Abstract
Significant progress has been made in understanding the roles of crucial residues/motifs in the channel function of P2X receptors during the pre-structure era. The recent structural determination of P2X receptors allows us to reevaluate the role of those residues/motifs. Residues Arg-309 and Asp-85 (rat P2X4 numbering) are highly conserved throughout the P2X family and were involved in loss-of-function polymorphism in human P2X receptors. Previous studies proposed that they participated in direct ATP binding. However, the crystal structure of P2X demonstrated that those two residues form an intersubunit salt bridge located far away from the ATP-binding site. Therefore, it is necessary to reevaluate the role of this salt bridge in P2X receptors. Here, we suggest the crucial role of this structural element both in protein stability and in channel gating rather than direct ATP interaction and channel assembly. Combining mutagenesis, charge swap, and disulfide cross-linking, we revealed the stringent requirement of this salt bridge in normal P2X4 channel function. This salt bridge may contribute to stabilizing the bending conformation of the β2,3-sheet that is structurally coupled with this salt bridge and the α2-helix. Strongly kinked β2,3 is essential for domain-domain interactions between head domain, dorsal fin domain, right flipper domain, and loop β7,8 in P2X4 receptors. Disulfide cross-linking with directions opposing or along the bending angle of the β2,3-sheet toward the α2-helix led to loss-of-function and gain-of-function of P2X4 receptors, respectively. Further insertion of amino acids with bulky side chains into the linker between the β2,3-sheet or the conformational change of the α2-helix, interfering with the kinked conformation of β2,3, led to loss-of-function of P2X4 receptors. All these findings provided new insights in understanding the contribution of the salt bridge between Asp-85 and Arg-309 and its structurally coupled β2,3-sheet to the function of P2X receptors.
Keywords: ATP, conformational change, ion channel, protein expression, protein stability, P2X receptors, salt bridge
Introduction
P2X receptors are ATP-gated non-selective cation channels that are widely distributed in various eukaryotic cells (1). They sense the elevation of extracellular ATP with Ca2+ and Na+ ions passing through activated channels into the cell, leading to diverse physiological and pathological activities (1, 2), including synaptic transmission, pain sensation, cardiovascular diseases, respiratory functions, gastrointestinal disorders, immune balance, and urinary regulations. So far, seven P2X (P2X1–P2X7) receptors have been cloned from eukaryotic cells, which can form homomeric or heteromeric trimers (3–7). Dysfunctions of those homomeric or heteromeric P2X channels are associated with numerous diseases (2) such as neuropathic pain (8), hypertension (9), and thrombosis (10). Knowledge of the gating mechanism of those channels will advance our understanding of the structure-function relationship of those trimeric receptors, which in turn will contribute to the development of new drugs targeting P2X receptors (6).
In the past 2 decades, the functional roles of the essential structural elements, such as conserved residues/motifs in P2X receptors activation, have been identified through sequence alignment, mutagenesis, and electrophysiological recording before the crystal structure was determined (3, 11–15). The recent structural determination of zebrafish P2X4 receptors (zfP2X4) at the resting and open states allowed us to study both conserved and non-conserved key residues/motifs and their allosteric transitions during channel activation at the atomic level (16, 17). Those two structures led to the discovery that the subunit embedded in the membrane resembles a leaping dolphin, which consists of intracellular N and C termini, two transmembrane helices, and a rigid extracellular body domain, covalently attached by a cysteine-rich head domain, a right flipper, a left flipper (LF),4 and a dorsal fin (DF) domain. The resolved crystal structures will benefit an in-depth investigation of the conformational transition during the channel gating of P2X receptors (18, 19). Significant progress has been made (19–27) because the structural determination of P2X receptors (18, 19, 28), mainly including the downward motion of the head domain, coordinated tightening of the ATP-binding site jaw, the relative motions of the LF and DF domains, and the following pore-opening motions. Most importantly, the resolved crystal structures would also benefit intensive studies on the key structural elements with uncertain or distorted roles in the activation of P2X receptors, due to the previous lack of crystal structures.
It is known that the genetic polymorphism at the highly conserved residue (R307Q) causes loss-of-function of the human P2X7 (hP2X7) receptor and is associated with the lack of ATP-stimulated phospholipase D activity in lymphocytes (29). Before the structural determination of zfP2X4, it was thought that the polymorphism interrupted ATP binding of hP2X7 receptors, resulting in dysfunction of hP2X7. However, the crystal structures of the zfP2X4 receptor revealed that the shortest distance between the guanidinium group of Arg-312 (equivalent to the Arg-307 of hP2X7) and the triphosphate group of ATP is 12.7 Å (Fig. 1A), which is too far to produce direct interactions between the residues and the ATP. Instead, the guanidinium (RNHC(NH2)2+) of Arg-312 interacts with the anionic carboxylate (R-COO−) of Asp-88, another highly conserved residue throughout the P2X family, forming a salt bridge in both resting and open states of zfP2X4 receptors (Fig. 1B). Salt bridges, commonly formed by a combination of non-covalent interactions (hydrogen bonding and electrostatic interactions) between acidic and alkaline amino acids, play important roles in maintaining the structures and functions of enzymes, receptors, and other proteins (30–38). Previous studies have demonstrated important roles of salt bridges in channel functions, such as channel gating (31) and protein expression (34), in a wide variety of ion channels. Therefore, the reason why the polymorphism of one residue of this salt bridge induced loss-of-function in human P2X receptors should be reevaluated. Also, the functional roles of this highly conserved salt bridge in P2X receptors also need to be elucidated. Combining mutagenesis, disulfide cross-linking, electrophysiology, Western blotting, and molecular modeling, we suggest here the critical role of this highly conserved salt bridge in protein stability and channel gating via a mechanism of stabilizing the kinked conformation of the β2,3-sheet, a structural element covalently coupled with one residue of this salt bridge.
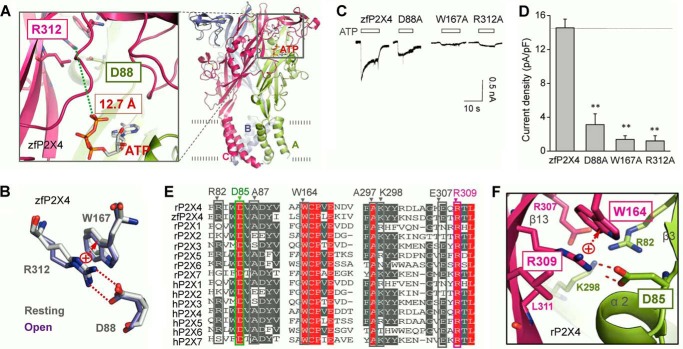
FIGURE 1.
Salt bridge between Arg-312 and Asp-88 shows high conservation in different subtypes of P2X receptors. A, three-dimensional structure of zfP2X4 in open state. Green broken line shows closest distance between Arg-312 and ATP. B, superposition of residues Arg-312, Asp-88, and Trp-167 at resting and open states. C and D, representative maximum current traces at a saturated concentration ATP (10 mm) (C) and pooled data (D) of zfP2X4 and its mutants. All data are expressed as mean ± S.E. (error bars) from 8 to 10 experiments. **, p < 0.01 versus WT (dashed line), Student's t test. E, multiple -sequence alignment of P2X family. F, zoom-in view of rP2X4 at open state shows details of the salt bridge and its surrounding residues. Red broken lines indicate hydrogen bonding between Arg-309 and Asp-85. Red arrow indicates cation π-electronic interactions between Arg-309 and Trp-164. All figures in this paper were drawn with PyMOL.
Experimental Procedures
Cell Culture
As we described previously (39), human embryonic kidney 293 cells (HEK-293) were cultured in DMEM containing 10% FBS and antibiotics at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. Transfections of plasmids were performed with Hilymax (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer's protocol.
Drugs and Mutagenesis
Most of the drugs were purchased from Sigma. The plasmids pcDNA3-rP2X4, pcDNA3-rP2X2, and pcDNA3-rP2X7 were kindly gifted by Drs. Alan North and Lin-Hua Jiang. The cDNA of zfP2X4.1 was obtained from Dr. Eric Gouaux and was then subcloned into the pcDNA3.1 vector. The cDNA of hP2X1 was purchased from JiKai Gene and was subcloned into the pcDNA3.1 vector. All mutants were obtained using a QuikChange mutagenesis kit and subsequently verified by DNA sequencing.
Electrophysiology
As we previously described (39), whole-cell patch clamp recordings were performed 24–48 h after transient transfection at room temperature (23 ± 2 °C) using Axon 200B under the voltage clamp. The recording electrodes were filled with a pipette solution, and the resistance was between 3 and 5 megohms. Membrane current signals were amplified by using a patch clamp amplifier (Axon 200B, Axon Instruments, Foster City, CA). Currents were low-pass filtered at 2 kHz. All currents were sampled and analyzed using a Digidata 1440 interface and a computer running the Clampex and Clampfit 10.0 software (Molecular Devices). Solutions were applied using a “Y-tube” method throughout the experiments. The external solution contained (in mm) 150 NaCl, 5 KCl, 10 glucose, 2 CaCl2·2H2O, 10 HEPES, and 1 MgCl2·6H2O; the pH was adjusted to 7.2–7.4. Pipette solutions contained (in mm) 120 KCl, 30 NaCl, 1 MgCl2·6H2O, 0.1 CaCl2·2H2O, and 5 EGTA; the pH was adjusted to 7.2 using Tris-base. Cells were patched and held at −60 mV throughout the experiment under voltage clamp conditions. Drugs were dissolved in external solutions. By using nystatin (0.1 μg/μl) in high potassium pipette solutions, perforated patch clamp was employed when ATP was administered repeatedly to avoid the desensitization of P2X receptors. High potassium pipette solutions contained (in mm) 75 K2SO4, 55 KCl, 5 MgSO4, 10 HEPES. The interval time between ATP delivery was 8–15 min to avoid receptor desensitization. The maximal currents of WT and mutated channels were measured using standard whole-cell recordings, whereas the nystatin-perforated patch clamp was used for recordings of dose-dependent response curves and the current comparison before and after DTT application.
Western Blotting
Surface biotinylation and Western blotting were performed according to our previous description (39). In brief, the transfected HEK-293 cells were washed in chilled PBS and then incubated with sulfo-NHS-LC-biotin (Pierce) dissolved in chilled PBS+/+ (containing 1 mm MgCl2 and 0.1 mm CaCl2, pH 8.0). The reaction was terminated by incubating the cells with glycine (20 mm) in PBS and washed three times with PBS. The cells were then collected and lysed with RIPA buffer (Thermo Scientific). With centrifugation, 10% of the volume of the supernatant was diluted with SDS loading buffer and used as the total protein fraction. The remaining biotinylated proteins were adsorbed to agarose resin linked to NeutrAvidin by 3–5 h of incubation at 4 °C and subsequently washed with chilled PBS 3–5 times. All of the Western blotting protein samples were boiled with SDS loading buffer with (for reducing experiment) or without (for non-reducing experiments) β-mercaptoethanol (1%) for 5 min. Bound proteins were used as surface proteins. Protein samples were then analyzed by Western blotting. The samples were separated by SDS-PAGE using an 8–10% main gel and a 4% stacking gel and transferred to a polyvinylidene difluoride (PVDF) membrane. The membrane was blocked with 5% milk at room temperature for 1–2 h and then incubated overnight at 4 °C with anti-EE tag (1:1,000; Abcam) or anti-GAPDH (1:2,000; Sungene Biotech) antibodies dissolved in 5% milk. Then the membrane was washed and incubated with appropriate HRP-conjugated secondary antibodies for EE tag (25 °C, 1 h, 1:1,000; goat anti-rabbit IgG(H+L)-HRP; Sungene Biotech) or GAPDH (25 °C, 1 h, 1:3,000, goat anti-mouse IgG(H+L)-HRP; Sungene Biotech) and finally visualized using an ECL solution (Thermo) and exposure with ImageQuant RT ECL system (GE Healthcare) for 1–3 min. The results were then analyzed using ImageQuant software.
Homology Modeling
The homology models of rP2X4 and mutants were created based on the crystal structures (Protein Data Bank entries 4DW0 and 4DW1) using Modeler 9.9 following our previous description (39). The alignments of the target sequences were made by Modeler 9.9 and manually adjusted to match published alignments (16). The constructed model was checked and validated by ProCheck (40).
Molecular Dynamic Simulations
According to our previous description (39), all MD simulations were performed by using the program DESMOND 3.0 (41) with a constant number of particles, pressure (1 bar), and temperature (300 K) and periodic boundary conditions, which uses a particular “neutral territory” method called the midpoint method to efficiently exploit a high degree of computational parallelism (41). A default OPLS_2005 force field, following the functional form of the OPLS-AA family of force fields, was used for the protein and ATP molecules. The energy-minimized structures by DESMOND 3.0 were used as the starting structures for MD simulations (41). A large dimyristoylphosphatidylcholine bilayer was constructed to generate a suitable membrane system in which the transmembrane domain of the rP2X4 could be embedded. The protein/dimyristoylphosphatidylcholine system was subsequently dissolved in simple point charge water molecules. Counter ions were then added to compensate for the net negative charge of the system. NaCl (150 mm) was added into the simulation box that represented background salt at physiological conditions. All of the MD simulations were run on the DAWNING TC2600 system. Preparation, analysis, and visualization were performed on a DELL T7500 graphic working station.
Data Analysis
All static results were expressed as means ± S.E. Data were analyzed with Clampfit 10.2 (Molecular Devices). Statistical comparisons were performed using Student's t test. The significant levels were considered statistically as follows: *, p < 0.05, and **, p < 0.01. Concentration-response relationships for ATP activation of wild-type or mutant receptors were performed by administration of ATP with different concentrations, and data were obtained from 4–12 cells in at least three different batches of transfections. All of the results used to generate a concentration-response relationship were from the same group. Currents induced by each concentration were normalized by the maximal current, and then data were pooled for statistical analysis. The data were fitted to the Hill equation, I/Imax = 1/(1 + (EC50/[ATP])n), where I is the normalized current at a given concentration of ATP; Imax is the maximum normalized current; EC50 is the concentration of ATP yielding a current that is half of the maximum, and nH is the Hill coefficient.
Results
Highly Conserved Salt Bridge between Arg-312 and Asp-88 Is Essential for the Channel Function of the zfP2X4 Receptor
The crystal structure of the zfP2X4 receptor revealed that the guanidinium (RNHC(NH2)2+) of Arg-312 in one subunit interacts with the anionic carboxylate (R-COO−) of Asp-88 in another subunit, with the N—O bond distances of 1.79 and 1.74 Å, respectively, forming a salt bridge, both at resting and open states (Fig. 1B). Additionally, a cation-π interaction was also observed between Arg-312 and Trp-167 (Fig. 1B). Alanine substitutions on these three residues (R312A, D88A, and W167A) led to significantly decreased maximal currents (Imax) of zfP2X4 receptors induced by saturated ATP (10 mm), indicating the important roles of those residues in P2X4 function (Imax = 1.2 ± 0.6, 3.12 ± 1.3, 1.37 ± 0.44 pA/pF, for R312A, D88A, and W167A, respectively, versus Imax = 14.6 ± 1.0 pA/pF for WT, respectively, n = 7, p < 0.01; Fig. 1, C and D). However, the shortest atomic distance between ATP and the residue Arg-312 is longer than 12.0 Å (Fig. 1A), suggesting that Arg-312 indirectly influences the ATP-induced effect. The contribution of those three residues in channel functions remains to be clarified.
The residues Asp-88, Arg-312, and Trp-167 of zfP2X4 receptors are highly conserved across the P2X family (Fig. 1E). Moreover, the spatially adjacent residues surrounding them are also conserved in zfP2X4 and rat P2X4 (rP2X4) receptors (Fig. 1E), indicating a comparatively highly conserved architecture in P2X4 proteins among different species. Because zfP2X4 showed low apparent ATP affinity (EC50, the concentration of yielding one-half of maximal currents, >1 mm) (17) and poorer expression efficiency in HEK-293 cells (Imax = 14.6 ± 1.0, n = 8, and 313 ± 36 pA/pF, n = 25 for zfP2X4 and rP2X4, respectively), when compared with rP2X4 receptors, all the following procedures of mutagenesis, electrophysiological recordings, and protein expression measurements were carried out on equivalent residues of rP2X4 to facilitate the further study into this interaction cluster.
To examine the locations and orientations of equivalent amino acids of this salt bridge and adjacent residues intuitively, we built a homology model of rP2X4 based on the crystal structures of zfP2X4, at both resting (PDB entry code 4DW0) and open (PDB entry code 4DW1) states. The homology model revealed that Arg-309 (located at β13, corresponding to Arg-312 of zfP2X4), interacted with Asp-85 (located at the loop between α2 and β3, corresponding to Asp-88 of zfP2X4), forming a salt bridge (Fig. 1F). Meanwhile, a strong cation-π interaction was also observed between Arg-309 and Trp-164 (corresponding to Trp-167 of zfP2X4) (Fig. 1F). Adjacent residues that are highly conserved between the zfP2X4 and rP2X4 receptors, including Arg-82, Ala-87, Asp-88, Ala-297, and Glu-307 (rP2X4 numbering), which are shown to be highly conserved between the zfP2X4 and rP2X4 receptors, may contribute to supporting the interactions between Asp-85, Arg-309, and Trp-164 (Fig. 1E).
Salt Bridge between Arg-309 and Asp-85 Is a Stringent Structural Element Required for Normal rP2X4 Function
To elucidate the functional roles of this salt bridge in rP2X4, we substituted these three amino acids and their adjacent residues with alanines. The substitutions at the residues almost abolished ATP-induced rP2X4 currents (Imax = 12.7 ± 3.1, 33.9 ± 6.0, 43.7 ± 9.2 versus 312.8 ± 36 pA/pF; R309A, D85A, and W164A versus WT, n = 6–25, p < 0.01, Fig. 2, A and B), which are consistent with identical mutations in zfP2X4 (Fig. 1, C and D). In comparison, mutations in adjacent residues (K298A, R82A, and E307A) displayed no significant effects on maximal responses of rP2X4 (Fig. 2, A and B).
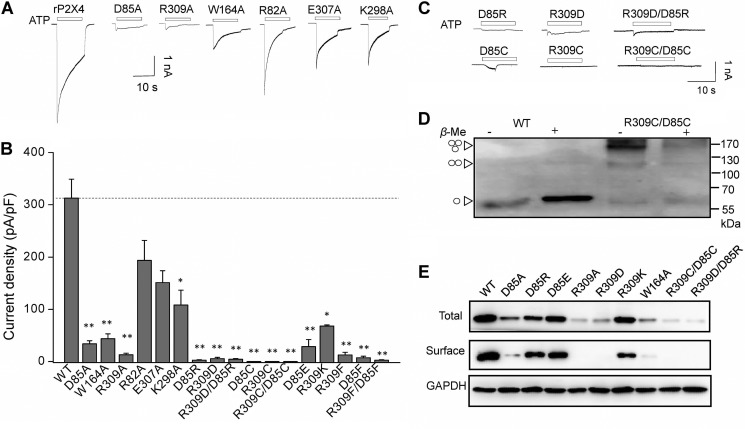
FIGURE 2.
Essential role of salt bridge between Arg-309 and Asp-85 in normal rP2X4 functions. A–C, representative maximum current traces and pooled data of rP2X4 and its mutants induced by saturated ATP (1–10 mm). All data are expressed as mean ± S.E. from 6 to 25 experiments. *, p < 0.05; **, p < 0.01 versus WT (dashed line), Student's t test. D, rP2X4 with double mutation R309C/D85C formed homologous oligomers, mainly trimers in non-reducing Western blotting indicated by arrowheads (left). Western blotting results were observed in at least three independent experiments for each receptor. E, representative Western blotting with anti-EE of total and surface-biotinylated proteins from HEK-293 expressing WT and various mutants of rP2X4. The protein expression experiments were conducted independently at least three times.
Previous studies demonstrated that the ion channel function could be restored by charge swapping or disulfide trapping of the salt bridge in some receptors (31, 32, 35). Here, disruption of the salt bridge by charge reversal at one amino acid of the salt bridge attenuated ATP-induced maximum currents (Imax = 5.46 ± 2.6 and 2.69 ± 0.7 pA/pF, for R309D and D85R, respectively; n = 6–10; Fig. 2, B and C), indicating an essential role of this structural element to normal channel function. However, charge swapping (R309D/D85R) could not restore the currents induced by saturated ATP (10 mm) to the WT level (R309D/D85R, Imax = 4.32 ± 1.5 pA/pF, n = 6; Fig. 2, B and C). We then examined the role of the salt bridge by replacing it with an inter-domain/intersubunit disulfide bond. Cysteine substitutions were introduced to the salt bridge pair. As a result, single or double substitutions resulted in complete loss of channel function (Fig. 2, B and C). The Western blotting result revealed that R309C/D85C mutants mainly existed as trimers and, to a lesser degree, as dimers, which can be separated into monomers by the reducing reagent β-mercaptoethanol (1%) (Fig. 2D). As a control, the WT receptor was expressed as a monomer in both conditions (Fig. 2D). These results suggested the formation of intersubunit disulfide bonds in R309C/D85C mutant, although the expression of mutated channels was much lower compared with that of the WT. Therefore, the loss of function of R309C/D85C was not due to its failure to form intersubunit disulfide bonds; instead, it may have resulted from the cross-linking-induced conformational change that could not support the normal channel function.
The fact that charge swapping and disulfide bond trapping could not restore the channel function shows a stringent structural requirement of the salt bridge between Arg-309 and Asp-85 for normal rP2X4 function. In addition even minor alterations, such as D85E and R309K, could significantly reduce the ATP-induced maximal current (Imax = 28.6 ± 13 and 67.8 ± 2.3 pA/pF for D85E and R309K, respectively; n = 4–5; Fig. 2B), indicating that any subtle change in this position may lead to a significant impact in channel function, let alone changes with bulkier sizes of the aromatic side chains in D85F, R309F, and D85F/R309F mutants (Imax = 7.21 ± 3.1, 12.7 ± 5.1, and 2.7 ± 0.8 pA/pF for D85F, R309F, and D85F/R309F, respectively; n = 4–5; Fig. 2, B and C). Taken together, the salt bridge between Arg-309 and Asp-85 is essential for the functional rP2X4 receptors. Any subtle changes of charge or size at the side chains could result in a loss of channel function.
Salt Bridge between Arg-309 and Asp-85 Is Essential for rP2X4 Stability and Channel Gating Rather than Direct ATP Interaction and Channel Assembly
Given that the decreased channel response is generally attributed to lower protein expression or impaired channel gating, we measured the protein expression level of the rP2X4 receptor and its mutants, at both total and membrane expression levels. Protein expression levels of mutants D85A, R309A, and W164A decreased significantly, especially for their surface expression levels (Figs. 2E and 3, A and B). The mutation D85R reduced the channel expression level to half of the WT expression. Also, R309D and R309D/D85R caused a larger reduction of the channel expression of P2X4 receptors (Figs. 2E and 3, A and B). In addition, introducing a disulfide bond between Arg-309 and Asp-85 by double cysteine substitutions (R309C/D85C) could not rescue the decreased protein expression level by single mutations (Figs. 2E and 3, A and B).
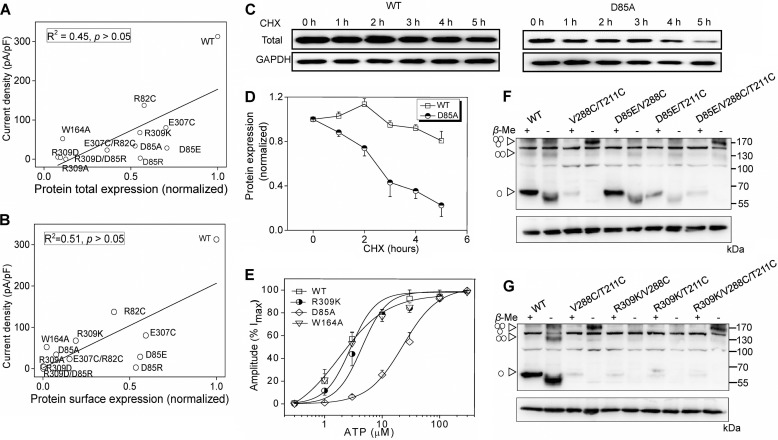
FIGURE 3.
Salt bridge between Arg-309 and Asp-85 is essential for protein stability and channel gating rather than ATP binding and channel assembly. A and B, correlation map between current density and total (A) or surface (B) protein expression of different mutants. C, representative Western blotting of rP2X4 protein for WT (left) and D85A (right). Cells were treated with 20 μg/ml CHX in a time course experiment as indicated. The results were observed in at least three independent experiments for statistical analysis. D, time-response curves (mean ± S.E., n = 4) showing protein expression levels at different time points following the incubation of 20 μg/ml CHX. E, ATP concentration-response curve of rP2X4 and its mutants. All data are expressed as means ± S.E. from 4 to 12 experiments. Data point was fitted with Hill equation to the ATP-dependent activation. F and G, protein samples extracted from transfected HEK-293 cells separated by non-reducing Western blotting. Monomeric, dimeric, and trimeric receptors are indicated by triangle arrows on the left. Molecular weight markers are shown on the right. Similar results were observed in at least three independent experiments.
Multiple factors can influence the protein expression level, for example, altered protein stability, channel assembly, and surface trafficking. To determine whether the disruption of the salt bridge affected the stability of protein, we incubated the transfected HEK-293 cells with cycloheximide (CHX, 20 μg/ml), an inhibitor of protein biosynthesis, in time scale experiments. The negligible protein expression of R309A rendered the mutant expression difficult to be quantified. We chose D85A as an example to investigate the protein stability. After incubation with CHX for 5 h, the protein level of D85A decreased by 77 ± 11% and that of the WT decreased by 19 ± 8% (Fig. 3, C and D), suggesting a profound instability of the D85A mutant when compared with the WT (p < 0.05).
In contrast to the profound reduction in maximum currents, little to no change was observed in EC50 values for the functional mutants, namely R309K (EC50 = 4.75 ± 1.6 μm, nh = 1.4 ± 0.5, n = 12) and W164A (EC50 = 2.3 ± 0.3 μm; nh = 1.2 ± 0.4, n = 9), compared with WT channels (EC50 = 2.3 ± 0.7 μm, nh = 1.6 ± 0.6, n = 4). The EC50 value of the channel was partly enhanced in D85A (EC50 = 41.9 ± 7.1 μm, nh = 1.5 ± 0.4, n = 10, Fig. 3E), which is located in the loop between the α2-helix and the β3-sheet (Fig. 1F). The increased EC50 may originate from the fact that the motion of this domain was associated with the downward movement of the head domain, which is coupled with the ATP binding and channel gating activity (20, 24, 42). However, compared with the mutations at positively charged residues, which interact directly with the negative triphosphate of ATP, such as K71A and K69R (the EC50 of ATP >20 mm) (43, 44), the rightward shift of the concentration-response curve for D85A was nearly negligible.
Taking into account that D85E and R309K displayed significantly decreased ATP-induced currents (Fig. 2B) but with less influence in the protein expression (Fig. 2E), we proposed that the impaired channel gating of rP2X4 may be the main cause of the functional decrease of those mutants, in addition to their poor expression. To further test this hypothesis, we analyzed the relevance between protein expression and current density of all mutants. The current densities showed poor correlation with total (R2 = 0.45, p > 0.05) or surface protein expressions of ion channels (R2 = 0.51, p > 0.05), verifying that the reduction of protein expression cannot fully account for the decreased channel activity from the disrupted salt bridge between Arg-309 and Asp-85 (Fig. 3, A and B). To further exclude the possibility that this intersubunit salt bridge between Arg-309 and Asp-85 participates in channel assembly, we introduced additional mutations, D85E and R309K, into mutant V288C/T211C, which could form trimeric receptors in non-reduction SDS-PAGE, due to the formation of intersubunit disulfide bonds as we previously demonstrated (39). Trimeric bands of mutants, D85E/V288C/T211C and R309K/V288C/T211C, were observed in the non-reducing Western blotting, indicating that the alterations of the salt bridge between Arg-309 and Asp-85 did not render a significant deficiency in channel assembly of P2X4 receptors (Fig. 3, F and G). Thus, given the normal surface expression, channel assembly, and EC50 of D85E and R309K, the significantly reduced maximal currents induced by saturated ATP might be due to changes in some essential or constituent elements of P2X4 channel gating, for example, the decreased rate of channel opening and/or mean channel open time or an increased rate of channel desensitization. Taken together, the salt bridge together with cation-π interactions among Arg-309 and Asp-85 and Trp-164 plays important roles in channel function by participating in protein expression and stability, as well as channel gating, rather than direct ATP interaction and channel assembly.
Molecular Simulations Indicate the Essential Role of the Salt Bridge between Arg-309 and Asp-85 in Stabilizing the Kinked Conformation of the β2,3-Sheet
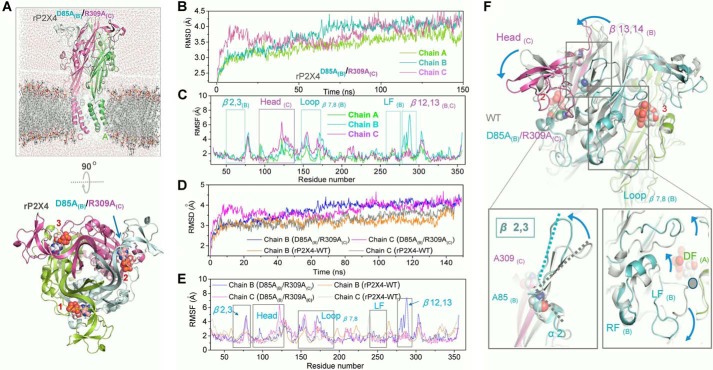
Although it is clear that the Arg-309 and Asp-85 salt bridge is possibly involved in protein stability and the channel gating function of rP2X4 receptors, the underlying mechanism still remains unclear. To gain insights into the conformational change induced by the absence of this salt bridge, we constructed a homology model, named rP2X4D85A(B)/R309A(C), substituting alanine for Asp-85 in chain B and Arg-309 in chain C of rP2X4 at open state. Therefore, the salt bridge that formed between chain B and chain C was absent in the model, whereas chain A performed as a WT subunit (Fig. 4A). During the 150-ns time scale of the MD simulations, root mean square deviations of the backbone atoms in the WT chain A were obviously smaller than the mutated chains B and C, suggesting that notably changed conformations in chains B and C occurred when the constraint derived from the intersubunit salt bridge was lost (Fig. 4B). In addition, root mean squared fluctuation analysis of rP2X4D85A(B)/R309A(C) showed profound fluctuation of residues in mutated chains (chains B and C) when compared with the WT chain (chain A), especially for the residues in the head domain, the loops β7,8, β12,13, and β2,3, and the DF domain (Figs. 4, C and F, and 5A). This point was further confirmed by a structural flexibility comparison between the mutated chains (chains B and C) of rP2X4D85A(B)/R309A(C) and the unmutated chains of WT rP2X4 (Fig. 4, D and E).
FIGURE 4.
Interrupting salt bridge between Arg-309 and Asp-85 causes conformational changes in rP2X4 receptors. A, built simulation system of rP2X4D85A(B)/R309A(C) at the open state viewed parallel (upper) and perpendicular (lower) to the membrane, with different subunit displayed in green, cyan, and magenta cartoons, respectively. The three ATP molecules are highlighted by spheres and marked by numbers. The blue arrow indicates the broken salt bridge. B, time dependence of the root mean square deviation of the Cα from the initial structure of rP2X4D85A(B)/R309A(C) receptor during 150-ns MD simulation. C, comparison of simulated root mean squared fluctuations (RMSF) of each residue in three subunits. D, time dependence of the root mean square deviation of the Cα from initial homology models of WT and mutated channel rP2X4D85A(B)/R309A(C). E, root mean square deviation of WT and mutated channels rP2X4D85A(B)/R309A(C) during MD simulations. F, zoom-in view of the constructed model of rP2X4D85A(B)/R309A(C) based on the open structure, showing the movements of different domains (blue arrows) after MD simulations.
FIGURE 5.
Illustration showing salt bridge Arg-309 and Asp-85 stabilizes the kinked conformation of β2,3 and multiple-domain stacking interactions. A, illustration of alternations in the multiple domains interactions. Dashed line represents the mutated receptors rP2X4D85A(B)/R309A(C), and solid line represents the conformation of WT receptors. Different colors represent different regions of adjacent subunits. Arrows indicate the movement of different domains. B, superposition of the structure of β2,3 of WT (gray) and the representative conformation of R309A/D85A (azure) after MD simulations. Gray and azure dashed lines represent the bending angle formed between β2,3 and α2 of WT and R309A/D85A, respectively. Azure arrow represents the movement of β2,3 in the R309A/D85A mutant when compared with the original conformation of WT.
Asp-85 and Arg-309 were located in the linker region between α2 and β3 in one subunit and β13 in another subunit, respectively (Fig. 1F). Because of the stretching force derived from the formation of the intersubunit salt bridge between Arg-309 and Asp-85, together with the repelling force from the head domain, β2,3 was forced to form a kinked conformation with an ∼90° angle relative to the α2-helix in the WT channel (Fig. 5B). Additionally, a certain angle also exists between β2 and β3 (like an “X” shape) due to the existence of those constraints (Figs. 4F, 5B, and 6A). Once the salt bridge was interrupted, β2,3 moved to the upper left side and exhibited an extended conformation (Figs. 4F, lower panel, and 5). The β2,3-sheet was located at the interface among the head domain, the loop β7,8, and the loop β13,14 (Figs. 4F and 6B), where β2,3 stacked with those domains together through electronic and/or hydrophobic interactions. Thus, the conformations of those physically coupled domains were dramatically changed after the loss of the salt bridge constraint, such as the downward movement of the head domain, the upward movement of loop β7,8, and the simultaneous downward movement of the LF domain (Figs. 4F, lower panel, and 5A). Therefore, weakening of the stacking interaction among various domains from three distinct subunits may reduce the protein stability.
FIGURE 6.
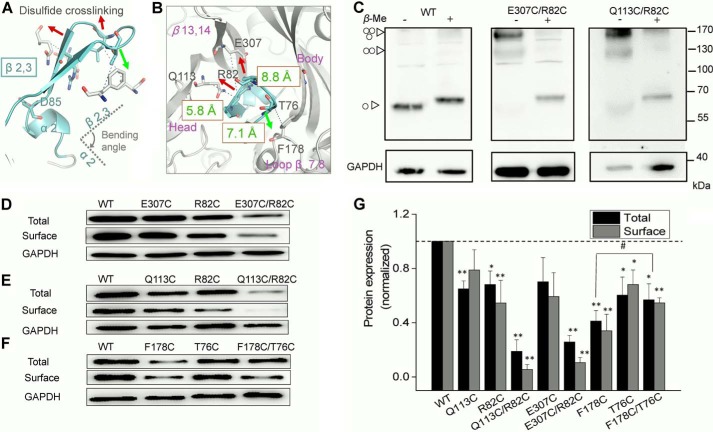
Changed conformation of β2,3 leads to a decreased protein expression of rP2X4. A and B, zoom-in view of the bending angle of β2,3 relative to the α2-helix. Red and green arrows indicate the direction to alter the orientation of β2,3 from three different directions. Residues selected to generate disulfide bond are highlighted with sticks. Green text shows the Cβ-Cβ distance between the three paired residues. Purple text indicates the domains surrounding β2,3. C, homo-oligomeric proteins, mainly trimeric rat P2X4 receptors formed by introduction of disulfide bond, were separated by Western blotting. Molecular weight markers are shown on the right. The symbols at left represent the monomeric, dimeric, and trimeric rP2X4 receptor, respectively. Similar results were observed in at least three independent experiments. D–G, representative Western blotting of total and surface expressions, their pooled data of rP2X4, and the mutants with cysteine substitution extracted from the transiently transfected HEK-293 cells. Bars represent mean ± S.E. (n = 3) of the total or surface levels normalized to the WT protein; #, p < 0.05 versus control; *, p < 0.05; **, p < 0.01 versus WT (dashed line), Student's t test.
Meanwhile, given that β2,3 is physically coupled with head and LF domains, two domains proved to be crucial for channel gating of the P2X4 receptors. It is reasonable to assume that the salt bridge has a role in affecting the channel gating of P2X4 receptors through stabilizing the kinked conformation of β2,3. Therefore, we proposed that the salt bridge between Arg-309 and Asp-85 functioned as the fulcrum of multiple region interactions to stabilize the protein structure. The loss of constraint from this salt bridge would change the kinked conformation of β2,3, which consequently interferes with the regions physically coupled with β2,3, leading to the instability of the protein or a deficiency in channel gating (Fig. 5).
Kinked Conformation of β2,3 Is Essential for Protein Stability and Channel Gating of P2X4 Receptors
To verify our hypothesis, we engineered the disulfide bond trapping in three directions that modify the bending angle of β2,3 without destroying the salt bridge (Fig. 6, A and B). The Cβ-Cβ distances of Gln-113 and Arg-82 and of Glu-307 and Arg-82 were 5.8 and 8.8 Å, respectively (Fig. 6B), whereas the general Cβ-Cβ distance of the disulfide bond was ∼4.5–5 Å. Therefore, the kinked conformation of β2,3 would be altered by shortening Cβ-Cβ distances of Q113C/R82C and E307C/R82C, rendering a transition of β2,3 from a kinked conformation to an extended conformation (Fig. 6, A and B). In contrast, the introduction of the disulfide bond in F178C/T76C will roughly hold the original direction of β2,3 (Fig. 6, A and B). By non-reducing Western blotting, the presence of trimetric receptors shown in Q113C/R82C and E307C/R82C confirmed the establishment of intersubunit disulfide cross-linking (Fig. 6C).
Consistent with our hypothesis, the introduction of double cysteine mutations Q113C/R82C and E307C/R82C significantly reduced the total protein expressions and surface protein expressions of rP2X4 when compared with those of WT channels (Fig. 6, D, E, and G). Single cysteine mutations (Q113C, R82C, and E307C) also decreased both total and surface protein expression levels, but to a lesser extent, when compared with the double cysteine mutations (Fig. 6, D, E, and G, p < 0.05). Moreover, generated by F178C/T76C, the downward movement of β2,3 had a slighter effect on protein expression than the other two disulfide bonds (Fig. 6, F and G, p < 0.05). The single mutations F178C and T76C rendered about half a decrease in protein expression level, especially for F178C (42.1 ± 7.8% of WT). Yet interestingly, the double cysteine mutation F178C/T76C could partially rescue the protein loss caused by F178C (56.9 ± 11% versus 42.1 ± 7.8%, F178C/T76C versus F178C, p < 0.05; Fig. 6, F and G), indicating the essential role of the kinked conformation of β2,3 in maintaining protein expression of rP2X4.
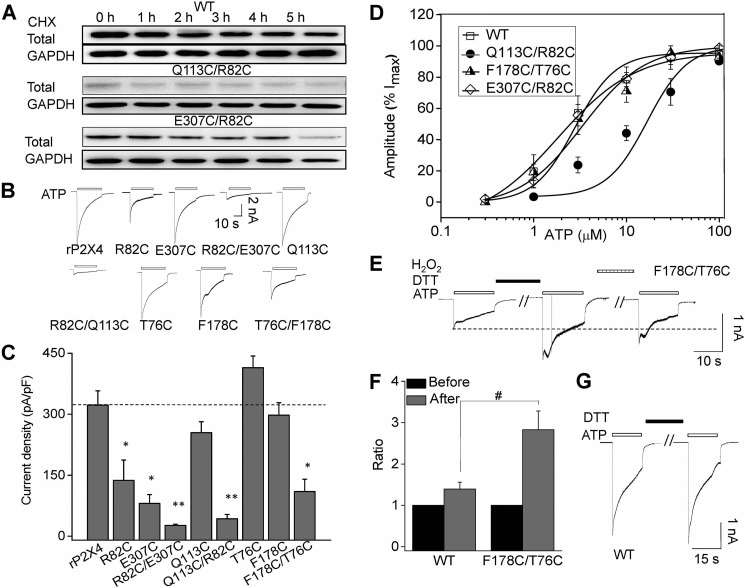
We then examined protein stability using a CHX assay. Channel proteins were almost completely degraded when Q113C/R82C- or E307C/R82C-transfected cells were incubated with CHX for 1 or 5 h, respectively, whereas CHX had little impact on the WT proteins (Fig. 7A), indicating that alternations of the kinked conformation of β2,3 rendered the instability of rP2X4 receptors. Meanwhile, the maximal currents deceased dramatically in double cysteine mutants Q113C/R82C and E307C/R82C (Fig. 7, B and C). Additionally, double cysteine mutants caused little to no shift in ATP-response curves (Fig. 7D, EC50 = 15.6 ± 7.4 μm, nH = 0.79 ± 0.05, n = 4; 2.8 ± 0.8 μm, nH = 1.6 ± 0.2, n = 7; 2.01 ± 0.4 μm, nH = 1.4 ± 0.1, n = 4 versus 2.3 ± 0.7 μm, nH = 1.6 ± 0.6, n = 4 for Q113C/R82C, E307C/R82C, F178C/T76C, and WT, respectively).
FIGURE 7.
Changed conformations of β2,3 render the instability and gating deficiency of P2X4 receptors. A, representative Western blotting of rP2X4 receptors expressing WT or mutants in HEK-293 incubated with CHX in time course experiments. Similar results were observed in at least three independent experiments. B and C, representative current traces and pooled data for WT and mutant receptors evoked by applications of ATP. Average current density induced by saturated ATP (1 mm). All data are expressed as mean ± S.E. from 6 to 25 experiments. *, p < 0.05; **, p < 0.01 versus WT (dashed line), Student's t test. D, concentration-response curve of rP2X4 and its mutants (n = 4). Data points were fitted with the Hill equation. E, representative current traces recorded from cells expressing F178C/T76C in response to application of 100 μm ATP. Cells were perfused with DTT (10 mm) for 8 min and H2O2 (0.3%) for 5 min. F, pooled data for the effects of DTT on saturated ATP-induced currents of WT and F178C/T76C. y axis represents the ratio of current induced by ATP after DTT application normalized to the current before DTT administration. All data are expressed as mean ± S.E. (error bars) from six cells in three independent experiments. #, p < 0.05 versus control, Student's t test. G, representative current traces recorded from cells expressing WT rP2X4.
Interestingly, although the protein expression level of F178C/T76C was higher than that of F178C, the amplitudes of ATP-induced currents were smaller (Imax = 297 ± 30, 414 ± 29, and 110 ± 31 pA/pF, for F178C, T76C, and F178C/T76C, respectively, p < 0.01). The disproportional current density and protein expression level in F178C/T76C indicated a possible role of β2,3 in P2X4 channel gating. The current amplitude of F178C/T76C increased 2–4-fold after dithiothreitol (DTT, 10 mm) application (Fig. 7, E and F); the current declined again after treatment with H2O2 (0.3%), indicating that the disulfide bridge may impair the channel gating of F178C/T76C. Because DTT application displayed no significant effect on the current of WT rP2X4 (Fig. 7, F and G), the breaking of disulfide between F178C/T76C may be responsible for DTT application-evoked increases in the current amplitude of F178C/T76C. Taken together, alterations of β2,3 decreased both maximal channel responses and the stability of the channel protein, whereas preserving the natural kinked conformation of β2,3 exhibited little effect on channel function and protein expression. These data confirmed the important role of the kinked conformation of β2,3 in maintaining normal P2X4 function.
Additional Strategies to Alter the Kinked Conformation of β2,3 Lead to a Deficiency in Normal P2X4 Functions
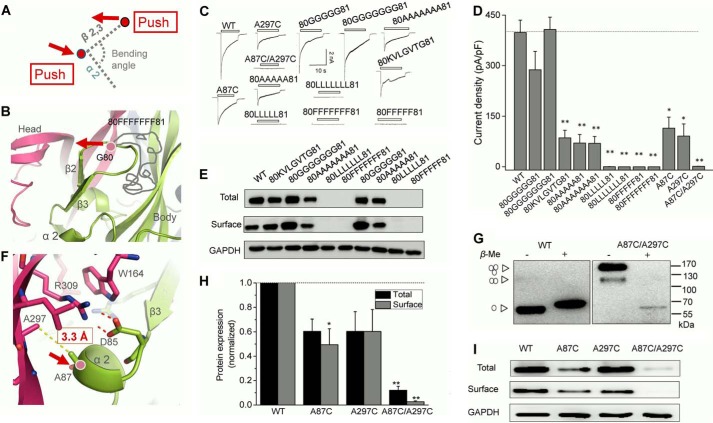
To further confirm the important role of the kinked conformation of β2,3, we put inserts with different amino acid sequences into the loop between β2 and β3 (between Gly-80 and Phe-81), which would push β2,3 toward the upper left direction (Fig. 8A). Little to no change was observed in both expression and current densities of the mutants, which were inserted with five or seven glycines (Fig. 8, B and C). In contrast, insertion of a random sequence (KVLGVTG) or five or seven alanines between Gly-80 and Phe-81 all caused obvious decreases in currents and expressions of channels (Fig. 8, B–E). Moreover, complete loss of channel current and protein expression was observed in the mutants with inserts composed of leucines or phenylalanines (five or seven), suggesting that elongated aliphatic side chains and the bulky steric hindrance of inserted amino acids could push β2,3 to move further toward the upper left side and consequently induce a more extending conformation of β2,3 and finally cause a profound influence in channel function.
FIGURE 8.
Additional strategies to alter the conformation of β2,3 lead to similar changes in rP2X4. A, illustration of our strategies to alter the bending angle formed by β2,3 toward α2. The red arrows represent the directions induced by the alteration in the adjacent area. B, zoom-in view of insertion site in the middle loop between β2 and β3. The gray curve represents the amino acids inserted. Red arrow shows the left motion of β2,3 driven by long amino acid insertion. C and D, typical currents and pooled data from HEK-293 cells expressing rP2X4 and its mutants. All data are expressed as mean ± S.E. from 8 to 25 experiments. *, p < 0.05; **, p < 0.01 versus WT (dashed line), Student's t test. E, Western blotting analysis of protein expressions of rP2X4 and its mutants in HEK-293 cells. Similar results were observed in at least three independent experiments. F, zoom-in view of salt bridge region. The Cβ-Cβ distance of Ala-297 and Ala-87 is indicated by yellow dashed line. G, rP2X4A87C/A297C formed homologous oligomers, mainly trimers as indicated by arrowheads on the left. H and I, Western blotting and pooled data of rP2X4 and the cysteine substitution mutants A87C, A297C, and A87C/A297C. Bars represent mean ± S.E. (n = 3) of the total or surface levels normalized to the WT protein. *, p < 0.05 versus WT (dashed line), Student's t test. **, p < 0.01.
Furthermore, we selected two residues adjacent to the salt bridge between Asp-85 and Arg-309, namely Ala-297 and Ala-87 (situated in α2 helix), with a 3.3-Å Cβ-Cβ distance between the two alanines (Fig. 8F). Given that Cβ-Cβ distance between the formed disulfide bond is about 4.5–5 Å, the distance between Ala-297 and Ala-87 was lengthened when they were replaced with cysteines. Therefore, the cross-linking by mutation A297C/A87C could slightly push the α2 helix to the right and consequently alter the interaction between Asp-85 and Arg-309 and finally change the kinked conformation of β2,3 (Fig. 8, A and F). In non-reducing SDS-PAGE analysis, obvious high molecular mass proteins, mainly trimers, were observed, indicating successful introduction of disulfide bonds (Fig. 8G). The subtle distance changes caused by double mutation A297C/A87C resulted in a profound reduction in protein expression and maximal currents, indicating the rigidity of the conformation in this region (Fig. 8, C, D, H, and I).
Taken together, we concluded that the salt bridge formed between Arg-309 and Asp-85 and the kinked conformation of β2,3 mainly functioned as a fulcrum to stabilize the interactions among various domains. The destruction of the salt bridge will render conformational changes in β2,3 as well as other physically coupled domains, and it will finally influence protein expression and channel gating.
Substitution of Identical Salt Bridges in Different P2X Subtypes Causes a Similar Deficiency in Normal Channel Functions
In consideration of the significance of the salt bridge and its high conservation across the P2X family, we chose P2X1, P2X2, and P2X7 to further study the function of the salt bridge in different subtypes. Similar to what we observed in rP2X4, alanine substitution of the three conserved core residues leads to the deficiency of channel functions, reflected by electrophysiological study (Fig. 9, A and B). Among the conserved residues, Arg-309 (rP2X4 numbering) was the most stringent in the four P2X family members we studied, with the substitution of it causing a complete loss of channel function. Western blotting data showed that the protein expression, especially at the cell surface level, was conspicuously decreased in the equivalent mutants of Arg-309 (rP2X4 numbering) in P2X7, P2X1, and P2X2 (Fig. 9, C and D). These data indicated that the loss of function in polymorphism of R307Q may have been due to the deficiency of surface protein expression (Fig. 9, C and D), rather than impaired direct ATP contacts, as reported previously (29). In addition, although part of the surface expression was partially retained in P2X7D89A, P2X7R307A, P2X1R305A, and P2X2R304A, none of these mutants produced currents in response to saturated ATP. These results were in accordance with the data of corresponding mutants in rP2X4, suggesting that the conserved salt bridge was not only involved in protein expression but also in channel gating. It should also be noted that those mutants had less impact in total protein expression compared with P2X4, suggesting a conserved role of this salt bridge in influencing the normal channel function but somewhat different from those existing in various subtype P2X receptors.
FIGURE 9.
Substitution of salt bridge in different P2X subtypes caused similar deficiency. A and B, saturated ATP-induced representative current traces (A) and pooled data (B) for different P2X subtypes and their mutants. All data are expressed as mean ± S.E. from 4 to 10 experiments. *, p < 0.05; **, p < 0.01 versus WT, Student's t test. C and D, representative Western blotting of total and surface protein expressions and pooled data of P2X receptors and its mutants extracted from transiently transfected HEK-293 cells. Bars represent mean ± S.E. (n = 3–5) of the total or surface levels normalized to the WT protein.
Discussion
A salt bridge is a strong noncovalent interaction, with a combination of hydrogen bond and electrostatic interactions (45). This interaction is the most commonly observed interaction to contribute to the stability of the conformation of proteins (46–48). Previous studies have demonstrated that a salt bridge is implicated in many protein functions, such as the ligand binding of the GABAA receptor (33), the stabilization of the activation state of Kir1.1 (37), the folding of the serotonin transporter (38), the surface expression of ASIC channels (34), and the protein thermostability of enzymes (36). Here, we suggest that the salt bridge formed between Arg-309 and Asp-85 played an essential role in the protein stability and channel gating of P2X4 receptors. Losing the constraints from this salt bridge will lead to the instability of the channel protein. Located at the intersubunit interface near the ATP-binding pocket, the salt bridge between Arg-309 and Asp-85 may function as a fulcrum of the stacking interactions among the head domain, the right flipper domain, and the LF domain by stabilizing the conformation of β2,3. Once the salt bridge is interrupted, significant conformational changes will take place like the collapse of dominoes, in domains including β2,3, DF, loop β7,8, loop β12,13, and the head domain (Fig. 5A). Among these regions, some domains have been proven to be associated with channel gating, such as the head and LF domains (20–25, 42). Additionally, even a minor change in size of the side chain of those two residues may cause a loss-of-function of P2X receptors. Notably, the significant decreases in current density have also been observed in P2X1, P2X2, P2X4, and P2X7 with mutations in the residues of the salt bridge, without dramatically lower express levels, such as the P2X4D85K, P2X7R307Q, P2X7D89A, P2X7R307A, P2X1R305A, P2X1W164A, and P2X2R304A (Figs. 2 and 9). Because the EC50 values and channel assembly were proven to be comparable with the WT channels in some mutants (Fig. 3), the decreased current density in those mutations might be attributed to altered channel gating. Previous studies demonstrated that the head, the DF, and the LF domains have participated in nearly every essential or constituent element of P2X channel gating (20–22, 24, 39, 42), including channel desensitization (24). Given that the changed salt bridge possibly resulted in the conformational changes in β2,3 and its physically coupled head and LF domains, the decreased current density in those mutants may be attributed to the combination of the affecting factors mentioned above. Taken together, this salt bridge is important for the normal channel function of P2X receptors. Loss of the constraint of this strong interaction will lead to the instability of channel protein, whereas a minor alteration in this interaction would render deficiency of the channel gating, although the expression level is comparable with WT channels.
Previous studies have suggested that the residue of this salt bridge is implicated in the ATP-sensing of P2X receptors (29, 43, 44). However, the crystal structures exclude this possibility, because the shortest atomic distance between ATP and the residue of the salt bridge is over 12 Å (Fig. 1A) (16, 17). It has been demonstrated that channel activation was associated with the downward motion of the head domain that is coordinated with ATP binding (20, 25). As revealed in MD simulations, we observed alterations of movement in the head domain after losing this salt bridge (Figs. 4F and 5A). It is speculated that the salt bridge may partially contribute to stabilize the shape of the ATP-binding pocket rather than a direct involvement in ATP recognition. This may be the reason why some mutants that interrupt this salt bridge as well as its physically coupled β2,3 could change the apparent EC50 value of the P2X4 receptor, for example, the slightly increased EC50 values of rP2X4D85A and rP2X4Q113C/R82C. Thus, this salt bridge only plays an indirect and negligible role in the ATP sensing of P2X receptors.
Although both Asp-85 and Arg-309 are required to form the salt bridge, their contributions to P2X4 functions are not equal (Fig. 2). Being the more stringent component, the replacement of Arg-309 with the hydrophobic amino acid (rP2X4R309A) caused complete loss of function in protein expression and channel function. The mutant rP2X4D85A showed a relatively smaller effect on the protein expression when compared with R309A despite the obvious reduced ATP currents. The same results were also observed in P2X1, P2X2, and P2X7 (Fig. 9, A–C). This unequal feature has also been reported in other ion channels, such as the Glu-153 and Lys-196 of the GABAA receptor (31) and the Asp-107 and Arg-153 (34) interaction pair of the ASIC3 channel. Until now, there is no proven explanation for this phenomenon. It is possible that that one residue of the salt bridge pair bears more strain energy than the other residue does in maintaining protein conformation and stability. Nevertheless, other possible functions of the residue before the protein folding cannot be ruled out, which rendered one residue different from the other.
Additionally, as an indispensable membrane ion channel, P2X has exhibited its important role in disease intervention and treatment (49–52). In recent years, many P2X-targeted drugs have been developed in clinical trials (50–52). In-depth knowledge of the dynamics and interplay among different structural elements during channel gating will improve our understanding of the structure-function relationship of the P2X receptor and aid future new drug designs. For example, elucidation of subtle differences in the channel gating of distinct subtypes will boost subtype-specific drug designs targeting P2X receptors. Although the salt bridge identified here is highly conserved through the evolution, the sequences of the surrounding regions (head domain, LF, DF, and β2,3) showed a high diversity across the P2X family, which may contribute to subtype-specific differences in the physical coupling between those domains and the salt bridge. As an important functional element, the salt bridge and its surrounding structures are closely related to many ion channel diseases. In the other three subtypes we have identified, P2X7, P2X2, and P2X1, alternation of Asp-85 and Arg-309 and Trp-164 resulted in a similar reduction of channel currents to P2X4, but they showed some difference in total expression as well as channel activation, suggesting some differences underlying the channel dysfunction by mutations in different subtypes. The future study of variability between subunits is helpful for drug design and disease intervention.
In summary, our study identified the essential roles of an intersubunit salt bridge in the normal function of P2X receptors. As a fulcrum, the salt bridge functions by maintaining the interactions of surrounding domains and is indispensable for protein stability and channel gating. These findings enrich our understanding of the role of this salt bridge in P2X receptors and will aid in exploring the mechanisms behind the site mutation-related diseases as well as developing new therapeutic approaches.
Author Contributions
Ye Yu conceived and designed the project. W.-S. Z., M.-Y. S., L.-F. S. Y. L., and L.-D. H. performed the experiments. Ye Yu, W.-S. Z., and Y. T. analyzed the data. Yang Yang, W.-S. Z., Ye Yu, X.-Y. C., and L.-Y. L. wrote the manuscript. Ye Yu, Y.-Z. F., Y.-M. H., and P. C. devised the protein homology model. R. W. and Ye Yu supervised Wen-Shan Zhao.
Acknowledgments
We are grateful to Drs. Lin-Hua Jiang, Alan North, and Eric Gouaux for their kind gifts of plasmids pCDNA3-rP2X4, rP2X2, rP2X7, and pFastBac OP-1-zfP2X4.1.
This work was supported by National Excellent Young Scientist Foundation of China Grant 31222018, National Program on Key Basic Research Project of China Grant 2014CB910302, National Natural Science Foundation of China Grants 31570832, 31170787, 31400707, 81473377 and 81302694, Key National S&T Program “Major New Drug Development” Grant 2012ZX09504001-003, “Shanghai Jiao Tong University-SMC Mutual Funds” for Excellent Young Scholar, Science and Technology Fund of Shanghai Jiao Tong University School of Medicine Grant 13XJ10042, Shanghai Jiao Tong University School of Medicine-Key Incubation Project, and the Opening Project of National Key Laboratory of Receptors Grant SIMM1601KF-02. The authors declare that they have no conflicts of interest with the contents of this article.
- LF
- left flipper
- DF
- dorsal fin
- h
- human
- zf
- zebrafish
- CHX
- cycloheximide
- pF
- picofarad
- MD
- molecular dynamics.
References
- 1.Surprenant A., and North R. A. (2009) Signaling at purinergic P2X receptors. Annu. Rev. Physiol. 71, 333–359 [DOI] [PubMed] [Google Scholar]
- 2.Khakh B. S., and North R. A. (2006) P2X receptors as cell-surface ATP sensors in health and disease. Nature 442, 527–532 [DOI] [PubMed] [Google Scholar]
- 3.Coddou C., Yan Z., Obsil T., Huidobro-Toro J. P., and Stojilkovic S. S. (2011) Activation and regulation of purinergic P2X receptor channels. Pharmacol. Rev. 63, 641–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis C., Neidhart S., Holy C., North R. A., Buell G., and Surprenant A. (1995) Coexpression of P2X2 and P2X3 receptor subunits can account for ATP-gated currents in sensory neurons. Nature 377, 432–435 [DOI] [PubMed] [Google Scholar]
- 5.Nicke A., Bäumert H. G., Rettinger J., Eichele A., Lambrecht G., Mutschler E., and Schmalzing G. (1998) P2X1 and P2X3 receptors form stable trimers: a novel structural motif of ligand-gated ion channels. EMBO J. 17, 3016–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.North R. A., and Surprenant A. (2000) Pharmacology of cloned P2X receptors. Annu. Rev. Pharmacol. Toxicol. 40, 563–580 [DOI] [PubMed] [Google Scholar]
- 7.North R. A. (2002) Molecular physiology of P2X receptors. Physiol. Rev. 82, 1013–1067 [DOI] [PubMed] [Google Scholar]
- 8.Chessell I. P., Hatcher J. P., Bountra C., Michel A. D., Hughes J. P., Green P., Egerton J., Murfin M., Richardson J., Peck W. L., Grahames C. B., Casula M. A., Yiangou Y., Birch R., Anand P., and Buell G. N. (2005) Disruption of the P2X7 purinoceptor gene abolishes chronic inflammatory and neuropathic pain. Pain 114, 386–396 [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto K., Sokabe T., Matsumoto T., Yoshimura K., Shibata M., Ohura N., Fukuda T., Sato T., Sekine K., Kato S., Isshiki M., Fujita T., Kobayashi M., Kawamura K., Masuda H., et al. (2006) Impaired flow-dependent control of vascular tone and remodeling in P2X4-deficient mice. Nat. Med. 12, 133–137 [DOI] [PubMed] [Google Scholar]
- 10.Hechler B., Lenain N., Marchese P., Vial C., Heim V., Freund M., Cazenave J. P., Cattaneo M., Ruggeri Z. M., Evans R., and Gachet C. (2003) A role of the fast ATP-gated P2X1 cation channel in thrombosis of small arteries in vivo. J. Exp. Med. 198, 661–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adriouch S., Bannas P., Schwarz N., Fliegert R., Guse A. H., Seman M., Haag F., and Koch-Nolte F. (2008) ADP-ribosylation at R125 gates the P2X7 ion channel by presenting a covalent ligand to its nucleotide binding site. FASEB J. 22, 861–869 [DOI] [PubMed] [Google Scholar]
- 12.Zemkova H., Yan Z., Liang Z., Jelinkova I., Tomic M., and Stojilkovic S. S. (2007) Role of aromatic and charged ectodomain residues in the P2X(4) receptor functions. J. Neurochem. 102, 1139–1150 [DOI] [PubMed] [Google Scholar]
- 13.Roberts J. A., Valente M., Allsopp R. C., Watt D., and Evans R. J. (2009) Contribution of the region Glu181 to Val200 of the extracellular loop of the human P2X1 receptor to agonist binding and gating revealed using cysteine scanning mutagenesis. J. Neurochem. 109, 1042–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werner P., Seward E. P., Buell G. N., and North R. A. (1996) Domains of P2X receptors involved in desensitization. Proc. Natl. Acad. Sci. U.S.A. 93, 15485–15490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clyne J. D., Wang L. F., and Hume R. I. (2002) Mutational analysis of the conserved cysteines of the rat P2X2 purinoceptor. J. Neurosci. 22, 3873–3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawate T., Michel J. C., Birdsong W. T., and Gouaux E. (2009) Crystal structure of the ATP-gated P2X(4) ion channel in the closed state. Nature 460, 592–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hattori M., and Gouaux E. (2012) Molecular mechanism of ATP binding and ion channel activation in P2X receptors. Nature 485, 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young M. T. (2010) P2X receptors: dawn of the post-structure era. Trends Biochem. Sci. 35, 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Browne L. E., Jiang L. H., and North R. A. (2010) New structure enlivens interest in P2X receptors. Trends Pharmacol. Sci. 31, 229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang R., Taly A., Lemoine D., Martz A., Cunrath O., and Grutter T. (2012) Tightening of the ATP-binding sites induces the opening of P2X receptor channels. EMBO J. 31, 2134–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang R., Lemoine D., Martz A., Taly A., Gonin S., Prado de Carvalho L., Specht A., and Grutter T. (2011) Agonist trapped in ATP-binding sites of the P2X2 receptor. Proc. Natl. Acad. Sci. U.S.A. 108, 9066–9071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jie Y., Zhang L., Xu H., Gao C., Ma W., and Li Z. (2014) Involvement of the left-flipper-to-dorsal-fin interface of the zebrafish P2X4 receptor in ATP binding and structural rearrangement. Neurosci. Lett. 582, 1–5 [DOI] [PubMed] [Google Scholar]
- 23.Jiang R., Taly A., and Grutter T. (2013) Moving through the gate in ATP-activated P2X receptors. Trends Biochem. Sci. 38, 20–29 [DOI] [PubMed] [Google Scholar]
- 24.Lörinczi É., Bhargava Y., Marino S. F., Taly A., Kaczmarek-Hájek K., Barrantes-Freer A., Dutertre S., Grutter T., Rettinger J., and Nicke A. (2012) Involvement of the cysteine-rich head domain in activation and desensitization of the P2X1 receptor. Proc. Natl. Acad. Sci. U.S.A. 109, 11396–11401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kowalski M., Hausmann R., Dopychai A., Grohmann M., Franke H., Nieber K., Schmalzing G., Illes P., and Riedel T. (2014) Conformational flexibility of the agonist binding jaw of the human P2X3 receptor is a prerequisite for channel opening. Br. J. Pharmacol 171, 5093–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Habermacher C., Martz A., Calimet N., Lemoine D., Peverini L., Specht A., Cecchini M., and Grutter T. (2016) Photo-switchable tweezers illuminate pore-opening motions of an ATP-gated P2X ion channel. Elife 5, e11050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J., and Yu Y. (2016) Insights into the channel gating of P2X receptors from structures, dynamics and small molecules. Acta Pharmacol. Sin. 37, 44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasuya G., Fujiwara Y., Takemoto M., Dohmae N., Nakada-Nakura Y., Ishitani R., Hattori M., and Nureki O. (2016) Structural insights into divalent cation modulations of ATP-gated P2X receptor channels. Cell Rep. 14, 1–13 [DOI] [PubMed] [Google Scholar]
- 29.Gu B. J., Sluyter R., Skarratt K. K., Shemon A. N., Dao-Ung L. P., Fuller S. J., Barden J. A., Clarke A. L., Petrou S., and Wiley J. S. (2004) An Arg307 to Gln polymorphism within the ATP-binding site causes loss of function of the human P2X7 receptor. J. Biol. Chem. 279, 31287–31295 [DOI] [PubMed] [Google Scholar]
- 30.Dibb K. M., Rose T., Makary S. Y., Claydon T. W., Enkvetchakul D., Leach R., Nichols C. G., and Boyett M. R. (2003) Molecular basis of ion selectivity, block, and rectification of the inward rectifier Kir3.1/Kir3.4 K(+) channel. J. Biol. Chem. 278, 49537–49548 [DOI] [PubMed] [Google Scholar]
- 31.Venkatachalan S. P., and Czajkowski C. (2008) A conserved salt bridge critical for GABA(A) receptor function and loop C dynamics. Proc. Natl. Acad. Sci. U.S.A. 105, 13604–13609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang R., Martz A., Gonin S., Taly A., de Carvalho L. P., and Grutter T. (2010) A putative extracellular salt bridge at the subunit interface contributes to the ion channel function of the ATP-gated P2X2 receptor. J. Biol. Chem. 285, 15805–15815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laha K. T., and Wagner D. A. (2011) A state-dependent salt bridge interaction exists across the β/α intersubunit interface of the GABAA receptor. Mol. Pharmacol. 79, 662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y., Yu Y., Cheng J., Liu Y., Liu D. S., Wang J., Zhu M. X., Wang R., and Xu T. L. (2012) Highly conserved salt bridge stabilizes rigid signal patch at extracellular loop critical for surface expression of acid-sensing ion channels. J. Biol. Chem. 287, 14443–14455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hausmann R., Günther J., Kless A., Kuhlmann D., Kassack M. U., Bahrenberg G., Markwardt F., and Schmalzing G. (2013) Salt bridge switching from Arg290/Glu167 to Arg290/ATP promotes the closed-to-open transition of the P2X2 receptor. Mol. Pharmacol. 83, 73–84 [DOI] [PubMed] [Google Scholar]
- 36.Lam S. Y., Yeung R. C., Yu T. H., Sze K. H., and Wong K. B. (2011) A rigidifying salt bridge favors the activity of thermophilic enzyme at high temperatures at the expense of low-temperature activity. PLoS Biol. 9, e1001027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sackin H., Nanazashvili M., Li H., Palmer L. G., and Walters D. E. (2009) An intersubunit salt bridge near the selectivity filter stabilizes the active state of Kir1.1. Biophys. J. 97, 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koban F., El-Kasaby A., Häusler C., Stockner T., Simbrunner B. M., Sitte H. H., Freissmuth M., and Sucic S. (2015) A salt bridge linking the first intracellular loop with the C terminus facilitates the folding of the serotonin transporter. J. Biol. Chem. 290, 13263–13278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao W. S., Wang J., Ma X. J., Yang Y., Liu Y., Huang L. D., Fan Y. Z., Cheng X. Y., Chen H. Z., Wang R., and Yu Y. (2014) Relative motions between left flipper and dorsal fin domains favour P2X4 receptor activation. Nat. Commun. 5, 4189. [DOI] [PubMed] [Google Scholar]
- 40.Laskowski R. A., MacArthur M. W., Moss D. S., and Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 41.Shaw D. E. (2005) A fast, scalable method for the parallel evaluation of distance-limited pairwise particle interactions. J. Comput. Chem. 26, 1318–1328 [DOI] [PubMed] [Google Scholar]
- 42.Huang L. D., Fan Y. Z., Tian Y., Yang Y., Liu Y., Wang J., Zhao W. S., Zhou W. C., Cheng X. Y., Cao P., Lu X. Y., and Yu Y. (2014) Inherent dynamics of head domain correlates with ATP-recognition of P2X4 receptors: insights gained from molecular simulations. PLoS ONE 9, e97528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang L. H., Rassendren F., Surprenant A., and North R. A. (2000) Identification of amino acid residues contributing to the ATP-binding site of a purinergic P2X receptor. J. Biol. Chem. 275, 34190–34196 [DOI] [PubMed] [Google Scholar]
- 44.Ennion S., Hagan S., and Evans R. J. (2000) The role of positively charged amino acids in ATP recognition by human P2X1 receptors. J. Biol. Chem. 275, 35656. [PubMed] [Google Scholar]
- 45.Musafia B., Buchner V., and Arad D. (1995) Complex salt bridges in proteins: statistical analysis of structure and function. J. Mol. Biol. 254, 761–770 [DOI] [PubMed] [Google Scholar]
- 46.Makhatadze G. I., Loladze V. V., Ermolenko D. N., Chen X., and Thomas S. T. (2003) Contribution of surface salt bridges to protein stability: guidelines for protein engineering. J. Mol. Biol. 327, 1135–1148 [DOI] [PubMed] [Google Scholar]
- 47.Waldburger C. D., Schildbach J. F., and Sauer R. T. (1995) Are buried salt bridges important for protein stability and conformational specificity? Nat. Struct. Biol. 2, 122–128 [DOI] [PubMed] [Google Scholar]
- 48.Takano K., Tsuchimori K., Yamagata Y., and Yutani K. (2000) Contribution of salt bridges near the surface of a protein to the conformational stability. Biochemistry 39, 12375–12381 [DOI] [PubMed] [Google Scholar]
- 49.Honore P., Donnelly-Roberts D., Namovic M. T., Hsieh G., Zhu C. Z., Mikusa J. P., Hernandez G., Zhong C., Gauvin D. M., Chandran P., Harris R., Medrano A. P., Carroll W., Marsh K., Sullivan J. P., et al. (2006) A-740003 [N-(1-{[(Cyanoimino)(5-quinolinylamino) methyl]amino}-2,2-dimethylpropyl)-2-(3,4-dimethoxyphenyl)acetamide], a novel and selective P2X7 receptor antagonist, dose-dependently reduces neuropathic pain in the rat. J. Pharmacol. Exp. Ther. 319, 1376–1385 [DOI] [PubMed] [Google Scholar]
- 50.Keystone E. C., Wang M. M., Layton M., Hollis S., McInnes I. B., and D1520C00001 Study Team. (2012) Clinical evaluation of the efficacy of the P2X7 purinergic receptor antagonist AZD9056 on the signs and symptoms of rheumatoid arthritis in patients with active disease despite treatment with methotrexate or sulphasalazine. Ann. Rheum. Dis. 71, 1630–1635 [DOI] [PubMed] [Google Scholar]
- 51.Abdulqawi R., Dockry R., Holt K., Layton G., McCarthy B. G., Ford A. P., and Smith J. A. (2015) P2X3 receptor antagonist (AF-219) in refractory chronic cough: a randomised, double-blind, placebo-controlled phase 2 study. Lancet 385, 1198–1205 [DOI] [PubMed] [Google Scholar]
- 52.Turner R. D., Rajakulasingam R. K., Bhowmik A., and Bothamley G. H. (2015) P2X3 receptor antagonist in chronic cough. Lancet 386, 244. [DOI] [PubMed] [Google Scholar]