Abstract
Tetracyclines possess many properties considered ideal for antibiotic drugs, including activity against Gram-positive and -negative pathogens, proven clinical safety, acceptable tolerability, and the availability of intravenous (IV) and oral formulations for most members of the class. As with all antibiotic classes, the antimicrobial activities of tetracyclines are subject to both class-specific and intrinsic antibiotic-resistance mechanisms. Since the discovery of the first tetracyclines more than 60 years ago, ongoing optimization of the core scaffold has produced tetracyclines in clinical use and development that are capable of thwarting many of these resistance mechanisms. New chemistry approaches have enabled the creation of synthetic derivatives with improved in vitro potency and in vivo efficacy, ensuring that the full potential of the class can be explored for use against current and emerging multidrug-resistant (MDR) pathogens, including carbapenem-resistant Enterobacteriaceae, MDR Acinetobacter species, and Pseudomonas aeruginosa.
Antibiotic-resistance mechanisms limit the usefulness of tetracyclines in treating bacterial infections. But ongoing optimization has resulted in derivatives (e.g., eravacycline) with improved potency.
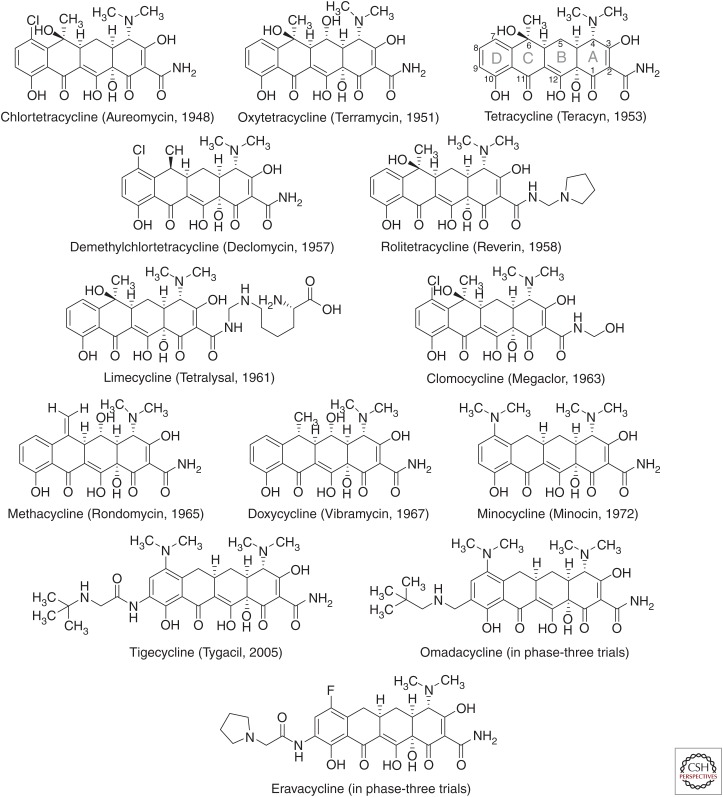
Tetracycline antibiotics are well known for their broad spectrum of activity, spanning a wide range of Gram-positive and -negative bacteria, spirochetes, obligate intracellular bacteria, as well as protozoan parasites. The first tetracyclines were natural products derived from the fermentations of actinomycetes. Chlortetracycline, produced by Streptomyces aureofaciens, and marketed as Aureomycin, was first reported by Benjamin Duggar at Lederle Laboratories in 1948 and approved for clinical use that same year (Duggar 1948). Soon after, Pfizer (New York) scientists isolated oxytetracycline, approved by the U.S. Food and Drug Administration (FDA) in 1950 and marketed as Terramycin (Finlay et al. 1950). Other tetracyclines that followed over the next two decades were also natural products produced by streptomycetes (tetracycline, demethylchlortetracycline) or semisynthetic derivatives with improved antibacterial potency, spectrum, resistance coverage, solubility, and/or oral bioavailability (methacycline, rolitetracycline, lymecycline, doxycycline, and minocycline) (Jarolmen et al. 1970; Cunha et al. 1982; Nelson and Levy 2011). Several of these “legacy” tetracyclines remain in clinical use for the treatment of uncomplicated respiratory, urogenital, gastrointestinal, and other rare and serious infections; however, the dissemination of tetracycline-resistant mechanisms has narrowed their utility, limiting use to only infections with confirmed susceptibility (Fig. 1).
Figure 1.
Chemical structures of clinically used tetracyclines and development candidates. Tetracycline structures are labeled with generic names; trade names and year of discovery are indicated within parentheses. The core structure rings (A–D) and carbons (1–12) are labeled in the chemical structure of tetracycline using the convention for tetracycline carbon numbering and ring letter assignments.
After a long pause in the advancement of the tetracycline class, renewed interest in optimization of tetracyclines during the late 1980s led to the discovery of semisynthetic derivatives with improved potency against difficult-to-treat emerging multidrug-resistant (MDR) Gram-negative and -positive pathogens, including bacteria with tetracycline-specific resistance mechanisms. Tigecycline, a semisynthetic parenteral glycylcycline, was discovered in 1993 by scientists at Lederle (which later became Wyeth, New York), and introduced into clinical use in 2005 (Sum and Petersen 1999; Zhanel et al. 2004). Tigecycline continues to be an important treatment option for serious infections caused by pathogens resistant to other antibiotic classes. In recent years, two new tetracyclines have entered clinical development: omadacycline, a semisynthetic aminomethylcycline derivative of minocycline discovered at Paratek Pharmaceuticals (Boston, MA) (Draper et al. 2014), and eravacycline, a fully synthetic fluorocycline discovered at Tetraphase Pharmaceuticals (Watertown, MA) (Clark et al. 2012; Xiao et al. 2012). In addition to efficacy against MDR infections, an important feature of these two new antibiotics is their oral formulations. This review will focus on recent developments in the understanding of tetracycline-resistance mechanisms and their potential impact on the clinical utility of tetracycline-class antibiotics.
MECHANISM, UPTAKE, AND TETRACYCLINE-SPECIFIC RESISTANCE
In recent surveillance studies, the prevalence of tetracycline resistance in selected European countries was found to be 66.9% and 44.9% for extended-spectrum β-lactamase (ESBL)-producing Escherichia coli and Klebsiella species (spp.), respectively (Jones et al. 2014), and global tetracycline-resistance percentages were 8.7% and 24.3% for methicillin-resistant Staphylococcus aureus (MRSA) and Streptococcus pneumoniae, respectively (Mendes et al. 2015). Resistance to tetracyclines is usually attributed to one or more of the following: the acquisition of mobile genetic elements carrying tetracycline-specific resistance genes, mutations within the ribosomal binding site, and/or chromosomal mutations leading to increased expression of intrinsic resistance mechanisms. Three general class-specific mechanisms have been well described: efflux, ribosomal protection, and enzymatic inactivation of tetracycline drugs. As there are several recent reviews on the topics of tetracycline-specific resistance determinants and their prevalence in clinical and environmental settings (Roberts 2005, 2011; Jones et al. 2008; Thaker et al. 2010), only a limited discussion of these areas will be covered here.
Uptake and Mechanism of Action
Tetracyclines preferentially bind to bacterial ribosomes and interact with a highly conserved 16S ribosomal RNA (rRNA) target in the 30S ribosomal subunit, arresting translation by sterically interfering with the docking of aminoacyl-transfer RNA (tRNA) during elongation (Maxwell 1967; Brodersen et al. 2000; Pioletti et al. 2001). Tetracyclines are usually considered bacteriostatic antibiotics; however, organism- and isolate-specific bactericidal activity in vitro has been described (Norcia et al. 1999; Petersen et al. 2007; Bantar et al. 2008; Noviello et al. 2008), and, recently, the bactericidal activity of tigecycline against E. coli and K. pneumoniae in a mouse model suggests that in vitro bactericidal assessments may not necessarily predict in vivo outcomes (Tessier and Nicolau 2013).
The mechanism of tetracycline uptake has been reviewed by Nikaido and Thanassi (1993). Briefly, in Gram-negative cells such as E. coli, tetracycline passively diffuses through the outer membrane porins OmpF and OmpC (Mortimer and Piddock 1993; Thanassi et al. 1995), most likely as a Mg2+ chelate, and this is consistent with the finding that outer membrane porin mutants show decreased susceptibility to tetracyclines (Pugsley and Schnaitman 1978). Accumulation of tetracycline in the periplasm is driven by the Donnan potential across the outer membrane. The dissociation of tetracycline from Mg2+ enables the weakly lipophilic, uncharged form to diffuse through the inner membrane to the cytoplasm where it may be complexed with magnesium and reach its ribosomal target. Uptake into the cytoplasm is partially energy dependent, involving passive diffusion, proton motive force, and phosphate bond hydrolysis (McMurry and Levy 1978; Smith and Chopra 1984; Yamaguchi et al. 1991).
Ribosomal Interactions
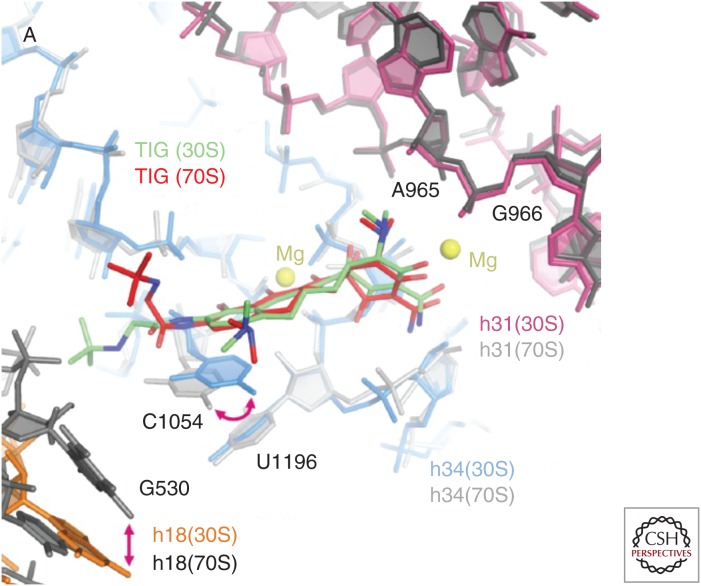
Crystallographic studies with the Thermus thermophilus 30S ribosomal subunit have revealed at least one high-occupancy tetracycline-binding site (Tet-1) and five other minor binding sites in 16S rRNA (Brodersen et al. 2000; Pioletti et al. 2001). Tetracycline most likely binds complexed with two Mg2+ ions at the Tet-1 site located in a pocket formed between helices h34 and h31, near the A-site where aminoacyl-tRNA docks onto the 30S subunit, consistent with the known mechanism of action (Jenner et al. 2013). The significance of the other five tetracycline-binding sites located elsewhere within the 30S subunit is unclear, and recent crystallographic studies with tigecycline and tetracycline binding to the T. thermophilus 70S ribosome (Jenner et al. 2013) and tigecycline binding to the 30S ribosome (Schedlbauer et al. 2015) showed that tigecycline was bound only to the Tet-1 site, and secondary binding sites were not observed (Fig. 2). Additional interactions made between the 9-tert-butylglycylamido moiety of tigecycline and C1054 in h34 are consistent with the higher binding affinity and greater antitranslational potency of tigecycline compared with tetracycline (Olson et al. 2006). Interestingly, a different orientation of this tigecycline side chain was observed in the 30S versus the 70S structure (Fig. 2), suggesting that tigecycline must accommodate conformation changes in the primary binding site that occur during decoding (Schedlbauer et al. 2015). Consistent with this recent finding, earlier work by Bauer et al. (2004) showed that tigecycline and tetracycline produced slightly different patterns of Fe2+-mediated RNA cleavage and dimethylsulfate modification, suggesting that both antibiotics bind at the same binding site, but in somewhat different orientations. Ribosome-binding competition experiments with [3H]tetracycline show relative IC50 values as follows: eravacycline, 0.22 µm; tigecycline, 0.22 µm; minocycline, 1.63 µm; omadacycline, 1.96 µm; and tetracycline, 4 µm; and results were consistent with these and other novel tetracycline derivatives binding at a single major site (Olson et al. 2006; Grossman et al. 2012; Jenner et al. 2013; Draper et al. 2014).
Figure 2.
Alternative binding modes of tigecycline at the primary ribosomal-binding site. Alternative tigecycline-binding modes in the 30S (green) and 70S (red) structures are shown, superimposed within the primary tetracycline-binding site. Key nucleotides (G530, A965, G966, C1054, U1196) and helices (h18, h31, h34) are shown in both structures. (From Schedlbauer et al. 2015; reprinted, with permission, from the American Society for Microbiology © 2015.)
Binding-Site Mutations
Because most bacteria have multiple rRNA copies, target-based mutations in rRNA conferring tetracycline resistance are usually found in bacteria with low rRNA gene copy numbers. Mutations in 16S rRNA have been reported in Propionibacterium acnes (2–3 16S rRNA copies), Helicobacter pylori (1–2 16S rRNA copies), Mycoplasma bovis (1–2 16S rRNA copies), and S. pneumoniae (4 16S rRNA copies), and the effects of these mutations on tetracycline binding can generally be explained by crystallographic or biophysical data. In H. pylori, a triple mutation AGA 965-967 TTC in the h31 loop, and a deletion of G942 (E. coli numbering), each conferred tetracycline resistance (Trieber and Taylor 2002). Residues 965–967 are located in the primary, or Tet-1, binding site, whereas G942 is located in the Tet-4 secondary binding site (Brodersen et al. 2000; Pioletti et al. 2001). Mutations in h34 of 16S rRNA were associated with increased tetracycline resistance in P. acnes (G1058C) and M. bovis (G1058A/C), and tetracycline-resistance mutations A965T, A967T/C, and U1199C (which base pairs with G1058 in h34) were also found in M. bovis (Ross et al. 1998; Amram et al. 2015). Although G1058 does not directly interact with tetracycline, mutation to cytosine likely causes a conformational change in the binding site, reducing the affinity of tetracycline for the 30S ribosomal subunit. Preexisting G1058C mutations in P. acnes reduced the antibacterial activities of tetracycline, doxycycline, eravacycline, and tigecycline, consistent with all of these tetracycline antibiotics having common interactions with rRNA in bacteria (Grossman et al. 2012). In S. pneumoniae, mutations in 16S RNA C1054T and T1062G/A conferred tigecycline resistance when present in the four genomic copies of 16S rRNA (Lupien et al. 2015). Whereas resistance caused by a mutation in C1054 can be explained by the interaction of this residue with tigecycline, a more indirect effect on tigecycline binding may be conferred by mutations in T1062. Nonsense mutations in a gene encoding a 16S rRNA methyltransferase in S. pneumoniae were also found to confer reduced tigecycline susceptibility in the study by Lupien et al. (2015). This enzyme methylates position N(2) of G966 in h31 of 16S rRNA in E. coli and the alterations in this activity may reduce tigecycline binding to the ribosome.
Unlike rRNA genes, genes encoding ribosomal proteins are single copy and mutations in these genes can confer antibiotic resistance. Mutations in the rpsJ, encoding changes or deletions in residues 53–60 in the 30S ribosomal subunit protein S10, have been linked to tetracycline or tigecycline resistance in in vitro studies with Gram-positive bacteria Bacillus subtilis, Enterococcus faecium, E. faecalis, and S. aureus (Williams and Smith 1979; Wei and Bechhofer 2002; Beabout et al. 2015a; Cattoir et al. 2015), in clinical isolates of Gram-negative bacteria Neisseria gonorrhoeae and K. pneumoniae (Hu et al. 2005; Villa et al. 2014), and in in vitro studies with E. coli and Acinetobacter baumannii (Beabout et al. 2015a). Identification of a tigecycline-resistant K. pneumoniae strain with an rpsJ mutation encoding Val57Leu in S10 was the first description of tetracycline resistance attributable, at least in part, to a target site mutation in Enterobacteriaeceae (Villa et al. 2014). In the T. thermophilus crystal structure, these S10 residues map to a loop projecting toward the aminoacyl-tRNA-binding site in the 30S structure (Brodersen et al. 2000; Carter et al. 2000). Although located ∼8.5 Å from tetracycline in the structure, it has been proposed that this region of the S10 protein may alter the interaction of tetracyclines and 16S rRNA in this region (Hu et al. 2005). Mutations in rpsC encoding Lys4Arg and His175Asp variations in ribosomal protein S3 were associated with reduced tigecycline susceptibility in S. pneumoniae (Lupien et al. 2015). Ribosomal protein S3 has been shown to be important for tetracycline binding to the ribosome (Buck and Cooperman 1990).
Tetracycline-Specific Ribosomal Protection
Tetracycline ribosomal protection proteins (RPPs), originally described in Campylobacter jejuni and Streptococcus spp., are GTPases with significant sequence and structural similarity to elongation factors EF-G and EF-Tu (Burdett 1986; Taylor et al. 1987; Sanchez-Pescador et al. 1988; Kobayashi et al. 2007). According to a nomenclature list maintained at the University of Washington (faculty.washington.edu/marilynr), there are currently 12 reported ribosomal protection genes. These genes are disseminated through bacterial populations on mobile genetic elements, and many of the genes are found in both Gram-negative and Gram-positive organisms (Roberts 2011). The most common and best characterized RPPs are Tet(O) and Tet(M), with 75% sequence similarity to each other. These proteins catalyze the GTP-dependent release of tetracycline from the ribosome (Connell et al. 2003a,b). Cryoelectron microscopic structural studies indicate that RPPs compete with EF-G for an overlapping binding site, and it is thought that RPPs dissociate tetracycline from its binding site by directly interfering with the stacking interaction of the tetracycline D-ring and 16S rRNA base C1054 within h34 (Donhofer et al. 2012; Li et al. 2013). Conformational changes induced by RPPs promote rapid binding of the EF-Tu • GTP • aa-tRNA ternary complex, enabling translation to continue in the presence of tetracycline (Donhofer et al. 2012). RPP mechanisms confer resistance to tetracycline, minocycline, and doxycycline; however, other tetracyclines containing side chains at the C-9 position of the D-ring, such as tigecycline and other glycylcyclines, eravacycline and other fluorocyclines, and omadacycline, generally retain translational inhibitory and antibacterial activities in the presence of RPPs (Table 1) (Rasmussen et al. 1994; Bergeron et al. 1996; Grossman et al. 2012; Jenner et al. 2013). The 9-t-butylglycylamido moiety at the C-9 position in tigecycline was shown to improve binding affinity and translational inhibition by >100-fold and 20-fold, respectively, over that of tetracycline; however, the mechanism of RPP evasion could not be fully explained (Olson et al. 2006). Recently, using a set of novel synthetic tetracycline derivatives containing C-9 side chains with different degrees of bulkiness, Jenner et al. (2013) showed that, in addition to conferring enhanced interactions with C1054 (Schedlbauer et al. 2015), steric interference by the bulk of the C-9 side chain is also a significant factor in maintaining ribosome binding in the presence of RPPs. Although earlier reports have shown the relative immunity of tigecycline to RPP mechanisms, a recent study by Beabout et al. (2015b) has linked Tn916-associated constituitive overexpression and increased copy number of tet(M) to tigecycline resistance in E. faecalis.
Table 1.
The activities of tetracyclines against recombinant E. coli expressing major tetracycline-specific resistance mechanisms
| MIC (μg/mL) | ||||||
|---|---|---|---|---|---|---|
| E. coli lacZ | E. coli tet(M) | E. coli tet(K) | E. coli tet(A) | E. coli tet(B) | E. coli tet(X) | |
| Eravacycline | 0.063 | 0.063 | 0.031 | 0.25 | 0.063 | 4 |
| Tigecycline | 0.063 | 0.13 | 0.063 | 1 | 0.063 | 2 |
| Doxycycline | 2 | 64 | 4 | 32 | 32 | 16 |
| Minocycline | 0.5 | 64 | 1 | 8 | 16 | 4 |
| Tetracycline | 2 | 128 | 128 | >128 | >128 | 128 |
| Ceftriaxone | 0.063 | 0.13 | 0.063 | 0.13 | 0.13 | 0.13 |
Genes were overexpressed in E. coli DH10B from a recombinant expression vector under the control of an arabinose promoter. Standardized MIC assays were performed according to CLSI methodology as previously described.
MIC, minimal inhibitory concentration; tet(M), ribosomal protection; tet(K), Gram-positive tetracycline efflux; tet(A) and tet(B), Gram-negative efflux; tet(X), flavin-dependent monooxygenase.
Data is reprinted, with permission, from Grossman et al. (2012).
Tetracycline-Specific Efflux
The most common tetracycline-specific efflux pumps are members of the major facilitator superfamily (MFS) of transporters (Chopra and Roberts 2001); however, there have been rare reports of non-MFS pumps (Teo et al. 2002; Warburton et al. 2013). The latest tally shows that 30 distinct tetracycline-specific efflux pumps reported in bacteria (faculty.washington.edu/marilynr; updated August 6, 2015). These pumps extrude tetracycline antibiotics from the inside of cells at the expense of a proton, and have been assigned to seven different groups according to amino acid sequence similarities and the number of times they traverse the inner membrane (9–14 times) (Guillaume et al. 2004; Thaker et al. 2010). The most clinically prevalent pumps are members of either group 1 or group 2. The group 1 drug−H+ antiporters contain 12 transmembrane segments organized into α and β domains connected by a large interdomain cytoplasmic loop. This group includes Tet(A) and Tet(B), the most commonly found tetracycline pumps in Gram-negative clinical isolates. The group 2 pumps possess 14 transmembrane segments and include Tet(K) and Tet(L), the most common tetracycline-specific efflux pumps in Gram-positive clinical isolates. In addition to their role in conferring tetracycline resistance, group 2 pumps are also monovalent cation–H+ antiporters, and may play a role in coping with sodium stress, alkali stress, and potassium insufficiency (Guay et al. 1993; Krulwich et al. 2001). Pumps assigned to group 3–7 include pumps that are less prevalent clinically (Guillaume et al. 2004).
The order of substrate preference across all tetracycline efflux pump types can be shown in recombinant E. coli strains overexpressing representative pumps in an isogenic background: tetracycline > minocycline, doxycycline > tigecycline, eravacycline (Table 1) (Grossman et al. 2012). It should be noted, however, that it is likely that multiple strain-specific factors in clinical isolates affecting uptake and intrinsic efflux systems, in addition to the level of expression of tetracycline-specific pumps, play a coordinated role in the overall susceptibility to tetracyclines. Tet(A), Tet(B), and Tet(K) pumps are all able to recognize tetracycline, minocycline, and doxycycline. Whereas Tet(B) and Tet(K) overexpression had no effect on tigecycline and eravacycline, overexpression of Tet(A) produced a fourfold increase in eravacycline minimal inhibitory concentration (MIC) and a 16-fold increase in tigecycline MIC versus the negative control strain, indicating that these newer tetracyclines are recognized to differing extents by the Tet(A) pump (Table 1) (Grossman et al. 2012). Earlier characterizations of the substrate specificity of Tet pumps in nonisogenic strain backgrounds led to the conclusion that tigecycline was not a substrate for Tet(A) (Petersen et al. 1999), and that a naturally occurring amino acid sequence variation (Ser-Phe-Val→Ala-Ser-Phe) in the interdomain loop sequence at residues 201–203 affected recognition of tigecycline and minocycline (Tuckman et al. 2000). More recent work has shown that recombinant expression of either Tet(A) pump variation in E. coli produced similar tigecycline and minocycline susceptibility, confirming that these amino acid residues do not appear to be involved in substrate recognition (Fyfe et al. 2013). The notion that mutations in tetracycline pumps can alter substrate specificity is, however, supported by studies with tet(B) in which mutations encoding residues in transmembrane domains had opposing effects on tetracycline versus glycylcycline susceptibility (Guay et al. 1994), and site-directed mutations in the interdomain loop had opposing effects on tetracycline versus minocycline and doxycycline susceptibility (Sapunaric and Levy 2005). These studies suggest the possibility that tetracyclines could select for resistance mutations within tetracycline pump genes during clinical use; however, this has not yet been reported in clinical isolates.
Enzymatic Inactivation of Tetracyclines
Evidence of a tetracycline-modifying enzyme mechanism was first described as an activity encoded by a Bacteroides plasmid expressed in E. coli (Speer and Salyers 1988, 1989). This activity was subsequently characterized as a flavin-dependent monooxygenase, encoded by an expanding family of tet(X) orthologs, capable of covalently inactivating all tetracyclines with the addition of a hydroxyl group to the C-11a position located between the C and B rings of the tetracycline core (Fig. 1) (Speer et al. 1991; Yang et al. 2004; Moore et al. 2005; Grossman et al. 2012; Aminov 2013). Because Bacteroides species are obligate aerobes, it is not surprising that the oxidoreductases encoded by tet(X) and its orthologs tetX1 and tetX2 do not confer resistance in the isolates in which they were originally found (Whittle et al. 2001). The environmental origin of tet(X) is suggested by its identification in Sphinogbacterium spp., a Gram-negative soil bacterium that expresses a functional Tet(X) (Ghosh et al. 2009). Further, the presence of tet(X) and genes encoding similar tetracycline-inactivating activities, also known as “tetracycline destructases,” in agricultural and aquacultural bacteria ensures the persistence of this resistance mechanism in the food chain, facilitating crossover into human pathogens (Aminov 2013; Forsberg et al. 2015). Because of the conjugative nature of tet(X)-containing plasmids and transposons, recent reports of tet(X) in Enterobacteriaceae and Pseudomonadaceae hospital urinary tract infection (UTI) isolates in Sierra Leone, and tet(X) in A. baumannii in a Chinese hospital, are of concern with regard to the spread of this mechanism (Leski et al. 2013; Deng et al. 2014).
Other less well-characterized tetracycline-modifying mechanisms have also been described. An NADP-requiring tetracycline-modifying activity similar to that of Tet(X) was expressed from the metagenomic DNA of uncultivatable oral microflora; however, there is no homology between the deduced amino acid sequence of Tet(37) from the oral metagenome and Tet(X) from Bacteroides (Diaz-Torres et al. 2003). Another gene, tet(34), has been cloned from the chromosome of Vibrio spp. and encodes a xanthine-guanine phosphoribosyltransferase capable of conferring resistance to oxytetracycline (Nonaka and Suzuki 2002). The clinical relevance of these two tetracycline-modifying enzymes remains to be determined.
INTRINSIC MULTIDRUG-RESISTANCE MECHANISMS AFFECTING TETRACYCLINES
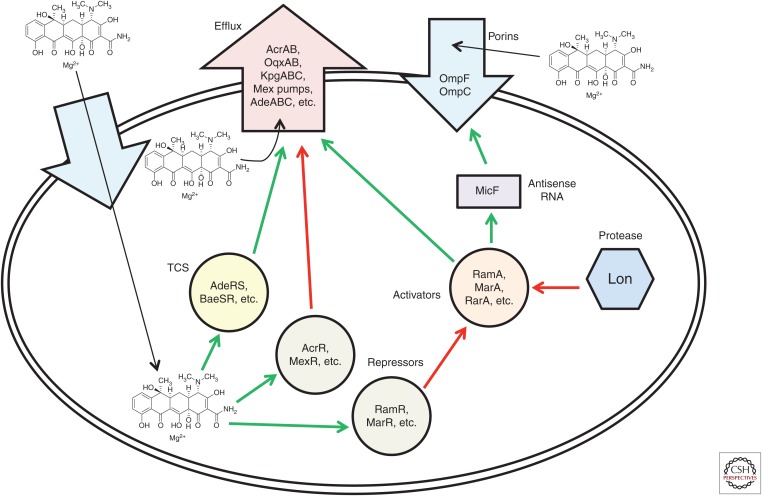
Complex intrinsic regulatory networks in bacteria modulate the uptake and intracellular accumulation of most antibiotics, including tetracyclines. Mutations affecting expression and/or function of one or more key repressor, activator, pump, or porin can simultaneously impact the susceptibility to a broad range of antibiotic classes (Fig. 2).
AraC Transcriptional Activators in Gram-Negative Bacteria
MarA, RamA, SoxS, RobA, and the newly described RarA are members of the “AraC-family” of bacterial transcriptional activators that enable Gram-negative bacteria to respond to different types of environmental stress, including antibiotic exposure (Fig. 3) (Martin and Rosner 2001; Grkovic et al. 2002; De Majumdar et al. 2013). Each activator regulates a set of genes in response to a specific type of stress (Martin et al. 2008; Martin and Rosner 2011); for instance, MarA regulates more than 60 genes collectively referred to as the “mar regulon,” for multiple antibiotic resistance (Barbosa and Levy 2000). AraC-family activators bind to a consensus 20 base pair sequence via two helix–turn–helix motifs that comprise the DNA-binding domain. The DNA-binding site is known as the “box” and is located in the promoter region of stress-responsive genes (i.e., “marbox” for MarA, etc.). Further, AraC-family activators can bind their own promoters and autoactivate their own expression (Alekshun and Levy 1997; Rosenblum et al. 2011). Mutations promoting constituitive expression of AraC-family regulons are now known to be common mechanisms contributing to multidrug resistance.
Figure 3.
Regulation of expression of Gram-negative intrinsic multidrug-resistance mechanisms affecting tetracyclines. A summary of known regulatory mechanisms affecting tetracycline susceptibility are shown. Green arrows indicate interactions in which tetracycline resistance is “increased,” and red arrows indicate interactions in which tetracycline resistance is “reduced.” See the text for details. TCS, Two-component signal transduction system.
The first description of mar in E. coli by George and Levy in 1983 showed that amplifiable resistance to tetracyclines, as well as structurally and mechanistically unrelated antibiotics, including chloramphenicol, penicillins, cephalosporins, puromycin, nalidixic acid, and rifampin, was caused by an energy-dependent efflux system (George and Levy 1983a,b). The Mar regulon is now known to be widespread in enteric Gram-negative species, including E. coli, K. pneumoniae, Salmonella spp., Shigella spp., Citrobacter spp., Enterobacter spp., and Yersinia spp. (Cohen et al. 1993; Alekshun and Levy 1997). The mar locus encodes two divergent operons regulated by a repressor MarR that binds to an operator MarO (Martin and Rosner 1995; Seoane and Levy 1995). Induction of MarR triggers the expression of marC in one direction and marRAB in the other direction. Whereas tetracycline has been shown to be an inducer of marA expression (Hachler et al. 1991), induction appears to be via an indirect mechanism because direct binding of tetracycline to MarR could not be shown (Martin and Rosner 1995). MarA is a key activator of stress-responsive genes, and its role in promoting overexpression of the major multidrug efflux pump, AcrAB (Li and Nikaido 2009), is central to conferring an MDR phentoype in enteric bacteria (Gambino et al. 1993; Alekshun and Levy 1997). MarA also controls the expression of the major Gram-negative porin OmpF through the up-regulation of micF. MicF is an antisense RNA regulator of ompF expression, acting by reducing the levels of ompF mRNA (Cohen et al. 1988; Andersen and Delihas 1990; Gambino et al. 1993). Reduction in ompF expression contributes to reduced accumulation of tetracycline and other antibiotics (Mortimer and Piddock 1993; Thanassi et al. 1995). The roles of MarC and MarB in multidrug resistance are less well defined; MarC has been shown to encode a periplasmic protein, which appears to indirectly affect the transcription of marA (Vinue et al. 2013). Whereas first-step mar mutants may not confer clinically relevant resistance to some classes of antibiotics, it is possible that first-step mutants can achieve clinically relevant resistance to tetracycline (George and Levy 1983b); however, this has not yet been shown in clinical isolates. Reduced susceptibility to tigecycline in E. coli clinical isolates has been attributed to increased overexpression of AcrAB correlating with mutations in marR and increased transcription of marA (Keeney et al. 2008). In a study by Linkevicius et al. (2013), targeted sequencing of loci suspected to be involved in tigecycline resistance found a deletion in marR in one of eight E. coli clinical isolates with reduced tigecycline susceptibility; however, MICs were still well below the resistance breakpoint (MIC = 0.19 µg/mL).
RamA, another AraC-family activator, was first identified in K. pneumoniae showing reduced susceptibility to a range of unrelated antibiotics, including tetracycline (George et al. 1995). Expression of the ramA gene is repressed by RamR, encoded by the ramR gene that is divergently transcribed from the nearby ramA gene. Similar to MarR, tetracycline does not directly bind RamR; thus, induction of ramA appears indirect (Yamasaki et al. 2013). Analogous to regulation by marA in E. coli, overexpression of ramA was shown to also reduce porin expression and up-regulate AcrAB efflux in K. pneumoniae (George et al. 1995; Ruzin et al. 2005b), S. enterica (van der Straaten et al. 2004a,b; Nikaido et al. 2008), and Enterobacter cloacae (Keeney et al. 2007). RamA function appears to be independent of MarA, as RamA-mediated increases in AcrAB expression were not associated with increases in MarA expression (Ruzin et al. 2005b). Although, heterologously expressed ramA from bacteria, including K. pneumoniae, Salmonella, Citrobacter, and Enterobacter spp., is functional in E. coli (Chollet et al. 2004; van der Straaten et al. 2004b; Ruzin et al. 2005b; Reinhardt 2014), a ramA gene has not been identified in several enteric species, notably E. coli and Shigella spp.
In K. pneumoniae, a survey of recent literature suggests an emerging theme that AraC-family activators, especially ramA, play a prominent role in clinically relevant resistance to tetracycline antibiotics. A study by Bratu et al. (2009) showed that reduced tigecycline susceptibility in K. pneumoniae clinical isolates from New York City correlated with ramA and soxS expression, but not with marA or acrAB expression (Bratu et al. 2009). However, in the same study, K. pneumoniae mutants selected in vitro for reduced tigecycline susceptibility showed increases in marA and acrB expression, but not soxS and ramA, so the interplay of regulators appears complicated. In another study, analysis of 72 demographically and geographically diverse K. pneumoniae clinical isolates from the tigecycline phase 3 clinical trials showed that isolates with tigecycline MIC values >2 µg/mL had a statistically significant correlation with elevated expression of ramA and a less significant trend with acrA expression (Ruzin et al. 2008). The association of tigecycline resistance with mutations in genes encoding MDR repressors (ramR, acrR) and/or increased expression of genes encoding AraC-family activators (rarA, marA, ramA) and efflux pump subunits (acrB, oqxB) has been described in several studies with geographically diverse isolates from Germany (Hentschke et al. 2010); Turkey, Singapore, Chile, and Pakistan (Rosenblum et al. 2011); Italy (Villa et al. 2014); and China (Bratu et al. 2009; Sheng et al. 2014; Zhong et al. 2014; He et al. 2015).
Additional pathways to tigecycline resistance in K. pneumoniae are suggested by the identification of tigecycline-resistant K. pneumoniae clinical isolates that do not overexpress ramA (Rosenblum et al. 2011) and by the isolation of low-level tigecycline-resistant strains from a K. pneumoniae ramA deletion mutant (Veleba and Schneiders 2012). Bioinformatic scanning of the K. pneumoniae genome for new AraC-family regulators identified rarA (Veleba et al. 2012). Expression of rarA and the nearby operon oqxAB encoding an MDR efflux pump were found to be elevated in geographically diverse K. pneumoniae MDR clinical isolates (Veleba et al. 2012) and E. cloacae isolates (Veleba et al. 2013) with reduced tigecycline susceptibility. The presence of rarA was confirmed in the genomes of Enterobacter and Serratia spp., and similar to other AraC-family regulators, overexpression of rarA produced a low-level MDR phenotype, including tigecycline resistance in K. pneumoniae and E. cloacae (Veleba et al. 2012, 2013). RarA is thought to be the activator of oqxAB (Kim et al. 2009), and has been linked to the regulation of acrAB and ompF expression (De Majumdar et al. 2013).
Two-Component Systems
Two-component signal transduction systems (TCSs) in bacteria are the most common form of bacterial signal transduction, and are generally composed of a membrane-bound histidine kinase and a response regulator, to which a phoshoryl group is transferred, allowing it to function as a transcription factor affecting the expression of responsive genes (Bem et al. 2015). Several TCSs have been implicated in modulating susceptibility to tetracycline-class antibiotics in Gram-negative and -positive bacteria, presumably by affecting permeability and/or expression of intrinsic multidrug efflux systems.
In A. baumannii, the AdeRS TCS controls expression of the major multidrug efflux pump AdeABC (Marchand et al. 2004). Mutations in the AdeR regulator and/or AdeS sensor affecting the normal phosphotransfer process can lead to the constituitive expression of the AdeABC efflux pump (Marchand et al. 2004). Ruzin et al. (2007) were the first to show that elevated tigecycline MICs (4 µg/mL) were associated with constituitive overexpression of AdeABC in two clinical isolates, and coincided with an insertion element in the adeS gene in both isolates. In more recent studies, characterizing 81 genetically diverse XDR and 38 carbapenem-resistant MDR A. baumannii clinical isolates from Taiwan (Sun et al. 2012, 2014b) reported that tigecycline resistance (MIC ≥8 µg/mL) correlated with overexpression of AdeABC in the majority of isolates, likely resulting from mutations in adeR and adeS encoding changes in conserved amino acid residues, or an insertion sequence (IS) in adeS producing a truncated constituitively “on” form of AdeS. Although a more detailed understanding of how these mutations impact AdeRS signaling and AdeABC expression remains to be elucidated, the recurrence of genetic alterations in adeR and adeS genes strongly implicates the involvement of AdeRS and the AdeABC efflux system in tigecycline resistance. The existence of multiple mechanisms affecting tigecycline susceptibility in A. baumannii is suggested by cases of tigecycline-resistant isolates in which either no mutations in adeR and adeS were found (Hornsey et al. 2010c; Yoon et al. 2013; Sun et al. 2014b) or additional mutations in rpsJ, rrf, msbA, and gna were associated with increasing the level of tigecycline resistance in adeS mutants; the possible roles of rrf, msbA, and gna in tigecycline resistance remains to be shown (Hammerstrom et al. 2015). Interestingly, reduced susceptibility to tigecycline, minocycline, and doxycycline was associated with a deletion in the trm (tigecycline-related methyltransferase) gene encoding an S-adenosyl-l-methionine-dependent methyltransferase in an A. baumannii isolate; this newly identified mechanism may be responsible for some resistance not attributable to AdeABC (Chen et al. 2014; Lomovskaya et al. 2015).
Other TCSs that have been associated with resistance to tetracyclines include BaeSR in A. baumannii (Lin et al. 2014), PhoBR in K. pneumoniae (Srinivasan et al. 2012), and RprXY in Bacteroides fragilis (Rasmussen and Kovacs 1993); however, the relevance of these systems in conferring clinical resistance to tetracyclines is not yet understood.
Lon Protease
Induction of multidrug resistance through AraC-family regulators in Gram-negative bacteria is posttranslationally regulated by the cytoplasmic ATP-dependent serine protease, Lon, which is involved in the degradation of unstable or misfolded proteins (Tsilibaris et al. 2006). In the absence of environmental stress, Lon promotes rapid reversion of stress phenotypes by binding at amino-terminal residues of activators MarA, RamA, SoxS and proteolytically degrading them (Griffith et al. 2004; Nicoloff et al. 2006; Ricci et al. 2014). It follows that mutations in lon may prolong the stability of these stress-responsive activators, increasing expression of acrAB and other resistance genes, leading to antibiotic resistance or reduced susceptibility.
The involvement of Lon protease in the development of antibiotic resistance was shown in a series of 13 E. coli cultures derived from a single inoculum in which a significant subpopulation (∼3.7 × 10−4) contained a lon::IS186 mutation, or deletion in lon, and was capable of growing in low-level tetracycline and chloramphenicol (Nicoloff et al. 2007). Most mutants characterized in this study also contained IS elements in marR or acrR, or tandem amplifications of the acrAB region, and the antibiotic-resistance phenotype, at least in part, could be attributed to these mutations. Because Lon protease is also involved in the stability of transposase enzymes from IS elements and transposons, E. coli lon mutant strains show higher transposition rates and greater genome instability (Derbyshire et al. 1990; Nagy and Chandler 2004; Rouquette et al. 2004). Further, the lon gene itself is a hotspot for IS insertions (SaiSree et al. 2001). Thus, the potential to select for early steps in drug resistance in vitro appears to be much higher in lon mutants, and this is supported by the finding that genomic duplications of the region encoding the major efflux pump, AcrAB, can be readily isolated in an E. coli lon mutant (Nicoloff et al. 2006; Nicoloff and Andersson 2013).
Whether lon mutations increase the potential to select for resistance to tetracyclines, or any other antibiotic class, in clinical isolates during infection is not entirely clear (Butler et al. 2006). There has been at least one report of an E. coli acrR (A191V), lon::IS186 mutant isolated from a UTI, and this mutant showed reduced susceptibility to tigecycline (MIC = 0.25 µg/mL), but still did not reach a level considered clinically significant (Linkevicius et al. 2013). A tigecycline-resistant K. pneumoniae clinical isolate containing a frameshift within the coding region of lon was reported by Fyfe et al. (2015); however, this mutant also had a deletion in ramR, which presumably also contributed to the tigecycline-resistant phenotype (MIC = 8 µg/mL). In this same study, K. pneumoniae lon mutants generated by transposon mutagenesis showed 8- to 32-fold increases in the parental tigecycline MIC (0.5 µg/mL), suggesting that mutation in lon can contribute to clinically significant resistance levels. Additional studies are needed to clarify the contribution of lon to the development of resistance to tetracyclines and other antibiotics in clinical isolates.
Intrinsic Efflux of Tetracyclines
A large component of the intrinsic antibiotic-resistance response in bacteria is due to increased expression of intrinsic efflux pumps (Piddock 2006; Li et al. 2015). As described earlier, expression of these genes can be modulated by locally or distally encoded negative and positive regulators, and mutations up-regulating or down-regulating expression of the regulators themselves, or the efflux pumps they regulate, can impact antibiotic susceptibility. Susceptibility to tetracycline-class antibiotics has been linked to a wide variety of intrinsic efflux systems in Gram-negative and -positive bacteria summarized in Table 2.
Table 2.
Intrinsic bacterial multidrug efflux mechanisms conferring resistance to tetracycline drugs
| Pathogen | Efflux pump family | Known tetracycline specificity | References |
|---|---|---|---|
| A. baumannii | RND | AdeABC: tetracycline, tigecycline, minocycline,a doxycyclinea AdeDE: tetracycline AdeFGH: tetracycline, minocycline, tigecycline AdeIJK: tetracycline, minocycline, doxycycline, tigecycline |
Chau et al. 2004; Ruzin et al. 2007; Damier-Piolle et al. 2008; Coyne et al. 2010, 2011; Ruzin et al. 2010; Lomovskaya et al. 2014, 2015 |
| B. fragilis | RND | BmeABC: tetracycline | Pumbwe et al. 2006 |
| E. coli | RND | AcrAB: tetracycline, tigecycline, minocycline, doxycycline AcrEF: tetracycline, tigecycline, minocycline, doxycycline |
Hirata et al. 2004 |
| Enterobacter spp. | RND | AcrAB: tetracycline, tigecycline, minocycline OqxAB: tigecycline |
Keeney et al. 2007; Hornsey et al. 2010b; Veleba et al. 2013 |
| E. faecalis | ABC | EfrAB: doxycycline (not tetracycline) | Lee et al. 2003 |
| K. pneumoniae | RND | AcrAB: tetracycline, tigecycline, minocycline OqxAB: tigecycline, tetracycline KpgABC: tigecycline |
Ruzin et al. 2005b; Ruzin et al. 2008; Veleba and Schneiders 2012; Nielsen et al. 2014; Zhong et al. 2014; He et al. 2015 |
| P. aeruginosa | RND | MexAB-OprM: tetracycline, minocycline, doxycycline, chlortetracycline, oxytetracycline, tigecycline MexCD-OprJ: tetracycline, chlortetracycline, oxytetracycline, tigecycline MexJK: tetracycline MexXY-OprM: tetracycline, minocycline, doxycycline, chlortetracycline, oxytetracycline, tigecycline |
Masuda et al. 2000; Morita et al. 2001; Chuanchuen et al. 2002; Dean et al. 2003; Schweizer 2003 |
| P. mirabilis | RND | AcrAB: tigecycline, minocycline | Visalli et al. 2003 |
| S. aureus | MATE | MepA: tigecycline, eravacycline (not tetracycline) NorB: tetracycline (not minocycline) |
McAleese et al. 2005; Truong-Bolduc et al. 2005; Sutcliffe et al. 2013 |
| Stenotrophomonas maltophilia | ABCRND | SmrA: tetracycline SmeDEF: tetracycline, doxycycline, minocycline, tigecycline |
Alonso and Martinez 2001; Zhang et al. 2001; Al-Hamad et al. 2009 |
| Serratia marcescens | RNDABC | SdeXY-HasF: tetracycline, tigecycline SmdAB: tetracycline |
Chen et al. 2003; Matsuo et al. 2008; Hornsey et al. 2010a |
aPoorer substrates for AdeABC as compared with other tetracycline drugs.
RND, Resistance-nodulation-division superfamily; MATE, multidrug and toxic compound extrusion family; ABC, ATP-binding cassette transporter family.
Overexpression of AcrAB, the major pump found in Enterobacteriaceae, and a member of the resistance-nodulation-division (RND) superfamily, has been implicated in resistance to tigecycline, in E. coli (Hirata et al. 2004), Enterobacter spp. (Keeney et al. 2007), K. pneumoniae (Ruzin et al. 2005b), Morganella morganii (Ruzin et al. 2005a), and Proteus mirabilis (Visalli et al. 2003). Two recently identified pumps, OqxAB and KpgABC, in K. pneumoniae also appear to have some association with tigecycline resistance, but their clinical significance is uncertain (Nielsen et al. 2014; Bialek-Davenet et al. 2015; He et al. 2015). In Serratia marcescens, pumps with specificity for tetracycline and/or tigecycline include SdeXY-HasF and SmdAB (Chen et al. 2003; Matsuo et al. 2008; Hornsey et al. 2010a).
RND-type pumps are also implicated in conferring reduced susceptibility in nonfermenter and anaerobic Gram-negative bacteria. In A. baumannii clinical isolates, as discussed earlier, reduced susceptibility to tigecycline and eravacycline has been correlated with AdeABC pump expression (Ruzin et al. 2010; Abdallah et al. 2015). Interestingly, the AdeABC pump appears to show some selectivity among the tetracyclines, as minocycline is reported to be a weaker substrate than other tetracyclines (Coyne et al. 2011; Lomovskaya et al. 2014). Pseudomonas aeruginosa strains are generally less susceptible to tetracycline antibiotics, including tigecycline, and this is largely because of expression of the MexAB-OprM, MexCD-OprJ, MexXY-OprM pumps (Dean et al. 2003). In Stenotrophomonas maltophilia, SmrA and SmeDEF (Alonso and Martinez 2001; Zhang et al. 2001; Al-Hamad et al. 2009), and in B. fragilis, BmeABC (Pumbwe et al. 2006), have been reported to recognize tetracycline, but their clinical significance remains to be shown.
Much less is known about the regulation of intrinsic resistance to tetracyclines in Gram-positive bacteria. The best-characterized intrinsic Gram-positive pump with demonstrated specificity for tetracyclines is the multidrug and toxic compound extrusion (MATE)-family pump, MepA, in S. aureus. Although this pump does not appear to recognize tetracycline as a substrate, fourfold and 64-fold increases in MIC for eravacycline and tigecycline, respectively, were observed for a MepA overexpressing strain versus the isogenic parent, indicating a distinct tetracycline substrate specificity for this pump (McAleese et al. 2005; Sutcliffe et al. 2013). The NorB pump, negatively regulated by MgrA in S. aureus, has also been reported to recognize tetracycline (Truong-Bolduc et al. 2005).
THE PRESENT AND FUTURE FOR TETRACYCLINES
Minocycline
Historically, minocycline has been available in both oral and intravenous dosage formulations. As options for the treatment of MDR A. baumannii are limited, the recent approval of a new intravenous (IV) formulation, Minocin IV, for treatment of Acinetobacter spp. and other difficult-to-treat Gram-positive and -negative pathogens, is a valuable repurposing of an old antibiotic for targeted use (The Medicines Company 2015). In the 2004–2013 Tigecycline Evaluation and Surveillance Trial (TEST) report, the highest level of in vitro susceptibility against A. baumannii isolates was reported for minocycline (84.5%), and 70.3% susceptibility was observed against MDR A. baumannii (Hoban et al. 2015). In the global 2007–2011 SENTRY surveillance program, minocycline was the second most active antibiotic against A. baumannii (79.1% susceptible) (Castanheira et al. 2014). This might be explained to some extent by the ability of minocycline to thwart AdeABC efflux, and a lower rate of minocycline-resistance development in A. baumannii (Lomovskaya et al. 2014). Clinical responses to Minocin IV used as a monotherapy or in combination for the treatment of MDR A. baumannii infections appear encouraging (Goff et al. 2014; Ritchie and Garavaglia-Wilson 2014; Falagas et al. 2015), but this therapy will likely be a stop-gap as the spread of RPPs should increase minocycline resistance.
Tigecycline
Tigecycline has a broad spectrum of coverage, including activity against MRSA, vancomycin-resistant Enterococcus spp. (VRE), MDR A. baumannii, and ESBL-producing and carbapenem-resistant Enterobacteriaceae (CRE), supporting the currently approved indications of complicated skin and skin structure infections, complicated intra-abdominal infections (cIAI), and community-acquired bacterial pneumonia (CABP) (Stein and Babinchak 2013; Wyeth Pharmaceuticals 2016). The administration of tigecycline is limited to IV only. Given its activity against MDR pathogens, tigecycline was evaluated for use in hospital-acquired pneumonia, ventilator-associated pneumonia, and diabetic foot infections; however, clinical studies showed lower cure rates versus comparator drugs (Wyeth Pharmaceuticals 2016). Broader usage and alternative dosing regimens for serious infections continue to be explored in clinical studies (Ramirez et al. 2013; Stein and Babinchak 2013). Based on meta-analyses of clinical trial data, the FDA issued a safety alert in 2010 and a black box warning in 2013 because of an observed increase in mortality risk in patients treated with tigecycline, as compared with other drugs (U.S. Food and Drug Administration 2010, 2013). Whereas the cause of death during tigecycline treatment remains uncertain, mortality appeared to occur in patients with complicated worsening infections or underlying medical conditions.
Since its approval in 2005, tigecycline maintains high levels of susceptibility in global surveillance studies despite sporadic reports of resistance during use: E. coli and K. pneumoniae in the United Kingdom (Stone et al. 2011; Spanu et al. 2012); K. pneumoniae in Greece (Neonakis et al. 2011), Saudi Arabia (Al-Qadheeb et al. 2010), Spain (Rodriguez-Avial et al. 2012), the United States (Nigo et al. 2013); E. hormaechei in France (Daurel et al. 2009); A. baumannii in the United States (Peleg et al. 2007; Reid et al. 2007; Anthony et al. 2008); E. faecalis in the United Kingdom (Cordina et al. 2012) and Germany (Werner et al. 2008); and B. fragilis in the United States (Sherwood et al. 2011). There are also reports in which tigecycline resistance actually predated the use of tigecycline in institutions in which resistance was detected (Rosenblum et al. 2011; Zhong et al. 2014) or arose during treatment with another antibiotic (Hornsey et al. 2010b).
In the 2004–2013 TEST report, among the Enterobacteriaceae (n = 118,899), enterococci (n = 20,782), methicillin-resistant S. aureus (n = 14,647), and S. pneumoniae (n = 14,562), susceptibility to tigecycline was 97%, >99%, ≥99.9%, and ≥99.9%, respectively (Hoban et al. 2015). Against MDR Enterobacteriaceae (n = 9372), defined as resistant to more than three different classes of antibiotics, susceptibility to tigecycline was 83.2%. And, among the carbapenem-resistant Enterobacter spp. (n = 578), E. coli (n = 181), and K. pneumoniae (n = 1330), susceptibility to tigecycline was 83%, 97.2%, and 92%, respectively. Although there are no breakpoints available against A. baumannii, tigecycline maintained an MIC90 (MIC inhibiting 90% of the isolates) of 2 µg/mL against all A. baumannii (n = 16,778), as well as an MDR subset (n = 6743). The gap in coverage of P. aeruginosa is evident by an MIC90 of 16 µg/mL against all collected isolates (n = 28,413) and ≥32 µg/mL against the MDR subset (n = 3496). Similar findings were observed in recent reports from the SENTRY (Sader et al. 2013, 2014), Regional Resistance Surveillance (Jones et al. 2014), and CANWARD (Zhanel et al. 2013) programs.
Omadacycline
The aminomethylcycline derivative of minocycline, omadacycline, has completed a phase 2 trial for safety and efficacy in skin and skin structure infections (SSSI) and is being developed for use in SSSI, CABP, and UTIs with IV and oral formulations (Noel et al. 2012). Omadacycline was shown to have MIC90 values against MRSA (n = 39), VRE (n = 19), Streptococcus pyogenes (n = 30), penicillin-resistant S. pneumoniae (n = 23), and Haemophilus influenzae (n = 53) of 0.5, 0.5, 0.25, ≤0.06, and 2 µg/mL, respectively (Macone et al. 2014). Against E. coli (n = 23) and K. pneumoniae (n = 14), MIC90 values were 2 µg/mL and 4 µg/mL, respectively. The 9-alkylaminomethyl modification of minocycline endows omadacycline with activity against ribosomal protection mechanisms (Draper et al. 2014).
Eravacycline
Eravacycline, a broad spectrum, fully synthetic fluorocycline with novel C-9 pyrrolidinoacetamido and C-7 fluoro modifications, completed a phase 2 trial for cIAI and has completed pivotal phase 3 trials for cIAI and complicated UTI (Solomkin et al. 2014), with future indications expected to include other serious infections. Both IV and oral formulations are in development. In evaluations against large panels of aerobic and anaerobic Gram-negative and -positive bacteria, eravacycline showed MIC90 values ranging from ≤0.008 to 2 µg/mL for all species, except P. aeruginosa and Burkholderia cenocepacia (MIC90 values of 16–32 µg/mL) (Sutcliffe et al. 2013; McDermott et al. 2015; Morrissey et al. 2015a,b,c). In the study by Sutcliffe et al. (2013), eravacycline showed activity against tetracycline-resistant E. coli (MIC50/90 = 0.25/0.5 µg/mL; n = 157), E. cloacae (MIC50/90 = 2/4 µg/mL; n = 25), and P. mirabilis (MIC50/90 = 1/2 µg/mL; n = 109). In a recent study with more than 4000 contemporary Gram-negative pathogens from New York City hospitals, eravacycline MIC50/90 values were 0.12/0.5 µg/mL for E. coli, 0.25/1 µg/mL for K. pneumoniae, 0.25/1 µg/mL for Enterobacter aerogenes, 0.5/1 µg/mL for E. cloacae, and 0.5/1 µg/mL for A. baumannii (Abdallah et al. 2015). Eravacycline also shows good activity against MDR bacteria, including Enterobacteriaceae and A. baumannii expressing extended spectrum β-lactamases, carbapenem resistance, and mechanisms conferring resistance to other antibiotic classes (Sutcliffe et al. 2013; Grossman et al. 2014b; Abdallah et al. 2015).
CONCLUSION
Tetracycline-class antibiotics have treated serious life-threatening infections for more than 60 years; however, as with every other antibiotic class, use has led to resistance development. Historically, potency, spectrum, and tetracycline-resistance hurdles have been addressed semisynthetically with chemical modifications of earlier natural product derivatives. The most successful examples of this approach include minocycline, doxycycline, tigecycline, and omadacycline. More recently, a fully synthetic chemistry approach has led to the discovery of eravacycline, which shows promise in the treatment of serious infections caused by a broad range of bacterial pathogens. Ongoing exploration of synthetic tetracycline derivatives has enabled improvements in potency against P. aeruginosa and other difficult-to-treat MDR Gram-negative pathogens (Deng et al. 2012; O’Brien et al. 2012; Xiao et al. 2013; Grossman et al. 2014a; Sun et al. 2014a, 2015). The ability to synthesize completely novel, “unnatural” tetracyclines opens new opportunities to more fully explore the potential of this familiar and clinically validated antibiotic class.
ACKNOWLEDGMENTS
I thank Joyce Sutcliffe, Patricia Bradford, Kathy Kerstein, and Corey Fyfe for reading this manuscript, and Charlie Xiao for helping to prepare Figure 1.
Footnotes
Editors: Lynn L. Silver and Karen Bush
Additional Perspectives on Antibiotics and Antibiotic Resistance available at www.perspectivesinmedicine.org
REFERENCES
- Abdallah M, Olafisoye O, Cortes C, Urban C, Landman D, Quale J. 2015. Activity of eravacycline against Enterobacteriaceae and Acinetobacter baumannii, including multidrug-resistant isolates, from New York City. Antimicrob Agents Chemother 59: 1802–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekshun MN, Levy SB. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: The mar regulon. Antimicrob Agents Chemother 41: 2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hamad A, Upton M, Burnie J. 2009. Molecular cloning and characterization of SmrA, a novel ABC multidrug efflux pump from Stenotrophomonas maltophilia. J Antimicrob Chemother 64: 731–734. [DOI] [PubMed] [Google Scholar]
- Alonso A, Martinez JL. 2001. Expression of multidrug efflux pump SmeDEF by clinical isolates of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 45: 1879–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Qadheeb NS, Althawadi S, Alkhalaf A, Hosaini S, Alrajhi AA. 2010. Evolution of tigecycline resistance in Klebsiella pneumoniae in a single patient. Ann Saudi Med 30: 404–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminov RI. 2013. Evolution in action: Dissemination of tet(X) into pathogenic microbiota. Front Microbiol 4: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amram E, Mikula I, Schnee C, Ayling RD, Nicholas RA, Rosales RS, Harrus S, Lysnyansky I. 2015. 16S rRNA gene mutations associated with decreased susceptibility to tetracycline in Mycoplasma bovis. Antimicrob Agents Chemother 59: 796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J, Delihas N. 1990. micF RNA binds to the 5′ end of ompF mRNA and to a protein from Escherichia coli. Biochemistry 29: 9249–9256. [DOI] [PubMed] [Google Scholar]
- Anthony KB, Fishman NO, Linkin DR, Gasink LB, Edelstein PH, Lautenbach E. 2008. Clinical and microbiological outcomes of serious infections with multidrug-resistant Gram-negative organisms treated with tigecycline. Clin Infect Dis 46: 567–570. [DOI] [PubMed] [Google Scholar]
- Bantar C, Schell C, Posse G, Limansky A, Ballerini V, Mobilia L. 2008. Comparative time-kill study of doxycycline, tigecycline, sulbactam, and imipenem against several clones of Acinetobacter baumannii. Diagn Microbiol Infect Dis 61: 309–314. [DOI] [PubMed] [Google Scholar]
- Barbosa TM, Levy SB. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J Bacteriol 182: 3467–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer G, Berens C, Projan SJ, Hillen W. 2004. Comparison of tetracycline and tigecycline binding to ribosomes mapped by dimethylsulphate and drug-directed Fe2+ cleavage of 16S rRNA. J Antimicrob Chemother 53: 592–599. [DOI] [PubMed] [Google Scholar]
- Beabout K, Hammerstrom TG, Perez AM, Magalhaes BF, Prater AG, Clements TP, Arias CA, Saxer G, Shamoo Y. 2015a. The ribosomal S10 protein is a general target for decreased tigecycline susceptibility. Antimicrob Agents Chemother 59: 5561–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beabout K, Hammerstrom TG, Wang TT, Bhatty M, Christie PJ, Saxer G, Shamoo Y. 2015b. Rampant parasexuality evolves in a hospital pathogen during antibiotic selection. Mol Biol Evol 32: 2585–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bem AE, Velikova N, Pellicer MT, Baarlen P, Marina A, Wells JM. 2015. Bacterial histidine kinases as novel antibacterial drug targets. ACS Chem Biol 10: 213–224. [DOI] [PubMed] [Google Scholar]
- Bergeron J, Ammirati M, Danley D, James L, Norcia M, Retsema J, Strick CA, Su WG, Sutcliffe J, Wondrack L. 1996. Glycylcyclines bind to the high-affinity tetracycline ribosomal binding site and evade Tet(M)- and Tet(O)-mediated ribosomal protection. Antimicrob Agents Chemother 40: 2226–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek-Davenet S, Lavigne JP, Guyot K, Mayer N, Tournebize R, Brisse S, Leflon-Guibout V, Nicolas-Chanoine MH. 2015. Differential contribution of AcrAB and OqxAB efflux pumps to multidrug resistance and virulence in Klebsiella pneumoniae. J Antimicrob Chemother 70: 81–88. [DOI] [PubMed] [Google Scholar]
- Bratu S, Landman D, George A, Salvani J, Quale J. 2009. Correlation of the expression of acrB and the regulatory genes marA, soxS and ramA with antimicrobial resistance in clinical isolates of Klebsiella pneumoniae endemic to New York City. J Antimicrob Chemother 64: 278–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen DE, Clemons WM Jr, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103: 1143–1154. [DOI] [PubMed] [Google Scholar]
- Buck MA, Cooperman BS. 1990. Single protein omission reconstitution studies of tetracycline binding to the 30S subunit of Escherichia coli ribosomes. Biochemistry 29: 5374–5379. [DOI] [PubMed] [Google Scholar]
- Burdett V. 1986. Streptococcal tetracycline resistance mediated at the level of protein synthesis. J Bacteriol 165: 564–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SM, Festa RA, Pearce MJ, Darwin KH. 2006. Self-compartmentalized bacterial proteases and pathogenesis. Mol Microbiol 60: 553–562. [DOI] [PubMed] [Google Scholar]
- Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407: 340–348. [DOI] [PubMed] [Google Scholar]
- Castanheira M, Mendes RE, Jones RN. 2014. Update on Acinetobacter species: Mechanisms of antimicrobial resistance and contemporary in vitro activity of minocycline and other treatment options. Clin Infect Dis 59: S367–S373. [DOI] [PubMed] [Google Scholar]
- Cattoir V, Isnard C, Cosquer T, Odhiambo A, Bucquet F, Guerin F, Giard JC. 2015. Genomic analysis of reduced susceptibility to tigecycline in Enterococcus faecium. Antimicrob Agents Chemother 59: 239–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau SL, Chu YW, Houang ET. 2004. Novel resistance-nodulation-cell division efflux system AdeDE in Acinetobacter genomic DNA group 3. Antimicrob Agents Chemother 48: 4054–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kuroda T, Huda MN, Mizushima T, Tsuchiya T. 2003. An RND-type multidrug efflux pump SdeXY from Serratia marcescens. J Antimicrob Chemother 52: 176–179. [DOI] [PubMed] [Google Scholar]
- Chen Q, Li X, Zhou H, Jiang Y, Chen Y, Hua X, Yu Y. 2014. Decreased susceptibility to tigecycline in Acinetobacter baumannii mediated by a mutation in trm encoding SAM-dependent methyltransferase. J Antimicrob Chemother 69: 72–76. [DOI] [PubMed] [Google Scholar]
- Chollet R, Chevalier J, Bollet C, Pages JM, Davin-Regli A. 2004. RamA is an alternate activator of the multidrug resistance cascade in Enterobacter aerogenes. Antimicrob Agents Chemother 48: 2518–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I, Roberts M. 2001. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 65: 232–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuanchuen R, Narasaki CT, Schweizer HP. 2002. The MexJK efflux pump of Pseudomonas aeruginosa requires OprM for antibiotic efflux but not for efflux of triclosan. J Bacteriol 184: 5036–5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RB, Hunt DK, He M, Achorn C, Chen CL, Deng Y, Fyfe C, Grossman TH, Hogan PC, O’Brien WJ, et al. 2012. Fluorocyclines. 2: Optimization of the C-9 side-chain for antibacterial activity and oral efficacy. J Med Chem 55: 606–622. [DOI] [PubMed] [Google Scholar]
- Cohen SP, McMurry LM, Levy SB. 1988. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J Bacteriol 170: 5416–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SP, Yan W, Levy SB. 1993. A multidrug resistance regulatory chromosomal locus is widespread among enteric bacteria. J Infect Dis 168: 484–488. [DOI] [PubMed] [Google Scholar]
- Connell SR, Tracz DM, Nierhaus KH, Taylor DE. 2003a. Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrob Agents Chemother 47: 3675–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell SR, Trieber CA, Dinos GP, Einfeldt E, Taylor DE, Nierhaus KH. 2003b. Mechanism of Tet(O)-mediated tetracycline resistance. EMBO J 22: 945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordina C, Hill R, Deshpande A, Hood J, Inkster T. 2012. Tigecycline-resistant Enterococcus faecalis associated with omeprazole use in a surgical patient. J Antimicrob Chemother 67: 1806–1807. [DOI] [PubMed] [Google Scholar]
- Coyne S, Rosenfeld N, Lambert T, Courvalin P, Perichon B. 2010. Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob Agents Chemother 54: 4389–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne S, Courvalin P, Perichon B. 2011. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother 55: 947–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha BA, Sibley CM, Ristuccia AM. 1982. Doxycycline. Ther Drug Monit 4: 115–135. [DOI] [PubMed] [Google Scholar]
- Damier-Piolle L, Magnet S, Bremont S, Lambert T, Courvalin P. 2008. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob Agents Chemother 52: 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daurel C, Fiant AL, Bremont S, Courvalin P, Leclercq R. 2009. Emergence of an Enterobacter hormaechei strain with reduced susceptibility to tigecycline under tigecycline therapy. Antimicrob Agents Chemother 53: 4953–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean CR, Visalli MA, Projan SJ, Sum PE, Bradford PA. 2003. Efflux-mediated resistance to tigecycline (GAR-936) in Pseudomonas aeruginosa PAO1. Antimicrob Agents Chemother 47: 972–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Majumdar S, Veleba M, Finn S, Fanning S, Schneiders T. 2013. Elucidating the regulon of multidrug resistance regulator RarA in Klebsiella pneumoniae. Antimicrob Agents Chemother 57: 1603–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Grossman T, Clark RB, Xiao XY, Sutcliffe J. 2012. The intravenous pharmacokinetics (PK) and efficacy of TP-433 in murine infection models with Pseudomonas aeruginosa. In Abstr 25th ECCMID, Abstract AbsP1426 London. [Google Scholar]
- Deng M, Zhu MH, Li JJ, Bi S, Sheng ZK, Hu FS, Zhang JJ, Chen W, Xue XW, Sheng JF, et al. 2014. Molecular epidemiology and mechanisms of tigecycline resistance in clinical isolates of Acinetobacter baumannii from a Chinese university hospital. Antimicrob Agents Chemother 58: 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire KM, Kramer M, Grindley ND. 1990. Role of instability in the cis action of the insertion sequence IS903 transposase. Proc Natl Acad Sci 87: 4048–4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Torres ML, McNab R, Spratt DA, Villedieu A, Hunt N, Wilson M, Mullany P. 2003. Novel tetracycline resistance determinant from the oral metagenome. Antimicrob Agents Chemother 47: 1430–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donhofer A, Franckenberg S, Wickles S, Berninghausen O, Beckmann R, Wilson DN. 2012. Structural basis for TetM-mediated tetracycline resistance. Proc Natl Acad Sci 109: 16900–16905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper MP, Weir S, Macone A, Donatelli J, Trieber CA, Tanaka SK, Levy SB. 2014. Mechanism of action of the novel aminomethylcycline antibiotic omadacycline. Antimicrob Agents Chemother 58: 1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggar BM. 1948. Aureomycin; a product of the continuing search for new antibiotics. Ann NY Acad Sci 51: 177–181. [DOI] [PubMed] [Google Scholar]
- Falagas ME, Vardakas KZ, Kapaskelis A, Triarides NA, Roussos NS. 2015. Tetracyclines for multidrug-resistant Acinetobacter baumannii infections. Int J Antimicrob Agents 45: 455–460. [DOI] [PubMed] [Google Scholar]
- Finlay AC, Hobby GL, P’an SY, Regna PP, Routien JB, Seeley DB, Shull GM, Sobin BA, Solomons IA, Vinson JW, et al. 1950. Terramycin, a new antibiotic. Science 111: 85. [DOI] [PubMed] [Google Scholar]
- Forsberg KJ, Patel S, Wencewicz TA, Dantas G. 2015. The tetracycline destructases: A novel family of tetracycline-inactivating enzymes. Chem Biol 22: 888–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyfe C, Sutcliffe JA, Grossman TH. 2013. Susceptibility of tetracyclines to Tet(A) resistance is independent of interdomain loop sequence. Antimicrob Agents Chemother 57: 2430–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyfe C, Norris D, Sutcliffe JA, Grossman TH. 2015. Identification of Klebsiella pneumoniae genes involved in tigecycline-resistance using transposon mutagenesis. In Abstr 25th ECCMID, Abstract P1022 Copenhagen, Denmark. [Google Scholar]
- Gambino L, Gracheck SJ, Miller PF. 1993. Overexpression of the MarA positive regulator is sufficient to confer multiple antibiotic resistance in Escherichia coli. J Bacteriol 175: 2888–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George AM, Levy SB. 1983a. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: Involvement of a non-plasmid-determined efflux of tetracycline. J Bacteriol 155: 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George AM, Levy SB. 1983b. Gene in the major cotransduction gap of the Escherichia coli K-12 linkage map required for the expression of chromosomal resistance to tetracycline and other antibiotics. J Bacteriol 155: 541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George AM, Hall RM, Stokes HW. 1995. Multidrug resistance in Klebsiella pneumoniae: A novel gene, ramA, confers a multidrug resistance phenotype in Escherichia coli. Microbiology 141: 1909–1920. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Sadowsky MJ, Roberts MC, Gralnick JA, LaPara TM. 2009. Sphingobacterium sp. strain PM2-P1–29 harbours a functional tet(X) gene encoding for the degradation of tetracycline. J Appl Microbiol 106: 1336–1342. [DOI] [PubMed] [Google Scholar]
- Goff DA, Bauer KA, Mangino JE. 2014. Bad bugs need old drugs: A stewardship program’s evaluation of minocycline for multidrug-resistant Acinetobacter baumannii infections. Clin Infect Dis 1: S381–S387. [DOI] [PubMed] [Google Scholar]
- Griffith KL, Shah IM, Wolf RE Jr, 2004. Proteolytic degradation of Escherichia coli transcription activators SoxS and MarA as the mechanism for reversing the induction of the superoxide (SoxRS) and multiple antibiotic resistance (Mar) regulons. Mol Microbiol 51: 1801–1816. [DOI] [PubMed] [Google Scholar]
- Grkovic S, Brown MH, Skurray RA. 2002. Regulation of bacterial drug export systems. Microbiol Mol Biol Rev 66: 671–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman TH, Starosta AL, Fyfe C, O’Brien W, Rothstein DM, Mikolajka A, Wilson DN, Sutcliffe JA. 2012. Target- and resistance-based mechanistic studies with TP-434, a novel fluorocycline antibiotic. Antimicrob Agents Chemother 56: 2559–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman T, Fyfe C, Kerstein K, Sun C, Clark R, Xiao XY, Sutcliffe J. 2014a. In vivo efficacy of novel, fully synthetic tetracyclines in a murine lung infection model challenged with KPC-producing Klebsiella pneumonia, Abstract P0300. In Abstr 24th ECCMID, Barcelona, Spain. [Google Scholar]
- Grossman T, O’Brien W, Fyfe C, Sutcliffe J. 2014b. Eravacycline is potent against third generation cephalosporin- and carbapenem-resistant Enterobacteriaceae, carbapenem-resistant Acinetobacter baumannii and has isolate-specific bactericidal activity. In Abstr 54th Intersci Conf Antimicrob Agents Chemother, Abstract C-1374 Washington, DC. [Google Scholar]
- Guay GG, Tuckman M, McNicholas P, Rothstein DM. 1993. The tet(K) gene from Staphylococcus aureus mediates the transport of potassium in Escherichia coli. J Bacteriol 175: 4927–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guay GG, Tuckman M, Rothstein DM. 1994. Mutations in the tetA(B) gene that cause a change in substrate specificity of the tetracycline efflux pump. Antimicrob Agents Chemother 38: 857–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaume G, Ledent V, Moens W, Collard JM. 2004. Phylogeny of efflux-mediated tetracycline resistance genes and related proteins revisited. Microb Drug Resist 10: 11–26. [DOI] [PubMed] [Google Scholar]
- Hachler H, Cohen SP, Levy SB. 1991. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J Bacteriol 173: 5532–5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerstrom TG, Beabout K, Clements TP, Saxer G, Shamoo Y. 2015. Acinetobacter baumannii repeatedly evolves a hypermutator phenotype in response to tigecycline that effectively surveys evolutionary trajectories to resistance. PLoS ONE 10: e0140489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Fu Y, Chen Q, Ruan Z, Hua X, Zhou H, Yu Y. 2015. Tigecycline susceptibility and the role of efflux pumps in tigecycline resistance in KPC-producing Klebsiella pneumoniae. PLoS ONE 10: e0119064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschke M, Wolters M, Sobottka I, Rohde H, Aepfelbacher M. 2010. ramR mutations in clinical isolates of Klebsiella pneumoniae with reduced susceptibility to tigecycline. Antimicrob Agents Chemother 54: 2720–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata T, Saito A, Nishino K, Tamura N, Yamaguchi A. 2004. Effects of efflux transporter genes on susceptibility of Escherichia coli to tigecycline (GAR-936). Antimicrob Agents Chemother 48: 2179–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban DJ, Reinert RR, Bouchillon SK, Dowzicky MJ. 2015. Global in vitro activity of tigecycline and comparator agents: Tigecycline Evaluation and Surveillance Trial 2004–2013. Ann Clin Microbiol Antimicrob 14: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornsey M, Ellington MJ, Doumith M, Hudson S, Livermore DM, Woodford N. 2010a. Tigecycline resistance in Serratia marcescens associated with up-regulation of the SdeXY-HasF efflux system also active against ciprofloxacin and cefpirome. J Antimicrob Chemother 65: 479–482. [DOI] [PubMed] [Google Scholar]
- Hornsey M, Ellington MJ, Doumith M, Scott G, Livermore DM, Woodford N. 2010b. Emergence of AcrAB-mediated tigecycline resistance in a clinical isolate of Enterobacter cloacae during ciprofloxacin treatment. Int J Antimicrob Agents 35: 478–481. [DOI] [PubMed] [Google Scholar]
- Hornsey M, Ellington MJ, Doumith M, Thomas CP, Gordon NC, Wareham DW, Quinn J, Lolans K, Livermore DM, Woodford N. 2010c. AdeABC-mediated efflux and tigecycline MICs for epidemic clones of Acinetobacter baumannii. J Antimicrob Chemother 65: 1589–1593. [DOI] [PubMed] [Google Scholar]
- Hu M, Nandi S, Davies C, Nicholas RA. 2005. High-level chromosomally mediated tetracycline resistance in Neisseria gonorrhoeae results from a point mutation in the rpsJ gene encoding ribosomal protein S10 in combination with the mtrR and penB resistance determinants. Antimicrob Agents Chemother 49: 4327–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarolmen H, Hewel D, Kain E. 1970. Activity of minocycline against R factor-carrying Enterobacteriaceae. Infect Immun 1: 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner L, Starosta AL, Terry DS, Mikolajka A, Filonava L, Yusupov M, Blanchard SC, Wilson DN, Yusupova G. 2013. Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis. Proc Natl Acad Sci 110: 3812–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CH, Murphy E, Bradford PA. 2008. Genetic determinants of tetracycline resistance and their effect on tetracycline and glycylcycline antibiotics. Anti-Infect Agents Med Chem 7: 84–96. [Google Scholar]
- Jones RN, Flonta M, Gurler N, Cepparulo M, Mendes RE, Castanheira M. 2014. Resistance surveillance program report for selected European nations (2011). Diagn Microbiol Infect Dis 78: 429–436. [DOI] [PubMed] [Google Scholar]
- Keeney D, Ruzin A, Bradford PA. 2007. RamA, a transcriptional regulator, and AcrAB, an RND-type efflux pump, are associated with decreased susceptibility to tigecycline in Enterobacter cloacae. Microb Drug Resist 13: 1–6. [DOI] [PubMed] [Google Scholar]
- Keeney D, Ruzin A, McAleese F, Murphy E, Bradford PA. 2008. MarA-mediated overexpression of the AcrAB efflux pump results in decreased susceptibility to tigecycline in Escherichia coli. J Antimicrob Chemother 61: 46–53. [DOI] [PubMed] [Google Scholar]
- Kim HB, Wang M, Park CH, Kim EC, Jacoby GA, Hooper DC. 2009. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob Agents Chemother 53: 3582–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nonaka L, Maruyama F, Suzuki S. 2007. Molecular evidence for the ancient origin of the ribosomal protection protein that mediates tetracycline resistance in bacteria. J Mol Evol 65: 228–235. [DOI] [PubMed] [Google Scholar]
- Krulwich TA, Jin J, Guffanti AA, Bechhofer H. 2001. Functions of tetracycline efflux proteins that do not involve tetracycline. J Mol Microbiol Biotechnol 3: 237–246. [PubMed] [Google Scholar]
- Lee EW, Huda MN, Kuroda T, Mizushima T, Tsuchiya T. 2003. EfrAB, an ABC multidrug efflux pump in Enterococcus faecalis. Antimicrob Agents Chemother 47: 3733–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leski TA, Bangura U, Jimmy DH, Ansumana R, Lizewski SE, Stenger DA, Taitt CR, Vora GJ. 2013. Multidrug-resistant tet(X)-containing hospital isolates in Sierra Leone. Int J Antimicrob Agents 42: 83–86. [DOI] [PubMed] [Google Scholar]
- Li XZ, Nikaido H. 2009. Efflux-mediated drug resistance in bacteria: An update. Drugs 69: 1555–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Atkinson GC, Thakor NS, Allas U, Lu CC, Chan KY, Tenson T, Schulten K, Wilson KS, Hauryliuk V, et al. 2013. Mechanism of tetracycline resistance by ribosomal protection protein Tet(O). Nat Commun 4: 1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Plesiat P, Nikaido H. 2015. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 28: 337–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Lin YY, Yeh HW, Lan CY. 2014. Role of the BaeSR two-component system in the regulation of Acinetobacter baumannii adeAB genes and its correlation with tigecycline susceptibility. BMC Microbiol 14: 1471–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkevicius M, Sandegren L, Andersson DI. 2013. Mechanisms and fitness costs of tigecycline resistance in Escherichia coli. J Antimicrob Chemother 68: 2809–2819. [DOI] [PubMed] [Google Scholar]
- Lomovskaya O, Sun D, King P, Dudley MN. 2014. Tigecycline (TIG) but not minocycline (MINO) selects for clinically relevant efflux-mediated resistance (R) in Acinetobacter spp. (ACB). In Abstr 54th Intersci Conf Antimicrob Agents Chemother, Abstract C1-1087. [Google Scholar]
- Lomovskaya O, Sun D, Rubio-Aparicio D, Dudley MN. 2015. Accumulation of several chromosomal mutations have limited impact on the sensitivity of Acinetobacter baumannii (ACB) to minocycline (MINO). In Abstr Intersci Conf Antimicrob Agents Chemother/Internat Cong Chemother Infect, Abstract C-1009. [Google Scholar]
- Lupien A, Gingras H, Leprohon P, Ouellette M. 2015. Induced tigecycline resistance in Streptococcus pneumoniae mutants reveals mutations in ribosomal proteins and rRNA. J Antimicrob Chemother 70: 2973–2980. [DOI] [PubMed] [Google Scholar]
- Macone AB, Caruso BK, Leahy RG, Donatelli J, Weir S, Draper MP, Tanaka SK, Levy SB. 2014. In vitro and in vivo antibacterial activities of omadacycline, a novel aminomethylcycline. Antimicrob Agents Chemother 58: 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand I, Damier-Piolle L, Courvalin P, Lambert T. 2004. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob Agents Chemother 48: 3298–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RG, Rosner JL. 1995. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc Natl Acad Sci 92: 5456–5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RG, Rosner JL. 2001. The AraC transcriptional activators. Curr Opin Microbiol 4: 132–137. [DOI] [PubMed] [Google Scholar]
- Martin RG, Rosner JL. 2011. Promoter discrimination at class I MarA regulon promoters mediated by glutamic acid 89 of the MarA transcriptional activator of Escherichia coli. J Bacteriol 193: 506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RG, Bartlett ES, Rosner JL, Wall ME. 2008. Activation of the Escherichia coli marA/soxS/rob regulon in response to transcriptional activator concentration. J Mol Biol 380: 278–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-oprM efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother 44: 3322–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Chen J, Minato Y, Ogawa W, Mizushima T, Kuroda T, Tsuchiya T. 2008. SmdAB, a heterodimeric ABC-type multidrug efflux pump, in Serratia marcescens. J Bacteriol 190: 648–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell IH. 1967. Partial removal of bound transfer RNA from polysomes engaged in protein synthesis in vitro after addition of tetracycline. Biochim Biophys Acta 138: 337–346. [DOI] [PubMed] [Google Scholar]
- McAleese F, Petersen P, Ruzin A, Dunman PM, Murphy E, Projan SJ, Bradford PA. 2005. A novel MATE family efflux pump contributes to the reduced susceptibility of laboratory-derived Staphylococcus aureus mutants to tigecycline. Antimicrob Agents Chemother 49: 1865–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott L, Jacobus NV, Snydman DR, Kerstein K, Grossman TH, Sutcliffe JA. 2015. Evaluation of the in vitro activity of eravacycline against a broad spectrum of recent clinical anaerobic isolates. In Abstr Intersci Conf Antimicrob Agents Chemother/Internat Cong Chemother Infect, Abstract C-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry L, Levy SB. 1978. Two transport systems for tetracycline in sensitive Escherichia coli: Critical role for an initial rapid uptake system insensitive to energy inhibitors. Antimicrob Agents Chemother 14: 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes RE, Farrell DJ, Sader HS, Streit JM, Jones RN. 2015. Update of the telavancin activity in vitro tested against a worldwide collection of Gram-positive clinical isolates (2013), when applying the revised susceptibility testing method. Diagn Microbiol Infect Dis 81: 275–279. [DOI] [PubMed] [Google Scholar]
- Moore IF, Hughes DW, Wright GD. 2005. Tigecycline is modified by the flavin-dependent monooxygenase TetX. Biochemistry 44: 11829–11835. [DOI] [PubMed] [Google Scholar]
- Morita Y, Komori Y, Mima T, Kuroda T, Mizushima T, Tsuchiya T. 2001. Construction of a series of mutants lacking all of the four major mex operons for multidrug efflux pumps or possessing each one of the operons from Pseudomonas aeruginosa PAO1: MexCD-OprJ is an inducible pump. FEMS Microbiol Lett 202: 139–143. [DOI] [PubMed] [Google Scholar]
- Morrissey I, Sutcliffe J, Hackel M, Hawser S. 2015a. Assessment of eravacycline against 3,467 recent gram-positive bacteria, including multidrug-resistant isolates collected from 2013–2014. In Abstr Intersci Conf Antimicrob Agents Chemother/Internat Cong Chemother Infect, Abstract C-563. [Google Scholar]
- Morrissey I, Sutcliffe J, Hackel M, Hawser S. 2015b. Assessment of Eravacycline against a recent global collection of 4,462 Enterobacteriaceae clinical isolates (2013–2014). In Abstr Intersci Conf Antimicrob Agents Chemother/Internat Cong Chemother Infect, Abstract C-619. [Google Scholar]
- Morrissey I, Sutcliffe J, Hackel M, Hawser S. 2015c. Assessment of eravacycline against non-fermenting gram-negative clinical isolates isolated in 2013–2014. Abstr Intersci Conf Antimicrob Agents Chemother/Internat Cong Chemother Infect, Abstract C-095. [Google Scholar]
- Mortimer PG, Piddock LJ. 1993. The accumulation of five antibacterial agents in porin-deficient mutants of Escherichia coli. J Antimicrob Chemother 32: 195–213. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Chandler M. 2004. Regulation of transposition in bacteria. Res Microbiol 155: 387–398. [DOI] [PubMed] [Google Scholar]
- Nelson ML, Levy SB. 2011. The history of the tetracyclines. Ann NY Acad Sci 1241: 17–32. [DOI] [PubMed] [Google Scholar]
- Neonakis IK, Stylianou K, Daphnis E, Maraki S. 2011. First case of resistance to tigecycline by Klebsiella pneumoniae in a European University Hospital. Indian J Med Microbiol 29: 78–79. [DOI] [PubMed] [Google Scholar]
- Nicoloff H, Andersson DI. 2013. Lon protease inactivation, or translocation of the lon gene, potentiate bacterial evolution to antibiotic resistance. Mol Microbiol 90: 1233–1248. [DOI] [PubMed] [Google Scholar]
- Nicoloff H, Perreten V, McMurry LM, Levy SB. 2006. Role for tandem duplication and lon protease in AcrAB-TolC- dependent multiple antibiotic resistance (Mar) in an Escherichia coli mutant without mutations in marRAB or acrRAB. J Bacteriol 188: 4413–4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoloff H, Perreten V, Levy SB. 2007. Increased genome instability in Escherichia coli lon mutants: Relation to emergence of multiple-antibiotic-resistant (Mar) mutants caused by insertion sequence elements and large tandem genomic amplifications. Antimicrob Agents Chemother 51: 1293–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen LE, Snesrud EC, Onmus-Leone F, Kwak YI, Aviles R, Steele ED, Sutter DE, Waterman PE, Lesho EP. 2014. IS5 element integration, a novel mechanism for rapid in vivo emergence of tigecycline nonsusceptibility in Klebsiella pneumoniae. Antimicrob Agents Chemother 58: 6151–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigo M, Cevallos CS, Woods K, Flores VM, Francis G, Perlman DC, Revuelta M, Mildvan D, Waldron M, Gomez T, et al. 2013. Nested case-control study of the emergence of tigecycline resistance in multidrug-resistant Klebsiella pneumoniae. Antimicrob Agents Chemother 57: 5743–5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H, Thanassi DG. 1993. Penetration of lipophilic agents with multiple protonation sites into bacterial cells: Tetracyclines and fluoroquinolones as examples. Antimicrob Agents Chemother 37: 1393–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido E, Yamaguchi A, Nishino K. 2008. AcrAB multidrug efflux pump regulation in Salmonella enterica serovar Typhimurium by RamA in response to environmental signals. J Biol Chem 283: 24245–24253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel GJ, Draper MP, Hait H, Tanaka SK, Arbeit RD. 2012. A randomized, evaluator-blind, phase 2 study comparing the safety and efficacy of omadacycline to those of linezolid for treatment of complicated skin and skin structure infections. Antimicrob Agents Chemother 56: 5650–5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka L, Suzuki S. 2002. New Mg2+-dependent oxytetracycline resistance determinant tet 34 in Vibrio isolates from marine fish intestinal contents. Antimicrob Agents Chemother 46: 1550–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norcia LJ, Silvia AM, Hayashi SF. 1999. Studies on time-kill kinetics of different classes of antibiotics against veterinary pathogenic bacteria including Pasteurella, Actinobacillus and Escherichia coli. J Antibiot 52: 52–60. [DOI] [PubMed] [Google Scholar]
- Noviello S, Ianniello F, Leone S, Fiore M, Esposito S. 2008. In vitro activity of tigecycline: MICs, MBCs, time-kill curves and post-antibiotic effect. J Chemother 20: 577–580. [DOI] [PubMed] [Google Scholar]
- O’Brien W, Fyfe C, Grossman T, Chen CL, Clark R, Deng Y, He M, Hunt D, Sun C, Xiao XY, et al. 2012. In vitro potency of novel tetracyclines against Pseudomonas aeruginosa and other major Gram-negative pathogens. In Abstr 22th ECCMID, Abstract P1448 London, United Kingdom. [Google Scholar]
- Olson MW, Ruzin A, Feyfant E, Rush TS III, O’Connell J, Bradford PA. 2006. Functional, biophysical, and structural bases for antibacterial activity of tigecycline. Antimicrob Agents Chemother 50: 2156–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg AY, Potoski BA, Rea R, Adams J, Sethi J, Capitano B, Husain S, Kwak EJ, Bhat SV, Paterson DL. 2007. Acinetobacter baumannii bloodstream infection while receiving tigecycline: A cautionary report. J Antimicrob Chemother 59: 128–131. [DOI] [PubMed] [Google Scholar]
- Petersen PJ, Jacobus NV, Weiss WJ, Sum PE, Testa RT. 1999. In vitro and in vivo antibacterial activities of a novel glycylcycline, the 9-t-butylglycylamido derivative of minocycline (GAR-936). Antimicrob Agents Chemother 43: 738–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen PJ, Jones CH, Bradford PA. 2007. In vitro antibacterial activities of tigecycline and comparative agents by time-kill kinetic studies in fresh Mueller–Hinton broth. Diagn Microbiol Infect Dis 59: 347–349. [DOI] [PubMed] [Google Scholar]
- Piddock LJ. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev 19: 382–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioletti M, Schlunzen F, Harms J, Zarivach R, Gluhmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, et al. 2001. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J 20: 1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley AP, Schnaitman CA. 1978. Outer membrane proteins of Escherichia coli. VII: Evidence that bacteriophage-directed protein 2 functions as a pore. J Bacteriol 133: 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumbwe L, Ueda O, Yoshimura F, Chang A, Smith RL, Wexler HM. 2006. Bacteroides fragilis BmeABC efflux systems additively confer intrinsic antimicrobial resistance. J Antimicrob Chemother 58: 37–46. [DOI] [PubMed] [Google Scholar]
- Ramirez J, Dartois N, Gandjini H, Yan JL, Korth-Bradley J, McGovern PC. 2013. Randomized phase 2 trial to evaluate the clinical efficacy of two high-dosage tigecycline regimens versus imipenem-cilastatin for treatment of hospital-acquired pneumonia. Antimicrob Agents Chemother 57: 1756–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen BA, Kovacs E. 1993. Cloning and identification of a two-component signal-transducing regulatory system from Bacteroides fragilis. Mol Microbiol 7: 765–776. [DOI] [PubMed] [Google Scholar]
- Rasmussen BA, Gluzman Y, Tally FP. 1994. Inhibition of protein synthesis occurring on tetracycline-resistant, TetM-protected ribosomes by a novel class of tetracyclines, the glycylcyclines. Antimicrob Agents Chemother 38: 1658–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid GE, Grim SA, Aldeza CA, Janda WM, Clark NM. 2007. Rapid development of Acinetobacter baumannii resistance to tigecycline. Pharmacotherapy 27: 1198–1201. [DOI] [PubMed] [Google Scholar]
- Reinhardt A, Kuhn F, Heisig A, Heisig P. 2014. Overexpression of ramA from Citrobacter freundii mediates MDR phenotype in various Citrobacter species and also in Escherichia coli. In Abstr 24th ECCMID, Abstract P1102 Barcelona, Spain. [Google Scholar]
- Ricci V, Blair JM, Piddock LJ. 2014. RamA, which controls expression of the MDR efflux pump AcrAB-TolC, is regulated by the Lon protease. J Antimicrob Chemother 69: 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie DJ, Garavaglia-Wilson A. 2014. A review of intravenous minocycline for treatment of multidrug-resistant Acinetobacter infections. Clin Infect Dis 59: S374–S380. [DOI] [PubMed] [Google Scholar]
- Roberts MC. 2005. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett 245: 195–203. [DOI] [PubMed] [Google Scholar]
- Roberts MC. 2011. Mechanisms of bacterial antibiotic resistance and lessons learned from environmental tetracycline-resistant bacteria. In Antimicrobial Resistance in the Environment (ed. Keen P, Montforts MHMM). Wiley, Hoboken, NJ. [Google Scholar]
- Rodriguez-Avial C, Rodriguez-Avial I, Merino P, Picazo JJ. 2012. Klebsiella pneumoniae: Development of a mixed population of carbapenem and tigecycline resistance during antimicrobial therapy in a kidney transplant patient. Clin Microbiol Infect 18: 61–66. [DOI] [PubMed] [Google Scholar]
- Rosenblum R, Khan E, Gonzalez G, Hasan R, Schneiders T. 2011. Genetic regulation of the ramA locus and its expression in clinical isolates of Klebsiella pneumoniae. Int J Antimicrob Agents 38: 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JI, Eady EA, Cove JH, Cunliffe WJ. 1998. 16S rRNA mutation associated with tetracycline resistance in a gram-positive bacterium. Antimicrob Agents Chemother 42: 1702–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquette C, Serre MC, Lane D. 2004. Protective role for H-NS protein in IS1 transposition. J Bacteriol 186: 2091–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin A, Keeney D, Bradford PA. 2005a. AcrAB efflux pump plays a role in decreased susceptibility to tigecycline in Morganella morganii. Antimicrob Agents Chemother 49: 791–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin A, Visalli MA, Keeney D, Bradford PA. 2005b. Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob Agents Chemother 49: 1017–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin A, Keeney D, Bradford PA. 2007. AdeABC multidrug efflux pump is associated with decreased susceptibility to tigecycline in Acinetobacter calcoaceticus–Acinetobacter baumannii complex. J Antimicrob Chemother 59: 1001–1004. [DOI] [PubMed] [Google Scholar]
- Ruzin A, Immermann FW, Bradford PA. 2008. Real-time PCR and statistical analyses of acrAB and ramA expression in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother 52: 3430–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin A, Immermann FW, Bradford PA. 2010. RT-PCR and statistical analyses of adeABC expression in clinical isolates of Acinetobacter calcoaceticus–Acinetobacter baumannii complex. Microb Drug Resist 16: 87–89. [DOI] [PubMed] [Google Scholar]
- Sader HS, Flamm RK, Jones RN. 2013. Tigecycline activity tested against antimicrobial resistant surveillance subsets of clinical bacteria collected worldwide (2011). Diagn Microbiol Infect Dis 76: 217–221. [DOI] [PubMed] [Google Scholar]
- Sader HS, Farrell DJ, Flamm RK, Jones RN. 2014. Variation in potency and spectrum of tigecycline activity against bacterial strains from U.S. medical centers since its approval for clinical use (2006 to 2012). Antimicrob Agents Chemother 58: 2274–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SaiSree L, Reddy M, Gowrishankar J. 2001. IS186 insertion at a hot spot in the lon promoter as a basis for lon protease deficiency of Escherichia coli B: Identification of a consensus target sequence for IS186 transposition. J Bacteriol 183: 6943–6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pescador R, Brown JT, Roberts M, Urdea MS. 1988. Homology of the TetM with translational elongation factors: Implications for potential modes of tetM-conferred tetracycline resistance. Nucleic Acids Res 16: 1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapunaric FM, Levy SB. 2005. Substitutions in the interdomain loop of the Tn10 TetA efflux transporter alter tetracycline resistance and substrate specificity. Microbiology 151: 2315–2322. [DOI] [PubMed] [Google Scholar]
- Schedlbauer A, Kaminishi T, Ochoa-Lizarralde B, Dhimole N, Zhou S, Lopez-Alonso JP, Connell SR, Fucini P. 2015. Structural characterization of an alternative mode of tigecycline binding to the bacterial ribosome. Antimicrob Agents Chemother 59: 2849–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer HP. 2003. Efflux as a mechanism of resistance to antimicrobials in Pseudomonas aeruginosa and related bacteria: Unanswered questions. Genet Mol Res 2: 48–62. [PubMed] [Google Scholar]
- Seoane AS, Levy SB. 1995. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J Bacteriol 177: 3414–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng ZK, Hu F, Wang W, Guo Q, Chen Z, Xu X, Zhu D, Wang M. 2014. Mechanisms of tigecycline resistance among Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother 58: 6982–6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood JE, Fraser S, Citron DM, Wexler H, Blakely G, Jobling K, Patrick S. 2011. Multi-drug resistant Bacteroides fragilis recovered from blood and severe leg wounds caused by an improvised explosive device (IED) in Afghanistan. Anaerobe 17: 152–155. [DOI] [PubMed] [Google Scholar]
- Smith MC, Chopra I. 1984. Energetics of tetracycline transport into Escherichia coli. Antimicrob Agents Chemother 25: 446–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomkin JS, Ramesh MK, Cesnauskas G, Novikovs N, Stefanova P, Sutcliffe JA, Walpole SM, Horn PT. 2014. Phase 2, randomized, double-blind study of the efficacy and safety of two dose regimens of eravacycline versus ertapenem for adult community-acquired complicated intra-abdominal infections. Antimicrob Agents Chemother 58: 1847–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu T, De Angelis G, Cipriani M, Pedruzzi B, D’Inzeo T, Cataldo MA, Sganga G, Tacconelli E. 2012. In vivo emergence of tigecycline resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli. Antimicrob Agents Chemother 56: 4516–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer BS, Salyers AA. 1988. Characterization of a novel tetracycline resistance that functions only in aerobically grown Escherichia coli. J Bacteriol 170: 1423–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer BS, Salyers AA. 1989. Novel aerobic tetracycline resistance gene that chemically modifies tetracycline. J Bacteriol 171: 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer BS, Bedzyk L, Salyers AA. 1991. Evidence that a novel tetracycline resistance gene found on two Bacteroides transposons encodes an NADP-requiring oxidoreductase. J Bacteriol 173: 176–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan VB, Venkataramaiah M, Mondal A, Vaidyanathan V, Govil T, Rajamohan G. 2012. Functional characterization of a novel outer membrane porin KpnO, regulated by PhoBR two-component system in Klebsiella pneumoniae NTUH-K2044. PLoS ONE 7: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein GE, Babinchak T. 2013. Tigecycline: An update. Diagn Microbiol Infect Dis 75: 331–336. [DOI] [PubMed] [Google Scholar]
- Stone NR, Woodford N, Livermore DM, Howard J, Pike R, Mushtaq S, Perry C, Hopkins S. 2011. Breakthrough bacteraemia due to tigecycline-resistant Escherichia coli with New Delhi metallo-β-lactamase (NDM)-1 successfully treated with colistin in a patient with calciphylaxis. J Antimicrob Chemother 66: 2677–2678. [DOI] [PubMed] [Google Scholar]
- Sum PE, Petersen P. 1999. Synthesis and structure-activity relationship of novel glycylcycline derivatives leading to the discovery of GAR-936. Bioorg Med Chem Lett 9: 1459–1462. [DOI] [PubMed] [Google Scholar]
- Sun JR, Perng CL, Chan MC, Morita Y, Lin JC, Su CM, Wang WY, Chang TY, Chiueh TS. 2012. A truncated AdeS kinase protein generated by ISAba1 insertion correlates with tigecycline resistance in Acinetobacter baumannii. PLoS ONE 7: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Hunt D, Clark R, Fyfe C, Kerstein K, Grossman T, Sutcliffe J, Xiao XY. 2014a. In vitro potency of novel, fully synthetic tetracyclines against MDR Gram-negative pathogens including carbapenem-resistant Enterobacteriaceae. In 24th ECCMID, Abstract P0299 Barcelona, Spain. [Google Scholar]
- Sun JR, Perng CL, Lin JC, Yang YS, Chan MC, Chang TY, Lin FM, Chiueh TS. 2014b. AdeRS combination codes differentiate the response to efflux pump inhibitors in tigecycline-resistant isolates of extensively drug-resistant Acinetobacter baumannii. Eur J Clin Microbiol Infect Dis 33: 2141–2147. [DOI] [PubMed] [Google Scholar]
- Sun C, Hunt DK, Chen CL, Deng Y, He M, Clark RB, Fyfe C, Grossman TH, Sutcliffe JA, Xiao XY. 2015. Design, synthesis, and biological evaluation of hexacyclic tetracyclines as potent, broad spectrum antibacterial agents. J Med Chem 20: 20. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JA, O’Brien W, Fyfe C, Grossman TH. 2013. Antibacterial activity of eravacycline (TP-434), a novel fluorocycline, against hospital and community pathogens. Antimicrob Agents Chemother 57: 5548–5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DE, Hiratsuka K, Ray H, Manavathu EK. 1987. Characterization and expression of a cloned tetracycline resistance determinant from Campylobacter jejuni plasmid pUA466. J Bacteriol 169: 2984–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo JW, Tan TM, Poh CL. 2002. Genetic determinants of tetracycline resistance in Vibrio harveyi. Antimicrob Agents Chemother 46: 1038–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier PR, Nicolau DP. 2013. Tigecycline displays in vivo bactericidal activity against extended-spectrum-β-lactamase-producing Enterobacteriaceae after 72-hour exposure period. Antimicrob Agents Chemother 57: 640–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaker M, Spanogiannopoulos P, Wright GD. 2010. The tetracycline resistome. Cell Mol Life Sci 67: 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanassi DG, Suh GS, Nikaido H. 1995. Role of outer membrane barrier in efflux-mediated tetracycline resistance of Escherichia coli. J Bacteriol 177: 998–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Medicines Company. 2015. Prescribing Information, Minocin; Parsippany, NJ. [Google Scholar]
- Trieber CA, Taylor DE. 2002. Mutations in the 16S rRNA genes of Helicobacter pylori mediate resistance to tetracycline. J Bacteriol 184: 2131–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong-Bolduc QC, Dunman PM, Strahilevitz J, Projan SJ, Hooper DC. 2005. MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J Bacteriol 187: 2395–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsilibaris V, Maenhaut-Michel G, Van Melderen L. 2006. Biological roles of the Lon ATP-dependent protease. Res Microbiol 157: 701–713. [DOI] [PubMed] [Google Scholar]
- Tuckman M, Petersen PJ, Projan SJ. 2000. Mutations in the interdomain loop region of the tetA(A) tetracycline resistance gene increase efflux of minocycline and glycylcyclines. Microb Drug Resist 6: 277–282. [DOI] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration. 2010. FDA Drug Safety Communication: Increased risk of death with Tygacil (tigecycline) compared to other antibiotics used to treat similar infections. FDA, Silver Spring, MD. [Google Scholar]
- U.S. Food and Drug Administration. 2013. FDA Drug Safety Communication: FDA warns of increased risk of death with IV antibacterial Tygacil (tigecycline) and approves new Boxed Warning. FDA, Silver Spring, MD. [Google Scholar]
- van der Straaten T, Janssen R, Mevius DJ, van Dissel JT. 2004a. Salmonella gene rma (ramA) and multiple-drug-resistant Salmonella enterica serovar typhimurium. Antimicrob Agents Chemother 48: 2292–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Straaten T, Zulianello L, van Diepen A, Granger DL, Janssen R, van Dissel JT. 2004b. Salmonella enterica serovar Typhimurium RamA, intracellular oxidative stress response, and bacterial virulence. Infect Immun 72: 996–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veleba M, Schneiders T. 2012. Tigecycline resistance can occur independently of the ramA gene in Klebsiella pneumoniae. Antimicrob Agents Chemother 56: 4466–4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veleba M, Higgins PG, Gonzalez G, Seifert H, Schneiders T. 2012. Characterization of RarA, a novel AraC family multidrug resistance regulator in Klebsiella pneumoniae. Antimicrob Agents Chemother 56: 4450–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veleba M, De Majumdar S, Hornsey M, Woodford N, Schneiders T. 2013. Genetic characterization of tigecycline resistance in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J Antimicrob Chemother 68: 1011–1018. [DOI] [PubMed] [Google Scholar]
- Villa L, Feudi C, Fortini D, Garcia-Fernandez A, Carattoli A. 2014. Genomics of KPC-producing Klebsiella pneumoniae sequence type 512 clone highlights the role of RamR and ribosomal S10 protein mutations in conferring tigecycline resistance. Antimicrob Agents Chemother 58: 1707–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinue L, McMurry LM, Levy SB. 2013. The 216-bp marB gene of the marRAB operon in Escherichia coli encodes a periplasmic protein which reduces the transcription rate of marA. FEMS Microbiol Lett 345: 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visalli MA, Murphy E, Projan SJ, Bradford PA. 2003. AcrAB multidrug efflux pump is associated with reduced levels of susceptibility to tigecycline (GAR-936) in Proteus mirabilis. Antimicrob Agents Chemother 47: 665–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PJ, Ciric L, Lerner A, Seville LA, Roberts AP, Mullany P, Allan E. 2013. TetAB46, a predicted heterodimeric ABC transporter conferring tetracycline resistance in Streptococcus australis isolated from the oral cavity. J Antimicrob Chemother 68: 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Bechhofer DH. 2002. Tetracycline induces stabilization of mRNA in Bacillus subtilis. J Bacteriol 184: 889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner G, Gfrorer S, Fleige C, Witte W, Klare I. 2008. Tigecycline-resistant Enterococcus faecalis strain isolated from a German intensive care unit patient. J Antimicrob Chemother 61: 1182–1183. [DOI] [PubMed] [Google Scholar]
- Whittle G, Hund BD, Shoemaker NB, Salyers AA. 2001. Characterization of the 13-kilobase ermF region of the Bacteroides conjugative transposon CTnDOT. Appl Environ Microbiol 67: 3488–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G, Smith I. 1979. Chromosomal mutations causing resistance to tetracycline in Bacillus subtilis. Mol Gen Genet 177: 23–29. [DOI] [PubMed] [Google Scholar]
- Wyeth Pharmaceuticals. 2016. Prescribing information, Tygacil; Philadelphia, PA. [Google Scholar]
- Xiao XY, Hunt DK, Zhou J, Clark RB, Dunwoody N, Fyfe C, Grossman TH, O’Brien WJ, Plamondon L, Ronn M, et al. 2012. Fluorocyclines. 1: 7-fluoro-9-pyrrolidinoacetamido-6-demethyl-6-deoxytetracycline: A potent, broad spectrum antibacterial agent. J Med Chem 55: 597–605. [DOI] [PubMed] [Google Scholar]
- Xiao XY, Deng Y, Sun C, Hunt D, Clark R, Fyfe C, Grossman T, Sutcliffe J. 2013. Novel 7-CF3-8-heterocyclyl tetracyclines with promising antibacterial activity against Gram-positive and Gram-negative pathogens, including Pseudomonas aeruginosa. In Abstr 53th Intersci Conf Antimicrob Agents Chemother, Abstract F-632 Denver, CO. [Google Scholar]
- Yamaguchi A, Ohmori H, Kaneko-Ohdera M, Nomura T, Sawai T. 1991. ΔpH-dependent accumulation of tetracycline in Escherichia coli. Antimicrob Agents Chemother 35: 53–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S, Nikaido E, Nakashima R, Sakurai K, Fujiwara D, Fujii I, Nishino K. 2013. The crystal structure of multidrug-resistance regulator RamR with multiple drugs. Nat Commun 4: 2078. [DOI] [PubMed] [Google Scholar]
- Yang W, Moore IF, Koteva KP, Bareich DC, Hughes DW, Wright GD. 2004. TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics. J Biol Chem 279: 52346–52352. [DOI] [PubMed] [Google Scholar]
- Yoon EJ, Courvalin P, Grillot-Courvalin C. 2013. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: Major role for AdeABC overexpression and AdeRS mutations. Antimicrob Agents Chemother 57: 2989–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhanel GG, Homenuik K, Nichol K, Noreddin A, Vercaigne L, Embil J, Gin A, Karlowsky JA, Hoban DJ. 2004. The glycylcyclines: A comparative review with the tetracyclines. Drugs 64: 63–88. [DOI] [PubMed] [Google Scholar]
- Zhanel GG, Adam HJ, Baxter MR, Fuller J, Nichol KA, Denisuik AJ, Lagace-Wiens PR, Walkty A, Karlowsky JA, Schweizer F, et al. 2013. Antimicrobial susceptibility of 22746 pathogens from Canadian hospitals: Results of the CANWARD 2007-11 study. J Antimicrob Chemother 68: 7–22. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li XZ, Poole K. 2001. SmeDEF multidrug efflux pump contributes to intrinsic multidrug resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 45: 3497–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Xu H, Chen D, Zhou H, Hu X, Cheng G. 2014. First emergence of acrAB and oqxAB mediated tigecycline resistance in clinical isolates of Klebsiella pneumoniae pre-dating the use of tigecycline in a Chinese hospital. PLoS ONE 9: e115185. [DOI] [PMC free article] [PubMed] [Google Scholar]