Masuda et al. show that Arid5a regulates the fate of naive CD4+ T cells to pro- or antiinflammatory T cells through selective stabilization of Stat3 mRNA under Th17-polarizing conditions.
Abstract
Balance in signal transducer and activator of transcription (STAT) activation is a key factor in regulating the fate of naive CD4+ T cells. Here, we demonstrate that AT-rich interactive domain-containing protein 5a (Arid5a) in T cells directs naive CD4+ T cells to differentiate into inflammatory CD4+ T cells, especially Th17 cells, through selective stabilization of Stat3 (but not Stat1 and Stat5) mRNA in an IL-6–dependent manner. Loss of Arid5a in T cells led to reduction of STAT3 level under Th17-polarizing conditions, whereas STAT1 and STAT5 in Arid5a-deficient T cells were highly activated compared with those of WT T cells under the same conditions. These cells displayed the feature of antiinflammatory (Il10-expressing) CD4+ T cells. Thus, we show a T cell–intrinsic role of Arid5a on fate decisions of naive CD4+ T cells through selective stabilization of Stat3 mRNA.
Activation of signal transducer and activator of transcription 3 (STAT3) is associated with cancer and autoimmunity in the inflammatory milieu (Yu et al., 2009; Flanagan et al., 2014). In particular, STAT3 activation in naive CD4+ Th cells directs the development of inflammatory CD4+ T cells, such as IL-17–producing T (Th17) cells and follicular helper T cells (Harris et al., 2007; Crotty, 2014; Ma and Deenick, 2014). Although STAT3 has been reported to also be essential for the development of Th2 cells, STAT6 (in cooperation with STAT3) is required for Th2 cell development (Stritesky et al., 2011). Signaling via cytokine receptors on CD4+ T cells promotes the development of a distinct lineage of helper T cells through activation of STAT family members (Murphy and Reiner, 2002; Zhu et al., 2010). Cytokines such as IL-6 and -21 have been shown to activate STAT3 in CD4+ T cells, and the development of Th17 cells via the IL-6–STAT3 axis has been reported to be critically involved in numerous autoimmune diseases, such as rheumatoid arthritis (RA) and multiple sclerosis (Muranski and Restifo, 2013; O’Reilly et al., 2013; Dong, 2014; Masuda and Kishimoto, 2014). Upon binding of IL-6 to a complex of the receptor for IL-6 (IL-6R) and gp130, STAT3 is mainly recruited and activated via the Janus kinase (JAK)–STAT pathway (Kishimoto, 2005), whereas IL-6 was reported to activate other STAT family, including STAT1 and STAT5, in T cells (Tormo et al., 2012). Activated STAT3 regulates the transcriptional activity of the genes including Rorc, Rorα, Batf, and Maf, resulting in promoted Th17 cell development (Zhou et al., 2007; Durant et al., 2010; Ciofani et al., 2012; Muranski and Restifo, 2013; Yosef et al., 2013). In contrast, although IL-27 also activates STAT3 and STAT1 in CD4+ T cells, naive CD4+ T cells are differentiated into type 1 regulatory T (Tr1) cells with immunosuppressive functions under the control of IL-27 and TGF-β signaling (Awasthi et al., 2007). A recent study has shown that STAT1 activation forces the feature of IL-27–driven CD4+ T cells with immunosuppressive aspects, whereas STAT3 is also necessary for the development of immunosuppressive CD4+ T cells (Zhu et al., 2015). Thus, a balance in STATs activation is an important factor in controlling the fate of naive CD4+ T cells.
Overproduction of IL-6 in vivo triggers constitutive STAT3 activation (Kishimoto, 2010; Jeltsch et al., 2014; Nagashima et al., 2014). Our group has shown that AT-rich interactive domain (Arid)–containing protein 5a (Arid5a) contributes to selective elevation of IL-6 level in vivo through posttranscriptional regulation of the Il6 gene (Masuda et al., 2013). Thus far, the Arid family is categorized as a 15 member superfamily that possesses different cellular functions, such as cell proliferation, cell growth, and progression (Lin et al., 2014), whereas the immunological function of Arid5a, which is also known as modulator recognition factor 1–like (MRF1-like), remains to be understood (Masuda et al., 2013). It was first demonstrated by our group that Arid5a is one of the mRNA-stabilizing proteins that associates with the 3′ untranslated region (3′UTR) of Il6 mRNA (the IL-6 3′UTR), but not the TNF 3′UTR (Masuda et al., 2013). Il6 mRNA is stabilized by Arid5a, whereby binding to the IL-6 3′UTR inhibits the function of destabilizing proteins with access to its 3′UTR, such as an endoribonuclease Zc3h12a, which is also called protein regulatory RNase 1 (Regnase-1; Matsushita et al., 2009; Masuda et al., 2013). Accordingly, IL-6 level in serum is dramatically attenuated in Arid5a-deficient mice after LPS shock or experimental autoimmune encephalomyelitis induction, which, in turn, results in the reduction of IL-17–producing T cell population in draining lymph nodes (Masuda et al., 2013).
Both Arid5a and Zc3h12a (which encodes Regnase-1 protein) are TLR-inducible genes (Matsushita et al., 2009; Iwasaki et al., 2011; Masuda et al., 2013). Expression of mRNA and proteins of Arid5a and Regnase-1 is tightly regulated in macrophages under the control of TLR4 signaling (Matsushita et al., 2009; Iwasaki et al., 2011; Masuda et al., 2013). A recent study has shown that a T cell–intrinsic role of Regnase-1 is essential for suppression of systemic autoimmunity, in which Regnase-1 in T cells destabilized several inflammatory mRNAs, including mRNAs of Il6, c-Rel, Il2, Ox40, Ctla-4, and Icos (Uehata et al., 2013). Regnase-1 protein levels in T cells are also controlled by the cleavage of the paracaspase MALT1 under TCR signaling strength (Uehata et al., 2013). Thus, control of mRNAs of inflammatory genes by Regnase-1 in T cells, as well as macrophages, has been shown to be essential for immune homeostasis.
T cell–intrinsic functions of Arid5a, however, have not been elucidated. Here, we demonstrate that Arid5a in T cells is a key molecule, which regulates the fate of naive CD4+ T cells to pro- or antiinflammatory T cells through selective stabilization of Stat3 mRNA under Th17-polarizing conditions.
RESULTS
Arid5a expression is specifically enhanced under Th17-polarizing conditions in an IL-6–dependent manner, but not in other distinct T cell subsets, including Th1, Th2, and regulatory T cells
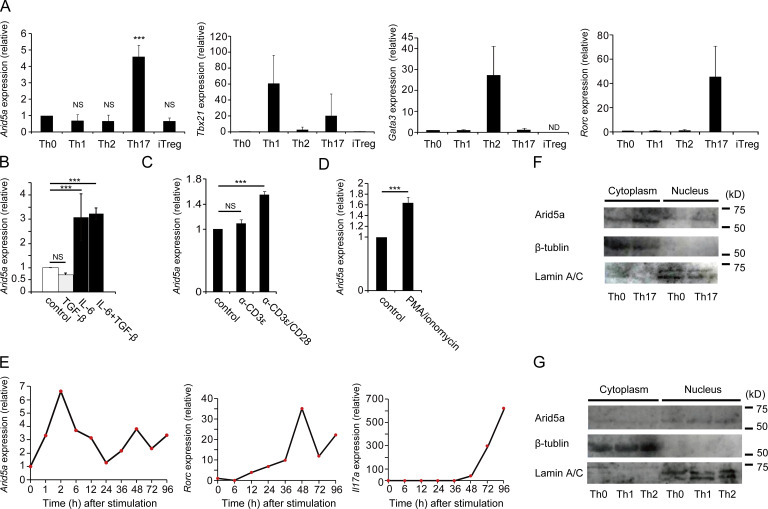
We previously identified a unique Il6 mRNA-stabilizing protein, Arid5a, which was involved in inflammation and autoimmunity through specific elevation of IL-6 level in vivo (Masuda et al., 2013). Expression of Arid5a mRNA (Arid5a expression) was enhanced under the control of TLR4 signaling in LPS-treated macrophages (Masuda et al., 2013). In this study, we found that Arid5a expression was also specifically augmented in CD4+ T cells under Th17-polarizing conditions, whereas Arid5a expression in CD4+ T cells under Th1, Th2, or regulatory T (T reg) cell conditions was not significantly enhanced compared with that of Th0 cells (Fig. 1 A). In addition, Arid5a expression was augmented in CD4+ T cells stimulated with anti-CD3ε and anti-CD28 antibodies in the presence of IL-6, whereas its expression was not drastically enhanced in CD4+ T cells stimulated with anti-CD3ε or anti-CD28 antibodies in the absence of IL-6 or PMA and ionomycin (Fig. 1, B–D). To investigate during which stages Arid5a mRNA is highly expressed, we next analyzed time-course expression of Arid5a mRNA under Th17 cell–inducing conditions. Arid5a expression was rapidly increased in CD4+ T cells polarized toward Th17 cells at an early stage, 2 h after stimulation (Fig. 1 E). Notably, Arid5a proteins in CD4+ T cells were augmented in the cytoplasm under Th17 cell conditions (Fig. 1 F). In contrast, levels of Arid5a protein in the cytoplasm were not increased under Th1 or Th2 cell conditions (Fig. 1 G). Collectively, Arid5a expression was specifically elevated in CD4+ T cells polarized toward Th17 cells (in an IL-6–dependent manner) but not under Th1, Th2, or T reg cell conditions, and might play a role as a stability protein in the early step of Th17 cell development.
Figure 1.
Specific elevation of Arid5a expression in CD4+ T cells under Th17-polarizing conditions. (A) Quantitative real-time PCR analysis of mRNAs of Arid5a, Tbx21, Gata3, and Rorc in CD4+ T cells differentiated for 48 h under Th0, Th1, Th2, Th17, or T reg cell conditions, normalized to the expression of Gapdh mRNA. (B) Quantitative real-time PCR analysis of Arid5a expression in CD4+ T cells stimulated for 48 h with TGF-β or IL-6, normalized to the expression of Gapdh mRNA. (C) Quantitative real-time PCR analysis of Arid5a expression in CD4+ T cells stimulated for 48 h with anti-CD3ε or anti-CD28 antibodies, normalized to the expression of Gapdh mRNA. (D) Quantitative real-time PCR analysis of Arid5a expression in CD4+ T cells stimulated for 48 h with PMA and ionomycin, normalized to the expression of Gapdh mRNA. (E) Time course expression of mRNAs of Arid5a, Rorc, and Il17a in CD4+ T cells differentiated for the indicated times under Th17 cell conditions. Data are representative from two independent experiments with similar results. (F) Immunoblot analysis of Arid5a, β-tubulin, and Lamin A/C in the cytoplasm or the nucleus of CD4+ T cells differentiated for 12 h toward Th17 cells. (G) Immunoblot analysis of Arid5a, β-tubulin, and Lamin A/C in the cytoplasm or the nucleus in CD4+ T cells differentiated for 12 h into Th0, Th1, or Th2 cells. Data are representative of three independent experiments (A–D, F, and G). Error bars show mean ± SD (A–D). ***, P < 0.001 (Student’s t test).
Arid5a in T cells selectively stabilizes Stat3 (but not Stat1 and Stat5) mRNA under Th17-polarizing conditions
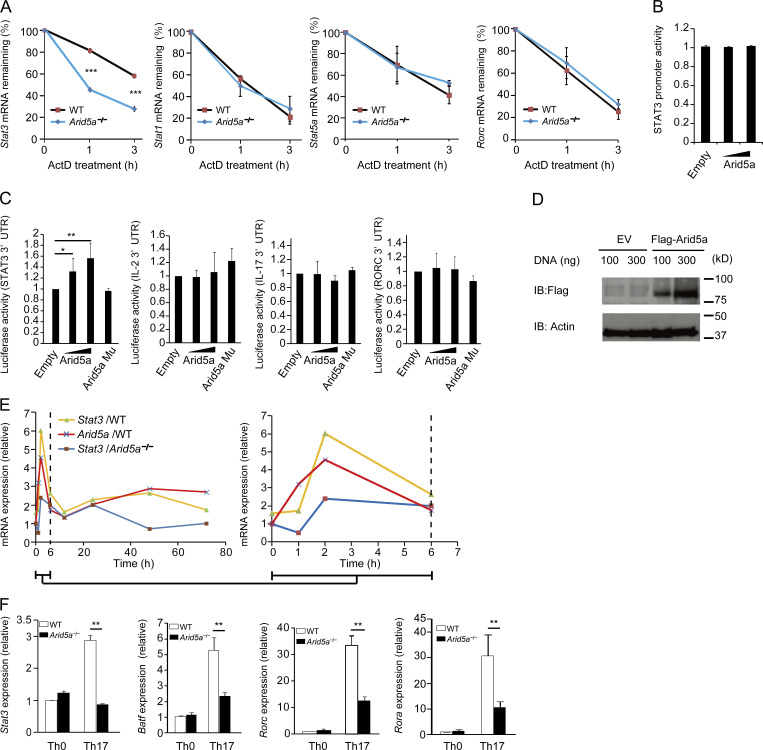
Because Arid5a expression in T cells had increased specifically under Th17-polarizing conditions (in an IL-6–dependent manner), we next examined whether Arid5a affects mRNA stability of STAT family genes under Th17 cell–inducing conditions. After actinomycin D treatment, mRNA half-life of Stat3 gene in Arid5a-deficient CD4+ T cells under Th17-polarizing conditions became significantly shorter than that in WT T cells (Fig. 2 A). In contrast, mRNA half-lives of Stat1, Stat5a, and Rorc in Arid5a-deficient T cells were similar to those in WT T cells under Th17-inducing conditions (Fig. 2 A). These results suggest that Arid5a selectively stabilizes Stat3 mRNA but not mRNAs of other STAT family genes, including Stat1 and Stat5, in CD4+ T cells under Th17-inducing conditions. Moreover, Arid5a did not affect STAT3 promoter activity (Fig. 2 B). Next, we investigated whether Arid5a controls Stat3 mRNA stability via the total STAT3 3′UTR using HEK293T cells. Overexpression of Arid5a enhanced the luciferase activity of pGL3 vector encoding the STAT3 3′UTR in a dose-dependent manner (Fig. 2 C). In contrast, mutant Arid5a lacking the Arid domain did not have any influence on the luciferase activity (Fig. 2 C). In addition, overexpression of Arid5a did not affect the luciferase activity of pGL3 vector encoding the total IL-2, IL-17, or RORC 3′UTR (Fig. 2 C). We also confirmed that level of Flag-tagged Arid5a protein in HEK293T cells was elevated in a dose-dependent manner (Fig. 2 D). These results indicate that Arid5a is able to specifically influence stabilization of Stat3 mRNA via the STAT3 3′UTR. Furthermore, we compared the kinetics of expression of Stat3 mRNA between WT and Arid5a-deficient T cells under Th17-polarizing conditions. Interestingly, the kinetics of Stat3 expression were similar to that of Arid5a expression in WT CD4+ T cells under Th17-polarizing conditions (Fig. 2 E), and Stat3 mRNA level in Arid5a-deficient T cells was remarkably lower than that of WT T cells at 2 or 48 h after stimulation (Fig. 2 E). Consequently, the expression levels of STAT3-regulated genes, including Batf, Rorc, and Rora in Arid5a-deficient T cells under Th17 cell conditions, were impaired compared with WT T cells (Fig. 2 F). Collectively, Arid5a selectively stabilizes Stat3 (but not Stat1 and Stat5) mRNA in CD4+ T cells under Th17-polarizing conditions, which in turn resulted in enhancement of the expression of STAT3-controlled genes.
Figure 2.
Selective stabilization of Stat3 mRNA by Arid5a in CD4+ T cells under Th17-polarizing conditions. (A) Quantitative real-time PCR analysis of mRNAs of Stat1, Stat3, Stat5a, and Rorc in CD4+ T cells differentiated for 12 h under Th17 cell-inducing conditions, followed by treatment for 0–180 min with actinomycin D (ActD). (B) Luciferase activity of HEK 293T cells transfected for 48 h with the luciferase reporter vector (100 ng) encoding the STAT3 human promoter in combination with Arid5a expression vector (100–300 ng) or empty vector (100–300 ng). (C) Luciferase activity of HEK 293T cells transfected for 48 h with pGL3 vector encoding the total STAT3 3′UTR, IL-2 3′UTR, IL-17 3′UTR, or RORc 3′UTR in combination with Arid5a expression vector, empty vector, or Arid5a mutant (lacking Arid domain) vector. (D) Immunoblot analysis of Arid5a expression in HEK293T cells transfected for 48 h with Flag-Arid5a expression vector or empty vector (EV). (E) Quantitative real-time PCR analysis of mRNAs of Stat3 and Arid5a in either WT or Arid5a-deficient CD4+ T cells differentiated for 0, 1, 2, 6, 12, 24, 48, or 72 h under Th17 cell conditions. Time-course expression (0–6 h) of Stat3 and Arid5a in WT or Arid5a-deficient CD4+ T cells was enlarged (right). Data are representative of two independent experiments with similar results. (F) Quantitative real-time PCR analysis of mRNAs of Stat3, Batf, Rorc, or Rora in either WT or Arid5a-deficient CD4+ T cells differentiated for 48 h under Th17 cell conditions. Data are representative of three independent experiments (A–D and F). Error bars show mean ± SD (A–C and F). *, P < 0.05; **, P < 0.02; ***, P < 0.001 (Student’s t test).
The 3′UTR of Stat3 mRNA contains the responsive elements of both Arid5a and an RNase Regnase-1
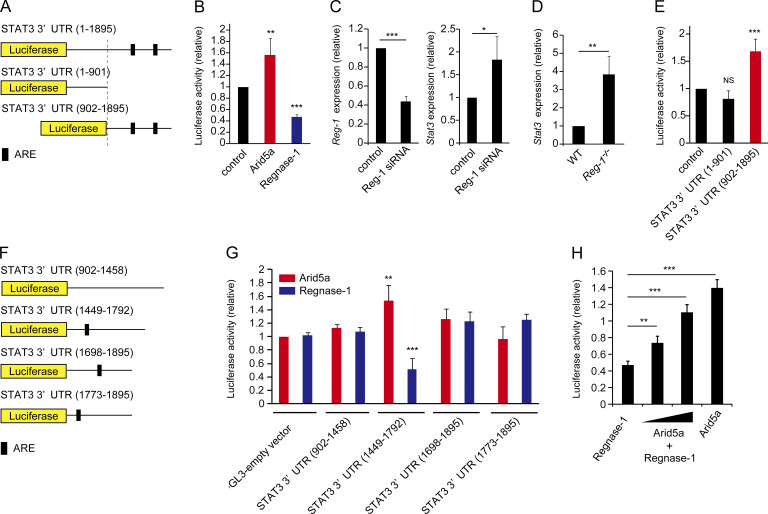
Having found that Arid5a controls Stat3 mRNA stability via the STAT3 3′UTR, we next attempted to identify which elements in the STAT3 3′UTR are critical for stabilization of Stat3 mRNA by Arid5a. We initially prepared the constructs of pGL3 luciferase vector encoding the total STAT3 3′UTR (1–1895), STAT3 3′UTR (1–901), or STAT3 3′UTR (902–1895; Fig. 3 A). Interestingly, overexpression of Regnase-1, which was reported to degrade Il6 mRNA on the IL-6 stem-loop region (Matsushita et al., 2009), reduced the luciferase activity of pGL3 encoding the total STAT3 3′UTR (1–1895), whereas overexpression of Arid5a augmented the activity of the total STAT3 3′UTR vector (Fig. 3 B). This result suggests that Stat3 mRNA could be a target of Regnase-1. In support of this hypothesis, knockdown of Regnase-1 in T cells led to the augmentation of Stat3 mRNA level (Fig. 3 C). Stat3 mRNA level in Regnase-1 deficient MEFs was also higher than WT MEFs (Fig. 3 D). Moreover, overexpression of Arid5a enhanced the luciferase activity of pGL3 encoding the Stat3 3′UTR (902–1895), but not the STAT3 3′UTR (1–901; Fig. 3 E). These results suggest that Arid5a is able to function as a stability protein in the region of the STAT3 3′UTR (902–1895). We also designed the constructs of pGL3 luciferase vector encoding the fragments (902–1458, 1449–1792, 1698–1895, and 1773–1895) of the STAT3 3′UTR (Fig. 3 F). Notably, overexpression of Arid5a increased the luciferase activity of pGL3 vector encoding the fragment (1449–1792) of the STAT3 3′UTR, whereas overexpression of Regnase-1 reduced the luciferase activity of the same region (1449–1792) of the STAT3 3′UTR (Fig. 3, F and G). In contrast, overexpression of either Arid5a or Regnase-1 did not affect the luciferase activity of pGL3 vector encoding the fragments (902–1458, 1698–1895, and 1773–1895) of the STAT3 3′UTR(Fig. 3, F and G). Next, we investigated whether Arid5a neutralizes the inhibitory effect of Regnase-1 on the luciferase activity of pGL3 vector encoding the total STAT3 3′UTR. As a result, overexpression of Arid5a rescued the luciferase activity of the total STAT3 3′UTR impaired by Regnase-1 in a dose-dependent manner (Fig. 3 H). Interestingly, an in silico secondary structure prediction showed that the region of the STAT3 3′UTR (1449–1792) included a stem-loop sequence (1738–1765) that had the potential to associate with Arid5a (Fig. S1). Thus, a stem-loop region (1738–1765) in the STAT3 3′UTR might play an important role as a responsive element for both Arid5a and Regnase-1 proteins.
Figure 3.
The STAT3 3′UTR (1449–1792) is the responsive sites of Arid5a and an endoribonuclease Regnase-1. (A) Diagram of the region of pGL3 vector encoding STAT3 3′UTR (1–1895), STAT3 3′UTR (1–901), or STAT3 3′UTR (902–1895). The black bar shows AU-rich elements (ARE). (B) Luciferase activity of the vector encoding the STAT3 3′UTR (1–1895) in HEK293T cells transfected for 48 h together with Arid5a expression vector, Regnase-1 expression vector, or empty vector. (C) Quantitative real-time PCR analysis of mRNAs of Regnase-1 (Reg-1) and Stat3 in Jurkat cells electroporated with Reg-1 siRNA or control siRNA, normalized to human Gapdh mRNA expression. (D) Quantitative real-time PCR analysis of Regnase-1 (Reg-1) mRNA in Regnase-1–deficient MEFs, normalized to Gapdh mRNA expression. (E) Luciferase activity of the pGL3-vector encoding the STAT3 3′UTR (1–901) or STAT3 3′UTR (902–1895), or the pGL3-empty vector cotransfected for 48 h with Arid5a expression vector. (F) Diagram of the region of pGL3 vector encoding the STAT3 3′UTR (902–1458), STAT3 3′UTR (1449–1792), STAT3 3′UTR (1698–1895), or STAT3 3′UTR (1773–1895). The black bar shows AU-rich elements (ARE). (G) Luciferase activity of the vector encoding each area of the STAT3 3′UTR as in F cotransfected with either Arid5a expression vector or Regnase-1 expression vector. (H) Luciferase activity of the vector encoding the total STAT3 3′UTR (1–1895) in HEK293T cells for 48 h cotransfected with either Regnase-1 vector or Arid5a expression vector. Data are representative of three independent experiments (B–E, G, and H). Error bars show mean ± SD (B–E, G, and H). *, P < 0.05; **, P < 0.02; ***, P < 0.001 (Student’s t test).
The stem structure of the STAT3 3′UTR (1738–1765) is critical for Arid5a binding
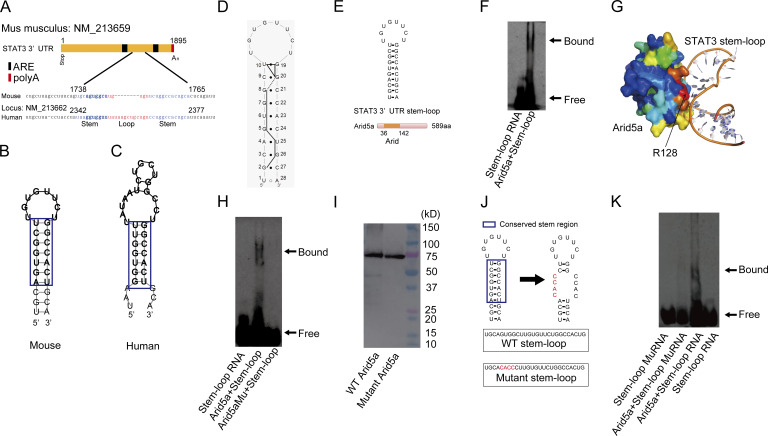
Because we have identified a critical responsive elements of the STAT3 3′UTR for mRNA stability, we next investigated the detailed mechanism of how Arid5a associates with the STAT3 3′UTR (1738–1765). Interestingly, the stem structure of the identified elements (1738–1765) of the STAT3 3′UTR is highly conserved between human and mouse (Fig. 4, A–C). We also confirmed that the predicted secondary structure of the STAT3 3′UTR (1738–1765) matched the structure determined by NMR (Fig. 4 D). Moreover, an in silico analysis showed that the stem region of the STAT3 3′UTR (1738–1765) exhibited high affinity with Arid5a protein (unpublished data). Therefore, we tried to confirm the interaction of recombinant mouse Arid5a protein with the possible binding site (5′-UGCAGUGGCUUGUGUUCUGGCCACUGCA-3′) of the STAT3 3′UTR using electrophoresis mobility shift assay (EMSA; Fig. 4 E). As a result, Arid5a physically bound to the stem-loop region (1738–1765) of the STAT3 3′UTR (Fig. 4 F). Next, we investigated which residues of Arid5a protein are essential for its binding to the stem-loop sequence (1738–1765) of the STAT3 3′UTR. We searched for potential RNA-binding residues on Arid5a by means of in silico analysis. Consequently, we identified residue R128 as likely candidate to interact with the stem-loop element of the STAT3 3′UTR (Fig. 4 G). Accordingly, we next prepared mutant Arid5a recombinant protein with an alanine substitution at its residue 128. As predicted, the mutant Arid5a protein did not bind to the sequence (1738–1765) consisting of the STAT3 stem-loop structure (Fig. 4 H). Mouse recombinant Arid5a and mutant Arid5a proteins were detected by anti-Arid5a antibody (Fig. 4 I). Because we predicted the docking mode of Arid5a-the STAT3 3′UTR (Fig. 4 G), we examined whether Arid5a physically binds to the mutant stem-loop STAT3 3′UTR. As a result, Arid5a did not interact with the mutant stem-loop STAT3 3′UTR (Fig. 4, J and K). These results indicate that Arid5a physically associates with the stem region of the STAT3 3′UTR (1738–1765) via its residue R128, which might prevent RNA-destabilizing proteins such as Regnase-1 from binding the STAT3 stem-loop region.
Figure 4.
Arid5a physically associates with the stem structure of the STAT3 3′UTR (1738–1765) via its residue R128. (A) A predicted conserved stem-loop region in the STAT3 3′UTR between human and mouse. Blue and red colored regions indicate stem and loop regions, respectively. (B and C) Secondary structure models of the stem-loop regions of mouse (B) and human (C) as in A. The conserved elements between human and mice are highlighted in blue. (D) Identification of the secondary structure of the STAT3 3′UTR (1738–1765) by NMR analysis. The trace of the NOESY connectivity is indicated by bold solid lines. Closed circles indicate base pairs that were experimentally identified. (E) Diagram of the stem-loop structure (1738–1765) of the STAT3 3′UTR and recombinant mouse Arid5a proteins. (F) Gel mobility shift assay (EMSA) of interaction of Arid5a recombinant protein with 3′-biotinated nucleotides as in E. (G) Docking model of Arid5a-STAT3 3′UTR (1738–1765) in silico. (H) EMSA of noninteraction of mutant Arid5a recombinant protein or interaction of Arid5a recombinant protein with 3′-biotinated nucleotides as in E. (I) Immunoblot analysis of recombinant Arid5a protein or mutant recombinant Arid5a by anti-Arid5a antibody, respectively. (J) Diagram of the stem-loop structure of the STAT3 3′UTR (1738–1765) and the mutant stem-loop structure. (K) EMSA of noninteraction of the 3′-biotinated mutant stem-loop or interaction of the 3′-biotinated stem-loop with Arid5a recombinant protein as in J. Data are representative of three independent experiments (D, F, H, I, and K).
Arid5a impairs the binding of Regnase-1 on the stem-loop STAT3 3′UTR (1738–1765)
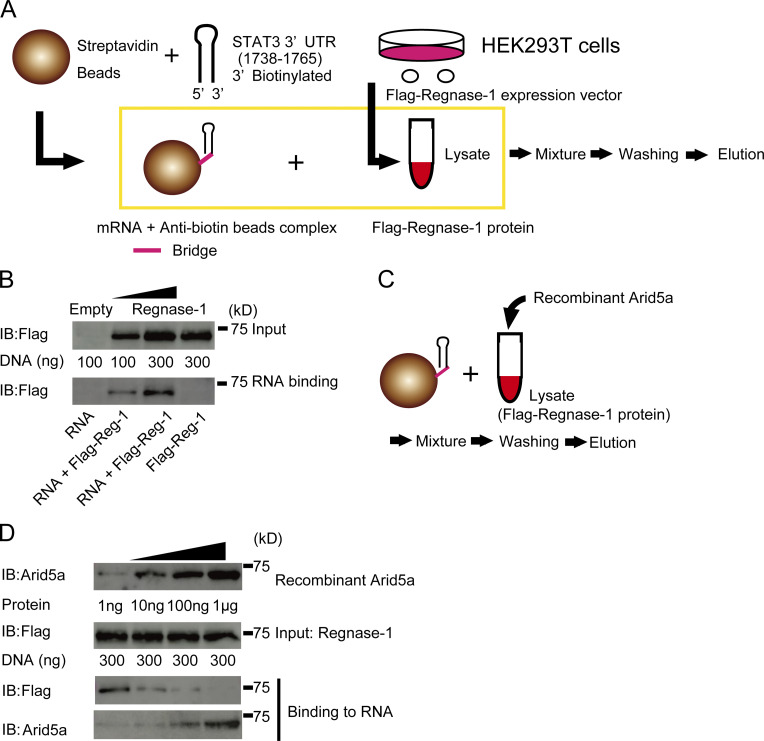
Because we have shown that Arid5a physically associates with the stem of the STAT3 3′UTR (1738–1765), and inhibits the biological function of Regnase-1 on the STAT3 3′UTR, we next examined if Arid5a physically prevents Regnase-1 from binding to the stem-loop of the STAT3 3′UTR (1738–1765). To confirm the physical binding of Regnase-1, we initially prepared the RNA-binding beads and the lysates from HEK293T cells transfected with Flag-Regnase-1 expression or control plasmid (Fig. 5 A). Following the protocol shown in Fig. 5 A, we found that Regnase-1 physically binds to the stem-loop of STAT3 3′UTR (1738–1765; Fig. 5 B). It is intriguing that the loop region of the STAT3 3′UTR (1738–1765) includes a unique sequence of UGU for interaction of Regnase-1 (Mino et al., 2015). Next, we investigated whether Arid5a interferes with the binding of Regnase-1 on the stem-loop region STAT3 3′UTR (1738–1765) following the protocol (Fig. 5 C). As a result, Arid5a blocked the binding of Regnase-1 on the STAT3 3′UTR (1738–1765) in a dose-dependent manner (Fig. 5 D).
Figure 5.
Arid5a inhibits the binding of Regnase-1 to the stem-loop STAT3 3′UTR (1738–1765). (A) Schematic diagram of an RNA-protein binding assay. The lysates were prepared from HEK293T cells transfected with Flag-Regnase-1 expression vector or empty vector for 48 h. The 3′-biotinated nucleotides (5′-UGCAGUGGCUUGUGUUCUGGCCACUGCA-3′) of the STAT3 3′UTR (1738–1765) was conjugated with Streptavidin beads. Next, the lysates and the beads were mixed, and then washed three times. Finally, the binding proteins with the beads were eluted. (B) Immunoblot analysis of Flag-Regnase-1 protein in HEK293T cells for 48 h transfected with Flag-Regnase-1 expression vector or Flag-Regnase-1 protein binding to the stem-loop STAT3 3′UTR (1738–1765). (C) Schematic diagram of a competitive assay of RNA-protein binding. The lysates with or without Arid5a recombinant protein were prepared from HEK293T cells transfected with Flag-Regnase-1 expression vector for 48 h. Next, the lysates were mixed with the RNA-binding beads as in A, and washed three times. Finally, the binding proteins were eluted. (D) Immunoblot analysis of recombinant Arid5a protein and Flag-Regnase-1 protein in the lysates or the eluted samples, as in C. Data are representative of three independent experiments (B and D).
Attenuation of STAT3 activation by loss of Arid5a in T cells prolongs the activation of STAT1 under Th17-polarizing conditions
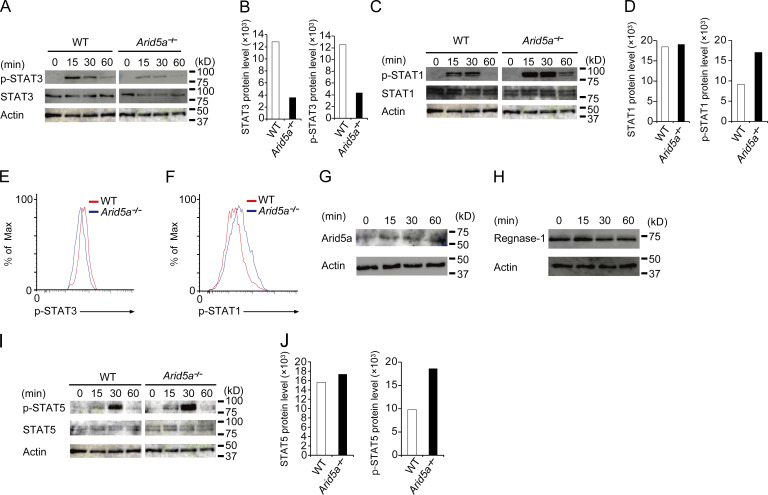
We next compared levels of STAT family proteins (including STAT1, STAT3, and STAT5) or its phosphorylation in either WT and Arid5a-deficient CD4+ T cells under Th17 cell-inducing conditions. We found that both levels of phosphorylated STAT3 and STAT3 proteins in Arid5a-deficient T cells were significantly less than those of WT T cells under Th17-polarizing conditions (Fig. 6, A and B). Conversely, level of phosphorylated STAT1 protein in Arid5a-deficient T cells was higher than that of WT T cells, whereas level of STAT1 protein in Arid5a-deficient T cells was similar to that of WT T cells (Fig. 6, C and D). Thus, reduction of STAT3 level by Arid5a deficiency was associated with imbalance of phosphorylated levels of STAT1 and STAT3 proteins between WT and Arid5a-deficient T cells (Fig. 6, E and F). We next examined the levels of Arid5a and Regnase-1 proteins, whereas naive CD4+ T cells were differentiated into Th17 cells (0–60 min). Expression of Arid5a protein was enhanced under Th17-polarizing conditions after stimulation (Fig. 6 G). Regnase-1 protein was also constitutively expressed under the same conditions (Fig. 6 H). These data suggest that Arid5a is able to compete with Regnase-1, whereas naive CD4+ T cells were differentiated into Th17 cells (0–60 min). STAT5 phosphorylation was also strengthened compared with that of WT T cells under Th17-polarizing conditions, whereas the level of STAT5 in Arid5a-deficient T cells was not altered under the same conditions (Fig. 6, I and J). Collectively, Arid5a deficiency led to reduction of STAT3 level, and also contributed to the activation of other STAT family proteins including STAT1 and STAT5 in CD4+ T cells under Th17 cell–inducing conditions.
Figure 6.
Reduction of STAT3 level in T cells lacking Arid5a impairs a balance among STAT1, STAT3, and STAT5 activation under Th17-polarizing conditions. (A) Immunoblot analysis of STAT3, phosphorylated STAT3, and β-actin in WT or Arid5a-deficient CD4+ T cells differentiated for the indicated times toward Th17 cells. (B) Intensity of STAT3 or phosphorylated STAT3 level as in A. (C) Immunoblot analysis of STAT1, phosphorylated STAT1, and β-actin in WT or Arid5a-deficient CD4+ T cells differentiated for the indicated times toward Th17 cells. (D) Intensity of STAT1 or phosphorylated STAT1 level as in C, respectively. (E and F) Phosphorylation of STAT3 or STAT1 in WT or Arid5a-deficient CD4+ T cells for 30 min differentiated toward Th17 cells by FACS analysis. (G and H) Immunoblot analysis of Arid5a or Regnase-1 protein in CD4+ T cells differentiated for the indicated times into Th17 cells. (I) Immunoblot analysis of STAT5, phosphorylated STAT5, and β-actin in WT or Arid5a-deficient CD4+ T cells differentiated for the indicated times toward Th17 cells. (J) Intensity of STAT5 or phosphorylated STAT5 level as in I. Data are representative of three independent experiments (A–J). Data are representative from three independent experiments with similar results (B, D, and J).
Naive CD4+ T cells in the absence of Arid5a are differentiated into CD4+ T cells with an immunosuppressive phenotype under Th17-polarizing conditions
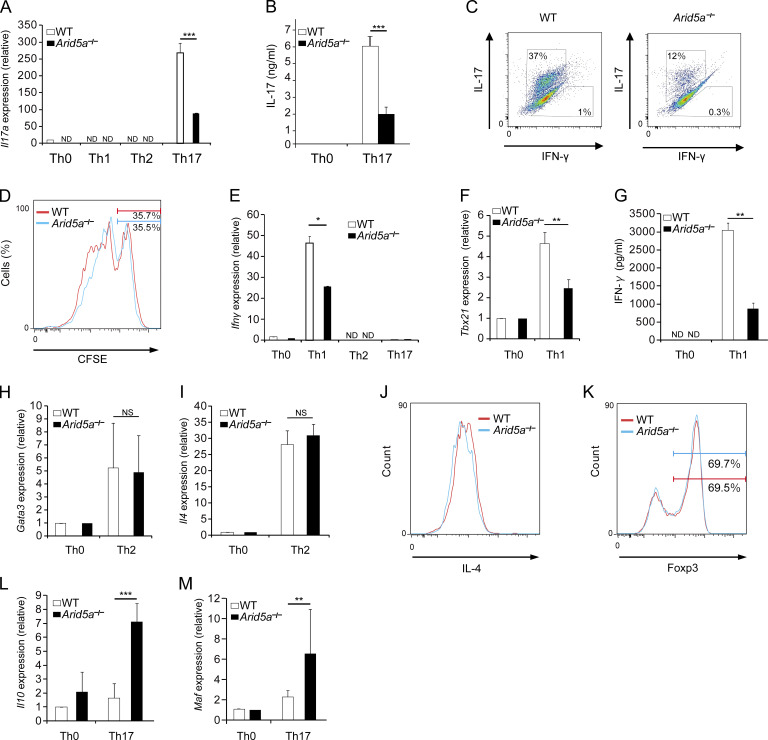
As we have shown that reduction of STAT3 level in Arid5a-deficient T cells leads to imbalance among activation of STAT1, STAT3, and STAT5, we next compared the differentiation of naive WT and Arid5a-deficient CD4+ T cells toward Th17 cells. The differentiation of naive CD4+ T cells toward Th17 cells was impaired in the absence of Arid5a, in which mRNA and protein levels of IL-17A in Arid5a-deficient T cells were significantly lower than those of WT T cells (Fig. 7, A and B). Moreover, the frequency of IL-17–producing T cell population was decreased in Arid5a-deficient T cells compared with WT T cells in vitro (Fig. 7 C), although cell proliferation in CD4+ T cells under Th17 cell conditions did not alter between WT and Arid5a-deficient T cells (Fig. 7 D). Unexpectedly, IFN-γ production was also abrogated in Arid5a-deficient T cells under Th1 cell conditions, compared with WT T cells, in which expression of both Tbx21 and Ifnγ mRNAs was inhibited (Fig. 7, E–G). In contrast, loss of Arid5a in T cells did not affect Gata3 and Il4 mRNA levels under Th2 cell conditions, and also Th2 cell populations (Fig. 7, H–J). In addition, Arid5a deficiency did not significantly influence Foxp3 expression in CD4+ T cells under T reg cell conditions (Fig. 7 K). Notably, the level of Il10 mRNA in Arid5a-deficient T cells was significantly higher than WT T cells under Th17 cell condition (Fig. 7 L), although Foxp3 expression was not detected in Arid5a-deficient T cells under the same conditions (unpublished data). Because c-Maf (which is encoded by Maf) was reported to control Il10 expression in CD4+ T cells under Th17 cell conditions (Xu et al., 2009), we next examined the expression level of Maf in WT and Arid5a-deficient T cells. As a result, Maf expression in Arid5a-deficient T cells was significantly higher than WT T cells under Th17 cell conditions (Fig. 7 M). It is conceivable that it is a result of imbalance of STAT1 and STAT3 activation. Indeed, the pattern of STAT1- or STAT3-controlling gene expression in Arid5a-deficient T cells was different from that in WT T cells under Th17 cell-inducing conditions (Table S1). These results suggest that loss of Arid5a in T cells failed to direct the development of canonical Th17 cells and changed the phenotype of Th17 cells.
Figure 7.
Loss of Arid5a in T cells alters the character of inflammatory CD4+ T cells under Th17-polarizing conditions. (A) Quantitative real-time PCR analysis of Il17a mRNA in CD4+ WT or Arid5a-deficient T cells differentiated for 48 h in Th0, Th1, Th2, or Th17 cell conditions, normalized to the expression of Gapdh mRNA. (B) Level of IL-17 in the supernatants in CD4+ WT or Arid5a-deficient T cells differentiated for 72 h in Th0 or Th17 cell conditions by ELISA. (C) Frequency of IL-17–producing or IFN-γ–producing T cell population in WT or Arid5a-deficient CD4+ T cells differentiated for 72 h toward Th17 cells. (D) Proliferation of naive WT or Arid5a-deficient CD4+ T cells differentiated for 48 h toward Th17 cells. (E) Quantitative real-time PCR analysis of Ifnγ mRNA in CD4+ WT or Arid5a-deficient T cells differentiated for 48 h in Th0, Th1, Th2, or Th17 cell conditions, normalized to the expression of Gapdh mRNA. (F) Quantitative real-time PCR analysis of Tbx21 mRNA in CD4+ WT or Arid5a-deficient T cells differentiated for 48 h into Th0 or Th1 cells, normalized to the expression of Gapdh mRNA. (G) Level of IFN-γ in the supernatants in CD4+ WT or Arid5a-deficient T cells differentiated for 48 h in Th0 or Th17 cell conditions by ELISA. (H and I) Quantitative real-time PCR analysis of Gata3 or Il4 mRNA in CD4+ WT or Arid5a-deficient T cells differentiated for 72 h into Th0 or Th2 cells, normalized to the expression of Gapdh mRNA. (J) FACS analysis of IL-4 in CD4+ WT or Arid5a-deficient T cells differentiated for 96 h into Th2 cells. (K) FACS analysis of FoxP3 in CD4+ WT or Arid5a-deficient T cells differentiated for 48 h into T reg cells. (L and M) Quantitative real-time PCR analysis of Il10 or Maf mRNA in CD4+ WT or Arid5a-deficient T cells differentiated for 48 h in Th0 or Th17 cell conditions, normalized to the expression of Gapdh mRNA. Data are representative of three independent experiments (A–M). Error bars show mean ± SD (A, B, E–I, L, and M). *, P < 0.05; **, P < 0.02; ***, P < 0.001 (Student’s t test).
The attenuation of Th17 cell differentiation in Arid5a-deficient T cells is mainly caused by reduction of STAT3 level in an IL-6–dependent manner
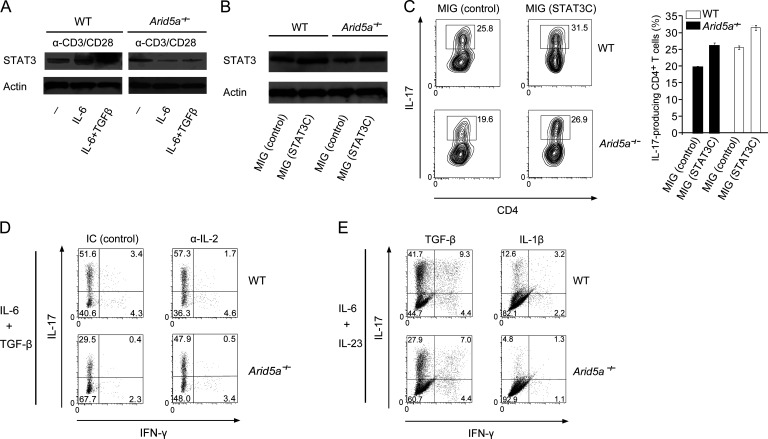
We next examined if reduction of STAT3 level in T cells lacking Arid5a had a dominant effect on the impairment of Th17 cell differentiation in an IL-6–dependent manner. First, we confirmed whether the level of STAT3 was reduced in an IL-6–dependent manner in Arid5a-deficient T cells. As a result, STAT3 level was decreased in Arid5a-deficient T cells activated by anti-CD3ε and anti-CD28 antibodies together with IL-6 stimulation alone or the combination of IL-6 and TGF-β (Fig. 8 A). Next, we investigated the influence of the decrease of STAT3 levels in Arid5a-deficient T cells on the attenuation of Th17 cell differentiation by means of a rescue experiment in which retrovirus vectors were used, including MSCV-IRES-GFP (MIG) as a control and MIG-STAT3C overexpressing a hyperactive STAT3C form, as previously described (Bromberg et al., 1999). Retrovirus-induced overexpression of STAT3C complemented the STAT3 level decrease in Arid5a-deficient T cells under Th17-polarizing conditions (Fig. 8 B), which resulted in the recovery of Th17 cell populations lost by Arid5a deficiency (Fig. 8 C). These results suggest that reduction of STAT3 level in T cells lacking Arid5a critically contributes to the impairment of Th17 cell differentiation. Because it has been reported that elevated IL-2 production from STAT3-deficient CD4+ T cells restrains Th17 cell differentiation through STAT5 activation (Laurence et al., 2007), we next examined the effect of anti–IL-2 treatment on Th17 cell populations in Arid5a-deficient T cells. The Th17 cell populations attenuated by Arid5a deficiency were partially recovered compared with WT Th17 cell populations (Fig. 8 D). Furthermore, to investigate the effect of TGF-β signaling on the frequency of Th17 cells in Arid5a-deficient T cells, naive CD4+ WT or Arid5a-deficient T cells were differentiated into Th17 cells under IL-6, IL-23, and TGF-β or IL-1β stimulation (without TGF-β). As a result, the induction of Th17 cell differentiation was impaired in Arid5a-deficient T cells even without the control of TGF-β signaling (Fig. 8 E). These results suggest that Arid5a contributes to the promotion of Th17 cell differentiation mainly through posttranscriptional control of Stat3 gene via the pathway of IL-6 signaling.
Figure 8.
Decreased STAT3 level in Arid5a-deficient T cells critically contributes to the attenuation of Th17 cell populations. (A) Immunoblot analysis of STAT3 and β-actin in WT or Arid5a-deficient CD4+ T cells for 48 h stimulated by anti-CD3ε and anti-CD28 antibodies with or without IL-6 plus TGF-β or IL-6 alone. (B and C) WT and Arid5a-deficient CD4+ T cells were transfected with MIG or MIG-STAT3C retrovirus vector for 24 h, respectively, and then cultured for 4 d under Th17-polarizing conditions. STAT3 or β-actin level in these cells was analyzed by immunoblot analysis, respectively (B). The frequency of Th17 cells in WT or Arid5a-deficient T cells by FACS analysis (C). Error bars show mean ± SD (C). (D) FACS analysis of WT or Arid5a-deficient T cells differentiated for 5 d under Th17-polarizing conditions with anti-IL-2 (JES6-1A12) treatment or isotype control (IC). (E) FACS analysis of CD4+ WT or Arid5a-deficient T cells differentiated for 5 d into Th17 cells after the stimulation of anti-CD3ε and anti-CD28 antibodies with IL-6 and IL-23 plus TGF-β (left) or IL-1β (right). Data are representative of three independent experiments (A–E).
DISCUSSION
A recent study reported that elevated expression of Arid5a was associated with peripheral CD4+ T cells of RA patients compared with healthy controls (Saito et al., 2014). Interestingly, a humanized anti–IL-6 receptor antibody, tocilizumab, efficiently suppressed elevation of Arid5a expression in peripheral CD4+ T cells of RA patients, in which it was reported that Arid5a negatively regulated the function of RORgt (known as a regulator of Th17 cell differentiation), independent of the control of IL-6 signaling (Saito et al., 2014). In contrast, our group has demonstrated that Arid5a regulates Stat3 mRNA half-life as a stability protein under Th17-polarizing conditions (in an IL-6–dependent manner), which resulted in the promotion of Th17 cell differentiation.
IL-6 activates STAT3 via gp130 and IL-6 receptor complex (Kishimoto, 2010). It is known that IL-6 is a key cytokine that directs the fate of naive CD4+ T cells through activation of STAT3 (O’Shea et al., 2011; Hunter and Jones, 2015). Notably, STAT3 activation was reported to be important for the induction of Arid5a expression (Saito et al., 2014). Our data has also shown that STAT3 level is reduced in Arid5a-deficient CD4+ T cells activated by anti-CD3ε and anti-CD28 antibodies in an IL-6–dependent manner. Therefore, a positive feedback loop between STAT3 and Arid5a might facilitate Arid5a to stabilize Stat3 mRNA in Th17 cell conditions (in an IL-6–dependent manner). Ultimately, further clarification is needed to reveal the mechanisms of Arid5a activation for stabilization of Stat3 mRNA under the control of IL-6 signaling.
It is important to ascertain how Arid5a selectively stabilizes Stat3 mRNA via the STAT3 3′UTR. We previously discussed stabilization of Il6 mRNA by Arid5a. Although we demonstrated that Arid5a counteracts the destabilizing effect of Regnase-1 on the IL-6 stem-loop site (Masuda et al., 2013), the detailed mechanism of competition between Arid5a and Regnase-1 on the stem-loop structure was not elucidated. Interestingly, we have shown that Stat3 mRNA stability is also controlled by both Arid5a and Regnase-1, in which Arid5a associated with the stem region of the STAT3 3′UTR (1738–1765) via its residue R128, as well as by Regnase-1 bound to the STAT3 3′UTR (1738–1765). Moreover, the loop region of the mouse STAT3 3′UTR (1738–1765) or the human STAT3 3′UTR (2342–2377) harbored a unique sequence UGU (mouse) or UAU (human) for the association of Regnase-1 or Roquin (Mino et al., 2015). Collectively, these findings indicate that the binding of Arid5a to the stem structure prevents Regnase-1 from binding to the loop region of the STAT3 3′UTR (1738–1765). In contrast, Regnase-1 did not function as a destabilizing protein in the region of the STAT3 3′UTR (1698–1895), which also includes the stem-loop elements (1738–1765). It is conceivable that some cooperators such as UPF-1, which has been reported to be necessary for Regnase-1 function (Mino et al., 2015), might fail to act on the region of the STAT3 3′UTR (1698–1895).
A recent study has also shown that STAT3 is highly activated in the lung of T cell–conditional Roquin-deficient mice associated with loss of Regnase-1 function (Jeltsch et al., 2014). Notably, Regnase-1 in corporation with Roquin has been shown to impair the development of Th17 cells in which suppression of STAT3 activation was associated (Jeltsch et al., 2014). Moreover, we have shown that knockdown of Regnase-1 in T cells enhances Stat3 mRNA level. These results indicate that level of Stat3 mRNA and its activation in T cells are controlled by a balance of activation of Arid5a and Regnase-1 (possibly also Roquin) in CD4+ T cells during IL-6 signaling, which could regulate the differentiation of Th17 cells.
Unexpectedly, Arid5a-deficient T cells impaired the differentiation of naive T cells into Th1 cells during IL-12 stimulation, in which Tbx21 mRNA expression was inhibited compared with WT T cells, whereas Stat4 mRNA level was not altered (unpublished data). Moreover, Arid5a expression levels in T cells were not augmented under the control of IL-12 signaling. Thus, although further investigation will be needed to reveal a role of Arid5a in Th1 cell conditions, Arid5a might be able to control Tbx21 expression under the control of IL-12 signaling in a differently than an IL-6–dependent manner.
It is intriguing that a balance among phosphorylated levels of STAT1, STAT3, and STAT5 was impaired between WT and Arid5a-deficient T cells under Th17-polarizing conditions, possibly due to the reduction of STAT3 level. Recently, the asymmetry of STAT1 and STAT3 activation has been argued, in which STAT1 activation was prolonged by IL-6 in the absence of STAT3 in MEFs (Costa-Pereira et al., 2002), and imbalance of suppressor of cytokine signaling 1 and 3 also influenced phosphorylated levels of STAT1 and STAT3 (Hong et al., 2002). Thus, although there could be several factors that decide a balance between STAT1 and STAT3 activation, further investigation will be needed to uncover how STAT1 activation is prolonged in Arid5a-deficient T cells.
Consequently, prolonged activation of STAT1 in Arid5a-deficient T cells might change the feature of canonical Th17 cells with inflammatory aspects into antiinflammatory CD4+ T cells with immunosuppressive aspects. Arid5a-deficient T cells highly stimulated the expression of Il10 under Th17-polarizing conditions, independent of Foxp3 expression, compared with WT T cells. Although the character of Arid5a-deficient T cells under the control of IL-6 and TGF-β signaling needs to be further clarified, Arid5a-deficient T cells under Th17 cell conditions had a similar phenotype to Tr1 cells, in which the level of Maf expression was significantly higher than WT CD4+ T cells. Maf expression was shown to be dependent on STAT3 activation in CD4+ T cells under Th17 cell conditions (Xu et al., 2009), whereas its expression was also enhanced by STAT1 in CD4+ T cells under Tr1 cell conditions, which contributed to the elevation of Il10 expression in CD4+ T cells (Ng et al., 2013; Zhu et al., 2015). Moreover, enhancement of STAT5 phosphorylation could also limit canonical Th17 cell differentiation, possibly due to high levels of IL-2 (Laurence et al., 2007; Yang et al., 2011). Indeed, anti–IL-2 treatment had a partial effect on the frequency of Th17 cells in Arid5a-deficient T cells. This suggests that IL-2 secreted from Arid5a-deficient T cells could partly restrain the differentiation of Th17 cells through activation of STAT5. Because our data also showed that retrovirus-induced overexpressing STAT3C recovered the decreased Th17 cell populations in Arid5a-deficient T cells, both enhanced phosphorylation of STAT1 and STAT5 associated with reduction of STAT3 levels might secondarily contribute to the impairment of Th17 cell differentiation in Arid5a-deficient T cells.
In this study, we have shown a T cell–intrinsic function of Arid5a as a stability protein of Stat3 mRNA. Arid5a bound to the stem-loop region (1738–1765) of Stat3 mRNA via its residue R128, which in turn resulted in inhibition of Regnase-1 binding to the same region of the Stat3 3′UTR. In T cells, levels of Arid5a expression are augmented via the IL-6–dependent pathway. Loss of Arid5a in T cells led to reduction of STAT3 protein, which correlated with an imbalance in STAT1, STAT3, and STAT5 activation under Th17-polarizing conditions. Activation of STAT1 in T cells lacking Arid5a could be a key factor in the alteration of the feature of inflammatory CD4+ T cells under Th17 cell conditions into that of antiinflammatory CD4+ T cells like Tr-1 cells. Thus, given that Arid5a functions as a director of the development of inflammatory CD4+ T cells, it is strongly implicated that it is an effective therapeutic strategy in IL-6–dependent autoimmune disease to specifically inhibit the function of Arid5a not only in macrophages but also in T cells.
MATERIALS AND METHODS
Mice
C57BL/6J wild-type mice (8–10 wk) were obtained from CLEA Japan, Inc. Arid5a−/− mice were generated as previously described (Masuda et al., 2013). Rag2−/− mice were purchased from Charles River. Mice were maintained under specific pathogen–free conditions. All animal experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committees of the Graduate School of Frontier Bioscience (Osaka University, Osaka, Japan).
In vitro T cell differentiation
Naive CD4+ T cells (CD4+CD44lowCD62LhiCD25−) were isolated by MACS beads cell isolation kits (Miltenyi Biotec) or flow cytometry from the spleen using FACS ARIAII (BD). Naive CD4+ T cells were stimulated with plate-bound anti-CD3ε (5 µg/ml; 145-2C11; BioLegend) and anti-CD28 (2 µg/ml; 37.51; BioLegend), in the presence of IL-12 (20 ng/ml) and anti–IL-4 (10 µg/ml; 11B11; BioLegend) for the generation of Th1 cells; or IL-4 (50 ng/ml) and anti–IFN-γ (10 µg/ml; XMG1.2; BioLegend) for the generation of Th2 cells; or IL-6 (20 ng/ml), TGF-β1 (4 ng/ml), anti-IL4, and anti-IFN-γ for the generation of Th17 cells; or IL-2 (20 U/ml) and hTGF-β1 (4 ng/ml) for the generation of regulatory T cells. Mouse IL-4, IL-6, IL-12, IL-2, and human TGF-β1 were obtained from R&D Systems.
Measurement of cytokines
Cytokines were measured by ELISA kits provided by R&D Systems. Naive CD4+ WT or Arid5a-deficient T cells were differentiated under Th1 or Th17 cell–inducing conditions. The supernatants were collected for the measurement of IL-17 or IFN-γ.
Quantitative real-time PCR analysis
Total RNA was extracted with RNeasy columns (QIAGEN). Reverse transcription of mRNA was performed in a thermal cycler (Applied Biosystems). Real-time PCR was performed in the ABI PRISM 7900 HT (Applied Biosystems) using the following primers for TaqMan gene expression (Applied Biosystems): Arid5a (Mm00524454_m1), Gata3 (Mm00484683_m1), Maf (Mm02581355_s1), Tbx21 (Mm00450960_m1), Rorc (Mm01261022_m1), Rora (Mm01173766_m1), Il17a (Mm00439618_m1), Il10 (Mm00439614_m1), Il4 (Mm00445259_m1), Zc3h12a (Mm00462535_g1), Stat3 (Mm01219775_m1), Stat4 (Mm00448890_m1), Stat1 (Mm00439531_m1), Stat5a (Mm03053818_s1), Stat6 (Mm01160477_m1), Batf (Mm00479410_m1), Il2 (Mm00434256_m1), Gapdh (Mm99999915_g1), Icos (Mm00497600_m1), Irf1 (Mm01288580_m1), and Ifng (Mm01168134_m1). The following primers for FastStart Universal SYBR Green Master (Rox; Roche) were also used: human GAPDH forward, 5′-AGGGCTGCTTTTAACTCTGGT-3′, reverse, 5′-CCCCACTTGATTTTGGAGGGA-3′; human STAT3 forward, 5′-AAGAGGCGGCAACAGATT-3′, and reverse, 5′-CGGTCTTGATGACGAGGG-3′; human ZC3H12A forward, 5′-AACTGGAGAAGAAGAAGATCCTGG-3′, and reverse, 5′-ATTGACGAAGGAGTACATGAGCAG-3′.
Stability assay of mRNA in T cells
Naive T cells were cultured for 12 h under Th1-, Th2-, or Th17-polarizing conditions. After actinomycin D treatment, total mRNA was collected for the measurement of real-time PCR analysis.
Intracellular cytokine staining
T cells were stimulated with 50 ng/ml PMA (Sigma-Aldrich) and 800 ng/ml ionomycin (EMD Millipore) for 5 h, with GolgiStop (BD) added for the final 2 h, followed by fixation and permeabilization with Cytofix/Cytoperm (BD). Cells were stained intracellularly with phycoerythrin-conjugated anti–IL-17 (BioLegend) and FITC-labeled anti–IFN-γ (eBioscience). For Foxp3 staining, T cells were fixed and permeabilized with the Fixation/Permeabilization buffer (eBioscience) for 2 h at 4°C before intracellular staining with FITC-conjugated anti-Foxp3 (eBioscience). Flow cytometric analysis was performed with FACS ARIAII (BD).
Flow cytometric analysis of phospho-STAT1 (Y701) and phospho-STAT3 (Y705)
Naive T cells were cultured for 30 min in the presence of IL-6 and TGF-β. Cells were fixed with 4% paraformaldehyde (Thermo Fisher Scientific) for 10 min at 37°C, and then permeabilized in 90% methanol for 30 min on ice. Cells were washed twice by Stain Buffer (BD), and stained with Alexa Fluor 488–conjugated phospho-STAT1 (Y701) antibody or phycoerythrin-conjugated phospho-STAT3 (Y705) antibody for 1 h at room temperature (BD). Flow cytometric analysis was performed with FACS ARIAII (BD).
RNA EMSA
EMSA was performed according to the protocol of LightShift Chemiluminescent RNA EMSA kit (Thermo Fisher Scientific). The RNA was synthesized as single strand and 3′-end labeled by biotin (Hokkaido System Science). The sequences used were 5′-UGCAGUGGCUUGUGUUCUGGCCACUGCA-3′, and 5′-UGCACACCCUUGUGUUCUGGCCACUGCA-3′. The mouse Arid5a recombinant protein or mutant Arid5a protein with an alanine substitution at residue 128 was prepared by Chugai Pharmaceutical Co.
Retroviral infection
Naive CD4+ T cells were transduced with retroviruses, as previously described (Chen et al., 2005). Murine stem cell virus retroviral DNA plasmids (MSCV-IRES-GFP, MIG) were transfected into a potent retrovirus packaging cell line named Platinum-E, Plat-E (Morita et al., 2000). After 3 d, retrovirus-containing supernatants were collected. CD4+ T cells purified by magnetic-activated cell sorting were activated for 24 h with plate-bound anti-CD3 and anti-CD28, and then infected by centrifugation (45 min at 2,000 rpm) with retrovirus-containing supernatant supplemented with polybrene (8 µg/ml; Sigma-Aldrich) and recombinant human IL-2 (25 U/ml).
Plasmids
The plasmid expressing Arid5a or Regnase-1 was prepared as previously described, respectively (Masuda et al., 2013). The luciferase vector of pGL3 encoding full-length STAT3 3′UTR (1–1895) and a fragment of STAT3 3′UTR (1–901), STAT3 3′UTR (902–1895), STAT3 3′UTR (902–1458), STAT3 3′UTR (1449–1792), STAT3 3′UTR (1698–1895), or STAT3 3′UTR (1773–1895) was prepared in-house. The luciferase vector of pGL3 encoding IL-2 3′UTR, IL-17 3′UTR, or RORc 3′UTR was prepared as previously described (Uehata et al., 2013).
STAT3 promoter assay
HEK293T cells were transfected with the luciferase vector encoding human STAT3 promoter (Switchgear Genomics), together with either empty vector alone or human Arid5a expression vector. Cells were lysed after 48 h and analyzed using LightSwitch Luciferase Assay Reagent (Switchgear Genomics).
Proliferation assays
Cell proliferation assay was followed by means of CFSE assay kit (Invitrogen). Naive WT or Arid5a-deficient T cells were stimulated with plate-coated anti-CD3ε and anti-CD28 antibodies in the presence of IL-6 and TGF-β. Cells were stained by CFSE for 48 h, and then analyzed by flow cytometry.
Preparation of recombinant mouse Arid5a
Recombinant mouse Arid5a protein and mutant Arid5a protein were provided by Chugai Pharmaceutical Co., Ltd. FLAG-tagged mouse Arid5a cDNA was cloned into pcDNA 3.4 vector (Thermo Fisher Scientific). Freestyle 293-F cells were transfected with the FLAG-mArid5a plasmid according to the manufacturer’s instruction and cultured for 48 h. Cells were harvested and then disrupted by sonication in suspension buffer (50 mM Hepes-OH, pH 7.5, 500 mM NaCl, and 10% glycerol) supplemented with protease inhibitor cocktail and 12.5 U/ml benzonase (EMD Chemicals Inc.). Cell lysate was clarified by centrifugation and subjected to anti-FLAG M2 affinity resin (Sigma-Aldrich). After 2-h incubation at 4°C, the resin was washed with suspension buffer containing 5 mM ATP and 10 mM MgCl2, and then elution was performed with suspension buffer containing 0.1 mg/ml FLAG peptide. Eluate was collected, supplemented with DTT to give a final concentration of 2 mM, and then concentrated by ultrafiltration. The concentrated eluate was loaded onto a Superdex200 10/300 GL column (GE Healthcare) equilibrated with GPC buffer (50 mM Hepes-OH, pH 7.5, 250 mM NaCl, 0.1% CHAPS, 5 mM DTT, and 1 mM EDTA), which was also used as a buffer for the elution. Fractions containing monomeric FLAG-mArid5a were collected as purified mArid5a. Arid5a mutant (R128 mutant) protein was prepared following the method for the purification of recombinant mouse Arid5a protein.
NMR sample preparation
The synthesized STAT3 3′UTR (1738–1765), (r[UGCAGUGGCUUGUGUUCUGGCCACUGCA]), was purified by HPLC (Japan Bio Services Co., Ltd.), and then desalted. The lyophilized STAT3 3′UTR was dissolved in 10 mM sodium phosphate buffer (pH 6.5) containing 30 mM NaCl, 2 mM MgCl2, and 0.01 mM 2,2-dimethylsilapentane-5-sulfonic acid (DSS). The concentration of the STAT3 3′UTR was 450 µM. The sample was heated at 95°C for 5 min, followed by gradual cooling to room temperature.
NMR spectroscopy
NMR spectra were recorded with a Bruker AVANCE III HD 600 spectrometers equipped with a cryogenic probe with a Z-gradient. NOESY experiment was acquired at 5°C with a mixing time of 400 ms. Chemical shift was calibrated with a DSS resonance. NMR data were processed and analyzed using TopSpin/XWIN-NMR (Bruker), NMRPipe (Delaglio et al., 1995), and an NMR Assignment Program (Sparky).
Arid5a-STAT3 modeling
A model of Arid5a was constructed by first building 3D models of the protein using HHPred (Söding, 2005) and Spanner (Lis et al., 2011) with MRF-2 (Protein Data Bank identifier 1ig6; sequence identity was 75%) as a template. Stem-loops in the STAT3 3′UTR were identified using RNA RNAstructure (Bellaousov et al., 2013) and rendered in 3D using iFoldRNA program (Sharma et al., 2008; Ding et al., 2012). Protein-RNA docking was performed using surFit server. To consider the flexibility of the RNA structure, we ran 10 × 10 ns MD simulations using Gromacs (Hess et al., 2008) with the AMBER ff12SB force field (Salomon-Ferrer et al., 2013). From the docking results, we picked the top 100 clusters and investigated these manually. Here, we used residue-level RNA binding propensity from the program aaRNA (Li et al., 2014) and the DNA-binding mode of MRF2 to identify putative RNA binding residues.
Statistical analysis
Student’s t test (two tailed) was used to analyze data for statistically significant differences. Values of P < 0.05 were regarded as statistically significant.
Online supplemental material
Fig. S1 shows the sequence of the mouse STAT3 3′UTR (902-1895). Table S1 is a comparison of STAT3- or STAT1-controlling gene expression between Arid5a-deficient T cells and WT T cells under Th17 cell conditions. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151289/DC1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Y. Kabumoto (Chugai Pharmaceutical Co., Ltd., Tokyo, Japan) for cell sorting analysis and for recombinant Arid5a protein and mutant Arid5a protein.
This work was supported by the Japan Society for the Promotion of Science Grants-in-Aid for Young Scientists (B; 26860328) and by Scientific Research on Innovative Areas (15H01256); the Japanese Science and Technology Agency and the Japanese Ministry of Education, Culture, Sports, Science and Technology for integrated promotion of social system reform and research and development; and the Kishimoto Foundation.
The authors declare no competing financial interests.
Author contributions: K. Masuda did most of the experiments; K. Yamashita and D. Standley performed computational analysis; T. Mashima and M. Katahira performed NMR analysis; K. Masuda, B. Ripley, K. Nyati, P. Dubey, M. Zaman, H. Hanieh, K. Yamashita, D. Standley, O. Takeuchi, and T. Kishimoto analyzed data; M. Higa and O. Takeuchi contributed reagents and analytical tools; T. Okamoto and Y. Matsuura performed virus infection experiments; T. Kishimoto supervised the project; K. Masuda and T. Kishimoto designed the experiments; K. Masuda, B. Ripley, and T. Kishimoto wrote the paper.
Footnotes
Abbreviations used:
- 3′UTR
- 3′ untranslated region
- Arid
- AT-rich interactive domain
- Arid5a
- Arid-containing protein 5a
- EMSA
- electrophoresis mobility shift assay
- MIG
- MSCV-IRES-GFP
- MRF
- modulator recognition factor
- RA
- rheumatoid arthritis
- Regnase-1
- protein regulatory RNase 1
References
- Awasthi A., Carrier Y., Peron J.P., Bettelli E., Kamanaka M., Flavell R.A., Kuchroo V.K., Oukka M., and Weiner H.L.. 2007. A dominant function for interleukin 27 in generating interleukin 10-producing anti-inflammatory T cells. Nat. Immunol. 8:1380–1389. 10.1038/ni1541 [DOI] [PubMed] [Google Scholar]
- Bellaousov S., Reuter J.S., Seetin M.G., and Mathews D.H.. 2013. RNAstructure: Web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 41:W471–W474 10.1093/nar/gkt290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg J.F., Wrzeszczynska M.H., Devgan G., Zhao Y., Pestell R.G., Albanese C., and Darnell J.E. Jr. 1999. Stat3 as an oncogene. Cell. 98:295–303. 10.1016/S0092-8674(00)81959-5 [DOI] [PubMed] [Google Scholar]
- Chen L., Willis S.N., Wei A., Smith B.J., Fletcher J.I., Hinds M.G., Colman P.M., Day C.L., Adams J.M., and Huang D.C.. 2005. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell. 17:393–403. 10.1016/j.molcel.2004.12.030 [DOI] [PubMed] [Google Scholar]
- Ciofani M., Madar A., Galan C., Sellars M., Mace K., Pauli F., Agarwal A., Huang W., Parkurst C.N., Muratet M., et al. 2012. A validated regulatory network for Th17 cell specification. Cell. 151:289–303. 10.1016/j.cell.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Pereira A.P., Tininini S., Strobl B., Alonzi T., Schlaak J.F., Is’harc H., Gesualdo I., Newman S.J., Kerr I.M., and Poli V.. 2002. Mutational switch of an IL-6 response to an interferon-gamma-like response. Proc. Natl. Acad. Sci. USA. 99:8043–8047. 10.1073/pnas.122236099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. 2014. T follicular helper cell differentiation, function, and roles in disease. Immunity. 41:529–542. 10.1016/j.immuni.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., and Bax A.. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 6:277–293. 10.1007/BF00197809 [DOI] [PubMed] [Google Scholar]
- Ding F., Lavender C.A., Weeks K.M., and Dokholyan N.V.. 2012. Three-dimensional RNA structure refinement by hydroxyl radical probing. Nat. Methods. 9:603–608. 10.1038/nmeth.1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C. 2014. Targeting Th17 cells in immune diseases. Cell Res. 24:901–903. 10.1038/cr.2014.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant L., Watford W.T., Ramos H.L., Laurence A., Vahedi G., Wei L., Takahashi H., Sun H.W., Kanno Y., Powrie F., and O’Shea J.J.. 2010. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 32:605–615. 10.1016/j.immuni.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan S.E., Haapaniemi E., Russell M.A., Caswell R., Lango Allen H., De Franco E., McDonald T.J., Rajala H., Ramelius A., Barton J., et al. 2014. Activating germline mutations in STAT3 cause early-onset multi-organ autoimmune disease. Nat. Genet. 46:812–814. 10.1038/ng.3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T.J., Grosso J.F., Yen H.R., Xin H., Kortylewski M., Albesiano E., Hipkiss E.L., Getnet D., Goldberg M.V., Maris C.H., et al. 2007. Cutting edge: An in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J. Immunol. 179:4313–4317. 10.4049/jimmunol.179.7.4313 [DOI] [PubMed] [Google Scholar]
- Hess B., Kutzner C., van der Spoel D., and Lindahl E.. 2008. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 4:435–447. 10.1021/ct700301q [DOI] [PubMed] [Google Scholar]
- Hong F., Jaruga B., Kim W.H., Radaeva S., El-Assal O.N., Tian Z., Nguyen V.A., and Gao B.. 2002. Opposing roles of STAT1 and STAT3 in T cell-mediated hepatitis: regulation by SOCS. J. Clin. Invest. 110:1503–1513. 10.1172/JCI0215841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C.A., and Jones S.A.. 2015. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 16:448–457. 10.1038/ni.3153 [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Takeuchi O., Teraguchi S., Matsushita K., Uehata T., Kuniyoshi K., Satoh T., Saitoh T., Matsushita M., Standley D.M., and Akira S.. 2011. The IκB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR-IL-1R by controlling degradation of regnase-1. Nat. Immunol. 12:1167–1175. 10.1038/ni.2137 [DOI] [PubMed] [Google Scholar]
- Jeltsch K.M., Hu D., Brenner S., Zöller J., Heinz G.A., Nagel D., Vogel K.U., Rehage N., Warth S.C., Edelmann S.L., et al. 2014. Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote T(H)17 differentiation. Nat. Immunol. 15:1079–1089. 10.1038/ni.3008 [DOI] [PubMed] [Google Scholar]
- Kishimoto T. 2005. Interleukin-6: from basic science to medicine--40 years in immunology. Annu. Rev. Immunol. 23:1–21. 10.1146/annurev.immunol.23.021704.115806 [DOI] [PubMed] [Google Scholar]
- Kishimoto T. 2010. IL-6: from its discovery to clinical applications. Int. Immunol. 22:347–352. 10.1093/intimm/dxq030 [DOI] [PubMed] [Google Scholar]
- Laurence A., Tato C.M., Davidson T.S., Kanno Y., Chen Z., Yao Z., Blank R.B., Meylan F., Siegel R., Hennighausen L., et al. 2007. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 26:371–381. 10.1016/j.immuni.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Li S., Yamashita K., Amada K.M., and Standley D.M.. 2014. Quantifying sequence and structural features of protein-RNA interactions. Nucleic Acids Res. 42:10086–10098. 10.1093/nar/gku681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Song W., Bi X., Zhao J., Huang Z., Li Z., Zhou J., Cai J., and Zhao H.. 2014. Recent advances in the ARID family: focusing on roles in human cancer. Onco Targets Ther. 7:315–324. 10.2147/OTT.S57023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis M., Kim T., Sarmiento J., Kuroda D., Dinh H., Kinjo A.R., Devadas S., Nakamura H., and Standley D.M.. 2011. Bridging the gap between single-template and fragment based protein structure modeling using Spanner. Immunome Res. 7:1. [Google Scholar]
- Ma C.S., and Deenick E.K.. 2014. Human T follicular helper (Tfh) cells and disease. Immunol. Cell Biol. 92:64–71. 10.1038/icb.2013.55 [DOI] [PubMed] [Google Scholar]
- Masuda K., and Kishimoto T.. 2014. Th17 cells: inflammation and regulation. Atlas Genet. Cytogenet. Oncol. Haematol. 18:611–623. 10.4267/2042/54019 [DOI] [Google Scholar]
- Masuda K., Ripley B., Nishimura R., Mino T., Takeuchi O., Shioi G., Kiyonari H., and Kishimoto T.. 2013. Arid5a controls IL-6 mRNA stability, which contributes to elevation of IL-6 level in vivo. Proc. Natl. Acad. Sci. USA. 110:9409–9414. 10.1073/pnas.1307419110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita K., Takeuchi O., Standley D.M., Kumagai Y., Kawagoe T., Miyake T., Satoh T., Kato H., Tsujimura T., Nakamura H., and Akira S.. 2009. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 458:1185–1190. 10.1038/nature07924 [DOI] [PubMed] [Google Scholar]
- Mino T., Murakawa Y., Fukao A., Vandenbon A., Wessels H.H., Ori D., Uehata T., Tartey S., Akira S., Suzuki Y., et al. 2015. Regnase-1 and Roquin Regulate a Common Element in Inflammatory mRNAs by Spatiotemporally Distinct Mechanisms. Cell. 161:1058–1073. 10.1016/j.cell.2015.04.029 [DOI] [PubMed] [Google Scholar]
- Morita S., Kojima T., and Kitamura T.. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063–1066. 10.1038/sj.gt.3301206 [DOI] [PubMed] [Google Scholar]
- Muranski P., and Restifo N.P.. 2013. Essentials of Th17 cell commitment and plasticity. Blood. 121:2402–2414. 10.1182/blood-2012-09-378653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K.M., and Reiner S.L.. 2002. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2:933–944. 10.1038/nri954 [DOI] [PubMed] [Google Scholar]
- Nagashima H., Okuyama Y., Asao A., Kawabe T., Yamaki S., Nakano H., Croft M., Ishii N., and So T.. 2014. The adaptor TRAF5 limits the differentiation of inflammatory CD4(+) T cells by antagonizing signaling via the receptor for IL-6. Nat. Immunol. 15:449–456. 10.1038/ni.2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T.H., Britton G.J., Hill E.V., Verhagen J., Burton B.R., and Wraith D.C.. 2013. Regulation of adaptive immunity; the role of interleukin-10. Front. Immunol. 4:129 10.3389/fimmu.2013.00129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly S., Cant R., Ciechomska M., and van Laar J.M.. 2013. Interleukin-6: a new therapeutic target in systemic sclerosis? Clin. Transl. Immunology. 2:e4 10.1038/cti.2013.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea J.J., Lahesmaa R., Vahedi G., Laurence A., and Kanno Y.. 2011. Genomic views of STAT function in CD4+ T helper cell differentiation. Nat. Rev. Immunol. 11:239–250. 10.1038/nri2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y., Kagami S., Sanayama Y., Ikeda K., Suto A., Kashiwakuma D., Furuta S., Iwamoto I., Nonaka K., Ohara O., and Nakajima H.. 2014. AT-rich-interactive domain-containing protein 5A functions as a negative regulator of retinoic acid receptor-related orphan nuclear receptor γt-induced Th17 cell differentiation. Arthritis Rheumatol. 66:1185–1194. 10.1002/art.38324 [DOI] [PubMed] [Google Scholar]
- Salomon-Ferrer R., Case D.A., and Walker R.C.. 2013. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 3:198–210. 10.1002/wcms.1121 [DOI] [Google Scholar]
- Sharma S., Ding F., and Dokholyan N.V.. 2008. iFoldRNA: three-dimensional RNA structure prediction and folding. Bioinformatics. 24:1951–1952. 10.1093/bioinformatics/btn328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics. 21:951–960. 10.1093/bioinformatics/bti125 [DOI] [PubMed] [Google Scholar]
- Stritesky G.L., Muthukrishnan R., Sehra S., Goswami R., Pham D., Travers J., Nguyen E.T., Levy D.E., and Kaplan M.H.. 2011. The transcription factor STAT3 is required for T helper 2 cell development. Immunity. 34:39–49. 10.1016/j.immuni.2010.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormo A.J., Letellier M.C., Sharma M., Elson G., Crabé S., and Gauchat J.F.. 2012. IL-6 activates STAT5 in T cells. Cytokine. 60:575–582. 10.1016/j.cyto.2012.07.002 [DOI] [PubMed] [Google Scholar]
- Uehata T., Iwasaki H., Vandenbon A., Matsushita K., Hernandez-Cuellar E., Kuniyoshi K., Satoh T., Mino T., Suzuki Y., Standley D.M., et al. 2013. Malt1-induced cleavage of regnase-1 in CD4(+) helper T cells regulates immune activation. Cell. 153:1036–1049. 10.1016/j.cell.2013.04.034 [DOI] [PubMed] [Google Scholar]
- Xu J., Yang Y., Qiu G., Lal G., Wu Z., Levy D.E., Ochando J.C., Bromberg J.S., and Ding Y.. 2009. c-Maf regulates IL-10 expression during Th17 polarization. J. Immunol. 182:6226–6236. 10.4049/jimmunol.0900123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.P., Ghoreschi K., Steward-Tharp S.M., Rodriguez-Canales J., Zhu J., Grainger J.R., Hirahara K., Sun H.W., Wei L., Vahedi G., et al. 2011. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat. Immunol. 12:247–254. 10.1038/ni.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef N., Shalek A.K., Gaublomme J.T., Jin H., Lee Y., Awasthi A., Wu C., Karwacz K., Xiao S., Jorgolli M., et al. 2013. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 496:461–468. 10.1038/nature11981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Pardoll D., and Jove R.. 2009. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer. 9:798–809. 10.1038/nrc2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Ivanov I.I., Spolski R., Min R., Shenderov K., Egawa T., Levy D.E., Leonard W.J., and Littman D.R.. 2007. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 8:967–974. 10.1038/ni1488 [DOI] [PubMed] [Google Scholar]
- Zhu C., Sakuishi K., Xiao S., Sun Z., Zaghouani S., Gu G., Wang C., Tan D.J., Wu C., Rangachari M., et al. 2015. An IL-27/NFIL3 signalling axis drives Tim-3 and IL-10 expression and T-cell dysfunction. Nat. Commun. 6:6072 10.1038/ncomms7072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Yamane H., and Paul W.E.. 2010. Differentiation of effector CD4 T cell populations (*). Annu. Rev. Immunol. 28:445–489. 10.1146/annurev-immunol-030409-101212 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.