Abstract
Background
Circadian clocks are endogenous timing systems that regulate various aspects of mammalian metabolism, physiology and behavior. Traditional chronotherapy refers to the administration of drugs in a defined circadian time window to achieve optimal pharmacokinetic and therapeutic efficacies. In recent years, substantial efforts have been dedicated to developing novel small-molecule modulators of circadian clocks.
Methods
Here, we review the recent progress in the identification of molecular targets of small-molecule clock modulators and their efficacies in clock-related disorders. Specifically, we examine the clock components and regulatory factors as possible molecular targets of small molecules, and we review several key clock-related disorders as promising venues for testing the preventive/therapeutic efficacies of these small molecules. Finally, we also discuss circadian regulation of drug metabolism.
Results
Small molecules can modulate the period, phase and/or amplitude of the circadian cycle. Core clock proteins, nuclear hormone receptors, and clock-related kinases and other epigenetic regulators are promising molecular targets for small molecules. Through these targets small molecules exert protective effects against clock-related disorders including the metabolic syndrome, immune disorders, sleep disorders and cancer. Small molecules can also modulate circadian drug metabolism and response to existing therapeutics.
Conclusion
Small-molecule clock modulators target clock components or diverse cellular pathways that functionally impinge upon the clock. Target identification of new small-molecule modulators will deepen our understanding of key regulatory nodes in the circadian network. Studies of clock modulators will facilitate their therapeutic applications, alone or in combination, for clock-related diseases.
Keywords: Circadian clock, small-molecule modulator, molecular target, nuclear receptor, metabolic syndrome, drug metabolism
Introduction
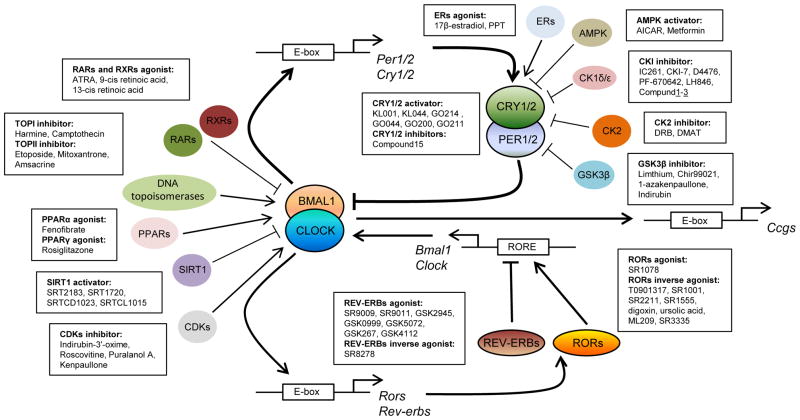
Circadian clocks are endogenous timing systems that drive daily rhythms in a variety of metabolic, physiological and behavioral processes [1–3]. In mammals, the cell-autonomous molecular oscillator is the basic unit of the clock system, comprised of transcriptional-translational feedback loops including a core loop and a secondary stabilization loop (Fig. 1) [2, 4]. Briefly, heterodimers of transcription factors CLOCK/BMAL1 and NPAS2/BMAL1 bind to E-box DNA elements on the promoters of Period1/2 (Per1/2) and Cryptochrome1/2 (Cry1/2) genes and activate their expression. PER1/2 and CRY1/2 proteins in turn form complexes and inhibit CLOCK/BMAL1 and NPAS2/BMAL1 activities and thus their own expression, completing one cycle and priming for the next. In the stabilization loop, CLOCK/BMAL1 and NPAS2/BMAL1 drive the expression of genes encoding the nuclear receptor REV-ERBs and RORs. REV-ERBs and RORs respectively repress and activate transcription of Bmal1 and several other clock genes, through direct binding to the RORE elements on their promoters. In addition, the transcriptional-translational feedback loops are subjected to regulation at various levels including post-translational modifications such as phosphorylation, ubiquitination and acetylation [5, 6]. Post-translational modifications of clock proteins provide the necessary plasticity to fine-tune the clockwork and respond to environmental changes. These cell-autonomous circadian oscillators regulate oscillatory expression of clock-controlled genes (CCGs) in a tissue-specific manner, a process coordinated by the SCN (suprachiasmatic nuclei) master pacemaker in the hypothalamus.
Fig. 1. Circadian clock loops and possible molecular targets of small-molecule modulators.
The circadian clock oscillator is composed of transcriptional–translational feedback loops including a core loop (BMAL1/CLOCK and PERs/CRYs) and a stabilization loop (REV-ERBs and RORs). In addition to core clock components, various cellular pathways that modulate the molecular feedback loops can be targeted by small molecules leading to changes in the amplitude, phase, and period of circadian rhythms. Results from circadian phenotype-based screening or target-based rational design will continue to expand potential druggable pathways, which could lead to novel therapeutic strategies for clock-related disorders.
Importantly, circadian misalignment in humans has been shown to contribute to sleep disorders, metabolic syndrome, cancer and other pathologies [7–9]. For example, human subjects who lived on an enforced 28-hr rhythm for 10 days were found to exhibit impaired glucose tolerance and hyperinsulinemia [8]. These experimental data are well aligned with epidemiological evidence showing increased disease risk in shift workers [10]. Furthermore, circadian mutant mice with genetic disruption in clock genes displayed a variety of physiological deficits [11–13]. For example, Bmal1−/−, Cry1/2−/− and ClockΔ19/Δ19 mutant mice harboring disruptive mutations in the core loop have been reported to exhibit a spectrum of metabolic disorders including obesity, hyperlipidemia, hepatic steatosis and hyperglycemia [14–18]. These findingstogether indicate thata robust circadian clock is necessary for health .
The therapeutic relevance of circadian clocks has traditionally been limited to the domain of chronotherapy, which entails selecting the best time during the daily cycle for drug administration to achieve the optimal therapeutic index and pharmacokinetic profile [19]. For example, standard weekly treatments of a broad-spectrum anti-cancer drug gemcitabine often lead to hematologic toxicity, which could be significantly diminished by treating patients at 9AM as opposed to 3PM [20]. More recently, increasing attention has turned to the discovery and exploitation of small molecule modulators which can manipulate the circadian timing to prevent or alleviate clock-related pathological deficits [21]. Several endogenous molecules such as fibroblast growth factor (FGF), epidermal growth factor (EGF), insulin, heme, melatonin and cAMP are known to modulate the circadian clock [22], providing proof-of-principle for the identification of novel natural or synthetic small-molecule modulators. Indeed, a number of small molecule modulators have been successfully discovered through circadian phenotype-based screening or target-based design [21, 23, 24]. In this review, we will enumerate modulators of key components or regulatory factors of the mammalian circadian clock and review their efficacies in manipulating circadian rhythms and ameliorating clock-related diseases.
Targets of small-molecule clock modulators
Below we describe small molecule modulators which directly interact with core clock proteins or key regulatory factors of the circadian clock.
1. Core clock proteins
The core clock proteins include components of the transcriptional-translational feedback loops, such as BMAL1, NPAS2, CLOCK, PER1/2, CRY1/2, REV-ERBs and RORs. Synthetic or natural ligands for CRY1/2, REV-ERBs and RORs have been reported (Table 1 and Fig. 1).
Table 1.
Small-molecule modulators targeting core clock proteins
| Molecular target | Target activity | Compound | Circadian phenotype | Reference |
|---|---|---|---|---|
| CRY1/2 | activator | KL001 | Period lengthening Amplitude reducing |
[25] |
| KL044 | Period lengthening | [26, 27] | ||
| GO214 | Period lengthening | [26, 27] | ||
| GO044 | Period shortening | [26, 27] | ||
| GO200 | Period shortening | [26, 27] | ||
| GO211 | Period shortening | [26, 27] | ||
| Inhibitor | 2-ethoxypropanoic acid | Period shortening Amplitude reducing |
[28] | |
| REV-ERBs | Agonist | SR9009 | Amplitude reducing | [30] |
| SR9011 | Amplitude reducing | [30] | ||
| GSK2945 | Amplitude reducing | [31, 32] | ||
| GSK0999 | Amplitude reducing | [31, 32] | ||
| GSK5072 | Amplitude reducing | [31, 32] | ||
| GSK267 | Amplitude reducing | [31, 32] | ||
| GSK4112 | N/A | [31] | ||
| Inverse agonist | SR8278 | N/A | [33] | |
| RORs | Agonist | SR1078 | N/A | [37] |
| Inverse agonist | T0901317 | N/A | [31] | |
| SR1001 | N/A | [36] | ||
| SR2211 | N/A | [31] | ||
| SR1555 | N/A | [31] | ||
| Digoxin | N/A | [31] | ||
| Ursolic acid | N/A | [31] | ||
| ML209 | N/A | [31] | ||
| SR3335 | N/A | [38] |
1.1 CRY1/2 proteins
CRY1/2 proteins function as thenegative regulator sof the core circadian feedback loop. In an unbiased cell-based circadian phenotypic screen, a carbazole compound KL001 was shown to specifically interact with CRY proteins. KL001 stabilized CRY1/2 by preventing ubiquitin-dependent CRY degradation, and consequently lengthened the circadian period and reduced the circadian amplitude [25]. Among KL001 derivatives directly targeting CRY, KL044 and GO214 also lengthened the circadian period, whereas GO044, GO200 and GO211 showed the opposite effects [26, 27]. In another study, compound 15, a derivative of 2-ethoxypropanoic acid, was reported to bind to the C-terminal tail of CRY1/2 and inhibit their repressive function, resulting in slight shortening of the circadian period accompanied by significant reduction of the circadian amplitude [28]. These studies describe CRY-interacting small molecules showing distinct biochemical mechanisms and functional outcome, yet interestingly dampening the rhythms in both cases. Further studies are required to fully understand the mechanisms of these compounds withinthe molecular oscillator .
1.2 REV-ERB receptors
The REV-ERB nuclear receptors (consisting of REV-ERBα and REV-ERBβ) repress Bmal1 transcription through binding to the promoter RORE elements [13, 29]. Synthetic REV-ERB agonists SR9009 and SR9011, optimized from the lead compound GSK4112, reduced the circadian amplitude in SCN slices from Per2::Luciferase reporter mice (PER2::LUC) without affecting the circadian period [30, 31]. SR9011 also disrupted wheel-running activity immediately after single administration [30]. Moreover, several other derivative compounds, including GSK2945, GSK0999, GSK5072 and GSK267, activated REV-ERBα and diminished the oscillation of the Bmal1-Luciferease reporter in U2OS cells. Paradoxically however, a more recent study showed that SR9009 and SR9011 displayed greater selectivity for LXRα than REV-ERBα [31, 32]. Further experiments are required to resolve this discrepancy. In addition to agonists, structure-activity analysis focusing on the GSK4112 tertiary amine scaffold also identified an REV-ERB antagonist SR8278 displaying inhibitory activities in a luciferase-based cellular assay [33]. These findings reveal that tertiary amine derivatives are rich sources for the identification of REV-ERB agonists or antagonists.
1.3 ROR receptors
The retinoic acid receptor-related orphan receptors (RORs; consisting of RORα, RORβ and RORγ) enhance Bmal1 transcription through direct competition with REV-ERBs for binding to RORE elements [34, 35]. A number of RORs ligands have been identified via circadian phenotype-based screening or target-based design [31, 36–38]. For example, T0901317 and SR1001 act as inverse agonists for RORα and RORγ [36]. Among isoform-specific ligands, SR2211, SR1555, digoxin, ursolic acid and ML209 function as RORγ inverse agonists [31], whereas SR3335 is a RORα inverse agonist [38]. Ligands activating RORs are also emerging in recent literature; for example, SR1078 was shown to activate both RORα and RORγ [37]. How these RORs agonists or inverse agonists modulate circadian clocks remains to be characterized.
2. Kinases
Phosphorylation of core clock proteins is a prevalent regulatory mechanism for the circadian clock [39]. Therefore, ligands targeting cognate kinases can profoundly alter circadian characteristics(Table 2 and Fig. 1).
Table 2.
Small-molecule modulators targeting regulators of circadian clocks
| Molecular target | Target activity | Compound | Circadian phenotype | Reference |
|---|---|---|---|---|
| ALK | Inhibitor | SB432542 | Attenuated phase delay | [54] |
| AMPK | Activator | AICAR | Period lengthening Amplitude reducing |
[49] |
| Metformin | Induced circadian expression | [50, 51] | ||
| CDKs | Inhibitor | Indirubin-3'-oxime | Period shortening | [40] |
| Kenpaullone | Period shortening | [40] | ||
| Roscovitine | Period lengthening | [40] | ||
| Puralanol A | Period lengthening | [40] | ||
| CKI | Inhibitor | IC261 | Period lengthening | [40, 41] |
| CKI-7 | Period lengthening | [40–42] | ||
| D4476 | Period lengthening | [40–42] | ||
| PF-670642 | Period lengthening | [43, 44] | ||
| LH846 | Period lengthening | [43, 44] | ||
| Compund1–3 | Period lengthening | [46] | ||
| CK2 | Inhibitor | DRB | Period lengthening | [40, 47] |
| DMAT | Period lengthening | [40, 47] | ||
| ERs | Agonist | 17β-estradiol | Period shortening | [57] |
| PPT | Period lengthening | [42] | ||
| GSK3β | Inhibitor | Lithium | Period lengthening | [40] |
| Chir99021 | Period shortening | [40] | ||
| 1-azakenpaullone | Period shortening | [40] | ||
| Indirubin | Period shortening | [40] | ||
| PPARs | Agonist | Fenofibrate | Induced circadian rhythm | [58] |
| Rosiglitazone | Induced circadian expression | [59] | ||
| RARs and RXRs | Agonist | ATRA | Phase shift | [61] |
| 9-cis retinoic acid | Induced circadian rhythm | [61] | ||
| 13-cis retinoic acid | Induced circadian rhythm | [61] | ||
| SIRT1 | Activator | SRT2183 | Reduced circadian expression | [64] |
| SRT1720 | Reduced circadian expression | [64] | ||
| SRTCD1023 | Period lengthening Amplitude reducing |
[64] | ||
| SRTCL1015 | Period lengthening Amplitude reducing |
[64] | ||
| TOPI | Inhibitor | Harmine | Period lengthening | [66, 67] |
| Camptothecin | Period lengthening | [65] | ||
| TOPII | Inhibitor | Etoposide | Period shortening | [40] |
| Mitoxantrone | Period shortening | [40] | ||
| Amsacrine | Period lengthening | [40] |
2.1 Casein kinaseI (CKI)
In mammals, several genes encode CKI isoforms (α, β, γ1, γ2, γ3, δ and ε), among which α, δ and ε have been implicated in the regulation of BMAL1, PERs, and CRYs [39]. CK1δ/ε inhibitors IC261, CKI-7 and D4476 previously showed period lengthening effects in cultured cells [40–42]. Moreover, PF-670642 and LH846, as potent CKIδ inhibitors, also significantly lengthened molecular oscillations in U2OS cells and mouse tissue explants in vitro and circadian locomotor activity rhythms in vivo [43, 44]. Interestingly, PF-4800567 strongly inhibited CKIε yet displayed only a minimal effect on the circadian clock [45]. In our previous study, compounds 1–3 showed robust CKIε inhibitory activities and markedly lengthened the period in cultured cells and mouse tissue explants; it is possible that these compounds also inhibit CK1δ [46]. These studies together indicate that among the several CKI isoforms implicated in clock regulation, CKIδ, and perhaps CKIε to a lesser degree, may bepromising targetsfor pharmacological modulation of circadian clocks.
2.2 Casein kinase 2 (CK2)
Previous biochemical studies revealed that CK2 regulates circadian clocks by phosphorylating BMAL1 and PER proteins [39]. Downregulating CK2 expression by RNAi has been shown to lengthen the circadian period [47]. Pharmacological inhibition of CK2 by DRB and DMAT also resulted in period lengthening [40, 47], indicating a strong correlation between clock lengthening and CK2 inhibition .
2.3 Glycogen synthase kinase3 (GSK3)
Among the two mammalian GSK3 isoforms (GSK3α and GSK3β), GSK3β has been reported to regulate the circadian clock by phosphorylating CLOCK, PER, REV-ERBα and CRY proteins [39]. Lithium, a weak GSK3β inhibitor, lengthened the circadian period in cultured cells and in vivo. However, GSK3β-selective inhibitors including Chir99021, 1-azakenpaullone and indirubin shortened the circadian period [40]. Because lithium broadly inhibits inositol monophosphatase, GSK3 and other phosphomonoesterases [48], the period-lengthening effect of lithium might be mediated by target protein(s) other than GSK3β .
2.4 5' AMP-activated protein kinase (AMPK)
AMPK, a heterotrimeric protein kinase consisting of a catalytic (α) subunit and two regulatory (β , γ) subunits, has been shown to phosphorylate and destabilize PER and CRY proteins [39, 49]. AICAR, an AMPK activating metabolite, reduced the oscillatory amplitude and lengthened the period [49]. Furthermore, metformin, a broadly used diabetes drug and an AMPK activator, potentiated the degradation of PER2 and altered the expression of clock genes in mouse muscle, liver and heart [50, 51]. In the diabetic db/db mice, metformin also restored the expression pattern of clock genes in white fat via the AMPK-Nampt-Sirt1 pathway [52]. The precise role of AMPK in the clock, via PER or CRY phosphorylation or another target pathway, requiresfurther study .
2.5 Cyclin-dependent kinases (CDKs)
Recent studies revealed that CDK5, one of 11 CDK isoforms, regulates E-box dependent transcriptional activation by directly phosphorylating CLOCK [53]. Indirubin-3’-oxime and kenpaullone, inhibitors of both CDK and GSK3, were found to shorten the circadian period [40]. In contrast, CDK inhibitors with varying target specificity such as roscovitine (for CDK1, CDK2 and CDK5) and puralanol A (for CDK2, CDK4 and CDK5) significantly lengthened the circadian period [40]. NU6102, a selective inhibitor of CDK1 and CDK2, was incapable of lengthening the period [40]. Therefore, additional studies are necessary to clarify the complex roles of CDK inhibitors and determine relative contributions of CDK isoforms tocircadian regulation .
2.6 other kinases
A number of other kinase inhibitors, including the p38 inhibitors SB203580 and PD169316, the JNK inhibitor SP600125, and the CLK inhibitor TG003, were identified as period-lengthening small molecules in NIH-3T3 and U2OS cells [42]. However, these compounds also seemed to inhibit CK1δ/ε other than their primary targets [42] and the exact role of p38, JNK and CLK, if any, in period lengthening is unclear. SB432542, an inhibitor of activinreceptor -like kinase (ALK), was found to attenuate phase delays through acute induction of the circadian transcriptional regulator DEC1 [54]. Furthermore, a kinase inhibitor screen established inhibitors against CK1δ/ε (IC261), CK2 (DMAT and DRB) and p38 (SB202190) as period-lengthening compounds, and also revealed similar effects by those of PI3-kinase (LY294002) and Akt (BML-297) [55]. In general, however, the prevalent promiscuity of kinase inhibitors presents a significant challenge to pinpoint the causal relationship regarding circadian clockeffects .
3. Nuclear receptorsother than REV -ERBs and RORs
A growing number ofnuclear receptors have been reportedto play important regulatory roles in circadian clocks. In addition to REV-ERBs and RORs acting in the stabilization loop of the core oscillator, glucocorticoid receptor (GR), estrogen receptors (ERs), peroxisome proliferator activated receptors (PPARs), retinoic acid receptors (RARs) and retinoid X receptors (RXRs) have also been shown to regulate the clock [56], therefore implicating their ligands as clock modulators (Table 2 and Fig. 1). An in-depth understanding of the roles of nuclear receptor ligands in the clock system will enhance their therapeutic potential for clock-related disorders.
3.1 Estrogen receptors(ER s)
In tissue explant cultures from PER2::LUC reporter mice, 17β-estradiol, an ERα/β agonist, shortened the period of rhythmic PER2::LUC expression in the uterus but not in the SCN, possibly through regulation of the Per2 promoter [57]. However, PPT, a specific ERα agonist, lengthened the circadian period [42]. Since PPT also acts on CK1δ/ε [42], determining the exact function of ERα/β on circadian period requires improved pharmacological and genetic tools.
3.2 Peroxisome proliferator activated receptors (PPARs)
PPARα regulates the expression of Bmal1 and Rev-erbα via binding to the PPRE promoter elements [58]. Fenofibrate, a synthetic PPARα agonist, was shown to induce circadian clock gene expression in vitro and to up-regulate hepatic Bmal1 in vivo [58]. In addition, PPARγ also directly regulates the expression of Bmal1 in the vascular system and thus participates in the circadian regulation of blood pressure and heart rate [59]. In accordance, administration of the PPARγ agonist rosiglitazone induced aortic expression of Bmal1 [59]. However, the natural PPARγ ligand 15-deoxy-Delta12, 14-prostaglandin J2 entrained cellular circadian clocks in a PPARγ-independent manner, suggesting an off-target effect[ 60].
3.3 Retinoic acid receptors (RARs) and retinoid X receptors (RXRs)
In the vasculature, RARα and RXRα were shown to negatively regulate the BMAL1/CLOCK- mediated transcriptional activation of clock gene expression through direct interaction with CLOCK [61]. All-trans retinoic acid (ATRA), a ligand for both RAR and RXR, can phase-shift Per2 mRNA rhythm in serum-induced smooth muscle cells and in vivo [61]. In addition, 9-cis retinoic acid and 13-cis retinoic acid, ligands for both RAR and RXR, are also able to entrain circadian rhythms in vitro and in vivo [61]. These studies collectively suggest that ligands for both RAR and RXR modulate circadian clocks by altering the interaction between RAR/RXR and CLOCK. This mode of action contrasts with that of other nuclear receptor ligands which function primarily by altering binding of nuclear receptors to cognate promoterelements of clock genes.
4. Silent Information Regulator 1 (SIRT1)
SIRT1 is a sirtuin family deacetylase, directly recruited to the CLOCK/BMAL1 chromatin complex to suppress circadian transcription [62, 63]. Apart from histones, BMAL1 has also been identified as a SIRT1 substrate. Previously, several SIRT1 activators were characterized. For example, SRT2183 and SRT1720 decrease d circadian gene expression, and SRTCD1023 and SRTCL1015 also reduced the amplitude and lengthened the period of circadian rhythms. In contrast, SRTCE1022 did not activate SIRT1 in biochemical or cell-based assays, and showed no effects on circadian amplitude and period [64]. These results demonstrate a negative role of SIRT1 incircadian gene expression.
5. DNA topoisomerases( TOPs)
Topoisomerases, including type I (TOPI) and type II (TOPII), are key enzymes for DNA replication, transcription, recombination and chromatin remodeling [65]. In a recent study, it was shown that TOPI suppressed Bmal1 transcription via binding to an intermediate region between the two RORE elements in the promoter. Camptothecin, a TOPI inhibitor, enhanced Bmal1 transcription and lengthened the circadian period [65]. In addition, the TOPI inhibitor harmine also lengthened the circadian period by enhancing the trans-activating function of RORα in vitro and in vivo [66, 67]. In comparison, three TOPII inhibitors including etoposide, mitoxantrone and amsacrine have been shown to shorten the circadian period [40], suggesting differential functions of TOPI and TOPII in the circadian clock.
Therapeutic potentials of small-molecule clock modulators
In this section, we firstdescribe in vitro and mouse studies demonstrating the therapeutic potential of clock-modulating small molecules in several clock-related disorders. We will then consider their possible application in traditional chronotherapy where drug metabolism is a predominant factordetermining efficacy and toxicity .
1. Clock-related disorders
1.1 Metabolic syndrome
To combat the current global epidemic of metabolic disorders, circadian research has demonstrated novel interventions via directly manipulating circadian rhythms [30, 68]. For example, nighttime-only intake of high-fat diet (HFD) was shown to protect mice against metabolic disease and improve their motor coordination compared with unrestricted feeding [68]. It has been hypothesized that restricting caloric intake during the rest phase when energy expenditure is suppressed prevents storage of excess nutrients and thus reduces body weight gain. Interestingly, the oscillatory amplitude of clock and metabolic gene expression was significantly enhanced in mice fed with time-restricted intake of HFD, raising the possibility that small molecules inducing amplitude enhancement may also protect against metabolic diseases. On the other hand, the REV-ERB agonist SR9009 has been shown to elicit beneficial metabolic effects in diet-induced obese mice, yet acutely repressed circadian behavior and oscillatory amplitude [30]. These data on the one hand suggest that targeting the clock machinery may be efficacious against metabolic disorders, on the other also reveal mechanistic complexity, particularly regarding the functional relationship between clock amplitude and metabolic output.
1.2 Immune disorders
The clock is known to regulate the expression of various pro-inflammatory cytokines such as IL-6, TNF, IL-17 and CXCL1 [69]. In a recent study, the REV-ERB ligand GSK4112 was found to inhibit expression of IL6, CXCL11, CXCL6 and CCL2 in primary human macrophages with LPS stimulation [70]. In addition, digoxin and ursolic acid, as RORγ inverse agonists, ameliorated autoimmune disorders including arthritis and encephalomyelitis via suppression of Th17 differentiation [71–74]. These studies indicate that targeting clock componentsmay constitute a valid strategy against inflammation and autoimmune diseases.
1.3 Sleepdisorders
In addition to acute sleep disturbance such as jetlag, familial advanced sleep phase syndrome (FASPS) or delayed sleep phase syndrome (DSPS) are among the circadian sleep disorders with a genetic basis [75]. FASPS is characterized by circadian period shortening due to a T44A mutation in the CK1δ protein [75]. Interestingly, the CK1δ inhibitor PF-670462 has been shown to induce behavioral rhythms in mice rendered arrhythmic by constant light exposure or the Vipr2−/− mutation [43], indicating pharmacological targeting of CK1δ constitutes a therapeutic modulation of perturbed circadian behavior. It will be interesting to investigate whether PF-670462 can prolong the period in FASPS patients or model animals and alleviate the sleep symptom. Likewise, other small molecule modulators capable of altering circadian period and/or phase may be efficacious for the treatment of circadian sleep disorders.
1.4 Cancer
Circadian clocks have been postulated to serve tumor suppressor functions. Epidemiological and genetic studies have provided evidence that circadan disruption increases cancer risk and clock gene expression is altered in various tumors [9, 76]. For example, several studies revealed that the incidence of breast cancer is higher among women who work night shifts [77]. It has also been reported that REV-ERBβ is over-expressed in various breast cancer cell lines, as well as in tumor cell lines derived from skin, liver and prostate [78]. Interestingly, treatment of breast cancer cell lines with the REV-ERB agonist SR9011 resulted in a significant decrease in Cyclin A2 levels and caused a dose-dependent reduction in cell viability [79]. Although not directly targeting core clock proteins, L-methyl selenocysteine (MSC) is a clock-modulating compound that upregulates Bmal1 transcription and importantly protects against toxicity induced by the chemotherapeutic agent cyclophosphamide in mice [80]. Further efforts should be dedicated to exploiting pharmacological modulation of circadian clocksas an anti -cancer strategy.
2. Drug metabolism
Circadian pharmacokinetics and pharmacodynamics studies have revealed administration time-dependent efficacy and toxicity for a growing number of drugs, facilitating chronotherapeutic applications for various diseases such as rheumatoid arthritis, asthma, ulcer, cardiovascular disease, metabolic disease and cancer [19]. In accordance, transcriptome analysis of mouse liver indicated that hepatic drug-processing genes involving in absorption, biotransformation, and excretion of exogenous and endogenous compounds are rhythmically expressed in liver [81]. Genetic studies further demonstrate a regulatory role of the circadian clock in the expression of drug metabolism genes. For example, RORα and RORγ deficient mice displayed perturbed expression of genes encoding several phase I and phase II metabolic enzymes, including 3β-hydroxysteroid dehydrogenases, cytochrome P450 enzymes, and sulfotransferases [82]. Likewise, mice deficient for the PAR-bZip family circadian transcription factors (DBP, TEF and HLF) showed reduced basal expression of genes involved in drug metabolism and detoxification, including cytochrome P450 enzymes, carboxylesterases, and constitutive androstane receptor (CAR); furthermore, the mice are hypersensitive to xenobiotic challenges [83]. These studies together indicate that circadian expression of cytochrome P450s (CYPs) and other xenobiotic metabolism enzymes regulates drug absorption, distribution, metabolism and elimination (ADME), contributing to chronotherapeutic effects[ 84].
In a mouse genetic study, deficiency in Clock or Bmal1 rendered mice highly sensitive to the toxic effect of the anticancer agent cyclophosphamide, whereas mice with Cry1/2 double knockout were more resistant to the same treatment [85]. These results suggested that the functional interplay between the positive and negative circadian components impactsdrug toxicity by regulating expression of genes encoding either enzymes important for drug metabolism or targets of the drug. In that regard, it is interesting to note that 56 of the top 100 best-selling drugs are known to target proteins encoded by rhythmically expressed genes [86]. Clock-modulating small molecules can be exploited to fine-tune the functional state of the clock, thereby optimizing both drug metabolism and response, i.e., pharmacokinetics and pharmacodynamics. In essence, these clock modulators can serve in combination therapies to enhance the therapeutic index of existing drugs(see also below) .
Future directions and conclusions
Small molecule modulators are useful probes targeting a diverse array of clock components including core clock proteins and other regulatory factors [21, 87]. Several important points should be reiterated. First, compounds with known primary targets may act on other targets to exert circadian effects. Whereas small-molecule modulators targeting core clock proteins such as CRYs may be expected to be more specific, cross-reactivity among kinases or nuclear receptors is not uncommon. For example, the effects of the p38 inhibitors SB203580 and PD169316, JNK inhibitor SP600125 and CLK inhibitor TG003 on circadian clocks may be mainly attributable to inhibition of CK1δ/ε rather than their primary targets [42]. For REV-ERB and ROR receptors with ligand binding domains sharing sequence homology with other receptors, selectivity of derived ligands toward related receptors should be experimentally evaluated [32]. Second, regulatory proteins of the circadian loops may also possess non-clock functions, suggesting clock-independent effects of their pharmacological agents. The aforementioned kinases again exemplify this point as they are known to regulate various cellular pathways in addition to the clock. Finally, given the multilayer feedback regulation embedded in the oscillator, significant challenge remains in discerning the precise biochemical mechanism and oscillator-wide function of these compounds. For example, the clock gene expression and overall amplitude depends on the stoichiometric ratio of positive and negative factors [88]. Conceivably, excessive activation of the positive factors or suppression of the negative factors will likely disrupt the balance within the oscillator, causing the cyclic pattern to collapse or dampen.
Despite these challenges, clock-targeting pharmaceuticals show preventive and therapeutic potentials against clock-related disorders. Continuing efforts can be dedicated to development of modulators for other clock proteins and their functional complexes, such as PERs, BMAL1, CLOCK and NPAS2 proteins, and PER/CRY, BMAL1/NPAS2 and BMAL1/CLOCK complexes. Two areas of studies should be conducted to fully exploit the potential of small-molecule modulators for therapeutic application. First, in vivo studies are required to characterize the effects of small-molecule modulators on the circadian clock and clock-related pathologies, e.g., using animal models for sleep disorders, cancer, obesity and diabetes. Synthetic REV-ERB agonists were previously found to alter circadian and metabolic gene expression and improve metabolic homeostasis in high-fat diet induced obese mice but not in ob/ob mice [30]; it will be interesting to investigate why REV-ERB agonists display distinct effects on these two models and whether other REV-ERB agonists also improve the metabolic syndrome. Second, as mentioned above, small molecule modulators may improve efficacy and reduce toxicity of existing therapeutic agents. For example, the expression of MCT1, a lactate acid transporter, is required for tumor cell survival and known to be regulated by the clock [89, 90]. It is possible that small molecules modulating MCT1 circadian expression could enhancethe th erapeutic index of the MCT1 inhibitor AZD3965. In addition, given the demonstrated circadian control of CYP expression [81–84], small-molecule modulators may reduce the toxicity of drugs when co-administeredwith other drugs.
In conclusion, small-molecule clock modulators target clock components or diverse cellular pathways functionally impinging upon the clock. Target identification of new small-molecule modulators will deepen our understanding of key regulatory nodes in the circadian network. Importantly, studies of clock modulators will also facilitate their therapeutic applications, alone or in combination, for clock-related diseases.
Acknowledgments
We thank Seung-Hee (Sally) Yoo for critical reading. This work was in part supported by the Robert A. Welch Foundation (AU-1731), American Heart Association (11SDG7600045), and NIH/NIA (R01 AG045828) to Z.C.
ABBREVIATIONS
- ALK
Activin receptor-like kinase
- AMPK
5' AMP-activated protein kinase
- ATRA
All-trans retinoic acid
- CCGs
Clock controlled genes
- CDKs
Cyclin-dependent kinases
- CK1
Casein kinase 1
- CK2
Casein kinase 2
- Cry
Cryptochrome
- CYPs
cytochrome P450s
- DSPS
Delayed sleep phase syndrome
- EGF
Epidermal growth factor
- ERs
Estrogen receptors
- FASPS
Familial advanced sleep phase syndrome
- FGF
Fibroblast growth factor
- GR
Glucocorticoid receptor
- GSK3
Glycogen synthase kinase 3
- Per
Period
- PPARs
Peroxisome proliferator activated receptors
- RARs
Retinoic acid receptors
- RORs
Retinoic acid receptor-related orphan receptors
- RXRs
Retinoid X receptors
- SIRT1
Silent information regulator 1
- TOP
DNA topoisomerase
Footnotes
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
References
- 1.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9(10):764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu AC, Lewis WG, Kay SA. Mammalian circadian signaling networks and therapeutic targets. Nat Chem Biol. 2007;3(10):630–639. doi: 10.1038/nchembio.2007.37. [DOI] [PubMed] [Google Scholar]
- 3.Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6(7):544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu W, Hardin PE. Circadian oscillators of Drosophila and mammals. J Cell Sci. 2006;119(Pt 23):4793–4795. doi: 10.1242/jcs.03174. [DOI] [PubMed] [Google Scholar]
- 5.Kojima S, Shingle DL, Green CB. Post-transcriptional control of circadian rhythms. J Cell Sci. 2011;124(Pt 3):311–320. doi: 10.1242/jcs.065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8(2):139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 7.Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A. 2009;106(11):4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parsons MJ, Moffitt TE, Gregory AM, Goldman-Mellor S, Nolan PM, Poulton R, Caspi A. Social jetlag, obesity and metabolic disorder: investigation in a cohort study. Int J O bes. 2015;39(5):842–848. doi: 10.1038/ijo.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3(5):350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 10.Arendt J. Shift work: coping with the biological clock. Occup Med (Lond) 2010;60(1):10–20. doi: 10.1093/occmed/kqp162. [DOI] [PubMed] [Google Scholar]
- 11.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134(5):728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bass J. Circadian topology of metabolism. Nature. 2012;491(7424):348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 13.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13(2):125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Shi SQ, Ansari TS, McGuinness OP, Wasserman DH, Johnson CH. Circadian disruption leads to insulin resistance and obesity. Curr Biol. 2013;23(5):372–381. doi: 10.1016/j.cub.2013.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, Lopez JP, Philipson LH, Bradfield CA, Crosby SD, JeBailey L, Wang X, Takahashi JS, Bass J. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466(7306):627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308(5724):1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barclay JL, Shostak A, Leliavski A, Tsang AH, Johren O, Muller-Fielitz H, Landgraf D, Naujokat N, van der Horst GT, Oster H. High-fat diet-induced hyperinsulinemia and tissue-specific insulin resistance in Cry-deficient mice. Am J Physiol Endocrinol Metab. 2013;304(10):E1053–1063. doi: 10.1152/ajpendo.00512.2012. [DOI] [PubMed] [Google Scholar]
- 18.Jeong K, He B, Nohara K, Park N, Shin Y, Kim S, Shimomura K, Koike N, Yoo SH, Chen Z. Dual attenuation of proteasomal and autophagic BMAL1 degradation in Clock(Delta19/+) mice contributes to improved glucose homeostasis. Sci Rep. 2015;5:12801. doi: 10.1038/srep12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levi F, Schibler U. Circadian rhythms: mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 20.Iwata K, Aizawa K, Sakai S, Jingami S, Fukunaga E, Yoshida M, Hamada A, Saito H. The relationship between treatment time of gemcitabine and development of hematologic toxicity in cancer patients. Biol Pharm Bull. 2011;34(11):1765–1768. doi: 10.1248/bpb.34.1765. [DOI] [PubMed] [Google Scholar]
- 21.Wallach T, Kramer A. Chemical chronobiology: Toward drugs manipulating time. FEBS Lett. 2015;589(14):1530–1538. doi: 10.1016/j.febslet.2015.04.059. [DOI] [PubMed] [Google Scholar]
- 22.Antoch MP, Kondratov RV. Pharmacological modulators of the circadian clock as potential therapeutic drugs: focus on genotoxic/anticancer therapy. Handb Exp Pharmacol. 2013;(217):289–309. doi: 10.1007/978-3-642-25950-0_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Z, Yoo SH, Takahashi JS. Small molecule modifiers of circadian clocks. Cell Mol Life Sci. 2013;70(16):2985–2998. doi: 10.1007/s00018-012-1207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirota T, Kay SA. High-throughput screening and chemical biology: new approaches for understanding circadian clock mechanisms. Chem Biol. 2009;16(9):921–927. doi: 10.1016/j.chembiol.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirota T, Lee JW, St John PC, Sawa M, Iwaisako K, Noguchi T, Pongsawakul PY, Sonntag T, Welsh DK, Brenner DA, Doyle FJ, 3rd, Schultz PG, Kay SA. Identification of small molecule activators of cryptochrome. Science. 2012;337(6098):1094–1097. doi: 10.1126/science.1223710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JW, Hirota T, Kumar A, Kim NJ, Irle S, Kay SA. Development of Small-Molecule Cryptochrome Stabilizer Derivatives as Modulators of the Circadian Clock. ChemMedChem. 2015;10(9):1489–1497. doi: 10.1002/cmdc.201500260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oshima T, Yamanaka I, Kumar A, Yamaguchi J, Nishiwaki-Ohkawa T, Muto K, Kawamura R, Hirota T, Yagita K, Irle S, Kay SA, Yoshimura T, Itami K. C-H activation generates period-shortening molecules that target cryptochrome in the mammalian circadian clock. Angew Chem. 2015;54(24):7193–7197. doi: 10.1002/anie.201502942. [DOI] [PubMed] [Google Scholar]
- 28.Chun SK, Jang J, Chung S, Yun H, Kim NJ, Jung JW, Son GH, Suh YG, Kim K. Identification and validation of cryptochrome inhibitors that modulate the molecular circadian clock. ACS Chem Biol. 2014;9(3):703–710. doi: 10.1021/cb400752k. [DOI] [PubMed] [Google Scholar]
- 29.Gerhart-Hines Z, Lazar MA. Rev-erbalpha and the circadian transcriptional regulation of metabolism. Diabetes Obes Metab. 2015;17(Suppl 1):12–16. doi: 10.1111/dom.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R, Yoo SH, Takahashi JS, Butler AA, Kamenecka TM, Burris TP. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485(7396):62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kojetin DJ, Burris TP. REV-ERB and ROR nuclear receptors as drug targets. Nat Rev Drug Discov. 2014;13(3):197–216. doi: 10.1038/nrd4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trump RP, Bresciani S, Cooper AW, Tellam JP, Wojno J, Blaikley J, Orband-Miller LA, Kashatus JA, Boudjelal M, Dawson HC, Loudon A, Ray D, Grant D, Farrow SN, Willson TM, Tomkinson NC. Optimized chemical probes for REV-ERBalpha. J Med Chem. 2013;56(11):4729–4737. doi: 10.1021/jm400458q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kojetin D, Wang Y, Kamenecka TM, Burris TP. Identification of SR8278, a synthetic antagonist of the nuclear heme receptor REV-ERB. ACS Chem Biol. 2011;6(2):131–134. doi: 10.1021/cb1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43(4):527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 35.Jetten AM, Kang HS, Takeda Y. Retinoic acid-related orphan receptors alpha and gamma: key regulators of lipid/glucose metabolism, inflammation, and insulin sensitivity. Front Endocrinol (Lausanne) 2013;4:1. doi: 10.3389/fendo.2013.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solt LA, Kumar N, Nuhant P, Wang Y, Lauer JL, Liu J, Istrate MA, Kamenecka TM, Roush WR, Vidovic D, Schurer SC, Xu J, Wagoner G, Drew PD, Griffin PR, Burris TP. Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature. 2011;472(7344):491–494. doi: 10.1038/nature10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Kumar N, Nuhant P, Cameron MD, Istrate MA, Roush WR, Griffin PR, Burris TP. Identification of SR1078, a synthetic agonist for the orphan nuclear receptors RORalpha and RORgamma. ACS Chem B iol. 2010;5(11):1029–1034. doi: 10.1021/cb100223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar N, Kojetin DJ, Solt LA, Kumar KG, Nuhant P, Duckett DR, Cameron MD, Butler AA, Roush WR, Griffin PR, Burris TP. Identification of SR3335 (ML-176): a synthetic RORalpha selective inverse agonist. ACS Chem Biol. 2011;6(3):218–222. doi: 10.1021/cb1002762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reischl S, Kramer A. Kinases and phosphatases in the mammalian circadian clock. FEBS Lett. 2011;585(10):1393–1399. doi: 10.1016/j.febslet.2011.02.038. [DOI] [PubMed] [Google Scholar]
- 40.Hirota T, Lewis WG, Liu AC, Lee JW, Schultz PG, Kay SA. A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3beta. Proc Natl Acad Sci U S A. 2008;105(52):20746–20751. doi: 10.1073/pnas.0811410106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eide EJ, Woolf MF, Kang H, Woolf P, Hurst W, Camacho F, Vielhaber EL, Giovanni A, Virshup DM. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25(7):2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Isojima Y, Nakajima M, Ukai H, Fujishima H, Yamada RG, Masumoto KH, Kiuchi R, Ishida M, Ukai-Tadenuma M, Minami Y, Kito R, Nakao K, Kishimoto W, Yoo SH, Shimomura K, Takao T, Takano A, Kojima T, Nagai K, Sakaki Y, Takahashi JS, Ueda HR. CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci U S A. 2009;106(37):15744–15749. doi: 10.1073/pnas.0908733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meng QJ, Maywood ES, Bechtold DA, Lu WQ, Li J, Gibbs JE, Dupre SM, Chesham JE, Rajamohan F, Knafels J, Sneed B, Zawadzke LE, Ohren JF, Walton KM, Wager TT, Hastings MH, Loudon AS. Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc Natl Acad Sci U S A. 2010;107(34):15240–15245. doi: 10.1073/pnas.1005101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee JW, Hirota T, Peters EC, Garcia M, Gonzalez R, Cho CY, Wu X, Schultz PG, Kay SA. A small molecule modulates circadian rhythms through phosphorylation of the period protein. Angew Chem. 2011;50(45):10608–10611. doi: 10.1002/anie.201103915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walton KM, Fisher K, Rubitski D, Marconi M, Meng QJ, Sladek M, Adams J, Bass M, Chandrasekaran R, Butler T, Griffor M, Rajamohan F, Serpa M, Chen Y, Claffey M, Hastings M, Loudon A, Maywood E, Ohren J, Doran A, Wager TT. Selective inhibition of casein kinase 1 epsilon minimally alters circadian clock period. J Pharmacol Exp Ther. 2009;330(2):430–439. doi: 10.1124/jpet.109.151415. [DOI] [PubMed] [Google Scholar]
- 46.Chen Z, Yoo SH, Park YS, Kim KH, Wei S, Buhr E, Ye ZY, Pan HL, Takahashi JS. Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc Natl Acad Sci U S A. 2012;109(1):101–106. doi: 10.1073/pnas.1118034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maier B, Wendt S, Vanselow JT, Wallach T, Reischl S, Oehmke S, Schlosser A, Kramer A. A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 2009;23(6):708–718. doi: 10.1101/gad.512209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quiroz JA, Gould TD, Manji HK. Molecular effects of lithium. Mol Interv. 2004;4(5):259–272. doi: 10.1124/mi.4.5.6. [DOI] [PubMed] [Google Scholar]
- 49.Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, Thompson CB, Evans RM. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326(5951):437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Um JH, Yang S, Yamazaki S, Kang H, Viollet B, Foretz M, Chung JH. Activation of 5'-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2. J Biol Chem. 2007;282(29):20794–20798. doi: 10.1074/jbc.C700070200. [DOI] [PubMed] [Google Scholar]
- 51.Barnea M, Haviv L, Gutman R, Chapnik N, Madar Z, Froy O. Metformin affects the circadian clock and metabolic rhythms in a tissue-specific manner. Biochim Biophys Acta. 2012;1822(11):1796–1806. doi: 10.1016/j.bbadis.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 52.Caton PW, Kieswich J, Yaqoob MM, Holness MJ, Sugden MC. Metformin opposes impaired AMPK and SIRT1 function and deleterious changes in core clock protein expression in white adipose tissue of genetically-obese db/db mice. Diabetes Obes Metab. 2011;13(12):1097–1104. doi: 10.1111/j.1463-1326.2011.01466.x. [DOI] [PubMed] [Google Scholar]
- 53.Kwak Y, Jeong J, Lee S, Park YU, Lee SA, Han DH, Kim JH, Ohshima T, Mikoshiba K, Suh YH, Cho S, Park SK. Cyclin-dependent kinase 5 (Cdk5) regulates the function of CLOCK protein by direct phosphorylation. J Biol Chem. 2013;288(52):36878–36889. doi: 10.1074/jbc.M113.494856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kon N, Hirota T, Kawamoto T, Kato Y, Tsubota T, Fukada Y. Activation of TGF-beta/activin signalling resets the circadian clock through rapid induction of Dec1 transcripts. Nat Cell Biol. 2008;10(12):1463–1469. doi: 10.1038/ncb1806. [DOI] [PubMed] [Google Scholar]
- 55.Yagita K, Yamanaka I, Koinuma S, Shigeyoshi Y, Uchiyama Y. Mini screening of kinase inhibitors affecting period-length of mammalian cellular circadian clock. Acta Histochem Cytochem. 2009;42(3):89–93. doi: 10.1267/ahc.09015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang X, Lamia KA, Evans RM. Nuclear receptors, metabolism, and the circadian clock. Cold Spring Harb Symp Quant Biol. 2007;72:387–394. doi: 10.1101/sqb.2007.72.058. [DOI] [PubMed] [Google Scholar]
- 57.Nakamura TJ, Sellix MT, Menaker M, Block GD. Estrogen directly modulates circadian rhythms of PER2 expression in the uterus. Am J Physiol Endocrinol Metab. 2008;295(5):E1025–1031. doi: 10.1152/ajpendo.90392.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Canaple L, Rambaud J, Dkhissi-Benyahya O, Rayet B, Tan NS, Michalik L, Delaunay F, Wahli W, Laudet V. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor alpha defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol. 2006;20(8):1715–1727. doi: 10.1210/me.2006-0052. [DOI] [PubMed] [Google Scholar]
- 59.Wang N, Yang G, Jia Z, Zhang H, Aoyagi T, Soodvilai S, Symons JD, Schnermann JB, Gonzalez FJ, Litwin SE, Yang T. Vascular PPARgamma controls circadian variation in blood pressure and heart rate through Bmal1. Cell Metab. 2008;8(6):482–491. doi: 10.1016/j.cmet.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakahata Y, Akashi M, Trcka D, Yasuda A, Takumi T. The in vitro real-time oscillation monitoring system identifies potential entrainment factors for circadian clocks. BMC Mol Biol. 2006;7:5. doi: 10.1186/1471-2199-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McNamara P, Seo SB, Rudic RD, Sehgal A, Chakravarti D, FitzGerald GA. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: a humoral mechanism to reset a peripheral clock. Cell. 2001;105(7):877–889. doi: 10.1016/s0092-8674(01)00401-9. [DOI] [PubMed] [Google Scholar]
- 62.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134(2):317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 64.Bellet MM, Nakahata Y, Boudjelal M, Watts E, Mossakowska DE, Edwards KA, Cervantes M, Astarita G, Loh C, Ellis JL, Vlasuk GP, Sassone-Corsi P. Pharmacological modulation of circadian rhythms by synthetic activators of the deacetylase SIRT1. Proc Natl AcadSci U S A. 2013;110(9):3333–3338. doi: 10.1073/pnas.1214266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Onishi Y, Kawano Y. Rhythmic binding of Topoisomerase I impacts on the transcription of Bmal1 and circadian period. Nucleic Acids Res. 2012;40(19):9482–9492. doi: 10.1093/nar/gks779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Onishi Y, Oishi K, Kawano Y, Yamazaki Y. The harmala alkaloid harmine is a modulator of circadian Bmal1 transcription. Biosci Rep. 2012;32(1):45–52. doi: 10.1042/BSR20110002. [DOI] [PubMed] [Google Scholar]
- 67.Kondoh D, Yamamoto S, Tomita T, Miyazaki K, Itoh N, Yasumoto Y, Oike H, Doi R, Oishi K. Harmine lengthens circadian period of the mammalian molecular clock in the suprachiasmatic nucleus. Biol Pharm Bull. 2014;37(8):1422–1427. doi: 10.1248/bpb.b14-00229. [DOI] [PubMed] [Google Scholar]
- 68.Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, Ellisman MH, Panda S. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15(6):848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scheiermann C, Kunisaki Y, Frenette PS. Circadian control of the immune system. Nat Rev Immunol. 2013;13(3):190–198. doi: 10.1038/nri3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, Boyce SH, Farrow SN, Else KJ, Singh D, Ray DW, Loudon AS. The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Nucleic Acids Res. 2012;109(2):582–587. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee J, Baek S, Lee DG, Park MK, Cho ML, Park SH, Kwok SK. Digoxin ameliorates autoimmune arthritis via suppression of Th17 differentiation. Int Immunopharmacol. 2015;26(1):103–111. doi: 10.1016/j.intimp.2015.03.017. [DOI] [PubMed] [Google Scholar]
- 72.Baek SY, Lee J, Lee DG, Park MK, Kwok SK, Cho ML, Park SH. Ursolic acid ameliorates autoimmune arthritis via suppression of Th17 and B cell differentiation. Acta Pharmacol Sin. 2014;35(9):1177–1187. doi: 10.1038/aps.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huh JR, Leung MW, Huang P, Ryan DA, Krout MR, Malapaka RR, Chow J, Manel N, Ciofani M, Kim SV, Cuesta A, Santori FR, Lafaille JJ, Xu HE, Gin DY, Rastinejad F, Littman DR. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORgammat activity. Nature. 2011;472(7344):486–490. doi: 10.1038/nature09978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu T, Wang X, Zhong B, Nurieva RI, Ding S, Dong C. Ursolic acid suppresses interleukin-17 (IL-17) production by selectively antagonizing the function of RORgamma t protein. J Biol Chem. 2011;286(26):22707–22710. doi: 10.1074/jbc.C111.250407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jones CR, Huang AL, Ptacek LJ, Fu YH. Genetic basis of human circadian rhythm disorders. Exp Neurol. 2013;243:28–33. doi: 10.1016/j.expneurol.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sahar S, Sassone-Corsi P. Regulation of metabolism: the circadian clock dictates the time. Trends Endocrinol Metab. 2012;23(1):1–8. doi: 10.1016/j.tem.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jia Y, Lu Y, Wu K, Lin Q, Shen W, Zhu M, Huang S, Chen J. Does night work increase the risk of breast cancer? A systematic review and meta-analysis of epidemiological studies. Cancer Epidemiol. 2013;37(3):197–206. doi: 10.1016/j.canep.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 78.De Mei C, Ercolani L, Parodi C, Veronesi M, Lo Vecchio C, Bottegoni G, Torrente E, Scarpelli R, Marotta R, Ruffili R, Mattioli M, Reggiani A, Wade M, Grimaldi B. Dual inhibition of REV-ERBbeta and autophagy as a novel pharmacological approach to induce cytotoxicity in cancer cells. Oncogene. 2015;34(20):2597–2608. doi: 10.1038/onc.2014.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Y, Kojetin D, Burris TP. Anti-proliferative actions of a synthetic REV-ERBalpha/beta agonist in breast cancer cells. Biochem Pharmacol. 2015;96(4):315–322. doi: 10.1016/j.bcp.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hu Y, Spengler ML, Kuropatwinski KK, Comas-Soberats M, Jackson M, Chernov MV, Gleiberman AS, Fedtsova N, Rustum YM, Gudkov AV, Antoch MP. Selenium is a modulator of circadian clock that protects mice from the toxicity of a chemotherapeutic drug via upregulation of the core clock protein, BMAL1. Oncotarget. 2011;2(12):1279–1290. doi: 10.18632/oncotarget.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang YK, Yeager RL, Klaassen CD. Circadian expression profiles of drug-processing genes and transcription factors in mouse liver. Drug Metab Dispos. 2009;37(1):106–115. doi: 10.1124/dmd.108.024174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kang HS, Angers M, Beak JY, Wu X, Gimble JM, Wada T, Xie W, Collins JB, Grissom SF, Jetten AM. Gene expression profiling reveals a regulatory role for ROR alpha and ROR gamma in phase I and phase II metabolism. Physiol Genomics. 2007;31(2):281–294. doi: 10.1152/physiolgenomics.00098.2007. [DOI] [PubMed] [Google Scholar]
- 83.Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4(1):25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 84.Kosir R, Spaninger K, Rozman D. Circadian events in human diseases and in cytochrome P450-related drug metabolism and therapy. IUBMB Life. 2013;65(6):487–496. doi: 10.1002/iub.1160. [DOI] [PubMed] [Google Scholar]
- 85.Gorbacheva VY, Kondratov RV, Zhang R, Cherukuri S, Gudkov AV, Takahashi JS, Antoch MP. Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the CLOCK/BMAL1 transactivation complex. Proc Natl Acad Sci U S A. 2005;102(9):3407–3412. doi: 10.1073/pnas.0409897102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A. 2014;111(45):16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nohara K, Yoo SH, Chen ZJ. Manipulating the circadian and sleep cycles to protect against metabolic disease. Front Endocrinol (Lausanne) 2015;6:35. doi: 10.3389/fendo.2015.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee Y, Chen R, Lee HM, Lee C. Stoichiometric relationship among clock proteins determines robustness of circadian rhythms. J Biol Chem. 2011;286(9):7033–7042. doi: 10.1074/jbc.M110.207217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Polanski R, Hodgkinson CL, Fusi A, Nonaka D, Priest L, Kelly P, Trapani F, Bishop PW, White A, Critchlow SE, Smith PD, Blackhall F, Dive C, Morrow CJ. Activity of the monocarboxylate transporter 1 inhibitor AZD3965 in small cell lung cancer. Clin Cancer Res. 2014;20(4):926–937. doi: 10.1158/1078-0432.CCR-13-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stearns AT, Balakrishnan A, Rhoads DB, Ashley SW, Tavakkolizadeh A. Diurnal rhythmicity in the transcription of jejunal drug transporters. J Pharmacol Sci. 2008;108(1):144–148. doi: 10.1254/jphs.08100sc. [DOI] [PMC free article] [PubMed] [Google Scholar]