Abstract
Rabies is a rapidly progressive lyssavirus encephalitis that is statistically 100% fatal. There are no clinically effective antiviral drugs for rabies. An immunologically naïve teenager survived rabies in 2004 through improvised supportive care; since then, 5 additional survivors have been associated with use of the so-called Milwaukee Protocol (MP). The MP applies critical care focused on the altered metabolic and physiologic states associated with rabies. The aim of this study was to examine the metabolic profile of cerebrospinal fluid (CSF) from rabies patients during clinical progression of rabies encephalitis in survivors and nonsurvivors and to compare these samples with control CSF samples. Unsupervised clustering algorithms distinguished three stages of rabies disease and identified several metabolites that differentiated rabies survivors from those who subsequently died, in particular, metabolites related to energy metabolism and cell volume control. Moreover, for those patients who survived, the trajectory of their metabolic profile tracked toward the control profile and away from the rabies profile. NMR metabolomics of human rabies CSF provide new insights into the mechanisms of rabies pathogenesis, which may guide future therapy of this disease.
Keywords: rabies, metabolomics, NMR spectroscopy, cerebrospinal fluid
Graphical Abstract
INTRODUCTION
Rabies is a rapidly progressive encephalomyelitis caused by RNA viruses in the family Rhabdoviridae, genus Lyssavirus, with a very high fatality rate.1 While preventable by vaccine, more than 55000 cases occur annually worldwide, mostly in children and resource-limited settings.2 Rabies virus infection is notable for being minimally cytolytic and poorly inflammatory.3 For this reason, experts have long postulated that rabies is a disorder of neurotransmission; however, a clear candidate neurotransmitter or signaling pathway has not been identified. Since the advent of critical care in the 1970s, five rare survivors were reported among persons developing rabies; all had received rabies vaccine prior to development of rabies symptoms.4–8 The first vaccine-naïve survivor was treated in 2004 using an approach later named the Milwaukee Protocol (MP).9 The protocol was improvised around ketamine, a neuroprotective anesthetic with putative activity against the rabies virus in vivo.10,11 Treatment requires sedating the patient for prevention of dysautonomia until the patient generates a natural immune response to the rabies virus. The MP has been applied 43 times, with five additional young survivors and a significant increase in survival times.12
Extended survival times have permitted systematic observations in the clinic and laboratory of patients treated with the MP.9,13–17 It has been shown that rabies infection results in disordered metabolism, including a reproducible and reversible deficiency of tetrahydrobiopterin (BH4) and monoamine neurotransmitters, and an increase in lactic acid in cerebrospinal fluid (CSF).17,18 Vasospasm has been noted in some reports but not in others.16,19–22 CSF lactic acidosis >4 mM has been incorporated in the MP as a component of the ad hoc criteria for medical futility in rabies.12
There are no antiviral drugs that show clinical benefit in rabies. Improved survival in human rabies remains dependent on critical care and anticipation of complications. Given that rabies includes several acquired disorders of metabolism, metabolomics is a promising approach to confirm and expand our understanding of the natural history of rabies during critical care. Metabolomics measures a range of small molecules present in a biological system, usually by mass spectrometry or nuclear magnetic resonance. When combined with multivariate statistical analysis, metabolomics characterizes the physiologic state of different biofluids.23–25 Metabolomics has contributed greatly to our understanding of the metabolic actions of pharmaceutical agents26 and the diagnosis of chronic and infectious diseases.27,28 Recently, metabolomic technologies have been applied to the analysis of CSF from normal and diseased cohorts.24,29 CSF is partially derived from interstitial fluid in the central nervous system (CNS); therefore, the composition of the CSF can be expected to reflect the biological processes of the brain. Indeed, metabolomic investigations have described metabolite profiles characteristic of CNS diseases including meningitis,30 multiple sclerosis,29 and amyotrophic lateral sclerosis.31
The present study set out to test whether metabolomics could track the progression of metabolic changes over time in survivors and nonsurvivors. The application of metabolomics in this case may contribute to a deeper understanding of rabies pathogenesis and may thereby help to refine future rabies therapy. We anticipate that proton nuclear magnetic resonance (1H NMR) based metabolomics may be useful for guiding appropriate treatment.
MATERIALS AND METHODS
Sample Collection
For this study, rabies CSF was acquired either from residual material collected during treatment, or additional volumes (1 mL) collected during standard care and obtained with informed consent. In total, 44 CSF samples were collected from 11 patients with laboratory confirmed rabies. Control CSF volumes were collected from the Clinical Chemistry laboratory at Children’s Hospital of Wisconsin from 25 patients aged 5–25 years without corresponding microbiological assessment. All samples were deidentified and analyzed in this state. For rabies samples, select information restricted to a unique anonymous identifier (for time-series analysis), age (years), gender, country and source of rabies were retained for rabies subjects to properly interpret the data. Clinical information on rabies patients in this study was abstracted from detailed correspondence during the medical care of each patient, but formal charts were not available. The study was approved by the Institutional Review Boards at the Children’s Hospital of Wisconsin, and UC Davis. Once collected, CSF was centrifuged to discard cellular elements and refrigerated or frozen (−20 °C) until the time that they were transferred to the coordinating research center where they were stored at −80 °C until analysis. Control samples retained from residual CSF samples at the Children’s Hospital of Wisconsin were collected, centrifuged and stored at −80 °C until analysis.
1H NMR Spectroscopy
Sample Preparation
CSF samples were removed from −80 °C storage and allowed to thaw. Once defrosted, samples were centrifuged using a filter with a cutoff of 3000 MW (Pall, Ann Arbor, MI) to remove lipids and proteins. The filtrate volume was adjusted to 585 µL with Type I ultrapure water from Millipore Synergy UV system (Millipore, Billerica, MI). Samples were prepared for analysis by the addition of 65 µL of internal standard containing approximately 5 mmol/L of DSS-d6 [3-(trimethylsilyl)-1-propanesulfonic acid-d6], 0.2% NaN3, in 99.8% D2O to 585 µL of CSF. The pH of each sample was adjusted to 6.8 ± 0.1 by adding small amounts of NaOH or HCl. A 600 µL aliquot was subsequently transferred to 5 mm Bruker NMR tubes and stored at 4 °C until NMR acquisition (within 24 h of sample preparation).
Data Acquisition and Metabolite Quantification
NMR spectra were acquired as previously described23 on a Bruker Avance 600-MHz NMR equipped with a SampleJet autosampler using a NOESY-presaturation pulse sequence (noesypr) at 25 °C. Spectra were acquired with 8 dummy scans and 32 transients over a spectral width of 12 ppm with a total acquisition time of 2.5 s. Water saturation was achieved during the prescan delay (2.5 s) and mixing time (100 ms). Once acquired, all spectra were zero-filled to 128k data points and Fourier transformed with a 0.5-Hz line broadening applied. Spectra were manually phased and baseline corrected using NMR Suite v6.1 Processor (Chenomx Inc., Edmonton, Canada). Metabolite quantification was achieved using the 600-MHz library from Chenomx NMR Suite v6.1 Profiler (Chenomx Inc., Edmonton, Canada), which uses the concentration of a known reference signal (in this case DSS) to determine the concentration of individual compounds as previously described.32 Identification and quantitation of compounds using Chenomx software has previously been validated and shown to be both accurate and precise.23 Metabolites were quantified in micromolar (µM) units and exported from Chenomx. The drug vehicle propylene glycol was removed from the list of metabolites prior to analysis to eliminate impact on metabolic interpretations.
Data Analysis and Visualization
Metabolite concentrations were log10-transformed and imported into SIMCA-P software (version 11.0; Umetrics, Umeå, Sweden) for analysis. Data were mean centered and unit variance scaled. Unsupervised principal component analysis (PCA) was applied to all CSF samples and scores plots were visually inspected for trends or outliers in the data. Partial least-squares discriminant analysis (PLS-DA) was then used to explore variations in metabolite concentrations between different classes within the data, e.g. control samples versus rabies samples, stages of rabies disease and survivors versus nonsurvivors of rabies. A scores plot was created to visualize the PLS-DA model, and the corresponding loadings provided information on the contribution of metabolites to the separation of classes. The variable importance in the projection (VIP) value of each metabolite in the model was calculated to indicate its contribution to the classification of samples. The VIP is a weighted sum of squares of the PLS weights with the weights calculated from the amount of dependent variable variance of each PLS component. Since the average of squared VIP values equals 1, we chose a cut off of >1.5 to identify metabolites that were important in discriminating between groups. The quality of all models was judged by the goodness-of-fit parameter (R2) and the predictive ability parameter (Q2), which is calculated by an internal cross-validation of the data and the predictability calculated on a leave-out basis. In addition, all PLS-DA models were validated externally by randomly selecting three-quarters of the samples to be used as a training data set with the remaining samples acting as a test data set. The class of each sample in the test data set was predicted on the basis of the model built from the training set. This was repeated until all samples were predicted. Differences in metabolite concentrations between groups were validated in PASW Statistics version 18.0 for Windows (IBM SPSS Inc. Chicago, IL) using independent t tests. P-values were adjusted for multiple testing using the false discovery rate (FDR) correction method.33 Significance was assumed at p < 0.05.
Identification of Stages of Rabies Disease
Unsupervised cluster analysis was performed using the k-means cluster algorithm in PASW Statistics version 18.0 for Windows (IBM SPSS Inc., Chicago, IL) to identify different stages of rabies disease from CSF metabolite data. Before clustering, all metabolite concentrations were log10-transformed. The k-means method assumes a certain number of clusters, k, fixed a priori, and produces a separation of the objects into nonoverlapping groups coming from Euclidean distances. Cluster membership is exclusive and dependent on minimizing the Euclidean distance within each cluster and maximizing differences between clusters at each step of an iterative procedure. Thus, a cluster represents a group of individuals with similar metabolic characteristics. We examined a split according to three clusters and a maximum of ten iterations were used. Differences in the metabolite concentrations across clusters were evaluated using ANOVA. Where statistically different effects were identified (p < 0.05), comparisons were made between clusters using the Bonferroni post hoc multiple comparison test. k-means cluster determined stages of rabies disease were visualized using PLS-DA scores plots as described in the previous section.
RESULTS
Twenty-five control CSF samples and 44 CSF samples from 11 patients with laboratory confirmed rabies were analyzed using 1H NMR spectroscopy. A total of 56 metabolites were identified in the CSF profiles from patients and controls with complete overlap. Initial data inspection identified extreme outlying samples from two rabid patients. Nine samples from one patient included extreme concentrations of ethylene glycol suggesting intoxication from local remedies or counterfeit medications. This patient’s clinical course was atypical, with early seizures and cardiopulmonary arrests. One more sample from a different patient was also associated with a cardiopulmonary arrest hours earlier. These extreme outliers were removed and the remaining 34 samples from nine rabies patients formed the basis of the primary analysis.
CSF samples from rabies patients were collected at time points determined by medical treatment, ranging from hospital day (HD) 1 to 59; the number of samples collected ranged from 2 to 8 samples per patient. Six of the patients were children aged between 4 and 11, 5 males and 1 female; and three were adults aged 37–42, 2 males and 1 female. Three patients contracted rabies from a bat while six contracted rabies from a dog. Countries of origin included Colombia, Dominican Republic, Equatorial Guinea, Ireland (ex-South Africa), Peru, and cases imported into the United States from India and the Philippines. Clinical characteristics for the rabies patients included in this analysis are listed in Table 1. The 25 control samples were single sample collections from anonymous patients ranging in age from 5 to 25 years.
Table 1.
Clinical Characteristics of Rabies Patients
| case | age (years) |
gender | contry of origin/ treatment site |
animal | CSF samples |
clinical outcome |
|---|---|---|---|---|---|---|
| 1 | 11 | Male | Philippines/United States | Dog | 3 | Deceased |
| 2 | 5 | Male | Equatorial Guinea | Dog | 5 | Deceased |
| 3 | 8 | Female | Columbia | Bat | 6 | Survival |
| 4 | 37 | Female | South Africa/Ireland | Dog | 2 | Deceased |
| 5 | 4 | Male | Dominican Republic | Dog | 2 | Deceased |
| 6 | 7 | Male | Dominican Republic | Dog | 3 | Deceased |
| 7 | 42 | Male | India/United States | Dog | 8 | Deceased |
| 8 | 42 | Male | Peru | Bat | 2 | Survival |
| 9 | 9 | Male | Peru | Bat | 3 | Deceased |
Discrimination of Rabies from Controls
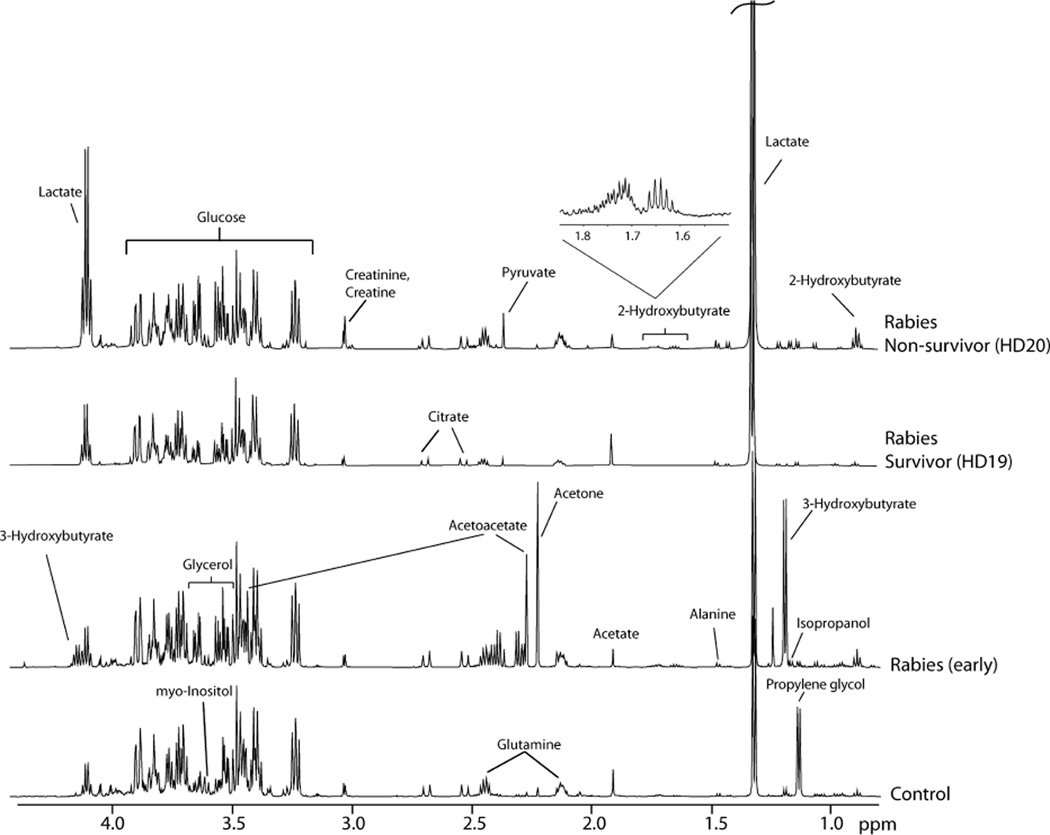
Typical 1H NMR spectra obtained for CSF samples from rabies patients and controls are shown in Figure 1. A principal component analysis (PCA) scores plot of all 1H NMR CSF data was created; the first three components accounted for 43% of the variation in the data. PCA illustrated a clear separation between rabies patients and control samples. To investigate this further, PLS-DA was applied resulting in a 2 component model (R2X = 0.34, R2Y = 0.87, Q2 = 0.81). External validation of the model, as described in the Materials and Methods section, indicated a robust model. Analysis of variable importance in the projection (VIP) values identified metabolites responsible for the separation of patients and controls (Table 2). Independent t tests confirmed that the concentrations of the metabolites identified from multivariate analysis were significantly different between control and rabies samples. The most significant discriminating metabolites (VIP >1.5) included quinolinate, malate, lactate, glycerol, and isobutyrate.
Figure 1.
1H NMR spectra obtained for CSF samples from controls, early stage rabies patients, and late stage (hospital day 19 and 20) rabies patients (survivor vs nonsurvivor).
Table 2.
Concentrations and VIP Values of the Metabolites Responsible for the Separation of Control Patients and Rabies Patientsa
| control (µM) | rabies (µM) | ||||||
|---|---|---|---|---|---|---|---|
| n = 25 | n = 34 | ||||||
| metabolite | pathway | VIP | mean | SD | mean | SD | P value |
| Quinolinate | Excitotoxic | 2.06 | 0.19b | 0.61b | 15.15 | 14.03 | <0.0001 |
| Malate | Cell volume/TCA cycle | 1.62 | 3.29b | 3.78b | 17.59 | 13.28 | <0.0001 |
| Lactate | Glycolysis | 1.58 | 1590.08 | 344.14 | 4539.63 | 3114.26 | <0.0001 |
| Glycerol | Lipid turnover | 1.53 | 249.84 | 111.64 | 563.83 | 246.67 | <0.0001 |
| Isobutyrate | Organic acid | 1.50 | 7.64 | 2.22 | 13.23 | 4.52 | <0.0001 |
| 2-Hydroxybutyrate | Oxidative stress | 1.45 | 32.91 | 13.17 | 69.96 | 33.50 | <0.0001 |
| Fructose | Carbohydrate | 1.41 | 138.78 | 47.67 | 71.09 | 44.32 | <0.0001 |
| N-Acetylaspartate | Cell volume/TCA cycle | 1.34 | 2.22 | 0.66 | 7.00 | 5.43 | <0.0001 |
| Arginine | Protein catabolism/Urea cycle/NO | 1.30 | 25.85 | 7.15 | 41.94 | 13.92 | <0.0001 |
| Formate | Organic acid | 1.28 | 26.80 | 7.47 | 68.82 | 60.45 | <0.0001 |
| 2-Oxoglutarate | Cell volume/TCA cycle/Transamination | 1.27 | 1.82 | 2.01 | 14.38 | 21.28 | <0.0001 |
| 2-Hydroxyisovalerate | Organic acid | 1.24 | 6.84 | 6.35 | 18.80 | 15.29 | <0.0001 |
| Pyroglutamate | Organic acid | 1.23 | 21.97 | 8.46 | 51.75 | 46.77 | <0.0001 |
| Methionine | Protein catabolism | 1.22 | 3.19 | 0.76 | 5.91 | 2.41 | <0.0001 |
| Urea | Urea cycle | 1.19 | 2113.74 | 1245.24 | 5548.26 | 4861.16 | <0.0001 |
| Ornithine | Urea cycle | 1.18 | 4.20 | 1.64 | 12.46 | 13.00 | <0.0001 |
| Choline | Lipid turnover | 1.15 | 1.82 | 0.48 | 5.15 | 4.24 | <0.0001 |
| Succinate | Cell volume/TCA cycle | 1.13 | 0.64 | 0.58 | 4.06 | 6.39 | <0.0001 |
| NAAG | Cell volume | 1.12 | 1.46 | 0.67 | 0.91 | 0.39 | <0.0001 |
| 2-Oxoisocaproate | Organic acids | 1.10 | 1.36 | 0.41 | 2.14 | 0.84 | <0.0001 |
| Fucose | Carbohydrate | 1.09 | 4.66 | 3.26 | 8.30 | 4.19 | <0.0001 |
| Alanine | Protein catabolism/Transamination | 1.06 | 28.05 | 9.04 | 53.34 | 35.49 | <0.0001 |
| Serine | Protein catabolism | 1.04 | 44.97 | 10.52 | 33.34 | 15.22 | <0.0001 |
| Glucose | Glycolysis | 1.02 | 3394.95 | 464.15 | 4396.57 | 1376.40 | 0.048 |
Metabolite concentrations were quantified from NMR spectra of CSF from control samples and rabies samples. Data are presented as mean and standard deviation (SD). P values are based on independent samples t-tests, adjusted for multiple testing using the FDR correction. VIP, Variable importance in the projection; NAAG, N-acetylaspartylglutamate; NO, nitric oxide.
Mean concentrations are not accurate due to concentrations for many samples being below the detection limit.
Identification of Three Stages of Rabies Pathogenesis
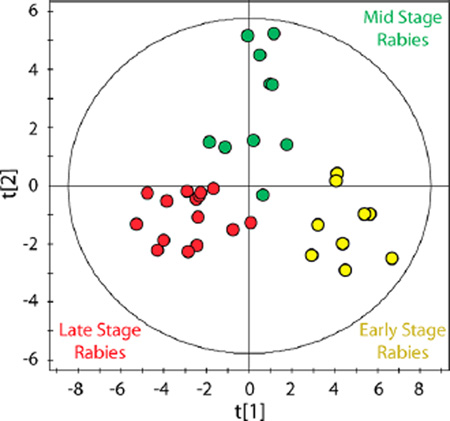
k-means cluster analysis was employed to probe inherent patterns in human rabies CSF. This unsupervised approach identified 3 distinct groups of samples. These clusters of metabolites were found to segregate temporally, as early stage disease (HD 0–7), mid stage disease (HD 8–12) and late stage disease (HD 12+). Partitioning between mid stage and late stage disease was imperfect because samples collected from two rabies survivors did not progress through the three stages of disease. A survivor was defined a priori as a patient with compatible illness and laboratory-confirmed rabies, who was discharged from an intensive care unit to a rehabilitation unit, further documented to lack detectable rabies virus in skin or saliva in the presence of neutralizing antibody (>0.5 IU/mL) to the rabies virus. CSF samples from survivors, collected at HD 19, 39, 48, and 59 and expected to cluster with the late stage group, were instead clustered with midstage samples. For this reason, the mid stage rabies group will be referred to as mid/survivor in future analyses and discussion.
Metabolites that differentiated the three disease stage groups defined by cluster analysis are highlighted in Table 3 and Supporting Information Figure S1. Each stage of rabies is characterized by a unique metabolite profile. Early stage samples have significantly higher concentrations of 3-hydroxybutyrate, acetoacetate and acetone. Mid/survivor samples were characterized by lower concentrations of isopropanol and pyruvate, and higher concentrations of formate, acetate, propionate and glycolate. Late stage samples, associated with nonsurvivors, had significantly higher concentrations of many metabolites. Certain metabolites appear to follow a specific trend over time. For example, lactate, quinolinate, glutamate, malate, 2-oxoglutarate (alpha-ketoglutarate), ornithine, as well as many amino acids and lipid metabolites show an increasing trend across stages (Table 3).
Table 3.
Metabolites that Differentiate the Three Stages of Rabies Disease: Early, Mid, and Late Stage Rabiesa
| early stage (µM) | mid/survivor (µM) | late stage (µM) | |||||
|---|---|---|---|---|---|---|---|
| n = 9 | n = 10 | n = 15 | |||||
| metabolite | mean | SD | mean | SD | mean | SD | P value |
| Energy metabolism | |||||||
| Lactate | a b 2123.60 | 540.93 | a c3358.46 | 674.4 | b c 6776.70 | 3505.78 | <0.001 |
| Pyruvate | 73.37 | 43.98 | a 43.60 | 37.3 | a 132.45 | 63.04 | 0.025 |
| Creatine | a 41.43 | 7.2 | b 54.50 | 26.29 | a b 99.97 | 46.76 | 0.001 |
| Ketosis | |||||||
| 3-Hydroxybutyrate | a b 244.54 | 305.41 | a 10.75 | 8.73 | b 10.11 | 3.48 | <0.001 |
| Acetoacetate | a b 135.28 | 167.67 | a 4.73 | 2.91 | 6.49 | 3.25 | <0.001 |
| Acetone | a b 156.83 | 173.97 | a 31.94 | 79.52 | b 9.65 | 9.93 | <0.001 |
| Cell volume/TCA cycle | |||||||
| Malate | a 9.14 | 3.39 | 19.56 | 16.39 | a 21.35 | 13.14 | 0.052 |
| Succinate | a b 0.76 | 0.59 | a 5.68 | 8.46 | b 4.94 | 6.35 | 0.029 |
| 2-Oxoglutarate | a 2.34 | 1.39 | 9.42 | 12.89 | a 24.92 | 27.01 | 0.012 |
| Glutamate | a 7.22 | 3.17 | 10.34 | 5.29 | a 14.09 | 7.19 | 0.032 |
| Excitotoxic | |||||||
| Glycine | a 16.89 | 18.37 | 16.48 | 18.05 | a 39.33 | 41.18 | 0.08 |
| Quinolinate | a 3.10 | 2.5 | 17.05 | 15.49 | a 21.10 | 13.13 | <0.001 |
| Protein catabolism | |||||||
| Lysine | a 20.65 | 7.16 | 27.94 | 15.71 | a 43.78 | 23.02 | 0.024 |
| Methionine | a 4.43 | 1.6 | 5.99 | 2.54 | a 6.74 | 2.44 | 0.083 |
| Phenylalanine | a 12.93 | 3.16 | 17.78 | 10.93 | a 23.06 | 10.45 | 0.044 |
| Proline | a 9.06 | 3.66 | 10.77 | 6.9 | a 21.20 | 10.77 | 0.012 |
| Tyrosine | a 11.47 | 2.71 | b 17.57 | 8.26 | a b 26.71 | 11.49 | 0.003 |
| Alanine | a b 24.29 | 7.21 | a 47.40 | 18.16 | b 74.74 | 40.86 | <0.001 |
| Arginine | a 33.49 | 8.89 | 37.81 | 10.81 | a 49.76 | 14.60 | 0.044 |
| Urea Cycle | |||||||
| Ornithine | a 6.68 | 7.58 | 9.31 | 3.83 | a 18.03 | 17.16 | 0.007 |
| Urea | a 3374.30 | 2025.69 | 3784.26 | 2143.44 | a 8028.64 | 6208.90 | 0.054 |
| Lipid turnover | |||||||
| Choline | a 2.38 | 1.63 | 3.92 | 1.81 | a 7.64 | 5.1 | 0.005 |
| Ethanolamine | a 15.05 | 10.64 | 25.68 | 14.46 | a 29.36 | 13.45 | 0.043 |
| Other | |||||||
| 2-Hydroxybutyrate | a 85.57 | 29.67 | a 49.77 | 15.94 | 74.05 | 38.89 | 0.063 |
| Ascorbate | 30.81 | 23.88 | a 16.01 | 14.5 | a 49.09 | 41.65 | 0.025 |
| Fructose | a b 108.05 | 64.68 | a 55.96 | 25.53 | b 59.00 | 24.94 | 0.045 |
| Isopropanol | a 9.39 | 7.57 | a b 2.27 | 1.96 | b 16.77 | 19.65 | 0.003 |
Metabolite concentrations were quantified from NMR spectra of CSF samples. Data are presented as mean and standard deviation (SD). The highest mean concentration for each metabolite across stages of disease is highlighted in boldface, the lowest concentration is underlined. P values are based on independent samples t-tests, adjusted for multiple testing using the FDR correction. Superscript letters identify stages with significantly different (p < 0.05) metabolite concentrations.
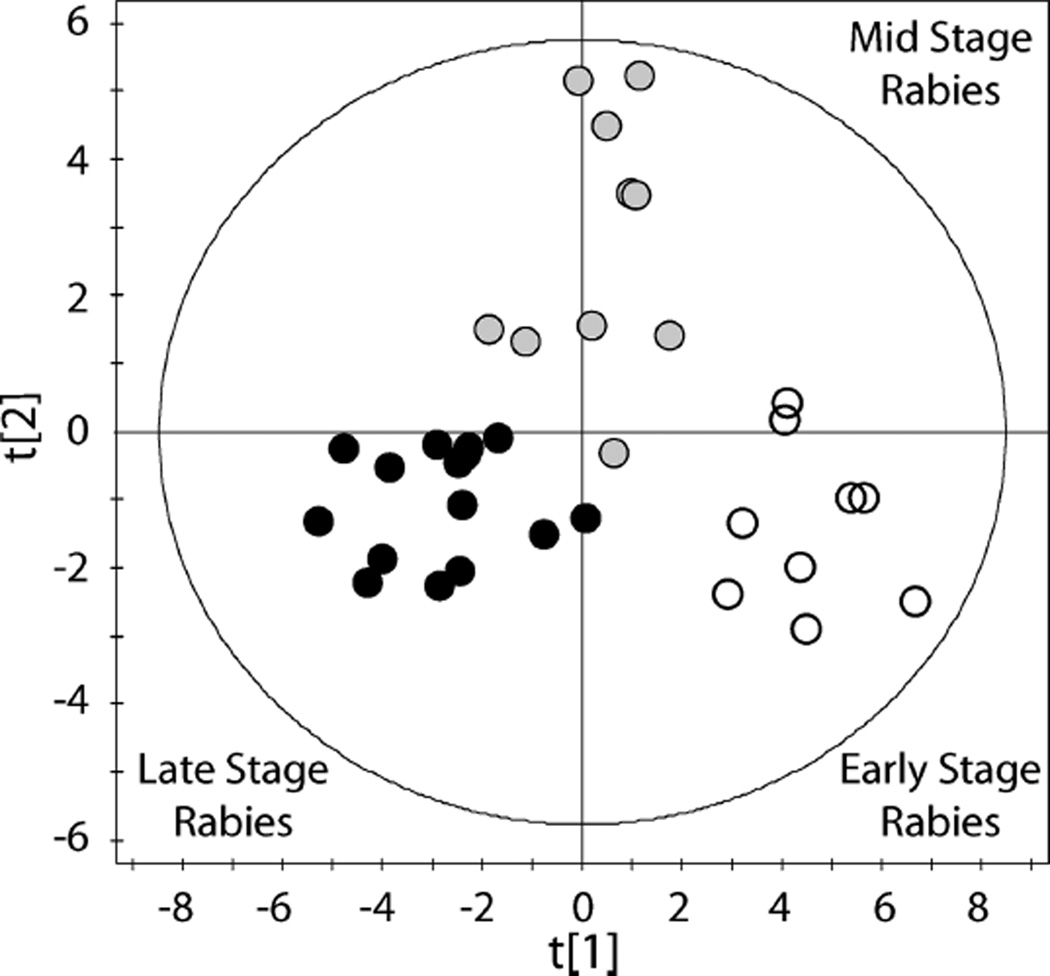
To visualize these phenotypic disease stage profiles, cluster membership information was used to classify samples by PLS-DA. This supervised analysis resulted in a 2 component model describing the metabolomic data (Figure 2: R2X = 0.30, R2Y = 0.77, Q2 = 0.64). PLS-DA also allowed the predictability of the model to be determined; external validation indicated a robust model. To further probe the metabolic differences between cluster groups and to determine the most influential metabolites driving the separation between groups, pairwise comparisons were performed using PLS-DA. Three robust models were created comparing early and mid/survivor samples (R2X = 0.21, R2Y = 0.85, Q2 = 0.68), mid/survivor and late stage samples (R2X = 0.16, R2Y = 0.74, Q2 = 0.56) and early and late stage samples (R2X = 0.26, R2Y = 0.89, Q2 = 0.82). In order to determine the metabolites that were most influential in differentiating rabies stages, a conservative cut off was applied (VIP ≥ 1.5). Increased acetoacetate, 3-hydroxybutyrate, acetone, isopropanol and 2-hydroxybutyrate and lower alanine, lactate and quinolinate separated early samples from mid/survivor samples. Lower concentrations of isopropanol, pyruvate, lactate, creatine and proline and higher concentrations of acetate, propionate and glycolate in mid/survivor samples separated these from late stage rabies samples. As expected, these findings by PLS-DA identified the same metabolites that defined cluster analysis-determined disease stages presented in Table 3.
Figure 2.
Identification of three stages of Rabies pathogenesis. Partial least-squares discriminant analysis (PLS-DA) of 1H NMR CSF metabolomic data comparing three stages of rabies (R2X = 0.30, R2Y = 0.77, Q2 = 0.64). Disease stage groups were determined from metabolomic data using k-means cluster analysis. Open circles represent early stage disease, gray filled circles represent mid stage disease, and black filled circles represent late stage disease. Hotelling’s T2 95% confidence ellipse is indicated on the plot.
Rabies Survivors
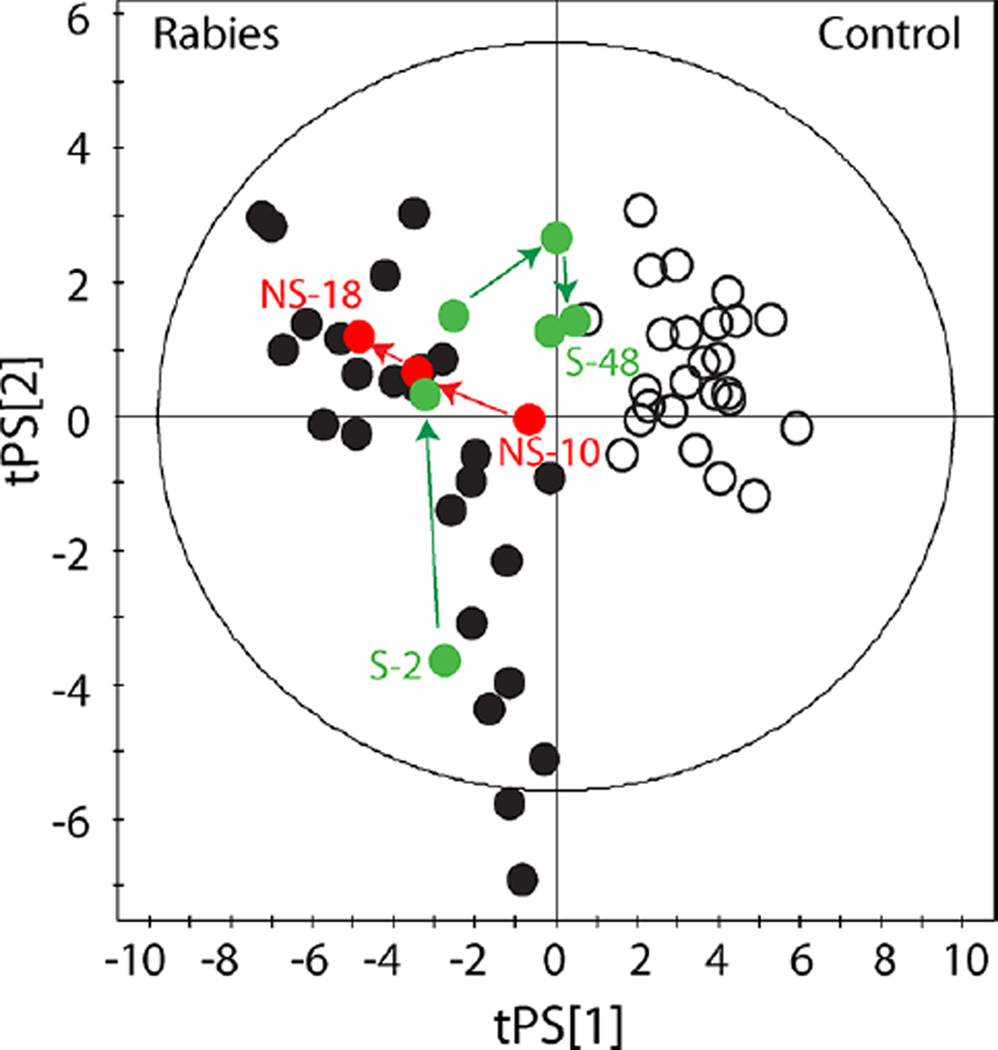
It was evident from cluster analysis that the metabolomic profiles of CSF samples collected from the two rabies survivors differed from those for whom rabies was fatal. PLS-DA scores plots illustrate how survivor profiles return toward controls, while nonsurvivors diverge incrementally (Figure 3). To determine which metabolites discriminate survivors from persons ultimately dying of rabies, the metabolomic profiles of samples collected on HD 19, 39, 48, and 59 from the two surviving patients were compared to four randomly selected late stage CSF samples taken from patients who died. PLS-DA resulted in a 2-component model (R2X = 0.48, R2Y = 0.99, Q2 = 0.82). Concentrations of ascorbate, isopropanol, 2-hydroxyisovalerate, lactate, 3-hydroxyisovalerate, N-acetylaspartate, 2-oxoglutarate, isobutyrate and pyruvate were higher in non-survivors (Table 4).
Figure 3.
Tracking metabolic profiles of survivors and nonsurvivors. Partial least-squares discriminant analysis (PLS-DA) of 1H NMR CSF metabolomic data comparing samples collected from rabies patients and controls (R2X = 0.34, R2Y = 0.87, Q2 = 0.81). Open circles represent control samples; black filled circles represent rabies patient samples. Red filled circles track a nonsurviving (NS) patient from hospital day 10 to day 18, as disease progresses this patient diverges further from control samples. The green filled circles track a survivor (S) who changes from a diseased metabolic profile to reflect a normal metabolic profile as the patient recovers from rabies disease. Hotelling’s T2 95% confidence ellipse is indicated on the plot.
Table 4.
Metabolite Concentrations and VIP Values of the Discriminating Metabolites for Comparison of Rabies Survivors and Persons Dying of Rabiesa
| rabies survivors (µM) | irreversible rabies (µM) | ||||||
|---|---|---|---|---|---|---|---|
| n = 4 | n = 4 | ||||||
| Metabolite | Pathway | VIP | mean | SD | mean | SD | P value |
| Ascorbate | Vitamin | 1.69 | 7.01 | 4.93 | 49.82 | 23.19 | 0.018 |
| Isopropanol | Acetone metabolism | 1.66 | 0.66 | 0.28 | 17.36 | 22.60 | 0.015 |
| 2-Hydroxyisovalerate | Organic acid | 1.53 | 4.87 | 0.94 | 14.06 | 8.27 | 0.038 |
| Lactate | Glycolysis | 1.49 | 3948.63 | 614.75 | 9736.11 | 4121.83 | 0.039 |
| 3-Hydroxyisovalerate | Organic acid | 1.49 | 2.38 | 0.60 | 4.49 | 1.07 | 0.032 |
| N-Acetylaspartate | Cell volume/TCA cycle | 1.49 | 3.21 | 1.61 | 10.00 | 3.62 | 0.027 |
| 2-Oxoglutarate | Cell volume/TCA cycle/Transamination | 1.49 | 3.81 | 1.74 | 36.99 | 23.96 | 0.024 |
| Isobutyrate | Organic acid | 1.47 | 10.41 | 3.72 | 20.63 | 4.83 | 0.023 |
| Pyruvate | Glycolysis | 1.47 | 20.09 | 21.62 | 131.39 | 71.96 | 0.021 |
Metabolite concentrations were quantified from NMR spectra of CSF samples. Data are presented as mean and standard deviation (SD). P values are based on independent samples t-tests, adjusted for multiple testing using the FDR correction. VIP, Variable Importance in the Projection.
DISCUSSION
Despite the heterogeneity of patients, samples, treatment conditions, and sources of rabies virus, 1H NMR spectroscopy combined with multivariate statistical procedures clearly distinguished rabies cases from controls and also distinguished three stages of rabies in patients treated with the Milwaukee Protocol. Fifty-six metabolites were identified and quantified using 1H NMR based metabolomics techniques. This is in accordance with a previous study employing NMR methods that identified fifty one metabolites in CSF of normal samples taken from patients receiving spinal anesthesia before non-neurological surgery.24 The majority of metabolites detected in the present study were identified in all CSF samples, except for quinolinate and malate which did not reach the levels of detection in some control samples. The biological variation (calculated as relative standard deviation) ranged from 14 to 286% depending on the metabolite in question and the sample group (rabies patients versus controls). As expected, variation in metabolite concentrations across samples was generally lower in the control group 14–128% (except for phenylalanine 249%). Stoop and colleagues (2010) reported similar ranges in normal samples (12–143%).24 Moreover, almost identical variation in glucose concentrations was reported in both studies highlighting the analytical reproducibility of NMR measurements. The tight clustering of the anonymized CSFs and the low variation in their metabolite concentrations which closely reflect published normal data,34 supports their use as normal controls in this study.
Unsupervised clustering techniques using solely metabolite concentrations clustered rabies CSF samples into 3 groups that corresponded closely with times of sampling after hospitalization. Rabies was associated with metabolite markers for inflammation, excitotoxicity, increased mitochondrial flux, lactic acidosis and increased cerebral volume. One striking observation was that rabies patients presented with ketoacidosis on time of admission. Mild ketoacidosis is frequently encountered among febrile, anorectic patients when first arriving for medical care. In the case of rabies, it is assumed to be related to hydrophobia and reduced food and water intake. Another key feature of rabies disease was elevated concentrations of quinolinate from the day of admission and throughout. Increased concentrations of quinolinate reflect activation of the tryptophan–kynurenine pathway in invading macrophages and activated microglia.35 Concentrations of quinolinate in human rabies CSF exceeded those measured in most cases of meningitis,36 cerebral malaria37,38 or pediatric HIV encephalopathy.39 Quinolinate is a potent agonist of the of N-methyl-d-aspartate (NMDA) receptor, and is capable of promoting excitotoxic damage of the brain.40 Ketamine, one of the drugs used in the MP, inhibits quinolinate mediated agonism of the NMDA receptor.41 Quinolinate concentrations in one survivor decreased as their metabolomic profile approached the controls. We cannot comment on quinolinate concentrations from our second survivor at early time points because no sample was collected before HD 20 (quinolinate 28 µM); however, later quinolinate concentrations decreased with time. In contrast, quinolinate concentrations increased exponentially in CSF samples collected from patients who later die from rabies infection.
Malate, 2-oxoglutarate and N-acetylaspartate (NAA), three metabolites associated with maintenance of cell volume and the aspartate-malate shuttle were also elevated in rabies compared to controls.42 NAA is a specialized amino acid almost exclusively synthesized by neurons, where it may serve as a molecular water pump.43 Elevated concentrations of NAA are also seen in patients with Canavan disease44 which is caused by inactive aspartyl acylase and is characterized by enlarged head, radiologically subtle cerebral edema, developmental delay, and vacuolization of the white matter.45 NAA synthesis in neuronal mitochondria is tied to neuronal energy metabolism.46 In particular, experimental data suggest that as concentrations of pyruvate increase, the rate of NAA efflux from mitochondria also increases.47 Elevated NAA, 2-oxoglutarate, and malate in rabies CSF treated with the MP may suggest increased mitochondrial flux and export of NAA, but it is unclear whether this is causal of or responsive to changes in cerebral volume. When we compare late stage samples from rabies survivors to nonsurvivors, pyruvate and NAA are higher in individuals that die, suggesting deregulated cerebral volume control and an increased energy demand, progressing over time.
CSF lactate concentrations were higher in rabies patients compared to controls and increased over time. Increased concentrations of lactate have been shown previously in patients with bacterial meningitis, severe encephalitis, and other metabolic disorders.48–51 Lactate is typically interpreted as a marker of anaerobic metabolism, but this may not be true in rabies. Asphyxia was not encountered clinically except in two patients with arrests who were excluded from the final analyses. We consider mitochondrial dysfunction unlikely. In human CSF, ratios of lactate to pyruvate greater than 2552 and 4053 have been used to diagnose hypoxic-ischemia or mitochondrial disorders. In this study, lactate to pyruvate ratios greater than 40 were rarely observed, and without any clear trend (data not shown). The CSF metabolic profile for human rabies differed from that of myotubes intoxicated by mitochondrial complex I or III inhibitors. Glucose (increased), formate (increased) and serine (decreased) in rabies CSF moved inversely to the same metabolites measured in media from myotubes intoxicated with rotenone or antimycin A.54 ATP deficiency from mitochondrial dysfunction can be partially offset by increased glycolysis. Cerebral hyperglycolysis, believed to be the result of ionic imbalances and neurochemical-induced excitotoxicity, has been characterized in patients with traumatic brain injury,55,56 but CSF glucose concentrations were normal in our series. Alternatively, increased lactate may reflect decreased neuronal metabolism of lactate as a consequence of rabies infection or the deep sedation used in the MP.
In this study, metabolomics was able to detect adverse effects of medical therapy. From our data, it appears that alcohol dehydrogenase activity of the patient may contribute to adverse outcomes during rabies treatment. This highly complex enzyme system varies genetically, with high multiplicity and variability in both structure and function.57 Propylene glycol was detected in CSF samples in our series. This was expected because it is a commonly used solvent for intravenous benzodiazepines used in the MP. However, propylene glycol is metabolized by hepatic alcohol dehydrogenase to form lactate. This might suggest that the progressive CSF lactic acidosis we measured in rabies is related to treatment rather than rabies virus infection itself, particularly given the lack of correlation with CSF pyruvate and glucose levels.58 Furthermore, high concentrations of isopropanol were a strong predictor of demise from rabies in our series. Acetone is metabolized to isopropanol by alcohol dehydrogenase.59,60 This suggests that late ketogenesis or other elaboration of acetone during rabies may be detrimental. Thus, interindividual and inter-racial differences in alcohol dehydrogenase could impact response to rabies infection and protocolized treatment; pharmacogenomic studies of rabies patients are planned. One clear therapeutic intervention is to minimize sedation during treatment of rabies – this is already recommended after the first week of treatment when the risk of fatal dysautonomia abates. Alternate sedatives might be considered. Perfusion studies in animal livers show that the addition of insulin prevents the increase in lactate associated with propylene glycol administration.61 Insulin also prevents ketogenesis. Low dose insulin combined with adequate nutrition during rabies treatment could therefore attenuate sedation-related toxicity as well as modulate protein and lipid catabolism62 characteristic of late stage (unsurvivable) rabies.
Metabolomics of CSF was able to perfectly differentiate patients with rabies from controls, and define biochemical stages of rabies that closely corresponded to the temporal progression of disease. Metabolomic studies comparing rabies with other CNS diseases are underway and will help to clarify the metabolic abnormalities that are unique to rabies or shared with other CNS infections or intoxications. Indeed, this study did not examine CSF from other brain disorders, so no comment can be made on the specificity of the metabolomics findings for rabies. Our analysis of survivors might be confounded by the fact that both survivors had rabies of bat phylogeny. Sample collections in newly diagnosed rabies patients are ongoing and future studies will likely include survivors of cosmopolitan dog phylogeny.
Survival from rabies was previously extremely rare. For the two survivors in our series, trajectory plots reverted toward the control region suggestive of a return to metabolic homeostasis. Results from this study therefore suggest a role for metabolomics in real time monitoring during metabolically targeted therapy. Moreover, since current ante-mortem diagnosis of rabies uses a composite of laboratory tests that require up to several days to complete at highly skilled reference laboratories, metabolomics should be further investigated for its potential to improve the rapidity and accuracy of the diagnosis of human rabies. Currently, the MP targets excitotoxicity, altered neurotransmission, and cerebral perfusion. Novel target metabolic pathways were identified in this study that may prove useful in guiding future experimental therapy, while identifying interventions as simple as changes in sedation and incorporation of low dose insulin with adequate nutrition to mitigate adverse effects of alcohol dehydrogenase metabolism during use of the MP. The ultimate goal is to develop the MP as a metabolically targeted supportive therapy of rabies, previously considered incurable. Similar approaches may serve as a basic platform for treatment of other untreatable neurotropic infections.
Supplementary Material
Acknowledgments
This work was supported by the Zach Jones Foundation. The authors thank all of the patients and patients' families that were involved in this study. We gratefully acknowledge A. Jansen (Medical College of Wisconsin), J. Blanton, F. Jackson, I. Kuzmin, N. Kuzmina, X. Ma, M. Niezgoda, L. Orciari, O. Urazova, and P. Yager (CDC) for laboratory support and J. Rubin (Centro Medico La Paz, Equatorial Guinea), Y. Caicedo (Hospital Universidad del Valle, Columbia), A. Roy-Berman (Children’s Hospital & Research Center, Oakland, CA), C. Glaser (California Department of Public Health, Richmond, CA), Y. Oba (University of Missouri), G. Arroyo Sanchez (Hospital Nacional Cayetano Heredia, Lima, Peru), and S. Lo (Children’s Hospital of Wisconsin) for providing CSF samples.
ABBREVIATIONS
- HD
hospital day
- MP
Milwaukee Protocol
- PCA
principal components analysis
- PLS-DA
partial least-squares–discriminant analysis
- CSF
cerebrospinal fluid
- CNS
central nervous system
- DSS-d6
3-(trimethylsilyl)-1-propanesulfonic acid-d6
- VIP
variable importance in the projection
- FDR
false discovery rate
- NAA
N-acetyl aspartate
- NMDA
N-methyl-d-aspartate
Footnotes
ASSOCIATED CONTENT
Supporting Information
This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Committee on Infectious Diseases. Rabies-prevention policy update: new reduced-dose schedule. Pediatrics. 2011;127(4):785–787. doi: 10.1542/peds.2011-0095. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. WHO expert consultation on rabies. Geneva: World Health Organization; 2005. [PubMed] [Google Scholar]
- 3.Faber M, Pulmanausahakul R, Hodawadekar SS, Spitsin S, McGettigan JP, Schnell MJ, Dietzschold B. Overexpression of the rabies virus glycoprotein results in enhancement of apoptosis and antiviral immune response. J. Virol. 2002;76(7):3374–3381. doi: 10.1128/JVI.76.7.3374-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hattwick MA, Weis TT, Stechschulte CJ, Baer GM, Gregg MB. Recovery from rabies. A case report. Ann. Intern. Med. 1972;76(6):931–942. doi: 10.7326/0003-4819-76-6-931. [DOI] [PubMed] [Google Scholar]
- 5.Porra SC, Barboza JJ, Fuenzalida E, Adaros HL, Oviedo AM, Furst J. Recovery from rabies in man. Ann. Intern. Med. 1976;85(1):44–48. doi: 10.7326/0003-4819-85-1-44. [DOI] [PubMed] [Google Scholar]
- 6.Tillotson JR, Axelrod D, Lyman DO. Rabies in a laboratory worker - New York. MMWR. 1977;26:183–184. [Google Scholar]
- 7.Alvarez L, Fajardo R, Lopez E, Pedroza R, Hemachudha T, Kamolvarin N, Cortes G, Baer GM. Partial recovery from rabies in a nine-year-old boy. Pediatr. Infect. Dis. J. 1994;13(12):1154–1155. doi: 10.1097/00006454-199412000-00020. [DOI] [PubMed] [Google Scholar]
- 8.Madhusudana SN, Nagaraj D, Uday M, Ratnavalli E, Kumar MV. Partial recovery from rabies in a six-year-old girl. Int. J. Infect. Dis. 2002;6(1):85–86. doi: 10.1016/s1201-9712(02)90144-x. [DOI] [PubMed] [Google Scholar]
- 9.Willoughby RE, Tieves KS, Hoffman GM, Ghanayem NS, Amlie-Lefond CM, Schwabe MJ, Chusid MJ, Rupprecht CE. Survival after treatment of rabies with induction of coma. N. Engl. J. Med. 2005;352(24):2508–2514. doi: 10.1056/NEJMoa050382. [DOI] [PubMed] [Google Scholar]
- 10.Lockhart BP, Tordo N, Tsiang H. Inhibition of rabies virus transcription in rat cortical neurons with the dissociative anesthetic ketamine. Antimicrob. Agents Chemother. 1992;36(8):1750–1755. doi: 10.1128/aac.36.8.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weli S, Scott C, Ward C, Jackson A. Rabies virus infection of primary neuronal cultures and adult mice: failure to demonstrate evidence of excitotoxicity. J. Virol. 2006;80(20):10270–10273. doi: 10.1128/JVI.01272-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willoughby RE. Rabies treatment protocol and registry. 2011 wwwmcw edu/rabies. [Google Scholar]
- 13.Willoughby RE. ″Early death″ and the contraindication of vaccine during treatment of rabies. Vaccine. 2009;27(51):7173–7177. doi: 10.1016/j.vaccine.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 14.Hunter M, Johnson N, Hedderwick S, McCaughey C, Lowry K, McConville J, Herron B, McQuaid S, Marston D, Goddard T, Harkess G, Goharriz H, Voller K, Solomon T, Willoughby RE, Fooks AR. Immunovirological correlates in human rabies treated with therapeutic coma. J. Med. Virol. 2010;82(7):1255–1265. doi: 10.1002/jmv.21785. [DOI] [PubMed] [Google Scholar]
- 15.Rubin J, David D, Willoughby RE, Rupprecht CE, Garcia C, Guarda DC, Zohar Z, Stamler A. Applying the Milwaukee Protocol to treat canine rabies in Equatorial Guinea. Scand. J. Infect. Dis. 2009;41(5):372–375. doi: 10.1080/00365540902798333. [DOI] [PubMed] [Google Scholar]
- 16.Willoughby RE, Roy-Burman A, Martin KW, Christensen JC, Westenkirschner DF, Fleck JD, Glaser C, Hyland K, Rupprecht CE. Generalised cranial artery spasm in human rabies. Dev. Biol. (Basel) 2008;131:367–375. [PubMed] [Google Scholar]
- 17.Willoughby RE, Opladen T, Maier T, Rhead W, Schmiedel S, Hoyer J, Drosten C, Rupprecht C, Hyland K, Hoffmann G. Tetrahydrobiopterin deficiency in human rabies. J. Inherit. Metab. Dis. 2009;32(1):65–72. doi: 10.1007/s10545-008-0949-z. [DOI] [PubMed] [Google Scholar]
- 18.Warrell DA, Davidson NM, Pope HM, Bailie WE, Lawrie JH, Ormerod LD, Kertesz A, Lewis P. Pathophysiologic studies in human rabies. Am. J. Med. 1976;60(2):180–190. doi: 10.1016/0002-9343(76)90427-7. [DOI] [PubMed] [Google Scholar]
- 19.Grattan-Smith PJ, O’Regan WJ, Ellis PS, O’Flaherty SJ, McIntyre PB, Barnes CJ. Rabies, a second Australian case, with a long incubation period. Med. J. Aust. 1992;156:651–654. doi: 10.5694/j.1326-5377.1992.tb121465.x. [DOI] [PubMed] [Google Scholar]
- 20.Lamas CC, Martinez AJ, Baraff R, Bajwa J. Rabies encephaloradiculomyelitis. Acta Neuropathol. (Berl.) 1980;51(3):245–247. doi: 10.1007/BF00687393. [DOI] [PubMed] [Google Scholar]
- 21.Sing TM, Soo MY. Imaging findings in rabies. Australas. Radiol. 1996;40(3):338–341. doi: 10.1111/j.1440-1673.1996.tb00415.x. [DOI] [PubMed] [Google Scholar]
- 22.Roine RO, Hillbom M, Valle M, Haltia M, Ketonen L, Neuvonen E, Luimio J, Lähdevirta J. Fatal encephalitis caused by a bat-borne rabies-related virus. Brain. 1988;111(6):1505–1516. doi: 10.1093/brain/111.6.1505. [DOI] [PubMed] [Google Scholar]
- 23.Slupsky CM, Rankin KN, Wagner J, Fu H, Chang D, Weljie AM, Saude EJ, Lix B, Adamko DJ, Shah S, Greiner R, Sykes BD, Marrie TJ. Investigations of the effects of gender, diurnal variation, and age in human urinary metabolomic profiles. Anal. Chem. 2007;79(18):6995–7004. doi: 10.1021/ac0708588. [DOI] [PubMed] [Google Scholar]
- 24.Stoop MP, Coulier L, Rosenling T, Shi S, Smolinska AM, Buydens L, Ampt K, Stingl C, Dane A, Muilwijk B, Luitwieler RL, Sillevis Smitt PAE, Hintzen RQ, Bischoff R, Wijmenga SS, Hankemeier T, van Gool AJ, Luider TM. Quantitative proteomics and metabolomics analysis of normal human cerebrospinal fluid samples. Mol. Cell. Proteomics. 2010;9(9):2063–2075. doi: 10.1074/mcp.M110.000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walsh MC, Brennan L, Malthouse JPG, Roche HM, Gibney MJ. Effect of acute dietary standardization on the urinary, plasma, and salivary metabolomic profiles of healthy humans. Am. J. Clin. Nutr. 2006;84:531–539. doi: 10.1093/ajcn/84.3.531. [DOI] [PubMed] [Google Scholar]
- 26.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. 2009;106(34):14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slupsky CM, Steed H, Wells TH, Dabbs K, Schepansky A, Capstick V, Faught W, Sawyer MB. Urine metabolite analysis offers potential early diagnosis of ovarian and breast cancers. Clin. Cancer Res. 2010;16(23):5835–5841. doi: 10.1158/1078-0432.CCR-10-1434. [DOI] [PubMed] [Google Scholar]
- 28.Slupsky CM, Rankin KN, Fu H, Chang D, Rowe BH, Charles PGP, McGeer A, Low D, Long R, Kunimoto D, Sawyer MB, Fedorak RN, Adamko DJ, Saude EJ, Shah SL, Marrie TJ. Pneumococcal pneumonia: potential for diagnosis through a urinary metabolic profile. J. Proteome Res. 2009;8(12):5550–5558. doi: 10.1021/pr9006427. [DOI] [PubMed] [Google Scholar]
- 29.Sinclair A, Viant M, Ball A, Burdon M, Walker E, Stewart P, Rauz S, Young S. NMR-based metabolomic analysis of cerebrospinal fluid and serum in neurological diseases - a diagnostic tool? NMR Biomed. 2010;23(2):123–132. doi: 10.1002/nbm.1428. [DOI] [PubMed] [Google Scholar]
- 30.Coen M, O’Sullivan M, Bubb WA, Kuchel PW, Sorrell T. Proton nuclear magnetic resonance-based metabonomics for rapid diagnosis of meningitis and ventriculitis. Clin. Infect. Dis. 2005;41(11):1582–1590. doi: 10.1086/497836. [DOI] [PubMed] [Google Scholar]
- 31.Blasco H, Corcia P, Moreau C, Veau S, Fournier C, Vourc’h P, Emond P, Gordon P, Pradat PF, Praline J, Devos D, Nadal-Desbarats L, Andres CR. 1H-NMR-based metabolomic profiling of CSF in early amyotrophic lateral sclerosis. PLoS ONE. 2011;5(10):e13223. doi: 10.1371/journal.pone.0013223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal. Chem. 2006;78(13):4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 33.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 34.Wishart DS, Lewis MJ, Morrissey JA, Flegel MD, Jeroncic K, Xiong Y, Cheng D, Eisner R, Gautam B, Tzur D, Sawhney S, Bamforth F, Greiner R, Li L. The human cerebrospinal fluid metabolome. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2008;871(2):164–173. doi: 10.1016/j.jchromb.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 35.Heyes MP, Saito K, Lackner A, Wiley CA, Achim CL, Markey SP. Sources of the neurotoxin quinolinic acid in the brain of HIV-1-infected patients and retrovirus-infected macaques. FASEB J. 1998;12(10):881–896. doi: 10.1096/fasebj.12.10.881. [DOI] [PubMed] [Google Scholar]
- 36.Heyes MP, Saito K, Milstien S, Schiff SJ. Quinolinic acid in tumors, hemorrhage and bacterial infections of the central nervous system in children. J. Neurol. Sci. 1995;133(1–2):112–118. doi: 10.1016/0022-510x(95)00164-w. [DOI] [PubMed] [Google Scholar]
- 37.Medana IM, Day NPJ, Salahifar-Sabet H, Stocker R, Smythe G, Bwanaisa L, Njobvu A, Kayira K, Turner GDH, Taylor TE, Hunt NH. Metabolites of the kynurenine pathway of tryptophan metabolism in the cerebrospinal fluid of malawian children with malaria. J. Infect. Dis. 2003;188(6):844–849. doi: 10.1086/377583. [DOI] [PubMed] [Google Scholar]
- 38.Medana IM, Hien TT, Day NP, Nguyen HP, Nguyen THM, Chu’ong LV, Tran THC, Taylor A, Salahifar H, Stocker R, Smythe G, Turner GDH, Farrar J, White NJ, Hunt NH. The clinical significance of cerebrospinal fluid levels of kynurenine pathway metabolites and lactate in severe malaria. J. Infect. Dis. 2002;185(5):650–656. doi: 10.1086/339009. [DOI] [PubMed] [Google Scholar]
- 39.Brouwers P, Heyes MP, Moss HA, Wolters PL, Poplack DG, Markey SP, Pizzo PA. Quinolinic acid in the cerebrospinal fluid of children with symptomatic human immunodeficiency virus type 1 disease: relationships to clinical status and therapeutic response. J. Infect. Dis. 1993;168(6):1380–1386. doi: 10.1093/infdis/168.6.1380. [DOI] [PubMed] [Google Scholar]
- 40.Stone TW. Neuropharmacology of quinolinic and kynurenic acids. Pharmacol. Rev. 1993;45(3):309–379. [PubMed] [Google Scholar]
- 41.Keilhoff G, Wolf G, Stastný F. Effects of MK-801, ketamine and alaptide on quinolinate models in the maturing hippocampus. Neuroscience. 1991;42(2):379–385. doi: 10.1016/0306-4522(91)90382-x. [DOI] [PubMed] [Google Scholar]
- 42.Surendran S, Matalon KM, Szucs S, Tyring SK, Matalon R. Metabolic changes in the knockout mouse for Canavan’s disease: implications for patients with Canavan’s disease. J. Child. Neurol. 2003;18(9):611–615. doi: 10.1177/08830738030180090701. [DOI] [PubMed] [Google Scholar]
- 43.Baslow MH, Hrabe J, Guilfoyle DN. Dynamic relationship between neurostimulation and N-acetylaspartate metabolism in the human visual cortex: evidence that NAA functions as a molecular water pump during visual stimulation. J. Mol. Neurosci. 2007;32(3):235–245. doi: 10.1007/s12031-007-0049-9. [DOI] [PubMed] [Google Scholar]
- 44.Al-Dirbashi O, Rashed M, Al-Qahtani K, Al-Mokhadab M, Kurdi W, Al-Sayed M. Quantification of N-acetylaspartic acid in urine by LC-MS/MS for the diagnosis of Canavan disease. J. Inherit. Metab. Dis. 2007;30(4):612. doi: 10.1007/s10545-007-0635-6. [DOI] [PubMed] [Google Scholar]
- 45.Janson CG, McPhee SWJ, Francis J, Shera D, Assadi M, Freese A, Hurh P, Haselgrove J, Wang DJ, Bilaniuk L, Leone P. Natural history of Canavan disease revealed by proton magnetic resonance spectroscopy (1H-MRS) and diffusion-weighted MRI. Neuropediatrics. 2006;37(04):209–221. doi: 10.1055/s-2006-924734. [DOI] [PubMed] [Google Scholar]
- 46.Moffett JR, Ross B, Arun P, Madhavarao CN, Namboodiri AMA. N-Acetylaspartate in the CNS: from neurodiagnostics to neurobiology. Prog. Neurobiol. 2007;81(2):89–131. doi: 10.1016/j.pneurobio.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel TB, Clark JB. Synthesis of N-acetyl-L-aspartate by rat brain mitochondria and its involvement in mitochondrial/cytosolic carbon transport. Biochem. J. 1979;184:539–546. doi: 10.1042/bj1840539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komorowski RA, Farmer SG, Hanson GA, Hause LL. Cerebrospinal fluid lactic acid in diagnosis of meningitis. J. Clin. Microbiol. 1978;8(1):89–92. doi: 10.1128/jcm.8.1.89-92.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hiraoka A, Miura I, Hattori M, Tominaga I, Kushida K, Maeda M. Proton magnetic resonance spectroscopy of cerebrospinal fluid as an aid in neurological diagnosis. Biol. Pharm. Bull. 1994;17(1):1–4. doi: 10.1248/bpb.17.1. [DOI] [PubMed] [Google Scholar]
- 50.Hansen EL, Kristensen HS, Brodersen P, Paulson OB, Möllertz S, Jessen O. Acid-base pattern of cerebrospinal fluid and arterial blood in bacterial meningitis and in encephalitis. Acta Med. Scand. 1974;196(1–6):431–437. doi: 10.1111/j.0954-6820.1974.tb01035.x. [DOI] [PubMed] [Google Scholar]
- 51.Chow SL, Rooney ZJ, Cleary MA, Clayton PT, Leonard JV. The significance of elevated CSF lactate. Arch. Dis. Child. 2005;90(11):1188–1189. doi: 10.1136/adc.2005.075317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Debray FG, Mitchell GA, Allard P, Robinson BH, Hanley JA, Lambert M. Diagnostic accuracy of blood lactate-to-pyruvate molar ratio in the differential diagnosis of congenital lactic acidosis. Clin. Chem. 2007;53(5):916–921. doi: 10.1373/clinchem.2006.081166. [DOI] [PubMed] [Google Scholar]
- 53.Vespa P, Bergsneider M, Hattori N, Wu H-M, Huang S-C, Martin NA, Glenn TC, McArthur DL, Hovda DA. Metabolic crisis without brain ischemia is common after traumatic brain injury: a combined microdialysis and positron emission tomography study. J. Cereb. Blood Flow Metab. 2005;25(6):763–774. doi: 10.1038/sj.jcbfm.9600073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Q, Vu H, Liu L, Wang T-C, Schaefer WH. Metabolic profiles show specific mitochondrial toxicities in vitro in myotube cells. J. Biomol. NMR. 2011;49(3–4):207–219. doi: 10.1007/s10858-011-9482-8. [DOI] [PubMed] [Google Scholar]
- 55.Andersen BJ, Marmarou A. Post-traumatic selective stimulation of glycolysis. Brain Res. 1992;585(1–2):184–189. doi: 10.1016/0006-8993(92)91205-s. [DOI] [PubMed] [Google Scholar]
- 56.Bergsneider M, Hovda DA, Shalmon E, Kelly DF, Vespa PM, Martin NA, Phelps ME, McArthur DL, Caron MJ, Kraus JF, Becker DP. Cerebral hyperglycolysis following severe traumatic brain injury in humans: a positron emission tomography study. J. Neurosurg. 1997;86(2):241–251. doi: 10.3171/jns.1997.86.2.0241. [DOI] [PubMed] [Google Scholar]
- 57.Jörnvall H, Höög JO, Persson B, Parés X. Pharmacogenetics of the alcohol dehydrogenase system. Pharmacology. 2000;61(3):184–191. doi: 10.1159/000028399. [DOI] [PubMed] [Google Scholar]
- 58.Ruddick JA. Toxicology, metabolism, and biochemistry of 1,2-propanediol. Toxicol. Appl. Pharmacol. 1972;21(1):102–111. doi: 10.1016/0041-008x(72)90032-4. [DOI] [PubMed] [Google Scholar]
- 59.Davis PL, Cortivo LAD, Maturo J. Endogenous isopropanol: forensic and biochemical implications. J. Anal. Toxicol. 1984;8(5):209–212. doi: 10.1093/jat/8.5.209. [DOI] [PubMed] [Google Scholar]
- 60.Lewis GD, Laufman AK, McAnalley BH, Garriott JC. Metabolism of acetone to isopropyl alcohol in rats and humans. J. Forensic Sci. 1984;29(2):541–549. [PubMed] [Google Scholar]
- 61.Newman HW, Van Winkle W, Kennedy NK, Morton MC. Comparative effects of propylene glycol, other glycols, and alcohol on the liver directly. J. Pharmacol. Exp. Ther. 1940;68(1):194–200. [Google Scholar]
- 62.Aoki TT, Assal J-P, Manzano FM, Kozak GP, Cahill GF. Plasma and cerebrospinal fluid amino acid levels in diabetic ketoacidosis before and after corrective therapy. Diabetes. 1975;24(5):463–467. doi: 10.2337/diab.24.5.463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.