Abstract
The mismatch repair (MMR) family is a highly conserved group of proteins that function in correcting base–base and insertion–deletion mismatches generated during DNA replication. Disruption of this process results in characteristic microsatellite instability (MSI), repair defects, and susceptibility to cancer. However, a significant fraction of MSI-positive cancers express MMR genes at normal levels and do not carry detectable mutation in known MMR genes, suggesting that additional factors and/or mechanisms may exist to explain these MSI phenotypes in patients. To systematically investigate the MMR pathway, we conducted a proteomic analysis and identified MMR-associated protein complexes using tandem-affinity purification coupled with mass spectrometry (TAP-MS) method. The mass spectrometry data have been deposited to the ProteomeXchange with identifier PXD003014 and DOI 10.6019/PXD003014. We identified 230 high-confidence candidate interaction proteins (HCIPs). We subsequently focused on MSH2, an essential component of the MMR pathway and uncovered a novel MSH2-binding partner, WDHD1. We further demonstrated that WDHD1 forms a stable complex with MSH2 and MSH3 or MSH6, i.e. the MutS complexes. The specific MSH2/WDHD1 interaction is mediated by the second lever domain of MSH2 and Ala1123 site of WDHD1. Moreover, we showed that, just like MSH2-deficient cells, depletion of WDHD1 also led to 6-thioguanine (6-TG) resistance, indicating that WDHD1 likely contributes to the MMR pathway. Taken together, our study uncovers new components involved in the MMR pathway, which provides candidate genes that may be responsible for the development of MSI-positive cancers.
Cells are equipped with a number of repair mechanisms to correct various types of DNA lesions. At least five major complimentary, but partially overlapping, multistep damage repair pathways are known to operate in mammals: mismatch repair (MMR)1, nucleotide excision repair, base excision repair, and double-strand break repair, which includes both homologous recombination repair and nonhomologous end joining (see review: (1, 2)). In particular, MMR is a major repair pathway that prevents both base substitution and insertion–deletion mismatches due to replication errors (3–5).
MMR is a highly conserved biological pathway that exists from bacteria to mammals. MMR process can be divided into three key steps: mismatch recognition, excision, and resynthesis (5, 6). The initial mismatch recognition step is fulfilled by MutS protein complexes, either MutSα (the MSH2-MSH6 heterodimer) or MutSβ (the MSH2-MSH3 heterodimer). The MutSα is primarily responsible for repairing base–base mismatches and small insertion–deletion loops of 1–2 nucleotides (7–9), while MutSβ preferentially recognizes larger insertion–deletion loops containing up to 14 extra nucleotides (10–12). Binding to mispaired DNA primes MutS to undergo a conformational change and recruitment of MutL to form an ATP-dependent ternary complex (13). Three different MutL heterodimeric complexes, MutLα, MutLβ, and MutLγ have been identified in the mammalian system. MLH1 heterodimerizes with PMS2, PMS1, or MLH3 to form MutLα, MutLβ, or MutLγ, respectively. MutLα plays a crucial role in MMR, as cells that lack either protein inactivate MMR in human cells, while loss of MutLβ or MutLγ heterodimers leads to minor defects in MMR. MutL is able to recognize and excise the lagging strand from the mismatch both distally and proximally (14, 15). Moreover, MutL interacts physically with MutS, enhances mismatch recognition, and recruits and activates exonuclease1 (EXO1) (16, 17). Exonuclease1 (EXO1) is the only enzyme with capabilities to excise nucleotide in 5′-3′ direction (18). In the case of 3′ excision, proliferating cell nuclear antigen (PCNA)/replication factor C-dependent endonuclease activity plays a critical role in 3′-5′ excision involving EXO1. EXO1 then excises nascent DNA from the nick toward and beyond the mismatch to generate a single-strand gap, which is filled by DNA polymerases δ (lagging strand) or ε (leading strand) using the parental DNA strand as a template. Finally, the nick is sealed by DNA ligase I (19, 20). In addition, two MutS homologues, MSH4 and MSH5, share similar structure and sequence features with the other members of the MutS family. Recent evidence suggests that they function beyond MMR and are involved in processes such as recombinant repair, DNA damage signaling, and immunoglobulin class switch recombination (21, 22).
It has been well documented that impairment of MMR genes, especially MSH2 and MLH1, cause susceptibility to certain types of cancer, including human nonpolyposis colorectal cancer. At the cellular level, deficient MMR results in a strong mutator phenotype known as microsatellite instability (MSI), which is a hallmark of MMR deficiency (3–5). However, a significant fraction of MSI-positive colorectal cancers express MMR genes at normal levels and do not carry detectable mutation or hypermethylation in known MMR genes (23). Similarly, certain noncolorectal cancer cells with MSI also appear to have normal expression of known MMR protein (24, 25). These observations suggest that additional factors and/or mechanisms may exist to explain these MSI phenotypes in patients.
To address this question, we performed tandem affinity purification coupled with mass spectrometry analysis (TAP-MS) to uncover MMR-associated protein complexes. Our proteomics study of the MMR family led to the discovery of many novel MMR-associated proteins, and gene ontology analysis expanded the roles of MMR in multiple biological processes. Specifically for MSH2, we uncovered a novel MutS binding partner WDHD1, which associates with both MutSα (MSH2-MSH6 heterodimer) and MutSβ (MSH2-MSH3 heterodimer). We provide additional evidence suggesting that WDHD1 is involved in the MMR pathway, which can be used as potential biomarker for MSI phenotypes in cancer patients.
EXPERIMENTAL PROCEDURES
Cell Culture, Plasmids and Transfection
HEK293T cells were maintained in DMEM medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in a humidified incubator with 5% CO2. The Gateway entry pDONR201-MMR genes MutS homolog 2 (MSH2), MutS homolog 3 (MSH3), MutS homolog 4 (MSH4), MutS homolog 5 (MSH5), MutS homolog 6 (MSH6), EXO1, Postmeiotic Segregation Increased 1 (PMS1), Postmeiotic Segregation Increased 2 (PMS2), MutL homolog 1 (MLH1), MutL homolog 3 (MLH3), and WD Repeat And HMG-Box DNA Binding Protein 1 (WDHD1), SWI/SNF-Related, Matrix-Associated Actin-Dependent Regulator Of Chromatin, Subfamily A, Containing DEAD/H Box 1 (SMARCAD1), KIAA1671, Stromal Cell Derived Factor 4 (SDF4), and Methyl CpG Binding Protein 2 (MeCP2) plasmids (obtained from Harvard PlasmID database and The ORFeome Collaboration) were recombined into gateway-compatible destination vector for the expression of SFB-tagged or Myc-tagged fusion proteins as described previously (26). All truncated WDHD1 and MSH2 constructs were generated by polymerase chain reaction and subcloned into pDNR201 vector with use of Gateway Technology (Invitrogen) as the entry clones. Deletions or point mutants of WDHD1 and MSH2 were generated using the Quick-change site directed mutagenesis kit (Stratagene) using pDNR201-WDHD1 or pDNR201-MSH2 as a template and then cloned into an SFB tag or Myc tag destination vectors. All mutations were verified by DNA sequencing. Plasmid transfection was carried out using polyethylenimine as described previously (27).
Antibodies
The anti-MSH2 antibody was obtained from Cell Signaling Technology. The monoclonal anti-FLAG M2, anti-β-actin, and anti-WDHD1 antibodies were purchased from Sigma-Aldrich. The anti-Myc (9E10) antibody was obtained from Covance.
Coprecipitation and Western blotting
Cells were lysed with NETN buffer (100mM NaCl; 1mM EDTA; 20mM Tris HCl; and 0.5% Nonidet P-40) containing protease inhibitors on ice for 20 min. The soluble fractions were collected after centrifugation and incubated with protein A agarose beads coupled with anti-MSH2 antibody, or S-protein beads for 4 h at 4 °C. The precipitates were then washed and boiled in 2 × sodium dodecyl sulfate (SDS) loading buffer. Samples were resolved on SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene fluoride membrane, and immunoblotting was carried out with antibodies as indicated.
Clonogenic Survival Assays
Briefly, a total of 1 × 103 HeLa cells were seeded onto 60 mm dish in triplicates. Twenty-four hours after seeding, cells were treated with different concentrations of 6-TG (0, 1 μm, 3 μm, 8 μm) for 3 days, washed, and cultured in the medium. After 14 days, cells were stained with crystal violet and colonies counted. Numbers of colonies were expressed as a percentage of the colonies formed in the absence of the drug. Results were the averages of data obtained from three independent experiments.
Tandem Affinity Purification
HEK293T cells were transfected with plasmids encoding various SFB-tagged MMR proteins. Stable cell lines were selected with media containing 2 μg/ml puromycin and confirmed by immunostaining and Western blotting. HEK293T cells stably expressing SFB-tagged MMR proteins were lysed with NETN buffer on ice for 20 min. After removal of cell debris by centrifugation, crude lysates were incubated with streptavidin Sepharose beads for 1 h at 4 °C. The bead-bound proteins were washed three times with NETN buffer and eluted twice with 2 mg/ml biotin (Sigma) for 1 h at 4 °C. The eluates were combined and then incubated with S-protein agarose (Novagen) for 1 h at 4 °C. The S beads were washed three times with NETN buffer. The proteins bound to S-protein agarose beads were separated by SDS-PAGE and visualized by Coomassie Blue staining. The eluted proteins were identified by mass spectrometry analysis, performed by the Taplin Biological Mass Spectrometry Facility (Harvard Medical School).
Mass Spectrometry Analysis
Gel bands were excised into small pieces, destained completely, disulfide bonds were reduced with 5 mm tris(2-carboxyethyl)phosphine (TCEP), cysteines were alkylated with 10 mm IAA, and then subjected to trypsin digestion at 37 °C for overnight. The peptides were extracted with acetonitrile and vacuum dried. Samples were reconstituted in HPLC solvent A (2.5% acetonitrile, 0.1% formic acid), delivered onto a Proxeon EASY-nLC II liquid chromatography pump (Thermo Fisher, Waltham, MA), and eluted with acetonitrile gradient by increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid) from 6% to 30% in 30 mins. The eluates directly entered Orbitrap Elite MS (Thermo Fisher), setting in positive ion mode and data-dependent manner with full MS scan from 350–1250 m/z, resolution at 60,000, automatic gain control target at 1 × 106. The top 10 precursors were then selected for MS2 analysis.
The MS/MS spectra were used to search SEQUEST (ver. 28) (Thermo Fisher). Spectra were converted to mzXML using a modified version of the ReAdW.exe. Database searching included all entries from the human Uniprot database (March 11, 2014). This database was concatenated with one composed of all protein sequences in the reverse order. The number of entries in the database was 141,456. Searches were performed using a 50 ppm precursor ion tolerance for total protein level analysis. The product ion tolerance was set to 1 Da. Enzyme specificity was set to partially tryptic with two missed cleavages. Carboxyamidomethyl for cysteine residues (+57.021 Da) was set as static modifications, and oxidation for methionine residues (+15.995) was set as a variable modification. The identified peptides were filtered by false discovery rate < 1% based on the target–decoy method. The parameters XCorr, ΔCn, missed cleavages, peptide length, charge state, and precursor mass accuracy were considered for the peptide-spectrum match (PSM) filtering using a linear discriminant analysis (28, 29). Single peptide identifications were removed. The identified proteins and peptides are shown in Supplemental Tables S1 and S2.
Mismatch Repair Protein Interactome Analysis
For the evaluation of potential protein–protein interactions, identified proteins and the corresponding PSM numbers (10 baits of mismatch proteins with one biological repeat for each) were subjected to assessment using the CRAPome methodology. The CRAPome scoring strategy is based on quantitative comparison of abundance (spectral counts) of coprecipitating proteins in purifications with bait against the distribution of prey abundances across a set of negative controls. This fold change (FC) score includes primary score FC-A and more stringent score FC-B. The FC-A calculation averages the counts across all control, whereas the FC-B score takes the average of the top three highest spectral counts for the abundance estimate (30). In this study, we used 233 TAP-MS data with randomly selected baits as the control group. An FC-B score higher than two was taken as the threshold for potential binding proteins. To further select for HCIPs, we chose the proteome profiling data of HEK293T whole-cell lysis as the background to assess the specificity of protein–protein interaction. The spectra number for the identified proteins was normalized by total spectra counts. By comparing with this global expression background, only proteins that were enriched above the average enrichment fold following the TAP-MS procedure were included in the HCIP lists.
The HCIPs of MMR proteins were analyzed by Cytoscape (31). We analyzed the network and created custom styles then applied yFiles organic layout with minor adjustments when necessary. The principal component analysis of the interactomes was studied with R statistical computing software. The HCIPs with normalized spectra number for each MMR protein were analyzed. The gene ontology annotations with p value were performed based on the Knowledge Base provided by Ingenuity Pathway Analysis software (IPA, Ingenuity Systems), which contains findings and annotations from multiple sources, including the Gene Ontology Database. False discovery rate correction of p value was used to correct for multiple testing to get the significantly enriched function with R statistical computing (32).
Experimental Design and Statistical Rationale
All the TAP-MS experiments of MMR proteins were performed with two biological replicates in HEK293T cells. These biological replicates came from two independent stable clones. The purified protein lysis from TAP were digested with trypsin and analyzed by MS. The raw data were calculated with SEQUEST and filtered by false discovery rate < 1% as we described in the methods. These identified proteins were filtered by combining CRAPome analysis and background enrichment strategy considering the two biological replicates. Function enrichment of the HCIPs was analyzed by IPA. False discovery rate correction of p value was used to correct for multiple testing. Clonogenic survival assays were performed with at least three biological replicates and statistical analysis was performed using the Student's test.
RESULTS
Proteomics Study of Mismatch Repair Protein Interactome Using TAP-MS Approach
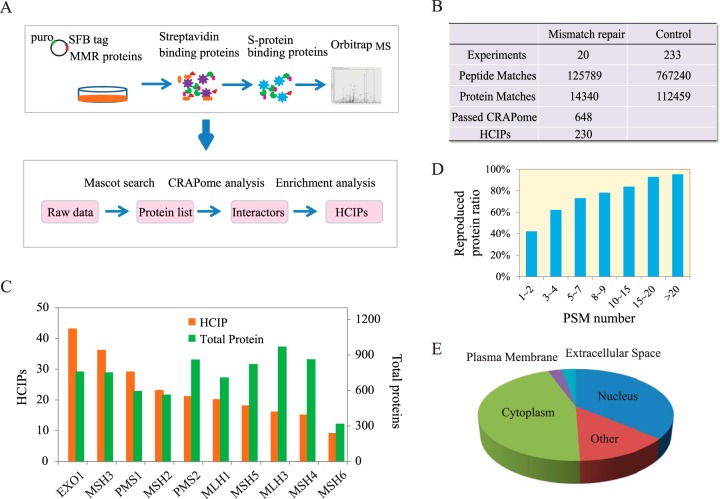
To build the interaction network of DNA MMR pathway, we used the well-established tandem affinity purification followed by mass spectrometry (TAP-MS) strategy (33–35), which was described in Fig. 1A, to identify the binding proteins. In humans, the DNA MMR pathway includes 10 proteins, MSH2, MSH3, MSH4, MSH5, MSH6, PMS1, PMS2, MLH1, MLH3, and EXO1. We established HEK293T derivative cell lines stably expressing each of the triple-tagged (S-protein, FLAG, and streptavidin-binding peptide) MMR proteins. TAP experiments were performed twice for each protein using independent stable clones, and the purified proteins were digested and delivered to mass spectrometry for identification. The identified protein numbers are shown in Figs. 1B and 1C. Details of the identification results are shown in Supplemental Tables S1 and S2. In total, 131,449 peptides and 20,001 proteins were acquired from the 20 TAP-MS experiments. Analysis of our repeat purifications verified strong reproducibility of our TAP-MS procedure (Fig. 1D), especially for the proteins identified with high PSMs, suggesting the high quality of our TAP-MS data.
Fig. 1.
Proteomics analysis of human DNA mismatch repair pathway. (A) Diagrams of interactome study pipeline using tandem affinity purification-mass spectrometry (TAP-MS) approach and HCIP filtration employed in this study. (B) Summary of TAP-MS results of MMR proteins and control experiments, including identified peptides, proteins, and HCIPs. The CRAPome score FC-B > 2 was used as the cutoff to identify HCIPs. Additional filtration using proteomic profile of HEK293T cell was also applied. (C) Total numbers of identified proteins and corresponding HCIPs for each DNA MMR protein were presented. (D) The data reproducibility distribution within TAP-MS experiment repeats. As expected, proteins with higher numbers of PSMs have higher reproducibility. (E) The localization analysis of HCIPs of DNA MMR proteins is presented.
To obtain the high-confidence candidate interacting proteins (HCIP) list, we submitted 20 TAP-MS results with spectra counts information for DNA MMR proteins and 233 controls with random selected unrelated control proteins for CRAPome analysis (30). FC-B score was used to filter our TAP-MS dataset for HCIPs. We obtained 648 proteins out of the total 14,340 identification list with the score higher than two. Furthermore, to improve the confidence of our interacting protein list, we adopted the proteome profiling data of input cell lysate as background for our protein–protein interaction study, which allowed us to remove background contaminants. Finally, we obtained 230 HCIPs as the “interactome” for all 10 MMR proteins with 36.1% of the proteins identified as nuclear proteins and 45.0% as cytoplasmic components (Fig. 1E). The details of the identified proteins and HCIPs for each bait protein are shown in Fig. 1C and Supplemental Table S3.
Overview of Protein–Protein Interaction Network of Human DNA Mismatch Repair Pathway
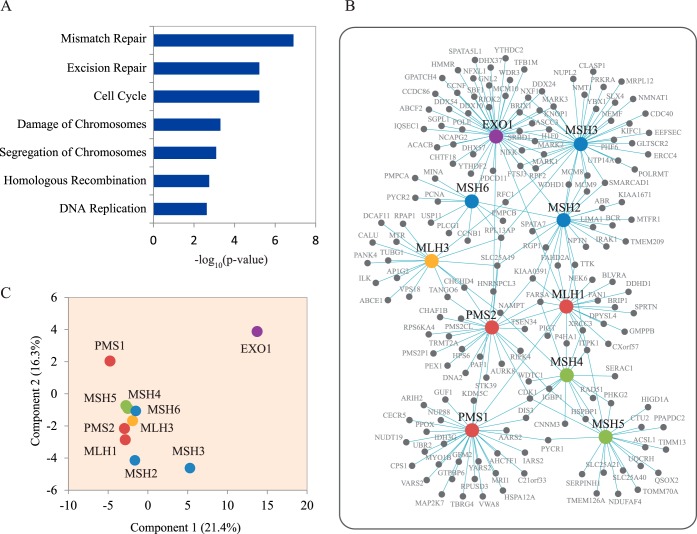
To understand the interactomes of MMR proteins, we first used IPA to reveal the function of all the identified HCIPs. The IPA analysis found that interactomes are highly enriched in proteins with reported roles in the MMR pathway, cell cycle, cellular growth and proliferation, DNA damage response, cellular development, cell morphology, and cellular assembly and organization (Fig. 2A). Our results are in agreement with many published reports, which not only further demonstrate the high reliability of our dataset and methodology but also provide us with clues on how these proteins function in the MMR pathway.
Fig. 2.
Interactomes of mismatch repair proteins. (A) Cellular function annotation of all HCIPs using gene ontology analysis. The function pathways with significant multiple testing p value were shown in the histogram. (B) Network integration and analysis of all HCIPs of DNA MMR proteins were conducted using cytoscape software. In this network, the cycles with blue color are MutS complexes, and the red ones are MutL complexes, and the green ones are MSH4-MSH5 complex. The purple cycle EXO1 interacts with both MutS and MutL complexes. The yellow one is MLH3, which indicates no strong binding between MLH3 and other MMR proteins found in this study. (C) Principal component analysis of the HCIPs and the corresponding PSMs of MMR protein interactomes. The ratios for x and y axis indicate the percentage of variance explained by these two principal components.
MMR proteins do not function in isolation. There are many interactions among these MMR proteins and their HCIPs. Therefore, we studied the interactome network of all the HCIPs using Cytoscape (Fig. 2B). From the interaction data among various DNA MMR proteins, we found there are strong bindings among some of these proteins, which are already known as functional complexes involved in MMR, for example, the MutS and MutL complexes. MSH2 forms heterodimers with MSH6 and MSH3, which are, respectively, called MutSα and MutSβ, while MLH1 forms heterodimers with PMS2 and PMS1, which are MutLα, and MutLβ. As some of the HCIPs are shared among several MMR proteins, the comparison of different identified spectra number for the common HCIPs were analyzed by unsupervised principal component analysis of the 10 TAP-MS results. We generated the principal component analysis plot with the top two principal components, which explained 21.4% and 16.3% of total data variation (Fig. 2C). As expected, our analysis validated the MutS and MutL complexes. A DNA exonuclease EXO1 has been reported to function in DNA MMR by excising mismatch-containing DNA tracts directed by strand breaks to the mismatch (36, 37). According to our HCIPs list, EXO1 interacts with both MutS and MutL complexes, supporting active interaction and coordination between MutS, MutL, and EXO1 in lesion recognition, incision, and excision steps during the MMR process. We also identified an interaction between MSH4 and MSH5, suggesting that they may function as a complex in MMR pathway and/or other cellular processes.
Subinteractome Network Study of MutS, MutL, and EXO1
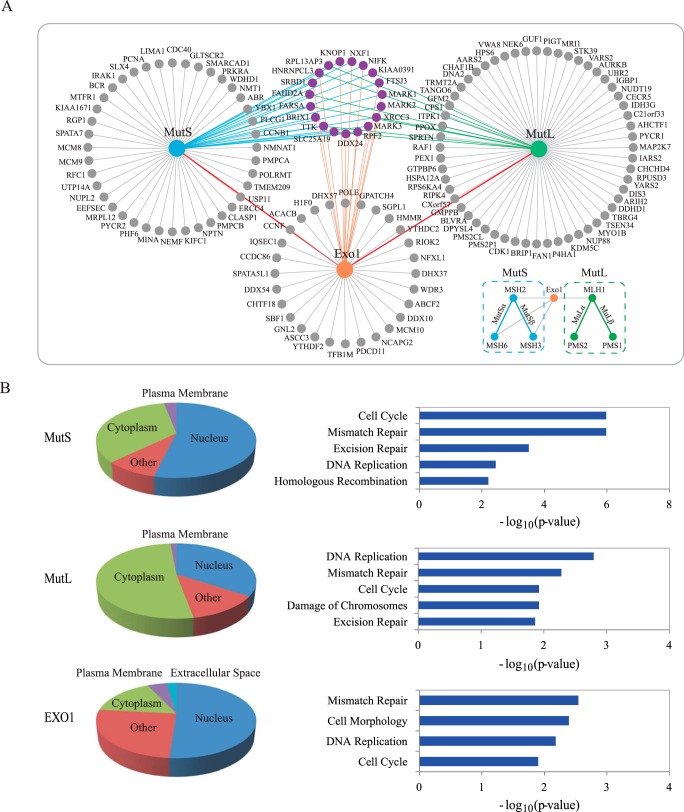
As EXO1 interacts with both MutS and MutL complexes, they may form a large “MMR repairsome” involved in the MMR process. Here, we further studied the subinteractome network. First, we integrated the HCIPs of MutS complexes (including MSH2, MSH3, and MSH6) and MutL complexes (including MLH1, PMS1, and PMS2) individually, and then built a subnetwork with HCIPs of these three components, MutS, MutL, and EXO1 (Fig. 3A). The proteins in the three cycles around the baits or complexes are the proteins only identified as HCIPs in the identical TAP-MS experiment. The proteins labeled in purple are the HCIPs identified by at least two baits. Some of the common identified HCIPs are involved in DNA repair pathways. For instance, x-ray repair cross-complementing protein 3 (XRCC3) is involved in the homologous recombination repair pathway (38). This protein was identified as HCIPs of MutS and MutL complexes, indicating that it may play a role in the DNA MMR pathway. Of course, it is also possible that MutS and MutL may associate with XRCC3 and function in homologous recombination repair pathway.
Fig. 3.
Subnetwork analysis of mismatch repair protein complexes. (A) Interaction maps of MutS (MSH2, MSH3, and MSH6), MutL (MLH1, PMS1, and PMS2), and exonuclease EXO1. HCIPs of two major MutS complexes, MSH2-MSH6 (MutSα) and MSH2-MSH3 (MutSβ), were combined. Similarly, HCIPs of two major MutL complexes, MLH1-PMS1 (MutLα) and MLH1-PMS2 (MutLβ) were also combined. The purple cycles are shared HCIPs among these three interactomes. (B) Gene ontology analysis of MutS, MutL, and EXO1 HCIPs, including localization and cellular function analysis.
To globally reveal the functions of HCIPs of MutS, MutL, and EXO1 identified in our TAP-MS study, we used the software IPA for the localization and function analyses (Fig. 3B). Many of the HCIPs localize in the nucleus, which include 53.73% HCIPs of MutS complex, 51.16% of EXO1, and 34.29% of MutL complex. The functional analysis illustrates that these HCIPs are highly enriched in several functional pathways, including the MMR pathway, homologous recombination repair, nucleotide excision repair, cell cycle, and DNA replication. The proteins with these functions may be involved in DNA MMR pathway and vice versa.
Validation of MSH2 Interactome Reveals a Novel MutS-Binding Partner WDHD1
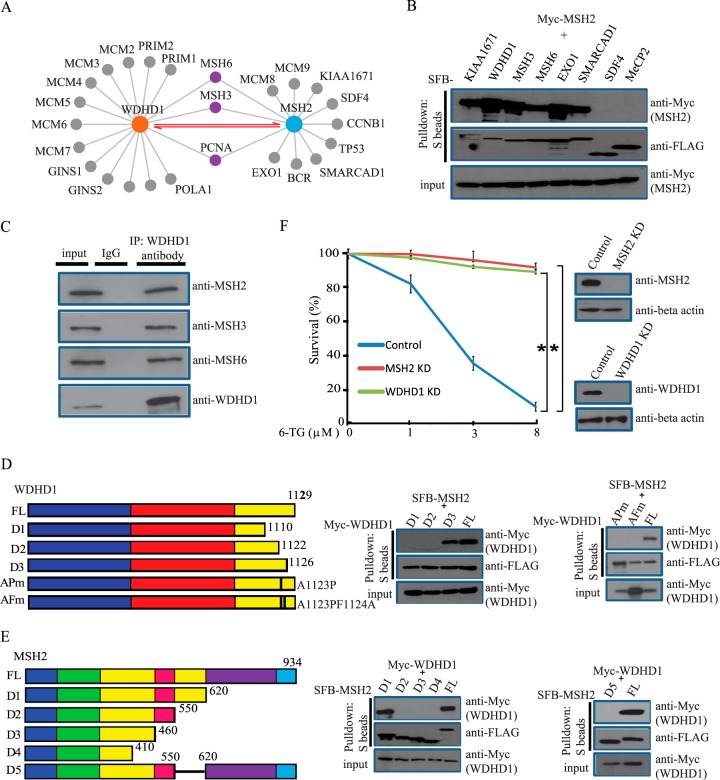
To further validate our proteomics data, we decided to perform an in-depth study of the MSH2 interactome. In this interactome, we identified several known MSH2-binding proteins, including MSH3, MSH6, and EXO1 (Fig. 4A). Excitingly, we uncovered WDHD1 as a major MSH2-associated protein (Fig. 4A). To confirm that WDHD1 exists in the same complex as MSH2, we performed reversal TAP-MS analyses using SFB-tagged WDHD1 as the bait protein and were excited to identify MSH2, MSH3, and MSH6 as WDHD1-associated proteins (Fig. 4A). These data suggest that WDHD1 is likely a novel component of MutS complexes and associates with both MutSα and MutSβ in vivo. To verify the MSH2 interactome revealed by our TAP-MS analysis, we coexpressed SFB-tagged MSH3, MSH6, EXO1, WDHD1, KIAA1671, SMARCAD1, SDF4, and MeCP2 (negative control) with Myc-tagged MSH2 in HEK293T cells. Results indicated that, besides the known interactions (i.e. MSH2-MSH3, MSH2-MSH6, MSH2-EXO1), several HCIPs such as WDHD1, KIAA1671, and SMARCAD1 also bind to MSH2, therefore validating the MSH2 interactome we identified (Fig. 4B). In addition, we confirmed the MSH2-WDHD1 interaction between endogenous proteins (Fig. 4C), suggesting that these two proteins indeed associate with each other in vivo.
Fig. 4.
MSH2 interacts with WDHD1. (A) Interactomes of MSH2 or WDHD1 identified by our TAP-MS approach. (B) Validation of MSH2 interactome. HEK293T cells were transfected with plasmids encoding SFB-tagged various proteins together with plasmids encoding Myc-tagged MSH2. Precipitation reactions were conducted using S-protein beads and then subjected to Western blotting using indicated antibodies. (C) Endogenous WDHD1 interacts with MutS complex. HeLa cell lysates were prepared and immunoprecipitated with WDHD1 antibody. Immunoprecipitates were blotted using antibodies as indicated. (D, E) Mapping of the corresponding regions required for the MSH2/WDHD1 interaction. (Left panel) Schematic presentation of wild type and mutants of WDHD1 and MSH2 used in this study. For WDHD1, the blue rectangle indicates WD domain, red rectangle indicates SepB domain, and yellow rectangle indicates HMG domain. WDHD1 D1 mutant (1–1110aa), D2 mutant (1–1122aa), D3 mutant (1–1126aa), AP mutant (Ala1123Pro), and AF mutant (Ala1123ProPhe1124Ala); MSH2 D1 mutant (1–620aa), D2 mutant (1–550aa), D3 mutant (1–460aa), D4 mutant (1–410aa), and D5 mutant (del 550–620aa) were indicated, respectively (left panel). For MSH2, the blue rectangle indicates mismatch domain, green rectangle indicates connector domain, yellow rectangles indicate lever domains, red rectangle indicates clamp domain, purple domain indicates ATPase domain, and light blue rectangle indicates helix-turn-helix domain. (Right panel) Immunoprecipitation reactions were performed using S-protein beads and then subjected to Western blot analyses using antibodies as indicated. (F) WDHD1 depletion confers an increased cellular resistance to 6-TG. Colony-formation assays were performed as described in the Experimental Procedures. Statistical analysis was performed using the Student's test. A p value less than 0.05 was considered significant. An asterisk (*) represents the p value. Data are presented as mean ± S.E.
Mapping the Interaction Domains of WDHD1 and MSH2
Wenext attempted to define the MSH2-binding region(s) on WDHD1. A series of truncation mutants of WDHD1 were coexpressed with SFB-tagged MSH2 in HEK293T cells. We were able to map the minimal MSH2-binding region to a small region at the C terminus of WDHD1 (residues 1122–1126). Interestingly, within this region, an Ala1123 to Pro missense mutant of WDHD1 was detected in a lung cancer patient Catalogue of Somatic Mutations in Cancer (COSMIC). We found that mutation of Ala1123 (Ala1123Pro) site alone or both Ala1123 and Phe1124 (Ala1123ProPhe1124Ala) sites abolished the interaction between MSH2 and WDHD1 (Fig. 4D), indicating that this interaction may contribute to cancer development.
Next, we sought to identify the region(s) of MSH2 that is responsible for its interaction with WDHD1. Again, we generated a series of truncation and internal deletion mutants of MSH2. As shown in Fig. 4E, the D5 mutant (the second lever domain, residues 550–620) of MSH2 dramatically reduced the MSH2-WDHD1 interaction, indicating that this domain of MSH2 is important for its binding to WDHD1.
WDHD1 Depletion Confers an Increased Cellular Resistance to 6-thioguanine (6-TG)
It is well documented that the levels of MSH2 inversely correlate with 6-TG and N-methyl-N'-nitro-N-nitrosoguanidine (MNNG) resistance (39). Consistently, there were fewer colonies formed in parental HeLa cells upon 6-TG treatment, while knockdown of MSH2 or WDHD1 in these cells resulted in resistance to 6-TG, i.e. more colonies formed after 6-TG treatment (Fig. 4F). These results indicate that WDHD1 may not only bind to MSH2 but also function with MSH2 in the MMR pathway.
DISCUSSION
This work provides an extensive analysis of MMR protein–protein interaction network, identifies over 230 HCIPs, and therefore greatly broadens our current understanding of the MMR pathway. We uncovered several uncharacterized partners for MMR proteins like MutS, MutL, and EXO1 (Fig. 3). The biological significance of these interactions remains to be determined. Given that the MMR pathway is a critical genome maintenance pathway and MMR deficiency leads to MSI and cancer development, we speculate that some of the MMR-binding proteins discovered in this study may be mutated or downregulated in cancer and therefore contribute to cancer development and MSI phenotypes identified in cancer patients. This possibility warrants further investigation.
In our proteomics analysis of the MMR pathway, we built a subnetwork with HCIPs for three MMR components, MutS, MutL, and Exo1 (Fig. 3A). It is known that MMR is implicated in other repair processes, including DNA damage signaling, homologous recombination, interstrand cross-link repair, and meiotic DNA recombination (40). Indeed, HCIPs of three MMR components clearly indicate the connections between MMR and other DNA repair pathways. For example, XRCC3 was identified as HCIPs of MutS and MutL complexes. This protein is implicated in homologous recombination repair (41, 42), indicating that the MMR pathway may participate in homologous recombination repair through interactions with multiple factors involved in homologous recombination. MutL complexes have also been shown to participate in the repair of interstrand cross-links, with the evidence that MutLα interacts specifically with Fanconi anemia protein FANCJ (Fanconi Anemia Group J Protein, BRCA1-Interacting Protein 1) to facilitate interstrand cross-link repair (43). As a matter of fact, BRIP1 was repeatedly identified in our purifications of MutL complexes (Fig. 3A). Moreover, another Fanconi anemia protein FAN1 was also identified in the MutL complexes (Fig. 3A). The specific interaction between FAN1 (FANCD2/FANCI-Associated Nuclease 1), and MutL was further confirmed by reverse purification conducted by us and others (44, 45), suggesting that MutL may participate in interstrand cross-link repair through its interaction with several Fanconi anemia proteins.
MSH2 is a central component of the MMR pathway that recognizes mismatches arising during DNA replication. The analysis of MSH2 interactome revealed not only several known components of MutS complex, including MSH3 and MSH6 but also several previously unidentified partners, such as WDHD1, KIAA1671, and SMARCAD1 (Fig. 4A). In particular, WDHD1 protein containing an amino-terminal WD40 domain (tryptophan-aspartic acid (W-D) dipeptide repeat) and a carboxyl-terminal HMG High-mobility group motif. It has been shown that WDHD1 acts as a component of the replisome to regulate DNA replication and S phase progression (46–49). It is also well documented that MMR corrects DNA mismatches generated during DNA replication. Thus, it is reasonable to speculate that WDHD1 may function to recruit MutS complex to chromatin during DNA replication and thus facilitate the MMR pathway in removing mismatches after ongoing DNA replication forks. In this study, we not only validated the interaction between MSH2 and WDHD1 but also showed that a missense mutant of WDHD1, Ala1123-to-Pro, detected in a lung cancer patient (COSMIC), abolished the WDHD1-MSH2 interaction, indicating that this mutation in WDHD1 may be functionally important for lung cancer development. Of note, we also checked cBioPortal and found 163 WDHD1 mutations in colorectal cancer, endometrial cancer, bladder cancer, and others, indicating that WDHD1 may be mutated in multiple types of cancers and contribute to tumorigenesis. Moreover, similar to MSH2, knockdown of WDHD1 confers cellular resistant to 6-TG, indicating that WDHD1 likely participates in the MMR pathway. Future studies will be directed at defining whether and how WDHD1 may facilitate the loading of MSH2 during DNA replication and promote MMR.
In conclusion, our proteomics analysis of the MMR pathway provides a rich resource for further exploration of MMR functions in various DNA repair pathways, which will offer new ideas and therapeutic approaches for cancer patients.
Supplementary Material
Acknowledgments
We would like to thank all the colleagues in Dr. Chen's laboratory for insightful discussion and technical assistance, especially Dr. Xu Li. We also want to thank Dr. Steven Gygi and Dr. Ross Tomaino (Taplin Mass Spectrometry Facility, Harvard Medical School) for their help with mass spectrometry analysis and providing raw data for the submission. We thank Dr. Alma Faust for critical reading of the manuscript.
Footnotes
Author contributions: Z.C., Z.G., and J.C. designed the research; Z.C., M. Tran, M. Tang, W.W., and Z.G. performed the research; Z.C., Z.G., and J.C. analyzed data; and Z.C., Z.G., and J.C. wrote the paper.
* This work was supported in part by the Department of Defense (DOD) Era of Hope research scholar award to J.C. (W81XWH-09-1-0409) and an R21 grant to Z.G. (CA192052). This work was also in part supported by the NIH/NCI Cancer Center Support Grant P30 CA016672. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental material Supplemental Tables S1-Se.
This article contains supplemental material Supplemental Tables S1-Se.
1 The abbreviations used are:
- MMR
- mismatch repair
- HCIP
- high-confidence interacting proteins
- CRAPome
- Contaminant Repository for Affinity Purification
- SFB
- S tag-Flag tag-SBP
- IPA
- Ingenuity Pathway Analysis.
REFERENCES
- 1.Altieri F., Grillo C., Maceroni M., and Chichiarelli S. (2008) DNA damage and repair: From molecular mechanisms to health implications. Antiox. Redox. Signal. 10, 891–937 [DOI] [PubMed] [Google Scholar]
- 2.Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., and Linn S. (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 3.Kolodner R. (1996) Biochemistry and genetics of eukaryotic mismatch repair. Genes Develop. 10, 1433–1442 [DOI] [PubMed] [Google Scholar]
- 4.Kunkel T. A., and Erie D. A. (2005) DNA mismatch repair. Annu. Rev. Biochem. 74, 681–710 [DOI] [PubMed] [Google Scholar]
- 5.Li G. M. (2008) Mechanisms and functions of DNA mismatch repair. Cell Res. 18, 85–98 [DOI] [PubMed] [Google Scholar]
- 6.Hsieh P., and Yamane K. (2008) DNA mismatch repair: Molecular mechanism, cancer, and ageing. Mechanisms Ageing Develop. 129, 391–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Acharya S., Wilson T., Gradia S., Kane M. F., Guerrette S., Marsischky G. T., Kolodner R., and Fishel R. (1996) hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc. Natl. Acad. Sci. U.S.A. 93, 13629–13634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marsischky G. T., Filosi N., Kane M. F., and Kolodner R. (1996) Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Develop. 10, 407–420 [DOI] [PubMed] [Google Scholar]
- 9.Marsischky G. T., and Kolodner R. D. (1999) Biochemical characterization of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 complex and mispaired bases in DNA. J. Biol. Chem. 274, 26668–26682 [DOI] [PubMed] [Google Scholar]
- 10.Habraken Y., Sung P., Prakash L., and Prakash S. (1996) Binding of insertion/deletion DNA mismatches by the heterodimer of yeast mismatch repair proteins MSH2 and MSH3. Curr. Biol. 6, 1185–1187 [DOI] [PubMed] [Google Scholar]
- 11.Harrington J. M., and Kolodner R. D. (2007) Saccharomyces cerevisiae Msh2-Msh3 acts in repair of base–base mispairs. Mol. Cell Biol. 27, 6546–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palombo F., Iaccarino I., Nakajima E., Ikejima M., Shimada T., and Jiricny J. (1996) hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr. Biol. 6, 1181–1184 [DOI] [PubMed] [Google Scholar]
- 13.Gradia S., Subramanian D., Wilson T., Acharya S., Makhov A., Griffith J., and Fishel R. (1999) hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol. Cell 3, 255–261 [DOI] [PubMed] [Google Scholar]
- 14.Kadyrov F. A., Dzantiev L., Constantin N., and Modrich P. (2006) Endonucleolytic function of MutLalpha in human mismatch repair. Cell 126, 297–308 [DOI] [PubMed] [Google Scholar]
- 15.Kadyrov F. A., Holmes S. F., Arana M. E., Lukianova O. A., O'Donnell M., Kunkel T. A., and Modrich P. (2007) Saccharomyces cerevisiae MutLalpha is a mismatch repair endonuclease. J. Biol. Chem. 282, 37181–37190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tishkoff D. X., Boerger A. L., Bertrand P., Filosi N., Gaida G. M., Kane M. F., and Kolodner R. D. (1997) Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc. Natl. Acad. Sci. U.S.A. 94, 7487–7492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran P. T., Simon J. A., and Liskay R. M. (2001) Interactions of Exo1p with components of MutLalpha in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 98, 9760–9765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran P. T., Erdeniz N., Symington L. S., and Liskay R. M. (2004) EXO1-A multi-tasking eukaryotic nuclease. DNA Repair 3, 1549–1559 [DOI] [PubMed] [Google Scholar]
- 19.Constantin N., Dzantiev L., Kadyrov F. A., and Modrich P. (2005) Human mismatch repair: Reconstitution of a nick-directed bidirectional reaction. J. Biol. Chem. 280, 39752–39761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Yuan F., Presnell S. R., Tian K., Gao Y., Tomkinson A. E., Gu L., and Li G. M. (2005) Reconstitution of 5′-directed human mismatch repair in a purified system. Cell 122, 693–705 [DOI] [PubMed] [Google Scholar]
- 21.Sekine H., Ferreira R. C., Pan-Hammarström Q., Graham R. R., Ziemba B., de Vries S. S., Liu J., Hippen K., Koeuth T., Ortmann W., Iwahori A., Elliott M. K., Offer S., Skon C., Du L., Novitzke J., Lee A. T., Zhao N., Tompkins J. D., Altshuler D., Gregersen P. K., Cunningham-Rundles C., Harris R. S., Her C., Nelson D. L., Hammarström L., Gilkeson G. S., and Behrens T. W. (2007) Role for Msh5 in the regulation of Ig class switch recombination. Proc. Natl. Acad. Sci. U.S.A. 104, 7193–7198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tompkins J. D., Wu X., Chu Y. L., and Her C. (2009) Evidence for a direct involvement of hMSH5 in promoting ionizing radiation induced apoptosis. Experiment. Cell Res. 315, 2420–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peltomäki P. (2003) Role of DNA mismatch repair defects in the pathogenesis of human cancer. J. Clin. Oncol. 21, 1174–1179 [DOI] [PubMed] [Google Scholar]
- 24.Gu L., Cline-Brown B., Zhang F., Qiu L., and Li G. M. (2002) Mismatch repair deficiency in hematological malignancies with microsatellite instability. Oncogene 21, 5758–5764 [DOI] [PubMed] [Google Scholar]
- 25.Wang K., Kan J., Yuen S. T., Shi S. T., Chu K. M., Law S., Chan T. L., Kan Z., Chan A. S., Tsui W. Y., Lee S. P., Ho S. L., Chan A. K., Cheng G. H., Roberts P. C., Rejto P. A., Gibson N. W., Pocalyko D. J., Mao M., Xu J., and Leung S. Y. (2011) Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat. Genet. 43, 1219–1223 [DOI] [PubMed] [Google Scholar]
- 26.Gong Z., Cho Y. W., Kim J. E., Ge K., and Chen J. (2009) Accumulation of Pax2 transactivation domain interaction protein (PTIP) at sites of DNA breaks via RNF8-dependent pathway is required for cell survival after DNA damage. J. Biol. Chem. 284, 7284–7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Longo P. A., Kavran J. M., Kim M. S., and Leahy D. J. (2013) Transient mammalian cell transfection with polyethylenimine (PEI). Meth. Enzymol. 529, 227–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elias J. E., and Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 29.Elias J. E., and Gygi S. P. (2010) Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol. Biol. 604, 55–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellacheruvu D., Wright Z., Couzens A. L., Lambert J. P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., Al-Hakim A., Bouchard A., Faubert D., Fermin D., Dunham W. H., Goudreault M., Lin Z. Y., Badillo B. G., Pawson T., Durocher D., Coulombe B., Aebersold R., Superti-Furga G., Colinge J., Heck A. J., Choi H., Gstaiger M., Mohammed S., Cristea I. M., Bennett K. L., Washburn M. P., Raught B., Ewing R. M., Gingras A. C., and Nesvizhskii A. I. (2013) The CRAPome: A contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benjamini Y., and Hochberg Y. (1995) Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. Royal Stat. Soc. B Met. 57, 289–300 [Google Scholar]
- 33.Li X., Wang W., Wang J., Malovannaya A., Xi Y., Li W., Guerra R., Hawke D. H., Qin J., and Chen J. (2015) Proteomic analyses reveal distinct chromatin-associated and soluble transcription factor complexes. Mol. Syst. Biol. 11, 775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang W., Li X., Huang J., Feng L., Dolinta K. G., and Chen J. (2014) Defining the protein-protein interaction network of the human hippo pathway. Mol. Cell Proteomics 13, 119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu S., Li X., Gong Z., Wang W., Li Y., Nair B. C., Piao H., Yang K., Wu G., and Chen J. (2014) Proteomic analysis of the human cyclin-dependent kinase family reveals a novel CDK5 complex involved in cell growth and migration. Mol. Cell. Proteomics 13, 2986–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Genschel J., and Modrich P. (2003) Mechanism of 5′-directed excision in human mismatch repair. Mol. Cell 12, 1077–1086 [DOI] [PubMed] [Google Scholar]
- 37.Lee Bi B. I., Nguyen L. H., Barsky D., Fernandes M., and Wilson D. M. 3rd (2002) Molecular interactions of human Exo1 with DNA. Nucleic Acids Res. 30, 942–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chun J., Buechelmaier E. S., and Powell S. N. (2013) Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway. Mol. Cell Biol. 33, 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karran P. (2006) Thiopurines, DNA damage, DNA repair and therapy-related cancer. Brit. Med. Bull. 79–80, 153–170 [DOI] [PubMed] [Google Scholar]
- 40.Jiricny J. (2006) The multifaceted mismatch-repair system. Nature Rev. Mol. Cell. Biol. 7, 335–346 [DOI] [PubMed] [Google Scholar]
- 41.Brenneman M. A., Wagener B. M., Miller C. A., Allen C., and Nickoloff J. A. (2002) XRCC3 controls the fidelity of homologous recombination: Roles for XRCC3 in late stages of recombination. Mol. Cell 10, 387–395 [DOI] [PubMed] [Google Scholar]
- 42.Peng G., Dai H., Zhang W., Hsieh H. J., Pan M. R., Park Y. Y., Tsai R. Y., Bedrosian I., Lee J. S., Ira G., and Lin S. Y. (2012) Human nuclease/helicase DNA2 alleviates replication stress by promoting DNA end resection. Cancer Res. 72, 2802–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng M., Litman R., Xie J., Sharma S., Brosh R. M. Jr., and Cantor S. B. (2007) The FANCJ/MutLalpha interaction is required for correction of the cross-link response in FA-J cells. EMBO J. 26, 3238–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu T., Ghosal G., Yuan J., Chen J., and Huang J. (2010) FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science 329, 693–696 [DOI] [PubMed] [Google Scholar]
- 45.Smogorzewska A., Desetty R., Saito T. T., Schlabach M., Lach F. P., Sowa M. E., Clark A. B., Kunkel T. A., Harper J. W., Colaiácovo M. P., and Elledge S. J. (2010) A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol. Cell 39, 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bermudez V. P., Farina A., Tappin I., and Hurwitz J. (2010) Influence of the human cohesion establishment factor Ctf4/AND-1 on DNA replication. J. Biol. Chem. 285, 9493–9505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gambus A., van Deursen F., Polychronopoulos D., Foltman M., Jones R. C., Edmondson R. D., Calzada A., and Labib K. (2009) A key role for Ctf4 in coupling the MCM2–7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 28, 2992–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Im J. S., Ki S. H., Farina A., Jung D. S., Hurwitz J., and Lee J. K. (2009) Assembly of the Cdc45-Mcm2–7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc. Natl. Acad. Sci. U.S.A. 106, 15628–15632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu W., Ukomadu C., Jha S., Senga T., Dhar S. K., Wohlschlegel J. A., Nutt L. K., Kornbluth S., and Dutta A. (2007) Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Develop. 21, 2288–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.