Despite widespread vernalization responsiveness in the grass subfamily Pooideae, the flowering repressor VERNALIZATION2 evolved more recently in core members of this subfamily.
Abstract
Flowering of many plant species is coordinated with seasonal environmental cues such as temperature and photoperiod. Vernalization provides competence to flower after prolonged cold exposure, and a vernalization requirement prevents flowering from occurring prior to winter. In winter wheat (Triticum aestivum) and barley (Hordeum vulgare), three genes VRN1, VRN2, and FT form a regulatory loop that regulates the initiation of flowering. Prior to cold exposure, VRN2 represses FT. During cold, VRN1 expression increases, resulting in the repression of VRN2, which in turn allows activation of FT during long days to induce flowering. Here, we test whether the circuitry of this regulatory loop is conserved across Pooideae, consistent with their niche transition from the tropics to the temperate zone. Our phylogenetic analyses of VRN2-like genes reveal a duplication event occurred before the diversification of the grasses that gave rise to a CO9 and VRN2/Ghd7 clade and support orthology between wheat/barley VRN2 and rice (Oryza sativa) Ghd7. Our Brachypodium distachyon VRN1 and VRN2 knockdown and overexpression experiments demonstrate functional conservation of grass VRN1 and VRN2 in the promotion and repression of flowering, respectively. However, expression analyses in a range of pooids demonstrate that the cold repression of VRN2 is unique to core Pooideae such as wheat and barley. Furthermore, VRN1 knockdown in B. distachyon demonstrates that the VRN1-mediated suppression of VRN2 is not conserved. Thus, the VRN1-VRN2 feature of the regulatory loop appears to have evolved late in the diversification of temperate grasses.
The initiation of flowering is a major developmental transition in the plant life cycle. When flowering initiates, shoot apical meristems shift from forming vegetative organs such as leaves to forming flowers. In many plant species, flowering occurs at a particular time of year in response to the sensing of seasonal cues such as changes in day length and temperature. In some plants adapted to temperate climates, exposure to the prolonged cold of winter (vernalization) results in the ability to flower in the next growing season (Chouard, 1960; Amasino, 2010). Although vernalization ultimately enables flowering, vernalization responsiveness is typically an adaptation to ensure that flowering does not occur prematurely in the fall season. This has obvious adaptive value; for example, many vernalization-responsive plants become established in the fall season (during which flowering would not lead to successful reproduction) and then rapidly flower in the spring when conditions for reproduction and seed maturation are optimal.
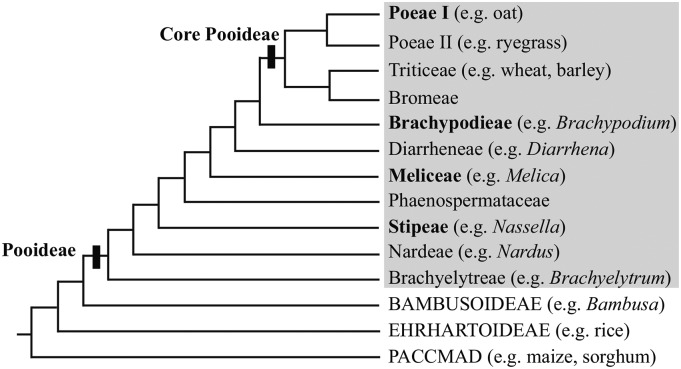
The grass family (Poaceae) originated approximately 70 million years ago as part of the tropical forest understory. However, grasses have since diversified across the globe occupying a variety of ecological niches (Kellogg, 2001). Exemplifying this, the ∼3,800 species of grass subfamily Pooideae, including the economically important cereals wheat (Triticum aestivum, Triticeae), barley (Hordeum vulgare, Triticeae), oat (Avena sativa, Poeae), and rye (Lolium perenne, Poeae), have adapted to cool climates of both northern and southern hemispheres (Hartley, 1973; Grass Phylogeny Working Group, 2001; Edwards and Smith, 2010). Phylogenetic analyses indicate the above-mentioned species within Triticeae and Poeae are a closely related group often referred to as crown or core pooid grasses (Schneider et al., 2009; Grass Phylogeny Working Group II, 2012). The remaining noncore pooids are in tribes consecutively sister to the core pooids, including Brachypodieae that contains the emerging plant model Brachypodium distachyon (Meliceae and Stipeae; Brkljacic et al., 2011; Grass Phylogeny Working Group II, 2012; Fig. 1).
Figure 1.
Pooideae phylogeny showing the eleven major tribes (gray box) and delimitation of the core pooids based on Schneider et al. (2009) and Grass Phylogeny Working Group II (2012). Outgroups are the closely related grass subfamilies, Bambusoideae and Ehrhartoideae, which together with Pooideae form the BEP clade. Sister to the BEP clade is the PACCMAD clade that contains tropical cereals such as maize and sorghum (Sorghum bicolor). Focal tribes in this study are highlighted in bold.
It is hypothesized that vernalization responsiveness evolved early during the diversification of Pooideae, as a key adaptation allowing for their transition into the temperate zone (Preston and Sandve, 2013; Fjellheim et al., 2014). Within core Pooideae, many species have been characterized as vernalization responsive (Heide, 1994; Grass Phylogeny Working Group, 2001; Grass Phylogeny Working Group II, 2012). However, it is unclear how widespread vernalization responsiveness is outside core pooids and whether pooids with this trait share a conserved ancestral vernalization pathway. To explore the extent to which the vernalization pathway is conserved in Pooideae, we characterized the expression and function of vernalization pathway homologs in B. distachyon and other pooids.
The current molecular model of vernalization responsiveness in wheat and barley involves a leaf-specific regulatory loop among VERNALIZATION1 (VRN1), VRN2, and VRN3 (Dennis and Peacock, 2009; Distelfeld et al., 2009a; Greenup et al., 2009; Sasani et al., 2009), the latter of which is homologous to Arabidopsis (Arabidopsis thaliana) FT, which encodes a small protein that moves from leaves to the shoot apical meristem to promote flowering (Yan et al., 2006; Turck et al., 2008). During growth of vernalization-requiring cereals in the fall season, the CONSTANS-like gene VRN2 represses FT to prevent flowering, and the FRUITFULL-like gene VRN1 is transcribed at very low levels (Yan et al., 2004a, 2006; Hemming et al., 2008; Sasani et al., 2009). During winter, VRN1 transcript levels increase, causing the repression of VRN2 and the derepression of VRN3/FT (Yan et al., 2004b; Trevaskis et al., 2006; Sasani et al., 2009). Although vernalization alleviates FT repression, FT also requires long days to become activated; thus, flowering only occurs during the lengthening days of spring and summer (Yan et al., 2006; Hemming et al., 2008; Sasani et al., 2009). In wheat and barley, VRN2 is necessary for the vernalization requirement because deletions of the entire locus or point mutations in the CCT domain result in spring varieties, which do not require vernalization (Yan et al., 2004; Dubcovsky et al., 2005; Karsai et al., 2005; von Zitzewitz et al., 2005; Distelfeld et al., 2009b).
In wheat, there is a negative correlation between VRN1 and VRN2 expression in leaves. VRN1 levels increase during cold and remain elevated following cold (Trevaskis et al., 2003; Yan et al., 2003; Sasani et al., 2009), and this correlates with the stable reduction of VRN2 during and after cold exposure (Yan et al., 2004). Recently, it was shown that VRN1 binds to the VRN2 promoter and thus directly regulates VRN2 expression (Deng et al., 2015). Furthermore, mutations in the wheat VRN1 locus result in elevated VRN2 expression and delayed flowering (Chen and Dubcovsky, 2012). The delayed flowering phenotype in the vrn1 mutants is largely due to the presence of VRN2 because wheat vrn1 vrn2 double mutants flower significantly earlier than vrn1 single mutants (Chen and Dubcovsky, 2012).
Expression of VRN1 in the noncore pooid B. distachyon is consistent with it being conserved as floral promoter involved in vernalization (Ream et al., 2014; for review on flowering in B. distachyon, see Woods and Amasino 2015). As in wheat and barley, B. distachyon VRN1 (BdVRN1) mRNA levels increase quantitatively during increasing durations of cold exposure and remain elevated post cold (Ream et al., 2014; Woods et al., 2014). Furthermore, overexpression of BdVRN1 results in rapid flowering and is correlated with elevated BdFT and reduced BdVRN2 expression (Ream et al., 2014). However, contrary to VRN2 behavior in core pooids, BdVRN2 mRNA levels increase rather than decrease during cold, despite a simultaneous increase in BdVRN1 expression (Ream et al., 2014). Moreover, after cold exposure, BdVRN2 expression levels return to prevernalization expression levels. Lastly, rapid-flowering accessions of B. distachyon, which have elevated BdVRN1 and BdFT mRNA levels without cold exposure, do not have correspondingly lower levels of BdVRN2 compared to delayed-flowering accessions (Ream et al., 2014). Thus, the BdVRN2 expression patterns are not consistent with BdVRN2 acting as a floral repressor that is down regulated by vernalization through BdVRN1 (Ream et al., 2014).
Here, we conduct extensive phylogenetic analyses that infer a gene duplication event occurred before the divergence of grasses, giving rise to a CONSTANS9 (CO9) and a VRN2/Ghd7 clade. Analyses across representative pooids suggest that VRN2/GhD7-like gene expression is only repressed by vernalization in core pooids, including oats. Furthermore, although functional data in B. distachyon demonstrates that BdVRN2 is indeed a conserved repressor of flowering, BdVRN1 does not negatively regulate the expression of BdVRN2. Thus, the incorporation of vernalization-mediated repression of VRN2 as part of the vernalization system was likely to have occurred after the divergence of Brachypodieae and core pooids.
RESULTS
Phylogenetic Analyses of VRN2-Like Genes Suggest a Duplication Event before the Base of Grasses
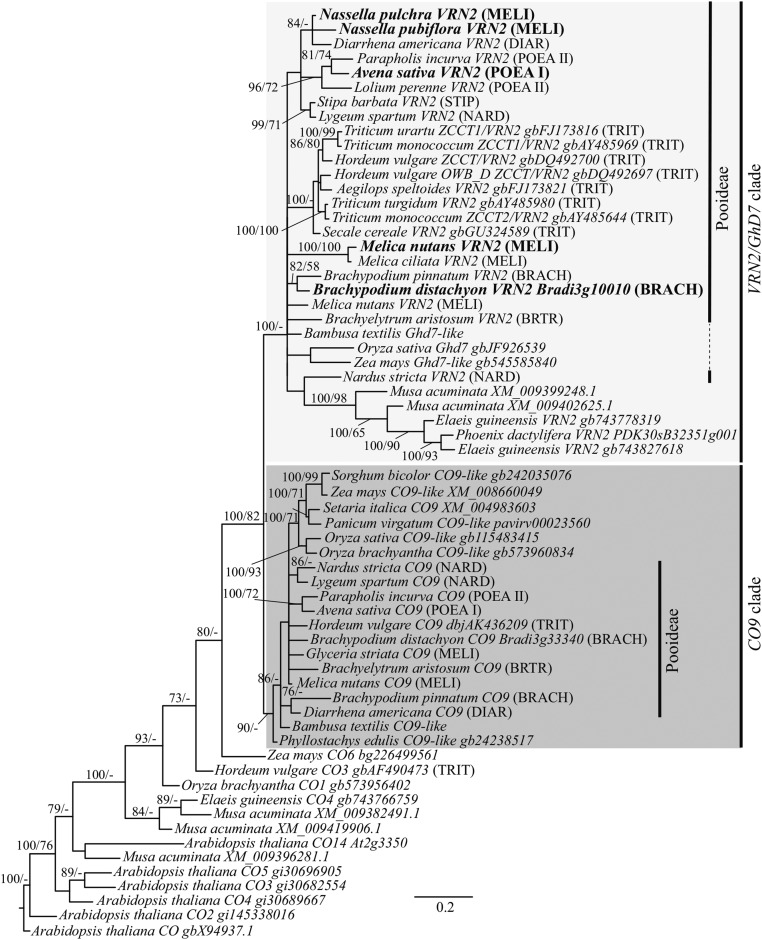
Genes from representatives within the cereal grass subfamily Pooideae, in addition to representatives from other grass subfamilies, including Bambusoideae, Panicoideae, and Ehrhartoideae, were included in our phylogenetic analyses of VRN2-like genes. Banana (Musa acuminata, Musaceae), date palm (Phoenix dactylifera, Arecaceae), and oil palm (Elaeis guineensis, Arecaceae) genes were also included to sample monocots outside of the grass family. Bayesian and maximum likelihood analyses on an alignment of the highly conserved CCT and 3′ coding domain of VRN2-like genes infer two major clades containing cereal VRN2- and CO9-like genes, respectively (Fig. 2). The best tree topology supports the inclusion of rice (Oryza sativa) Grain number, plant height, and heading date7 (Ghd7) and sequences from banana, date palm, and oil palm within the VRN2 clade (100% posterior probability), which is sister to the supported CO9 clade (90% posterior probability). The Shimodaira-Hasegawa topology test also supports the position of rice Ghd7 within the VRN2 clade (−ln L of 3,554) as the most likely topology; the topologies of Ghd7 outside of VRN2 and CO9, and CO9 sister to Ghd7 were equally less likely (–ln L of 3,562). Together, these data support a gene duplication before the diversification of commelinid monocots, giving rise to the VRN2/Ghd7 and CO9 clades. For most Pooideae and Ehrhartoideae (e.g. rice) species sampled, at least two VRN2-like genes were isolated, one falling within the VRN2/Ghd7 clade and the other within the CO9 clade. Interestingly, two sequences from the banana genome (M. acuminata) were supported within the VRN2 clade in addition to oil palm (E. guineensis) and date palm (P. dactylifera). CO9 sequences from the banana and palm genomes were not identified, suggesting a loss of CO9 took place within those lineages. Alternatively, CO9 sequences are present but were not recovered in the protein blast search. Relationships within each clade did not track the species phylogeny closely, although there was little support for tribal-level relationships. All newly sequenced VRN2- and CO9-like genes possessed a conserved CCT domain and a conserved nine-amino acid motif upstream of the stop codon. However, the two Melica nutans and one Melica ciliata (Meliceae) VRN2 clade genes had a frameshift mutation in the 3′ end of the coding region, leading to a premature stop codon seven amino acids upstream of the usual position. Species used for further analyses were Nassella pulchra (Stipeae), Nassella pubiflora (Stipeae), A. sativa (Poeae I), and M. nutans (Meliceae). Members of the Stipeae and Meliceae tribe are the earliest diverging Pooideae representatives included in this study and provided insight into the early diverging lineages of the Pooideae. The Poeae representative (A. sativa) gave an expanded picture of the evolution of cold-mediated VRN2 expression in the core Pooideae. Together, the sampling of different tribes within the Pooideae enabled a diverse look at the evolution of VRN2 across the Pooideae clade.
Figure 2.
Bayesian inference of the phylogenetic relationships among VRN2- and CO9-like genes and rice Ghd7 based on a nucleotide alignment of the conserved CCT domain and 3′ coding region. The presence of rice Ghd7 and M. acuminata (wild ancestor of banana) genes in the VRN2 clade suggests that the gene duplication giving rise to the VRN2/Ghd7 and CO9 clades occurred prior to the diversification of commelinid monocots. Bayesian posterior probabilities (left) and maximum likelihood bootstrap (right) support values above 70% are indicated at each branch; dashes denote lower than 70% where applicable. Scale bar indicates substitutions per site. Focal genes are labeled in large bold font. Abbreviated tribal names are indicated for members of Pooideae: BRTR, Brachyelytreae; NARD, Nardeae; STIP, Stipeae; MELI, Meliceae; DIAR, Diarrheneae; BRACH, Brachypodieae; TRIT, Triticeae; Poea I, Poeae I; and Poea II, Poeae II.
Vernalization Responsiveness Is Widespread in Pooideae
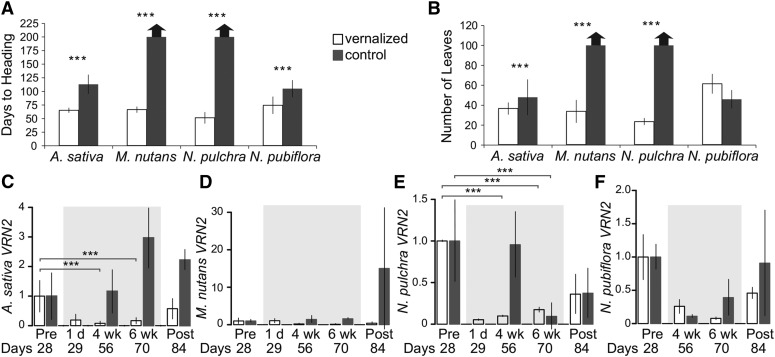
Our data, based on days to heading (after subtraction of six weeks in cold), tiller number at heading, and leaf number at heading, indicate that the core pooid winter oat ‘Norline’ and the noncore pooids M. nutans and N. pulchra (Stipeae) are responsive to vernalization. In contrast, although N. pubiflora headed sooner in cold than those grown without cold exposure with the conservative subtraction of 6 weeks of the time in cold (Fig. 3A), we consider it nonresponsive to vernalization because it had a similar number of leaves (P = 0.068; Fig. 3B) and tillers (P = 0.017) in the cold versus warm treatment. Consistent with previous work, winter oat flowered an average of 44 d later without versus with vernalization (P = 0.025), with an average of six extra tillers (P < 0.001) and 17 extra leaves (P < 0.001; Preston and Kellogg, 2008; Fig. 3, A and B). Under warm conditions, M. nutans and N. pulchra plants failed to flower after 200 d with at least 100 leaves, whereas vernalization resulted in M. nutans flowering after an average of 66 d with 10 tillers and 33 leaves, and N. pulchra flowering after an average of 51 d with seven tillers and 23 leaves (Fig. 3, A and B). Together with previous studies from B. distachyon (Higgins et al., 2010), these results show that vernalization responsiveness is phylogenetically widespread in Pooideae and that there is variation for the presence of this trait in both core and noncore pooids.
Figure 3.
VRN2 regulation in noncore pooids differs from wheat and barley. A, Vernalization causes rapid flowering (days to heading with 6 weeks vernalization subtracted) in A. sativa, M. nutans, N. pulchra, and N. pubiflora relative to control conditions. B, Vernalization decreases leaf number at heading in A. sativa, M. nutans, and N. pulchra, but not N. pubiflora, relative to control conditions. C, As predicted, winter oat VRN2 is negatively regulated as a function of cold exposure. D, M. nutans VRN2 mRNA levels are unaffected by cold and time. E, N. pulchra VRN2 expression is negatively regulated by time in both cold and control conditions. F, N. pubiflora VRN2 expression is not significantly affected by cold or time. Thick arrows in A and B denote nonflowering individuals. Error bars in C to F show standard deviations for three biological replicates. Experimental replicates to control for chamber effects showed similar results. Asterisks above bars indicate statistically significant contrasts (*P < 0.05, **P < 0.01, and ***P < 0.001).
The VRN2 Expression Pattern during and after Cold Is Different in Core and Noncore Pooids
As predicted based on the model in wheat and barley, VRN2-like gene expression in the core pooid winter oat (P = 0.006; Fig. 3C) and the noncore pooid N. pulchra (P < 0.001; Fig. 3E) showed a significant time point by growth temperature interaction. Furthermore, pairwise contrasts between pretreatment (28 d) and 56 or 70 d time points indicate a significant down-regulation of VRN2 transcription in winter oat (P = 0.001) and N. pulchra (P = 0.001) with versus without cold (Fig. 3, C and E). However, although expression of VRN2 in N. pulchra dropped after 4 and 6 weeks of cold exposure, a similar decrease in expression was also observed in the warm treatment at the 70 d time point (P = 0.017). This suggests that regulation of VRN2 differs between N. pulchra and winter wheat, barley, and oat.
Contrary to predictions, no significant time point by treatment interaction was found for the noncore pooid M. nutans VRN2 expression (P = 0.558; Fig. 3D). Indeed, for this species, VRN2 expression was similar across both treatments and across all available time points. In the case of the vernalization nonresponsive noncore pooid N. pubiflora, we did not expect an effect of time point, treatment, or their interaction on VRN2 expression; indeed, there was no significant effect of time point by treatment (P = 0.312). However, there was a significant effect of time point (P = 0.002), with a decrease in expression from 28 to 56 d, followed by an increase from 70 to 84 d in both vernalized and control treatments (Fig. 3F).
Reduction of BdVRN2 Expression Results in Rapid Flowering and Elevated Expression of BdFT and BdVRN1
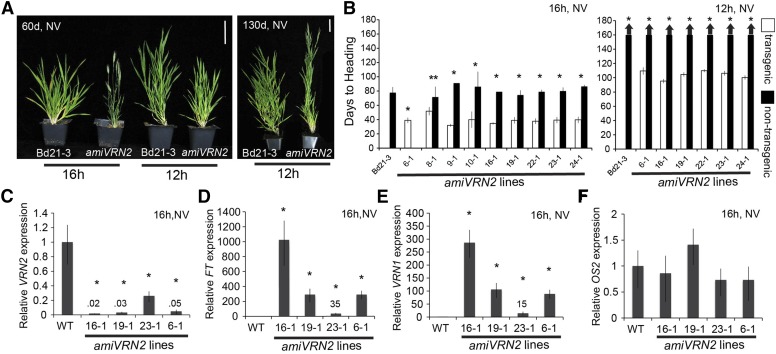
To investigate BdVRN2’s role in flowering time, we transformed the vernalization-responsive B. distachyon accession Bd21-3 (Ream et al., 2014) with an artificial microRNA (amiVRN2; Warthmann et al., 2008) that posttranscriptionally down-regulates BdVRN2 mRNA levels. Eight independent transgenic lines segregated for the amiVRN2 transgene, and one independent line (Bd6-1) had the transgene fixed (Fig. 4B). Under a 16-h photoperiod without vernalization, wild-type Bd21-3 and segregating nontransgenic control plants flowered between 78 and 90 d with an average of 17 leaves (Fig. 4B; leaf data not shown). In contrast, transgenic plants harboring amiVRN2 flowered between 31 and 52 d and produced an average of five to seven leaves (Fig. 4, A and B; leaf data not shown). This indicates that BdVRN2 acts as a repressor of flowering under inductive 16-h photoperiods.
Figure 4.
VRN2 knockdown causes rapid flowering. A, Representative photos of Bd21-3 wild-type and rapid flowering amiVRN2 knockdown plants grown in a 16- or 12-h photoperiods without vernalization (NV). Pictures were taken 60 and 130 d after germination as indicated. Bar = 5 cm. B, Flowering times of Bd21-3 wild-type and segregating nontransgenic (black bars) compared with independent amiVRN2 transgenic lines (white bars). Lines with no nontransgenic plants are fixed for the transgene. Bars represent the average of 6 plants ± sd. The experiment was repeated with similar results (data not shown). Arrows above bars indicate that none of the plants flowered at the end of the experiment (120 d). C to F, Quantitative RT-PCR expression data from the upper leaf of nonvernalized Bd21-3 wild-type (WT) and amiVRN2 plants at the three-leaf stage grown in a 16-h photoperiod. Average relative BdVRN2 (C), BdFT (D), BdVRN1 (E), and BdOS2 expression (F) is shown for three biological replicates ± sd. Expression analyses were repeated with similar results. Single asterisks indicate P-values < 0.01, and two asterisks indicate P-values < 0.005. Primers for BdVRN1, BdVRN2, and BdFT were previously optimized by Ream et al. (2014), and BdOS2 primers were optimized by Ruelens et al. (2013).
In 12- and 8-h photoperiods, wild-type and segregating nontransgenic control plants failed to flower by 160 d without vernalization (Fig. 4B; 8 h data not shown). However, the flowering of the wild type in 12-h photoperiods (but not in 8- or 10-h photoperiods) could be accelerated with 4 weeks vernalization (Supplemental Fig. S1). Even though amiVRN2 plants flowered more rapidly than the wild type in 12-h photoperiods without vernalization (Fig. 4B), they did not flower as rapidly as 4 week vernalized wild-type plants or lines overexpressing BdVRN1 (BdVRN1 overexpression lines flowered around 40 d; Supplemental Fig. S1). Under noninductive 8-h photoperiods, in which BdVRN2 expression was previously demonstrated to be low in Bd21-3 (Ream et al., 2014), none of the nontransgenic or transgenic plants flowered after 160 d, consistent with previous findings that 8 h is a noninductive photoperiod (Ream et al., 2014; data not shown). This is consistent with VRN2 acting as a repressor of flowering only under inductive long days.
As expected, BdVRN2 expression levels in leaves of four independent amiVRN2 transgenic lines grown in 16-h photoperiods without vernalization were significantly lower (P < 0.01) than expression in wild-type Bd21-3 plants, confirming efficiency of the amiVRN2 transgene (Fig. 4C). Moreover, expression levels of BdFT, BdVRN1, and the paralog of BdVRN1 (BdFUL2) were proportionally significantly elevated in leaves of amiVRN2 transgenic compared with wild-type plants (P < 0.05), consistent with their rapid flowering and indicating a role for BdVRN2 in the repression of BdFT, BdVRN1, and BdFUL2 (Fig. 4, D and E; Supplemental Fig. S2). None of the recently identified FLOWERING LOCUS C-like (FLC-like) genes ODDSOC1 (OS1), OS2, and MADS37 (Ruelens et al., 2013) showed differences in expression in the amiVRN2 lines compared with the wild type (Fig. 4F; Supplemental Fig. S2). In Arabidopsis, FLC is a potent floral repressor turned off by cold conferring a vernalization requirement in Brassicaceae (Amasino, 2010).
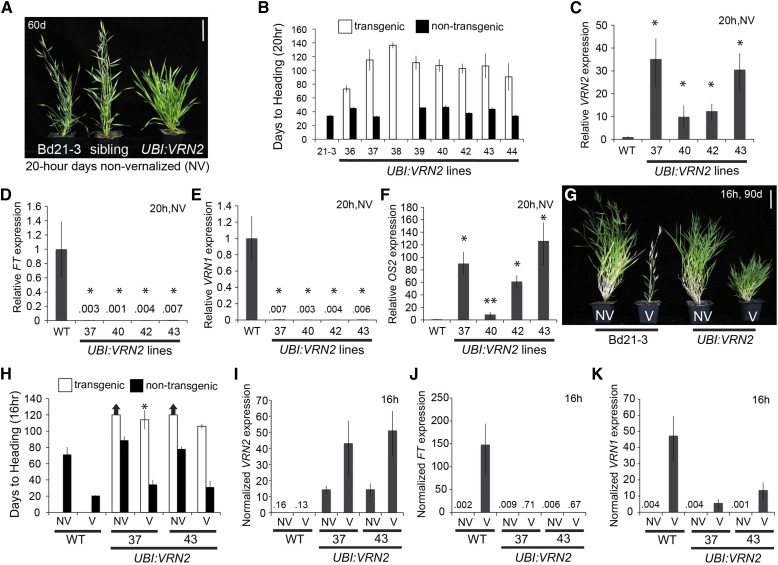
Overexpression of BdVRN2 Delays Flowering, Resulting in Reduced BdFT and BdVRN1 Expression
To further investigate the role of VRN2 as a repressor of flowering, we generated 10 independent transgenic Bd21-3 plants constitutively expressing BdVRN2 under control of the maize (Zea mays) ubiquitin promoter. All of the T0 transgenic plants were delayed in flowering compared to control plants lacking the transgene, when grown in a normally highly inductive 20-h photoperiod without prior vernalization (data not shown). For eight of the transgenic lines that segregated for the transgene in the T1 generation, flowering was significantly delayed by roughly 70 d relative to siblings lacking the transgene and wild-type plants in 20-h photoperiods without prior vernalization (Fig. 5, A and B). Furthermore, vernalized (4 weeks) transgenic plants overexpressing VRN2 flowered roughly 100 d later than vernalized wild-type and nontransgenic sibling plants when grown under inductive 16-h photoperiods (Fig. 5, G and H). Vernalized UBI:VRN2 lines were even more delayed than nonvernalized Bd21-3 plants; however, they did flower within 120 d, whereas nonvernalized UBI:VRN2 lines failed to flower and were larger than vernalized plants (Fig. 5, G and H). Thus, overexpression of BdVRN2 delays flowering and is able to suppress the vernalization response.
Figure 5.
VRN2 overexpression delays flowering. A, Representative photo of Bd21-3 wild-type, nontransgenic sibling, and delayed flowering UBI:VRN2 plants grown in a 20-h photoperiod without vernalization (NV) 60 d postgermination. Bar = 5 cm. B, Days to heading for Bd21-3 wild type (WT) and segregating nontransgenic (black bars) compared with independent UBI:VRN2 transgenic (white bars) plants. Lines with no nontransgenic plants are fixed for the transgene. Bars represent the average of six plants ± sd. The experiment was repeated with similar results (data not shown). C to F, Quantitative RT-PCR expression data from the third leaf of Bd21-3 wild type and UBI:VRN2 plants at the three-leaf stage grown in 20-h photoperiods without vernalization. Average relative BdVRN2 (C), BdFT (D), BdVRN1 (E), and BdOS2 expression (F) is shown for three biological replicates ± sd. Expression analyses were repeated with similar results. G, Representative photo of nonvernalized and 4-week 5°C vernalized (V) Bd21-3, and UBI:VRN2 plants grown for 90 d under a 16-h photoperiod. Bar = 5 cm. H, Days to heading for Bd21-3 wild type and segregating nontransgenic (black bars) compared with independent UBI:VRN2 transgenic (white bars) plants. Bars represent the average of six plants ± sd. Arrows above bars indicate that none of the plants flowered at the end of the experiment (120 d), and asterisks indicate that only some plants in the treatment did not flower after 120 d. I to K, Average relative qRT-PCR data from the upper leaf of Bd21-3 wild type and UBI:VRN2 plants at the three-leaf stage grown in a 16-h photoperiod with and without vernalization. BdVRN2 (I), BdFT (J), and BdVRN1 expression (K) is shown for three biological replicates ± sd.
As expected, leaf BdVRN2 expression levels in four independent UBI:VRN2 transgenic lines grown in 20-h photoperiods were significantly elevated (P < 0.05) compared with wild-type Bd21-3 plants (Fig. 5C). Conversely, BdFT and BdVRN1 expression was significantly lower in leaves of UBI:VRN2 transgenic compared with wild-type plants (P < 0.05), consistent with the delayed-flowering phenotype of UBI:VRN2 plants (Fig. 5, B, D, and E). Interestingly, BdOS2 expression levels were significantly elevated in the UBI:VRN2 lines compared with wild-type plants; however, BdOS1 and BdMADS37 were unaffected by elevated BdVRN2 levels (Fig. 5F; data not shown).
VRN2 expression levels were also significantly elevated in leaves of UBI:VRN2 lines grown in 16-h photoperiods compared with Bd21-3 either with or without vernalization. The newly expanded third leaf was harvested for both the nonvernalized and vernalized samples when the third leaf was reached. Surprisingly, VRN2 expression levels were higher in vernalized UBI:VRN2 lines than nonvernalized UBI:VRN2 lines (Fig. 5I). As was the case in 20-h photoperiods without vernalization, BdFT and BdVRN1 were significantly lower in the UBI:VRN2 lines regardless of vernalization treatment, consistent with the delayed-flowering phenotype of the UBI:VRN2 lines (Fig. 5, J and K). However, BdVRN1 expression levels were still elevated in the UBI:VRN2 vernalized lines compared with nonvernalized UBI:VRN2 lines (Fig. 5K).
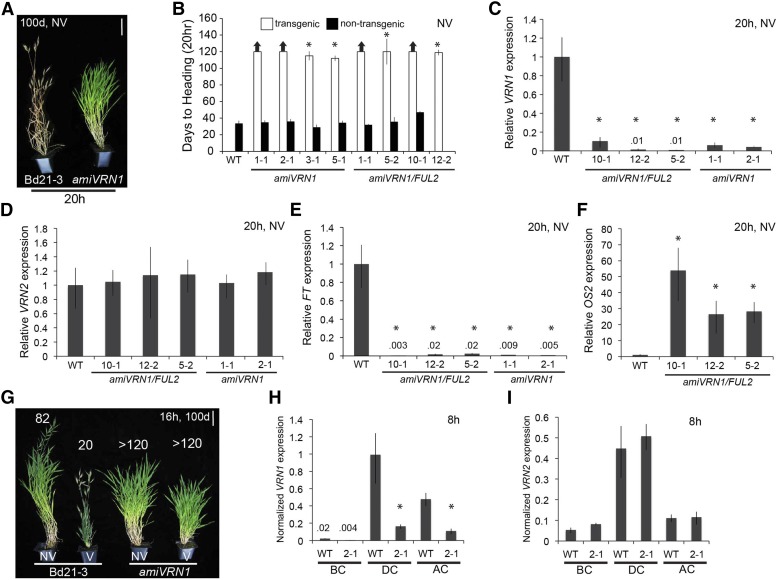
Reduction of BdVRN1 Expression Results in Delayed Flowering but Does Not Affect the Expression of BdVRN2
To evaluate if BdVRN1 and/or BdFUL2 (a paralog of VRN1; Preston and Kellogg, 2006) affect the expression of BdVRN2, and to better define their roles in flowering, we developed artificial microRNAs (amiRNAs) to silence both BdVRN1 and BdFUL2 or BdVRN1 alone (Fig. 6, A and B). Of the eight BdVRN1 and 18 BdVRN1/FUL2 independent T0 transgenic lines, none flowered within 220 d when grown in inductive 20-h photoperiods without vernalization (data not shown). Furthermore, seven segregating and one fixed T1 independent transgenic line flowered more than 100 d later than sibling plants lacking the transgene and wild-type plants with 20-h photoperiods (Fig. 6, A and B). However, there was no significant difference in flowering time between BdVRN1 and BdVRN1/FUL2 plants, suggesting a redundant function for BdVRN1 and BdFUL2 in flowering or that BdFUL2 does not affect flowering. Unlike wild-type plants that flowered after an average of 20 d, amiVRN1 plants did not respond to vernalization and failed to flower within 120 d (Fig. 6G; Supplemental Fig. S3). The nonvernalized amiVRN1 plants also failed to flower after 120 d, whereas nonvernalized wild-type plants flowered after 82 d on average (Fig. 6G; Supplemental Fig. S3).
Figure 6.
VRN1 knockdown delays flowering. A, Representative image of nonvernalized (NV) Bd21-3 wild type and amiVRN1 plants grown in a 20-h photoperiod 100 d postgermination. Bar = 5 cm. B, Days to heading for Bd21-3 wild type (WT) and segregating nontransgenic controls (black bars) compared with independent amiVRN1 transgenic (white bars) plants. Lines with no nontransgenic plants are fixed for the transgene. Bars represent the average of six plants ± sd. The experiment was repeated with similar results. Arrows above bars indicate that none of the plants flowered at the end of the experiment (120 d), and asterisks indicate that only some plants did not flower after 120 d. C to F, Quantitative RT-PCR expression data for the upper leaf of Bd21-3 and amiVRN1 plants grown in a 20-h photoperiod without vernalization. Average relative BdVRN1 (C), BdFT (D), BVRN2 (E), and BdOS2 expression (F) is shown for three biological replicates ± sd. Expression analysis was repeated with similar results. G, Representative photo of Bd21-3 and amiVRN1 plants grown without or with (V) 4 weeks 5°C vernalization with a 16-h photoperiod. Numbers above plants represent average days to heading of six plants per line (for details, see Supplemental Fig. S3). H and I, qRT-PCR expression data for 8-h photoperiod grown Bd21-3 and amiVRN1 plants before cold (BC), during 4 weeks cold (DC), and 7 d after cold (AC). Average relative BdVRN1 (H) and BdVRN2 expression (I) is shown for three biological replicates ± sd.
BdVRN1 and BdFUL2 transcript levels in leaves of three independent amiVRN1/FUL2 and two amiVRN1 only transgenic lines were significantly lower (P < 0.05) than wild-type Bd21-3 plants grown in 20-h photoperiods without vernalization (Fig. 6C; Supplemental Fig. S4C). BdFT expression was also significantly lower in leaves of amiVRN1 or amiVRN1/FUL2 versus wild-type plants consistent with the delayed flowering phenotype of the former (Fig. 6, B and E). Despite the reduction of BdVRN1 in amiVRN1 transgenic plants, the expression levels of BdVRN2 were not significantly different from wild-type plants (Fig. 6D). However, for the FLC-like genes BdOS2, but not BdOS1 and BdMADS37, expression was significantly elevated (P < 0.05) in amiVRN1 plants (Fig. 6F; Supplemental Fig. S4). Recently, VRN1 has been shown to directly bind to the OS2 promoter in barley (Deng et al., 2015), and OS2 expression is reduced in barley lines with highly expressed VRN1; however, vrn1 mutants in barley do not effect OS2 expression before vernalization (Greenup et al., 2010). In contrast with barley, OS2 levels are elevated in B. distachyon with reduced VRN1 mRNA levels but do not change relative to the wild type when VRN1 is up-regulated in amiVRN2 lines (Fig. 4F) or in UBI:VRN1 lines (data not shown). It will be interesting to determine if OS2 contributes to the delayed-flowering phenotype of amiVRN1 by generating vrn1/os2 double mutants.
To corroborate the results from above that indicate that VRN1 does not affect VRN2 expression, we analyzed the expression of both genes in leaves of wild-type and amiVRN1 plants harvested before, during, and after cold (Fig. 6, H and I). As expected, BdVRN1 expression was significantly lower (P < 0.05) in amiVRN1 versus wild-type leaves (Fig. 6H). BdVRN1 expression significantly increased during and after cold in both wild-type and amiVRN1 plants (Fig. 6H). However, despite the significantly lower BdVRN1 expression levels in amiVRN1 lines, BdVRN2 expression was not significantly different from Bd21-3 wild-type plants (Fig. 6I). Similar results were observed in the amiVRN1/FUL2 transgenic lines (data not shown). Thus, reduction of BdVRN1 or BdFUL2 mRNA expression does not affect the expression of BdVRN2 as is the case in wheat, and BdVRN2 does not appear to contribute to the delayed-flowering phenotype of the amiVRN1 or amiVRN1/FUL2 transgenic lines. However, it will be interesting to determine if BdVRN2 contributes to the delayed-flowering amiVRN1 phenotype by generating vrn1 vrn2 double mutants.
DISCUSSION
The attainment of flowering competence in response to vernalization has evolved multiple times independently across major lineages of angiosperms and is hypothesized to be a key adaptation facilitating niche shifts from the tropics to the temperate zone (Ream et al., 2012; Preston and Sandve, 2013). One such niche transition occurred in the grass subfamily Pooideae, members of which are distributed primarily in the northern temperate zone (Hartley, 1973; Edwards and Smith, 2009) and are heavily relied upon for grain, turf, and fodder. To determine the likelihood that the known cereal vernalization gene network was established early in the diversification of pooids, we identified vernalization-responsive species outside core Pooideae and tested whether the vernalization-mediated repression of VRN2 is conserved. Functional analyses in the noncore pooid species B. distachyon strongly support conservation of VRN2 as a repressor of flowering. However, unlike the network in wheat and barley, BdVRN1 does not negatively regulate BdVRN2, and noncore pooid VRN2 genes are not responsive to vernalization. Together, these data support a model in which orthologous VRN2/Ghd7 genes have retained a repressive flowering function during the diversification of pooids, but that co-option of VRN2 into the network of genes regulated during vernalization occurred after the divergence of Brachypodieae and core Pooideae.
Evolutionary History of Pooid VRN2/Ghd7 and CO9 Genes
Two hypotheses about the evolutionary history of VRN2-like genes have been previously proposed (Cockram et al., 2012; Ream et al., 2012). One posits a single gene duplication event before the base of grasses, giving rise to a VRN2/Ghd7- and a CO9-containing clade and implying an orthologous relationship between pooid VRN2 genes and rice Ghd7 (Ream et al., 2012). The other postulates two duplication events before the base of grasses, with the first giving rise to the VRN2 and Ghd7/CO9 clades and the second producing the Ghd7 and CO9 clades, followed by loss of Ghd7 genes at the base of pooids (Cockram et al., 2012). Our phylogenetic results based on VRN2/Ghd7/CO9-like sequences from multiple grass and other monocot species indicate that VRN2 is orthologous to rice Ghd7, supporting a single duplication event before the base of grasses that gave rise to Ghd7/VRN2 and CO9 clade genes.
Conservation of VRN2 as a Flowering Repressor
Functional data from wheat, barley, B. distachyon, and rice, combined with our gene tree topology, strongly support conservation of VRN2/Ghd7-clade genes as repressors of flowering under long days. In winter wheat and barley, loss-of-function mutations in VRN2 are associated with early flowering, and silencing of VRN2 reduces heading time (Yan et al, 2004a; Dubcovsky et al., 2005). Similarly, we found that B. distachyon Bd21-3 amiVRN2 knockdown and BdVRN2-overexpressing lines flower significantly earlier and later than the wild type, respectively. Functional alleles of the rice VRN2 ortholog Ghd7 likewise delay heading date under long days (Xue et al., 2008). Similar to many subtropical grasses, rice is a short-day plant, and the requirement for short days to flower in rice is augmented by the long-day repression conferred by Ghd7 (Xue et al., 2008; Weng et al., 2014). Although little is known about members of the VRN2/Ghd7 sister CO9 clade, barley CO9 has also been shown to prevent precocious flowering, but in this case under noninductive short-day conditions that accompany winter (Kikuchi et al., 2012). Thus, we infer that the ancestor of VRN2/Ghd7 and CO9 clades repressed flowering but that photoperiod regulation of these genes evolved following their duplication.
Co-Option of VRN2 in Core Pooid Vernalization Responsiveness
Previous work demonstrated that BdVRN1 and BdFT in the noncore pooid B. distachyon interact in a positive feedback circuit (Ream et al., 2014), as present in the core pooids wheat and barley (Yan et al., 2006; Shimada et al., 2009; Distelfeld and Dubcovsky, 2010). Although BdVRN2 appears to act as a repressor of flowering, VRN1 amiRNA knockdown lines do not show the predicted increase in VRN2 expression. This indicates that BdVRN2 does not interact with BdVRN1. A second piece of evidence supporting the absence of cold and VRN1-mediated regulation of VRN2 outside core Pooideae comes from gene expression analyses. Rather than decreasing in response to cold, as in winter wheat, barley, and oat (Yan et al., 2004a; Dubcovsky et al., 2005; von Zitzewitz et al., 2005; Distelfeld et al., 2009b) (Fig. 2), VRN2 expression actually increases transiently in B. distachyon Bd21-3 (Ream et al., 2014) or is unaffected by cold in vernalization-responsive M. nutans and N. pulchra. Thus, despite vernalization responsiveness being widespread throughout subfamily Pooideae, cold-regulated VRN2 expression appears to have evolved after the major niche transition of Pooideae from the tropics to the temperate zone. This either suggests that pooid vernalization responsiveness evolved multiple times independently or, more likely, that VRN2 later became subject to VRN1 regulation, possibly allowing further diversification of core pooids into even colder, more seasonal climates, of the temperate north (Edwards and Smith, 2010).
MATERIAL AND METHODS
Plant Growth and Flowering Time Measurements
Seeds of Parapholis incurva, winter oat ‘Norline’, Brachypodium pinnatum, Melica nutans, Melica ciliata, Stipa barbata, Glyceria striata, Nassella pubiflora, Nassella pulchra, Diarrhena americana, Lygeum spartum, Nardus stricta, and Brachyelytrum aristosum were germinated on 1% agar plates for 1 week in the dark, planted in soil, and grown at 20 to 22°C in long days (16 h light:8 h dark) in a greenhouse at the University of Vermont. Bambusa textilis rhizomes were acquired from the U.S. Department of Agriculture (PI 80872) and grown under the same greenhouse conditions. Brachypodium distachyon Bd21-3 wild-type and transgenic plants were grown at the University of Wisconsin, Madison as previously described (Ream et al., 2014).
For the flowering-time experiments, at least 40 germinated seedlings of Avena sativa, M. nutans, N. pulchra, and N. pubiflora were planted in soil and each individual was randomly assigned to one of two growth treatments. Plants in both treatments were initially grown for 28 d at 20°C, followed by 42 d at 4°C (vernalization treatment) or 42 d at 20°C (control treatment). All plants were then given an additional 14 d at 20°C before being transferred to a common 20 to 22°C greenhouse to monitor for flowering time. Experiments were conducted under long-day photoperiods, and treatments were replicated two or three times. Heading time was measured as days from germination to overtopping of the flag leaf by the inflorescence in warm-treated plants. To correct for inhibitory effects of cold on growth in A. sativa, M. nutans, N. pulchra, and N. pubiflora, 6 weeks of cold exposure was not counted in the final heading date. In the case of wild-type and transgenic B. distachyon 21-3, seeds were imbibed with water and for vernalization treatments exposed to 5°C for 4 weeks; note that the time in cold was not included in the final heading date as there is limited to no growth during the cold. Nonvernalization temperatures averaged 21°C during the light period and 18°C the dark period.
Tissue Sampling, RNA Extraction, and cDNA Synthesis
For experiments in A. sativa, M. nutans, N. pulchra, and N. pubiflora, leaves for RNA extraction were collected from the youngest expanded leaf of four individuals without repeated measures at 28 (pretreatment), 29 (cold exposure), 56 (4 weeks with or without vernalization), 70 (6 weeks with or without vernalization), and 84 (posttreatment) d postgermination. RNA was extracted using TRI Reagent (Ambion) followed by DNase treatment with Turbo DNA-free DNase (Ambion) according to the manufacturer’s instructions. cDNA was synthesized using 0.5 μg of RNA in an iScript cDNA synthesis reaction (Bio-Rad). RNA extraction from the upper leaves of B. distachyon wild-type and transgenic plants followed Ream et al. (2014).
Cloning, Sequencing, and Phylogenetic Analysis
VRN2-like genes were amplified from leaf-derived cDNA using degenerate primers based on the CCT domain and 3′ coding region of barley (Hordeum vulgare) ZCCT1, ZCCT2, and CO9; B. distachyon VRN2 and CO9; and rice (Oryza sativa) Ghd7 and CO9 (Supplemental Table S1). Longer VRN2-like sequences were also obtained from a few species using nested gene-specific forward primers in combination with a polyT reverse primer (Supplemental Table S1). Each amplicon was cloned into pGEM-T (Promega), and eight colonies were picked for Sanger sequencing at Beckman Coulter Genomics. Nucleotide sequences were initially aligned with existing VRN2-like genes from GenBank and Phytozome 10.3 using MAFFT (Yan et al., 2003, 2004; von Zitzewitz et al., 2005; Cockram et al., 2007; Pidal et al., 2009; Katoh and Standley, 2013), before manual alignment of amino acid sequences in Mesquite (Maddison and Maddison, 2011; Supplemental Fig. S5). Unalignable regions were pruned from the analysis in order to minimize random noise in the data. A Bayesian analysis was run on the final nucleotide alignment using MrBayes on the CIPRES XSEDE server with 10 million generations, using the GTR+G model as determined by Mr. ModelTest version 2.3 (Ronquist and Huelsenbeck, 2003; Nylander, 2004). Following stationarity, 25% of samples were discarded as burn-in. Maximum likelihood analyses were conducted using RaxML Blackbox on CIPRES XSEDE. The Shimodaira-Hasegawa topology test was done using PAUP4 (Swofford, 2003) on three topologies that were manipulated in Mesquite to differ in the position of rice Ghd7 (sister to the VRN2 clade, sister to the CO9 clade, or sister to both).
Gene Expression Analyses
VRN2 qRT-PCR primers for A. sativa, M. nutans, N. pulchra, and N. pubiflora were designed in Primer3 (Rozen and Skaletsky, 2000) based on results of our phylogenetic analyses (Supplemental Table S1). Primer efficiencies were checked using the dilution series method (Scoville et al., 2011), and amplicons were sequence-verified. Critical threshold values were normalized using the geomean of two reference housekeeping genes, UBIQUITIN5 (UBQ5) and ELONGATION FACTOR 1a (EF1α; Supplemental Table S1) as previously described (Scoville et al., 2011). Three technical replicates were used per biological replicate, and three biological replicates were used per two to three experimental replicates for a total of six to nine individual replicates per time point/treatment. For B. distachyon, qRT-PCR for BdVRN1, BdVRN2, BdFT, and BdUBC18 followed Ream et al. (2014) and for BdOS1, BdOS2, and BdMADS37 followed Ruelens et al. (2013) with three biological replicates and two experimental replicates. BdFUL2 primers were optimized as described previously (Supplemental Table S1).
Statistical Analyses
For A. sativa, M. nutans, N. pulchra, and N. pubiflora, linear mixed effects models were employed in R (v3.1.2; multcomp and nlme packages) to test for the effect of time point, treatment, and their interaction on VRN2 expression. Replicate and time were accounted for as random effects, and data for which there were no a priori predictions (29 d cold shock and 84 d posttreatment time points) were omitted from analyses to reduce heteroscedasticity. Data were subjected to log transformation to increase normality. Pairwise comparisons of expression were done between pretreatment (28 d) and 4 (56 d) or 6 (70 d) weeks of vernalization and between 70 d minus pretreatment expression for vernalization and control treatments. When no time point by treatment interaction was significant, models were simplified by removing the interaction term, and contrasts were done exclusively within time point and treatment. For B. distachyon, differences in heading date and gene expression between wild-type, nontransgenic, and transgenic plants were assessed using the Student’s t test and deemed significant if P < 0.05.
Generation of UBI:VRN2 Transgenic Lines
BdVRN2 cDNAs were amplified from Bd1-1 cDNA pooled from vernalized and nonvernalized leaf tissue. cDNAs were gel extracted (Qiagen) and were cloned into pENTR-D-TOPO (Life Technologies) using the manufacturer’s protocol. Clones were verified by sequencing. pENTR-cDNAs were recombined into pANIC10a (Mann et al., 2012) using Life Technologies LR Clonase II following the manufacturer’s protocol. Clones were verified by sequencing in pANIC10a and then transformed into chemically competent Agrobacterium tumefaciens strain Agl-1. Plant callus transformation was performed as previously described (Vogel and Hill, 2008). Independent transgenic lines were genotyped for the transgene using a cDNA specific forward and pANIC vector AcV5 tag reverse primer (Supplemental Table S1). Primer pairs used to clone each cDNA are listed in Supplemental Table S1. The UBI:VRN1 lines were previously published by Ream et al. (2014).
Generation of amiVRN2 and amiVRN1 Transgenic Lines
amiRNA sequences targeting either BdVRN2 or BdVRN1 transcripts were designed using the amiRNA designer tool at wmd3.weigelworld.org based on MIR528 from rice in the pNW55 vector. Parameters were set such that amiRNAs would be specific to the desired target gene, except for DW4 that was designed to target both BdVRN1 and BdFUL2. To increase the chances of obtaining successful knockdown of BdVRN1 and BdVRN2 expression, three amiRNAs were developed targeting the 5′, middle, and 3′ ends of the coding region. amiRNAs targeting the 3′ end of both BdVRN1 and BdVRN2 were the most efficient for knocking down expression; thus, lines with these constructs were used in all the experiments. Gateway-compatible amiVRN1 and amiVRN2 PCR products were recombined into pDONR221 using Life Technologies BP Clonase II following the manufacturer’s protocol. Clones were verified by sequencing. The pDONR221 vector containing the desired amiRNA in combination with another vector containing the maize ubiquitin promoter were both recombined into destination vector p24GWI (designed by Devin O’Connor at the Plant Gene Expression Center, Albany, CA) using Life Technologies LR clonase II plus following the manufacturer’s protocol. Clones were verified by sequencing to ensure that the maize ubiquitin promoter was upstream from the developed amiRNA in order for the amiRNA to be continually expressed. The generated constructs were transformed into A. tumefaciens strain Agl-1. Plant callus transformation was performed as previously described (Vogel and Hill, 2008). Independent transgenic lines were genotyped for the transgene using an amiRNA forward primer specific for the targeted transcript and a reverse primer derived from the pNW55 backbone sequence (Supplemental Table S1). Primers used to generate the amiRNAs are listed in Supplemental Table S1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers KT354940 to KT354963.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Days to heading of Bd21-3 and OX:VRN1 lines grown in 20,16,15,14,12,10, and 8 hour days.
Supplemental Figure S2. BdOS1,BdMADS37 and BdFUL2 gene expression in Bd21-3 and amiVRN2.
Supplemental Figure S3. Days to heading of Bd21-3 and amiVRN1 plants non-vernalized and vernalized.
Supplemental Figure S4. BdOS1 and BdMADS37 gene expression in amiVRN1 and OXVRN1 and FUL2 expression in amiVRN1/FUL2 lines.
Supplemental Figure S5. Alignment of VRN2/Ghd7/CO9 genes and outgroups used for phylogenetic analysis.
Supplementary Material
Acknowledgments
We thank Devin O’Connor for kindly providing and allowing use of unpublished amiRNA constructs that he developed while a graduate student in Sarah Hake’s lab. The Amasino lab thanks Thomas Ream for useful discussions and the cloning of the VRN2 cDNA for overexpression analyses, and Jill Mahoy and Heidi Kaeppler for performing B. distachyon transformations. We also thank two anonymous reviewers of this article for their valuable comments.
Glossary
- amiRNA
artificial microRNA
Footnotes
J.C.P. was supported by USDA-HATCH and by the National Science Foundation (IOS-1353056). R.M.A. was supported by the National Science Foundation (Grant IOS-1258126) and by the Great Lakes Bioenergy Research Center (Department of Energy Biological and Environmental Research Office of Science Grant DE-FCO2-07ER64494). D.P.W. was supported in part by a National Institutes of Health-sponsored predoctoral training fellowship to the University of Wisconsin Genetics Training Program. Y.D. was funded by the China Scholarship Council.
Articles can be viewed without a subscription.
References
- Amasino R. (2010) Seasonal and developmental timing of flowering. Plant J 61: 1001–1013 [DOI] [PubMed] [Google Scholar]
- Brkljacic J, Grotewold E, Scholl R, Mockler T, Garvin DF, Vain P, Brutnell T, Sibout R, Bevan M, Budak H, et al. (2011) Brachypodium as a model for the grasses: today and the future. Plant Physiol 157: 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Dubcovsky J (2012) Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genet 8: e1003134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouard P. (1960) Vernalization and its relations to dormancy. Annu Rev Plant Physiol 11: 191–238 [Google Scholar]
- Cockram J, Mackay IJ, O’Sullivan DM (2007) The role of double-stranded break repair in the creation of phenotypic diversity at cereal VRN1 loci. Genetics 177: 2535–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockram J, Thiel T, Steuernagel B, Stein N, Taudien S, Bailey PC, O’Sullivan DM (2012) Genome dynamics explain the evolution of flowering time CCT domain gene families in the Poaceae. PLoS One 7: e45307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Casao MC, Wang P, Sato K, Hayes PM, Finnegan EJ, Trevaskis B (2015) Direct links between the vernalization response and other key traits of cereal crops. Nat Commun 6: 5882. [DOI] [PubMed] [Google Scholar]
- Dennis ES, Peacock WJ (2009) Vernalization in cereals. J Biol 8: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A, Dubcovsky J (2010) Characterization of the maintained vegetative phase deletions from diploid wheat and their effect on VRN2 and FT transcript levels. Mol Genet Genomics 283: 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A, Li C, Dubcovsky J (2009a) Regulation of flowering in temperate cereals. Curr Opin Plant Biol 12: 178–184 [DOI] [PubMed] [Google Scholar]
- Distelfeld A, Tranquilli G, Li C, Yan L, Dubcovsky J (2009b) Genetic and molecular characterization of the VRN2 loci in tetraploid wheat. Plant Physiol 149: 245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Chen C, Yan L (2005) Molecular characterization of the allelic variation at the VRN-H2 vernalization locus in barley. Mol Breed 15: 395–407 [Google Scholar]
- Edwards EJ, Smith SA (2010) Phylogenetic analyses reveal the shady history of C4 grasses. Proc Natl Acad Sci USA 107: 2532–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjellheim S, Boden S, Trevaskis B (2014) The role of seasonal flowering responses in adaptation of grasses to temperate climates. Front Plant Sci 5: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group (2001) Phylogeny and subfamilial classification of the grasses (Poaceae). Ann Mo Bot Gard 88: 373–457 [Google Scholar]
- Grass Phylogeny Working Group II (2012) New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytol 193: 304–312 [DOI] [PubMed] [Google Scholar]
- Greenup A, Peacock WJ, Dennis ES, Trevaskis B (2009) The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Ann Bot (Lond) 103: 1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenup AG, Sasani S, Oliver SN, Talbot MJ, Dennis ES, Hemming MN, Trevaskis B (2010) ODDSOC2 is a MADS box floral repressor that is down-regulated by vernalization in temperate cereals. Plant Physiol 153: 1062–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley W. (1973) Studies on origin, evolution, and distribution of Gramineae. V. Subfamily Festucoideae. Aust J Bot 21: 201–234 [Google Scholar]
- Heide OM. (1994) Control of flowering and reproduction in temperate grasses. New Phytol 128: 347–362 [DOI] [PubMed] [Google Scholar]
- Hemming MN, Peacock WJ, Dennis ES, Trevaskis B (2008) Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiol 147: 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JA, Bailey PC, Laurie DA (2010) Comparative Genomics of Flowering Time Pathways Using Brachypodium distachyon as a Model for the Temperate Grasses. PLoS ONE 5: e10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsai I, Szűcs P, Mészáros K, Filichkina T, Hayes PM, Skinner JS, Láng L, Bedő Z (2005) The Vrn-H2 locus is a major determinant of flowering time in a facultative × winter growth habit barley (Hordeum vulgare L.) mapping population. Theor Appl Genet 110: 1458–1466 [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg EA. (2001) Evolutionary history of the grasses. Plant Physiol 125: 1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi R, Kawahigashi H, Oshima M, Ando T, Handa H (2012) The differential expression of HvCO9, a member of the CONSTANS-like gene family, contributes to the control of flowering under short-day conditions in barley. J Exp Bot 63: 773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR (2011) Mesquite: a modular system for evolutionary analysis, Version 3.03. http://mesquiteproject.org
- Mann DGJ, Lafayette PR, Abercrombie LL, King ZR, Mazarei M, Halter MC, Poovaiah CR, Baxter H, Shen H, Dixon RA, Parrott WA, Neal Stewart C Jr (2012) Gateway-compatible vectors for high-throughput gene functional analysis in switchgrass (Panicum virgatum L.) and other monocot species. Plant Biotechnol J 10: 226–236 [DOI] [PubMed] [Google Scholar]
- Nylander JAA. (2004) MrModeltest v2 (program distributed by the author). Evolutionary Biology Centre, Uppsala University, Sweden [Google Scholar]
- Pidal B, Yan L, Fu D, Zhang F, Tranquilli G, Dubcovsky J (2009) The CArG-box located upstream from the transcriptional start of wheat vernalization gene VRN1 is not necessary for the vernalization response. J Hered 100: 355–364 [DOI] [PubMed] [Google Scholar]
- Preston JC, Kellogg EA (2008) Discrete developmental roles for temperate cereal grass VERNALIZATION1/FRUITFULL-like genes in flowering competency and the transition to flowering. Plant Physiol 146: 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston JC, Kellogg EA (2006) Reconstructing the evolutionary history of paralagous APETALA1/FRUITFULL-like genes in grasses. Genetics 174: 421–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston JC, Sandve SR (2013) Adaptation to seasonality and the winter freeze. Front Plant Sci 4: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ream TS, Woods DP, Amasino RM (2012) The molecular basis of vernalization in different plant groups. Cold Spring Harb Symp Quant Biol 77: 105–115 [DOI] [PubMed] [Google Scholar]
- Ream TS, Woods DP, Schwartz CJ, Sanabria CP, Mahoy JA, Walters EM, Kaeppler HF, Amasino RM (2014) Interaction of photoperiod and vernalization determines flowering time of Brachypodium distachyon. Plant Physiol 164: 694–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Ruelens P, de Maagd RA, Proost S, Theißen G, Geuten K, Kaufmann K (2013) FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat Commun 4: 2280. [DOI] [PubMed] [Google Scholar]
- Sasani S, Hemming MN, Oliver SN, Greenup A, Tavakkol-Afshari R, Mahfoozi S, Poustini K, Sharifi HR, Dennis ES, Peacock WJ, Trevaskis B (2009) The influence of vernalization and daylength on expression of flowering-time genes in the shoot apex and leaves of barley (Hordeum vulgare). J Exp Bot 60: 2169–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J, Döring E, Hilu KW, Röser M (2009) Phylogenetic structure of the grass subfamily Pooideae based on comparison of plastid matK gene-3′trnK exon and nuclear ITS sequences. Taxon 58: 405–424 [Google Scholar]
- Scoville AG, Barnett LL, Bodbyl-Roels S, Kelly JK, Hileman LC (2011) Differential regulation of a MYB transcription factor is correlated with transgenerational epigenetic inheritance of trichome density in Mimulus guttatus. New Phytol 191: 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada S, Ogawa T, Kitagawa S, Suzuki T, Ikari C, Shitsukawa N, Abe T, Kawahigashi H, Kikuchi R, Handa H, Murai K (2009) A genetic network of flowering-time genes in wheat leaves, in which an APETALA1/FRUITFULL-like gene, VRN1, is upstream of FLOWERING LOCUS T. Plant J 58: 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. (2003) PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sinauer Associates, Sunderland, MA [Google Scholar]
- Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES (2003) MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci USA 100: 13099–13104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Peacock WJ, Dennis ES (2006) HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol 140: 1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Vogel J, Hill T (2008) High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Rep 27: 471–478 [DOI] [PubMed] [Google Scholar]
- Warthmann N, Chen H, Ossowski S, Weigel D, Hervé P (2008) Highly specific gene silencing by artificial miRNAs in rice. PLoS One 3: e1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng X, Wang L, Wang J, Hu Y, Du H, Xu C, Xing Y, Li X, Xiao J, Zhang Q (2014) Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response. Plant Physiol 164: 735–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods DP, Amasino RM (2015) Dissecting the Control of Flowering Time in Grasses Using Brachypodium distachyon. In J Vogel, ed, Genetics and Genomics of Brachypodium. Springer International Publishing, Cham, pp 259–273 [Google Scholar]
- Woods DP, Ream TS, Amasino RM (2014) Memory of the vernalized state in plants including the model grass Brachypodium distachyon. Front Plant Sci 5: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, Zhang Q (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40: 761–767 [DOI] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103: 19581–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J (2004b) Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor Appl Genet 109: 1677–1686 [DOI] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004a) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100: 6263–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zitzewitz J, Szűcs P, Dubcovsky J, Yan L, Francia E, Pecchioni N, Casas A, Chen THH, Hayes PM, Skinner JS (2005) Molecular and structural characterization of barley vernalization genes. Plant Mol Biol 59: 449–467 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.