Abstract
The nuclear immunophilin FKBP25 interacts with chromatin-related proteins and transcription factors and is suggested to interact with nucleic acids. Currently the structural basis of nucleic acid binding by FKBP25 is unknown. Here we determined the nuclear magnetic resonance (NMR) solution structure of full-length human FKBP25 and studied its interaction with DNA. The FKBP25 structure revealed that the N-terminal helix-loop-helix (HLH) domain and C-terminal FK506-binding domain (FKBD) interact with each other and that both of the domains are involved in DNA binding. The HLH domain forms major-groove interactions and the basic FKBD loop cooperates to form interactions with an adjacent minor-groove of DNA. The FKBP25–DNA complex model, supported by NMR and mutational studies, provides structural and mechanistic insights into the nuclear immunophilin-mediated nucleic acid recognition.
INTRODUCTION
Immunophilins are a versatile family of chaperone proteins that control cis-trans isomerization of peptidyl-prolyl bonds in proteins and also serve as molecular receptors for several immunosuppressive drugs (1). FK506 binding proteins (FKBPs) belong to the immunophilin family of proteins, known to bind the immunosuppressive drugs, such as FK506 and rapamycin (2). FKBPs are conserved from yeast to animals, and several of their members have been discovered and studied in detail. Most of the FKBPs are multi-domain proteins and harbor the much conserved FK506-binding domain (FKBD), also known as the PPIase domain (3). FKBPs are involved in several biological processes including protein folding, receptor signaling and protein trafficking, and are also implicated in several diseases, such as Alzheimer's, Parkinson's, Refsum, malaria and cancer (2,4–9). However, recent studies have shown novel functions of FKBPs in the nucleus, including histone chaperone activity, gene regulation and chromatin remodeling .
Human FKBP25 (also known as FKBP3), a member of the FKBP family, is a 25-kDa protein that binds with both FK506 and rapamycin drugs (10). Most of the FKBPs discovered to-date are known to be distributed in the cytoplasm. Interestingly, FKBP25 was the first FKBP family member that was identified to mainly reside in the nucleus (11). FKBP25 contains an N-terminal helix-loop-helix (HLH) domain and a C-terminal FKBD, which are linked by a flexible loop that is 35 amino acids in length (Figure 1A and Supplementary Figure S1a). The unique N-terminal domain of FKBP25, which was predicted to interact with DNA (12), does not show a significant sequence similarity to any mammalian protein.
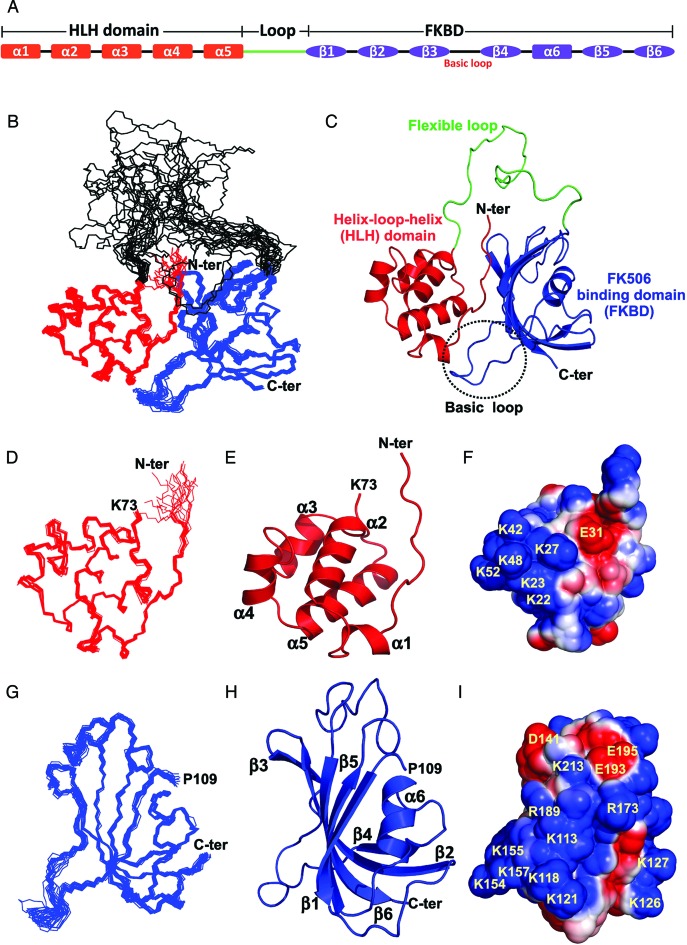
Figure 1.
Nuclear magnetic resonance (NMR) solution structure of the FKBP25. (A) The domain organization and secondary structures of human FKBP25. (B) Superposition of the backbone traces from the final ensembles of 20 solution structures of the full-length FKBP25 determined by NMR spectroscopy. The N-terminal HLH (residue M1-K73) and C-terminal FKBD (residue P109-D224) are highlighted in red and blue respectively. (C) A ribbon representation of FKBP25 is displayed, using the same color scheme and the central flexible loop (G74-P108) linking the two domains is highlighted in green. Ensembles of 20 solution structures, a ribbon representation, and the electrostatic potential surface of the N-terminal HLH (D–F) and C-terminal FKBD of FKBP25 (G–I) are displayed for brevity. The secondary structural elements are indicated as labeled. The positively charged surface is in blue and the negatively charged surface is in red. Refer online version for color coding.
Functionally FKBP25, a downstream gene of p53 (13), interacts with the mouse double minute 2 homolog (MDM2) and increases the auto-ubiquitination of MDM2, which in turn leads to the activation of the p53 signaling pathway (14). Human FKBP25 also interacts with histone deacetylase (HDAC1/HDAC2) and the transcription factor, Yin Yang 1 (YY1) (15), which increases its transcriptional repressive activity by increasing YY1-mediated DNA binding. It also interacts with other nucleic acid-associating proteins, such as high mobility group II and nucleolin (16,17). Fpr3 and Fpr4, the yeast homologs of human FKBP25, and AtFKBP53 from Arabidopsis thaliana are known to possess histone chaperone activity and play an important role in the repression of ribosomal RNA gene expression (18–21). Taken together, these studies underscore the role of FKBP25 in nuclear events, such as transcription regulation, p53 activation, chromatin regulation and DNA repair (22).
While the structures of two the individual domains of human FKBP25 are known (23,24), the structure of full-length FKBP25 and the molecular mechanism of its interaction with nucleic acid remain unknown. To this end, we have determined the nuclear magnetic resonance (NMR) solution structure of the full-length human FKBP25, characterized its DNA binding and proposed a model to show the DNA-binding mechanism of FKBP25. Further we have also characterized the role of FKBP25 in YY1-mediated DNA recognition.
MATERIALS AND METHODS
Sample preparation
Human FKBP25 was expressed and purified as described previously (25). FKBP25 mutants were generated by polymerase chain reaction-based site-directed mutagenesis from the full-length human FKBP25 and confirmed by DNA sequencing. For purification of the mutants, the same protocol was used as that for wild-type human FKBP25. Gene for DNA binding domain of YY1 (referred to as YY1–DBD; spanning residues 286–414), was amplified by polymerase chain reaction and cloned into pET29b. Clones were transformed into BL21 (DE3) Escherichia coli cells. Cells were induced by 1 mM IPTG and the LB broth media was supplemented with 0.1 mM ZnCl2. Further YY1–DBD protein was purified by Ni-NTA column using the same buffer as used for FKBP25 except for the addition of 0.1 mM ZnCl2 in all the buffers used in purification. YY1–DBD was finally exchanged with 25 mM Bis-Tris, 150 mM NaCl and 0.1 mM ZnCl2 at pH 7.0. YY1 peptide (residues 300-333) was purchased from GL Biochem (Shanghai) Ltd and it was dissolved into buffer containing 25 mM Bis-Tris, 150 mM NaCl and 0.1 mM ZnCl2 at pH 7.0. For NMR experiments, uniformly 15N- and 15N/13C-labeled FKBP25 was purified as previously described (25) and protein sample was prepared in a buffer containing 20 mM sodium phosphate, 50 mM NaCl, 0.01% NaN3, 90% H2O and 10% D2O at pH 7. For FKBP25–DNA titration, labeled protein was prepared in 20 mM Bis-Tris, pH 7.0, with 50 or 150 mM NaCl, 90% H2O and 10% D2O.
The 23-bp oligonucleotide (DNAYY1) and ethylenediaminetetraacetic acid-derivatized deoxythymidine (dT-EDTA)-conjugated oligonucleotides were purchased from Sigma-Aldrich (Singapore). The sequence of the DNAYY1 oligo was 5′AGGGTCTCCATTTTGAAGCATGC3′. Double-stranded DNA (dsDNA) was prepared by mixing an equal amount of two complementary oligonucleotides in 25 mM Tris and 150 mM NaCl at pH 7.0, heating to 95°C for 5 min and cooling slowly to room temperature. The concentration of DNA was measured by ultraviolet absorbance at 260 nm with an ND-100 UV-Vis spectrophotometer (NanoDrop Technologies, Inc.)
NMR spectroscopy data acquisition and assignments
Samples for NMR experiments contained 0.5–0.6 mM protein in 20 mM sodium phosphate, pH 7.0, 50 mM NaCl, 1 mM DTT, 0.01% NaN3 and 10 or 100% D2O. Conventional two-dimensional (2D) and three-dimensional (3D) heteronuclear NMR data were collected using uniformly 15N- and 13C/15N-labeled samples (26). All NMR experiments were performed on Bruker Avance 600 and 700 MHz spectrometers equipped with a cryoprobe at 298 K. Assignments of the protein backbone 15N, 1HN, 13Cα, 13Cβ, 13C′ and Hα chemical shifts of FKBP25 were carried out based on HNCACB, CBCA(CO)NH, HNCA, HN(CO)CA, HNCO, HN(CA)CO and HNHA spectra. The chemical shifts of side-chain resonances were obtained from a combination of (H)CC(CO)NH, H[(CC(CO)]NH, 1H-13C-HCCH-TOCSY, 1H-15N-NOESY-HSQC (τm 100 ms) and 1H-13C-NOESY-HSQC (τm 100 ms) spectra. 1H resonances of free DNAYY1 oligomer were assigned using a combination of homonuclear 2D TOCSY and NOESY (τm 200 ms) recorded in 90% H2O/10% D2O or 99.9% D2O conditions. DNA assignments in the FKBP25–DNAYY1 complex were obtained from 2D-F1, F2-[13C/15N]-filtered NOESY spectra (27–30). Intermolecular NOEs between FKBP25 and DNAYY1 were assigned from 3D-13C F1-filtered F3-edited NOESY spectra recorded in D2O (27–30). All spectra were processed with NMRPipe (31) and analyzed using SPARKY. The assigned chemical shift values of FKBP25 have been deposited in the Biological Magnetic Resonance Bank (BMRB ID: 19551) (25). To measure residual dipolar coupling (RDC) constants, poly (ethylene glycol)/alcohol mixtures were used as alignment media for the preparation of anisotropic sample condition as described previously (32). Briefly, 50 μl of C12E5 were mixed in 530 μl of NMR buffer containing 90% H2O and 10% D2O for stock solution preparation. 1-Hexanol was gradually added in 2 μl increments with vigorous shaking to a final molar C12E5:1-hexanol ratio of 0.96. After 1 h of resting at room temperature, air bubbles were removed by centrifugation at 5000 × g for several minutes. For the measurement of RDC, 300 μl of the C12E5:1-hexanol stock solution was added to 200 μl of protein solution. The final concentration of the C12E5:1-hexanol mixture in the NMR sample was about 5% (wt/wt). One-bond N-NH RDC constants were measured using 2D 1H-coupled IPAP 1H-15N-HSQC spectra (33) with 512 complex t1 (15N) points and 128 scans per t1 increment for both isotropic and anisotropic conditions. The data analysis and calculation of the alignment tensor were performed using REDCAT software (34).
Structure calculation and refinement of human FKBP25
Solution structures of the human FKBP25 were calculated by simulated annealing in torsion angle space with a combination of the programs CYANA 2.1(35) and CNS 1.2 (36). Nuclear overhauser effect (NOE) distance constraints were derived by analyzing 1H-15N-NOESY-HSQC (100 ms mixing time) and 13C-edited NOESY-HSQC (100 ms mixing time) spectra of uniformly 15N- or 13C/15N-labeled samples of FKBP25. The secondary structure was predicted by the TALOS+ program (37) based on the results of the analysis of chemical shifts of the main-chain N, HA, CA and C atoms and sequential (|i-j| = 1), short range (|i-j| < 5) NH–NH and NH-aliphatic contacts on a 1H-15N-NOESY-HSQC spectrum. Dihedral angle (phi, psi) restraints were also calculated from chemical shifts using TALOS+ and hydrogen bond restraints were obtained based on the protein structure during structure calculations. NOE cross-peaks on NOESY spectra were classified based on their intensities and were applied with an upper distance limit of 3.0 Å (strong), 3.5 Å (medium), 5.0 Å (weak) and 6.0 Å (very weak), respectively. An additional 0.5 Å was added for NOEs that involved methylene and methyl groups. Upper distance limit for the inter-domain NOE contacts between the N-terminal HLH and C-terminal FKBD were set at 6.5 Å. A total of 200 conformers were generated as initial structures by CYANA 2.1 from 5836 NOE and 281 backbone dihedral angle constraints. After calculation, initial conformers were sorted by target function values and the lowest 100 conformers were selected for further refinement using CNS 1.2. One hundred and thirty-six backbone hydrogen bonds were identified on the basis of initial structures and 128 1DNH RDC constraints were included in the final stage of the calculation. The final structure was refined using a simulated annealing protocol with a combination of torsion angle space and cartesian coordinate dynamics as described previously (38). Finally, 20 structures were selected by their total energy values for display and structural analysis. MOLMOL (39) and PyMOL (40) programs were used for structure visualization and PROCHECK-NMR and Protein Structure Validation Software suite were used for structure validation (41,42). The 20 NMR ensemble structures have been deposited in the PDB ID: 2MPH.
NMR relaxation measurement
To study dynamic behavior of FKBP25, 15N relaxation NMR data were recorded at 298 K for 0.5 mM of 15N-labeled FKBP25 on a 700 MHz NMR spectrometer. Amide 15N relaxation data of R1, R2 and heteronuclear 1H-15N-NOE data experiments were performed as described (43). R1 data were measured with eight different relaxation delays 5, 65, 145, 246, 366, 527, 757 and 1148 ms, and R2 data were obtained by using 10 different relaxation delays of 8.4, 25.1, 41.8, 58.6, 75.3, 100, 120 and 142 ms. Duplicated time points were used for estimation of the error. Steady state heteronuclear 1H-15N -NOE spectra were recorded with and without 3 s of 1H proton saturation. The relaxation rates and error estimation were determined by using Sparky and the correlation time (τc) for each domain was calculated TENSOR2 (44).
NMR titration of FKBP25 with DNA or peptide
NMR samples for DNAYY1 was prepared in buffer containing 25 mM Tris, pH 7.0, 150 mM NaCl and 10% D2O while for YY1 peptide or YY1–DBD we used same buffer supplemented with 0.1 mM ZnCl2. Molecular interactions of full-length FKBP25 with DNAYY1, YY1 peptide or YY1–DBD were studied by 2D TROSY-HSQC spectroscopy (45) using 15N-labeled FKBP25 on a Bruker Avance 700 spectrometer at 298 K. The weighted chemical shift perturbations (CSPs) for backbone 15N and 1HN were calculated by the formula Δδ = [(Δ1HN)2 + (Δ15N/5)2]0.5 where Δ1H and Δ15N are change in chemical shift of 1H and 15N respectively, upon DNA or YY1 peptide binding.
Paramagnetic relaxation enhancement (PRE) experiment
To observe the intermolecular interaction between FKBP25 and DNA, oligonucleotides with dT-EDTA at positions 5 and 27 were purchased from Sigma-Aldrich (Singapore). The oligonucleotides were annealed with their respective unlabeled strand to generate two types of dsDNA denoted as DNA-1 (labeled at dT5) and DNA-2 (labeled at dT27) (Figure 6C). The dT-EDTA-labeled dsDNAs were mixed with equal amounts of Mn2+ or Ca2+ to achieve a paramagnetic or diamagnetic state respectively, and free Mn2+ or Ca2+ was subsequently removed on a PD-10 column. For paramagnetic relaxation enhancement (PRE) measurements, NMR samples (0.15 mM) were prepared by mixing 15N-labeled FKBP25 with a DNA-Mn2+ or DNA-Ca2+complex in a 1:1 molar ratio. 15N TROSY-HSQC spectra were acquired on a Bruker 600 MHz NMR spectrometer equipped with a cryoprobe. The peak intensities of paramagnetic and diamagnetic states were measured and the intensity ratios of the paramagnetic to diamagnetic state (Ipar/Idia) were calculated.
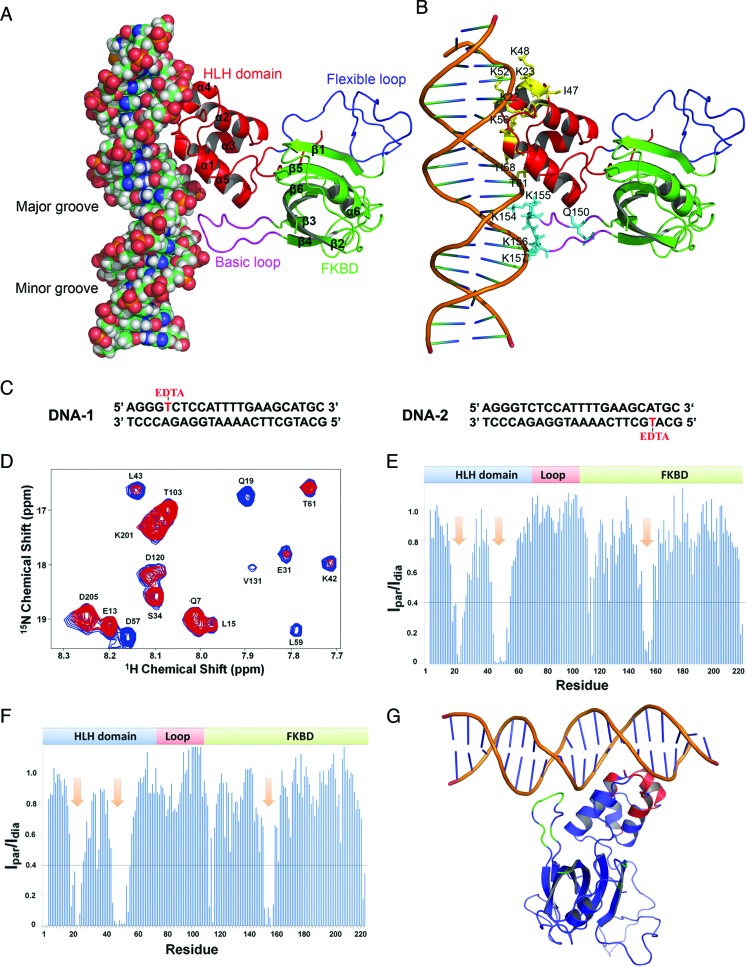
Figure 6.
Structural model of the FKBP25–DNA complex. (A) FKBP25–DNA complex structure model obtained from HADDOCK docking. The HLH domain, flexible loop, FKBD and basic loop are shown in red, blue, cyan and purple, respectively). (B) The FKBP25–DNA model with the residues of FKBP25 important for the interaction with DNA. DNA and FKBP25 are presented in a cartoon representation and residues are shown in a stick representation in yellow for HLH and cyan for basic loop. (C) Sequences of two modified DNAs with one strand having EDTA-labeled thymine either at position 5 (denoted as DNA-1) or at position 27 (denoted as DNA-2). Thymines with EDTA labeling are shown in red. (D) A section of overlaid 1H 15N TROSY-HSQC spectra of the FKBP25–DNA-1 complex obtained in paramagnetic (in red) and diamagnetic (in blue) states by chelating labeled EDTA with Mg2+ or Ca2+, respectively. (E and F) The plot depicts the ratio of peak intensities of all residues from paramagnetic to diamagnetic states for FKBP25 in complex with DNA-1 or DNA-2 respectively. The arrows in the plot show a stretch of residues that are important for DNA binding. (G) The effect of PRE on the FKBP25–DNA model; the residues with an intensity ratio <0.4 are colored blue or red for the HLH domain and FKBD, respectively. Refer online version for color coding.
Isothermal titration calorimetry (ITC) experiment
The binding of the 23-bp double-stranded DNAYY1 with FKBP25 and YY1–DBD were analyzed by isothermal titration calorimetry (ITC) experiments carried out on a MicroCal iTC200 (MicroCal Inc., Northampton) at 25°C. For FKBP25–DNAYY1 binding study, 100 μM DNAYY1 was titrated into 25 μM FKBP25 or into the ITC buffer containing 25 mM Tris and 150 mM NaCl at pH 7.0. For each titration, 1 μl of DNA was injected 24 times with a time interval of 180 s and stirring speed was maintained at 500 rpm. The reference power was 7 μcals−1. For YY1–DBD and DNAYY1 interaction, 200 μM dsDNAYY1 was titrated into 25 μM YY1–DBD by mixing 2.5 μl of DNA per injection for 16 injections. The time interval for each injection was 150 s and the stirring speed was fixed to 500 rpm. The YY1–DBD and DNAYY1 were prepared by dialyzing samples into aliquots from a common stock buffer containing 25 mM Bis-Tris, pH 7.0, 150 mM NaCl and 0.1 mM ZnCl2 to avoid any signal obtained from buffer mismatch. For the buffer blank experiments, DNAYY1 was titrated into the same buffer. The data collected from all of the ITC experiments were integrated using MicroCal Origin 5.0 and signals from blank were subtracted. The data was fitted to a one-site binding model (due to mixing artifacts, the heat associated with the first peak was excluded from the data analysis).
Tryptophan quenching experiment
To confirm the binding of 23-bp DNAYY1 with FKBP25 and its mutants, change in the intrinsic fluorescence of the tryptophan moieties of FKBP25 were recorded using a Cary Eclipse fluorescence spectrophotometer (Varian, Inc.). A total of 5 μM of human FKBP25, K22A, Q150E or K157A was titrated with DNAYY1 and fluorescence intensity was recorded until the binding reached to saturation. The sample was excited at 290 nm and emission spectra were recorded between 300 and 420 nm at 25°C. Relative fluorescence intensity, which was obtained by [(F0-F)/F0] where F0 and F is the fluorescence intensity in absence and presence of DNA, was plotted against the DNA concentrations. The binding affinity Kd was determined by Origin pro software using ligand depletion model.
Gel retardation assay
A total of 300 ng of supercoiled plasmid DNA (pSUMO) was mixed with increasing concentrations of FKBP25 in 20 mM phosphate buffer and 150 mM NaCl, pH7.0. The used DNA:FKBP25 molar ratios were 1:0, 1:25, 1:125 and 1:250. The FKBP25–DNA mixture was also prepared with different salt concentrations ranging from 0 to 1600 mM NaCl. The FKBP25–DNA mixture was incubated at room temperature for 30 min and loaded onto a 1% agarose gel. ssDNA was prepared by linearizing pSUMO plasmid DNA (by digesting with BamH1 restriction enzyme) followed by heating at 100°C and immediate cooling to 0°C as previously described (46).
HADDOCK docking
With the inputs of NMR titration, mutational analysis and intermolecular interactions between DNA and FKBP25 derived from isotope filtered NOESY experiment, we performed docking on a HADDOCK web server (47) and built a model for the FKBP25–DNA complex. The sequence of first 20 residues of 23-bp DNAYY1 used in this study were same as the sequence of 20-bp dsDNA used in crystal structure of YY1–DNA complex (PDB ID: 1UBD). We used the structure of DNA from the YY1–DNA complex and the solution structure of the full-length human FKBP25 for docking experiments. The active residues of FKBP25 were defined by combinations as those showing CSPs larger than the averages (Δδ > 0.05), the presence of intermolecular NOEs between FKBP25 and DNA and mutagenesis data with solvent accessible surface area larger than 30% for either main-chain or side chain atoms calculated with NACCESS and MOLMOL program. We defined Lys22, Lys23, Lys42, Lys48, Gln150, Lys156 and Lys157 as active residues for protein. In the case of DNAYY1, based on ambiguous or unambiguous intermolecular NOE information, we selected nucleotides A10, C23, T24, T25, C26, G32, A34 and G35 as active residues. Passive residues for both FKBP25 and DNAYY1 were automatically picked by the HADDOCK server program. These experimental restraints were used as input for the HADDOCK program and default parameters were used for docking. The resulting structures were clustered by a default cutoff value (7.5 Å). HADDOCK clustered 153 structures in 10 clusters, which represent 76.5% of the water-refined models HADDOCK generated. Finally, based on the HADDOCK score and total energy, the best model from the first cluster was selected as the final model of the FKBP25–DNA complex. The quality of the HADDOCK-derived complex models was checked using PROCHECK (48).
RESULTS
Solution structure of full-length human FKBP25
The solution structure of the full-length human FKBP25 was determined based on a total of 5972 NMR-derived distance restraints, 281 dihedral angle restraints and 128 RDC constraints. The ensemble of 20 low-energy structures is shown in Figure 1B. Excluding the 35 residues (Gly74-Pro108) in the flexible loop between the N-terminal HLH domain and C-terminal FKBD, the root-mean-square deviation (RMSD) values relative to the mean coordinate of 20 conformers were 0.67 ± 0.25 Å for the backbone atoms and 1.01 ± 0.26 Å for all heavy atoms (Table 1). The topology of FKBP25 showed that the HLH and FKBD are connected by a long and unstructured flexible linker from Gly74-Pro108 (Figure 1B and C). The N-terminal domain consists of five α-helices (Figure 1D–F). The C-terminal domain shows a canonical FKBD fold which consists of six anti-parallel β-strands and a short central α-helix (Figure 1G–I). The calculated RMSD values for the Cα trace between our NMR and previously reported structures were 1.40 Å and 1.87 Å for HLH and FKBD, respectively (Supplementary Figure S1b and S1c).
Table 1. Structural statistics for FKBP25.
| Number of NOE constraints | |
|---|---|
| All | 5835 |
| Intra residues |i-j| = 0 | 1008 |
| Sequential, |i-j| = 1 | 1568 |
| Medium-range, 1<|i-j|<5 | 1011 |
| Long-range, |i-j|> = 5 | 2248 |
| Number of hydrogen bond constraints | 136 |
| Number of dihedral angle constraints | 281 |
| Number of RDC constraints (1DHN) | 128 |
| Number of constraint violations (>0.5Å) | 0 |
| Number of angle violations (>5°) | 0 |
| Number of RDC violations (>1.0Hz) | 0 |
| CNS energy (kcal.mol-1) | |
| Etotal | 133.58 ± 1.58 |
| ENOE | 6.73 ± 0.53 |
| Ecdih | 0.62 ± 0.14 |
| Ebond + Eangle + Eimproper | 91.86 ± 1.40 |
| Evdw | 33.29 ± 1.41 |
| RMSD for Residue 2–73,109–223 to mean (excluding flexible loop 74–108)1 | |
| Backbone | 0.67 ± 0.25Å |
| Heavy Atoms | 1.01 ± 0.26Å |
| RMSD for N-terminal helix-loop-helix domain (2–73)1 | |
| Backbone | 0.55 ± 0.13Å |
| Heavy atoms | 0.86 ± 0.16Å |
| RMSD for C-terminal FK506 binding domain (109–223)1 | |
| Backbone | 0.60 ± 0.32Å |
| Heavy atoms | 1.00 ± 0.34Å |
| Ramachandran plot (excluding flexible loop 74–108)2 | |
| Most favored regions | 83.1% |
| Additionally allowed regions | 14.5% |
| Generously allowed regions | 2.4% |
| Disallowed regions | 0.0% |
1Number of structures used in RMSD calculation was 20.
2Ramachandran analysis was performed using PROCHECK-NMR program.
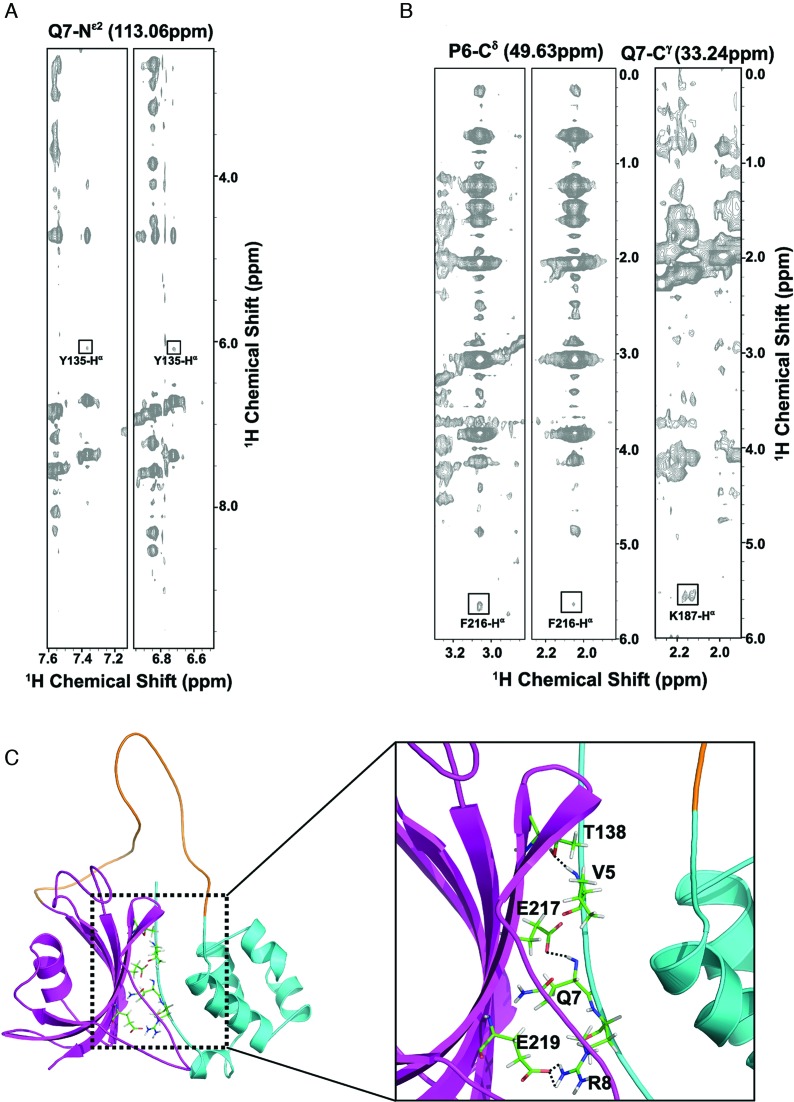
As there are no known FKBP family proteins with structural features showing inter-domain interaction when interacting with their substrates or ligands, the domain–domain contact between the N-terminal HLH and C-terminal FKBD of FKBP25 is an interesting structural feature among the family. The FKBP25 structure revealed that residues located in the N-terminal HLH domain (Val5-Arg8) interact with residues of the C-terminal β2 and β6, which we confirmed by observing a series of inter-domain NOEs between HLH and FKBD from 15N-edited NOESY-HSQC and 13C-edited NOESY-HSQC spectra (Figure 2A and B). The backbone amides of residues Val5 and Gln7 at the N-terminus form hydrogen bonds with the side-chain oxygen atoms of Thr138 of β2 and Glu217 of β6, respectively (Figure 2C). In addition, an ionic interaction between Arg8 and Glu219 was also observed (Figure 2C). Furthermore, truncation of the C-terminal FKBD caused slight perturbations of the chemical shifts on the N-terminal HLH domain, indirectly indicating the presence of domain–domain interactions between the HLH and C-terminal FKBD (Supplementary Figure S2). The connecting loop between these two domains is unstructured in solution as demonstrated by the lack of medium and long-range NOEs and also random coil secondary chemical shifts and dynamic properties determined in a heteronuclear NOE experiment. These data suggested that the N-terminal HLH and C-terminal FKBD cooperate with each other to perform their molecular functions.
Figure 2.
Molecular interaction between the N-terminal HLH and C-terminal FKBD domains. (A) Representative strips of the 15N-edited three-dimensional (3D) NOESY spectrum of FKBP25 showing NOE cross-peaks between residues on the N-terminal (Gln7) and the C-terminal FKBD (Tyr135). (B) Representative strips of the 13C-edited 3D NOESY spectrum of FKBP25, with NOE cross-peaks between residues on the N-terminal HLH (Pro6, Gln7) and the C-terminal FKBD (Phe216, Lys187). (C) A ribbon representation of the interaction between the HLH and FKBD. The HLH domain and FKBD are highlighted in cyan and pink respectively. Crucial residues forming hydrogen bonds (Val5, Gln7, Thr138 and Glu217) and electrostatic interactions (Arg8, Glu219) are shown as sticks and hydrogen bonds are indicated by dashed lines. Refer online version for color coding.
To study internal motion and the overall domain dynamics, we measured 15N relaxation data for FKBP25 (Supplementary Figure S3). R1, R2 and heteronuclear data corroborate that FKBP25 comprises of well ordered N- terminal HLH and C-terminal FKBD connected by a flexible linker. The average tumbling correlation time for overall residues in the N-terminal HLH domain is τc ≈ 7.5 ns, which is significantly larger than the estimated value of τc ≈ 5.9 ns, using HYDRONMR software (49). For C-terminal FKBD, the average τc of 11 ns is slightly higher than the theoretical value, τc ≈ 10 ns. These values suggest that the rotational diffusion of both domains are coupled and do not tumble independently. However 15N R2/R1 ratios and different correlation times of the HLH and FKBD are significantly different, indicating that the interaction between both domains is not strong. These results suggest that HLH and FKBD of FKBP25 are in between the two extreme dynamics models. The first model is single rigid tumbling of both domains, and the second one is each two domain is dynamically independent. 15N relaxation data and observation of weak interdomain NOEs support FKBP25's interdomain flexibility and weak domain–domain interaction between both HLH domain and FKBD.
FKBP25 binds to DNA
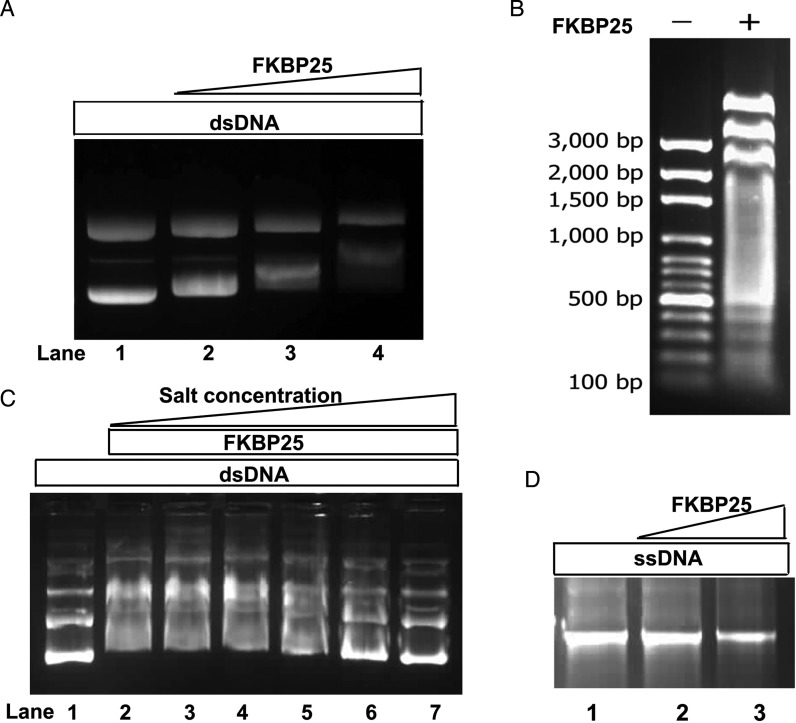
The electrostatic surface potential enabled us to visualize a patch of positively charged residues on both of the domains of FKBP25 (Figure 1F and I), which indicated that FKBP25 is a potential nucleic acid-binding protein. In order to study the interaction of FKBP25 with nucleic acid we first examined the DNA-binding ability of FKBP25 in a gel retardation assay by employing supercoiled plasmid DNA (pSUMO). A significant retardation of the migration of plasmid DNA was observed on an agarose gel when DNA was mixed with human FKBP25 (Figure 3A). The retardation was clearly detectable when the molar ratio of DNA:FKBP25 was close to 1:125. We obtained similar results with different DNA sequences (Figure 3B), suggesting that FKBP25 binds to double-stranded DNA (dsDNA) in a sequence-independent manner in vitro. As sequence-independent protein–DNA binding is mainly mediated by ionic interactions, we examined the effect of increase in salt concentration on FKBP25-mediated DNA binding, which revealed that the gel retardation decreases with an increase in salt concentration (Figure 3C). However, no gel retardation was observed when single-stranded DNA (ssDNA) was incubated with FKBP25 (Figure 3D). These findings suggest that FKBP25 has a preferred DNA binding ability to dsDNA over ssDNA.
Figure 3.
Gel mobility shift experiments showing binding of FKBP25 with DNA. (A) A gradual increase in band shift in purified plasmid DNA (300 ng) mixed with increasing concentrations of FKBP25 (molar ratio 1:0, 1:25, 1:125 and 1:250; lane 1–4 respectively) is shown. (B) The retardation of DNA fragment migration after FKBP25 binding is shown; DNA fragments of all sizes (ranging from 3 kb to 100 bp) display reduced migration. (C) Gel retardation assay of the FKBP25-plasmid complex in the presence of increasing concentrations of NaCl shows that the interaction of FKBP25 with plasmid DNA decreases with increasing concentration of NaCl. Lane 1 has DNA alone while lane 2–7 had DNA incubated with FKBP25 in 1:125 molar ratio and increasing concentration of NaCl (0, 100, 200, 400, 800 and 1600 mM NaCl respectively). (D) Gel shift assay of single-stranded plasmid DNA alone (lane 1) and with FKBP25 (lane 2, molar ratio 1:1 and lane 3, molar ratio 1:250) showing no binding to ssDNA.
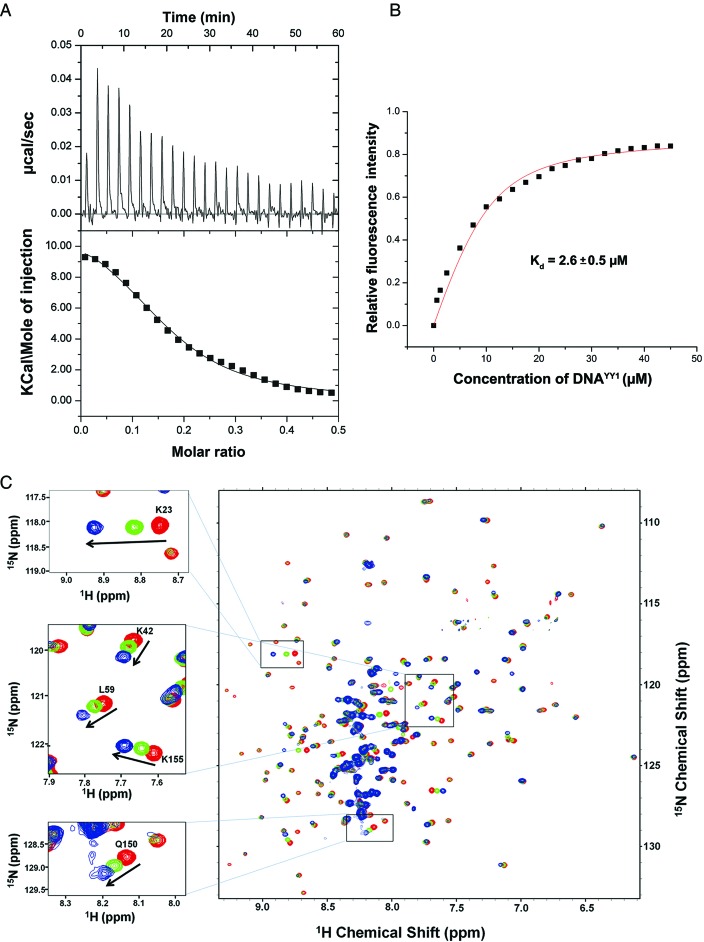
Since FKBP25 recognizes large dsDNA segments (plasmid DNA or linear DNA fragments), we wanted to test FKBP25's ability to recognize oligonucleotides. To perform biophysical characterization of FKBP25–nucleic acid interaction and also to study the possible ternary complex formation with YY1 protein, a transcription factor known to bind FKBP25 (15), we decided to use a 23-bp dsDNA oligonucleotide (referred to as DNAYY1). The sequence of DNAYY1 used here is the same (except for the three additional nucleotides at its 3′ end) as that which has been shown to bind YY1 earlier (15), which helps us to make more reliable observations on the ternary complex formation. To this end, we quantitatively examined the molecular interaction between FKBP25 and DNAYY1 using ITC, which indicated an upward trend of the titration peaks and the positive resultant integrated heats (Figure 4A), confirming the physical interaction between these two. The ΔS were observed to be positive which indicates that it's entropy driven interaction. The estimated Kd value for the DNA binding was 1.23 ± 0.15 μM and the thermodynamic parameters of interaction have been summarized in Supplementary Table S1. We also examined the FKBP25–nucleic acid interaction by tryptophan fluorescence quenching experiment and showed that the fluorescence emission reduced with increasing concentrations of DNA. The estimated binding affinity of FKBP25 with DNA was 2.6 ± 0.5 μM which is comparable with the value obtained by ITC (Figure 4A and B).
Figure 4.
Characterization of FKBP25–DNA binding by isothermal titration calorimetry (ITC), fluorescence quenching and NMR titration. (A) ITC results of the FKBP25 titrated with DNA; the raw data of heat changes (upper panels) and the process curve fit (lower panel) are shown for 0.1 mM of 23 bp dsDNAYY1. (B) The relative fluorescence intensity [(Fo-F)/Fo] of 5 μM FKBP25 is shown as function of DNAYY1 concentration, F and F0 are the fluorescence intensities at 342 nm in absence and presence of DNA respectively. Plot was used to estimate binding constant of FKBP25–DNAYY1 interaction. (C) The overlaid TROSY-HSQC spectra of FKBP25 without (red), with 1:1 (green) or 1:2 (blue) dsDNAYY1; the spectra show a gradual shift in some cross-peaks on DNA binding. CSPs of some residues are zoomed and shown in a box. Refer online version for color coding.
Both the N-terminal HLH domain and C-terminal FKBD interact with DNA
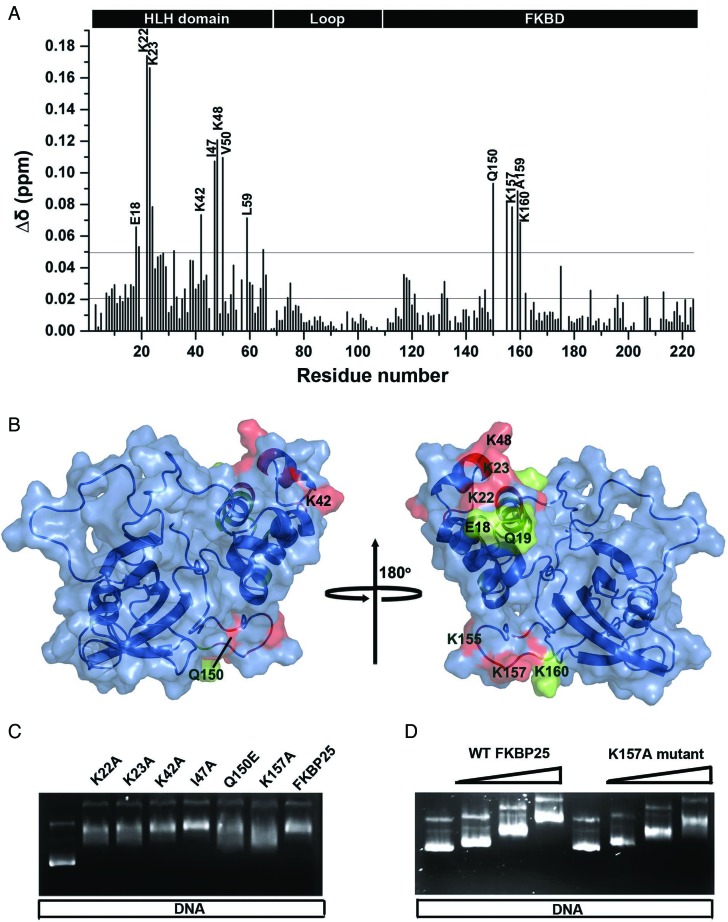
The gel retardation, ITC and trytophan fluorescence quenching results indicated that FKBP25 can interact with dsDNA. To confirm this binding and to further probe the molecular interface of FKBP25–DNA complex, we performed NMR titration of FKBP25 with different molar ratios of DNAYY1. As seen in the Figure 4C, the titration caused shifting and broadening of some of the cross peaks in the 1H-15N HSQC spectrum confirming the binding of FKBP25 with DNA. The significant changes (>0.05) in chemical shift were observed for the backbone resonances of Glu18, Gln19, Lys22, Lys23, Asp24, Lys42, Ile47, Lys48, Val50, Leu59, Gln150, Lys155, Lys157, Ala159 and Lys160 (Figure 5A and B). These residues are either directly involved in the interaction with DNA or their chemical shifts are affected indirectly by the interaction. Except for five residues (Gln150, Lys155, Lys157, Ala159 and Lys160) from FKBD, all of the other residues showing notable chemical shift changes belonged to the N-terminal HLH domain, which signifies the role of the HLH domain in FKBP25–DNA interactions. Interestingly, the five residues from the C-terminal FKBD are located in its 40s loop (referred as basic loop), suggesting an involvement of this basic loop in these interactions.
Figure 5.
DNA-binding surface in FKBP25 revealed by NMR. (A) Weighted CSPs for the 15N and 1H resonance of FKBP25 after DNA binding; the lower black line represents the average CSP while upper black line is the sum of average CSPs and standard deviation. (B) DNA-binding surface of FKBP25 mapped by CSP results represented in the surface representation; the residues having CSP more than 0.07 are represented in red while those showing CSP between 0.07 and 0.05 are shown in green. (C) Gel shift assay of the wild-type FKBP25 and mutants. Lane 1 had pSUMO plasmid DNA alone while other lanes had plasmid DNA incubated with wild-type or mutant FKBP25 in 1:250 molar ratio. (D) Gel retardation of the plasmid DNA in increasing concentrations of wild-type or K157A mutant FKBP25 in DNA to protein molar ratio 1:0, 1:25, 1:125 and 1:250. Refer online version for color coding.
The mapping of the CSP's from the NMR titration onto the surface of FKBP25 revealed two major patches, one on HLH domain and the other on the basic loop of FKBD (Figure 5B). We also performed NMR titration of FKBP25 with DNAYY1 at 1:1 molar ratio at two different salt concentrations (50 and 150 mM). The overlay spectra of free FKBP25 with FKBP25–DNAYY1 complex, either in the buffer with 50 or 150 mM salt concentration, showed that CSPs of the cross peaks reduced when salt concentration was increased, confirming that FKBP25–DNA interaction is salt dependent (Supplementary Figure S4). In order to investigate the interaction of FKBP25 with ssDNA, we also performed a HSQC experiment in the absence and presence of ssDNAYY1. The results from NMR titration showed that there were negligible CSPs in any of the residues. These data confirm that the FKBP25-ssDNA interaction is feeble (Supplementary Figure S5), which corroborates the gel shift assay results (Figure 3D).
Mutational studies reveal critical amino acids for the FKBP25–DNA interaction
NMR titration indicated that some residues of FKBP25 show significant CSPs upon DNA binding. Anyhow few of these residues could be directly involved in binding while others could be indirectly involved through conformational changes brought about in FKBP25. To determine which interaction provides the greatest contribution to the observed affinity, we site-specifically mutated residues (K22A, K23A, K42A, I47F, Q150E and K157A) that showed significant CSPs, and examined their role in DNA binding. Analysis of the DNA migration patterns in the presence of the mutants indicated that all mutants, except for I47F, were critical for DNA interactions (Figure 5C and D). To further validate the result, we also performed tryptophan fluorescence quenching experiments using these mutants. FKBP25, K22A, Q150E and K157A were titrated with increasing concentration of DNAYY1. The change in fluorescence intensity at 342 nm was plotted against different DNA concentrations and then the Kd were estimated for wild-type and mutant proteins. Binding affinities of DNA with FKBP25 and its mutant have been summarized in Supplementary Table S2. Consistent with the gel shift assay result (Figure 5C and D), the mutants showed almost 4-fold reduced affinities to DNA with respect to the wild-type FKBP25, confirming that residues K22, Q150 and K157 are critical in the DNA recognition by FKBP25 (Supplementary Figure S6a and S6b).
Structural basis of nucleic acid recognition by FKBP25
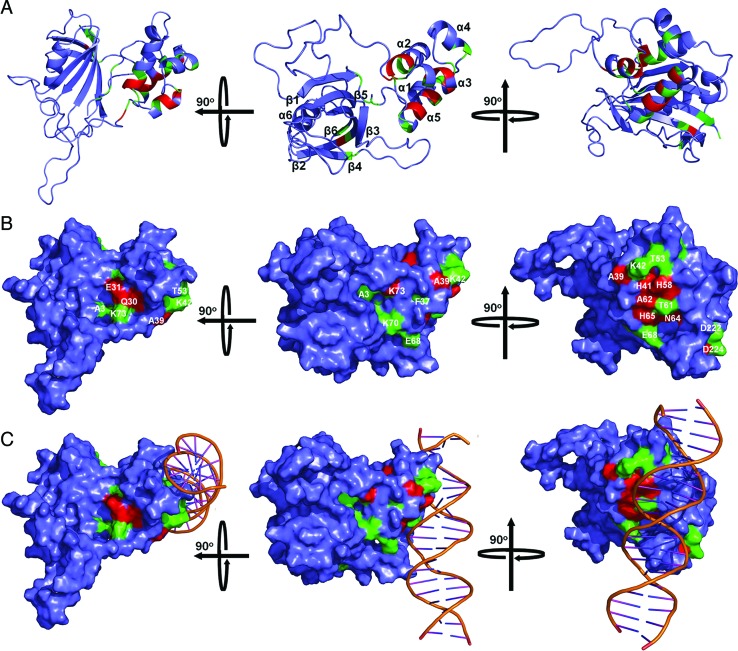
To understand the structural basis of nucleic acid recognition by FKBP25, we collected NMR data of FKBP25 in the presence of 23-bp DNAYY1, which revealed overlapping resonances and lack of sufficient unambiguous intermolecular NOE constraints. Despite the severely overlapped DNA resonances in the protein–DNA complex, a few unambiguous intermolecular NOEs were observed in 3D-13C F1-filtered F3-edited NOESY spectra taken with 13C/15N labeled protein and unlabeled DNAYY1: Lys22 CϵH to H2’/H4’ of T31 and H5 of C32 (Supplementary Figure S7 and Supplementary Table S3). Thus, in this study we used selected number of intermolecular NOEs, HSQC titration and mutational data, and generated an FKBP25–DNA complex model by HADDOCK (Supplementary Table S3). The FKBP25–DNAYY1 structure model revealed that both the N-terminal HLH domain and the C-terminal FKBD bind directly to DNA (Figure 6A). Consistent with the mutational study, residues Lys22, Lys23 and Lys42 interact with DNA through salt bridges formed with the phosphate back bone of DNA indicating that these side chains are required for interaction with DNA (Figure 6B). From the C-terminal FKBD, residues like Lys154, Lys155 and Lys156 makes direct salt bridge contact with the phosphate backbone of DNAYY1 (Figure 6B). Other residues that showed significant CSPs on DNA binding were present in close proximity to the DNA, suggesting that the FKBP25–DNA structural model was consistent with the NMR titration data. The structure also confirmed that the basic loop of FKBD plays an important role in DNA-binding. Sequence alignment of human FKBP25 with its homologs from yeasts and plants or other human FKBPs showed that the highly charged basic loop with a patch of lysine residues is exclusively present only in human FKBP25 (Supplementary Figure S8). The structure of the full-length FKBP25 showed that this long and unique basic loop is present in close proximity to the N-terminal domain (Figure 6A) and serves as a novel motif to aid nucleic acid binding by FKBP25.
Paramagnetic relaxation enhancement (PRE) measurements
To validate the model of the FKBP25–DNA complex generated by HADDOCK, we performed PRE experiments. We prepared two modified DNAs, which have the same sequences as DNAYY1, each DNA having one thymine labeled with EDTA. DNA-1 had EDTA labeling at position 5 while DNA-2 had EDTA labeling at position 27 (Figure 6C). Paramagnetic and diamagnetic states were obtained by generating DNA in complex with either Mn2+ or Ca2+, respectively. Then 15N TROSY-HSQC spectra were acquired for FKBP25–DNA-1 and FKBP25–DNA-2 complexes in paramagnetic and diamagnetic states. To check whether spin labeling had an effect on FKBP25–DNA binding, the spectra from the complex with or without the spin label were superimposed. Because these superimposed spectra fit well, we concluded that spin labeling did not cause any changes in FKBP25–DNA binding (data not shown). Moreover, the overlaid spectra of the FKBP25–DNA-1 complex in paramagnetic and diamagnetic states showed that the intensities of some of the peaks decreased in the paramagnetic state (Figure 6D). Later, the ratios of the peak intensities for paramagnetic to those for diamagnetic states (Ipar/Idia) were estimated and plotted for all residues (Figure 6E and F). PRE measurements for the FKBP25–DNA-1 complex showed that the Ipar/Idia ratio was significantly attenuated for residues Glu18-Ile26 from α1/α2 loop and α2 helix, Gly45-Asp57 from α3/α4 loop, α4/α5 loop and α4 helix, Gln150-Lys160 from β3/ β4 loop, the basic loop of FKBD (Figure 6E). In the paramagnetic state, the peaks of some residues, such as Lys22, Lys23, Lys48, Lys52, Thr53 and Ala159, completely disappeared, indicating that these residues are located close to Mn2+ bound to DNA-1(Figure 6G). The PRE results very well supported the FKBP25–DNA model, because all of the residues that showed PRE effects were positioned in close proximity to DNA in the FKBP25–DNA complex model (Figure 6B). The Lys22, Lys23, Lys42 and Lys48 residues showed maximum CSPs on DNA binding (Figure 5A). These residues showed ionic interactions with the DNA phosphate backbone, and in support of that model, the PRE experiments also showed maximum PRE effects for these residues. The peaks of FKBD residues Gln150-Lys160 were significantly attenuated, which confirmed that FKBD, through its basic loop, was also involved in binding with DNA, thus supporting the FKBP25–DNA model. Some residues from FKBD, but not those belonging to the basic loop, such as Lys118, Ile223 and Asp224, also showed significantly reduced peaks under paramagnetic conditions, which is logical because these residues are located close to the basic loop of FKBD. Surprisingly, the results of PRE experiments we obtained for DNA-1 and DNA-2 were found to be same (Figure 6E and F). This observation could be explained by the fact that FKBP25 binds to DNA in a sequence-independent manner and any dT-EDTA-labeled end of DNA (either DNA-1 or DNA-2) could bind to any side of protein (either HLH domain or FKBD) and hence no difference was seen in the PRE results for binding of DNA-1 and DNA-2 to FKBP25. Thus, we could conclude that any end of DNA could bind with the HLH domain while the other end could bind with the FKBD of FKBP25.
Transcription factor YY1-binding surface on FKBP25 revealed by NMR titration
YY1 belongs to the zinc finger family possessing four zinc fingers that form the DNA binding domain (referred to as YY1–DBD) (Supplementary Figure S1d). YY1 interacts with the HLH domain of FKBP25 through residues 300–333, which resides in the first zinc finger of YY1 (15). In order to map the YY1 binding site on FKBP25, NMR TROSY-HSQC experiment of 15N-labeled FKBP25 with either YY1–DBD or YY1 peptide (amino acid residues 300–333) was performed. Both YY1–DBD or YY1 peptide showed similar binding to FKBP25 as the binding shifted the same set of residues on FKBP25 (data not shown), thus we decided to use the YY1 peptide for further NMR titrations. After confirming YY1 peptide binding to FKBP25, we performed titration of FKBP25 with YY1 peptide to find residues gradually shifting upon YY1 peptide binding. The overlaid HSQC spectra of FKBP25 showed increase in CSPs of some of the residues with increasing concentrations of the YY1 peptide (Supplementary Figure S9a). Most of residues that shifted on YY1 peptide binding (Gln30-His32, Leu38-Ala39, Asn64-Leu66 and G74) belonged to the HLH domain (Figure 7A and B; Supplementary Figure S9b), which was consistent with previous report (23). Interestingly, we also found that some residues (His132, Cys133, Leu162 and Asp222) from the C-terminal FKBD also showed CSPs after YY1 peptide binding (Supplementary Figure S9b), making a patch located on the rear side of the FK506-binding pocket facing toward the HLH domain (Figure 7A, middle and right panels). Mapping of YY1-binding site on the FKBP25–DNA complex model suggested that the YY1–FKBP25 interaction may affect the FKBP25-mediated DNA binding or vice versa (Figure 7C). To further understand the relationship between YY1 and DNAYY1 binding to FKBP25, HSQC spectra for FKBP25–DNA–YY1 peptide (1:1:1, 1:1:2, 1:1:5 and 1:1:10 molar ratios) were obtained and compared with the HSQC spectra of free FKBP25, FKBP25–DNA (1:1) and FKBP25-YY1 peptide (1:1). The FKBP25–DNA complex and the FKBP25–YY1 peptide complex showed a shift in residues specific to DNA or YY1 binding respectively. Although the overlaid spectra of FKBP25 and FKBP25–DNA–YY1 (1:1:1) revealed that YY1 could not abolish the binding of FKBP25 with DNAYY1 at a 1:1 molar ratio of DNAYY1 to YY1 peptide. Gradually increase in YY1 molar ratio from 1:1 to 1:10, resulted in a gradual decrease and increase in the CSP of the DNA-binding residues and YY1 binding residues of FKBP25, respectively. At 1:10 molar ratio of DNAYY1 to YY1 peptide, YY1 peptide was able to completely abolish the FKBP25–DNAYY1 interaction by forming an FKBP25–YY1 peptide complex (Supplementary Figure S10). Taken together, the NMR data points to a model where high concentration of YY1 competes with DNA for FKBP25 binding. To further probe the dynamics of FKBP25, YY1–DBD and DNAYY1 we performed several ITC experiments. The result of ITC experiment performed to investigate binding of YY1–DBD to DNAYY1 showed that the interaction is exothermic and enthalpy driven with the estimated Kd of 0.39 ± 0.6 μM (Supplementary Table S1 and Supplementary Figure S11). In this direction we also tried to measure the affinity of FKBP25 with YY1–DBD, but the Kd was not measurable probably due to relatively minimal heat release upon interaction, indicating feeble binding (data not shown). The comparison of binding affinity suggests that DNAYY1 has stronger affinity for YY1–DBD than FKBP25 (almost 3-fold higher binding affinity), while binding affinity of FKBP25 with YY1–DBD domain is weakest among all three binary complexes.
Figure 7.
Mapping of the YY1-binding site in FKBP25. The mapped YY1-binding site in FKBP25 is presented either in a cartoon (A) or in a surface representation (B). The model (in the middle panel) has been rotated 90° either on the horizontal axis (left panel) or on the vertical axis (right panel) to show the complete binding site. The YY1-binding site is indicated and the residues having CSP more than 0.15 are represented in red while those showing CSP between 0.07 and 0.15 are shown in green. (C) Model of the FKBP25–DNA complex showing the YY1-binding sites. A portion of the YY1-binding site overlaps with the DNA-binding site (middle panel) but the major section of the YY1-binding site does not overlap with the DNA-binding site of FKBP25 (left panel and right panel). Refer online version for color coding.
DISCUSSION
The NMR solution structure of FKBP25 displays molecular characteristics with inter-domain interaction between the N-terminal HLH and C-terminal FKBD, which allows nucleic acid recognitions. These structural features could play an important role in the biological actions of the immunophilin in the nucleus, especially in the regulation of epigenetic functions through associations with various types of nucleic acids and chromatin modification enzymes and transcription factors, such as HDAC and YY1.
The full-length FKBP25 structure presented in this study revealed that the N- and C-terminal domains interact with each other through the formation of hydrogen bonds and electrostatic interactions. Further we have shown that FKBP25 interacts with DNA, in a sequence independent manner, mainly through electrostatic forces between the two macromolecules. In particular residues in the α2 (Lys22-His32) and α4 (Ile47-Thr53) helices of the N-terminal HLH domain and the basic loop (Gln150-Ala159) of the C-terminal FKBD are involved in DNA recognition, suggesting that both domains are required for DNA recognition by FKBP25. Our structural model suggests that fourth helix of the HLH domain might form major groove interactions and the basic loop of FKBD cooperate to form interactions with an adjacent minor groove. Interestingly, the key DNA recognition helix α4 was shown to interact with DNA in a tilted angle. Moreover, the basic loop, also known as 40s loop of canonical FKBP family, was found to be notably larger than that of other FKBPs and bears a stretch of lysine residues (KKKKNAK) that appeared to be important for FKBP25-mediated DNA binding and to be a novel molecular function of FKBP family proteins. Because the inter-domain interaction has been shown to be weak, the long and flexible loop between the N- and C-terminal domains might provide structural flexibility for the formation of complexes with their binding partners. The long loop could allow a level of flexibility with regard to DNA binding: maybe the basic loop could bind to the minor/major groove that's not only one turn away from the HLH binding site, but also bind to the groove that is 2 or 3 turns away.
FKBP25 was shown to interact with the transcription factor YY1 (15). Here we confirmed the previous findings (23) in our NMR study and also mapped the YY1-binding site on FKBP25. We showed that higher concentrations of YY1 peptide were required to abolish FKBP25–DNA interaction and form a binary complex with FKBP25, suggesting that FKBP may have differential binding affinities to nucleic acids and YY1. The binding affinity (Kd) of DNAYY1 with YY1–DBD was estimated to be 0.39 μM using ITC, which is comparable with the previously reported values of 0.56 and 0.80 μM, determined by ITC (50) and fluorescence anisotropy measurements (51) respectively. The binding affinity of YY1 to DNA is relatively low (in micro molar range), in comparison to other zinc finger transcription factors which usually show binding affinity in nanomolar range (51). Based on the low binding affinity of YY1, it was suggested that YY1 may require other co-regulator proteins to increase its affinity to DNA. For example, it was shown that the transcriptional activation mediated by YY1 was enhanced by binding to INO80, an adenosine triphosphate-dependent chromatin-remodeling complex (52). Similarly YY1 was also found to interact with FKBP25 and this interaction was shown to be important for increased repression activity of YY1 (15). The increased repression activity was a result of the increased binding affinity of YY1 to DNA by an unknown mechanism. It was also showed that YY1 mainly binds to N-terminal HLH domain through its first zinc finger (residues 300–333). The co-crystal structure of YY1–DNA complex suggested that the second and third zinc fingers are important for DNA sequence recognition and binding, while the first zinc finger is relatively loosely bound to DNA (the first zinc finger makes a single base contact to DNA while other zinc-fingers of YY1 makes multiple contacts). Our structural studies suggest that FKBP25 can also bind both DNA or YY1 zinc finger 1 through different residues of N-terminal HLH domain while the basic loop from FKBD also provides additional binding to DNA and thus FKBP25 could serve as a co-regulator of YY1. In addition, it was also noted that the interacting DNA regions are almost mutually exclusive for FKBP25 (Supplementary Figure S12b) and YY1. Taken all these information together, we proposed a model for the FKBP25–DNA–YY1 ternary complex. YY1 binds to DNA through the zinc fingers 2 and 3, while the zinc finger 1 binds with the N-terminal HLH domain of FKBP25 near the major groove of DNA and both HLH domain and basic loop of FKBD could bind to major and minor groove of the same DNA and thus this ternary complex could stabilize and increase binding affinity of YY1 to the DNA. Similar ternary complex formation was shown before where Pax-5, a paired box family protein, interacts with Ets-1, a transcription regulator and both of these proteins bind with same DNA to form a ternary complex (53).
Previous study has shown that YY1 can bind FKBP25 even in the absence of DNA. Here we propose that FKBP25 could bind to YY1 first (Supplementary Figure S12a) and then this binary complex searches for the DNA binding sequence for YY1 (as both YY1 and FKBP25 has comparable affinity to DNA of 0.39 and 1.20 μM respectively) where it forms a ternary complex in the above mentioned fashion (Supplementary Figure S12b). This ternary complex formation could be necessary for increasing the affinity of YY1 for DNA. Finally, FKBP25 can release itself from the DNA and not necessarily from YY1 as suggested by our NMR titration (Supplementary Figure S10), implying that an increase in YY1 concentration abolishes FKBP25–DNA interaction (Supplementary Figure S12c). Apart from this it is also possible that these two proteins can independently interact with DNA and then form a ternary complex. However, given the limitations of these predicted models more studies are required to further understand the molecular mechanism governing the interactome among these macromolecules forming the ternary complex.
In addition, knockdown of FKBP25 resulted in a notable increase in MDM2 and decrease in p53 and p21 levels (14), suggesting a potential role of FKBP25 in homeostatic regulation of p53–MDM2 interaction. Human FKBP25 interacts with HDAC1/2 (15) and its yeast homolog Fpr3 plays a regulatory role in meiotic recombination checkpoint (18). A recent proteomic study indicated the potential roles of FKBP25 in DNA packaging, pre-messenger RNA splicing and chromatin remodeling (54,55), suggesting a possible link between human FKBP25 and the recognition of nucleic acids. Several proteins, which are known to interact with DNA in a sequence-independent manner, were shown to be involved in DNA repair or have a role in chromatin remodeling (17,46,56–58). Human FKBP25 interacted with the high mobility group II and it serves as a substrate for poly [adenosine phosphate-ribose] polymerase 1, which led to the adenosine phosphate-ribosylation of FKBP25 in response to DNA damage stress (59), suggesting a possible role of FKBP25 in DNA repair and also in the regulation of gene expression. Interestingly, a structural motif characterized by a positively charged surface patch centered on the tilted helix α4 in its N-terminal HLH domain was also present in human HectD1, which is homologous to the E6AP carboxyl terminus domain containing E3 ubiquitin ligase1 (23). Taken together, recent data suggest the involvement of FKBP25 in various nuclear events. Considering its functional versatility, chromatin immunoprecipitation sequencing (Chip-Seq) on FKBP25-bound nucleic acid complexes might provide fundamental insights into molecular regulatory mechanism of nucleic acid recognition by FKBP25 at a genome-wide level.
In conclusion, in this study we present a structural basis of DNA binding by FKBP25 and provide evidence for functional domain–domain cross-talk of FKBP25 and an emerging role of its FKBD in the molecular recognition of nucleic acids. More studies are needed to fully understand the physiological significance of these immunophilin-mediated nuclear interactions.
ACCESSION NUMBER
Twenty NMR ensemble structures of FKBP25 have been deposited in the Protein Data Bank with the code 2MPH.
Supplementary Material
Acknowledgments
We thank Sirano Dhe-Paganon and Michael Sattler for critical reading of the manuscript.
Footnotes
Present address: Ho Sup Yoon, Division of Structural Biology & Biochemistry, School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, 637551, Singapore.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Singapore Ministry of Education AcRF Tier 1 Grant [RG 47/15]; Singapore Ministry of Health NMRC IRG Grant [NMRC/1245/2010]. Funding for open access charge: Singapore Ministry of Education AcRF Tier 1 Grant [RG 47/15]; Singapore Ministry of Health NMRC IRG Grant [NMRC/1245/2010].
Conflict of interest statement. None declared.
REFERENCES
- 1.Ivery M.T. Immunophilins: switched on protein binding domains? Med. Res. Rev. 2000;20:452–484. doi: 10.1002/1098-1128(200011)20:6<452::aid-med2>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 2.Kang C.B., Hong Y., Dhe-Paganon S., Yoon H.S. FKBP family proteins: immunophilins with versatile biological functions. Neurosignals. 2008;16:318–325. doi: 10.1159/000123041. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn E.A., Walkinshaw M.D. Targeting FKBP isoforms with small-molecule ligands. Curr. Opin. Pharmacol. 2011;11:365–371. doi: 10.1016/j.coph.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Cao W., Konsolaki M. FKBP immunophilins and Alzheimer's disease: a chaperoned affair. J. Biosci. 2011;36:493–498. doi: 10.1007/s12038-011-9080-7. [DOI] [PubMed] [Google Scholar]
- 5.Chambraud B., Radanyi C., Camonis J.H., Rajkowski K., Schumacher M., Baulieu E.E. Immunophilins, Refsum disease, and lupus nephritis: the peroxisomal enzyme phytanoyl-COA alpha-hydroxylase is a new FKBP-associated protein. Proc. Natl. Acad. Sci. U.S.A. 1999;96:2104–2109. doi: 10.1073/pnas.96.5.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nilsson A., Skold K., Sjogren B., Svensson M., Pierson J., Zhang X., Caprioli R.M., Buijs J., Persson B., Svenningsson P., et al. Increased striatal mRNA and protein levels of the immunophilin FKBP-12 in experimental Parkinson's disease and identification of FKBP-12-binding proteins. J. Proteome Res. 2007;6:3952–3961. doi: 10.1021/pr070189e. [DOI] [PubMed] [Google Scholar]
- 7.Yao Y.L., Liang Y.C., Huang H.H., Yang W.M. FKBPs in chromatin modification and cancer. Curr. Opin. Pharmacol. 2011;11:301–307. doi: 10.1016/j.coph.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Chattopadhaya S., Harikishore A., Yoon H.S. Role of FK506 binding proteins in neurodegenerative disorders. Curr. Med. Chem. 2011;18:5380–5397. doi: 10.2174/092986711798194441. [DOI] [PubMed] [Google Scholar]
- 9.Romano S., Sorrentino A., Pace A.L., Nappo G., Mercogliano C., Romano M.F. The emerging role of large immunophilin FK506 binding protein 51 in cancer. Curr. Med. Chem. 2011;18:5424–5429. doi: 10.2174/092986711798194333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin Y.J., Burakoff S.J., Bierer B.E. Molecular cloning of a 25-kDa high affinity rapamycin binding protein, FKBP25. J. Biol. Chem. 1992;267:10942–10945. [PubMed] [Google Scholar]
- 11.Jin Y.J., Burakoff S.J. The 25-kDa FK506-binding protein is localized in the nucleus and associates with casein kinase II and nucleolin. Proc. Natl. Acad. Sci. U.S.A. 1993;90:7769–7773. doi: 10.1073/pnas.90.16.7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riviere S., Menez A., Galat A. On the localization of FKBP25 in T-lymphocytes. FEBS Lett. 1993;315:247–251. doi: 10.1016/0014-5793(93)81173-w. [DOI] [PubMed] [Google Scholar]
- 13.Ahn J., Murphy M., Kratowicz S., Wang A., Levine A.J., George D.L. Down-regulation of the stathmin/Op18 and FKBP25 genes following p53 induction. Oncogene. 1999;18:5954–5958. doi: 10.1038/sj.onc.1202986. [DOI] [PubMed] [Google Scholar]
- 14.Ochocka A.M., Kampanis P., Nicol S., Allende-Vega N., Cox M., Marcar L., Milne D., Fuller-Pace F., Meek D. FKBP25, a novel regulator of the p53 pathway, induces the degradation of MDM2 and activation of p53. FEBS Lett. 2009;583:621–626. doi: 10.1016/j.febslet.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 15.Yang W.M., Yao Y.L., Seto E. The FK506-binding protein 25 functionally associates with histone deacetylases and with transcription factor YY1. EMBO J. 2001;20:4814–4825. doi: 10.1093/emboj/20.17.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leclercq M., Vinci F., Galat A. Mammalian FKBP-25 and its associated proteins. Arch. Biochem. Biophys. 2000;380:20–28. doi: 10.1006/abbi.2000.1883. [DOI] [PubMed] [Google Scholar]
- 17.Stros M. DNA bending by the chromosomal protein HMG1 and its high mobility group box domains: effect of flanking sequences. J. Biol. Chem. 1998;273:10355–10361. [PubMed] [Google Scholar]
- 18.Hochwagen A., Tham W.H., Brar G.A., Amon A. The FK506 binding protein Fpr3 counteracts protein phosphatase 1 to maintain meiotic recombination checkpoint activity. Cell. 2005;122:861–873. doi: 10.1016/j.cell.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Nelson C.J., Santos-Rosa H., Kouzarides T. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell. 2006;126:905–916. doi: 10.1016/j.cell.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 20.Xiao H., Jackson V., Lei M. The FK506-binding protein, Fpr4, is an acidic histone chaperone. FEBS Lett. 2006;580:4357–4364. doi: 10.1016/j.febslet.2006.06.093. [DOI] [PubMed] [Google Scholar]
- 21.Li H., Luan S. AtFKBP53 is a histone chaperone required for repression of ribosomal RNA gene expression in Arabidopsis. Cell Res. 2010;20:357–366. doi: 10.1038/cr.2010.22. [DOI] [PubMed] [Google Scholar]
- 22.Hanes S.D. Prolyl isomerases in gene transcription. Biochim. Biophys. Acta. 2014;10:2017–2034. doi: 10.1016/j.bbagen.2014.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helander S., Montecchio M., Lemak A., Fares C., Almlof J., Li Y., Yee A., Arrowsmith C.H., Dhe-Paganon S., Sunnerhagen M. Basic tilted helix bundle—a new protein fold in human FKBP25/FKBP3 and HectD1. Biochem. Biophys. Res. Commun. 2014;447:26–31. doi: 10.1016/j.bbrc.2014.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang J., Hung D.T., Schreiber S.L., Clardy J. Structure of the human 25 kDa FK506 binding protein complexed with rapamycin. J. Am. Chem. Soc. 1996;118:1231–1232. [Google Scholar]
- 25.Prakash A., Shin J., Yoon H.S. H, C and N resonance assignments of human FK506 binding protein 25. Biomol. NMR Assign. 2015;9:43–46. doi: 10.1007/s12104-014-9541-7. [DOI] [PubMed] [Google Scholar]
- 26.Kay L.E. Pulsed field gradient multi-dimensional NMR methods for the study of protein structure and dynamics in solution. Prog. Biophys. Mol. Biol. 1995;63:277–299. doi: 10.1016/0079-6107(95)00007-0. [DOI] [PubMed] [Google Scholar]
- 27.Ogura K., Terasawa H., Inagaki F. An improved double-tuned and isotope-filtered pulse scheme based on a pulsed field gradient and a wide-band inversion shaped pulse. J. Biomol. NMR. 1996;8:492–498. doi: 10.1007/BF00228150. [DOI] [PubMed] [Google Scholar]
- 28.Zwahlen C., Legault P., Vincent S.J.F., Greenblatt J., Konrat R., Kay L.E. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: application to a bacteriophage lambda N-peptide/boxB RNA complex. J. Am. Chem. Soc. 1997;119:6711–6721. [Google Scholar]
- 29.Breeze A.L. Isotope-filtered NMR methods for the study of biomolecular structure and interactions. Prog. Nucl. Mag. Res. Sp. 2000;36:323–372. [Google Scholar]
- 30.Iwahara J., Wojciak J.M., Clubb R.T. Improved NMR spectra of a protein-DNA complex through rational mutagenesis and the application of a sensitivity optimized isotope-filtered NOESY experiment. J. Biomol. NMR. 2001;19:231–241. doi: 10.1023/a:1011296112710. [DOI] [PubMed] [Google Scholar]
- 31.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 32.Rückert M., Otting G. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J. Am. Chem. Soc. 2000;122:7793–7797. [Google Scholar]
- 33.Cordier F., Dingley A.J., Grzesiek S. A doublet-separated sensitivity-enhanced HSQC for the determination of scalar and dipolar one-bond J-couplings. J. Biomol. NMR. 1999;13:175–180. doi: 10.1023/a:1008301415843. [DOI] [PubMed] [Google Scholar]
- 34.Valafar H., Prestegard J.H. REDCAT: a residual dipolar coupling analysis tool. J. Magn. Reson. 2004;167:228–241. doi: 10.1016/j.jmr.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 35.Guntert P. Automated structure determination from NMR spectra. Eur. Biophys. J. 2009;38:129–143. doi: 10.1007/s00249-008-0367-z. [DOI] [PubMed] [Google Scholar]
- 36.Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Shen Y., Delaglio F., Cornilescu G., Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brunger A.T. Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 39.Koradi R., Billeter M., Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 40.DeLano W.L. PyMOL molecular viewer: updates and refinements. Abstr. Pap. Am. Chem. S. 2009;238 [Google Scholar]
- 41.Laskowski R.A., Rullmannn J.A., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 42.Bhattacharya A., Tejero R., Montelione G.T. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 43.Farrow N.A., Muhandiram R., Singer A.U., Pascal S.M., Kay C.M., Gish G., Shoelson S.E., Pawson T., Forman-Kay J.D., Kay L.E. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 44.Dosset P., Hus J.C., Blackledge M., Marion D. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J. Biomol. NMR. 2000;16:23–28. doi: 10.1023/a:1008305808620. [DOI] [PubMed] [Google Scholar]
- 45.Pervushin K., Riek R., Wider G., Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hua Q., He R.Q., Haque N., Qu M.H., del Carmen Alonso A., Grundke-Iqbal I., Iqbal K. Microtubule associated protein tau binds to double-stranded but not single-stranded DNA. Cell. Mol. Life Sci. 2003;60:413–421. doi: 10.1007/s000180300034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Vries S.J., van Dijk M., Bonvin A.M. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 48.Laskowski R.A., Rullmannn J.A., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 49.Garcia de la Torre J., Huertas M.L., Carrasco B. HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J. Magn. Reson. 2000;147:138–146. doi: 10.1006/jmre.2000.2170. [DOI] [PubMed] [Google Scholar]
- 50.Houbaviy H.B., Burley S.K. Thermodynamic analysis of the interaction between YY1 and the AAV P5 promoter initiator element. Chem. Biol. 2001;8:179–187. doi: 10.1016/s1074-5521(00)90066-8. [DOI] [PubMed] [Google Scholar]
- 51.Golebiowski F.M., Gorecki A., Bonarek P., Rapala-Kozik M., Kozik A., Dziedzicka-Wasylewska M. An investigation of the affinities, specificity and kinetics involved in the interaction between the Yin Yang 1 transcription factor and DNA. FEBS J. 2012;279:3147–3158. doi: 10.1111/j.1742-4658.2012.08693.x. [DOI] [PubMed] [Google Scholar]
- 52.Cai Y., Jin J., Yao T., Gottschalk A.J., Swanson S.K., Wu S., Shi Y., Washburn M.P., Florens L., Conaway R.C., et al. YY1 functions with INO80 to activate transcription. Nat. Struct. Mol. Biol. 2007;14:872–874. doi: 10.1038/nsmb1276. [DOI] [PubMed] [Google Scholar]
- 53.Garvie C.W., Hagman J., Wolberger C. Structural studies of Ets-1/Pax5 complex formation on DNA. Mol. Cell. 2001;8:1267–1276. doi: 10.1016/s1097-2765(01)00410-5. [DOI] [PubMed] [Google Scholar]
- 54.Galat A., Thai R. Rapamycin-binding FKBP25 associates with diverse proteins that form large intracellular entities. Biochem. Biophys. Res. Commun. 2014;450:1255–1260. doi: 10.1016/j.bbrc.2014.06.105. [DOI] [PubMed] [Google Scholar]
- 55.Gudavicius G., Dilworth D., Serpa J.J., Sessler N., Petrotchenko E.V., Borchers C.H., Nelson C.J. The prolyl isomerase, FKBP25, interacts with RNA-engaged nucleolin and the pre-60S ribosomal subunit. RNA. 2014;20:1014–1022. doi: 10.1261/rna.042648.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tarsounas M., Davies A.A., West S.C. RAD51 localization and activation following DNA damage. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004;359:87–93. doi: 10.1098/rstb.2003.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paull T.T., Haykinson M.J., Johnson R.C. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- 58.Sultan A., Nesslany F., Violet M., Begard S., Loyens A., Talahari S., Mansuroglu Z., Marzin D., Sergeant N., Humez S., et al. Nuclear tau, a key player in neuronal DNA protection. J. Biol. Chem. 2011;286:4566–4575. doi: 10.1074/jbc.M110.199976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tao Z., Gao P., Liu H.W. Studies of the expression of human poly(ADP-ribose) polymerase-1 in Saccharomyces cerevisiae and identification of PARP-1 substrates by yeast proteome microarray screening. Biochemistry. 2009;48:11745–11754. doi: 10.1021/bi901387k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.