A NADPH dehydrogenase is critical for balancing electron flow between energy-dissipating and energy-generating pathways during the acclimation to photoautotrophic nitrogen deprivation.
Abstract
When photosynthetic organisms are deprived of nitrogen (N), the capacity to grow and assimilate carbon becomes limited, causing a decrease in the productive use of absorbed light energy and likely a rise in the cellular reduction state. Although there is a scarcity of N in many terrestrial and aquatic environments, a mechanistic understanding of how photosynthesis adjusts to low-N conditions and the enzymes/activities integral to these adjustments have not been described. In this work, we use biochemical and biophysical analyses of photoautotrophically grown wild-type and mutant strains of Chlamydomonas reinhardtii to determine the integration of electron transport pathways critical for maintaining active photosynthetic complexes even after exposure of cells to N deprivation for 3 d. Key to acclimation is the type II NADPH dehydrogenase, NDA2, which drives cyclic electron flow (CEF), chlororespiration, and the generation of an H+ gradient across the thylakoid membranes. N deprivation elicited a doubling of the rate of NDA2-dependent CEF, with little contribution from PGR5/PGRL1-dependent CEF. The H+ gradient generated by CEF is essential to sustain nonphotochemical quenching, while an increase in the level of reduced plastoquinone would promote a state transition; both are necessary to down-regulate photosystem II activity. Moreover, stimulation of NDA2-dependent chlororespiration affords additional relief from the elevated reduction state associated with N deprivation through plastid terminal oxidase-dependent water synthesis. Overall, rerouting electrons through the NDA2 catalytic hub in response to photoautotrophic N deprivation sustains cell viability while promoting the dissipation of excess excitation energy through quenching and chlororespiratory processes.
Oxygenic photosynthesis involves the conversion of light energy into chemical bond energy by plants, green algae, and cyanobacteria and the use of that energy to fix CO2. The photosynthetic electron transport system, located in thylakoid membranes, involves several major protein complexes: PSII (water-plastoquinone oxidoreductase), cytochrome b6f (cyt b6f; plastoquinone-plastocyanin oxidoreductase), PSI (plastocyanin-ferredoxin oxidoreductase), and the ATP synthase (CFoCF1). Light energy absorbed by the photosynthetic apparatus is used to establish both linear electron flow (LEF) and cyclic electron flow (CEF), which drive the production of ATP and NADPH, the chemical products of the light reactions needed for CO2 fixation in the Calvin-Benson-Bassham (CBB) cycle.
With the absorption of light energy by pigment-protein complexes associated with PSII, energy is funneled into unique chlorophyll (Chl) molecules located in the PSII reaction center (RC), where it can elicit a charge separation that generates a large enough oxidizing potential to extract electrons from water. In LEF, electrons from PSII RCs are transferred sequentially along a set of electron carriers, initially reducing the plastoquinone (PQ) pool, then the cyt b6f complex, and subsequently the lumenal electron carrier plastocyanin (PC). Light energy absorbed by PSI excites a special pair of Chl molecules (P700), causing a charge separation that generates the most negative redox potential in nature (Nelson and Yocum, 2006). The energized electron, which is replaced by electrons from PC, is sequentially transferred to ferredoxin and ferredoxin NADP+ reductase, generating reductant in the form of NADPH.
Electron transport from water to NADPH in LEF is accompanied by the transport of H+ into the thylakoid lumen. For each water molecule oxidized, two H+ are released in the thylakoid lumen. In addition, H+ are moved into the lumen by the transfer of electrons through cyt b6f (Q cycle). H+ accumulation in the thylakoid lumen dramatically alters the lumenal pH, and the transmembrane H+ gradient (ΔpH) together with the transmembrane ion gradient constitute the proton motive force (pmf), which drives ATP formation by ATP synthase (Mitchell, 1961, 1966, 2011). This pmf also promotes other cellular processes, including the dissipation of excess absorbed excitation energy as heat in a photoprotective process (see below; Li et al., 2009; Erickson et al., 2015). The NADPH and ATP molecules generated by LEF and CEF fuel the synthesis of reduced carbon backbones (in the CBB cycle) used in the production of many cellular metabolites and fixed carbon storage polymers.
A basic role for CEF is to increase the ATP-NADPH ratio, which can satisfy the energy requirements of the cell and augment the synthesis of ATP by LEF, which is required to sustain CO2 fixation by the CBB cycle (Allen, 2003; Kramer et al., 2004; Iwai et al., 2010; Alric, 2014). There are two distinct CEF pathways identified in plants and algae. In both pathways, electrons flow from the PQ pool through cyt b6f to reduce the oxidized form of P700 (P700+). In one CEF pathway, electrons are transferred back to the PQ pool prior to the formation of NADPH. This route involves the proteins PGR5 and PGRL1 (DalCorso et al., 2008; Tolleter et al., 2011; Hertle et al., 2013) and is termed PGR5/L1-dependent CEF. A second route for CEF includes an NADPH dehydrogenase that oxidizes NADPH (product of LEF) to NADP+, simultaneously reducing the PQ (Allen, 2003; Kramer et al., 2004; Rumeau et al., 2007). The reduced PQ pool is then oxidized by cyt b6f, causing H+ translocation into the thylakoid lumen, followed by the transfer of electrons to P700+ via PC. In the green alga Chlamydomonas reinhardtii, this second route for CEF involves a type II NADPH dehydrogenase (NDA2; Jans et al., 2008; Desplats et al., 2009).
Oxygenic photosynthetic organisms have inhabited the planet for approximately 3 billion years and have developed numerous strategies to acclimate to environmental fluctuations. These acclimation processes confer flexibility to the photosynthetic machinery, allowing it to adjust to changes in conditions that impact the metabolic/energetic state of the organism and, most importantly, the formation of reactive oxygen species that may damage the photosynthetic apparatus and other cellular components (Li et al., 2009). Several ways in which the photosynthetic apparatus adjusts to environmental fluctuations have been established. A well-studied acclimation process, nonphotochemical quenching (NPQ), reduces the excitation pressure on PSII when oxidized downstream electron acceptors are not available (Eberhard et al., 2008; Li et al., 2009; Erickson et al., 2015). Several processes constitute NPQ, as follows. (1) qT, which involves the physical movement of light-harvesting complexes (LHCs) from one photosystem to another (this is also designated state transitions; Rochaix, 2014). (2) qE, which involves thermal dissipation of the excitation energy. This energy-dependent process requires an elevated ΔpH and involves an LHC-like protein, LHCSR3 (in C. reinhardtii) or PSBS (in plants), as well as the accumulation of specific xanthophylls (mainly lutein in C. reinhardtii and zeaxanthin in plants; Niyogi et al., 1997b; Li et al., 2000, 2004; Peers et al., 2009). (3) qZ, which is energy independent and involves the accumulation of zeaxanthin (Dall’Osto et al., 2005; Nilkens et al., 2010). (4) qI, which promotes quenching following physical damage to PSII core subunits (Aro et al., 1993). Additional mechanisms that can impact LEF and CEF are the synthesis and degradation of pigment molecules, changes in levels of RC and antenna complexes, and the control of electron distribution between LEF and CEF as the energetic demands of the cell change (Allen, 2003; Kramer et al., 2004). In addition, electrons can be consumed by mitochondrial and chlororespiratory activities (Bennoun, 1982; Peltier and Cournac, 2002; Johnson et al., 2014; Bailleul et al., 2015). The latter mainly involves the plastid terminal oxidase PTOX2, which catalyzes the oxidation of the PQ pool and the reduction of oxygen and H+ to form water molecules (Houille-Vernes et al., 2011; Nawrocki et al., 2015).
Photosynthetic processes also must be modulated as organisms experience changes in the levels of available nutrients (Grossman and Takahashi, 2001). The macronutrient nitrogen (N), which represents 3% to 5% of the dry weight of photosynthetic organisms, is required to synthesize many biological molecules (e.g. amino acids, nucleic acids, and various metabolites) and also participates in posttranslational modifications of proteins (e.g. S-nitrosylation; Romero-Puertas et al., 2013). Importantly, N is highly abundant in chloroplasts in the form of DNA, ribosomes, Chl, and polypeptides (e.g. Rubisco and LHCs; Evans, 1989; Raven, 2013). Furthermore, there is a strong integration between N and carbon assimilation. During N limitation under photoautotrophic conditions, the inability of the organism to synthesize amino acids and other N-containing molecules necessary for cell growth and division can feed back to inhibit both carbon fixation by the CBB cycle and electron transport processes and also can negatively impact the expression of genes encoding key CBB cycle enzymes (Terashima and Evans, 1988; Huppe and Turpin, 1994; Nunes-Nesi et al., 2010).
C. reinhardtii is a well-established model organism in which to study photosynthesis and acclimation processes, including acclimation to nutrient limitation (Wykoff et al., 1998; Grossman and Takahashi, 2001; Moseley et al., 2006; Grossman et al., 2009; Terauchi et al., 2010; Aksoy et al., 2013). This unicellular alga grows rapidly as a photoheterotroph (on fixed carbon in the light) or as a heterotroph (on fixed carbon in the dark), has completely sequenced nuclear, chloroplast, and mitochondrial genomes, can be used for classical genetic analyses, and is haploid, which makes some aspects of molecular manipulation (e.g. the generation of knockout mutants) easier (Merchant et al., 2007; Blaby et al., 2014). In the past few years, there have been many studies on the ways in which C. reinhardtii responds to N deprivation (Bulté and Wollman, 1992; Blaby et al., 2013; Goodenough et al., 2014; Schmollinger et al., 2014; Wei et al., 2015; Juergens et al., 2015). Cells deprived of N under photoheterotrophic conditions (i.e. acetate as an external carbon source) minimize the use of N (referred to as N sparing) and induce mechanisms associated with scavenging N from both external and internal pools, all of which eventually lead to proteome modifications and an elevated carbon-N ratio (Schmollinger et al., 2014). Acclimation under photoheterotrophic conditions also causes dramatic modifications of cellular metabolism and energetics: photosynthesis is down-regulated at multiple levels, with a portion of its N content recycled (mainly Chl and polypeptides of the photosynthetic apparatus), while there is enhanced accumulation of mitochondrial complexes leading to increased respiratory activity (Schmollinger et al., 2014; Juergens et al., 2015). Additionally, while fixed carbon cannot be used for growth in the absence of N, it may be stored as starch and triacylglycerol (Work et al., 2010; Siaut et al., 2011; Davey et al., 2014; Goodenough et al., 2014).
In contrast to the acclimation of photoheterotrophically grown C. reinhardtii to N deprivation, little is known about how the photosynthetic machinery in this alga adjusts in response to N deprivation under photoautotrophic conditions, when the cells absolutely require photosynthetic energy generation for maintenance. Specifically, we sought to understand how photosynthesis adjusts to metabolic restrictions that slow down the CBB cycle, which in turn could cause the accumulation of photoreductant, particularly NADPH, as the demand for electrons declines (Peltier and Schmidt, 1991; Rumeau et al., 2007). Based on analyses of mutants and the use of spectroscopic and fluorescence measurements, we established a critical role for NDA2 in the acclimation of C. reinhardtii to N deprivation under photoautotrophic conditions, including (1) an augmented capacity for alternative routes of electron utilization (which decrease the NADPH-NADP+ ratio) based on increased NDA2-dependent CEF and chlororespiration, and (2) elevated qE, which relies on the H+ gradient generated by NDA2-dependent CEF.
RESULTS
Photoautotrophically Grown Cells Retain Their Capacity to Harvest Light Energy during N Deprivation
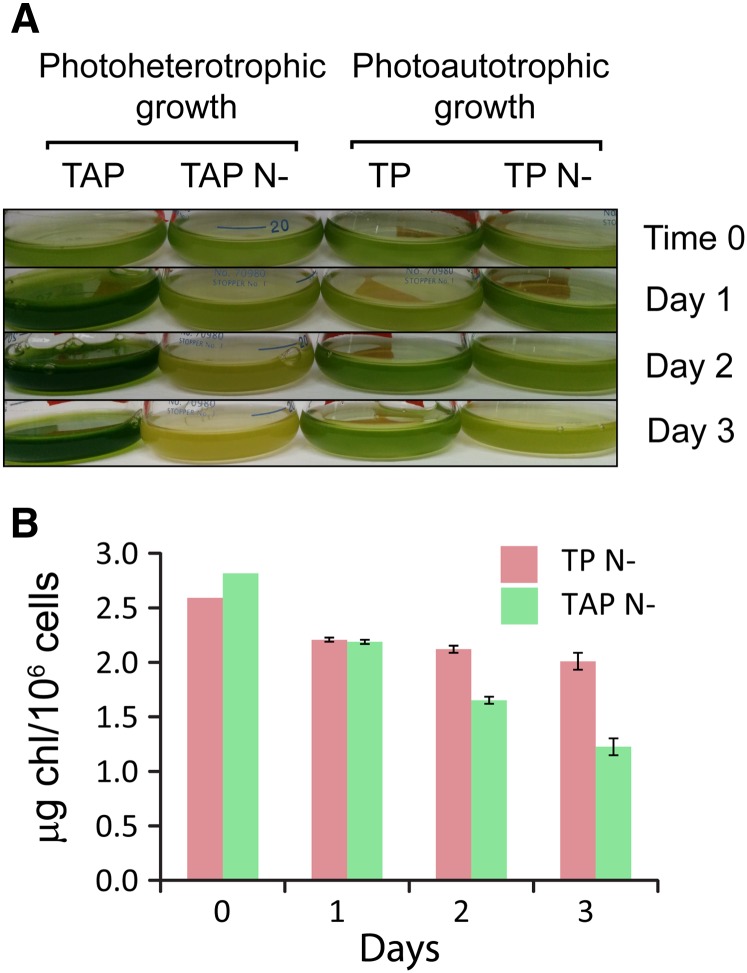
In order to understand photosynthesis-related acclimation mechanism(s) in C. reinhardtii during N deprivation, we first analyzed the Chl content of cells grown under photoautotrophic conditions and compared it with that of cells grown photoheterotrophically (Fig. 1). While photosynthesis is essential under the former growth conditions (where all cellular energy would be derived from photosynthesis), it is not essential under the latter conditions, since photoheterotrophic medium includes a fixed carbon source (acetate). Following 2 d of N deprivation, photoheterotrophically grown (TAP N−) wild-type [D66+ (CC-4425)] cells lost their dark-green pigmentation compared with cells deprived of N for 2 d during photoautotrophic growth (Fig. 1A). Following 3 d of N deprivation, cells grown in TAP N− became yellowish while cells grown in TP N− retained their light green pigmentation (Fig. 1A). Hence, while TP N− cells retain most of their Chl, the level of Chl declines dramatically in TAP N−-grown cells (which is likely associated with a decrease in photosynthetic complexes, as shown by others; Bulté and Wollman, 1992; Work et al., 2010; Schmollinger et al., 2014; Wei et al., 2014), and the cells appear yellow. The difference in this pigmentation phenotype became more pronounced following 7 d of N deprivation (Supplemental Fig. S1). This was confirmed by the quantification of Chl per cell (Fig. 1B). After 1 d of N deprivation, the Chl per cell of both photoautotrophically and photoheterotrophically grown cells decreased to approximately the same extent (15% and 23%, respectively); however, following 3 d of deprivation, cells grown photoautotrophically retained most of their Chl (approximately 80% of their initial Chl per cell), while in photoheterotrophically grown cells, the Chl per cell declined to approximately 50% of the level in nutrient-replete cells.
Figure 1.
Cells deprived of N under photoautotrophic conditions maintain most of their pigmentation. A, Visual representation of cells grown on Tris-acetate-phosphate (TAP) medium, N-deplete Tris-acetate-phosphate medium (TAP N−), Tris-phosphate (TP) medium, and N-deplete Tris-phosphate medium (TP N−) for 0 to 3 d. B, Chl levels (µg Chl 10−6 cells) after growth on TAP N− and TP N− for 0, 1, 2, and 3 d.
N-Deprived Cells Do Not Degrade Their Photosynthetic Complexes
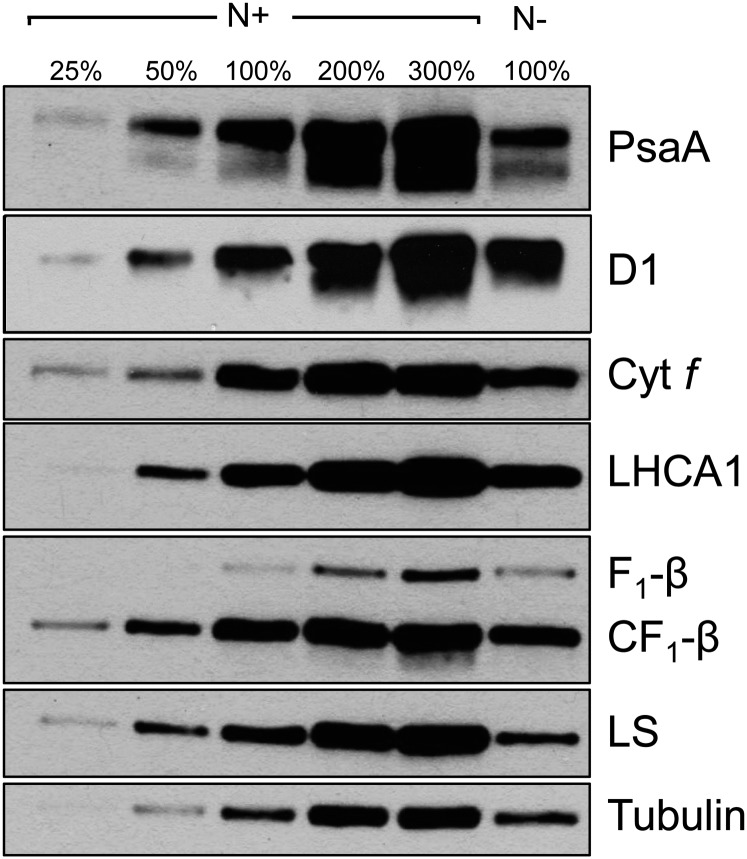
To explore whether the photosynthetic machinery was retained during photoautotrophic growth in the absence of N, we immunologically analyzed the levels of subunits of the major photosynthetic complexes following 3 d of N deprivation and compared them with those in N-replete cells (Fig. 2). All proteins involved in photochemistry that were analyzed, as well as subunits of the chloroplast and mitochondrial ATP synthases, accumulated to approximately the same level in N-replete and N-deprived cells under photoautotrophic conditions. The polypeptides analyzed were from PSI, PSII, LHC, cyt b6f, and ATP synthase (Fig. 2). Hence, during photoautotrophic N deprivation, the photosynthetic complexes do not experience massive degradation, suggesting that the cells may still have the potential to perform photochemistry. However, the levels of Rubisco large subunit declined to approximately 50% of the level observed in N-replete cells. This suggests a decrease in the capacity to assimilate CO2 via the CBB cycle, potentially associated with the scarcity of N and an inability to synthesize amino acids and nucleotides de novo (Terashima and Evans, 1988). Interestingly, we observed a slight decrease in the major PSII-related light-harvesting protein LHCBM1 as well as a 2-fold increase in the light-harvesting-like protein LHCSR3 (see below), which is associated with the dissipation of excitation energy from PSII LHCs as heat (Peers et al., 2009; Bonente et al., 2011). This phenomenon may reflect the need to reduce LEF in photoautotrophic N-deprived cells and is discussed below.
Figure 2.
Levels of polypeptide subunits of the major complexes of the photosynthetic apparatus in photoautotrophically grown N-deprived and N-replete cells. PsaA, Core subunit of PSI; D1, core subunit of PSII; Cyt f, subunit of cyt b6f; LHCA1, light-harvesting subunit of PSI; F1-β, catalytic subunit of the mitochondrial ATP synthase; CF1-β, catalytic subunit of the chloroplast ATP synthase; LS, large subunit of Rubisco. The samples representing 100% nutrient-replete growth conditions (N+) and N-deplete growth conditions (N−) were 0.125 and 0.15 μg of Chl, respectively.
C. reinhardtii Cells Generate NPQ during Photoautotrophic N Deprivation
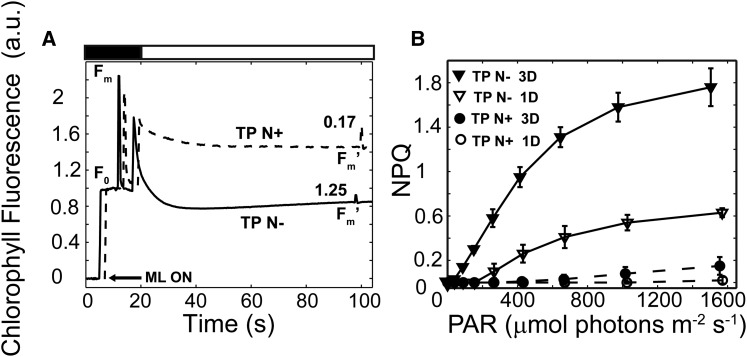
Chl fluorescence measurements were performed 3 d following the transfer of C. reinhardtii wild-type cells to photoautotrophic N-replete TP medium and TP N− in low light (LL; 40 μmol photons m−2 s−1). To examine the ability of nutrient-replete and N-deprived cells to quench Chl fluorescence, the cells were exposed to high-intensity actinic light (HL; 750 μmol photons m−2 s−1). The relative difference between the dark-acclimated level of maximum fluorescence (Fm) and the maximum fluorescence following an actinic light period (Fm′) can be used to quantify NPQ elicited in the light. As shown in Figure 3A, nutrient-replete cells exhibited a low level of NPQ (0.17 ± 0.07), while cells deprived of N for 3 d exhibited pronounced NPQ (1.25 ± 0.24).
Figure 3.
C. reinhardtii cells generate high levels of NPQ during photoautotrophic N deprivation. A, Representative Chl fluorescence traces of photoautotrophically grown, nutrient-replete (TP N+; dashed line) and N-deprived (TP N−; solid line) cells after growth in LL for 3 d. Prior to the analyses, the cells were dark adapted for 30 min followed by a brief exposure (20 s) to dark in which a saturating flash was applied (Fm) and then to HL (750 µmol photons m−2 s−1; white bar) for 80 s. The average (n = 3) NPQ values are presented above the Fm′ fluorescence peaks. Chl fluorescence traces were normalized to minimum fluorescence (F0). For convenient visualization, the kinetic traces for the nutrient-replete and N-deprived cells are shifted by 2 s. The arrow indicates activation of the measuring light (ML ON). a.u., Arbitrary units. B, NPQ at different light intensities was calculated from Chl fluorescence measurements of TP N+ (circles) and TP N− (triangles) cells grown in LL for 1 d (white symbols) and 3 d (black symbols). n = 3 ± sd. PAR, Photosynthetically active radiation.
We also determined the effect of different actinic light intensities on NPQ of photoautotrophic nutrient-replete and N-deprived (for 1 and 3 d) cells (Fig. 3B). Nutrient-replete wild-type cells exhibited very low levels of NPQ (Fig. 3B, white and black circles). After 1 d of N deprivation, the cells developed increased levels of NPQ at increasing light intensities up to 1,600 μmol photons m−2 s−1. However, after 3 d of N starvation, the cells exhibited a dramatic light intensity-dependent increase in NPQ, which, at the highest intensities, was approximately 3 times that of 1-d N-deprived cells (Fig. 3B, compare black and white triangles). Furthermore, N-deprived cells initiated NPQ as the actinic light saturated photochemical quenching and electron transport (Ik values of 263 ± 14 µmol photons m−2 s−1 [n = 3 ± sd] and 220 ± 5 µmol photons m−2 s−1 [n = 3 ± sd] for 1 and 3 d of N deprivation, respectively). Interestingly, cells grown in LL under photoheterotrophic conditions (either N+ or N−) exhibited very low levels of Chl fluorescence quenching and NPQ (Supplemental Fig. S2; note the difference in scale between Fig. 3B and Supplemental Fig. S2B). These findings suggest that the capacity to generate high levels of NPQ during N deprivation is unique to photoautotrophic growth and/or is specifically inhibited in the presence of acetate. Finally, these responses in our wild-type strain were essentially identical to those of another frequently used laboratory strain, 4A+ (CC-4051) (Supplemental Fig. S3).
C. reinhardtii Cells Generate qE When Deprived of N
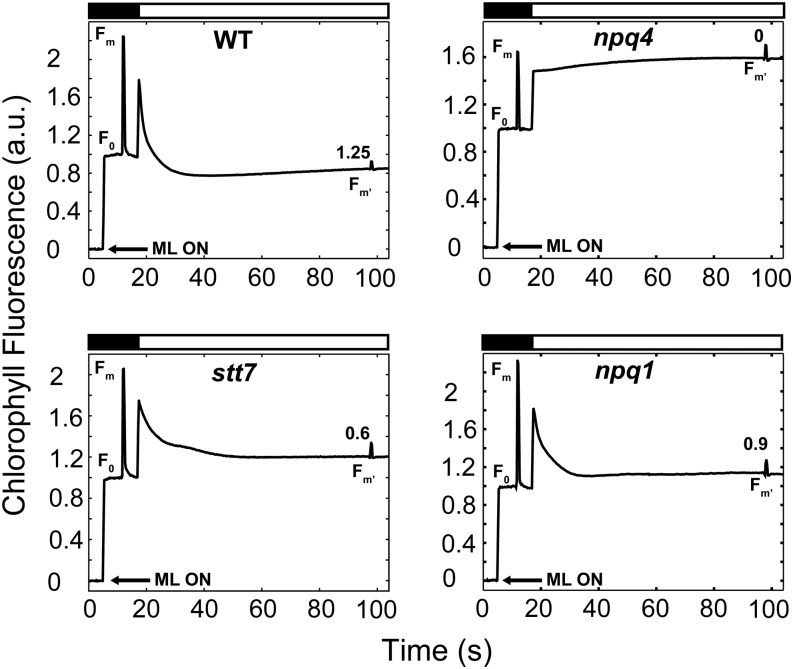
In photosynthetic organisms, NPQ can reflect a number of different photoprotective mechanisms that dissipate excess absorbed light energy. To explore the nature of NPQ in N-deprived cells under photoautotrophic conditions, we performed similar experiments on strains disrupted in specific components required for normal NPQ. The npq4 mutant lacks LHCSR3, a member of the light-harvesting protein family in C. reinhardtii, and cannot perform energy-dependent quenching (qE) despite having a normal xanthophyll cycle (Peers et al., 2009). Under N-deplete, photoautotrophic conditions, the npq4 mutant exhibited essentially no NPQ (less than 0.1; Fig. 4, compare top right and top left). These results demonstrated that most NPQ generated during photoautotrophic N deprivation is LHCSR3 dependent. We also assayed the stt7 strain of C. reinhardtii, which lacks the protein kinase involved in LHCII phosphorylation during state transitions (qT component of NPQ; Lemeille et al., 2009). In contrast to npq4, stt7 exhibited NPQ that was only somewhat less than that of wild-type cells (Fig. 4, bottom left). Therefore, the contribution of qT to NPQ in N-deprived, photoautotrophically grown cells appears to be relatively minor. The difference in the level of NPQ observed also could reflect the different genetic backgrounds of the wild-type and stt7 strains. Lastly, we assessed the impact of N deprivation on quenching in the npq1 mutant; this mutant is unable to catalyze the deepoxidation of violaxanthin to zeaxanthin (Niyogi et al., 1997a, 1997b). The N-deprived, photoautotrophically grown npq1 mutant displayed a high level of NPQ during exposure to HL. The quenching ability of npq1, despite the absence of zeaxanthin, rules out the formation of high levels of qZ, a zeaxanthin-dependent form of NPQ that is independent of an H+ gradient (Nilkens et al., 2010; Erickson et al., 2015) and also supports the idea that qE in C. reinhardtii is mediated by lutein (Niyogi et al., 1997b). A fourth component of NPQ, qI, represents physical damage to PSII core subunits (Aro et al., 1993; Eberhard et al., 2008; Li et al., 2009). To determine whether PSII damage occurred during photoautotrophic N deprivation, we analyzed the recovery of Fm′ during the relaxation of Chl fluorescence in the dark. Following exposure to HL, 30 s of darkness resulted in complete recovery of the Fm′ to the initial Fm values of wild-type cells (102% ± 2.9% [n = 3 ± sd]). Our data also suggest that not more than 10% of the Chl fluorescence quenching of the stt7 and npq1 strains can be contributed by photodamage to PSII (95.2% ± 4.9% [n = 3 ± sd] and 92% ± 3.5% [n = 3 ± sd], respectively). These results lend further support to the hypothesis that the NPQ observed during photoautotrophic N deprivation is primarily qE dependent.
Figure 4.
Evidence for a dominant role of the qE component of NPQ during N deprivation based on analysis of the various mutants. Representative Chl fluorescence traces show the wild type (WT; top left), npq4 (top right), stt7 (bottom left), and npq1 (bottom right) following 3 d of N deprivation. Black bars at the top of the graphs indicate dark periods, and white bars indicate light periods (HL; 750 µmol photons m−2 s−1). NPQ values (average from biological replicates) generated during HL are indicated above the Fm′ fluorescence peaks near the end of the light period. All Chl fluorescence traces were normalized to F0. Arrows indicate activation of the measuring light (ML ON). a.u., Arbitrary units.
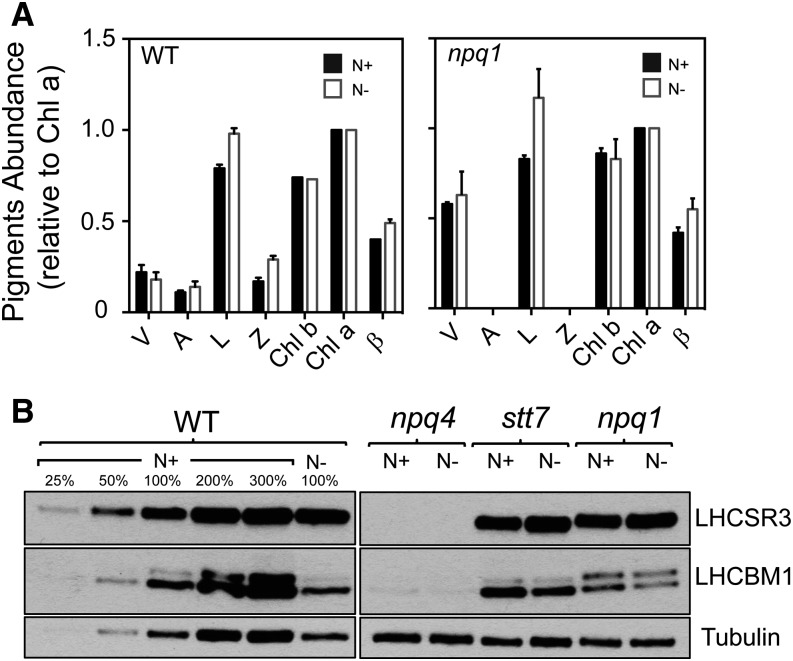
Molecular requirements for triggering qE in C. reinhardtii include the accumulation of xanthophyll pigments (mostly lutein but zeaxanthin to some extent), the LHCSR3 protein, and the establishment of a transmembrane H+ gradient (Li et al., 2009; Erickson et al., 2015). Therefore, we monitored pigment composition following 3 d of N deprivation in wild-type and npq1 cells (Fig. 5A; Supplemental Table S1). Wild-type cells exposed to photoautotrophic N deprivation in LL accumulated more zeaxanthin and antheraxanthin (42% and 24%, respectively) and less violaxanthin (21%) than cells grown in nutrient-replete medium (Fig. 5A, left; Supplemental Table S1), leading to a 20% increase in the deepoxidation state in N-deprived versus nutrient-replete cells (Student’s t test, P < 0.05; Supplemental Fig. S4). In addition, 3-d N-deprived wild-type cells also accumulated approximately 20% more lutein and β-carotene (Student’s t test, P < 0.05; Fig. 5A, left; Supplemental Table S1). Although the deepoxidation state does not change between 1 and 3 d of deprivation (Supplemental Fig. S4), the amount of NPQ generated following 3 d of N deprivation is significantly higher than the levels of NPQ generated following 1 d of N deprivation (Fig. 3B), and this rise in NPQ parallels the accumulation of lutein and β-carotene (Fig. 5A, left; Supplemental Table S1). Moreover, N-deprived cells unable to synthesize zeaxanthin (npq1) still generate substantial NPQ (Fig. 4, bottom right) and accumulate significantly more lutein and β-carotene than cells that were not deprived (Student’s t test, P < 0.05; Fig. 5A, right; Supplemental Table S1). Together, these results support a dominant role of lutein in qE in C. reinhardtii cells during photoautotrophic N deprivation (similar to its role in high light; Niyogi et al., 1997b).
Figure 5.
Pigment and protein analyses are in accord with qE as the major form of NPQ during N deprivation. A, Pigment accumulation (n = 3 ± sd) relative to Chl a in the presence and absence of N in the growth medium following 3 d of N deprivation (black and white bars, respectively). A, Antheraxanthin; L, lutein; V, violaxanthin; Z, zeaxanthin; β, β-carotene. B, LHCSR3 and LHCBM1 accumulation following 3 d of N deprivation assayed by immunoblots under nutrient-replete (N+) and N-deplete (N−) growth conditions for the wild type (WT) and NPQ mutant strains. The samples representing 100% nutrient-replete growth conditions and N-deplete growth conditions were 0.125 and 0.15 μg of Chl, respectively. Different amounts of wild-type membranes were analyzed, as indicated.
We also monitored the accumulation of LHCSR3 under N deprivation conditions. Cells grown photoautotrophically in LL under nutrient-replete conditions accumulated basal levels of LHCSR3 (Peers et al., 2009; Bonente et al., 2011). Interestingly, wild-type cells maintained for 3 d in medium lacking N exhibited an approximately 2-fold increase in LHCSR3 accumulation (Fig. 5B), while LHCSR3 accumulation in stt7 and npq1 did not show a major change following 3 d of N deprivation (Fig. 5B). These results suggest that relatively low levels of LHCSR3 are sufficient to support elevated qE. We also analyzed the possibility that the observed NPQ was a consequence of the accumulation of the LHCBM1 protein (Elrad et al., 2002). According to immunoblot analyses, 3 d of N deprivation caused a small decrease in LHCBM1 levels (the accumulation in N-deprived wild-type cells is approximately 80% of that of N-replete cells; Fig. 5B). Similar results were observed when comparing the LHCBM1 level for the stt7 and npq1 mutants, suggesting that this major LHC may not be acting as a quencher (or at least may be limiting for quenching) during N deprivation. Lastly, we examined the effect of the ΔpH on NPQ by adding the uncoupler nigericin to the assay. Nigericin caused a gradual relaxation of the quenched state in N-deprived cells (Supplemental Fig. S5A). Moreover, the levels of NPQ were reduced by greater than 50% following a 2-min incubation with nigericin (Supplemental Fig. S5B). Together, the physiological and biochemical evidence support the hypothesis that N deprivation under photoautotrophic conditions allows C. reinhardtii cells to generate extremely high levels of qE as the cells experience an increase in excitation pressure.
Additional Quenching Mechanisms during N Deprivation
The dissipation of absorbed light energy by qE is a strategy that allows cells to cope with light intensities that saturate photochemistry. However, cells often exploit multiple mechanisms to ameliorate the impact of excess absorbed light (Li et al., 2009; Erickson et al., 2015). During photoautotrophic N deprivation, we observed that the total Chl per cell was reduced relative to nutrient-replete conditions (Table I), although the Chl a:Chl b ratio remained approximately the same. Therefore, it is likely that the number of PSII RCs in the cells decreased to a similar extent as the LHCs. In addition, spectroscopic analysis of the carotenoid electrochromic shift (ECS) revealed that the ratio of PSI to PSII after 3 d of N deprivation increased by more than 20% relative to N-replete cells (Student’s t test, P < 0.05; Table I). This information supports the findings that N-deprived cells have reduced Chl per cell and that the cells reduced their rate of LEF and probably increased their rate of CEF (discussed below). Interestingly, the Fv/Fm (Table I) remains the same as in N-replete growing cells (i.e. PSII is fully active, and its level [quantified by immunoblots for D1 protein; Fig. 2] was comparable to that of N-replete cells). Finally, it was suggested previously that photoautotrophically grown cells deprived of N experienced a transition from state 1 to state 2 even under LL conditions (Peltier and Schmidt, 1991). Indeed, we observed an increase in the LHCs associated with PSI supercomplexes (a transition to state 2) throughout the deprivation period (from 1 to 3 d of N deprivation in LL), based on analyses of solubilized thylakoid membranes, in which the photosynthetic complexes were resolved by Suc gradient centrifugation coupled with SDS-PAGE (Supplemental Fig. S6).
Table I. F0, maximum PSII efficiency (Fv/Fm), and Chl measurements of N-replete (N+) and N-deplete (N−) wild-type cells over a 3-d period.
Values represent averages and sd of at least three biological replications.
| Day | N | F0 (Arbitrary Units) | Fv/Fm | PSI:PSII Ratio | Chl per Cell (μg 1026 cells) | Chl a:Chl b Ratio |
|---|---|---|---|---|---|---|
| 1 | N+ | 0.09 (0.01) | 0.60 (0.00) | 2.64 (0.18) | 2.53 (0.02) | 1.77 (0.03) |
| N− | 0.10 (0.00) | 0.62 (0.00) | 2.78 (0.06) | 2.21 (0.02) | 1.80 (0.01) | |
| 2 | N+ | 0.54 (0.03) | 2.77 (0.04) | 1.83 (0.07) | ||
| N− | 0.60 (0.01) | 2.12 (0.03) | 1.78 (0.01) | |||
| 3 | N+ | 0.12 (0.02) | 0.57 (0.01) | 2.86 (0.24) | 2.53 (0.04) | 1.83 (0.02) |
| N− | 0.17 (0.01) | 0.61 (0.00) | 3.60 (0.34) | 2.01 (0.08) | 1.84 (0.01) |
N-Deprived Cells Have a Reduced PQ Pool and Elevated Chlororespiration
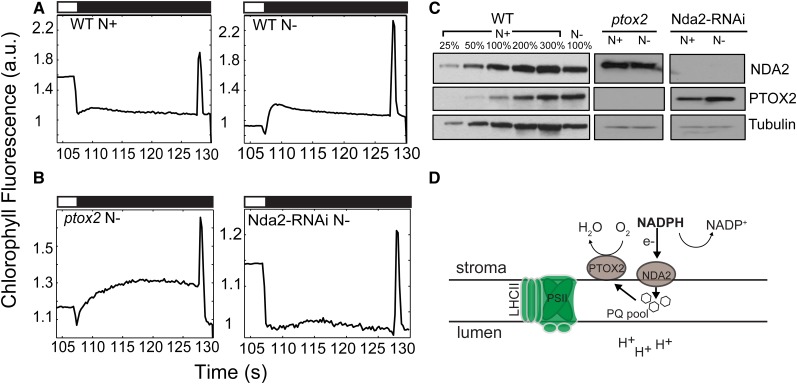
Fluorescence analyses revealed that N-deprived, photoautotrophically grown wild-type cells displayed a dramatic Chl fluorescence rise during the dark period immediately following exposure to HL (Fig. 6A, right). Upon light extinction (black bars in Fig. 6), there was a small dip in Chl fluorescence immediately followed by a rapid increase (hereafter designated dark fluorescence) that was sustained for 3 to 5 s before it gradually relaxed over an approximately 20-s period. Cells maintained in N-replete medium did not exhibit a similar dark fluorescence rise under the same experimental conditions (Fig. 6A, left). The increase in dark fluorescence may be the consequence of hyperreduction of the PQ pool during the dark period (Jans et al., 2008; Gotoh et al., 2010; Houyoux et al., 2011). This hypothesis is supported by the higher F0 values measured in 3-d N-deprived cells relative to nutrient-replete cells (Student’s t test, P < 0.05; Table I). The major enzyme involved in reducing the PQ pool in the dark is the chloroplast type II NAD(P)H dehydrogenase NDA2 in C. reinhardtii (Jans et al., 2008). Furthermore, the plastid terminal oxidases PTOX1 and PTOX2 oxidize the PQ pool by catalyzing the transfer of electrons to oxygen to generate water (Houille-Vernes et al., 2011; Nawrocki et al., 2015). These redox reactions from NADPH to oxygen are collectively referred to as chlororespiration (Bennoun, 1982; Peltier and Cournac, 2002). To determine whether the dark fluorescence observed in N-deprived cells is a consequence of PQ pool reduction by NDA2 and whether its relaxation is facilitated by PTOX2 (the major PTOX in C. reinhardtii chloroplasts), we analyzed Chl fluorescence kinetics in strains that are unable to accumulate NDA2 and PTOX2, Nda2-RNAi and ptox2, respectively (Fig. 6B). After 3 d of photoautotrophic N deprivation, the ptox2 strain exhibited a pronounced increase in dark fluorescence relative to the wild-type strain; dark fluorescence was sustained for 20 s with essentially no relaxation (Fig. 6, A and B). A similar impact on dark fluorescence was observed previously in C. reinhardtii wild-type cells subjected to HL and treated with the PTOX inhibitor propyl gallate (Houyoux et al., 2011). Therefore, we concluded that the dark fluorescence observed following N deprivation reflected a reduction of the PQ pool (by NDA2) and that its relaxation was a consequence of chlororespiratory activity facilitated by PTOX2, as depicted in the model in Figure 6D. In accord with this hypothesis, the Nda2-RNAi strain, which has essentially no NDA2 protein (Fig. 6C) and would be unable to reduce the PQ pool in the dark, did not exhibit dark fluorescence under photoautotrophic N deprivation conditions (Fig. 6B, right). Furthermore, 3 d of photoautotrophic N deprivation of wild-type cells led to an approximately 3-fold increase in the accumulation of PTOX2 and a slight increase in the accumulation of NDA2 (Fig. 6C). These results suggest that a high level of chlororespiration accompanies photoautotrophic N deprivation, which becomes apparent during the light-to-dark transitions, and that this respiratory pathway acts as an alternative electron valve (Fig. 6D) as N deprivation limits the major electron sinks.
Figure 6.
Chlororespiration is induced during N deprivation under photoautotrophic conditions. Representative Chl fluorescence traces are shown following light extinction (black bars) after 3 d of photoautotrophic growth. A, Wild-type (WT) cells under nutrient-replete (top left) and N-deplete (top right) conditions. B, ptox2 (bottom left) and Nda2-RNAi (bottom right) mutant strains under N-deplete conditions. All cells were exposed to HL (750 µmol photons m−2 s−1; white bars) prior to a dark period, as indicated in “Materials and Methods.” All Chl fluorescence traces were normalized to F0. a.u., arbitrary units. C, Immunoblot analysis for PTOX2 and NDA2 in wild-type, ptox2, and Nda2-RNAi strains under nutrient-replete (N+) and N-deplete (N−) growth conditions. Each lane contains protein from solubilized cells containing 2 μg of Chl. Different amounts of wild-type membranes were analyzed, as indicated. D, Schematic representation of the chlororespiratory pathway in darkness (or after the transition from light to darkness) as described in the text.
NDA2-Dependent CEF Regenerates NADP+ in the Light
Chlororespiration is a mechanism to oxidize NADPH in the dark, relieving elevated redox pressure generated by photosynthetic electron transport. In the light, however, PTOX2 is not as efficient as cyt b6f at oxidizing the PQ pool (Peltier and Cournac, 2002; Houille-Vernes et al., 2011; Nawrocki et al., 2015); therefore, other processes that accept electrons from NADPH may allow for the regeneration of NADP+ during the light period. In addition to its role in chlororespiration, NDA2 participates in CEF around PSI (Jans et al., 2008; Desplats et al., 2009) that is independent of the PGR5/L1-dependent CEF pathway (DalCorso et al., 2008; Hertle et al., 2013; Alric, 2014). In the former pathway, NADP+ is reduced to NADPH by ferredoxin NADP+ reductase, which is then oxidized by NDA2 with concomitant reduction of PQ. In the latter CEF pathway, however, electrons are cycled from reduced ferredoxin back to cyt b6f (likely via a PQ molecule), with no involvement of NADPH oxidation (Hertle et al., 2013; Alric, 2014). To explore whether CEF is activated during N deprivation, we measured the dark rereduction kinetics of P700+ after its complete oxidation in the light. After 1 d of N deprivation, wild-type cells exhibited a greater than 2-fold decrease in the half-time (t1/2) for P700+ rereduction (66.3 ± 7.4 ms compared with 142 ± 9.1 ms [n = 3 ± sd]; Student’s t test, P < 0.05), suggesting a doubling in the rate of CEF. To determine whether the NDA2- and/or PGR5/L1-dependent pathways were active during photoautotrophic N deprivation, we analyzed P700+ rereduction kinetics in C. reinhardtii mutants lacking components of one or the other of the two CEF pathways; these strains are Nda2-RNAi and a pgrl1 null mutant. P700+ rereduction rates for pgrl1 under nutrient-replete or nutrient-deprived conditions were similar to those of the wild-type strain (t1/2 of 57 ± 6.3 ms for N-deprived and 147 ± 30.7 ms for nutrient-replete conditions [n = 3 ± sd]; Student’s t test, P > 0.05). In contrast, the rate of P700+ rereduction for the N-deprived Nda2-RNAi strain was half as fast as that for the N-deprived wild type and similar to those of the wild-type and pgrl1 strains maintained under nutrient-replete conditions (t1/2 of 166.5 ± 10.3 ms and 160.7 ± 4 ms, respectively; Student’s t test, P > 0.05). The findings that NDA2 accumulation increased slightly during N deprivation (Fig. 6C) and that the Nda2-RNAi strain did not generate dark fluorescence (Fig. 6B, right) or allow for an elevated rate of CEF, likely because it was unable to efficiently reduce the PQ pool through the oxidization of NADPH, suggest that N-deprived cells regenerate NADP+ in the light primarily by an increase in the rate of reduction of the PQ pool and CEF via the NDA2-dependent pathway, with little or no contribution of the PGR5/L1-dependent pathway.
NDA2-Dependent CEF Establishes a Proton Gradient Necessary for qE
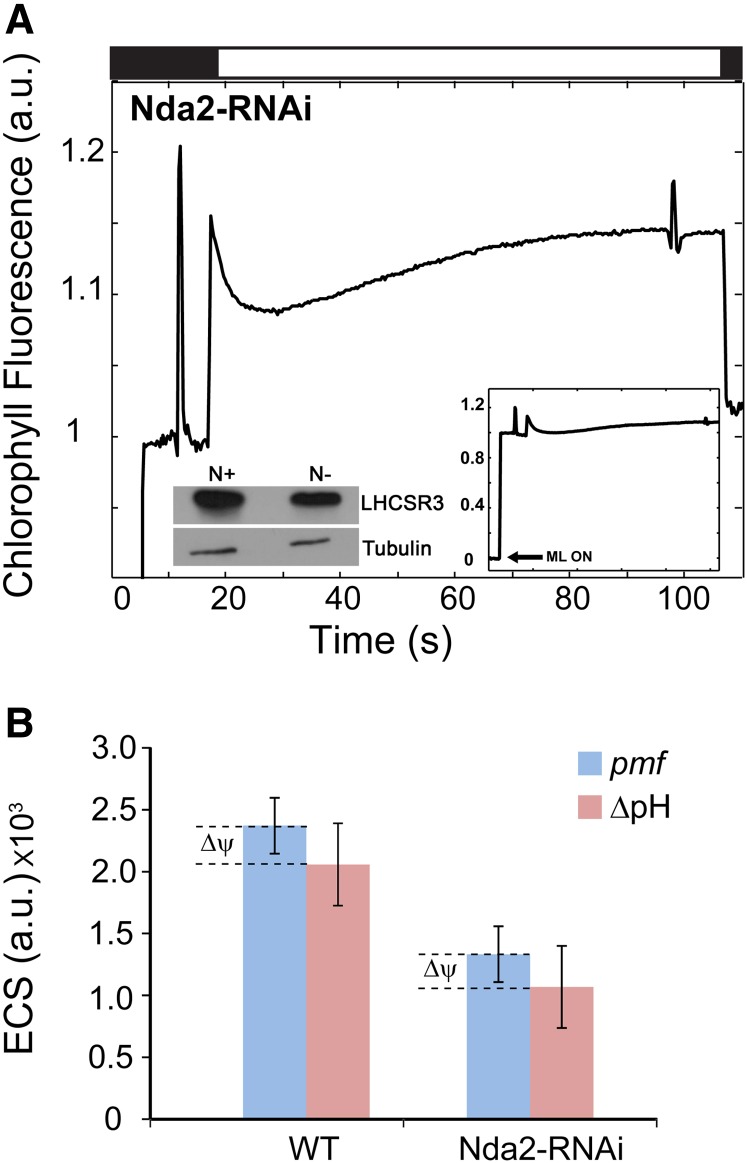
To determine whether NDA2 activity is required for Chl fluorescence quenching (qE) in photoautotrophic N-deprived cells, we monitored the fluorescence kinetics of the N-deprived Nda2-RNAi strain. Similar to npq4 (Fig. 4A), the Nda2-RNAi strain exhibited little quenching after exposure to HL (Fig. 7A). This finding highlights a key connection between NDA2-dependent CEF and qE under N deprivation conditions (where electron sinks are limited). To generate qE, LHCSR3 undergoes conformational changes triggered by acidification of the thylakoid lumen (Bonente et al., 2011). Accordingly, we hypothesized that under photoautotrophic N deprivation, NDA2-dependent CEF is necessary to generate the ΔpH required to establish qE. To further explore this hypothesis, we compared LHCSR3 accumulation and pigment content in Nda2-RNAi cells grown in nutrient-replete and N-deprived conditions. Following 3 d of N deprivation, Nda2-RNAi cells maintained a level of LHCSR3 similar to that of nutrient-replete wild-type cells (left inset in Fig. 7A). Also, like wild-type cells and the npq1 mutant strain, N deprivation of the Nda2-RNAi strain elicited an approximately 20% increase in the relative amount of lutein (Student’s t test, P < 0.05; Supplemental Table S1). To further explore the link between NDA2-dependent CEF and qE following 3 d of N deprivation, we probed the relaxation kinetics of the ECS signal as a proxy for changes in pmf (pmf = ΔpH + Δψ [the transmembrane ion gradient]) and the ΔpH, during 300 ms and 70 s in darkness, respectively. The relaxation kinetics of the ECS signal, in darkness, were monitored after the ECS attained a steady-state signal during exposure of the cells to 525 μmol photons m−2 s−1 (Baker et al., 2007). The total pmf generated by wild-type cells was 56% higher than the pmf generated by the Nda2-RNAi strain (Student’s t test, P < 0.05; Fig. 7B). Similarly, the calculated ΔpH was 51% higher for N-deprived wild-type cells than for N-deprived Nda2-RNAi cells (Student’s t test, P < 0.05; Fig. 7B). Therefore, the low level of qE generated in the N-deprived Nda2-RNAi strain (despite the accumulation of LHCSR3 and lutein) likely reflects the inability of this strain to generate a sufficient ΔpH because it is unable to achieve adequate rates of NDA2-dependent CEF.
Figure 7.
NDA2-dependent CEF provides the necessary energy for qE. A, Representative Chl fluorescence trace during HL (750 µmol photons m−2 s−1; white bar) for the Nda2-RNAi strain (for better visualization, the relevant part of the trace is shown in the main figure). Left inset, LHCSR3 accumulation assayed by immunoblots under nutrient-replete (N+) and N-deplete (N−) growth conditions for the Nda2-RNAi strain. Right inset, The same trace as in the main figure; the activation of measuring light (ML ON; arrow) is shown. The Chl fluorescence trace was normalized to F0. Each lane contains protein from solubilized cells containing 1 μg of Chl. B, pmf and ΔpH of wild-type (WT) and Nda2-RNAi strains calculated from ECS relaxation kinetics following 3 d of N deprivation (n = 3 ± sd). a.u., arbitrary units.
DISCUSSION
In this study, we examined the dynamics of the photosynthetic apparatus during the acclimation of C. reinhardtii to the absence of N under photoautotrophic growth conditions (Fig. 8 presents a model showing these dynamics). Under these conditions, the CBB cycle slows down as a consequence of feedback inhibition caused by reduced anabolic metabolism, including the reduced synthesis of amino acids and nucleotides, which in turn can lead to an imbalance in the chloroplast energetic state (i.e. changes in NADP+:NADPH and NADPH:ATP ratios).
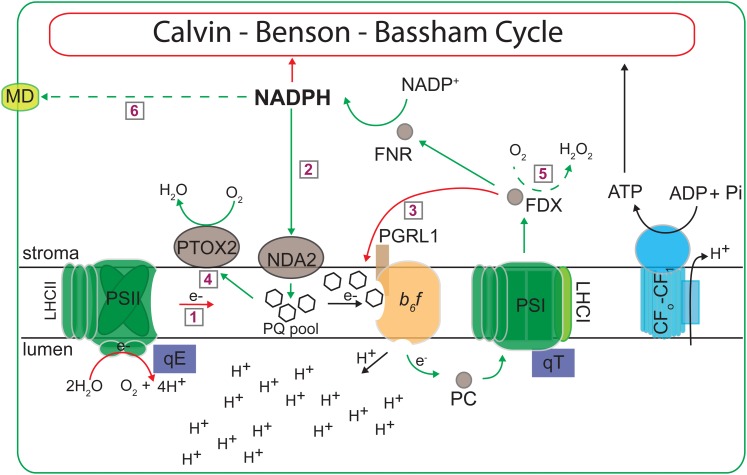
Figure 8.
Dynamics of the photosynthetic machinery during photoautotrophic N deprivation. Electron flow pathways during N-deplete growth conditions are shown. Red arrows indicate processes that appear to be down-regulated (as well as the red circle around CBB), while green arrows indicate processes that appear to be up-regulated, during N deprivation (relative to nutrient-replete cells). Dashed green arrows represent pathways identified by others. Pathways are as follows: 1, LEF; 2, NDA2-dependent CEF; 3, PGR5/L1-dependent CEF; 4, chlororespiration; 5, Mehler reaction; 6, movement of reductant out of the chloroplast through the formation of malate by malate dehydrogenase (MD). FDX, Ferredoxin; FNR, ferredoxin NADP+ reductase; Pi, inorganic phosphate.
The ways in which C. reinhardtii acclimates to N deprivation depend on the energy source used for growth and maintenance. When the cells are supplemented with an external fixed carbon source, Chl level declines (Fig. 1) and many of the complexes of the photosynthetic apparatus are degraded (Bulté and Wollman, 1992; Schmollinger et al., 2014; Wei et al., 2014; Juergens et al., 2015). A large pool of N is released during the degradation of proteins of the photosynthetic machinery, allowing the cells to transition to a new metabolic organization that sustains their viability. Furthermore, in the absence of photosynthesis and the presence of acetate, N-starved cells are dominated by respiratory processes (Schmollinger et al., 2014). In contrast, when external fixed carbon is not available, photosynthesis serves as the dominant mode of energy generation; therefore, the photosynthetic machinery is protected and maintained even when the number of electron sinks is severely reduced, as is the case for N-deprived cells. In this work, we showed that the major photosynthetic complexes, PSI, PSII, LHCs, cyt b6f, and ATP synthase, all were retained following 3 d of N deprivation (Figs. 2 and 5C). Retaining the photosynthetic machinery involves dynamic interactions among a range of bioenergetics processes that include, on the one hand, the activation of NPQ and the minimization of input of excitation energy that is directed to PSII RCs, and on the other hand, maximizing the use of alternative electron outlets (i.e. oxygen as an electron acceptor in chlororespiration) while generating sufficient energy for cell maintenance (by CEF). It should kept in mind that some aspects of the N deprivation response may be different among the different strains used in this study, because of their genetic background. Some strains (D66+, 4A+, ptox2, pgrl1, Nda2-RNAi) are defective for the regulator NIT2 while others (npq1, npq4, stt7) are not. Although N deprivation in photoheterotrophic conditions causes photosynthetic complex degradation in a NIT1- and nitric oxide-dependent manner, this response is much less important in photoautotrophic conditions and NIT2 does not seem to play a crucial role in either condition (Wei et al., 2014). Indeed, as noted above, we observed that N deprivation in photoautotrophic conditions did not greatly affect the abundance of electron transport components (Fig. 2). Despite this, photosynthetic electron flow was appreciably modified in response to N deprivation, as we discuss further below. Concerning our analysis of chlororespiratory mutants, the ptox2 and Nda2-RNAi strains both lack NIT2 and are therefore in a similar genetic background as the wild-type D66+.
N-deprived C. reinhardtii cells exhibited rapid, extensive, and reversible NPQ (qE; Fig. 3) that depended on LHCSR3 (Figs. 4 and 5B) and was accompanied by the accumulation of xanthophylls, mainly lutein (Fig. 5A). These results suggest that various cellular responses are elicited in photoautotrophic N-deprived cells in order to minimize LEF (Fig. 8, pathway 1). This conclusion is supported by the finding that the onset of qE in N-deprived cells occurs at moderate light intensities (200–300 μmol photons m−2 s−1; Fig. 3B) and coincides with the saturation of photochemical quenching. Additional quenching mechanisms associated with photoautotrophic N deprivation include a decrease in Chl per cell, an increase in the PSI-PSII ratio (Table I), and qT (Supplemental Fig. S6; Peltier and Schmidt, 1991). The decrease in Chl reflects a decline in both the level of RCs and associated LHCs, as the N-deprived cells exhibit little change in their Chl a:Chl b ratio, which also has been shown for other photosynthetic organisms (Terashima and Evans, 1988). Overall, in order to reduce the excitation pressure from PSII and LEF during prolonged N deprivation (for 3 d), the cells exhibited a range of quenching mechanisms that are affected on different time scales. On the one hand, a reduced level of Chl per cell, a decline in the PSI:PSII ratio, as well as translocation of the mobile light-harvesting antenna from PSII to PSI (during exposure to LL) take place over a time scale of several hours to days. These processes (with the exception of state transition) will also require a relatively long time to reverse once N levels in the environment increase. State transitions, which usually can occur in a few minutes upon exposure of C. reinhardtii cells to high light, can also reverse relatively rapidly (Eberhard et al., 2008; Erickson et al., 2015). At this point, however, we do not understand the nature of the state transition during N deprivation and the specific antenna proteins associated with the process. On the other hand, the cells also developed very high levels of NPQ, which were primarily a consequence of qE. As N-deprived cells grown in LL experience a limited number of electron sinks (as cells lose the major pathway represented by the CBB cycle), qE, which usually develops in response to HL exposure (Li et al., 2009), is established. This mechanism of quenching allows for rapid and extensive NPQ when N-deprived cells experience sudden increases in light intensity and allows for a rapid recovery as the excitation pressure declines.
The localization of lutein accumulation during the acclimation of cells to N deprivation and the specific role of this pigment in quenching are open questions. When C. reinhardtii cells acclimate to HL, lutein accumulates in both the PSII core and LHC complexes (Pineau et al., 2001). In addition, a high level of lutein does not appear to be tightly associated with photosynthetic complexes but may be loosely bound to photosynthetic polypeptides (Pineau et al., 2001). Moreover, reconstitution assays have suggested that both lutein and violaxanthin are bound to the L1 and L2 binding sites of the LHCSR3 polypeptide; the selectivity of these binding sites was less strict than the selectivity of the xanthophyll-binding sites on the LHC proteins (Bonente et al., 2011). A third binding site on LHCSR3 was partially occupied by a xanthophyll (not necessarily lutein; Bonente et al., 2011). Since the localization of xanthophylls within the photosynthetic membranes of C. reinhardtii during acclimation to photoautotrophic N deprivation has not been examined, we speculate that the pattern of localization of lutein in N-deprived cells may be similar to that of cells acclimated to HL (Pineau et al., 2001), especially since, as in HL, N-deprived cells experience a rise in the cellular reduction state and accumulate LHCSR3 (Fig. 5B). The slight decline in the level of violaxanthin during N deprivation (in contrast to the accumulation of lutein; Fig. 5A, left; Supplemental Table S1) may leave more LHCSR3 sites available for lutein binding. However, lutein also may be loosely bound to proteins or remain free in the membranes; additional biochemical and genetic studies specifically focused on the localization of lutein in membranes and photosynthetic complexes during N deprivation would be required to delineate the lutein-binding sites.
A critical question raised in these studies is what mechanisms promote lumen acidification for eliciting qE, especially considering the suppression of LEF during photoautotrophic N deprivation? LEF establishes a transmembrane ΔpH, but it can only be sustained if downstream electron acceptors, such as NADP+, are available. On the other hand, CEF (Fig. 8, pathways 2 and 3) is a mechanism to alleviate the potentially high NADPH:NADP+ ratio that would form during N deprivation by cycling electrons around PSI and also would function to generate a trans-thylakoid ΔpH that could sustain qE while at the same time providing ATP to fuel cellular processes (Allen, 2003). In this work, we have demonstrated that wild-type cells doubled their rate of CEF during N deprivation and that this CEF is NDA2 dependent (see “Results”). An Nda2-RNAi strain, which does not accumulate detectable levels of NDA2 (Fig. 6C), was unable to increase its rate of CEF (see “Results”). In contrast, the rate of CEF for pgrl1, a strain incapable of performing PGR5/L1-dependent CEF, was comparable to that of the wild type (see “Results”; Fig. 8, up-regulation for pathway 2 and suppression of pathway 3). The Nda2-RNAi strain, grown under photoautotrophic, N deprivation conditions, also was unable to elicit the dark rise in fluorescence associated with reduction of the PQ pool (Fig. 6B) or establish qE, even though it maintained LHCSR3 and accumulated elevated lutein levels (Fig. 7A; Supplemental Table S1). Moreover, the Nda2-RNAi strain generated approximately half of the wild-type pmf and ΔpH levels during 3 d of N deprivation (Fig. 7B), which may be insufficient to sustain qE. These results strongly support our hypothesis that there is an essential connection between qE and NDA2-dependent CEF, with the latter providing the energy (ΔpH) required to establish rapid and reversible, energy-dependent NPQ when the cells experience N deprivation (and potentially in response to other deprivation conditions).
While the processes described above would suppress LEF and the reduction of NADP+, sustained N deprivation could still lead to the accumulation of NADPH (and potentially other reduced molecules) and limit the amount for oxidized electron acceptors, which in turn could feed back on electron transport and other cellular processes (Eberhard et al., 2008). To regenerate NADP+, N-deprived cells would have to oxidize NADPH, which likely would involve the chlororespiratory proteins NDA2 (also involved in CEF, as mentioned above) and PTOX (Fig. 6; Peltier and Schmidt, 1991; Peltier and Cournac, 2002). NDA2 would couple the oxidation of NADPH to the reduction of the PQ pool (Fig. 8, pathway 2), followed by the oxidation of PQH2 by PTOX; this latter reaction is coupled to the reduction of oxygen and H+ to form water (Fig. 8, pathway 4). Chlororespiration is thought to occur primarily in the dark, since, in the light, PTOX is outcompeted by cyt b6f for PQH2 (Peltier and Cournac, 2002; Houille-Vernes et al., 2011; Nawrocki et al., 2015). However, we cannot exclude the possibility that, under some conditions, oxidation of the PQ pool by PTOX can occur in the light. This would become more likely when the photosynthetic electron transfer chain is hyperreduced (e.g. when terminal electron acceptors are scarce) and the oxidation rate of cyt b6f by PC is slow (relative to the oxidation rate of the PQ pool by PTOX). While the fluorescence phenotypes of the Nda2-RNAi and ptox2 strains support the hypothesis that chlororespiration is critical for the reoxidation of NADPH, our results cannot rule out the possibility that other mechanisms, including the donation of electrons by PSI to oxygen via a Mehler-type reaction (Asada, 1999), also may be exploited by the cells during photoautotrophic N deprivation. This latter possibility is suggested by recent work in which the elimination of chloroplast Fru-1,6-bisphosphate (FBPase) in Arabidopsis thaliana stimulated the production of high levels of hydrogen peroxide and increased the activity of the NADPH dehydrogenase-dependent CEF pathway (Strand et al., 2015), which is analogous to CEF catalyzed by C. reinhardtii NDA2. FBPase is an anabolic enzyme involved in the CBB cycle and gluconeogenesis. Its expression in C. reinhardtii is down-regulated during photoheterotrophic N deprivation (Blaby et al., 2013; Park et al., 2015), which would lead to reduced rates of CO2 fixation, the accumulation of NADPH, and hyperreduction of PSI. This raises the possibility that photoautotrophic N deprivation may elicit superoxide and hydrogen peroxide production through a Mehler-type reaction (Asada, 1999), which would help reoxidize the hyperreduced electron transport chain (Fig. 8, pathway 5). In addition, electrons may be transported from the chloroplast to the mitochondria, where they could be used for mitochondrial respiration (Johnson et al., 2014; Bailleul et al., 2015). This pathway was not explored in this work.
Our results strongly suggest a critical role for NDA2 in the acclimation of C. reinhardtii to N deprivation. This key enzyme integrates a range of processes that help balance the energetic state of the cell and are essential for survival as the cells become nutrient deprived. NDA2 can relieve the redox pressure through oxidizing NADPH and also promote energy generation by functioning in CEF and/or in routing electrons to the chlororespiratory pathways. NDA2-driven CEF also would provide the H+ gradient that would elicit high levels of NPQ (qE).
CONCLUSION
Environmental conditions can markedly impact the metabolic state and energetic balance of photosynthetic organisms. In response to environmental cues (in this work, N deprivation), the eukaryotic unicellular green alga C. reinhardtii employs a range of strategies to sustain viability when electron sinks become limited while allowing rapid recovery and growth once conditions improve. Photosynthetic organisms may achieve such plasticity by activating mechanisms to dissipate excess absorbed light energy (through NPQ and chlororespiration) while maintaining sufficient energy for cell maintenance (through CEF around PSI). These processes are maintained through the activity of NDA2, a type II NAD(P)H dehydrogenase. Furthermore, this analysis highlights major shifts in the functioning of the photosynthetic apparatus as the cells respond to dynamic fluctuations in their nutrient environment (and probably other abiotic factors); overall, the chloroplast appears to be transformed from an organelle that performs photosynthesis and fixes CO2 at high rates to one in which quenching and respiratory processes become more active, allowing for the dissipation of much of the absorbed light energy as heat.
MATERIALS AND METHODS
Strains and Growth Conditions
D66+ (CC-4425) was used as the wild-type strain of Chlamydomonas reinhardtii for all experiments unless indicated otherwise. Mutants used in this study include npq1 (CC-4100) and npq4 (CC-4614), both obtained from the Chlamydomonas Resource Center, ptox2 and pgrl1, provided by X. Johnson, and the Nda2-RNAi and stt7 (CC-4178) strains, provided by G. Peltier and F.-A. Wollman, respectively. General propagation of the strains was performed using TAP solid and liquid media. For photoautotrophic growth, TP medium (no acetate) was used. For N deprivation, cells were grown in liquid TAP medium, pelleted by centrifugation, and washed at least twice prior to resuspension in TP N−. For N deprivation under photoheterotrophic conditions, a similar procedure was used except that the cells were resuspended in TAP N−. For all experiments, unless noted otherwise, strains were grown under constant LL (40 µmol photons m−2 s−1) and aerated by shaking.
Chl Concentration and Cell Count
Chl concentrations were determined by methanol extraction according to Porra et al. (1989). Cell densities were determined using a particle counter/size analyzer (Z1 Coulter Counter; Beckman).
Chl Fluorescence Analysis
For Chl fluorescence measurements, cells were pelleted by centrifugation (1,200g, 5 min, and 23°C) and resuspended to a concentration of 10 µg mL−1 Chl in fresh medium. Fluorescence measurements were performed using a DUAL-PAM-100 (Walz). Prior to assaying NPQ, the cells were acclimated in the dark for at least 30 min. Following dark acclimation, we first measured the Fv/Fm and then exposed the cells to approximately 100 s of high actinic light (HL; approximately 750–800 µmol photons m−2 s−1), followed by 30 s of darkness and a saturating light pulse. Light curves were generated by exposing cells to increasing light intensities, with the time at each intensity step lasting 1 min. NPQ was calculated as (Fm − Fm′)/Fm, while Ik values were calculated by fitting the light curve according to Platt et al. (1980). To probe the dissipation of ΔpH during the analysis, 8.5 μm nigericin (dissolved in methanol) was added to the reaction during the light period. Alternatively, cells were incubated in the dark for 2 min with 8.5 μm nigericin prior to exposing them to actinic light.
P700 Activity Measurements
Absorbance changes associated with P700 oxidation-reduction were monitored at 705 nm using a JTS10 spectrophotometer (Biologic) according to Alric (2014). Cells were pelleted by centrifugation (as above) and resuspended to a concentration of 30 µg mL−1 Chl in 20 mm HEPES-KOH, pH 7.2, and 10% (w/v) Ficoll. Twenty micromolar 3-(3,4-dichlorophenyl)-1,1-dimethylurea and 1 mm hydroxylamine were added to the suspension to inhibit LEF prior to the measurement. Dark-adapted cells were exposed to 285 µmol photons m−2 s−1 during PSI oxidation followed by a saturating pulse. P700+ rereduction rates were measured as t1/2, calculated from the rereduction curves.
Spectroscopic Measurements of the Carotenoid ECS and ΔpH
Wild-type and Nda2-RNAi strains were grown on nutrient-replete and N-deplete media for 3 d, as described above. Cells containing 15 µg mL−1 Chl were pelleted by centrifugation (1,000g, 5 min, and 23°C) and resuspended in 20 mm HEPES-KOH, pH 7.2, and 10% (w/v) Ficoll. The cells were exposed to light (either a pulse or extended exposure, as indicated), and absorbance changes associated with thylakoid membrane carotenoids (designated the ECS) were monitored at 520 and 546 nm using a JTS10 spectrophotometer (Biologic; Joliot and Joliot, 2008). The PSI-PSII ratio was calculated as the ratio of the level of the phase of the ECS that was induced by a laser flash in untreated and 3-(3,4-dichlorophenyl)-1,1-dimethylurea-treated cells. Total pmf and ΔpH were estimated according to Baker et al. (2007). Briefly, cells were exposed to 525 μmol photons m−2 s−1 for 80 s, at which time the ECS signal reached a steady state, followed by extinction of the light and relaxation of the ECS signal over a 300-ms period in the dark. A proxy for the ΔpH was calculated as the difference between the lowest point in the decay of the ECS signal following the extinction of the light and the signal observed after complete relaxation over the period of 70 s in the dark.
Protein Extraction and Immunoblot Analysis
Total cell protein was obtained by pelleting cells by centrifugation (1,200g, 5 min, and 23°C), resuspending the pellets in a protein extraction buffer containing 5 mm HEPES-KOH, pH 7.5, 100 mm dithiothreitol, 100 mm Na2CO3, 2% (w/v) SDS, and 12% (w/v) Suc, and boiling the samples for 1 min. Samples used for SDS-PAGE (12% acrylamide gels) were normalized to Chl content (and similar levels of tubulin), and the resolved polypeptides were transferred to polyvinylidene difluoride membranes using a semidry transfer apparatus (Bio-Rad) according to the manufacturer’s directions. For immunoblot analysis, membranes were blocked for 1h at room temperature in Tris-buffered saline-0.1% (v/v) Tween containing 5% milk followed by an overnight incubation of the membranes at 4°C with the primary antibodies in Tris-buffered saline-0.1% (v/v) Tween containing powdered milk (3% [w/v]). Primary antibodies were diluted according to the manufacturer’s suggestions. Proteins were detected by enhanced chemiluminescence (GE Healthcare).
HPLC Analysis of Chl and Carotenoids
Briefly, cells were collected by centrifugation (16,100g, 5 min, and 23°C), and cell pellets were frozen in liquid N2 and stored at −80°C. Pigments were extracted by vortexing the pellets in 200 µL of acetone, and following a brief centrifugation (16,100g, 1 min, and 23°C), the supernatant was passed through a 0.45-µm nylon filter spin column (Costar). Samples were stored in HPLC vials in the dark at 4°C prior to analysis. Extracted pigments (25 µL) were separated on a C18 Spherisorb ODS1 4.6-mm × 250-mm cartridge column (Waters) at 23°C using the method described (Müller-Moulé et al., 2002). The deepoxidation state was calculated as (A+Z)/(V+A+Z).
Thylakoid Membrane Purification, Solubilization, and Separation of Pigment-Protein Complexes by Suc Gradient Centrifugation
Thylakoid membranes were purified according to Takahashi et al. (2006). Purified thylakoids were pelleted and resuspended in a buffer containing 5 mm HEPES-KOH, pH 7.5, 10 mm EDTA, and protease inhibitors. n-Dodecyl β-d-maltoside was added to the membranes (300 µg of Chl) to a final concentration of 0.75% (w/v), and solubilization was on ice in the dark for 40 min. Solubilized membranes were overlaid on a continuous 0.1 to 1.3 m Suc gradient and centrifuged at 188,000g for 16 h at 4°C. Fractions of equal volume (approximately 500 µL) were collected and prepared for SDS-PAGE. Resolved proteins were visualized by silver staining.
Statistics
The data were analyzed by Student’s t tests, and significant differences are defined as those with P < 0.05.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Chl degradation after 7 days of nitrogen deprivation.
Supplemental Figure S2. Chl fluorescence and NPQ analyses of Chlamydomonas cells growing under photoheterotrophic N deprivation.
Supplemental Figure S3. NPQ analysis for 4A+ WT strain.
Supplemental Figure S4. NPQ generation by N-deprived WT cells under photoautotrophic conditions involves de-epoxidation of xanthophylls.
Supplemental Figure S5. The effect of nigericin on Chl fluorescence quenching of N-deprived WT cells under photoautotrophic growth conditions.
Supplemental Figure S6. N-deprived cells promote a transition to state 2.
Supplemental Table S1. N-deprived cells promote a transition to state 2.
Supplementary Material
Acknowledgments
We thank Francis-André Wollman, Gilles Peltier, and Xenie Johnson for kindly providing the stt7, ptox2, and pgrl1 strains and the Nda2-RNAi line as well as antibodies; Marilyn Kobayashi for assistance with pigment analysis; David Kramer, Francis-André Wollman, and Fabrice Rappaport for extensive and fruitful discussions; and Fabrice Rappaport for helping with the interpretation of the ECS data.
Glossary
- cyt b6f
cytochrome b6fp
- LEF
linear electron flow
- CEF
cyclic electron flow
- Chl
chlorophyll
- RC
reaction center
- PQ
plastoquinone
- PC
plastocyanin
- pmf
proton motive force
- CBB
Calvin-Benson-Bassham
- NPQ
nonphotochemical quenching
- N
nitrogen
- TAP
Tris-acetate-phosphate
- TP
Tris-phosphate
- TP N−
N-deplete Tris-phosphate medium
- TAP N−
N-deplete Tris-acetate-phosphate medium
- LL
low light
- HL
high light
- Fm
dark-acclimated level of maximum fluorescence
- Fm′
maximum fluorescence following an actinic light period
- ECS
electrochromic shift
- F0
minimum fluorescence
- Fv/Fm
maximum PSII efficiency
- t1/2
half-time
- ΔpH
transmembrane H+ gradient
Footnotes
This work was supported by a Vaadia-BARD Postdoctoral Fellowship, United States-Israel Binational Agriculture Research and Development Fund (grant no. FI–485–2013 to S.I.S.), the Carnegie Institution for Science (to S.I.S.), and the National Science Foundation (grant no. MCB 0951094 to A.R.G.).
Articles can be viewed without a subscription.
References
- Aksoy M, Pootakham W, Pollock SV, Moseley JL, González-Ballester D, Grossman AR (2013) Tiered regulation of sulfur deprivation responses in Chlamydomonas reinhardtii and identification of an associated regulatory factor. Plant Physiol 162: 195–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF. (2003) Cyclic, pseudocyclic and noncyclic photophosphorylation: new links in the chain. Trends Plant Sci 8: 15–19 [DOI] [PubMed] [Google Scholar]
- Alric J. (2014) Redox and ATP control of photosynthetic cyclic electron flow in Chlamydomonas reinhardtii. II. Involvement of the PGR5-PGRL1 pathway under anaerobic conditions. Biochim Biophys Acta 1837: 825–834 [DOI] [PubMed] [Google Scholar]
- Aro EM, Virgin I, Andersson B (1993) Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta 1143: 113–134 [DOI] [PubMed] [Google Scholar]
- Asada K. (1999) The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol 50: 601–639 [DOI] [PubMed] [Google Scholar]
- Bailleul B, Berne N, Murik O, Petroutsos D, Prihoda J, Tanaka A, Villanova V, Bligny R, Flori S, Falconet D, et al. (2015) Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524: 366–369 [DOI] [PubMed] [Google Scholar]
- Baker NR, Harbinson J, Kramer DM (2007) Determining the limitations and regulation of photosynthetic energy transduction in leaves. Plant Cell Environ 30: 1107–1125 [DOI] [PubMed] [Google Scholar]
- Bennoun P. (1982) Evidence for a respiratory chain in the chloroplast. Proc Natl Acad Sci USA 79: 4352–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby IK, Blaby-Haas CE, Tourasse N, Hom EFY, Lopez D, Aksoy M, Grossman A, Umen J, Dutcher S, Porter M, et al. (2014) The Chlamydomonas genome project: a decade on. Trends Plant Sci 19: 672–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby IK, Glaesener AG, Mettler T, Fitz-Gibbon ST, Gallaher SD, Liu B, Boyle NR, Kropat J, Stitt M, Johnson S, et al. (2013) Systems-level analysis of nitrogen starvation-induced modifications of carbon metabolism in a Chlamydomonas reinhardtii starchless mutant. Plant Cell 25: 4305–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonente G, Ballottari M, Truong TB, Morosinotto T, Ahn TK, Fleming GR, Niyogi KK, Bassi R (2011) Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLoS Biol 9: e1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulté L, Wollman FA (1992) Evidence for a selective destabilization of an integral membrane protein, the cytochrome b6/f complex, during gametogenesis in Chlamydomonas reinhardtii. Eur J Biochem 204: 327–336 [DOI] [PubMed] [Google Scholar]
- DalCorso G, Pesaresi P, Masiero S, Aseeva E, Schünemann D, Finazzi G, Joliot P, Barbato R, Leister D (2008) A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132: 273–285 [DOI] [PubMed] [Google Scholar]
- Dall’Osto L, Caffarri S, Bassi R (2005) A mechanism of nonphotochemical energy dissipation, independent from PsbS, revealed by a conformational change in the antenna protein CP26. Plant Cell 17: 1217–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MP, Horst I, Duong GH, Tomsett EV, Litvinenko ACP, Howe CJ, Smith AG (2014) Triacylglyceride production and autophagous responses in Chlamydomonas reinhardtii depend on resource allocation and carbon source. Eukaryot Cell 13: 392–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplats C, Mus F, Cuiné S, Billon E, Cournac L, Peltier G (2009) Characterization of Nda2, a plastoquinone-reducing type II NAD(P)H dehydrogenase in Chlamydomonas chloroplasts. J Biol Chem 284: 4148–4157 [DOI] [PubMed] [Google Scholar]
- Eberhard S, Finazzi G, Wollman FA (2008) The dynamics of photosynthesis. Annu Rev Genet 42: 463–515 [DOI] [PubMed] [Google Scholar]
- Elrad D, Niyogi KK, Grossman AR (2002) A major light-harvesting polypeptide of photosystem II functions in thermal dissipation. Plant Cell 14: 1801–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson E, Wakao S, Niyogi KK (2015) Light stress and photoprotection in Chlamydomonas reinhardtii. Plant J 82: 449–465 [DOI] [PubMed] [Google Scholar]
- Evans JR. (1989) Photosynthesis and nitrogen relationships in leaves of C3 plants. Oecologia 78: 9–19 [DOI] [PubMed] [Google Scholar]
- Goodenough U, Blaby I, Casero D, Gallaher SD, Goodson C, Johnson S, Lee JH, Merchant SS, Pellegrini M, Roth R, et al. (2014) The path to triacylglyceride obesity in the sta6 strain of Chlamydomonas reinhardtii. Eukaryot Cell 13: 591–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh E, Matsumoto M, Ogawa K, Kobayashi Y, Tsuyama M (2010) A qualitative analysis of the regulation of cyclic electron flow around photosystem I from the post-illumination chlorophyll fluorescence transient in Arabidopsis: a new platform for the in vivo investigation of the chloroplast redox state. Photosynth Res 103: 111–123 [DOI] [PubMed] [Google Scholar]
- Grossman A, Takahashi H (2001) Macronutrient utilization by photosynthetic eukaryotes and the fabric of interactions. Annu Rev Plant Physiol Plant Mol Biol 52: 163–210 [DOI] [PubMed] [Google Scholar]
- Grossman AR, Gonzalez-Ballester D, Shibagaki N, Pootakham W, Moseley J (2009) Responses to macronutrient deprivation. In Pareek A, Sopory SK, Bohnert HJ, Govindjee, eds, Abiotic Stress Adaptation in Plants: Physiological, Molecular and Genomic Foundation. Springer, The Netherlands, Dordrecht pp 307–348 [Google Scholar]
- Hertle AP, Blunder T, Wunder T, Pesaresi P, Pribil M, Armbruster U, Leister D (2013) PGRL1 is the elusive ferredoxin-plastoquinone reductase in photosynthetic cyclic electron flow. Mol Cell 49: 511–523 [DOI] [PubMed] [Google Scholar]
- Houille-Vernes L, Rappaport F, Wollman FA, Alric J, Johnson X (2011) Plastid terminal oxidase 2 (PTOX2) is the major oxidase involved in chlororespiration in Chlamydomonas. Proc Natl Acad Sci USA 108: 20820–20825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houyoux PA, Ghysels B, Lecler R, Franck F (2011) Interplay between non-photochemical plastoquinone reduction and re-oxidation in pre-illuminated Chlamydomonas reinhardtii: a chlorophyll fluorescence study. Photosynth Res 110: 13–24 [DOI] [PubMed] [Google Scholar]
- Huppe HC, Turpin DH (1994) Integration of carbon and nitrogen metabolism in plant and algal cells. Annu Rev Plant Physiol Plant Mol Biol 45: 577–607 [Google Scholar]
- Iwai M, Takizawa K, Tokutsu R, Okamuro A, Takahashi Y, Minagawa J (2010) Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature 464: 1210–1213 [DOI] [PubMed] [Google Scholar]
- Jans F, Mignolet E, Houyoux PA, Cardol P, Ghysels B, Cuiné S, Cournac L, Peltier G, Remacle C, Franck F (2008) A type II NAD(P)H dehydrogenase mediates light-independent plastoquinone reduction in the chloroplast of Chlamydomonas. Proc Natl Acad Sci USA 105: 20546–20551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X, Steinbeck J, Dent RM, Takahashi H, Richaud P, Ozawa S, Houille-Vernes L, Petroutsos D, Rappaport F, Grossman AR, et al. (2014) Proton gradient regulation 5-mediated cyclic electron flow under ATP- or redox-limited conditions: a study of ΔATpase pgr5 and ΔrbcL pgr5 mutants in the green alga Chlamydomonas reinhardtii. Plant Physiol 165: 438–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot P, Joliot A (2008) Quantification of the electrochemical proton gradient and activation of ATP synthase in leaves. Biochim Biophys Acta 1777: 676–683 [DOI] [PubMed] [Google Scholar]
- Juergens MT, Deshpande RR, Lucker BF, Park JJ, Wang H, Gargouri M, Holguin FO, Disbrow B, Schaub T, Skepper JN, et al. (2015) The regulation of photosynthetic structure and function during nitrogen deprivation in Chlamydomonas reinhardtii. Plant Physiol 167: 558–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer DM, Avenson TJ, Edwards GE (2004) Dynamic flexibility in the light reactions of photosynthesis governed by both electron and proton transfer reactions. Trends Plant Sci 9: 349–357 [DOI] [PubMed] [Google Scholar]
- Lemeille S, Willig A, Depège-Fargeix N, Delessert C, Bassi R, Rochaix JD (2009) Analysis of the chloroplast protein kinase Stt7 during state transitions. PLoS Biol 7: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XP, Björkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK (2000) A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Li XP, Gilmore AM, Caffarri S, Bassi R, Golan T, Kramer D, Niyogi KK (2004) Regulation of photosynthetic light harvesting involves intrathylakoid lumen pH sensing by the PsbS protein. J Biol Chem 279: 22866–22874 [DOI] [PubMed] [Google Scholar]
- Li Z, Wakao S, Fischer BB, Niyogi KK (2009) Sensing and responding to excess light. Annu Rev Plant Biol 60: 239–260 [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. (1961) Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191: 144–148 [DOI] [PubMed] [Google Scholar]
- Mitchell P. (1966) Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biol Rev Camb Philos Soc 41: 445–502 [DOI] [PubMed] [Google Scholar]
- Mitchell P. (2011) Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. 1966. Biochim Biophys Acta 1807: 1507–1538 [DOI] [PubMed] [Google Scholar]
- Moseley JL, Chang CW, Grossman AR (2006) Genome-based approaches to understanding phosphorus deprivation responses and PSR1 control in Chlamydomonas reinhardtii. Eukaryot Cell 5: 26–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Moulé P, Conklin PL, Niyogi KK (2002) Ascorbate deficiency can limit violaxanthin de-epoxidase activity in vivo. Plant Physiol 128: 970–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki WJ, Tourasse NJ, Taly A, Rappaport F, Wollman FA (2015) The plastid terminal oxidase: its elusive function points to multiple contributions to plastid physiology. Annu Rev Plant Biol 66: 49–74 [DOI] [PubMed] [Google Scholar]
- Nelson N, Yocum CF (2006) Structure and function of photosystems I and II. Annu Rev Plant Biol 57: 521–565 [DOI] [PubMed] [Google Scholar]
- Nilkens M, Kress E, Lambrev P, Miloslavina Y, Müller M, Holzwarth AR, Jahns P (2010) Identification of a slowly inducible zeaxanthin-dependent component of non-photochemical quenching of chlorophyll fluorescence generated under steady-state conditions in Arabidopsis. Biochim Biophys Acta 1797: 466–475 [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Bjorkman O, Grossman AR (1997a) Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell 9: 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi KK, Björkman O, Grossman AR (1997b) The roles of specific xanthophylls in photoprotection. Proc Natl Acad Sci USA 94: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Fernie AR, Stitt M (2010) Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol Plant 3: 973–996 [DOI] [PubMed] [Google Scholar]
- Park JJ, Wang H, Gargouri M, Deshpande RR, Skepper JN, Holguin FO, Juergens MT, Shachar-Hill Y, Hicks LM, Gang DR (2015) The response of Chlamydomonas reinhardtii to nitrogen deprivation: a systems biology analysis. Plant J 81: 611–624 [DOI] [PubMed] [Google Scholar]
- Peers G, Truong TB, Ostendorf E, Busch A, Elrad D, Grossman AR, Hippler M, Niyogi KK (2009) An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462: 518–521 [DOI] [PubMed] [Google Scholar]
- Peltier G, Cournac L (2002) Chlororespiration. Annu Rev Plant Biol 53: 523–550 [DOI] [PubMed] [Google Scholar]
- Peltier G, Schmidt GW (1991) Chlororespiration: an adaptation to nitrogen deficiency in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 88: 4791–4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau B, Gerard-Hirne C, Selve C (2001) Carotenoid binding to photosystem I and II of Chlamydomonas reinhardtii cells grown under weak light or exposed to intense light. Plant Physiol Biochem 39: 73–85 [Google Scholar]
- Platt T, Gallegos C, Harrison W (1980) Photoinhibition of photosynthesis in natural assemblages of marine phytoplankton. J Mar Res 38: 687–701 [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Raven JA. (2013) Rubisco: still the most abundant protein of Earth? New Phytol 198: 1–3 [DOI] [PubMed] [Google Scholar]
- Rochaix JD. (2014) Regulation and dynamics of the light-harvesting system. Annu Rev Plant Biol 65: 287–309 [DOI] [PubMed] [Google Scholar]
- Romero-Puertas MC, Rodríguez-Serrano M, Sandalio LM (2013) Protein S-nitrosylation in plants under abiotic stress: an overview. Front Plant Sci 4: 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumeau D, Peltier G, Cournac L (2007) Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ 30: 1041–1051 [DOI] [PubMed] [Google Scholar]
- Schmollinger S, Mühlhaus T, Boyle NR, Blaby IK, Casero D, Mettler T, Moseley JL, Kropat J, Sommer F, Strenkert D, et al. (2014) Nitrogen-sparing mechanisms in Chlamydomonas affect the transcriptome, the proteome, and photosynthetic metabolism. Plant Cell 26: 1410–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaut M, Cuiné S, Cagnon C, Fessler B, Nguyen M, Carrier P, Beyly A, Beisson F, Triantaphylidès C, Li-Beisson Y, et al. (2011) Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand DD, Livingston AK, Satoh-Cruz M, Froehlich JE, Maurino VG, Kramer DM (2015) Activation of cyclic electron flow by hydrogen peroxide in vivo. Proc Natl Acad Sci USA 112: 5539–5544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Iwai M, Takahashi Y, Minagawa J (2006) Identification of the mobile light-harvesting complex II polypeptides for state transitions in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 103: 477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima I, Evans JR (1988) Effects of light and nitrogen nutrition on the organization of the photosynthetic apparatus in spinach. Plant Cell Physiol 29: 143–155 [DOI] [PubMed] [Google Scholar]
- Terauchi AM, Peers G, Kobayashi MC, Niyogi KK, Merchant SS (2010) Trophic status of Chlamydomonas reinhardtii influences the impact of iron deficiency on photosynthesis. Photosynth Res 105: 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolleter D, Ghysels B, Alric J, Petroutsos D, Tolstygina I, Krawietz D, Happe T, Auroy P, Adriano JM, Beyly A, et al. (2011) Control of hydrogen photoproduction by the proton gradient generated by cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell 23: 2619–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Derrien B, Gautier A, Houille-Vernes L, Boulouis A, Saint-Marcoux D, Malnoë A, Rappaport F, Vitry C, de, Vallon O, et al. (2014) Nitric Oxide–Triggered Remodeling of Chloroplast Bioenergetics and Thylakoid Proteins upon Nitrogen Starvation in Chlamydomonas reinhardtii. Plant Cell. 26: 353–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Work VH, Radakovits R, Jinkerson RE, Meuser JE, Elliott LG, Vinyard DJ, Laurens LML, Dismukes GC, Posewitz MC (2010) Increased lipid accumulation in the Chlamydomonas reinhardtii sta7-10 starchless isoamylase mutant and increased carbohydrate synthesis in complemented strains. Eukaryot Cell 9: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff DD, Davies JP, Melis A, Grossman AR (1998) The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol 117: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.