Abstract
Negative feedback regulation of cholesterol metabolism in mammalian cells ensures a proper balance of cholesterol with other membrane lipids, principal among these being the major phospholipid phosphatidylcholine (PC). Processes such as cholesterol biosynthesis and efflux, cholesteryl ester storage in lipid droplets, and uptake of plasma lipoproteins are tuned to the cholesterol/PC ratio. Cholesterol-loaded macrophages in atherosclerotic lesions display increased PC biosynthesis that buffers against elevated cholesterol levels and may also facilitate cholesterol trafficking to enhance cholesterol sensing and efflux. These same mechanisms could play a generic role in homeostatic responses to acute changes in membrane free cholesterol levels. Here, I discuss the established and emerging roles of PC metabolism in promoting intracellular cholesterol trafficking and membrane lipid homeostasis.
Keywords: cholesterol, phosphatidylcholine, lipid droplets, cholesterol efflux, Niemann, Pick C disease
Introduction
Cholesterol is an essential lipid component of mammalian cell membranes. Due to its structural properties and assembly characteristics, cholesterol modulates the fluidity of membrane bilayers and partitions into specific lipid domains that are vital to overall cell organization.1,2 The unique rigid ring structure and the small hydroxy headgroup of cholesterol allow it to readily flip-flop between leaflets of a membrane bilayer and to be absorbed and extracted from membranes with relative ease by protein carriers.3,4 Cholesterol is synthesized in the endoplasmic reticulum (ER) and peroxisomes in a multistep process involving more than 20 enzymes and is also released from cholesteryl ester (CE) in cytoplasmic lipid droplets (LDs) or lipoproteins internalized in lysosomes/late endosomes (L/LEs).3 To maintain membrane cholesterol homeostasis and prevent toxic cholesterol over-accumulation, the cell is equipped with negative feedback mechanisms involving crosstalk between the following two families of sterol-responsive transcription factors: the sterol regulatory element-binding proteins (SREBPs) that control the expression of proteins involved in cholesterol biosynthesis and CE-rich low-density lipoprotein (LDL) uptake5 and liver-X receptors (LXRs) that increase the expression of cholesterol transporters and promote the efflux of excess free cholesterol to extracellular acceptors.6 Homeostatic responses to acute changes in cellular cholesterol levels occur mainly in the ER, a cholesterol-poor organelle containing ~1% of total cellular cholesterol. Therefore, interorganelle cholesterol trafficking plays a critical role in cholesterol sensing, yet these mechanisms remain poorly understood.7,8
The heterogeneous distribution of cholesterol in cells is hypothesized to be largely dictated by its affinity for sphingomyelin (SM) and highly saturated phospholipids enriched in lateral membrane nanodomains or rafts in the plasma membrane (PM), trans-Golgi network, and endocytic membranes.9–11 A recent study using high-resolution secondary ion mass spectrometry revealed that micrometer-scale sphingolipid-enriched domains in the PM of cultured fibroblasts were not enriched in cholesterol.12 This observation should be examined further in other cell types and under different metabolic conditions. It should also be noted that membrane lipid nanodomain size is considered to be in the scale of 10–100 nm,13,14 highlighting the technical challenges in assessing their role in cellular cholesterol distribution. Other phospholipids could also play an important role, such as phosphatidylserine that has recently been shown to affect enrichment of cholesterol in the cytoplasmic leaflet of the PM.15 It has been postulated that membrane cholesterol in excess of the complexing activity of phospholipids represents the active or free cholesterol capable of dynamic movement, including transport between organelles, conversion to regulatory oxysterols, and efflux to extracellular acceptors.16 Consistent with this idea, treatment of cells with agents that disrupt membrane domains, such as SMase and oxysterols, results in a redistribution of PM cholesterol to the ER.17,18 The rapid transfer of cholesterol between cell compartments in part involves nonvesicular transport at membrane contact sites (MCSs) that allow cholesterol to freely equilibrate between donor and acceptor membranes.19 The efficiency of this mode of transport is presumably limited by free cholesterol concentrations in acceptor membranes. Therefore, membrane-driven processes that act as sinks for excess ER cholesterol have the potential to drive cholesterol transport to the ER from other locations. These cholesterol sinks include (1) ER membrane phospholipid biogenesis to absorb free cholesterol, (2) CE storage in LDs, and (3) cholesterol efflux (Fig. 1). In this commentary, I discuss the influence of the major membrane phospholipid phosphatidylcholine (PC) on these processes and thus the potential role of PC metabolic enzymes in promoting intracellular cholesterol transport and membrane cholesterol homeostasis.
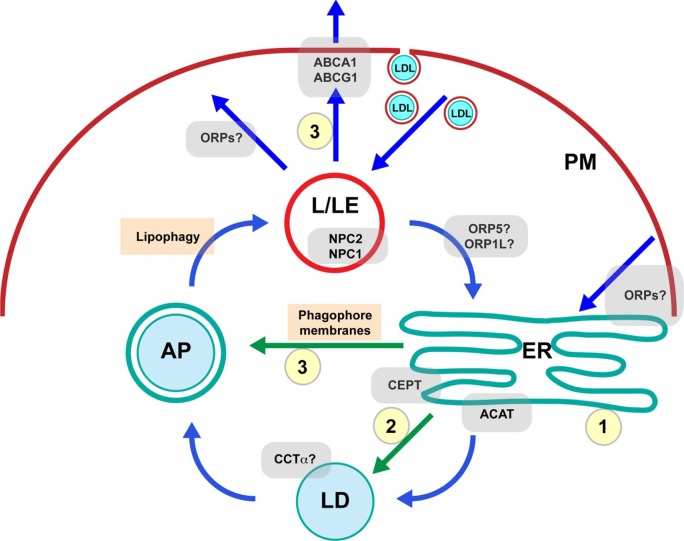
Figure 1.
A simplified model of intracellular cholesterol trafficking. PC metabolism potentially regulates several important cholesterol sinks (yellow circles) that maintain low ER cholesterol concentrations. These in turn improve cholesterol sensing and facilitate interorganelle cholesterol trafficking (blue arrows), including pathways to the ER from sites of high cholesterol concentration, such as L/LEs and PM. Trafficking of newly synthesized PC relevant to cholesterol sinks is indicated by green arrows. Cholesterol sinks regulated by PC metabolic enzymes include (1) ER membrane biogenesis and expansion, (2) CE storage in cytoplasmic LDs, and (3) cholesterol efflux involving LD turnover by lipophagy, which may involve PC synthesis in the ER to supply phagophore membranes for autophagosome (AP) biogenesis.
Abbreviations: CEPT, choline/ethanolamine phosphotransferase; CCTα, CTP:phosphocholine cytidylyltransferase α; ACAT, acyl-CoA:cholesterol acyltransferase; LDL, low-density lipoprotein; NPC1, Niemann–Pick C1; ORP, OSBP-related protein.
PC Buffers Against Excess Cholesterol in Cells
PC is the most abundant phospholipid in eukaryotic cells and has a positive influence on the incorporation of cholesterol in membranes.20,21 PC is also a precursor for the synthesis of SM by the enzyme SM synthase 1 in the Golgi apparatus22 and, thus, can indirectly affect SM-enriched nanodomains that may affect intracellular cholesterol distribution.9 PC and cholesterol contents are maintained within narrow ratios in mammalian cells, reflective of the counter-balancing effects of these lipids on membrane fluidity.2,23 As an example, cholesterol-laden macrophage foam cells within atherosclerotic lesions have highly upregulated PC biosynthesis that fuels ER membrane biogenesis to absorb excess free cholesterol.24,25 Failure in this adaptive response results in apoptotic cell death due to toxic concentrations of cholesterol in ER membranes and the resultant ER stress.26 Under basal conditions, excess free cholesterol is converted to CE by the ER-localized enzyme acyl-CoA:cholesterol acyltransferase 1/2 for storage in cytoplasmsic LDs.8 LD CE is continuously hydrolyzed and reesterified, which cycles the excess cholesterol through membrane pools for cholesterol sensing and efflux pathways.27 LDs are dynamic organelles consisting of a neutral lipid core surrounded by a membrane monolayer composed largely of PC.28 PC biosynthetic enzymes have been shown to regulate the LD size and number in a variety of cell types.29–32 There-fore, PC metabolism regulates not only the capacity of cell membranes to absorb free cholesterol but also the storage of excess cholesterol in LDs.
Enzymes of PC Metabolism
The Kennedy pathway for de novo PC synthesis consists of three enzymatic reactions for the conversion of the soluble metabolite choline to membrane PC. First, choline kinase phosphorylates choline, followed by the formation of the nucleotide intermediate cytidine diphosphate (CDP)-choline catalyzed by CTP:phosphocholine cytidylyltransferase (CCT), and finally the transfer of phosphocholine from CDP-choline to diacylglycerol (DAG) catalyzed by the ER-localized enzyme choline/ethanolamine phosphotransferase 1 (CEPT1).33,34 The rate-limiting reaction under most conditions is the formation of CDP-choline catalyzed by CCT.35 CCTα, the major ubiquitously expressed isoform, cycles between the nucleus and cytoplasm in response to cellular demands for PC.30,36,37 For instance, CCTα is cytosolic in type II alveolar cells that require large amounts of PC for secretion of the lung surfactant38 but is predominantly nuclear localized in a variety of other cells and tissues.30,39,40 CCTα is an amphitropic enzyme, existing as both soluble/inactive and membrane-bound/active forms.35,39,41 Membrane translocation occurs via an amphipathic α-helical regulatory domain (domain M) and is triggered by high levels of lipid activators, typically substrates for phospholipid biosynthesis, such as fatty acids and DAG.42 CCTα also translocates to membrane surfaces that exhibit lipid-packing defects, thus signaling the need for increased PC content.42,43 Binding of CCTα to artificial liposomes causes membrane nanotubule formation, signifying a membrane-deforming ability similar to several other lipid-binding proteins containing amphipathic helical domains.44,45 Within cells, nuclear CCTα membrane translocation induces the formation of tubular invaginations of the double membrane nuclear envelope, collectively termed the nucleoplasmic reticulum, implicated in intranuclear calcium signaling and nucleo/cytoplasmic transport.44,46 The membrane-deforming ability of CCTα could also impact extranuclear sites of membrane translocation, but this possibility remains unexplored.
The Kennedy pathway is responsible for de novo PC production in all cells and tissues, with the methylation of phosphatidylethanolamine (PE) to PC catalyzed by PE methyltransferase also playing a major role in liver and adipocytes.47 Further metabolism of de novo synthesized PC through the Lands cycle involves fatty acid removal from the sn-2 position by phospholipase A2 activity followed by reacylation by lysophosphatidylcholine acyltransferase (LPCAT) that modifies the fatty acid composition of PC to the mature form found in membranes.48,49 Stimulated PC turnover by various phospholipases produces bioactive lipids, such as phosphatidic acid, lysophosphatidylcholine, DAG, and unsaturated fatty acids.33 PC synthesis and turnover are closely regulated with the cell cycle, thus ensuring the coordination of cell growth and membrane biogenesis.33 In other conditions of stimulated membrane growth, such as ER stress-induced ER expansion, increased flux through the Kennedy pathway can produce an overshoot response.50,51 However, studies of CCTα overexpression or increased activation indicate that excess PC in cells is efficiently degraded to maintain steady-state membrane levels.52,53
The Role of PC in Cholesterol Storage in LDs
Due to the ubiquitous role of PC in membrane structure and as a source of bioactive lipids, defining the role(s) of PC metabolism in cholesterol trafficking is complex. An emerging area of interest involves the role of PC in regulating the function of LDs, metabolic organelles that control membrane lipid storage and trafficking.54 The esterification of excess cholesterol and fatty acids for storage in LDs occurs in all cell types and plays a critical role in membrane homeostasis.55 LDs consist of a neutral lipid core of triacylglycerol (TAG) and CE surrounded by a membrane monolayer composed mainly of PC with numerous associated proteins.54,56 LDs are dynamic organelles that rapidly change their size, number, and position in the cytoplasm depending on the nutritional status of the cell.28 A genome-wide RNAi screen conducted in oleate-loaded Drosophila S2 cells identified the Kennedy pathway enzymes among genes that when knocked down affected the LD morphology.57 CCTα deficiency leading to decreased PC synthesis results in the formation of large LDs in cells. This may result from the shunting of DAG to TAG synthesis58 or decreased PC on LD surfaces where it acts as a surfactant to suppress the LD fusion.29 The surfactant effect of the phospholipid monolayer lowers the exposed surface area, decreases the surface tension, and increases the stability of LDs.59 In oleate-loaded Drosophila S2 cells, the CCTα homolog CCT1 shuttled from the nucleus to the cytoplasm and localized to the surface of growing LDs.29 This suggests that CCTα can directly monitor PC content on LD surfaces in some cell types. In contrast, CCTα remained nuclear localized and translocated to the nuclear envelope during LD expansion in differentiating preadipocytes but nevertheless regulated the LD size and number by controlling the PC supply.30 It remains unknown how newly synthesized PC is transferred to the LD surface following the terminal reaction in the Kennedy pathway catalyzed by the ER-localized transmembrane enzyme CEPT1. This may involve phospholipid transfer proteins or membrane bridges between the ER and LD surface.28 Other PC metabolic enzymes have also been shown to affect the LD PC content and regulate the LD size, including PEMT31 and the Lands cycle enzymes LPCAT-1 and LPCAT-2 that localize to LDs in some cell types.32,60 Collectively, these studies support an important role of PC metabolic enzymes in regulating neutral lipid storage capacity and the hydrolytic release of stored fatty acids and cholesterol.
PC and Cholesterol Coregulation by the SREBP Pathway
The central regulators of membrane lipid homeostasis are the SREBPs, a family of transcription factors that regulate the expression of genes involved in cholesterol and fatty acid synthesis and their uptake in plasma lipoproteins.5 The SREBP isoforms have the following separable but overlapping gene targets: SREBP-1a controls genes in both cholesterol and fatty acid biosynthetic pathways, while SREBP-1c, the major SREBP-1 isoform expressed in tissues, and SREBP-2 are relatively specific for fatty acid and cholesterol synthesis, respectively.61 SREBPs are initially synthesized as inactive membrane-bound precursors localized in the ER. Vesicle-mediated transport to the Golgi apparatus results in sequential cleavage by two Golgi-resident proteases, releasing cytosolic mature forms that traffic to the nucleus and activate target genes.5 Processing of SREBPs is regulated by the cholesterol/phospholipid ratio in ER membranes, where a switch-like change in the chemical activity of cholesterol occurs above a threshold of ~5 mol%.62 The cholesterol sensor is SREBP-cleavage-activating protein (SCAP) that binds to and escorts SREBPs from the ER to the Golgi.63 Direct binding of cholesterol to SCAP induces a conformation change and binding to insulin-induced gene protein (Insig).64 This results in retention of the SCAP/SREBP complex in the ER, thus preventing the proteolytic activation of SREBPs at the Golgi.65
Differential regulation of SREBP-1 and SREBP-2 isoforms occurs in cultured cells and in liver where SREBP-1c is highly upregulated as compared to SREBP-2 at both the mRNA and processing levels in response to insulin signaling.66,67 Unsaturated fatty acids were shown to be required to fully suppress the processing of SREBP-1 isoforms but not SREBP-2 in the presence of sterols in HEK 293 cells.68 Recently, it was demonstrated that PC synthesis via the Kennedy pathway negatively affects the proteolytic activation of SREBP-1 isoforms in C. elegans, human HepG2 cells, and mouse liver.69 SREBP-1c exerts significant control of PC metabolism in an indirect manner by controlling the production of unsaturated fatty acids, which serve as membrane lipid activators of CCTα and as well as Kennedy pathway substrate.70 This suggests a negative feedback loop, whereby increased mature SREBP-1c stimulates unsaturated fatty acid and PC synthesis that in turn suppresses the precursor SREBP-1c processing. The requirement of a phospholipid-derived signal for the complete suppression of SREBP-1c processing could be important for the maintenance of phospholipid synthesis and membrane balance under conditions of elevated cellular cholesterol levels. In Drosophila, the phospholipid PE controls proteolytic processing of the single SREBP ortholog.71 Drosophila, like other insects, are auxotrophic for sterols, and PE comprises >50% of the total phospholipids.72 Interestingly, PE and cholesterol share the physical property of promoting the hexagonal HII-phase formation in artificial bilayer systems.73 Thus, it is possible that SREBP processing is affected by perturbations in the membrane environment surrounding SCAP. This could in turn be regulated by PE/PC balance in ER membranes.69
Regulation of PC Synthesis by Oxysterols and the LXR Pathway
A portion of excess cholesterol in cells is channeled to mitochondria for the synthesis of various oxysterols with altered physiochemical properties compared to cholesterol, such as enhanced solubility and spontaneous diffusion across cellular membranes.74,75 The majority of cholesterol turnover in the body takes place in the liver where 7α-hydroxylated sterols are substrates for the synthesis of bile acids.76 In the brain, turnover of excess cholesterol involves its conversion to 24-hydroxycho-lesterol, followed by secretion and clearance from the circulation by the liver for incorporation into the bile acid biosynthetic pathway.77,78 In all cells and tissues, oxysterols serve as potent signaling molecules that improve cell and whole body membrane homeostasis through several mechanisms.74 For instance, oxysterols induce degradation of 3-hydroxy 3-methylglutaryl (HMG)-CoA reductase, the rate-limiting enzyme in cholesterol biosynthesis, and suppress cholesterol biosynthetic gene expression by inhibiting SREBP-2 proteolytic activation.79,80 Although cholesterol itself is a poor membrane activator of CCTα, the oxysterol 25-hydroxycholesterol directly stimulates CCTα activity in vitro and induces CCTα membrane translocation and activation in cultured cells.81
A primary signaling function of oxysterols is as ligand activators of LXRs (LXR-α and LXR-β), transcription factors that increase the expression of cholesterol transporters, such as the ATP-binding cassette transporter ABCA1, along with other proteins involved in cholesterol efflux.82 Through a process known as reverse cholesterol transport (RCT), excess cholesterol from peripheral tissues is effluxed to high-density lipoprotein acceptors and transported to the liver for eventual excretion in the bile. The protective effects of RCT are especially vital for the removal of cholesterol from the intimal space of the vasculature, where macrophages are prone to becoming atherogenic foam cells through the scavenging of oxidized LDL particles.83 Accordingly, LXRs have emerged as important therapeutic targets to inhibit atherosclerosis by promoting RCT.84 However, a caveat is that SREBP-1c is also an LXR target gene. As a result, treatment with synthetic LXR agonists leads to a detrimental increase in liver lipogenesis accompanied by hypertriglyceridemia and hepatic steatosis in animal models tested.85 The role of SREBP-1c induction in the LXR gene regulatory program remains unclear. As noted earlier, SREBP-1c activity produces membrane activators of CCTα along with substrates for de novo PC synthesis.70 Thus, one possibility is that increased lipogenesis under conditions of excess cholesterol improves the ratio of free cholesterol with membrane phospholipids.86 The increased fatty acid and TAG production could also detoxify the effect of elevated cholesterol by promoting its storage as CE in cytoplasmic LDs.87
Potential Role of Membrane PC Synthesis in Lipophagy
Autophagy is a constitutive degradative pathway conserved from yeast to humans that plays an essential role in the maintenance of cellular energy and metabolic homeostasis. Autophagic (self-eating) degradation involves engulfment of targeted cytosolic components by phagocytic membranes to form an autophagosome (AP) that fuses with L/LEs where the contents are degraded, releasing nutrients and other building blocks for use in cellular metabolism.88 A specialized form of autophagy known as lipophagy degrades cytoplasmic LDs, thus liberating stored fatty acids for energy production during starvation.89 Recently, lipophagy was shown to play a critical role in cholesterol efflux from macrophage foam cells, whereby lysosomal acid lipase hydrolyzes LD CE releasing free cholesterol for ABCA1-mediated efflux.90 Membranes required for AP formation can arise from various sources depending on the cellular context. In starvation-induced macroautophagy, this may involve physical remodeling of existing membranes originating at the ER,91 ER–mitochondria contact sites,92,93 Golgi apparatus,94 or recycling endosomes.95,96 Under nutrient replete conditions, AP biogenesis has been shown to involve hydrolysis of TAG stores in LDs, releasing fatty acids to supply de novo PC synthesis in the ER.97 Formation of APs to engulf large CE-rich LDs may also require lipogenesis to provide the needed substrate for new membrane lipid synthesis. Thus, SREBP-1c induction leading to CCTα activation and increased de novo PC synthesis could support lipophagy, identified as an essential early step in LXR-stimulated cholesterol efflux from macrophage foam cells.90 While LD degradation by lipophagy has been demonstrated in extreme conditions, such as starvation89 and cholesterol-laden macrophage foam cells,90 a generic role in membrane homeostasis remains undefined. Importantly, a recent report showed that SREBP-2 regulates the expression of several protein factors involved in autophagy.98 This points to a potential role of lipophagy in mobilizing LD CE stores to augment membrane free cholesterol pools under cholesterol-depleted conditions, allowing a more rapid response as compared to CE hydrolysis by a cytoplasmic neutral CE hydrolase. If so, the potential roles of PC metabolism in AP formation and lipophagy may not only apply to extreme situations of cholesterol loading in cells but also to cholesterol sensing and membrane cholesterol homeostasis under basal conditions.
Transport of Cholesterol From L/LEs
An important cholesterol trafficking pathway originates in L/LEs where hydrolytic breakdown of the CE component of internalized LDL releases free cholesterol for use in cell metabolism.3 PM cholesterol internalized in endocytic vesicles is also processed in the L/LE compartment for recycling to intracellular membranes.99 An initial step is the binding of free cholesterol by the soluble luminal protein Niemann–Pick C (NPC) 2, followed by handoff of the cholesterol molecule to the polytopic membrane protein NPC1 for transfer to unknown cytosolic binding proteins or insertion into the L/LE-limiting membrane.100 NPC disease is a progressive fatal disorder associated with neurodegeneration and liver dysfunction caused by loss-of-function mutation in either NPC1 (~95% of cases) or NPC2. NPC disease is characterized morphologically in cells and tissues by the accumulation of free cholesterol and glycosphingolipids in the luminal L/LE compartment.101 Interestingly, transport of cholesterol from lysosomes to mitochondria was found to be not defective in NPC1-deficient Chinese hamster ovary cells.102 Instead, the endolysosomal membrane protein meta-static lymph node protein 64, a cholesterol-binding member of the steroidogenic acute regulatory-related lipid transfer domain-containing protein family, participated with NPC2 to transfer LDL-derived free cholesterol to mitochondria.102 In vitro studies have also shown that recombinant NPC2 can directly transfer cholesterol to phospholipid membranes, independent of NPC1.103,104 The reason for the additional requirement of NPC1 in cells remains unknown, although one likely aspect is to chaperone the hydrophobic cholesterol molecule across a glycocalyx that lines the interior of lysosomes.99,105 The handoff from NPC2 to NPC1 may also improve the kinetics of cholesterol insertion into membranes against a concentration gradient.104 In this scenario, the requirement of NPC1 would be partially dependent on the downstream export of cholesterol that effectively lowers the cholesterol concentration in the L/LE-limiting membrane.106
The cholesterol trafficking pathways from L/LEs to other organelles remain largely unknown but involve both vesicular and nonvesicular mechanisms.3,107,108 Potentially important players are members of the oxysterol-binding-protein (OSBP)-related protein (ORP) family, known to participate in nonvesicular transfer of sterols and other lipids at interorganelle MCSs.109,110 OSBP along with several other cytoplasmic ORPs binds to oxysterols, which induces membrane translocation in conjunction with specific organelle-targeting signals.111 Several ORPs have recently been implicated in the nonvesicular export of cholesterol from L/LEs. In one study, ORP1L was shown to interact with the ER resident protein, vesicle-associated membrane protein-associated protein, to regulate L/LE positioning in response to cholesterol depletion and L/LE-to-ER MCSs in human MelJuSo melanoma cells.112 Knockdown of ORP5, a membrane-anchored ORP localized in the ER, caused L/LE accumulation of LDL-derived free cholesterol in HeLa cells, mimicking the NPC phenotype.113 Detailed confocal microscopy analysis revealed that accumulation of free cholesterol occurred in the L/LE-limiting membrane in addition to the luminal compartment.113 Therefore, ORP5 may function downstream of NPC1 to mediate the exit of LDL-derived cholesterol from L/LEs to the ER. A recent study showed that L/LE-to-peroxisome transport of cholesterol occurs via a MCS involving the L/LE membrane protein synaptotagmin VII binding to peroxisomal PI(4,5)P2.108 Knock-down of critical peroxisomal genes in HEK 293 cells blocked L/LE cholesterol trafficking and caused L/LE accumulation of LDL-derived cholesterol. In addition, L/LE cholesterol accumulation occurred in cell and animal models of various peroxisomal disorders.108 Importantly, these results point to a previously unappreciated role for peroxisomes as an important conduit for L/LE cholesterol export and, further, that this transport is mediated through MCSs.108
Potential Roles of Cholesterol Sinks in NPC Disease Reversal
Various manipulations in lipid metabolism or membrane trafficking have been shown to reverse L/LE cholesterol accumulation in NPC1-deficient cells.114–119 Several of these mechanisms appear to act at extra-lysosomal sites and, thus, may relieve bottlenecks in the intracellular movement of cholesterol. Conversely, the accumulation of lipoprotein-derived cholesterol in L/LEs has been shown to occur in cells overloaded with free cholesterol in the absence of NPC protein defects.120,121 This suggests that the export of free cholesterol from L/LEs becomes rate limiting upon saturation of cholesterol levels in downstream membranes. If, as recent studies suggest, a substantial amount of intracellular cholesterol trafficking involves transport down a chemical concentration gradient at interorganelle MCSs,19,110,122 then lower cholesterol concentrations in acceptor membranes would facilitate cholesterol movement. In this model, mechanisms that lower ER cholesterol concentrations would drive cholesterol trafficking to the ER from distal sites, leading to improved cholesterol sensing and homeostasis. As detailed herein, PC metabolism is intimately linked with several cholesterol sinks that may facilitate cholesterol trafficking to the ER by this mechanism, specifically (1) expansion of ER membranes to absorb free cholesterol, (2) LD CE storage, and (3) cholesterol efflux (Fig. 1). Increased flux in these pathways could conceivably alleviate the cholesterol trafficking block in NPC1-deficient cells by improving the kinetics of NPC2-mediated cholesterol transfer. Indeed, one study showed that ABCA1 overexpression reversed the NPC disease phenotype in human fibroblasts deficient in NPC1 but not in NPC2.117 This effect was highest upon addition of the cholesterol efflux acceptor apoA-I to the culture medium, suggesting that increased ABCA1-mediated cholesterol efflux provided the needed cholesterol sink to bypass the requirement for NPC1. However, it is also possible that the overexpressed ABCA1 acted directly in cholesterol transfer at the L/LE membrane in cooperation with NPC2.117 Clearly, further studies are needed to investigate whether reversal of the cholesterol trafficking defect in NPC1-deficient cells and tissues requires treatments that compensate directly within the L/LE compartment or whether downstream cholesterol sinks can shift the rate-limiting step and improve the kinetics of NPC2-dependent cholesterol transfer. In this regard, the role of PC metabolism in L/LE cholesterol export could be tested in cell models in which PC synthesis is upregulated and/or PC turnover is inhibited, leading to an increase in the cellular PC/cholesterol ratio of downstream membranes. Conversely, knockdown of key enzymes in the Kennedy pathway, such as CCTα and CEPT1, could lead to decreased trafficking of LDL-derived cholesterol from L/LEs and mimic the NPC phenotype.
Conclusion
The maintenance of cholesterol/PC ratios in cell membranes is vital to membrane integrity and fluidity and promotes stability and function of transmembrane proteins. Critical increases in ER cholesterol concentrations result in depletion of ER calcium stores and activation of ER stress pathways that can lead to apoptotic cell death.26 Sequestration of excess free cholesterol in L/LEs could protect the cell against cholesterol toxicity and ER stress in the short term, but the long-term consequence is pathological L/LE dysfunction, as seen in various neurodegenerative lipid storage disorders.123 L/LE cholesterol accumulation in tissue macrophages in the vessel wall and liver has also been linked to detrimental inflammatory responses in cardiovascular disease and nonalcoholic steatohepatitis, respectively.124,125 PC plays a critical role in cellular cholesterol sinks that buffer against cholesterol-induced ER stress and assist in the maintenance of cellular cholesterol gradients that drive interorganelle cholesterol transport. Therefore, the enzymes of PC metabolism potentially represent control points affecting cholesterol trafficking and storage and, ultimately, cholesterol homeostasis.
Footnotes
ACADEMIC EDITOR: Tim Levine, Editor in Chief
PEER REVIEW: One peer reviewer contributed to the peer review report. Reviewers’ reports totaled 414 words, excluding any confidential comments to the academic editor.
FUNDING: TAL is supported by operating grants from the Canadian Institutes of Health Research and the Heart and Stroke Foundation of Canada and in part by a Heart and Stroke Foundation of Canada New Investigator Award. The author confirms that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Author discloses no potential conflicts of interest.
Paper subject to independent expert single-blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived the concepts: TAL. Wrote the first draft of the manuscript: TAL. Developed the structure and arguments for the paper: TAL. Made critical revisions: TAL. The author reviewed and approved of the final manuscript.
REFERENCES
- 1.Maxfield FR, van Meer G. Cholesterol, the central lipid of mammalian cells. Curr Opin Cell Biol. 2010;22(4):422–429. doi: 10.1016/j.ceb.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yeagle PL. Lipid regulation of cell membrane structure and function. FASEB J. 1989;3(7):1833–1842. [PubMed] [Google Scholar]
- 3.Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nat Rev Mol Cell Biol. 2008;9(2):125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- 4.Iaea DB, Maxfield FR. Cholesterol trafficking and distribution. Essays Biochem. 2015;57:43–55. doi: 10.1042/bse0570043. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124(1):35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 6.Repa JJ, Mangelsdorf DJ. The liver X receptor gene team: potential new players in atherosclerosis. Nat Med. 2002;8(11):1243–1248. doi: 10.1038/nm1102-1243. [DOI] [PubMed] [Google Scholar]
- 7.Maxfield FR, Wustner D. Intracellular cholesterol transport. J Clin Invest. 2002;110(7):891–898. doi: 10.1172/JCI16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang TY, Chang CC, Ohgami N, Yamauchi Y. Cholesterol sensing, trafficking, and esterification. Annu Rev Cell Dev Biol. 2006;22:129–157. doi: 10.1146/annurev.cellbio.22.010305.104656. [DOI] [PubMed] [Google Scholar]
- 9.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327(5961):46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 10.Klemm RW, Ejsing CS, Surma MA, et al. Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J Cell Biol. 2009;185(4):601–612. doi: 10.1083/jcb.200901145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gagescu R, Demaurex N, Parton RG, Hunziker W, Huber LA, Gruenberg J. The recycling endosome of Madin-Darby canine kidney cells is a mildly acidic compartment rich in raft components. Mol Biol Cell. 2000;11(8):2775–2791. doi: 10.1091/mbc.11.8.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frisz JF, Klitzing HA, Lou K, et al. Sphingolipid domains in the plasma membranes of fibroblasts are not enriched with cholesterol. J Biol Chem. 2013;288(23):16855–16861. doi: 10.1074/jbc.M113.473207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan C, Furlong J, Burgos P, Johnston LJ. The size of lipid rafts: an atomic force microscopy study of ganglioside GM1 domains in sphingomyelin/DOPC/cholesterol membranes. Biophys J. 2002;82(5):2526–2535. doi: 10.1016/S0006-3495(02)75596-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eggeling C, Ringemann C, Medda R, et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457(7233):1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 15.Maekawa M, Fairn GD. Complementary probes reveal that phosphatidylserine is required for the proper transbilayer distribution of cholesterol. J Cell Sci. 2015;128(7):1422–1433. doi: 10.1242/jcs.164715. [DOI] [PubMed] [Google Scholar]
- 16.Steck TL, Lange Y. Cell cholesterol homeostasis: mediation by active cholesterol. Trends Cell Biol. 2010;20(11):680–687. doi: 10.1016/j.tcb.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slotte JP, Hedstrom G, Rannstrom S, Ekman S. Effects of sphingomyelin degradation on cell cholesterol oxidizability and steady-state distribution between the cell surface and the cell interior. Biochim Biophys Acta. 1989;985(1):90–96. doi: 10.1016/0005-2736(89)90108-9. [DOI] [PubMed] [Google Scholar]
- 18.Lange Y, Ye J, Rigney M, Steck TL. Regulation of endoplasmic reticulum cholesterol by plasma membrane cholesterol. J Lipid Res. 1999;40(12):2264–2270. [PubMed] [Google Scholar]
- 19.Baumann NA, Sullivan DP, Ohvo-Rekila H, et al. Transport of newly synthesized sterol to the sterol-enriched plasma membrane occurs via nonvesicular equilibration. Biochemistry. 2005;44(15):5816–5826. doi: 10.1021/bi048296z. [DOI] [PubMed] [Google Scholar]
- 20.Lange Y, Tabei SM, Ye J, Steck TL. Stability and stoichiometry of bilayer phospholipid-cholesterol complexes: relationship to cellular sterol distribution and homeostasis. Biochemistry. 2013;52(40):6950–6959. doi: 10.1021/bi400862q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ali MR, Cheng KH, Huang J. Assess the nature of cholesterol-lipid interactions through the chemical potential of cholesterol in phosphatidylcholine bilayers. Proc Natl Acad Sci U S A. 2007;104(13):5372–5377. doi: 10.1073/pnas.0611450104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huitema K, van den Dikkenberg J, Brouwers JF, Holthuis JC. Identification of a family of animal sphingomyelin synthases. EMBO J. 2004;23(1):33–44. doi: 10.1038/sj.emboj.7600034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ridgway ND, Byers DM, Cook HW, Storey MK. Integration of phospholipid and sterol metabolism in mammalian cells. Prog Lipid Res. 1999;38(4):337–360. doi: 10.1016/s0163-7827(99)00007-7. [DOI] [PubMed] [Google Scholar]
- 24.Tabas I. Phospholipid metabolism in cholesterol-loaded macrophages. Curr Opin Lipidol. 1997;8(5):263–267. doi: 10.1097/00041433-199710000-00004. [DOI] [PubMed] [Google Scholar]
- 25.Tabas I. Consequences of cellular cholesterol accumulation: basic concepts and physiological implications. J Clin Invest. 2002;110(7):905–911. doi: 10.1172/JCI16452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng B, Yao PM, Li Y, et al. The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat Cell Biol. 2003;5(9):781–792. doi: 10.1038/ncb1035. [DOI] [PubMed] [Google Scholar]
- 27.Brown MS, Ho YK, Goldstein JL. The cholesteryl ester cycle in macrophage foam cells. Continual hydrolysis and re-esterification of cytoplasmic cholesteryl esters. J Biol Chem. 1980;255(19):9344–9352. [PubMed] [Google Scholar]
- 28.Walther TC, Farese RV., Jr The life of lipid droplets. Biochim Biophys Acta. 2009;1791(6):459–466. doi: 10.1016/j.bbalip.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krahmer N, Guo Y, Wilfling F, et al. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 2011;14(4):504–515. doi: 10.1016/j.cmet.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aitchison AJ, Arsenault DJ, Ridgway ND. Nuclear-localized CTP: phosphocholine cytidylyltransferase alpha regulates phosphatidylcholine synthesis required for lipid droplet biogenesis. Mol Biol Cell. 2015;26(16):2927–2938. doi: 10.1091/mbc.E15-03-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horl G, Wagner A, Cole LK, et al. Sequential synthesis and methylation of phosphatidylethanolamine promote lipid droplet biosynthesis and stability in tissue culture and in vivo. J Biol Chem. 2011;286(19):17338–17350. doi: 10.1074/jbc.M111.234534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moessinger C, Kuerschner L, Spandl J, Shevchenko A, Thiele C. Human lysophosphatidylcholine acyltransferases 1 and 2 are located in lipid droplets where they catalyze the formation of phosphatidylcholine. J Biol Chem. 2011;286(24):21330–21339. doi: 10.1074/jbc.M110.202424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fagone P, Jackowski S. Phosphatidylcholine and the CDP-choline cycle. Biochim Biophys Acta. 2013;1831(3):523–532. doi: 10.1016/j.bbalip.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henneberry AL, Wright MM, McMaster CR. The major sites of cellular phospholipid synthesis and molecular determinants of fatty acid and lipid head group specificity. Mol Biol Cell. 2002;13(9):3148–3161. doi: 10.1091/mbc.01-11-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cornell RB, Ridgway ND. CTP:phosphocholine cytidylyltransferase: function, regulation, and structure of an amphitropic enzyme required for membrane biogenesis. Prog Lipid Res. 2015;59:147–171. doi: 10.1016/j.plipres.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Northwood IC, Tong AH, Crawford B, Drobnies AE, Cornell RB. Shuttling of CTP:phosphocholine cytidylyltransferase between the nucleus and endoplasmic reticulum accompanies the wave of phosphatidylcholine synthesis during the G(0) –. G(1) transition. J Biol Chem. 1999;274(37):26240–26248. doi: 10.1074/jbc.274.37.26240. [DOI] [PubMed] [Google Scholar]
- 37.Houweling M, Cui Z, Anfuso CD, Bussiere M, Chen MH, Vance DE. CTP: phosphocholine cytidylyltransferase is both a nuclear and cytoplasmic protein in primary hepatocytes. Eur J Cell Biol. 1996;69(1):55–63. [PubMed] [Google Scholar]
- 38.Ridsdale R, Tseu I, Wang J, Post M. CTP:phosphocholine cytidylyltransferase alpha is a cytosolic protein in pulmonary epithelial cells and tissues. J Biol Chem. 2001;276(52):49148–49155. doi: 10.1074/jbc.M103566200. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Sweitzer TD, Weinhold PA, Kent C. Nuclear localization of soluble CTP:phosphocholine cytidylyltransferase. J Biol Chem. 1993;268(8):5899–5904. [PubMed] [Google Scholar]
- 40.DeLong CJ, Qin L, Cui Z. Nuclear localization of enzymatically active green fluorescent protein- CTP:phosphocholine cytidylyltransferase alpha fusion protein is independent of cell cycle conditions and cell types. J Biol Chem. 2000;275(41):32325–32330. doi: 10.1074/jbc.M004644200. [DOI] [PubMed] [Google Scholar]
- 41.Huang HK, Taneva SG, Lee J, Silva LP, Schriemer DC, Cornell RB. The membrane-binding domain of an amphitropic enzyme suppresses catalysis by contact with an amphipathic helix flanking its active site. J Mol Biol. 2013;425(9):1546–1564. doi: 10.1016/j.jmb.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 42.Cornell RB, Taneva SG. Amphipathic helices as mediators of the membrane interaction of amphitropic proteins, and as modulators of bilayer physical properties. Curr Protein Pept Sci. 2006;7(6):539–552. doi: 10.2174/138920306779025675. [DOI] [PubMed] [Google Scholar]
- 43.Cornell RB, Northwood IC. Regulation of CTP:phosphocholine cytidylyltransferase by amphitropism and relocalization. Trends Biochem Sci. 2000;25(9):441–447. doi: 10.1016/s0968-0004(00)01625-x. [DOI] [PubMed] [Google Scholar]
- 44.Lagace TA, Ridgway ND. The rate-limiting enzyme in phosphatidylcholine synthesis regulates proliferation of the nucleoplasmic reticulum. Mol Biol Cell. 2005;16(3):1120–1130. doi: 10.1091/mbc.E04-10-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taneva SG, Lee JM, Cornell RB. The amphipathic helix of an enzyme that regulates phosphatidylcholine synthesis remodels membranes into highly curved nanotubules. Biochim Biophys Acta. 2012;1818(5):1173–1186. doi: 10.1016/j.bbamem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 46.Goulbourne CN, Malhas AN, Vaux DJ. The induction of a nucleoplasmic reticulum by prelamin A accumulation requires CTP:phosphocholine cytidylyltransferasealpha. J Cell Sci. 2011;124(pt 24):4253–4266. doi: 10.1242/jcs.091009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vance DE, Walkey CJ, Cui Z. Phosphatidylethanolamine N-methyltransferase from liver. Biochim Biophys Acta. 1997;1348(1–2):142–150. doi: 10.1016/s0005-2760(97)00108-2. [DOI] [PubMed] [Google Scholar]
- 48.Lands WE. Metabolism of glycerolipides; a comparison of lecithin and triglyceride synthesis. J Biol Chem. 1958;231(2):883–888. [PubMed] [Google Scholar]
- 49.Lands WE. Stories about acyl chains. Biochim Biophys Acta. 2000;1483(1):1–14. doi: 10.1016/s1388-1981(99)00177-8. [DOI] [PubMed] [Google Scholar]
- 50.Maiuolo J, Bulotta S, Verderio C, Benfante R, Borgese N. Selective activation of the transcription factor ATF6 mediates endoplasmic reticulum proliferation triggered by a membrane protein. Proc Natl Acad Sci U S A. 2011;108(19):7832–7837. doi: 10.1073/pnas.1101379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Higgins JA. Studies on the biogenesis of smooth endoplasmic reticulum membranes in hepatocytes of phenobarbital-treated rats. II. The site of phospholipid synthesis in the initial phase of membrane proliferation. J Cell Biol. 1974;62(3):635–646. doi: 10.1083/jcb.62.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baburina I, Jackowski S. Cellular responses to excess phospholipid. J Biol Chem. 1999;274(14):9400–9408. doi: 10.1074/jbc.274.14.9400. [DOI] [PubMed] [Google Scholar]
- 53.Lagace TA, Storey MK, Ridgway ND. Regulation of phosphatidylcholine metabolism in Chinese hamster ovary cells by the sterol regulatory element-binding protein (SREBP)/SREBP cleavage-activating protein pathway. J Biol Chem. 2000;275(19):14367–14374. doi: 10.1074/jbc.275.19.14367. [DOI] [PubMed] [Google Scholar]
- 54.Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RG. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem. 2004;279(5):3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- 55.Listenberger LL, Han X, Lewis SE, et al. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci U S A. 2003;100(6):3077–3082. doi: 10.1073/pnas.0630588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J Biol Chem. 2002;277(46):44507–44512. doi: 10.1074/jbc.M207712200. [DOI] [PubMed] [Google Scholar]
- 57.Guo Y, Walther TC, Rao M, et al. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453(7195):657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jackowski S, Wang J, Baburina I. Activity of the phosphatidylcholine biosynthetic pathway modulates the distribution of fatty acids into glycerolipids in proliferating cells. Biochim Biophys Acta. 2000;1483(3):301–315. doi: 10.1016/s1388-1981(99)00203-6. [DOI] [PubMed] [Google Scholar]
- 59.Thiam AR, Farese RV, Jr, Walther TC. The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol. 2013;14(12):775–786. doi: 10.1038/nrm3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moessinger C, Klizaite K, Steinhagen A, et al. Two different pathways of phosphatidylcholine synthesis, the Kennedy pathway and the lands cycle, differentially regulate cellular triacylglycerol storage. BMC Cell Biol. 2014;15:43. doi: 10.1186/s12860-014-0043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horton JD. Sterol regulatory element-binding proteins: transcriptional activators of lipid synthesis. Biochem Soc Trans. 2002;30(pt 6):1091–1095. doi: 10.1042/bst0301091. [DOI] [PubMed] [Google Scholar]
- 62.Sokolov A, Radhakrishnan A. Accessibility of cholesterol in endoplasmic reticulum membranes and activation of SREBP-2 switch abruptly at a common cholesterol threshold. J Biol Chem. 2010;285(38):29480–29490. doi: 10.1074/jbc.M110.148254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nohturfft A, DeBose-Boyd RA, Scheek S, Goldstein JL, Brown MS. Sterols regulate cycling of SREBP cleavage-activating protein (SCAP) between endoplasmic reticulum and Golgi. Proc Natl Acad Sci U S A. 1999;96(20):11235–11240. doi: 10.1073/pnas.96.20.11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Radhakrishnan A, Goldstein JL, McDonald JG, Brown MS. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 2008;8(6):512–521. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun LP, Seemann J, Goldstein JL, Brown MS. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc Natl Acad Sci U S A. 2007;104(16):6519–6526. doi: 10.1073/pnas.0700907104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sheng Z, Otani H, Brown MS, Goldstein JL. Independent regulation of sterol regulatory element-binding proteins 1 and 2 in hamster liver. Proc Natl Acad Sci U S A. 1995;92(4):935–938. doi: 10.1073/pnas.92.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Owen JL, Zhang Y, Bae SH, et al. Insulin stimulation of SREBP-1c processing in transgenic rat hepatocytes requires p70 S6-kinase. Proc Natl Acad Sci U S A. 2012;109(40):16184–16189. doi: 10.1073/pnas.1213343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hannah VC, Ou J, Luong A, Goldstein JL, Brown MS. Unsaturated fatty acids downregulate SREBP isoforms 1a and 1c by two mechanisms in HEK-293 cells. J Biol Chem. 2001;276(6):4365–4372. doi: 10.1074/jbc.M007273200. [DOI] [PubMed] [Google Scholar]
- 69.Walker AK, Jacobs RL, Watts JL, et al. A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell. 2011;147(4):840–852. doi: 10.1016/j.cell.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ridgway ND, Lagace TA. Regulation of the CDP-choline pathway by sterol regulatory element binding proteins involves transcriptional and post-transcriptional mechanisms. Biochem J. 2003;372(pt 3):811–819. doi: 10.1042/BJ20030252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dobrosotskaya IY, Seegmiller AC, Brown MS, Goldstein JL, Rawson RB. Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila. Science. 2002;296(5569):879–883. doi: 10.1126/science.1071124. [DOI] [PubMed] [Google Scholar]
- 72.Jones HE, Harwood JL, Bowen ID, Griffiths G. Lipid composition of subcellular membranes from larvae and prepupae of Drosophila melanogaster. Lipids. 1992;27(12):984–987. doi: 10.1007/BF02535576. [DOI] [PubMed] [Google Scholar]
- 73.Hayakawa E, Naganuma M, Mukasa K, Shimozawa T, Araiso T. Change of motion and localization of cholesterol molecule during L(alpha)-H(II) transition. Biophys J. 1998;74(2 pt 1):892–898. doi: 10.1016/S0006-3495(98)74012-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brown AJ, Jessup W. Oxysterols: sources, cellular storage and metabolism, and new insights into their roles in cholesterol homeostasis. Mol Aspects Med. 2009;30(3):111–122. doi: 10.1016/j.mam.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 75.Bauman DR, Bitmansour AD, McDonald JG, Thompson BM, Liang G, Russell DW. 25-Hydroxycholesterol secreted by macrophages in response to toll-like receptor activation suppresses immunoglobulin A production. Proc Natl Acad Sci U S A. 2009;106(39):16764–16769. doi: 10.1073/pnas.0909142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Russell DW. Fifty years of advances in bile acid synthesis and metabolism. J Lipid Res. 2009;50(Suppl):S120–S125. doi: 10.1194/jlr.R800026-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lund EG, Xie C, Kotti T, Turley SD, Dietschy JM, Russell DW. Knockout of the cholesterol 24-hydroxylase gene in mice reveals a brain-specific mechanism of cholesterol turnover. J Biol Chem. 2003;278(25):22980–22988. doi: 10.1074/jbc.M303415200. [DOI] [PubMed] [Google Scholar]
- 78.Russell DW, Halford RW, Ramirez DM, Shah R, Kotti T. Cholesterol 24-hydroxylase: an enzyme of cholesterol turnover in the brain. Annu Rev Biochem. 2009;78:1017–1040. doi: 10.1146/annurev.biochem.78.072407.103859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sever N, Yang T, Brown MS, Goldstein JL, DeBose-Boyd RA. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol Cell. 2003;11(1):25–33. doi: 10.1016/s1097-2765(02)00822-5. [DOI] [PubMed] [Google Scholar]
- 80.Radhakrishnan A, Ikeda Y, Kwon HJ, Brown MS, Goldstein JL. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: oxysterols block transport by binding to Insig. Proc Natl Acad Sci U S A. 2007;104(16):6511–6518. doi: 10.1073/pnas.0700899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gehrig K, Lagace TA, Ridgway ND. Oxysterol activation of phosphatidylcholine synthesis involves CTP:phosphocholine cytidylyltransferase alpha translocation to the nuclear envelope. Biochem J. 2009;418(1):209–217. doi: 10.1042/BJ20081923. [DOI] [PubMed] [Google Scholar]
- 82.Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294(5548):1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 83.Levin N, Bischoff ED, Daige CL, et al. Macrophage liver X receptor is required for antiatherogenic activity of LXR agonists. Arterioscler Thromb Vasc Biol. 2005;25(1):135–142. doi: 10.1161/01.ATV.0000150044.84012.68. [DOI] [PubMed] [Google Scholar]
- 84.Joseph SB, McKilligin E, Pei L, et al. Synthetic LXR ligand inhibits the development of atherosclerosis in mice. Proc Natl Acad Sci U S A. 2002;99(11):7604–7609. doi: 10.1073/pnas.112059299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schultz JR, Tu H, Luk A, et al. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14(22):2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Repa JJ, Liang G, Ou J, et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRa and LXRb. Genes Dev. 2000;14(22):2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ignatova ID, Angdisen J, Moran E, Schulman IG. Differential regulation of gene expression by LXRs in response to macrophage cholesterol loading. Mol Endocrinol. 2013;27(7):1036–1047. doi: 10.1210/me.2013-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol. 2015;16(8):461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- 89.Singh R, Kaushik S, Wang Y, et al. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13(6):655–667. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11(12):1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 92.Hamasaki M, Furuta N, Matsuda A, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495(7441):389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 93.Hailey DW, Rambold AS, Satpute-Krishnan P, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141(4):656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yen WL, Shintani T, Nair U, et al. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol. 2010;188(1):101–114. doi: 10.1083/jcb.200904075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 2013;154(6):1285–1299. doi: 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Knaevelsrud H, Soreng K, Raiborg C, et al. Membrane remodeling by the PX-BAR protein SNX18 promotes autophagosome formation. J Cell Biol. 2013;202(2):331–349. doi: 10.1083/jcb.201205129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dupont N, Chauhan S, Arko-Mensah J, et al. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr Biol. 2014;24(6):609–620. doi: 10.1016/j.cub.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Seo YK, Jeon TI, Chong HK, Biesinger J, Xie X, Osborne TF. Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell Metab. 2011;13(4):367–375. doi: 10.1016/j.cmet.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gallala HD, Breiden B, Sandhoff K. Regulation of the NPC2 protein-mediated cholesterol trafficking by membrane lipids. J Neurochem. 2011;116(5):702–707. doi: 10.1111/j.1471-4159.2010.07014.x. [DOI] [PubMed] [Google Scholar]
- 100.Kwon HJ, Abi-Mosleh L, Wang ML, et al. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 2009;137(7):1213–1224. doi: 10.1016/j.cell.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Karten B, Peake KB, Vance JE. Mechanisms and consequences of impaired lipid trafficking in Niemann-Pick type C1-deficient mammalian cells. Biochim Biophys Acta. 2009;1791(7):659–670. doi: 10.1016/j.bbalip.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 102.Charman M, Kennedy BE, Osborne N, Karten B. MLN64 mediates egress of cholesterol from endosomes to mitochondria in the absence of functional Niemann-Pick type C1 protein. J Lipid Res. 2010;51(5):1023–1034. doi: 10.1194/jlr.M002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Babalola JO, Wendeler M, Breiden B, et al. Development of an assay for the intermembrane transfer of cholesterol by Niemann-Pick C2 protein. Biol Chem. 2007;388(6):617–626. doi: 10.1515/BC.2007.063. [DOI] [PubMed] [Google Scholar]
- 104.Infante RE, Wang ML, Radhakrishnan A, Kwon HJ, Brown MS, Goldstein JL. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc Natl Acad Sci U S A. 2008;105(40):15287–15292. doi: 10.1073/pnas.0807328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li J, Deffieu MS, Lee PL, Saha P, Pfeffer SR. Glycosylation inhibition reduces cholesterol accumulation in NPC1 protein-deficient cells. Proc Natl Acad Sci U S A. 2015;112(48):14876–14881. doi: 10.1073/pnas.1520490112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Subramanian K, Balch WE. NPC1/NPC2 function as a tag team duo to mobilize cholesterol. Proc Natl Acad Sci U S A. 2008;105(40):15223–15224. doi: 10.1073/pnas.0808256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Urano Y, Watanabe H, Murphy SR, et al. Transport of LDL-derived cholesterol from the NPC1 compartment to the ER involves the trans-Golgi network and the SNARE protein complex. Proc Natl Acad Sci U S A. 2008;105(43):16513–16518. doi: 10.1073/pnas.0807450105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chu BB, Liao YC, Qi W, et al. Cholesterol transport through lysosome-peroxisome membrane contacts. Cell. 2015;161(2):291–306. doi: 10.1016/j.cell.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 109.Schulz TA, Choi MG, Raychaudhuri S, et al. Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J Cell Biol. 2009;187(6):889–903. doi: 10.1083/jcb.200905007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Prinz WA. Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics. J Cell Biol. 2014;205(6):759–769. doi: 10.1083/jcb.201401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ngo MH, Colbourne TR, Ridgway ND. Functional implications of sterol transport by the oxysterol-binding protein gene family. Biochem J. 2010;429(1):13–24. doi: 10.1042/BJ20100263. [DOI] [PubMed] [Google Scholar]
- 112.Rocha N, Kuijl C, van der Kant R, et al. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 glued and late endosome positioning. J Cell Biol. 2009;185(7):1209–1225. doi: 10.1083/jcb.200811005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Du X, Kumar J, Ferguson C, et al. A role for oxysterol-binding protein-related protein 5 in endosomal cholesterol trafficking. J Cell Biol. 2011;192(1):121–135. doi: 10.1083/jcb.201004142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Choudhury A, Dominguez M, Puri V, et al. Rab proteins mediate Golgi transport of caveola-internalized glycosphingolipids and correct lipid trafficking in Niemann-Pick C cells. J Clin Invest. 2002;109(12):1541–1550. doi: 10.1172/JCI15420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Linder MD, Uronen RL, Holtta-Vuori M, van der Sluijs P, Peranen J, Ikonen E. Rab8-dependent recycling promotes endosomal cholesterol removal in normal and sphingolipidosis cells. Mol Biol Cell. 2007;18(1):47–56. doi: 10.1091/mbc.E06-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Narita K, Choudhury A, Dobrenis K, et al. Protein transduction of Rab9 in Niemann-Pick C cells reduces cholesterol storage. FASEB J. 2005;19(11):1558–1560. doi: 10.1096/fj.04-2714fje. [DOI] [PubMed] [Google Scholar]
- 117.Boadu E, Nelson RC, Francis GA. ABCA1-dependent mobilization of lysosomal cholesterol requires functional Niemann-Pick C2 but not Niemann-Pick C1 protein. Biochim Biophys Acta. 2012;1821(3):396–404. doi: 10.1016/j.bbalip.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 118.Devlin C, Pipalia NH, Liao X, Schuchman EH, Maxfield FR, Tabas I. Improvement in lipid and protein trafficking in Niemann-Pick C1 cells by correction of a secondary enzyme defect. Traffic. 2010;11(5):601–615. doi: 10.1111/j.1600-0854.2010.01046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cianciola NL, Carlin CR. Adenovirus RID-alpha activates an autonomous cholesterol regulatory mechanism that rescues defects linked to Niemann-Pick disease type C. J Cell Biol. 2009;187(4):537–552. doi: 10.1083/jcb.200903039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Frolov A, Srivastava K, Daphna-Iken D, Traub LM, Schaffer JE, Ory DS. Cholesterol overload promotes morphogenesis of a Niemann-Pick C (NPC)-like compartment independent of inhibition of NPC1 or HE1/NPC2 function. J Biol Chem. 2001;276(49):46414–46421. doi: 10.1074/jbc.M108099200. [DOI] [PubMed] [Google Scholar]
- 121.Dove DE, Su YR, Zhang W, et al. ACAT1 deficiency disrupts cholesterol efflux and alters cellular morphology in macrophages. Arterioscler Thromb Vasc Biol. 2005;25(1):128–134. doi: 10.1161/01.ATV.0000148323.94021.e5. [DOI] [PubMed] [Google Scholar]
- 122.Du X, Brown AJ, Yang H. Novel mechanisms of intracellular cholesterol transport: oxysterol-binding proteins and membrane contact sites. Curr Opin Cell Biol. 2015;35:37–42. doi: 10.1016/j.ceb.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 123.Platt FM, Boland B, van der Spoel AC. The cell biology of disease: lysosomal storage disorders: the cellular impact of lysosomal dysfunction. J Cell Biol. 2012;199(5):723–734. doi: 10.1083/jcb.201208152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol. 2013;13(10):709–721. doi: 10.1038/nri3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hendrikx T, Walenbergh SM, Hofker MH, Shiri-Sverdlov R. Lysosomal cholesterol accumulation: driver on the road to inflammation during atherosclerosis and non-alcoholic steatohepatitis. Obes Rev. 2014;15(5):424–433. doi: 10.1111/obr.12159. [DOI] [PubMed] [Google Scholar]