A heat stress transcription factor is involved in tomato pollen thermotolerance, contributing to stress responses as well as the abundance of chaperones in the priming program activated during microsporogenesis.
Abstract
Male reproductive tissues are more sensitive to heat stress (HS) compared to vegetative tissues, but the basis of this phenomenon is poorly understood. Heat stress transcription factors (Hsfs) regulate the transcriptional changes required for protection from HS. In tomato (Solanum lycopersicum), HsfA2 acts as coactivator of HsfA1a and is one of the major Hsfs accumulating in response to elevated temperatures. The contribution of HsfA2 in heat stress response (HSR) and thermotolerance was investigated in different tissues of transgenic tomato plants with suppressed HsfA2 levels (A2AS). Global transcriptome analysis and immunodetection of two major Hsps in vegetative and reproductive tissues showed that HsfA2 regulates subsets of HS-induced genes in a tissue-specific manner. Accumulation of HsfA2 by a moderate HS treatment enhances the capacity of seedlings to cope with a subsequent severe HS, suggesting an important role for HsfA2 in regulating acquired thermotolerance. In pollen, HsfA2 is an important coactivator of HsfA1a during HSR. HsfA2 suppression reduces the viability and germination rate of pollen that received the stress during the stages of meiosis and microspore formation but had no effect on more advanced stages. In general, pollen meiocytes and microspores are characterized by increased susceptibility to HS due to their lower capacity to induce a strong HSR. This sensitivity is partially mitigated by the developmentally regulated expression of HsfA2 and several HS-responsive genes mediated by HsfA1a under nonstress conditions. Thereby, HsfA2 is an important factor for the priming process that sustains pollen thermotolerance during microsporogenesis.
Survival upon exposure to heat stress (HS) and acquisition of thermotolerance depend on the activation of heat stress response (HSR). HSR requires a complex network of distinct and interconnected pathways involved in maintenance of protein homeostasis, reduction of cellular damages, and readjustment of metabolic activities. Genes encoding for heat shock proteins (Hsps), such as molecular chaperones, are the best studied HS-induced genes, but a plethora of other genes are up-regulated to contribute to HSR as shown by transcriptome analysis in several plant species (Finka et al., 2011; Jung et al., 2012; Liu and Charng, 2013; Sarkar et al., 2014; Fragkostefanakis et al., 2015b). The rapid and strong transcriptional activation of the majority of genes is controlled by the activity of the heat stress transcription factors (Hsfs). Plants have evolved a large repertoire of Hsfs, ranging from 21 in Arabidopsis (Arabidopsis thaliana) to 27 in tomato (Solanum lycopersicum), reaching up to 52 in soybean (Glycine max; Scharf et al., 2012).
The role of individual Hsfs in HSR and thermotolerance has been shown in several genetic studies, mainly in Arabidopsis, but also in some crop plants (reviewed in Fragkostefanakis et al., 2015a; Scharf et al., 2012). Despite the high similarity among Hsf orthologs in different plant species, discrepancies regarding a specific role of individual Hsfs in HSR have been reported. In tomato, HsfA1a, one of the four A1 Hsfs, is considered as master regulator required for the induction of HSR. In Arabidopsis, the master regulator function is shared among HsfA1a, HsfA1b, and HsfA1d as concluded from thermotolerance assays in single and multiple gene knockout mutants (Liu et al., 2011; Yoshida et al., 2011). In addition, Arabidopsis HsfB1 acts as transcriptional repressor during the attenuation phase of HSR, while tomato HsfB1 possesses both coactivator and repressor functions (Bharti et al., 2004; Ikeda et al., 2011).
In tomato, the fundamental Hsf network for control of HSR depends on the activities of HsfA1a, HsfA2, and HsfB1 (Hahn et al., 2011). HsfA1a and HsfB1 are constitutively expressed with mRNA steady state levels of low abundance. The transcripts of HsfA2 are hardly detectable under control temperature conditions, but upon HS, the accumulation of HsfA2 mRNA and protein is strongly induced. Consequently, HsfA2 becomes the most abundant Hsf during recovery or after several cycles of repeated HS (Hahn et al., 2011; Scharf et al., 1998). During recovery from HS, the availability of HsfA2 is controlled at the protein level by interactions with class CI and CII small Hsps (sHsps) and the activity of higher molecular weight chaperones such as Hsp70 and Hsp101 (Port et al., 2004). HsfA2 forms hetero-oligomeric complexes with HsfA1a. These “superactivator” complexes activate downstream genes more strongly than the two individual factors alone (Chan-Schaminet et al., 2009). Ectopic expression or suppression of members of HsfA2 subclass in different plant species have been related to increased or reduced basal and acquired thermotolerance (Charng et al., 2007; Chauhan et al., 2013; Nishizawa et al., 2006; Ogawa et al., 2007; Yokotani et al., 2008).
Tomato HsfA2 requires interaction with HsfA1a for efficient nuclear retention and transcriptional activity (Mishra et al., 2002; Scharf et al., 1998). Overexpression of Arabidopsis HsfA2 in the quadruple HsfA1a/b/d/e knockout mutant restored thermotolerance, indicating that Arabidopsis HsfA2 is functional in the absence of the master regulator (Liu and Charng, 2013). Moreover, a preferential regulation of genes involved in metabolism and redox homeostasis by HsfA2 was shown, while genes preferentially controlled by HsfA1s are involved in transcription (Liu and Charng, 2013). Therefore, beyond the expected redundancy, diverse specificity or preferential functions are shared among different members of the Hsf family.
Genetic studies have highlighted the involvement of Hsfs in various growth and developmental processes in addition to their well established role in HSR. In some cases, homologous or heterologous ectopic expression of HsfA2 co-orthologs resulted in growth and developmental alterations, including growth retardation and dwarfism as well as accelerated plant and callus growth in the absence of HsfA1s in Arabidopsis (Liu and Charng, 2013; Ogawa et al., 2007). In addition, HsfA2 and other HS-induced Hsps that are minimally expressed in vegetative tissues under physiological conditions are strongly expressed at early stages of anther and pollen development (Frank et al., 2009; Giorno et al., 2010). However, despite the increased levels of HsfA2, pollen at meiotic and early mitotic phases exhibit only weak HSR at the transcriptional level, which is assumed to be related with the increased sensitivity of these stages at higher temperatures (Frank et al., 2009; Giorno et al., 2010). A tomato cultivar with higher basal expression of HsfA2 and some Hsps accumulating at early microspore stage exhibited higher pollen thermotolerance compared to another cultivar with lower Hsf-Hsp levels (Frank et al., 2009). This might suggest that the accumulation of chaperones during progression from meiocytes to mitotic microspores enhances protection and priming of HSR for more advanced developmental stages (Chaturvedi et al., 2013; Frank et al., 2009).
Through microarray meta-analysis, a putative HsfA2 regulatory network was proposed for tomato (Fragkostefanakis et al., 2015b). This network suggests a potential HsfA2 function in different abiotic stress responses. In addition to classical Hsps, this network includes other HS-induced genes with diverse functions, linking the activity of HsfA2 to various aspects of HSR. Here, the relevance of HsfA2 for HSR and thermotolerance of different tissues is examined using transgenic plants with enhanced or suppressed HsfA2 expression. In addition, the transcriptional networks controlled by HsfA2 are examined in young leaves and anthers to dissect the functional relation of HsfA2 in different tissues by transcriptome analysis. These two tissues were selected as leaves mostly represent photosynthesizing (metabolically active) cells, while anthers mostly consist of sporophytic cells supporting the development of male gametophytes. This analysis revealed the existence of tissue-specific regulatory networks controlled by HsfA2. HsfA2 suppression also causes reduced pollen viability and germination after exposure to a moderate HS treatment. We show that under physiological conditions, HsfA2 controls the abundance of genes with significant developmental functions and propose the implication of HsfA2 in pollen thermotolerance.
RESULTS
The Role of HsfA2 in Heat Stress Response of Tomato Seedlings
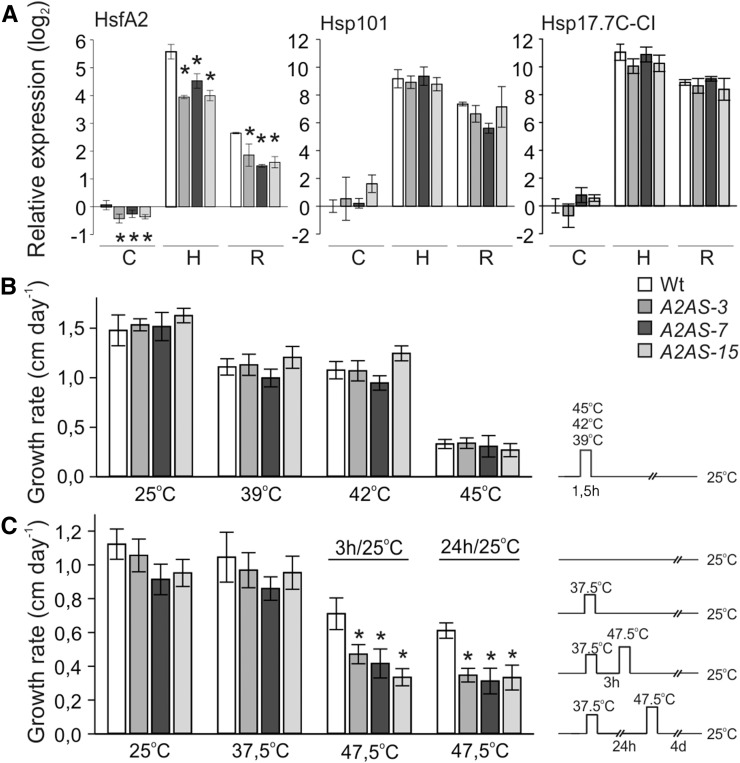
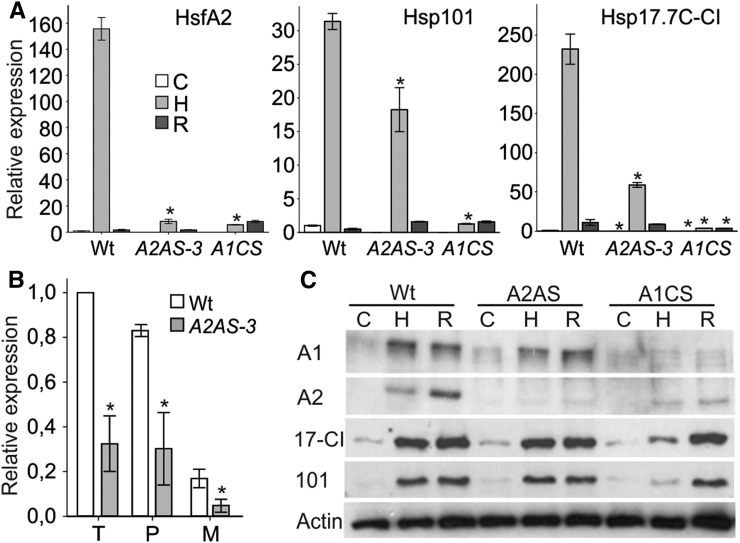
We generated transgenic plants expressing a cDNA cassette of tomato HsfA2 either in the sense (A2S) or in antisense orientation (A2AS) under the control of the Cauliflower mosaic virus (CaMV) 35S promoter as previously described for HsfA1a (Mishra et al., 2002). We selected three independent transgenic lines (A2AS-3, -7, and -15) with significantly reduced HsfA2 levels as confirmed by transcript analysis using qRT-PCR in seedlings (Fig. 1A) exposed for 1 h to 39°C (sample H) and after subsequent recovery for 1.5 h at 25°C (sample R). The effect of HsfA2 suppression on the transcript abundance of heat induced molecular chaperones was examined for Hsp101 and Hsp17.7C-CI in control and stressed seedlings (Fig. 1A). Transient suppression of HsfA2 in tomato mesophyll protoplasts resulted in reduced levels of these two Hsps (Fragkostefanakis et al., 2015b). As expected, in seedlings, both Hsp101 and Hsp17.7C-CI followed the expression profile of the HS-induced HsfA2 transcript levels reaching their maximum after HS treatment, while further increase in the corresponding protein levels was observed during the subsequent recovery period at 25°C (Fig. 1A). Surprisingly, HsfA2 suppression in transgenic A2AS seedlings did not significantly affect the expression profile of these two Hsps, which was similar to the wild type (Fig. 1A).
Figure 1.
Basal and acquired thermotolerance of etiolated seedlings of tomato wild-type and HsfA2 transgenic lines. A, Relative gene expression (2–ΔΔCt) of HsfA2, Hsp17.7C-CI, and Hsp101 in wild-type and A2AS transgenic seedlings kept at 25°C (control, sample C) or exposed to 1 h HS at 39°C (sample H), which was followed by recovery for 1.5 h at 25°C (sample R). The Ct value for each gene was normalized to Ct values for EF1α and UBI housekeeping genes and to expression in wild-type control. Vertical bars are the average ± sd of three replicates. B, Four-day-old etiolated tomato wild-type and A2AS-3, -7, and -15 transgenic seedlings were exposed to 39, 42, or 45°C for 90 min. Hypocotyl length was recorded for 5 d after stress and growth rate was calculated. C, Same seedlings as in B were exposed to 37.5°C for 90 min and then allowed to recover at 25°C for 3 or 24 h. Following recovery, a severe HS at 47.5°C for 90 min was applied. Hypocotyl length was recorded for 5 d after stress, and growth rate was calculated. Pictographs on the right in B and C indicate the applied HS regimes, respectively. Each data point is the mean ± se of at least 10 seedlings for each genotype and treatment. For A and C, asterisk denotes significant difference (P < 0.05) compared to the wild type for a given sample or stress treatment as shown by pairwise t test analyses.
This result obviously indicates some discrepancy in comparison to the situation observed in leaf mesophyll protoplasts. Therefore, we analyzed the expression of these genes and accumulation of the corresponding proteins in seedlings of the A2S line to verify this phenomenon in more detail.
As expected, HsfA2 was ectopically expressed in A2S seedlings under all tested conditions as judged from transcript abundance determined by qRT-PCR (Supplemental Fig. S1A). However, similar to the wild type, HsfA2 protein was not detectable in nonstressed A2S seedlings, although the protein abundance was higher after HS or recovery when compared to the wild type (Supplemental Fig. S1B). Consequently, A2S seedlings accumulated higher levels of Hsp101 and Hsp17.7C-CI transcripts (Supplemental Fig. S1A), and the abundance of Hsp101 protein was strongly enhanced compared to wild-type seedlings (Supplemental Fig. S1A). Thus, HsfA2 is not essential for the induction of Hsp101 and Hsp17.7C-CI in heat-stressed tomato seedlings, but increasing the levels by ectopically expressed HsfA2 strongly enhances the accumulation of these Hsps.
The role of HsfA2 for basal thermotolerance was examined by exposing 4-d-old dark-grown seedlings to 25, 39, 42, or 45°C for 90 min (Fig. 1B). Following HS, the hypocotyl elongation rate of each seedling was determined. We observed a reduction in the growth rate of the wild-type and transgenic seedlings already after treatment at 39°C, while the impact of the stress was more severe in seedlings exposed to 45°C (Fig. 1B). However, the stress had similar effect on the growth rate of all genotypes, suggesting that HsfA2 is not essential for basal thermotolerance (Fig. 1B). This result is consistent with the finding that HsfA2 suppression does not influence the accumulation of Hsps (Fig. 1A). However, A2S seedlings showed increased basal thermotolerance compared to the wild type when the relative hypocotyl growth was monitored after HS treatment at 42 or 45°C (Supplemental Fig. S1C). This result suggests that HsfA2 is not required for maintenance of basal thermotolerance, but increased levels of HsfA2 enhance the capacity of seedlings to cope with acute stress.
We further examined the contribution of HsfA2 in short- and long-term acquired thermotolerance. Seedlings were kept at 37.5°C for 90 min and were then allowed to recover at 25°C for 3 or 24 h (Fig. 1C). Following recovery for the indicated time, seedlings were exposed to a sever HS at 47.5°C for 90 min (Fig. 1C). The 37.5°C pretreatment does not affect the hypocotyl elongation rate but yields a strong accumulation of HsfA2 protein. The protein was still detectable after 48 h of recovery in wild-type seedlings (Supplemental Fig. S2). In contrast, there was no HsfA2 or Hsp17.7C-CI protein accumulating in response to a direct HS treatment at 47.5°C, which eventually was lethal for both wild-type and A2AS seedlings. A2AS-3 seedlings exhibited slightly reduced Hsp101 and significantly lower Hsp17-CI protein levels in the short- and long-term acquired thermotolerance tests (Supplemental Fig. S2). The lower protein accumulation seems to correlate with a significantly reduced growth rate compared to the wild type, which was observed for seedlings from all three A2AS lines in both short- and long-term acquired thermotolerance tests (Fig. 1C). Worth mentioning here, there were no differences between seedlings from different genotypes in the loss of acquired thermotolerance after 48 h recovery, which might mark the time limit for maintenance of the acquired protection (data not shown). Collectively these results show that accumulation of HsfA2 by a mild stress can increase the capacity of the seedlings to induce Hsp synthesis at higher temperatures and to recover from otherwise lethal HS treatments, while it is not essential for manifestation of the HSR at moderate HS conditions.
The Role of HsfA2 in Heat Stress Response of Young Leaves and Developing Anthers
The absence of a significant effect of HsfA2 suppression on basal thermotolerance in seedlings raised the question whether this holds true for other tissues with different physiological functions as well. On the one hand, we focused on metabolically highly active, photosynthesizing young leaves and, on the other hand, on anthers containing male sporophytic and gametophytic cells. For direct comparison of potential HsfA2 functions in the two tissues, we used 8-week-old flowering tomato plants. For HS treatment, plants were either exposed to 39°C for 1 h (H) and then allowed to recover for 1.5 h at 25°C (R) or kept all the time at 25°C as control (C).
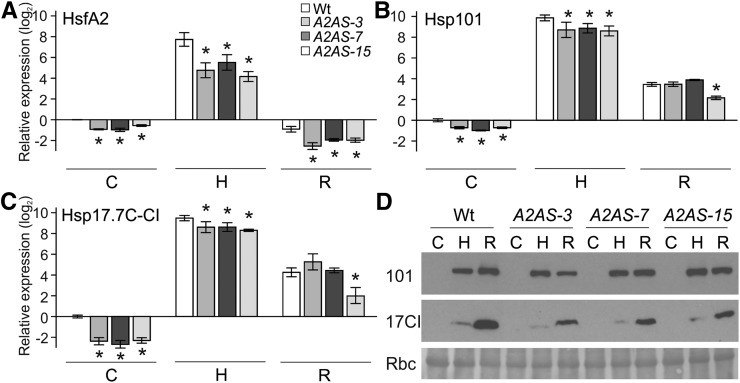
HsfA2 transcript levels were lower in control, HS, and recovery samples from leaves of all A2AS lines compared to the wild type (Fig. 2A). This yielded a lower protein abundance when compared to the wild type as exemplified for A2AS-3 (Supplemental Fig. S3D). HsfA2 suppression resulted in reduced Hsp101 and Hsp17.7C-CI transcript levels after 1 h HS but showed similar levels to the wild type after recovery with the exception of A2AS-15 line (Fig. 2, B and C). In turn, the reduced transcript abundance was followed by significantly lower accumulation of Hsp17.7C-CI protein but had only minimal effect on Hsp101 (Fig. 2D). Hsp101 protein has been also previously shown to accumulate in leaves of A1CS plants, suggesting efficient translation and Hsp101 protein accumulation even at reduced transcript levels (Mishra et al., 2002). The reduced accumulation of Hsps in heat-stressed leaves of A2AS lines compared to the wild type indicates that HsfA2 plays an important role as coactivator during induction of the HSR.
Figure 2.
HSR of young leaves of wild-type and A2AS transgenic plants. A to C, Relative gene expression (2–ΔΔCt) of HsfA2 (A), Hsp17.7C-CI (B), and Hsp101 (C) in leaves from wild-type and A2AS transgenic plants exposed to HS at 39°C for 1 h (sample H) and then allowed to recover for 1.5 h (sample R) or kept under control conditions for the same period (sample C). The expression was determined and represented as in Fig. 1A. Vertical bars are the average ± sd of three replicates. D, Leaves of wild-type and A2AS-3, -7, and -15 plants were treated as in A, and equal amounts of total protein extract (20 μg, controlled by large subunit of Rubisco [Rbc] after Ponceau S staining) were subjected to immunoblot analysis with Hsp17-CI and Hsp101 antibodies.
The conclusion is further supported by the observation that leaves from heat-stressed A2S plants accumulate higher HsfA2 transcript and protein levels, which resulted in an increased expression of Hsp101 and Hsp17.7C-CI as well (Supplemental Fig. S3, A–D). Similar to seedlings, we could not detect HsfA2 protein in control leaves of A2S plants, while HsfA2 protein levels were enhanced under HS but similar to the wild type in recovery samples (Supplemental Fig. S3D). We further confirmed the stimulatory effect of HsfA2 overexpression on additional HS-induced genes, including several Hsps but also for Hsa32, APX3, and MBF1c (Supplemental Fig. S3E). Based on these observations, we conclude that the higher accumulation of HsfA2 in the A2S plants leads to a stronger HSR.
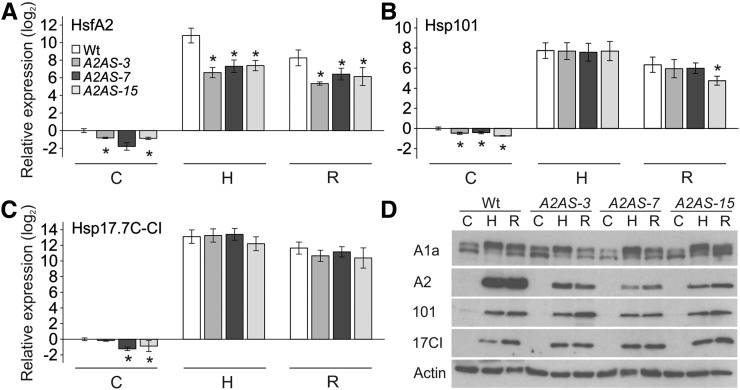
For investigation of the role of HsfA2 in HSR of male reproductive tissues, we collected anthers containing meiocytes and tetrads (flower bud length <4 mm), unicellular microspores (flower bud length 4–8 mm), and binucleate mature pollen (flower bud length >8 mm) after exposure to HS. The different anther stages were pooled and samples were used for analyses of transcript and protein levels (Fig. 3). Remarkably, HsfA2 transcript and protein abundance was significantly reduced in control and stressed anthers in all A2AS lines compared to the wild type (Fig. 3, A and D). However, partially similar to seedlings, but different to leaves, suppression of HsfA2 does not cause a significant reduction of Hsp101 or Hsp17-CI expression (Fig. 3, B–D). In contrast to the impairment of HsfA2 expression reported for HsfA1 cosuppression plants (Mishra et al., 2002), knockdown of HsfA2 seems not to affect the expression level of HsfA1a in anthers of the A2AS plants (Fig. 3D). Transcript analysis of HsfA2, Hsp101, and Hsp17.7C-CI in A1CS anthers confirmed that HsfA1a is required for the induction of these three genes in response to HS (Supplemental Fig. S4). In view of these findings, it is tempting to speculate that the proposed coactivator function of HsfA2 might be tissue or cell type specific and seems not to be required for efficient induction of Hsp expression in anthers. Moreover and in contrast to our observation in seedlings and leaves, in pooled anther stages from nonstressed or stressed A2S plants, we were unable to detect any increased accumulation of HsfA2 transcript or protein compared to the wild type (data not shown). This might be due to the weak CaMV 35S promoter activity in reproductive tissues, which might be sufficient for suppression but not for detectable increases by ectopic expression.
Figure 3.
HSR of anthers of wild-type and HsfA2 antisense plants. A to C, Relative gene expression (2–ΔΔCt) of HsfA2 (A), Hsp101 (B), and Hsp17.7C-CI (C) in anther from wild-type and A2AS transgenic plants exposed to HS at 39°C for 1 h (sample H) and then allowed to recover for 1.5 h (sample R) or kept under control conditions (sample C) for the same period. The expression was determined and represented as in Fig. 1A. Vertical bars are the average ± sd of three replicates. D, Anthers of different developmental stages from wild-type and A2AS-3, -7, and -15 plants were treated as in A, and equal amounts of total protein extract (20 μg, controlled by actin levels) were subjected to immunoblot analysis with specific antibody against HsfA1a, HsfA2, Hsp101, and Hsp17-CI. HsfA1a antibody identified two bands, probably representing different modification states of the protein.
Effect of HsfA2 Suppression on Pollen Thermotolerance
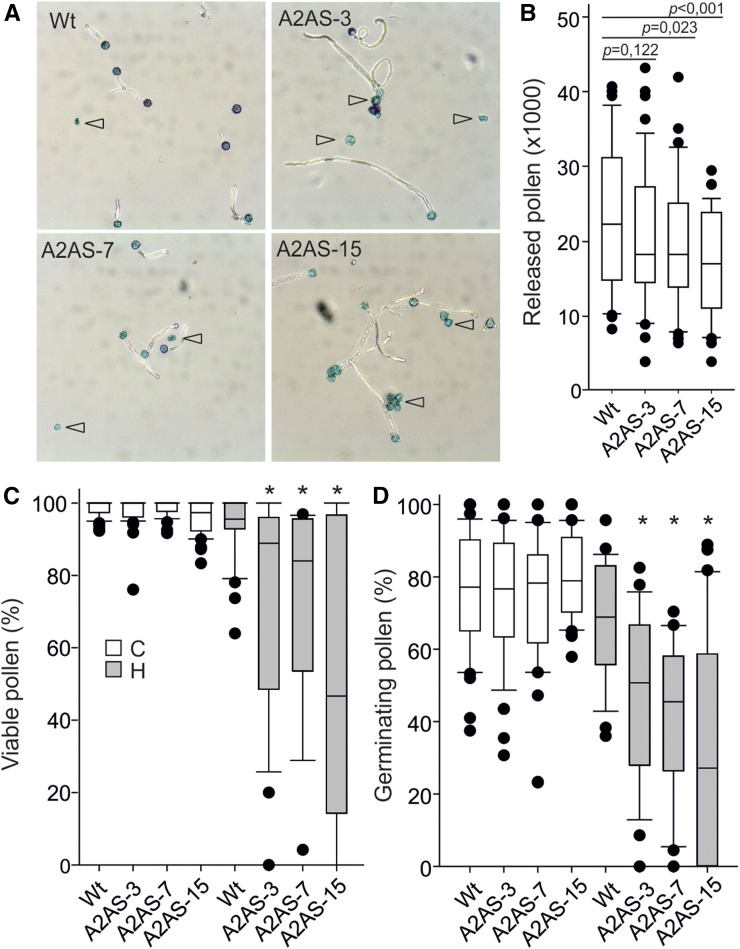
We investigated the impact of HsfA2 knockdown on thermotolerance of pollen (Fig. 4A). Pollen meiosis in tomato occurs approximately 10 d before anthesis and is considered as the developmental stage most sensitive to HS (Iwahori, 1966). Flowering wild-type and A2AS plants from all three transgenic lines were exposed to 3 h HS at 39°C or kept under control conditions. For individual flowers, the time until anthesis was monitored and the number of released pollen grains (Fig. 4B), pollen viability (Fig. 4C), and germination rate (Fig. 4D) were determined. We realized changes in these attributes only for flowers reaching anthesis 9 to 12 d after performing the HS treatment. Flowers that received the stress in more advanced stages did not yield significant differences between wild-type and A2AS plants (data not shown).
Figure 4.
Effect of HsfA2 knockdown on pollen release and quality. A, Representative light microscopy pictures of mature pollen grains isolated from plants exposed to HS 9 to 12 d before anthesis. Pollen grains were allowed to germinate for 3 h in germination solution and then stained with Alexander dye. Open arrowheads indicate dead pollen (blue colored, nongerminated). B, Number of released pollen isolated from anthers of wild-type and A2AS-3, -7, and -15 transgenic plants kept under control conditions. The P values determined by pairwise comparison between wild-type and A2AS lines are given on top of the box plots. C and D, Percentage of viable (C) and germinating (D) pollen released from control and stressed wild-type and A2AS transgenic plants. Asterisks denote significant differences between pollen from A2AS plants and the wild type as determined by t test (P < 0.05). The results are derived from quality analysis of pollen grains isolated from 25 to 47 individual flowers for each genotype and treatment.
Interestingly, anthers from nontreated A2AS plants released lower number of pollen grains than the wild type (Fig. 4B). The difference was statistically significant for A2AS-7 and A2AS-15 lines but not for A2AS-3 (P = 0.122) which had approximately 20% decreased number of released pollen grains compared to the wild type (Fig. 4B). The effect of HsfA2 suppression on pollen release has been confirmed in independent experiments for all transgenic lines. HS caused a reduction in pollen release from wild-type and A2AS anthers; however, the differences were not significant among the genotypes.
Nontreated wild-type and A2AS anthers released pollen of similar viability and germination rate, suggesting that HsfA2 is not involved in pollen development under nonstress conditions (Fig. 4, C and D). In the same line, untreated wild-type and A2AS pollen did not show any morphological abnormalities and pollen development was not affected as confirmed by staining with 4′,6-diamino-phenylindole (data not shown). However, both viability and germination rate of heat-stressed pollen grains were significantly lower in all A2AS lines compared to the wild type (Fig. 4, C and D). These results suggest that HsfA2 is involved in both release of pollen from anthers and in the regulation of thermotolerance during early stages (pollen mother cells/tetrads) of pollen development.
The higher sensitivity of A2AS pollen is most likely not a consequence of physiological alterations in vegetative tissues. On the one hand, the HS treatment did not cause significant alterations of the quantum yield of photochemical energy conversion in photosystem II [Φ(II)], the electron transport rate, the coefficient of photochemical quenching, or the nonphotochemical quenching (Supplemental Fig. S5). On the other hand, HS did not cause any significant alterations in the growth of wild-type or transgenic plants. These results indicate that HsfA2 suppression does neither influence the photosynthetic activity of young leaves nor the general plant growth. Therefore, we conclude that the observed pollen sensitivity can be attributed to direct effects on anthers and pollen.
Genome-Wide Identification of HsfA2-Dependent HS-Induced Genes in Leaves and Anthers
The differences obtained by expression analysis in leaves and anthers compared to the thermotolerance results (Figs. 2 and 3) prompted us to investigate the global transcriptional alterations caused by HsfA2 suppression in the two tissues. Leaf and anther samples were collected at the same time from wild-type and A2AS-3 plants either kept at control temperature (25°C) or exposed to a 39°C HS for 1 h followed by 1.5 h at 25°C to recover (HR). Transcriptome analysis was performed using massive analysis of 3′-cDNA ends (MACE), a digital RNA-seq gene expression profiling technique that allows the identification and quantification of low abundant transcripts (Fragkostefanakis et al., 2015b; Simm et al., 2015; Zawada et al., 2014). We were able to monitor the expression of 24.874 genes (Supplemental Table S1). The identification and verification of differentially expressed genes between tissues (anther versus leaf), genotypes (wild type versus A2AS-3), and conditions (control versus heat stressed) was performed via NOISeq using a PNOI-value > 0.95 for differential expression (Tarazona et al., 2011).
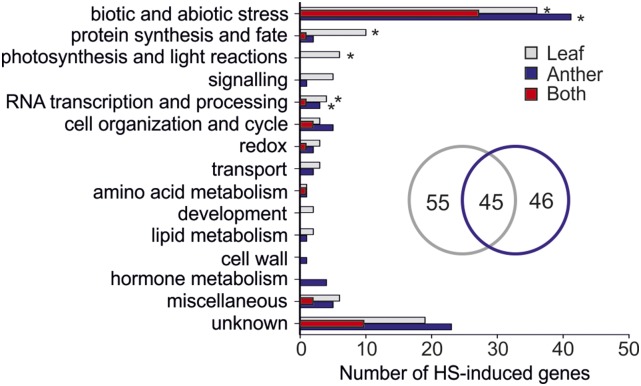
Higher expression after HS in either one or both wild-type tissues was observed for 146 genes in comparison to nontreated plants (Fig. 5; Supplemental Table S2). Only 45 genes were common for both tissues, while 55 and 46 appeared to be upregulated specifically in leaves and anthers, respectively (Fig. 5, inset). Although upregulated in both tissues, HsfA2 was found to sustain significantly increased levels specifically in anthers during recovery (Supplemental Table S2), which is in agreement with our qRT-PCR analysis (Figs. 2 and 3). The majority of up-regulated genes in both tissues are related to biotic and abiotic stress responses. Moreover, genes encoding for proteins involved in RNA transcription and processing are significantly up-regulated in at least one of the two tissues, while genes involved in protein synthesis and photosynthesis are specifically induced in leaves (Fig. 5). Approximately half of the genes induced in both leaves and anthers encode for Hsps (Supplemental Table S2).
Figure 5.
Classification of HS-induced genes in leaves and anthers of wild-type tomato plants. HS-induced genes (PNOI > 0.95) were categorized based on their putative functions. Each bar shows the total number of genes induced by HS in leaves (gray), anthers (blue), or in both tissues (red). The number of leaf- and anther-specific HS-induced genes are shown in a Venn diagram. Asterisks denote significant enrichment of the particular category in the tissues (P < 0.05) based on comparison against the whole genome.
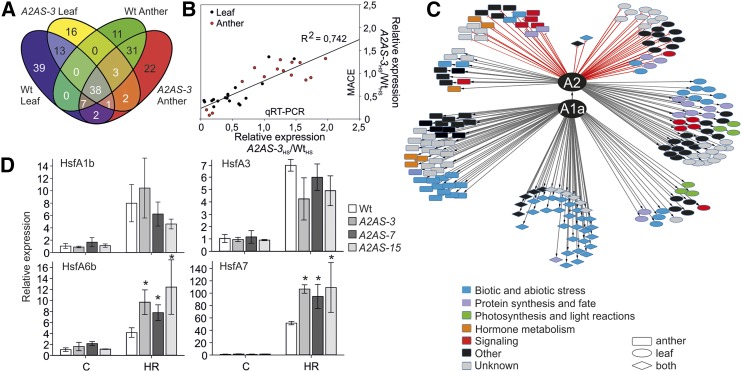
Out of the 100 genes upregulated in wild-type leaves (Fig. 5; PNOI > 0.95), 48 did not show a significant accumulation in A2AS-3 leaves, suggesting an HsfA2-dependent expression (Fig. 6A; Supplemental Table S3A). These genes are mainly involved in protein synthesis and degradation, photosynthesis, signal transduction, and amino acid and lipid metabolic pathways (Fig. 6C; Supplemental Table S3A).
Figure 6.
Differential gene expression in wild-type and A2AS leaves and anthers in response to heat stress. A, Venn diagram depicting the number of genes with significantly enhanced transcript abundance (PNOI > 0.95) in response to HS in wild-type and A2AS-3 leaves and anthers. B, Correlation between MACE and qRT-PCR results (Supplemental Fig. S1) on expression of different genes in HS samples. Expression values of A2AS-3 heat-stressed samples were normalized to the values of stressed samples in wild-type leaves and anthers. Each point is the average of three independent experiments. R2 shows the correlation between MACE and qRT-PCR. C, Regulatory network of HsfA2 in HSR of leaves and anthers. In the interaction scheme, genes are depicted as nodes and interactions as edges. Gray and red lines indicate positive and negative relation, respectively. The shape of each node denotes tissue specificity and the color functional categorization. D, Relative expression (2–ΔΔCt) of four Hsfs in anthers of wild-type and A2AS transgenic tomato plants exposed at 39°C for 1 h followed by 1.5 h recovery (gray bar). The Ct value for each gene was normalized to EF1α and UBI and to the expression under control conditions (white bar) individually for the wild type and A2AS. Vertical bars are the average ± se of three independent experiments.
Only 11 out of 90 genes found to be upregulated in wild-type anthers need HsfA2 for higher expression upon HS (Fig. 6A; Supplemental Table S3B). Thus, many genes do not require HsfA2 for induction upon HS in anthers (Supplemental Table S3, G and H). Anther-specific HsfA2-dependent HS targets include among others a putative peptide transporter, a dehydration-responsive family protein, and a type-C DnaJ protein. Remarkably, none of the HsfA2-regulated genes (genes that are only upregulated in the wild type but not in A2AS-3 plants) were common for leaves and anthers, pointing to tissue-specific functions of HsfA2 regarding the regulation of target genes (Fig. 6A).
The results observed by the global transcriptome analysis were confirmed by qRT-PCR on 20 selected genes identified as differentially regulated in leaves or anthers from wild-type and A2AS-3 plants (Fig. 6B; Supplemental Fig. S6). Comparison of the HS samples of A2AS-3 versus the wild type for each tissue confirmed the MACE analysis and also revealed a high correlation of expression levels between MACE and qRT-PCR (R2 = 0.742; Fig. 6B). Expression analysis was further extended for leaves and anthers from A2AS-15 plants, which confirmed the results obtained from the A2AS-3 line (Supplemental Fig. S6).
We also identified 27 genes showing significant induction in A2AS-3, but not in wild-type anthers, representing a response induced by impaired HsfA2 expression (Supplemental Table S3, D and E). Among those genes two were significantly upregulated in A2AS-3 leaves as well, namely, an acid phosphatase and Hsp90-4 (Supplemental Table S3E). However, the chloroplast ClpB/Hsp100 gene, two chaperonins, and a late embryogenesis abundant protein were specifically induced in A2AS-3 anthers (Supplemental Table S3D). In a similar way, we found 21 genes specifically induced in A2AS-3 leaves (Fig. 6C; Supplemental Table S3, C and E).
The majority of the genes identified as differentially regulated comprise heat stress elements (HSEs) in their putative promoter regions, indicating that they are directly regulated by HsfA2 or other Hsfs (Supplemental Table S4). From a total of 198 genes, 191 have at least one dimeric HSE within the −1,000 upstream promoter/5′ untranslated region, which is required for binding of Hsfs.
Expression of Class A Hsfs in Stressed A2AS Anthers
Many genes do not require HsfA2 for transcriptional induction upon HS in anthers (Fig. 6C). This might result from the activation of other Hsfs, which can compensate for HsfA2 suppression. In this direction, we analyzed the transcript abundance of HS-induced Hsfs, such as HsfA1b, HsfA6b, HsfA7, and the highly expressed HsfA3 in anthers from the wild type and three transgenic lines by qRT-PCR (Supplemental Table S5). For all four Hsfs, we observed increased expression after heat treatment irrespective of the genotype analyzed (Fig. 6D); however, the induction of HsfA6b and HsfA7 was much stronger in all three transgenic lines compared to the wild type. Thus, it is likely that HsfA1b and HsfA3 contribute to the regulation of HS response in anthers in general, while the strongly enhanced expression of HsfA7 and HsfA6b in A2AS plants appears to compensate for the suppression of HsfA2.
HsfA2-Dependent Gene Regulation in Nonstressed Anthers
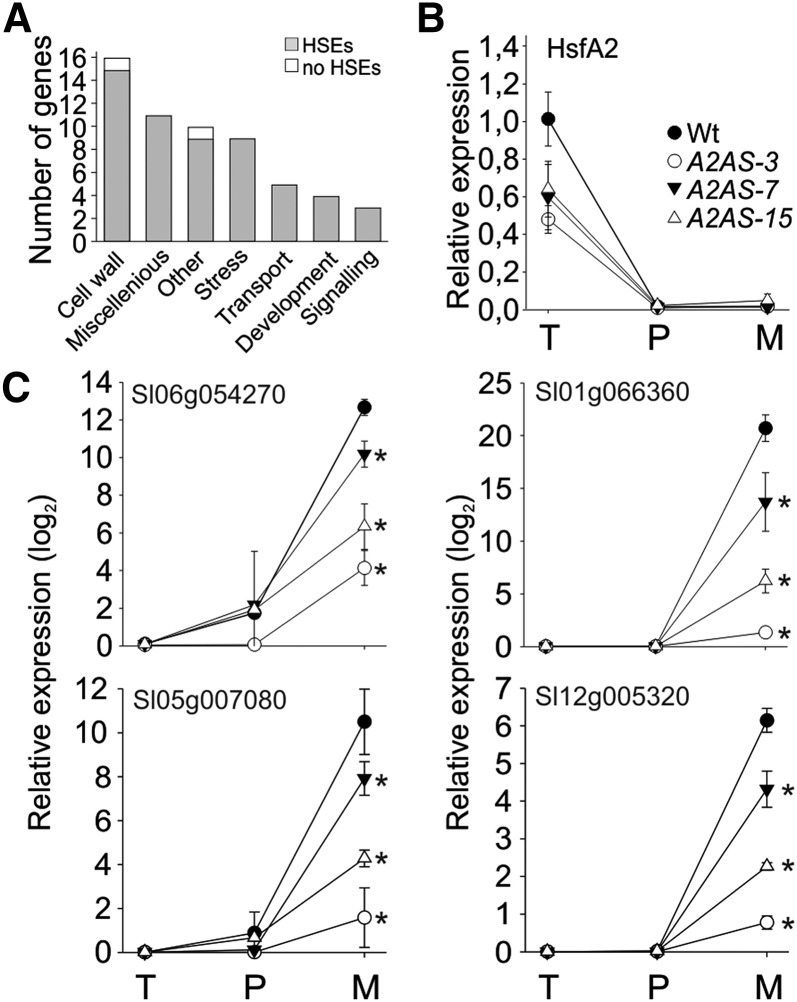
A2AS anthers from nontreated plants release lower number of pollen grains, which points to a function of HsfA2 in sporophytic tissues. Thus, we identified the genes with lower expression in nonstressed anthers of A2AS-3 plants compared to the wild type. Seventy-six genes show a significantly lower transcript abundance in A2AS anthers (PNOI > 0.9) and are therefore considered as developmentally regulated HsfA2-dependent genes (Fig. 7A; Supplemental Table S6). Approximately 25% of the genes with assigned function code for cell wall-modifying proteins, including several pectin methylesterases, pectin acetylesterases, pectate lyases, and an endo-1,4-β-xylanase. In addition, nine of the 76 HsfA2-dependent genes code for stress-related genes, while two of them encoded for desiccation-related proteins. Another five genes possess putative functions in transport of solutes and peptides, suggesting that HsfA2 affects various relevant aspects of anther development.
Figure 7.
Genes affected by HsfA2 suppression in nonstressed anthers. A, Genes expressed at significantly higher levels (PNOI > 0.90) in the wild type than A2AS-3 anthers under control conditions as indicated by MACE analysis. Categorization was based on the putative functions of the genes according to MapMan analysis (see “Materials and Methods”). Genes with unknown functions are not shown. The number of genes belonging to each category is shown. Gray bars indicate the number of genes having at least one dimeric or trimeric HSE in their putative promoter regions. B, Relative expression (2–ΔΔCt) of HsfA2 under control conditions in wild-type and A2AS-3 anthers of different developmental stages corresponding to pollen at tetrad (T), postmeiotic microspore (P), and mature pollen (M) stage, respectively. C, Relative expression (2–ΔΔCt) of a solute carrier protein/Glc transporter (Solyc06g054270), a pectinesterase (Solyc01g066360), a pectate lyase (Solyc05g007080), and a pectinacetylesterase (Solyc12g005320) during development of anthers as described in B. In all cases the expression values were normalized against anthers containing pollen at tetrad stage. Each point is the average ± sd of four replicates. Asterisks indicate statistically significant differences (P < 0.05) as indicated by pairwise t test between wild-type and A2AS samples.
Surprisingly, none of these 76 genes is induced by HS in the wild type or A2AS, indicating that the developmental and the stress-related regulatory networks controlled by HsfA2 in anthers are distinct. Promoter analysis showed that only six of these genes do not comprise HSEs required for binding of Hsfs (Fig. 7A; Supplemental Table S4). We randomly selected four genes for expression analysis in A2AS and wild-type anthers of different developmental stages. We realized that these genes did not follow the expression profile of HsfA2 during anther development (Fig. 7B) but showed a very strong induction in mature anthers (Fig. 7C). Interestingly, in all cases the transcript levels of these genes were significantly lower in mature anthers of the A2AS lines (Fig. 7C).
HsfA2 Regulates the Transcript Abundance of HS-Induced Genes in Pollen
Transcriptome analysis revealed a low number of genes not induced in A2AS anthers in response to HS. We further asked if this holds true for pollen as well. The transcript and protein abundance of HsfA2 was tested in pollen derived from wild-type, A2AS-3, and A1CS plants. Pollen was collected from flower buds of different developmental stages as described for anthers, which were further pooled prior to RNA or protein extraction. We analyzed the expression of HsfA2, Hsp101, and Hsp17.7C-CI in pollen isolated from heat-treated plants as described for leaves and anthers (Fig. 8A). The transcript abundance of HsfA2 was strongly enhanced after HS treatment in wild-type pollen, but during recovery it rapidly decreased to the level of control (Fig. 8A). HsfA2 expression in pollen of A2AS-3 and A1CS plants was found to be suppressed under both control and HS conditions. The efficient suppression of HsfA2 was also confirmed in individual stages of pollen development in A2AS-3 plants (Fig. 8B). Similar to HsfA2, Hsp101 and Hsp17.7C-CI expression was detectable in nonstressed wild-type pollen and the accumulation of their transcripts was reduced in A2AS-3 and strongly repressed in A1CS during HS (Fig. 8A).
Figure 8.
Effect of HsfA2 suppression on expression of heat stress induced genes in pollen. A, Relative expression (2–ΔΔCt) of indicated genes in wild-type, A2AS-3, and A1CS pollen isolated from anthers of nontreated plants (sample C) or exposed to HS at 39°C for 1 h (sample H) and after recovery at 25°C for 1.5 h (sample R). Pollen for each control or stress sample were collected from pooled anthers of different developmental stages ranging from tetrads to mature pollen. Each point is the average ± sd of three replicates. Asterisks indicate statistically significant differences (P < 0.05) as indicated by pairwise t test between wild-type, A2AS-3, and A1CS samples. B, Relative expression (2–ΔΔCt) of HsfA2 in pollen at tetrad (T), postmeiotic (P), and mature binucleate (M) developmental stages isolated from wild-type and A2AS-3 plants. Transcript abundance of HsfA2 normalized to wild-type tetrads. C, Immunoblot analysis with specific antibodies against HsfA1a (A1), HsfA2 (A2), Hsp17-CI (17-CI), and Hsp101 (101) in cell lysates from wild-type, A2AS-3, and A1CS pollen isolated from anthers of plants treated as in A. Equal protein loading (15 μg) was controlled by immunodetection of actin.
Subsequently, we analyzed the protein abundance of HsfA1a, HsfA2, Hsp17-CI, and Hsp101. We observed the typical induction of HsfA2 protein accumulation during HS and recovery in the wild type (Fig. 8C), which was inhibited in pollen of A2AS-3 and A1CS plants. This suggests that HsfA1a is a central factor for the regulation of HSR in pollen as shown for anthers. In contrast, the increase of Hsp17-CI and Hsp101 at the protein level was comparable between the wild type and A2AS-3, while in HS-treated A1CS pollen, the induction was significantly delayed (Fig. 8C). Thus, the decrease of transcript abundance under HS in A2AS-3 pollen (Fig. 8A) does not result in a corresponding reduction at the protein level; therefore, HsfA2 action seems not to be required for the accumulation of these proteins in pollen in response to HS.
HsfA2 Is Involved in Developmental Regulation of HSR Genes in Anthers and Pollen
It is assumed that Hsps are synthesized during microsporogenesis and stored for recruitment in cases of stress occurring during later stages of pollen development (Chaturvedi et al., 2013). In addition, anther tissues and pollen are metabolically very active, as indicated by the increased expression of many ribosome biogenesis factors, which exhibit expression profiles similar to HsfA2 (Simm et al., 2015). In this direction, we asked whether HsfA2 regulates the abundance of HS-related genes such as Hsps under control conditions in anthers and pollen.
The higher expression of Hsps and other HS response genes has been shown for only a limited number of genes in early stages of tomato pollen development (Frank et al., 2009; Giorno et al., 2010). We extended these findings by monitoring the expression profile of several Hsps in tetrads, postmeiotic microspores, and mature binucleate pollen isolated from wild-type plants. For a direct comparison with vegetative tissues, the transcript levels were normalized against the expression of these genes in nonstressed young leaves of the same plants. In general, we observed a significantly higher expression in tetrads compared to young leaves that, however, was reduced again in postmeiotic microspores and mature pollen grains (Supplemental Fig. S7).
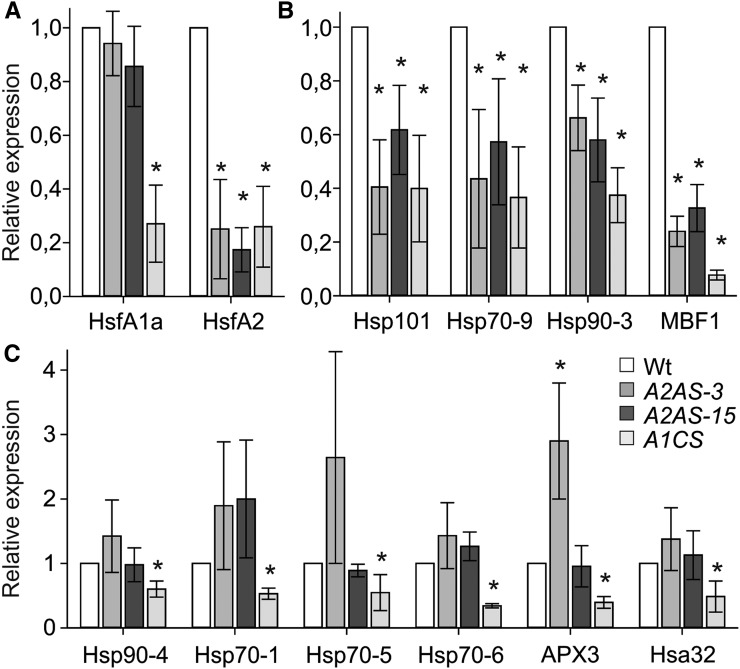
We checked whether HsfA2 is directly involved in the developmental regulation of these genes by qRT-PCR on RNA isolated from meiotic tetrad cells of wild-type, A2AS-3, and A2AS-15 plants (Fig. 9). Only Hsp101, Hsp90-3, Hsp70-9, and MBF1c were down-regulated in the A2AS background, while the same analysis in A1CS tetrads revealed that HsfA1a is required for higher expression of all analyzed Hsps (Fig. 9, B and C). This result suggests that the master regulator function of HsfA1a is not restricted to HSR but has a broader role for the function and regulatory activity of the Hsf network in male reproductive cells as well. In contrast, HsfA2 is required for increased expression of only a subset of developmentally regulated Hsp genes. Expression analysis for Hsp101, Hsp90-3, Hsp70-9, and MBF1c in wild-type, A2AS-3, -7, and -15 anthers yielded the same results (Supplemental Fig. S8B). The lower abundance of HsfA2 and Hsp101 proteins in A2AS-3, A2AS-7, A2AS-15, and A1CS anthers compared to the wild type was confirmed by immunoblot analysis of protein extracts from anthers corresponding to the three different developmental stages investigated (Supplemental Figs. S8A and S9). Collectively, we were able to show that HsfA2 is required for the increased expression of several HS-related genes in early stages of pollen and anther development.
Figure 9.
Regulation of HS-responsive genes in pollen at tetrad stage. A to C, Relative expression (2–ΔΔCt) is shown for HsfA1a and HsfA2 (A), genes depending on HsfA1a and HsfA2 for higher expression (B), and genes requiring HsfA1a but not HsfA2 (C) in tetrads from wild-type, A2AS-3, A2AS-15, and A1CS plants. Transcript abundance of HsfA2 normalized to wild-type tetrads. Asterisks indicate statistically significant differences (P < 0.05) based on pairwise t test between the wild type and A2AS-3, A2AS-15, or A1CS.
Metabolic Changes Influenced by HsfA2 in Control and Heat-Stressed Anthers
Disturbance of protein homeostasis by impaired accumulation of molecular chaperones is expected to influence different cellular activities including metabolite synthesis or uptake. In turn, alterations in the abundance of metabolites in anthers can influence both pollen development at physiological conditions and thermotolerance because developing pollen are highly dependent on supply of nutrients and metabolites from surrounding sporophytic tissues (Paupière et al., 2014). We performed nontargeted metabolomic profiling by gas chromatography coupled to mass spectrometry of control and stressed anthers from wild-type and A2AS-3 plants to examine whether HsfA2 suppression influences the primary metabolite composition of these tissues.
High-temperature stress caused the elevation of several metabolites that were shown previously to increase after HS treatment in Arabidopsis leaves (Kaplan et al., 2004), including putrescine, succinic acid, γ-amino butyric acid (GABA), and Val (Table I). From the metabolites identified here, 19 were increased in wild-type anthers by HS, while two were found to be reduced, namely, Fru and Glc. Interestingly, from the 19 induced metabolites, 13 did not show a similar increase in stressed A2AS anthers, indicating that HsfA2 suppression affects the accumulation of metabolites, which could further compromise thermotolerance. Such metabolites are GABA, putrescine, Fru-6-P and Glc-6-P, Fuc, galactonic acid, and succinic acid. Interestingly, we detected lower levels of several metabolites, including Fru-6-P and Glc-6-P, putrescine, and several amino acids such as Phe, Tyr, and Val already under control conditions, suggesting a developmental effect of HsfA2 suppression. Thus, HsfA2 is involved in the developmental priming of anthers as its suppression already affects the transcriptome, proteome, and metabolome of anthers in the absence of any stress.
Table I. Metabolites affected by HsfA2 suppression in control or heat-stressed anthers Letters indicate statistically significant differences (P < 0.05) as indicated by pairwise t test comparisons. The ±sd for each treatment is indicated (n = 5). WT, wild type.
| Class | Name | Retention Time (min) | Match Factor | ∆ Retention Index | WT Control | WT Heat Stress | A2AS Control | A2AS Heat Stress | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alcohol | 1,2,3-Butantriol | 13.90 | 655 | 56.68 | 3,780b | ±385 | 5,016a | ±823 | 2,778c | ±582 | 3,147bc | ±455 |
| Amino acid | γ- aminobutyric acid | 7.31 | 937 | 18.12 | 60,016b | ±9,755 | 82,222a | ±8,462 | 49,377b | ±8,036 | 60,699b | ±12,546 |
| l-Ser | 5.75 | 952 | −16.45 | 40,524ab | ±10,216 | 46,117a | ±15,824 | 25,392b | ±7,614 | 32,806ab | ±8,199 | |
| l-Phe | 8.60 | 727 | 82.53 | 3,475b | ±480 | 5,004a | ±629 | 2,457d | ±619 | 3,245bc | ±325 | |
| l-Tyr | 10.67 | 769 | 103.91 | 4,921c | ±942 | 9,788a | ±850 | 3,573d | ±790 | 6,952b | ±1,332 | |
| l-Val | 4.33 | 874 | −5.60 | 5,955c | ±583 | 9,496a | ±761 | 4,935d | ±572 | 7,270b | ±1,028 | |
| Organic acid | Glyceric acid-3-P | 9.29 | 726 | 12.57 | 2,418b | ±310 | 4,324a | ±773 | 2,403b | ±343 | 2,968b | ±427 |
| Citric acid | 9.60 | 910 | 52.72 | 20,623b | ±13,040 | 14,156b | ±3,034 | 32,393c | ±13,562 | 49,168a | ±7,144 | |
| 2,3-Dihydroxybutanedioic acid | 7.60 | 905 | −15.20 | 15,976b | ±1,146 | 19,963a | ±3,868 | 13,150b | ±1,753 | 16,491ab | ±3,481 | |
| Citramalic acid | 6.87 | 700 | 19.31 | 1,770a | ±124 | 2,086a | ±235 | 1,320b | ±232 | 1,814a | ±315 | |
| Pentonic acid | 8.51 | 857 | −45.11 | 8,626b | ±334 | 11,046a | ±1,928 | 7,573b | ±902 | 9,087b | ±982 | |
| Galactonic acid | 10.15 | 905 | −32.10 | 42,050b | ±5,056 | 50,100a | ±6,956 | 31,707c | ±3,658 | 38,715bc | ±6,816 | |
| Gluconic acid | 10.30 | 872 | −13.21 | 8,590b | ±988 | 10,692a | ±2,178 | 7,224b | ±797 | 8,197b | ±849 | |
| Putrescine | 8.38 | 904 | −52.92 | 34,036b | ±6,882 | 43,149a | ±6,009 | 23,797c | ±6,205 | 26,103c | ±6,307 | |
| Succinic acid | 5.88 | 884 | 32.02 | 28,793b | ±1,925 | 37,527a | ±7,699 | 23,408b | ±3,484 | 27,117b | ±4,641 | |
| Phosphate | Phosphoric acid | 5.35 | 934 | 27.46 | 63,899a | ±2,204 | 64,838a | ±2,333 | 59,068b | ±2,267 | 63,123a | ±3,625 |
| Sugar | Fru | 9.48 | 944 | −21.02 | 84,662a | ±2,847 | 77,245b | ±2,350 | 81,132a | ±2,867 | 69,483c | ±4,430 |
| Fru-6-P | 12.44 | 530 | 49.84 | 898b | ±70 | 1,056a | ±123 | 722c | ±105 | 827bc | ±151 | |
| Maltose | 13.97 | 625 | −107.40 | 7,107b | ±733 | 9,285a | ±2,248 | 5,810b | ±1,049 | 5751b | ±964 | |
| Melibiose | 15.97 | 740 | −12.76 | 4,343bc | ±452 | 5,559a | ±432 | 3,824c | ±372 | 4591b | ±567 | |
| Pentitol | 7.84 | 697 | −80.70 | 721bc | ±120 | 1,266a | ±266 | 652c | ±150 | 941b | ±169 | |
| Rhamnose | 8.35 | 642 | −33.39 | 2,792c | ±383 | 9,015a | ±1,943 | 2,425cd | ±473 | 6,093b | ±1,682 | |
| Xyl | 8.07 | 862 | −8.97 | 10,435a | ±2,804 | 10,080a | ±1,480 | 10,088a | ±2,034 | 7,004b | ±860 | |
| Glc | 9.57 | 941 | −21.30 | 80,963a | ±7,976 | 64,851b | ±7,848 | 90,324a | ±8,774 | 81,349a | ±6,638 | |
| Glc-6-P | 12.54 | 882 | 53.71 | 8,528ab | ±781 | 9403a | ±746 | 7,038c | ±849 | 7,618bc | ±1,403 | |
| β-d-Methylfructofuranoside | 9.04 | 843 | 10.31 | 10,992ab | ±1,823 | 11,825a | ±2,800 | 5,762c | ±3,038 | 8,245abc | ±2,928 | |
| Fuc | 8.46 | 578 | −26.55 | 7,332b | ±501 | 9,795a | ±1,495 | 6,278b | ±824 | 6,890b | ±708 | |
DISCUSSION
Tissue-Specific Regulation of Tomato HsfA2
HSR is characterized by the activation of Hsfs, which induce the expression of a cohort of selected genes to facilitate primarily the protection of protein homeostasis and adjustment of cellular metabolism (Mishra et al., 2002; Liu and Charng, 2013). Different cell types with diverse functions show different sensitivity to HS and are therefore characterized by distinct qualitative and quantitative transcriptional changes. Tomato HsfA2 belongs to the most strongly induced Hsfs and is shown here to be highly upregulated in response to HS in seedlings, leaves, anthers, and pollen. Analyzing HsfA2 transcript and protein levels after 1 h of HS or during subsequent recovery periods indicates that HsfA2 expression and stability undergo tissue-specific regulation. In leaves, the expression of HsfA2 was repressed during recovery and transcripts rapidly declined to the level found in the untreated samples, while in anthers, the decrease was only moderate (Figs. 2A and 3A). Hsp101 and Hsp17.7C-CI transcript levels were also reduced but remained present at higher abundance during recovery in all tissues. Therefore, we can assume that the observed tissue-specific behavior is an HsfA2-specific regulatory feature related to the attenuation phase of HSR. In contrast to the mRNA decline, HsfA2 protein was stabilized during recovery in all tissues, and in seedlings, HsfA2 was detectable even 48 h after the stress (Supplemental Fig. S2). The ongoing accumulation of HsfA2 protein and maintenance of high levels during recovery is in accordance with the increased expression of Hsps and points to the proposed function of HsfA2 as major coactivator of HSR in thermotolerant cells (Scharf et al., 1998; Schramm et al., 2006; Chan-Schaminet et al., 2009).
The existence of tissue-specific regulation of HsfA2 is further supported by the absence of any detectable amount of protein in nonstressed wild-type or A2S seedlings and leaves, which is in contrast to the strong accumulation of HsfA2 mRNA (Supplemental Figs. S1B and S3D). This indicates that HsfA2 in nonstressed cells is subjected to posttranscriptional regulation that prevents translation of the mRNA or rapid degradation of the newly synthesized protein is promoted. Although treatment of A2S leaf discs with the proteasome inhibitor MG132 did not result in detectable HsfA2 protein accumulation (data not shown), the contribution of both mechanisms cannot be ruled out and further investigations are needed. However, such a control mechanism seems to be tissue specific because HsfA2 protein was detectable in early stages of anther development under nonstress conditions (Giorno et al., 2010).
HsfA2 Is Required for Enhanced Activation of HSR and Increased Thermotolerance in Specific Tissues and Cell Types
Accumulating HsfA2 under ongoing HS conditions is recruited into cytoplasmic heat stress granules and is released during recovery to enhance the expression of proteins required for protection and restoration of cellular homeostasis during repeated HS periods (Scharf et al., 1998; Mishra et al., 2002). In this sense, the stability and activity of HsfA2 is controlled by interacting with HsfA1a and class CI and CII sHsps (Port et al., 2004), thereby HsfA2 becomes the major Hsf involved in regulation of acquired thermotolerance (Charng et al., 2007). Here, we show that the knockdown of tomato HsfA2 expression leads to significant reduction of the capacity of seedlings to acquire increased thermotolerance, i.e. to resist a challenging stress treatment at otherwise lethal conditions following an acclimation by sublethal HS pretreatment (Fig. 1C). Similar observations regarding loss of long-term acquired thermotolerance have been reported for knockout lines of Arabidopsis HsfA2 (Charng et al., 2007). However, in contrast to this report, we noticed a reduced acquired thermotolerance in the A2AS seedlings already after a short recovery period of 3 h (Fig. 1C). This is in agreement with the reduced accumulation of Hsp101 and Hsp17-CI in tomato seedlings, which was not observed in Arabidopsis and might indicate species-specific functional diversification between the two HsfA2 orthologs in the regulation of the initial HSR.
HsfA2 suppression, on the other hand, affected neither the accumulation of Hsps in seedlings exposed to 39°C for 1 h nor the basal thermotolerance of seedlings (Fig. 1). In contrast, A2AS leaves show lower accumulation of Hsps, suggesting that knockdown of HsfA2 hampers the full activation of HSR in leaves. Even more, transcriptome analysis revealed that in leaves, 48 genes require HsfA2 for increased expression during recovery from stress. However, no alteration in thermotolerance of leaves was detected, as indicated by the unchanged photosynthetic activity (Supplemental Fig. S5), suggesting that HsfA2 accumulation and subsequently a strong coactivation of HSR are not essential for protection of the photosynthetic activity in leaves under sublethal HS conditions in leaves.
In general, our transcriptome analysis revealed a lower number of stress-induced genes compared to other studies (Finka et al., 2011; Liu and Charng, 2013; Rizhsky et al., 2004). This on one hand can be attributed to the high stringency criteria we used here for the identification of differentially expressed genes and, on the other hand, on the fact that stressed samples were harvested after 1.5 h of recovery. Based on the results for Hsp101 and Hsp17.7C-CI in leaves, we cannot exclude the possibility that during HS a higher number of genes require HsfA2 for increased expression.
Anthers containing sporophytic cells and pollen showed different requirements for HsfA2 than leaves during HSR. Only 12 genes in anthers are dependent on HsfA2 for maintaining increased expression during recovery from heat stress. Remarkably, genes requiring HsfA2 were different in leaves and anthers, pointing to a tissue-specific HsfA2 function regarding the regulation of target genes. In general, the majority of the genes identified as HsfA2 dependent in the two tissues (i.e. 56 out of 59 in total) comprise at least one HSE in the promoter region and can be therefore directly regulated by HsfA2 (Supplemental Table S4). The three genes that do not have HSEs are probably not controlled directly by HsfA2 or other Hsfs but rather by other transcription factors. Interestingly, functional prediction assigned a HSE in the promoter of a putative WRKY transcription factor, which is expressed in an HsfA2-dependent manner.
Several genes with increased expression were identified in A2AS stressed leaves and/or anthers, but not in the wild type. Considering that HsfA2 as a class A Hsf is predicted to possess only activator function, we assume that the elevated levels of those genes in A2AS plants might be either due to the reduced activity of a transcriptional repressor or the increased activity of other Hsfs or due to alterations in the stability of their transcripts in the A2AS background. Indeed, MACE analysis showed that the stress inducible HsfA6b and HsfA7 that share the highest sequence homology with HsfA2 (Scharf et al., 2012) sustained higher levels in A2AS stressed anthers compared to the wild type. This indicates a possible compensation for the suppressed HsfA2 function by HsfA6b and HsfA7 (Fig. 6D). In the same direction, the absence of a strong phenotype in stressed A2AS seedlings might also be due to the activation of other Hsfs.
The Contribution of HsfA2 in Pollen Thermotolerance
Despite the low number of genes affected by HsfA2 suppression in heat-stressed anthers and the efficient protein accumulation of two major Hsps, we realized a significantly lower viability and germination rate for A2AS pollen compared to the wild type (Fig. 4). In contrast to vegetative tissues, HsfA2 and other HS-induced genes are expressed at earlier stages of pollen development under nonstress conditions (Supplemental Fig. S7). The fact that the increased thermosensitivity was apparent only for pollen derived from flowers that received the stress 9 to 12 d prior to anthesis suggests that the function of HsfA2 is developmental stage specific. In fact, HsfA2 knockdown resulted in lower accumulation of several Hsps and the transcriptional regulator MBF1c in pollen (Fig. 9). This suggests that the increased expression of HsfA2 in developing pollen leads to higher protection against thermal stress in a manner similar to acquired thermotolerance in seedlings (Fig. 1C). The expression of HsfA2 in nonstressed tetrads and microspores is under the control of HsfA1a. Therefore, during microsporogenesis, the stimulation of HsfA1a and HsfA2 activates a priming process that enhances pollen fitness in case of environmental stress (Duck and Folk, 1994; Chaturvedi et al., 2013).
The implication of Hsf-Hsp networks in developmental programs has been shown for embryogenesis and seed maturation (Almoguera et al., 2009; Kotak et al., 2007; Prieto-Dapena et al., 2008; Wehmeyer and Vierling, 2000). Seed-specific overexpression of the sunflower (Helianthus annuus) HsfA9 in tobacco (Nicotiana tabacum) enhanced the accumulation of Hsps, improved the resistance of seeds against controlled deterioration, and increased seed longevity (Prieto-Dapena et al., 2006).
HsfA2 suppression in anthers resulted already under control conditions in reduced levels of several genes with diverse putative functions in anther and pollen development. Interestingly, none of these genes was HS induced, suggesting a function of HsfA2 beyond HS. The absence of Hsps can be attributed to the fact that the RNA used for transcriptome analysis derived from pooling of anthers of different stages while the HsfA2 suppression affects Hsps only at the tetrad stage. Nevertheless, the genes identified to be developmentally regulated by HsfA2 in anthers might be related to the lower number of released pollen in A2AS plants or with the reduced thermotolerance of pollen.
Anther dehiscence that leads to pollen release is a complex process requiring successive cell differentiation and degeneration of specific tissues (Sato et al., 2000). The cell wall was the highest represented functional category among the 76 HsfA2-regulated genes, including genes encoding for enzymes related to cell wall remodeling, such as pectin methylesterases, pectin acetylesterases, and pectate lyases (Supplemental Table S6). Pectin is very abundant in anther and pollen cell walls, and genetic studies in different species have pointed to the significance of pectin remodeling in various aspects of development of male reproductive tissues. For example, QUARTET1 in Arabidopsis codes for a pectin methylesterase, which is expressed in both pollen and surrounding anther tissues and is implicated in separation of tetrads (Francis et al., 2006). Furthermore, overexpression of cotton (Populus trichocarpa) pectin acetylesterase 1 (PtPAE1) in tobacco impaired the development of pollen grains resulting in anthers with fewer grains in pollen sacs (Gou et al., 2012).
Fifteen out of sixteen cell wall-related genes comprise HSEs in their promoter region, suggesting that they might be directly regulated by HsfA2 (Supplemental Table S4). However, stage-specific expression analysis of four genes revealed that they follow a different profile than HsfA2 by showing a gradual increase during anther development (Fig. 7, B and C). This suggests that these genes are probably not solely regulated by HsfA2 but also by other Hsfs or even other transcription factors.
Other genes with lower expression in A2AS anthers under control conditions are predicted to be involved in various metabolic processes, including synthesis of secondary metabolites. A gene encoding for a putative chalcone isomerase was found to be down-regulated in A2AS anthers. Chalcone isomerase is implicated in flavonoid biosynthesis and a self-fertile chalcone synthase null allele in morning glory (Ipomoea purpurea) exhibited reduced male fertilization success at high temperatures, suggesting that flavonoids can mitigate HS effects (Coberly and Rausher, 2003). Phenylpropanoids also accumulate in response to HS and enhanced activity of Phe ammonia-lyase, the key enzyme of phenylpropanoid pathway, promotes acclimation of cells to HS (Rivero et al., 2001). In this direction, reduced levels of hydroxycinnamoyl CoA quinate transferase, which is involved in phenylpropanoid biosynthesis, might be related to the lower capacity of anther and pollen to withstand HS.
Furthermore, the pleiotropic effects of HsfA2 suppression in anthers include also changes in the abundance of primary metabolites including sugars, carbohydrates, and amino acids, the levels of which are critical for thermotolerance (Table I). Interestingly, already under control conditions several metabolites such as the sugars Fru- and Glc-6-phosphate as well as the polyamine putrescine exhibited lower levels in A2AS anthers, which also led to decreased contents in stressed anthers. Similar to protein homeostasis, disturbance of the metabolic equilibrium can cause developmental defects and increase the sensitivity of cells against stress conditions. This also applies for pollen that is highly dependent on nutrient and metabolite supply from surrounding tissues. In addition, we also observed lower content of some metabolites in stressed A2AS anthers that are not affected in control, including Fru, maltose, and GABA, while Glc and citric acid are maintained at higher levels in A2AS stressed anthers compared to the wild type. The nonproteinogenic amino acid GABA accumulates during HS and is believed to have a protective role against oxidative stress (Kinnersley and Turano, 2000). Interestingly, the changes in the levels of metabolites were not directly correlated with the expression of genes involved in the respective metabolic pathways. We hypothesize that this can be an indirect effect of HsfA2 suppression, for example, because of disturbed protein homeostasis due to the lower levels of molecular chaperones in developing nonstressed anthers.
CONCLUSION
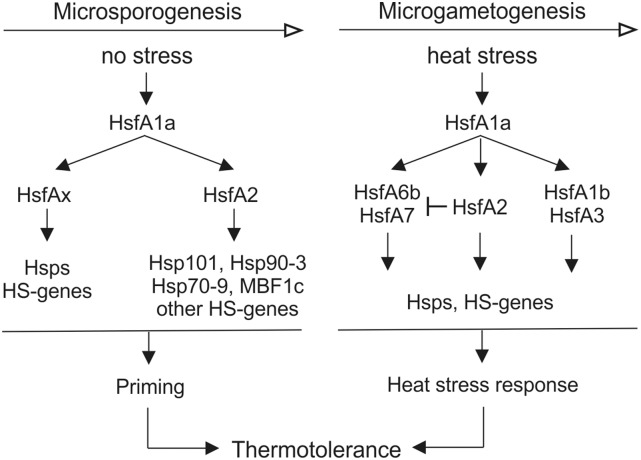
The capacity of male gametophytes to tolerate HS is dependent on developmentally and/or HS-responsive transcriptional cascades mediated by Hsfs (Fig. 10). Pollen mother cells, tetrads, and young microspores compensate for their reduced capacity to induce a strong HSR by activating a priming process that leads to the accumulation of Hsps and other HS-responsive genes (Fig. 10). In this process, HsfA1a acts as master regulator, while HsfA2 is required for the enhanced expression of HS-induced chaperones such as Hsp101 and other protective components characteristic for thermotolerant cells. Other Hsps might be directly regulated by HsfA1a alone or in cooperation with other class A Hsfs. Thermotolerance in more advanced stages of pollen development is mainly dependent on the activation of HSR mechanisms. Based on our results, HSR in maturing microspores is primarily mediated by HsfA1a and HsfA2. Induction of HsfA1b and HsfA3 is HsfA2 independent and cannot compensate for reduced HsfA2 activities. In contrast, the higher induction of HsfA6b and HsfA7 may compensate for decreased HsfA2 availability (Fig. 10).
Figure 10.
Proposed model for regulation of thermotolerance at different stages of pollen development. Thermotolerance of pollen at early stages of development is highly dependent on a priming program that leads to the accumulation of Hsps and other HS-induced genes under nonstress conditions. The induction of these genes is controlled by the master regulator HsfA1a, which further activates HsfA2 and probably other class A Hsfs (HsfAx) to induce downstream genes. HsfA2 is required for the higher expression of some HS genes, including Hsp101, Hsp90-3, Hsp70-9, and MBF1c. Following microspore formation and during microgametogenesis, the capacity of pollen to induce HSR is restored. Here, HsfA1a induces and subsequently interacts with HsfA1b, HsfA2, HsfA3, HsfA6b, and HsfA7 to activate HS genes. HsfA2 induction alleviates the up-regulation of HsfA6b and HsfA7, which in turn are strongly induced in case of limited HsfA2 availability caused by suppression.
MATERIALS AND METHODS
Generation of Transgenic Plants
Tomato (Solanum lycopersicum) plants (cv Moneymaker) were transformed as described by Koncz and Schell (1986) using the Agrobacterium tumefaciens strain GV3101(pMP90). The binary vector pGPTV-KAN (Becker et al., 1992) was modified by removing the β-glucuronidase (uidA) gene and insertion of the HindIII fragment containing the sense or antisense HsfA2 cDNA expression cassette from the corresponding pRT vectors under the control of CaMV 35S promoter to generate A2S and A2AS transgenic lines, respectively (Scharf et al., 1990). Leaf disc transformation and plant regeneration were done as described (Knapp et al., 1994). Calli were generated on Murashige and Skoog nutrient medium (Duchefa) containing 100 μg mL−1 kanamycin and 500 μg mL−1 carbenicillin. Shoot formation was induced by the addition of 2 μg mL−1 zeatin. After root formation, regenerated T0 plants were transferred to soil and further grown in the greenhouse under 16 h light/8 h dark at 24 and 18°C, respectively. Segregation of the T0 lines was followed for at least the next three generations (T1 to T3) by kanamycin resistance tests. Plants used in this study were all from T5 generation. HsfA1 cosuppression transgenic plant (A1CS2 here referred as A1CS) has been previously described by Mishra et al. (2002).
HS Treatments and Thermotolerance Assays
Eight-week-old wild-type and HsfA2 transgenic tomato plants (S. lycopersicum cv Moneymaker, lines A2AS-3, -7, and -15) grown in a glasshouse under a 16/8-h day/night cycle (25–20°C) were transferred to a growth chamber (120 μmol m−2 s−1 light intensity similar to greenhouse conditions) and exposed to HS conditions for 1 h at 39°C and then allowed to recover for 1.5 h at 25°C or kept for the same time at 25°C as control. Each biological replicate comprised young leaves and anthers, which were pooled from at least eight control and HS-treated plants and stored at −80°C until further processing.
The photosynthetic activity of control and stressed young leaves from wild-type and A2AS plants was estimated using a PAM fluorometer (Walz). Prior to PAM measurements, the plants were dark adapted for 30 min. Five areas of interest were chosen on the second youngest leaf of each plant. Φ(II), electron transport rate, nonphotochemical quenching, and the coefficient of photochemical quenching were determined according to the instructions of the manufacturer.
Thermotolerance assays on seedlings of wild-type and transgenic lines were performed similar to the hypocotyl elongation assay established for evaluation of thermotolerance in Arabidopsis (Arabidopsis thaliana) seedlings (Queitsch et al., 2000). Tomato seeds were surface sterilized using a quick wash with ethanol, a 15-min wash with 1% sodium hypochloride, and several washes with sterile water. Seeds were allowed to germinate in dark on wet paper towels in petri dishes at 25°C. Ten 4-d-old etiolated seedlings from each genotype grown vertically were exposed to either 42 or 45°C HS and then allowed to recover in the dark. Control seedlings were kept constantly at 25°C. Dark conditions allowed us to minimize the effect of oxidative stress, which is concomitantly generated with HS. For acquired thermotolerance, seedlings were preexposed to 37.5°C for 90 min, then allowed to recover for the indicated time and treated for another 90 min at 47.5°C. The seedlings were photographed before and after the HS treatment every day onwards, and hypocotyl length was determined by using ImageJ. Each experiment was repeated three times.
For evaluation of pollen thermotolerance, 8-week-old plants grown in a greenhouse under conditions as described above were exposed to 39°C for 3 h. Pollen number and viability were determined in flowers treated at early stages of pollen development (meiocytes and tetrads; ∼9–12 d prior to anthesis; ∼2–4-mm bud length at the time of treatment).
For pollen isolation, flowers at anthesis stage from control and treated plants were collected, anthers were excised, transferred to germination solution [1 mm KNO3, 3 mm Ca(NO3)2, 0.8 mm MgSO4, 1.6 mm H3BO3, 5% (w/v) Suc, and 25% (w/v) PEG4000], and vortexed for 30 s to allow the collection of released pollen fraction. After vortexing, anthers were transferred to new tubes and the number of the remaining pollen (unreleased fraction) was determined. Pollen grains that remained in the anthers after vortexing were released by mechanical disruption using micropestles in germination solution. Pollen number was determined using a Fuchs-Rosenthal cell counter and viability by staining with Alexander dye (Alexander, 1980). Released pollen were allowed to germinate at 25°C under gentle shaking for 3 h in germination solution and were considered as germinated when the pollen tube length had at least the diameter of the pollen grain. The experiment was conducted at least three times with 15 plants per genotype and treatment. Two to three flowers were harvested from each plant.
RNA Extraction and qRT-PCR
Total RNA was extracted using the E.Z.N.A. Plant RNA Kit (Omega Bio-Tek) following the manufacturer’s instructions. One microgram of total RNA was used for cDNA synthesis with Revert Aid reverse transcriptase (Thermo Scientific) following the manufacturer's protocol. Expression of selected genes was determined using real-time PCR on a Stratagene Mx3000P cycler (Agilent Technologies). The qRT-PCR reaction (10 μL) consisted of gene-specific primers (Supplemental Table S7), PerfeCTa SYBR Green FastMix Low ROX (Quanta Biosciences), and the template. Thermal cycling conditions were 95°C/3 min followed by 95°C/15 s, 60°C/30 s, and 72°C/30 s for 40 cycles. Gene primers (Supplemental Table S7) were designed using PRIMER3 (http://primer3.ut.ee/). Data were analyzed by standard methods (Livak and Schmittgen, 2001) and presented as relative levels of gene expression using ubiquitin (UBI; Solyc07g064130) and EF1α (Solyc06g005060) genes as internal standards. The selection of housekeeping genes was done using BestKeeper (Pfaffl et al., 2004).
Western-Blot Analysis
Frozen homogenized seedlings, leaf tissue, or pollen (50–100 mg) was used for protein extraction (Mishra et al., 2002). Extracts containing about 15 to 20 μg of protein were mixed with an equal volume of 2× SDS sample buffer and separated on 10 or 12% SDS-polyacrylamide gels. Alternatively, proteins from frozen homogenized anther tissues were isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. For immunoblot analysis, proteins were transferred to nitrocellulose membrane (Protran nitrocellulose transfer membrane; Whatman) and further processed for chemiluminescence detection following the manufacturer’s protocol (Perkin-Elmer). Antibodies used for detection of HsfA1a, HsfA2, and Hsp17-CI have been described previously (Mishra et al., 2002). Hsp101 antibody was purchased from Agrisera (AS07-253).
Transcriptome Analysis Using MACE
Total RNA was extracted using the E.Z.N.A. Plant RNA Kit (Omega Bio-Tek) following the manufacturer’s instructions. MACE (Zawada et al., 2014) libraries were prepared and sequenced by GenXPro (Yakovlev et al., 2014). Analysis of MACE read libraries was done using the pipeline by Simm et al. (2015), performing read quality control using FastQC (James et al., 2011), linker sequence trimming via BLAT (Kent, 2002), read-mapping onto the tomato genome (ITAG2.3; SGN; Bombarely et al., 2011) via SSAHA2 (Ning et al., 2001), and transcript counting considering that each MACE read derives from a single transcript (Supplemental Table S1). The number of transcripts per gene was normalized by the library size of mapped reads multiplied by one million. The MACE libraries are deposited in the Gene Expression Omnibus (GSE68500). NOISeq simulated five replicates for each MACE library and used the median to calculate the probability of differentially expressed genes in pairwise comparisons. Here, we consider as differentially expressed genes those with PNOI > 0.95 for HS and >0.9 for development related samples, respectively. The selected criteria are of high stringency because in a comparative study, PNOI > 0.8 for NOISeq showed high correlation with a probability of 0.999 for baySeq and an adjusted P value threshold of 0.001 for edgeR and DESeq (Tarazona et al., 2011). In addition, for PNOI > 0.9, the false positive rate for NOISeq is lower than 0.05 (Zheng and Moriyama, 2013). Functional categorization of selected genes was done using MapMan mapping file S. lycopersicum (http://mapman.gabipd.org/web/guest/mapmanstore). The network of HsfA2-regulated genes for leaf and anther tissues (anther, leaf, or both) was created using Cytoscape (Shannon et al., 2003). The identification of HSEs was performed as described by Fragkostefanakis et al. (2015b).
Metabolomics
Frozen anther material was grinded in liquid N2 with a pestle and mortar. Anther powder (10–45 mg) was subsequently weighed and transferred into a 1.5-mL Eppendorf tube. The extraction of primary metabolites was performed at room temperature as previously described by Carreno-Quintero et al. (2012). Seven hundred microliters of 100% methanol together with 300 μL of distilled water were added to each anther samples. Samples were sonicated for 20 min followed by a centrifugation at maximum speed (17,000g) for 10 min. Three hundred microliters of supernatant was transferred into a new 2-mL Eppendorf tube. Six hundred microliters of distilled water together with 400 μL of chloroform was added. The Eppendorf tube was manually shaken for 5 min followed by a centrifugation at maximum speed (17,000g) for 10 min. Forty microliters of supernatant was then transferred into a crimp vial with insert and dried overnight in a speed vacuum before being injected for the gas chromatography with time-of-flight mass spectrometer (GC-TOF-MS) analysis. Polar metabolites were analyzed with the 7890B gas chromatography system from Agilent Technologies coupled to a Leco Pegasus HAT GC-TOF-MS system with a Gerstel autosampler system. The data were recorded with Chroma TOF software version 4.51.6. Gas chromatography-mass spectrometry raw data were processed for baseline correction, noise determination, and spectral alignment using MetAlign software (Lommen and Kools, 2012). Estimated noise values were then subtracted per mass peak and low intensity values (<100 of mass spectrometry response) were replaced by random values between 80 and 100. Nonspecific masses (<85 D) were removed from the data. Compound mass spectra were extracted using MSClust software (Tikunov et al., 2012) and subsequently annotated using NIST MSSearch (National Institute of Standards and Technology) and the T_MSRI_ID database of GC-TOF-MS spectra (http://csbdb.mpimp-golm.mpg.de/csbdb/gmd/msri/gmd_msri.html). Compounds were annotated based on their match factor and the delta retention index between the library and the data. Data were further normalized by using total ion count.
Accession Numbers
Sequence data from this article can be found in the GenBank data libraries under accession numbers GSE68500.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Thermotolerance of etiolated tomato seedlings of wild-type and HsfA2 transgenic plants.
Supplemental Figure S2. Importance of HsfA2 for acquired thermotolerance.
Supplemental Figure S3. Heat stress response of young leaves of wild-type and HsfA2 transgenic plants.
Supplemental Figure S4. Heat stress response in anthers of wild-type and A1CS plants.
Supplemental Figure S5. Photosynthetic activity parameters in wild-type and HsfA2 transgenic leaves.
Supplemental Figure S6. Transcript levels of HS-induced genes in stressed leaves and anthers as determined by MACE and qRT-PCR.
Supplemental Figure S7. Expression of HS-responsive genes in pollen at tetrad and postmeiotic microspore stages.
Supplemental Figure S8. Regulation of HS-responsive genes during anther development
Supplemental Figure S9. Regulation of HsfA2 and Hsp101 during anther development by HsfA1a.
Supplemental Table S1. Overview of characteristics of the analyzed MACE libraries prepared from young leaves and anthers of control and heat-stressed tomato wild-type and A2AS plants.
Supplemental Table S2. Genes differentially expressed in response to heat stress in young leaves and anthers from wild-type flowering plants as determined by NOISeq analysis.
Supplemental Table S3. Genes differentially expressed in response to heat stress in young leaves and anthers from wild-type and A2AS flowering plants as determined by NOISeq analysis.
Supplemental Table S4. Heat stress elements identified in the region 1 kb upstream of the start codon of differentially expressed genes.
Supplemental Table S5. Expression (transcripts per million [TPM]) of Hsfs and Hsps in MACE libraries.
Supplemental Table S6. Genes with significantly lower expression in control A2AS anthers compared to the wild type.
Supplemental Table S7. List of primers used for qRT-PCR analysis.
Supplementary Material
Acknowledgments
We thank Lutz Nover for long-term endorsement and critical attendance of the work, Daniela Bublak, Lisa Vanessa Jahn, and Kerstin Pohl for excellent technical assistance, Sascha Röth and members of the SPOT-ITN consortium for helpful comments during the preparation of the manuscript, as well as Holger Schranz and Gerald Kircher for maintenance of plants in the greenhouse. We also thank Alexandra Florian and Alisdair Fernie for their help with metabolomics analysis. This manuscript is a contribution of SPOT-ITN.
Glossary
- HS
heat stress
- HSR
heat stress response
- CaMV
Cauliflower mosaic virus
- HSE
heat stress element
- GABA
γ-amino butyric acid
- MACE
massive analysis of 3′-cDNA ends
Footnotes
This work was supported by grants from the Bundesministerium für Bildung and Forschung to K.-D.S., from the Deutsche Forschungsgemeinschaft (SFB902 to E.S.), and from SPOT-ITN/Marie-Curie to A.B., E.S., and K.-D.S.
Articles can be viewed without a subscription.
References
- Alexander MP. (1980) A versatile stain for pollen fungi, yeast and bacteria. Stain Technol 55: 13–18 [DOI] [PubMed] [Google Scholar]
- Almoguera C, Prieto-Dapena P, Díaz-Martín J, Espinosa JM, Carranco R, Jordano J (2009) The HaDREB2 transcription factor enhances basal thermotolerance and longevity of seeds through functional interaction with HaHSFA9. BMC Plant Biol 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20: 1195–1197 [DOI] [PubMed] [Google Scholar]
- Bharti K, Von Koskull-Döring P, Bharti S, Kumar P, Tintschl-Körbitzer A, Treuter E, Nover L (2004) Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 16: 1521–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, Leto J, Gosselin J, Mueller LA (2011) The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res 39: D1149–D1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno-Quintero N, Acharjee A, Maliepaard C, Bachem CW, Mumm R, Bouwmeester H, Visser RG, Keurentjes JJ (2012) Untargeted metabolic quantitative trait loci analyses reveal a relationship between primary metabolism and potato tuber quality. Plant Physiol 158: 1306–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan-Schaminet KY, Baniwal SK, Bublak D, Nover L, Scharf K-D (2009) Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression. J Biol Chem 284: 20848–20857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, Wang TT (2007) A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol 143: 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi P, Ischebeck T, Egelhofer V, Lichtscheidl I, Weckwerth W (2013) Cell-specific analysis of the tomato pollen proteome from pollen mother cell to mature pollen provides evidence for developmental priming. J Proteome Res 12: 4892–4903 [DOI] [PubMed] [Google Scholar]
- Chauhan H, Khurana N, Agarwal P, Khurana JP, Khurana P (2013) A seed preferential heat shock transcription factor from wheat provides abiotic stress tolerance and yield enhancement in transgenic Arabidopsis under heat stress environment. PLoS One 8: e79577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coberly LC, Rausher MD (2003) Analysis of a chalcone synthase mutant in Ipomoea purpurea reveals a novel function for flavonoids: amelioration of heat stress. Mol Ecol 12: 1113–1124 [DOI] [PubMed] [Google Scholar]
- Duck NB, Folk WR (1994) Hsp70 heat shock protein cognate is expressed and stored in developing tomato pollen. Plant Mol Biol 26: 1031–1039 [DOI] [PubMed] [Google Scholar]
- Finka A, Mattoo RU, Goloubinoff P (2011) Meta-analysis of heat- and chemically upregulated chaperone genes in plant and human cells. Cell Stress Chaperones 16: 15–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkostefanakis S, Röth S, Schleiff E, Scharf KD (2015a) Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks. Plant Cell Environ 38: 1881–1895 [DOI] [PubMed] [Google Scholar]
- Fragkostefanakis S, Simm S, Paul P, Bublak D, Scharf K-D, Schleiff E (2015b) Chaperone network composition in Solanum lycopersicum explored by transcriptome profiling and microarray meta-analysis. Plant Cell Environ 38: 693–709 [DOI] [PubMed] [Google Scholar]
- Francis KE, Lam SY, Copenhaver GP (2006) Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiol 142: 1004–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank G, Pressman E, Ophir R, Althan L, Shaked R, Freedman M, Shen S, Firon N (2009) Transcriptional profiling of maturing tomato (Solanum lycopersicum L.) microspores reveals the involvement of heat shock proteins, ROS scavengers, hormones, and sugars in the heat stress response. J Exp Bot 60: 3891–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorno F, Wolters-Arts M, Grillo S, Scharf K-D, Vriezen WH, Mariani C (2010) Developmental and heat stress-regulated expression of HsfA2 and small heat shock proteins in tomato anthers. J Exp Bot 61: 453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou J-Y, Miller LM, Hou G, Yu X-H, Chen X-Y, Liu C-J (2012) Acetylesterase-mediated deacetylation of pectin impairs cell elongation, pollen germination, and plant reproduction. Plant Cell 24: 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn A, Bublak D, Schleiff E, Scharf K-D (2011) Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 23: 741–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M (2011) Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol 157: 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahori S. (1966) High temperature injuries in tomato. V. Fertilization and development of embryo with special reference to the abnormalities caused by high temperature. J Jpn Soc Hortic Sci 35: 55–62 [Google Scholar]
- Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- James LR, Andrews S, Walker S, de Sousa PR, Ray A, Russell NA, Bellamy TC (2011) High-throughput analysis of calcium signalling kinetics in astrocytes stimulated with different neurotransmitters. PLoS One 6: e26889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K-H, Ko H-J, Nguyen MX, Kim S-R, Ronald P, An G (2012) Genome-wide identification and analysis of early heat stress responsive genes in rice. J Plant Biol 55: 458–468 [Google Scholar]
- Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL (2004) Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol 136: 4159–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. (2002) BLAT--the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnersley AM, Turano FJ (2000) Gamma aminobutyric acid (GABA) and plant responses to stress. Crit Rev Plant Sci 19: 479–509 [Google Scholar]
- Knapp S, Larondelle Y, Rossberg M, Furtek D, Theres K (1994) Transgenic tomato lines containing Ds elements at defined genomic positions as tools for targeted transposon tagging. Mol Gen Genet 243: 666–673 [DOI] [PubMed] [Google Scholar]
- Kotak S, Vierling E, Bäumlein H, von Koskull-Döring P (2007) A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 19: 182–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HC, Charng YY (2013) Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol 163: 276–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HC, Liao HT, Charng YY (2011) The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ 34: 738–751 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lommen A, Kools HJ (2012) MetAlign 3.0: performance enhancement by efficient use of advances in computer hardware. Metabolomics 8: 719–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf K-D (2002) In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev 16: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Z, Cox AJ, Mullikin JC (2001) SSAHA: a fast search method for large DNA databases. Genome Res 11: 1725–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S (2006) Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J 48: 535–547 [DOI] [PubMed] [Google Scholar]
- Ogawa D, Yamaguchi K, Nishiuchi T (2007) High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J Exp Bot 58: 3373–3383 [DOI] [PubMed] [Google Scholar]
- Paupière MJ, van Heusden AW, Bovy AG (2014) The metabolic basis of pollen thermo-tolerance: perspectives for breeding. Metabolites 4: 889–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol Lett 26: 509–515 [DOI] [PubMed] [Google Scholar]
- Port M, Tripp J, Zielinski D, Weber C, Heerklotz D, Winkelhaus S, Bublak D, Scharf K-D (2004) Role of Hsp17.4-CII as coregulator and cytoplasmic retention factor of tomato heat stress transcription factor HsfA2. Plant Physiol 135: 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Dapena P, Castaño R, Almoguera C, Jordano J (2006) Improved resistance to controlled deterioration in transgenic seeds. Plant Physiol 142: 1102–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Dapena P, Castaño R, Almoguera C, Jordano J (2008) The ectopic overexpression of a seed-specific transcription factor, HaHSFA9, confers tolerance to severe dehydration in vegetative organs. Plant J 54: 1004–1014 [DOI] [PubMed] [Google Scholar]
- Queitsch C, Hong S-W, Vierling E, Lindquist S (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero RM, Ruiz JM, García PC, López-Lefebre LR, Sánchez E, Romero L (2001) Resistance to cold and heat stress: accumulation of phenolic compounds in tomato and watermelon plants. Plant Sci 160: 315–321 [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R (2004) When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar NK, Kim Y-K, Grover A (2014) Coexpression network analysis associated with call of rice seedlings for encountering heat stress. Plant Mol Biol 84: 125–143 [DOI] [PubMed] [Google Scholar]
- Sato S, Peet M, Thomas J (2000) Physiological factors limit fruit set of tomato (Lycopersicon esculentum Mill) under chronic, mild heat stress. Plant Cell Environ 23: 719–726 [Google Scholar]
- Scharf K-D, Berberich T, Ebersberger I, Nover L (2012) The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim Biophys Acta 1819: 104–119 [DOI] [PubMed] [Google Scholar]
- Scharf K-D, Heider H, Höhfeld I, Lyck R, Schmidt E, Nover L (1998) The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol Cell Biol 18: 2240–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf K-D, Rose S, Zott W, Schöffl F, Nover L (1990) Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. EMBO J 9: 4495–4501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm F, Ganguli A, Kiehlmann E, Englich G, Walch D, von Koskull-Döring P (2006) The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol Biol 60: 759–772 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm S, Fragkostefanakis S, Paul P, Keller M, Einloft J, Scharf K-D, Schleiff E (2015) Identification and expression analysis of ribosome biogenesis factor co-orthologs in Solanum lycopersicum. Bioinform Biol Insights 9: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A (2011) Differential expression in RNA-seq: a matter of depth. Genome Res 21: 2213–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikunov YM, Laptenok S, Hall RD, Bovy A, de Vos RC (2012) MSClust: a tool for unsupervised mass spectra extraction of chromatography-mass spectrometry ion-wise aligned data. Metabolomics 8: 714–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehmeyer N, Vierling E (2000) The expression of small heat shock proteins in seeds responds to discrete developmental signals and suggests a general protective role in desiccation tolerance. Plant Physiol 122: 1099–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovlev IA, Lee Y, Rotter B, Olsen JE, Skrøppa T, Johnsen Ø, Fossdal CG (2014) Temperature-dependent differential transcriptomes during formation of an epigenetic memory in Norway spruce embryogenesis. Tree Genet Genomes 10: 355–366 [Google Scholar]
- Yokotani N, Ichikawa T, Kondou Y, Matsui M, Hirochika H, Iwabuchi M, Oda K (2008) Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis. Planta 227: 957–967 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Ohama N, Nakajima J, Kidokoro S, Mizoi J, Nakashima K, Maruyama K, Kim JM, Seki M, Todaka D, et al. (2011) Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol Genet Genomics 286: 321–332 [DOI] [PubMed] [Google Scholar]
- Zawada AM, Rogacev KS, Müller S, Rotter B, Winter P, Fliser D, Heine GH (2014) Massive analysis of cDNA ends (MACE) and miRNA expression profiling identifies proatherogenic pathways in chronic kidney disease. Epigenetics 9: 161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Moriyama EN (2013) Comparative studies of differential gene calling using RNA-Seq data. BMC Bioinformatics (Suppl) 14: S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.