Abstract
Hypoxia was a major challenge faced by cetaceans during the course of secondary aquatic adaptation. Although physiological traits of hypoxia tolerance in cetaceans have been well characterized, the underlying molecular mechanisms remain unknown. We investigated the sequences of 17 hypoxia-tolerance-related genes in representative cetaceans to provide a comprehensive insight into the genetic basis of hypoxia tolerance in these animals. Genes involved in carrying and transporting oxygen in the blood and muscle (hemoglobin-α and β, myoglobin), and genes involved in the regulation of vasoconstriction (endothelin-1, -2, and -3; endothelin receptor type A and B; adrenergic receptor α-1D; and arginine vasopressin) appear to have undergone adaptive evolution, evidence for positive selection on their particular sites, and radical physiochemical property changes of selected condons. Interestingly, “long-diving” cetaceans had relatively higher ω (dN/dS) values than “short-diving” cetaceans for the hemoglobin β gene, indicating divergent selective pressure presented in cetacean lineages with different diving abilities. Additionally, parallel positive selection or amino acid changes (ADRA1D: P50A, A53G, AVPR1B: I/V270T) among animals exposed to different hypoxia habitats reflect functional convergence or similar genetic mechanisms of hypoxia tolerance. In summary, positive selection, divergent selective pressures, and parallel evolution at the molecular level provided some new insights into the genetic adaptation of hypoxia tolerance.
Keywords: Cetacean, hypoxia tolerance, positive selection, adaptive evolution
Introduction
Cetaceans, including whales, dolphins, and porpoises, have lived in an exclusively aquatic environment for approximately 53–56 Myr after their transition from land to water (Thewissen et al. 2007). This change in lifestyle means they have evolved a series of specializations for life in a fully aquatic habitat, such as lack of distal hindlimbs, loss of hair, and derivation of echolocation (Gatesy and O'Leary 2001). Several researchers have demonstrated adaptive evolution in candidate loci associated with aquatic features in cetaceans (McGowen et al. 2012; Yim et al. 2014), such as brain size (Xu et al. 2012), taste receptors (Feng et al. 2014; Zhu et al. 2014), osmoregulation (Xu et al. 2013), triacylglycerol metabolism (Wang et al. 2015), and innate immunity (Shen et al. 2012), supporting the idea that cetaceans have evolved adaptively to their aquatic environment (McGowen et al. 2014).
Cetaceans are distinguished by their excellent diving ability; for example, sperm whales (Physeter catodon) can dive down to 3,000 m and remain submerged for up to 73 min (Watkins et al. 1993). However, because mammals need to breathe air, hypoxia (reduction in convective oxygen delivery in blood and tissues; Elsner and Gooden 1983; Butler and Jones 1997) is recognized as one of the biggest challenges for diving cetaceans. Previous studies have revealed physiological changes in terms of respiration, circulation, and hematology in cetaceans compared with their terrestrial relatives (Ramirez et al. 2007; Panneton 2013). For example, cetaceans have high levels of hemoglobin (HB) and a high hematocrit (Hct, volume percentage of red blood cells in blood and its influence on oxygen transport; Birchard 1997), resulting in blood oxygen stores in bottlenose dolphins (Tursiops truncatus) that are more than three times higher than those in terrestrial species (Snyder 1983). In addition, neuroglobin (NGB) expression levels in whale brains were 4–15 times higher than in terrestrial cattle (Schneuer et al. 2012), while a recent study showed that myoglobin (MB) levels in whales were 2–7 times higher than in horses (Helbo and Fago 2012). The elevated concentrations of these oxygen-carrying proteins in different tissues provide the basis for increased intrinsic oxygen stores during diving, as seen in other animals that are habitually exposed to hypoxia. For example, an important adaptation of subterranean mole rats to environmental hypoxia is mediated by higher tissue globin expression (e.g., NGB) (compared with rat), high blood–oxygen affinity, and high Hct (Van Aardt et al. 2007; Avivi et al. 2010). In addition to the above physiological adaptations, adaptive changes in the cardiovascular system in response to hypoxia tolerance have been found in cetaceans, such as dramatic bradycardia (reduction in heart rate in response to apneic immersion; Manley 1990) and selective peripheral vasoconstriction, the latter being responsible for redistributing the blood circulation to maintain blood flow to the central nervous system and heart, and reducing flow to the skin, muscle, and splanchnic organs (e.g., spleen, kidney) (Ramirez et al. 2007; Panneton 2013). These adjustments are essential to achieve effective oxygen conservation. Vasoconstrictor tone plays an important role in regulating blood pressure and distributing blood flow in the body, which is mainly dependent on a complex interplay of vasoconstrictors and their receptors that stimulate ion channels and trigger the contraction by signaling cascade reaction (Jackson 2000). The circulating humoral factors, including endothelin, angiotensin, histamine, and so on, can induce a local vasoconstrictor response (Greer 2015). For example, the release of endothelin-1 (EDN1) can lead to vasoconstriction of arterial blood vessels by either acting on endothelin receptors (e.g. EDNRA) on vascular smooth muscle cells, or through the release of aldosterone and a reduction in renal blood flow (Sowerby 2009). Besides, α1-adrenoceptor-evoked contractions have been implicated in hypoxic sympatholysis (Marshall 2015). Furthermore, anatomical evidence suggests that extensive venous plexi in the heads of dolphins may promote hypoxia tolerance (Costidis and Rommel 2012).
Although physiological adaptions for hypoxia tolerance have been well-studied in cetaceans, its molecular basis has not been well addressed. In this study, we investigated the sequences of 17 candidate genes encoding proteins with important roles in oxygen storage and transport, and vasoconstriction. Comparison of the sequences of these genes in cetaceans with terrestrial mammals will allow us to reveal the genetic basis for the adaptation of cetaceans to hypoxia, and improve our understanding of the genetic and evolutionary architecture of cetaceans during the course of secondary aquatic adaptation.
Methods
Screening Candidate Genes
We screened a total of 17 candidate genes (supplementary table S1, Supplementary Material online). Among the studied genes, those encoding HB, MB, and NGB are responsible for oxygen storage and transport in the blood, muscle, and nerons, respectively (Dickerson and Geis 1983; Burmester et al. 2000; Ordway and Garry 2004). HB is a heterotetramer protein composed of two α (HBA) and two β chains (HBB). Given the significant role of vasoconstriction in hypoxia tolerance, we also screened vasoconstriction-related genes. Based on the vascular smooth muscle contraction signaling pathway (http://www.genome.jp/kegg/) (KO: 04270), several candidate vasoconstrictors and receptors have been identified, including endothelin-1, -2, and -3 (EDN1, EDN2, EDN3); endothelin receptors type A and B (EDNRA and EDNRB) (Masaki 2000); angiotensin II receptor type 1 and 2 (AGTR1 and AGTR2) (Ardaillou 1999); adrenergic receptor α-1A, B, and D (ADRA1A, ADRA1B, and ADRA1D) (Wright et al. 2012); arginine vasopressin (AVP), and arginine vasopressin receptor 1A and B (AVPR1A and AVPR1B) (Holmes et al. 2003). The information on gene function and related disease was searched from UniProtKB/Swiss-Prot (http://www.uniprot.org/) and GeneCards website (http://www.genecards.org/).
Species Coverage, Sequence Acquisition, and Compilation
A total of 11 cetacean species, representing 8 different families—Delphinidae (Bottlenose dolphin, T. truncatus; Long-beaked common dolphin, Delphinus capensis; Short-beaked common dolphin, Delphinus delphis; Chinese white dolphin, Sousa chinensis), Lipotidae (Baiji, Lipotes vexillifer), Phocoenidae (Finless porpoise, Neophocaena phocaenoides), Ziphiidae (Beaked whales, Mesoplodon densirostris), Physeteridae (Sperm whales, Physeter macrocephalus), Kogiidae (Dwarf sperm whale, Kogia sima), and Balaenopteridae (Minke whale, Balaenoptera acutorostrata; Omura’s whale, Banaenoptera omurai)—were sampled in our study (supplementary table S2, Supplementary Material online). Genomic DNA was extracted from muscle and blood samples using standard phenol/chloroform extraction protocols. Each sample used for this study was collected from dead individuals in the wild, and approved by the Nanjing Normal University Animal Care Committee. To amplify the open reading frames of candidate genes across the studied species, specific primers were designed using the homologous sequences from the genome of cetaceans and other mammals by using PrimerPrimer5 (Lalitha 2000). The polymerase chain reaction (PCR) primers have been listed in supplementary table S3, Supplementary Material online. All genes were amplified with 15 μl total volume PCR mixtures, containing 0.3 μl of each primer (10 μM), 0.3 μl of DNA, and 7.5 μl of 2× EasyTaq PCR SuperMix (TransGen Biotech). PCR reaction was performed on ABI 9700 with the following PCR conditions: 95 °C for 5 min, 35 times (94 °C for 30 s, primer-specific annealing temperature [50–55 °C] for 40 s, 72 °C for 40 s), and 72 °C elongation for 10 min. The PCR products were purified using DNA Gel Extraction Kit (Axygen) and sequenced directly or cloned into PMD-18 vector (Takara). Three to eight colonies randomly selected were sequenced using vector primers M13F and M13R. Sequencing in both directions is conducted by ABI 3730 automated DNA sequencer. Eleven hypoxia-tolerance-related genes, including HBA, HBB, NGB, MB, EDN1, EDN2, EDN3, EDNRA, EDNRB, AGTR1, and AGTR2, from major lineages of cetaceans were amplified here, and the successfully amplified exons for each gene were deposited in supplementary table S2, Supplementary Material online.The exons of each gene were compiled and concatenated according to the coding sequences of the known genomes of cetacean species. Novel sequences were deposited in GenBank under accession numbers KT191174–KT191292. In contrast, the remaining six genes including ADRA1A, ADRA1B, ADRA1D, AVP, AVPR1A, and AVPR1B were searched from draft genome sequences of relevant cetaceans, that is, baiji, killer whale (Orcinus orca), sperm whale, bowhead whale (Balaena mysticetus), and mink whale using bottlenose dolphin genes as query to conduct BLAST (Johnson et al. 2008). Gene sequences from other mammalian orders, including artiodactyls, carnivores, primates, lagomorphs, rodents, and afrotheria, were downloaded from GenBank (http://www.ncbi.nlm.nih.gov), the Ensemble Genome database (http://www.ensembl.org), or UCSC genome data (http://genome.ucsc.edu/) with accession numbers listed in supplementary table S4, Supplementary Material online. The nucleotide and deduced amino acid sequences of each gene were aligned using MUSCLE in MEGA6 (Tamura et al. 2013) and checked by eye.
Analyses of Evolutionary Selective Pressure
The selective pressures were estimated using the different codon substitution site models implemented in CodeML of the phylogenetic analysis with maximum-likelihood (ML) software (PAML release 4.7) (Yang 2007). Comparison of ω = dN/dS, the ratio of nonsynonymous (dN) to synonymous (dS) substitution rates, among sites and branches is an indication of the form and intensity of natural selection, with ω < 1, ω = 1, and ω > 1 indicating negative selection, neutral evolution, and positive selection, respectively. Both the species tree (supplementary fig. S1, Supplementary Material online; Ranwez et al. 2007) and gene tree reconstructed using Bayesian inference implemented in MrBayes 3.1.2 (Ronquist and Huelsenbeck 2003) were separately used as the guide tree in all the analyses.
We performed the site-specific models, M8a and M8 (Swanson et al. 2003), which allow for variable selection patterns among amino acid sites to test for the presence of sites under positive selection in cetaceans-only data set. To further test whether the positively selected sites were restricted to specific lineage, the branch-sites model was also compared with the fixed model to detect positive selection that may have affected specific sites along a specific branch in the all-mammals data set (Zhang et al. 2005) (supplementary fig. S1, Supplementary Material online). The likelihood ratio test (LRT) with a χ2 distribution was used to determine which models were statistically different from the null model at a threshold of P < 0.05. Bayes empirical Bayes (BEB) analysis was used to identify sites under positive selection with posterior probabilities ≥0.80 (Yang et al. 2005).
It was noted that ω was estimated by PAML models that only consider the variation in the nonsynonymous substitution rate whereas the synonymous rate fixed across the sequence. Thus, we further adopted a fixed-effect likelihood (FEL), random-effect likelihood (REL), and fast unconstrained Bayesian approximation (FUBAR) for detecting positive selection implemented in the Datamonkey website, which incorporated variation in the rate of synonymous substitution (Pond and Frost 2005a). Because a small number of sequences may lead to a high false-positive rate, we performed more stringent significance threshold than the ones suggested by simulation to correspond to true type I errors of ∼ 0.5% (Pond and Frost 2005b). Thus, sites with a significance level <0.2 for FEL, Bayes factor >50 for REL, or posterior probability >0.8 for FUBAR were regarded as candidates for selection.
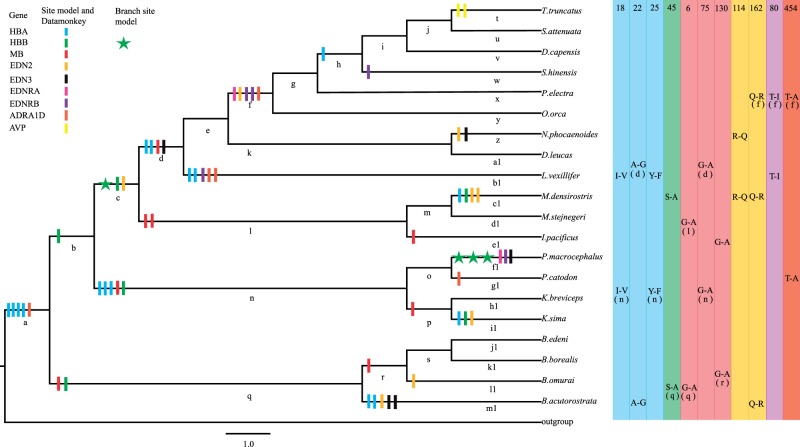
Although Codeml and Datamonkey assess the influence of natural selection at the codon level, selection for changes in amino acid physicochemical properties was analyzed using complementary protein-level approach implemented in TreeSAAP (Woolley et al. 2003), which measures the selective influences based on 31 structural and biochemical amino acid property changes by means of goodness-of-fit test. A series of z-score was also produced in each category and higher categories (6–8) indicating more radical substitution. The number of radical changes per codon in amino acid properties served as a proxy of positive selection and more radical changes might indicate adaptive evolution (Sunagar et al. 2012). For the sites identified to be under selection by more than one ML method, the radical amino acid changes were mapped onto the phylogeny (fig. 1).
Fig. 1.—
Radical amino acid changes in selected sites across the cetacean phylogeny. Radical amino acid changes of positively selected sites in selected genes marked on different colors were labeled on the cetaceans phylogeny from branch a to m1. Parallel amino acid changes which occurred in selected genes HBA (blue), HBB (green), MB (red), EDN2 (orange), EDN3 (black), EDNRA (pink), EDNRB (purple), ADRA1D (purplish red), and AVP (yellow) among different lineages of cetaceans are shown on the right of the corresponding terminal branches, while letter in parentheses stand for the internal branches. Amino acid positions (numbers) and parallel changes at each position were listed in the right part of figure 1 corresponding to genes marked in different colors. Selected sites are indicated separately by vertical lines and stars.
Structural Analysis
To gain insight into the functional significance of the putatively selected sites, we mapped selective sites onto the protein secondary and three-dimensional (3D) structures (supplementary figs. S2 and S3, Supplementary Material online). The secondary structures of each gene were identified using UniProt (http://www.uniprot.org/). Furthermore, the 3D structures of genes were either searched from Protein Data Bank, which is a database containing experimentally determined 3D structures of proteins, nucleic acids, and other biological macromolecules (http://www.rcsb.org/pdb/), or constructed by the I-TASSER (Threading ASSEmbly Refinement), a hierarchical protein structure modeling approach based on the secondary structure enhanced Profile–Profile threading Alignment and the iterative implementation of the TASSER program (supplementary table S1, Supplementary Material online) (Zhang 2008).
Selection Pressure Analysis in Cetacean Clades with Contrasting Diving Abilities
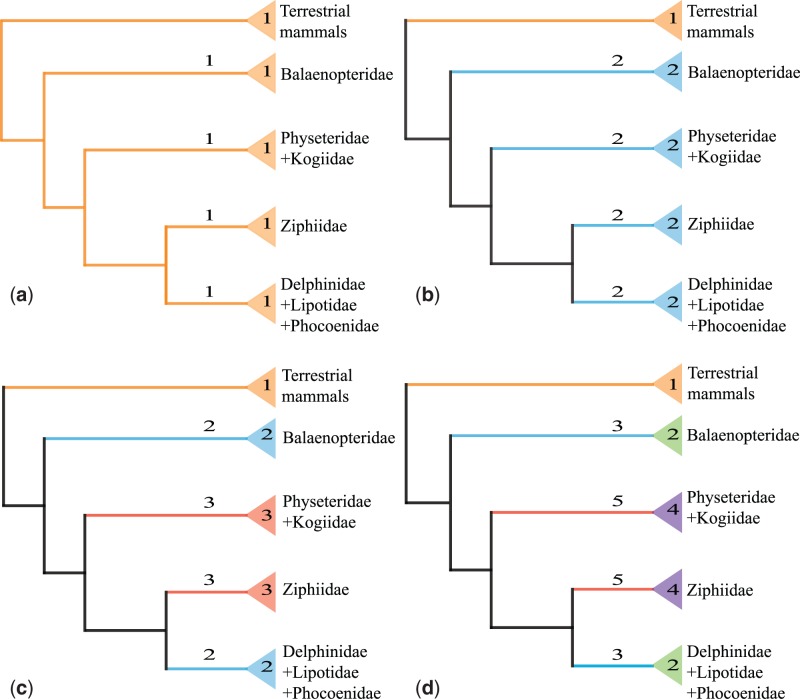
To investigate the possible role of positive Darwinian selection on the evolution of 17 genes in cetacean clades with contrasting diving abilities, the most diving talents like beaked whales, sperm whales, and dwarf sperm whale (the maximum dive duration > 40 min) were classified as the “long-diving” group, and the remaining families were regarded as the “short-diving” group according to the proposal by Nery et al. (2013a) (supplementary table S5, Supplementary Material online). We used branch models that allow different branch groups to have different ω, leading to the so-called “two ratio,” “three ratio,” “five ratio,” and so on (Yang 1998) models which were implemented in CODEML (fig. 2). Foreground (particular lineages of interest) and background lineages (the remaining lineages) were first set up. First, to test whether different selective pressure acting on the terrestrial mammals and cetacean lineages, the one ratio model that enforces the same ω ratio for all lineages (fig. 2a) was compared with the two ratio (2ω) model (fig. 2b) that allows one ω ratio for all terrestrial mammals branches (orange; labeled 1) and another for all branches of cetacean families (blue; labeled 2). Second, to explore the rate variation among cetacean clades with different diving abilities, the three ratio (3ω) model (fig. 2c) assumes that ancestral and descendant branches of long-diving (pink; labeled 3) and short-diving (blue; labeled 2) whales have independent rates, and a third ω value for all terrestrial mammals branches (orange; labeled 1). Finally, the five ratio (5ω) model (fig. 2d) was used to estimate whether descendant branches (labeled 2 and 4) have independent rates from their ancestral branches in the long-diving (branch 5) and short-diving (branch 3) families, which was compared with the 2ω model. The inference of positive selection was conducted by performing am LRT in which the following two pairs of models were compared (table 1). Three starting values (0.5, 1, and 2) were used to check the existence of multiple local optima.
Fig. 2.—
Graphical representation of nested branch models implemented to test the possible role of positive selection over cetacean lineages with different diving abilities. For details see Materials and Methods.
Table 1.
Log Likelihood and Omega Values Estimated under Different Branch Models according to Contracting Diving Abilities of Cetacean
| Gene | Model | −lnL | LRT | Comparisons | P Value | ω Values |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Terrestrial Mammals | Mysticetes | Delphinidae + Lipotidae + Phocoenidae | Physeteridae + Kogiidae | Ziphiidae | ||||||
| HBB | 1ω | 3,528.038 | 0.283 | 0.283 | 0.283 | 0.283 | 0.283 | |||
| 2ω | 3,525.152 | 5.772 | 2ω vs. 1ω | 0.016 | 0.262 | 0.513 | 0.513 | 0.513 | 0.513 | |
| 3ω | 3,522.168 | 5.968 | 3ω vs. 2ω | 0.015 | 0.262 | 0.255 | 0.255 | 1.007 | 1.007 | |
| 5ω | 3,521.819 | 6.666 | 5ω vs. 2ω | 0.036 | 0.262 | 0.369/0.207 | 0.369/0.207 | 1.563/0.917 | —/0.917 | |
NOTE.—For 5ω model, the omega value was estimated for the ancestral and descendant branches, the number at the left side refers to the ancestral branch omega value, whereas the number at the right side to the descendant branch omega. “—”denotes the absence of omega value estimated for the ancestral branch because of one species in Ziphiidae family.
Convergent Evolution Analyses in Hypoxia Species
To determine if similar patterns of evolution occurred in animals habitually exposed to hypoxia but living in diverse environments, we first constructed phylogenies using MrBayes 3.1.2 (Ronquist and Huelsenbeck 2003). For MrBayes analysis, four Markov chains were run for 2 × 106 generations and sampled every 1,000 generations. A consensus tree was estimated after removing the first 1,000 tree as burn-in. Modeltest (Posada and Crandall 1998) and ProtTest (Abascal et al. 2005) were used to determine the best-fit models for nucleotide and amino acid substitution, individually, under the Akaike information criterion. Then, we searched for parallel amino acid substitutions from the internal ancestor nodes to terminal branches along paraphyletic lineages of species exposed to hypoxic environment. The ancestral amino acids sequences were reconstructed using the Bayesian approach implemented in the BASEML program from the PAML package (Yang 2007). The software CONVERG 2 was used to test whether the observed parallel substitutions in focal branches were fixed randomly or by natural selection (Zhang and Kumar 1997).
Results
Positive Selection of Hypoxia-Tolerance-Related Genes in Cetaceans
Considering that selection analyses using species tree was basically the same as that using gene tree, only the former analyses were shown here. We used the one ratio model that only allows a single ω ratio for all branches. The ω values obtained for all 17 genes ranged from 0.044 to 0.460 and were significantly <1, suggesting that strong purifying selection played a major role in the evolution of hypoxia-related genes to keep its important functions in hypoxic adaptation (supplementary table S6, Supplementary Material online).
To determine if individual codons in each gene were subject to positive selection, we used a pair of site model (M8a vs. M8) in the cetaceans-only data set. The result showed that the M8 model that included positive selection fitted the data better than the neutral model M8a. Specially, M8 model detected 23 sites to be under positive selection at HBA, HBB, MB, EDN2, and ADRA1D genes (supplementary table S7, Supplementary Material online). Significant evidence of positive selection was further identified by other three ML methods, that is, FEL, REL, and FUBAR model implemented in Datamonkey. A total of ten hypoxia-tolerance-related genes (HBA, HBB, MB, EDN1, EDN2, EDN3, EDNRA, EDNRB, ADRA1D, and AVP) were subjected to positive selection, five of which were also detected by M8. The FEL methods found 23 positively selected codons in 9 genes at significance level of 0.2. Using the REL method, 25 codons in 7 genes were identified under positive selection at significant level of Bayes factor > 50. Additionally, FUBAR also identified 27 sites in 10 genes under diversifying selection with a posterior probability > 0.8 (supplementary table S8, Supplementary Material online).
The more stringent branch-site model was then used to investigate whether positive selection acting on specific sites in cetacean lineages (fig. 1: branch a-m1). Evidence for positive selection was identified along the lineages leading to the dwarf sperm whale (fig. 1: branch i1, P = 0.001), sperm whale (fig. 1: branch f1, P = 0.003), and the last common ancestor of Delphinidae, baiji, and beaked whale (fig. 1: branch c, P < 0.001) at HBB with three, seven, and one sites as possible targets of positive selection identified by BEB procedure, respectively (table 2).
Table 2.
Selective Pressure Analyses of Hypoxia-Exposed Animals in All-Mammals Data Set by Branch-Site Model
| Genes | Branch-Site Model | −lnL | 2Δ (lnL) | P Value | ω Values | Positively Selected Sites |
|---|---|---|---|---|---|---|
| HBA | Branch (terminal branch of Pantholops hodgsonii) | |||||
| Null | 3,551.894 | ω0 = 0.068, ω1 = 1, ω2 = 1 | ||||
| Alternative | 3,548.763 | 6.262 | 0.012 | ω0 = 0.068, ω1 = 1, ω2 = 270.801 | 21-0.993, 80-0.994, 105-0.999, 132-0.994 | |
| HBB | Branch (last common ancestral branch of Delphinidae, Lipotes vexillifer, and Mesoplodon densirostris) | |||||
| Null | 4,185.624 | ω0 = 0.047, ω1 = 1, ω2 = 1 | ||||
| Alternative | 4,177.163 | 16.922 | <0.001 | ω0 = 0.047, ω1 = 1, ω2 = 494.933 | 63-1.000 | |
| Branch (terminal branch of Kogia sima) | ||||||
| Null | 4,186.525 | ω0 = 0.048, ω1 = 1, ω2 = 1 | ||||
| Alternative | 4,181.318 | 10.414 | 0.001 | ω0 = 0.048, ω1 = 1, ω2 = 81.753 | 60-0.988, 88-0.933, 134-0.971 | |
| Branch (terminal branch of Physeter macrocephalus) | ||||||
| Null | 4,182.625 | ω0 = 0.044, ω1 = 1, ω2 = 1 | ||||
| Alternative | 4,178.154 | 8.942 | 0.003 | ω0 = 0.045, ω1 = 1, ω2 = 822.915 | 3-0.938, 11-0.928, 12-0.995, 51-0.943, 60-0.960, 62-0.997, 73-0.936 | |
| Branch (last common ancestral branch of Bos grunniens and Bos mutus) | ||||||
| Null | 4,189.876 | ω0 = 0.046, ω1 = 1, ω2 = 1 | ||||
| Alternative | 4,186.130 | 7.492 | 0.006 | ω0 = 0.048, ω1 = 1, ω2 = 999 | 136-0.956 | |
| NGB | Branch (terminal branch of Heterocephalus glaber) | |||||
| Null | 2,529.909 | ω0 = 0.059, ω1 = 1, ω2 = 1 | ||||
| Alternative | 2,526.051 | 7.716 | 0.005 | ω0 = 0.059, ω1 = 1, ω2 = 999 | 3-0.973, 13-0.976, 15-0.998, 67-0.858, 97-0.952, 67-0.969, 97-0.949 | |
Together, the combination of results from the above 5 ML methods showed that 42 codons from 10 genes (HBA: 9, HBB: 11, MB: 5, EDN1: 1, EDN2: 3, EDN3: 3, EDNRA: 3, EDNRB: 2, ADRA1D: 3, AVP: 2) were detected to be under positive selection in cetaceans by at least 2 ML methods (supplementary table S8, Supplementary Material online), which were regarded as strong candidate codons of positive selection. Moreover, 78.6% (33/42) of selected sites were also detected to be under radical changes by TreeSAAP (table 3), which provides additional evidence for the operation of positive selection on cetaceans. Furthermore, the 33 radical change sites were scattered throughout most of the cetacean phylogeny (fig. 1).
Table 3.
Radical Amino Acid Sites under Positive Selection Detected by PAML, Datamonkey, and TreeSAAP Simultaneously
| Gene | Site | PAML |
Datamonkey |
TreeSAAP Properties |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Position | Site Model (M8)a | Branch-Site Modela | FEL P < 0.2b | REL BF > 50b | FUBAR pp > 0.8b | AA Changes | Parallel Changes | Cladec | Radical Changes in AA Propertiesd | Total | |
| HBA | 16 | 0.912 | 0.106 | 450.038 | 0.820 | G-A | a | Pα, Pc, P | 3 | ||
| A-S | h | Pα, Pc, P | 3 | ||||||||
| A-G | c1 | Pα, Pc, P | 3 | ||||||||
| 18 | 0.897 | 0.134 | 406 | 0.805 | V-I | a | pK’ | 1 | |||
| I-V | Yes | b1, n | pK’ | 1 | |||||||
| 22 | 0.981 | 0.084 | 475.336 | 0.836 | A-G | Yes | d, m1 | Pα, Pc, P | 3 | ||
| 25 | 0.920 | 0.116 | 416.941 | 0.839 | Y-F | Yes | n,b1 | F | 1 | ||
| 52 | 0.843 | 176.704 | 0.859 | G-D | m1 | αc, Pr, Et, | 3 | ||||
| 74 | 0.802 | 134.561 | M-I | d | pK’ | 1 | |||||
| 76 | 0.864 | 146.049 | D-N | a | αc, | 1 | |||||
| T-A | i1 | Pα, | 1 | ||||||||
| 112 | 0.886 | 178.977 | A-V | a | Pβ | 1 | |||||
| V-A | n | Pβ | 1 | ||||||||
| HBB | 11 | 0.928 | A-G | f1 | Pα, Pc, P | 3 | |||||
| 45 | 0.953 | 0.092 | 442.775 | 0.963 | S-A | Yes | q, c1 | Pα, Pc, P | 3 | ||
| S-H | n | F | 1 | ||||||||
| 60 | 0.917 | 56.360 | K-N | b | pHi | 1 | |||||
| 0.960 | N-K | f1 | pHi | 1 | |||||||
| 62 | 0.997 | K-Q | f1 | pHi | 1 | ||||||
| 63 | 0.994 | 0.200 | 384.186 | 0.958 | A-S | i1 | Pα, Pc, P | 3 | |||
| 1.000 | A-K | c | Ko, Br, Esm, Et, h | 5 | |||||||
| 88 | 0.891 | 0.169 | 308.649 | 0.926 | T-A | c | Pα | 1 | |||
| MB | 6 | 0.861 | 0.140 | G-A | Yes | l, q | Pα, Pc, P | 3 | |||
| 36 | 0.921 | 0.102 | 0.896 | S-H | p | F | 1 | ||||
| 75 | 0.159 | 0.875 | G-A | Yes | d, n | Pα, Pc, P | 3 | ||||
| A-G | k | Pα, Pc, P | 3 | ||||||||
| 130 | 0.856 | 0.151 | G-A | Yes | r, e1 | Pα, Pc, P | 3 | ||||
| EDN2 | 12 | 0.039 | 761.565 | 0.965 | V-A | c | Pβ | 1 | |||
| V-I | i1 | pK’ | 1 | ||||||||
| 114 | 0.942 | 0.023 | 190.831 | 0.984 | R-Q | Yes | z, c1 | pHi | 1 | ||
| R-L | l1 | Ns, αn, pHi, Br, RF, h, Hnc, p, Esm, Et | 10 | ||||||||
| 162 | 0.937 | 0.057 | 70.988 | 0.977 | Q-R | Yes | f, c1, m1 | pHi | 1 | ||
| EDN3 | 34 | 0.144 | 0.838 | T-A | f1 | Pα | 1 | ||||
| T-R | m1 | pHi, Hnc, Esm, | 3 | ||||||||
| 152 | 0.155 | 0.843 | Q-L | z | Ns, Br, h, F, p, Et | 6 | |||||
| Q-R | m1 | pHi | 1 | ||||||||
| 167 | 0.169 | 0.842 | R-G | d | Ca, pHi, Mv, Mw, Hnc, V0, u, Esm, Et | 9 | |||||
| EDNRA | 19 | 0.196 | 63.872 | 0.912 | I-V | f | Pk’ | 1 | |||
| 22 | 0.173 | 73.036 | 0.916 | N-D | f1 | αc | 1 | ||||
| EDNRB | 80 | 10721.8 | 0.888 | T-I | Yes | f, b1 | Ns, Br, Pk’, Ra, Hp, Ht | 6 | |||
| T-A | f1 | Pα | 1 | ||||||||
| 347 | 0.195 | 11076.6 | L-R | f | Ns, Br, RF, h, pHi, Hnc, p, αn, Esm, Et | 10 | |||||
| R-H | w | Esm | 1 | ||||||||
| ADRA1D | 454 | 0.988 | 0.041 | 436.44 | 0.968 | P-T | a | Ko, Ht | 2 | ||
| T-A | Yes | f, g1 | Pα | 1 | |||||||
| 475 | 0.867 | G-R | b1 | Ca, pHi, Mv, Mw, Hnc, V0, µ, Esm, Et | 9 | ||||||
| 483 | 0.909 | Q-P | b1 | Pc, Ko, αM, Ht | 3 | ||||||
| AVP | 159 | 1111.96 | 0.852 | A-S | t | Pα, Pc, P | 3 | ||||
| 162 | 1271.98 | 0.885 | G-P | t | Bl, αc, Ht | 3 | |||||
Codons identified by PAML as under positive selection along with Bayesian (BEB) analysis PPs for sites with P > 80% under M8 and branch-site models.
Codons were estimated in Datamonkey.
Amino acid substitution occurred along clades, with detailed information marked in figure 1.
Radical changes in amino acid properties under categories 6–8 were detected in TreeSAAP. Physicochemical amino acid properties available in TreeSAAP are as follows: αc: power to be C-term α-helix; αn: power to be in the N-terminal of an α-helix; Br: buriedness; Ca: helical contact energy; El: long-range nonbonded energy; Esm: short- and medium-range nonbonded energy; Et: total nonbonding energy; F: mean r.m.s. fluctuation displacement; h: hydropathy; Hnc: normal consensus hydrophobicity; Hp: surrounding hydrophobicity; Ht: thermodynamic transfer hydrophobicity; Ko: compressibility; µ: refractive index; Mv: molecular volume;Mw: molecular weight; Ns: average number of surrounding residues; Pα: α- helical tendencies; Pβ: β-structure tendencies; Pc: coil tendencies; P: turn tendencies; p: polarity; pHi: isoelectric point; pK’: equilibrium constant of ionization for COOH; Pr: polar requirement; Ra: solvent accessible reduction ratio; RF: chromatographic index; V0: partial-specific volume.
Structural Links to Protein Function
To gain insight into the functional significance of the putatively selected sites, we mapped them onto the secondary and 3D structures of the corresponding proteins. The results showed that most sites were located within or close to the functional domain (supplementary figs. S2 and S3, Supplementary Material online). For HBA and HBB, six sites (HBA: 16, 18; HBB: 3, 60, 62, 63) were situated in or near the glycosylation and metal-binding domains (supplementary fig. S3A–C, Supplementary Material online), which are responsible for co- and posttranslational protein modification and oxygen binding, respectively. In contrast, positively selected amino acids in vasoconstriction-related proteins were located primarily in functional domains facilitating ligand–receptor interactions, such as propeptide (EDN1: 125; EDN2: 114, 162; EDN3: 34, 152, 167), peptide (AVP: 159, 162), transmembrane (EDNRA: 161, EDNRB: 347), and extracellular topological domains (EDNRA: 22; EDNRB: 80; ADRA1D: 454, 475, 483) (supplementary fig. S3D–J, Supplementary Material online).
Selective Patterns in Cetaceans with Contrasting Diving Abilities
Branch model with several ratios was used to explore whether cetacean groups with divergent diving abilities (classed into two groups, i.e., long-diving and short-diving cetaceans) evolved under different evolutionary pressure. The results revealed that the model that included positive selection, including 2ω, 3ω, and 5ω models, fitted the data better than their null model at HBB gene (2ω vs. 1ω: P = 0.016; 3ω vs. 2ω: P = 0.015; and 5ω vs. 2ω: P = 0.036, respectively, table 1). For example, under 2ω model (fig. 2b), the cetacean ω ratio was 0.513, whereas the background ω ratio for terrestrial mammals was 0.262. Interestingly, the ω ratio (1.007) for long-diving cetaceans was much higher than that of short-diving cetaceans (0.255; fig. 3). The five ratios (5 ω) model (fig. 2d) showed significant model improvement over the 2ω model (table 1), suggesting that ancestral and descendant branches had independent evolutionary ratios. In addition, ω values for the ancestral branches of both groups were always higher than that of the descendant branches. More importantly, significant sign of positive selection was identified along the ancestral branches of the long-diving group (ω = 1.563; table 1). Although the same analysis had been implemented, neither accelerated evolution nor positive selection was detected in other genes (data not shown).
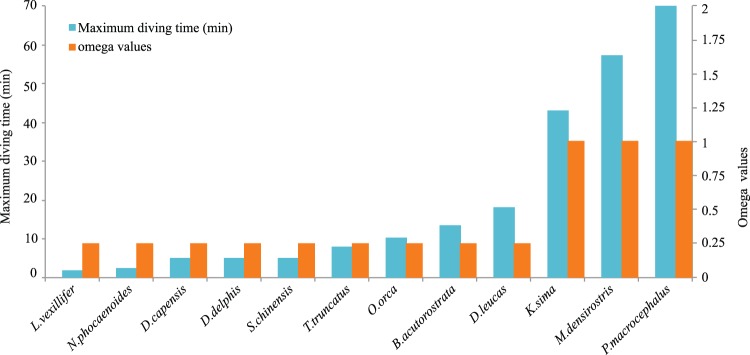
Fig. 3.—
Comparison of ω values estimated by the 3ω model in HBB among cetaceans according to different diving abilities. Compared with other lineages, long-diving cetaceans, including Physeter macrocephalus, Kogia sima, and Mesoplodon densirostris, show a greater ω values as well as longer diving time than other cetacean.
Convergent Evolution in Species Habitually Exposed to Hypoxia
To determine if similar patterns of evolution occurred in other species habitually exposed to hypoxia, we tested positive selection in other animals that inhabited hypoxia-exposed environments, such as burrowing and plateau species (supplementary table S9, Supplementary Material online). Branch-site model test had been used to detect positive selection along hypoxia species clades. A branch-site model showed evidence of positive selection along lineages such as the Tibetan antelope (Pantholops hodgsonii) in HBA (P = 0.012), naked mole rat (Heterocephalus glaber) in NGB (P = 0.005), and the last common ancestral branch of the yak (Bos mutus and Bos grunniens) in HBB (P = 0.006) (table 2). Four, one, and seven codons, respectively, were identified as being under positive selection in HBA, HBB, and NGB (posterior probability > 0.80). However, no sign of positive selection was detected in the remaining genes. Additionally, convergent/parallel amino acid changes (ADRA1D: P50A, A53G; AVPR1B: I/V270T) were identified specially in the lineages leading to naked mole rats and the ancestral lineage of cetaceans, although they were only distantly related in the phylogenetic tree (supplementary fig. S1, Supplementary Material online), and the three parallel/convergent substitutions deviated significantly from the random expectation at a significance level of 0.05 (supplementary table S10, Supplementary Material online). However, a phylogenetic tree based on nucleotide and amino acid data for ADRA1D and AVPR1B did not cluster hypoxia-exposed mammals into a single monophyletic clade (supplementary fig. S4, Supplementary Material online).
Discussion
Adaptive Evolution of Cetacean Hypoxia-Tolerance-Related Genes
Physiological adaptation of hypoxia tolerance in marine mammals has been widely investigated for decades (Kooyman et al. 1981). It was suggested that adaptive changes might have occurred in oxygen stores and transport, as well as cardiovascular system, however, exploring the genetic basis of these adaptations has just begun recently. In this study, we conducted comparative analyses of the selective pressure on 17 hypoxia-tolerance-related genes in cetaceans and terrestrial mammals, including not only oxygen store and transport-related genes but also vasoconstrictor-related genes, to comprehensively understand the mechanism of hypoxia tolerance during the origin and diversification of cetaceans.
Our analyses provide strong evidence that several hypoxia-tolerance-related genes have been subjected to adaptive evolution in cetacean. Selection analysis showed that significant evidence for positive selection was examined at 10 of 17 hypoxia-tolerance-related genes including HBA, HBB, MB, EDN1, EDN2, EDN3, EDNRA, EDNRB, ADRA1D, and AVP in cetacean. Particularly, 42 specific codons identified repeatedly under selection by several ML methods made this conclusion more conservative. Moreover, adaptive evolution was further supported by evidence that high proportion (78.6%) of the putatively selected sites was under radical changes in amino acid property, and several of them fall in regions important for function based on structural information, which suggested that the radical amino acid changes might have influenced protein properties and functions. Overall, positive selection may be the major driving force for the evolution of hypoxia-tolerance-related genes to evolve enhancing oxygen storage and transportation ability, as well as vasoconstriction in cetaceans.
The strongest evidences for positive selection were seen at HBA, HBB, and MB, in agreement with previous studies (Dasmeh et al. 2013; Mirceta et al. 2013; Nery et al. 2013b). Mirceta et al. (2013) reported an adaptive molecular signature of elevated MB net surface charge in diving species with maximal MB concentration by modeling the evolution history of MB (Mirceta et al. 2013). Significant higher evolutionary rate was also identified at MB in cetaceans, and several mutations of positively selected sites were suggested to contribute to a higher MB stabilization correlated with increased protein abundance (Dasmeh et al. 2013). Moreover, Nery et al. (2013b) identified several mutations in HBA and HBB genes in two cetacean species that overall contribute to their ability to cope with oxygen limitation. In our study, 25 codons were concordant between at least 2 ML methods and thus constitute strong candidates for positive selection. Some of the amino acid changes at these selected sites are radical in terms of their physicochemical properties strengthening the case for positive selection. Thus, adaptation of increased oxygen storage and transportation in cetacean may be explained by physiochemical property changes in particular selected codons. For example, position 60 in HBB is an N6-acetyllysine modified residue, which is characterized by changes in its “Isoelectric Point” (pHi) property in cetaceans (table 3). Protein isoelectric point plays an important role in the modulation of protein–surfactant interactions, especially the electrostatic contribution at low surfactant concentrations (Santiago et al. 2010). We therefore deduce that the unambiguous amino acid changes at this site are likely to be involved in regulation of protein interaction. Interestingly, some of the sites with evidence of positive selection detected here have been reported to be associated with functional adaptation in previous studies, which further supported our conclusions. For example, Nery et al. (2013b) found that the residue 62 of HBB gene falls into E helix of the beta chain, and seemed to be essential in the maintenance of the heme in the nonpolar pockets of the alpha and beta chains. In spite of the evidence for selection documented here, further investigation on the functional verification of these important sites in cetaceans is necessary in the future to determine their role in hypoxia adaptation.
Different with the above globins, vasoconstrictor tone plays an important role in regulating blood pressure and distributing blood flow in the body (Jackson 2000) that were typically exhibited in physiological adaptation of cetaceans during hypoxic diving. Seven genes (EDN1, EDN2, EDN3, EDNRA, EDNRB, ADRA1D, AVP) also showed extensive evidence of positive selection, which most likely explains the adaptation at molecular level of vasoconstriction observed in cetaceans. Of these genes, ADRA1D is known to be responsible for the contractile response, especially in the aorta, and ADRA1D-knockout mice showed dramatically decreased contractile responses in the aorta and in the mesenteric arterial beds (Tanoue et al. 2003). The observed positive selection in the ADRA1D gene in cetaceans thus suggests that it is important for enhanced arterial vasoconstriction ability. In addition, the AVP gene, which is one of the strongest vasoconstrictors in the skin (Dünser et al. 2003), was also subject to positive selection across cetaceans, suggesting that AVP seems to be essential in enhanced vasoconstriction abilities for reducing blood flow to the skin. Furthermore, the endogenous vasoconstrictor endothelins (EDN1, EDN2, and EDN3) also regulate vasoconstriction by binding to receptors (EDNRA and EDNRB) (Masaki 2000). In this case, 12 selected condons appear robust among analyses. Of these, site 347 in ENDRB is a charge-modifying amino acid located in a ligand interaction subdomain (Sakamoto et al. 1993), and ten properties including polarity (p) had been detected (table 3). Hence, it is reasonable to deduce that shift on this site may affect binding affinity of endothelin. It should be noted that EDNRB knockout mice have a significant reduction in vascular contraction (Berthiaume et al. 1998), which suggests that EDNRB plays an important role in maintaining vasoconstriction effect. The structure of propeptide region has been shown to be associated with synthesis of endothelins (Moore et al. 1992), and several of the putative sites under selection (EDN1: 125; EDN2: 114, 162; EDN3: 34, 152, 167) are in this region. Hence, it appears that these sites under positive selection are important for endothelin synthesis. Again, the endothelin system has a basal vasoconstricting role in the vascular homeostasis (Kedzierski and Yanagisawa 2001), and its positive selection can thus suggest an essential role in regulating basal vascular tone in cetaceans. Additionally, the endothelin system also plays a key role in controlling renal blood flow and water and sodium reabsorption in the kidney (Kedzierski and Yanagisawa 2001), suggesting that its positive selection maybe associated with reduced blood flow in the kidney and ceased renal filtration to save oxygen during diving (Elsner et al. 1966). Taken collectively, consistent evidences at molecular level show a strong functional role for vasoconstrictor-related genes, which drive cetaceans to evolve effective strategies of vascular regulation to cope with the challenge of hypoxic conditions.
Selection Pressures in Cetaceans in Relation to Diving Abilities
Cetaceans have evolved a fully aquatic lifestyle and diversified into all major aquatic ecosystems. It was shown that different selective pressures presented in hypoxia-tolerance-related genes between long-diving with short-diving cetaceans, which were expected given the wide variety of niches and environments these cetaceans occupy. For short-diving cetaceans, they are usually coastal and inshore, and their foraging takes place in epipelagic or shallow waters (Barros and Randall 1998). In contrast, long-diving cetaceans are almost exclusively benthic or bathypelagic, and prefer to forage in benthic habitats primarily targeting cephalopods (Karczmarski et al. 2000; Watwood et al. 2006). We therefore hypothesized that divergent selection acted on cetaceans with different diving abilities. Additionally, our results demonstrated evidence for positive selection of HBB in long-diving cetaceans. It was suggested that deep-diving, pelagic species required more oxygen for metabolism (Ridgway and Johnston 1966). Notably, species-specific substitutions identified on long-diving cetaceans may be functional adaptations for the deep-diving foraging behavior. For example, a substitution (H3Q) (supplementary fig. S5, Supplementary Material online) was identified in the HBB gene in the sperm whale, a species famous for its exceptional long-diving ability. A functional study demonstrated that this mutation corresponded to a 2, 3-bisphosphoglycerate-binding site that plays a key role in oxygen affinity (Harano et al. 1983). Thus, positive selection at this site suggests further improvements in oxygen storage and transport in the long-diving whales, consistent with their outstanding diving abilities.
Convergent Evolution in Hypoxia-Exposed Animals
Convergence is an evolutionary phenomenon whereby a similar-appearing trait evolves independently in two or more lineages as a result of adaptation to similar environmental pressures, for example, echolocation in bats and dolphins (Parker et al. 2013). Interestingly, the different hypoxia-exposed animals studied here, including plateau, burrowing, and marine species of mammals, also shared a number of convergent phenotypic adaptations to increased oxygen utilization, regulation of cardiovascular function, and reduced energy metabolism (Ramirez et al. 2007). However, the genetic bases of these convergent phenotypic traits have not been well studied. We revealed two lines of genetic evidence underlying convergent phenotypic traits in these animals. First, some positively selected genes (i.e., HBA and HBB) with known functional associations overlapped among different hypoxia-exposed animals, consistent with the convergent phenotypic evolution in these species. HBA and HBB are indispensable for the formation of HB that plays important roles in storing and transporting oxygen in the blood (Hardison 1996). Physiological studies reported that hypoxia-exposed animals show increased oxygen storage and/or more effective oxygen-loading and diffusion strategies in order to sustain their oxygen supply and utilization (Ramirez et al. 2007; Storz et al. 2010). HBA and HBB were positively selected in plateau and marine mammals, and may in turn increase their oxygen storage and transport abilities, which represent the main phenotypic selection targets in a hypoxic environment. Second, we identified three parallel nonsynonymous changes in two vasoconstrictor genes (ADRA1D, AVPR1B) along more than one hypoxia-resistant lineage, representing significant convergent evolution. Vasoconstrictors play important roles in regulating contractile activity in the systemic circulation (Jackson 2000). Convergent evolution of these vasoconstrictor genes could therefore be related to convergent phenotypic traits, such as changes in the regulation of vascular tone. For example, vasoconstriction is thought to cause hypoxia-induced pulmonary arterial hypertension (Humbert et al. 2004), which often occurs in humans unused to high altitudes but not in hypoxia-resistant populations from high altitudes, such as Tibetans (Beall 2007) and yaks (Qiu et al. 2012). In contrast, peripheral vasoconstriction represents an efficient strategy for overcoming hypoxia during diving in marine mammals (Ramirez et al. 2007). Convergent evolution of vasoconstrictor genes thus reflects the similar evolutionary pattern of vascular-tone regulation in response to increased oxygen demands in different hypoxia-exposed animals. In summary, the observed genetic and phenotypic convergence suggests that different hypoxia-exposed animals have followed similar evolutionary paths in order to adapt to hypoxic conditions.
Supplementary Material
Supplementary tables S1–S10 and figures S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the National Outstanding Young Scientists Foundation of China to G.Y. (grant no. 31325025); the National Natural Science Foundation of China (grant no. 31172069 to G.Y., grant no. 31370401 to WH Ren, grant no. 31570379 to S.X.); the Priority Academic Program Development of Jiangsu Higher Education Institutions to G.Y. and S.X.; the Natural Science Foundation of Jiangsu Province to S.X. (grant no. BK20141449); and the Cultivation Program for Doctoral New Academic Researcher granted by Nanjing Normal University to R.T. (grant no. 1812000002115). We are grateful to Dr Anli Gao, Mr Xinrong Xu, and Dr Bingyao Chen for their contributions to sample collection, and to Dr Wenhua Ren, Weijing Zhang, and Yanyan Liang for helping in data edition and bioinformatic analyses.
Literature Cited
- Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105. [DOI] [PubMed] [Google Scholar]
- Ardaillou R. 1999. Angiotensin II receptors. J Am Soc Nephrol. 10:S30–S39. [PubMed] [Google Scholar]
- Avivi A, et al. 2010. Neuroglobin, cytoglobin, and myoglobin contribute to hypoxia adaptation of the subterranean mole rat Spalax. Proc Natl Acad Sci U S A.. 107:21570–21575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros NB, Randall SW. 1998. Prey and feeding patterns of resident bottlenose dolphins (Tursiops truncatus) in Sarasota Bay, Florida. J Mammal. 79:1045–1059. [Google Scholar]
- Beall CM. 2007. Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc Natl Acad Sci U S A. 104:8655–8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthiaume N, et al. 1998. Pharmacology of endothelins in vascular circuits of normal or heterozygous endothelin-A or endothelin-B knockout transgenic mice. J Cardiovasc Pharm. 31:S561–S564. [DOI] [PubMed] [Google Scholar]
- Birchard GF. 1997. Optimal hematocrit: theory, regulation and implications. Am Zool. 37:65–72. [Google Scholar]
- Burmester T, Weich B, Reinhardt S, Hankeln T. 2000. A vertebrate globin expressed in the brain. Nature 407:520–523. [DOI] [PubMed] [Google Scholar]
- Butler PJ, Jones DR. 1997. Physiology of diving birds and mammals. Physiol Rev. 77:837–899. [DOI] [PubMed] [Google Scholar]
- Costidis A, Rommel SA. 2012. Vascularization of air sinuses and fat bodies in the head of the Bottlenose dolphin (Tursiops truncatus): morphological implications on physiology. Front Physiol. 3:243.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasmeh P, Serohijos AW, Kepp KP, Shakhnovich EI. 2013. Positively selected sites in cetacean myoglobins contribute to protein stability. PLoS Comput Biol. 9:760–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson RE, Geis I. 1983. Hemoglobin: structure, function, evolution, and pathology. Menlo Park (CA): Benjamin Cummings. [Google Scholar]
- Dünser MW, et al. 2003. Ischemic skin lesions as a complication of continuous vasopressin infusion in catecholamine-resistant vasodilatory shock: incidence and risk factors. Crit Care Med. 31:1394–1398. [DOI] [PubMed] [Google Scholar]
- Elsner R, Franklin DL, Van Citters RL, Kenney DW. 1966. Cardiovascular defense against asphyxia. Science 153:941–949. [DOI] [PubMed] [Google Scholar]
- Elsner R, Gooden B. 1983. Diving and asphyxia: a comparative study of animals and man. Monogr Physiol Soc. 40:1–168. [PubMed] [Google Scholar]
- Feng P, Zheng J, Rossiter SJ, Wang D, Zhao H. 2014. Massive losses of taste receptor genes in toothed and baleen whales. Genome Biol Evol. 6:1254–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatesy J, O'Leary MA. 2001. Deciphering whale origins with molecules and fossils. Trends Ecol Evol. 16:562–570. [Google Scholar]
- Greer JR. 2015. Pathophysiology of cardiovascular dysfunction in sepsis. BJA Educ. 15:316–321. [Google Scholar]
- Harano T, et al. 1983. Hemoglobin okayama [beta 2 (NA 2) His replaced by Gln]: a new ‘silent’ hemoglobin variant with substituted amino acid residue at the 2, 3-diphosphoglycerate binding site. FEBS Lett. 156:20–22. [DOI] [PubMed] [Google Scholar]
- Hardison RC. 1996. A brief history of hemoglobins: plant, animal, protist, and bacteria. Proc Natl Acad Sci U S A. 93:5675–5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbo S, Fago A. 2012. Functional properties of myoglobins from five whale species with different diving capacities. J Exp Biol. 215:3403–3410. [DOI] [PubMed] [Google Scholar]
- Holmes CL, Landry DW, Granton JT. 2003. Science review: vasopressin and the cardiovascular system part 1-receptor physiology. Crit Care. 7:427–434.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert M, Sitbon O, Simonneau G. 2004. Drug therapy: treatment of pulmonary arterial hypertension. New Engl J Med. 351:1425–1436. [DOI] [PubMed] [Google Scholar]
- Jackson WF. 2000. Ion channels and vascular tone. Hypertension 35:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M, et al. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res. 36:W5–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczmarski L, Cockcroft VG, Mclachlan A. 2000. Habitat use and preferences of Indo-Pacific humpback dolphins Sousa chinensis in Algoa Bay, South Africa. Mar Mammal Sci. 16:65–79. [Google Scholar]
- Kedzierski RM, Yanagisawa M. 2001. Endothelin system: the double-edged sword in health and disease. Annu Rev Pharmacol Toxicol. 41:851–876. [DOI] [PubMed] [Google Scholar]
- Kooyman G, Castellini M, Davis R. 1981. Physiology of diving in marine mammals. Annu Rev Physiol. 43:343–356. [DOI] [PubMed] [Google Scholar]
- Lalitha S. 2000. Primer Premier 5. Biotechnol Softw Intern Rep. 1(6):270–272. [Google Scholar]
- Manley L. 1990. Apnoeic heart rate responses in humans. Sports Med. 9:286–310. [DOI] [PubMed] [Google Scholar]
- Marshall JM. 2015. Interactions between local dilator and sympathetic vasoconstrictor influences in skeletal muscle in acute and chronic hypoxia. Exp Physiol. 100:1400–1411. [DOI] [PubMed] [Google Scholar]
- Masaki T. 2000. The endothelin family: an overview. J Cardiovasc Pharm. 35:S3–S5. [DOI] [PubMed] [Google Scholar]
- McGowen MR, Gatesy J, Wildman DE. 2014. Molecular evolution tracks macroevolutionary transitions in Cetacea. Trends Ecol Evol. 29:336–346. [DOI] [PubMed] [Google Scholar]
- McGowen MR, Grossman LI, Wildman DE. 2012. Dolphin genome provides evidence for adaptive evolution of nervous system genes and a molecular rate slowdown. Proc Biol Sci. 279:3643–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirceta S, et al. 2013. Evolution of mammalian diving capacity traced by myoglobin net surface charge. Science 340:1234192.. [DOI] [PubMed] [Google Scholar]
- Moore K, et al. 1992. Plasma endothelin immune reactivity in liver disease and the hepatorenal syndrome. New Engl J Med. 327:1774–1778. [DOI] [PubMed] [Google Scholar]
- Nery MF, Arroyo JI, Opazo JC. 2013a. Accelerated evolutionary rate of the myoglobin gene in long-diving whales. J Mol Evol. 76:380–387. [DOI] [PubMed] [Google Scholar]
- Nery MF, Arroyo JI, Opazo JC. 2013b. Genomic organization and differential signature of positive selection in the alpha and beta globin gene clusters in two cetacean species. Genome Biol Evol. 5:2359–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordway GA, Garry DJ. 2004. Myoglobin: an essential hemoprotein in striated muscle. J Exp Biol. 207:3441–3446. [DOI] [PubMed] [Google Scholar]
- Panneton WM. 2013. The mammalian diving response: an enigmatic reflex to preserve life? Physiology 28:284–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J, et al. 2013. Genome-wide signatures of convergent evolution in echolocating mammals. Nature 502:228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SLK, Frost SD. 2005a. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533. [DOI] [PubMed] [Google Scholar]
- Pond SLK, Frost SD. 2005b. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 22:1208–1222. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818. [DOI] [PubMed] [Google Scholar]
- Qiu Q, et al. 2012. The yak genome and adaptation to life at high altitude. Nat Genet. 44:946–949. [DOI] [PubMed] [Google Scholar]
- Ramirez JM, Folkow LP, Blix AS. 2007. Hypoxia tolerance in mammals and birds: from the wilderness to the clinic. Annu Rev Physiol. 69:113–143. [DOI] [PubMed] [Google Scholar]
- Ranwez V, et al. 2007. OrthoMaM: a database of orthologous genomic markers for placental mammal phylogenetics. BMC Evol Biol. 7:241.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway SH, Johnston DG. 1966. Blood oxygen and ecology of porpoises of three genera. Science 151:456–458. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. [DOI] [PubMed] [Google Scholar]
- Sakamoto A, et al. 1993. Distinct subdomains of human endothelin receptors determine their selectivity to endothelin A-selective antagonist and endothelin B-selective agonists. J Biol Chem. 268:8547–8553. [PubMed] [Google Scholar]
- Santiago PS, et al. 2010. Isoelectric point determination for Glossoscolex paulistus extracellular hemoglobin: oligomeric stability in acidic pH and relevance to protein-surfactant interactions. Langmuir 26:9794–9801. [DOI] [PubMed] [Google Scholar]
- Schneuer M, et al. 2012. Neuroglobin of seals and whales: evidence for a divergent role in the diving brain. Neuroscience 223:35–44. [DOI] [PubMed] [Google Scholar]
- Shen T, et al. 2012. Adaptive evolution and functional constraint at TLR4 during the secondary aquatic adaptation and diversification of cetaceans. BMC Evol Biol. 12:39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder GK. 1983. Respiratory adaptations in diving mammals. Respir Physiol. 54:269–294. [DOI] [PubMed] [Google Scholar]
- Sowerby A. 2009. An investigation of the properties of large-conductance Ca2+ activated K+ channels of rat arterial smooth muscle and their modulation by vasoconstrictors [phd dissertation]. [Leicester (UK)]: University of Leicester.
- Storz JF, Scott GR, Cheviron ZA. 2010. Phenotypic plasticity and genetic adaptation to high-altitude hypoxia in vertebrates. J Exp Biol. 213:4125–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagar K, Johnson WE, O'Brien SJ, Vasconcelos V, Antunes A. 2012. Evolution of CRISPs associated with toxicoferan-reptilian venom and mammalian reproduction. Mol Biol Evol. 29:1087–1822. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Nielsen R, Yang Q. 2003. Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol. 20:18–20. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue A, et al. 2003. Insights into α1 adrenoceptor function in health and disease from transgenic animal studies. Trends Endocrinol Metab. 14:107–113. [DOI] [PubMed] [Google Scholar]
- Thewissen JG, Cooper LN, Clementz MT, Bajpai S, Tiwari B. 2007. Whales originated from aquatic artiodactyls in the Eocene epoch of India. Nature 450:1190–1194. [DOI] [PubMed] [Google Scholar]
- Van Aardt WJ, Bronner G, Buffenstein R. 2007. Hemoglobin-oxygen-affinity and acid-base properties of blood from the fossorial mole-rat, Cryptomys hottentotu spretoriae. Comp Biochem Physiol A Mol Integr Physiol. 147:50–56. [DOI] [PubMed] [Google Scholar]
- Wang Z, et al. 2015. ‘Obesity’ is healthy for cetaceans? Evidence from pervasive positive selection in genes related to triacylglycerol metabolism. Sci Rep. 5:14187.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins WA, Daher MA, Fristrup KM, Howald TJ, Di Sciara GN. 1993. Sperm whales tagged with transponders and tracked underwater by sonar. Mar Mammal Sci. 9:55–67. [Google Scholar]
- Watwood SL, et al. 2006. Deep-diving foraging behaviour of sperm whales (Physeter macrocephalus). J Anim Ecol. 75:814–825. [DOI] [PubMed] [Google Scholar]
- Woolley S, Johnson J, Smith MJ, Crandall KA, McClellan DA. 2003. TreeSAAP: selection on amino acid properties using phylogenetic trees. Bioinformatics 19:671–672. [DOI] [PubMed] [Google Scholar]
- Wright CD, Wu SC, Dahl EF, Sazama AJ, O'Connell TD. 2012. Nuclear localization drives α1-adrenergic receptor oligomerization and signaling in cardiac myocytes. Cell Signal. 24:794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, et al. 2012. Positive selection at the ASPM gene coincides with brain size enlargements in cetaceans. Proc Biol Sci. 279:4433–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, et al. 2013. Adaptive evolution of the osmoregulation-related genes in cetaceans during secondary aquatic adaptation. BMC Evol Biol. 13:189.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 15:568–573. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WS, Nielsen R. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 22:1107–1118. [DOI] [PubMed] [Google Scholar]
- Yim HS, et al. 2014. Minke whale genome and aquatic adaptation in cetaceans. Nat Genet. 46:88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kumar S. 1997. Detection of convergent and parallel evolution at the amino acid sequence level. Mol Biol Evol. 14:527–536. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 22:2472–2479. [DOI] [PubMed] [Google Scholar]
- Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K, et al. 2014. The loss of taste genes in cetaceans. BMC Evol Biol. 14:218.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.