterial clone libraries of the same individual (Supplementary Table S5). In addition, the predominant Thiotrichaceae and Methylococcales phylotypes in the epibiotic 16S rRNA gene clone library tended to be found frequently in the gut bacterial 16S rRNA gene clone library (Supplementary Table S5).
All of the epsilonproteobacterial phylotypes from the intestine and setae were phylogenetically related to members of the genus Sulfurovum (Supplementary Table S5). The number of Sulfurovum-affiliated phylotypes in the epibiotic 16S rRNA gene clone library was slightly higher than that in the gut bacterial 16S rRNA gene clone library (seven and nine phylotypes, respectively). The Chao index showed that the species richness level of the Sulfurovum group in the epibiotic clone library (14 species) was twice that in the gut bacterial clone library (7 species) (Supplementary Table S7). The Sulfurovum phylotypes in the gut bacterial clone library were not always found in the epibiotic clone library and the predominant epibiotic Sulfurovum phylotype (setae1_78) was not found in the gut bacterial clone library (Supplementary Table S5).
13C-bicarbonate tracer experiments and stable carbon isotope analysis
13C-bicarbonate tracer experiments were performed using live S. crosnieri and dissected intestines (Supplementary Figure S1). The 13C enrichment level in setae with epibionts (3530‰) was clearly higher and the 13C enrichment level in the intestine (−19.5‰) was slightly higher compared with that in the muscle (−22.8‰) (Table 1). The 13C amounts assimilated in the whole setae including epibionts, the full-length intestine and the whole muscle of a living S. crosnieri individual for 48 h were calculated as 133 μmol, 0.22 μmol and 3.2 μmol, respectively (Table 1). The 13C amount assimilated in the dissected S. crosnieri intestine for 48 h was 0.48 μmol (Table 1). In addition, the natural stable carbon isotope composition was determined in the tissues, including setae, intestine and muscle, of three S. crosnieri individuals (Table 2). The δ13C value was slightly elevated in the muscles (−32.6‰) compared with that in the setae (−36.4‰) (Table 2) and these differences were significant (unpaired t-test, P<0.05). In contrast, the δ13C values were similar in the muscles (−32.6‰) and intestine (−33.4‰) (Table 2), and there was no significant difference (unpaired t-test, P>0.05).
Table 1. Stable carbon isotope compositions of different tissues of a living S. crosnieri individual and a dissected intestine before and after 13C-labelled bicarbonate tracer experiments.
| Labelled specimen | Tissue |
δ13C (‰) |
Enrichment of 13C/12C (%) | Total organic carbon/whole tissue (mmol) | Assimilated 13C/whole tissue (μmol) | |
|---|---|---|---|---|---|---|
| Natural abundancea | After incubationb | |||||
| S. crosnieri | Setae | −36.4 | 3530±78 | 370±8 | 3.3 | 133±3 |
| Intestine | −33.4 | −19.5±0.3 | 1.4±0.03 | 1.4 | 0.22±0.005 | |
| Muscle | −32.6 | −22.8±0.1 | 1.1±0.01 | 29.1 | 3.2±0.03 | |
| Dissected intestine | — | −33.4 | −0.5±0.6 | 3.4±0.06 | 1.3 | 0.48±0.009 |
Values are means of three independent determinations measured in Table 2.
Values were measured in triplicate and were expressed as mean±s.d.
Table 2. Natural stable isotopic composition of different tissues of S. crosnieri from a natural habitat of Iheya North.
| Specimen no. |
δ13C (‰) |
||
|---|---|---|---|
| Setae | Intestine | Muscle | |
| 1 | −35.9±0.3 | −33.3±0.2 | −32.1±0.1 |
| 2 | −36.0±0.9 | −34.0±0.7 | −32.6±0.1 |
| 3 | −37.2±0.5 | −33.0±0.1 | −33.2±0.1 |
| Mean | −36.4 | −33.4 | −32.6 |
The δ13C value of a sample was measured in triplicate. They are expressed as the mean±s.d.
Discussion
The deep-sea vent galatheid crabs, S. crosnieri and K. puravida, frequently exhibit a typical behaviour where they comb their setae, which are covered with dense epibionts, using the third maxillipeds and then bringing the maxillipeds to the mouths, although this behaviour has not been observed in R. exoculata (Watsuji et al., 2010; Thurber et al., 2011). Based on these observations, it has been hypothesized that S. crosnieri and K. puravida ingest their epibiotic microbial populations directly via this typical feeding behaviour, and digest them in their digestive organs (Watsuji et al., 2010; Thurber et al., 2011). In this study, we found setae fragments in almost every S. crosnieri intestine (Figure 1), which strongly suggests that the setae were ingested via the combing behaviour in S. crosnieri. In addition, it is likely that the epibiotic population on the setae, as well as the setae, are ingested during this behaviour given that S. crosnieri setae are always covered with dense epibiotic microbial communities in natural deep-sea vent habitats (Figure 1a; Watsuji et al., 2010, 2012). However, the setae found in the S. crosnieri intestinal tracts were not accompanied by obvious filamentous epibiotic assemblages (Figure 1c). Typical filamentous epibiotic cells were not observed among the intestinal components of S. crosnieri observed via the dye-stained epibiont tracer experiment, but apparent dye pigmentation and faintly coloured granules were identified in the intestinal components (Figures 2a and c). These results suggest that the epibiotic populations on the ingested S. crosnieri setae and the ingested dye-stained epibionts were digested in the gut. The in vitro digestion experiment clearly indicated that the S. crosnieri epibionts, but not the setae, were digested at significant levels by the intestinal extract (Figure 3). In addition, the intestinal extract exhibited digestive enzyme activities, such as α-amylase, lipase, protease and nuclease activities (Supplementary Table S4 and Supplementary Figure S2). Therefore, these results suggest that the epibionts were ingested and digested in the gut.
The sulphide- and methane-oxidizing activities of living S. crosnieri individuals have previously been identified in both atmospheric and elevated hydrostatic pressure conditions (Watsuji et al., 2012, 2014). In addition, thioautotrophic and methanotrophic productions have been detected in the epibiotic bacterial community (Watsuji et al., 2010, 2012). In this study, the sulphide- and methane-oxidizing activities of the potential gut microbial community in S. crosnieri were found to be negligible (Figures 4a and b and Supplementary Figure S1), which indicates that the viable gut microbial community exhibited little chemolithotrophic (sulphide- and methane-oxidizing) activity and productivity to support the host's nutrition. In addition, this result suggests that the ingested epibiotic microbial populations lost their indigenous chemolithotrophic (sulphide- and methane-oxidizing) activities in the intestine because of digestion by various intestinal enzymes.
The phylogenetic diversity of gut microbial communities in deep-sea vent invertebrates that harbour epibiotic microbial communities was characterized in R. exoculata (Zbinden and Cambon-Bonavita, 2003; Durand et al., 2009). These analyses revealed that the predominant phylotypes in the R. exoculata gut community were related to members of Deferribacteres, whereas they were not identified in the R. exoculata epibiotic community (Durand et al., 2009). Based on these results, it has been speculated that a gut-specific microbial community is present in the R. exoculata intestine. Meanwhile, we found that many of the bacterial 16S rRNA gene sequences retrieved from the S. crosnieri intestine were phylogenetically related to those of the S. crosnieri epibionts (Supplementary Table S5). In particular, eight phylotypes that represented 19% of the gut bacterial clones were very closely related to the S. crosnieri epibiont clones identified in previous studies and this study (>99% identity) (Supplementary Table S5). Thus, the similar bacterial phylotypes in the gut microbial DNA assemblages and the epibiotic microbial community of S. crosnieri provides molecular evidence that the epibiotic microbial population is ingested and digested by S. crosnieri as a nutritional source for the host.
Our analysis of the phylogenetic diversity in the epibiotic community and the gut bacterial assemblage of the same individual showed that a higher diversity of Thiotrichaceae and Methylococcales phylotypes was found in the epibiotic community compared with the gut bacterial assemblage (Supplementary Tables S5 and S7). All of the epsilonproteobacterial phylotypes from the intestine and setae were phylogenetically related to the Sulfurovum group and the epibiotic community exhibited a higher diversity of Sulfurovum phylotypes than the gut bacterial assemblage (Supplementary Tables S5 and S7). These results indicate that the phylogenetic diversity of the ingested epibionts may decline during their digestion in the intestine. Genetic microheterogeneity was also evident in the epibiotic community and the gut bacterial assemblage of the same individual, particularly in the Sulfurovum-affiliated phylotypes, that is, the predominant Sulfurovum-affiliated phylotypes in the gut bacterial assemblage were not detected in the epibiotic community (Supplementary Table S5). We cannot completely exclude the possibility that several distinctive Sulfurovum phylotypes from the epibiotic Sulfurovum phylotypes thrive specifically in the intestine, but we hypothesize that the genetic microheterogeneity in the epibiotic community and the gut bacterial assemblage may be attributable to the indigenous genetic microheterogeneity of the epibiotic community. The current analysis and previous phylogenetic analyses of the epibiotic communities in S. crosnieri individuals were conducted using the whole setae or different parts of setae samples, thus the phylotype compositions were represented as the average entities in an individual or several individuals (Watsuji et al., 2010, 2012, 2014). However, it is likely that different parts of the setae will host similar but slightly heterogeneous epibiont phylotype compositions. The feeding-like behaviour of living S. crosnieri should always transfer epibionts from different parts of the setae to the gut, thus the ingested epibiotic populations in the intestine may exhibit genetic microheterogeneity. Even if the gut bacterial phylotypes were derived from the gut-endemic microbial community of S. crosnieri, these bacterial components would not provide a nutritional source for S. crosnieri because they lose their chemolithotrophic activities (Figure 4).
The proportion of Sulfurovum-affiliated phylotype clones was 42.6% in the gut bacterial assemblage but only 14.7% in the epibiotic bacterial community, although the proportions of Thiotrichaceae- and Methylococcales-related phylotypes in the gut bacterial assemblage decreased as compared with those in the epibiotic bacterial community (Supplementary Tables S5 and S6). These results imply that the Sulfurovum-affiliated epibionts tend to reside in the intestine throughout the digestive process, in contrast to the Thiotrichaceae- and Methylococcales-related epibiotic populations. However, our in vitro digestion experiment using an epibiont cell suspension with the intestinal extract detected no apparent phylotype-specific digestion of epibionts by the intestinal enzymes (that is, significant resistance of the Sulfurovum group to digestion), although the total bacterial cells and DNA levels drastically decreased during digestion (Supplementary Tables S1, S2 and S3). In many decapods, it has been reported that foods are masticated and digested in the stomach, and finer food particles are absorbed in the hepatopancreas, whereas coarser particles pass through gland filters in the intestine (Schaefer 1970; Kunze and Anderson 1979). A previous fluorescence in situ hybridization of S. crosnieri epibionts clearly demonstrated that the Sulfurovum epibionts had the largest cell size as well as the thickest and longest filaments in the epibiotic community (Watsuji et al., 2010). Therefore, because the Sulfurovum epibionts have relatively larger cells and a filamentous morphology compared with other epibionts, the ingested and masticated Sulfurovum epibionts are more likely to be transported into the intestine via the gland filters rather than the hepatopancreas. Therefore, the Sulfurovum epibionts may be more likely to reside in the intestine throughout the digestive process as compared with the Thiotrichaceae- and Methylococcales-related epibiotic populations.
The phylotypes affiliated with Verrucomicrobiae and Mollicutes from the intestine were not related to the bacterial 16S rRNA gene sequences identified in the S. crosnieri epibionts (Supplementary Table S5). Phylotypes related to Verrucomicrobiae and Mollicutes were also detected in the gut of R. exoculata, where the gut-endemic microbial community would occur (Durand et al., 2009). The phylotype gut_h89 belonging to Gammaproteobacteria was also not related to previously known epibiotic 16S rRNA gene sequences, although this was the predominant phylotype in the gut bacterial assemblage. Thus, these bacterial phylotypes found in the S. crosnieri intestine DNA assemblages may represent viable components of the gut-endemic microbial community of S. crosnieri.
The hydrothermal vent mussels Bathymodiolus azoricus and B. puteoserpentis are nutritionally sustained by chemolithoautotrophic and methanotrophic endosymbionts (Duperron et al., 2006; Petersen et al., 2011). When 13C enrichment was evaluated in 13C-labelled bicarbonate tracer experiments using living B. azoricus individuals, the 13C enrichment level in the gill containing thioautotrophic endosymbionts was the highest among all tissues, whereas the 13C enrichment level of tissues, including the digestive system, was slightly higher compared with that of muscles (Riou et al., 2008). A similar 13C tracer experiment using live S. crosnieri showed that the 13C enrichment of setae with thioautotrophic epibionts was the highest, whereas the 13C enrichment of the intestine was slightly higher compared with that of the muscle (Table 1). Thus, as found in these deep-sea vent mussels, our results may indicate that the organic carbon assimilated by the epibiotic microbial population is supplied to the living S. crosnieri body via digestion in the digestive organs.
Finally, the nutritional transfer rate was estimated from the results of the 13C-labelled bicarbonate tracer experiments (Supplementary Figure S1). The total amount of 13C assimilated in the dissected S. crosnieri intestine (0.48 μmol), which was assumed to be the net 13C amount assimilated by functionally active gut microbial assemblages, was much lower than that of the whole muscle in a living S. crosnieri individual (3.2 μmol; Table 1). Thus, it is very unlikely that 13C assimilated in the intestine with gut microbial assemblages sustained the 13C incorporated into the muscle and the other tissues in the body of S. crosnieri. In contrast, the total amount of 13C assimilated in the whole setae of a living S. crosnieri individual (133 μmol) was much greater than that of the whole muscle (Table 1). Thus, only the 13C assimilated by setae epibionts can be the source of 13C supplied to muscle and other tissues in the body of S. crosnieri.
We determined the natural stable carbon isotope compositions of the tissues, including setae, intestine and muscle, of three S. crosnieri individuals (Table 2). As noted previously (Watsuji et al., 2010), the highly 13C-depleted values of the setae with epibionts indicated the significant contribution of the methanotrophic populations using highly 13C-depleted methane in the hydrothermal fluids, in addition to the primary production of thioautotrophic epibionts. In general, a small but significant elevation in the δ13C value in the animal body relative to the δ13C value in the available food is observed as compared with the stable carbon isotope compositions of natural animal communities, although the δ13C values of animal tissues resemble those in the animal's diet (Rau et al., 1983). According to the natural carbon isotope compositions of S. crosnieri tissues, a small elevation in the δ13C value of S. crosnieri muscle relative to the δ13C value for the setae with epibionts was also observed, although the δ13C values were almost equal in the muscles and intestines (Table 2). Thus, the natural carbon isotope compositions of different S. crosnieri tissues also suggest that the epibiotic microbial population on the setae is the primary diet of natural S. crosnieri populations in deep-sea vent habitats.
Concluding remarks
Our results indicate that the predominant deep-sea vent invertebrate S. crosnieri found in the Okinawa Trough hydrothermal systems is nutritionally sustained by host-associated chemosynthetic bacteria, as in the case of another deep-sea vent shrimp R. exoculata (Watsuji et al., 2010; Ponsard et al., 2012). Given that the typical feeding-like behaviour has been observed frequently in these deep-sea vent galatheid crabs in their natural habitats and under rearing conditions, it was predicted that deep-sea vent galatheid crabs such as S. crosnieri and K. puravida ingest epibiotic microbial populations and utilize them as their primary diet (Watsuji et al., 2010; Thurber et al., 2011). However, the nutritional transfer from the epibionts to the host has not been directly shown and its mechanism remains unclear. Therefore, in this study, our experiments and analyses demonstrated that S. crosnieri populations from the Okinawa Trough hydrothermal systems ingest thioautotrophic and methanotrophic epibionts via their mouth using the third maxillipeds, and digest and assimilate them as their primary nutritional source. This is a striking example, in which the manner of nutritional transfer in the ectosymbiosis between chemosynthetic bacteria and deep-sea invertebrates is clearly characterized.
Acknowledgments
We are grateful to the crews of RV/Natsushima and the operation teams of DSV/HyperDolphin for helping us collect the deep-sea hydrothermal samples.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Cary SC, Cottrell MT, Stein JL, Camacho F, Desbruyeres D. (1997). Molecular identification and localization of filamentous symbiotic bacteria associated with the hydrothermal vent annelid Alvinella pompejana. Appl Environ Microbiol 63: 1124–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh CM, Gardiner SL, Jones ML, Jannasch HW, Waterbury JB. (1981). Prokaryotic cells in the hydrothermal vent tube worm Riftia pachyptila: possible chemoautotrophic symbionts. Science 213: 340–342. [DOI] [PubMed] [Google Scholar]
- Cavanaugh CM, Levering PR, Maki JS, Mitchell R, Lidstrom ME. (1987). Symbiosis of methylotrophic bacteria and deep-sea mussels. Nature 325: 346–348. [Google Scholar]
- Childress JJ, Fisher CR, Brooks JM, Kennicutt MC, Bidigare R, Anderson AE. (1986). A methanotrophic marine molluscan (Bivalvia, Mytilidae) symbiosis: mussels fueled by gas. Science 233: 1306–1308. [DOI] [PubMed] [Google Scholar]
- Corliss JB, Dymond J, Gordon LI, Edmond JM, von Herzen RP, Ballard RD et al. (1979). Submarine thermal springs on the Galapagos rift. Science 203: 1073–1083. [DOI] [PubMed] [Google Scholar]
- Duperron S, Bergin C, Zielinski F, Blazejak A, Pernthaler A, McKiness ZP et al. (2006). A dual symbiosis shared by two mussel species, Bathymodiolus azoricus and Bathymodiolus puteoserpentis (Bivalvia: Mytilidae), from hydrothermal vents along the northern Mid-Atlantic Ridge. Environ Microbiol 8: 1441–1447. [DOI] [PubMed] [Google Scholar]
- Durand L, Zbinden M, Cueff-Gauchard V, Duperron S, Roussel EG, Shillito B et al. (2009). Microbial diversity associated with the hydrothermal shrimp Rimicaris exoculata gut and occurrence of a resident microbial community. FEMS Microbiol Ecol 71: 291–303. [DOI] [PubMed] [Google Scholar]
- Felbeck H. (1981). Chemoautotrophic potential of the hydrothermal vent tube worm, Riftia pachyptila Jones (Vestimentifera). Science 213: 336–338. [DOI] [PubMed] [Google Scholar]
- Goffredi SK, Jones WJ, Erhlich H, Springer A, Vrijenhoek RC. (2008). Epibiotic bacteria associated with the recently discovered Yeti crab, Kiwa hirsuta. Environ Microbiol 10: 2623–2634. [DOI] [PubMed] [Google Scholar]
- Grzymski JJ, Murray AE, Campbell BJ, Kaplarevic M, Gao GR, Lee C et al. (2008). Metagenome analysis of an extreme microbial symbiosis reveals eurythermal adaptation and metabolic flexibility. Proc Natl Acad Sci USA 105: 17516–17521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze J, Anderson DT. (1979). Functional morphology of the mouthparts and gastric mill in the hermit crabs Clibanarius taeniatus (Milne Edwards), Clibanarius virescens (Krauss), Paguristes squamosus (McCulloch) and Dardanus setifer (Milne-Edwards) (Anomura: Paguridae). Aust J Mar Freshwat Res 30: 683–722. [Google Scholar]
- Petersen JM, Zielinski FU, Pape T, Seifert R, Moraru C, Amann R et al. (2011). Hydrogen is an energy source for hydrothermal vent symbioses. Nature 476: 176–180. [DOI] [PubMed] [Google Scholar]
- Polz MF, Cavanaugh CM. (1995). Dominance of one bacterial phylotype at a Mid-Atlantic Ridge hydrothermal vent site. Proc Natl Acad Sci USA 92: 7232–7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polz MF, Robinson JJ, Cavanaugh CM, Van Dover CL. (1998). Trophic ecology of massive shrimp aggregations at a Mid-Atlantic Ridge hydrothermal vent site. Limnol Oceanography 43: 1631–1638. [Google Scholar]
- Ponsard J, Cambon-Bonavita MA, Zbinden M, Lepoint G, Joassin A, Corbari L et al. (2012). Inorganic carbon fixation by chemosynthetic ectosymbionts and nutritional transfers to the hydrothermal vent host-shrimp Rimicaris exoculata. ISME J 7: 96–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau GH, Mearns AJ, Young DR, Olson RJ, Schafer HA, Kaplan IR. (1983). Animal 13C/12C correlates with trophic level in pelagic food webs. Ecology 64: 1314–1318. [Google Scholar]
- Riou V, Halary S, Duperron S, Bouillon S, Elskens M, Bettencourt R et al. (2008). Influence of CH4 and H2S availability on symbiont distribution, carbon assimilation and transfer in the dual symbiotic vent mussel Bathymodiolus azoricus. Biogeosciences 5: 1681–1691. [Google Scholar]
- Schaefer N. (1970). The functional morphology of the foregut of three species of decapod Crustacea: Cyclograpsus punctatus Milne-Edwards, Diogenes brevirostris Stimpson, and Upogebia africana (Ortmann). Zool Afr 5: 309–326. [Google Scholar]
- Thurber AR, Jones WJ, Schnabel K. (2011). Dancing for food in the deep sea: bacterial farming by a new species of yeti crab. PloS One 6: e26243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watsuji TO, Nakagawa S, Tsuchida S, Toki T, Hirota A, Tsunogai U et al. (2010). Diversity and function of epibiotic microbial communities on the galatheid crab, Shinkaia crosnieri. Microb Environ 25: 288–294. [DOI] [PubMed] [Google Scholar]
- Watsuji TO, Nishizawa M, Morono Y, Hirayama H, Kawagucci S, Takahata N et al. (2012). Cell-specific thioautotrophic productivity of epsilon-proteobacterial epibionts associated with Shinkaia crosnieri. PloS One 7: e46282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watsuji TO, Yamamoto A, Takaki Y, Ueda K, Kawagucci S, Takai K. (2014). Diversity and methane oxidation of active epibiotic methanotrophs on live Shinkaia crosnieri. ISME J 8: 1020–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbinden M, Cambon-Bonavita MA. (2003). Occurrence of Deferribacterales and Entomoplasmatales in the deep-sea Alvinocarid shrimp Rimicaris exoculata gut. FEMS Microbiol Ecol 46: 23–30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.