Abstract
Schizophrenia (SCZ) is a serious psychiatric disorder that affects 1% of general population and places a heavy burden worldwide. The underlying genetic mechanism of SCZ remains unknown, but studies indicate that the disease is associated with a global gene expression disturbance across many genes. Next-generation sequencing, particularly of RNA sequencing (RNA-Seq), provides a powerful genome-scale technology to investigate the pathological processes of SCZ. RNA-Seq has been used to analyze the gene expressions and identify the novel splice isoforms and rare transcripts associated with SCZ. This paper provides an overview on the genetics of SCZ, the advantages of RNA-Seq for transcriptome analysis, the accomplishments of RNA-Seq in SCZ cohorts, and the applications of induced pluripotent stem cells and RNA-Seq in SCZ research.
Keywords: RNA sequencing (RNA-Seq), schizophrenia (SCZ), induced pluripotent stem cells (iPSCs)
Introduction
Schizophrenia (SCZ) is a common mental disorder that affects 21 million people worldwide. In the United States, the prevalence of SCZ is ∼1% among American adults and the estimated cost of the disease was $62.7 billion in 2002.1 It significantly interferes with the quality of life for the patients and their families. The risk of suicide for SCZ patients is eightfold higher than general population. Up to 40% of people with SCZ attempt suicide and 15% of them actually complete suicide. SCZ has an equal incidence in men and in women, with the delayed onset in women.2 The symptoms of SCZ include hallucinations, delusions, disorganized speech, abnormal motor behavior, flattened affect and social withdrawal.3 Medications can be used to relieve some symptoms, but there is no cure for SCZ and the underlying mechanisms remain elusive. While the causes of SCZ are still unknown, there is no doubt that the disease has a strong genetic basis. The family history is the most significant risk factor for SCZ.4 A number of genetic approaches were performed to identify the candidate variants and genes implicated in SCZ.5
In the past decade, the sequencing costs have fallen significantly with the development of next-generation sequencing (NGS). RNA sequencing (RNA-Seq) has become a powerful and common tool for genetic research. RNA-Seq can accurately quantify the gene expression levels and establish a global view of whole transcriptome.6 RNA-Seq has been used for transcriptome analysis to understand the genetic mechanism of the SCZ. In this review, we first introduce the genetic basis of SCZ with a focus on genetic studies using high-throughput technologies to identify sequence variants and genes implicated in SCZ. We describe the RNA-Seq approach and its advantages for transcriptome analysis of human complex disorders. We then summarize RNA-Seq studies published so far for analyzing the transcripts in the tissues of SCZ patients. Finally, we provide a synopsis of RNA-Seq studies using induced pluripotent stem cells (iPSCs) as a model for SCZ research.
SCZ Genetics
Although the causes of SCZ remain unknown, there is a clear genetic component to the disease. The risk for developing SCZ increases significantly with the history of family psychosis. SCZ is present in 1% of the general population, and it occurs in 13% of those with one parent who had SCZ. If one identical twin is diagnosed with SCZ, the rate of developing SCZ for the other is ∼50%. The twin and family studies suggest that the SCZ has a significant heritability at 81%.7 However, little is known about the roles of genetic factors in the development of SCZ. Identifying the variants and genes associated with SCZ could help elucidate the genetic mechanisms of the disease.
The genetic variants play a significant role in the nature of risk for SCZ, and a number of advanced high-throughput technologies were carried out to identify the sequence variants associated with SCZ. Genome-wide association studies (GWASs) have been widely used to identify the common single-nucleotide polymorphisms (SNPs) at novel loci implicated in SCZ.8–14 A GWAS of 3,322 European SCZ patients and 3,587 healthy controls showed convincing statistical evidence for major histocompatibility complex (MHC) region.8 Other GWASs with large sample sizes identified the common variants associated with SCZ on chromosomes 6p22.1,11 18q21.2,12 and 11q24.2,12 and microRNA 137.13 Most common SNPs identified from GWASs have modest effects on the disease risk, but the combined polygenetic effect of many genes may increase the liability to psychiatric disorders, rationalizing the large contribution of common variation to the heritability of SCZ.8 Psychiatric Genomics Consortium (PGC) has established a list of >100 common loci implicated in SCZ and other major mental disorders.14 The GWAS data sets can be accessed from the PGC website (https://www.med.unc.edu/pgc). However, common variants explain only a small proportion of the genetic contribution to SCZ and the remaining missing heritability is apparently unexplained. In the past decade, NGS has powerful capabilities for discovering genetic variations. Copy number variation and exome sequencing studies using NGS demonstrated the growing appreciation of rare genetic variants of large effect on the risk for SCZ.15 Rare copy number variants associated with SCZ have been identified at loci 1q21.1, 3q29, 15q13.3, 16p11.2, and 22q11.21,16 and structural rearrangements implicated in SCZ have been found in phosphodiesterase 4B (PDE4B) and nudE neurodevelopment protein 1 (NDE1) genes.17,18 The case–control exome sequencing studies of SCZ demonstrated that the rare damaging mutations were enriched in the genes involved in neural development or implicated in mental disorders. For example, a recent whole exome sequencing for 2,536 cases and 2,543 controls found an enrichment of rare disruptive variants in genes related to calcium channels, the postsynaptic activity-regulated cytoskeleton-associated protein (ARC) complex, and the fragile X mental retardation protein.19 Another deep sequencing project for a total of 980 cases and 955 controls observed an excess of rare nonsense variants in 101 candidate genes associated with SCZ.20
The sequencing studies provided the target genes that can be used to plan the next steps, and follow-up functional studies have established some candidate genes for developing SCZ.5 Disrupted in schizophrenia 1 (DISC1) gene is a significant candidate gene for SCZ. The risk gene is identified from the linkage analysis of a reciprocal chromosome (1;11) (q42;q14) translocation in a large Scottish pedigree highly burdened for major mental diseases.21 The people with the translocation present a reduced P300 event-related potential associated with SCZ.21 Several independent linkage and association studies also reported evidence for the association of DISC1 variants with various psychiatric disorders as well as multiple cognitive and neurophysiological traits.22–24 The gene expression of DISC1 was changed on SCZ patients, and some spliced transcripts were highly expressed in the hippocampus of these patients.25 The dysbindin (DTNBP1) is another SCZ susceptibility gene, with a reduced expression in the midbrain of SCZ patients.26 Interestingly, DISC1 can enhance the stability of DTNBP1 through forming a complex and the inter-action is important for the process of neurite outgrowth.27 In addition, the association studies have found common variants in susceptibility to major psychiatric disorders in DISC1-interacting partners, such as activating transcription factor 4, pericentriolar material 1, NDE1, and PDE4B.23 Therefore, it is believed that multiple genes can contribute to the development of SCZ by altering interaction networks required for normal brain functions. However, most of the candidate genes have not been well validated for their functional roles and more comprehensive approaches need to be performed to identify the susceptibility genes associated with SCZ. The transcriptome analysis can give functional sense to the genetic knowledge of SCZ.
RNA-Seq and Human Diseases
Over the past decade, the sequencing landscape was dramatically changed. The Sanger sequencing,28 referred as the first-generation sequencing, uses a chain termination and fragmentation approach to sequence DNA. It requires bacterial cloning and can only carry out hundreds of sequencing reaction. Human Genome Project29 costs a total of $3 billion, and it has taken 13 years to sequence the first reference genome using Sanger-based sequencing. It stimulated the development of massively parallel sequencing, called NGS, to reduce the sequence costs and speed up the process. NGS technologies can prepare libraries in a cell-free system and perform millions of sequencing reactions in parallel, which enable the sequencing of genomes at the remarkable price and speed.30,31 The latest NGS platforms can complete the sequencing of whole human genome at a few thousand dollars within one week. The NGS technologies enable scientists to address the biological questions at an unprecedented large scale, which revolutionize the SCZ research.32
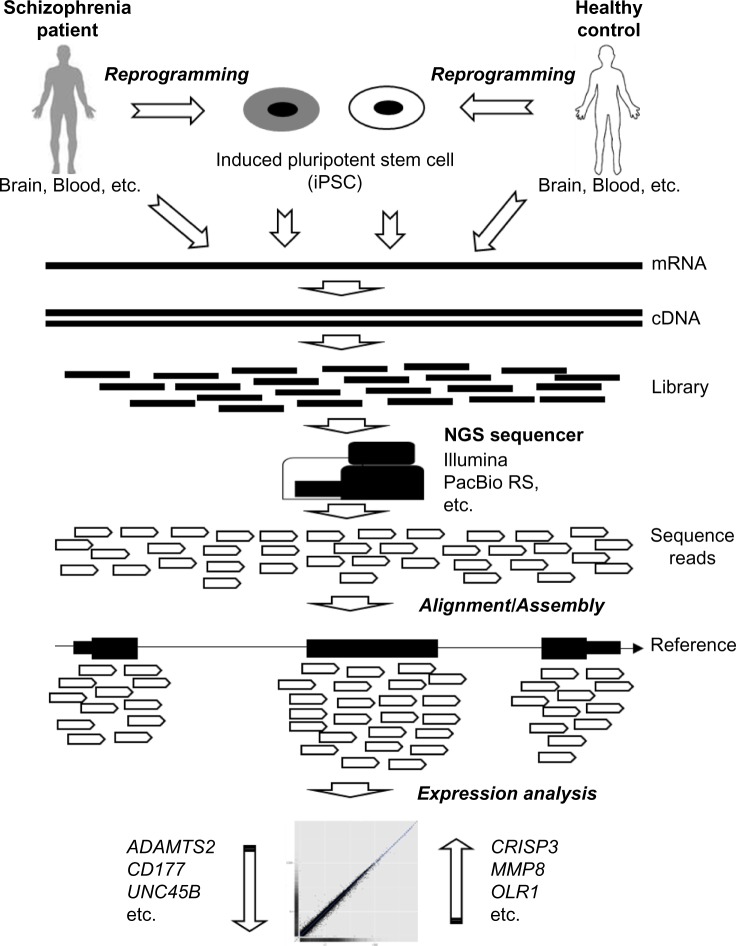
RNA-Seq uses the latest NGS technologies to sequence the transcriptome and conduct gene expression analysis for SCZ (Fig. 1). In general, the samples are collected from human tissues, such as brain, and blood or pluripotent stem cells, such as iPSCs, derived from the tissue cells in SCZ patients and/or healthy individuals. RNAs are extracted from the samples and converted to cDNA fragments with adaptors added for NGS library preparation. Each short cDNA fragment is sequenced using massively parallel sequencing on NGS sequencers, such as Illumina33 and PacBio RS34 systems. Then, the sequence reads are aligned to a known reference genome/transcriptome or assembled without the reference.6 This technology enables the scientists to discover, quantify, and profile RNAs in SCZ studies. RNA-Seq has several advantages over other high-throughput RNA detection technologies, such as DNA microarray hybridization technology (Table 1). It needs a small amount of RNA as inputs and does not require probes based on existing genomic sequence. RNA-Seq has low background noise and can measure the transcripts in a single base level.6 It can quantify the transcripts without limit and detect a dynamic range of gene expression levels. In addition, RNA-Seq can detect exon splicing for different isoforms, allelic expressions, rare transcripts, and noncoding RNAs, which are very difficult to be measured by microarray.6 Thus, RNA-Seq has been widely used for coding and non-coding RNA expression profiling, coding gene annotation, and genome rearrangement detection,35 which provide useful information to elucidate the genetic mechanism of SCZ.
Figure 1.
Schematic overview of RNA-Seq analysis in SCZ.
Table 1.
Comparison of microarray hybridization and RNA-Seq.
| MICROARRAY | RNA-Seq | |
|---|---|---|
| Technology details6 | ||
| Input RNA amount | High | Low |
| Probes | Yes | No |
| Resolution | Relatively Low | High; Single base |
| Background noise | High | Low |
| Cost | High | Relatively Low |
| Application35 | ||
| Discovered gene range | Limited; Only on array | Wide; Whole transcriptome |
| Different isoform | Limited | Yes |
| Allelic expression | Limited | Yes |
| Rare/New transcript | Limited | Yes |
| Noncoding RNA | Limited | Yes |
RNA-Seq provides an unbiased way to investigate the gene expression, and it has been widely used to research the human complex disorders, including cancers as well as neurodegenerative and neuropsychiatric diseases.36 RNA-Seq can be used to investigate transcripts in cancer cells and may help us understanding the carcinogenesis in human. Scientists found that the gene fusions or alternative splicing events generated by genome rearrangements are involved in the pathogenesis of many types of common cancers, such as prostate cancer,37 ovarian cancer, and sarcoma.38 In addition, RNA-Seq can be used to investigate the roles of noncoding RNAs in the pathogenic mechanisms of cancers. For instance, Prensner et al found 121 novel prostate cancer-associated noncoding RNAs and identified prostate cancer associated transcript 1 (PCAT1) as a transcriptional repressor implicated in prostate cancer.39 RNA-Seq has been used to sequence the brain transcriptome of human and animal models, which provided information about the transcriptional profiles in pathological states of neurodegenerative and psychiatric disorders.40 For example, the RNA-Seq studies described the differing gene expression levels and splicing isoforms in the brain regions of the Alzheimer’s patients.41 Martin et al42 carried out RNA-Seq on the medial prefrontal cortex of rats treated with lysergic acid diethylamide and found that the differentially expressed genes are enriched in processes implicated in SCZ. In the next section, we summarize the recent accomplishments of RNA-Seq in human individuals with SCZ and other neuropsychiatric disorders.
RNA-Seq in SCZ Cohorts
RNA-Seq allows investigators to better tailor the gene expression changes in SCZ patients compared to normal people (Table 2). Researchers applied RNA-Seq to sequence transcripts derived from the brain tissues of SCZ patients and unaffected controls to survey gene expression changes and identify novel splicing isoforms. Wu et al.43 used RNA-Seq to sequence the mRNAs collected from postmortem superior temporal gyrus in nine SCZ patients and nine matched controls. They demonstrated that the differentially expressed genes were enriched in energy production, neurotransmission, presynaptic function, and neural development. In addition, they observed that the genes that showed significant differences in alternative splicing and promoter usage were exemplified by doublecortin like kinase 1 (DCLK1) gene, which implicated the critical roles of aberrant RNA processing in the pathophysiology of SCZ. A follow-up study used the differentially expressed genes derived from the same RNA-Seq data and SCZ candidate genes collected from different literature data sets to establish an SCZ-mediator network.44 The network analysis of mitochondrial and coagulation pathways suggested that hemostatic process and energy metabolism play critical roles in the pathogenesis of SCZ. RNA-Seq has been used to investigate genes associated with antipsychotic mechanisms in SCZ. Crespo-Facorro et al45 analyzed the blood mRNAs collected from 22 SCZ patients before and after atypical antipsychotic medication. They identified that the expression levels of six genes with the evidence in a previous study of drug-naive SCZ patients were significantly changed in the patients after medication.
Table 2.
Summary of RNA-Seq studies in SCZ cohorts.
| SAMPLE SIZE* | TISSUE | PLATFORMS† | CANDIDATE GENES | REFERENCE |
|---|---|---|---|---|
| 18 (9S, 9C) | Superior temporal gyrus | Illumina GA | DCLK1; TP53, PRKACA, STAT3, SP1, DRD2, DDR3, HTR2A | Wu et al. (2012)43; Huang et al. (2014)44 |
| 22 (22S) | Blood | Illumina GA | ADAMTS2, CD177, CNTNAP3, ENTPD2, RFX2, UNC45B | Crespo-Facorro et al. (2015)45 |
| 29 (14S, 15C) | Hippocampus | Illumina GA | IFITM1, IFITM2, IFITM3, APOL1, ADORA2A, IGFBP4, CD163 | Hwang et al. (2013)46 |
| 40 (20S, 20C) | Dorsolateral prefrontal cortex | SOLiD | IL-6, IL-8, IL-1β, SERPINA3 | Fillman et al. (2013)47; Catts et al. (2014)48 |
| 6 (3S, 3C) | Blood | Illumina GA | S100A8, TYROBP | Xu et al. (2012)49 |
| 76 (36S, 40C) | Blood | Illumina GA | ADAMTS2, CSMD1, EHF, RFX2, GRIK3, LPL, S100B, SNCA, SYN2, TUBB2A, SELENBP1, CSMD1 | Sainz et al. (2013)50 |
| 79 (17S, 16B, 17M, 40C) | Dentate gyrus (Hippocampus) | SOLiD | miR-182 | Kohen et al. (2014)53 |
| 144 (55S, 34B, 55C) | Prefrontal cortex | SOLiD | FKBP5, PTGES3, BAG1 | Sinclair et al. (2013)54 |
| 82 (31S, 25B, 26C) | Cingulate cortex | Illumina GA and HiSeq | FER, FGF13, IDH1, DYNLT3, TRPV1, CBFA2T2,MTA2, IGSF11, NAP1L5 | Zhao et al. (2015)55 |
| 22 (11B, 11C) | Dorsolateral prefrontal cortex | Illumina GA and HiSeq | ABCG2, SRSF5, RFX4 | Akula et al. (2015)56 |
| 59 (30M, 29C) | Dorsolateral prefrontal cortex | Illumina MiSeq | SAT1 | Pantazatos et al. (2015)57 |
Notes:
Schizophrenia (S); bipolar disorder (B); recurrent major depressive disorder (M); and healthy control (C).
Illumina Genome Analyzer (GA), HiSeq, and MiSeq sequencers and Applied Biosystems SOLiD™ sequencer.
The most significant association was identified between SCZ and locus on chromosome 6p in the MHC region,8,12 which strongly implicates that the immune system contributes to the pathophysiology of SCZ. The RNA-Seq studies for the brain tissues demonstrated that the differentially expressed genes in SCZ patients were enriched in the process associated with immune/inflammation system. Hwang et al.46 identified 144 differentially expressed genes from the hippocampus of SCZ patients and healthy controls. The co-expression network analysis indicated that these genes were highly enriched in the immune/inflammation responses. The transcriptome analysis for dorsolateral prefrontal cortex of 20 SCZ patients and 20 controls found 798 differentially expressed genes, which are overrepresented in the process corresponding to inflammatory response.47 A follow-up study suggested that the expressions of astrocyte marker genes were increased in SCZ patients with neuroinflammation.48 The RNA-Seq analyses of blood samples also support the important roles of inflammatory responses in the development of SCZ. The gene expression profiling in blood samples of six individuals (three SCZ patients and three controls) indicated that the differentially expressed genes are enriched in the processes correlated with immune/inflammatory response.49 Another study of a larger cohort (36 SCZ patients and 40 controls) also supported that immune/inflammatory response is the major biological process presented in the genes with significant differential expressions in SCZ patients.50 These RNA-Seq studies indicated that immune response may underlie the pathophysiology of SCZ with gene expression alterations occurring in both brain and blood tissues. These findings could be used to elucidate the molecular mechanisms of SCZ, identify target genes for the disease, and develop antipsychotic drugs.
The strong evidence based on neuropathological and genetic studies suggested that SCZ has a clear and overlapping genetic component with other common psychiatric disorders, including bipolar disorder (BD) and recurrent major depressive disorder (rMDD).51,52 RNA-Seq was used to identify the shared expression change patterns and investigate the etiological mechanisms for these common psychiatric disorders. The transcriptome analysis of human hippocampus dentate gyrus granule cells from 79 individuals with psychiatric disorders and healthy controls showed that microRNA 182 (miR-182) was involved in shaping the dentate gyrus and the disrupted signaling was found in controls and BD patients, but not in patients with rMDD and SCZ.53 Sinclair et al.54 analyzed prefrontal cortex of 144 subjects (55 SCZ patients, 34 BD patients, and 55 controls) and found that the expressions of FK506 binding protein 5 (FKBP5), prostaglandin E synthase 3 (PTGES3), and BCL2-associated athanogene 1 (BAG1) were significantly changed in SCZ and/or BD patients. The whole transcriptome analysis of cingulate cortex from 82 samples (31 SCZ patients, 25 BD patients, and 26 controls) suggested that the differentially expressed genes were enriched in signals from GWASs for both SCZ and BD.55 The transcriptome analyses of dorsolateral prefrontal cortex identified significant gene expression changes in ATP-binding cassette sub-family G-member 2 (ABCG2), serine/arginine-rich splicing factor 5 (SRSF5), and regulatory factor X4 (RFX4) in BD56 patients and spermidine/spermine N1-acetyltransferase (SAT1) in rMDD57 patients. The microRNA expression study of the prefrontal cortex, hippocampus, and cerebellum showed that the differentially expressed microRNA targets were highly enriched for gene sets associated with major psychiatric disorders, such as SCZ, BD, rMDD, and autism, which suggest that microRNAs play important roles in the transcriptional networks of developing major psychiatric and neurodevelopmental disorders.58
In addition to investigating altered gene expression, RNA-Seq can be used to analyze the important cellular mechanisms, such as RNA editing.59 The most prevalent type of RNA editing in human beings is adenosine-to-inosine (A-to-I) RNA editing catalyzed by the adenosine deaminases acting on RNA (ADAR), which has been implicated in SCZ.60 RNA-Seq provides an accurate and quantitative approach to detect the A-to-I RNA editing sites at genome level.61 Li et al.62 applied the parallel DNA capturing and sequencing to analyze the transcripts in seven tissues from a single individual and identified several hundred potential human RNA editing sites. However, Zhu et al.63 examined these putative RNA editing sites using an ultra-high-throughput sequencing and showed that the variability of RNA editing at the sites may be overestimated in normal human brain. Interestingly, they analyzed 29 confirmed RNA editing sites in SCZ and rMDD patients but did not identify any significant alterations of RNA editing for these sites. Silberberg et al.64 applied high-throughput sequencing to analyze the editing levels within neuroreceptor in postmortem brain samples obtained from SCZ patients, BD patients, and healthy individuals. In SCZ samples, they observed that the expression of specific ADAR variants was increased but the catalytic activity was decreased. In addition, they found that the RNA editing sites in the glutamate receptor, ionotropic, kainate 2 (GRIK2) transcript are edited frequently in BD patients.
A number of resources have been developed to collect and analyze the RNA-Seq data involved in these common psychiatric disorders. PD_NGSAtlas65 (http://210.46.85.200/pd_ngsatlas/) includes 37 expression profiles collected by RNA-Seq in blood and brain regions of SCZ, BD, and healthy individuals. Users can download gene expression data for their research or view multidimensional data under a given genomic context from its own genome browser. Wang and Cairns developed an open-source software, called SeqGSEA66 (https://www.bioconductor.org/packages/release/bioc/html/SeqGSEA.html), for differential expression and splicing analysis as well as functional gene set analysis in RNA-Seq data. They showed that this approach can be used to carry out the analysis on SCZ candidate gene set, with high statistical significance and biological relevancy.67
RNA-Seq Analysis of iPSCs for SCZ Modeling
A challenging problem of SCZ research is the inaccessibility of the human materials, such as brain tissues. In particular, the studies for investigating pathological progression of SCZ have been hampered by the brain tissues of SCZ patients, which are usually collected from a particular stage of the disease. Fortunately, iPSC technology provides reliable patient-derived cellular model systems to solve this challenging issue in psychiatric research. iPSCs are a type of adult mammalian cells that can be genetically reprogramed to an embryonic stem cell (ESC)-like state by being forced to express genes important for maintaining the essential properties of ESCs. The iPSC technology not only provides the tools to investigate the development and epigenetic reprograming but also can be used to investigate the human neuropsychiatric disorders, using the disease-specific neurons derived from iPSCs of patients.68 For instance, the iPSCs can be used to prepare brain cells that contain the actual genetic information of the SCZ patients. In addition, the in vitro differentiation tool makes it possible to construct an accurate SCZ disease model.69
The advent of iPSC technology allows scientists to investigate the cellular and molecular effects of SCZ.70 Chiang et al.71 generated the first integration-free iPS cell line from the skin biopsies of SCZ patients with a DISC1 mutation. Then, Brennand et al.72 and Tran et al.73 established human iPSCs from fibroblasts of SCZ patients and differentiated them to SCZ-specific iPSC neurons. The neurons had defects in neuronal connectivity and reduced expression levels of PSD95 dendritic protein and glutamate receptor.72 Interestingly, the SCZ-specific iPSC neurons showed the differential expressions in genes implicated in AMP and WNT signaling pathways, which were consistent with the previous GWAS of SCZ.72 These efforts make it possible to perform in vitro genetic and pharmacological analyses to study the underlying mechanisms of SCZ.
RNA-Seq has been used to analyze the transcripts of patient-specific neurons from iPSCs for testing the expression alterations in SCZ candidate genes. The gene expression profiling on neurons from iPSCs programed from dental pulp showed that the high expressed genes were enriched for the genes associated with SCZ.74 Zeng et al.75 used the neural stem cells differentiated from iPSCs as a model to study the cellular effects of exonic deletions in an SCZ candidate gene neurexin 1 (NRXN1), which suggested that iPSC is a reliable model for studying the functional genetic link between sequence variation and neurodevelopment. Lin et al used the iPSC and RNA-Seq to investigate imprinting and stochastic processes in human neurons and observed that the SCZ candidate genes have allele-biased expressions.76 They observed dramatic expression changes in SCZ candidate genes, including NRXN1 and neuroligin 1 (NLGN1).77 In addition, they found that the long noncoding RNAs associated with a GWAS of SCZ have increased expression during the transition from iPSCs to differentiating neurons. The environmental factors, such as maternal immune activation, play important roles in the development of SCZ. The transcriptome analysis of neuronal aggregates derived from iPSC showed that the SCZ candidate genes have dramatic expression changes after heat shock.78 RNA-Seq has also been applied to characterize the iPSCs derived from the patients with other major psychiatric disorders. Madison et al.79 built 12 iPSC lines from two affected brothers with BD and their two unaffected parents. They utilized RNA-Seq to analyze the transcripts of the cell lines and observed that the differentially expressed transcripts are enriched in the biological process associated with neuron differentiation and development.
Differentiating neurons derived from iPSCs provide an ideal system for RNA-Seq to study the expression profiling of biological processes in the pathophysiology of SCZ. However, only a few applications of iPSC technology have been used for SCZ research. There are still several major obstacles that must be overcome in this revolution technology. First, the iPSC neurons are closer to fetal neurons rather than adult ones,80 and the poor maturation may be an issue for studying the late-onset SCZ. Second, most iPSCs contain multiple gene disruptions due to reprograming process, which may result in genetic dysfunction. The reprograming transgenes make iPSCs prone to tumor formation and may lead to abnormal differentiation. Finally, there are some variations in gene expressions between ESCs and iPSCs,81 which could be particularly problematic in RNA-Seq analysis for SCZ research. Despite these limitations, iPSC provides a good cellular model system to accurately reflect disease conditions. By combining RNA-Seq technology, iPSC can be used to identify the candidate genes associated with SCZ and customize the cell therapies of the disease in the future.82
Conclusions
The pathophysiology of SCZ remains largely obscure. It is considered to be a complex genetic disorder with multiple genes that contribute to the risk of the disease. The main direction of SCZ genetic research is to identify genes conferring risk or protection. RNA-Seq is a powerful tool to investigate the disease-related gene expression alterations at the level of mRNA, with high resolution and reduced cost. It has been fruitfully applied to elucidate the involvement of multiple genes in the pathogenesis of SCZ. The RNA-Seq analyses for both brain and blood tissues of SCZ patients demonstrated that the immune system plays critical roles in the pathophysiology of SCZ. In addition, RNA-Seq can be used to investigate the shared genetic factors between SCZ and other psychiatric disorders. The patient-specific neurons derived from iPSCs provide an ideal system for disease modeling and gene expression profiling using RNA-Seq, which provides useful information for further analyses to characterize the functional effects of candidate genes and design antipsychotic drugs. RNA-Seq studies of SCZ improve our understanding of the roles of multiple genes in the causation of common psychiatric disorders. The advents of RNA-Seq will help biomedical scientists and doctors to develop the better diagnostic and treatment methods for SCZ patients.
Footnotes
ACADEMIC EDITOR: Thomas Dandekar, Associate Editor
PEER REVIEW: Four peer reviewers contributed to the peer review report. Reviewers’ reports totaled 856 words, excluding any confidential comments to the academic editor.
FUNDING: Authors disclose no external funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert single-blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived the idea of the manuscript: ST. Wrote the manuscript: XL, ST. Reviewed and approved the final manuscript: XL, ST.
REFERENCES
- 1.Wu EQ, Birnbaum HG, Shi L, et al. The economic burden of schizophrenia in the United States in 2002. J Clin Psychiatry. 2005;66:1122–9. doi: 10.4088/jcp.v66n0906. [DOI] [PubMed] [Google Scholar]
- 2.Hafner H. Gender differences in schizophrenia. Psychoneuroendocrinology. 2003;28(suppl 2):17–54. doi: 10.1016/s0306-4530(02)00125-7. [DOI] [PubMed] [Google Scholar]
- 3.Schultz SH, North SW, Shields CG. Schizophrenia: a review. Am Fam Physician. 2007;75:1821–9. [PubMed] [Google Scholar]
- 4.Mortensen PB, Pedersen CB, Westergaard T, et al. Effects of family history and place and season of birth on the risk of schizophrenia. N Engl J Med. 1999;340:603–8. doi: 10.1056/NEJM199902253400803. [DOI] [PubMed] [Google Scholar]
- 5.Kim Y, Zerwas S, Trace SE, Sullivan PF. Schizophrenia genetics: where next? Schizophr Bull. 2011;37:456–63. doi: 10.1093/schbul/sbr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Gerstein M, Snyder M. RNA-seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–92. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 8.Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SH, DeCandia TR, Ripke S, et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44:247–50. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ripke S, O’Dushlaine C, Chambert K, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–9. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi J, Levinson DF, Duan J, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–7. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefansson H, Ophoff RA, Steinberg S, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–7. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ripke S, Sanders A, Kendler K, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–76. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ripke S, Neale BM, Corvin A, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schreiber M, Dorschner M, Tsuang D. Next-generation sequencing in schizophrenia and other neuropsychiatric disorders. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:671–8. doi: 10.1002/ajmg.b.32156. [DOI] [PubMed] [Google Scholar]
- 16.Levinson DF, Duan J, Oh S, et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168:302–16. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Millar JK, Pickard BS, Mackie S, et al. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310:1187–91. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- 18.Ingason A, Rujescu D, Cichon S, et al. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol Psychiatry. 2011;16:17–25. doi: 10.1038/mp.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell SM, Moran JL, Fromer M, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–90. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu X, Zhang B, Liu W, et al. A survey of rare coding variants in candidate genes in schizophrenia by deep sequencing. Mol Psychiatry. 2014;19:857–8. doi: 10.1038/mp.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders – cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69:428–33. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carless MA, Glahn DC, Johnson MP, et al. Impact of DISC1 variation on neuroanatomical and neurocognitive phenotypes. Mol Psychiatry. 2011;16(1063):1096–104. doi: 10.1038/mp.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bradshaw NJ, Porteous DJ. DISC1-binding proteins in neural development, signalling and schizophrenia. Neuropharmacology. 2012;62:1230–41. doi: 10.1016/j.neuropharm.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnstone M, Thomson PA, Hall J, McIntosh AM, Lawrie SM, Porteous DJ. DISC1 in schizophrenia: genetic mouse models and human genomic imaging. Schizophr Bull. 2011;37:14–20. doi: 10.1093/schbul/sbq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakata K, Lipska BK, Hyde TM, et al. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc Natl Acad Sci U S A. 2009;106:15873–8. doi: 10.1073/pnas.0903413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weickert CS, Straub RE, McClintock BW, et al. Human dysbindin (DTNBP1) gene expression in normal brain and in schizophrenic prefrontal cortex and midbrain. Arch Gen Psychiatry. 2004;61:544–55. doi: 10.1001/archpsyc.61.6.544. [DOI] [PubMed] [Google Scholar]
- 27.Lee SA, Kim SM, Suh BK, et al. Disrupted-in-schizophrenia 1 (DISC1) regulates dysbindin function by enhancing its stability. J Biol Chem. 2012;290:7087–96. doi: 10.1074/jbc.M114.614750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–7. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adekoya E, Ait-Zahra M, Allen N, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 30.van Dijk EL, Auger H, Jaszczyszyn Y, Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30:418–26. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 31.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–45. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 32.Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER. The next-generation sequencing revolution and its impact on genomics. Cell. 2013;155:27–38. doi: 10.1016/j.cell.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bentley D, Balasubramanian S, Swerdlow H, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–9. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eid J, Fehr A, Gray J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–8. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 35.Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–64. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Costa V, Aprile M, Esposito R, Ciccodicola A. RNA-Seq and human complex diseases: recent accomplishments and future perspectives. Eur J Hum Genet. 2013;21:134–42. doi: 10.1038/ejhg.2012.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pflueger D, Terry S, Sboner A, et al. Discovery of non-ETS gene fusions in human prostate cancer using next-generation RNA sequencing. Genome Res. 2011;21:56–67. doi: 10.1101/gr.110684.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McPherson A, Hormozdiari F, Zayed A, et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol. 2011;7:e1001138. doi: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prensner JR, Iyer MK, Balbin OA, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–9. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hitzemann R, Darakjian P, Walter N, Dan Iancu O, Searles R, McWeeney S. Introduction to sequencing the brain transcriptome. Int Rev Neurobiol. 2014;116:1–19. doi: 10.1016/B978-0-12-801105-8.00001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Twine NA, Janitz K, Wilkins MR, Janitz M. Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by Alzheimer’s disease. PLoS One. 2011;6:e16266. doi: 10.1371/journal.pone.0016266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin DA, Marona-Lewicka D, Nichols DE, Nichols CD. Chronic LSD alters gene expression profiles in the mPFC relevant to schizophrenia. Neuropharmacology. 2014;83:1–8. doi: 10.1016/j.neuropharm.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu JQ, Wang X, Beveridge NJ, et al. Transcriptome sequencing revealed significant alteration of cortical promoter usage and splicing in schizophrenia. PLoS One. 2012;7:e36351. doi: 10.1371/journal.pone.0036351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang KC, Yang KC, Lin H, Tsao TT, Lee SA. Transcriptome alterations of mitochondrial and coagulation function in schizophrenia by cortical sequencing analysis. BMC Genomics. 2014;15(suppl 9):S6. doi: 10.1186/1471-2164-15-S9-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crespo-Facorro B, Prieto C, Sainz J. Schizophrenia gene expression profile reverted to normal levels by antipsychotics. Int J Neuropsychopharmacol. 2015;18:yu066. doi: 10.1093/ijnp/pyu066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hwang Y, Kim J, Shin JY, et al. Gene expression profiling by mRNA sequencing reveals increased expression of immune/inflammation-related genes in the hippocampus of individuals with schizophrenia. Transl Psychiatry. 2013;3:e321. doi: 10.1038/tp.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fillman SG, Cloonan N, Catts VS, et al. Increased inflammatory markers identified in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol Psychiatry. 2013;18:206–14. doi: 10.1038/mp.2012.110. [DOI] [PubMed] [Google Scholar]
- 48.Catts VS, Wong J, Fillman SG, Fung SJ, Shannon Weickert C. Increased expression of astrocyte markers in schizophrenia: association with neuroinflammation. Aust N Z J Psychiatry. 2014;48:722–34. doi: 10.1177/0004867414531078. [DOI] [PubMed] [Google Scholar]
- 49.Xu J, Sun J, Chen J, et al. RNA-Seq analysis implicates dysregulation of the immune system in schizophrenia. BMC Genomics. 2012;13(suppl 8):S2. doi: 10.1186/1471-2164-13-S8-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sainz J, Mata I, Barrera J, et al. Inflammatory and immune response genes have significantly altered expression in schizophrenia. Mol Psychiatry. 2013;18:1056–7. doi: 10.1038/mp.2012.165. [DOI] [PubMed] [Google Scholar]
- 51.Gottesman II, Laursen TM, Bertelsen A, Mortensen PB. Severe mental disorders in offspring with 2 psychiatrically ill parents. Arch Gen Psychiatry. 2010;67:252–7. doi: 10.1001/archgenpsychiatry.2010.1. [DOI] [PubMed] [Google Scholar]
- 52.Lichtenstein P, Yip BH, Bjork C, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–9. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kohen R, Dobra A, Tracy JH, Haugen E. Transcriptome profiling of human hippocampus dentate gyrus granule cells in mental illness. Transl Psychiatry. 2014;4:e366. doi: 10.1038/tp.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sinclair D, Fillman SG, Webster MJ, Weickert CS. Dysregulation of glucocorticoid receptor co-factors FKBP5, BAG1 and PTGES3 in prefrontal cortex in psychotic illness. Sci Rep. 2013;3:3539. doi: 10.1038/srep03539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao Z, Xu J, Chen J, et al. Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. Mol Psychiatry. 2015;20:563–72. doi: 10.1038/mp.2014.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akula N, Barb J, Jiang X, et al. RNA-sequencing of the brain transcriptome implicates dysregulation of neuroplasticity, circadian rhythms and GTPase binding in bipolar disorder. Mol Psychiatry. 2014;19:1179–85. doi: 10.1038/mp.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pantazatos SP, Andrews SJ, Dunning-Broadbent J, et al. Isoform-level brain expression profiling of the spermidine/spermine N1-acetyltransferase1 (SAT1) gene in major depression and suicide. Neurobiol Dis. 2015;79:123–34. doi: 10.1016/j.nbd.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ziats MN, Rennert OM. Identification of differentially expressed microRNAs across the developing human brain. Mol Psychiatry. 2014;19:848–52. doi: 10.1038/mp.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng Z, Cheng Y, Tan BC-M, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol. 2012;30:253–60. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- 60.Akbarian S, Smith MA, Jones EG. Editing for an AMPA receptor subunit RNA in prefrontal cortex and striatum in Alzheimer’s disease, Huntington’s disease and schizophrenia. Brain Res. 1995;699:297–304. doi: 10.1016/0006-8993(95)00922-d. [DOI] [PubMed] [Google Scholar]
- 61.Ramaswami G, Zhang R, Piskol R, et al. Identifying RNA editing sites using RNA sequencing data alone. Nat Methods. 2013;10:128–32. doi: 10.1038/nmeth.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li JB, Levanon EY, Yoon J-K, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–3. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 63.Zhu H, Urban DJ, Blashka J, et al. Quantitative analysis of focused a-to-I RNA editing sites by ultra-high-throughput sequencing in psychiatric disorders. PLoS One. 2012;7:e43227. doi: 10.1371/journal.pone.0043227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Silberberg G, Lundin D, Navon R, Öhman M. Deregulation of the A-to-I RNA editing mechanism in psychiatric disorders. Hum Mol Genet. 2012;21:311–21. doi: 10.1093/hmg/ddr461. [DOI] [PubMed] [Google Scholar]
- 65.Zhao Z, Li Y, Chen H, et al. PD_NGSAtlas: a reference database combining next-generation sequencing epigenomic and transcriptomic data for psychiatric disorders. BMC Med Genomics. 2014;7:512. doi: 10.1186/s12920-014-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang X, Cairns MJ. SeqGSEA: a bioconductor package for gene set enrichment analysis of RNA-Seq data integrating differential expression and splicing. Bioinformatics. 2014;30:1777–9. doi: 10.1093/bioinformatics/btu090. [DOI] [PubMed] [Google Scholar]
- 67.Wang X, Cairns MJ. Understanding complex transcriptome dynamics in schizophrenia and other neurological diseases using RNA sequencing. Int Rev Neurobiol. 2014;116:127–52. doi: 10.1016/B978-0-12-801105-8.00006-0. [DOI] [PubMed] [Google Scholar]
- 68.Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–63. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Imaizumi Y, Okano H. Modeling human neurological disorders with induced pluripotent stem cells. J Neurochem. 2014;129:388–99. doi: 10.1111/jnc.12625. [DOI] [PubMed] [Google Scholar]
- 70.Brennand KJ, Gage FH. Concise review: the promise of human induced pluripotent stem cell-based studies of schizophrenia. Stem Cells. 2011;29:1915–22. doi: 10.1002/stem.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chiang C-H, Su Y, Wen Z, et al. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Mol Psychiatry. 2011;16:358–60. doi: 10.1038/mp.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brennand KJ, Simone A, Jou J, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–5. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tran NN, Ladran IG, Brennand KJ. Modeling schizophrenia using induced pluripotent stem cell-derived and fibroblast-induced neurons. Schizophr Bull. 2013;39:4–10. doi: 10.1093/schbul/sbs127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen J, Lin M, Foxe JJ, et al. Transcriptome comparison of human neurons generated using induced pluripotent stem cells derived from dental pulp and skin fibroblasts. PLoS One. 2013;8:e75682. doi: 10.1371/journal.pone.0075682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zeng L, Zhang P, Shi L, Yamamoto V, Lu W, Wang K. Functional impacts of NRXN1 knockdown on neurodevelopment in stem cell models. PLoS One. 2013;8:e59685. doi: 10.1371/journal.pone.0059685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin M, Hrabovsky A, Pedrosa E, Wang T, Zheng D, Lachman HM. Allele-biased expression in differentiating human neurons: implications for neuropsychiatric disorders. PLoS One. 2012;7:e44017. doi: 10.1371/journal.pone.0044017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin M, Pedrosa E, Shah A, et al. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS One. 2011;6:e23356. doi: 10.1371/journal.pone.0023356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin M, Zhao D, Hrabovsky A, Pedrosa E, Zheng D, Lachman HM. Heat shock alters the expression of schizophrenia and autism candidate genes in an induced pluripotent stem cell model of the human telencephalon. PLoS One. 2014;9:e94968. doi: 10.1371/journal.pone.0094968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Madison JM, Zhou F, Nigam A, et al. Characterization of bipolar disorder patient-specific induced pluripotent stem cells from a family reveals neurodevelopmental and mRNA expression abnormalities. Mol Psychiatry. 2015;20:703–17. doi: 10.1038/mp.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Patani R, Lewis PA, Trabzuni D, et al. Investigating the utility of human embryonic stem cell-derived neurons to model ageing and neurodegenerative disease using whole-genome gene expression and splicing analysis. J Neurochem. 2012;122:738–51. doi: 10.1111/j.1471-4159.2012.07825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10:678–84. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 82.Kim KS. Induced pluripotent stem (iPS) cells and their future in psychiatry. Neuropsychopharmacology. 2010;35:346–8. doi: 10.1038/npp.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]