The isolation of bovine kidney Na+,K+-ATPase is presented, allowing the successful crystallization and crystal structure determination of an ouabain-bound E2–BeF3 − complex.
Keywords: P-type ATPase; native source isolation; cardiotonic steroids; Na+,K+-ATPase; ouabain
Abstract
Na+,K+-ATPase is responsible for the transport of Na+ and K+ across the plasma membrane in animal cells, thereby sustaining vital electrochemical gradients that energize channels and secondary transporters. The crystal structure of Na+,K+-ATPase has previously been elucidated using the enzyme from native sources such as porcine kidney and shark rectal gland. Here, the isolation, crystallization and first structure determination of bovine kidney Na+,K+-ATPase in a high-affinity E2–BeF3 −–ouabain complex with bound magnesium are described. Crystals belonging to the orthorhombic space group C2221 with one molecule in the asymmetric unit exhibited anisotropic diffraction to a resolution of 3.7 Å with full completeness to a resolution of 4.2 Å. The structure was determined by molecular replacement, revealing unbiased electron-density features for bound BeF3 −, ouabain and Mg2+ ions.
1. Introduction
Sodium potassium adenosine triphosphatase (Na+,K+-ATPase; EC 3.6.3.9) is a plasma-membrane protein that is present in all animal cells. In each ATPase cycle, three sodium ions are extruded out of and two potassium ions are transported into the cell (Post & Jolly, 1957 ▸). This activity is critical for fundamental biological processes such as maintenance of the resting membrane potential, cell excitability and activity of secondary transporters (Rudnick, 1998 ▸; Rose et al., 2009 ▸). Na+,K+-ATPase consists of a catalytic α-subunit and a supporting β-subunit and is often associated with a tissue-specific regulatory FXYD protein. Each subunit exists in several isoforms, and different combinations of isoforms are found in different tissue types (Blanco & Mercer, 1998 ▸; Sweadner & Rael, 2000 ▸). In kidney tissue, the α1β1 dimer is associated with FXYD2, also known as the γ-subunit.
The Na+,K+-ATPase α-subunit consists of ten transmembrane (TM) helices and three cytoplasmic subdomains: the actuator (A) domain, the nucleotide-binding (N) domain and the phosphorylation (P) domain. During the catalytic cycle, autophosphorylation of a conserved aspartate in the P domain (Asp374 in bovine Na+,K+-ATPase) takes place immediately after the binding of intracellular Na+, followed by occlusion. Subsequent events are extracellular Na+ release, K+ binding, autodephosphorylation and K+ release to the intracellular side. The conformational transitions between Na+-bound and K+-bound states, also named the E1 and E2 states, respectively, are accompanied by large domain movements. The supporting β-subunit and the FXYD protein both have one TM helix. In addition, the β-subunit has a large, glycosylated C-terminal extracellular domain of about 240 amino acids (Miller & Farley, 1988 ▸).
Na+,K+-ATPase can interact with a variety of substances ranging from small molecules such as cardiotonic steroids (CTSs) to proteins (Reinhard et al., 2013 ▸). CTSs, such as the plant-derived ouabain and digoxin, are specific Na+,K+-ATPase inhibitors which have been used as drugs in the treatment of congestive heart failure for hundreds of years. They inhibit the enzyme by binding to the TM domain of the α-subunit from the extracellular side and by stabilization of the outward-open phosphorylated E2 state (E2P state), thus antagonizing K+ binding (Lingrel et al., 1997 ▸). CTSs are also proposed to affect intracellular kinase signalling pathways using Na+,K+-ATPase as a receptor (Mohammadi et al., 2001 ▸).
To date, crystal structures representing both the E2 (Morth et al., 2007 ▸; Shinoda et al., 2009 ▸; Ogawa et al., 2015 ▸) and E1 (Kanai et al., 2013 ▸; Nyblom et al., 2013 ▸) states of the enzyme have been determined. In the abovementioned cases the cytoplasmic domains are stabilized by different phosphate analogues, while the transmembrane domain has K+ or Na+ occluded. Another group of structures represent CTS-stabilized forms in which varying CTSs are bound with low affinity (ouabain; Ogawa et al., 2009 ▸) as well as with high affinity (ouabain, bufalin and digoxin; Yatime et al., 2011 ▸; Laursen et al., 2013 ▸, 2015 ▸). All of the above structures were based on preparations of native Na+,K+-ATPase from either porcine kidney or shark rectal glands (Jørgensen, 1988 ▸; Skou & Esmann, 1988 ▸; Klodos et al., 2002 ▸).
To date, heterologous expression systems for Na+,K+-ATPase have remained challenging for the quantitative purification of fully active enzyme. Therefore, attempts to crystallize the enzyme from alternative native sources are important, as even small changes in the primary structure or the native lipid composition can have a large effect on crystallogenesis. Here, we explore bovine kidney as a novel native source of Na+,K+-ATPase suitable for subsequent crystallization and structure-determination experiments; the α/β-subunits are about 98/93% identical in sequence to those of the porcine enzyme and 87/65% identical in sequence to those of the shark enzyme. We present the first crystal structure of the bovine Na+,K+-ATPase enzyme (α1β1γ), which is locked in an E2–BeF3 − state forming a high-affinity complex with the cardiotonic steroid ouabain.
2. Experimental methods
2.1. Membrane preparation and solubilization
The purification of bovine kidney Na+,K+-ATPase followed the rationale described for the purification of the porcine kidney enzyme (Klodos et al., 2002 ▸). Unless stated otherwise, all steps were performed at 277 K.
For the purification, approximately 2 l buffer A (25 mM imidazole, 250 mM sucrose, 1 mM ethylenediaminetetraacetic acid, pH adjusted to 7.3 using HCl) was required. Using rongeurs (KLS Martin), approximately 110 g of tissue was dissected from the red outer medulla of nine fresh bovine kidneys obtained from the production line of a dairy-cow slaughterhouse (Skare Beef, Aarhus Slagtehus, Denmark). The dissection was performed on-site within 10–120 min post mortem. The tissue fragments were stored in buffer A on ice. Excess blood cells were rinsed away by adding fresh buffer A to a total volume of 300–400 ml, followed by incubation for 30–60 min before removal of the buffer. The rinsing step was repeated five times, whereas the final incubation interval was overnight. The tissue was then suspended in fresh, ice-cold buffer A to a total volume of 230 ml and disintegrated using an ordinary kitchen blender. The blender was rinsed with 100 ml buffer A, which was combined with the disintegrated sample. The combined fractions were then homogenized in batches of approximately 40 ml. All homogenization steps were performed using glass homogenizers (Fisher Scientific) with the piston attached to a drilling machine operating at 1000–2000 rev min−1.
The homogenized sample was centrifuged for 40 min at 6000g in a JLA-8.1000 rotor. The supernatant was collected and stored on ice. The pellet was resuspended in buffer A to a volume of 300 ml and the homogenization and centrifugation procedure was repeated. The two supernatants were combined and centrifuged for 40 min at 11 750g in a JLA-8.1000 rotor. The resulting supernatant was subjected to a subsequent step of centrifugation for 50 min at 48 000g in a Beckman Ti45 rotor. The resulting microsome pellets (about 20 g in total) were each resuspended in 10 ml buffer A, pooled and homogenized in small batches of 10 ml. The homogenate was diluted to 76 ml (about 0.26 g microsomes per millilitre). The protein concentration was estimated by the Bradford protein assay supplemented with NaOH (Bradford, 1976 ▸; Stoscheck, 1990 ▸) and adjusted to 10 mg ml−1 with buffer A. The Na+,K+-ATPase-rich microsomes were stored at 253 K in aliquots of 12 ml and a 1 ml aliquot was used for activity assays.
Microsomes were purified by mild sodium dodecyl sulfate (SDS) treatment, removing peripheral and contaminating proteins (Forbush, 1982 ▸; Jørgensen, 1988 ▸). The optimal SDS concentration was determined as described by Klodos et al. (2002 ▸). In brief, the diluted microsomes (the separately stored 1 ml aliquot) were incubated for 1 h at room temperature with different concentrations of SDS (ranging from 0 to 1.3 mg ml−1). In a 96-well SpectraPlate-96 MB (PerkinElmer) the Na+,K+-ATPase activity was analyzed by quantification of the ouabain-sensitive release of inorganic phosphate from ATP in the presence of Na+, K+ and Mg2+ (Baginski & Zak, 1960 ▸; Cariani et al., 2004 ▸). Absorbance measurements at 715 nm were performed using a VICTOR3 photometer (PerkinElmer). The specific Na+,K+-ATPase activity was calculated from the difference in absorbance in the absence and presence of 1 mM ouabain. Maximal activity was observed after incubation with 0.8 mg ml−1 SDS.
For the purposes of large-scale purification, the incubation time with the optimal SDS concentration was increased to 12 h. The membranes were then ultracentrifuged for 50 min at 127 000g and 284 K in a Beckman Ti45 rotor and the pellets were resuspended in fresh buffer A to a total volume of 30 ml followed by homogenization. The centrifugation and homogenization procedure was repeated four times to remove SDS, resulting in pellets with a total mass of 4.56 g. The collected pellets were resuspended in 10 ml buffer A and homogenized. Subsequently, the sample was diluted with buffer A to a protein concentration of about 8 mg ml−1. The whole preparation yielded approximately 155 mg of Na+,K+-ATPase in microsomes and was stored in 450 µl aliquots at 253 K.
Membrane solubilization was based on previously described procedures (Hansen et al., 2011 ▸). 450 µl bovine Na+,K+-ATPase-enriched membranes were thawed on ice, supplemented with 500 µl solubilization buffer (30 mM K HEPES pH 7.0, 400 µM BeSO4, 1.6 mM KF, 3 mM MgCl2, 0.5 mM ouabain, 5 mM DTT) and incubated on ice for 45 min. The sample was ultracentrifuged for 30 min at 140 000g in a JLA-100 rotor and the resulting pellet was resuspended in 190 µl solubilization buffer and incubated for a further 45 min on ice. The Na+,K+-ATPase was then solubilized by the addition of 13.1 µl 350 mg ml−1 octaethylene glycol monododecyl ether (C12E8), aiming at a protein:detergent ratio of 1:1(w:w), incubated for 5 min at room temperature and then centrifuged for 30 min at 140 000g and 277 K in a JLA-100 rotor. This procedure typically resulted in a 200 µl supernatant of solubilized Na+,K+-ATPase at a concentration of 7–9 mg ml−1.
2.2. Crystallization
The purified and solubilized protein was set up for crystallization at a concentration of 7–9 mg ml−1 using the hanging-drop vapour-diffusion method, mixing 1 µl precipitant solution and 1 µl solubilized protein sample and equilibrating against 400 µl precipitant solution in the well. Initial crystallization conditions were identified in the PEG/Ion Screen from Hampton Research [condition No. 5; 200 mM MgCl2, 20%(w/v) PEG 3350]. Further crystal optimization was obtained by grid and additive screening and was based on continual diffraction-potential evaluation. The final crystallization conditions were 14%(w/v) PEG 3350, 100 mM MOPS/N-methyl-d-glucamine pH 7.0, 210 mM MgCl2, 25 mM β-mercaptoethanol. Prior to mounting the crystals in LithoLoops (Molecular Dimensions), 0.75 µl precipitant solution supplemented with 0.5 mM ouabain and 30%(v/v) glycerol was added to the drops. The crystals were flash-cooled in liquid nitrogen and stored for transport to the synchrotron.
2.3. Diffraction data collection and processing
Screening for X-ray diffraction and data collection were performed on beamlines I911-2 and I911-3 at MAX-lab, Lund, Sweden (Ursby et al., 2013 ▸), on the European Molecular Biology Laboratory (EMBL) beamline P14 at the Deutsches Elektronen-Synchrotron (DESY) in Hamburg, Germany or on the MX BL 14.2 beamline at the Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung (BESSY) at the Helmholz Center in Berlin, Germany (Mueller et al., 2012 ▸). The data were processed and scaled using XDS (Kabsch, 2010 ▸). Subsequently, the data were anisotropically scaled and R free flags were randomly assigned using the Diffraction Anisotropy Server (Strong et al., 2006 ▸).
2.4. Structure determination and refinement
The structure was determined by molecular replacement with Phaser (McCoy et al., 2007 ▸) using the protein chains of porcine E2P–ouabain–Na+,K+-ATPase (PDB entry 4hyt; Laursen et al., 2013 ▸) as a search model. Rigid-body and restrained refinement was performed with PHENIX (Adams et al., 2010 ▸) and model building with Coot (Emsley et al., 2010 ▸). Rigid bodies and translation–libration–screw (TLS) groups were based on six separate domains: the A (α27–α85), N (α382–α593) and P (α354–α381 + α594–α751) domains, the β-ectodomain (β63–β299) and one domain containing all 12 TM helices [αTM1–10 (α86–α159 + α281–α353 + α752–α1021), βTM (β19–β62) and γ-subunit (γ5–γ36)]. For refinement, grouped atomic displacement parameters (ADPs) were applied using two parameter groups per amino-acid residue, as well as individual ADP refinement on all nonprotein features, i.e. ions and ligands. Tight geometrical restraints were applied in order to stabilize the refinement. The composite OMIT map was calculated using PHENIX (Adams et al., 2010 ▸). Geometric parameters were analysed using MolProbity (Chen et al., 2010 ▸) and all structure representations depicted were prepared with PyMOL (Schrödinger). The atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank (Berman et al., 2000 ▸) as PDB entry 4xe5.
3. Results and discussion
The Na+,K+-ATPase (α1β1γ) from bovine kidney was isolated, providing a new animal source which yields enzyme preparations that are suitable for crystallization and structure-solution experiments. The purification procedure includes dissection of the red outer medulla, differential centrifugation, an SDS wash in order to remove peripheral bound membrane proteins and subsequent solubilization using the detergent C12E8. About 155 mg of protein embedded in microsomes was obtained from nine bovine kidneys and approximately half of the protein could be solubilized using C12E8 in a 1:1(w:w) molar ratio. Based on SDS–PAGE analysis, the purity of the protein in the supernatant was around 90%. The total yield of solubilized Na+,K+-ATPase which could be obtained from one batch of purification was thus about 70 mg.
Beryllium fluoride (BeFx) is a well characterized phosphate analogue which mimics phosphorylation within the cytoplasmic P domain of P-type ATPases. As a consequence, the P-type ATPases can be fixed in the functional state analogous to E2P with inhibited ion transport (Murphy & Coll, 1992 ▸; Olesen et al., 2007 ▸). In order to produce a homogenous sample for crystallization purposes, we locked the Na+,K+-ATPase by the addition of BeFx and the Na+,K+-ATPase-specific inhibitor ouabain, resulting in an E2–BeF3 −–ouabain complex. The protein was crystallized using standard vapour-diffusion techniques (Table 1 ▸). Plate-like crystals appeared after 2–3 d at 293 K and grew to a full size of 120 × 80 × 30 µm in 2–3 weeks (Fig. 1 ▸). Single crystals were mounted in LithoLoops, cryoprotected with about 30% glycerol and 0.5 mM ouabain added directly to the crystallization droplet and stored in liquid nitrogen. Assessment of the X-ray diffraction potential and subsequent data collections were performed on several European synchrotron beamlines and the data-collection statistics of the best diffracting crystal are summarized in Table 2 ▸. The crystals belonged to the orthorhombic space group C2221, with one molecule in the asymmetric unit.
Table 1. Crystallization.
| Method | Hanging-drop vapour diffusion |
| Plate type | 24-well VDX (Hampton Research) |
| Temperature (K) | 293 |
| Protein concentration (mg ml−1) | 7–9 |
| Buffer composition of protein solution | 30 mM K HEPES pH 7.0, 400 µM BeSO4, 1.6 mM KF, 3 mM MgCl2, 0.5 mM ouabain, 5 mM DTT, 22 mM C12E8 |
| Composition of reservoir solution | 14%(w/v) PEG 3350, 100 mM MOPS/N-methyl-D-glucamine pH 7.0, 210 mM MgCl2, 25 mM β-mercaptoethanol |
| Volume and ratio of drop | 2 µl, 1:1 ratio |
| Volume of reservoir (µl) | 400 |
Figure 1.
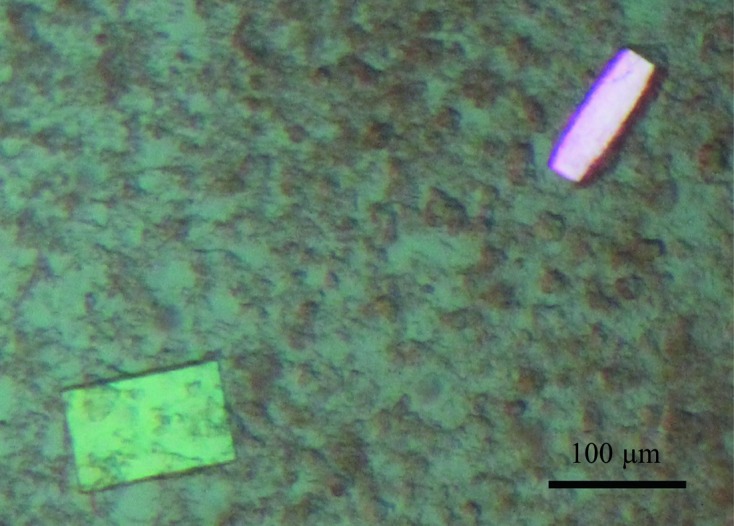
Orthorhombic crystals of bovine Na+,K+-ATPase in an E2–BeF3 −–ouabain complex.
Table 2. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Diffraction source | I911-3, MAX-lab |
| Wavelength (Å) | 1.0 |
| Temperature (K) | 100 |
| Detector | MAR Mosaic 225 |
| Crystal-to-detector distance (mm) | 385 |
| Rotation range per image (°) | 0.25 |
| Total rotation range (°) | 120 |
| Exposure time per image (s) | 10 |
| Space group | C2221 |
| a, b, c (Å) | 64.7, 301.1, 242.3 |
| Mosaicity (°) | 0.32 |
| Resolution range† | |
| Upper limit along a*, b*, c* (Å) | 6.1, 3.7, 3.9 (3.8–3.7) |
| Lower limit (Å) | 30.0 |
| Total No. of reflections | 70244 |
| No. of unique reflections | 15157 |
| Completeness | |
| Low-resolution cutoff‡ (%) | 99.4 (99.7) |
| High-resolution cutoff† (%) | 59.2 (4.2) |
| Multiplicity | 4.6 (3.0) |
| 〈I/σ(I)〉 | |
| Low-resolution cutoff‡ (%) | 8.17 (2.16) |
| High-resolution cutoff† (%) | 9.60 (2.97) |
| R r.i.m. | 0.177 (0.455) |
| Overall B factor from Wilson plot (Å2) | 91.5 |
Elliptical truncation (Strong et al., 2006 ▸): 30–3.7 Å, with values for 3.8–3.7 Å given in parentheses.
Spherical truncation: 30–4.2 Å, with values for 4.3–4.2 Å given in parentheses.
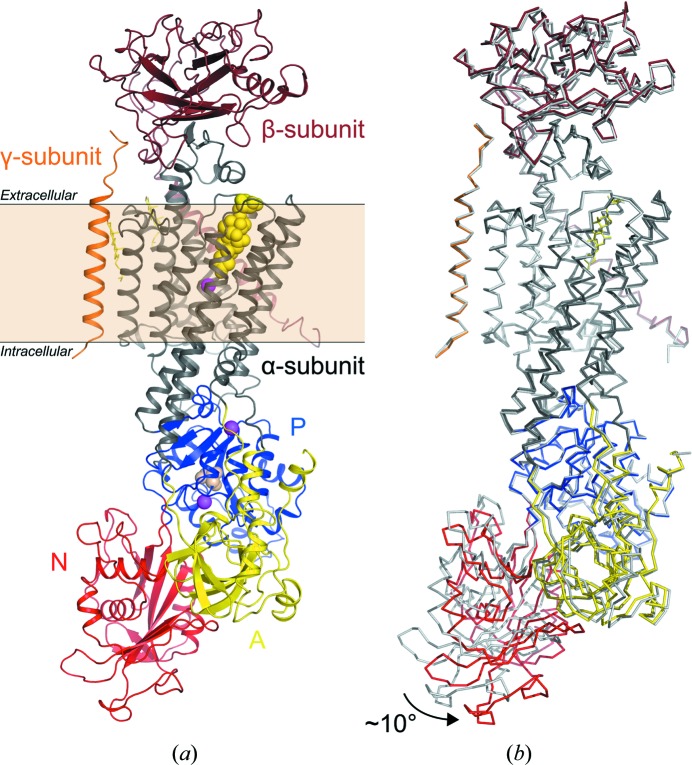
The structure of bovine Na+,K+-ATPase in the E2–BeF3 −–ouabain form was solved by molecular replacement using the protein chains of the E2P–ouabain complex of the homologous porcine Na+,K+-ATPase as a search model (PDB entry 4hyt; Laursen et al., 2013 ▸). One complete molecule (α1β1γ) was placed in the asymmetric unit (Fig. 2 ▸ a). The bovine Na+,K+-ATPase forms type I membrane-protein crystals (Michel, 1983 ▸), which are characterized by stacking of two-dimensional membrane layers. After rigid-body refinement, in which the individual domains were defined as moving groups, a movement of about 10° of the N domain towards the A domain became obvious, with no effect on the overall conformation of the other domains (Fig. 2 ▸ b).
Figure 2.
(a) Structure of bovine Na+,K+-ATPase E2–BeF3 −–ouabain with the α-subunit in dark grey, the β-subunit in brown and the FXYD protein (γ-subunit) in orange. The cytoplasmic domains of the α-subunit are the A (yellow), N (red) and P (blue) domains. Ouabain, BeF3 − and Mg2+ are shown as yellow, wheat and pink spheres, respectively. Cholesterol molecules are shown as yellow sticks. The membrane layer is indicated. (b) Superposition of the bovine Na+,K+-ATPase E2–BeF3 −–ouabain and the porcine Na+,K+-ATPase E2P–ouabain (PDB entry 4hyt; Laursen et al., 2013 ▸; depicted in light grey) structures. The 10° displacement of the N domain is indicated by an arrow.
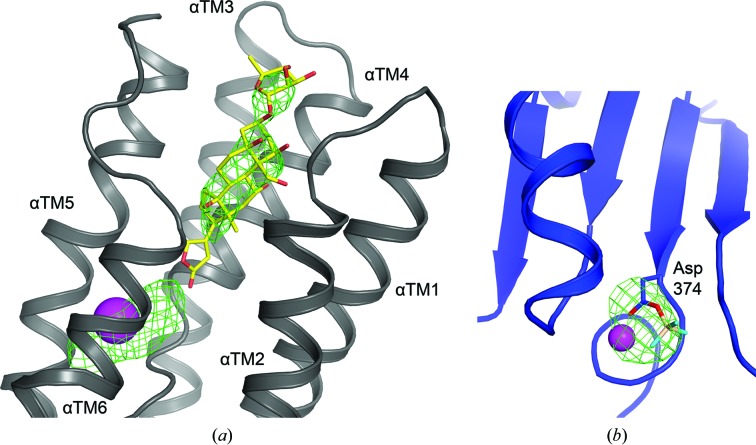
During the course of structure refinement, unbiased F o − F c electron densities were observed both at the CTS binding pocket, showing the presence of bound ouabain and magnesium (Fig. 3 ▸ a), and next to the side-chain carboxylate of the conserved Asp374 phosphorylation site within the cytoplasmic P domain, indicating bound beryllium fluoride (Fig. 3 ▸ b). The composite OMIT map also clearly shows the presence of ouabain and beryllium fluoride (Supplementary Fig. S1). In addition, extra density features at the periphery of the extracellular side of the TM domain were attributed to two cholesterol molecules, as also reported for the crystal structures of Na+,K+-ATPase in various functional states (Ogawa et al., 2009 ▸; Shinoda et al., 2009 ▸; Kanai et al., 2013 ▸; Laursen et al., 2013 ▸, 2015 ▸; Nyblom et al., 2013 ▸). The final refinement statistics are summarized in Table 3 ▸.
Figure 3.
Unbiased electron density (F o − F c map at 3 r.m.s.d., green mesh) after rigid-body refinement (a) around the ouabain-binding site located at the extracellular side of the αTM domain (in grey) and (b) at the phosphorylation site of Asp374 located in the P domain (in blue) attributed to BeF3 − occupancy (in wheat/cyan sticks). Magnesium ions are shown as pink spheres.
Table 3. Structure-refinement statistics.
Values in parentheses are for the highest resolution shell.
| Resolution range (Å) | 30–3.9 (4.2–3.9) |
| Completeness (%) | 64.93 |
| σ Cutoff | 2.0 |
| No. of reflections, working set | 14398 (1087) |
| No. of reflections, test set | 731 (63) |
| Final R cryst | 0.322 (0.434) |
| Final R free | 0.336 (0.458) |
| No. of non-H atoms | |
| Protein | 10257 |
| Ion | 3 |
| Ligand | 97 |
| Water | 5 |
| Total | 10362 |
| R.m.s. deviations | |
| Bonds (Å) | 0.006 |
| Angles (°) | 0.906 |
| Average B factors (Å2) | |
| Protein | 112.0 |
| Ion | 90.8 |
| Ligand | 90.7 |
| Water | 91.6 |
| Ramachandran plot (%) | |
| Favoured regions | 94.93 |
| Additionally allowed | 4.92 |
| Outliers | 0.15 |
The presented structure of bovine Na+,K+-ATPase is the first structure of an Na+,K+-ATPase in an E2–BeF3 − state, which is analogous to an E2P state. The conformation has high affinity for ouabain (Forbush, 1983 ▸) and the obtained structure is therefore a high-affinity CTS complex. As mentioned above, the only significant structural variation compared with the previously determined high-affinity E2P–ouabain structure of the porcine kidney enzyme (Laursen et al., 2013 ▸) is the location of the N domain, which is rotated away by about 10°. However, the same kind of domain movement was previously observed for the high-affinity porcine E2P–bufalin Na+,K+–ATPase structure (Laursen et al., 2015 ▸). This indicates that the movement of the N domain is not induced by the presence of BeF3 −. Instead, it is reasonable to assume that the position of the N domain is highly sensitive to the crystal packing of the different crystal forms, as all three samples were crystallized in different space groups.
Our enzyme preparation contained potassium and magnesium ions, and from a crystallographic point of view alone it is challenging to address which metal ion is bound close to the ouabain-binding site, which would lead to the low- or high-affinity ouabain complex, respectively. Although we have no explicit proof of the absence of potassium, we rationalize that the structure presented here is the high-affinity E2–BeF3 −–ouabain Na+,K+-ATPase complex because (i) BeF3 − is known to form an E2P analogous state which binds ouabain with high affinity (Forbush, 1983 ▸), (ii) crystals suitable for X-ray diffraction were only obtained with high amounts of magnesium (optimized at 210 mM MgCl2) in the crystallization buffer, hence facilitating Mg2+-dependent binding of ouabain to the Na+,K+-ATPase, which results in a high-affinity complex (Laursen et al., 2013 ▸), and (iii) the overall domain arrangement is compatible with magnesium and ouabain binding to the bovine Na+,K+-ATPase E2–BeF3 −–ouabain structure, i.e. similar to the previously determined high-affinity E2P–CTS structures of the porcine kidney enzyme (Laursen et al., 2013 ▸, 2015 ▸).
4. Conclusions
All previously available structural information on Na+,K+-ATPase was obtained from preparations of the porcine and shark enzymes. We have developed a protocol for the isolation of bovine Na+,K+-ATPase that is suitable for crystallization using standard vapour-diffusion techniques. We have solved the crystal structure of bovine Na+,K+-ATPase in a high-affinity E2–BeF3 −–ouabain conformation. Despite the moderate resolution reported, the diffraction potential of the bovine preparation is comparable to recently reported CTS complex structures of the porcine kidney enzyme (Laursen et al., 2015 ▸), and the bovine preparation can therefore be considered to be a useful resource for future protein crystallization.
Supplementary Material
PDB reference: Na+,K+-ATPase from bovine kidney, 4xe5
Supplementary Figure S1.. DOI: 10.1107/S2053230X1600279X/pg5057sup1.pdf
Acknowledgments
We would like to thank Skare Beef, Aarhus Slagtehus, Jægergårdsgade 154, DK-8000 Aarhus C for bovine kidneys and assistance at the slaughterhouse. Temitope Ayeotan, Lotte Thue Pedersen, Peter Aasted Paulsen, Hanne Poulsen and Miroslav Huličiak helped to collect the tissue. A PhD fellowship to JLG was co-financed by the Danish Council for Independent Research and The Graduate School of Science and Technology, Aarhus University. A PhD fellowship to DM was co-financed by the Graduate School of Science and Technology, Aarhus University. PN was supported by the European Research Council through the Advanced Research Grant BIOMEMOS. LR was supported by a postdoctoral fellowship from The Danish Council for Independent Research – Medical Science (FSS) . The authors wish to thank the beamline staff at MAX-lab, Lund, Deutsches Elektronen-Synchrotron (DESY), Hamburg, BESSY, Berlin and the European Synchrotron Radiation Facility (ESRF), Grenoble for their help at the synchrotrons.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Baginski, E. & Zak, B. (1960). Clin. Chim. Acta, 5, 834–838. [DOI] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Blanco, G. & Mercer, R. W. (1998). Am. J. Physiol. 275, F633–F650. [DOI] [PubMed]
- Bradford, M. M. (1976). Anal. Biochem. 72, 248–254. [DOI] [PubMed]
- Cariani, L., Thomas, L., Brito, J. & del Castillo, J. R. (2004). Anal. Biochem. 324, 79–83. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Forbush, B. (1982). J. Biol. Chem. 257, 12678–12684. [PubMed]
- Forbush, B. (1983). Curr. Top. Membr. Trans. 19, 167–201.
- Hansen, A. S., Kraglund, K. L., Fedosova, N. U. & Esmann, M. (2011). Biochem. Biophys. Res. Commun. 406, 580–583. [DOI] [PubMed]
- Jørgensen, P. L. (1988). Methods Enzymol. 156, 29–43.
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kanai, R., Ogawa, H., Vilsen, B., Cornelius, F. & Toyoshima, C. (2013). Nature (London), 502, 201–206. [DOI] [PubMed]
- Klodos, I., Esmann, M. & Post, R. L. (2002). Kidney Int. 62, 2097–2100. [DOI] [PubMed]
- Laursen, M., Gregersen, J. L., Yatime, L., Nissen, P. & Fedosova, N. U. (2015). Proc. Natl Acad. Sci. USA, 112, 1755–1760. [DOI] [PMC free article] [PubMed]
- Laursen, M., Yatime, L., Nissen, P. & Fedosova, N. U. (2013). Proc. Natl Acad. Sci. USA, 110, 10958–10963. [DOI] [PMC free article] [PubMed]
- Lingrel, J. B., Argüello, J. M., Van Huysse, J. & Kuntzweiler, T. A. (1997). Ann. N. Y. Acad. Sci. 834, 194–206. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Michel, H. (1983). Trends Biochem. Sci. 8, 56–59.
- Miller, R. P. & Farley, R. A. (1988). Biochim. Biophys. Acta, 954, 50–57. [DOI] [PubMed]
- Mohammadi, K., Kometiani, P., Xie, Z. & Askari, A. (2001). J. Biol. Chem. 276, 42050–42056. [DOI] [PubMed]
- Morth, J. P., Pedersen, B. P., Toustrup-Jensen, M. S., Sørensen, T. L., Petersen, J., Andersen, J. P., Vilsen, B. & Nissen, P. (2007). Nature (London), 450, 1043–1049. [DOI] [PubMed]
- Mueller, U., Darowski, N., Fuchs, M. R., Förster, R., Hellmig, M., Paithankar, K. S., Pühringer, S., Steffien, M., Zocher, G. & Weiss, M. S. (2012). J. Synchrotron Rad. 19, 442–449. [DOI] [PMC free article] [PubMed]
- Murphy, A. J. & Coll, R. J. (1992). J. Biol. Chem. 267, 5229–5235. [PubMed]
- Nyblom, M., Poulsen, H., Gourdon, P., Reinhard, L., Andersson, M., Lindahl, E., Fedosova, N. & Nissen, P. (2013). Science, 342, 123–127. [DOI] [PubMed]
- Ogawa, H., Cornelius, F., Hirata, A. & Toyoshima, C. (2015). Nature Commun. 6, 8004. [DOI] [PMC free article] [PubMed]
- Ogawa, H., Shinoda, T., Cornelius, F. & Toyoshima, C. (2009). Proc. Natl Acad. Sci. USA, 106, 13742–13747. [DOI] [PMC free article] [PubMed]
- Olesen, C., Picard, M., Winther, A. M., Gyrup, C., Morth, J. P., Oxvig, C., Møller, J. V. & Nissen, P. (2007). Nature (London), 450, 1036–1042. [DOI] [PubMed]
- Post, R. L. & Jolly, P. C. (1957). Biochim. Biophys. Acta, 25, 118–128. [DOI] [PubMed]
- Reinhard, L., Tidow, H., Clausen, M. J. & Nissen, P. (2013). Cell. Mol. Life Sci. 70, 205–222. [DOI] [PMC free article] [PubMed]
- Rose, E. M., Koo, J. C. P., Antflick, J. E., Ahmed, S. M., Angers, S. & Hampson, D. R. (2009). J. Neurosci. 29, 8143–8155. [DOI] [PMC free article] [PubMed]
- Rudnick, G. (1998). J. Bioenerg. Biomembr. 30, 173–185. [DOI] [PubMed]
- Shinoda, T., Ogawa, H., Cornelius, F. & Toyoshima, C. (2009). Nature (London), 459, 446–450. [DOI] [PubMed]
- Skou, J. C. & Esmann, M. (1988). Methods Enzymol. 156, 43–46. [DOI] [PubMed]
- Stoscheck, C. M. (1990). Methods Enzymol. 182, 50–68. [DOI] [PubMed]
- Strong, M., Sawaya, M. R., Wang, S., Phillips, M., Cascio, D. & Eisenberg, D. (2006). Proc. Natl Acad. Sci. USA, 103, 8060–8065. [DOI] [PMC free article] [PubMed]
- Sweadner, K. J. & Rael, E. (2000). Genomics, 68, 41–56. [DOI] [PubMed]
- Ursby, T., Unge, J., Appio, R., Logan, D. T., Fredslund, F., Svensson, C., Larsson, K., Labrador, A. & Thunnissen, M. M. G. M. (2013). J. Synchrotron Rad. 20, 648–653. [DOI] [PMC free article] [PubMed]
- Yatime, L., Laursen, M., Morth, J. P., Esmann, M., Nissen, P. & Fedosova, N. U. (2011). J. Struct. Biol. 174, 296–306. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Na+,K+-ATPase from bovine kidney, 4xe5
Supplementary Figure S1.. DOI: 10.1107/S2053230X1600279X/pg5057sup1.pdf