Abstract
In the developing and mature brain, mitochondria act as central hubs for distinct but interwined pathways, necessary for neural development, survival, activity, connectivity and plasticity. In neurons, mitochondria assume diverse functions, such as energy production in the form of ATP, calcium buffering and generation of reactive oxygen species. Mitochondrial dysfunction contributes to a range of neurodevelopmental and neurodegenerative diseases, making mitochondria a potential target for pharmacological-based therapies. Pathogenesis associated with these diseases is accompanied by an increase in mitochondrial mass, a quantitative increase to overcome a qualitative deficiency due to mutated mitochondrial proteins that are either nuclear- or mitochondrial-encoded. This compensatory biological response is maladaptive, as it fails to sufficiently augment the bioenergetically functional mitochondrial mass and correct for the ATP deficit. Since regulation of neuronal mitochondrial biogenesis has been scantily investigated, our current understanding on the network of transcriptional regulators, co-activators and signaling regulators mainly derives from other cellular systems. The purpose of this review is to present the current state of our knowledge and understanding of the transcriptional and signaling cascades controlling neuronal mitochondrial biogenesis and the various therapeutic approaches to enhance the functional mitochondrial mass in the context of neurodevelopmental disorders and adult-onset neurodegenerative diseases.
Keywords: Mitochondrial DNA maintenance, adaptive mitochondrial biogenesis, mitochondrial fusion-fission, mitophagy, maternally inherited mitochondrial diseases, Parkinson’s disease, Huntington’s disease, small-molecule-based therapies
1. INTRODUCTION
Due to their eubacterial ancestry, mitochondria contain their own genome characterized by limited encoding capacity, making these organelles dependent on the nuclear genome for their functions. Even though mitochondria are ubiquitous organelles, their mitochondrial proteome is complex and exhibits tissue-heterogeneity to meet the metabolic and energy needs specific to each cell type, to assume diverse functions tailored to specific organs, and to adapt a myriad of mitochondrial pathways to specific metabolic state of the cell [1]. Half of the mitochondrial proteins are tissue-specific, while the other half makes up the core protein components [2]. Using a combination of in-depth mass spectrometry, microscopy and computation, the mitochondrial compendium has been estimated at 1098 genes with their expression profiles analyzed across 14 distinct mouse tissues [3]. This mitochondrial protein atlas, which is constantly expanded and updated on the MitoCarta inventory available at http://www.broad.mit.edu/publications/MitoCarta, provides a platform to decipher tissue-specific programs and pathogenesis of mitochondrial diseases. This inventory highlights a lack of tissue specificity for most of the subunits of the oxidative phosphorylation (OXPHOS) system and the Krebs cycle, whereas many of the mitochondrial ribosomes, half of the subunits of complex IV, enzymes involved in the ketogenesis and urea cycle pathways exhibit some degree of tissue-specificity [3].
Mitochondrial dysfunction causes a range of diseases spanning from incurable neonatal neurodevelopmental diseases to adult-onset neurodegenerative diseases [4]. Maintenance of an adequate and functional mitochondrial population during the lifetime of neurons is critical and involves a balance between mitochondrial biogenesis and turnover. Mitochondrial quality control occurs via the process of mitophagy through the autophagy machinery, a process that is often dysregulated in many neurological diseases [5, 6]. Mitochondrial biogenesis is a complex process requiring a coordinated bigenomic regulation to execute several distinct processes: 1) inner and outer mitochondrial membrane synthesis; 2) synthesis of mitochondrial-encoded proteins; 3) synthesis and import of nuclear-encoded mitochondrial proteins; and 4) replication of mitochondrial DNA (mtDNA). Since neuronal mitochondrial biogenesis by itself has been poorly explored, the identity of the neuronal-specific regulators responsible to coordinate mitochondrial biogenesis with neural development remains unknown. In the case of the mitochondrial respiratory disorders and adult-onset neurodegenerative diseases, neurons exhibit a mixed population of healthy and defective mitochondria whose overall functionality depends on the equilibrium between mitochondrial biogenesis and mitophagy. As a result of bioenergetic deficit and sub-optimal ATP synthesis, a compensatory biological response is triggered as an attempt to produce more mitochondria- a quantitative increase to overcome a qualitative deficiency due to mutated mitochondrial proteins that are either nuclear- or mitochondrial-encoded.
This review will concentrate on mitochondrial biogenesis in the context of neuronal differentiation, its timing and regulation, the signaling pathways responsible to enhance mitochondrial mass in response to neuronal stimuli. We will discuss potential small molecule-based therapeutic strategies to boost the mitochondrial biogenic response in various animal models for mitochondrial encephalomyopathies and their implications for mitochondrial respiratory disorders and adult-onset neurodegenerative diseases (Table 1).
Table 1.
Discussed Neurodevelopmental and Neurodegenerative Diseases
| Disease | Clinical features | Age of onset |
Mt or nuclear mutations |
Pattern of Inheritance |
Primary or secon- dary mitochon- drial disorders |
Impaired mitochondrial biogenesis |
Impaired OXPHOS activities |
Aberrant mitochondrial dynamics |
|---|---|---|---|---|---|---|---|---|
| MELAS | stroke-like, seizures, cortical blindness, migraines, lactic acidosis, dystonia, progressive mental retardation, diabetes, hemiparesis, cardiomyopathy | Childhood or early adulthood | Mt | M | Primary | Yes | Yes | ND |
| MERRF | myoclonus, myopathy with ragged-red fibers, spasticity, cardiomyopathy, seizures, ataxia, dementia, peripheral neuropathy | Childhood or early adulthood | Mt | M | Primary | Yes | Yes | ND |
| AdPEO | Cataract, deafness, depression, myopathy with ragged-red fibers, parkinsonism, sensory ataxic neuropathy, ptosis, | Childhood | Nuclear | AD | Primary | Yes | Yes | ND |
| Leigh | Ataxia, intellectual retardation, hypotonia, nystagmus, cardiomyopathy, brainstem dysfunction, demylelination, vascular proliferation | Childhood | Mt or nuclear | M or AR or X-linked | Primary | Yes | Yes | ND |
| COX deficiency | Developmental regression, ataxia, nystagmus, hypotonia, lactic acidosis, kidney problems, respiratory failure | Childhood | Mt or nuclear | M or AR | Primary | Yes | Yes | ND |
| CMT2A | Progressive motor and sensory neuropathy in lower limbs, hypotonia | Childhood or adulthood | Nuclear | AD | Secondary | No | No | Yes |
| Parkinson’s disease | Bradykinesia, rigidity, resting tremor, balance problems, cognitive and speech impairment, degeneration of DA neurons of the substantia nigra | Adulthood | Nuclear | AR | Secondary | Yes | Yes | Yes |
| Huntington’s disease | Cognitive impairment, choreoathetotic movements, dementia, selective degeneration of striatal neurons, atrophy of caudate and putamen | Adulthood | Nuclear | AD | Secondary | Yes | Yes | Yes |
| Alzheimer’s disease | Selective memory impairment, dementia | Adulthood | Nuclear | Sporadic or AD | Secondary | Yes | Yes | Yes |
Abbreviations: AD: Autonomic Dominant; AdPEO: Autonomic dominant Progressive external Ophtalmoplaegia; AR: Autonomic Recessive; COX: Cytochrome c Oxidase; CMT2A: Charcot-Marie-Tooth Type 2A; DA: Dopaminergic; M: Maternal; MELAS: Mitochondrial Encephalopathy Lactic Acidosis Stroke-like episode; MERRF: Myoclonic Epilepsy Ragged-Red Fibers; Mt; Mitochondrial; ND: Not Determined.
2. MITOCHONDRIA ARE ESSENTIAL ORGANELLES FOR DEVELOPING AND MATURE NEURONS
It has long been known that the brain is a highly energy-demanding organ that consumes more than 20% of the total energy produced by the organism, even though it only makes 2–3% of the whole body weight [7, 8]. Most of the energy produced in the form of ATP derives from OXPHOS, since glycolysis generates a small proportion of cellular ATP [9]. Thus, developing neurons are highly dependent on mitochondrial biogenesis to assume their energy need associated with cytoskeletal remodeling, axonal and dendritic growth, axonal transport of synaptic vesicles and synaptic transmission, which consume about 50% of generated ATP [10–13]. Similarly, a mature brain requires more than 50% of the brain’s energy consumption to maintain synaptic homeostasis and plasticity. Therefore, a dysfunctional mitochondrial biomass and/or disturbed mitochondrial trafficking, two pathological hallmarks of neurodegenerative diseases, lead to altered neurometabolic coupling, neural processing and circuitry, and functional connectivity in mature neurons [14–16].
Aside from producing ATP, mitochondria regulate calcium (Ca2+) homeostasis by modulating Ca2+ uptake in an energy-dependent and pulsatile manner via the mitochondrial Ca2+ uniporter (mCU) and by providing ATP to stimulate the plasma membrane Ca2+-ATPase, thereby influencing synaptic transmission, cellular survival and metabolism [17–21]. Moreover, Ca2+ sequestration dictates mitochondrial motility to specific sub-cellular domains of neurons to regulate synaptic function, with defective mitochondrial movements causing neurological disorders [22, 23]. Under pathological conditions, mitochondrial Ca2+ overload compromises the integrity of the mitochondrial inner membrane, thereby triggering mitochondrial permeability transition (MPT), a process when sustained results in cessation of ATP production, permeability of the outer mitochondrial membrane, cytochrome c leakage and subsequent neuronal cell death [24, 25]. Thus, the mitochondrion functions as a major hub for several distinct but interconnected pathways necessary for neural development, survival, activity, connectivity and plasticity.
3. MAINTENANCE OF MITOCHONDRIAL HOMEOSTASIS
A hallmark of brain metabolism is the tight coupling between energy demand and supply, which involves constant regulation of mitochondrial homeostasis and correct localization of mitochondria in the distinct subcellular compartments of the extremely polarized neurons to address their unique regional metabolic needs in response to signaling cues [23]. Thus, mitochondria are not uniformly distributed among the subcellular domains, the soma, axon and dendrites [26]. They occupy up to 40% of the cytoplasmic volume of neuronal cells with their number oscillating from a few hundred to thousands. They are transported to specific compartments of neurons characterized by high ATP consumption and calcium dynamics, such as active growth cones, axonal branching points [27–31], nodes of Ranvier [32], myelination boundaries and demyelinated regions [33, 34].
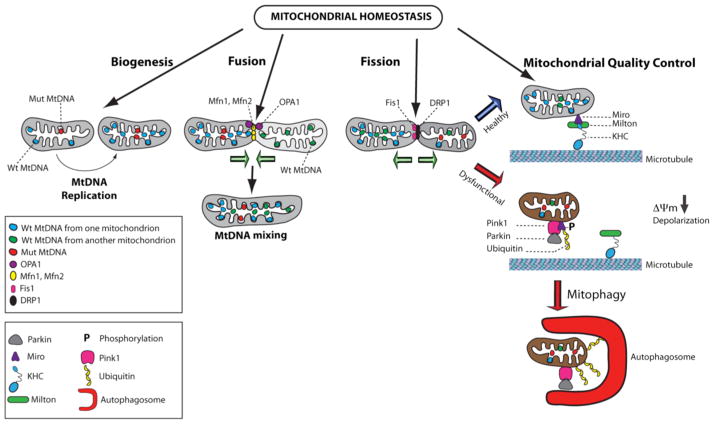
Although mitochondria have a longer half-life in neurons than in other post-mitotic somatic cells, their renewal is essential for proper neural development and survival [35–39]. The question of how to define mitochondrial biogenesis has been addressed in many different cellular systems, nonpathological and pathological contexts and distinct developmental stages, which led to further questions. Is this a self-sufficient or partially independent or dependent process of preexisting mitochondria? The most likely response is that the mitochondrial mass is under the control of three distinct mechanisms: fission of pre-existing mitochondria, biogenesis of newly generated mitochondria and mitochondrial quality control to dispose of dysfunctional mitochondria via two pathways, mitophagy and the ubiquitin proteasome system (UPS) (Fig. 1). While fission does not necessitate replication of mtDNA and newly synthesized mitochondrial membrane, mitochondrial biogenesis generates new mitochondria via replication of mtDNA and synthesis of mitochondrial lipids for the genesis of outer and inner mitochondrial membranes [40]. Cardiolipin is a unique phospholipid that is almost exclusively present in mitochondrial membranes and required for mitochondrial biogenesis and maintenance of the mitochondrial membrane composition [41, 42].
Fig. 1.
Maintenance of mitochondrial homeostasis. Mitochondrial number and health are under the control of three pathways: biogenesis, fusion-fission cycles and mitochondrial quality control. In healthy cells, these pathways are well balanced. Mitochondrial biogenesis involves mtDNA replication, while fusion promotes mtDNA mixing. Wild type mtDNA is illustrated in blue or green, while mutated mtDNA is indicated in red. Execution of the fusion process requires merging of the outer and inner mitochondrial membranes via the sequential action of the GTPase proteins, OPA1 (purple circles) and the mitofusins, Mfn1 and Mfn2 (yellow circles). Mitochondrial mass is also influenced by the process of fission, a process during which one mitochondrion gives rise to two healthy mitochondria. Fission is controlled by the dynamin-related protein DRP1 protein (black circles) and its receptors, such as Fis1 protein (pink circles), which together constrict the membranes to cause separation of mitochondria. The mitochondrial quality control prevents accumulation of dysfunctional mitochondria exhibiting mitochondrial membrane depolarization, which are targeted by the autophagic machinery for clearance. Healthy mitochondria are attached to microtubules via the kinesin heavy chain (KHC in blue) and the Miro-Milton adaptor complex illustrated by purple and green symbols, respectively. Following mitochondrial membrane depolarization provoked by various insults or increased mutated mtDNA population, PINK1 (pink symbol) accumulates in the outer mitochondrial membrane to recruit Parkin (grey symbol). Subsequently, Miro is phosphorylated resulting in detachment from Milton and microtubules. Parkin promotes ubiquitination (yellow symbol) of Miro and additional mitochondrial proteins, thereby inducing mitophagy with the assistance of autophagosomes (red symbol).
In neurons, mitochondrial biogenesis mainly occurs in the soma although it has been observed locally within the axon, albeit with low frequency [43]. Its regulation and timing will be discussed in subsequent sections of this review. Local increase of the mitochondrial density can be modulated by repeated cycles of fusion-fission and mitochondrial trafficking on actin and microtubules – both processes involved in maintaining functional integrity of the mitochondrial network. Mitochondria are dynamic and mobile organelles, which shuttle between “individual ” and “network ” states characterized by distinct morphologies, discrete spheres or interconnected tubules, respectively [44–46]. Mitochondrial fusion results in elongated and interconnected mitochondrial network via coordinated merging of the outer and inner mitochondrial membranes under the control of three conserved transmembrane GTPases proteins, mitofusin1 (Mfn1), mitofusin 2 (Mfn2), and optic atrophy 1 protein (OPA1) [47, 48]. Fusion allows exchanging matrix content and mtDNA molecules between mitochondria, which favors optimal mitochondrial physiology by diluting mutated mtDNA and rescuing damaged mitochondria via acquisition of key components from healthy mitochondria (Fig. 1) [49]. Mfn1 or Mfn2 knockout mice exhibit early embryonic lethality due to a placental defect [50]. However, conditional Mfn2 knockout mice display cerebellar neurodegeneration, while Mfn1 is not essential for cerebellar development [51]. Although mitochondrial fusion is a ubiquitous process, its dysregulation provokes specific neurodegenerative diseases, including autosomal-dominant optic atrophy and the Charcot-Marie-Tooth neuropathy type 2A (CMT2A) [52–54]. It is believed that certain neurons are particularly sensitive to defective mitochondrial fusion-fission, as it impacts mitochondrial distribution in neurons with long axons or extensive dendritic branching. Cerebellar Purkinje cells of Mfn2 conditional knockout mice display aggregated mitochondria in dendrites thereby preventing microtubule-dependent transport of organelles [51]. A similar occlusion in neuritic branches has been observed in mouse models of Alzheimer’s disease (AD) and is considered as a potential mechanism for neurodegeneration (Table 1) [55]. Finally, abnormal trafficking has been reported in the longest axons of sensory neurons affected in CMT2A (Table 1) [56]. In contrast, fission of preexisting mitochondria generates new mitochondria in the absence of mtDNA replication under the control of the master mediator dynamin-related protein Drp1, whose activity is regulated through post-translational modifications and interactions with specific receptor proteins, such as the Fission 1 protein (Fis1) (Fig. 1) [57]. While mitochondrial fusion-fission influences many cellular functions, neurons critically rely on this dynamic process to ensure adequate distribution of mitochondria for supporting synaptic activity via the formation of synapses and dendritic spines [58, 59]. Moreover, imbalance of mitochondrial dynamics is predicted to lead to neurodegenerative diseases, as continuous mitochondrial fission can induce a neurodegenerative cascade [60–63].
The mitochondrial quality control system is essential to maintain a functional mitochondrial population in neurons, which occurs via UPS and mitophagy to clear aged and dysfunctional mitochondria (Fig. 1) [64–65]. The question of whether induction of mitochondrial biogenesis triggers the mitophagic process of older mitochondria to keep constant the overall mitochondrial content in healthy neurons remains to be answered. In contrast, dysregulation of either process has been the subject of intense studies in the context of adult-onset neurodegenerative diseases. The mitochondrial quality control system has mainly been investigated as an intrinsic mechanism to selectively sequester aberrant mitochondria as a consequence of bioenergetic deficit, accumulation of reactive oxygen species or high levels of Ca2+ influx, followed by degradation via the autophagy-lysosomal pathway, also referred to as mitophagy [66]. Although direct evidence for compartmentally restricted mitophagy in neurons is scant, it is speculated that the process of mitophagy occurs predominantly in the soma where mature lysosomes reside, and therefore coupled to mitochondrial retrograde movement [67]. However, the notion of retrograde movement of depolarized mitochondria is controversial given that axonal mitochondrial transport depends on the strength of the mitochondrial membrane potential [29, 30]. Recent time-lapse imaging provides new insight on neuronal mitophagy, as depolarized mitochondria were observed undergoing local mitophagy in the somatodentritic regions where lysosomes are mostly positioned [68]. Furthermore, recent studies have revealed the colocalization of lysosomal markers and autophagosomes in the distal axons of dorsal root ganglion neurons, congruent with local mitochondrial clearance [69, 70]. The surveillance pathway under the control of the serine/threonine PTEN-Induced putative Kinase 1 (PINK1) and the E3 ubiquitin ligase Parkin regulates mitophagy in various cellular contexts and systems [65, 71]. Following depolarization, PINK1 moves from the inner mitochondrial membrane (IMM) to the outer mitochondrial membrane (OMM) and phosphorylates the molecular/adaptor molecule, Mitochondrial Rho (Miro), which connects mitochondria to microtubules via its binding partner Milton and kinesin heavy chain (KHC). PINK1 also recruits Parkin to the OMM of compromised mitochondria, setting the stage for Parkin-mediated ubiquitination and degradation of Miro causing mitochondrial motility arrest and clearance via mitophagy [72, 73]. In addition, Parkin functions as a molecular link between the UPS and mitophagy pathways by ubi-quinating numerous proteins from the mitochondrial proteome, among them Mfn1 and Mfn2, to favor mitochondrial fission and fragmentation, making mitochondria more susceptible to mitophagy [74–76]. The PINK1-Parkin pathway is particularly relevant to the nervous system, as mutations in the Parkin and PINK1 genes are linked to familial forms of Parkinson’s disease (PD) [77, 78]. Furthermore, compelling evidence is congruent with the notion of a dysregulated mitochondrial quality control system leading to mitochondrial dysnfunction and neuronal loss, two hallmarks of numerous neurodegenerative diseases, such as AD and Huntington’s disease (HD) as discussed in the section 7 of the review [5, 64; 79–82].
4. HIGHLIGHTS OF NEURONAL MITOCHONDRIAL BIOGENESIS AND ITS REGULATORS
Little is known about neuronal mitochondrial biogenesis, as most studies have been conducted in the context of adipocyte and muscle cell differentiation [83]. However, the high degree of conservation among key transcription factors and co-activators suggests that a similar regulatory cascade should modulate the mitochondrial content in neurons [84]. Since mitochondria cannot be made de novo, the formation of new mitochondria occurs from preexisting mitochondria via mitochondrial biogenesis [85]. Mitochondrial biogenesis is an extremely complex process requiring replication of the mtDNA, the synthesis, import, and incorporation of proteins and lipids to the existing mitochondrial reticulum, and a certain threshold of mitochondrial membrane potential as the result of proton gradient generated during oxidative phosphorylation. Mitochondrial biogenesis requires a coordinated regulation of two distinct genomes, the nuclear and mitochondrial genomes given that the majority of mitochondrial proteins are encoded by nuclear genes. This is potentially challenging for post-mitotic neurons due to the extremely polarized architecture, forcing the majority of mitochondrial biogenesis to be confined to the soma and requiring mitochondrial renewal in the distal axons to rely on axonal transport via microtubules.
Findings over the last decade have revealed several converging signaling pathways intersecting with the basic mitochondrial regulatory machinery via a mitochondrial-nuclear crosstalk to replicate the mitochondrial genome in parallel with increase in mitochondrial mass. The circular double stranded mtDNA (16,569 bp) encodes a total of 37 genes, 13 of which encode essential protein subunits of the OXPHOS respiratory complexes, while the other genes are dedicated for synthesis of mitochondrial proteins by encoding 22 transfer RNAs and 2 ribosomal RNAs [86]. None of the mitochondrial genes encodes for proteins directly involved in mitochondrial DNA replication and maintenance. Therefore, the functional mitochondrial mass is influenced by the nuclear-mitochondrial cross talk and cellular context.
Mitochondria contain between 800 to 1000 copies of mtDNA, which are maternally inherited and packaged in high-ordered nucleo-protein structures called nucleoids [87–91]. Although nucleoids are distributed throughout the mitochondrial matrix, they are often located in proximity of the cristae, which carry the OXPHOS system. In vitro studies have revealed that nucleoids are packed in a sphere consisting of two components, the core center and the peripheral layer [92–94]. While the core center is composed of two to ten mtDNA copies bound to proteins responsible for mtDNA transcription, replication and maintenance, the peripheral layer contains proteins involved in diverse functions, such as protein folding and metabolism, which tether the nucleoids to the inner mitochondrial membrane of the cristae [93, 95, 96]. Although the in vivo structure and composition of nucleoids remain to be established, alteration in nucleoid architecture is detrimental to mitochondrial functions, as bitransgenic mice overexpressing Tfam and Twinkle, two essential regulators of mtDNA copy number, exhibited nucleoid enlargement accompanied by mtDNA deletions due to high mtDNA copy number interfering with mtDNA replication and transcription [97].
A compilation of recent studies has provided key evidence in support of the concept of the nucleoid functioning as the mitochondrial genetic inheritance unit [98]. Our understanding of how nucleoid numbers are regulated remains elusive and the question of whether their copy number is specific to cell fate and/or the mitotic status remains to be answered. Two models for the mode of nucleoid-mediated mtDNA propagation have been postulated: the static or “faithful nucleoid” model and the “dynamic nucleoid” [99]. The faithful model implies that nucleoids do not exchange mtDNA between each other, while the “dynamic nucleoid” model states that nucleoids under go dynamic reorganization allowing mtDNA exchange. A recent study has demonstrated that the “faithful nucleoid” model appears to be the predominant mode of propagation in rapidly dividing cells [94]. By fusing two cybrid cell lines, each containing a homoplastic population of mtDNA with two distinct non-overlapping Δ mtDNA deletions, the authors found that the two distinct nucleoid populations did not exchange mtDNAs after many cell divisions, congruent with the notion of nucleoids being essentially autonomous. The packaging of mtDNA into nucleoids is believed to be the limiting factor preventing the mixing of two distinct mtDNAs populations. More importantly, the two-nucleoid populations complemented each other to restore bioenergetic functions, suggesting that mtDNA-derived transcripts and/or polypeptides involved in the assembly of OXPHOS complexes diffuse locally within the matrix. This is in agreement with the notion of the relative threshold of heteroplasmic burden observed in patients affected with maternally-inherited mitochondrial respiratory diseases. However, it remains to be determined whether in vivo nucleoids may exchange mtDNA between each other during specific cellular events.
The regulatory circuitry responsible to modulate mitochondrial biogenesis in response to developmental and physiological cues is composed of transcriptional coactivators, such as members of the peroxisome proliferator-activated receptor γ coactivator-1 (PGC-1) family, and transcription factors, including the nuclear respiratory factors 1 (NRF-1) and 2 (NRF-2) (Fig. 2) [100]. PGC-1α is a ubiquitously expressed transcriptional co-activator that was initially identified as the co-activator of the nuclear receptor PPARγ, a regulator of adaptive thermogenesis in brown adipose tissue [101]. It belongs to the PGC-1 family, which is composed of three members, PGC-1α, PGC-1β, and PGC-1-related co-activator (PRC), all of them known to interact with the family of nuclear receptors PPARα, PPARβ/γ, and PPARγ [102]. PGC-1α is expressed at high levels in tissues with high metabolic demands, such as heart, skeletal muscle, kidney and brain [103, 104]. PGC-1α is expressed in the developing brain, from embryonic day 15 to postnatal day 3, with accentuated expression levels in GABAergic neurons to regulate their metabolism and survival capacity [105]. Consistent with PGC-1α critical role in the brain is the phenotype of PGC-1α null mice exhibiting striatal degeneration and small lesions in the cortical layer V/VI of the motor cortex, nucleus accumbens, substantia nigra, hippocampus and the mammalliary body, reminiscent of the neurodegenerative HD [106]. Although PGC-1α remains expressed in the mature brain, its function and activities remain elusive.
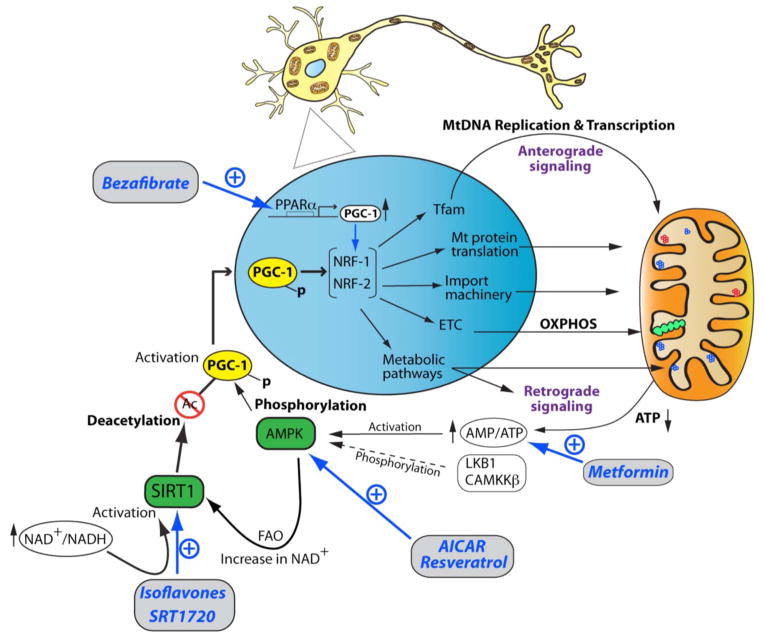
Fig. 2.
An integrated view of therapeutic strategies targeting the transcriptional network and signaling pathways promoting neuronal mitochondrial biogenesis. Diagrammed are the anterograde and retrograde signaling crosstalks between the nucleus (blue) and mitochondria (orange). The OXPHOS system is depicted with green circles, while wild type and mutated mtDNA molecules are illustrated with blue and red circles, respectively. The signaling and transcriptional cascades are limited to the PGC-1α-NRF-1-NRF-2 axis, given its important regulatory role for neuronal mitochondrial biogenesis. Pharmacological manipulations of key regulators for mitochondrial biogenesis and bioenergetics are illustrated in blue in the diagrammatic summary. PGC-1α expression can be pharmacologically enhanced by bezafibrate, an agonist of PPARα, which leads to increased expression of the NRF-1 and NRF-2 genes. In addition, PGC-1α directly stimulates the transcriptional activity of the NRF-1 and NRF-2 transcription factors via protein-protein interactions. Consequently, PGC-1α-mediated increased expression levels and activity of NRF-1 and NRF-2 lead to stimulation of gene expression relevant to mtDNA replication via the TFAM protein, OXPHOS activity (ETC), import of nuclear-encoded proteins, the mitochondrial translation machinery, and diverse mitochondrial and cellular metabolic pathways. The major retrograde signaling pathway is under the control of the AMP/ATP ratio, which augments upon decreased ATP levels. Increased AMP/ATP ratio activates AMPK, which subsequently phosphorylates the PGC-1α protein. In neuronal cells, activation of AMPK is also under the control of the LKB1 and CAMMKβ kinase. Phosphorylated PGC-1α migrates to the nucleus, where it stimulates the expression levels of the NRF-1 and NRF-2 genes as well as their transcriptional activities. In a neuronal context, AMPK activity can be modulated via several pharmacological means, such as AICAR, resveratrol, and metformin, which are depicted in blue in the diagrammatic summary. AMPK also influences the activity of the NAD+-dependent deacetylase SIRT1 via fatty acid oxidation (FAO), resulting in increased NAD+ levels. Subsequently, SIRT1 is activated via phosphorylation, which can occur independently of AMPK upon increase of the NAD+/NADH ratio. Isoflavones and the pharmacological agent SRT1720 also activate SIRT1, which in turn deacetylate PGC-1α, thereby linking the cellular metabolic status to a network of gene expression relevant for mitochondrial biogenesis.
PGC-1α is a highly versatile transcriptional co-activator that behaves as the master regulator of mitochondrial biogenesis by increasing the expression of various relevant transcription factors and cooperating with them to potentiate their transcriptional activity. In addition to mitochondrial biogenesis, PGC-1α regulates mitochondrial pathways critical for neuronal metabolism, such as fatty acid oxidation (FAO) and OXPHOS metabolism (Fig. 2) [83]. PGC-1α not only induces the expression levels of the two key transcription factors, NRF-1 and NRF-2, but also interacts with them to stimulate their transcriptional activity, resulting increased expression of genes responsible for mtDNA replication and transcription, such as the essential regulator of mtDNA copy number, mitochondrial transcription factor A (TFAM) (Fig. 2) [107]. Subsequently, TFAM stimulates the biogenesis of mitochondria via increased expression of mitochondrial-encoded polypeptides, OXPHOS respiration and intracellular ATP concentrations [108–110]. In parallel, PGC-1α stimulates the expression levels of numerous nuclear-encoded subunits of the mitochondrial respiratory complexes of the OXPHOS system via the NRF-1/NRF-2 pathway, thereby directly stimulating the mitochondrial bioenergetic output (Fig. 2). Thus, PGC-1α governs mitochondrial biogenesis, by making mitochondria more bioenergetically competent, which in turn favors biogenesis due to increased import of key nuclear-encoded mitochondrial proteins, a process that is dependent on the mitochondrial membrane potential [111–113]. Illustrating the importance of PGC-1α to control mitochondrial functions essential for neuronal survival is the decreased expression levels of PGC-1α associated with the neurodegenerative diseases, HD, PD and AD [114].
In various cellular contexts, NRF-1 and NRF-2 control multiple mitochondrial functions, most notably the oxidative phosphorylation metabolism by binding to promoter regions of nuclear genes encoding subunits of the five respiratory complexes of the OX-PHOS system (Fig. 2) [100]. Since their properties have been extensively described in numerous reviews [100, 102, 107, 108, 115–117], this review focusses on the impact of NRF-1 and NRF-2 on mitochondrial functions vis-à-vis neurogenesis. Both NRF-1 and NRF-2 regulate genes involved in assembly of the respiratory apparatus, the heme biosynthetic pathway, import of nuclear-encoded mitochondrial proteins, and mtDNA replication and transcription, most notably the Tfam gene. Therefore, NRF-1 and NRF-2 are transcription factors critical for modulating the nuclear-mitochondrial crosstalk in order to adapt the mitochondrial biomass and oxidative metabolism to a specific cellular context or developmental stage. Their critical roles are illustrated by the early embryonic lethality of the NRF-1 and NRF-2 null mice. NRF-1 null mice die between embryonic day E 3.5 and E6.5, as a consequence of dramatic decrease in mtDNA content and mitochondrial membrane potential in blastocysts [118]. NRF-2, which is the human homolog of the transcription factor GA-binding protein (GABP), is essential for early embryonic development in light of the peri-implantation lethal phenotype of NRF-2α homozygous null mice [119].
Unlike for other organs, such as the liver and skeletal muscle, our knowledge on the coupling signaling mechanisms between cellular activity and mitochondrial biogenesis in the brain is far from comprehensive. Moreover, most mitochondrial nuclear-encoded genes are expressed in both a pan- and neural-specific manner as demonstrated by comprehensive mitochondrial proteomic studies [2]. Our current knowledge about the integrative roles of NRF-1 and NRF-2 in linking neural activity to mitochondrial bioenergetics is derived from extensive studies performed on the rat visual cortex [120]. The cytochrome c oxidase (COX) respiratory complex of the OXPHOS system is a critical energy-generating enzyme in glutamatergic visual cortical neurons [121]. Both NRF-1 and NRF-2 regulate the ten nuclear-encoded COX subunits by binding to regulatory elements that are conserved among the rat, mouse and human species [116–119]. Additionally, NRF-1 and NRF-2 indirectly regulate the expression of the three mitochondrial-encoded subunits of COX by binding to the Tfam promoter to activate mtDNA transcription [115]. Furthermore, NRF-1 couples mitochondrial bioenergetics to neural activity induced by KCl depolarization of visual cortical neurons via the concomitant transcriptional regulation of the 13 subunits of COX and the N-methyl-D-aspartate (NMDA) glutamate receptor subunits, thereby promoting an efficient intersection between synaptic transmission and energy metabolism [126].
TFAM, which is a direct target of NRF-1 and NRF-2, behaves as an essential regulator of embryonic development in light of the embryonic lethality of Tfam null mice coinciding with onset of neurogenesis, as a result of mtDNA depletion and severe respiratory defects [127]. The fact that this phenotype is similar to that of the DNA polymerase γ null mice infers a requirement of mtDNA replication and maintenance for early organogenesis [128]. The TFAM protein is a member of the high mobility transcription factor group that is essential for regulating transcription and replication of mtDNA, two interlinked processes given that transcription of mtDNA controls mtDNA gene expression and provides the RNA primers necessary for initiation of mtDNA replication at the origin of replication of the heavy chain [129]. TFAM binds to mitochondrial promoter sequences, the two heavy chain-specific promoters (HSP1 and HSP2) and the light chain-specific promoter (LSP) in a sequence-specific manner to initiate transcription together with the two basal mitochondrial transcription factors, TFB1M and TFB2M, and the mitochondrial RNA polymerase (POLRMT) [130, 131]. In addition, TFAM possesses histone-like properties by binding to mtDNA molecules in a non-specific manner and packaging mtDNA into nucleoids [95, 132, 133]. TFAM is a limiting determinant factor for mtDNA copy number given the tight correlation between TFAM expression levels and mtDNA copy number observed in vitro and in vivo using Tfam+/− heterozygous mice [127, 134–136]. Several animal models were generated to investigate the functional consequences of decreased Tfam expression levels and mtDNA copy number in neuronal cells. The mitochondrial late-onset neurodegeneration (MILON) mice were generated by crossing Tfam-loxP/TfamloxP mice with mice heterozygous for a transgene expressing cre recombinase from the calcium-dependent calmodulin kinase II promoter (+/CaMKII-cre) to disrupt postnatal Tfam expression in forebrain neurons [137]. Surprisingly, MILON mice exhibited adult corticohippocampal neurodegeneration associated with 40% decrease in mtDNA copy number. Since patients with PD display impaired respiratory functions due to high levels of somatic mtDNA mutations, the effect of decreased mtDNA copy number in dopaminergic (DA) neurons was investigated in the MitoPark mouse model in which Tfam is specifically deleted in DA neurons by crossing TfamloxP/TfamloxP mice with mice heterozygous for a transgene expressing cre recombinase from the DA transporter (DAT) promoter (+/DAT-cre) [138]. Low mtDNA copy number in DA neurons mimics parkinsonian symptoms associated with disrupted OXPHOS [138, 139]. Collectively, these studies demonstrate the pivotal function of TFAM in maintaining mtDNA copy number via induced mitochondrial biogenesis in developing and mature brain.
Further strengthening the functional relationship between critical mtDNA content and initiation of organogenesis, and therefore neurogenesis, is the early embryonic lethality of the Twinkle (T7 gp4-like protein with intramitochondrial nucleoid localization) null mice at E8.5 due to severe mtDNA depletion [140]. Twinkle is a nuclear-encoded mtDNA helicase with structural similarity to the bacteriophage T7 primase-helicase (T7gp4), which catalyzes hydrolysis of nucleoside triphosphates to unwind the mtDNA duplex with a 5′ to 3′ orientation [141, 142]. It is an essential component of the minimal replisome necessary for the mtDNA replication, along with the heterotrimeric mtDNA polymerase (POLG) and the tetrameric single-stranded DNA-binding protein (mtSSB) [143]. Bromodeoxyuridine (BrdU) labeling has demonstrated that Twinkle specifically regulates de novo mtDNA synthesis [97]. The critical role of Twinkle in regulating mtDNA copy number has been validated by its colocalization with TFAM and mtSSB in nucleoids [95, 140] and the phenotype of conditional Twinkle knockout mice, which display severe mtDNA depletion and reduced respiration functions in skeletal muscle and heart [140]. A similar correlation between expression levels of TWINKLE protein and mtDNA copy number was observed in human osteosarcoma (143B) cells transfected with siRNAs to reduce Twinkle expression, which caused decreased mitochondrial nucleoids [144]. Conversely, modest over-expression of Twinkle in transgenic mice under the control of the β-actin promoter or endogenous regulatory elements increased mtDNA copy number in skeletal muscle, heart and brain without provoking abnormal mitochondrial morphology [140, 144]. Interestingly, mutations in the human Twinkle (C10orf2) gene induce mtDNA deletions rather than mtDNA depletion, causing autosomal dominant progressive external opthalmoplegia (adPEO) characterized by a late-onset myopathy affecting the extraocular, limb, and facial muscles [141, 145]. These mtDNA deletions and symptoms of human PEO were recapitulated in the mouse model “Deletor” that ubiquitously expresses a mutant Twinkle cDNA carrying a dominant patient PEO mutation against a background of wild type TWINKLE [146]. More than 31 dominant mutations and 3 recessive mutations in the Twinkle gene cause inherited mitochondrial diseases, such as infantile-onset spinocerebellar ataxia, epileptic encephalopathy with mtDNA depletion [147, 148].
5. SIGNALING PATHWAYS MODULATING NEURONAL MITOCHONDRIAL BIOGENESIS
Mitochondrial biogenesis is tightly regulated by distinct signaling pathways in response to various physiological stimuli, such as oxygen supply, calcium levels, AMP/ATP ratio, and NAD+/NADH (Fig. 2). The AMP-activated kinase (AMPK) and Sirtuin-1 (SIRT1) are key cellular sensors to tailor the functional mitochondrial mass to the energy needs required for sustaining brain functions by interfacing with the major transcriptional regulator of mitochondrial biogenesis PGC-1α to form the pivotal regulatory AMPK-SIRT1-PGC-1α axis (Fig. 2).
AMPK is a heterotrimeric serine/threonine protein kinase composed of a catalytic subunit (α1 or α2) and two β (β1 or β2) and γ (γ1, γ2, or γ3) regulatory subunits [149, 150]. AMPK activation is triggered by events increasing ATP consumption, such as rising cytoplasmic calcium levels, and pathological stresses often associated with neurodevelopmental disorders and neurodegenerative diseases, such as hypoxia, ischemia, energy crisis and glucose deprivation [151, 152]. In the nervous system, AMPK activation is controlled by two kinases, the tumor suppressor liver serine/threonine kinase B1 (LKB1, also called STK11 or Par4) and the calcium/calmodulin-dependent protein kinase β, (CAMKKβ, also known as CAMKK2) depending on the cell type (Fig. 2) [153, 154]. In immature neurons, AMPK is activated by CAMKKβ rather than by LKB1 [155–157]. The activated AMPK not only down-regulates ATP-consuming (anabolic) pathways, such as synthesis of lipids, carbohydrates, and proteins, but also up-regulates ATP-generating (catabolic) pathways, including mitochondrial biogenesis to maintain energy homeostasis [158, 159]. More specifically, mitochondrial biogenesis is enhanced via AMPK-mediated phosphorylation of PGC-1α, which is subsequently translocated into the nucleus to upregulate genes involved in mitochondrial biogenesis, FAO and ATP synthesis (Fig. 2) [160, 161]. In visual cortical neurons, AMPK integrates signaling cues to regulate the PGC-1α-NRF-1 axis to adapt the mitochondrial bioenergetic metabolism to neural activity [162]. Using KCl depolarization, the authors demonstrated that AMPK is quickly activated to increase expression levels of PGC-1α and subsequently NRF-1 and TFAM expression levels, resulting in augmented ATP levels in cultured primary visual cortical neurons. Maintenance of this expression pattern requires the presence of neural cue. Conversely, deprivation of neural activity in the in vivo model for monocular visual deprivation leads to a substantial reduction in AMPK activity followed by reduced PGC-1α and NRF-1 expression levels and therefore decreased mitochondrial mass and ATP levels [162]. Recent studies have suggested that activation of AMPK plays a role in the neurodegenerative disease AD [163]. In fact, amyloid-β 1-42 (Aβ42) oligomers activate AMPK in a CAMKKβ-dependent manner in neurons with activated AMPK enriched in tangle- and pretangle-bearing neurons in patients with AD [164–166]. Although the role of the CAMKKβ-AMPK pathway in the pathophysiology of AD remains unknown, some studies suggest an AMPK-mediated protective effect by diminishing Aβ production/APP cleavage or boosting Aβ clearance [167, 168]. In an animal model for PD, AMPK activation by 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) was also reported, which promoted neuronal survival by an unknown molecular mechanism [169]. However, it is tempting to speculate that activated AMPK may alleviate the mitochondrial complex I deficiency and altered ATP synthesis found in PD patients and inhibited upon MPTP exposure [170–174]. Given that disruption of the Tfam gene in dopaminergic neurons of the conditional knockout “MitoPark” mice provokes a parkinsonian phenotype, it is plausible that neuronal survival triggered by MPTP-mediated activation of AMPK may in part result from sustained mtDNA replication and therefore mitochondrial biogenesis [138, 175].
SIRT1, a NAD+-dependent deacetylase, is another energy sensor that modulates mitochondrial biogenesis as a result of increased NAD+/NADH ratio (Fig. 2) [176]. SIRT1 is the mammalian homolog of the yeast Silent information regulator (Sir2) belonging to the class III of histone deacetylases (HDAC), which include seven members (SIRT1-7) [177, 178]. Since brain-specific deletion of SIRT1 fails to impact brain development, SIRT1 functions in the central nervous system remain elusive [179]. However, SIRT1-deficient mice display abnormal retinal development and sporadic exencephaly, possibly due to interference with the Hairy and Enhancer of Split-1 (Hes1)/Hairy and Enhancer of Split-related with YRPW motif protein-2 (Hey2) pathways [180]. Congruent with these observations is the SIRT1-mediated differentiation of neural precursor cells via repression of Notch1-Hes1 signaling pathway [181]. Recent studies have revealed SIRT1 impact on cognitive function and synaptic plasticity, two processes in which mitochondria play a critical role [182, 183]. In addition, emerging evidence suggests that SIRT1 confers neuroprotection in two neurodegenerative diseases, amyotrophic lateral sclerosis (ALS) and AD [184–187]. In response to increased NAD+ levels provoked by energy stress, SIRT1 activity augments to deacetylate many protein clients, PGC-1 α being the most relevant for enhancing mitochondrial biogenesis [188–190]. However, the mechanism by which SIRT1 is activated has remained elusive until recently. Upon cellular metabolic stress causing increased NAD+/NADH ratio, SIRT1 is phosphorylated at Thr 522 residue, which subsequently modulates its oligomeric status by favoring a monomeric state and increased activity [191–195]. SIRT1 regulates the transcriptional activity of PGC-1α by deacetylating at least one of the 13 acetylated lysine residues, thereby reversing the effects of the acetyltransferase GCN5 responsible to repress PGC-1α activity via acetylation of specific lysine residues [196, 197]. Consequently, deacetylated PGC-1α cooperates with key transcription factors to stimulate expression levels of genes involved in OXPHOS, mitochondrial biogenesis, and mitochondrial metabolic pathways, including the tri-carboxylic acid (TCA) cycle and FAO [189]. Finally, the two critical energy sensors, AMPK and SIRT1, are part of an integrated signaling pathway, in which AMPK acts as the initial sensor to increase intracellular levels of NAD+ as a consequence of fatty acid oxidation, leading to SIRT1 activation and subsequent deacetylation of PGC-1α (Fig. 2) [190, 198–200].
6. TIMING OF MITOCHONDRIAL BIOGENESIS DURING NEUROGENESIS
During pre-implantation stages of embryonic development, mitochondrial biogenesis does not occur in actively dividing pluri-potent blastomeres, resulting in fewer and fewer mtDNA molecules in the blastocyst in keeping with its reliance on anaerobic respiration for ATP production (Fig. 3) [201]. This progressive reduction in mitochondrial number is referred to as the bottleneck theory [202]. At the onset of organogenesis, increase in the number of mitochondria coincides with loss of pluripotency and cellular differentiation to promote greater levels of ATP via a metabolic shift from glycolytic to oxidative respiration (Fig. 3) [8]. During neurogenesis, the transition from neural stem cells to differentiated neurons, astrocytes, or oligodendrocytes is a high-energy demand process consuming about 50% of cellular ATP to execute key differentiation processes, including plasmalemmal biogenesis, cytoskeletal assembly associated with axonal and dendritic growth, growth cone development, synaptic functions, and organelle transport [10].
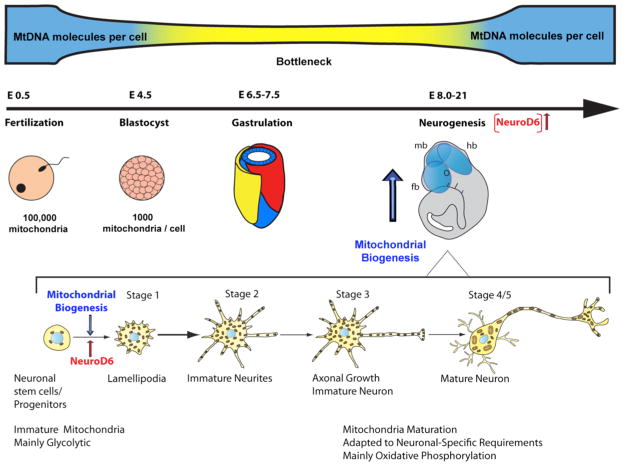
Fig. 3.
Timing of mitochondrial biogenesis during embryogenesis and neuronal differentiation. Depicted are key developmental stages during embryogenesis during which the mitochondrial genetic bottleneck occurs. Initiation of mitochondrial biogenesis coincides with increased expression of NeuroD6 and precedes neurogenesis as indicated by a blue or red arrow, respectively. The early stages of neuronal differentiation are illustrated as defined by Dotti et al., 1988. Onset of mitochondrial biogenesis coincides with the transition from neural/progenitor cells to the lamellipodial stage. The degree of mitochondrial maturity and metabolism is indicated throughout neuronal differentiation. (fb) forebrain; (mb) midbrain; (hb) hindbrain.
Early biochemical studies in developing neurons have revealed a biphasic augmentation in total mitochondrial proteins coinciding with growth cone formation and establishment of the neuronal network with synaptic activity [203, 204]. Despite the vital dependence of neurons on mitochondria and aerobic metabolism, little is known about the neuronal regulators responsible to tailor mitochondrial biogenesis to the onset of neuronal differentiation. And this is particularly critical given that half of the mammalian mitochondrial proteome is tissue-specific [1]. Most studies have focused on the ubiquitously expressed transcription factors and co-factors, such as NRF-1, NRF-2 and members of the PGC-1 family, in the context of physiological adaptations and muscle and adipocyte differentiation [116].
Our studies have addressed this gap in our knowledge by demonstrating a direct link between the mitochondrial mass and the neurogenic basic helix-loop-helix (bHLH) transcription factor NeuroD6 (previously known as Nex1/MATH-2) during the early stages of neuronal differentiation [31]. NeuroD6, which contributes to the specification of multipotential progenitors towards a glutamatergic pyramidal fate during corticogenesis [205–207], links neuritogenesis to mitochondrial biogenesis by coordinating several distinct gene networks, such as cytoskeletal proteins involved in axonal outgrowth and mitochondrial trafficking, molecular chaperones essential for translocation of nuclear-encoded mitochondrial proteins, and determinants for mtDNA replication [208–211]. NeuroD6 stimulates maximal mitochondrial mass at the earliest stage of neuronal differentiation, lamellipodia stage, as defined by Dotti et al [212], thereby preceding axonal outgrowth (Fig. 3) [31]. Moreover, NeuroD6 synchronizes mitochondrial bioenergetics with neuritogenesis by promoting accumulation of mitochondria in axonal branching and growth cone, thereby tailoring the bioenergetic needs of these subcellular domains, an essential process for proper establishment of neural circuitry during neurogenesis [31, 208–210]. NeuroD6 modulates the mitochondrial membrane potential and expression of key subunits of OXPHOS components involved in ATP synthesis, thereby generating a bioenergetic reserve [31]. Subsequently, NeuroD6 confers cellular tolerance to mitochondrial stressors and oxidative stress, known to compromise neurogenesis and cause specific neurodevelopmental disorders and neurodegenerative diseases, including autism spectrum disorder and PD [211, 213]. Coordination of mitochondrial bioenergetics and neuritogenesis has also been demonstrated in immortalized hippocampal neuroblasts and upon local nerve groth factor (NGF) signaling in axons of cultured embryonic chick sensory neurons [30, 214]. Our collective finding of NeuroD6-mediated regulation of mitochondrial biogenesis and bioenergetics at the outset of neuronal differentiation is congruent with the concept that the network of immature mitochondria must expand via biogenesis and undergo an active process of bioenergetic maturation to fuel the needs associated with the developing brain by promoting the synthesis of greater levels of ATP and maintenance of mitochondrial homeostasis.
The link between mitochondrial biogenesis and cell fate has also been investigated in the context of neuronal differentiation of embryonic stem cells (ESCs). In undifferentiated mouse and human ESCs, mitochondria exhibit a spherical and immature morphology with a predominantly perinuclear localization, as observed in blastomeres during the early stages of mammalian embryonic development [215]. Furthermore, ESCs have low numbers of mitochondria and mtDNA copy number coinciding with low levels of regulators of mtDNA replication and transcription, such as PolG and TFAM, and high expression levels of pluripotent markers, like Oct-4, Sox2, and nanog [216–220]. Moreover, ESCs display low levels of ATP production, low oxygen consumption and modest levels of antioxidant enzymes, which are in keeping with high expression levels of glycolytic enzymes and low expression levels of TCA enzymes and specific subunits of respiratory complexes for the OXPHOS system [221, 222]. These collective observations have led to the hypothesis that mitochondrial properties might be critical for the maintenance of pluripotency and self-renewal, supporting the notion of mitochondrial regulators being potential ESCs markers [223]. In support of this concept is the retinoic acid (RA)-induced reprogramming of mitochondria in ESCs committed to a neural fate. RA-treated ESCs cells harbor increased number of mitochondria with a tubular morphology and numerous elongated cristae, consistent with activation of a mitochondrial aerobic metabolism, OXPHOS activity, and increased mitochondrial membrane potential and ATP synthesis [215]. Consistent with this RA-mediated change in mitochondrial status is the increased number of mtDNA molecules and expression levels of regulators pivotal for mitochondrial biogenesis and bioenergetics, such as PolgA, PolgB, Tfam, and PGC-1α [219]. Expression of neural fate regulators, like the transcription factors Mash1 and Pax6 and the signaling molecule Wnt1, might also contribute to mitochondrial maturation in RA-induced ESCs [224–227]. Our studies revealed a similar mitochondrial remodelling during the transition from progenitors to neuronal-like PC12 cells in the presence of constitutive expression of the differentiation transcription factor NeuroD6 or NGF signaling [31, 211].
Further evidence of the importance of mitochondrial biogenesis for neurogenesis arises from the timing of embryonic lethality of homozygous Tfam and PolgA null mice. PolgA−/− knockout embryos display developmental arrest at the onset of organogenesis, causing early embryonic lethality between embryonic day E7.5 and E8.5 due to severe mtDNA depletion and loss of cytochrome oxidase activity [128]. Similarly, Tfam−/− knockout embryos show early embryonic lethality at the onset of neurogenesis between E8.5 and E11.5 due to mtDNA depletion and severe respiratory deficiency [127]. Such timing of embryonic lethality illustrates that mtDNA replication and maintenance are required for development beyond late gastrulation and early organogenesis to ensure adequate mitochondrial biogenesis and bioenergetics upon activation of the aerobic mitochondrial metabolism associated with differentiation of neural stem cells during neurogenesis.
7. COMPENSATORY MITOCHONDRIAL BIOGENESIS ASSOCIATED WITH SPECIFIC NEURODEVELOPMENTAL AND NEURODEGENERATIVE DISEASES
Mitochondrial dysfunction contributes to a range of neurodevelopmental and neurodegenerative diseases, making mitochondria a potential target for pharmacological-based therapies [228–230]. More specifically, disruption of mitochondrial biogenesis, turnover and functions can lead to primary or secondary type of mitochondrial diseases. Primary mitochondrial diseases are caused by mutation(s) or deletion in a mitochondrial or nuclear gene encoding a mitochondrial protein, while secondary mitochondrial diseases result from pathological events initiated outside mitochondria (Table 1).
The first pathogenic mtDNA mutations were discovered in 1988, which led to the classification of mitochondrial diseases [231, 232]. Maternally inherited mutations affect either mitochondrial protein synthesis when mapped in a mt-tRNA or mt-rRNA gene, or the OXPHOS system when mapped in one of the 13 genes encoding a subunit of the respiratory complexes [233]. In contrast, mutations in nuclear genes exhibit Mendelian inheritance patterns and compromise mitochondrial bioenergetics or mitochondrial biogenesis [234]. Most maternally-inherited pathogenic mtDNA mutations only affect a subset of mtDNA copies, resulting in heteroplasmy, a mixture of mutant and wild type mtDNA copies [235, 236]. Nevertheless, there are rare mtDNA mutations that solely exist in a homoplasmic state, as exemplified in Leber’s hereditary optic neuropathy (LHON) [237]. In a heteroplasmic state, there is a threshold effect, which ranges from 60 to 90% of mutated mtDNA depending on the tissue and mutation, resulting in OXPHOS defect, insufficient ATP levels and a diseased phenotype [238, 239]. In fact, one of the prevalent therapeutic strategies involves shifting herero-plasmy toward wild type mtDNA via increased mitochondrial biogenesis and/or turnover of mutated mitochondria to attenuate the OXPHOS deficit and therefore somatic manifestations in patients [240, 241].
The mitochondrial respiratory disorders represent the largest subset of primary mitochondrial diseases, with a frequency of 1:5000, making them the most common inborn error of metabolism [242]. They are incurable, progressive and multisystemic diseases with clinical heterogeneity and phenotypic variability, which share a defective oxidative phosphorylation and disruption of ATP synthesis. Although the age of onset defines the severity of the disease, most patients face significant disability, poor prognosis and premature death [243]. The mitochondrial respiratory disorders are commonly referred to as mitochondrial encephalomyoptahies, as they affect both the skeletal muscle and central nervous system with manifestations often including epilepsy, stroke-like episodes, cognitive impairment, ataxia, hearing loss, and progressive dementia [244]. To illustrate the concept of adaptive mitochondrial biogenesis in response to mitochondrial dysfunction, we will focus on the two most prevalent maternally inherited mitochondrial respiratory disorders, mitochondrial encephalopathy lactic acidosis and stroke-like episodes (MELAS) and myoclonic epilepsy and ragged-red fiber disease (MERRF) (Table 1).
MELAS is caused by a common A to G substitution at position 3243 of the mitochondrial genome, corresponding to the mitochondrial gene encoding the tRNALeu(UUR) [245, 246]. This A3243G MELAS mutation is present in a heteroplasmic state, causing a severe respiratory chain deficiency with complex I being the most affected [247]. The MELAS patients also present hypertrophic cardiomyopathy, myopathy and diabetes (Table 1) [248, 249]. They exhibit a high glycolytic rate, increased lactate production, impaired NADH response, a reduced mitochondrial membrane potential and ATP production [250]. As an attempt to overcome this chronic bioenergetic crisis, MELAS patients display a compensatory biological response of producing more mitochondria, albeit maladaptive, as it fails to sufficiently augment the bioenergetically functional mitochondrial mass and compensate for the ATP deficit [242]. Accumulation of abnormal mitochondria in dendrites of Purkinje cells and endothelial and smooth muscle cells of cerebral small arteries is suspected to contribute to ataxia and stroke-like episodes [251].
MERRF is another example of maternally inherited mitochondrial respiratory disorder provoked by a common A to G substitution in the mitochondrial genome at position 8344, corresponding to the mitochondrial gene encoding the tRNALys (Table 1) [232, 252]. As an attempt to mitigate the energy deficit, enlarged and abnormal mitochondria proliferate and accumulate in the sub-sarcolemmal region of muscle cells, resulting in ragged-red fibers visualized by the Gomori trichrome stain [253]. Abnormal large mitochondria are also present in the dentate nucleus leading to severe neuronal loss [254]. Collectively, this pattern of aggregated mitochondria may in part contribute to the diverse symptoms displayed by MERFF patients, which include myoclonus, myopathy with spasticity, cardiomyopathy, recurrent seizures, ataxia, peripeheral neuropathy, dementia and potential hearing loss or optic atrophy [255, 256].
Recent studies have provided new insight on a major contributor of neuropathogenesis for several neurodegenerative diseases with an underlying mitochondrial dysfunction, including HD, PD, and AD (Table 1) [114]. The common denominator among these neurodegenerative diseases is impaired function of the master regulator for mitochondrial biogenesis PGC-1α. PGC-1α null and conditional knock-out mice display neurological abnormalities usually associated with HD mouse models, such as myoclonus, dystonia and neurodegeneration in the cortex, thalamus, basal ganglia, hippocampus and striatum, the latter exhibiting the highest degree of degeneration [106, 257, 258]. Autoptic HD tissue samples revealed significant OXPHOS deficit due to decreased enzymatic activities of the respiratory complexes II, III, and IV, in two regions of the striatum, caudate and putamen, known to be involved in the uncontrollable dance-like movement, a hallmark of HD [259, 260]. Further evidence of the underlying mitochondrial dysfunction associated with HD was provided by studies on HD patient-derived lymphoblastoid cell lines revealing a reduced ATP/ADP ratio, a bioenergetic response inversely proportional to the length of the CAG repeat tract in the huntingtin (htt) protein [261]. Several lines of evidence support the concept that the link between transcriptional abnormalities and mitochondrial dysfunction is the central node responsible for HD neuropathology. Both expression levels and transcriptional activities of PGC-1α are reduced in medium spiny striatal neurons from HD patients and knock-in HD mice [262]. Furthermore, genome-wide microarray studies using human striatum samples documented a down-regulation of PGC-1α target genes involved in mitochondrial biogenesis and bioenergetics [263]. The authors could prevent striatal atrophy in HD-like mice by restoring PGC-1α expression using a lentiviral approach. Finally, recent studies have highlighted a link between sequence variants of PGC-1α and severity of the symptoms for HD [264, 265]. PGC-1α dysfunction extends beyond the neuronal lineage to oligodendrocytes where PGC-1α regulates the expression of several genes required for proper myelination, such as myelin basic protein (MBP), in keeping with the observed deficient myelination in HD transgenic mice [266].
Abnormal PGC-1α-mediated mitochondrial biogenesis also interfaces with PD, the second most common neurodegenerative disease [267, 268]. A meta-analysis of independent microarray analyses using microdissected human DA neurons from PD patients has documented a down-regulation of PGC-1α-regulated target genes, congruent with the concept of altered PGC-1α expression being the cause rather than the consequence of PD pathogenesis [267]. In support of this notion is the discovery of the molecular link between PGC-1α-mediated mitochondrial dysregulation, parkin inactivation and neurodegeneration of dopaminergic neurons in the substantia nigra of PD human samples and PD mouse models [268]. The authors discovered a substrate of parkin, called PARIS (ZNF746), which represses the expression of PGC-1α and its target gene NRF-1, both regulating mitochondrial biogenesis and OX-PHOS [116]. PARIS functions as a Kruppel-associated box (KRAB) zing finger transcriptional repressor [269]. It accumulates in DA neurons upon Parkin inactivation as a result of a mutation associated with the familial form of PD, or in the presence of excessive reactive oxygen species (ROS) production, or upon dopaminergic stress, which caused reduced expression levels of PGC-1α and NRF-1. Considering that conditional knockout of parkin results in a PARIS-dependent loss of DA neurons, the authors argue that the Parkin-PARIS-PGC-1α pathway is one probable causal mechanism to PD pathogenesis.
Recent studies have demonstrated that reduction in mitochondrial number and impaired mitochondrial gene expression contribute to mitochondrial dysfunction associated with AD [270]. Morphometric analyses performed on hippocampal neurons of autoptic brains from patients with AD have revealed a significant decrease in intact mitochondria [271]. The correlation between the mitochondrial mass and expression of the amyloid precursor protein carrying the Swedish mutation (APPswe) was confirmed in the in vitro M17 cellular model [272]. Decreased expression levels of PGC-1α in AD brains imply abnormal mitochondrial biogenesis as a key event for reduction of mitochondrial mass and bioenergetic functions [273]. A recent study has demonstrated that the network of transcription factors regulating mitochondrial biogenesis, including NRF-1, NRF-2 and TFAM, was altered in AD brains suggesting a deficiency in the process of mitochondrial biogenesis [274]. The causal relationship between deficient mitochondrial biogenesis and AD was confirmed in APPswe M17 cells overexpressing PGC-1α, which displayed restored expression levels of NRF-1, NRF-2 and TFAM along with increased mtDNA content and mitochondrial mass [274]. Thus, the authors postulate that induction of mitochondrial biogenesis may alleviate mitochondrial dysfunction in AD patients. However, the pharmacological approach to improve mitochondrial biogenesis remains elusive. Activation of the protein kinase A (PKA)-CREB pathway may enhance mitochondrial biogenesis via increased expression of the PGC-1α, as the PGC-1α promoter is upregulated by the transcription factor CREB phosphorylated at Ser133 by PKA [275]. A functional correlation between CREB activity, PGC-1α expression and PKA activity was observed in the in vitro M17 cellular model for the familial form of AD [274]. Moreover, postmortem AD brains exhibit decreased CREB protein and phosphorylation levels as a result of inactivation of PKA [276].
8. PUTATIVE THERAPEUTIC OPTIONS TO MODULATE NEURONAL MITOCHONDRIAL BIOGENESIS
Currently, mitochondrial respiratory disorders are a group of incurable, genetically and clinically heterogeneous diseases. Most often, they are caused by inherited mutations in the mitochondrial or nuclear genome that induce defects in the mitochondrial OX-PHOS system, resulting in impaired ATP synthesis. Treatment of these mitochondrial diseases has remained challenging due to polyploidy of the mitochondrial genome, heteroplasmy, and the influence of the nuclear background of the patients. Thus far, palliative therapies available to patients, which consist of various cocktails of vitamins, anti-oxidants and nutrient supplements, have been ineffective further underscoring the need to design novel pharmacological interventions [277].
One of the main therapeutic avenues to overcome maladaptive mitochondrial biogenesis is to efficiently boost mitochondrial biogenesis via pharmacological means to compensate for the OXPHOS deficit associated with mitochondrial respiratory disorders and neurodegenerative diseases, with the objective to enrich the wild type mitochondrial population. This shift in heteroplasmy would optimize OXPHOS functions and alleviate symptomatic manifestations of these diseases [240, 278]. In support of this strategy are studies demonstrating that induction of mitochondrial biogenesis via transgenic overexpression of PGC-1α using a genetic approach was able to rescue ATP levels in cybrid cell lines harboring nuclear or mitochondrial mutations derived from patients with oxidative phosphorylation defects [279, 280]. Endurance exercise has also mimicked the beneficial effect of PGC-1α overexpression in terms of mitochondrial biogenesis, thereby improving OXPHOS functions and slowing down progression of mitochondrial myopathy due to cytochrome c oxidase deficiency [281]. Similar improvement has also been observed in patients affected with mitochondrial diseases [282, 283].
To develop novel and efficient pharmacological approaches for inducing mitochondrial biogenesis, several animal models were engineered to mimic distinct human mitochondrial encephalopathies [284, 285]. The “deletor” mouse carries dominant mutations in the nuclear gene encoding the mitochondrial helicase, Twinkle, responsible for human adult-onset progressive external ophtalmoplegia due to multiple mtDNA deletions [141, 145]. This mouse exhibits progressive OXPHOS deficit in the skeletal muscle and in specific neuronal populations, including cerebellar Purkinje cells and hippocampal neurons, making it a useful model to investigate therapeutic strategies to treat adult-onset mitochondrial myopathy and neurodegeneration [146]. Another widely used mouse model to investigate mitochondrial myopathy is the muscle-specific knockout of the assembly factor of the respiratory complex IV, COX10 [286], which mimics human infantile fatal COX deficiency [287, 288]. The conditional knockout mouse for COX15, a key enzyme for the synthesis of heme A, exhibits severe myopathy in skeletal muscle, making it another useful mouse model for the fatal infantile hypertrophic cardiomyopathy and myopathy [289–292]. However, not all mouse models closely mimic human mitochondrial diseases, as exemplified by the Surf1 null mice. Although mutations of Surf1, another assembly factor for COX, cause Leigh syndrome in humans, Surf1 knockout mice fail to exhibit histo-pathological hallmarks of Leigh syndrome, such as neurodegeneration, even though they display mild COX deficiency [293].
The current therapeutic modalities for patients with mitochondrial respiratory disorders aim at increasing expression levels of PGC-1α via increased activity of the nuclear PPAR receptors and/or boosting its activity via modulation of the energy sensors AMPK and SIRT1 [116, 190, 294, 295]. Bezafibrate, an effective cholesterol-lowering drug [296], and rosiglitazone, a thiazolodinedione drug used to reduce insulin resistance in type II diabetes [297, 298], act as pharmacological activators of PGC-1 α expression levels by functioning as pan-agonists of the three PPAR nuclear receptor isoforms α, β/δ, and γ (Fig. 2). Given the limited blood-brain barrier permeability of rosiglitazone, its efficacy for neurodegenerative disorders and neurodegenerative diseases is limited [289]. Therefore, most studies investigated the impact of bezafibrate on mitochondrial biogenesis. Exposure to bezafibrate yielded conflicting results for mitochondrial biogenesis in various animal models. In skeletal muscle of the Cox10−/− mouse model, bezafibrate increased levels of PGC-1α, mtDNA and ATP levels, thereby slowing down the progression of mitochondrial myopathy [300]. However, this response could not be achieved in other mouse models for different mitochondrial myopathies. In the Surf1−/− mouse model, bezafibrate failed to modulate the levels of PGC-1α and mtDNA as well as the enzymatic activities of OXPHOS respiratory complexes despite increased expression levels of PPARs α, β/δ, and γ [292]. Similarly, a lack of bezafibrate-induced mitochondrial biogenesis was observed in the muscle-specific Cox15−/− mouse model [292]. The “deletor” mice treated with bezafibrate failed to alter expression levels of PGC-1α while down-regulating mtDNA copy number and expression levels of OXPHOS genes [301]. Interestingly, a recent study showed that bezafibrate improved the phenotype and survival in the R6/2 transgenic mouse model of Huntington’s disease by increasing expression levels of PGC-1α, OXPHOS genes and PPARs α, β/δ, and γ, along with increased mitochondrial density in medium spiny striatal neurons [302]. In contrast, in vitro studies using cultured fibroblasts isolated from patients affected with mitochondrial respiratory disorders due to complex I, complex III, or complex IV deficiency showed that bezafibrate concomitantly induced PGC-1α expression levels and rescued OXPHOS activities [303]. In cybrid cells carrying the human A3243G MELAS mutation or the A8344G MERFF mutation, bezafibrate also induced expression levels of PGC-1 and mitochondrial biogenesis accompanied by enhanced OXPHOS activity [304]. The underlying mechanisms responsible for these conflicting results on mitochondrial biogenesis remain unclear, which may favor a personalized therapeutic approach for patients with mitochondrial respiratory diseases. Furthermore, bezafibrate efficacy remains to be established in humans.
Another pharmacological mean to induce mitochondrial biogenesis is to modulate the activity of PGC-1α, by posttranslational modifications, such as phosphorylation or deacetylation under the control of AMPK or NAD-dependent deacetylase Sirt1, respectively (Fig. 2) [176]. As a consequence of low energy reserve resulting in inverse ATP and AMP levels, PGC-1α is phosphorylated by activated AMPK and subsequently translocated to the nucleus to upregulate genes involved in mitochondrial biogenesis and FAO [166]. Mimicking increased AMP levels is 5-aminoimidazole-4-carboxamide ribonucleoside (AICAR), which acts as an AMPK agonist and therefore activates PGC-1α (Fig. 2) [305, 306]. A recent in vitro study has demonstrated the potential benefit of AICAR to correct the ATP deficit due to congenital deficiency of the mitochondrial respiratory complex I by inducing mitochondrial biogenesis rather than stimulating the mitochondrial membrane potential [307]. Moreover, AICAR activated the AMPK-PGC-1α axis in diverse genotypic and nuclear backgrounds from patients’ fibroblasts with different complex I defects. Surprisingly, AICAR ameliorated OXPHOS activity by increasing transcription and translation of key OXPHOS genes instead of increased mitochondrial biogenesis in three COX-defective mouse models, Cox15−/−, Surf1−/− and Sco2KO/KI [292]. These conflicting results between human- and murine-based studies imply that the nuclear background most likely plays an important role in dictating the efficacy and the gene pathways triggered by pharmacological drugs.
Metformin, an anti-type 2 diabetes mellitus drug, activates AMPK and modulates PGC-1 α activity [308]. However, metformin-mediated activation of AMPK is not direct as in the case of AICAR, but rather linked to the inhibition of mitochondrial respiratory complex I, thereby decreasing ATP synthesis while increasing ADP levels and subsequently AMP levels [309, 310]. A recent study showed that metformin also activates AMPK, independently of inhibition of complex I, via repression of the AMP deaminase enzyme, resulting in increased AMP levels [311]. Thus, metformin enhances the mitochondrial retrograde pathway controlling AMPK activity (Fig. 2). Due to its indesirable inhibition of complex I, the therapeutic potential of metformin cannot be exploited in the context of mitochondrial respiratory disorders, which are characterized by severe complex I deficiency [242]. However, studies in the roundworm Caenorhabditis elegans and the fruitfly Drosophila melanogaster may suggest otherwise given their extended lifespan upon RNA-interference-mediated knockdown of specific components of the OXPHOS system [312–314].
In light of the fact that sirtuins, more specifically SIRT1 and SIRT3, play a central role in regulating mitochondrial biogenesis, a strong emphasis has been placed on developing pharmacological strategies to activate the NAD-SIRT1-mitochondrial axis [315]. Upon SIRT1 activation in response to energy depletion, PGC-1α is deacetylated and activated in a NAD+-dependent manner (Fig. 2) [188, 196, 316]. Manipulation of the SIRT1-mediated pathway via overexpression successfully improved energy expenditure in several animal models for neurodegenerative diseases associated with mitochondrial dysfunction by unknown molecular mechanisms [317]. For example, SIRT1 transgenic expression in distinct HD mouse models attenuated brain atrophy and ameliorated brain and motor functions through activation of several SIRT1 targets in a manner similar to that of calorie restriction [318–320]. In the context of PD, SIRT1 overexpression suppressed the formation of α-synuclein by activating molecular chaperones involved in mitochondrial functions [321]. Likewise, modulation of SIRT1 expression levels and activity conferred neuroprotection and improved learning and memory deficits in various AD models [186, 322]. Based on these collective findings, it is reasonable to speculate that altering SIRT1 activity could be a valid therapeutic strategy to alleviate somatic manifestations in patients affected with inherited metabolic diseases, such as mitochondrial respiratory disorders.
The recently identified small molecule SIRT1 agonists, SRT1720 (N-[2-[3-(piperazin-1-ylmethyl)imidazo[2,1-b][1,3]thiazol-6-yl]phenyl]quinoxaline-2-carboxamide) and isoflavone-derived compounds, such as daidzein, formononetin, DCHC (3-(2′,4′-dicholorphenyl)-7-hydroxy-4H-chromen-4-one) and 7-C (7-hydroxy-4H-chromen-4-one) have been tested in non-neuronal cellular paradigms for their impact on mitochondrial biogenesis [323–327]. Mice treated with the pharmacological activator of SIRT1, SRT1720, which is structurally unrelated to resveratrol, exhibited enhanced SIRT1 activity and mitochondrial biogenesis via direct deacetylation of PGC-1α and indirect stimulation of AMPK (Fig. 2) [328]. In vitro studies using primary cultures of renal proximal tubule cells, SRT1720 rapidly induced mitochondrial biogenesis in a SIRT1-dependent manner via deacetylation of PGC-1α, causing increased mtDNA content and ATP levels [327]. However, SRT1720 direct activation of SIRT1 remains disputed due to artifacts derived from the use of the nonphysiological fluorescent “Fluor de Lys” substrate for measuring in vitro SIRT1 activity [329].
The plant polyphenol, resveratrol, may be another potential pharmacological tool given that it confers neuroprotection in mouse model of AD [186]. The in vivo molecular mechanism, by which it induces mitochondrial biogenesis, remains controversial. Although resveratrol was identified in a screen for Sir2 deacetylase activity as a potent agonist of Sir2 activity, it appears not to act as a direct activator of Sir2 protein [323, 330]. In cultured primary dorsal root ganglion neurons, cortical neurons and Neuro2a neuronal cells, resveratrol rapidly induces mitochondrial biogenesis via increased expression levels of TFAM and PGC-1α in a SIRT1-independent pathway [331]. Within two hours, it triggered neuritogenesis and mitochondrial biogenesis via AMPK activation under the influence of the LKB1 and CaMKKβ kinases. Such rapid response is therapeutically advantageous in the context of CNS injury and ischemic or hypoxic insults [332]. Other studies confirmed that SIRT1 is not essential for immediate resveratrol-induced AMPK activity and stimulation of PGC-1α expression, but rather behaves as a downstream mediator of AMPK actions to activate PGC-1α via deacetylation [200, 333]. Moreover, it is postulated that resveratrol may promote mitochondrial biogenesis by initially creating an energy stress via its mild inhibition of the OXPHOS system. Subsquently, it activates AMPK and stimulates SIRT1 through enhanced NAD+ levels as a result of increased FAO (Fig. 2) [190, 198, 199, 334, 335]. However, this regulatory link between resveratrol, AMPK and SIRT1 may be adapted to cellular environment, physiological conditions and stressors, which may explain the current conflicting findings on the dependence of SIRT1 and AMPK upon resveratrol exposure in skeletal muscle versus neurons [200, 331, 336, 337].
8. CONCLUSION AND PERSPECTIVES
Mitochondrial biogenesis and functions are intertwined, as they influence each other and impact diverse neuronal functions in a developing or mature brain, thereby making mitochondrial dysfunction the prime source for specific neurodevelopmental disorders or adult neurodegenerative diseases. Although mitochondrial biogenesis is a key determinant of neuronal differentiation, its regulation has not been extensively studied. Therefore, a comprehensive and integrated analysis of the molecular mechanisms controlling mitochondrial biogenesis in the context of a developing or mature neuron is worth pursuing to develop novel therapeutic approaches tailored to the nervous system. Furthermore, it is essential to elucidate the dynamic balance between mitochondrial biogenesis, fusion-fission cycles and mitophagy, as it is a key determinant of the overall mitochondrial health and density, thereby influencing the overall energetic homeostasis within the cell. In addition, the crosstalk between these distinct but interdependent biological processes is constantly tuned by shared effectors and regulators [338–340]. During establishment of CNS connectivity, fusion-fission combined with mitochondrial trafficking is critical to locally modulate the mitochondrial population to sites with increased energy needs in response to growth factors, guidance cues, synaptic activity and axonal branching [341]. This functional interlink between mitochondrial distribution and synaptic activity is well documented at the presynaptic and the postsynaptic elements of dendritic spines of hippocampal neurons [58, 342]. In a mature brain, maintenance of an adequate functional mitochondrial mass is vital to ensure synaptic activity and plasticity and relies on a balance between the rate of biogenesis and clearance of dysfunctional or old mitochondria. Thus, adult neurodegenerative diseases may result from either impaired mitochondrial biogenesis, or inability to compensate for declined impaired mitochondrial functions, or a defective mitophagic process. In neurodegenerative diseases with a childhood onset, increased mitochondrial abundance is observed in tissues of patients affected with specific mitochondrial respiratory disorders, a quantitative increase to overcome a qualitative deficiency due to mutated mitochondrial proteins that are either nuclear- or mitochondrial-encoded.
Thus, the strategy for designing small molecules to manipulate the endogenous pathways controlling mitochondrial content and activity has been explored with the objective to potentiate the endogenous mitochondrial biogenic response. Promising results from mouse models have been obtained with various pharmacological tools acting as effectors of the AMPK-SIRT1-PGC-1α axis. However, these pharmacological interventions produced mixed results depending on the animal model for mitochondrial respiratory disorders. This mixed response may in part result from the role played by the nuclear background, a key modulator of mitochondrial biogenesis. Finally, it is important to highlight that none of the existing small molecules drugs have been tested in patients affected with mitochondrial respiratory diseases. In conclusion, improving our understanding of neuronal mitochondrial biogenesis and generating animal models mimicking diverse human mitochondrial diseases are essential to aggressively pursue the development of novel therapeutic strategies and pharmacological agents, as the field of mitochondrial pharmacology is nascent but auspicious.
Acknowledgments
We are grateful to the anonymous reviewers, who provided insightful suggestions and feedback to strengthen the review. This work was supported by the National Institute of Health, National Institute of Neurological Disorders and Stroke (Grant Number R01-NS041391 to AC).
ABBREVIATIONS
- AD
Alzheimer Disease
- adPEO
Autosomal Dominant Progressive External Opthalmoplegia
- AICAR
5-Aminoimidazole-4-carboxamide ribonucleoside
- ALS
Amyotrophic Lateral Sclerosis
- AMPK
AMP-activated Kinase
- APP
Amyloid Precursor Protein
- APPswe
Amyloid Precursor Protein carrying the Swedish mutation
- bHLH
Basic Helix-Loop-Helix
- BrdU
Bromodeoxyuridine
- CaMMK
Calcium/calmodulin-dependent protein Kinase
- CMT2A
Charcot-Marie-Tooth neuropathy type 2A
- COX
Cytochrome c Oxidase
- DA
Dopaminergic
- DAT
Dopaminergic Transporter
- DRP1
Dynamin Related Protein 1
- ETC
Electron Transfer Chain
- ESC
Embryonic Stem Cells
- FAO
Fatty Acid Oxidation
- GABP
GA-Binding Protein
- HD
Huntington Disease
- HDAC
Histine Deacetylase
- HES1
Hairy and Enhancer of Split-1
- HEY2
Hairy and Enhancer of split-related with YRPW motif protein-2
- HSP
Heavy Strand Promoter
- Htt
Huntingtin
- IMM
Inner Mitochondrial Membrane
- KRAB
Kruppel-Associated Box
- LHON
Leber’s Hereditary Optic Neuropathy
- LSP
Light Strand Promoter
- MBP
Myelin Basic Protein
- MELAS
Mitochondrial Encephalopathy Lactic Acidosis and Stroke-like episodes
- MERRF
Myoclonic Epilepsy and Ragged-Red Fiber
- Mfn
Mitofusin
- MILON
Mitochondrial Late-Onset Neurodegeneration
- Miro
Mitochondrial Rho
- MPTP
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- mtDNA
Mitochondrial DNA
- mtSSB
Mitochondrial Single Stranded Protein
- NAD
Nicotinamide Adenine Dinucleotide
- NGF
Nerve Growth Factor
- NMDA
N-methyl-D-aspartate
- NRF
Nuclear Respiratory Factor
- OMM
Outer Mitochondrial Membrane
- OPA1
Optic Atrophy 1
- OXPHOS
Oxidative Phosphorylation
- PARIS
Parkin Interacting Substrate
- PD
Parkinson Disease
- PINK1
PTEN INducing Kinase 1
- PKA
Protein Kinase A
- PGC1
Peroxisome Proliferator-Activated Receptor γ Coactivator-1
- POLG
Polymerase Gamma
- POLRMT
Mitochondrial RNA Polymerase
- PPAR
Peroxisome Proliferator-Activated Receptor
- PRC
PGC-1-Related Co-activator
- RA
retinoic acid
- ROS
Reactive Oxygen Species
- Sir
Silent Information Regulator
- SIRT1
Sirtuin 1
- TCA
Tricarboxylic Acid
- TFAM
Mitochondrial Transcription Factor A
- TFB1M
Mitochondrial Basal Transcription Factor 1
- Twinkle
T7 gp4-like protein With Intramitochondrial Nucleoid Llocalization
- UPS
Ubiquitin Proteasome System
Footnotes
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
References
- 1.Calvo SE, Mootha VK. The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mootha VK, Bunkenborg J, Olsen JV, et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–40. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 3.Pagliarini DJ, Calvo SE, Chang B, et al. A mitochondrial protein compedium elucidates complex I disease biology. Cell. 2008;134:112–23. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DiMauro S. A history of mitochondrial diseases. J Inherit Metab Dis. 2011;34:261–76. doi: 10.1007/s10545-010-9082-x. [DOI] [PubMed] [Google Scholar]
- 5.De Castro IP, Martins ML, Tufi R. Mitochondrial quality control and neurological disease: an emerging connection. Expert Rev Mol Med. 2010;12:e12. doi: 10.1017/S1462399410001456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stetler RA, Leak R, Gao Y, Chen J. The dynamics of the mitochondrial organelle as a potential therapeutic organelle. J Cereb Blood Flow Metab. 2013;33:22–32. doi: 10.1038/jcbfm.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Attwell D, Laughlin SB. An energy budget for signaling in the grey matter of the brain. J Cereb Blood Flow Metab. 2001;21:1133–45. doi: 10.1097/00004647-200110000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Erecinska M, Cherian S, Silver IA. Energy metabolism in mammalian brain during development. Prog Neurobiol. 2004;73:397–445. doi: 10.1016/j.pneurobio.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Gunter TE, Gunter KK, Sheu SS, Gavin CE. Mitochondrial calcium transport: physiological and pathological relevance. Am J Physiol. 1994;267:C313–39. doi: 10.1152/ajpcell.1994.267.2.C313. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein BW, Bamburg JR. Actin-ATP hydrolysis is a major energy drain for neurons. J Neurosci. 2003;23:1–6. doi: 10.1523/JNEUROSCI.23-01-00002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verstreken P, Ly CV, Venken KJ, Koh TW, Zhou Y, Bellen HJ. Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron. 2005;47:365–78. doi: 10.1016/j.neuron.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 12.Kann O, Kovács R. Mitochondria and neuronal activity. Am J Physiol Cell Physiol. 2007;292:C641–57. doi: 10.1152/ajpcell.00222.2006. [DOI] [PubMed] [Google Scholar]
- 13.Lee CW, Peng HB. The function of mitochondria in presynaptic development at the neuromuscular junction. Mol Biol Cell. 2008;19:150–8. doi: 10.1091/mbc.E07-05-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Attwell D, Gibb A. Neuroenergetics and the kinetic design of excitatory synapses. Nat Rev Neurosci. 2005;6:841–9. doi: 10.1038/nrn1784. [DOI] [PubMed] [Google Scholar]
- 15.Korf J, Gramsbergen JB. Timing of potential and metabolic brain energy. J Neurochem. 2007;103:1697–708. doi: 10.1111/j.1471-4159.2007.04909.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhang D, Raichle ME. Disease and the brain’s dark energy. Nat Rev Neurol. 2010;6:15–28. doi: 10.1038/nrneurol.2009.198. [DOI] [PubMed] [Google Scholar]
- 17.Zenisek D, Matthews G. The role of mitochondria in presynaptic calcium handling at a ribbon synapse. Neuron. 2000;25:229–37. doi: 10.1016/s0896-6273(00)80885-5. [DOI] [PubMed] [Google Scholar]
- 18.Kirichok Y, Krapivinsky G, Clapman DE. The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 2004;427:360–4. doi: 10.1038/nature02246. [DOI] [PubMed] [Google Scholar]
- 19.Baughman JM, Perocchi F, Girgis HS, et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–5. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Stefani D, Raffaello A, Teardo E, Szabo I, Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476:336–40. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rizzuto R, De Stefani D, Raffaello A, Mammucar C. Mitochondria as sensors and regulators of calcium signalling. Nature Rev. 2012:566–78. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- 22.Chang K, Niescier RF, Min K-T. Mitochondrial matrix Ca2+ as an intrinsic signal regulating mitochondrial motility in axons. Proc Natl Acad Sci USA. 2011;108:15456–61. doi: 10.1073/pnas.1106862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheng Z-H, Cai Q. Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nat Rev Neurosci. 2012;13:77–93. doi: 10.1038/nrn3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong Z, Saikumar P, Weinberg JM, Venkatachalan MA. Calcium in cell injury and death. Annu Rev Pathol Mech Dis. 2006;1:405–34. doi: 10.1146/annurev.pathol.1.110304.100218. [DOI] [PubMed] [Google Scholar]
- 25.Pivovarova NB, Andrews SB. Calcium-dependent mitochondrial function and dysfunction in neurons. FEBS J. 2010;277:3622–36. doi: 10.1111/j.1742-4658.2010.07754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollenbeck PJ, Saxton WM. The axonal transport of mitochondria. J Cell Sci. 2005;118:5411–9. doi: 10.1242/jcs.02745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris RL, Hollenbeck PJ. The regulation of bidirectional mitochondrial transport is coordinated with axonal outgrowth. J Cell Sci. 1993;104:917–27. doi: 10.1242/jcs.104.3.917. [DOI] [PubMed] [Google Scholar]
- 28.Ruthel G, Hollenbeck PJ. Response of mitochondrial traffic to axon determination and differential branch growth. J Neurosci. 2003;23:8618–24. doi: 10.1523/JNEUROSCI.23-24-08618.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller KE, Sheetz MP. Axonal mitochondrial transport and potential are correlated. J Cell Sci. 2004;117:2791–804. doi: 10.1242/jcs.01130. [DOI] [PubMed] [Google Scholar]
- 30.Verberg J, Hollenbeck PJ. Mitochondrial membrane potential in axons increases with local nerve growth factor or semaphoring signaling. J Neurosci. 2008;28:8306–15. doi: 10.1523/JNEUROSCI.2614-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baxter KK, Uittenbogaard M, Yoon J, Chiaramello A. The neurogenic basic helix-loop-helix transcription factor NeuroD6 concomitantly increases mitochondrial mass and regulates cytoskeletal organization in the early stages of neuronal differentiation. ASN Neuro. 2009;1:e00016. doi: 10.1042/AN20090036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berthold CH, Fabricius C, Rydmark M, Andersen B. Axoplasmic organelles at nodes of Ranvier. I. Occurrence and distribution in large myelinated spinal root axons of the adult cat. J Neurocytol. 1993;22:925–40. doi: 10.1007/BF01218351. [DOI] [PubMed] [Google Scholar]
- 33.Mutsaers SE, Carroll WM. Focal accumulation of intra-axonal mitochondria in demyelination of the cat optic nerve. Acta Neuropathol. 1998;96:139–43. doi: 10.1007/s004010050873. [DOI] [PubMed] [Google Scholar]
- 34.Bristow EA, Griffiths PG, Andrews RM, Johnson MA, Turnbull DM. The distribution of mitochondrial activity in relation to optic nerve structure. Arch Ophtalmol. 2002;120:791–6. doi: 10.1001/archopht.120.6.791. [DOI] [PubMed] [Google Scholar]
- 35.Gross NJ, Getz GS, Rabinowitz M. Apparent turnover of mitochondrial deoxyribonucleic acid and mitochondrial phospholipids in the tissues of the rat. J Biol Chem. 1969;244:1552–62. [PubMed] [Google Scholar]
- 36.Menzies RA, Gold PH. The turnover of mitochondria in a variety of tissues of young adult and aged rats. J Biol Chem. 1971;246:2425–9. [PubMed] [Google Scholar]
- 37.Korr H, Kurz C, Seidler TO, Sommer D, Schmitz C. Mitochondrial DNA synthesis studied autoradiographically in various cell types in vivo. Braz J Med Biol Res. 1998;31:289–98. doi: 10.1590/s0100-879x1998000200012. [DOI] [PubMed] [Google Scholar]
- 38.Miwa S, Lawless C, von Zglinicki T. Mitochondrial turnover in liver is fast in vivo and is accelerated by dietary restriction: application of a simple dynamic model. Aging Cell. 2008;7:920–23. doi: 10.1111/j.1474-9726.2008.00426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Toole M, Latham R, Baqri RM, Miller KE. Modeling mitochondrial dynamics during in vivo axonal elongation. J Theor Biol. 2008;255:369–77. doi: 10.1016/j.jtbi.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 40.Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- 41.Chicco AJ, Sparagna GC. Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am J Physiol Cell Physiol. 2007;292:C33–44. doi: 10.1152/ajpcell.00243.2006. [DOI] [PubMed] [Google Scholar]
- 42.Schlame M, Ren M. The role of cardiolipin in the structural organization of mitochondrial membranes. Biochem Biophys Acta. 2009;1788:2080–83. doi: 10.1016/j.bbamem.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amiri M, Hollenbeck JP. Mitochondrial biogenesis in the axons of vertebrate peripheral neurons. Dev Neurobiol. 2008;68:1348–61. doi: 10.1002/dneu.20668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoppins S, Lackner L, Nunnari J. The machines that divide and fuse mitochondria. Annu Rev Biochem. 2007;76:751–80. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- 45.Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–87. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 46.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–5. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Escobar-Henriques M, Anton F. Mechanistic perspective of mitochondrial fusion: Tubulation vs. fragmentation. Biochem Biophys Acta. 2013;1833:162–75. doi: 10.1016/j.bbamcr.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 48.Belenguer P, Pellegrini L. The dynamin GTPase OPA1: More than mitochondria. Biochem Biophys Acta. 2013;1833:176–83. doi: 10.1016/j.bbamcr.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 49.McBride H, Scorrano L. Mitochondrial dynamics and physiology. Biochem Byiophys Acta. 2013;1833:148–9. doi: 10.1016/j.bbamcr.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 50.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen H, McCaffery M, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–62. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 52.Delettre C, Lenaers G, Griffoin JM, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–10. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 53.Alexander C, Votruba M, Pesch UE, et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet. 2000;26:211–15. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- 54.Züchner S, Mersiyanova LV, Muglia M, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–51. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 55.Stokin GB, Lillo C, Falzone TL, et al. Axonopathy and transport deficits early in the pathogenesis of Alzheimer’s disease. Science. 2005;307:1282–8. doi: 10.1126/science.1105681. [DOI] [PubMed] [Google Scholar]
- 56.Baloh RH, Schmidt RE, Pestronk A, Milbrandt J. Altered axonal mitochondrial transport in the pathogenesis of Charcot-Marie-Tooth disease from mitofusin 2 mutations. J Neurosci. 2007;27:422–30. doi: 10.1523/JNEUROSCI.4798-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elgass K, Pakay J, Ryan MT, Palmer CS. Recent advances into the understanding of mitochondrial fission. Biochem Biophys Acta. 2013;1833:150–61. doi: 10.1016/j.bbamcr.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 58.Li Z, Okamoto K, Hayashi Y, Sheng M. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell. 2004;119:873–87. doi: 10.1016/j.cell.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 59.Verstreken P, Ly CV, Venken KJ, Koh TW, Zhou Y, Bellen HJ. Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron. 2005;4:365–78. doi: 10.1016/j.neuron.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 60.Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E. Mitochondrial fragmentation in neurodegeneration. Nature Rev Neurosci. 2008;9:505–18. doi: 10.1038/nrn2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen H, Chan DC. Mitochondrial dynamics-fusion, fission, movement, and mitophagy-in neurodegenerative diseases. Hum Mol Genet. 2009;18:169–76. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cho DH, Nakamura T, Lipton SA. Mitochondrial dynamics in cell death and neurodegeneration. Cell Mol Life Sci. 2010;67:3435–47. doi: 10.1007/s00018-010-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Narendra DP, Youle RJ. Neurodegeneration: Trouble in the cell’s powerhouse. Nature. 2012;483:418–9. doi: 10.1038/nature10952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rugarli EI, Langer T. Mitochondrial quality control: a matter of life and death for neurons. EMBO J. 2012;31:1336–49. doi: 10.1038/emboj.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Diff. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Holtzman E, Novikoff AB. Lysosomes in the rate sciatic nerve following crush. J Cell Biol. 1965;27:651–69. doi: 10.1083/jcb.27.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cai Q, Zakaria HM, Simone A, Sheng ZH. Spatial Parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr Biol. 2012;22:545–52. doi: 10.1016/j.cub.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maday S, Wallace KE, Holzbaur EL. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol. 2012;196:407–11. doi: 10.1083/jcb.201106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maday S, Holzbaur EL. Autophagosome assembly and cargo capture in the distal axon. Autophagy. 2012;8:858–60. doi: 10.4161/auto.20055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin SM, Youle RJ. PINK1- and Parkin-mediated mitophagy at a glance. J Cell Sci. 2012;125:795–9. doi: 10.1242/jcs.093849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X, Winter D, Ashrafi G, et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kane LA, Youle RJ. PINK1 and Parkin flag Miro to direct mitochondrial traffic. Cell. 2011;147:721–3. doi: 10.1016/j.cell.2011.10.028. [DOI] [PubMed] [Google Scholar]
- 74.Gegg ME, Cooper JM, Chau KY, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PIK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–70. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Glauser L, Sonnay S, Stafa K, Moore DJ. Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J Neurochem. 2011;118:636–45. doi: 10.1111/j.1471-4159.2011.07318.x. [DOI] [PubMed] [Google Scholar]
- 76.Sarraf SA, Raman M, Guarani-Pereira V, et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–6. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kitada T, Asakawa S, Hattori N, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–8. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 78.Valente EM, Abou-Sleiman PM, Caputo V, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–60. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 79.Rubinsztein DC, DiFiglia M, Hintz N, et al. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy. 2005;1:11–22. doi: 10.4161/auto.1.1.1513. [DOI] [PubMed] [Google Scholar]
- 80.Ventruti A, Cuervo AM. Autophagy and neurodegeneration. Curr Neurol Neurosci Rep. 2007;7:443–51. doi: 10.1007/s11910-007-0068-5. [DOI] [PubMed] [Google Scholar]
- 81.Schapira AHV. Mitochondrial pathology in Parkinson’s disease. Mount Sin J Med. 2011;78:872–81. doi: 10.1002/msj.20303. [DOI] [PubMed] [Google Scholar]
- 82.Battevi Y, La Spada AR. Mitochondrial autophagy in neural function, neurodegenerative disease, neural cell death, and aging. Neurobiol Dis. 2011;43:46–51. doi: 10.1016/j.nbd.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–38. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 84.Onyango IG, Lu J, Rodova M, Lezi E, Crafter AB, Swerdlow RH. Regulation of neuronal mitochondrial biogenesis and relevance to brain health. Biochem Biophys Acta. 2010;1802:228–34. doi: 10.1016/j.bbadis.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 85.Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007;8:870–9. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- 86.Wallace DC. The mitochondrial genome in human adaptive radiation and disease: on the road to therapeutics and performance enhancement. Gene. 2005;354:169–80. doi: 10.1016/j.gene.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 87.Kaneda H, Hayashi J, Takahama S, Taya C, Lindahl KF, Yonekawa H. Elimination of paternal mitochondrial DNA in intraspecific crosses during early mouse embryogenesis. Proc Natl Acad Sci USA. 1995;92:4542–6. doi: 10.1073/pnas.92.10.4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shoubridge EA. Mitochondrial DNA segregation in the developing embryo. Hum Reprod. 2000;15:229–34. doi: 10.1093/humrep/15.suppl_2.229. [DOI] [PubMed] [Google Scholar]
- 89.Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat Rev Genet. 2005;6:815–25. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- 90.Spelbrink JN. Functional organization of mammalian mitochondrial DNA in nucleoids: history, recent developments, and future challenges. IUBMB Life. 2010;62:19–32. doi: 10.1002/iub.282. [DOI] [PubMed] [Google Scholar]
- 91.Bogenhagen DF. Mitochondrial DNA nucleoid structure. Biochem Biophys Acta. 2012;1819:914–20. doi: 10.1016/j.bbagrm.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 92.Iborra F, Kimura H, Cook P. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2004;2:9. doi: 10.1186/1741-7007-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang Y, Bogenhagen DF. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J Bio Chem. 2006;281:25791–802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- 94.Gilkerson RW, Schon EA, Hernandez E, Davidson MM. Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. J Cell Biol. 2008;181:1117–28. doi: 10.1083/jcb.200712101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Garrido N, Griparic L, Jokitalo E, Wartiovaara J, van der Bliek AM, Spelbrink JN. Composition and dynamics of human mitochondrial nucleoids. Mol Biol Cell. 2003;14:1583–96. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J Biol Chem. 2008;283:3665–75. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- 97.Ylikallio E, Tyynismaa H, Tsutsui H, Ide T, Suomalainen A. High mitochondrial DNA copy number has detrimental effects in mice. Hum Mol Genet. 2010;19:2695–705. doi: 10.1093/hmg/ddq163. [DOI] [PubMed] [Google Scholar]
- 98.Gilkerson RW. Mitochondrial DNA nucleoids determine mitochondrial genetics and dysfunction. Int J Biochem Cell Biol. 2009;41:1899–906. doi: 10.1016/j.biocel.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 99.Jacobs HT, Lehtinen SK, Spelbrink JN. No sex please, we’re mitochondria: a hypothesis on the somatic unit of inheritance of mammalian mtDNA. Bioessays. 2000;22:564–72. doi: 10.1002/(SICI)1521-1878(200006)22:6<564::AID-BIES9>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 100.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–68. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 101.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–39. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 102.Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α): Transcriptonal coactivator and metabolic regulator. Endocrine Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 103.Estesbauer H, Oberkofler H, Krempler F, Patsch W. Human peroxisome proliferator activated receptor gamma coactivator 1 (PPARGC1) gene: cDNA sequence, genomic organization, chromosomal localization, and tissue expression. Genomics. 1999;62:98–102. doi: 10.1006/geno.1999.5977. [DOI] [PubMed] [Google Scholar]
- 104.Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta), a novel PGC-1-related transcription coactivator associated with host cell factor. J Biol Chem. 2002;277:1645–8. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- 105.Cowell R, Blake KR, Russel JW. Localization of the transcriptional coactivator PGC-1alpha to GABAergic neurons during maturation of the rat brain. J Comp Neurol. 2007;502:1–18. doi: 10.1002/cne.21211. [DOI] [PubMed] [Google Scholar]
- 106.Lin J, Wu PH, Tarr PT, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–35. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 107.Scarpulla RC. Transcriptional activators and coactivators in the nuclear control of mitochondrial function in mammalian cells. Gene. 2002;286:81–9. doi: 10.1016/s0378-1119(01)00809-5. [DOI] [PubMed] [Google Scholar]
- 108.Wu Z, Puigserver P, Andersson U, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–24. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 109.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–56. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rasbach KA, Schnellmann RG. PGC-1alpha over-expression promotes recovery from mitochondrial dysfunction and cell injury. Biochem Biophys Res Comm. 2007;355:734–9. doi: 10.1016/j.bbrc.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 111.Geissler A, Krimmer T, Bömer U, Guiard B, Rassow J, Pfanner N. x Membrane potential-driven protein import into mitochondria. Mol Biol Cell. 2007;11:3977–91. doi: 10.1091/mbc.11.11.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wiedermann N, Frazier AE, Pfanner N. The protein import machinery of mitochondria. J Biol Chem. 2004;279:14473–6. doi: 10.1074/jbc.R400003200. [DOI] [PubMed] [Google Scholar]
- 113.Neupert W, Herrmann JH. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–49. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 114.Tsunemi T, La Spada AR. PGC-1α at the intersection of bioenergetics regulation and neuronal function: From Huntington’s disease to Parkinson’s disease and beyond. Prog Neurobiol. 2012;97:142–51. doi: 10.1016/j.pneurobio.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Scarpulla RC. Nuclear control of respiratory gene expression in mammalian cells. J Cell Biochem. 2006;97:673–83. doi: 10.1002/jcb.20743. [DOI] [PubMed] [Google Scholar]
- 116.Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochem Biophys Acta. 2011;1813:1269–78. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Scarpulla RC, Vega RB, Kelly DP. Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol Metab. 2012;23:459–66. doi: 10.1016/j.tem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Huo L, Scarpulla RC. Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol Cell Biol. 2001;21:644–54. doi: 10.1128/MCB.21.2.644-654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ristevski S, O’Leary DA, Thornell AP, Owen MJ, Kola I, Hertzog PJ. The ETS transcription factor GABPα is essential for early embryogenesis. Mol Cell Biol. 2004;24:5844–9. doi: 10.1128/MCB.24.13.5844-5849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wong-Riley MT. Bigenomic regulation of cytochrome c oxidase in neurons and the tight coupling between neuronal activity and energy metabolism. Adv Exp Med Biol. 2012;748:283–304. doi: 10.1007/978-1-4614-3573-0_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wong-Riley MT. Cytcohrome oxidase: an endogenous metabolic marker for neuronal activity. Trends Neurosci. 1989;12:94–101. doi: 10.1016/0166-2236(89)90165-3. [DOI] [PubMed] [Google Scholar]
- 122.Ongwijitwat S, Wong-Riley MT. Is nuclear respiratory factor 2 a master transcriptional coordinator for all ten nuclear-encoded cytochrome c oxidase subunits in neurons? Gene. 2005;360:65–77. doi: 10.1016/j.gene.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 123.Ongwijitwat S, Liang HL, Graboyes EM, Wong-Riley MT. Nuclear respiratory factor 2 senses changing cellular energy demands and its silencing down-regulates cytochrome oxidase and other target gene mRNAs. Gene. 2006;374:39–49. doi: 10.1016/j.gene.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 124.Yang SJ, Liang HL, Wong-Riley MT. Activity-dependent transcriptional regulation of nuclear respiratory factor-1 in cultured rat visual cortical neurons. Neuroscience. 2006;141:1181–92. doi: 10.1016/j.neuroscience.2006.04.063. [DOI] [PubMed] [Google Scholar]
- 125.Dhar SS, Ongwijitwat S, Wong-Riley MT. Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. J Biol Chem. 2008;283:3120–9. doi: 10.1074/jbc.M707587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dhar SS, Wong-Riley MT. Coupling of energy metabolism and synaptic transmission at the transcriptional level: role of nuclear respiratory factor 1 I regulating both cytochrome c oxidase and NMDA glutamate receptor subunit genes. J Neurosci. 2009;29:483–92. doi: 10.1523/JNEUROSCI.3704-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Larsson NG, Wang JM, Wilhelmsson H, et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nature Genet. 1998;18:231–6. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 128.Hance N, Ekstrand MI, Trifunovic A. Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum Mol Genet. 2005;14:1775–83. doi: 10.1093/hmg/ddi184. [DOI] [PubMed] [Google Scholar]
- 129.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–99. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 130.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet. 2002;31:289–94. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 131.Gaspari M, Falkenberg M, Larsson NG, Gustafsson CM. The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. EMBO J. 2004;23:4606–14. doi: 10.1038/sj.emboj.7600465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Alam TI, Kanki T, Muta T, et al. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003;31:1640–5. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kaufman BA, Durisic N, Mativetsky JM, et al. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell. 2007;18:3225–36. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ekstrand M, Falkenberg M, Rantanen A, et al. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–44. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 135.Pohjoismaki JL, Wanrooij S, Hyvarinen AK, et al. Alterations to the expression level of mitochondrial transcription factor A, TFAM, modify the mode of mitochondrial DNA replication in cultured human cells. Nucleic Acids Res. 2006;34:5815–28. doi: 10.1093/nar/gkl703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kang D, Kim SH, Hamasaki N. Mitochondrial transcription factor A (TFAM): roles in maintenance of mtDNA and cellular functions. Mitochondrion. 2007;7:39–44. doi: 10.1016/j.mito.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 137.Sörensen L, Ekstrand M, Silva JP, et al. Late-onset corticohippocampal neurodepletion attributable to catastrophic failure of oxidative phosphorylation in MILON mice. J Neurosci. 2001;21:8082–90. doi: 10.1523/JNEUROSCI.21-20-08082.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ekstrand M, Terzioglu M, Galter D, et al. Progressive parkinsonism in mice with respiration-chain-deficient dopamine neurons. Proc Natl Acad Sci USA. 2007;104:1325–30. doi: 10.1073/pnas.0605208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Galter D, Pernold K, Yoshitake T, et al. MitoPark mice mirror the slow progression of key symptoms and L-DOPA response in Parkinson’s disease. Genes Brain Behav. 2010;9:173–81. doi: 10.1111/j.1601-183X.2009.00542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Milenkovic D, Matic S, Kühl I, et al. Twinkle is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Spelbrink JN, Li FY, Tiranti V, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet. 2001;28:223–31. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 142.Korhonen JA, Gaspari M, Falkenberg M. TWINKLE has 5′->3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J Biol Chem. 2003;278:48627–32. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 143.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23:2423–29. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tyynismaa H, Sembongi H, Bokori-Brown M, et al. Twinkle helicase is essential for mtDAN maintenance and regulates mtDNA copy number. Hum Mol Genet. 2004;13:3219–27. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- 145.Suomalainen A, Majander A, Wallin M, et al. Autosomal dominant progressive external ophtalmoplegia with multiple deletions of mtDNA: clinical, biochemical, and molecular genetic features of the 10q-linked disease. Neurology. 1997;48:1244–53. doi: 10.1212/wnl.48.5.1244. [DOI] [PubMed] [Google Scholar]
- 146.Tyynismaa H, Mjosund KP, Wanrooij S, et al. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc Natl Acad Sci USA. 2005;102:17687–92. doi: 10.1073/pnas.0505551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wanrooij S, Falkenberg M. The human mitochondrial replication fork in health and disease. Biochem Biophys Acta. 2010;1797:1378–88. doi: 10.1016/j.bbabio.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 148.Copeland WC. Defects in mitochondrial DNA replication and human disease. Crit Rev Biochem Mol Biol. 2012;47:64–74. doi: 10.3109/10409238.2011.632763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–85. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 150.Fogarty S, Hawley SA, Green KA, Saner N, Mustard KJ, Hardie DG. Calmodulin-dependent protein kinase-β activates AMPK without forming a stable complex: synergistic effects of Ca2+ and AMP. Biochem J. 2010;426:109–18. doi: 10.1042/BJ20091372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bierbaum M. Activating AMP-activated protein kinase without AMP. Mol Cell. 2005;19:289–90. doi: 10.1016/j.molcel.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 152.Viollet B, Horman S, Leclerc J, et al. AMPK inhibition in health and disease. Crit Rev Biochem Mol Biol. 2010;45:276–95. doi: 10.3109/10409238.2010.488215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Steinberg GR, Kemp BE. AMPK in health and disease. Physiol Rev. 2009;89:1025–78. doi: 10.1152/physrev.00011.2008. [DOI] [PubMed] [Google Scholar]
- 154.Cantó C, Auwerx J. AMP-activated protein kinase and its downstream transcriptional pathways. Cell Mol Life Sci. 2010;67:3407–23. doi: 10.1007/s00018-010-0454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Barnes AP, Lilley BN, Pan YA, et al. LKB1 and SAD kinases define a pathway required for the polarization of cortical neurons. Cell. 2007;129:549–63. doi: 10.1016/j.cell.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 156.Anderson KA, Ribar TJ, Lin F, et al. Hypothalamic CAMKK2 contributes to the regulation of energy balance. Cell Metab. 2008;7:377–88. doi: 10.1016/j.cmet.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 157.Green MF, Anderson KA, Means AR. Characterization of the CaMKKβ-AMPK signaling complex. Cell Signal. 2011;23:2005–12. doi: 10.1016/j.cellsig.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zong H, Ren JM, Young LH, et al. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc Natl Acad Sci USA. 2002;25:15983–7. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bergeron R, Ren JM, Cadman KS, et al. Chronic activation of AMP kinase results in NRF-1 activation and mitochondrial biogenesis. Am J Physiol Endocrinol Metab. 2001;281:E1340–6. doi: 10.1152/ajpendo.2001.281.6.E1340. [DOI] [PubMed] [Google Scholar]
- 160.Reznick RM, Shulman GI. The role of AMP-activated protein kinase in mitochondrial biogenesis. J Physiol. 2006;574:33–39. doi: 10.1113/jphysiol.2006.109512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jäger S, Handschin C, St-Pierre J, Spiegelman BM. AMP-actiavted protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci USA. 2007;104:12017–22. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yu L, Yang SJ. AMP-activated protein kinase mediates activity-dependent regulation of peroxisome proliferator-activated receptor γ coactivator-1α and nuclear respiratory factor 1 expression in rat visual cortical neurons. Neuroscience. 2010;169:23–38. doi: 10.1016/j.neuroscience.2010.04.063. [DOI] [PubMed] [Google Scholar]
- 163.Salminen A, Kaarniranta K, Haapasalo A, Soininen H, Hiltunen M. AMP-activated protein kinase: a potential player in Alzheimer’s disease. J Neurochem. 2011;118:460–74. doi: 10.1111/j.1471-4159.2011.07331.x. [DOI] [PubMed] [Google Scholar]
- 164.Thornton C, Bright NJ, Sastre M, Muckett PJ, Carling D. AMP-activated protein kinase (AMPK) is a tau kinase, activated in response to amyloid β-peptide exposure. Biochem J. 2011;434:503–12. doi: 10.1042/BJ20101485. [DOI] [PubMed] [Google Scholar]
- 165.Vingtdeux V, Davies P, Dickson DW, Marambaud P. AMPK is abnormally activated in tangle- and pre-tangle-bearing neurons in Alzheimer’s disease and other tauopathies. Acta Neuropathol. 2011;121:337–49. doi: 10.1007/s00401-010-0759-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yoon SO, Park DJ, Ryu JC, et al. JNK3 perpetuates metabolic stress induced by Aβ peptides. Neuron. 2012;75:824–37. doi: 10.1016/j.neuron.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Vingtdeux V, Giliberto L, Zhao H, et al. AMP-activated protein kinase signaling activation by resveratrol modulates amyloid-beta peptide metabolism. J Biol Chem. 2010;285:9100–9113. doi: 10.1074/jbc.M109.060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Vingtdeux V, Chadakkar P, Zhao H, d’Abramo C, Davies P, Marambaud P. Novel synthetic small-molecule activators of AMPK as enhancers of autophagy and amyloid-β peptide degradation. FASEB J. 2011;25:219–31. doi: 10.1096/fj.10-167361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Choi JS, Park C, Jeong JW. AMP-activated proteine kinase is activated in Parkinson’s disease models mediated by 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. Biochem Biophys Res Commun. 2010;391:147–51. doi: 10.1016/j.bbrc.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 170.Langston JW, Ballard P, Tetrud JW, Irwin I. Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science. 1983;219:979–80. doi: 10.1126/science.6823561. [DOI] [PubMed] [Google Scholar]
- 171.Dauer W, Przedborski S. Parkinson’s disease: mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 172.Parker WD, Jr, Boyson SJ, Parks JK. Abnormalities of the electron transport chain in idiopathic Parkinson’s disease. Ann Neurol. 1989;26:719–23. doi: 10.1002/ana.410260606. [DOI] [PubMed] [Google Scholar]
- 173.Schapira AHV, Cooper JM, Dexter D, Clark JB, Jenner P, Marsden CD. Mitochondrial complex I deficiency in Parkinson’s disease. Lancet. 1989;8649:1269. doi: 10.1016/s0140-6736(89)92366-0. [DOI] [PubMed] [Google Scholar]
- 174.Parker WD, Jr, Parks JK, Swerdlow RH. Complex I deficiency in Parkinson’s disease frontal cortex. Brain Res. 2008;1189:215–18. doi: 10.1016/j.brainres.2007.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Liang CL, Wang TT, Luby-Phelps K, German DC. Mitochondria mass is low in mouse substantia nigra dopamine neurons: implications for Parkinson’s disease. Exp Neurol. 2007;203:370–80. doi: 10.1016/j.expneurol.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 176.Cantó C, Auwerx J. PGC-1α, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoojans K, Auwerx J. Sirtuins: the “magnificent seven”, function, metabolism and longevity. Ann Med. 2007;39:335–45. doi: 10.1080/07853890701408194. [DOI] [PubMed] [Google Scholar]
- 178.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Cohen DE, Supinski AM, Bonkowski MS, Domnez G, Guarente LP. Neuronal SIRT1 regulates endocrine and behavioral responses to calorie restriction. Genes Dev. 2009;23:2812–7. doi: 10.1101/gad.1839209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cheng HL, Mostoslavsky R, Saito S, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–9. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hisahara S, Chiba S, Matsumoto H, et al. Histone deacetylase SIRT1 modulates neuronal differentiation by its nuclear translocation. Proc Natl Acad Sci USA. 2008;105:15599–604. doi: 10.1073/pnas.0800612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gao J, Wang WY, Mao YW, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–9. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Michán S, Li Y, Chou MM, et al. SIRT1 is essential for normal cognitive function and synaptic plasticity. J Neurosci. 2010;30:9695–07. doi: 10.1523/JNEUROSCI.0027-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRt1 activation prevent axonal degeneration. Science. 2004;305:1010–3. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 185.Chen J, Zhou Y, Mueller-Steiner S, et al. SIRT1 protects against microglia-dependent amyloid-beta toxicity through inhibiting NF-kappaB signaling. J Biol Chem. 2005;280:40364–74. doi: 10.1074/jbc.M509329200. [DOI] [PubMed] [Google Scholar]
- 186.Kim D, Nguyen MD, Dobbin MM, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic sclerosis. EMBO J. 2007;26:3169–79. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell. 2010;142:320–32. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 188.Rodgers JT, Lerin C, Haas W, Gysi SP, Spiegelman BM, Puig-server P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–8. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 189.Gerhart-Hines Z, Rodgers JT, Bare O, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–23. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Cantó C, Gerhart-Hines Z, Feige JN, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–60. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sasaki T, Maier B, Koclega KD, et al. Phosphorylation regulates SIRT1 function. PLoS One. 2008;3:e4020. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kang H, Jung JW, Kim MK, Chung JH. CK2 is the regulator of SIRT1 substrate-binding affinity, deacetylase activity and cellular response to DNA damage. PLoS One. 4:e6611. doi: 10.1371/journal.pone.0006611. 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Nasrin N, Kaushik VK, Fortier E, et al. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS One. 2009;4:e8414. doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Guo X, Williams JG, Schug TT, Li X. DYRRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J Biol Chem. 2010;285:13223–32. doi: 10.1074/jbc.M110.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Guo X, Kesimer M, Tolun G, et al. The NAD+-dependent protein deacetylase activity of SIRT1 is regulated by its oligomeric status. Sci Rep. 2012;2:640. doi: 10.1038/srep00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J Biol Chem. 2005;280:16456–60. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 197.Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1α and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fulco M, Cen Y, Zhao P, et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–73. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Costford SR, Bajpeyi S, Pasarica M, et al. Skeletal muscle NAMPT is induced by exercise in humans. Am J Physiol Endocrinol Metab. 2010;298:E117–26. doi: 10.1152/ajpendo.00318.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Cantó C, Jiang LQ, Deshmukh AS, et al. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 2010;11:213–9. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Van Blerkom J, Davies P, Mathwig V, Alexander S. Domains of high-polarized and low-polarized mitochondria may occur in mouse and human oocytes and early embryos. Human Rep. 2002;17:393–406. doi: 10.1093/humrep/17.2.393. [DOI] [PubMed] [Google Scholar]
- 202.Cree LM, Samuels DC, de Sousa Lopes SC, et al. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat Genet. 2008;40:249–54. doi: 10.1038/ng.2007.63. [DOI] [PubMed] [Google Scholar]
- 203.Cordeau-Lossouam L, Vayssière JL, Larcher JC, Gros F, Croizat B. Mitochondrial maturation during neuronal differentiation in vivo and in vitro. Biol Cell. 1991;1:57–65. doi: 10.1016/0248-4900(91)90051-n. [DOI] [PubMed] [Google Scholar]
- 204.Vayssière JL, Cordeau-Lossouam L, Larcher JC, Baseville M, Gros F, Croizat B. Participation of the mitochondrial genome in the differentiation of neuroblastoma cells In vitro. Cell Dev Biol. 1992;28A:763–72. doi: 10.1007/BF02631065. [DOI] [PubMed] [Google Scholar]
- 205.Schwab MH, Druffel-Augustin S, Gass P, et al. Neuronal basic helix-loop-helix proteins (NEX, NeuroD, NDRF): spatiotemporal expression and targeted disruption of the Nex gene in transgenic mice. J Neurosci. 1998;18:1408–18. doi: 10.1523/JNEUROSCI.18-04-01408.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Schwab MH, Bartholomae A, Heimrich B, et al. Neuronal basic helix-loophelix proteins (NEX and BETA/NeuroD) regulate terminal granule cell differentiation in the hippocampus. J Neurosci. 2000;20:3714–24. doi: 10.1523/JNEUROSCI.20-10-03714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wu SX, Goebbels S, Nakamura K, et al. Pyramidal neurons of upper cortical layers generated by NEX-positive progenitor cells in the subventricular zone. Proc Natl Acad Sci USA. 2005;102:17172–7. doi: 10.1073/pnas.0508560102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Uittenbogaard M, Chiaramello A. Constitutive overexpression of the basic helix-loop-helix Nex/MATH-2 transcription factor promotes neuronal differentiation of PC12 cells and neurite regeneration. J Neurosci Res. 2002;67:235–45. doi: 10.1002/jnr.10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Uittenbogaard M, Chiaramello A. Expression profiling upon Nex1/MATH-2-mediated neuritogenesis in PC12 cells and its implication in regeneration. J Neurochem. 2004;91:1332–43. doi: 10.1111/j.1471-4159.2004.02814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Uittenbogaard M, Baxter KK, Chiaramello A. NeuroD6 genomic signature bridging neuronal differentiation to survival via the molecular chaperone network. J Neurosci. 2010a;88:33–54. doi: 10.1002/jnr.22182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Baxter KK, Uittenbogaard M, Chiaramello A. The neurogenic basic helix-loop-helix transcription factor NeuroD6 enhances mitochondrial biogenesis and bioenergetics to confer tolerance of neuronal PC12-NeuroD6 cells to the mitochondrial stressor rotenone. Exp Cell Res. 2012;318:2200–14. doi: 10.1016/j.yexcr.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8:1454–68. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Uittenbogaard M, Baxter KK, Chiaramello A. The neurogenic basic helix-loop-helix transcription factor NeuroD6 confers tolerance to oxidative stress by triggering an antioxidant response and sustaining the mitochondrial biomass. ASN Neuro. 2010b;2:e00034. doi: 10.1042/AN20100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Voccoli V, Colombaioni L. Mitochondrial remodeling in differentiating neuroblasts. Brain Res. 2009;152:15–29. doi: 10.1016/j.brainres.2008.11.026. [DOI] [PubMed] [Google Scholar]
- 215.Facucho-Oliveira JM, St John JC. The relationship between pluri-potency and mitochondrial DNA proliferation during early embryo development and embryonic stem cell differentiation. Stem Cell Rev Rep. 2009;5:140–58. doi: 10.1007/s12015-009-9058-0. [DOI] [PubMed] [Google Scholar]
- 216.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–6. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 217.Chambers I, Colby D, Robertson M, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–55. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 218.St John JC, Ramalho-Santos J, Gray HL, et al. The expression of mitochondrial DNA transcription factors during early cardiomyocyte in vitro differentiation from human embryonic stem cells. Cloning Stem Cells. 2005;7:141–53. doi: 10.1089/clo.2005.7.141. [DOI] [PubMed] [Google Scholar]
- 219.Facucho-Oliveira JM, Alderson J, Spikings EC, Egginton S, St John JC. Mitochondrial DNA replication during differentiation of murine embryonic stem cells. J Cell Sci. 2007;120:4025–34. doi: 10.1242/jcs.016972. [DOI] [PubMed] [Google Scholar]
- 220.Li J, Pan G, Cui K, Liu Y, Xu S, Pei D. A dominant-negative form of mouse SOX2 induces trophectoderm differentiation and progressive polyploidy in mouse embryonic stem cells. J Biol Chem. 2007;282:19481–92. doi: 10.1074/jbc.M702056200. [DOI] [PubMed] [Google Scholar]
- 221.Chung S, Dzeja PP, Faustino RS, Perrez-Terzic C, Behfar A, Terzic A. Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nat Clin Pract Cardiov Med. 2007;4:S60–7. doi: 10.1038/ncpcardio0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Kondoh H, Lleonart ME, Nakashima Y, et al. A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid Redox Signal. 2007;9:293–9. doi: 10.1089/ars.2006.1467. [DOI] [PubMed] [Google Scholar]
- 223.Parket GC, Acsadi G, Brenner CA. Mitochondria: Determinants of stem cell fate? Stem Cells Dev. 2009;18:803–6. doi: 10.1089/scd.2009.1806.edi. [DOI] [PubMed] [Google Scholar]
- 224.Bain G, Ray WJ, Yao M, Gottlieb DI. Retinoic acid promotes neural and represses mesodermal gene expression in mouse embryonic stem cells in culture. Biochem Biophys Res Comm. 1996;223:691–4. doi: 10.1006/bbrc.1996.0957. [DOI] [PubMed] [Google Scholar]
- 225.Gajovic S, St-Onge L, Yokota Y, Gruss P. Retinoic acid mediates Pax6 expression during in vitro differentiation of embryonic stem cells. Differentiation. 1997;62:187–92. doi: 10.1046/j.1432-0436.1998.6240187.x. [DOI] [PubMed] [Google Scholar]
- 226.Bibel M, Richter J, Schrenk K, et al. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat Neurosci. 2004;7:1003–9. doi: 10.1038/nn1301. [DOI] [PubMed] [Google Scholar]
- 227.Nordin N, Li M, Mason JO. Expression profiles of Wnt genes during neural differentiation of mouse embryonic stem cells. Cloning Stem Cells. 2008;10:37–48. doi: 10.1089/clo.2007.0060. [DOI] [PubMed] [Google Scholar]
- 228.Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol Mech Dis. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Duchen MR, Szabadkai G. Roles of mitochondria in human disease. Essays Biochem. 2010;47:115–37. doi: 10.1042/bse0470115. [DOI] [PubMed] [Google Scholar]
- 230.Smith RAJ, Hartley RC, Cochemé, Murphy MP. Mitochondrial pharmacology. Trends Pharmacol Sci. 2012;33:341–52. doi: 10.1016/j.tips.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 231.Wallace DC, Singh G, Lott MT, et al. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988a;242:1427–30. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 232.Wallace DC, Zheng X, Lott MT, et al. Familial mitochondrial encephalopathy (MERRF): genetic, pathophysiological, and biochemical characterization of a mitochondrial DNA disease. Cell. 1988b;55:601–10. doi: 10.1016/0092-8674(88)90218-8. [DOI] [PubMed] [Google Scholar]
- 233.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–8. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 234.Wallace DC, Brown MD, Lott MT. Mitochondrial DNA variation in human evolution and disease. Gene. 1999;238:211–30. doi: 10.1016/s0378-1119(99)00295-4. [DOI] [PubMed] [Google Scholar]
- 235.Holt IJ, Harding AE, Petty RK, Morgan-Hughes JA. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am J Hum Genet. 1990;46:428–33. [PMC free article] [PubMed] [Google Scholar]
- 236.Wallace DC. Mitochondrial DNA sequence variation inhuman evolution and disease. Proc Natl Acad Sci USA. 1994;91:8739–46. doi: 10.1073/pnas.91.19.8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Man PY, Tunrbull DM, Chinnery PF. Leber hereditary optic neuropathy. J Med Genet. 2002;39:162–9. doi: 10.1136/jmg.39.3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Sciacco M, Bonilla E, Schon EA, DiMauro S, Moraes CT. Distribution of wild type and common deletion forms of mtDNA in normal and respiration-deficient muscle fibers from patients with mitochondrial myopathy. Hum Mol Genet. 1994;3:13–9. doi: 10.1093/hmg/3.1.13. [DOI] [PubMed] [Google Scholar]
- 239.Larsson NG, Clayton DA. Molecular genetic aspects of human mitochondrial disorders. Annu Rev Genet. 1995;29:151–78. doi: 10.1146/annurev.ge.29.120195.001055. [DOI] [PubMed] [Google Scholar]
- 240.Wenz T, Willaims SL, Bacman SR, Moraes CT. Emerging therapeutic approaches to mitochondrial diseases. Dev Dis Res Rev. 2010;16:219–29. doi: 10.1002/ddrr.109. [DOI] [PubMed] [Google Scholar]
- 241.Schon EA, Przedborski Mitochondria: the next (neurode)generation. Neuron. 2011;70:1033–53. doi: 10.1016/j.neuron.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.DiMauro S, Schon EA. Mitochondrial disorders in the nervous system. Annu Rev Neurosci. 2008;31:91–123. doi: 10.1146/annurev.neuro.30.051606.094302. [DOI] [PubMed] [Google Scholar]
- 243.Schapira AH. Mitochondrial disease. Lancet. 2006;368:70–82. doi: 10.1016/S0140-6736(06)68970-8. [DOI] [PubMed] [Google Scholar]
- 244.Finsterer J. Inherited mitochondrial neuropathies. J Neurol Sci. 2011;304:9–16. doi: 10.1016/j.jns.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 245.Goto Y, Nonaka I, Horai S. A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalopathies. Nature. 1990;348:651–3. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- 246.Kobayashi Y, Momoi MY, Tominaga K, et al. A point mutation in the mitochondrial tRNA(Leu)(UUR) gene in MELAS (mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes) Biochem Biophys Res Commun. 1990;173:816–22. doi: 10.1016/s0006-291x(05)80860-5. [DOI] [PubMed] [Google Scholar]
- 247.Schon EA, Koga Y, Davidson M, Moraes CT, King MP. The mitochondrial tRNA(Leu)(UUR) mutation in MELAS: a model for pathogenesis. Biochem Biophys Acta. 1992;1101:206–9. [PubMed] [Google Scholar]
- 248.Kobayashi Y, Momoi MY, Tominaga K, et al. Respiration-deficient cells are caused by a single point mutation in the mitochondrial tRNLeu (UUR) gene in mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS) Am J Hum Genet. 1991;49:590–9. [PMC free article] [PubMed] [Google Scholar]
- 249.Hirano M, Pavlakis SG. Mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS): current concepts. J Child Neurol. 1994;9:4–13. doi: 10.1177/088307389400900102. [DOI] [PubMed] [Google Scholar]
- 250.Sproule DM, Kaufmann P. Mitochondrial encephalopathy, lactic acidosis, and stroke-like episodes: basic concepts, clinical phenotype, and therapeutic management of MELAS syndrome. Ann NY Acad Sci. 2008;1142:133–58. doi: 10.1196/annals.1444.011. [DOI] [PubMed] [Google Scholar]
- 251.Turnbull HE, Lax NZ, Diodato D, Ansorge O, Turnbull DM. The mitochondrial brain: From mitochondrial genome to neurodegeneration. Biochem Byophys Acta. 2010;1802:111–21. doi: 10.1016/j.bbadis.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Shoffner JM, Lott MT, Lezza AM, Seibel P, Ballinger SW, Wallace DC. Myoclonic epilepsy and ragger-red fiber disease (MERRF) is associate with a mitochondrial DNA tRNA(Lys) mutation. Cell. 1990;61:931–7. doi: 10.1016/0092-8674(90)90059-n. [DOI] [PubMed] [Google Scholar]
- 253.Naumann M, Kiefer R, Toyka KV, Sommer C, Seibel P, Reichmann H. Mitochondrial dysfunction with myoclonus epilepsy and ragged-red fibers point mutation in nerve, muscle, and adipose tissue of a patient with multiple symmetric lipomatosis. Muscle Nerve. 1997;20:833–9. doi: 10.1002/(sici)1097-4598(199707)20:7<833::aid-mus7>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 254.Tanji K, Kunimatsu T, Vu TH, Bonilla E. Neuropathological features of mitochondrial disorders. Semin Cell Dev Biol. 2001;12:429–39. doi: 10.1006/scdb.2001.0280. [DOI] [PubMed] [Google Scholar]
- 255.Chinnery PF, Howell N, Lightowlers RN, Turnbull DM. Molecular pathology of MELAS and MERRF. The relationship between mutation load and clinical phenotypes. Brain. 1997;120:1713–21. doi: 10.1093/brain/120.10.1713. [DOI] [PubMed] [Google Scholar]
- 256.DiMauro S, Hirano M, Kaufman P, et al. Clinical features and genetics of myoclonic epilepsy with ragged red fibers. Adv Neurol. 2002;89:217–29. [PubMed] [Google Scholar]
- 257.Leone TC, Lehman JJ, Finck BN, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Ma D, Li S, Lucas EK, Cowell RM, Lin JD. Neuronal inactivation of peroxisome proliferator-activated receptor gamma coactivator 1alpha (PGC-1alpha) protects mice from diet-induced obesity and leads to degenerative lesions. J Biol Chem. 2010;285:39087–95. doi: 10.1074/jbc.M110.151688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Browne SE, Bowling AC, MacGarvey U, et al. Oxidative damage and metabolic dysfunction in Huntington’s disease: selective vulnerability of the basal ganglia. Annu Neurol. 1997;41:646–53. doi: 10.1002/ana.410410514. [DOI] [PubMed] [Google Scholar]
- 260.Gu M, Gash MT, Mann VM, Javoy-Agid F, Cooper JM, Schapir AH. Mitochondrial defect in Huntington’s disease caudate nucleus. Annu Neurol. 1996;39:385–9. doi: 10.1002/ana.410390317. [DOI] [PubMed] [Google Scholar]
- 261.Seong IS, Ivanova E, Lee JM, et al. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum Mol Genet. 2005;14:2871–80. doi: 10.1093/hmg/ddi319. [DOI] [PubMed] [Google Scholar]
- 262.Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression by PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 263.Weydt P, Pineda VV, Torrence AE, et al. Thermoregulatory and metabolic defects in Huntington’s disease transgenic mice implicate PGC-1alpha in Huntington’s disease neurodegeneration. Cell Metab. 2006;4:349–62. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 264.Chaturvedi RK, Adhihetty P, Shukla S, et al. Impaired PGC-1alpha function in muscle in Huntington’s disease. Hum Mol Genet. 2009;18:3048–65. doi: 10.1093/hmg/ddp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Weydt P, Soyal SM, Gellera C, et al. The gene coding for PGC-1alpha modifies age at onset in Huntington’s disease. Mol Neurodegener. 2009;4:3. doi: 10.1186/1750-1326-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Xiang Z, Valenza M, Cui L, et al. Peroxisome-proliferator-activated receptor gamma coactivator 1 {alpha} contributes to dysmyelination in experimental models of Huntington’s disease. J Neurosci. 2011;31:9544–53. doi: 10.1523/JNEUROSCI.1291-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Zheng B, Liao Z, Locascio JJ, et al. PGC-1alpha, a potential therapeutic target for early intervention in Parkinson’s disease. Sci Transl Med. 2010;2:52ra73. doi: 10.1126/scitranslmed.3001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Shin JH, Ko HS, Kang H, et al. PARIS (ZNF746) repression of PGC-1α contributes to neurodegeneration in Parkinson’s disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Looman C, Abrink M, Mark C, Hellman L. KRAB zinc finger proteins: an analysis of the molecular mechanisms governing their increase in numbers and complexity during evolution. Mol Biol Evol. 2002;19:2118–30. doi: 10.1093/oxfordjournals.molbev.a004037. [DOI] [PubMed] [Google Scholar]
- 270.Zhu X, Perry G, Moreira PL, et al. Mitochondrial abnormalities and oxidative imbalance in Alzheimer disease. J Alzheimers Dis. 2006;9:147–53. doi: 10.3233/jad-2006-9207. [DOI] [PubMed] [Google Scholar]
- 271.Hirai K, Aliev G, Nunomura A, et al. Mitochondrial abnormalities in Alzheimer’s disease. J Neurosci. 2001;21:3017–23. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Wang X, Su B, Siedlack SL, et al. Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc Natl Acad Sci USA. 2008;105:19318–23. doi: 10.1073/pnas.0804871105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Qin W, Haroutunian V, Katsel P, et al. PGC-1alpha expression decreases in the Alzheimer disease brain as a function of dementia. Arch Neurol. 2009;66:352–61. doi: 10.1001/archneurol.2008.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Sheng B, Wang X, Su B, et al. Impaired mitochondrial biogenesis contributes to mitochondrial dysfunction in Alzheimer’s disease. J Neurochem. 2011;120:419–29. doi: 10.1111/j.1471-4159.2011.07581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Herzig S, Long F, Jhala US, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–83. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 276.Yamamoto-Sasaki M, Ozawa H, Saito T, Rosler M, Riederer P. Impaired phosphorylation of cyclic AMP response element binding protein in the hippocampus of dementia of the Alzheimer type. Brain Res. 1999;824:300–3. doi: 10.1016/s0006-8993(99)01220-2. [DOI] [PubMed] [Google Scholar]
- 277.Suomalainen A. Therapy for mitochondrial disorders: Little proof, high research activity, some promise. Sem Fetal Neonatal Med. 2011;16:236–40. doi: 10.1016/j.siny.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 278.Moraes CT. Making the most of what you’ve got: optimizing residual OXPHOS function in mitochondrial diseases. EMBO Mol Med. 2009;1:357–9. doi: 10.1002/emmm.200900049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Srivastava S, Barrett JN, Moraes CT. PGC-1α/β upregulation is associated with improved oxidative phosphorylation in cells harboring nonsense mtDNA mutations. Hum Mol Genet. 2007;16:993–1005. doi: 10.1093/hmg/ddm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Srivastava S, Diaz F, Iommarini L, Aure K, Lombes A, Moraes CT. PGC-1α/β induced expression partially compensates for respiratory chain defects in cells from patients with mitochondrial disorders. Hum Mol Genet. 2009;18:1805–12. doi: 10.1093/hmg/ddp093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Wenz T, Diaz F, Hernandez D, Moraes CT. Endurance exercise is protective for mice with mitochondrial myopathy. J Appl Physiol. 2009;106:1712–9. doi: 10.1152/japplphysiol.91571.2008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 282.Taivassalo T, Haller RG. Implications of exercise training in mtDNA defects-use it or lose it? Biochem Biophys Acta. 2004;1659:221–31. doi: 10.1016/j.bbabio.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 283.Taivassalo T, Haller RG. Exercise and training in mitochondrial myopathies. Med Sci Sports Exerc. 2005;37:2094–101. doi: 10.1249/01.mss.0000177446.97671.2a. [DOI] [PubMed] [Google Scholar]
- 284.Torraco A, Diaz F, Vempati UD, Moraes CT. Mouse models of oxidative phosphorylation defects: Powerful tools to study the pathobiology of mitochondrial diseases. Biochem Biophys Acta. 2009;1793:171–80. doi: 10.1016/j.bbamcr.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Tyynismaa H, Suomalainen A. Mouse models of mitochondrial DNA defects and their relevance for human disease. EMBO Reports. 2009;10:137–43. doi: 10.1038/embor.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Diaz F, Thomas CK, Garcia S, Hernandez D, Moraes CT. Mice lacking COX10 in skeletal muscle recapitulate the phenotype of progressive mitochondrial myopathies associated with cytochrome c oxidase deficiency. Hum Mol Genet. 2005;14:2737–48. doi: 10.1093/hmg/ddi307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.DiMauro S, Mendell JR, Sahenk Z, et al. Fatal infantile mitochondrial myopathy and renal dysfunction due to cytochrome c oxidase deficiency. Neurology. 1980;30:795–804. doi: 10.1212/wnl.30.8.795. [DOI] [PubMed] [Google Scholar]
- 288.Minchom PE, Dormer RL, Hughes IA, et al. Fatal infantile mitochondrial myopathy due to cytochrome c oxidase deficiency. J Neurol Sci. 1983;60:453–63. doi: 10.1016/0022-510x(83)90156-9. [DOI] [PubMed] [Google Scholar]
- 289.Barros MH, Carlson CG, Glerum DM, Tzagoloff A. Involvement of mitochondrial ferredoxin and Cox15p in hydroxylation of heme O. FEBS Lett. 2001;492:133–38. doi: 10.1016/s0014-5793(01)02249-9. [DOI] [PubMed] [Google Scholar]
- 290.Antonicka H, Mattman A, Carlson CG, et al. Mutations in COX15 produce a defect in the mitochondrial heme biosynthetic pathway, causing early-onset fatal hypertrophic cardiomyopathy. Am J Hum Genet. 2003;72:101–14. doi: 10.1086/345489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Khalimonchuck O, Rödel G. Biogenesis of cytochrome c oxidase. Mitochondrion. 2005;5:363–88. doi: 10.1016/j.mito.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 292.Viscomi C, Bottani E, Civiletto G, et al. In vivo correction of COX deficiency by activation of the AMPK/PGC-1α. Cell Metab. 2011;14:80–90. doi: 10.1016/j.cmet.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Dell’Agnello C, Leo S, Agostino A, et al. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Hum Mol Genet. 2007;16:4310–444. doi: 10.1093/hmg/ddl477. [DOI] [PubMed] [Google Scholar]
- 294.Wenz T. PGC-1α activation as a therapeutic approach in mitochondrial disease. IUBMB Life. 2009;6:1051–62. doi: 10.1002/iub.261. [DOI] [PubMed] [Google Scholar]
- 295.Nunnari J, Suomalainen A. Mitochondria: In sickness and in health. Cell. 2012;148:1145–59. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Tenenbaum A, Motro M, Fisman EZ. Dual and pan-peroxisome proliferator-activated receptors (PPAR) co-agonism: the bezafibrate lessons. Cardiovasc Diabetol. 2005;4:14. doi: 10.1186/1475-2840-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Lehrke M, Lazar M. The many faces of PPARgamma. Cell. 2005;123:993–9. doi: 10.1016/j.cell.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 298.Semple R, Chatterjee V, O’Rahilly S. PPAR gamma and human metabolic disease. J Clin Invest Ophtalmol Vis Sci. 2006;16:581–9. doi: 10.1172/JCI28003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Risner M, Saunders AM, Altman JF, et al. Efficacy of rosiglitazone in a genetically defined population with mild-to-moderate Alzheimer’s disease. Pharmacogenomics J. 2006;6:246–54. doi: 10.1038/sj.tpj.6500369. [DOI] [PubMed] [Google Scholar]
- 300.Wenz T, Diaz F, Spiegelman BM, Moraes CT. Activation of the PPAR/PGC-1alpha pathway prevents a bioenergetic deficit and effectively improves a mitochondrial myopathy phenotype. Cell Metab. 2008;8:249–56. doi: 10.1016/j.cmet.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 301.Yatsuga S, Suomalainen A. Effect of bezafibrate treatment on late-onset mitochondrial myopathy in mice. Hum Mol Genet. 2012;21:526–35. doi: 10.1093/hmg/ddr482. [DOI] [PubMed] [Google Scholar]
- 302.Johri A, Calingasan NY, Hennessey TM, et al. Pharmacological activation of mitochondrial biogenesis exerts widespread beneficial effects in a transgenic mouse model of Huntington’s disease. Hum Mol Genet. 2012;21:1124–37. doi: 10.1093/hmg/ddr541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Bastin J, Aubey F, Rötig A, Munnich A, Djouadi F. Activation of peroxisome proliferator-activated receptor pathway stimulates the mitochondrial respiratory chain and can correct deficiencies in patients’ cells lacking its components. J Clin Endocrinol Metab. 2008;93:1433–41. doi: 10.1210/jc.2007-1701. [DOI] [PubMed] [Google Scholar]
- 304.Wenz T, Wang X, Marini M, Moraes CT. A metabolic shift induced by a PPAR panagonist markedly reduces the effects of pathogenic mitochondrial tRNA mutations. J Cell Mol Med. 2011;15:2317–25. doi: 10.1111/j.1582-4934.2010.01223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Corton JM, Gillepsie JG, Hawley SA, Hardie DG. 5-amino-imidazole-4-carboxamide ribonucleoside. A specific method for activating AMP-activating protein kinase in intact cell? Eur J Biochem. 1995;229:558–65. doi: 10.1111/j.1432-1033.1995.tb20498.x. [DOI] [PubMed] [Google Scholar]
- 306.Merrill GF, Kurth EJ, Hardie DG, Winder WW. AICA riboside increases AMP-activated protein kinase, fatty acid oxidation, and glucose uptake in rat muscle. Am J Physiol. 1997;273:1107–12. doi: 10.1152/ajpendo.1997.273.6.E1107. [DOI] [PubMed] [Google Scholar]
- 307.Golubitzky A, Dan P, Weissman S, Link G, Wikstrom JD, Saada A. Screening for active small molecules in mitochondrial complex I deficient patient’s fibroblasts, reveal AICAR as the most beneficial compound. PLoS One. 2011;6:e26883. doi: 10.1371/journal.pone.0026883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Zhou G, Myers R, Li Y, et al. Role of AMK-activated protein kinase in mechanism of metfomin action. J Clin Invest. 2001;108:1167–74. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J. 2000;348:607–14. [PMC free article] [PubMed] [Google Scholar]
- 310.Brunmair B, Staniek K, Gras F, et al. Thiazolidinediones, like metformin, inhibit respiratory complex I: A common mechanism contributing to their antidiabetic actions? Diabetes. 2004;53:1052–9. doi: 10.2337/diabetes.53.4.1052. [DOI] [PubMed] [Google Scholar]
- 311.Ouyang J, Parakhia RA, Ochs RS. Metformin activates AMP kinase through inhibition of AMP deaminase. J Biol Chem. 2011;286:1–11. doi: 10.1074/jbc.M110.121806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans. Nat Genet. 2003;33:40–8. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 313.Copeland JM, Cho J, Lo T, Jr, et al. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr Biol. 2009;19:1591–8. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 314.Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–83. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- 315.Houtkooper RH, Auwerx J. Exploring the therapeutic space around NAD+ J Cell Biol. 2012;199:205–9. doi: 10.1083/jcb.201207019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Yu J, Auwerx J. The role of sirtuins in the control of metabolic homeostasis. Annu NY Acad Sci. 2009;(Suppl 1):E10–19. doi: 10.1111/j.1749-6632.2009.04952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Domnez G, Outeiro TF. SIRT1 and SIRT2: emerging targets in neurodegeneration. EMBO Mol Med. 2013;5:344–52. doi: 10.1002/emmm.201302451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Duan W, Guo Z, Jiang H, Ware M, Li XJ, Mattson MP. Dietary restriction normalizes glucose metabolism and BDNF levels, slows disease progression, and increases survival in huntingtin mutant mice. Proc Natl Acad Sci USA. 2003;100:2911–6. doi: 10.1073/pnas.0536856100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Cohen DE, Cui L, Supinski A, et al. Sirt1 mediates neuroprotection from mutant huntingtin by activation of TORC1 and CREB transcriptional pathway. Nat Med. 2012;18:159–65. doi: 10.1038/nm.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Jiang M, Wang J, Fu J, et al. Neuroprotective role of Sirt1 in mammalian models of Huntington’s disease through activation of multiple Sirt1 targets. Nat Med. 2012;18:153–9. doi: 10.1038/nm.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Domnez G, Arun A, Chung CY, McLean PJ, Lindquist S, Guarente L. SIRT1 protects against alpha-synuclein aggregation by activating molecular chaperones. J Neurosci. 2012;32:124–32. doi: 10.1523/JNEUROSCI.3442-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 322.Domnez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell. 2010;142:320–32. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 323.Howitz KT, Bitterman KJ, Cohen HY, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–6. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 324.Milne JC, Lambert PD, Schenk S, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–6. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Feige JN, Lagouge M, Cantó C, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2006;8:347–58. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 326.Rasbach KA, Schnellmann RG. Isoflavones promote mitochondrial biogenesis. J Pharmacol Exp Ther. 2008;325:536–43. doi: 10.1124/jpet.107.134882. [DOI] [PubMed] [Google Scholar]
- 327.Funk JA, Odejinmi S, Schnellmann RG. SRT1720 induces mitochondrial biogenesis and rescues mitochondrial function after oxidant injury in renal proximal tubule cells. J Pharm Exp Ther. 2010;333:593–601. doi: 10.1124/jpet.109.161992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Feige JN, Lagouge M, Cantó C, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–58. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 329.Pacholec M, Bleasdale JE, Chrunyk B, et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 2010;285:8340–51. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Kaeberlein M, McDonagh T, Heltweg B, et al. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005;280:17038–45. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 331.Dasgupta B, Milbrandt J. Resveratrol stimulates AMP kinase activity in neurons. Proc Natl Acad Sci USA. 2007;104:7217–22. doi: 10.1073/pnas.0610068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- 333.Um JH, Park SJ, Kang H, et al. AMP-activated protein kinase-deficient mice are resistant to the metabolic effects of resveratrol. Diabetes. 2010;59:554–63. doi: 10.2337/db09-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Zini R, Morin C, Bertelli A, Bertelli AA, Tillement JP. Effects of resveratrol on the rat brain respiratory chain. Drugs Exp Clin Res. 1999;25:87–97. [PubMed] [Google Scholar]
- 335.Cantó C, Auwerx J. Targeting sirtuin 1 to improve metabolism: all you need is NAD+? Pharm Rev. 2012;64:166–87. doi: 10.1124/pr.110.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Lagouge M, Argmann C, Gerhart-Hines Z, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell. 2006;127:1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 337.Menzies KJ, Singh K, Saleem A, Hood DA. Sirtuin 1-mediated effects of exercise and resveratrol on mitochondrial biogenesis. J Biol Chem. 2013;288:6968–79. doi: 10.1074/jbc.M112.431155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Ehses S, Raschke I, Mancuso G, et al. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J Cell Biol. 2009;187:1023–36. doi: 10.1083/jcb.200906084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Head B, Griparic L, Amiri M, Gandre-Babbe S, van der Bliek AM. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J Cell Biol. 2009;187:959–66. doi: 10.1083/jcb.200906083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–98. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Lovas JR, Wang X. The meaning of mitochondrial movement to a neuron’s life. Biochem Biophys Acta. 2013;1833:184–94. doi: 10.1016/j.bbamcr.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Stowers RS, Megeath LJ, Gorska-Andrzejak J, Meinertzhagen IA, Schwarz TL. Axonal transport of mitochondria to synapses depends on Milton, a novel Drosophila protein. Neuron. 2002;36:1063–77. doi: 10.1016/s0896-6273(02)01094-2. [DOI] [PubMed] [Google Scholar]