Abstract
Intestinal dysbiosis, characterized by a reduced Firmicutes/Bacteroidetes ratio, has been reported in systemic lupus erythematosus (SLE) patients. In this study, in vitro cultures revealed that microbiota isolated from SLE patient stool samples (SLE-M) promoted lymphocyte activation and Th17 differentiation from naïve CD4+ lymphocytes to a greater extent than healthy control-microbiota. Enrichment of SLE-M with Treg-inducing bacteria showed that a mixture of two Clostridia strains significantly reduced the Th17/Th1 balance, whereas Bifidobacterium bifidum supplementation prevented CD4+ lymphocyte over-activation, thus supporting a possible therapeutic benefit of probiotics containing Treg-inducer strains in order to restore the Treg/Th17/Th1 imbalance present in SLE. In fact, ex vivo analyses of patient samples showed enlarged Th17 and Foxp3+ IL-17+ populations, suggesting a possible Treg-Th17 trans-differentiation. Moreover, analyses of fecal microbiota revealed a negative correlation between IL-17+ populations and Firmicutes in healthy controls, whereas in SLE this phylum correlated directly with serum levels of IFNγ, a Th1 cytokine slightly reduced in patients. Finally, the frequency of Synergistetes, positively correlated with the Firmicutes/Bacteroidetes ratio in healthy controls, tended to be reduced in patients when anti-dsDNA titers were increased and showed a strong negative correlation with IL-6 serum levels and correlated positively with protective natural IgM antibodies against phosphorylcholine.
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease triggered by a combination of environmental and genetic factors that result in a breakdown in tolerance towards self-antigens1. The subsequent production of autoantibodies by autoreactive B cells constitutes a key pathological factor in SLE, since it leads to the formation and deposition of immune-complexes that cause tissue damage2. Likewise, naïve CD4+ cells activated by recognition of such self-antigens can be differentiated into several subsets based on the pattern of cytokines present in the local environment3. In addition to the well-known paradigm of Th1/Th2 cell immune response, nowadays much evidence reveals the presence of alterations in Th17 and regulatory T (Treg) cells in SLE disease4,5,6. With regard to Th17cells, some studies support their pivotal role as primary drivers of autoimmune responses in SLE through the secretion of proinflammatory cytokines involved in local inflammation and tissue destruction, including IL-17, IL-22 andIL-237,8. Accordingly, increased circulating levels of IL-17 and IL-17-producing T cells have been recently reported in SLE9,10,11. Moreover, IL-17-producing T cells have also been shown to infiltrate the lungs, skin and kidneys in lupus patients, contributing to organ damage10,12. Conversely, Treg cells are essential for preventing autoimmune and inflammatory diseases, since they present a suppressive activity on aberrant effector responses13. Naturally occurring Treg cells emerge from the thymus and are primarily characterized by the presence of high levels of CD25 (IL2Rα chain) and FOXP3, a transcription factor required for the development and function of Treg cells14. In addition, Treg cells could be expanded or induced in peripheral tissues in response to diverse antigens15. Most studies report either reduced numbers or impaired function of circulating Treg cells in SLE patients16,17,18.
Increasing evidence suggests that the composition of the commensal microbiota colonizing the gut affects the differentiation of immune cells present in the gut-associated lymphoid tissues (GALTs)19. Specifically, plasmatic cells in the lamina propria are involved in the production of T cell-independent antibodies against components of both commensal and pathogenic bacteria as well as apoptotic cells, named natural IgM antibodies20. Interestingly, several studies have reported immunoregulatory functions of natural IgM antibodies inhibiting the inflammatory signaling in innate immune cells and suppressing autoimmune disease21,22. On the other hand, after the recognition of bacterial antigens, gut dendritic cells (DCs) may induce the differentiation of naïve CD4+ T cells into different types of effector or regulatory T cells23,24,25,26. Under physiologic conditions, the normal microbiota presented in healthy individuals favors the maintenance of the intestinal immune homeostasis27. Conversely, several studies suggest that alterations in the gut microbiota composition, known as dysbiosis, may be a critical factor in the development of numerous immune-mediated pathologies, probably in disease-susceptible hosts, through the generation of an imbalance between Th and Treg cells19,28,29,30,31. In this sense, intestinal dysbiosis has been associated with the development of several autoimmune diseases, including inflammatory bowel disease, type 1 diabetes, rheumatoid arthritis and multiple sclerosis32,33,34,35,36,37,38. In this regard, we have recently described a SLE-associated intestinal dysbiosis characterized by a significantly lower Firmicutes to Bacteroidetes ratio, the most abundant phyla in the human gut39 that has been previously described as imbalanced in other disorders37,38,40. Since these studies suggest that microbiota could control the Th/Treg axis outside the gut, the immune stimulation by specific bacteria could have a beneficial effect on inflammatory diseases33. Thus, it is known that some bacterial strains might induce the generation of Treg cells (iTreg) from naïve precursors23,41,42,43. Specifically, accumulating evidence supports the role of commensal strains of Bifidobacterium and Clostridium spp. belonging to clusters IV and XIVa in the induction of Treg cells23,41,42,43.
The study aims to evaluate the influence of fecal microbiota obtained from SLE patients and healthy controls in the in vitro differentiation of Th and Treg populations as well as the possible effect of enriching SLE gut microbiota with bifidobacteria and Clostridia strains known to be inducers of Treg cells. Then, we analyzed the possible relationship between the SLE-associated gut dysbiosis and the presence of immune parameters characteristic of these patients, such as the Treg/Th populations, cytokine levels, disease activity and the production of both pathogenic anti-dsDNA and protective natural IgM anti-phosphoryl choline antibodies.
Results
In vitro effect of SLE fecal microbiota on Treg/Th differentiation
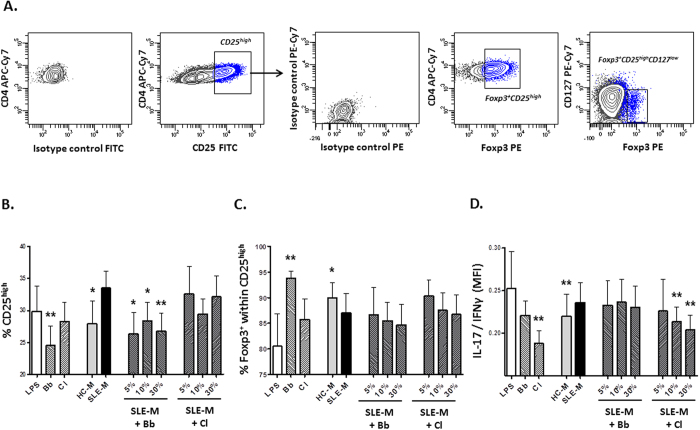
Given the gut dysbiosis recently reported in SLE patients39, we aimed to evaluate the influence of fecal microbiota obtained from SLE patients (SLE-M) and healthy controls (HC-M) in the in vitro differentiation of regulatory T cells (Treg), as well as Th1 and Th17 effector populations from naïve CD4+ T lymphocytes. In addition, to estimate the effect of enriching the gut microbiota with strains able to increase the Treg subset, 5, 10 or 30% of SLE-M were replaced with the same amounts of Bifidobacterium bifidum LMG13195 (Bb), a strain known to induce Foxp3 expression23,41, or with a mixture of two Clostridia strains (Cl) with a putative Treg-inducer effect (Ruminococcus obeum DSM25238 and Blautia coccoides DSM935)42,43. Thus, immature monocyte-derived DCs were treated for 48 h with LPS, as a maturation control, or with the different bacterial preparations and then used to prime CD4+CD45RA+ naïve T lymphocytes. After 12 days of culture in the presence of IL-2, Treg (CD4+CD25highCD127lowFoxp3+), Th17 and Th1(IFNγ- and IL-17-expresing cells, respectively) populations were determined by flow cytometry in CD4+ lymphocytes.
As expected, stimulation and expansion with IL-2 induced IL-2Rα (CD25) expression in most CD4+ lymphocytes (Fig. 1A), however the amount of cells presenting elevated CD25 levels (CD25high) showed differences among treatments (Fig. 1B). Specifically, SLE-M cultures tended to generate more CD25high cells than HC-M, whereas the lowest levels of this population were obtained after stimulation with the Bb strain. In fact, the upregulatory effect of SLE-M on CD25 expression was reverted after Bb supplementation. Regarding Treg cells (CD4+CD25high CD127lowFoxp3+), no significant differences were observed between HC-M or SLE-M stimulation. However, the proportion of Foxp3+ cells included within the CD25high population was lower in SLE- than in HC-M derived cultures (Fig. 1C), thus suggesting that part of the CD25high cells generated in the presence of SLE-M were activated lymphocytes rather than Treg cells. Unexpectedly, although Bb increased Foxp3+ cells notably, supplementation of SLE-M with this strain did not increase the generation of Foxp3+ cells within the CD25high subset.
Figure 1. Fecal microbiota isolated from SLE patients influences Treg/Th differentiation.
Naïve CD4+CD45RA+ lymphocytes were co-cultured for 12 days with DC previously maturated with LPS (maturation control), fecal microbiota isolated from healthy controls (HC-M) or SLE patients (SLE-M) as well as with Bifidobacterium bifidum LMG13195 (Bb), Clostridia strains (Cl) or SLE-M containing 5, 10 or 30% Bb or Cl. Cultured CD4+ T cells were recovered, stained for Treg markers, IL-17 and IFNγ and analyzed by flow cytometry. (A) Sequential gating strategy used to identify Treg cells (CD4+CD25highCD127lowFoxp3+ ). Positive cells for each marker were determined using fluorescence of cells labelled with the corresponding isotype-matched conjugated irrelevant MAb as a negative control. CD4+ T cells showing the highest CD25 expression were identified as CD4+CD25high. Then, CD25high population was analyzed to determine the proportion of cells expressing Foxp3 (Foxp3+ within CD25high) as well as the amount of Treg cells (CD25highCD127lowFoxp3+). Density plots correspond to a representative experiment. Analyses of (B) CD25highpopulation, (C) Foxp3+ cells within CD25high population, and (C) IL-17/IFNγ expression. Bars represent the mean and SEM of seven independent experiments performed with different blood donors. Statistical differences between SLE-M and the different treatments were evaluated by the Wilcoxon test for paired data. *p < 0.1; **p < 0.05.
Finally, although no significant differences were detected in IL-17 or IFNγ expression between cells treated with both fecal microbiotas, the IL-17/IFNγ ratio was significantly higher in SLE- than in HC-M cultures, whereas the lowest ratio was induced by Cl-conditioned DCs. Moreover, SLE-M supplementation with Cl, but not with Bb, induced a significant dose dependent reduction of the IL-17/IFNγ balance (Fig. 1D). Therefore, in vitro stimulation of naïve CD4+ T cells with SLE-M seems to promote lymphocyte activation and Th17 differentiation to a greater extent than HC-M treatment, thus supporting Th17/Treg disturbances in SLE. In addition, enrichment of SLE microbiota with Bb may prevent lymphocyte activation whereas Cl supplementation restores Th17/Th1 balance.
Relationship between immune parameters and fecal microbiota in SLE patients
To know whether the Treg/Th responses elicited in vitro by SLE-M could reflect the characteristic immune features of SLE patients, we analyzed Foxp3, CD25, CD127, IL-17 and IFNγ expression in fresh peripheral blood CD4+ lymphocytes as well as the serum levels of a battery of cytokines in 37 SLE patients and 36 HC (Table 1). In addition, since 40 of these individuals (20 SLE and 20 HC) have been previously included in a metagenomic study of fecal microbiota39 (Table 2), we determined the possible relationship between these immune parameters and the SLE-associated gut dysbiosis.
Table 1. Demographic and clinical features of SLE patients.
| SLE patients (N = 37) | |
|---|---|
| Sex (female/male) (n) | 36/1 |
| Age, years (mean ± SD) | 48.40 ± 12.99 |
| Age at diagnosis, years (mean ± SD) | 38.32 ± 13.64 |
| Disease duration, years (mean ± SD) | 8.48 ± 6.65 |
| SLEDAI score (mean ± SD) | 4.45 ± 3.28 |
| Clinical manifestations, n (%) | |
| Malar rash | 17 (45.95) |
| Discoid lesions | 8 (21.62) |
| Photosensitivity | 21 (56.76) |
| Oral ulcers | 16 (43.24) |
| Arthritis | 22 (59.46) |
| Serositis | 6 (16.22) |
| Cytopenia | 24 (64.86) |
| Renal disorder | 11 (29.73) |
| Neurological disorder | 2 (5.40) |
| Autoantibodies, n (%) | |
| ANAs | 37 (100.00) |
| Anti-dsDNA/titer, U/ml (mean ± SD) | 18 (48.65)/41.50 ± 61.31 |
| Anti-SSa | 19 (51.35) |
| Anti-SSb | 3 (9.68) |
| Anti-Sm | 4 (10.81) |
| Anti-RNP | 4 (10.81) |
| Treatment, n (%) | |
| None or NSAIDs | 4 (10.81) |
| Antimalarial drugs | 30 (81.08) |
| Glucocorticoids | 10 (27.03) |
| Immunosuppressive drugsa | 3 (8.11) |
dsDNA: double stranded DNA; NSAID: non-steroidal anti-inflammatory drug.
aMethotrexate and/or mycophenolate mophetil.
Table 2. SLE patients and healthy controls included in the intestinal microbiota analysis.
| SLE patients (N = 20) | Healthy controls (N = 20) | |
|---|---|---|
| Sex (female/male) (n) | 20/0 | 20/0 |
| Age (years) | 49.21 ± 10.70 | 46.92 ± 8.63 |
| BMI (Kg/m2) | 26.17 ± 5.34 | 25.20 ± 4.20 |
| Weigth (Kg) | 64.73 ± 11.23 | 67.28 ± 14.03 |
| Blood lipids (mg/dL) | ||
| Total cholesterol | 195.85 ± 37.22 | 198.32 ± 34.34 |
| HDL cholesterol | 64.80 ± 15.52 | 63.00 ± 9.09 |
| LDL cholesterol | 116.35 ± 38.52 | 120,32 ± 31.38 |
| Triglycerides | 71.55 ± 31.02 | 73.73 ± 37.33 |
| Disease parameters | ||
| Age at diagnosis (years) | 38.24 ± 11.85 | |
| Disease duration (years) | 10.40 ± 7.31 | |
| SLEDAI score | 4.83 ± 2.89 | |
| Anti-dsDNA titer (U/ml) | 22.01 ± 30.37 | |
| Treatment, n | ||
| None or NSAIDs | 2 | |
| Antimalarial drugs | 18 | |
Values represent means ± SD. BMI: body mass index; HDL: high-density lipoprotein; LDL: low-density lipoproteins; dsDNA: double stranded DNA; NSAID: non-steroidal anti-inflammatory drug.
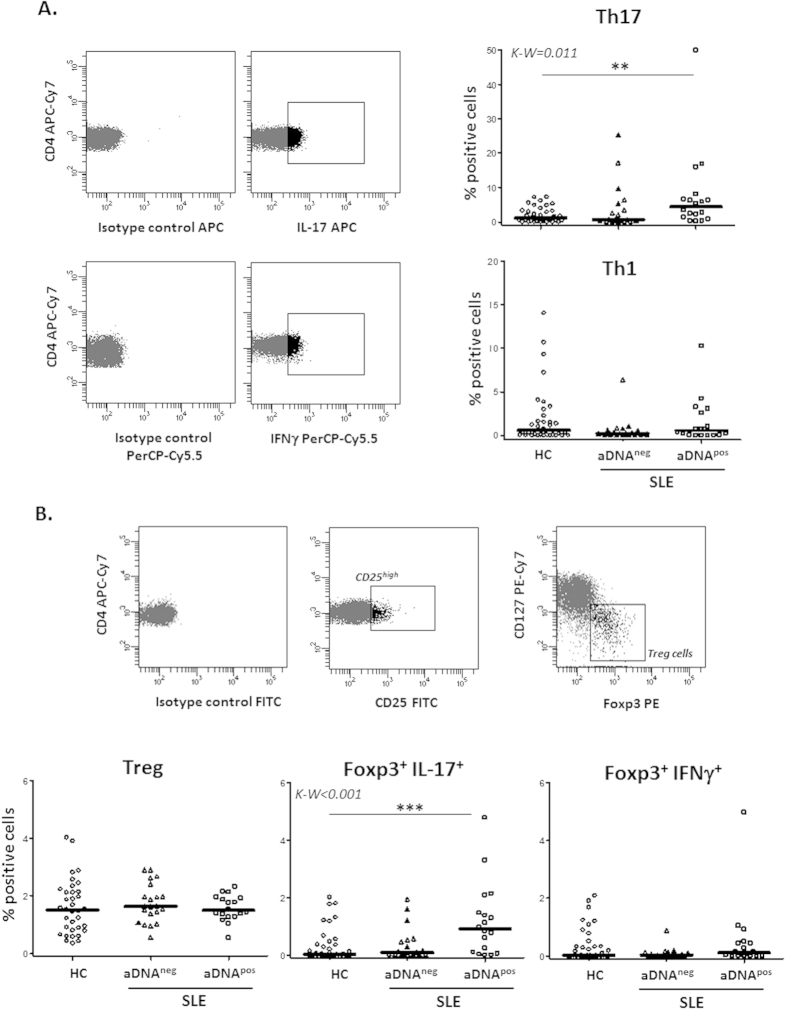
Results showed an increased frequency of Th17 cells in SLE patients compared to HC, especially in those presenting anti-dsDNA antibodies, whereas no significant differences were observed in the Th1 subset (Fig. 2A). Moreover, Foxp3+ IL-17+ double positive cells were significantly increased in SLE patients compared with HC, the highest levels of such cells being observed again in those patients presenting anti-dsDNA antibodies (Fig. 2B). No significant differences were detected in the percentage of Treg cells (CD4+ CD25high CD127lowFoxp3+), suggesting that a Treg-Th17 trans-differentiation process could be involved in the development of Foxp3+ cells without regulatory activity in SLE patients.
Figure 2. Increased IL-17 producing cells in SLE patients with anti-dsDNA antibodies.
Foxp3, CD25, CD127, IL-17 and IFNγ expression was analyzed in fresh peripheral blood CD4+ lymphocytes from SLE patients and HC. (A) Dot-plots show cells positive for IL-17 or IFNγ expression, determined attending to the fluorescence of cells labelled with the corresponding isotype-matched conjugated irrelevant MAb as a negative control. Scatter plots represent the percentage of IL-17+ (Th17) and IFNγ+ (Th1) CD4+ cells in HC and SLE patients presenting (pos) or not (neg) anti-dsDNA antibodies (aDNA). Horizontal bars show the median. (B) Treg cells were sequentially identified as CD4+CD25highCD127lowFoxp3+ cells. Scatter plots represent the quantity of Treg, Foxp3+ IL-17+ and Foxp3+ IFNγ+ cells in HC and SLE patients in function of their anti-dsDNA status, and horizontal bars show the median. Statistical differences among groups were evaluated by Kruskal-Wallis test and Dunn’s post test was conducted to determine which groups’ pairs had different means. **p < 0.01; ***p < 0.001.
On the other hand, the analysis of fecal microbiota in the HC group revealed a negative correlation between the frequency of Firmicutes and the size of the Th17 subset, most notably in Foxp3+ IL-17+ cells, whereas the opposite occurred with Bacteroidetes (Table 3); all these correlations were confirmed by multivariate linear regression analyses adjusted by weight, BMI and blood lipids (*p < 0.05, R2 > 0.6). However, such associations were completely absent in patients, in agreement with our previously described reduction of the Firmicutes/Bacteroidetes ratio in SLE.
Table 3. Associations of Firmicutes, Bacteroidetes and the Firmicutes to Bacteroidetes ratio with different CD4+ T cell subsets in healthy controls.
| CD4+ T subsets | Firmicutes | Bacteroidetes | Firmicutes/Bacteroidetes |
|---|---|---|---|
| Th1 | r = 0.040 | r = 0.183 | r = −0.119 |
| p = 0.862 | p = 0.427 | p = 0.606 | |
| Th17 | r = −0.444* | r = 0.294 | r = −0.329 |
| p = 0.044 | p = 0.196 | p = 0.145 | |
| Foxp3+ IL-17+ | r = −0.561* | r = 0.465* | r = −0.487* |
| p = 0.008 | p = 0.034 | p = 0.025 | |
| Foxp3+ IFNγ+ | r = 0.054 | r = 0.156 | r = −0.089 |
| p = 0.816 | p = 0.500 | p = 0.702 |
Correlation analyses were evaluated by Spearman test and confirmed by multivariate linear regression analyses adjusted by weight, BMI and blood lipids (*p < 0.05, R2 > 0.6).
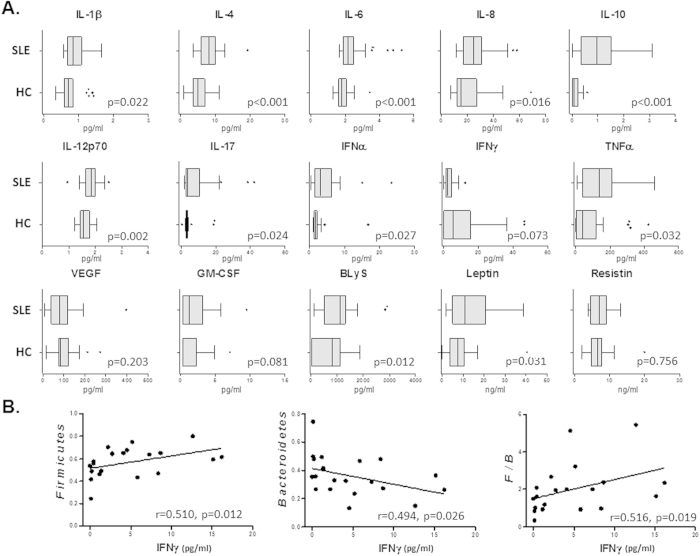
Regarding cytokine serum levels, SLE patients presented higher amounts of IL-1β, IL-4, IL-6, IL-8, IL-10, IL-12p70, IL-17A, IFNα, TNFα, GM-CSF, BLyS and leptin than HC, whereas IFNγ showed a clear tendency to reduction (Fig. 3A). Interestingly, none of the increased or unchanged cytokines in SLE displayed significant associations with Firmicutes or Bacteroidetes, however, IFNγ levels correlated negatively with Bacteroidetes and positively with Firmicutes and the Firmicutes/Bacteroidetes ratio in patients (Fig. 3B). These associations were not detected in HC.
Figure 3. IFNγ serum levels in SLE patients were associated with Firmicutes to Bacteroidetes ratio.
(A) Box and whiskers represent median and interquartile range of circulating amounts of cytokines in SLE patients compared to controls. Differences between both groups were assessed by the non-parametric Mann-Whitney U test. (B) Correlations between serum levels of IFNγ and the frequency of Firmicutes, Bacteroidetes or the Firmicutes to Bacteroidetes ratio (F/B) in SLE patients were evaluated using Spearman test.
Association between the frequency of Synergistetes and the presence of natural antibodies
As we previously reported39, the proportion of fecal Synergistetes did not show significant differences between SLE patients and healthy controls. However, we found a negative correlation trend between this bacterial group and the titer of anti-dsDNA antibodies (r = −0.386, p = 0.084), not detected with any other of the previously analyzed microbial groups. Likewise, serum levels of IL-6 associated negatively with the amount of Synergistetes in SLE patients (r = −0.738, p < 0.001), thus suggesting a possible relationship between this bacterial group and disease activity or antibody production. Therefore, aiming to expand the knowledge about the possible role played by intestinal Synergistetes in the development of humoral immune responses, we quantified serum levels of anti-PC IgM, natural protective antibodies, and anti-PC IgG (lacking this effect) as well as total circulating IgM and IgG. No significant differences were detected in anti-PC antibodies (IgM and IgG) and total IgM between patients and controls, but the amount of total IgG was increased in patients compared to controls (p = 0.027). Interestingly, anti-dsDNA titer correlated negatively with both isotype anti-PC antibodies (IgM: r = −0.508, p = 0.019; IgG: r = −0.460, p = 0.036) and with total IgM (r = −0.501, p = 0.021) but not IgG (r = 0.065, p = 0.780) levels, thus suggesting a detrimental effect of SLE disease activity on naturally occurring protective IgM antibodies.
On the other hand, Synergistetes exhibited a positive correlation with total and anti-PC IgM in SLE patients and with total and anti-PC IgM/IgG ratio in patients and controls (Table 4). Interestingly, serum levels of IL-6 in patients showed the opposite associations. All these results suggest that intestinal Synergistetes may promote the development of protective natural IgM antibodies, an effect particularly relevant in SLE patients since the increased levels of anti-dsDNA and/or IL-6 could downregulate this bacterial group.
Table 4. Relationship of fecal Synergistetes and IL-6 serum levels with anti-PC and total IgM and IgG antibodies.
|
Synergistetes |
IL-6 |
|||
|---|---|---|---|---|
| HC | SLE | HC | SLE | |
| IgM: | ||||
| anti-PC | r = 0.404 | r = 0.624* | r = −0.004 | r = −0.473 |
| p = 0.062 | p = 0.002 | p = 0.986 | p = 0.030 | |
| total | r = 0.101 | r = 0.824* | r = 0.081 | r = −0.642* |
| p = 0.662 | p < 0.001 | p = 0.729 | p = 0.002 | |
| IgG: | ||||
| anti-PC | r = −0.156 | r = 0.239 | r = −0.049 | r = −0.038 |
| p = 0.523 | p = 0.286 | p = 0.842 | p = 0.286 | |
| total | r = −0.301 | r = −0.078 | r = 0.133 | r = 0.332 |
| p = 0.211 | p = 0.737 | p = 0.586 | p = 0.142 | |
| IgM/IgG ratio: | ||||
| anti-PC | r = 0.670 | r = 0.495* | r = −0.005 | r = −0.488 |
| p = 0.002 | p = 0.023 | p = 0.983 | p = 0.025 | |
| total | r = 0.496 | r = 0.597* | r = −0.082 | r = −0.610* |
| p = 0.031 | p = 0.004 | p = 0.737 | p = 0.003 | |
Correlation analyses were evaluated by Spearman test and confirmed by multivariate linear regression analyses adjusted by weight, BMI and blood lipids (*p < 0.05, R2 > 0.6).
Finally, it is noteworthy that Synergistetes correlated negatively with Bacteroidetes (r = −0.486, p = 0.022) and positively with the Firmicutes/Bacteroidetes ratio (r = 0.443, p = 0.039) in controls, but not in SLE patients (r = 0.075, p = 0.745 and r = −0.136, p = 0.556, respectively), supporting the role of these bacterial groups in SLE.
Discussion
Intestinal dysbiosis has been associated with several immune mediated diseases, including SLE39 and other autoimmune conditions32,33,34,35,36,37,38, pointing out a possible role of the microbiome in the balance between inflammatory and regulatory responses, either in the intestine or systemically. Therefore, understanding how the gut microbiota shapes immune responses seems to be critical for human health, particularly in chronic inflammatory disorders such as autoimmune diseases. However, to the best of our knowledge, this is the first study evaluating the effect of intestinal microbiota isolated from SLE stool samples on the induction of effector or regulatory T cell responses.
Our findings revealed that the differences in the composition of the fecal microbiota between SLE patients and HC elicited different in vitro immune responses. Specifically, DCs stimulated with SLE-M promoted Th17 differentiation from naïve CD4+ T lymphocytes to a greater extent than HC-M conditioned DCs. No differences were detected in the generation of Treg cells, but the enlarged CD25high population obtained with SLE-M treatment tended to include a lower proportion of Foxp3+ cells, supporting their activated rather than regulatory status. Hence, the altered SLE gut microbiota could enhance lymphocyte activation and Th17 differentiation, thus sustaining the preexisting inflammation.
These findings are in accordance with the results of the ex vivo analyses, since the frequency of Th17 cells was increased in fresh peripheral blood from SLE patients compared to HC, especially in those presenting anti-dsDNA antibodies, whereas no differences were observed in Treg cells. Interestingly, SLE patients exhibited a higher proportion of Foxp3+ cells producers of IL-17, thus suggesting a possible Treg-Th17 trans-differentiation process that could be responsible for the reduced regulatory activity of Treg cells reported in SLE patients18. Similarly, an increased prevalence of circulating IL-17 and Foxp3 double-expressing CD4+ lymphocytes, together with a reduced Treg suppressor ability, have been found in patients with inflammatory bowel disease44. The elevated frequency of Treg cells present in the gut compared with other organs45 is noteworthy, but they have an increased Treg/Th plasticity which could be influenced by inflammatory mediators, specific bacterial strains and other micro-environmental factors46. In line with this, the analyses of fecal microbiota revealed a strong negative correlation between the size of IL-17+ Foxp3+, and to a lesser extend Th17, populations and the frequency of Firmicutes in healthy controls, suggesting that they could counteract Th17 differentiation. On the other hand, serum levels of IFNγ, the prototypic Th1 cytokine and slightly reduced in SLE, correlated directly in patients with the amount of Firmicutes and with the Firmicutes to Bacteroidetes ratio, this imbalance being the main feature of SLE dysbiosis and independent to disease duration, lifestyle and dietary-related factors39. Therefore, we hypothesize that bacterial strains belonging to the Firmicutes phylum could be involved in the generation and/or maintenance of functional Treg cells in the gut, avoiding the trans-differentiation into effector Th17 cells in physiological conditions, whereas a diminished proportion of these bacteria in pathological situations may promote Th17 vs Th1 and Treg bias and generate IL-17+ Foxp3+ cells. Thus, enrichment of gut microbiota in Treg-inducing bacteria could be a desirable goal for SLE patients. However, although Firmicutes/Bacteroidetes ratio represents the main difference between healthy controls and SLE patients, the specific bacterial groups implicated in the observed Th17 immune response associated to SLE microbiota could not be determined by our approach, even more having into account that Firmicutes phyla includes both Th1747 and Treg23,41,42,43,44 inducing bacteria. Hence, the confirmation of such hypothesis would benefit from additional experiments using germ-free mice or fecal microbiota transplant procedures in animal models.
Certain commensal microorganisms have demonstrated a Treg-inducer capability, and therefore have been proposed as potential probiotic strains appropriate for the modulation of an excessive inflammatory response to restore the immune homeostasis at the intestinal mucosa. Hence, in this study we performed in vitro analyses aiming to determine the possible effect of enriching SLE gut microbiota with strains known to be able to differentiate naïve T lymphocytes into Treg cells. Firmicutes phylum contains several Clostridium spp., belonging to clusters IV and XIVa, known to induce Treg cells23,41,42,43. Similarly, bifidobacteria have been also demonstrated to promote the Treg polarization and Th17 reduction in vivo using murine models of colitis48. Therefore, we used a mixture of two of these Clostridia strains, as well as a Bifidobacterium strain with previously demonstrated in vitro ability to generate functional Treg cells23,24. Unexpectedly, neither was able to increase this population, but results showed other beneficial effects, different for each bacterial treatment. Supplementation of SLE-M with Clostridia induced a significant dose-dependent reduction of the IL-17/IFNγ balance, thus restoring Th1 bias, whereas bifidobacteria enrichment prevented the over-activation of CD4+ lymphocytes, as detected by the reduction of CD25 expression. These effects, however, could be the consequence of an active suppression of effector Th cells. In fact, the observed down regulation of Th17 cells may be explained by a preferential promotion of Treg cells by Clostridia strains, in accordance with the results obtained by Atarashi et al.43 using germ-free mice colonized with different fractions of healthy human microbiota. Likewise, suppressive function of Treg cells generated with the Bifidobacterium strain used here results in downregulation of CD25 expression on effector cells23. Therefore, in view of these results and the previously reported SLE dysbiosis, it seems reasonable to consider the possible therapeutic benefit of the supplementation with probiotics containing Treg-inducer strains in order to restore the Treg/Th17/Th1 balance in SLE patients.
Another remarkable result is the suggested role played by Synergistetes, a little known intestinal bacterium, in SLE. This bacterial group correlated negatively with Bacteroidetes and positively with the Firmicutes to Bacteroidetes ratio in healthy controls, whereas in SLE patients a strong negative correlation was shown with serum levels of IL-6, a proinflammatory and Th17 promoting cytokine increased in SLE patients. Moreover, the amount of Synergistetes tended to be reduced when the titer of anti-dsDNA antibodies were increased, characteristic of SLE. These findings led us to evaluate the possible role of intestinal Synegistetes in the development of protective humoral immune responses. Natural IgM antibodies against commensal bacteria or neoantigens from apoptotic cells are commonly present in the human circulation from birth and have protective and immunoregulatory functions. Among them, IgM antibodies that recognize phosphorylcholine (PC) are valuable components of the immune system known to increase the phagocytosis of apoptotic cells and inhibit inflammatory pathways in autoimmunity and atherosclerosis20,22,49. However, anti-PC IgG did not seem to possess these protective effects. Results in our SLE cohort revealed that the proportion of Synergistetes correlated positively with total and anti-PC IgM and IgM/IgG ratio in SLE patients, whereas serum levels of IL-6 exhibited the opposite associations. These data suggest a protective role of intestinal Synergistetes promoting the generation of natural IgM antibodies, which could be hampered in SLE patients, especially those with high IL-6 levels. Interestingly, it has been reported that anti-PC IgM antibodies can counteract IL-6 upregulation in vitro and in vivo22, probably by the inhibition of MAPK responses to TLR agonists, including lupus immune complexes50, thus explaining the opposite associations of these natural antibodies with Synergistetes and IL-6. Moreover, B1 cells and anti-PC antibodies are increased in IL-6 knockout mice whereas the opposite occurs with B2 cells and IgG levels51. In line with these results, the anti-dsDNA titer correlated negatively with anti-PC levels, thus supporting a deleterious effect of disease activity on the amount of natural protective antibodies. In fact, higher anti-PC IgM levels have been associated with lower SLE disease activity20. Unfortunately, active patients were not included in our study of the intestinal microbiota to avoid interference with the treatments, so no significant associations were detected between SLEDAI score and Synergistetes or anti-PC IgM antibodies.
The involvement of Synergistetes in the promotion of natural antibodies, and especially those with an atheroprotective role, could be of clinical relevance for patients with SLE and other autoimmune diseases, since they usually develop premature atherosclerosis and have an increased cardiovascular risk not explained by classical factors. It has been reported that diminished levels of anti-PC IgM antibodies in SLE patients could predict a subclinical cardiovascular disease21. Moreover, patients that have suffered a cardiovascular event presented significantly lower levels of these antibodies compared to the rest of patients21,49. In fact, therapy based on passive transfer of anti-PC has been used to inhibit atherosclerosis development52. Therefore, the identification of intestinal microorganisms able to expand natural humoral responses through the promotion of protective IgM antibodies, as may be Synergistetes, could be valuable tools to design clinical interventions in order to improve both disease activity and prevention of cardiovascular complications. In this sense, the analysis of fecal Synergistetes in SLE patients with and without cardiovascular events should be of interest.
In summary, our in vitro cultures with fecal microbiota isolated from SLE patients suggested that immune responses against intestinal bacteria could be involved in the lymphocyte over-activation as well as in the Treg-Th17 trans-differentiation observed in SLE patients, with the reduced Firmicutes to Bacteroidetes ratio probably having a role in this process. The altered immune responses associated with the intestinal dysbiosis could be reestablished, at least in part, by the supplementation with beneficial bacterial strains able to induce suppressor responses. In addition, our results revealed a possible role of intestinal Synergistetes in the development of natural protective anti-PC IgM antibodies. This could be of special relevance in patients with high IL-6 and/or anti-dsDNA levels, since they present low frequency of these bacteria.
Methods
Ethics approval
Ethics approval for this study was obtained from the Bioethics Committee of CSIC (Consejo Superior de Investigaciones Científicas) and from the Regional Ethics Committee for Clinical Research (Servicio de Salud del Principado de Asturias), according to the Declaration of Helsinki. All methods were carried out in accordance with the approved guidelines and signed informed written consent was collected from all participants prior to participation in the study.
Generation of monocyte-derived DCs
Human peripheral blood mononuclear cells (PBMCs) were obtained from standard buffy-coat preparations from 7 healthy blood donors (Asturian Blood Transfusion Center, Oviedo, Spain) by centrifugation over Ficoll-Hypaque gradients (Lymphoprep, Nycomed, Oslo, Norway). Monocytes (CD14+ ≥ 95%) were isolated from previously obtained PBMCs by negative selection using the Human Monocyte enrichment kit (EasySep, Stem Cell Technologies, Canada).
Immature DCs were obtained from isolated monocytes by standard procedures. Thus, monocytes were cultured in 24-wellplates at 5 × 105 cells/ml for 7 days at 37 °C and 5% CO2 in complete RPMI medium (RPMIc) [RPMI 1640 containing 2 mML-glutamine and 25 mM Hepes (Bio Whitaker, Verviers, Belgium), supplemented with 10% heat-inactivated fetal calf serum (FCS) and the antibiotics streptomycin and ampicillin at 100 mg/ml] in the presence of recombinant human (rh) IL-4 (35 ng/ml) and rhGM-CSF (70 ng/ml) (R&D Systems, Abingdon, UK). At days 2 and 5, 0.5 ml of the medium was removed without disturbing the clusters of developing DC and 0.5 ml of freshly made GM-CSF- and IL-4-containing medium was added to the wells, restoring the final volume in each well to 1 ml. At day 7, immature DCs were recovered, washed and resuspended in RPMIc medium at 5 × 105 cells/ml for subsequent maturation.
Stimulation of monocyte-derived DCs with isolated fecal microbiota
Immature monocyte-derived DCs were cultured in RPMIc medium with 1 mg/ml LPS from E. coli 0111:B4 (Sigma, St. Louis, MO), as a positive control of maturation, or with different bacterial preparations at a DC:bacteria ratio of 1:10. To this end, a pooled fecal microbiota (M) isolated from either four healthy controls (HC-M) or five SLE patients (SLE-M), previously shown to be representative of both conditions by metagenomics studies39, were prepared and used in DC cultures. Also, SLE-M were enriched with different proportions of Bifidobacterium bifidum LMG13195 (Bb), a strain known to induce Foxp3 expression23,41, or with a mixture of two Clostridia strains (Cl: Ruminococcus obeum DSM25238 and Blautia coccoides DSM935, ratio 1:1), which have a putative Treg-inducer effect42,43. Thus, 5, 10 or 15% SLE-M were replaced with the same proportions of Bb or Cl and used for DCs stimulation. Cultures with Bb and Cl bacterial preparations were performed as controls. After 48 hours, DCs from the 11 different treatments were harvested, the mature phenotype tested and used to stimulate naïve CD4+ T cells.
Naïve CD4+ T cell stimulation with microbial conditioned-DCs
CD4+ T cells were isolated from PBMCs by negative selection using the Human CD4+ T Cell Enrichment Kit (EasySep), following the manufacturer’s instructions, and then naïve CD45RA+ T cells were separated after depletion of CD45RO+ cells (Miltenyi Biotec, Germany). Monocyte-derived DCs maturated with LPS or with the different bacterial preparations were cocultured with purified CD4+ CD45RA+ T cells in 48-well plates at DC:T cell ratio of 1:10. At day 5 and 8, cells incubated with all treatments were expanded with IL-2 (30 U). After 12 days, cells were collected and washed twice before analysis of the Treg/Th17/Th1 phenotype by flow cytometry.
Bacterial strains and growth conditions
Bifidobacterium bifidum LMG13195 was grown in de MRS broth (Difco, Detroit, MI) supplemented with 0.05% (w/v) L-cysteine (Sigma). Ruminococcus obeum DSM25238 and Blautia coccoides DSM935 were cultivated in a combination of Reinforced Clostridial Broth (Merck, Darmstadt, Germany) and Brain-Heart Infusion (Difco), supplemented with 5% (v/v) heat-inactivated FCS (LabClinics, Barcelona, Spain). Cultures were grown at 37 °C in a MG500 anaerobic chamber (Don Whitley Scientific, West Yorkshire, UK) with an atmosphere of 10% (v/v) H2, 10% CO2, and 80% N2. Cultures were harvested by centrifugation, washed three times in phosphate buffered saline (PBS) and resuspended in the same buffer to a concentration of 108 bacteria/ml. Bacterial were counted by using a Thoma cell counting chamber (Marienfeld Superior, Germany). Bacterial cells were killed by exposing them to three consecutive cycles of 30 minutes under radiation in a UV chamber (15 W, Selecta, Barcelona, Spain). Plate counting was carried out after UV treatment to corroborate the absence of bacteria able to recover in the proper medium. UV-killed bacterial suspensions were distributed in aliquots, and stored at −80 °C until use. The identity of the strains was confirmed by sequencing the V1 and V2 variable regions of the 16 S rRNA gene using primers plb16 (5′-AGAGTTTGATCCTGGCTCAG-3′) and mlb16 (5′-GGCTGCTGGCACGTAGTTAG-3′)53.
Stool samples and microbiota separation
One portion of stool sample of 4 healthy controls and 5 SLE patients, previously used in a metagenomic study39, was submitted to a density gradient centrifugation to separate the microbiota from the rest of the fecal material, according to the method of Courtois et al.54. Feces were homogenized in sterile NaCl 0.9% (1:9; w/v) for 1 min in a homogenizer (Stomacher Lab Blender 400, WVR, Barcelona, Spain). A solution of Nycodenz® 80% (w/v) (PROGEN Biotechnik GmbH, Heidelberg, DE) was prepared in ultrapure water, and sterilized at 121 °C for 15 min. A volume of 10.5 ml of the homogenized fecal samples was placed on the top of 3.5 ml of the Nycodenz® solution, and centrifuged at 10,000 g for 40 min at 4 °C. The upper phase, containing soluble debris, was discarded and the layers corresponding to the microbiota were kept in ice for 5 min, in order to allow non-soluble debris to precipitate, washed twice in PBS, and stored in the same buffer in aliquots of 1 ml of 108 microorganisms/ml at −80 °C. UV-inactivated microbiota was obtained as described in the previous section.
SLE patients and healthy controls
Thirty-seven SLE patients fulfilling at least four American College of Rheumatology (ACR) revised criteria for the classification of SLE55 were selected from the updated Asturian Register of Lupus56,57. Information on clinical features during the disease course was obtained by reviewing clinical histories (Table 1). Thirty-six sex and age-matched healthy blood donors were used as controls (mean age ± SD: 42.56 years ± 11.39). At the time of sampling, anti-dsDNA titer, SLE disease activity index (SLEDAI) and/or weight, BMI (body mass index) and blood lipids [Triglycerides, HDL (high-density lipoprotein), LDL (low-density lipoproteins) and total cholesterol] were evaluated and patients were asked precise questions regarding the treatment received over the previous 6 months.
Intestinal microbiota analysis
Metagenomic analysis of fecal microbiota were performed in 20 non active SLE patients without antibiotic or immunosuppressive treatment in the last 6 months and 20 age-matched healthy controls as previously described39. Faecal DNA extraction, 16 S rRNA amplification sequencing of 16 S rRNA gene-based amplicons and the sequence-based microbiota analysis were reported elsewhere39. The raw sequences reported in this article are deposited in the NCBI Short Read Archive (SRA) (study accession number: SRP028162).
Flow cytometric analysis
Phenotypic studies of cultured cells and blood samples were performed after staining with the appropriate monoclonalantibody (mAb). Maturation of cultured DCs was verified after staining for 30 min at 4 °C with anti -CD86 fluorescein isothiocyanate (FITC), -CD80 phycoerythrin (PE), -HLA-DR PE-Cy5, -CD1a FITC mAb, or with the corresponding isotype matched conjugated irrelevant mAb as a negative control (all mAb were supplied by Pharmingen). To determine Treg/Th17/Th1 phenotype both in cultured cells and blood samples [previously lysed with 2 ml BD Lysing Solution (BD Biosciences, San Diego, CA) for 5 minutes and washed twice with PBS], CD4+ lymphocytes were first extracellularly stained with anti-CD4 allophycocyanin-Cy7 (APC-Cy7), anti-CD25 FITC and anti-CD127 PE-Cy7mAb or with the corresponding isotype-matched conjugated irrelevant mAb (all from eBiosciences, San Diego, CA). Then, cells were fixed, permeabilized and intracellularly stained with anti-FOXP3 PE, IFNγ PerCP-Cy5.5 and IL-17 A APC (Foxp3/transcription factor staining buffer set; eBiosciences) or with the corresponding isotype-matched conjugated irrelevant mAb, following the manufacturer’s instructions. A minimum of 10,000 CD4+ lymphocytes were acquired on a FACSCanto II flow cytometer (BD) and analyzed using the FlowJo software (Tree Star Inc). The specific fluorescence intensity was quantified as the mean fluorescence intensity (MFI) calculated by subtracting the background of isotype matched control staining from the total fluorescence.
Cytokine determination
Serum samples from SLE patients and HC were collected and maintained at −80 °C until cytokine determination was carried out. IL-1β, IL-4, IL-6, IL-8, IL-10, IL-12p70, IL-17 A, IFNα, VEGF and GM-CSF amounts were analyzed by Cytometric Bead Arrays Flex Set using a FACS Canto II flow cytometer (BD Biosciences). For IL-1β, IL-6, IL-10 and IL-12p70, an Enhanced Sensitivity Flex Set was needed. ELISA kits were used for the quantification of TNFα, leptin, resistin (Mini EDK kit, PeproTech), BLyS (Human BAFF Instant ELISA, eBioscience) and IFNγ (OptEIA kit, BD) following the manufacturer’s instructions. The lower limits of detection were 48.4 fg/ml for IL-1β, 1.4 pg/ml for IL-4, 68.4 fg/ml for IL-6, 1.2 pg/ml for IL-8, 13.7 fg/ml for IL-10, 12.6 fg/ml for IL- 12p70, 0.3 pg/ml for IL-17 A, 1.25 pg/ml for IFNα, 4.5 pg/ml for VEGF, 0.2 pg/ml for GM-CSF, 3.9 pg/ml for TNFα, 63 pg/ml for leptin, 24 pg/ml for resistin, 130 pg/ml for BLyS and 0.58 pg/ml for IFNγ.
Anti-Phosphorylcholine antibodies quantification
IgG and IgM antibodies against Phosphorylcholine (anti-PC) were quantified in serum samples from patients and controls by an in-house ELISA test, as follows. Microtiter wells (Maxisorp, Nunc) were coated overnight with phosphorylcholine conjugated to bovine serum albumin (PC-BSA) (Biosearch Technologies, Petaluma) and blocked with PBS 2% BSA for 2 hours at 37 °C. A sera pool of healthy controls was used as anti-PC Ab standard. Serum samples and anti-PC standard were diluted in Tris Buffered Saline (TBS) and incubated for 2 hours at room temperature (RT). After washing with TBS/Tween 20 (0.05%), wells were incubated for 2 hours at RT with alkaline phosphatase-conjugated anti-human IgG or IgM (Immunostep, Salamanca, Spain). Finally, plates were washed twice and revealed using p-nitrophenylphosphate as substrate. Absorbance was determined at a wavelength of 405 nm. Quantities of serum anti-PC arbitrary units were calculated for each sample according to the standard curves. Similarly, total IgG or IgM were quantified by conventional ELISA techniques.
Statistical analysis
The Kolmogorov-Smirnov test was used to assess the normal distribution of the data. In vitro experiments data were represented by mean ± SEM and differences between culture conditions were assessed by the paired Wilcoxon test. SLE patients and healthy controls cytokine serum levels were expressed as the median value (interquartile range) and non-parametric Mann-Whitney U-test was used to determine differences between both groups. Percentages of Treg/Th1/Th17 cells were compared by using the Kruskal-Wallis test; when a significant test was obtained, Dunn’s post hoc tests were conducted to determine statistical differences in pairs of groups. Associations of fecal microbial group’s frequencies with T cell subsets, cytokine serum levels and autoantibodies were examined by Spearman’s rank correlation test and confirmed by multivariate linear regression analyses adjusted by weight, BMI and blood lipids (triglycerides, HDL, LDL and total cholesterol]. GraphPad Prism 5 software (GraphPad Software, USA) and SPSS 22 statistical software package (SPSS Inc.) were used for all determinations, and a p-value < 0.05 was considered significant.
Additional Information
How to cite this article: Lopez, P. et al. Th17 responses and natural IgM antibodies are related to gut microbiota composition in systemic lupus erythematosus patients. Sci. Rep. 6, 24072; doi: 10.1038/srep24072 (2016).
Acknowledgments
This work was supported by European Union FEDER funds, Fondo de Investigación Sanitaria (PI12/00523), Spanish Plan Nacional de I+D (AGL2010-14952 and AGL2013-44039-R) and Fundación para el Fomento en Asturias de la Investigación Científica Aplicada y la Tecnología (EQUIP09-19). J.R.-C. is a recipient of a FPU grant from the Spanish Ministerio de Educación, Cultura y Deporte. B.J. and A.H. are recipients of a Ramón y Cajal postdoctoral contract and a FPI grant, respectively, both from the Spanish Ministerio deEconomía y Competitividad. We thank SLE patients and ALAS (Asociación Lúpicos de Asturias) for their continuous encouragement.
Footnotes
Author Contributions P.L. participated in the study conduct, sample collection, review of clinical manifestations, disease activity and therapy of patients, experimental procedures, data analysis/interpretation and manuscript preparation. B.P. and J.R.-C. performed some experimental procedures and data analysis. B.S., A.H. and A.M. participated in microbiota related design study, experiments and analysis. A.S. contributed to study design/conduct, data interpretation and manuscript preparation.
References
- Rahman A. & Isenberg D. A. Systemic lupus erythematosus. N. Engl. J. Med. 358, 929–939 (2008). [DOI] [PubMed] [Google Scholar]
- Sanz I. & Lee F. E.-H. B cells as therapeutic targets in SLE. Nat. Rev. Rheumatol. 6, 326–337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. & Paul W. E. CD4+ T cells: fates, functions, and faults. Blood 112, 1557–1569 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alunno A. et al. Balance between Regulatory T and Th17 Cells in Systemic Lupus Erythematosus: The Old and the New. J. Immunol. Res. 2012, e823085 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. et al. The imbalance between regulatory and IL-17-secreting CD4+ T cells in lupus patients. Clin. Rheumatol. 29, 1251–1258 (2010). [DOI] [PubMed] [Google Scholar]
- Talaat R. M., Mohamed S. F., Bassyouni I. H. & Raouf A. A. Th1/Th2/Th17/Treg cytokine imbalance in systemic lupus erythematosus (SLE) patients: Correlation with disease activity. Cytokine 72, 146–153 (2015). [DOI] [PubMed] [Google Scholar]
- Pan H.-F. et al. Expression profiles of Th17 pathway related genes in human systemic lupus erythematosus. Mol. Biol. Rep. 40, 391–399 (2013). [DOI] [PubMed] [Google Scholar]
- Qu N. et al. Pivotal Roles of T-Helper 17-Related Cytokines, IL-17, IL-22, and IL-23, in Inflammatory Diseases. J. Immunol. Res. 2013, e968549 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. Q. et al. Plasma IL-17A is Increased in New-Onset SLE Patients and Associated with Disease Activity. J. Clin. Immunol. 30, 221–225 (2010). [DOI] [PubMed] [Google Scholar]
- Crispín J. C. et al. Expanded double negative T cells in patients with systemic lupus erythematosus produce IL-17 and infiltrate the kidneys. J. Immunol. Baltim. Md 1950 181, 8761–8766 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doreau A. et al. Interleukin 17 acts in synergy with B cell–activating factor to influence B cell biology and the pathophysiology of systemic lupus erythematosus. Nat. Immunol. 10, 778–785 (2009). [DOI] [PubMed] [Google Scholar]
- Wang Y. et al. Laser microdissection-based analysis of cytokine balance in the kidneys of patients with lupus nephritis. Clin. Exp. Immunol. 159, 1–10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevach E. M. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity 30, 636–645 (2009). [DOI] [PubMed] [Google Scholar]
- Fontenot J. D., Gavin M. A. & Rudensky A. Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 (2003). [DOI] [PubMed] [Google Scholar]
- Feuerer M., Hill J. A., Mathis D. & Benoist C. Foxp3+ regulatory T cells: differentiation, specification, subphenotypes. Nat. Immunol. 10, 689–695 (2009). [DOI] [PubMed] [Google Scholar]
- Lyssuk E. Y., Torgashina A. V., Soloviev S. K., Nassonov E. L. & Bykovskaia S. N. Reduced number and function of CD4+CD25highFoxP3+ regulatory T cells in patients with systemic lupus erythematosus. Adv. Exp. Med. Biol. 601, 113–119 (2007). [PubMed] [Google Scholar]
- Valencia X., Yarboro C., Illei G. & Lipsky P. E. Deficient CD4+CD25high T Regulatory Cell Function in Patients with Active Systemic Lupus Erythematosus. J. Immunol. 178, 2579–2588 (2007). [DOI] [PubMed] [Google Scholar]
- Gómez J., Prado C., López P., Suárez A. & Gutiérrez C. Conserved anti-proliferative effect and poor inhibition of TNFalpha secretion by regulatory CD4+CD25+ T cells in patients with systemic lupus erythematosus. Clin. Immunol. Orlando Fla 132, 385–392 (2009). [DOI] [PubMed] [Google Scholar]
- Round J. L. & Mazmanian S. K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönwall C. & Silverman G. J. Natural IgM: beneficial autoantibodies for the control of inflammatory and autoimmune disease. J. Clin. Immunol. 34 Suppl 1, S12–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönwall C. et al. Relation of carotid plaque with natural IgM antibodies in patients with systemic lupus erythematosus. Clin. Immunol. 153, 1–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Park Y.-B., Patel E. & Silverman G. J. IgM Antibodies to Apoptosis-Associated Determinants Recruit C1q and Enhance Dendritic Cell Phagocytosis of Apoptotic Cells. J. Immunol. 182, 6031–6043 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- López P., González-Rodríguez I., Gueimonde M., Margolles A. & Suárez A. Immune response to Bifidobacterium bifidum strains support Treg/Th17 plasticity. PloS One 6, e24776 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- López P., Gueimonde M., Margolles A. & Suárez A. Distinct Bifidobacterium strains drive different immune responses in vitro. Int. J. Food Microbiol. 138, 157–165 (2010). [DOI] [PubMed] [Google Scholar]
- Rescigno M. et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2, 361–367 (2001). [DOI] [PubMed] [Google Scholar]
- Joffre O., Nolte M. A. & Spörri R. & Reis e Sousa, C. Inflammatory signals in dendritic cell activation and the induction of adaptive immunity. Immunol. Rev. 227, 234–247 (2009). [DOI] [PubMed] [Google Scholar]
- Mazmanian S. K., Round J. L. & Kasper D. L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 (2008). [DOI] [PubMed] [Google Scholar]
- Ivanov I. I. et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V. et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689 (2009). [DOI] [PubMed] [Google Scholar]
- Yurkovetskiy L. et al. Gender bias in autoimmunity is influenced by microbiota. Immunity 39, 400–412 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markle J. G. M. et al. Sex Differences in the Gut Microbiome Drive Hormone-Dependent Regulation of Autoimmunity. Science 339, 1084–1088 (2013). [DOI] [PubMed] [Google Scholar]
- Garrett W. S. et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 8, 292–300 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D. N. et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. 104, 13780–13785 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh N., Burton J. P., Suppiah P., Reid G. & Stebbings S. The role of the microbiome in rheumatic diseases. Curr. Rheumatol. Rep. 15, 314 (2013). [DOI] [PubMed] [Google Scholar]
- Wen L. et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 455, 1109–1113 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. K., Menezes J. S., Umesaki Y. & Mazmanian S. K. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. 108, 4615–4622 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N. et al. Gut Microbiota in Human Adults with Type 2 Diabetes Differs from Non-Diabetic Adults. PLoS ONE 5, e9085 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S. M., Kaakoush N. O. & Mitchell H. M. The role of bacteria and pattern-recognition receptors in Crohn’s disease. Nat. Rev. Gastroenterol. Hepatol. 8, 152–168 (2011). [DOI] [PubMed] [Google Scholar]
- Hevia A. et al. Intestinal dysbiosis associated with systemic lupus erythematosus. mBio 5, e01548–01514 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006). [DOI] [PubMed] [Google Scholar]
- López P. et al. Interaction of Bifidobacterium bifidum LMG13195 with HT29 cells influences regulatory-T-cell-associated chemokine receptor expression. Appl. Environ. Microbiol. 78, 2850–2857 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K. et al. Induction of Colonic Regulatory T Cells by Indigenous Clostridium Species. Science 331, 337–341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 (2013). [DOI] [PubMed] [Google Scholar]
- Ueno A. et al. Increased prevalence of circulating novel IL-17 secreting Foxp3 expressing CD4+ T cells and defective suppressive function of circulating Foxp3+ regulatory cells support plasticity between Th17 and regulatory T cells in inflammatory bowel disease patients. Inflamm. Bowel Dis. 19, 2522–2534 (2013). [DOI] [PubMed] [Google Scholar]
- Hall J. A. et al. Commensal DNA Limits Regulatory T Cell Conversion and is a Natural Adjuvant of Intestinal Immune Responses. Immunity 29, 637–649 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai M., Krause P., Cheroutre H. & Kronenberg M. Regulatory T-cell stability and plasticity in mucosal and systemic immune systems. Mucosal Immunol. 3, 443–449 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas A. M. et al. Induction of Th17 cells by segmented filamentous bacteria in the murine intestine. J. Immunol. Methods 421, 104–111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B. et al. Bifidobacterium breve attenuates murine dextran sodium sulfate-induced colitis and increases regulatory T cell responses. PloS One 9, e95441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönwall C. et al. MAPK phosphatase-1 is required for regulatory natural autoantibody-mediated inhibition of TLR responses. Proc. Natl. Acad. Sci. USA 109, 19745–19750 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S., Husband A. J. & Beagley K. W. B1 B cell numbers and antibodies against phosphorylcholine and LPS are increased in IL-6 gene knockout mice. Cell. Immunol. 198, 139–142 (1999). [DOI] [PubMed] [Google Scholar]
- Su J. et al. Natural antibodies against phosphorylcholine as potential protective factors in SLE. Rheumatology 47, 1144–1150 (2008). [DOI] [PubMed] [Google Scholar]
- Faria-Neto J. R. et al. Passive immunization with monoclonal IgM antibodies against phosphorylcholine reduces accelerated vein graft atherosclerosis in apolipoprotein E-null mice. Atherosclerosis 189, 83–90 (2006). [DOI] [PubMed] [Google Scholar]
- Milani C. et al. Assessing the Fecal Microbiota: An Optimized Ion Torrent 16S rRNA Gene-Based Analysis Protocol. PLoS ONE 8, e68739 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois S. et al. Quantification of bacterial subgroups in soil: comparison of DNA extracted directly from soil or from cells previously released by density gradient centrifugation. Environ. Microbiol. 3, 431–439 (2001). [DOI] [PubMed] [Google Scholar]
- Arnett F. C. et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 31, 315–324 (1988). [DOI] [PubMed] [Google Scholar]
- Gómez J. et al. Systemic lupus erythematosus in Asturias, Spain: clinical and serologic features. Medicine (Baltimore) 85, 157–168 (2006). [DOI] [PubMed] [Google Scholar]
- López P., Mozo L., Gutiérrez C. & Suárez A. Epidemiology of systemic lupus erythematosus in a northern Spanish population: gender and age influence on immunological features. Lupus 12, 860–865 (2003). [DOI] [PubMed] [Google Scholar]