Abstract
The MCM DNA helicase is a central regulatory target during genome replication. MCM is kept inactive during G1 and activated in S phase to initiate replication. During this transition, the only known chemical change on MCM is the gain of multi-site phosphorylation that promotes cofactor recruitment. As replication initiation is intimately linked to multiple biological cues, additional changes on MCM can provide further regulatory points. Here, we describe a yeast MCM sumoylation cycle that negatively regulates replication. MCM subunits undergo sumoylation upon loading at origins in G1 prior to MCM phosphorylation. MCM sumoylation levels then decline as MCM phosphorylation levels rise, suggesting an inhibitory role in replication. Indeed, increasing MCM sumoylation impairs replication initiation through promoting the recruitment of a phosphatase that reduces MCM phosphorylation and activation. MCM sumoylation thus counterbalances kinase-based regulation to ensure accurate control of replication initiation.
Introduction
The initiation of DNA replication is tightly controlled to ensure that duplication of every locus occurs once and only once per cell cycle, and to establish specific replication programs unique to an organism or cell type. Impairment in regulating replication initiation can lead to various forms of genomic changes and instability, and consequently human diseases and cancers1–5. Previous studies have revealed multiple forms of regulation at both local and global levels, with several of them targeting a key replicative enzyme, the DNA helicase MCM4,6–12.
The MCM complex is composed of Mcm2–7 subunits and is highly conserved from yeast to humans. Among its many roles during replication, MCM is critical for replisome assembly. It is the first replisome component to arrive at replication initiation sites (or origins). In budding yeast, MCM loading at origins is mediated by Cdc6 and the origin recognition complex (ORC, comprising Orc1–6) in late mitosis and G1 phase in a process called origin licensing (Fig. 1a)13,14. A subset of loaded MCM then initiates stepwise replisome assembly in a process termed origin firing. This begins with the recruitment of two co-factors, namely Cdc45 and the heterotetrameric GINS complex (Fig. 1a). Recruitment of both factors requires kinases: Dbf4-dependent kinase (DDK, composed of Cdc7 and Dbf4) phosphorylation of primarily Mcm4 recruits Cdc45, and subsequent S-cyclin-dependent kinase (S-CDK) phosphorylation of non-MCM proteins recruits GINS (Fig. 1a)15,16. The complex formed by Cdc45, MCM, and GINS (or CMG) serves as the replicative helicase13,14. Following CMG formation, more than a dozen additional replisome members assemble in a highly ordered yet still poorly understood manner before replication is initiated17,18. Throughout this intricate replisome assembly process, MCM and CMG are kept inactive to prevent premature DNA unwinding.
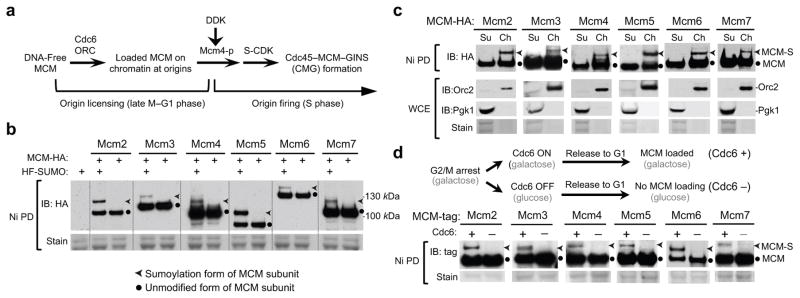
Figure 1. Sumoylation of six MCM subunits occurs on chromatin and depends on MCM loading at replication origins.
a. Schematic of key events for MCM loading and activation (see text for details). Phosphorylation of Mcm4 is indicated as “Mmc4-P”. CDK: S-cyclin-dependent kinase; DDK: Dbf4-dependent kinase.
b. Mono-sumoylation of each MCM subunit occurs under normal growth conditions. HF-Smt3 denotes His6-Flag-tagged SUMO, which allows the enrichment of sumoylated proteins on Ni-NTA beads, a method indicated as “Ni PD”. MCM subunits were tagged with HA. Unmodified and sumoylated bands are indicated by dots and arrowheads, respectively. Equal protein loading is shown by Ponceau S stain (stain). Similar methods and annotations are used in subsequent figure panels.
c. Only chromatin-bound MCM subunits show sumoylation. MCM subunits were tagged with HA and examined in chromatin fractionation and Ni PD tests. Ch and Su indicate chromatin-bound and supernatant fractions. Orc2 and Pgk1 are markers for chromatin and supernatant fractions in cell extracts (WCE), respectively.
d. Sumoylation of MCM subunits in G1 phase depends on Cdc6. Top, experimental scheme for Cdc6 depletion that prevents MCM loading, as described previously37. Mcmc2–4, 6, and 7 were tagged with HA, while Mcm5 was tagged with Strep tag II to be compatible with the Gal-Cdc6 construct. Successful Cdc6 depletion is shown in Supplementary Figure 2a.
The precision of many biological processes depends on a balanced act between positive and negative regulation. It is conceivable that the tightly controlled transition from inactive to active MCM states also requires additional regulation besides the known kinase-based positive regulation. Recent studies have indeed revealed other chemical modifications of MCM. In particular, proteomic screens in yeast, humans, and plants have shown that MCM subunits are sumoylated, revealing another highly conserved MCM modification19–21. Sumoylation entails the conjugation of the small protein modifier SUMO to lysine residue(s) on target proteins. This modification is reversible through desumoylation, and the cycle of sumoylation and desumoylation is highly dynamic in cells. The addition and removal of SUMO exert a range of effects on protein function, such as altering protein-protein interactions or enzymatic activities, and impact a variety of cellular processes22,23. While SUMO is known to affect genome maintenance, its roles in this arena have been examined mostly under genome damaging situations19,24–28; how SUMO influences the normal replication program is largely not answered.
To understand how sumoylation of MCM subunits pertains to normal replication programs, we first examined spatial and temporal patterns of this modification in budding yeast. We found that sumoylation of the six MCM subunits occurs exclusively on chromatin. Moreover, MCM sumoylation levels oscillate during the cell cycle in a manner opposite to those of MCM phosphorylation, suggesting that MCM sumoylation is an inhibitory marker for replication. The MCM sumoylation cycle depends on key MCM loaders and activators, suggesting that it is integral to MCM functions. Importantly, increased MCM sumoylation impairs replication initiation and decreases CMG levels. Mechanistically, these effects are linked to an enhanced recruitment of the phosphatase PP1 that counteracts DDK functions. Taken together, our findings suggest that MCM sumoylation enables a form of negative regulation during replication initiation. We propose that the dual control of MCM by two modifications ensures precise replication initiation and enables flexible control of genome duplication under different cellular contexts.
Results
Detection of MCM subunit sumoylation during normal growth
In search of additional means of MCM regulation, we investigated its sumoylation, which has been found in proteomic screens in multiple organisms19–21. To detect sumoylation, we followed a well-established method wherein denaturing conditions throughout protein preparation minimizes desumoylation29. In this method, the endogenous yeast SUMO (Smt3) is replaced with a His6-Flag-tagged version (HF-Smt3) at its genomic locus, such that sumoylated forms containing HF-Smt3 are enriched by Ni-NTA resin (referred to as Ni PD). Although unmodified forms of proteins show nonspecific histidine-mediated binding to Ni-NTA, they are distinguished from sumoylated forms upon western blotting for a particular protein by two criteria: 1) sumoylated forms are detected only in the presence of HF-Smt3, but not untagged Smt3, whereas unmodified forms are detectable regardless of the presence of HF-Smt3; 2) mono-sumoylated forms show ~20 kDa upshift compared to the unmodified forms.
In our tests, Mcm2–7 proteins were tagged with HA at their endogenous loci and fully supported cell growth. In the presence of HF-Smt3, but not untagged Smt3, a modified form of each MCM subunit was detected by the anti-HA antibody on western blots (Fig. 1b). In each case, the modified form exhibited a ~20 kDa upshift from the unmodified form of the protein (Fig. 1b). Consistent with being the sumoylated species, these modified forms showed smaller upshift when SUMO was tagged with a smaller tag (Supplementary Fig. 1a). Sumoylated form(s) of each MCM subunit were also seen when immunoprecipitated protein was examined by western blotting using SUMO-specific antibody (Supplemental Fig. 1b). In all cases, the detected levels of MCM sumoylation were not abundant, consistent with the dynamic nature of this modification. Taken together, two different approaches both showed that a fraction of each MCM subunit was sumoylated during normal growth. For our subsequent tests, we used the Ni PD method.
MCM sumoylation on chromatin requires its loading at origins
As MCM is subject to tight spatial regulation and can only function upon chromatin loading, we asked whether the chromatin-bound or soluble fraction of MCM was sumoylated. As shown in Figure 1c, sumoylated MCM subunits were detected exclusively in the chromatin-bound fraction. Consistent with this finding, MCM subunit sumoylation was absent when MCM loading onto chromatin was prevented by Cdc6 depletion in G1 cells (Fig. 1d and Supplementary Fig. 2a). These results suggest that MCM subunit sumoylation occurs on chromatin after the complex is loaded at origins.
MCM sumoylation levels peak in G1 and decline in S phase
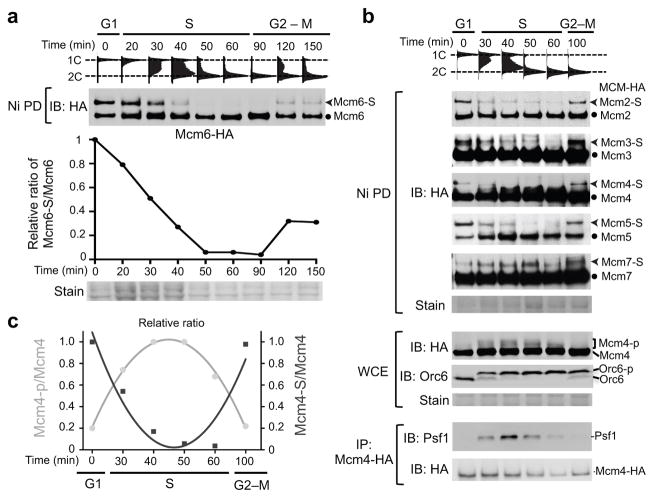
Next, we examined the temporal regulation of MCM sumoylation during the cell cycle. Cells were arrested in G1 and then allowed to synchronously progress through the cell cycle (Fig. 2a, FACS). Using Mcm6 as an example, we observed an oscillation in its sumoylation levels: sumoylation of Mcm6 peaked in G1 phase coinciding with its chromatin loading, declined during S phase, and reappeared in M phase, concurrent with the next loading cycle (Fig. 2a). Examination of chromatin-bound Mcm6 also showed a high sumoylation level in G1 and a low level in S phase (Supplementary Fig. 2b).
Figure 2. Sumoylation levels of MCM subunits oscillate during the cell cycle.
a. Mcm6 sumoylation level peaks in G1 phase, declines during S phase, and increases in late M phase. Flow cytometry (top) and immunoblotting (middle) show cell cycle progression and Mcm6 sumoylation status from cells arrested in G1 and indicated time points after release from G1. The graph depicts the relative ratio of sumoylated/unmodified Mcm6 on the western blot with the ratio in G1 cells set to 1. The ratio presented here and in other figures is not the absolute sumoylation level, as sumoylated forms were enriched; rather it is used to index changes in sumoylation levels. Staining at the bottom shows loading.
b. Dynamic MCM sumoylation and phosphorylation during the cell cycle. As in (a), FACS profiles and immunoblotting after Ni PD show sumoylation status of HA-tagged MCM subunits at indicated time points. Middle: examination of phosphorylation levels of Mcm4 and Orc6. Bottom: examination of CMG levels after immunoprecipitation of Mcm4 and probing for Psf1, a subunit of GINS.
c. The relative ratio of sumoylated or phosphorylated Mcm4 to total Mcm4 level. The ratio was calculated based on results in (b), the ratio in G1 cells was set to 1.
Similar patterns were seen for Mcm2–5 (Fig. 2b). Their sumoylation was detected in G1 when DDK activity, as indicated by Mcm4 phosphorylation, and CDK activity, as indicated by Orc6 phosphorylation, were low (Fig. 2b). Expectedly, CMG levels were also low in G1, as evidenced by the low amount of the GINS subunit Psf1 associated with Mcm4 (Fig. 2b). As cells entered S phase, DDK and CDK activities, as well as CMG levels, rose (Fig. 2b), while Mcm2–5 sumoylation levels diminished (Fig. 2b). In G2-M phase when DDK and CDK activities, as well as CMG levels fell, sumoylation levels of Mcm2–5 rose again (Fig. 2b). The Mcm7 sumoylation pattern was somewhat different. Like other MCM subunits, Mcm7 sumoylation was detected in G1; but unlike others, Mcm7 sumoylation persisted throughout most of S phase, then decreased in late S phase and reappeared in G2-M phase (Fig. 2b). The difference in sumoylation patterns between Mcm2–6 and Mcm7 likely reflect distinct regulation and functions.
Taken together, sumoylation levels of six MCM subunits oscillate during the cell cycle in a pattern opposite to that of DDK and CDK-mediated phosphorylation events. This was also visualized by plotting the relative ratio of sumoylated or phosphorylated Mcm4 vs. unmodified Mcm4 proteins (Fig, 2c). This temporal pattern suggests that contrary to phosphorylation, MCM sumoylation is an inhibitory marker for replication initiation.
MCM sumoylation loss in S phase requires DDK and GINS
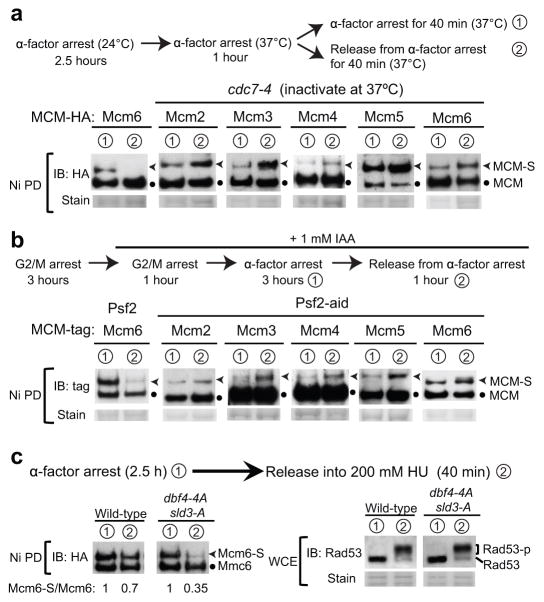
To gain a detailed understanding of the changes in Mcm2–6 sumoylation at the G1-S transition, we examined the roles of two key MCM regulators, namely DDK and GINS. To test the role of DDK, cells containing the temperature sensitive cdc7-4 allele and HA-tagged MCM subunits were arrested in G1 at permissive temperature (24°C), and then shifted to non-permissive temperature (37°C) before release into S phase (Fig. 3a). cdc7-4 inactivation upon temperature shift was confirmed by the lack of Mcm4 phosphorylation (Supplementary Fig. 2c). We found that sumoylation of Mcm2–6 still occurred in G1 when DDK was inactivated, and that the Mcm6 sumoylation level was similar to that in wild-type cells (Fig. 3a). Thus, bulk MCM sumoylation in G1 does not require DDK. On the other hand, DDK inactivation prevented the loss of Mcm2–6 sumoylation when G1 cells were released into S phase (Fig. 3a). This effect was not due to indirect alteration of CDK activity (monitored by Orc6 phosphorylation) or DNA damage checkpoint activity (monitored by Rad53 phosphorylation) (Supplementary Fig. 2c). Thus, DDK is required for loss of Mcm2–6 sumoylation when cells enter S phase.
Figure 3. Mcm2–6 sumoylation loss at the G1-S transition requires DDK, GINS, and replication initiation.
a. Mcm2–6 sumoylation in G1 phase does not require DDK, but its decrease in S phase depends on DDK. Top: experimental schemes for Cdc7 inactivation as described previously38 (see Methods). The sumoylation status of each HA-tagged MCM at indicated time points is shown.
b. Sumoylation of Mcm2–6 in G1 phase does not require GINS, but its decrease in S phase depends on the GINS subunit Psf2. Top: experimental schemes for Psf2 depletion (see Methods). Mcm5 was tagged with Strep tag II to be compatible with the Psf2–aid construct, and other MCM subunits are tagged with HA.
c. Mcm6 sumoylation loss coincides with firing of both early and late origins. Top: experimental schemes for G1 arrest and release. Immunoblots show Mcm6 sumoylation status at indicated time points. Note that dbf4-4A sld3-A cells allow late origin firing under this condition12. Rad53 phosphorylation (bottom) shows the effectiveness of HU treatment.
To test the role of GINS, we used an IAA-inducible degron (or aid) method to acutely deplete the GINS subunit Psf230,31. Psf2-aid and control cells were arrested in G2-M phase and Psf2-aid was degraded after IAA addition (Fig. 3b; Supplementary Fig. 2d–2e). Cells were next released into G1 arrest in the presence of IAA, and subsequently allowed to enter S phase (Fig. 3b). Efficient Psf2-aid degradation, proper cell cycle arrest and release, and expected lethality caused by Psf2 degradation are shown in Supplementary Figure 2d–2f. As in the case for DDK, Psf2 loss did not affect Mcm2–6 sumoylation in G1, but prevented Mcm2–6 sumoylation loss upon S phase entry (Fig. 3b). The observed effects were not due to a lack of CDK or abnormal checkpoint activity (Supplementary Fig. 2e).
Taken together, these results show that DDK and GINS are not required for Mcm2–6 sumoylation in G1; rather, they are required for the loss of this modification at the beginning of S phase. These findings, in conjunction with the cell cycle pattern of Mcm2–6 sumoylation, suggest that the modification takes place prior to DDK- and GINS-mediated events and then decreases in a DDK- and GINS-dependent manner upon S phase entry.
Mcm6 sumoylation loss coincides with origin firing
Next we addressed how the change in Mcm2–6 sumoylation status at the G1-S transition is related to early and late origin firing. It is known that releasing G1 cells into media containing 200 mM HU for a short time (e.g. 60 min) allows limited DNA synthesis from early origins32. The number of fired origins greatly increases when checkpoint-mediated inhibition of late origins is removed by mutating the phosphorylation sites on the DDK subunit Dbf4 and the replisome assembly factor Sld3 (dbf4-4A sld3-A)33. Thus, releasing G1 cells into HU media for a short time allows the assessment of the influence of firing only early origins (in wild-type cells) versus both early and late origins (in dbf4-4A sld3-A cells).
Using this experimental setup, we found that at 40 min post-release into HU, Mcm6 sumoylation level was reproducibly decreased by 30% in wild-type and 65% in dbf4-4A sld3-A cells, compared to those in G1-arrested cells (Fig. 3c). These results suggest that loss of Mcm6 sumoylation coincides with the firing of both early and late origins. As wild-type and dbf4-4A sld3-A cells do not experience replication termination under this treatment12, the observed Mcm6 sumoylation loss is not associated with replication termination.
Mcm6-SuOn increases MCM sumoylation
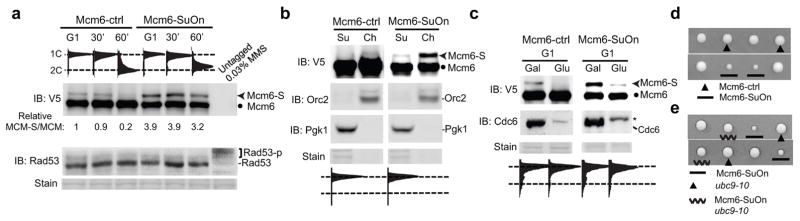
The correlation of Mcm2–6 sumoylation loss with origin firing led us to hypothesize that this modification is inhibitory to replication initiation. An important prediction of this hypothesis is that increasing MCM sumoylation should impair replication initiation. To test this, we employed a tagging strategy that utilizes the high affinity SUMO interaction region from a catalytically inactive SUMO protease domain to promote sumoylation of its fusion partner and subunits of the same complex, presumably by increasing local SUMO concentration34 (Supplementary Fig. 3a). This tag, referred to as SuOn (denoting SUMO on), was fused to Mcm6. As a control, Mcm6 was fused with the same tag that contains a point mutation of a key residue for SUMO interaction (Mcm6-ctrl)34,35. Both fusions were expressed from the endogenous MCM6 locus and tagged with the V5 epitope.
Compared with Mcm6-ctrl, Mcm6-SuOn showed ~4-fold increase in a modified form of the protein in both G1 and S phases (Fig. 4a). This form exhibited the characteristic ~20 kDa upshift of mono-SUMO modification, and was increased in Mcm6-SuOn cells, suggesting that it was the sumoylated form (Fig. 4a). We validated this conclusion using two additional tests. First, compared with untagged SUMO, HF-Smt3 led to a further upshift of this form (Supplementary Fig. 3b). Second, immunoprecipitation of Mcm6-SuOn or Mcm6-ctrl followed by western blotting showed that this form was recognized by a SUMO-specific antibody (Supplementary Fig. 3c). Taken together, these findings demonstrated that the modified form represented the sumoylated form. Mcm6-SuOn also increased sumoylation levels of three other MCM subunits (Mcm2, 4 and 7), thus is an effective tool to increase MCM sumoylation (Supplementary Fig. 3d).
Figure 4. Increasing sumoylation by Mcm6-SuOn slows growth and this defect is suppressed by a ubc9 mutant.
a. Mcm6-SuOn exhibits elevated sumoylation in G1 and S phases. Immunoblot at top shows Mcm6-ctrl and Mcm6-SuOn proteins (tagged with V5) from whole cell lysates. Equal loading is indicated by Ponceau stain, and lack of checkpoint activation is shown by the absence of Rad53 phosphorylation. MMS-treated sample without tagging shows no cross-reaction bands on α-V5 blot (top) and robust Rad53 phosphorylation (bottom).
b. Sumoylation of Mcm6-SuOn and Mcm6-ctrl occurs in the chromatin-bound fraction in G1 cells.
c. Sumoylation of Mcm6-SuOn in G1 requires Cdc6. Cell growth is described in Fig. 1d and Mcm6 sumoylation is detected as in (a). Cdc6 depletion is detected by immunoblotting. * indicates a cross-reaction band.
d. Mcm6-SuOn, but not Mcm6-ctrl, results in slow growth. Representative tetrads from Mcm6-SuOn/+ or Mcm6-ctrl/+ diploid strains are shown. Spores clones were grown at 30°C for 3–4 days and genotypes are indicated.
e. Slow growth of Mcm6-SuOn cells is suppressed by a SUMO E2 Ubc9 mutation (ubc9-10). Representative tetrads from dissection of Mcm6-SuOn/+ ubc9-10/+ diploid strains are shown as in (d).
Subsequent tests showed that Mcm6-SuOn did not affect the general behavior of MCM or modification of other replication factors: 1) Mcm6-SuOn did not change protein levels or MCM complex formation (Fig. 4a and Supplementary Fig. 3e); 2) the protein exhibited normal chromatin vs. cytosol distribution, suggesting proficient chromosomal loading (Fig. 4b); 3) its SUMO-form was detected in chromatin fraction in a Cdc6-dependent manner (Fig. 4b–4c); 4) Mcm6-SuOn did not affect sumoylation of several other DNA replication factors (Supplementary Fig. 3f); and 5) it did not show abnormal DNA damage checkpoint activation (Fig. 4a). These results suggest that the increased MCM sumoylation caused by Mcm6-SuOn obeys the rules of MCM sumoylation and does not perturb general MCM behavior or other replication and checkpoint protein modifications.
Mcm6-SuOn impairs replication initiation
Next, we examined how increased MCM sumoylation by Mcm6-SuOn affected replication and growth. Mcm6-SuOn grew slowly, while Mcm6-ctrl supported normal growth (Fig. 4d and Supplementary Fig. 4a). Importantly, this defect was ameliorated when the SUMO E2 Ubc9 was mutated (Fig. 4e). Thus, while tagging per se did not interfere with protein functions, Mcm6-SuOn impaired growth in a sumoylation-dependent manner.
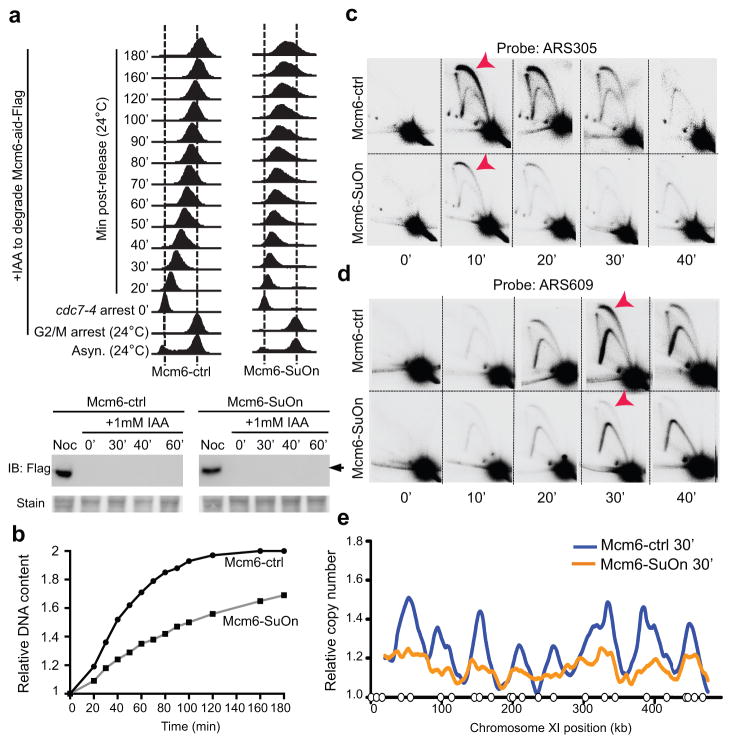
We next examined the kinetics of S phase progression. To avoid chronic defects caused by Mcm6-SuOn, we constructed diploid cells homozygous for cdc7-4, containing either Mcm6-SuOn or Mcm6-ctrl and an Mcm6-aid allele. As illustrated in Figure 5a, cells grown at the permissive temperature for cdc7-4 (24 °C) were synchronized in G2-M. Mcm6-aid was then depleted by IAA addition. Next, cells were released from G2-M arrest and synchronized at the G1-S boundary by raising the temperature to 37°C to inactivate cdc7-4. Finally, cells were released from this arrest by bringing the temperature back to 24°C to reactivate Cdc7-4. FACS profiles showed that cells containing Mcm6-SuOn or Mcm6-ctrl entered S phase after the final release, but Mcm6-SuOn cells exhibited a slower replication profile compared to Mcm6-ctrl cells (Fig. 5a). Quantification of DNA content from the FACS analyses suggested that the mutant moved through S phase at about half the speed as the control (Fig. 5b).
Figure 5. Mcm6-SuOn impairs replication initiation.
a. Mcm6-SuOn cells exhibit slower replication by flow cytometry analyses. Strains contained cdc7-4 to arrest cells before replication initiates. Experimental scheme and flow cytometry profiles show that cells were arrested in G2-M followed by IAA addition to degrade Mcm6-aid, then temperature shift to 37°C to inactivate cdc7-4 and achieve arrest before replication initiation, and cooling to 24°C to allow cells to progress into S phase. Mcm6-aid degradation is shown (bottom) and arrow denotes the Mcm6-aid band.
b. Quantification of DNA content from (a) is shown.
c. Mcm6-SuOn shows defective firing at the ARS305 locus. 2D gel results show different origin firing between Mcm6-ctrl and Mcm6-SuOn strains. Cells were from the experiment in (a). Bubble DNA structures representing origin firing events are labeled by red arrows.
d. Mcm6-SuOn shows defective firing at the ARS609 locus. As in (c), except ARS609-specific probe was used.
e. Copy number changes in a section of chromosome XI are shown based on genome sequencing of the 30 min post-release samples in (a). Open circles represent the confirmed replication origins according to OriBD.
To understand if the slow replication seen in Mcm6-SuOn cells was due to replication initiation defects, we subjected the samples collected in the above tests to two-dimensional agarose gel electrophoresis (2D gel). Using probes specific to an early origin (ARS305) and a late origin (ARS609), replication-firing events were detected as bubble DNA structures as shown previously (Fig. 5c–5d). While Mcm6-ctrl cells showed robust origin firing signals at both loci at expected times, Mcm6-SuOn cells showed weaker signals, indicating impaired replication initiation (Fig. 5c–5d).
Deep sequencing analysis of samples from cdc7-4 arrest (0 min) and 30 min after release was used to deduce copy number changes and genome-wide replication profiles. This analysis showed that Mcm6-SuOn cells exhibited decreased replication at nearly all the origins annotated in OriDB compared with Mcm6-ctrl (Fig. 4e and Supplementary Fig. 5). Taken together, these results demonstrate that increased MCM sumoylation levels impair genome-wide replication from both early and late origins.
Increasing MCM sumoylation levels compromises CMG formation
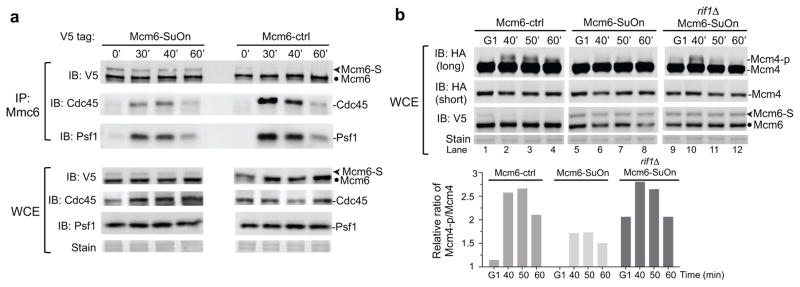
To gain insight into the molecular basis of the replication initiation defects associated with Mcm6-SuOn, we examined CMG formation, since it is critical for replisome assembly. CMG formation can be assessed by measuring the amount of Cdc45 or a GINS subunit (e.g. Psf1) that co-immunoprecipitates with Mcm6. Using the samples obtained from experiments depicted in Figure 5a, we found that Mcm6-SuOn co-purified lower amounts of both Cdc45 and Psf1, compared to Mcm6-ctrl, at 30 and 40 min after S phase entry (Fig. 6a). This finding suggested that Mcm6-SuOn interfered with CMG formation. This conclusion was substantiated by the observation that Mcm4 phosphorylation was reduced in Mcm6-SuOn cells, compared with Mcm6-ctrl in G1 and S phases (Fig. 6b, lane 1–8). In addition, we found that mimicking constitutive sumoylation by using an Mcm6-SUMO fusion led to similar defects as seen for Mcm6-SuOn, i.e., lower amounts of Cdc45 and Psf1 were associated with Mcm6-SUMO than with the control (Supplementary Fig. 6a). Consistent with this, Mcm6-SUMO fusion strains grew poorly and showed slower replication profiles (Supplementary Fig. 6b–6c). Our findings that increasing MCM sumoylation using two strategies resulted in similar molecular and phenotypic defects strongly support a negative role for MCM sumoylation in controlling replication initiation at a step involving CMG formation.
Figure 6. Mcm6-SuOn cells exhibit low levels of CMG and phosphorylation of Mcm4 that can be suppressed by rif1Δ.
a. Mcm6-SuOn cells have reduced CMG levels. Immunoblots at top show immunoprecipitated Mcm6 and associated Cdc45 and Psf1. Immunoblots at bottom show Mcm6, Cdc45, and Psf1 protein levels in the input. Indicated time points are the same as in experiments depicted in Fig. 5a.
b. The reduced Mcm4 phosphorylation in Mcm6-SuOn cells is restored by rif1Δ. As in Fig. 2b, immunoblots (top two) shows Mcm4 phosphorylation with long exposure for detecting Mcm4 phosphorylation and short exposures for detecting unmodified Mcm4 protein levels. Immunoblot at bottom shows Mcm6 protein levels and sumoylation status. Relative ratio of phosphorylated Mcm4 versus unmodified Mcm4 is plotted in the graph.
Removing a PP1 cofactor rescues defects of Mcm6-SuOn cells
As Mcm4 phosphorylation is a prerequisite for CMG formation, we determined if low Mcm4 phosphorylation could be responsible for the observed Mcm6-SuOn defects. We tested this idea genetically by removing the phosphatase PP1 (Glc7, essential in budding yeast) binding partner Rif1, as disruption of this complex increases Mcm4 phosphorylation in both G1 and S phases9–11. Rif1 loss in Mcm6-SuOn increased Mcm4 phosphorylation without affecting Mcm6 sumoylation level in both cell cycle phases (Fig. 6b, lanes 5–12).
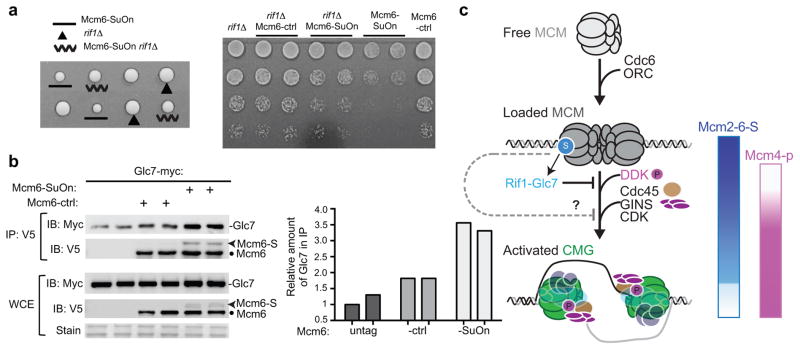
Importantly, Rif1 loss improved Mcm6-SuOn growth, as assessed by spore clone sizes and spotting assays (Fig. 7a). Control cells showed wild-type levels of Mcm4 phosphorylation, which were increased by rif1Δ as expected (Supplementary Fig. 6d). Also, no growth defects were seen for Mcm6-ctrl or Mcm6-ctrl rif1Δ cells (Fig. 7a). We also compared rif1Δ with two other mutations, mcm5-bob1 and mcm2-4-174Δ, known to improve replication when Mcm4 phosphorylation is dampened. While the lethality of Mcm6-ctrl mcm2-4-174Δ precluded testing this allele (Supplementary Fig. 4b), we found that mcm5-bob1 did not suppress cdc7-4 as potently as rif1Δ (Supplementary Fig. 4c) and failed to suppress Mcm6-SuOn growth defects (Supplementary Fig. 4d), suggesting that suppression of Mcm6-SuOn requires maximal bypass of Mcm4 phosphorylation defects. Taken together, the observed rif1Δ suppression of Mcm6-SuOn cells supports the notion that reduced Mcm4 phosphorylation is partly responsible for the replication defects in these cells.
Figure 7. rif1Δ suppresses Mcm6-SuOn growth defects and Mcm6-SuOn leads to enhanced association between Mcm6 and the Glc7 phosphatase.
a. Rif1 removal partially rescues the growth defects of Mcm6-SuOn. Left, representative tetrads from dissection of the Mcm6-SuOn/+ rif1Δ/+ diploid strain. Right, 5-fold serial dilutions of cells spotted on YPD plates.
b. Increased levels of Glc7 are associated with Mcm6-SuOn, compared to Mcm6-ctrl. Using G1-arrested cells, Mcm6 was immunoprecipitated and the associated Glc7 levels were examined. Quantification of the amount of Glc7 in the immunoprecipitated fraction is plotted, with the value of untagged control (first lane) set to 1.
c. A model depicts the spatial and temporal pattern of the MCM sumoylation cycle and a role of MCM sumoylation in negatively regulating replication initiation. For simplicity, only the replication factors used in this study are shown. After Cdc6-mediated MCM loading at replication origins, a fraction of Mcm2–7 subunits is sumoylated. This occurs prior to DDK-mediated Mcm4 phosphorylation and CMG formation. One function of MCM sumoylation is to aid Glc7 phosphatase recruitment to counteract Mcm4 phosphorylation in G1, preventing premature CMG formation. Roles for MCM sumoylation at an event after DDK activation are possible (“?”). As replication is initiated in waves from early and late origins, Mcm2–6 sumoylation decreases. The loss of Mcm2–6 sumoylation is concomitant with the appearance of DDK-mediated Mcm4 phosphorylation, both promoting replication initiation.
Mcm6-SuOn shows increased association with PP1
The above findings, in conjunction with a previously detected interaction between Glc7 and SUMO36, raised the possibility that the reduction in Mcm4 phosphorylation caused by MCM hypersumoylation may be due to increased recruitment of Glc7 to MCM. To test this, we examined the Glc7–Mcm6 association in G1 cells by co-immunoprecipitation. We detected a slight but reproducible enrichment of Glc7 in the immunoprecipitated fraction when Mcm6 was pulled down (Fig. 7b). Importantly, we found a 2-fold greater enrichment of Glc7 in the immunoprecipitated fraction of Mcm6-SuOn cells, compared to Mcm6-ctrl cells, both in G1 and upon release into S phase (Fig. 7b and Supplementary Fig. 6e). As the level of association of Glc7 with Mcm6-SuOn was not affected in cells lacking Rif1 (Supplementary Fig. 6f), Rif1 likely has additional roles in promoting Glc7 functions. Taken together, our results suggest that MCM sumoylation promotes Glc7 recruitment to MCM, thereby disfavoring MCM phosphorylation.
Discussion
Proper control of replication initiation is important for cell survival and for the prevention of human diseases. While positive regulation promotes origin licensing and replisome assembly, negative regulation is needed to prevent premature initiation or re-replication, and to ensure the proper sequence of events in the assembly of a functional replisome. Many of these regulatory targets are subunits of MCM due to its central role in multiple aspects of replication. While phosphorylation was the only known chemical modification on MCM that regulates replication initiation thus far, other modifications have recently been identified through studies such as proteomic screens. Here we examined a highly conserved MCM modification by SUMO and demonstrated its strict temporal and spatial regulation in cells (summarized in Fig. 7c). For each MCM subunit, a fraction of the protein showed sumoylation upon Cdc6-mediated MCM loading onto chromatin, prior to bulk Mcm4 phosphorylation (Fig. 1 and 2). As cells entered S phase, Mcm2–6 sumoylation levels greatly decreased (Fig. 2). This loss was associated with replication initiation from both early and late origins (Fig. 3c). That the pattern of MCM sumoylation was opposite to that of replication activity suggested that this modification played a negative role in replication. We tested this model by increasing MCM sumoylation through the use of Mcm6-SuOn or mimicking the modification by Mcm6-SUMO fusion (Fig. 4; Supplementary Fig. 6a–6c). In both cases, cell growth and replication were impeded and CMG levels were reduced, providing strong evidence for this model (Figs. 4d, 5a–5e; Supplementary Figs. 5, 6a–6c). In addition, the growth defect of Mcm6-SuOn was rescued either by reducing sumoylation or diminishing protein phosphatase 1 (PP1) function, which restored Mcm4-phosphorylation (Figs. 4e, 6b, 7a). Finally, our data suggest that MCM sumoylation promotes the association of PP1 with MCM (Fig. 7b; Supplementary Fig. 6e), providing a mechanism for targeting the phosphatase specifically to chromatin-loaded MCM in G1 phase (model in Fig. 7c).
Our results also suggest that the reversal of MCM sumoylation is important. The loss of Mcm2–6 sumoylation required DDK and GINS (Fig. 3a–3b), suggesting that an active desumoylation process may occur to remove this replication-inhibition marker. Consistent with an active role of DDK in MCM sumoylation loss, artificially increasing DDK activity by removing Rif1 reduced the sumoylation of Mcm2 and 6 proteins (Supplementary Fig. 7a), though this effect was masked in Mcm6-SuOn cells (Fig. 6b). DDK unlikely influences MCM sumoylation loss by altering substrate properties, since sumoylated forms of Mcm4 did not appear to be phosphorylated (Supplementary Fig. 7b). Rather, DDK could affect this process by modulating the desumoylation enzyme(s). Between the two desumoylation enzymes in yeast, acute depletion of Ulp2, but not Ulp1, increased the sumoylation levels of Mcm4 and 6 on chromatin (Supplementary Fig. 7c–7e), pointing to a potential role of this enzyme in DDK-mediated removal of MCM sumoylation. Thus, while our study focuses on MCM sumoylation and reveals a role for this modification in counteracting DDK-mediated phosphorylation in G1, the reverse is likely true at the junction of replication initiation. Such dual regulation could ensure the precise deployment of each regulatory module during replication initiation. The dynamic nature of sumoylation and desumoylation in cells and potential sumoylation loss during extraction likely explain the low levels of sumoylation observed for each MCM subunit, as for most other substrates. Our data suggest that even a low level of MCM sumoylation suffices to achieve a biological effect either because sumoylation promotes the recruitment of an enzyme, a small amount of which can catalyze multiple reactions, or because sumoylation of multiple MCM subunits may have redundant roles.
While our findings suggest one role of MCM sumoylation in replication regulation, they do not exclude other possible functions. For example, as Rif1 removal only partially rescues the Mcm6-SuOn growth defect (Fig. 7a), it is possible that MCM sumoylation has other roles in inhibiting replication. One probable scenario is that MCM sumoylation may block the recruitment of Cdc45 or GINS downstream of the DDK-mediated Mcm4 phosphorylation step (Fig. 7c). In addition, sumoylation of each MCM subunit may have distinct roles. For example, unlike Mcm2–6, Mcm7 sumoylation persisted throughout most of S phase (Fig. 2a–2b). Considering that Mcm7 is the only MCM subunit that undergoes ubiquitination to enable MCM removal during replication termination, its sumoylation may be relevant to this event. Our findings on one effect of MCM sumoylation will stimulate future studies to uncover the full scope of its biological effects.
In summary, our findings reveal that a novel SUMO-based regulation exerts a negative influence on MCM activation, adding to the known positive regulation conferred by MCM phosphorylation. This dual modulation can expand the range of regulation, allowing for flexible integration of multiple biological cues, ranging from chromatin structure to developmental stages, and for providing precision and flexibility in replication regulation. Considering that MCM sumoylation is highly conserved, our work will stimulate the elucidation of the range of effects of this important modification in higher organisms.
Online Methods
Yeast strains and techniques
Standard procedures were used in cell growth and medium preparation. Strains are isogenic to W1588-4C, a RAD5 derivative of W303, (MATa ade2-1 can1-100 ura3-1 his3-11,15 leu2-3,112 trp1-1 rad5-535)39. Strains and their usage in specific figure panels are listed in Supplementary Table. Proteins were tagged at their endogenous loci by standard methods and correct tagging was verified by sequencing. Each MCM subunit was tagged with 3HA at the C-terminus, except Mcm3, which was tagged at its N-terminus and expressed from the ADH1 promoter. As shown in Supplementary Figure 7f, PADH1-Mcm3-HA expression level was about half that of Mcm7-HA, consistent with their endogenous protein level ratio40 and indicating normal protein levels. Mcm5 was additionally tagged with the Strep tag II at its C-terminus in Figure 1d and 3b. Aid-tagging has been described previously30,31. In brief, Psf2 was fused with a 3V5 tag-IAA7 module at its C-terminus, and Mcm6 was fused with IAA17-3Flag at its C-terminus. Mcm6-SuOn and -ctrl were constructed as described with minor modifications34. Both tags and a 3V5 linker were fused to the C-terminus of Mcm6. SuOn is composed of the catalytically dead Ulp1 protease domain (418-621aa; with C580S abolishing the activity)34,35. The control tag has the F474A mutation that disrupts SUMO interaction34,35. We note that SuOn is different from the canonical SUMO-interacting motif (SIM) as it interacts with the C-terminal tail of SUMO through a distinct large interface and has strict orientation requirements35. Yeast dissection and spotting assays were performed using standard procedures. All genetic and biochemical experiments were performed using two different spore clones for each genotype.
Synchronization procedures
For experiments that entailed alpha factor arrest, cells were treated for 2.5 hours with 5 ug/ml (for BAR1+ cells) or 100 ng/ml (for bar1Δ cells) alpha factor. For experiments involving G2/M arrest, cells were grown in media containing 1% DMSO to early log phase and treated with 15 ug/ml nocodazole for 3 hours. In all cases, cell morphology was checked to confirm the arrest. Experiments in Figure 1d and 4c were performed as described previously37. In brief, cultures grown in YPGalactose at 24°C were arrested in G2-M and split into two, one of which received 2% glucose for 1 hour. Subsequently, cells were released into the same media containing alpha factor for 2.5 hours before harvesting. For the experiment in Figure 3a, cells were arrest by alpha factor at 24°C, then temperature was shifted to 37°C for 1 hour. Subsequently, cultures were split into two, only one of which was released from alpha factor into S phase. Samples were taken from both cultures 40 minutes afterwards. For experiments in Figure 3b, cells were arrested in G2-M phase at 24°C, IAA was added to a final concentration of 1 mM. After 1 hour, cells were released into media containing alpha factor and 1 mM IAA for 3 hours, and then released into media containing 1 mM IAA for 40 min. For the experiment in Figure 3c, alpha factor arrested cells were released into YPD media containing 300 ug/ml pronase and 200 mM HU at 24°C for 40 minutes. For experiments in Figures 5 and 6a, diploid cells containing Mcm6-SuOn or Mcm6-ctrl and a Mcm6-aid degron allele and homozygous for cdc7-4 were used. Cells were grown at the permissive temperature for cdc7-4 (24°C) and synchronized in G2-M. Then Mcm6-aid was depleted by the addition of 1 mM IAA for 1 hour. Subsequently, cells were released from G2-M arrest and synchronized at the G1-S boundary by raising the temperature to 37°C to inactivate cdc7-4. Finally, cells were released by rapid cooling to 24°C.
Detection of protein sumoylation
Unless otherwise indicated, standard Ni-NTA pull down method was used as described29. Smt3 was tagged with HF (6His-Flag) at its N terminus and expressed from its endogenous promoter41. Cell extracts prepared by 55% TCA precipitation were dissolved in Buffer A (6 M Guanidine HCl, 100 mM sodium phosphate, pH 8.0, 10 mM Tris-HCl, pH 8.0) with rotation at room temperature. Cleared supernatant was then incubated with Ni-NTA resin (Qiagen) after the addition of Tween 20 (0.05% final concentration) and imidazole (4.4 mM final concentration) overnight at room temperature. Beads were then washed twice with Buffer A containing 0.05% Tween 20 and four times with Buffer C (8 M urea, 100 mM sodium phosphate, pH 6.3, 10 mM Tris-HCl, pH 6.3) containing 0.05% Tween 20. HU buffer (8 M urea, 200 mM Tris-HCl, pH 6.8, 1 mM EDTA, 5% SDS, 0.1% bromophenol blue, 1.5% DTT, 200 mM imidazole) was used to elute proteins from the beads before loading on to a 3–8% gradient Tris-Acetate gel (Life Technologies). Western blots were probed with antibodies recognizing the tagged substrates detecting both sumoylated and unmodified bands. The latter is due to non-specific binding to the Ni-NTA beads and is not enriched when using HF-Smt3. Our previous work using a protein immunoprecipitation method showed sumoylation of two MCM subunits under normal growth conditions19 and Supplementary Fig. 1b demonstrates the sumoylation of additional MCM subunits by using this method. As desumoylation cannot be efficiently prevented during this procedure, low abundant sumoylation forms are difficult to detect. However, the use of both untagged and HF-SUMO allowed better assessment of sumoylation due to the different sizes of sumoylated forms in the two situations (Supplementary Fig. 1b).
Two-dimensional agarose gel electrophoresis (2D gel)
2D gel analyses were performed as previously described42. Genomic DNA was extracted from spheroplasts and purified by standard CsCl centrifugation at 90 krpm for 9 hours at 15°C. DNA was digested by EcoRI and separated by agarose gel electrophoresis in two dimensions. DNA was transferred to Hybond-XL membrane (GE Healthcare) and analyzed by Southern blot hybridization using probes specific for ARS305 or ARS609 as described previously43. Primer sequences used for amplification of the probes are available upon request.
Whole genome sequencing and copy number calculation
Both procedures were carried out as previously described38,44. Mcm6-ctrl and Mcm6-SuOn cells were collected at 0 and 30 min as described in Figure 5a. 1.5 ug genomic DNA from each sample was used to generate libraries using the KAPA’s library kit (iGO facility, MSKCC) and sequenced using HiSeq 2500 (Illumina). At least 10 million 50 bp paired-end reads were generated per sample. Reads were mapped to the S288c reference genome (SGD, SacCer2) excluding repetitive sequences, and summed in 1 kb bins using Genome Brower. Bins containing fewer than 600 reads were excluded. Custom R script was used to analyze the value for each locus. In brief, for each strain, the binned reads from the 30 min sample at a locus were divided by those from 0 min sample, and normalized to the ratio of total reads to give a genome-wide mean value of 1. This number was adjusted by the relative DNA content at 30 min in FACS fitting curve (Fig. 5b) to derive a relative copy number of the particular locus. The map of adjusted copy numbers were further smoothed with the “loess” function and shown in Figure 5e and Supplementary Figure 5. Detailed methods and data for calculating the relative copy number based on the whole genome sequencings are included in the GEO database (GSE70407).
Protein extraction and immunoprecipitation (IP)
For Figures 2b and 6a, cells were resuspended in IP buffer (100 mM HEPES/KOH pH 7.9, 100 mM KOAC, 2 mM MgOAC, 1 mM ATP, 1% Triton X-100, 2 mM NaF, 0.1 mM Na3VO4, 20 mM β-glycerophosphate, 1 mM PMSF, 10 mM Benzamidine HCl, 10 ug/ml leupeptin, 1 ug/ml pepstatin A) containing 1x protease inhibitor (EDTA-free, Roche) and 20 mM NEM. Cells were disrupted by bead beating. Benzonase was added to cell lysates, which were incubated for 1 hour at 4°C. After centrifugation for 20 min at 15,000 rpm at 4°C, the supernatant was collected and incubated with prewashed HA (26182, Fisher) or V5 conjugated beads (A7345, Sigma-Aldrich) for 2 hours at 4°C. For Figure 7b, minor changes were made to the IP buffer, 50 mM HEPES/KOH pH 7.9, 150 mM KOAC was used and glycerol was added at a final concentration of 10%.
Immunoblotting analysis and antibodies
Protein samples were resolved by 3–8% or 4–20% gradient gels (Life Technologies and Bio-Rad) and transferred to a 0.2 um nitrocellulose membrane (G5678144, GE). Antibodies used are: anti-HA (F-7, Sc-7392, Santa Cruz Biotechnology), anti-V5 (R960-25, Invitrogen), anti-myc (9E10, Bio X cell), PAP (P1291, Sigma), anti-flag (M2, Sigma), anti-Rad53 (yC-19, sc-6749, Santa Cruz Biotechnology), anti-Orc2 (SB46, Abcam), anti-Pgk1 (22C5D8, Invitrogen), anti-Cdc6 (9H8/5, Abcam), anti-Strep tag II (A01732, Genscript), anti-Psf1 and anti-Mcm6 (gifts from Karim Labib)17, anti-Orc6 and anti-Cdc45 (gifts from Bruce Stillman)4 and anti-SUMO39. Validation of these antibodies are either provided on the manufacturer’s website or from the cited references. For quantification purpose, membranes were scanned with Fujifilm LAS-3000 luminescent image analyzer, which has a linear dynamic range of 104. Quantification of blots and generation of figures were done using ImageGauge and Photoshop.
Lambda phosphatase treatment
After immunoprecipitation on HA (Fisher, 26182) or Ni-NTA beads (Qiagen), the beads were washed three times with wash buffer (50 mM K-HEPES pH7.9, 150 mM KOAC, 2 mM MgOAC, 10 ug/ml pepstatin, 10 ug/ml lepeptin, 0.5 mM PMSF, 1 mM benzamidine and 20 mM NEM). Beads were then resuspended in Lambda phosphatase reaction buffer (1x NEBuffer for PMP, 1 mM MnCl2 and 10 ug/ml pepstatin, 10 ug/ml lepeptin, 0.5 mM PMSF, 1 mM benzamidine and 20 mM NEM). In control tests, the beads were incubated with reaction buffer and phosphatase inhibitors (50 mM EDTA, 50 mM NaF and 10 mM Na3VO4). 80 U of lambda phosphatase (NEB, P0753S) was added, and incubated at 30°C for 30 min. Laemmili buffer was added to stop the reaction and the proteins were eluted by boiling at 95°C for 5 min before SDS-PAGE and western blotting analysis.
Other methods
Chromatin fractionation was performed as described previously with minor modifications45. In brief, spheroplasts were lysed in lysis buffer containing 1% Triton-X-100 and laid upon a 30% sucrose cushion and centrifuged at 13,000 rpm for 20 min to separate the supernatant and chromatin fractions. The chromatin-bound fraction was washed once with lysis buffer and re-suspended in the same buffer. Equal volumes of samples from lysate, supernatant and chromatin fractions were precipitated with 20% TCA, and resuspended in Laemmli buffer with the addition of 2M Tris to neutralize TCA. Flow cytometry was performed as previously described using FACSCalibur flow cytometer, and data were analyzed with FlowJo Software. To calculate the relative DNA content in Figure 5b, the distance between 1N and 2N DNA peaks was considered as 2 and the position of the 1N peak as 1. Then the distance between DNA peaks at each time point and the 1N DNA peak in G1 cells was measured and scaled between 1 and 2 and plotted. Original images of gels and blots used in this study can be found in Supplementary Data Set 1.
Supplementary Material
Acknowledgments
We are very grateful to T. Zhang and J. Xiang at the Genomics Resources Core Facility, Weill Cornell Medical College for their kind assistance in genome-wide sequencing analyses. We also thank K. Labib (University of Dundee), D. Shore (Institute of Genetics and Genomics in Geneva), B. Stillman (Cold Spring Harbor Laboratory), J. Diffley (Cancer Research UK London Research Institute), D. Remus (Memorial Sloan Kettering Cancer Center), J. Torres-Rosell (Universitat de Lleida), M. Kanemaki (Japan National Institute of Genetics), and D. Koshland (University of California, Berkeley, Berkeley) for providing strains, plasmids, and antibodies; Zhao lab members B. Wan for providing reagents and P. Sarangi for discussion. This study was supported by US National Institutes of Health grant GM080670 to X.Z..
Footnotes
Accession codes
Data files have been deposited in the Gene Expression Omnibus database under accession number GSE70407.
Author Contributions
L.W and X.Z. conceived the study and designed the experiments. L.W performed the experiments. L.W and X.Z analyzed the results and wrote the manuscript.
References
- 1.Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- 2.Zegerman P, Diffley JFX. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka S, et al. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 4.Sheu YJ, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463:113–117. doi: 10.1038/nature08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson AP, Laskey RA, Coleman N. Replication proteins and human disease. Cold Spring Harb Perspect Biol. 2014;6:327–342. doi: 10.1101/cshperspect.a013060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francis LI, Randell JCW, Takara TJ, Uchima L, Bell SP. Incorporation into the prereplicative complex activates the Mcm2-7 helicase for Cdc7-Dbf4 phosphorylation. Genes Dev. 2009;23:643–654. doi: 10.1101/gad.1759609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell. 2006;24:101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Randell JCW, et al. Mec1 is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7. Mol Cell. 2010;40:353–363. doi: 10.1016/j.molcel.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattarocci S, et al. Rif1 controls DNA replication timing in yeast through the PP1 phosphatase Glc7. Cell Rep. 2014;7:62–69. doi: 10.1016/j.celrep.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Davé A, Cooley C, Garg M, Bianchi A. Protein phosphatase 1 recruitment by Rif1 regulates DNA replication origin firing by counteracting DDK activity. Cell Rep. 2014;7:53–61. doi: 10.1016/j.celrep.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiraga SI, et al. Rif1 controls DNA replication by directing Protein Phosphatase 1 to reverse Cdc7-mediated phosphorylation of the MCM complex. Genes Dev. 2014;28:372–383. doi: 10.1101/gad.231258.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maric M, Maculins T, De Piccoli G, Labib K. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science. 2014;346:1253596. doi: 10.1126/science.1253596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Remus D, Diffley JFX. Eukaryotic DNA replication control: lock and load, then fire. Curr Opin Cell Biol. 2009;21:771–777. doi: 10.1016/j.ceb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Sclafani RA, Holzen TM. Cell Cycle Regulation of DNA Replication. Annu Rev Genet. 2007;41:237–280. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015;519:431–5. doi: 10.1038/nature14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heller RC, et al. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146:80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gambus A, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 18.Morohashi H, Maculins T, Labib K. The amino-terminal TPR domain of Dia2 tethers SCF(Dia2) to the replisome progression complex. Curr Biol. 2009;19:1943–1949. doi: 10.1016/j.cub.2009.09.062. [DOI] [PubMed] [Google Scholar]
- 19.Cremona CA, et al. Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the Mec1 checkpoint. Mol Cell. 2012;45:422–32. doi: 10.1016/j.molcel.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golebiowski F, et al. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 21.Elrouby N, Coupland G. Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc Natl Acad Sci USA. 2010;107:17415–20. doi: 10.1073/pnas.1005452107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 23.Sarangi P, Zhao X. SUMO-mediated regulation of DNA damage repair and responses. Trends Biochem Sci. 2015;40:233–42. doi: 10.1016/j.tibs.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Psakhye I, Jentsch S. Protein Group Modification and Synergy in the SUMO Pathway as Exemplified in DNA Repair. Cell. 2012;151:807–820. doi: 10.1016/j.cell.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 25.Morris JR, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–90. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 26.Galanty Y, et al. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung I, Zhao X. DNA break-induced sumoylation is enabled by collaboration between a SUMO ligase and the ssDNA-binding complex RPA. Genes Dev. 2015;29:1593–8. doi: 10.1101/gad.265058.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarangi P, et al. A versatile scaffold contributes to damage survival via sumoylation and nuclease interactions. Cell Rep. 2014;9:143–52. doi: 10.1016/j.celrep.2014.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulrich HD, Davies AA. In vivo detection and characterization of sumoylation targets in Saccharomyces cerevisiae. Methods Mol Biol. 2009;497:81–103. doi: 10.1007/978-1-59745-566-4_6. [DOI] [PubMed] [Google Scholar]
- 30.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 31.Havens KA, et al. A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol. 2012;160:135–142. doi: 10.1104/pp.112.202184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crabbe L, et al. Analysis of replication profiles reveals key role of RFC-Ctf18 in yeast replication stress response. Nat Struct Mol Biol. 2010;17:1391–7. doi: 10.1038/nsmb.1932. [DOI] [PubMed] [Google Scholar]
- 33.Zegerman P, Diffley JFX. Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature. 2010;467:474–478. doi: 10.1038/nature09373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Almedawar S, Colomina N, Bermudez-Lopez M, Pocino-Merino I, Torres-Rosell J. A SUMO-dependent step during establishment of sister chromatid cohesion. Curr Biol. 2012;22:1576–81. doi: 10.1016/j.cub.2012.06.046. [DOI] [PubMed] [Google Scholar]
- 35.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 36.Sung MK, et al. Genome-wide bimolecular fluorescence complementation analysis of SUMO interactome in yeast. Genome Res. 2013;23:736–46. doi: 10.1101/gr.148346.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desdouets C, et al. Evidence for a Cdc6p-independent mitotic resetting event involving DNA polymerase alpha. EMBO J. 1998;17:4139–46. doi: 10.1093/emboj/17.14.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hawkins M, et al. High-resolution replication profiles define the stochastic nature of genome replication initiation and termination. Cell Rep. 2013;5:1132–41. doi: 10.1016/j.celrep.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao X, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA. 2005;102:4777–82. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi Y, et al. Cooperation of Sumoylated Chromosomal Proteins in rDNA Maintenance. PloS Genet. 2008;4:e1000215. doi: 10.1371/journal.pgen.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedman KL, Brewer BJ. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 1995;262:613–27. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- 43.Hang LE, et al. Rtt107 Is a Multi-functional Scaffold Supporting Replication Progression with Partner SUMO and Ubiquitin Ligases. Mol Cell. 2015;60:268–79. doi: 10.1016/j.molcel.2015.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murakami H, Keeney S. Temporospatial coordination of meiotic DNA replication and recombination via DDK recruitment to replisomes. Cell. 2014;158:861–73. doi: 10.1016/j.cell.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schepers A, Diffley JF. Mutational analysis of conserved sequence motifs in the budding yeast Cdc6 protein. J Mol Biol. 2001;308:597–608. doi: 10.1006/jmbi.2001.4637. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.