Abstract
Objective:
To apply genetic analysis of genome-wide association data to study the extent and nature of a shared biological basis between migraine and coronary artery disease (CAD).
Methods:
Four separate methods for cross-phenotype genetic analysis were applied on data from 2 large-scale genome-wide association studies of migraine (19,981 cases, 56,667 controls) and CAD (21,076 cases, 63,014 controls). The first 2 methods quantified the extent of overlapping risk variants and assessed the load of CAD risk loci in migraineurs. Genomic regions of shared risk were then identified by analysis of covariance patterns between the 2 phenotypes and by querying known genome-wide significant loci.
Results:
We found a significant overlap of genetic risk loci for migraine and CAD. When stratified by migraine subtype, this was limited to migraine without aura, and the overlap was protective in that patients with migraine had a lower load of CAD risk alleles than controls. Genes indicated by 16 shared risk loci point to mechanisms with potential roles in migraine pathogenesis and CAD, including endothelial dysfunction (PHACTR1) and insulin homeostasis (GIP).
Conclusions:
The results suggest that shared biological processes contribute to risk of migraine and CAD, but surprisingly this commonality is restricted to migraine without aura and the impact is in opposite directions. Understanding the mechanisms underlying these processes and their opposite relationship to migraine and CAD may improve our understanding of both disorders.
Migraine affects 19% of women and 11% of men worldwide and causes more years lost to disability than any other neurologic disorder.1,2 In about one-third of patients, headache attacks are preceded by transient neurologic symptoms termed migraine aura, and migraine with and without aura (MA and MO, respectively) are believed to have a partially distinct pathogenic basis.3 It has long been assumed that the vascular system is involved in migraine pathogenesis, but little is known of the specific biological processes involved, and the relative importance of neuronal and vascular mechanisms remains controversial.3–6 Supporting a vascular basis, epidemiologic studies have found an increased risk for stroke among patients with migraine, most pronounced for MA.7 Some recent studies indicate a similar risk increase for coronary artery disease (CAD), the most common vascular disorder, although the association is less certain than for stroke.8–11 This raises the question of whether migraine and cardiovascular disease have a shared biological basis.
Both migraine and CAD have a strong genetic determination, and recent genome-wide association studies (GWAS) have identified risk variants for each. If migraine and CAD have a shared biological basis, one might anticipate that they will also share genetic variants that affect their risk. In this study, we utilized data from 2 large-scale nonoverlapping GWAS meta-analyses of migraine (the International Headache Genetics Consortium, IHGC)12 and CAD (Coronary ARtery DIsease Genome-Wide Replication And Meta-Analysis, CARDIoGRAM)13 to quantify shared genetic risk.
METHODS
Study cohorts.
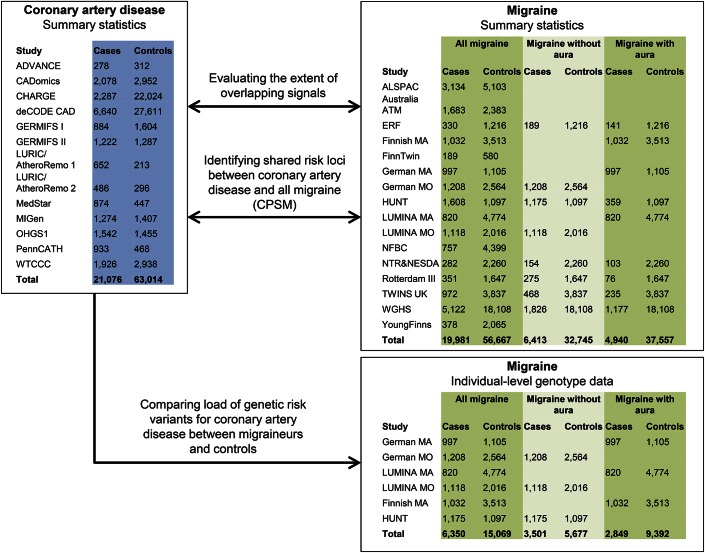
Summary statistics (p value and effect size) at single nucleotide polymorphism (SNP) level from 2 recently performed meta-analyses of genome-wide association data on migraine (IHGC)12 and CAD (CARDIoGRAM)13 were used in the present study. After excluding overlapping samples, the 2 studies consisted of 19,981 cases with migraine vs 56,667 controls, and 21,076 cases with CAD vs 63,014 controls. A proportion of the migraine cases were phenotyped in sufficient detail to allow subclassification into MO (6,413 cases, 32,745 controls) and MA (4,940 cases, 37,557 controls). In addition, individual-level genotype data were available for a proportion of the migraine cohorts (6,350 migraine cases vs 15,069 controls from the German MA and MO cohorts, Dutch LUMINA study, Finnish MA study, and the HUNT Study, Norway). All data sets were imputed by using the HapMap release 21 or 22 as reference. An overview of the study design and the included cohorts is given in figure 1. A detailed description of samples, genotyping, and association analyses is given in e-Methods, tables e-1 and e-2, and figure e-1 at Neurology.org/ng.
Figure 1. Study design and included cohorts.
CPSM = Cross-Phenotype Spatial Mapping.
Standard protocol approvals, registrations, and patient consents.
For all study cohorts, participation was based on informed consent. Each study was approved by local research ethics boards in the country where the study cohort was collected. See original publications of the 2 studies for full details of ethics and consent procedures.12,13
Analytic approach.
Evaluating extent of overlapping signals.
To assess whether more association signals were shared between the migraine and CAD studies than would be expected by chance, we used a set of 2,342,101 overlapping SNPs that were directly typed or imputed in both studies. Following the same procedure as described in a previous study,14 we first sorted the SNPs by association p value to migraine. Starting from the top of the list, all subsequent SNPs with linkage disequilibrium (LD) r2 > 0.05 (based on HapMap CEU release 27) were removed. This process was repeated until a set of 92,654 SNPs in approximate linkage equilibrium remained. For each of 5 separate p value cutoffs (1 × 10−2, 1 × 10−3, 1 × 10−4, 1 × 10−5, and 1 × 10−6), we counted the number of SNPs above and below the cutoff in each of the 2 studies, resulting in one 2 × 2 table for each p value cutoff. The Fisher exact test was used to estimate deviation from the expected distribution, and false discovery rate correction was performed on all 6 tests using the p.adjust function in R.15 A corrected Fisher p < 0.01 was taken to indicate an excess of overlapping signals. In order to obtain a more robust estimate of the significance of the observed overlap, this was also assessed through permutations. In each permutation cycle, the relation of p values to SNPs was randomized within each of the LD-pruned migraine and CAD data sets, and a Fisher p for overlap was calculated for each p value cutoff. We generated 100,000 permutations to produce an empirical null distribution of p values.
In an equivalent manner, secondary analyses were performed for MO (83,373 overlapping SNPs after LD pruning) and MA (88,031 overlapping SNPs after LD pruning).
Polygenic risk score analysis.
If shared genetic risk variants are in part or fully responsible for comorbidity between migraine and CAD, we would expect an accumulation of CAD risk alleles in migraineurs. To test this hypothesis, we used the 6 migraine cohorts in which individual-level genotype data were available for analysis (6,350 migraineurs vs 15,069 controls; figure 1). For each migraine case or control, we calculated a CAD polygenic risk score based on a previously published method.16 We first generated 3 sets of CAD risk SNPs by selecting SNPs with strong (p < 5 × 10−8; 149 SNPs), moderate (p < 1 × 10−4; 1,631 SNPs), or weak (p < 1 × 10−2; 36,384 SNPs) association to CAD among the 2,342,101 SNPs with information in both migraine and CAD studies. As suggested in the original description of the method,16 the analysis was based on non–LD-pruned SNP sets in order to optimize sensitivity. Using each set of CAD risk SNPs, we calculated a per-individual CAD polygenic risk score by summing the number of CAD risk alleles (or expected allele counts for imputed SNPs), each weighted by the log odds ratio from the CAD study. We subsequently assessed whether CAD polygenic risk score was associated with migraine status by applying a logistic regression model of the effect of CAD polygenic risk score (continuous) on migraine status (case, control), adjusted for sex and dummy-coded covariates representing the 6 individual migraine study cohorts.
Identifying shared risk loci.
In order to identify shared risk loci between migraine and CAD, we applied a novel method, Cross-Phenotype Spatial Mapping (CPSM; see e-Methods for an overview). This method compares 2 sets of p values from GWAS in order to find groups of SNPs at which they are correlated and thus identify shared patterns of association. We applied this method to the 2,342,101 overlapping SNPs from the migraine and CAD studies and selected genomic regions with signal above the 99.95th percentile of 1,000 permutations for further analysis. Potential effects of the shared association loci on regional gene expression (cis effect) were examined using an existing expression quantitative trait locus database from peripheral blood17 (e-Methods).
Lastly, we analyzed loci with previously reported genome-wide significant association to migraine or CAD (summarized in the original publications).12,13 The lead SNP at each locus was cross-analyzed for association to the other phenotype, Bonferroni correcting for the number of SNPs tested. All 13 of 13 reported migraine loci and 22 of 25 reported CAD loci were available in our data set and could be tested (excluding CAD risk SNPs rs17465637, rs1746048, and rs12413409).
RESULTS
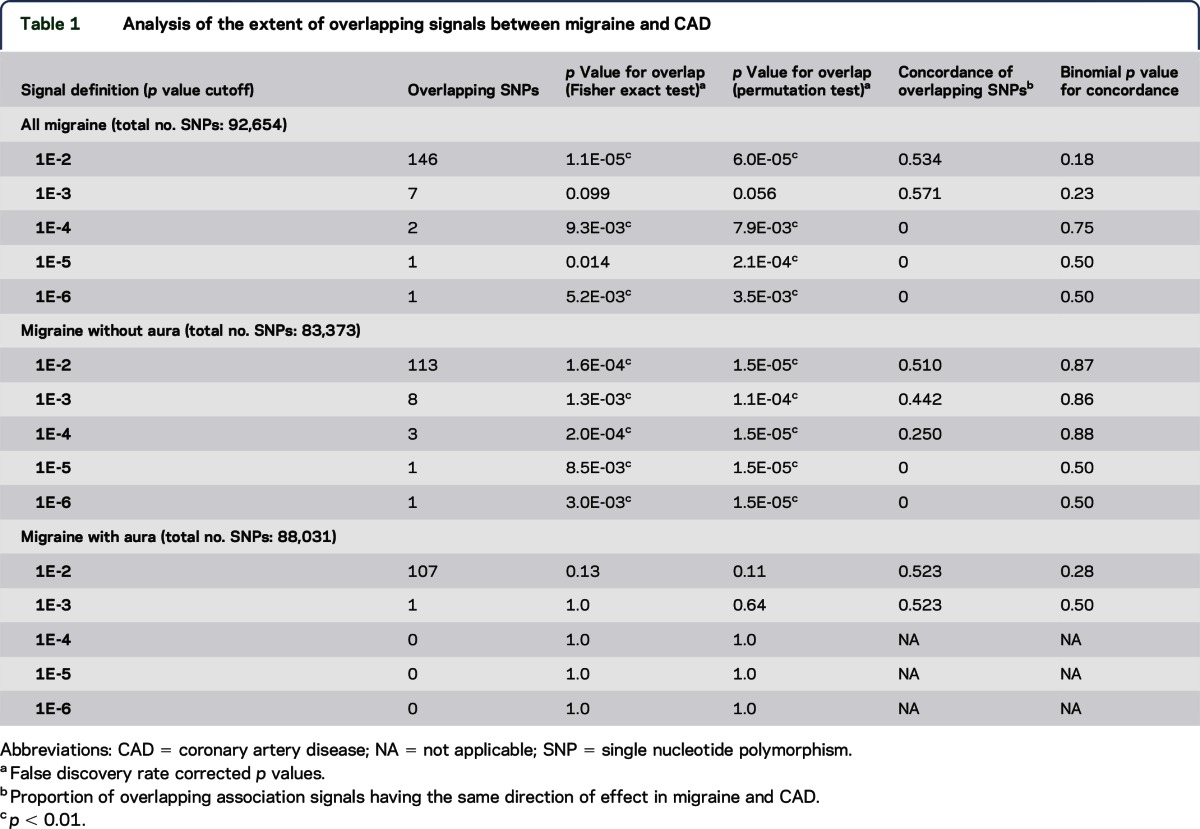
Comparing nominally significant SNPs from the migraine and CAD GWAS, we found an overlap of association signals in excess of what would be expected by chance (table 1). An overlap of signals was seen for SNPs with p values ≤1 × 10−2, 1 × 10−4, and 1 × 10−6. This was supported by permutation testing, which indicated a sharing of association signals at p value cutoff 1 × 10−5 as well. For reference, the full list of SNPs with association p value ≤1 × 10−2 to both CAD and migraine is given in table e-3. Secondary analyses by migraine subtype revealed an overlap of association signals between MO and CAD at all p value cutoffs (1 × 10−2, 1 × 10−3, 1 × 10−4, 1 × 10−5, and 1 × 10−6), while no overlap was seen between MA and CAD at any of the p value cutoffs. The direction of effect for overlapping association signals did not consistently agree between migraine and CAD, as evidenced by nonsignificant binomial p values for concordance (table 1).
Table 1.
Analysis of the extent of overlapping signals between migraine and CAD
To examine this further, the second analysis compared the load of genetic risk variants for CAD between migraineurs and controls, using individual-level data. The results indicated that a high CAD polygenic risk score was associated with a reduced risk of migraine (figure 2, further details in table e-4). For migraine overall, this association was seen for only the moderate CAD risk SNP set (p = 0.007). Secondary analyses revealed a similar, but more pronounced, association between CAD polygenic risk score and MO (p = 1.5 × 10−4 and 5.1 × 10−4 for the moderate and strong CAD risk SNP sets, respectively). No association was seen for MA. In the analysis of the weak CAD risk SNP set, there was no association to CAD genetic risk score for either migraine category, indicating that the observed associations were driven by a fairly limited number of loci that are at least moderately associated with CAD. These findings were consistent across men and women (figure e-2) and across individual independent cohorts within the same migraine subtype (figure e-3).
Figure 2. Association between coronary artery disease polygenic risk score and the presence of migraine.
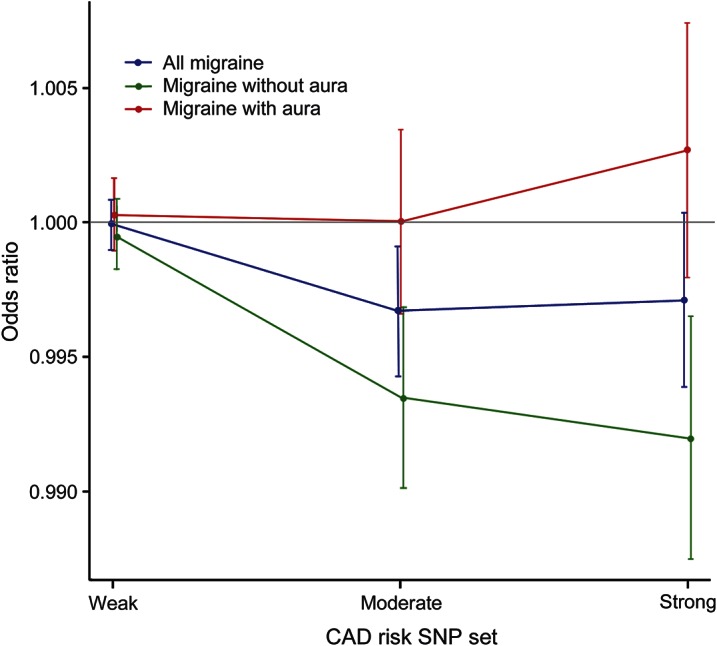
Results are given as odds ratios with 95% confidence intervals. Separate lines are shown for all migraine (blue), migraine without aura (green), and migraine with aura (red). The coronary artery disease (CAD) polygenic risk score was calculated based on single nucleotide polymorphisms (SNPs) with weak (p < 1 × 10−2), moderate (p < 1 × 10−4), or strong (p < 5 × 10−8) association to CAD in the Coronary ARtery DIsease Genome-Wide Replication And Meta-Analysis study.
CPSM yielded 16 loci that overlapped between migraine and CAD (table 2; figure e-4). Details of the most significant migraine and CAD SNPs at each locus are given in table e-5. The strongest evidence of shared association was seen at 6p24 (locus no. 1 of table 2), where both CAD and migraine showed genome-wide significant signals within the PHACTR1 gene (CAD: rs4714955, p = 9.8 × 10−11; migraine: rs9349379, p = 5.9 × 10−9). The second strongest overlapping signal was on 17q21 (locus no. 2), where the lead CAD SNP (rs46522, p = 2.6 × 10−7) was intragenic in UBE2Z, whereas the lead migraine SNP (rs11079844, p = 3.1 × 10−5) was intergenic between SNF8 and GIP. It is interesting that both lead SNPs are in high LD (r2 > 0.9) with 2 functional variants in GIP: Ser103Gly (rs2291725) and a splice site variant (rs2291726) that is predicted to lead to a prematurely truncated transcript18 (table e-6). The locus was also found to have a potential effect on the expression level of UBE2Z (table e-7). Lead SNPs in 5 loci were in high LD (r2 > 0.8) with nonsynonymous or splice site variants in nearby genes (table e-6). Ten of the 16 loci showed opposite direction of effect for migraine and CAD. In the secondary analyses, 12 of the 16 lead migraine SNPs had a lower association p value in MO than in MA (2-tailed binomial p = 0.08), and all 16 SNPs had the same effect direction in each of the 2 migraine subtypes. Local Manhattan plots and covariance plots for the identified loci are given in figure e-4.
Table 2.
Overlapping association loci between migraine and CAD as identified by CPSM analysis
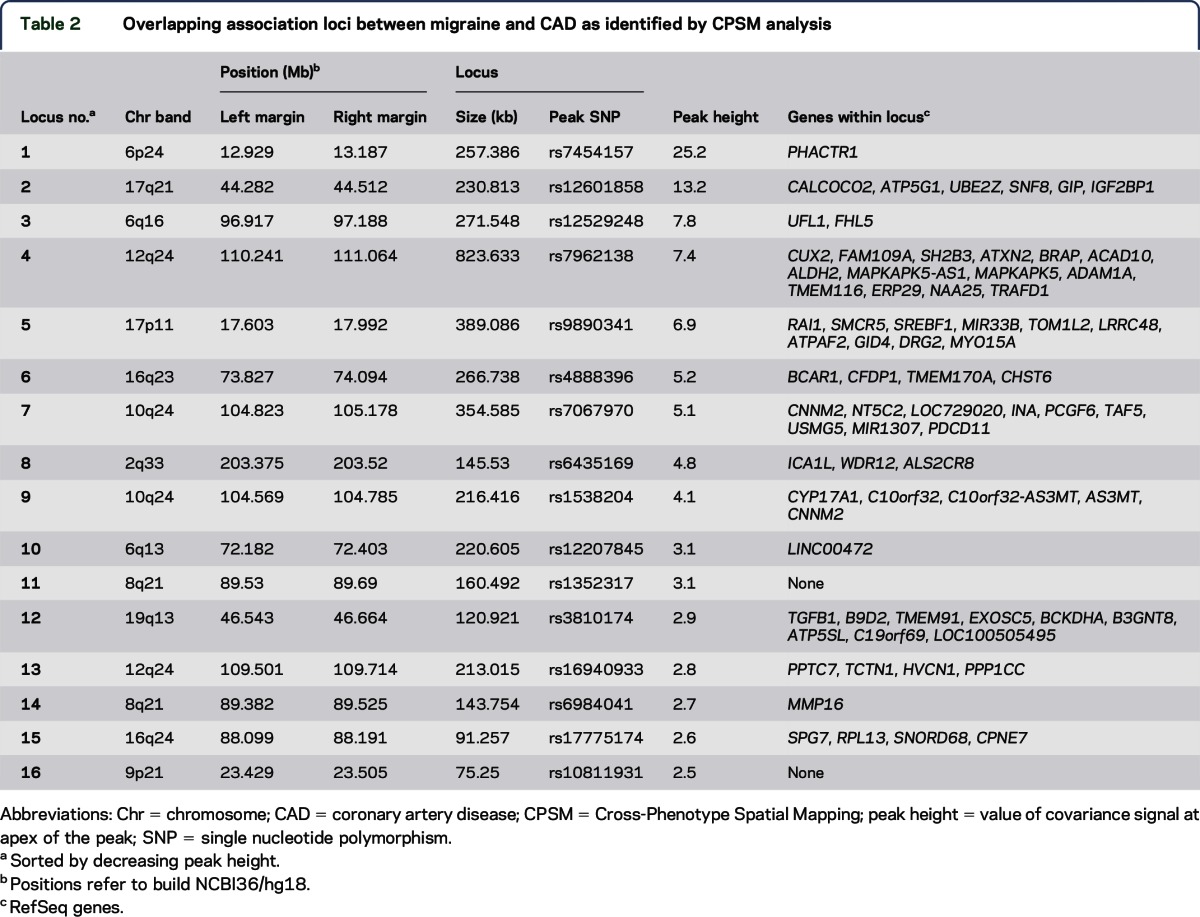
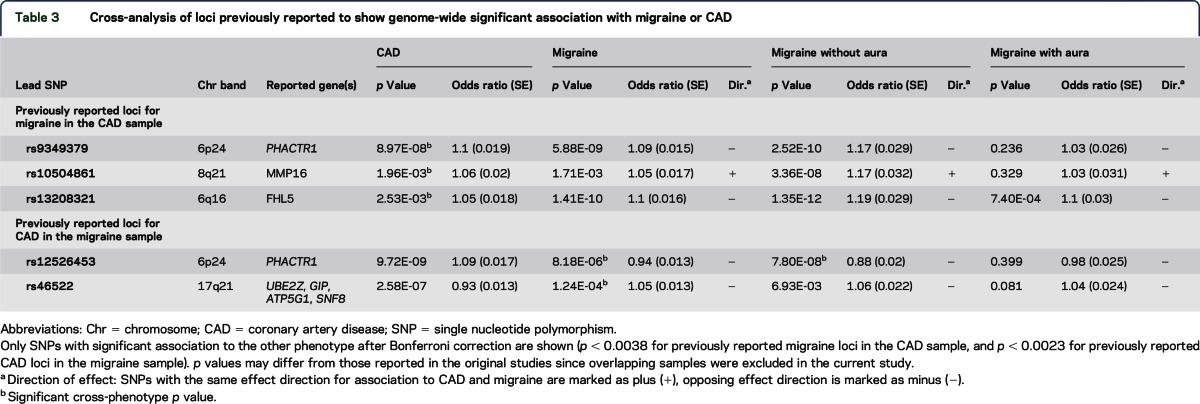
When considering previously reported risk loci for migraine and CAD, 3 CAD risk SNPs were associated to migraine at study-wide significance, and 2 migraine risk SNPs were associated to CAD (table 3). These correspond to loci no. 1, 2, 3, 11, and 14 as identified by the CPSM method and corroborate the evidence for shared genetic risk at these loci.
Table 3.
Cross-analysis of loci previously reported to show genome-wide significant association with migraine or CAD
DISCUSSION
In this study, we used data from 2 recently performed large-scale nonoverlapping GWAS to examine shared genetic risk between migraine and CAD. We found that association signals overlapped in excess of what would be expected by chance. Stratifying by migraine subtype further revealed that MO and MA behaved differently. MO had a genetic overlap with CAD, whereas MA did not. These results are unexpected, given the epidemiologic evidence that comorbidity with CAD is more common in MA than MO. Patients with MA were found to have a 2-fold increased risk for CAD8,10 and an increased risk for CAD-related mortality,9,11 although one cross-sectional study failed to find an association between CAD and any migraine subtype.19 Studies not differentiating on migraine subtype have been less conclusive, with some8,9,20,21 but not others19,22 indicating an increased risk of CAD related to migraine overall.
For MO, we found a clear overlap of association signals with CAD, whichever p value cutoff was used to define signals. Intriguingly, the impact was in the opposite direction, in that patients with MO had a lower load of CAD risk alleles than migraine-free controls. This association seemed to be driven by a limited number of loci. Only a proportion of the included migraine patients were phenotyped in sufficient detail to allow subclassification into MA or MO. When using the considerably larger set of all migraine patients, a similar association was seen as for MO, likely driven by this migraine subtype. While the results suggest that there are shared common risk variants between migraine and CAD, they do not indicate that these variants explain comorbidity between the 2 disorders.
The opposite direction of effect for some of the loci is consistent with a recent GWAS in which the migraine and CAD risk SNP rs9349379 (in PHACTR1) was associated with cervical artery dissection, with effect in the same direction as for migraine but opposite of CAD.23 Two further migraine SNPs showed evidence of association to cervical artery dissection with the same effect direction as for migraine (rs11172113 in LRP1 and rs13208321 in FHL5, the latter identified as locus 3 in the current study) but opposite direction for CAD.
The significant sharing of risk loci between migraine and CAD may reflect that they involve some of the same biological processes. Experimental studies will be needed to clarify this and whether the shared risk loci can give information on vascular mechanisms involved in migraine pathogenesis.
The lack of overlapping association signals between MA and CAD may indicate that the 2 disorders have separate and nonrelated genetic backgrounds. However, it may also result from insufficient power to detect shared common genetic risk factors for this migraine subtype. This is consistent with the relative failure so far in identifying common risk variants for MA; despite at least as high heritability and comparable study sample sizes, only one genome-wide significant locus has been identified for MA, compared to 9 for MO.12,24–27 It is possible that MA is a more heterogeneous disorder or is influenced by rare and low-frequency variants not captured by current imputation panels.12,28 Larger studies that also interrogate rare variants will be needed to determine the genetic basis of MA and its potential overlap with cardiovascular disease.
Six of the overlapping loci have previously been associated with CAD at genome-wide significant levels (loci 1, 2, 4, 7, 8, and 9 of table 2),13,29 and 2 with migraine (loci 1 and 3).12,25 The strongest overlapping region (locus 1) is entirely intragenic in PHACTR1 (which encodes phosphatase and actin regulator 1 protein). This locus is associated with both migraine and CAD at genome-wide significant levels in the current and previous studies13,25,30 and has also been associated with coronary artery calcification and stroke.31,32 PHACTR1 is highly expressed in the brain, and its transcript is an important regulator of synaptic activity and dendritic morphology through the control of protein phosphatase 1 and actin binding.33 More recently, PHACTR1 has been identified as a key regulator of endothelial function, including endothelial cell survival and angiogenesis,34 and it is associated with altered vasomotor tone.35 Both endothelial and vasomotor dysfunctions have been implicated in migraine,36,37 and this locus offers a potential focus for future studies. Alternatively, the pleiotropic effects of this gene on both synaptic and vascular functions may give rise to independent causal pathways for the 2 disorders.
The second strongest overlapping region (locus 2) is a previously identified risk locus for CAD.13 The lead CAD (rs46522) and migraine variants (rs11079844) are in strong LD (r2 = 0.94), and both are in strong LD (r2 > 0.90) with 2 potentially functional variants in GIP (which encodes gastric inhibitory polypeptide). GIP regulates glucose-induced insulin release from pancreatic β-cells and helps resensitize the insulin response.38 It is also expressed in the brain, where it may be involved in proliferation of neuronal progenitor cells.39 Whether GIP is involved in the observed tendency for insulin resistance and metabolic syndrome in migraineurs should be investigated.40
Strengths of this study include the use of large-scale nonoverlapping GWAS of migraine and CAD, stringent quality control measures, and sufficiently rich phenotyping to allow secondary analyses of the 2 migraine subtypes. Nevertheless, some limitations should also be acknowledged. First, only summary statistics and not individual-level genotype data were available for the majority of the samples included in this study. Second, in each included cohort, phenotype information was available on only migraine or CAD, not both. This prevented us from performing more in-depth analyses, including analysis for potential gene-gene interactions or identification of CAD risk loci specific to migraineurs. Third, considerable effort was devoted to the careful avoidance of shared controls between studies, and stringent quality control measures within each data set were enforced to reduce the risk of spurious effects resulting from biases within the data sets. Nevertheless, we cannot rule out subtle biases that could affect the current results. Two such concerns are the effects of migraine on survival and the possibility that migraineurs may be more likely to seek medical treatment and therefore be under closer surveillance with regards to other disorders. Future efforts should aim to replicate these findings in sufficiently large prospective data sets where both phenotypes are measured in the same individuals.
Our study provides novel insights into the relationship between migraine and CAD. Intriguingly, and unexpectedly, there was no genetic overlap between MA and CAD, for which epidemiologic studies suggest comorbidity, but there was compelling evidence for a genetic overlap between MO and CAD, where the impact of risk variants overall was in opposite direction for the 2 disorders. The results do not demonstrate that shared common genetic risk factors drive comorbidity between the 2 disorders. However, dissecting the mechanisms underlying the shared risk loci may improve our understanding of both disorders.
Supplementary Material
ACKNOWLEDGMENT
P.P. was supported by the European Commission FP7 project no. 261123 (gEUVADIS). C.K. and H.G. were funded by the German Federal Ministry of Education and Research (BMBF) within the framework of the National Genome Research Network (NGFN-Plus; grants 01GS08120 and 01GS1103 [to C.K.]) and the Deutsche Forschungsgemeinschaft (DFG). The Academy of Finland (grant 139795 to M.W.); the Folkhälsan Research Foundation (to M.W.); the Medicinska Understödsföreningen Liv & Hälsa (to M.W.); the Orion Farmos Research Foundation (to V. Anttila); and the Helsinki University Central Hospital (to M. Koiranen and V. Artto). The Women's Genome Health Study (WGHS) is supported by HL043851 and HL080467 from the National Heart, Lung, and Blood Institute and CA047988 from the National Cancer Institute, with collaborative scientific support and funding for genotyping provided by Amgen. Genetic analyses of migraine in WGHS have been supported by NS061836 from the National Institute of Neurological Disorders and Stroke. The Nord-Trøndelag Health Study (The HUNT Study) is a collaboration between HUNT Research Centre (Faculty of Medicine, Norwegian University of Science and Technology, NTNU), Nord-Trøndelag County Council, Central Norway Health Authority, and the Norwegian Institute of Public Institute of Public Health. The Young Finns Study has been financially supported by the Academy of Finland: grants 134309, 126925, 121584, 124282, 129378, 117787, and 41071, the Social Insurance Institution of Finland, Kuopio, Tampere and Turku University Hospital Medical Funds (grant 9N035 for T.L.), Juho Vainio Foundation, Paavo Nurmi Foundation, Finnish Foundation of Cardiovascular Research and Finnish Cultural Foundation, Tampere Tuberculosis Foundation, and Emil Aaltonen Foundation. The authors gratefully acknowledge the expert technical assistance in the statistical analyses by Irina Lisinen and Ville Aalto. TwinsUK: the study was funded by the Wellcome Trust and European Community's Seventh Framework Programme (FP7/2007–2013). The study also receives support from the National Institute for Health Research (NIHR) BioResource Clinical Research Facility and Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust and King's College London. SNP Genotyping was performed by The Wellcome Trust Sanger Institute and National Eye Institute via NIH/CIDR. The authors thank their funders, twin volunteers, and TwinsUK team. The LUMINA study is supported by grants obtained from the Netherlands Organization for the Health Research and Development (ZonMw) no. 90700217 and VIDI (ZonMw) no. 91711319 (to G.M.T.); the Netherlands Organisation for Scientific Research (NWO) VICI (918.56.602) and Spinoza (2009) grants (to M.D.F.); the 7th Framework EU project EUROHEADPAIN (no. 602633); and the Center for Medical Systems Biology (CMSB) established in the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research (NGI/NWO), project no. 050-060-409 (to M.D.F. and A.M.J.M.v.d.M.). Phenotype and genotype data collection in the Finnish Twin Cohort has been supported by the Wellcome Trust Sanger Institute, ENGAGE—European Network for Genetic and Genomic Epidemiology, FP7-HEALTH-F4-2007, grant 201413, National Institute of Alcohol Abuse and Alcoholism (grants AA-12502, AA-00145, and AA-09203 to R.J. Rose and AA15416 and K02AA018755 to D.M. Dick), and the Academy of Finland (grants 100499, 205585, 118555, 141054, 265240, 263278, and 264146 to J.K.). Phenotype and genotype data collection in NTR/NESDA was funded by the Netherlands Organization for Scientific Research (NWO: MagW/ZonMW grants 904-61-090, 985-10-002, 904-61-193, 480-04-004, 400-05-717, Addiction-31160008 Middelgroot-911-09-032, Spinozapremie 56-464-14192, Geestkracht program grant 10-000-1002), Centre for Medical Systems Biology (CMSB, NWO Genomics), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI—NL, 184.021.007), the VU University's Institute for Health and Care Research (EMGO+), Neuroscience Campus Amsterdam (NCA), ENGAGE (HEALTH-F4-2007-201413); European Science Council (ERC Advanced, 230374), Rutgers University Cell and DNA Repository (NIMH U24 MH068457-06), the Avera Institute for Human Genetics, Sioux Falls, SD, the National Institutes of Health (NIH, R01D0042157-01A), Genetic Association Information Network (GAIN) of the Foundation for the US National Institutes of Health (NIMH, MH081802), and by the Grand Opportunity grants 1RC2MH089951-01 and 1RC2MH089995-01 from the NIMH. NFBC1966 received financial support from the Academy of Finland (project grants 104781, 120315, 129269, 1114194, 24300796, Center of Excellence in Complex Disease Genetics and SALVE), Oulu University Hospital, Finland, Biocenter, University of Oulu, Finland 75617, 24002054, University of Oulu, Finland (grants 24000692 and 24500283: Well-being and health: Research in the Northern Finland Birth Cohorts 1966 and 1986, Phenotypic and Genomic analyses). NIH/NHLBI NHLBI grant 5R01HL087679-02 through the STAMPEED program (1RL1MH083268-01), NHLBI Consortium for Neuropsychiatric Phenomics Co-ordinating Center (1R01HL087679-01), and NIH/NIMH (5R01MH63706-02), United States. ENGAGE project and grant agreement HEALTH-F4-2007-201413. Medical Research Council (grant G1002319). The DNA extractions, sample quality controls, biobank upkeep, and aliquoting were performed in the National Public Health Institute, Biomedicum Helsinki, Finland and supported financially by the Academy of Finland and Biocentrum Helsinki. The authors thank Ms. Outi Tornwall and Ms. Minttu Jussila (DNA biobanking).
GLOSSARY
- CAD
coronary artery disease
- CARDIoGRAM
Coronary ARtery DIsease Genome-Wide Replication And Meta-Analysis
- CPSM
Cross-Phenotype Spatial Mapping
- GWAS
genome-wide association studies
- IHGC
International Headache Genetics Consortium
- LD
linkage disequilibrium
- MA
migraine with aura
- MO
migraine without aura
- SNP
single nucleotide polymorphism
Footnotes
Supplemental data at Neurology.org/ng
Contributor Information
Collaborators: Leonore Launer, George Davey Smith, George McMahon, Dale Nyholt, Alfons Macaya, Patricia Pozo-Rosich, Bru Cormand, Jessica Fernandez, Marta Vila-Pueyo, Celia Sintas, Jes Olesen, Anne Francke Christensen, Ann-Louise Esserlind, Najaf Amin, Tonu Esko, Aarno Palotie, Mikko Kallela, Maija Wessman, Ville Artto, Verneri Anttila, Eija Hämäläinen, Priit Palta, Padhraig Gormley, Ester Cuenca, Jaakko Kaprio, Martin Dichgans, Hartmut Göbel, Christian Kubisch, Tobias Freilinger, Rainer Malik, Bertram Muller-Myhsok, John-Anker Zwart, Bendik Winsvold, Line Jacobsen, Linda Pedersen, Alice Pressman, Arn van den Maagdenberg, Gisela Terwindt, Boukje de Vries, Rune R. Frants, Michel Ferrari, Dorret I. Boomsma, Lannie Ligthart, Brenda Penninx, Marjo-Riitta Jarvelin, Markku Koiranen, Cornelia van Duijn, M Arfan Ikram, Andrea Carmine Belin, Nancy Pedersen, Lynn Cherkas, Lydia Quaye, Daniel Chasman, Tobias Kurth, Markus Schuerks, Terho Lehtimaki, Olli Raitakari, Nick Eriksson, Devin Absher, Themistocles L. Assimes, Stephen Fortmann, Alan Go, Mark Hlatky, Carlos Iribarren, Joshua Knowles, Richard Myers, Thomas Quertermous, Steven Sidney, Neil Risch, Hua Tang, Stefan Blankenberg, Tanja Zeller, Arne Schillert, Philipp Wild, Andreas Ziegler, Renate Schnabel, Christoph Sinning, Karl Lackner, Laurence Tiret, Viviane Nicaud, Francois Cambien, Christoph Bickel, Hans J. Rupprecht, Claire Perret, Carole Proust, Thomas Münzel, Maja Barbalic, Joshua Bis, Eric Boerwinkle, Ida Yii-Der Chen, L. Adrienne Cupples, Abbas Dehghan, Serkalem Demissie-Banjaw, Aaron Folsom, Nicole Glazer, Vilmundur Gudnason, Tamara Harris, Susan Heckbert, Daniel Levy, Thomas Lumley, Kristin Marciante, Alanna Morrison, Christopher J. O'Donnell, Bruce M. Psaty, Kenneth Rice, Jerome I. Rotter, David S. Siscovick, Nicholas Smith, Albert Smith, Kent D. Taylor, Cornelia van Duijn, Kelly Volcik, Jaqueline Whitteman, Vasan Ramachandran, Albert Hofman, Andre Uitterlinden, Solveig Gretarsdottir, Jeffrey R. Gulcher, Hilma Holm, Augustine Kong, Kari Stefansson, Gudmundur Thorgeirsson, Karl Andersen, Gudmar Thorleifsson, Unnur Thorsteinsdottir, Jeanette Erdmann, Marcus Fischer, Anika Grosshennig, Christian Hengstenberg, Inke R. König, Wolfgang Lieb, Patrick Linsel-Nitschke, Michael Preuss, Klaus Stark, Stefan Schreiber, H.-Erich Wichmann, Andreas Ziegler, Heribert Schunkert, Zouhair Aherrahrou, Petra Bruse, Angela Doering, Jeanette Erdmann, Christian Hengstenberg, Thomas Illig, Norman Klopp, Inke R. König, Patrick Linsel-Nitschke, Christina Loley, Anja Medack, Christina Meisinger, Thomas Meitinger, Janja Nahrstaedt, Annette Peters, Michael Preuss, Klaus Stark, Arnika K. Wagner, H.-Erich Wichmann, Christina Willenborg, Andreas Ziegler, Heribert Schunkert, Bernhard O. Böhm, Harald Dobnig, Tanja B. Grammer, Eran Halperin, Michael M. Hoffmann, Marcus Kleber, Reijo Laaksonen, Winfried März, Andreas Meinitzer, Bernhard R. Winkelmann, Stefan Pilz, Wilfried Renner, Hubert Scharnagl, Tatjana Stojakovic, Andreas Tomaschitz, Karl Winkler, Benjamin F. Voight, Kiran Musunuru, Candace Guiducci, Noel Burtt, Stacey B. Gabriel, David S. Siscovick, Christopher J. O'Donnell, Roberto Elosua, Leena Peltonen, Veikko Salomaa, Stephen M. Schwartz, Olle Melander, David Altshuler, Sekar Kathiresan, Alexandre F. R. Stewart, Li Chen, Sonny Dandona, George A. Wells, Olga Jarinova, Ruth McPherson, Robert Roberts, Muredach P. Reilly, Mingyao Li, Liming Qu, Robert Wilensky, William Matthai, Hakon H. Hakonarson, Joe Devaney, Mary Susan Burnett, Augusto D. Pichard, Kenneth M. Kent, Lowell Satler, Joseph M. Lindsay, Ron Waksman, Christopher W. Knouff, Dawn M. Waterworth, Max C. Walker, Vincent Mooser, Stephen E. Epstein, Daniel J. Rader, Nilesh J. Samani, John R. Thompson, Peter S. Braund, Christopher P. Nelson, Benjamin J. Wright, Anthony J. Balmforth, Stephen G. Ball, and Alistair S. Hall
AUTHOR AFFILIATIONS
From the Department of Neurology (B.S.W., J.-A.Z.) and FORMI (B.S.W., L.M.J., L.M.P., J.-A.Z.), Oslo University Hospital, Oslo, Norway; Institute of Clinical Medicine (B.S.W., J.-A.Z.), University of Oslo, Norway; Wellcome Trust Sanger Institute (B.S.W., P.G., V. Anttila, P.P., E.H., A.P.), Wellcome Trust Genome Campus, Cambridge, United Kingdom; Department of Cardiovascular Sciences (C.P.N., N.J.S.), University of Leicester, Clinical Sciences Wing and National Institute for Health Research Leicester Biomedical Research Unit in Cardiovascular Disease (C.P.N., N.J.S), Glenfield Hospital, Leicester, United Kingdom; Institute for Stroke and Dementia Research (R. Malik, T.F., M.D.), Klinikum der Universität München, Ludwig-Maximilians-Universität, Munich, Germany; Munich Cluster for Systems Neurology (SyNergy) (R. Malik, M.D.), Munich, Germany; Program in Medical and Population Genetics (P.G., V. Anttila, H.-H.W., S.K., C.C., A.P.) and Stanley Center for Psychiatric Research (V. Anttila, A.P.), Broad Institute, Cambridge, MA; Psychiatric & Neurodevelopmental Genetics Unit (P.G., A.P.), Department of Psychiatry, Analytic and Translational Genetics Unit (V. Anttila, A.P.), Department of Medicine, Center for Human Genetic Research (H.-H.W., S.K.), Cardiovascular Research Center (H.-H.W., S.K.), and Department of Neurology (A.P.), Massachusetts General Hospital, Boston, MA; Division of Preventive Medicine (T. Kurth, D.I.C.), Brigham and Women's Hospital and Department of Medicine (V. Anttila, H.-H.W., S.K.), Harvard Medical School, Boston, MA; Department of Genetics (J.V.H., C.C.) and Department of Neurology (C.C.), Yale University School of Medicine, New Haven, CT; Wellcome Trust Centre for Human Genetics (K.S.E.), University of Oxford, United Kingdom; Department of Public Health (J.K.), Hjelt Institute and Institute for Molecular Medicine Finland (P.P., E.H., J.K., M.W., A.P.), University of Helsinki, Finland; Department of Epidemiology (N.A., M.A.I., C.v.D.), Department of Radiology (M.A.I.), and Department of Neurology (M.A.I.), Erasmus University Medical Centre, Rotterdam, the Netherlands; Department of Human Genetics (B.d.V., A.M.J.M.v.d.M.) and Department of Neurology (M.D.F., G.M.T., A.M.J.M.v.d.M.), Leiden University Medical Centre, Leiden, the Netherlands; Department of Neurology and Epileptology and Hertie-Institute for Clinical Brain Research (T.F.), University of Tübingen, Germany; Deutsches Herzzentrum München (T. Kessler, H.S.), Technische Universität München, Munich, Germany; DZHK (German Centre for Cardiovascular Research) (T. Kessler, H.S.), Partner Site Munich Heart Alliance, Munich, Germany; Institute of Health Sciences (M. Koiranen, M.-R.J.) and Biocenter Oulu (M.-R.J.), University of Oulu, Finland; Department of Biological Psychology (L.L., D.I.B.), VU University and EMGO+ Institute for Health and Care Research (L.L.), VU University Medical Centre, Amsterdam, the Netherlands; Medical Research Council (MRC) Integrative Epidemiology Unit at the University of Bristol (G.M., G.D.S.), United Kingdom; Institut für Integrative und Experimentelle Genomik (C.W., J.E.), Universität zu Lübeck, Lübeck, Germany; DZHK (German Research Centre for Cardiovascular Research) (C.W., J.E.), Partner Site Hamburg/Lübeck/Kiel, Lübeck, Germany; Department of Neurology (J.O.), Glostrup Hospital, University of Copenhagen, Denmark; Department of Neurology (V. Artto, M. Kallela), Helsinki University Central Hospital, Helsinki, Finland; Department of Medicine (T.L.A.), Stanford University School of Medicine, Stanford, CA; Medizinische Klinik und Poliklinik (S.B.), Universitätsmedizin Mainz, Johannes-Gutenberg Universität Mainz, Germany; Department of Twin Research and Genetic Epidemiology (L.C., L.Q.), King's College London, United Kingdom; MedStar Heart Institute (S.E.E.), Washington Hospital Center, Washington, DC; Kiel Pain and Headache Center (H.G.), Kiel, Germany; Division of Cardiovascular and Neuronal Remodelling (A.S.H.), Multidisciplinary Cardiovascular Research Centre, Leeds Institute of Genetics, Health and Therapeutics, University of Leeds, United Kingdom; Department of Children, Young People and Families (M.-R.J.) and Department of Mental Health and Substance Abuse Services (J.K.), National Institute for Health and Welfare, Helsinki, Finland; Department of Epidemiology and Biostatistics (M.-R.J.), School of Public Health, MRC–Health Protection Agency (HPA) Centre for Environment and Health, Faculty of Medicine, Imperial College, London, United Kingdom; Unit of Primary Care (M.-R.J.), Oulu University Hospital, Oulu, Finland; Department of Clinical Chemistry (T.L.), Fimlab Laboratories, Tampere, Finland; University of Tampere School of Medicine (T.L.), Finland; The John and Jennifer Ruddy Canadian Cardiovascular Genetics Centre (R. McPherson, R.R., A.F.R.S.) and Atherogenomics Laboratory (R. McPherson), University of Ottawa Heart Institute, Ontario, Canada; Synlab Center of Laboratory Diagnostics Heidelberg (W.M.), Heidelberg, Germany; Clinical Institute of Medical and Chemical Laboratory Diagnostics (W.M.), Medical University of Graz, Austria; Institute of Public Health (W.M.), Social and Preventive Medicine, Medical Faculty Manneim, University of Heidelberg, Germany; Institute of Health and Biomedical Innovation (D.R.N.), Queensland University of Technology, Brisbane, Australia; National Heart, Lung, and Blood Institute's Framingham Heart Study (C.J.O.), Framingham, MA; Department of Medicine, Institute for Translational Medicine and Therapeutics, and Cardiovascular Institute (D.J.R.), University of Pennsylvania, Philadelphia, PA; Department of Clinical Physiology and Nuclear Medicine (O.R.), Turku University Hospital, Turku, Finland; Research Centre of Applied and Preventive Cardiovascular Medicine (O.R.), University of Turku, Finland; Department of Neurology (M.S.), University Hospital Essen, Essen, Germany; deCODE genetics (U.T.), Reykjavik, Iceland; Faculty of Medicine (U.T.), University of Iceland, Reykjavik, Iceland; Institute of Genetics (M.W.), Folkhälsan Research Center, Helsinki, Finland; Institut National de la Santé et de la Recherche Médicale (INSERM) Research Center for Epidemiology and Biostatistics (U897) Team–Neuroepidemiology (T. Kurth), Bordeaux, France; University of Bordeaux (T. Kurth), France; and Institute of Human Genetics (C.K.), University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
AUTHOR CONTRIBUTIONS
Bendik S. Winsvold and Christopher P. Nelson: performed statistical analysis; conceived and designed the study; analyzed and interpreted the data; contributed data; drafted/revised the manuscript for content. Rainer Malik: conceived and designed the study; analyzed and interpreted the data; contributed data; drafted/revised the manuscript for content. Padhraig Gormley, Verneri Anttila, Jason Vander Heiden, Katherine S. Elliott, Line M. Jacobsen, and Priit Palta: analyzed and interpreted the data; contributed data; drafted/revised the manuscript for content. Najaf Amin, Boukje de Vries, Eija Hämäläinen, Tobias Freilinger, M. Arfan Ikram, Thorsten Kessler, Markku Koiranen, Lannie Ligthart, George McMahon, Linda M. Pedersen, Christina Willenborg, and Hong-Hee Won: contributed data; drafted/revised the manuscript for content. Jes Olesen: analyzed and interpreted the data; contributed data; drafted/revised the manuscript for content. Ville Artto, Themistocles L. Assimes, Stefan Blankenberg, Dorret I. Boomsma, Lynn Cherkas, George Davey Smith, Stephen E. Epstein, Jeanette Erdmann, Michel D. Ferrari, Hartmut Göbel, Alistair S. Hall, Marjo-Riitta Jarvelin, Mikko Kallela, Jaakko Kaprio, Sekar Kathiresan, Terho Lehtimäki, Ruth McPherson, Winfried März, Dale R. Nyholt, Christopher J. O'Donnell, Lydia Quaye, Daniel J. Rader, Olli Raitakari, Robert Roberts, Heribert Schunkert, Markus Schürks, Alexandre F.R. Stewart, Gisela M. Terwindt, Unnur Thorsteinsdottir, Arn M.J.M. van den Maagdenberg, Cornelia van Duijn, and Maija Wessman: contributed data; drafted/revised the manuscript for content. Tobias Kurth and Christian Kubisch: analyzed and interpreted the data; contributed data; drafted/revised the manuscript for content. Martin Dichgans: conceived and designed the study; analyzed and interpreted the data; contributed data; drafted/revised the manuscript for content. Daniel I. Chasman and Chris Cotsapas: analyzed and interpreted the data; contributed data; drafted/revised the manuscript for content. John-Anker Zwart, Nilesh J. Samani, and Aarno Palotie: conceived and designed the study; jointly supervised research; analyzed and interpreted the data; contributed data; drafted/revised the manuscript for content. All authors accept responsibility for conduct of research and will give final approval.
STUDY FUNDING
This work was supported by Academy of Finland (grant 251704 to A.P.), Sigrid Juselius Foundation (to A.P.), SynSys (to A.P.), the Wellcome Trust (grant 098051 to A.P.), EU FP7-242167 (to A.P.), NIH/RFA-HL-12-007, Genomic and Metabolomic Profiling of Finnish Familial Dyslipidemia Families (to A.P.), the South-Eastern Norway Regional Health Authority (grants 2010075 and 2011083 to B.S.W., L.M.J., and J.-A.Z.), the Research Council of Norway (grant 231187/F20 to B.S.W.), and the NIHR Leicester Cardiovascular Biomedical Research Unit and BHF (to C.P.N.). N.J.S. holds a Chair funded by the British Heart Foundation and is an NIHR Senior Investigator. Funding for study cohorts and remaining authors are listed in the acknowledgment. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
DISCLOSURE
Bendik S. Winsvold has received research support from the Research Council of Norway, the South-Eastern Norway Regional Health Authority, and Forsberg's and Auli Endowment. Christopher P. Nelson, Rainer Malik, and Padhraig Gormley report no disclosures. Verneri Anttila has received research support from Orion Farmos Research Foundation. Jason Vander Heiden has received funding for travel and/or speaker honoraria from New England Biolabs; and has received research support from National Library of Medicine and the United States-Israel Binational Science Foundation. Katherine S. Elliott reports no disclosures. Line M. Jacobsen is employed by AstraZeneca. Priit Palta has received research support from the Finnish Cultural Foundation. Najaf Amin has received research support from the Netherlands Brain Foundation. Boukje de Vries and Eija Hämäläinen report no disclosures. Tobias Freilinger has received funding for travel and/or speaker honoraria from Boehringer Ingelheim and Allergan; and has received research support from Deutsche Forschungsgemeinschaft (DFG). M. Arfan Ikram has received funding for travel and/or speaker honoraria from Kaizo, Ltd.; has served on the editorial boards of Neuroepidemiology, PLoS One, and Journal of Alzheimer Disease; and has received research support from Janssen Prevention Center, Netherlands Organization for Health Research and Development, the Netherlands Heart Foundation, Internationaal Parkinson Fonds, Internationale Stichting Alzheimer Onderzoek, and the Alzheimer Association. Thorsten Kessler and Markku Koiranen report no disclosures. Lannie Ligthart has received research support from EFIC-Grünenthal. George McMahon, Linda M. Pedersen, Christina Willenborg, and Hong-Hee Won report no disclosures. Jes Olesen has consulted for Jannsen Pharmaceutical Products; and has served on speakers' bureaus for Allergan. Ville Artto has received funding for travel and/or speaker honoraria from Boehringer Ingelheim, Orion, Menarini, Migraine Trust, and Bayer; and has received research support from the Finnish Medical Foundation and Helsinki University Central Hospital. Themistocles L. Assimes has served on the editorial board of Frontiers in Cardiovascular Medicine; has consulted and received research support from Telomere Diagnostics, Inc.; and has received research support from NHLBI. Stefan Blankenberg, Dorret I. Boomsma, Lynn Cherkas, and George Davey Smith report no disclosures. Stephen E. Epstein has received research support from MedStar Heart and Vascular Institute, MedStar WA Hospital Center; and holds stock/stock options and/or receives Board of Directors Compensation for CardioCell. Jeanette Erdmann reports no disclosures. Michel D. Ferrari has served on the editorial board of Cephalalgia; and has received research support from the Netherlands Organization for Scientific Research (NWO), European Community, ZonMW, and the Dutch Heart Foundation. Hartmut Göbel has served on Scientific Advisory Boards for Allergan, Bayer Vital, and St. Jude Medical; has received funding for travel and/or speaker honoraria from Amgen, Allergan, Hormosan, Klosterfrau, MSD, Mundipharma, St. Jude Medical, and Teva; has served on the editorial board of Der Schmerz, Pain Research and Treatment; has served on speakers' bureaus for Allergan, Hormosan, Klosterfrau, MSD, Mundipharma, St. Jude Medical, and Teva; and has received research support from St. Jude Medical. Alistair S. Hall and Marjo-Riitta Jarvelin report no disclosures. Mikko Kallela has served on Advisory Boards for MSD and Allergan; has received funding for travel and/or speaker honoraria from MSD, Allergan, TEVA, Novartis, and Genzyme; has received compensation for producing educational material from TEVA and Allergan; has received research support from Helsinki University Central Hospital; and holds stock/stock options and/or has received Board of Directors compensation from Helsinki Headache Center. Jaakko Kaprio has served on an Advisory Board for Copenhagen University; has received funding for travel and/or speaker honoraria; and has consulted for Pfizer Ltd. Sekar Kathiresan has served on scientific advisory boards for Regeneron, Merck, Eli Lilly, Aegerionm, Catabasis, Amarin, and Novartis; and has received research support from Regeneron, Aegerion, Merck, NIH, and Fondation Leducq. Terho Lehtimäki reports no disclosures. Ruth McPherson has served on the editorial board of Arteriosclerosis, Thrombosis & Vascular Biology; and has received research support from Canadian Institutes of Health Research and Heart & Stroke Foundation of Canada. Winfried März has served on Scientific Advisory Boards and speakers' bureaus and consulted for Aegerion Pharmaceuticals, AMGEN, Danone Research, Sanofi/Genzyme, Hoffmann LaRoche, MSD, Synageva, Eli Lilly, and BASF; has received funding for travel and/or speaker honoraria from Aegerion Pharmaceuticals, AMGEN, Danone Research, Sanofi/Genzyme, Hoffmann LaRoche, MSD, Synageva, Eli Lilly, and BASF; has served on the editorial boards of European Heart Journal and Journal of Laboratory Medicine; has been employed by and holds stock/stock options and/or Board of Directors compensation from Synlab Services GmbH; and has received research support from Aegerion Pharmaceuticals, AMGEN, Danone Research, Sanofi/Genzyme, Hoffmann LaRoche, MSD, Synageva, Eli Lilly, BASF, European Union, German Ministry of Research, German Ministry of Commerce, and Wissenschaftsinitiative Oberrhein. Dale R. Nyholt reports no disclosures. Christopher J. O'Donnell is employed by and has received research support from National Institutes of Health. Lydia Quaye reports no disclosures. Daniel J. Rader has served on the scientific advisory boards of Arteriosclerosis, Thrombosis & Vascular Biology; and has received research support from NIH and the Leducq Foundation. Olli Raitakari and Robert Roberts report no disclosures. Heribert Schunkert has served on scientific advisory boards, consulted for, received funding for travel and/or honoraria, and/or received research support from AstraZeneca, AMGEN, MSD SHARP & DOHME, Bayer Vital, Boehringer Ingelheim, Medtronic, Novartis, Pfizer, Sanofi-Aventis, St. Jude, Boston Scientific, and Daiichi Sankyo. Markus Schürks has served on the editorial boards of The Journal of Headache and Pain, and BMC Neurology; and consults Bayer HealthCare Pharmaceuticals. Alexandre F.R. Stewart has served on the editorial board of Frontiers in Cardiovascular Medicine—Cardiovascular Genetics. Gisela M. Terwindt has received research support from the Netherlands Organisation for Scientific Research (NWO). Unnur Thorsteinsdottir is an employee of deCODE genetics/Amgen. Arn M.J.M. van den Maagdenberg and Cornelia van Duijn report no disclosures. Maija Wessman has received research support from Folkhälsan Research Foundation, Academy of Finland, and Medicinska Understödsföreningen Liv och Hälsa. Tobias Kurth has served on editorial boards for BMJ and Cephalalgia; and has received research support from the French National Research Agency and the University of Bordeaux. Christian Kubisch reports no disclosures. Martin Dichgans has served on editorial boards for Stroke, the International Journal of Stroke, Cerebrovascular Diseases, and Journal of Neurochemistry; has consulted for Bayer Vital, Boehringer Ingelheim, Bristol-Myers Squibb, and Heel; and has received research support from Wellcome Trust, European Union, and German Federal Ministry of Education and Research. Daniel I. Chasman has served on the editorial board for Arteriosclerosis, Thrombosis, and Vascular Biology; and receives publishing royalties for Protein Structure: Determination, Analysis, and Applications for Drug Discovery (Marcel Dekker, 2003). Chris Cotsapas has served on the editorial board for PLoS Genetics; and has received research support from NINDS, NIAID, and RE Children's Consortium. John-Anker Zwart reports no disclosures. Nilesh J. Samani has served on editorial boards for Circulation: Cardiovascular Genetics and Heart; and has received research support from British Heart Foundation and National Institute for Health Research. Aarno Palotie has been a member of the Pfizer Genetics Scientific Advisory Panel; has received travel expenses and/or honoraria for lectures or educational activities not funded by industry; and has received research support from the Finnish Academy, European Union NIH, NINDS, Juselius Foundation, and the Finnish Foundation for Cardiovascular Research. Go to Neurology.org/ng for full disclosure forms.
REFERENCES
- 1.Vos T, Flaxman AD, Naghavi M, et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012;380:2163–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Headache Classification Committee of the International Headache Society (IHS). The International Classification of Headache Disorders, 3rd edition (beta version). Cephalalgia 2013;33:629–808. [DOI] [PubMed] [Google Scholar]
- 3.Charles AC, Baca SM. Cortical spreading depression and migraine. Nat Rev Neurol 2013;9:637–644. [DOI] [PubMed] [Google Scholar]
- 4.Charles A. Migraine is not primarily a vascular disorder. Cephalalgia 2012;32:431–432. [DOI] [PubMed] [Google Scholar]
- 5.Ashina M. Vascular changes have a primary role in migraine. Cephalalgia 2012;32:428–430. [DOI] [PubMed] [Google Scholar]
- 6.Noseda R, Burstein R. Migraine pathophysiology: anatomy of the trigeminovascular pathway and associated neurological symptoms, cortical spreading depression, sensitization, and modulation of pain. Pain 2013;154(suppl 1):S44–S53. [DOI] [PubMed] [Google Scholar]
- 7.Kurth T, Chabriat H, Bousser MG. Migraine and stroke: a complex association with clinical implications. Lancet Neurol 2012;11:92–100. [DOI] [PubMed] [Google Scholar]
- 8.Bigal ME, Kurth T, Santanello N, et al. Migraine and cardiovascular disease: a population-based study. Neurology 2010;74:628–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gudmundsson LS, Scher AI, Aspelund T, et al. Migraine with aura and risk of cardiovascular and all cause mortality in men and women: prospective cohort study. BMJ 2010;341:c3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurth T, Schurks M, Logroscino G, Gaziano JM, Buring JE. Migraine, vascular risk, and cardiovascular events in women: prospective cohort study. BMJ 2008;337:a636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liew G, Wang JJ, Mitchell P. Migraine and coronary heart disease mortality: a prospective cohort study. Cephalalgia 2007;27:368–371. [DOI] [PubMed] [Google Scholar]
- 12.Anttila V, Winsvold BS, Gormley P, et al. Genome-wide meta-analysis identifies new susceptibility loci for migraine. Nat Genet 2013;45:912–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schunkert H, Konig IR, Kathiresan S, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 2011;43:333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott KS, Chapman K, Day-Williams A, et al. Evaluation of the genetic overlap between osteoarthritis with body mass index and height using genome-wide association scan data. Ann Rheum Dis 2013;72:935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing [computer program]. Vienna, Austria; 2013. Available at: http://www.R-project.org/. Accessed September 20, 2013. [Google Scholar]
- 16.Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009;460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright FA, Sullivan PF, Brooks AI, et al. Heritability and genomics of gene expression in peripheral blood. Nat Genet 2014;46:430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nitz I, Fisher E, Weikert C, et al. Association analyses of GIP and GIPR polymorphisms with traits of the metabolic syndrome. Mol Nutr Food Res 2007;51:1046–1052. [DOI] [PubMed] [Google Scholar]
- 19.Le H, Tfelt-Hansen P, Russell MB, Skytthe A, Kyvik KO, Olesen J. Co-morbidity of migraine with somatic disease in a large population-based study. Cephalalgia 2011;31:43–64. [DOI] [PubMed] [Google Scholar]
- 20.Wang YC, Lin CW, Ho YT, Huang YP, Pan SL. Increased risk of ischemic heart disease in young patients with migraine: a population-based, propensity score-matched, longitudinal follow-up study. Int J Cardiol 2014;172:213–216. [DOI] [PubMed] [Google Scholar]
- 21.Sacco S, Ornello R, Ripa P, et al. Migraine and risk of ischaemic heart disease: a systematic review and meta-analysis of observational studies. Eur J Neurol 2015;22:1001–1011. [DOI] [PubMed] [Google Scholar]
- 22.Schurks M, Rist PM, Bigal ME, Buring JE, Lipton RB, Kurth T. Migraine and cardiovascular disease: systematic review and meta-analysis. BMJ 2009;339:b3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Debette S, Kamatani Y, Metso TM, et al. Common variation in PHACTR1 is associated with susceptibility to cervical artery dissection. Nat Genet 2015;47:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anttila V, Stefansson H, Kallela M, et al. Genome-wide association study of migraine implicates a common susceptibility variant on 8q22.1. Nat Genet 2010;42:869–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freilinger T, Anttila V, de Vries B, et al. Genome-wide association analysis identifies susceptibility loci for migraine without aura. Nat Genet 2012;44:777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chasman DI, Schurks M, Anttila V, et al. Genome-wide association study reveals three susceptibility loci for common migraine in the general population. Nat Genet 2011;43:695–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell MB, Olesen J. Increased familial risk and evidence of genetic factor in migraine. BMJ 1995;311:541–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nyholt DR, Anttila V, Winsvold BS, et al. Concordance of genetic risk across migraine subgroups: impact on current and future genetic association studies. Cephalalgia 2014;35:489–499. [DOI] [PubMed] [Google Scholar]
- 29.Ozaki K, Sato H, Inoue K, et al. SNPs in BRAP associated with risk of myocardial infarction in Asian populations. Nat Genet 2009;41:329–333. [DOI] [PubMed] [Google Scholar]
- 30.Coronary Artery Disease (C4D) Genetics Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet 2011;43:339–344. [DOI] [PubMed] [Google Scholar]
- 31.O'Donnell CJ, Kavousi M, Smith AV, et al. Genome-wide association study for coronary artery calcification with follow-up in myocardial infarction. Circulation 2011;124:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bevan S, Traylor M, Adib-Samii P, et al. Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke 2012;43:3161–3167. [DOI] [PubMed] [Google Scholar]
- 33.Allen PB, Greenfield AT, Svenningsson P, Haspeslagh DC, Greengard P. Phactrs 1-4: a family of protein phosphatase 1 and actin regulatory proteins. Proc Natl Acad Sci U S A 2004;101:7187–7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allain B, Jarray R, Borriello L, et al. Neuropilin-1 regulates a new VEGF-induced gene, Phactr-1, which controls tubulogenesis and modulates lamellipodial dynamics in human endothelial cells. Cell Signal 2012;24:214–223. [DOI] [PubMed] [Google Scholar]
- 35.Patel RS, Morris AA, Ahmed Y, et al. A genetic risk variant for myocardial infarction on chromosome 6p24 is associated with impaired central hemodynamic indexes. Am J Hypertens 2012;25:797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez-Osorio X, Sobrino T, Brea D, Martinez F, Castillo J, Leira R. Endothelial progenitor cells: a new key for endothelial dysfunction in migraine. Neurology 2012;79:474–479. [DOI] [PubMed] [Google Scholar]
- 37.Tietjen GE. Migraine as a systemic vasculopathy. Cephalalgia 2009;29:987–996. [DOI] [PubMed] [Google Scholar]
- 38.Baggio LL, Drucker DJ. Biology of incretins: GLP-1 and GIP. Gastroenterology 2007;132:2131–2157. [DOI] [PubMed] [Google Scholar]
- 39.Nyberg J, Anderson MF, Meister B, et al. Glucose-dependent insulinotropic polypeptide is expressed in adult hippocampus and induces progenitor cell proliferation. J Neurosci 2005;25:1816–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winsvold BS, Sandven I, Hagen K, Linde M, Midthjell K, Zwart JA. Migraine, headache and development of metabolic syndrome: an 11-year follow-up in the Nord-Trondelag Health Study (HUNT). Pain 2013;154:1305–1311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.