Significance
DNA-dependent RNA polymerases (RNAPs) are complex enzymes that synthesize RNA in a factor-dependent fashion. Like mechanical engines, RNAPs consist of rigid and flexible parts; the catalytic function of RNAPs critically relies on conformational changes. Based on single-molecule FRET measurements that directly report on the movements of the RNAP clamp of the archaeal 12-subunit RNAP, we show that the clamp domain exists in alternative states distinguishable by the width of the DNA binding channel. The conformation of the clamp is adjusted through the transcription cycle; more precisely, it varies as a function of (i) RNA subunits Rpo4/7, (ii) the presence of the DNA nontemplate strand, and (iii) transcription initiation and elongation factors TFE and Spt4/5, respectively.
Keywords: RNA polymerase, single-molecule FRET, transcription, archaea, clamp
Abstract
Transcription is an intrinsically dynamic process and requires the coordinated interplay of RNA polymerases (RNAPs) with nucleic acids and transcription factors. Classical structural biology techniques have revealed detailed snapshots of a subset of conformational states of the RNAP as they exist in crystals. A detailed view of the conformational space sampled by the RNAP and the molecular mechanisms of the basal transcription factors E (TFE) and Spt4/5 through conformational constraints has remained elusive. We monitored the conformational changes of the flexible clamp of the RNAP by combining a fluorescently labeled recombinant 12-subunit RNAP system with single-molecule FRET measurements. We measured and compared the distances across the DNA binding channel of the archaeal RNAP. Our results show that the transition of the closed to the open initiation complex, which occurs concomitant with DNA melting, is coordinated with an opening of the RNAP clamp that is stimulated by TFE. We show that the clamp in elongation complexes is modulated by the nontemplate strand and by the processivity factor Spt4/5, both of which stimulate transcription processivity. Taken together, our results reveal an intricate network of interactions within transcription complexes between RNAP, transcription factors, and nucleic acids that allosterically modulate the RNAP during the transcription cycle.
RNA polymerases (RNAPs) are at the heart of the transcription machinery and catalyze the synthesis of RNA in a DNA-dependent fashion. This intrinsically dynamic and highly regulated process can be divided into the initiation, elongation, and termination phases of transcription (1, 2). Variations in structures of different RNAPs have suggested that the RNAP clamp is a structurally mobile element that can adopt different degrees of closure over the DNA/RNA hybrid and the DNA binding channel (Fig. 1). Crystal structures of the complete 12-subunit Saccharomyces cerevisiae RNAPII (3, 4) and of the cognate transcription elongation complex (5–7) reveal the clamp in a closed state, with a concomitant narrowing of the DNA binding channel. By comparison, the clamp of a 10-subunit S. cerevisiae RNAPII that lacks the stalk consisting of subunits Rpb4/7 (8) is in an open state. Electron microscopy analysis of free RNAPII in solution further supported the notion of a conformational flexible clamp, suggesting that Rpb4/7 shifts the equilibrium between the closed and collapsed state of the clamp to the closed state (9). Moreover, the clamp of RNAPIII could recently be imaged in two distinct conformations that differ in the orientation of the stalk and the opening of the DNA cleft (10). These data have led to the theory that the stalk influences the clamp position. The archaeal RNAP is highly homologous to eukaryotic RNAPII, and X-ray structures show that the clamp adopts a closed conformation in the RNAPs of crenarchaea Sulfolobus shibatae and Sulfolobus solfataricus, whereas it is open in the RNAP of the euryarchaeon Thermococcus kodakarensis, all of which include the stalk (11, 12). A comparison of cryo-EM structures of the human initiation complexes also provides evidence that the clamp undergoes an opening-closing movement during initiation (13). The 4-subunit RNAP in bacteria is equivalent to the catalytic core of the 12-subunit enzymes in archaea and eukaryotes (1). It contains a clamp domain that is structurally not constrained by subunits such as the Rpo4/7 stalk, which is not present in bacteria. Single-molecule FRET studies of the bacterial RNAP have shown that the clamp closes during late initiation phase and remains closed in elongation complexes (14), whereas it is likely to partially open during pausing and termination.
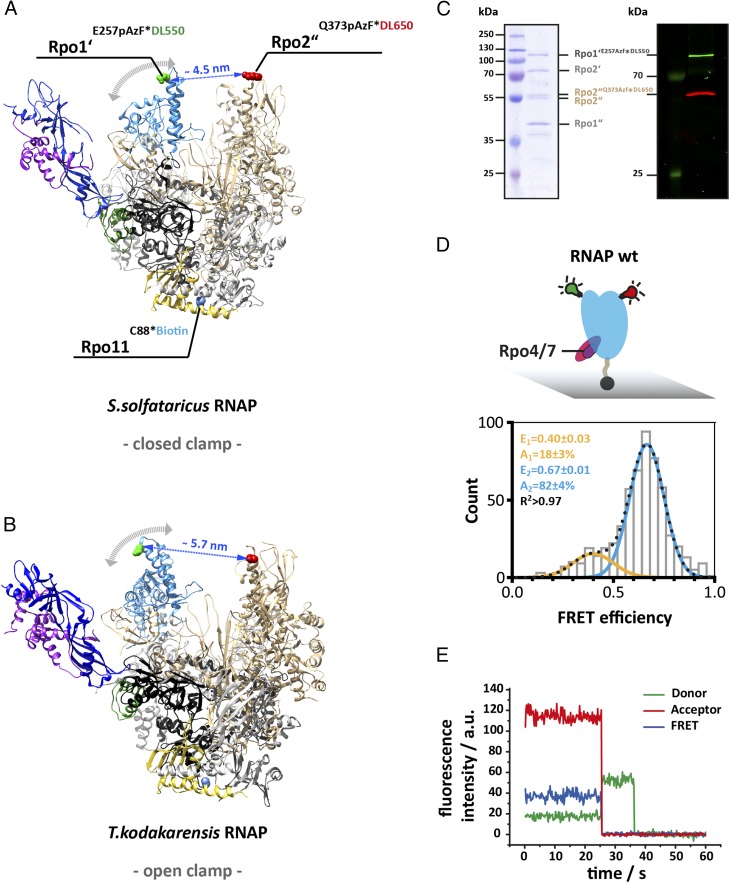
Fig. 1.
Accessing the conformation of the RNA polymerase clamp in solution using single-molecule FRET. Structure of the 12-subunit archaeal RNA polymerase with a (A) closed and (B) open clamp (Protein Data Bank ID codes 2PMZ and 4QIW, respectively). For FRET measurement, a donor (DyLight550) and acceptor (DyLight650) fluorophore were incorporated at the tip of the RNA polymerase clamp coiled coil (subunit Rpo1’ E257, highlighted in green) and the protrusion domain (subunit Rpo2” Q373, highlighted in red). For site-specific labeling the unnatural amino acid p-azidophenylalanine was incorporated at these sites followed by subsequent labeling via Staudinger-Bertozzi ligation. For immobilization purposes a biotin was coupled to a native cysteine in subunit Rpo11. Subunit coloring is as follows: Rpo1′ in dark gray, Rpo2″ in beige, Rpo11 in yellow, Rpo4 in purple, and Rpo7 in blue. (C) The Coomassie 10% SDS/PAGE shows the four large subunits (Left) and the fluorescence scan (Right) of a fully assembled RNAP after purification and demonstrates the specific labeling of subunit Rpo1′ and Rpo2″ with the donor or acceptor dye, respectively. (D) Conformation of the 12-subunit RNA polymerase clamp in the absence of nucleic acid substrates. Biotinylated RNAPs (symbolized by black sphere protruding from the RNAP core) were immobilized on a quartz slide (gray) for TIRF measurements. Histograms show FRET efficiencies determined for the 12-subunit RNAP (RNAP wt). The histogram was fitted with a double Gaussian function and the mean FRET efficiencies, E, the percentage distribution of the populations, A, and the coefficient of determination, R2, are given with SEs in the histograms. (E) Exemplary transients of an immobilized RNA polymerase show stable donor and acceptor fluorescence with no indication of multiple switching FRET states over several seconds and the resulting FRET transient until bleaching of the acceptor occurs (25 ms). Fluorescence transients were recorded using a TIRF microscopy setup with alternating laser excitation.
The RNAP clamp provides binding sites for the transcription initiation factor TFE (homologous to TFIIEα) and the elongation factor Spt4/5, both of which bind competitively to the clamp coiled coil motif (15–17) of archaeal-eukaryotic RNAPs. TFE facilitates open complex formation by interacting with the RNAP clamp and stalk and the nontemplate strand (NTS) (15, 18). The transcription elongation factor Spt4/5 bridges the gap across the DNA binding channel and stabilizes the elongation complex by preventing its dissociation, thereby stimulating the processivity of RNAP (15, 19, 20). Both the binding of TFE and Spt4/5 to RNAP and their modus operandi have been speculated to involve and depend on a repositioning of the clamp. In fact, much has been speculated about the involvement of the RNAP clamp during transcription initiation (13), elongation (21), pausing (22), and termination (23), but its impact on the movement of the clamp is mostly inferred from crystal structures and merits a much more careful study to elucidate the state of the clamp in solution. The conformation of the clamp in archaeal and eukaryotic RNAPs has only been captured in X-ray structures that are static by nature. In this paper, we overcome this limitation by combining a double-fluorescently labeled RNAP from the hyperthermophilic methanogen Methanocaldococcus jannaschii with single-molecule FRET measurements. This approach allowed us to characterize the conformational states of the clamp in solution in the context of transcription initiation and elongation complexes and to unravel the allosteric modulation of RNAP by TFE and Spt4/5.
Results
Experimental Design.
To determine the state of the RNAP clamp, we labeled the RNAP with a donor and acceptor fluorophore at the clamp’s coiled coil tip and at the protrusion domain of the second largest subunit Rpo2″ located opposite the clamp across the DNA binding channel (Fig. 1 A and B). The archaeal RNAP from M. jannaschii (MjRNAP) is composed of 12 subunits like its RNAPII counterpart. However, the large subunits Rpb1 and Rpb2 are split into Rpo1′/Rpo1″ and Rpo2′/Rpo2″, and the small eukaryotic subunits Rpb8 and Rpb9 are missing in the MjRNAP. The labeling of RNAP with dyes was achieved via Staudinger-Bertozzi ligation (24, 25) between the unnatural amino acid p-azido-l-phenylalanine (AzF) incorporated into recombinant RNAP subunits and a phosphine derivative of the donor and acceptor fluorophore (26, 27). For immobilization, a biotin moiety was incorporated into the Rpo11 subunit. RNAPs were assembled from the 12 subunits resulting in single-, double-, or triple-labeled RNAP variants (15, 28–30) (Fig. 1C). The labeled RNAPs were shown to respond to transcription factors and form transcription-competent RNAPs (Fig. S1). Labeled RNAPs were incubated with the respective transcription factors, nucleic acids, and/or nucleoside triphosphates (NTPs) at 65 °C to allow transcription complex formation (15, 19, 29, 31–33). The FRET efficiency between the donor and acceptor dye was measured at room temperature (21 °C) using a PRISM total internal reflection fluorescence (TIRF) microscope with alternating laser excitation (ALEX), which allowed us to spectroscopically select and analyze the complexes that contain both donor and acceptor dyes (34). In all configurations, we monitored stable fluorescence without any indication for dynamic changes in FRET efficiency until one of the dyes underwent photobleaching (Fig. 1E). The dyes did not influence the FRET efficiency, as samples with switched fluorescent probes showed no difference to the standard dye configuration (Fig. S2B). Neither did the direct immobilization of the RNAP via a biotin affect the conformation of the RNAP (Fig. S2C).
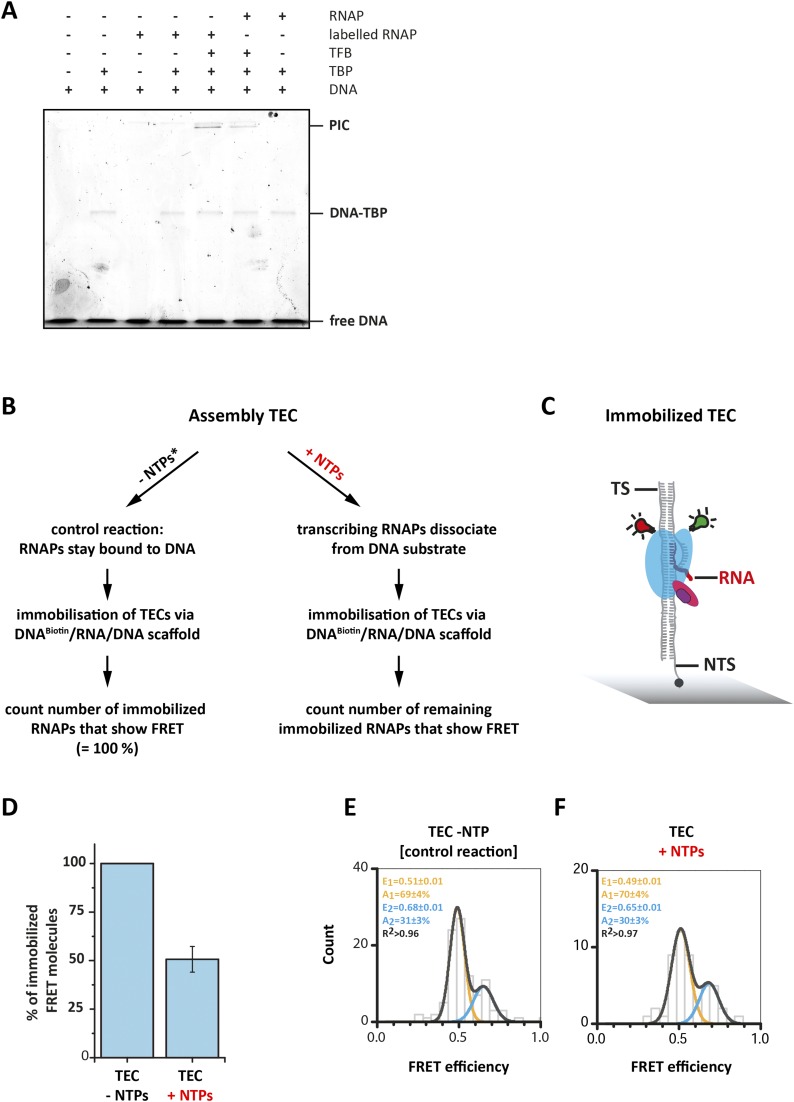
Fig. S1.
Modification of the RNAP does not interfere with preinitiation complex formation and transcriptional activity. (A) PIC formation was followed using an Alexa488-labeled SSV-T6 promoter DNA. Only if all components of the PIC are added is a slowly migrating fluorescent band corresponding to the PIC visible (0.4 µM RNAP, 8.7 µM TBP, 13 µM TFB, 133 nM DNA). The doubly labeled RNAP forms the PIC equally well as the unlabeled RNAP, indicating that the modified RNAP responds faithfully to the transcription initiation factors. (B) The transcriptional activity of donor/acceptor-labeled RNAPs. Double-labeled RNAPs were recruited to the elongation scaffold resulting in a TEC. Addition of NTPs and incubation of the sample at 65 °C starts transcription that ultimately leads to the dissociation of the RNAP when it reaches the end of the DNA substrate. Heparin was used as a scavenger that binds dissociated RNAPs, preventing reassociation of the released RNAP with the DNA substrate. These RNAPs will not be immobilized via the biotinylated elongation scaffold (biotin was positioned at the 5′-end of the NTS). (C) As a control, equal amounts of buffer instead of the NTP solution were added to the TEC, and all steps were performed as for the TEC+NTP sample. (D) The number of double-labeled RNAP molecules in the control (−NTP) vs. +NTP sample that are immobilized via the biotinylated elongation scaffold provides a direct readout about the number of double labeled RNAPs that were transcriptional active. NTP addition to the TEC led to a 51% reduction in the number of immobilized double-labeled RNAP molecules compared with the control sample providing evidence that the modified RNAPs are transcriptionally active. Other rare causes like spontaneous dissociation of the RNAP might occur but are not likely to contribute significantly to the total number of dissociated RNAPs. However, in this assay we are not able to differentiate between these processes. The FRET efficiency histograms derived for the control (E) and NTP incubated sample (F) are highly comparable exhibiting two FRET populations with the same relative amount of molecules in the corresponding populations. This demonstrated clearly that the transcriptionally active RNAPs are not depopulating a single “active” population but both RNAP populations are transcriptionally active. Data accumulated from three independent experiments.
Fig. S2.
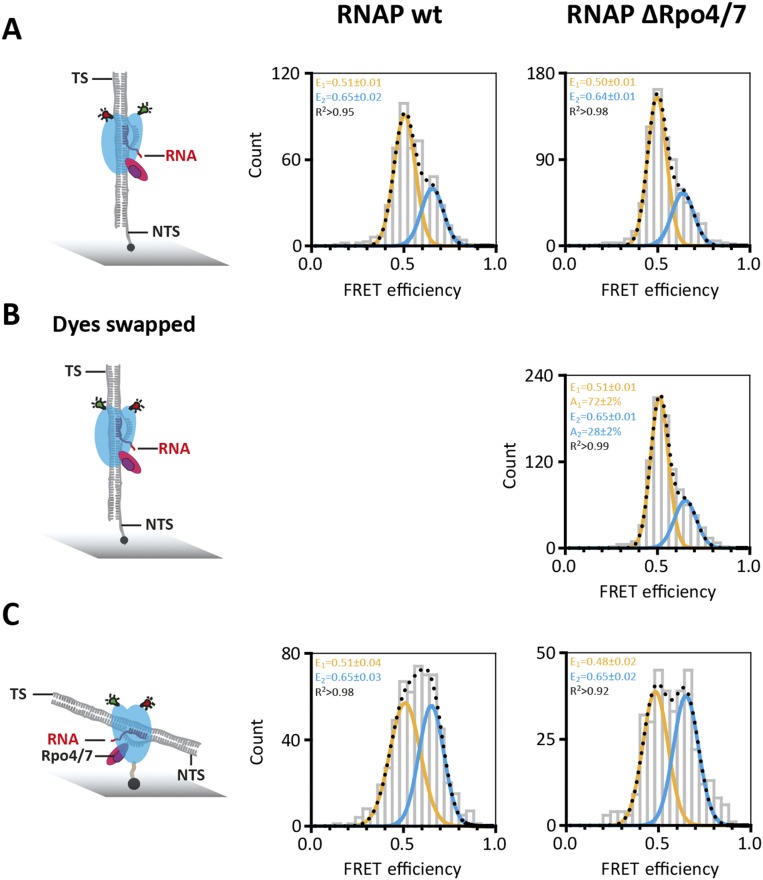
Conformation of the RNA polymerase with exchanged dyes and alternative immobilization strategy. (A) Standard dye configuration (subunit Rpo1′E257*DL550 and Rpo2″Q373*DL650) and immobilization strategy via the NTS of the elongation scaffold. (B) Donor (DyLight550) and acceptor (DyLight650) fluorophore were incorporated into Rpo2”Q373 and Rpo1’E257, respectively. Comparison with RNAP with the “standard” dye configuration shows essentially no difference, which demonstrates that the position of the dye does not influence the mean FRET efficiencies or the distribution between populations. These measurements were carried out when the RNAP was part of a transcription elongation complex (experimental conditions as in Fig. 4A). (C) Transcription elongation complexes were immobilized via a biotin incorporated directly into RNAP subunit 11 instead of a biotinylated nontemplate strand. In both cases, two populations are observed, indicating that the immobilization does not affect the functionality of the RNAP. Moreover, the mean FRET efficiencies values for the population remain unchanged. The difference in amplitude and width of the populations in A is most likely due to a subpopulation of directly immobilized RNAPs without a loaded elongation scaffold.
The RNAP Exhibits a Predominantly Closed Clamp.
We first investigated the conformation of the clamp of the “free” RNAP, i.e., not bound to nucleic acids, transcription factors, or substrates. The 12-subunit RNAP (RNAP wt) shows two populations: a major high FRET population (82 ± 4%) with a FRET efficiency (E) of 0.67 ± 0.03 and a minor low FRET population (18 ± 3%) at a FRET efficiency of 0.40 ± 0.03 (Fig. 1D). These data suggest that the clamp can adopt a closed and open conformation, respectively. Changes in FRET are a result of an altered interdye distance that, in principle, could also be caused by a pivoting motion of the core and shelf modules of the RNAP (35, 36). Please note that we refer to an open and closed state of the clamp if a low and high FRET population is present, which refers to the conformational state of the clamp and is not identical to the clamp states defined in crystal structures. We calculated the equivalent interdye distance to 4.7 and 5.7 nm (Materials and Methods). The distance between the labeled amino acids in the crenarchaeal RNAP structure is 4.5 nm, in good agreement with our solution data (37). We did not observe an interconversion between these two states in our measurements. However, it appears likely that dynamic switching occurs at the biological relevant temperature of 86 °C of the hyperthermophilic model organism. Due to the thermal instability of the immobilization and complex photophysical dye behavior, this idea could not be confirmed by measurements at higher temperature.
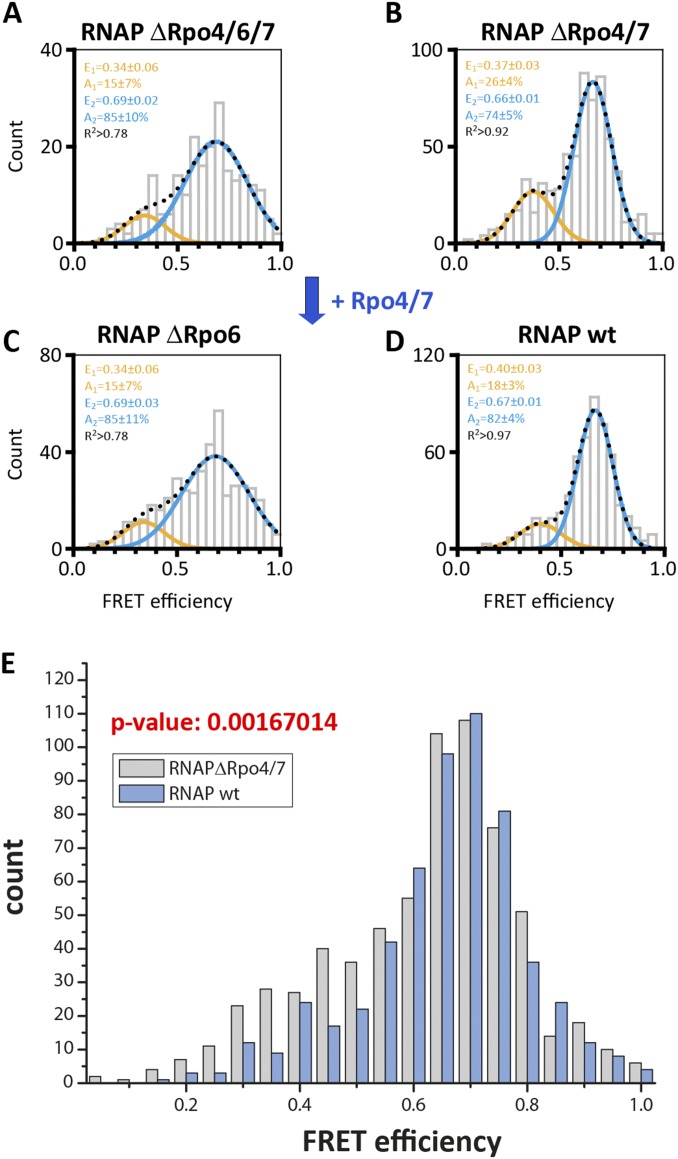
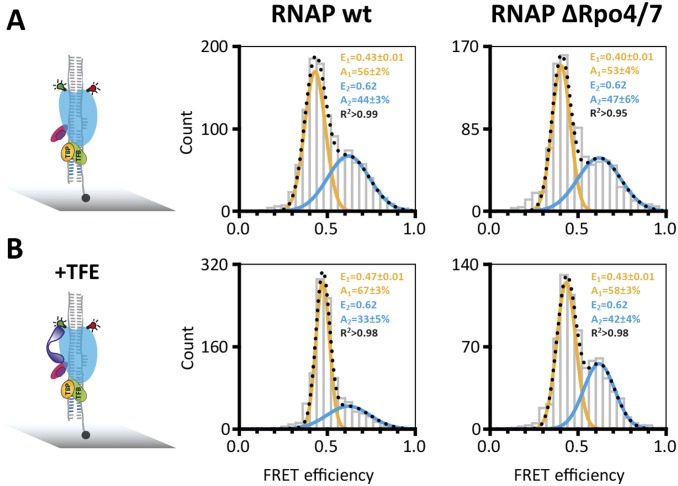
A comparison of X-ray structures suggest that the yeast RPB4/7 stalk biases the clamp toward the closed position. We prepared RNAP variants without the stalk subunits Rpo4/7. (RNAP ΔRpo4/7) and found two populations with mean FRET efficiencies of 0.37 ± 0.03 (26 ± 4%) and 0.66 ± 0.01 (74 ± 5%) (Fig. S3B). The slight shift toward the low FRET population indicates that the presence of the stalk slightly influences the conformation of the clamp, and statistical analysis using the Pearson χ2 test supported the fact that the difference between the dataset is statistically relevant (Fig. S3 B and E). To further verify that the small Rpo4/7-induced shift toward the low FRET population is real, we added Rpo4/7 to a ΔRpo4/6/7 RNAP lacking the Rpo6 docking site required for Rpo4/7 binding (29, 38) and observed no comparable shift (Fig. S3).
Fig. S3.
Conformation of the RNA polymerase clamp in the absence of subunit Rpo6 and Rpo4/7. Our recombinant archaeal model system allowed us to perturb the subunit composition of the labeled RNAPs. We prepared RNAP variants without the stalk subunits Rpo4/7 (RNAPΔRpo4/7) and without subunit Rpo6 that serves as docking site for Rpo4/7. Histograms showing the single-molecule FRET efficiencies determined for the 9-subunit RNAP (RNAP ΔRpo4/6/7; A) and 11-subunit RNAP (RNAP ΔRpo6; C) in comparison with the 10- and 12-subunit RNAP (RNAP ΔRpo4/7 and RNAP wt; B and D). RNAPs were immobilized via a biotin in subunit Rpo11 on a quartz slide for TIRF measurements. The histograms were fitted with a double Gaussian function and the mean FRET efficiencies, E, the percentage distribution of the populations, A, and the coefficient of determination, R2, are given with SEs in the histograms. Although addition of subunits Rpo4/7 to a RNAP ΔRpo4/7 enzyme slightly decreases the amount of low FRET population (E = 0.37), no shift was observed in an RNAP ΔRpo6 enzyme. Rpo6 is the docking site for subunits Rpo4/7, and hence, in its absence, Rpo4/7 cannot associate with the RNAP core. The absence of subunit Rpo6 leads to a very broad distribution, suggesting that Rpo6 might play a role in adjusting the position of the clamp. (E) Comparison of free RNAP wt vs. RNAP ΔRpo4/7 datasets: the null hypothesis that the datasets have the same distribution is rejected at the 5% level based on the Pearson χ2 test (P = 0.0016704 = 0.17%). In contrast, the null hypothesis that the datasets have the same distribution is not rejected when comparing and analyzing two RNAP wt or RNAP ΔRpo4/7 datasets. The Pearson χ2 test for RNAP wt datasets yielded P = 0.134966 (13.5%), and the Pearson χ2 test for RNAP ΔRpo4/7 datasets yielded P = 0.679731 (67.9%).
In conclusion, we observed that the clamp of the unliganded RNAP adopts two conformational states with the majority of the molecules found in the closed clamp state.
TFE Opens the RNAP Clamp in the PIC.
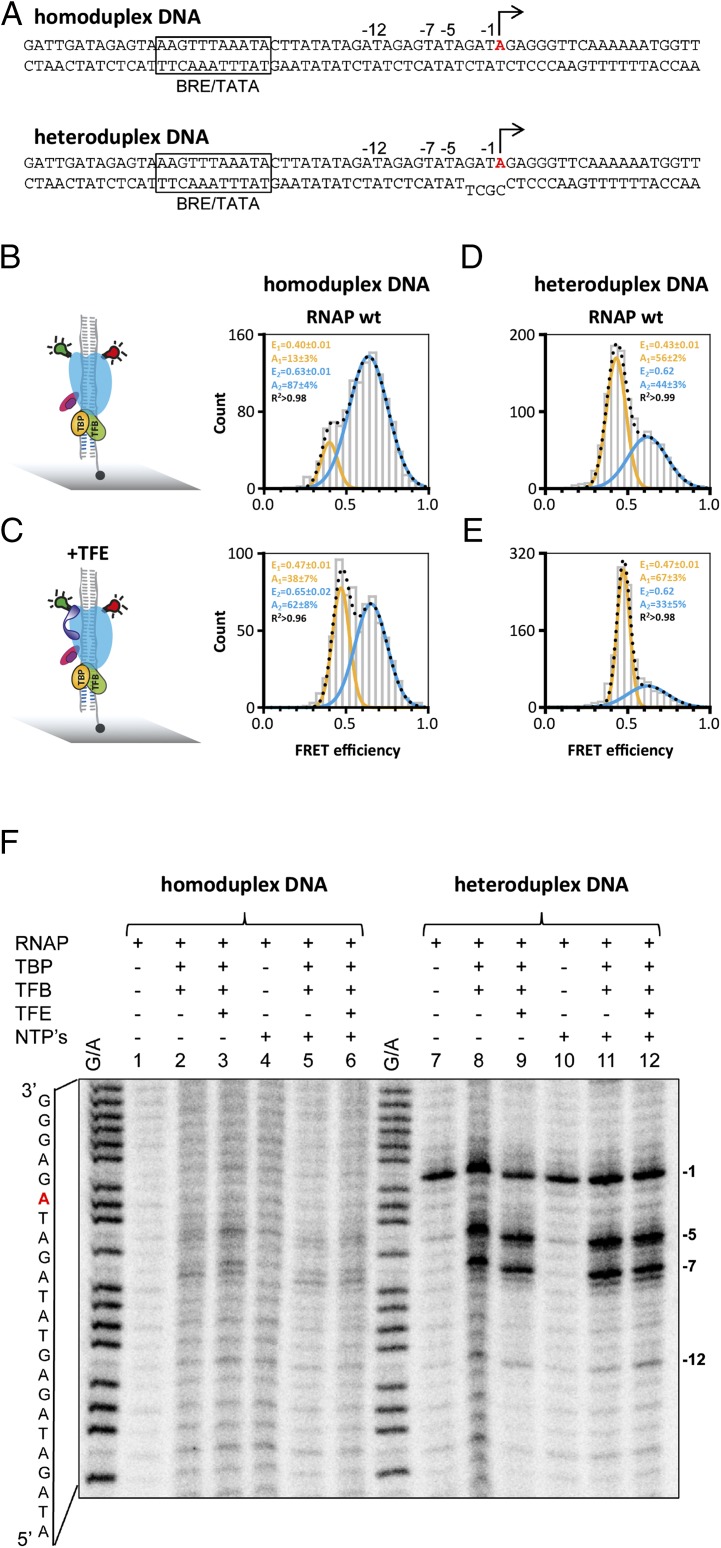
In archaea, two general factors, TATA-binding protein (TBP) and transcription factor B (TFB in archaea and TFIIB in eukaryotes), are necessary and sufficient to facilitate a basal level of transcription. Recruitment of the RNAP to the DNA-TBP-TFB complex leads to the formation of the preinitiation complex (PIC), the promoter DNA is melted, and the template strand is loaded into the active site of the RNAP, which is collectively referred to as open complex formation. This process is not very efficient but is stimulated by a third factor, TFE (TFIIEα) (15, 18, 31). It has not been possible to crystallize the open PIC for structure determination. However, single-molecule FRET mapping and cryo-EM approaches have provided high-confidence models of the PIC in archaea and eukaryotes (13, 33, 39). To characterize the conformational state of the RNAP clamp in the context of closed and open complexes, we assembled PICs using recombinant TBP, TFB, and RNAP on promoter templates that were either complementary (homoduplex, closed) or contained a short region of noncomplementarity (heteroduplex, open) proximal to the transcription start site (TSS) (Fig. 2A). The formation of both PICs is strictly dependent on TBP and TFB (Fig. S1). Permanganate footprinting experiments ascertained that the short region (4 bp) of noncomplementarity inherent in the oligonucleotide template is extended to form a complete transcription bubble. The exact bubble boundaries differ between promoters (40). In permanganate assays, the signal is dependent on the presence of T-residues in the promoter template and on the SSV1 T6 promoter the transcription bubble ranges at least from −12 to −1 (Fig. 2F).
Fig. 2.
Conformation of the RNA polymerase clamp in transcription initiation. (A) PICs were assembled by incubating transcription initiation factors TBP (orange), TFB (green), and a SSV T6 promoter DNA with a heteroduplex region of 4 bp surrounding the transcription start site (−3 to +1) or a fully complementary SSV T6 promoter DNA variant (homdoduplex) at 65 °C before measurement. The single transcription start site in residue +1 is highlighted in red; the BRE and TATA motifs are boxed. Complexes were formed using the doubly labeled RNAP wt. PICs were immobilized via a biotinylated template strand on a quartz glass slide (gray) for TIRF measurements. (B–D) Histograms showing the smFRET efficiencies determined for the PIC in the absence (B and D) and presence (C and E) of TFE (purple) (TFE was included in the PIC assembly reaction and incubated at 65 °C to allow complex formation). (F) Permanganate footprinting assay of the closed and open SSV T6 preinitiation complexes (Materials and Methods). The G+A marker lane allows orientation of the bands on the sequence in A. Using homoduplex templates, no permanganate reactivity was observed, congruent with the complexes predominantly being in the closed confirmation. In the heteroduplex promoter variant, the thymidine (T) residue at position −1 is located within the single-stranded region of the promoter and is permanganate reactive without any addition of TBP and TFB (lane 7, indicated with −1). The addition of TBP, TFB (lane 8) leads to two new reactive T residues at position −5 and −7 that reflect RNAP-directed transcription bubble formation. Addition of TFE gives rise to a new signal at register −12 (lane 9). The addition of nucleotides (NTPs) does not lead to additional signals.
Single-molecule FRET measurements demonstrate that the RNAP clamp in the PICs exists in two populations with mean FRET efficiencies of 0.40 ± 0.01 and 0.63 ± 0.01. This result demonstrates that the RNAP clamp in the PICs, similar to free RNAP, can adopt two conformational states (Fig. 2 B–D). A comparison of PICs assembled on homoduplex and heteroduplex templates demonstrates that the (pre)melting of the promoter shifts the equilibrium toward the open clamp state from 13 ± 3% to 56 ± 2% (Fig. 2 B and D). This result suggests that concomitant with DNA melting the RNAP clamp is opened. It is interesting to note that Rpo4/7 does not alter the relative distribution between the two populations nor the mean FRET efficiencies (E = 0.40 ± 0.01 and E = 0.62) (Fig. S4). This result implies that the tight interaction network between RNAP, TBP, TFB, and DNA overrides the effect of the stalk and controls the clamp conformation. TFE in archaea and TFIIE in the eukaryotic pol II system stabilizes the PIC and stimulates transcription by promoting DNA melting (13, 15, 16, 18, 39, 41, 42). TFE is composed of winged helix (WH) and zinc ribbon domains that are anchored to the RNAP clamp and stalk domains: these binding sites are ideally suited to manipulate clamp movements. In our experiments, the addition of TFE depopulates the high FRET state and increases the low FRET population (+25%), showing that TFE favors the opening of the RNAP clamp (Fig. 2 C and E). Notably, the width of the low FRET population is very narrow, indicative of a structurally highly defined state. The RNAP ΔRpo4/7 variant serves as negative control for the authenticity of TFE-induced clamp opening, because this variant cannot recruit TFE (15, 28), and the addition of TFE to ΔRpo4/7-based PICs does not lead to any significant change in main FRET efficiency values or the relative distribution between the two populations (Fig. S4).
Fig. S4.
Conformation of the RNA polymerase clamp in transcription initiation complex. Histograms showing the single-molecule FRET efficiencies determined for the PIC in the absence (A) and presence (B) of TFE (purple). PICs were assembled using transcription initiation factors TBP (orange); TFB (green), the heterdouplex SSV T6 promoter DNA and labeled RNAP wt. PICs were immobilized via a biotinylated template strand on a quartz slide (gray) for TIRF measurements. On addition of TFE, an increased number of RNAP molecules are found in the low FRET state (+11%) indicative of a TFE-induced opening of the RNAP clamp. RNA polymerases that lack subunits Rpo4/7 do not respond to TFE. Subunits Rpo4/7 are known to be important for TFE binding, and the RNAPΔRpo4/7 variant serves as control for the authenticity of the TFE effect. The datasets were fitted with a double Gaussian function, and the mean FRET efficiencies, E, the percentage distribution of the populations, A, and the coefficient of determination, R2, are given with SEs in the histograms.
In essence, both premelting of the promoter template (from 13% to 56% low FRET state without TFE) and TFE action (13–38% on homoduplex promoter) shift the equilibrium toward the open clamp state; the maximum effect is achieved by the combined action of TFE and the premelted promoter DNA (13–67%).
Spt4/5 and the Nontemplate Promoter Strand Affect the RNAP Clamp.
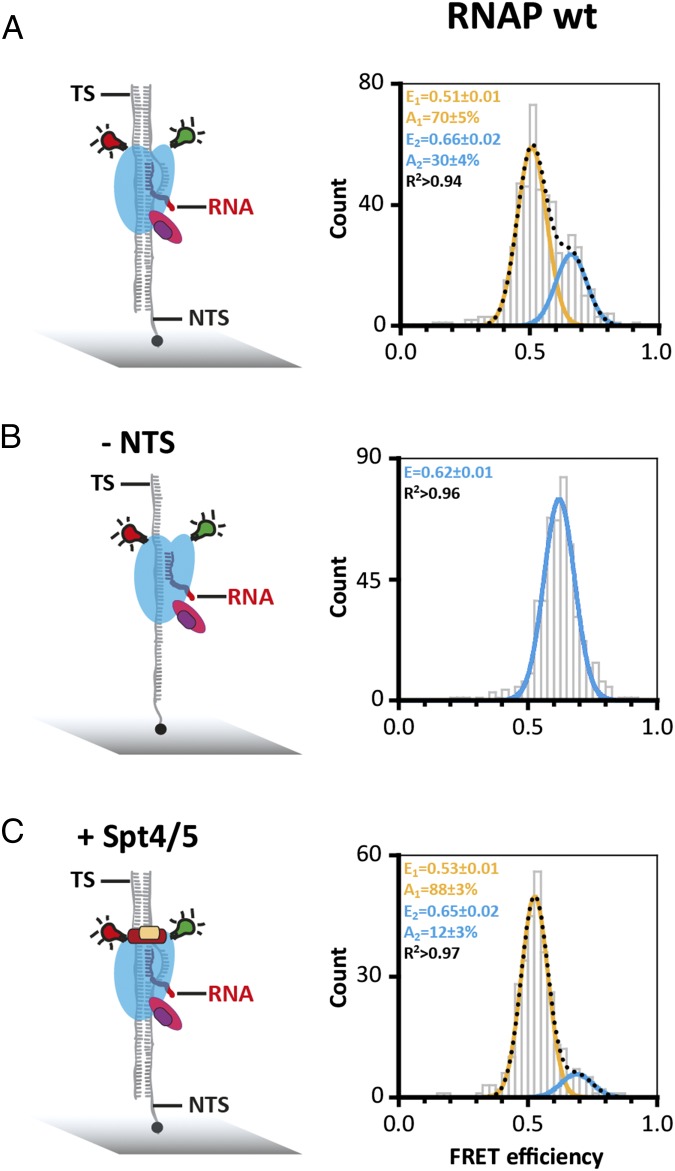
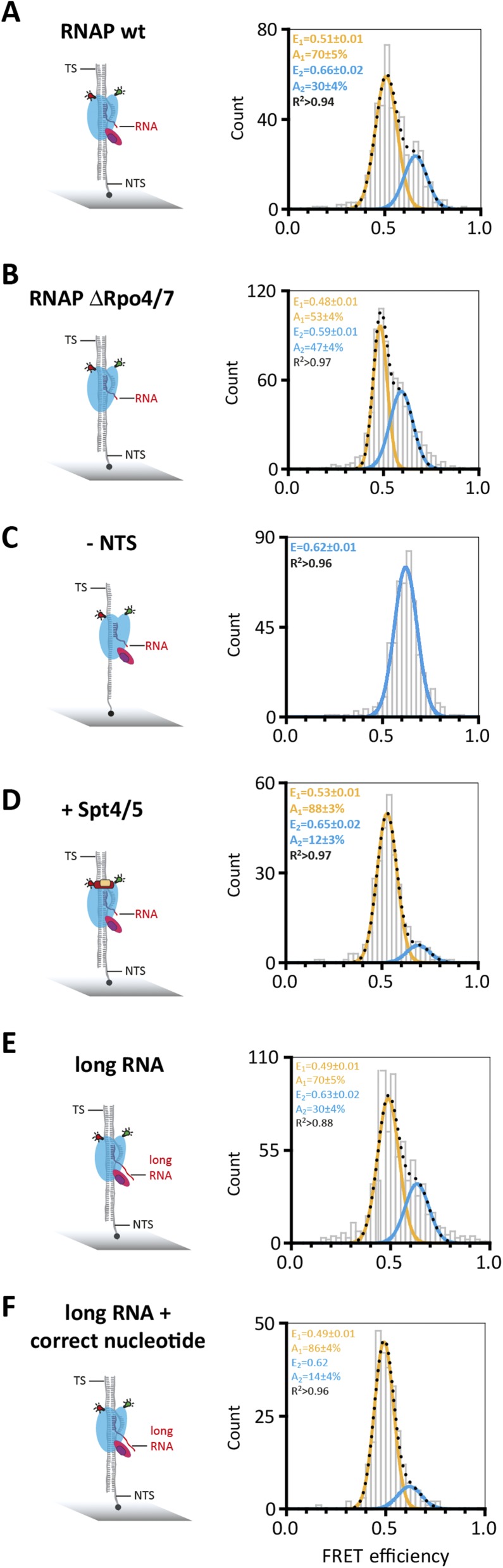
Having characterized the RNAP clamp state in free and promoter-bound RNAP, we next interrogated transcription elongation complexes (TECs). We assembled TECs using synthetic scaffolds made of the DNA template strand (TS), nontemplate strand (NTS), and an RNA primer. The formation of TECs was strictly dependent on the RNA primer, which is important to rule out that the RNAPs are engaged with the DNA template at its termini. Reminiscent of free RNAP and PICs, the TECs revealed two FRET populations centered at mean FRET efficiencies of a low FRET state of E = 0.51 ± 0.01 and a high FRET state of E = 0.66 ± 0.02 (Fig. 3A). It is important to note that the exact FRET values (and distances) are not identical to the two clamp states in free and promoter-bound RNAP. This difference in FRET values is likely due to structural adjustments of RNAP by TBP and TFB in the PICs. Bulk transcription assays have demonstrated the role of the elongation factor Spt4/5, the NTS, and the RNAP Rpo4/7 stalk for facilitating high transcription processivity (19, 32). To characterize the influence of Spt4/5 and the NTS on the RNAP clamp conformation, we added them to or omitted them from the TEC and recorded FRET efficiencies of the resulting complexes. The addition of Spt4/5 had a profound effect on the clamp as it abolished the high FRET state almost entirely (12% remaining in the high FRET population), i.e., fixed the clamp in a single state (Fig. 3, compare A and D). In contrast, omitting the NTS lead to the complete loss of the low FRET state, i.e., fixed the clamp in the alternative state. Because Spt4/5 and NTS are critical for high processivity in transcription assays, and both coincide with the low FRET state (E = 0.51) in our analysis, we infer that this low FRET open clamp state reflects a highly processive/active conformation of the RNAP in the context of the TEC. The Rpo4/7 stalk interacts with the nascent transcript and stimulates processivity (32). However, in line with the free and promoter-bound RNAP, the stalk did not lead to any changes of the clamp in the TEC or the relative distribution between the populations (Fig. S5), irrespective of the length of the RNA (Fig. S5E).
Fig. 3.
Conformation of the RNA polymerase clamp in transcription elongation. (A) Doubly labeled RNAP wt was specifically recruited to elongation scaffolds composed of double-stranded DNA with a 9-bp RNA/DNA heteroduplex to form an artificial transcription bubble. TECs were assembled at 65 °C before immobilization via a biotinylated NTS. (B) TECs were formed in the absence of the NTS and immobilization was achieved via a biotin on the template strand (TS). (C) FRET efficiency distribution for TECs containing the transcription elongation factor Spt4/5 (Spt4/5 was added to the TEC assembly reaction and incubated at 65 °C before immobilization of the TEC).
Fig. S5.
Conformation of the RNA polymerase clamp in transcription elongation. (A and C–F) Doubly labeled RNAP wt or (B) RNAP ΔRpo4/7 are specifically recruited to elongation scaffolds composed of double-stranded DNA with a partly complementary RNA sequence to form an artificial transcription bubble (nine bases). TECs were immobilized via a biotinylated NTS. (C) TECs were formed in the absence of the NTS, and immobilization was achieved via a biotin on the TS. (D) TECs containing the transcription elongation factor Spt4/5. (E and F) Doubly labeled RNAP wt was specifically recruited to elongation scaffolds composed of double-stranded DNA with a longer RNA (35 nt), which can potentially reach subunits Rpo4/7. (F) Incubation of the TEC complexes with the next correct nucleotide (UTP) before single-molecule interrogation resulted in a structural rearrangement to yield almost exclusively a single lower FRET population.
The Binding of Nucleotide Substrates and Analogs Affects the RNAP Clamp.
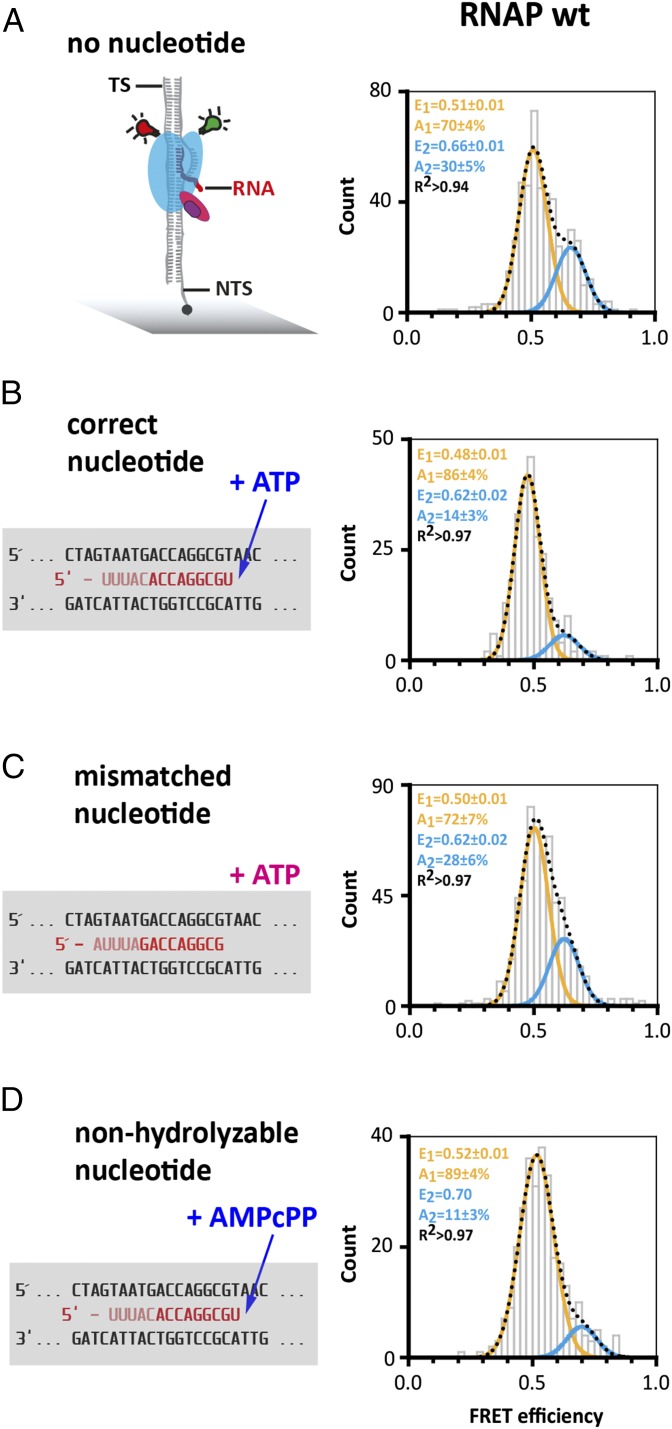
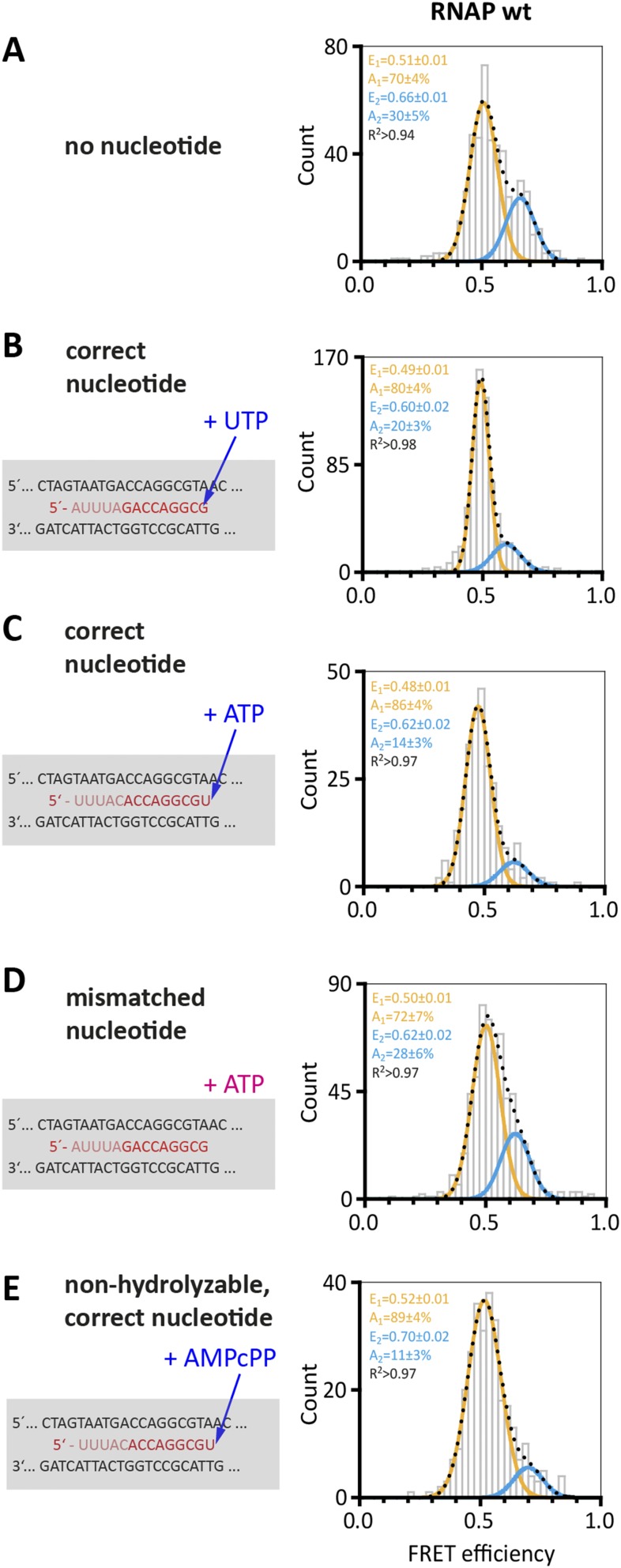
All complexes described above were recorded in the absence of NTP substrates. To test the influence of NTP binding on the conformation of the RNAP clamp, we added either the correct NTP that is complementary to +1 register in the TS, a mismatched NTP, or a nonhydrolyzable NTP analog (AMPcPP) to the TEC. The correct NTP abolished the high FRET population almost entirely (Fig. 4B)—i.e., 86% of the molecules adopt a conformation with low FRET efficiency (E = 0.48 ± 0.01)—that we interpret as clamp state directly comparable to the state induced by Spt4/5 or the NTS (E = 0.53 ± 0.01 and 0.51 ± 0.01), the processive elongation state. The fact that NTP incorporation leads to a quantitative change in the two populations serves as an internal control for the ability of RNAP to respond faithfully to substrates, and, in effect, ascertains correct folding of the in vitro reconstituted and fluorescently labeled RNAP complexes. The NTP-induced effect is dependent on precise Watson–Crick base-pairing because a mismatched nucleotide did not elicit the same response (Fig. 4C). The change is independent of the sequence context because it could be observed using different scaffold sequences (Fig. S6). A nonhydrolyzable nucleotide analog (AMPcPP) also shifts the equilibrium toward the low FRET population (E = 0.52 ± 0.01, +19%), which suggests that the binding of the nucleotide in the active center—rather than phosphodiester bond formation—triggers a conformational change that is translated to the RNAP clamp (Fig. 4D).
Fig. 4.
Conformation of the RNA polymerase clamp in transcription elongation in the presence of nucleotides. Transcription elongation complexes were assembled as described in Fig. 3 with doubly labeled RNAP wt (A) in the presence of nucleotides (B–D, the respective nucleotides were incubated with the TEC at 65 °C before immobilization of the TEC via the NTS). The RNA sequence was varied to test different nucleotides. In all cases, a 9-bp transcription bubble is formed despite the variation in RNA sequence. Shown is part of the standard elongation scaffold with the transcription bubble and the next correct nucleotide to be incorporated is either an ATP (B) or an UTP (C). Addition of a mismatched nucleotide (C) results in a FRET distribution with two populations comparable to the distribution of molecules determined for TECs in the absence of a nucleotide. TECs formed in the presence of a correct nucleotide (B) or a nonhydrolyzable nucleotide analog (D) exhibit only a minor high FRET population.
Fig. S6.
Addition of nucleotides to the TEC. TECs were assembled as described in Fig. 4 with doubly labeled RNAP wt (after incubation of the complexes with a series of nucleotides). The FRET efficiency distribution of the TEC in the absence (A) and presence of the next correct nucleotide (B and C) is shown for comparison. Addition of the correct nucleotide results in an almost complete shift to the low FRET population. The next correct nucleotide to be incorporated in the elongation scaffold (using the T40 RNA) in A, B, and D is an UTP. The elongation scaffold (T43 RNA) in C and E is modified and the next correct nucleotide to be incorporated is an ATP. (D) Addition of a mismatched nucleotide does not result in a significant redistribution of the populations. (E) Addition of a nonhydrolysable nucleotide (AMPcPP) triggers the conformational shift resulting in a clear increase in the low FRET conformation. All datasets were fitted with a single or double Gaussian function, and the mean FRET efficiencies, E, and the coefficient of determination, R2, are given with SEs in the histograms.
Discussion
Our results show that the RNAP clamp of the 12-subunit M. jannaschii RNAP can adopt closed and open conformations in response to (i) DNA melting, (ii) binding of the initiation factor TFE in the PIC, (iii) binding of the elongation factor Spt4/5 and the NTS in the TEC, and (iv) binding of the correct NTP substrate in the RNAP active center. Cryo-EM and X-ray crystallography that portray the RNAP, the PIC and EC strongly suggest that the opening and narrowing of the DNA binding channel is a consequence of the open-to-closed movement of the RNAP clamp. Several alternative mechanistic models for these conformational changes can be envisaged including a simple hinged movement of the clamp and a pivoting of the shelf and core modules; our measurements demonstrate that movements do occur but we cannot currently exclude any of these models (35, 36).
The Free RNAP Is Predominantly in a Conformational State with a Narrow DNA Cleft.
The free RNAP exists in two conformational states with the majority of the molecules found in a conformation with a very narrow DNA binding channel most likely due to a collapsed clamp (14). Inclusion of the Rpo4/7 stalk biases the distribution toward the collapsed state but no complete shift to the high FRET population was observed. In contrast to yeast RNAPII, X-ray structures of distinct archaeal RNAPs show that the clamp can adopt both closed and open conformations, whereas the Rpo4/7 stalk remains incorporated into the core enzyme (11, 12). Our data confirm that these structural clamp states exist in solution. Given the experimental conditions, we are confident that the vast majority of RNAP molecules contains the stalk (38); however, the low FRET state might reflect a minor subpopulation of core RNAP lacking Rpo4/7. Due to the absence of DNA, the clamp can close completely over the DNA binding channel resulting in this collapsed state. In analogy to the bacterial system, we term this state of free RNAP the collapsed state (E = 0.62).
DNA Melting Correlates with Clamp Opening.
Once the RNAP is recruited and bound to the promoter in a TBP- and TFB-dependent manner, the clamp can adopt conformational states with a narrow (E = 0.63) and widened (E = 0.40) DNA cleft, most likely as a result of a closed and open clamp. We interpret these two states as closed complexes, i.e., the DNA strands have not been separated yet, and open complexes, i.e., the template strand has been loaded into the active center, respectively. Dynamic loading, a flipping of the DNA in and out of the DNA binding channel, has been demonstrated for a minimal eukaryotic PIC (43). In our experiments, the equilibrium between the two states is shifted toward the open clamp conformation by using DNA templates that mimic the melted state corresponding to open promoter complexes. Even though we do not see switching between the open and closed states in our single-molecule FRET traces, it is likely that dynamic switching occurs at the biological relevant temperature of our hyperthermophilic model organism (optimal growth temperature at 86 °C). Single-molecule measurements at elevated temperatures, however, did not yield interpretable FRET traces due to the temperature-dependent complex photophysical behavior of the dyes. Measurements at room temperature might represent a “trapped” or “frozen” clamp of an otherwise dynamic process. Thermodynamically, the transition from a heteroduplex DNA containing a 4-bp mismatch to a fully melted (∼12 bp) transcription bubble is more favorable compared with the de novo 12-bp melting of homoduplex DNA, and this is reflected in the distribution of the FRET populations. In archaea, DNA melting is a spontaneous process that is stimulated by TFE but does not necessitate ATP hydrolysis and TFIIH-like helicase/translocase activities (31). In our setup, TFE-like artificial promoter melting favors the open RNAP clamp state, an observation that is supported by a recent EM study of the human PIC where loading of the DNA into the RNAP coincides with clamp opening (13). Archaeal TFE, eukaryotic TFIIE, and the paralogous RNAPI subunit A49 interact with single-stranded DNA (42, 44–46), and all are located proximal to or above the DNA binding channel (13, 15, 16, 35, 39, 47–49). We propose that TFE-like factors operate via two mechanisms: (i) they act as allosteric effectors by favoring RNAP clamp opening and (ii) simultaneously tether the nontemplate strand in the PIC. The outcome is a stimulation of DNA melting and stabilization of the PIC, both of which have been demonstrated using biochemical and functional assays.
Processive and Nonprocessive Transcription Elongation Complexes.
In the context of TEC, the M. jannaschii RNAP clamp adopts two states (E = 0.66 and 0.51). X-ray structures of eukaryotic and bacterial TECs show the RNAP clamp in a closed conformation (5, 6, 50, 51), which is likely to correspond to the E = 0.51 population in our measurements. The inclusion of the NTS in the TEC, the addition of Spt4/5, and the addition of the correct nucleotide eliminates the high FRET state almost entirely (14% of the molecules remaining in the high FRET population). Because a wealth of biochemical experiments have demonstrated that both Spt4/5 and the NTS improve the processivity of M. jannaschii RNAP (32, 52), it is tempting to speculate that there is a causal relationship between the low FRET state (E = 0.51) and the highly processive form of the RNAP TEC. This result implies that although the NTS is not strictly required for transcription, its presence in the TEC leads to conformational changes of the RNAP clamp that result in an increase in processivity. Spt4/5 and its homolog NusG sit astride the DNA binding channel of RNAP (12, 15, 53, 54), and the present data confirm that factor binding repositions the clamp. Interestingly, the correct, but not mismatched, NTP substrates induce the same allosteric response of the clamp; this effect only requires binding and not catalysis because a nonhydrolyzable NTP analog also elicits the clamp change. These results show that substrate binding in the active site triggers an allosteric communication that is translated a considerable distances to the RNAP clamp. What is the function of processive and nonprocessive conformations of RNAP in the TEC? We propose that these alternative states could provide a target for regulation and/or be part of the molecular mechanism that facilitates efficient TEC dissociation during termination. In bacteria, gene expression is abundantly regulated by promoting pausing (55–57) or overcoming it (54, 58), and the opening of the RNAP clamp has been predicted to precede termination (23).
What Are the Exact Conformational Changes of the RNAP?
Recent single-molecule data of RNAPII revealed that transition from the pretranslocated to a 1-bp backtracked state is thermodynamically favored (59). The −1 backtracked state is energetically neutral or even favorable as the first backtracked residue forms stacking interactions with a tyrosine in Rpb2 (60, 61). In addition, a transiently arrested TEC was demonstrated at register −6 (62); importantly, both convert into productive TECs on addition of nucleotides (62). It is conceivable that the high FRET state (E = 0.66) in our system represents a transiently arrested or −1 backtracked state. The addition of a correct nucleotide and subsequent incorporation allows the transition to an active TEC (E = 0.51; Fig. 4). Early models have suggested that the clamp hinges in and out via the switch motifs orthogonal to the direction of transcription (14, 63, 64). However, recent structural models have suggested that the conformational changes are more sophisticated than that (35, 36, 65); a widening of the cleft can alternatively be achieved by a pivoting motion of the two large RNAP subunits against each other. In the bacterial RNAP, a rotation of the core and shelf modules is induced by regulatory effectors such as Gfh1 that act via the NTP entry channel (36, 66). X-ray structures of a dimeric catalytically inactive state of eukaryotic RNA pol I show an expanded cleft, which distorts the active site and inhibits catalysis (35). Our experimental setup was designed to provide a single distance measurement across the DNA binding channel and the changes during the progression of RNAP through the transcription cycle; but, it is problematic to distinguish between the two different conformational models: the hinge and pivoting models. Future experiments using a wider range of derivatization positions will address this question. Our results show for the first time, to our knowledge, that the clamp of complex pol II-like enzymes undergoes conformational changes during its progression through the transcription cycle, in response to the binding of (i) initiation factor TFE and (ii) elongation factor Spt4/5 and (iii) the nucleic acid context of transcription initiation and elongation complexes (Fig. 5 and Fig. S7).
Fig. 5.
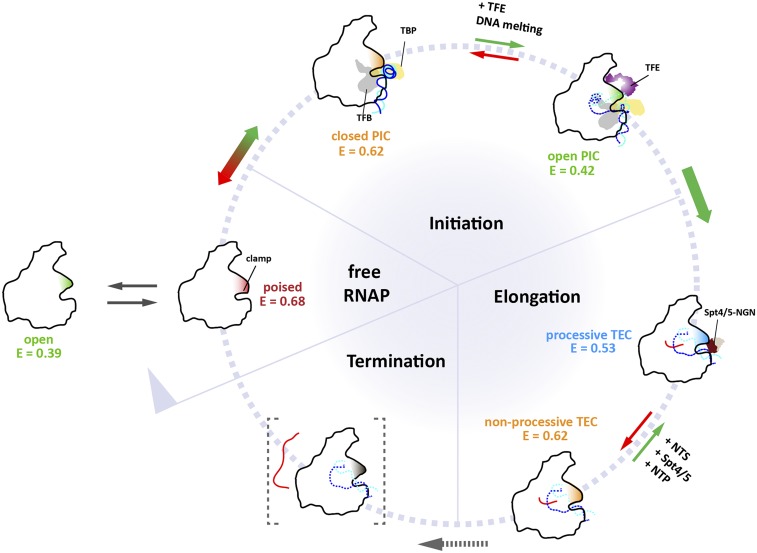
Model depicting the status of the archaeal RNA polymerase clamp at different stages of the transcription cycle. Single-molecule FRET measurements reveal the conformational space sampled by the RNAP clamp (for an overview of all clamp states see Fig. S7 and Tables S1 and S2). Different conformational states of the clamp that can be distinguished by the different mean FRET efficiencies, E, determined in this study are colored in red, green, orange, and blue. TBP is depicted in yellow, TFB in gray, TFE in purple, and Spt4/5 in dark and light brown. Green and red arrows represent productive and nonproductive pathways in the transcription cycle, respectively. In the free RNAP, the clamp completely closes over the binding channel resulting in a collapsed state. In the preinitiation complex RNAP can adopt two conformational states, termed open and closed PIC states. Transcription initiation factor TFE induces an opening of the clamp thereby stabilizing the open initiation complex with the template strand loaded into the active site. In the absence of TFE, the clamp is closed, and the template DNA is presumably not completely loaded into the RNAP resulting in a closed clamp state. Progression into the elongation phase of transcription results in a conformational change that leads to a closure of the clamp as seen in crystal structures of the TEC. However, a second conformational state is possible that reflects a less processive and presumably backtracked state of the RNAP (termed nonprocessive state). Addition of the elongation factor Spt4/5 and elongation of the RNA induced by nucleotide addition induce the transition to the closed TEC conformation that is characterized by high processivity. Termination of transcription is a state that cannot be mimicked in our model system but most likely requires the opening of the clamp for RNA strand release to commence.
Fig. S7.
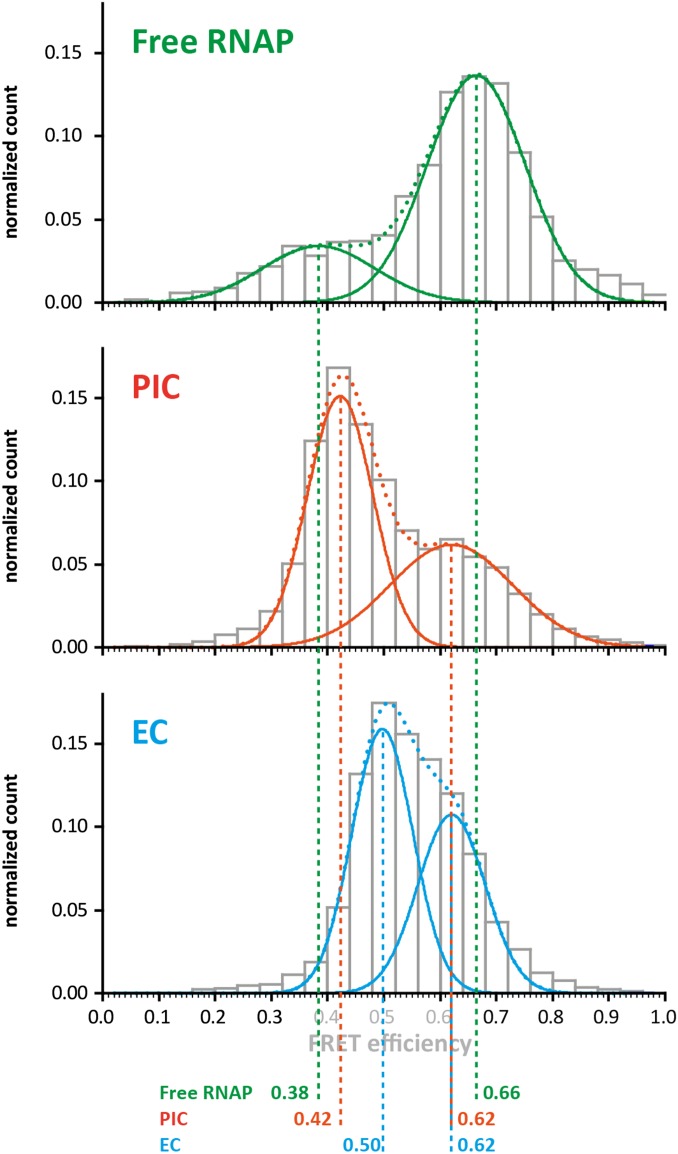
Overview over the conformations of the RNAP clamp at the different stages of the transcription cycle. All single-molecule datasets from the individual stages of the transcription cycle (free RNAP; preinitiation complex, PIC; transcription elongation complex, EC) acquired in the absence of transcription factors TFE and Spt4/5 in this study are summed up for this analysis. The mean FRET efficiencies determined from the histograms are indicated for clarity below. The histograms reflect the conformational space sampled by the RNA polymerase clamp in the different phases of the transcription cycle. In this study, we identified four populations with mean FRET efficiencies of 0.38/0.42, 0.50, 0.62, and 0.66. Whereas the widths of the populations are broadened in the combined analysis shown here, the mean FRET efficiencies correspond to the FRET efficiencies determined in the individual experiments. The E = 0.66 state (highlighted by the red bar) does only occur if the DNA is not loaded into the RNAP and the clamp can completely close over the DNA binding channel. However, some RNAP molecules adopt an open clamp state (E = 0.38, highlighted by the green bar). A comparable two-state situation is found in the initiation complex. TFE, a transcription initiation factor that stabilizes the initiation complex and leads to an increased activity of the RNAP, induces an opening of the clamp (E = 0.42). The fine-tuned adjustment of the clamp conformation can also be observed for the elongation complex. Here, the closed clamp (E = 0.5, highlighted by the blue bar) is in equilibrium with a closed clamp state (E = 0.62, highlighted by the orange bar) and addition of Spt4/5 or the next correct nucleotide results in a complete shift of the molecules into the closed clamp state. The compact state most likely represents the conformational state of a backtracked RNA polymerase.
Table S1.
Overview of the experimental data shown in the figures
| Transcription stage | Sample | No. of molecules | FRET efficiency | Relative percentage | ||
| E1 | E2 | A1 | A2 | |||
| RNAP only | RNAP ΔRpo4/7 | 667 | 0.37 ± 0.03 | 0.66 ± 0.01 | 26% | 74% |
| RNAP wt | 571 | 0.40 ± 0.03 | 0.67 ± 0.01 | 18% | 82% | |
| Initiation complex heteroduplex/homoduplex DNA | RNAP wt (homo) | 1,168 | 0.40 ± 0.01 | 0.63 ± 0.01 | 13% | 87% |
| RNAP wt (homo) + TFE | 670 | 0.47 ± 0.01 | 0.65 ± 0.02 | 38% | 62% | |
| RNAP wt (hetero) | 1,140 | 0.43 ± 0.01 | 0.62 | 56% | 44% | |
| RNAP wt (hetero) + TFE | 1,055 | 0.47 ± 0.01 | 0.62 | 67% | 33% | |
| Elongation complex | RNAP wt | 493 | 0.51 ± 0.01 | 0.66 ± 0.02 | 70% | 30% |
| RNAP wt (T40) + correct NTP | 713 | 0.48 ± 0.01 | 0.62 ± 0.02 | 86% | 14% | |
| RNAP wt − NTS | 479 | 0.62 ± 0.01 | 100% | |||
| RNAP wt + Spt4/5 | 305 | 0.53 ± 0.01 | 0.65 ± 0.02 | 88% | 12% | |
| RNAP wt (T40) + mismatch NTP | 611 | 0.50 ± 0.01 | 0.62 ± 0.02 | 72% | 28% | |
| RNAP wt (T43) +AmpCpp | 292 | 0.52 ± 0.01 | 0.70 ± 0.02 | 89% | 11% | |
The elongation scaffold type used in the measurements is indicated in the table, which shows the number of the RNA in the parentheses.
Table S2.
Overview of the experimental data shown in the SI figures
| Transcription stage | Sample | No. of molecules | FRET efficiency | Relative percentage | ||
| E1 | E2 | A1 | A2 | |||
| RNAP only | RNAP ΔRpo4/7 + ES | 348 | 0.48 ± 0.02 | 0.65 ± 0.02 | 55% | 45% |
| RNAP wt + ES | 551 | 0.51 ± 0.04 | 0.65 ± 0.03 | 51% | 49% | |
| RNAP ΔRpo4/6/7 | 448 | 0.34 ± 0.06 | 0.69 ± 0.02 | 15% | 85% | |
| RNAP ΔRpo6 | 231 | 0.34 ± 0.06 | 0.69 ± 0.03 | 15% | 85% | |
| Initiation complex (heteroduplex DNA) | RNAP ΔRpo4/7 | 943 | 0.40 ± 0.01 | 0.62 | 53% | 47% |
| RNAP ΔRpo4/7 + TFE | 750 | 0.43 ± 0.01 | 0.62 | 58% | 42% | |
| Elongation complex | RNAP ΔRpo4/7 dyes swapped | 915 | 0.51 ± 0.01 | 0.65 ± 0.01 | 72% | 28% |
| RNAP ΔRpo4/7 | 741 | 0.48 ± 0.01 | 0.59 ± 0.01 | 53% | 47% | |
| RNAP wt + long RNA | 745 | 0.49 ± 0.01 | 0.63 ± 0.02 | 70% | 30% | |
| RNAP wt +long RNA / + correct NTP | 256 | 0.49 ± 0.01 | 0.62 | 86% | 14% | |
| RNAP wt (T43) + correct NTP | 259 | 0.49 ± 0.01 | 0.60 ± 0.02 | 80% | 20% | |
The elongation scaffold type used in the measurements is indicated in the table, which shows the number of the RNA in the parentheses. ES, elongation scaffold; TS(Bio), biotinylated template strand.
Materials and Methods
Synthetic Oligonucleotides.
To mimic the different stages of the transcription cycle, suitable nucleic acid constructs were used (SI Materials and Methods). The oligonucleotides for the preinitiation complex consisted of a 110-nucleotide (nt)-long double-stranded DNA that contained the strong Sulfolobus spindle-shaped virus 1 (SSV) T6 gene promoter (31, 67). PICs were formed using either a promoter DNA heteroduplex that contained a 4-bp mismatch around the TSS [leading to open PIC formation (33) or a fully complementary promoter DNA (mimicking the closed PIC)]. The TS and NTS were incubated in a 1:1 molar ratio for 6 s at 95 °C and slowly cooled down to 4 °C. The TS carried a biotin at its 3′ end facilitating immobilization of the PIC. The elongation scaffold consisted of 83-nt-long double-stranded DNA with a preformed transcription bubble. A complementary RNA strand was annealed matching the template strand sequence in the transcription bubble. Depending on the experiment, either a short (14 nt) or long (35 nt) RNA was used. The DNA strands and the RNA primer were incubated in a 1.6:1:8 (NTS:TS:RNA) molar ratio for 6 s at 95 °C and slowly cooled down to 4 °C to promote annealing. Depending on the experiment, a biotin was coupled to either the NTS (attached to the 3′end) or the TS (attached to the 5′end) for surface immobilization. Oligonucleotides were purchased from IBA, MWG, or Sigma-Aldrich.
Proteins.
Unlabeled transcription factors TBP, TFB, TFE, and Spt4/5 from M. jannaschii were produced as described previously (15, 19, 29, 31).
Permanganate Footprinting Assay.
The DNA template encoding the SSV T6 promoter was prepared by annealing 5′-32P–labeled NTS (592: 5′-GATTGATAGAGTAAAGTTTAAATACTTATATAGATAGAGTATAGATAGAGGGTTCAAAAAATGGTT-3′) and unlabeled TS (593: 5′-AACCATTTTTTGAACCCTCTATCTATACTCTATCTATATAAGTATTTAAACTTTACTCTATCTATC-3′ for homoduplex; 600: 5′-AACCATTTTTTGAACCCTCCGCTTATACTCTATCTATATAAGTATTTAAACTTTACTCTATCTATC-3′ for hetero-duplex).
For the footprinting reactions, the components were combined in 23-μL reactions containing 1× HNME buffer [40 mM Hepes, (pH 7.3), 250 mM NaCl, 2.5 mM MgCl, 0.02 mM EDTA, and 1% glycerol], 8.3 nM template DNA, 0.14 μM RNAP, 2 μM TBP, 0.4 μM TFB, 1.2 μM TFE, and 30 μM NTP mix. The reaction was incubated at 65 °C for 15 min, followed by a 2-min incubation with 2 μL 16 mM KMnO4 and stopped with 1.5 μL 14 M β-mercaptoethanol. Protein was digested by addition of 0.25% SDS and 1 mg/mL proteinase K and incubation at 65 °C for 1 h. DNA was ethanol precipitated before treatment with 5% piperidine at 90 °C for 30 min, followed by one round of chloroform extraction and subsequent ethanol precipitation. To prepare the A+G ladder, the DNA was treated with formic acid for 5 min before DNA precipitation and piperidine treatment as described above. DNA was separated on 10% urea PAGE, exposed to a phosphor storage screen, and visualized on a Typhoon FLA 9500 bioimager.
Preparation of RNAP Derivatives.
Subunits Rpo1′ and Rpo2″ of the archaeal RNAP were expressed, purified, and labeled as described in ref. 15. Rpo11 was expressed in BL21(DE3)/Rosetta cells by inducing exponentially growing cultures with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at an optical density of 0.6–0.8. After induction, cultures were grown for 4 h at 37 °C. Bacterial cells were harvested (6,000 × g, 20 min), and the soluble fraction was extracted in P300 buffer (200 mM Tris/acetate, pH 7.9, 100 mM MgAc, 0.1 mM ZnSO4, 300 mM KAc, and 10% glycerol). Cells were lysed using sonication. Recombinant and heat stable Rpo11 could further be prepurified using a heat denaturation step (65 °C for 20 min). The heat-stable protein was precipitated with saturating amounts of ammonium sulfate followed by a centrifugation step (18,000 × g, 30 min). The precipitated protein was resuspended in P300/0.05% β-mercaptoethanol and further purified by size exclusion chromatography (HiPrep-Sephacryl, S100 16/60; GE Healthcare). Rpo11-containing fractions were combined and subsequently further purified by ion exchange chromatography (MonoQ 4.6/100 PE; GE Healthcare) using a gradient from 50 mM Tris, pH 7.5, and 50 mM NaCl to 50 mM Tris, pH 7.5, and 1 M NaCl. The labeling reaction was carried out using a 10-fold molar excess of biotin-maleimide (ThermoScientific) at 4 °C for 16 h, and the reaction was stopped by the addition of 0.5% β-mercaptoethanol. Subsequently, Rpo11 was combined with Rpo3 (molar ratio of 1:2 and denaturation of the proteins was reached by addition of 6 M urea). Dimerization of subunits Rpo3 and Rpo11 to form a stable heterodimer was achieved by stepwise dialysis of the sample to a urea-free buffer. Recombinant RNAP containing the modified subunits was reconstituted as described previously (29). Typical dye labeling efficiencies for Rpo1′ and Rpo2″ are 10–20%, resulting in a fraction of 1–4% of doubly labeled RNAPs.
Transcription Complex Preparation.
PIC complexes were assembled by incubating 0.13 µM promoter DNA, 5 µM TBP, 5 µM TFB, 0.1 µM RNAP, 6.7 mM DTT, and 0.07 mg/mL BSA in 1× TMNE buffer (40 mM Tris/HCl, pH 7.3, 250 mM sodium chloride, 2.5 mM magnesium chloride, 0.1 mM EDTA, and 5% glycerol) at 65 °C for 15 min (31, 33). To measure the PIC in the presence of the third basal transcription factor TFE, 15 µM TFE was added to the PIC and further incubated at 65 °C for 10 min (15, 33).
EC complexes were formed by incubating 0.1 µM labeled RNAP with 3 µM of the elongation scaffold and 6.7 mM DTT and 0.07 mg/mL BSA in 1× TMNE buffer (32). To measure the influence of the elongation factor Spt4/5 or NTPs on the RNAP clamp position, 20 µM Spt4/5 was added to the EC and further incubated for 10 min at 65 °C (15, 19). To reduce unspecific binding of the RNAP to the DNA, 0.8 mg/mL heparin was added to the PIC or TEC mixture and incubated for another 10 min at 65 °C before addition of TFE, Spt4/5, or NTPs.
Determination of the Förster Radius.
For the donor-acceptor pair DyLight550 and DyLight650, the Förster radius R0 was determined by calculating the overlap integrals from recorded donor emission spectra (excitation wavelength of 550 nm) and acceptor absorption spectra. Using the normalized spectra, the overlap integral was determined with the following equation:
with = normalized donor emission spectrum, = extinction coefficient of the acceptor, and λ = wavelength.
The Förster radius was further calculated using the formula
with , n = 1.33, ΦD = quantum yield of the donor, and J is the overlap integral.
The quantum yield of DyLight550 was calculated using ATTO532 as a reference and was determined to be 0.14. The Förster radius of the DyLight550-DyLight650 pair was calculated as 5.2 nm.
Rotational mobility of the dyes was checked using anisotropy measurements as published in Grohmann et al. (15). Addition of TFE and Spt4/5 to the labeled RNAPs did not lead to a change in anisotropy.
Surface Preparation.
Studies on immobilized molecules using a widefield setup were carried out on a polyethylene glycol (PEG) surface attached to a flow chamber for custom built PRISM TIRF microscope. Quartz slides were thoroughly cleaned with peroxymonosulfuric acid. After drying, the slides were silanized (Aminosilane A0700; amchro Hattersheim) and afterward PEGylized according to Roy et al. (68).
TIRF Immobilization Assay.
For TIRF measurements, the surface was incubated with 0.1 mg/mL neutravidin in 0.5× TMNE for 10 min and washed with 0.5× TMNE. PICs and ECs were immobilized via the biotinylated DNAs, and free RNAPs were immobilized with the internal biotin (subunits Rpo11) on the quartz glass slide via neutravidin. The quartz glass slide was rinsed with a 17-pM sample solution in 0.5× TMNE, and nonbound molecules were removed with 0.5× TMNE. This procedure yielded an immobilized RNAP density of approximately one molecule per 4 µm2. Afterward, the flow chamber was flushed with 0.5× TMNE containing 2 mM Trolox, 1% (wt/wt) glucose, 100 U/mL glucose oxidase, and 160 U/mL catalase (69). For experiments involving TFE or Spt4/5, high excess of these proteins was additionally added to the measuring buffer to ensure saturation of the RNAP with transcription factors during the measurement.
Widefield Single-Molecule Detection and Analysis.
All single-molecule FRET experiments were performed at room temperature (21 °C) on a homebuilt PRISM-TIRF setup based on an Olympus IX71 using ALEX. For two-color ALEX experiments, a yellow laser (568 nm; Coherent Sapphire 100 mW) was used for excitation of the donor, and a red laser (639 nm; Toptica iBeam Smart 150 mW) was used for direct excitation of the acceptor. Lasers were alternated in synchronization with the camera’s frame rate of 10 Hz using a polychromatic acousto-optic modulator (Crystal Technology). The fluorescence was collected by a 60× Olympus 1.20 NA water-immersion objective and split by wavelength with a dichroic mirror (640 DCXR; Chroma Technology) into two detection channels that were further filtered with two band-pass filters (Semrock BrightLine 582/75 and Semrock Brightline 609/54) in the orange channel and one long-pass filter (647-nm Semrock RazorEdge) in the near infrared detection range. Both detection channels were recorded by one electron multiplying charge-coupled device (EMCDD) camera (Andor IXon ×3, pregain 5.1, gain 250, frame rate 10 Hz) in a dual view configuration (TripleSplit; Cairn), and the acquired data were analyzed by custom-made software based on LabVIEW 2012 64bit (National Instruments). The fluorescent molecule spots were selected by an automated spot finder, and the fluorescence intensities were background corrected by subtracting the surrounding-pixel intensity. For the calculation of the FRET efficiency, the transients were corrected for leakage from the donor into the red detection channel (18%), the direct excitation of the acceptor by the 568-nm laser excitation (11%), and the gamma correction γ, which includes the fluorescence quantum yields of the fluorophores and the detection efficiencies of the two channels. FRET pairs, which did not show acceptor bleaching, were discarded for the analysis, because these transients do not allow an individual determination of the γ factor (70). The γ factor was determined on acceptor bleaching as follows:
with and as the intensities of the donor and acceptor before and and as the intensities of the donor and acceptor after acceptor bleaching. The γ factor was determined for each transient in a dataset, and overall γ factor for each dataset was derived by a Gaussian fit to a cumulative histogram of the γ factors (71).
FRET efficiency histograms shown are accumulated data from at least three independent measurements. The resulting histograms were fitted either with a single or double Gaussian fit, and the mean FRET efficiency and SE were determined from the fit. For statistical analysis of the data, we used the Pearson χ2 test.
From the FRET efficiencies, we calculated distances (r) using a Förster radius (R0) of 5.2 nm and the following equation:
Please note that we focused on FRET efficiencies rather than absolute distances, as the Förster radius R0 (e.g., the relative quantum yield of the fluorophores and κ2) might be slightly influenced due to a changed microenvironment of the dyes when the RNAP is part of the initiation or elongation complex.
SI Materials and Methods
EMSAs.
EMSAs were performed using Alexa488-labeled promoter DNA. The DNA (0.67 µM) was incubated with the reaction components (5 µM TBP, 5 µM TFB, 0.1 µM RNAP) in 6.7 mM DTT and 0.07 mg/mL BSA in 1× TMNE buffer for 20 min at 65 °C. Heparin (0.8 mg/mL) was added to prevent unspecific interaction with the DNA followed by an additional incubation step for 10 min at 65 °C. The reaction mixtures were separated on a native 12% Tris-glycine gel at 200 V for 50 min at room temperature, followed by visualizing of the fluorescently labeled complexes on a FLA7000 Scanner (GE Healthcare).
Single-Molecule Transcription Assay.
TECs were formed by incubating 0.1 µM of labeled RNAP with 3 µM TS/RNA and 6.7 mM DTT and 0.07 mg/mL BSA in 1× TMNE buffer for 15 min at 65 °C, followed by addition of the NTS and incubation for 10 min at 65 °C. To reduce unspecific binding of the RNAP to the DNA, 0.8 mg/mL heparin was added to the TEC mixture and incubated for another 10 min at 65 °C. Transcription was initiated by the addition of 0.75 mM ATP, GTP, UTP, and CTP. Equal amounts of water were added to the control reaction. The samples were incubated for another 5 min at 65 °C. Subsequently, the complexes were immobilized via a biotin coupled to the nontemplate strand (5′-end) of the elongation scaffold as described in Materials and Methods. For immobilization, the samples were incubated with the TEC samples for the exact same amount of time. After removal of the nonbound molecules, the number of immobilized RNAPs that showed stable FRET were counted. The number of molecules was accumulated from three independent experiments.
Anisotropy Measurements.
Steady-state fluorescence anisotropy measurements of donor and acceptor dyes were carried out using a fluorescence spectrometer (Varian; Cary Eclipse) with excitation at 562 nm (10-nm slit) and emission at 576 nm (5-nm slit) for studies of RNAP_Rpo1′E257AzF*DL550 and excitation at 652 nm (10-nm slit) and emission at 672 nm (5-nm slit) for studies of RNAP_Rpo2″Q373Azf*DL650 at an integration time of 15 s. The single labeled RNAPs were preincubated with either 20 µM Spt4/5 or 2 µM TFE at 65 °C for 10 min. The anisotropy values r were calculated by using the formula
where IVX corresponds to the vertically polarized excitation with either horizontally or vertically polarized emission. The G factor is given by , where IHX corresponds to the horizontally polarized excitation with either horizontally or vertically polarized emission.
Oligonucleotides Used in This Study.
Nontemplate strand homoduplex promoter DNA.
5′-GATTGATAGAGTAAAGTTTAAATACTTATATAGATAGAGTATAAGCGGAGGGTTGAGATGATGGTTAGGGTTGAGATGATGGTTATGGTTCATCTCGCAGCCACTCCGCA-3′
Template strand homoduplex promoter DNA.
5′-TGCGGAGTGGCTGCGAGATGAACCATAACCATCATCTCAACCCTAACCATCATCTCAACCCTCCGCTTATACTCTATCTATATAAGTATTTAAACTTTACTCTATCAATC-Biotin-3′
Nontemplate strand heteroduplex promoter DNA.
5′-GATTGATAGAGTAAAGTTTAAATACTTATAGAGATAGAGTATAGATAGAGGGTTGAGATGATGGTTAGGTTGAGATGATGGTTATGGTTCATCTCGCAGCCACTCCGCA-3′
Template strand heteroduplex promoter DNA.
5′-TGCGGAGTGGCTGCGAGATGAACCATAACCATCATCTCAACCCTAACCATCATCTCAACCCTCCGCTTATACTCTATCTCTATAAGTATTTAAACTTTACTCTATCAATC-Biotin-3′
Nontemplate strand elongation scaffold.
5′-CCGGCAGTACTAGTAATGACCAGGCGTAACTACTTCATCTTAACTACTCTAATGGATCTCCCATATGGTGGAGGTAAGGGTGG-Biotin-3′
Template strand elongation scaffold.
5′-CCACCCTTACCTCCACCATATGGGAGATCCATTAGAGTAGTTAAGATGAAGTAGTTACGCCTGGTCATTACTAGTACTGCCGG-3′
Template strand elongation scaffold (Biotin).
5′-Biotin-CCACCCTTACCTCCACCATATGGGAGATCCATTAGAGTAGTTAAGATGAAGTAGTTACGCCTGGTCATTACTAGTACTGCCGG-3′
RNA Oligonucleotides for the Assembly of the Elongation Complex.
T40.
5′-AUUUAGACCAGGCG-3′
T43.
5′-UUUACACCAGGCGU-3′
Long RNA.
5′-AACAUUACACGAAUAUAUAUGAUUUAGACCAGGCG-3′
Acknowledgments
We thank Richard Ebright for training in fluorescent labeling methods and comments; Jens Michaelis, Achillefs Kapanidis, Phil Holzmeister, and Alan Cheung for discussions and comments; Carsten Forthmann for assistance with programming of the analysis software; Jan Vogelsang and Sebastian Bange for assistance with the statistical analysis of the data; Bettina Wünsch for assistance with the determination of the Förster radius; and Peter Schultz for plasmids. D.G. was supported by the German Israel Foundation (Young Scientist Program 2292-2264.13/2011), the Fond der Chemischen Industrie, the Boehringer Ingelheim Fonds, and the Deutsche Forschungsgemeinschaft (Grants GR 3840/2-1 and SFB960). Research in the RNAP laboratory at University College London is funded by Wellcome Trust Investigator Award 079351/Z/06/Z and Biotechnology and Biological Sciences Research Council Grant BB/E008232/1 (to F.W.). P.T. was supported by a starting grant (SiMBA, EU 261162) of the European Research Council and the Volkswagen Foundation (86415).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515817113/-/DCSupplemental.
References
- 1.Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol. 2011;9(2):85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- 2.Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell. 2012;45(4):439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 3.Armache KJ, Kettenberger H, Cramer P. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc Natl Acad Sci USA. 2003;100(12):6964–6968. doi: 10.1073/pnas.1030608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bushnell DA, Kornberg RD. Complete, 12-subunit RNA polymerase II at 4.1-A resolution: Implications for the initiation of transcription. Proc Natl Acad Sci USA. 2003;100(12):6969–6973. doi: 10.1073/pnas.1130601100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16(6):955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 6.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292(5523):1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 7.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: Separation of RNA from DNA by RNA polymerase II. Science. 2004;303(5660):1014–1016. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- 8.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292(5523):1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 9.Craighead JL, Chang WH, Asturias FJ. Structure of yeast RNA polymerase II in solution: Implications for enzyme regulation and interaction with promoter DNA. Structure. 2002;10(8):1117–1125. doi: 10.1016/s0969-2126(02)00813-4. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann NA, et al. Molecular structures of unbound and transcribing RNA polymerase III. Nature. 2015;528(7581):231–236. doi: 10.1038/nature16143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirata A, Klein BJ, Murakami KS. The X-ray crystal structure of RNA polymerase from Archaea. Nature. 2008;451(7180):851–854. doi: 10.1038/nature06530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jun SH, et al. The X-ray crystal structure of the euryarchaeal RNA polymerase in an open-clamp configuration. Nat Commun. 2014;5:5132. doi: 10.1038/ncomms6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y, Fang J, Taatjes DJ, Nogales E. Structural visualization of key steps in human transcription initiation. Nature. 2013;495(7442):481–486. doi: 10.1038/nature11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakraborty A, et al. Opening and closing of the bacterial RNA polymerase clamp. Science. 2012;337(6094):591–595. doi: 10.1126/science.1218716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grohmann D, et al. The initiation factor TFE and the elongation factor Spt4/5 compete for the RNAP clamp during transcription initiation and elongation. Mol Cell. 2011;43(2):263–274. doi: 10.1016/j.molcel.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grünberg S, Warfield L, Hahn S. Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat Struct Mol Biol. 2012;19(8):788–796. doi: 10.1038/nsmb.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein BJ, et al. RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc Natl Acad Sci USA. 2011;108(2):546–550. doi: 10.1073/pnas.1013828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naji S, Grünberg S, Thomm M. The RPB7 orthologue E′ is required for transcriptional activity of a reconstituted archaeal core enzyme at low temperatures and stimulates open complex formation. J Biol Chem. 2007;282(15):11047–11057. doi: 10.1074/jbc.M611674200. [DOI] [PubMed] [Google Scholar]
- 19.Hirtreiter A, et al. Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res. 2010;38(12):4040–4051. doi: 10.1093/nar/gkq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werner F. A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J Mol Biol. 2012;417(1-2):13–27. doi: 10.1016/j.jmb.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grohmann D, Werner F. Cycling through transcription with the RNA polymerase F/E (RPB4/7) complex: Structure, function and evolution of archaeal RNA polymerase. Res Microbiol. 2011;162(1):10–18. doi: 10.1016/j.resmic.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Hein PP, et al. RNA polymerase pausing and nascent-RNA structure formation are linked through clamp-domain movement. Nat Struct Mol Biol. 2014;21(9):794–802. doi: 10.1038/nsmb.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weixlbaumer A, Leon K, Landick R, Darst SA. Structural basis of transcriptional pausing in bacteria. Cell. 2013;152(3):431–441. doi: 10.1016/j.cell.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287(5460):2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 25.Chakraborty A, Wang D, Ebright YW, Ebright RH. Azide-specific labeling of biomolecules by Staudinger-Bertozzi ligation phosphine derivatives of fluorescent probes suitable for single-molecule fluorescence spectroscopy. Methods Enzymol. 2010;472:19–30. doi: 10.1016/S0076-6879(10)72018-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chin JW, et al. Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J Am Chem Soc. 2002;124(31):9026–9027. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 27.Grohmann D, Werner F, Tinnefeld P. Making connections--strategies for single molecule fluorescence biophysics. Curr Opin Chem Biol. 2013;17(4):691–698. doi: 10.1016/j.cbpa.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ouhammouch M, Werner F, Weinzierl RO, Geiduschek EP. A fully recombinant system for activator-dependent archaeal transcription. J Biol Chem. 2004;279(50):51719–51721. doi: 10.1074/jbc.C400446200. [DOI] [PubMed] [Google Scholar]
- 29.Werner F, Weinzierl RO. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol Cell. 2002;10(3):635–646. doi: 10.1016/s1097-2765(02)00629-9. [DOI] [PubMed] [Google Scholar]
- 30.Schulz S, Kramm K, Werner F, Grohmann D. Fluorescently labeled recombinant RNAP system to probe archaeal transcription initiation. Methods. 2015;86:10–18. doi: 10.1016/j.ymeth.2015.04.017. [DOI] [PubMed] [Google Scholar]
- 31.Werner F, Weinzierl RO. Direct modulation of RNA polymerase core functions by basal transcription factors. Mol Cell Biol. 2005;25(18):8344–8355. doi: 10.1128/MCB.25.18.8344-8355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirtreiter A, Grohmann D, Werner F. Molecular mechanisms of RNA polymerase--the F/E (RPB4/7) complex is required for high processivity in vitro. Nucleic Acids Res. 2010;38(2):585–596. doi: 10.1093/nar/gkp928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagy J, et al. Complete architecture of the archaeal RNA polymerase open complex including DNA, TBP, TFB and TFE. Nat Commun. 2014;6:6161. doi: 10.1038/ncomms7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margeat E, et al. Direct observation of abortive initiation and promoter escape within single immobilized transcription complexes. Biophys J. 2006;90(4):1419–1431. doi: 10.1529/biophysj.105.069252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engel C, Sainsbury S, Cheung AC, Kostrewa D, Cramer P. RNA polymerase I structure and transcription regulation. Nature. 2013;502(7473):650–655. doi: 10.1038/nature12712. [DOI] [PubMed] [Google Scholar]
- 36.Tagami S, et al. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature. 2010;468(7326):978–982. doi: 10.1038/nature09573. [DOI] [PubMed] [Google Scholar]
- 37.Korkhin Y, et al. Evolution of complex RNA polymerases: The complete archaeal RNA polymerase structure. PLoS Biol. 2009;7(5):e1000102. doi: 10.1371/journal.pbio.1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grohmann D, Hirtreiter A, Werner F. RNAP subunits F/E (RPB4/7) are stably associated with archaeal RNA polymerase: Using fluorescence anisotropy to monitor RNAP assembly in vitro. Biochem J. 2009;421(3):339–343. doi: 10.1042/BJ20090782. [DOI] [PubMed] [Google Scholar]
- 39.Murakami K, et al. Architecture of an RNA polymerase II transcription pre-initiation complex. Science. 2013;342(6159):1238724. doi: 10.1126/science.1238724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blombach F, et al. Archaeal TFEα/β is a hybrid of TFIIE and the RNA polymerase III subcomplex hRPC62/39. eLife. 2015;4:e08378. doi: 10.7554/eLife.08378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen HT, Warfield L, Hahn S. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat Struct Mol Biol. 2007;14(8):696–703. doi: 10.1038/nsmb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grünberg S, Bartlett MS, Naji S, Thomm M. Transcription factor E is a part of transcription elongation complexes. J Biol Chem. 2007;282(49):35482–35490. doi: 10.1074/jbc.M707371200. [DOI] [PubMed] [Google Scholar]
- 43.Treutlein B, et al. Dynamic architecture of a minimal RNA polymerase II open promoter complex. Mol Cell. 2012;46(2):136–146. doi: 10.1016/j.molcel.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 44.Okuda M, et al. Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface. EMBO J. 2000;19(6):1346–1356. doi: 10.1093/emboj/19.6.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geiger SR, et al. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol Cell. 2010;39(4):583–594. doi: 10.1016/j.molcel.2010.07.028. [DOI] [PubMed] [Google Scholar]
- 46.Okamoto T, et al. Analysis of the role of TFIIE in transcriptional regulation through structure-function studies of the TFIIEbeta subunit. J Biol Chem. 1998;273(31):19866–19876. doi: 10.1074/jbc.273.31.19866. [DOI] [PubMed] [Google Scholar]
- 47.Wu CC, et al. RNA polymerase III subunit architecture and implications for open promoter complex formation. Proc Natl Acad Sci USA. 2012;109(47):19232–19237. doi: 10.1073/pnas.1211665109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jennebach S, Herzog F, Aebersold R, Cramer P. Crosslinking-MS analysis reveals RNA polymerase I domain architecture and basis of rRNA cleavage. Nucleic Acids Res. 2012;40(12):5591–5601. doi: 10.1093/nar/gks220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu CC, Lin YC, Chen HT. The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center. Mol Cell Biol. 2011;31(13):2715–2728. doi: 10.1128/MCB.05151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448(7150):157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 51.Vassylyev DG, et al. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448(7150):163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 52.Kireeva ML, Komissarova N, Kashlev M. Overextended RNA:DNA hybrid as a negative regulator of RNA polymerase II processivity. J Mol Biol. 2000;299(2):325–335. doi: 10.1006/jmbi.2000.3755. [DOI] [PubMed] [Google Scholar]
- 53.Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011;30(7):1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sevostyanova A, Belogurov GA, Mooney RA, Landick R, Artsimovitch I. The β subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Mol Cell. 2011;43(2):253–262. doi: 10.1016/j.molcel.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Farnham PJ, Greenblatt J, Platt T. Effects of NusA protein on transcription termination in the tryptophan operon of Escherichia coli. Cell. 1982;29(3):945–951. doi: 10.1016/0092-8674(82)90457-3. [DOI] [PubMed] [Google Scholar]
- 56.Ha KS, Toulokhonov I, Vassylyev DG, Landick R. The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase. J Mol Biol. 2010;401(5):708–725. doi: 10.1016/j.jmb.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma C, et al. RNA polymerase-induced remodelling of NusA produces a pause enhancement complex. Nucleic Acids Res. 2015;43(5):2829–2840. doi: 10.1093/nar/gkv108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Herbert KM, et al. E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J Mol Biol. 2010;399(1):17–30. doi: 10.1016/j.jmb.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dangkulwanich M, et al. Complete dissection of transcription elongation reveals slow translocation of RNA polymerase II in a linear ratchet mechanism. eLife. 2013;2:e00971. doi: 10.7554/eLife.00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471(7337):249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 61.Wang D, et al. Structural basis of transcription: Backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324(5931):1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states By translocating back and forth along the DNA and the RNA. J Biol Chem. 1997;272(24):15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 63.Mukhopadhyay J, et al. The RNA polymerase “switch region” is a target for inhibitors. Cell. 2008;135(2):295–307. doi: 10.1016/j.cell.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Srivastava A, et al. New target for inhibition of bacterial RNA polymerase: ‘Switch region’. Curr Opin Microbiol. 2011;14(5):532–543. doi: 10.1016/j.mib.2011.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sekine S, Murayama Y, Svetlov V, Nudler E, Yokoyama S. The ratcheted and ratchetable structural states of RNA polymerase underlie multiple transcriptional functions. Mol Cell. 2015;57(3):408–421. doi: 10.1016/j.molcel.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 66.Zuo Y, Wang Y, Steitz TA. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol Cell. 2013;50(3):430–436. doi: 10.1016/j.molcel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bell SD, Kosa PL, Sigler PB, Jackson SP. Orientation of the transcription preinitiation complex in archaea. Proc Natl Acad Sci USA. 1999;96(24):13662–13667. doi: 10.1073/pnas.96.24.13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5(6):507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ha T, Tinnefeld P. Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annu Rev Phys Chem. 2012;63:595–617. doi: 10.1146/annurev-physchem-032210-103340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McCann JJ, Choi UB, Zheng L, Weninger K, Bowen ME. Optimizing methods to recover absolute FRET efficiency from immobilized single molecules. Biophys J. 2010;99(3):961–970. doi: 10.1016/j.bpj.2010.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ha T, et al. Single-molecule fluorescence spectroscopy of enzyme conformational dynamics and cleavage mechanism. Proc Natl Acad Sci USA. 1999;96(3):893–898. doi: 10.1073/pnas.96.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]