A programmed cell death protein, by its interaction with histone acetyltransferases, has an important role during DNA damage responses induced by UV-B radiation in Arabidopsis.
Abstract
DNA damage responses have evolved to sense and react to DNA damage; the induction of DNA repair mechanisms can lead to genomic restoration or, if the damaged DNA cannot be adequately repaired, to the execution of a cell death program. In this work, we investigated the role of an Arabidopsis (Arabidopsis thaliana) protein, AtPDCD5, which is highly similar to the human PDCD5 protein; it is induced by ultraviolet (UV)-B radiation and participates in programmed cell death in the UV-B DNA damage response. Transgenic plants expressing AtPDCD5 fused to GREEN FLUORESCENT PROTEIN indicate that AtPDCD5 is localized both in the nucleus and the cytosol. By use of pdcd5 mutants, we here demonstrate that these plants have an altered antioxidant metabolism and accumulate higher levels of DNA damage after UV-B exposure, similar to levels in ham1ham2 RNA interference transgenic lines with decreased expression of acetyltransferases from the MYST family. By coimmunoprecipitation and pull-down assays, we provide evidence that AtPDCD5 interacts with HAM proteins, suggesting that both proteins participate in the same pathway of DNA damage responses. Plants overexpressing AtPDCD5 show less DNA damage but more cell death in root tips upon UV-B exposure. Finally, we here show that AtPDCD5 also participates in age-induced programmed cell death. Together, the data presented here demonstrate that AtPDCD5 plays an important role during DNA damage responses induced by UV-B radiation in Arabidopsis and also participates in programmed cell death programs.
DNA damage is a frequently occurring problem in living cells. For example, it was estimated that a mammalian genome undergoes approximately 100,000 modifications per day, each of which has some probability to cause mutations (Friedberg et al., 1995). DNA damage is one of the major consequences of exposure to high levels of UV-B radiation (Friedberg et al., 1995; Britt, 1996). Thus, a complex network of DNA damage responses has evolved to sense and respond to DNA damage (Rich et al., 2000). At the cellular level, the activation of DNA damage responses can have two outcomes: the induction of DNA repair mechanisms leading to genomic restoration or, if the damaged DNA cannot be adequately repaired, the execution of a cell death program (d’Adda di Fagagna, 2008; Zhang et al., 2009). However, little is known about the mechanisms by which a cell makes the decision to promote survival by repairing DNA damage or to die by triggering programmed cell death (PCD; Rich et al., 2000).
Chromatin remodeling, and histone acetylation in particular, are important epigenetic mechanisms involved in DNA damage repair induced by UV-B radiation in plants (Casati et al., 2008; Campi et al., 2012; Lario et al., 2013). After UV-B exposure, maize (Zea mays) plants deficient in chromatin-remodeling proteins show increased histone acetylation, in particular in the N-terminal domains of histones H3 and H4 (Casati et al., 2008). Moreover, maize and Arabidopsis (Arabidopsis thaliana) plants pretreated with an inhibitor of histone acetyltransferases (HATs) are deficient in DNA damage repair after UV-B exposure (Campi et al., 2012). In particular, two Arabidopsis HATs from the MYST family, AtHAM1 and AtHAM2, play important roles during DNA damage repair induced by UV-B, as Arabidopsis plants deficient in the expression of AtHAM1 and/or AtHAM2 show increased cyclobutane pyrimidine dimer (CPD) accumulation after UV-B exposure (Campi et al., 2012). The human homolog of AtHAM proteins is the acetyltransferase TAT-INTERACTIVE PROTEIN60 (TIP60). In humans, TIP60 is an essential player in different signaling pathways, including transcriptional regulation, chromatin remodeling, histone acetylation, and DNA repair (Sun et al., 2010). TIP60 acetylates the ε-amino groups of Lys residues on both histone and nonhistone proteins, including diverse targets such as histone H2A and H4, p53, the ATAXIA-TELANGIECTASIA MUTATED (ATM) kinase, among others (Sun et al., 2010; Reed et al., 2014). Different results suggest that TIP60 exerts diverse biological functions through mechanisms that are either dependent or independent of its intrinsic HAT activity, such as cellular signaling, DNA damage repair, cell cycle checkpoint control, and cell death induction (Sun et al., 2010). TIP60-mediated regulation typically involves the recruitment of TIP60 acetyltransferase activity to chromatin; in response to DNA damage, TIP60 is recruited to DNA lesions, where it participates in both the initial and the final stages of repair (Murr et al., 2006). The HAT activity of TIP60 is enhanced by PROGRAMMED CELL DEATH PROTEIN5 (PDCD5), a PCD-associated protein (Xu et al., 2009). PDCD5 regulates different types of PCD; for instance, HsPDCD5 regulates the translocation of Bax, a proapototic factor, from the cytosol to the mitochondria, causing cytochrome c release and an increase of CASPASE3 activity, which are early events of the onset of apoptosis (Chen et al., 2006). Also, HsPDCD5 is involved in the PCD pathway regulated by the tumor necrosis factor receptor TNFRSF19, a paraptosis-like cell death pathway (Wang et al., 2004). In response to DNA damage, HsPDCD5 is translocated to the nucleus, where it interacts with TIP60, improving its stability and promoting its HAT activity to repair DNA (Xu et al., 2009). Therefore, in humans, PDCD5 is involved both in the cytoplasmic pathway involved in the activation of PCD and in the nuclear pathway for DNA repair, where the cellular response to DNA damage is dependent on a signal threshold mechanism (Zhuge et al., 2011). In this way, depending on the level of DNA damage, PDCD5 regulates the switch of the cell response from DNA repair to apoptosis after UV-induced DNA damage in animals. Interestingly, model simulations suggest that PDCD5 nuclear translocation can attenuate cell apoptosis and that the PDCD5-TIP60 interaction can accelerate DNA damage-induced apoptosis, but the final cell fate decision is insensitive to the PDCD5-TIP60 interaction (Zhuge et al., 2011).
In plants, PDCD5 has only been identified and studied in the monocot plant rice (Oryza sativa; Attia et al., 2005; Su et al., 2006; Wang et al., 2010; Sun et al., 2012; Yang et al., 2013). OsPDCD5 overexpression causes many PCD symptoms, including leaf growth inhibition, DNA laddering, reduction of the total amount of proteins, and mitochondrial dysfunction (Attia et al., 2005). In addition, OsPDCD5 expression is up-regulated by UV-B radiation, low temperature, and high salinity (Su et al., 2006), and down-regulation of OsPDCD5 by antisense technology increased salt stress tolerance (Yang et al., 2013). Altogether, these results suggest an important role of PDCD5 in PCD and abiotic stress responses in rice and, probably, also in other plants.
Thus, the aim of this work was to investigate the role of Arabidopsis PDCD5 protein in the UV-B-induced DNA damage response and its possible participation in PCD programs. AtPDCD5 is induced by UV-B and is localized both in the nucleus and the cytosol. Moreover, pdcd5 mutants accumulate higher levels of DNA damage after UV-B exposure, similar to levels in ham1ham2 RNAi (for RNA interference) transgenic lines, and AtPDCD5 interacts with HAM proteins as detected by coimmunoprecipitation and pull-down experiments, suggesting that AtPDCD5 and HAM acetyltransferases participate in the same pathway of UV-B damage response. Consistent with a putative role of AtPDCD5 protein in PCD, pdcd5 mutants show an altered antioxidant metabolism. Plants overexpressing AtPDCD5 were found to be less sensitive to DNA damage but exhibit more cell death in root tips upon UV-B exposure. Finally, we here provide evidence that AtPDCD5 also may participate in age-induced PCD. Together, our results demonstrate that AtPDCD5 plays a role during the DNA damage response induced by UV-B radiation in Arabidopsis and also could participate in other PCD programs.
RESULTS
Identification of AtPDCD5 and Comparison with Other PDCD5 Proteins
In plants, PDCD5 has only been studied in rice. To find the Arabidopsis homolog of OsPDCD5 and HsPDCD5, we searched for a putative AtPDCD5 in the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) using protein sequence homology algorithms. This resulted in the identification of one putative AtPDCD5 protein (At1g29850) that shows 43% and 76% amino acid identity with HsPDCD5 and OsPDCD5, respectively (Supplemental Fig. S1, A and B).
AtPDCD5 has a predicted double-stranded DNA-binding domain of 112 amino acids, which is present in all the PDCD5 proteins identified, that extends from amino acids 9 to 119 and spans almost the entire coding region; it is predicted to bind about 20 nucleotides in DNA (pfam01984; Supplemental Fig. S2). The AtPDCD5 coding sequence shows three alternative splicing variants, which encode putative proteins of 129, 130, and 151 amino acids (Supplemental Fig. S1, A and C). The 129- and 130-amino acid variants only differ in one amino acid at position 23, while the 151-amino acid variant results from a change in the reading frame after position 119 (Supplemental Fig. S1B). At the DNA sequence level, the differences between the three variants occur in exon-exon junctions (Supplemental Fig. S3).
The expression of AtPDCD5 alternative splicing variants was analyzed in several tissues by quantitative reverse transcription (qRT)-PCR. Transcripts corresponding to the 129-amino acid protein were the most abundant in all the analyzed tissues (Supplemental Fig. S4). This isoform is also the most similar to all PDCD5s described from other organisms, including those from rice and humans; therefore, all the following experiments were conducted with this variant.
Regulation of AtPDCD5 Expression by UV-B Radiation and Subcellular Localization
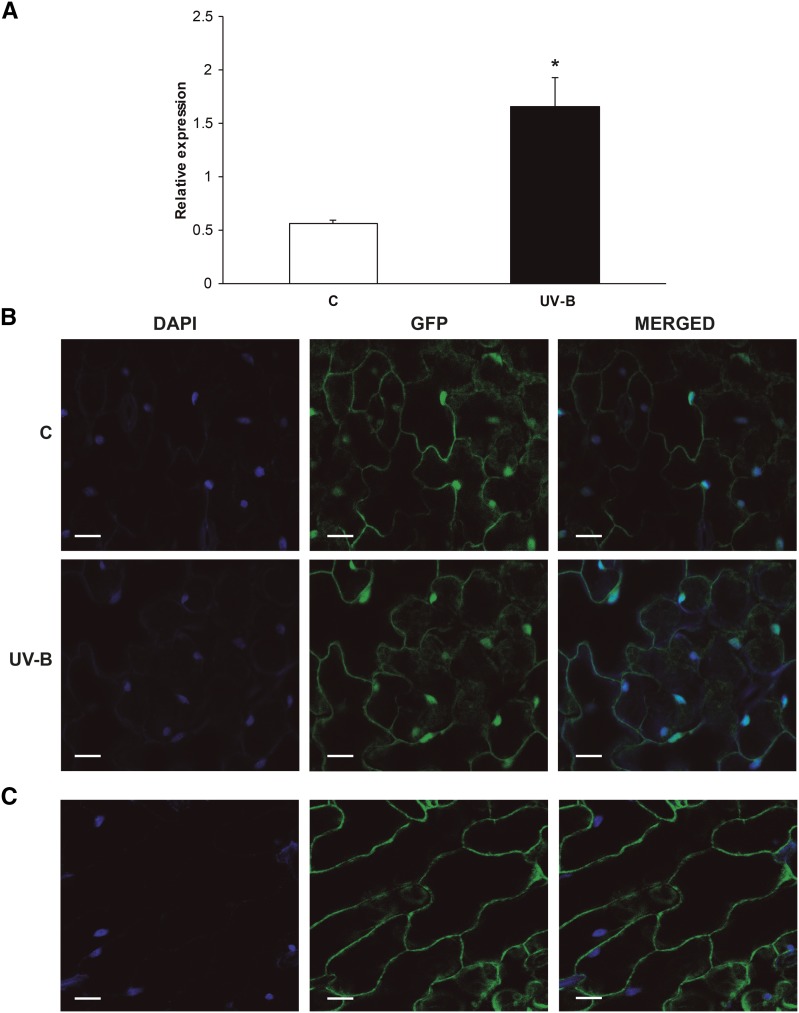
In humans, PDCD5 regulates the switch of the cell response from DNA repair to PCD after UV-induced DNA damage. In addition, as reported by Su et al. (2006), OsPDCD5 is UV-B regulated. Thus, it is possible that AtPDCD5 may have a role during DNA damage responses. To investigate if this is the case, we focused on the potential role of AtPDCD5 in the UV-B-induced DNA damage response pathway. We first analyzed its expression in 4-week-old Arabidopsis plants exposed under UV-B lamps for 4 h in growth chamber conditions. Immediately after the treatment, rosettes were collected for RNA extraction. AtPDCD5 transcript levels increased significantly (3-fold) after UV-B irradiation (Fig. 1A).
Figure 1.
Effects of UV-B radiation on PDCD5 expression and subcellular localization. A, Induction of AtPDCD5 expression by UV-B in Arabidopsis plants analyzed by qRT-PCR. Each reaction was normalized using the cycle threshold values corresponding to the CALCIUM PROTEIN KINASE3 mRNAs. The means of the results obtained using three independent RNAs as a template are shown, and error bars indicate the sd of the samples. The asterisk indicates a statistically significant difference applying Student’s t test (P < 0.05). C, Control. B, Subcellular localization of PDCD5 from Arabidopsis. Confocal images show 4′,6-diamino-phenylindole (DAPI) and GFP fluorescence in epidermal tissues of leaves from Pro-35S:AtPDCD5-GFP transgenic plants. DAPI staining indicates the positions of the nuclei in cells; GFP staining indicates the localization of AtPDCD5 in the cytoplasm and the nuclei of epidermal cells of transgenic plants. Images were merged to show signal overlap. Bars = 24 μm. C, Subcellular localization of GFP in Arabidopsis. Confocal images show DAPI and GFP fluorescence in epidermal tissues of leaves from Arabidopsis transgenic plants expressing GFP driven by the 35S promoter (Pro-35S:GFP). DAPI staining indicates the positions of the nuclei in cells; GFP staining indicates the localization of GFP in the cytoplasm of epidermal cells of transgenic plants. Images were merged to show signal overlap. Bars = 35 μm.
The presence of a putative DNA-binding domain in the AtPDCD5 structure and also the SUBAcon algorithm (http://suba.plantenergy.uwa.edu.au/; Hooper et al., 2014) predict that this protein could be localized to the nucleus. Moreover, HsPDCD5 is translocated to the nucleus after exposure to UV radiation, where it interacts with TIP60, promoting DNA repair (Xu et al., 2009). Therefore, we generated Arabidopsis transgenic plants that expressed AtPDCD5 as a fusion protein to the N-terminal domain of GFP driven by the cauliflower mosaic virus 35S promoter (Pro35S:AtPDCD5-GFP lines). Plants were grown 20 d under control conditions in the absence of UV-B. A subset was irradiated with UV-B, while a group was kept in the growth chamber as a control. Both in control and UV-B-exposed plants, GFP signal indicated that AtPDCD5-GFP fusion protein is localized both in the cytosol and in the nuclei (Fig. 1B). In Arabidopsis transgenic plants expressing GFP driven by the 35S promoter (Pro35S:GFP), the GFP fluorescence was detected only in the cytosol (Falcone Ferreyra et al., 2013; Fig. 1C); thus, the GFP signal in the nuclei shown in Figure 1B is due to AtPDCD5 localization in this organelle.
Mutant Plants with Decreased Levels of AtPDCD5 Accumulate More CPDs
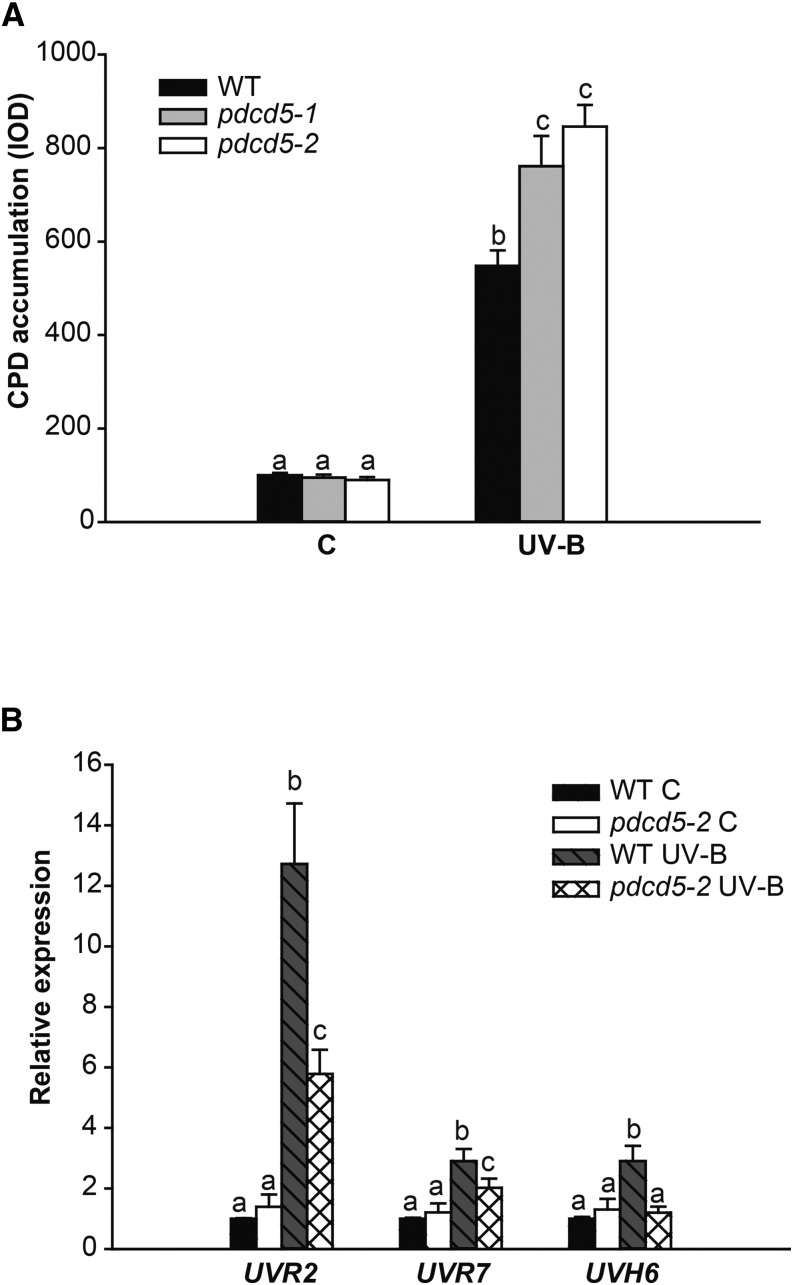
Absorption of UV-B by DNA induces the formation of covalent bonds between adjacent pyrimidines, giving rise to CPDs and, to a lesser extent, pyrimidine (6-4) pyrimidone photoproducts (Friedberg et al., 1995). We have demonstrated previously that Arabidopsis HAM1- and/or HAM2-deficient plants accumulate more CPDs than wild-type plants after UV-B exposure (Campi et al., 2012). Thus, we analyzed CPD accumulation after UV-B exposure in two AtPDCD5 knockdown mutant transfer DNA (T-DNA) insertion lines (pdcd5-1 and pdcd5-2; Supplemental Fig. S5), which have 2.7- and 5-fold decreases in AtPDCD5 transcripts levels, respectively, compared with wild-type plants. Supplemental Figure S6A shows that pdcd5-1 has a T-DNA insertion in the promoter, while pdcd5-2 has a T-DNA insertion in the 5′ untranslated region.
Arabidopsis wild-type and pdcd5 mutant plants were grown in a growth chamber in the absence of UV-B for 4 weeks. Plants were then exposed for 4 h to UV-B. As a control, different plants were irradiated with the same lamps covered with polyester plastic that absorbs UV-B. Leaf samples from control and UV-B-treated plants were collected immediately after the end of the treatment. DNA was extracted from pdcd5-1, pdcd5-2, and wild-type plants, and CPD accumulation was analyzed using monoclonal antibodies specifically raised against them. A comparison of CPD accumulation in samples from wild-type and mutant plants after the UV-B treatment and in control conditions in the absence of UV-B is shown in Figure 2A. In the absence of UV-B, the steady-state levels of CPDs in wild-type and mutant plants were similar (Fig. 2A). Nevertheless, after a 4-h UV-B exposure, more unrepaired lesions accumulated in the mutants than in wild-type plants (Fig. 2A). To analyze if the decreased expression of PDCD5 affects the expression of DNA repair enzymes, UVR2 (encoding a CPD photolyase), UVR7 (encoding ERCC1, a DNA excision repair protein of the nucleotide excision repair system), and UVH6 (encoding a DNA repair helicase of the nucleotide excision repair system) transcript levels were analyzed by qRT-PCR in wild-type and pdcd5-2 mutant plants. After a 4-h UV-B treatment under light conditions, UVR2 and UVR7 gene expression was induced in both the wild type and pdcd5 mutants, but the increase of both transcripts was significantly lower in pdcd5 mutants than in wild-type plants (Fig. 2B). Moreover, while UVH6 transcript levels were increased by UV-B in wild-type plants, UV-B induction of UVR6 was impaired in pdcd5 mutants. Therefore, the higher accumulation of CPDs in pdcd5 mutants by UV-B radiation can be due to lower expression levels of DNA repair enzymes in UV-B-exposed plants. Thus, our results demonstrate that, in the pdcd5 mutants, DNA repair mechanisms are impaired.
Figure 2.
CPD levels in wild-type (WT) and pdcd5 mutant plants exposed to UV-B radiation. A, CPD levels in the DNA of wild-type Columbia-0 (Col-0) and pdcd5-2 mutants under control conditions without UV-B (C) and immediately after a 4-h UV-B treatment (UV-B). CPD levels are indicated as integrated optical density (IOD) values. Data show means ± se of at least three independent experiments. Different letters over the bars indicate statically significant differences at P < 0.05 (ANOVA test). B, Relative expression of UVR2, UVR7, and UVH6 genes analyzed by qRT-PCR. Wild-type Col-0 and pdcd5-2 mutant plants were irradiated with UV-B for 4 h under light conditions or kept under control conditions without UV-B. Data are relativized to wild-type control values set at 1. For each transcript analyzed, different letters over the bars indicate statically significant differences at P < 0.05 (ANOVA test).
PDCD5-Deficient Plants Have an Altered Redox Metabolism
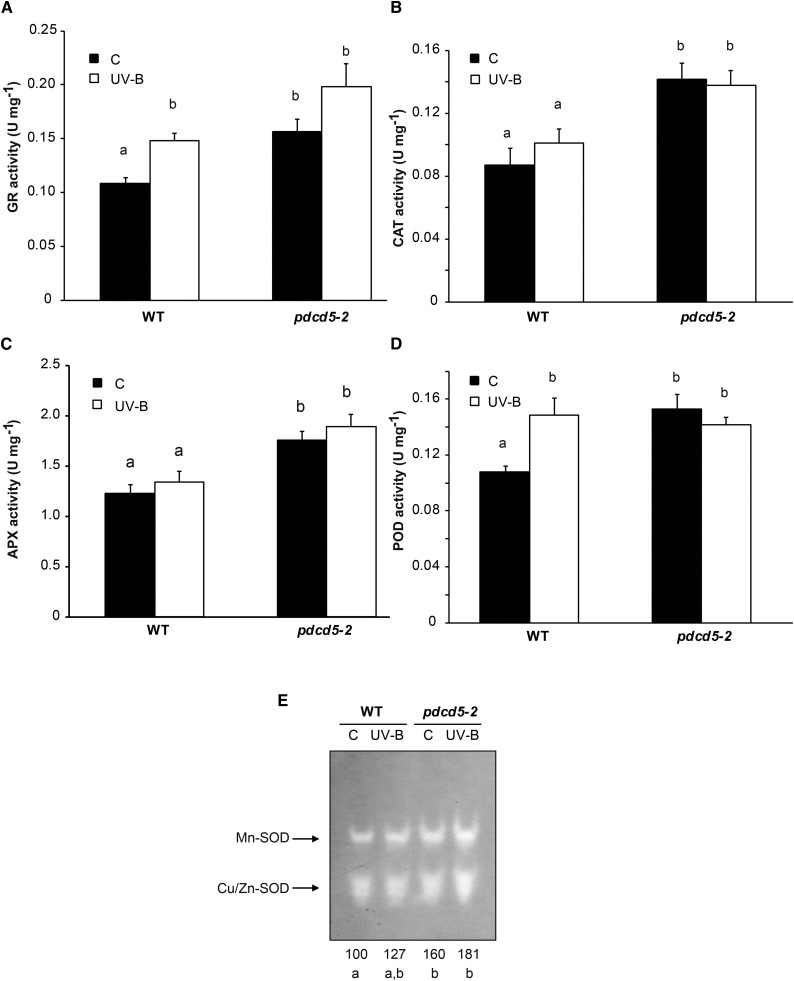
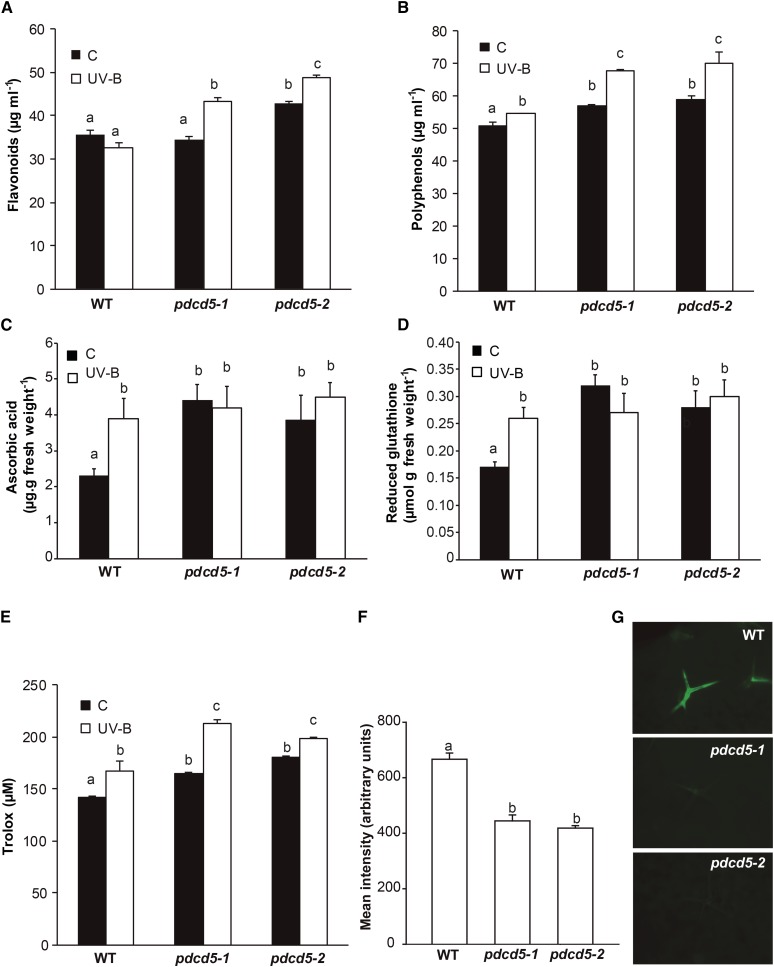
Reactive oxygen species (ROS) are not only toxic by-products of aerobic metabolism but also function as signaling agents, acting, for example, as important modulators of plant PCD (Gadjev et al., 2008). Therefore, we further evaluated the antioxidant metabolism in pdcd5 mutants, both under control conditions and after UV-B exposure. We first analyzed the activity of enzymes that participate in ROS detoxification: glutathione reductase (GR), catalase (CAT), peroxidase (POD), ascorbate peroxidase (APX), and superoxide dismutase (SOD). Under control conditions in the absence of UV-B, the activities of all five enzymes were higher in pdcd5-2 than in wild-type plants (Fig. 3). Interestingly, after a 4-h UV-B treatment at 2 W m−2, POD and GR activities increased in wild-type plants, showing similar levels to those detected in pdcd5 mutants under control conditions. On the other hand, the activities of the three other detoxifying enzymes did not increase in wild-type plants by UV-B but were significantly higher in pdcd5 than in wild-type plants under both control and UV-B conditions. This was also observed when plants were exposed to 10 times lower UV-B intensities (0.2 W m−2; Supplemental Fig. S6). Consistently, pdcd5 mutants also showed higher levels of antioxidant molecules, such as polyphenols, flavonoids, ascorbic acid, and reduced glutathione, in comparison with wild-type plants in control conditions (Fig. 4). After low-intensity UV-B exposure, the levels of ascorbic acid and reduced glutathione were similar in the wild type and pdcd5 mutants, while UV-B induced significant and higher accumulation of polyphenols and flavonoids in the mutants in comparison with wild-type plants. The overall antioxidant state of control and UV-B-irradiated plants was assayed using the ferric reducing ability of plasma (FRAP) assay, which is based on the ability of a certain biological sample to reduce ferric (Fe3+) to ferrous (Fe2+) ions (Benzie and Strain, 1996). Our results show that the antioxidant capacity (measured as Trolox concentration) is higher in pdcd5 mutants than in wild-type plants in control conditions and after UV-B exposure (Fig. 4). Finally, we analyzed direct ROS formation after a 4-h UV-B treatment at 2 W m−2. Both pdcd5 mutants exhibit lower levels of ROS than wild-type plants (Fig. 4, F and G), in correlation with the increased antioxidant capacity of the mutants shown in Figures 3 and 4. In summary, our results show that a deficiency in the expression of AtPDCD5 strongly affects the redox metabolism in Arabidopsis cells.
Figure 3.
Analysis of antioxidant enzyme activities in wild-type Col-0 (WT) and pdcd5 mutant plants under control conditions and after UV-B treatment. GR (A), CAT (B), APX (C), POD (D), and SOD (E) activities were determined under control conditions (C) and after a 4-h UV-B treatment (UV-B) at intensity of 2 W m−2. The numbers below the gel in E indicate the relative intensities corresponding to the sum of both reactive bands determined by densitometric analysis with respect to wild-type plants under control conditions (100%). Results represent averages ± se of three independent biological replicates. Statistical significance was analyzed using ANOVA; for each sample analyzed, different letters indicate significant differences at P < 0.01.
Figure 4.
Analysis of antioxidant metabolites and ROS formation in wild-type Col-0 (WT) and pdcd5 mutant plants under control conditions and after UV-B treatment. A to E, Levels of flavonoids (A), polyphenols (B), ascorbic acid (C), reduced glutathione (D), and Trolox (E), the product of the FRAP assay to measure antioxidant tissue capacity, are shown in control plants (C) and after a UV-B treatment (UV-B) at an intensity of 0.2 W m−2. Results represent averages ± se of three independent biological replicates. Statistical significance was analyzed using ANOVA; for each sample analyzed, different letters indicate significant differences at P < 0.05. F, Mean intensity of 2′7′-dichlorofluorescein diacetate (H2DCFDA) fluorescence in wild-type Col-0 and pdcd5 mutant plants after UV-B treatment at an intensity of 2 W m−2 to detect ROS formation. Results represent averages ± se of three independent biological replicates. Statistical significance was analyzed using ANOVA; for each sample analyzed, different letters indicate significant differences at P < 0.001. G, Representative images show ROS formation detected by H2DCFDA fluorescence (green) in leaves from wild-type Col-0 and pdcd5 mutant plants after UV-B exposure.
HAM1/HAM2 and AtPDCD5 Participate in the Same UV-B Response Pathway and Interact Physically in Vivo
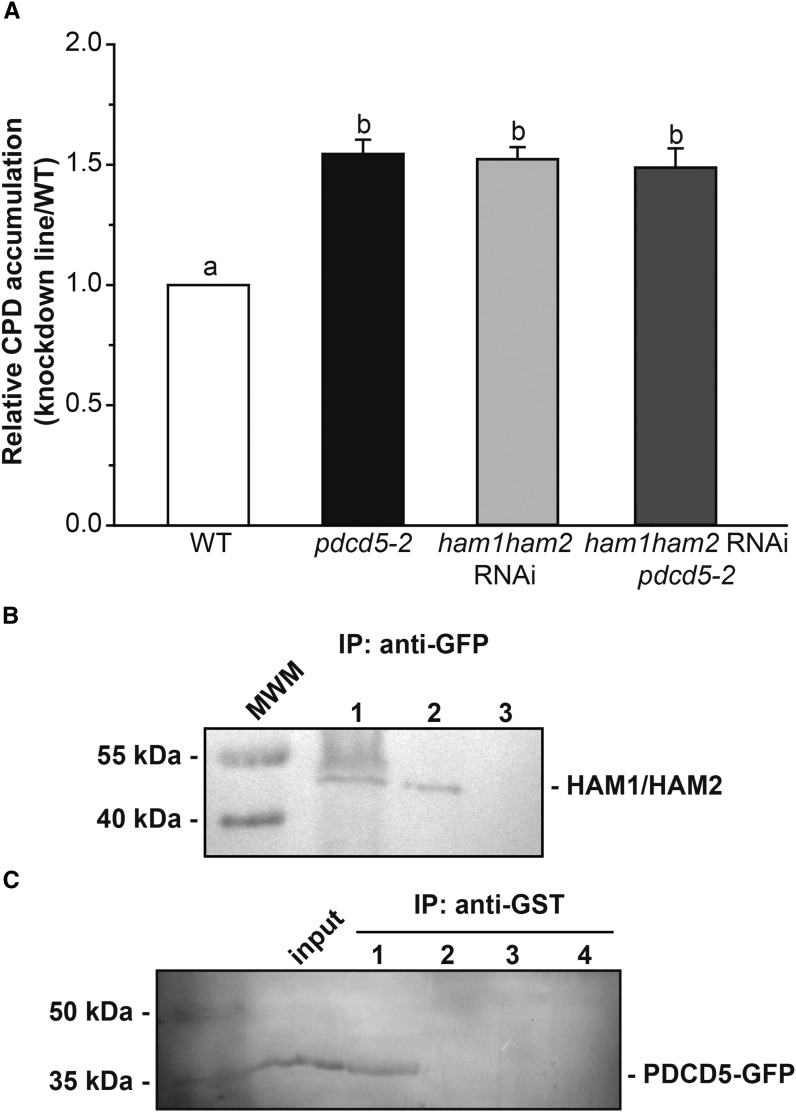
The human homolog of AtHAM1 and AtHAM2 is TIP60, and its HAT activity is regulated by HsPDCD5 under UV-B conditions (Xu et al., 2009). Similar to Arabidopsis pdcd5 plants, ham1ham2 RNAi plants showed an increased CPD accumulation after a 4-h UV-B treatment (Campi et al., 2012). Therefore, with the aim to elucidate if HAM1/HAM2 and AtPDCD5 are in the same UV-B response pathway, pdcd5-2 mutants were crossed with ham1ham2 RNAi transgenic plants. F2 progeny with reduced levels of the three transcripts (HAM1, HAM2, and PDCD5) from several independent F1 plants were obtained. Four-week-old triple knockdown plants (F3 progeny), together with single pdcd5, ham1ham2 RNAi, and wild-type plants, were irradiated with UV-B for 4 h, and CPD accumulation was assayed in samples collected at the end of the treatment. Figure 5A shows that DNA damage in the triple knockdown plants was similar to that of single pdcd5 mutants and the ham1ham2 RNAi transgenic plants, suggesting that AtPDCD5 and AtHAM proteins participate in the same UV-B-induced DNA damage response pathway.
Figure 5.
PDCD5 interacts with HAM1/HAM2 and participates in the same UV-B DNA damage response pathway. A, Relative CPD accumulation in the pdcd5-2 mutant, ham1ham2 RNAi, and ham1ham2 RNAi/pdcd5 plants versus wild-type (WT) plants immediately after a 4-h UV-B treatment under conditions that allowed photorepair in the light. B, Nuclear protein extracts from transgenic plants expressing Pro35S:AtPDCD5-GFP were used for coimmunoprecipitation (IP) experiments using anti-GFP antibodies (lane 1). As a negative control, a nuclear protein extract from wild-type Col-0 plants was incubated with anti-GFP antibodies (lane 3). Also, 30 μg of total nuclear protein extract before coimmunoprecipitation was included as a positive control (lane 2). Western blots were developed using anti-TIP60 antibodies (HAM1/HAM2). C, Nuclear protein extracts from transgenic plants expressing Pro35S:AtPDCD5-GFP were incubated with purified recombinant GST-AtHAM2 fusion protein and were used in pull-down experiments using anti-GST antibodies (5 μg; lane 1). As negative controls, nuclear extracts from transgenic plants were incubated with GST protein (5 μg; lane 2), and nuclear extracts from wild-type Col-0 plants were incubated with either GST-AtHAM2 (lane 3) or GST (lane 4). Western blots were developed using anti-GFP antibodies. Thirty micrograms of total nuclear protein extract before coimmunoprecipitation was included as a positive control (input). Prestained molecular weight markers (MWM) and their corresponding molecular masses are included at the left side of each gel.
Moreover, to further analyze if AtPDCD5 interacts in vivo with AtHAM1/HAM2, as reported in humans (Xu et al., 2009), we carried out coimmunoprecipitation experiments using nuclear lysates from Pro35S:AtPDCD5-GFP transgenic plants and anti-GFP antibodies. The immunoprecipitate was resolved by SDS-PAGE and analyzed by western blot using commercial antibodies against human TIP60 protein; these antibodies were shown previously to recognize AtHAM proteins (Lario et al., 2013). The analysis revealed that AtPDCD5-GFP coimmunoprecipitated with AtHAM1/HAM2 (Fig. 5B), demonstrating that HAM proteins and PDCD5 interact physically in the nucleus.
This interaction also was observed when pull-down experiments were carried out. For these experiments, nuclear extracts from Pro35S:AtPDCD5-GFP transgenic plants were incubated with purified recombinant glutathione S-transferase (GST)-AtHAM2, which is 92.5% similar in amino acid sequence to HAM1 (Campi et al., 2012; Supplemental Fig. S7), and this extract was then immunoprecipitated using anti-GST antibodies. As controls, GST protein or nuclear extracts from wild-type plants were used. Coimmunoprecipitates were analyzed by SDS-PAGE followed by western blot using anti-GFP antibodies, showing that AtHAM2 coimmunoprecipitates with AtPDCD5-GFP (Fig. 5C). Together, these results confirm the physical interaction between AtPDCD5 and HAM2, and probably with HAM1, in Arabidopsis nuclei (Fig. 5C), supporting the hypothesis that PDCD5’s role is conserved Arabidopsis.
AtPDCD5 Overexpression Increases Cell Death and DNA Damage Repair in UV-B-Exposed Plants
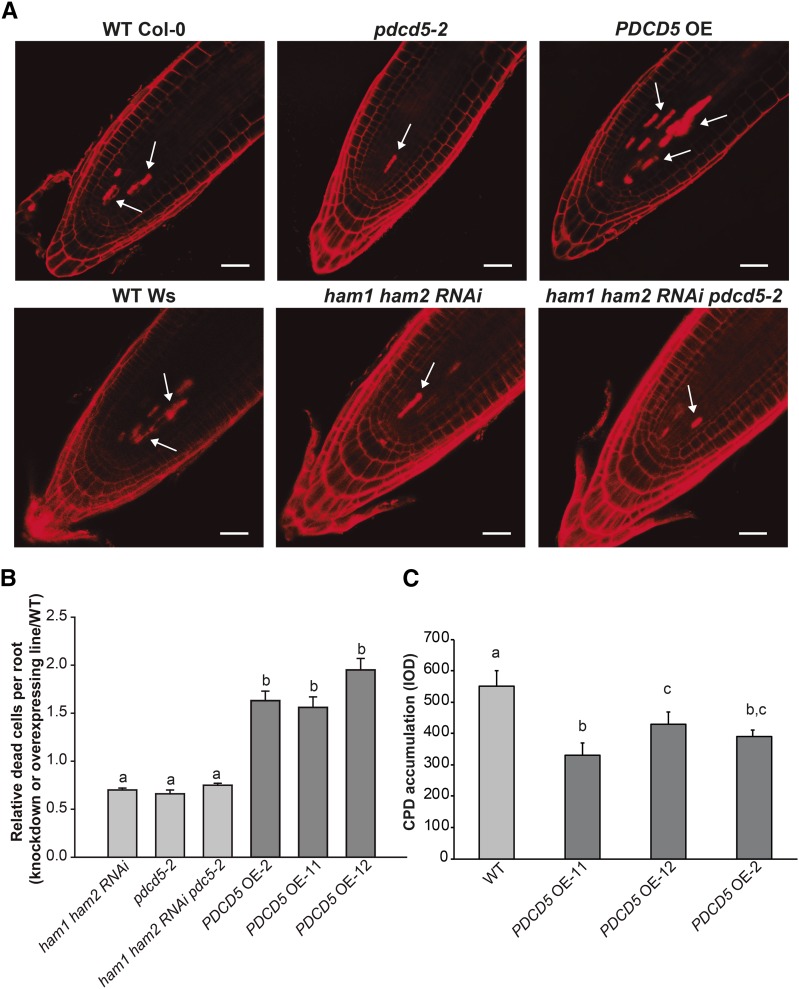
To assess if AtPDCD5 is involved in the induction of PCD in response to UV-B radiation in Arabidopsis, cell death was evaluated in root tips. This technique has been widely used to investigate the role of different proteins in PCD after DNA damage by UV-B and other genotoxic stresses. For example, in Arabidopsis, the role of ATM, ATR, and MPK1 in PCD after UV-B was analyzed by this technique (Furukawa et al., 2010; González Besteiro and Ulm, 2013); this technique also was used for the analysis of the role of several DNA translesion polymerases in PCD by UV-B (Curtis and Hays, 2011; Hays, 2011). To validate this hypothesis, we generated Arabidopsis transgenic plants that express AtPDCD5 driven by the cauliflower mosaic virus 35S promoter (PDCD5 OE lines; Supplemental Fig. S8). Five-day-old seedlings of the wild type (Col-0), pdcd5-2, and PDCD5 OE lines grown on Murashige and Skoog (MS) agar plates were exposed to UV-B radiation for 1 h (4 W m−2) and then moved to a growth chamber for 24 h in the absence of UV-B. Then, the roots were stained with propidium iodide (PI), as dead cells readily take up PI while cells that are alive actively exclude PI (Furukawa et al., 2010). pdcd5-2 root tips showed significantly fewer dead cells compared with the wild-type roots (Fig. 6, A and B). The opposite was observed in PDCD5 OE lines, which showed around 60% more dead cells than wild-type root tips (Fig. 6, A and B). In addition, we analyzed cell death in root tips of ham1ham2 RNAi double mutants and ham1ham2 RNAi pdcd5 triple mutants exposed to UV-B radiation. Both mutants have significantly fewer dead cells than wild-type roots but similar dead cells to pdcd5 mutants (Fig. 6, A and B). These results are consistent with the CPD accumulation measured in these plants after a 4-h UV-B treatment, strengthening the fact that AtPDCD5 and AtHAM proteins participate in the same UV-B-induced DNA damage response pathway. Interestingly, none of the analyzed lines showed dead cells when they were kept under control conditions in the absence of UV-B (Supplemental Fig. S9). These results suggest that the role of AtPDCD5 in PCD after UV-B exposure is conserved in Arabidopsis.
Figure 6.
PDCD5 overexpression induces cell death and increased DNA damage repair. A, Representative images of stem cells and adjacent daughter cells from wild-type (WT) Col-0 and Wassilewskija (Ws), pdcd5-2, ham1ham2 RNAi, and ham1ham2 RNAi/pdcd5 mutants, and PDCD5-overexpressing plants (PDCD5 OE) that were scored for intense PI staining to count dead stem cells per root after UV-B. Arrows indicate dead cells. Bars = 50 μm. B, Number of stem cells that are dead after UV-B exposure relative to those in wild-type plants (Col-0 and Ws). Three independent transgenic lines were analyzed (PDCD5 OE-2, PDCD5 OE-11, and PDCD5 OE-12). Statistical significance was analyzed using a one-way ANOVA test at P < 0.05; differences at P < 0.05 are marked with different letters. C, CPD levels in the DNA of wild-type Col-0 and PDCD5 OE plants (PDCD5 OE-2, PDCD5 OE-11, and PDCD5 OE-12) immediately after a 4-h UV-B treatment. CPD levels are indicated as integrated optical density (IOD) values. Results represent averages ± se of three independent biological replicates. Statistical significance was analyzed using a one-way ANOVA test at P < 0.05; differences from the control are marked with different letters.
Furthermore, we evaluated CPD accumulation after a 4-h UV-B treatment in plants overexpressing AtPDCD5 (PDCD5 OE lines). Interestingly, as shown in Figure 6C, plants that overexpress AtPDCD5 accumulate fewer CPDs after the UV-B treatment.
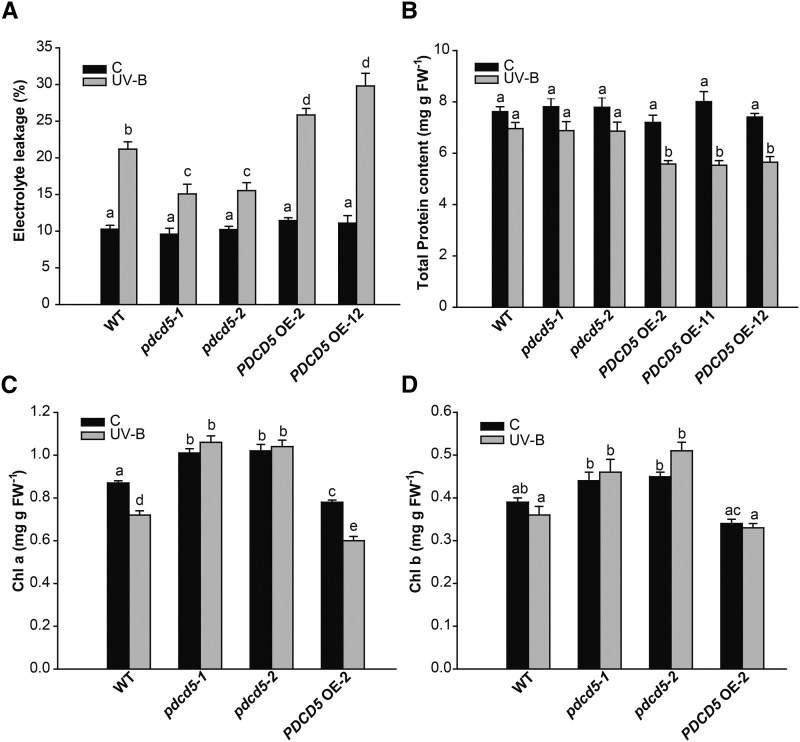
Finally, in order to confirm the participation of AtPDCD5 in PCD after UV-B exposure, we evaluated the integrity of the cells by measuring the electrolyte leakage of leaves. pdcd5 mutant plants exhibited a lower increase in electrolyte leakage than wild-type plants after the UV-B treatment, while the opposite was observed in PDCD5 OE lines (Fig. 7A). Moreover, while wild-type and pdcd5 mutant plants did not show any significant difference in total protein content after UV-B radiation or in control conditions, PDCD5 OE lines showed a significant reduction of total protein content after the UV-B treatment (Fig. 7B). We also measured chlorophyll a and b contents in mutant and overexpressing plants. Interestingly, under control conditions in the absence of UV-B, pdcd5 mutants showed higher levels of both chlorophyll a and b than wild-type plants (Col-0), while PDCD5 OE plants had lower chlorophyll levels than wild-type plants (Fig. 7, C and D). After a 4-h UV-B exposure, PDCD5 OE plants showed a pronounced decrease in chlorophyll content similar to wild-type plants; in contrast, pdcd5 mutants did not show any chlorosis. Taken together, our results demonstrate that AtPDCD5 overexpression increases DNA repair and cell death after UV-B exposure, while mutations in the PDCD5 gene cause the opposite effect.
Figure 7.
Plants with altered levels of PDCD5 show altered protein and chlorophyll contents and electrolyte leakage after UV-B exposure. Electrolyte leakage (A), total protein content (B), and chlorophyll content (Chl a and Chl b; C and D) are shown in wild-type Col-0 (WT), pdcd5 mutant plants, and PDCD5-overexpressing lines under control conditions (C) and after UV-B treatment (UV-B). Results represent averages ± se of three independent biological replicates. Statistical significance was analyzed using a two-way ANOVA test at P < 0.05; differences at P < 0.05 are marked with different letters. FW, Fresh weight.
AtPDCD5 Participates in Age-Induced PCD
Recently, it was reported that developmental and environmental PCD processes take place by separate regulatory pathways (Olvera-Carrillo et al., 2015). Thus, we finally investigated if AtPDCD5, in addition to its participation in PCD after UV-B exposure, could be involved in the natural senescence of plants. During early development, pdcd5 mutants looked similar to wild-type plants, except for having a smaller rosette area (Supplemental Fig. S10, A and B); however, pdcd5 mutants exhibited a remarkably delayed leaf senescence characterized by higher chlorophyll content compared with age-matched wild-type plants (Supplemental Fig. S10C). In contrast, PDCD5 OE plants showed lower levels of total chlorophyll in the leaves than wild-type plants (Supplemental Fig. S10C). Interestingly, at 40 days after sowing (DAS), while wild-type and PDCD5 OE plants exhibited mostly senescing leaves, pdcd5 leaves remained mainly green (Supplemental Fig. S10C). In pdcd5 plants, the onset of senescence occurred 5 d later than in wild-type or PDCD5 OE plants (36.5 DAS in pdcd5 mutants versus 31 DAS in wild-type and transgenic plants). The progress of leaf senescence also was delayed in pdcd5 mutants, which took on average 13 d for leaf 6 from pdcd5 plants (analyzing the progress of chlorosis from the leaf tip to the petiole) and 8.5 and 6 d for the same leaf from wild-type and PDCD5 OE plants, respectively. These results suggest that mutations in PDCD5 cause a delay in senescence, while its overexpression accelerates leaf senescence.
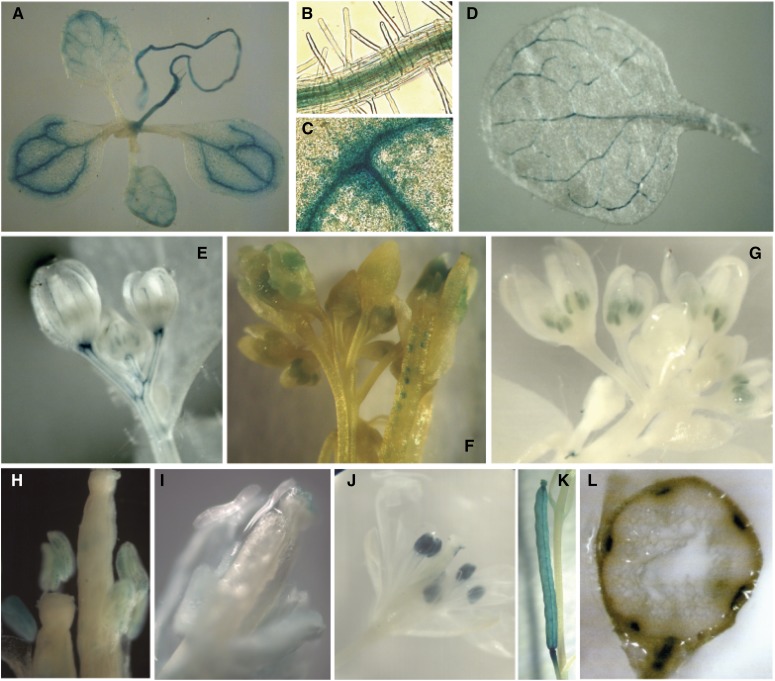
Finally, to further analyze if the spatiotemporal expression of AtPDCD5 correlates with its function in PCD during development, we constructed transgenic plants expressing the GUS gene under the control of the AtPDCD5 promoter as described in “Materials and Methods.” In all transgenic lines analyzed, the expression of AtPDCD5 was high in the vasculature of leaves, petals, stems, and roots (Fig. 8, A–E and L). In these tissues, PCD is essential during organ development (van Doorn and Woltering, 2005; Varnier et al., 2005; Gadjev et al., 2008). In addition, GUS activity was detected in anthers, pollen, stigma, and developing seeds (Fig. 8, F–J), while in mature green siliques, GUS staining was observed in the septum and valves (Fig. 8K).
Figure 8.
Histochemical analysis of GUS activity in transgenic Arabidopsis plants transformed with the ProAtPDCD5:GUS construct. A, Eight-day-old seedling. B, Magnification of A showing GUS staining in the root vasculature (light micrograph; 40×). C, Magnification of A showing GUS staining in the leaf vasculature (light micrograph; 40×). D, Mature leaf from a 28-d-old plant. E, Immature flower bud. F, Siliques with developing seeds and pollen grains in the silique tip. G, Flowers with immature anthers (light micrograph; 10×). H, Anthers with pollen grains (light micrograph; 10×). I, Gynoecium showing GUS staining in the stigma (light micrograph; 40×). J, Open flower. K, Mature green silique. L, Transverse cut of the stem.
Together, our data suggest that, besides its role in PCD after UV-B exposure, AtPDCD5 may also have a role in age-induced PCD, and also during other processes where PCD is essential during organ development, as described previously in humans and rice (Xu et al., 2009).
DISCUSSION
PCD is an essential component of plant development and an adaptation to adverse environmental conditions (Gadjev et al., 2008; Lord and Gunawardena, 2012). This process involves diverse molecular pathways, which execute the genetically controlled cellular suicide. Although in the last years several components of plant PCD pathways have been elucidated, many aspects remain unknown (Fendrych et al., 2014). In this work, we have identified and characterized an Arabidopsis protein, AtPDCD5, that is highly similar to rice and human PDCD5 proteins. In humans, PDCD5 expression increases during apoptosis (Liu et al., 1999); also, several studies have shown that HsPDCD5, when transiently or stably overexpressed in human cells, facilitates apoptosis triggered by certain stimuli, such as growth factor withdrawal (Liu et al., 1999). In addition, HsPDCD5 enhances TAJ/TROY-induced paraptotic cell death (Wang et al., 2004); paraptosis differs from apoptosis in the absence of caspase activation or typical nuclear changes, such as DNA fragmentation, and involves cytoplasmic vacuolation and mitochondrial swelling; this pathway exists in parallel with apoptosis (Sperandio et al., 2000; Wyllie and Golstein, 2001; Yoon et al., 2014). The presented evidence strongly suggests that PDCD5 may be a key molecule connecting the two cell death programs. Paraptosis seems to be an ancient form of programmed cell death, and because PDCD5 shares a high degree of primary sequence conservation between species (Supplemental Fig. S1), this protein may act as a core regulator of PCD in different species.
Our experiments strongly suggest that AtPDCD5 regulates PCD after UV-B irradiation. AtPDCD5 transcripts are increased after UV-B exposure, and transgenic plants overexpressing this protein show increased cell death in roots after UV-B exposure, while mutants in this gene are less affected by the treatment (Fig. 6). Moreover, AtPDCD5 locates both in the cytosol and the nucleus in transgenic plants that overexpress AtPDCD5 fused to GFP, similar to what happens in humans (Chen et al., 2001); this dual localization is observed both under control conditions and after UV-B exposure. In humans, depending on the UV treatment, PDCD5 can either regulate PCD in the cytosol or after movement to the nucleus, and together with the HAT TIP60, it participates in DNA damage repair (Zhuge et al., 2011). Those authors suggested that, in each cell, the ratio of PDCD5 in the cytosol and the nucleus may be the key factor that determines if the DNA damage repair or PCD response is triggered. However, it was recently reported that, in some type of cancer cells, PDCD5 does not interact with TIP60 and its participation in the DNA damage response is not rate limiting (Bock et al., 2015). The discrepancy between different published results remains unclear, but it could be due to experimental conditions and/or the type of cells analyzed. Interestingly, transgenic plants that overexpress AtPDCD5 show both increased cell death and reduced DNA damage accumulation after the UV-B treatment (Fig. 6). A possible explanation for this observation is that the decision to repair DNA damage or undergo PCD probably may differ between adjacent cells, depending on the level of damage that each cell accumulated after UV-B exposure. In this sense, it should be noted that, when DNA damage accumulation was measured, whole leaves were examined; so probably in those tissues, some cells suffered PCD and its DNA may have been degraded. Consequently, in our assay, we may have only measured DNA damage accumulation in cells in which PCD was not triggered. In agreement with this hypothesis, when stem cell death after UV-B exposure was examined, some cells underwent cell death but some neighboring cells did not. Nevertheless, like HsPDCD5, AtPDCD5 is localized in the cytosol and nucleus, and this dual localization may regulate DNA repair systems or PCD depending on the damage level after UV exposure. In a similar way to that described in humans, AtPDCD5 probably regulates the switch from DNA repair to PCD in a given cell when the damage cannot be properly repaired. However, it is also possible that overexpression of PDCD5 protein may lead to increased levels in cytoplasm and nucleus, triggering both responses in parallel.
Furthermore, our results demonstrate that AtPDCD5 interacts in vivo with AtHAM2 and possibly also with AtHAM1, the homolog acetyltransferases to human TIP60 (Fig. 5). Mutants in either or both AtPDCD5 and/or AtHAM proteins show similarly increased levels of CPD accumulation and decreased cell death after UV-B exposure compared with wild-type plants, suggesting that these proteins participate in the same pathway of UV-B damage response. In humans, PDCD5 is a positive regulator of TIP60 (Xu et al., 2009); in response to DNA double strand breaks, TIP60 is recruited to DNA lesions to participate in both the initial and final stages of repair (Murr et al., 2006). Also, in unstressed conditions, PDCD5 binds to TIP60 and enhances its stability; however, the amount of PDCD5 bound to TIP60 is increased significantly after UV irradiation. On the other hand, HsPDCD5 enhances the HAT activity of TIP60 and TIP60-dependent histone acetylation in both basal and UV-induced levels, and it increases TIP60-dependent K120 acetylation of p53 and participates in the p53-dependent expression of apoptosis-related genes (Xu et al., 2009). Similarly, if the interaction between AtPDCD5 and HAM proteins in Arabidopsis also regulates the expression of PCD genes, this also may explain why AtPDCD5-overexpressing lines show increased cell death after UV-B exposure. Moreover, the induction of DNA repair enzymes by UV-B radiation is affected in pdcd5 mutant plants (Fig. 3B), suggesting that their expression also may be regulated directly or indirectly by AtPDCD5. Together, our results suggest that, in Arabidopsis, AtPDCD5 may act as a regulator of HAM1 and/or HAM2 in UV-B DNA damage responses.
Our experiments show that AtPDCD5 responses may involve the participation of ROS, as plants deficient in AtPDCD5 expression show altered redox metabolism (Figs. 3 and 4; Supplemental Fig. S7). PCD can be initiated by different types of ROS, including hydrogen peroxide (H2O2), singlet oxygen, and superoxide radicals (Vranová et al., 2002; op den Camp et al., 2003). We also here describe that pdcd5 mutants have increased accumulation of DNA damage after UV-B exposure; so it is possible that this damage may activate different stress responses, including the accumulation of antioxidant molecules such as polyphenols, flavonoids, reduced glutathione, and ascorbate. Interestingly, under control conditions in the absence of UV-B, pdcd5 mutants also showed increased levels of some antioxidant molecules and enzymes when compared with wild-type plants. This metabolic state is frequently associated with increased stress tolerance; however, this does not seem to be the case. Similar results were reported previously in several plant species. For example, Creissen et al. (1999) showed that tobacco (Nicotiana tabacum) plants with increased levels of reduced glutathione were not stress tolerant but instead showed increased oxidative stress due to failure in redox sensing. Moreover, Xiang et al. (2001) demonstrated that transgenic Arabidopsis plants that overexpressed the enzyme γ-glutamyl-Cys synthetase, which also showed increased levels of reduced glutathione, were similarly tolerant to cadmium stress to wild-type plants. In this way, antioxidant molecules and enzymes may provide information on cellular redox state, and ROS may act as signals that could modulate the appropriate induction of acclimation processes or, alternatively, the execution of cell death programs (Foyer and Noctor, 2005). Therefore, a high level of antioxidant molecules is not necessarily conducive to increased stress tolerance.
Interestingly, we here provide evidence that AtPDCD5 also could have a role in age-induced PCD. While plants deficient in AtPDCD5 transcripts exhibit a delayed leaf senescence characterized by higher chlorophyll content compared with age-matched wild-type plants, PDCD5 OE plants show lower levels of total chlorophyll content in the leaves than wild-type plants (Supplemental Fig. S10C). Also, PDCD5 OE plants show a marked acceleration in the progression of senescence, suggesting that mutations in PDCD5 cause a delay in senescence, while its overexpression accelerates leaf senescence. Moreover, the spatiotemporal expression of AtPDCD5 is associated with tissues where PCD is essential during organ development, such as the vasculature of leaves, petals, stems, and roots, and also in anthers, pollen, and seeds (Fig. 8; van Doorn and Woltering, 2005; Varnier et al., 2005; Gadjev et al., 2008). Furthermore, these observations are supported by Arabidopsis eFP Browser data, which show high AtPDCD5 transcript levels in senescing leaves, sepals, petals, as well as the xylem of stems (Supplemental Fig. S11A; Winter et al., 2007) Thus, our data suggest that, besides its role in PCD after UV-B exposure, AtPDCD5 also may have a role in age-induced PCD. Finally, it is interesting that AtPDCD5 transcript levels also are increased after pathogen infection (Arabidopsis eFP Browser; Winter et al., 2007; Supplemental Fig. S11B). It was reported previously that PCD takes place during the hypersensitive response (Olvera-Carrillo et al., 2015); thus, AtPDCD5 also could be involved in PCD during biotic stress.
Together, the results presented here demonstrate that AtPDCD5 has an important role during DNA damage responses and participates in PCD under UV-B exposure in Arabidopsis. As described in humans, this role appears to be, at least in part, through its interaction with HAM acetyltransferases, and this interaction seems to be a vital component of the signaling pathways that regulate both DNA damage repair and PCD under UV-B.
MATERIALS AND METHODS
Plant Material, Growth Conditions and UV-B Treatments
Following a cold treatment (72 h at 4°C in the dark), Arabidopsis (Arabidopsis thaliana) ecotype Col-0 and Ws plants were grown in a growth chamber under light (100 μE m−2 s−1 provided by cool-white fluorescent tubes) with a 16-h-light/8-h-dark photoperiod at 22°C. The pdcd5-1 mutant (SAIL_880-E01) originated in the SAIL T-DNA insertion mutant collection and the pdcd5-2 mutant (SALK_024942) in the SALK T-DNA insertion mutant collection; both were obtained from the Arabidopsis Biological Resource Center. Both mutants are in the Col-0 ecotype. The ham1ham2 RNAi transgenic line (in ecotype Ws) was described previously (Campi et al., 2012). For PCD studies, Arabidopsis plants were germinated and grown on vertically oriented petri plates containing salt MS agar medium for 5 d. Previously, Arabidopsis seeds were sterilized and incubated at 4°C for 3 d. For subcellular localization analysis, plants were grown for 20 d on MS plates supplemented with kanamycin (50 mg L−1).
UV-B treatments were done in a growth chamber as described previously (Campi et al., 2012). Arabidopsis plants were treated for 4 h with a UV-B intensity of 2 W m−2 and a UV-A intensity of 0.65 W m−2. The UV-B bulbs were covered with cellulose acetate filters (100-mm extra-clear cellulose acetate plastic; Tap Plastics); the cellulose acetate sheeting does not remove any UV-B radiation from the spectrum but excludes wavelengths lower than 300 nm. This UV-B flux rate corresponds to UV-B levels in sunlight measured in summer at midday in Rosario, Argentina. Control plants without UV-B were exposed for the same period of time under the same UV-B lamps but covered with polyester filters (100-μm clear polyester plastic; Tap Plastics), which absorb both UV-B and wavelengths lower than 280 nm (UV-B, 0.02 W m−2; and UV-A, 0.45 W m−2). Samples were collected immediately after irradiation and stored at −80°C. The experiments were repeated at least three times. For subcellular localization and PCD studies, plants were irradiated for 1 h with a UV-B intensity of 4 W m−2 and 0.65 W m−2 UV-A, while control seedlings were treated under the same lamps covered with polyester film (UV-B at 0.02 W m−2 and UV-A at 0.45 W m−2). The experiments were repeated at least three times. For analysis of the antioxidant metabolism, plants also were irradiated for 4 h with a UV-B intensity of 0.2 W m−2.The experiments were repeated at least three times.
Subcellular Localization Analysis
Arabidopsis transgenic plants (Pro35S:AtPDCD5-GFP; T2 lines) were grown on MS agar plates for 20 d as described above, irradiated or not with UV-B, and analyzed immediately after the treatments. To visualize the nuclei, leaves were incubated with 2 µg mL−1 DAPI (Sigma) in phosphate-buffered saline (PBS) buffer (10 mm sodium phosphate and 130 mm NaCl, pH 7.2) with 0.1% (v/v) Tween for 15 min. The fluorescence of GFP and DAPI was visualized by confocal laser scanning microscopy (Nikon C1) under water with a 40× objective. GFP and DAPI were excited using an argon laser at 488 nm and a UV laser at 395 nm, respectively. GFP emission was collected between 515 and 530 nm to avoid any interference of chlorophyll autofluorescence. To analyze the percentage of GFP/DAPI colocalization, each nucleus identified by DAPI fluorescence was examined for GFP fluorescence, and whenever GFP fluorescence was detected, the nucleus was scored as showing colocalization. Thus, the relative intensity of nuclear GFP fluorescence was not considered in these measurements. The percentage of GFP/DAPI was calculated by the relation between the number of nuclei showing GFP localization and the total number of nuclei identified by DAPI fluorescence. For each condition, more than 100 nuclei were analyzed (20 images) from five different plants using four independent T2 transgenic lines with comparable GFP fluorescence. The experiments were repeated three times.
Nuclei Isolation, Coimmunoprecipitation, and Pull-Down Assays
Nuclei were isolated from 21-DAS transgenic Pro35S:AtPDCD5-GFP and wild-type (Col-0) Arabidopsis plants essentially as described by Gallagher and Ellis (1982).
For coimmunoprecipitation studies, 250 μL of nuclear extract was combined with 6 μL of anti-GFP antibody (Abcam ab290) and rotated end over end at 4°C for 3 h. Protein A-agarose beads (20 μL; Invitrogen) were added, and the incubation was continued for 1 h. Immunocomplexes were washed four times with 1 mL of ice-cold wash buffer (100 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% [v/v] Triton X-100, and 1 mm EDTA), resuspended in 40 µL of SDS-PAGE sample buffer (50 mm Tris-HCl, pH 6.8, 2% [w/v] SDS, 10% [v/v] glycerol, 2 mm 2-mercaptoethanol, 12.5 mm EDTA, and 0.02% [w/v] Bromophenol Blue) heated to 70°C for 5 min, and analyzed by SDS-PAGE followed by immunodetection according to Burnette (1981). Commercial IgG fractions were used for the detection of HAM1/HAM2 (anti-TIP60; Abcam ab23886).
For GST pull-down assays, nuclear extracts were incubated with the purified recombinant GST-AtHAM2 fusion protein (5 μg) or alternatively with purified GST protein. The extracts were rotated end over end at 4°C for 2 h. Anti-GST antibody (Abcam [3G10/1B3] ab92) was added (5 μL) and rotated end over end at 4°C for 3 h. Protein A-agarose beads (20 μL; Invitrogen) were added, and the incubation was continued for 1 h. After four washes, beads were resuspended in SDS sample buffer and analyzed by SDS-PAGE followed by western blot using anti-GFP antibodies. Bound antibodies were visualized by goat anti-rabbit or anti-mouse IgG conjugated to alkaline phosphatase according to the manufacturer’s instructions (Bio-Rad). The molecular masses of the polypeptides were estimated from a plot of the log of the molecular masses of marker standards (Thermo) versus migration distance.
Identification of Insertional T-DNA Mutants and Genetic Crosses
The genotype of the insertion lines was determined using a PCR-based approach. Basically, genomic DNA was isolated from leaves using a modified cetyl-trimethyl-ammonium bromide method (Sambrook et al., 1989). PCR on genomic DNA was done using specific primers for the AtPDCD5 gene (F-AtPDCD5-sc and R-AtPDCD5-sc) and one primer that hybridizes with the left border of the T-DNA. Three combinations of primers were used to identify homozygous, heterozygous, and wild-type plants for AtPDCD5. Primer sequences are listed in Supplemental Table S1.
pdcd5 homozygous (pdcd5-2) single mutants were crossed with ham1ham2 RNAi knockdown transgenic plants, and its resultant F2 population was screened for pdcd5 ham1ham2 RNAi double mutants. The genotypes were determined by PCR on genomic DNA using combinations of specific primers for PDCD5 and one primer that hybridizes with the left border of the T-DNA, by cosegregation of the hygromycin resistance of the RNA interference plants, and by the decreased expression of HAM1 and HAM2 by qRT-PCR on complementary DNA (cDNA) using specific primers (Supplemental Table S1; Campi et al., 2012). Finally, the F3 population was used for the experiments described.
DNA Damage Analysis
CPD accumulation was measured as described previously (Lario et al., 2013). Samples were collected from control and UV-B-treated plants. UV-B treatments were performed under light conditions; after the treatments, plant samples (0.1 g) were collected, immediately immersed in liquid nitrogen, and stored at −80°C. DNA was extracted by a modified cetyl-trimetyl-ammonium bromide method, denatured in 0.3 m NaOH for 10 min, and 6-fold dot blotted onto a nylon membrane (Perkin-Elmer Life Sciences). The membrane was incubated for 2 h at 80°C and then blocked in Tris-buffered saline (TBS; 20 mm Tris-HCl, pH 7.6, and 137 mm NaCl) containing 5% (w/v) dried milk for 1 h at room temperature. The blot was then washed with TBS and incubated with antibodies specific to CPDs (TDM-2; Cosmo Bio) overnight at 4°C with agitation. Unbound antibodies were washed away, and secondary antibody (Bio-Rad) conjugated to alkaline phosphatase in TBS was added. The blots were then washed several times and subsequently developed with 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium. Quantification was achieved by densitometry of the dot blots using ImageQuant software version 5.2. DNA concentration was fluorometrically determined using the Qubit dsDNA Assay Kit (Invitrogen) and checked on a 1% (w/v) agarose gel stained with SYBR Safe after quantification.
AtPDCD5 Promoter:GUS Expression in Transgenic Arabidopsis Plants
To make the AtPDCD5 promoter:GUS construct (ProAtPDCD5:GUS), the 5′ flanking DNA of the AtPDCD5 coding region (from −894 to +15) was amplified by PCR with specific primers for the gene, with HindIII and BamHI restriction sites in the forward and reverse primers, respectively (Supplemental Table S1). The PCR fragment (909 bp) was cloned into the pGEM-T Easy vector and sequenced. Then, this construct was digested with HindIII and BamHI restriction enzymes, and the product was purified from the gel and cloned into pBI101 binary vector, generating the pBI101-ProAtPDCD5:GUS construct. This plasmid was transformed into Col-0 plants as described above. Transgenic plants were selected on solid MS medium containing kanamycin (50 mg L−1). Five independent T3 transgenic lines were used for histochemical analysis. Samples were stained with 5-bromo-4-chloro-3-indolyl-d-glucuronide at 37°C for 24 h, followed by washes with ethanol, and kept in 50% (v/v) ethanol and 5% (v/v) acetic acid before being photographed. The experiments were repeated at least three times using five independent lines with similar results.
Analysis of PCD
After growing plants for 5 d on vertically oriented MS plates, seedlings were irradiated for 1 h with a UV-B intensity of 4 W m−2. UV-B-irradiated and nonirradiated control seedlings were then incubated for 24 h in the growth chamber in the dark, and PCD was analyzed as described by Furukawa et al. (2010). Root tips were stained using a modified pseudo-Schiff PI staining protocol and visualized by confocal laser scanning microscopy (Nikon C1) under water with a 40× objective. The excitation wavelength for PI-stained samples was 488 nm, and emission was collected at 520 to 720 nm. Dead (intensely PI staining) cells in the vicinity of the quiescent center were counted and scored as dead cells per root. Three independent transgenic lines (PDCD5 OE-2, PDCD5 OE-11, and PDCD5 OE-12) were analyzed.
Generation of Arabidopsis Transgenic Plants
Full-length open reading frames for AtPDCD5 with and without the stop codon were amplified from cDNA obtained from leaf tissues of Col-0 Arabidopsis plants. For the generation of Pro35S:AtPDCD5 plants, primers F-BamHI-PDCD5-OE and R-SalI-PDCD5-OE with the BamHI and SalI restriction sites, respectively, were used for further cloning, while for Pro35S:AtPDCD5-GFP plants, primers F-Kpn-PDCD5-GFP and R-BamHI-PDCD5-GFP with the KpnI and BamHI restriction sites, respectively, were used (Supplemental Table S1). PCRs were performed using GoTaq (Promega) and Pfu (Invitrogen) polymerases (10:1) under the following conditions: 1× GoTaq buffer, 1.5 mm MgCl2, 0.5 µm of each primer, and 0.5 mm of each deoxyribonucleotide triphosphate in a 25-μL final volume. The amplified products were purified, cloned into pGEM-T Easy vector (Promega), and sequenced. The KpnI-BamHI and BamHI-SalI fragments were further subcloned into pCS052_GFP_pCHF3 (a modified version of pCHF3; GFP coding sequence without the start codon is inserted into SalI-PstI sites), generating Pro35S:AtPDCD5-GFP and Pro35S:AtPDCD5 constructs.
The Pro35S:AtPDCD5 and Pro35S:AtPDCD5-GFP constructs were transformed into the Agrobacterium tumefaciens strain GV3101 by electroporation, and the transformation of Col-0 Arabidopsis with the resulting bacteria was performed by the floral dip method (Clough and Bent, 1998). Transformed seedlings (T1) were identified by selection on solid MS medium containing kanamycin (50 mg L−1), and then the plants were transferred to soil. The presence of Pro35S:AtPDCD5 and Pro35S:AtRPL10s-GFP transgenes in transformed plants was analyzed by PCR on the genomic DNA using the following combinations of primers: F-35Sprom and a reverse primer specific for AtPDCD5 (R-AtPDCD5-1), and F-AtPDCD5-1 and R-GFP, respectively Supplemental Table S1; Supplemental Fig. S8).
Cloning, Expression, and Purification of AtHAM2 and GST
A full-length cDNA corresponding to AtHAM2 was amplified from Col-0 Arabidopsis leaves (21 DAS) by PCR using the primers F-AtHAM2-EcoRI and R-AtHAM2-XhoI harboring the EcoRI and XhoI restriction sites, respectively, for further cloning. PCR was performed with GoTaq (Promega) and Pfu (Invitrogen) polymerases (10:1) under the following conditions: 1× buffer, 2 mm MgCl2, 0.5 µm of each primer, 0.5 mm of each deoxyribonucleotide triphosphate, 0.5 units of enzyme, and cDNA from Arabidopsis leaves in a 25-µL final volume under the following cycling conditions: 2 min of denaturation at 94°C; 35 cycles at 94°C for 20 s, 62°C for 30 s, and 72°C for 140 s; followed by 7 min at 72°C. The PCR product was purified from the gel, cloned in pGEM-T Easy vector (Promega), and sequenced. The pGEM-T-AtHAM2 construct was digested with the corresponding restriction enzymes, EcoRI and XhoI, and the insert was purified and cloned in pGEX-4T-1 vector, generating the construct pGEX-AtHAM2.
BL21 (DE3) pLys cells were transformed with the construct pGEX-AtHAM2 and the empty vector pGEX. Cell cultures (200 mL of Luria-Bertani medium containing 30 mg L−1 kanamycin and 35 mg L−1 chloramphenicol) were grown at 37°C until mid-log phase (optical density at 600 nm of 0.5–0.6), and the recombinant N-GST-AtHAM2 and GST expression were achieved by induction with 0.5 mm isopropylthio-β-galactoside for 5 h at 30°C. For the purification of GST-AtHAM2 and GST proteins, cells were harvested by centrifugation at 3,000g for 20 min at 4°C. Pellet was resuspended in PBS binding buffer (50 mm sodium phosphate, pH 7.5, and 140 mm NaCl) containing 0.1% (v/v) Tween 20, 1 mm phenylmethylsulfonyl fluoride, and complete EDTA-free protease inhibitor cocktail (Thermo). Cells were disrupted by sonication and then centrifuged at 12,000g for 20 min at 4°C to obtain soluble cell extracts. The protein was bound to a glutathione-agarose resin (GE) by rocking at 4°C for 1 h, and then the resin was washed three times with 15 volumes of binding buffer. Elution was carried out by four sequential additions of 1 mL of elution buffer (50 mm Tris-HCl, pH 8, 10 mm reduced glutathione, and 1 mm dithiothreitol). Finally, the recombinant protein was desalted in PBS buffer with 5% glycerol by four cycles of concentration and dilution using Amicon Ultra-50 30K and 3K (Millipore) and stored at −80°C. Protein concentration was estimated both by comparison with dilution series of bovine serum albumin on a Coomassie Blue-stained SDS-PAGE gel and also using the Bradford reagent (Bio-Rad; Bradford, 1976).
Protein Extraction
After UV-B treatments, Arabidopsis leaves were homogenized in an extraction buffer containing 50 mm potassium phosphate, pH 7.5, 1 mm EDTA, 15% (v/v) glycerol, 5 mm ascorbate, 1 mm phenylmethylsulfonyl fluoride, 2% (w/v) polyvinyl polypyrrolidone, and 1× complete protease inhibitor cocktail (Thermo). After centrifugation for 20 min at 20,000g and at 4°C, the supernatants recovered were desalted by the method of Penefsky (1977) using 3-mL columns of Sephadex G-25, and the elutes were used for activity assays.
Enzyme Activity Assays
The activities of APX, POX, CAT, and GR were determined spectrophotometrically in 50 mm potassium phosphate, pH 7.5, and 1 mm EDTA at 30°C in a 0.5-mL volume. APX activity was determined by the decrease in A290 of an assay mixture containing 1 mm ascorbate (extinction coefficient, 2.8 mm−1 cm−1) and 0.1 mm H2O2 (Gupta et al., 1993). Measurements were corrected for ascorbate autooxidation in the presence of 0.1 mm H2O2. GR activity was determined by following the decrease in A340 (extinction coefficient, 6.22 mm−1 cm−1) due to NADPH oxidation (Gupta et al., 1993) with 100 μm NADPH and 1 mm oxidized glutathione. CAT was determined by the decrease in A240 of an assay mixture containing 10 mm H2O2 (extinction coefficient, 39.4 mm−1 cm−1) as described previously (Kang et al., 1999). Peroxidase activity was determined with guaiacol as an electron donor by measuring the oxidation of guaiacol to tetraguiacol at 470 nm (extinction coefficient, 25.5 mm−1 cm–1) with 0.1 mm H2O2 and 17 mm guaiacol (Chance and Maehly, 1955). SOD activity was determined on nondenaturing polyacrylamide gels (8% acrylamide and 3% bis-acrylamide) as described by Beauchamp and Fridovich (1971). SOD isoforms were quantified by densitometric analysis. The gels were scanned, and the area of the bands was obtained by integration using ImageQuant software version 5.2. Total protein content was determined using Bradford reagent (Bio-Rad).
Determination of Antioxidant Metabolites
Arabidopsis leaves (0.1 g) were homogenized in 1 mL of 80% (v/v) ethanol. The homogenates were centrifuged for 10 min at 20,000g at 4°C. The supernatants were used for FRAP assays and polyphenol and flavonoid measurements.
For the FRAP assay, the reaction mix consisted of 0.25 m sodium acetate, 8.3 mm 2,4,6-Tris-(2 pyridyl)-S-triazin, and 16.7 mm FeCl3 (Benzie and Strain, 1996). A calibration curve was performed using 250 µm of a soluble analog of vitamin E, 6 hydroxy-2,5,7,8-tetramethylchrommane-2-carboxylic acid (Trolox; Sigma-Aldrich), in 80% (v/v) ethanol. Dilutions were performed and processed in parallel with samples. Then, 100 µL of the reaction mix was mixed with 100 µL of the samples on a microplate and incubated at 4°C for 20 min. Absorbance was measured at 600 nm.
Polyphenol and flavonoid determination was performed as described (Zhang et al., 2006). Samples (10 µL) were mixed with the Folin-Ciocalteu reagent (1×, 50 µL) on a microplate and incubated 5 min at room temperature. A saturated Na2CO3 solution was then added (40 µL) and incubated for 1 h at room temperature in the dark. For the calibration curves, gallic acid and quercetin were used for the determination of polyphenol and flavonoid contents, respectively. Polyphenols and flavonoids were quantified by absorbance measurements at 725 and 312 nm, respectively.
For the determination of ascorbate and glutathione, Arabidopsis leaves (0.2 g) were homogenized in 1 mL of 6% (w/v) meta-phosphoric acid. Homogenates were centrifuged at 20,000g for 15 min at 4°C. Supernatants (100 µL) were combined with 300 µL of 2 mm KCl, pH 2.5, and these mixtures were used for the determination of reduced ascorbate and glutathione. To quantify the total ascorbate and glutathione contents, 100 µL of supernatants was supplemented with dithiothreitol at a final concentration of 50 mm and incubated for 10 min at room temperature in the dark. The reduction reaction was finished by adding 250 µL of 2 mm KCl, pH 2.5. The measurements were performed three times using four independent biological samples. Ascorbate and glutathione contents were determined by HPLC using a Phenomenex LUNA C18 column (150 mm × 4.6 mm, 5 μm; LiChroSpher [Alltech] and LC-10ADVP [Shimadzu]). Absorbance was detected at 242 nm for ascorbate and at 196 nm for glutathione using a UV detector (SPD-M10Avp diode array detector; Shimadzu). Data were collected and analyzed using ClassVP 5.0 Shimadzu software.
Electrolyte Leakage and Chlorophyll Extraction
Three-week-old plants were exposed to 4 h of UV-B radiation. Leaves 4 and 5 were harvested and incubated in 1 mL of distilled water in a tube. Samples were shaken for 30 min, and electrolyte leakage from the leaves was measured using a conductivity meter (Twin Cond B.173; Horiba). Samples were boiled for 20 min, and total conductivity was determined. Data were expressed as (initial conductivity/total conductivity) × 100. Total chlorophylls were determined by standard procedures (Wintermans and de Mots, 1965).
ROS Detection and Image Analysis
After UV-B exposure, leaves 4 and 5 were harvested and incubated for 1 h with 10 μm H2DCFDA in the dark at room temperature. After incubation, leaves were washed with phosphate buffer solution and immediately visualized using a Nikon fluorescence microscope (Eclipse E800). ROS formation was quantified using ImageJ software. Experiments were carried out in triplicate using three plants, obtaining similar results.
Rosette Area Quantification
Plants were grown at 22°C under a 16-h/8-h light/dark regime. Every 3 to 4 d, photographs were taken, and the rosette area of each plant (cm2) was measured using Image Pro Plus 5.0 software.
qRT-PCR
Tissues from three independent biological replicates were frozen in liquid nitrogen and stored at −80°C. Total RNA was isolated from 100 mg of tissue using TRIzol reagent (Invitrogen) and treated with DNase (Promega). RNA was converted into first-strand cDNA using SuperScript II reverse transcriptase (Invitrogen) with oligo(dT) as a primer. The resultant cDNA was used as a template for quantitative PCR amplification in a MiniOPTICON2 apparatus (Bio-Rad) using the intercalation dye SYBR Green I (Invitrogen) as a fluorescent reporter and Platinum Taq Polymerase (Invitrogen). Primers were designed to generate unique 150- to 250-bp fragments using PRIMER3 software (Rozen and Skaletsky, 2000). Three replicates were performed for each sample plus a negative control (reaction without reverse transcriptase). To normalize the data of UV treatments, primers for a CALCIUM-DEPENDENT PROTEIN KINASE3 (CDPK3) transcript were used, while for tissue/stage-dependent expression studies and analysis of transgenic plants, primers for POLYUBIQUITIN10 (UBQ10) were used (Supplemental Table S1). Primers used for AtPDCD5 alternative splicing variants I (F-AtPDCD5-1 and R-AtPDCD5-1), II (F-AtPDCD5-2 and R-AtPDCD5-1), and III (F-AtPDCD5-1 and R-AtPDCD5-2) are listed in Supplemental Table S1. Amplification conditions were as follows: 2 min of denaturation at 94°C; 40 to 45 cycles at 94°C for 15 s, 57°C for 20 s, and 72°C for 20 s; followed by 10 min at 72°C. Melting curves for each PCR were determined by measuring the decrease of fluorescence with increasing temperature (from 65°C to 98°C). To confirm the size of the PCR products and to check that they corresponded to a unique and expected product, the final products were separated on a 2% (w/v) agarose gel. The PCR products were purified from the gel and sequenced to verify their identities.
Statistical Analysis
Data presented were analyzed using one-way and two-way ANOVA. Minimum significant differences were calculated by the Bonferroni, Tukey, Dunett, and Duncan tests (P < 0.05) using Sigma Plot 12.0 software. In some cases, data were compared using Student’s t test (n = 40 biological replicates in a single experiment; P < 0.05), and significant differences are indicated in the figures with asterisks.
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: PDCD5, At1g29850; HAM1, At5g64610; HAM2, At5g09740; AtUBQ10, At4g05320; CDPK3, At4g23650; UVR2, At1g12370; UVR7, At3g05210; and UVH6, At1g03190.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. PDCD5 proteins and different PDCD5 mRNA spliced forms from Arabidopsis.
Supplemental Figure S2. Representative scheme of the double-stranded DNA-binding domain conserved in PDCD5 proteins from eukaryotic species.
Supplemental Figure S3. Alignment of the AtPDCD5 genomic sequence with three AtPDCD5 mRNA spliced forms.
Supplemental Figure S4. AtPDCD5 spliced forms I, II, and III expression analysis in Arabidopsis.
Supplemental Figure S5. Characterization of pdcd5 mutant lines.
Supplemental Figure S6. Analysis of antioxidant enzyme activities in wild-type Col-0 and pdcd5 mutant plants under control conditions and after UV-B treatment.
Supplemental Figure S7. Recombinant expression of GST-AtHAM2 and GST proteins.
Supplemental Figure S8. Analysis of Arabidopsis transgenic plants expressing AtPDCD5-GFP and AtPDCD5.
Supplemental Figure S9. Images of root tips from wild-type Col-0 and Ws, pdcd5-2, ham1ham2 RNAi, and ham1ham2 RNAi/pdcd5 mutants, and PDCD5-overexpressing plants under control conditions without UV-B.
Supplemental Figure S10. Phenotypic analysis of pdcd5 mutants and transgenic PDCD5 OE plants.
Supplemental Figure S11. Relative expression levels of AtPDCD5 in different tissues and in leaves from plants infected with Pseudomonas syringae and Phytophthora infestans.
Supplemental Table S1. Primers used for cloning, qRT-PCR, and screening.
Supplementary Material
Acknowledgments
We thank Han Asard and Danny Huybrecht for assistance with analyses of antioxidant capacity and Els Prinsen for assistance with UV-B experiments.
Glossary
- CPD
cyclobutane pyrimidine dimer
- PCD
programmed cell death
- qRT
quantitative reverse transcription
- T-DNA
transfer DNA
- ROS
reactive oxygen species
- FRAP
ferric reducing ability of plasma
- Col-0
Columbia-0
- MS
Murashige and Skoog
- PI
propidium iodide
- H2O2
hydrogen peroxide
- Ws
Wassilewskija
- DAPI
4′,6-diamino-phenylindole
- PBS
phosphate-buffered saline
- cDNA
complementary DNA
- TBS
Tris-buffered saline
- H2DCFDA
2′7′-dichlorofluorescein diacetate
Footnotes
This work was supported by FONCyT (grant nos. PICT 2012–00267 and PICT 2013–268 to P.C.) and an Erasmus Mundus EADIC II Ph.D. grant to R.C.
Articles can be viewed without a subscription.
References
- Attia K, Ke-Gui L, Chun W, Guang-Ming H, Wei S, Jin-Shui Y (2005) Overexpression of the OsPDCD5 gene induces programmed cell death in rice. J Integr Plant Biol 47: 1115–1122 [Google Scholar]
- Beauchamp C, Fridovich I (1971) Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem 44: 276–287 [DOI] [PubMed] [Google Scholar]
- Benzie IF, Strain JJ (1996) The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: the FRAP assay. Anal Biochem 239: 70–76 [DOI] [PubMed] [Google Scholar]
- Bock FJ, Tanzer MC, Haschka MD, Krumschnabel G, Sohm B, Goetsch K, Kofler R, Villunger A (2015) The p53 binding protein PDCD5 is not rate-limiting in DNA damage induced cell death. Sci Rep 5: 11268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Britt AB. (1996) DNA damage and repair in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 75–100 [DOI] [PubMed] [Google Scholar]
- Burnette WN. (1981) “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem 112: 195–203 [DOI] [PubMed] [Google Scholar]
- Campi M, D’Andrea L, Emiliani J, Casati P (2012) Participation of chromatin-remodeling proteins in the repair of ultraviolet-B-damaged DNA. Plant Physiol 158: 981–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Campi M, Chu F, Suzuki N, Maltby D, Guan S, Burlingame AL, Walbot V (2008) Histone acetylation and chromatin remodeling are required for UV-B-dependent transcriptional activation of regulated genes in maize. Plant Cell 20: 827–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance B, Maehly AC (1955) Assay of catalase and peroxidases. Methods Enzymol 2: 764–775 [Google Scholar]
- Chen LN, Wang Y, Ma DL, Chen YY (2006) Short interfering RNA against the PDCD5 attenuates cell apoptosis and caspase-3 activity induced by Bax overexpression. Apoptosis 11: 101–111 [DOI] [PubMed] [Google Scholar]
- Chen Y, Sun R, Han W, Zhang Y, Song Q, Di C, Ma D (2001) Nuclear translocation of PDCD5 (TFAR19): an early signal for apoptosis? FEBS Lett 509: 191–196 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Creissen G, Firmin J, Fryer M, Kular B, Leyland N, Reynolds H, Pastori G, Wellburn F, Baker N, Wellburn A, et al. (1999) Elevated glutathione biosynthetic capacity in the chloroplasts of transgenic tobacco plants paradoxically causes increased oxidative stress. Plant Cell 11: 1277–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MJ, Hays JB (2011) Cooperative responses of DNA-damage-activated protein kinases ATR and ATM and DNA translesion polymerases to replication-blocking DNA damage in a stem-cell niche. DNA Repair (Amst) 10: 1272–1281 [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F. (2008) Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer 8: 512–522 [DOI] [PubMed] [Google Scholar]
- Falcone Ferreyra ML, Casadevall R, Luciani MD, Pezza A, Casati P (2013) New evidence for differential roles of L10 ribosomal proteins from Arabidopsis. Plant Physiol 163: 378–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M, Van Hautegem T, Van Durme M, Olvera-Carrillo Y, Huysmans M, Karimi M, Lippens S, Guérin CJ, Krebs M, Schumacher K, et al. (2014) Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Curr Biol 24: 931–940 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17: 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC [Google Scholar]
- Furukawa T, Curtis MJ, Tominey CM, Duong YH, Wilcox BWL, Aggoune D, Hays JB, Britt AB (2010) A shared DNA-damage-response pathway for induction of stem-cell death by UVB and by gamma irradiation. DNA Repair (Amst) 9: 940–948 [DOI] [PubMed] [Google Scholar]
- Gadjev I, Stone JM, Gechev TS (2008) Programmed cell death in plants: new insights into redox regulation and the role of hydrogen peroxide. Int Rev Cell Mol Biol 270: 87–144 [DOI] [PubMed] [Google Scholar]
- Gallagher TF, Ellis RJ (1982) Light-stimulated transcription of genes for two chloroplast polypeptides in isolated pea leaf nuclei. EMBO J 1: 1493–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González Besteiro MA, Ulm R (2013) ATR and MKP1 play distinct roles in response to UV-B stress in Arabidopsis. Plant J 73: 1034–1043 [DOI] [PubMed] [Google Scholar]
- Gupta AS, Webb RP, Holaday AS, Allen RD (1993) Overexpression of superoxide dismutase protects plants from oxidative stress: induction of ascorbate peroxidase in superoxide dismutase overexpressing plants. Plant Physiol 103: 1067–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays JB. (2011) New analytical methods for genetic dissection of biological responses to DNA lesions. DNA Repair (Amst) 10: 526–535 [DOI] [PubMed] [Google Scholar]
- Hooper CM, Tanz SK, Castleden IR, Vacher MA, Small ID, Millar AH (2014) SUBAcon: a consensus algorithm for unifying the subcellular localization data of the Arabidopsis proteome. Bioinformatics 30: 3356–3364 [DOI] [PubMed] [Google Scholar]
- Kang KS, Lim CJ, Han TJ, Kim JC, Jin CD (1999) Changes in the isozyme composition of antioxidant enzymes in response to aminotriazole in leaves of Arabidopsis thaliana. J Plant Biol 42: 187–193 [Google Scholar]
- Lario LD, Ramirez-Parra E, Gutierrez C, Spampinato CP, Casati P (2013) ANTI-SILENCING FUNCTION1 proteins are involved in ultraviolet-induced DNA damage repair and are cell cycle regulated by E2F transcription factors in Arabidopsis. Plant Physiol 162: 1164–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang Y, Zhang Y, Song Q, Di C, Chen G, Tang J, Ma D (1999) TFAR19, a novel apoptosis-related gene cloned from human leukemia cell line TF-1, could enhance apoptosis of some tumor cells induced by growth factor withdrawal. Biochem Biophys Res Commun 254: 203–210 [DOI] [PubMed] [Google Scholar]
- Lord CE, Gunawardena AH (2012) Programmed cell death in C. elegans, mammals and plants. Eur J Cell Biol 91: 603–613 [DOI] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z (2006) Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol 8: 91–99 [DOI] [PubMed] [Google Scholar]
- Olvera-Carrillo Y, Van Bel M, Van Hautegem T, Fendrych M, Huysmans M, Simaskova M, van Durme M, Buscaill P, Rivas S, Coll N, et al. (2015) A conserved core of programmed cell death indicator genes discriminates developmentally and environmentally induced programmed cell death in plants. Plant Physiol 169: 2684–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- op den Camp RGL, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Göbel C, Feussner I, et al. (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penefsky HS. (1977) Reversible binding of Pi by beef heart mitochondrial adenosine triphosphatase. J Biol Chem 252: 2891–2899 [PubMed] [Google Scholar]
- Reed SM, Hagen J, Tompkins VS, Thies K, Quelle FW, Quelle DE (2014) Nuclear interactor of ARF and Mdm2 regulates multiple pathways to activate p53. Cell Cycle 13: 1288–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich T, Allen RL, Wyllie AH (2000) Defying death after DNA damage. Nature 407: 777–783 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In Krawetz SA, Misener S, eds, Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 365–386 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sperandio S, de Belle I, Bredesen DE (2000) An alternative, nonapoptotic form of programmed cell death. Proc Natl Acad Sci USA 97: 14376–14381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Wu J, Wei C, Li K, He G, Attla K, Qian X, Yang J (2006) Interaction between programmed cell death 5 and calcineurin B-like interacting protein kinase 23 in Oryza sativa. Plant Sci 170: 1150–1155 [Google Scholar]
- Sun F, Qi W, Qian X, Wang Q, Yang M, Dong X, Yang J (2012) Investigating the role of OsPDCD5, a homolog of the mammalian PDCD5, in programmed cell death by inducible expression in rice. Plant Mol Biol Rep 30: 87–98 [Google Scholar]
- Sun Y, Jiang X, Price BD (2010) Tip60: connecting chromatin to DNA damage signaling. Cell Cycle 9: 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn WG, Woltering EJ (2005) Many ways to exit? Cell death categories in plants. Trends Plant Sci 10: 117–122 [DOI] [PubMed] [Google Scholar]
- Varnier AL, Mazeyrat-Gourbeyre F, Sangwan RS, Clément C (2005) Programmed cell death progressively models the development of anther sporophytic tissues from the tapetum and is triggered in pollen grains during maturation. J Struct Biol 152: 118–128 [DOI] [PubMed] [Google Scholar]
- Vranová E, Atichartpongkul S, Villarroel R, Van Montagu M, Inzé D, Van Camp W (2002) Comprehensive analysis of gene expression in Nicotiana tabacum leaves acclimated to oxidative stress. Proc Natl Acad Sci USA 99: 10870–10875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li X, Wang L, Ding P, Zhang Y, Han W, Ma D (2004) An alternative form of paraptosis-like cell death, triggered by TAJ/TROY and enhanced by PDCD5 overexpression. J Cell Sci 117: 1525–1532 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zha X, Zhang S, Qian X, Dong X, Sun F, Yang J (2010) Down-regulation of the OsPDCD5 gene induced photoperiod-sensitive male sterility in rice. Plant Sci 178: 221–228 [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermans JFGH, de Mots A (1965) Spectrophotometric characteristics of chlorophylls a and b and their pheophytins in ethanol. Biochim Biophys Acta 109: 448–453 [DOI] [PubMed] [Google Scholar]
- Wyllie AH, Golstein P (2001) More than one way to go. Proc Natl Acad Sci USA 98: 11–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Werner BL, Christensen EM, Oliver DJ (2001) The biological functions of glutathione revisited in Arabidopsis transgenic plants with altered glutathione levels. Plant Physiol 126: 564–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Chen Y, Song Q, Xu D, Wang Y, Ma D (2009) PDCD5 interacts with Tip60 and functions as a cooperator in acetyltransferase activity and DNA damage-induced apoptosis. Neoplasia 11: 345–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Sun F, Wang S, Qi W, Wang Q, Dong X, Yang J, Luo X (2013) Down-regulation of OsPDCD5, a homolog of the mammalian PDCD5, increases rice tolerance to salt stress. Mol Breed 31: 333–346 [Google Scholar]
- Yoon MJ, Lee AR, Jeong SA, Kim YS, Kim JY, Kwon YJ, Choi KS (2014) Release of Ca2+ from the endoplasmic reticulum and its subsequent influx into mitochondria trigger celastrol-induced paraptosis in cancer cells. Oncotarget 5: 6816–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhang J, Shen J, Silva A, Denis DA, Barrow CJ (2006) A simple 96-well microplate method for estimation of total polyphenol content in seaweeds. J Appl Phycol 18: 445–450 [Google Scholar]
- Zhang XP, Liu F, Cheng Z, Wang W (2009) Cell fate decision mediated by p53 pulses. Proc Natl Acad Sci USA 106: 12245–12250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuge C, Chang Y, Li Y, Chen Y, Lei J (2011) PDCD5-regulated cell fate decision after ultraviolet-irradiation-induced DNA damage. Biophys. J. 101: 2582–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.