Abstract
Background
Given the challenges of confirming prenatal alcohol exposure (PAE) during pregnancy using currently established biomarkers of alcohol consumption, we examined whether serum microRNAs (miRNAs) may serve as stable biomarkers for PAE. Alterations in the levels of specific circulating miRNAs have been associated with various disease states and in animal models of fetal alcohol spectrum disorder.
Methods
Pregnant women in this prospective study were recruited from substance abuse and general maternity clinics affiliated with the University of New Mexico. Serum was collected at the time of admission for delivery from 14 subjects who reported ≥1 binge‐drinking episode or ≥3 drinks/wk during pregnancy and 16 subjects who reported abstinence during pregnancy and tested negative for 5 ethanol biomarkers. Total RNA was isolated from serum and used for microarray analysis.
Results
False discovery rate‐corrected analyses of covariance revealed that 55 miRNAs were significantly altered between the 2 groups. Hierarchical clustering using only the significantly altered miRNAs grouped samples into alcohol‐consuming and non‐alcohol‐consuming individuals. Discriminant analysis then identified miRs‐122*, ‐126, ‐216b, ‐221*, ‐3119, ‐3942‐5p, ‐4704‐3p, ‐4743, ‐514‐5p, and ‐602 as the top 10 discriminators between the 2 groups. Ingenuity Pathway Analysis of putative miRNA targets illustrated that miRNAs identified in this study are involved in biological pathways that mediate the effects of alcohol, such as brain‐derived neurotrophic factor, ERK1/2, and PI3K/AKT signaling.
Conclusions
This is the first report of alterations in serum miRNA expression that are associated with alcohol use during human pregnancy. These results suggest that serum miRNAs could be useful as biomarkers of alcohol exposure.
Keywords: Serum miRNAs, Maternal Alcohol Consumption, Prenatal Alcohol Exposure, Biomarkers, Microarrays
Despite evidence that consuming alcohol during pregnancy is harmful for the developing fetus, more than 10% of women in the United States report at least some alcohol use during pregnancy (Tan et al., 2015). The prevalence of fetal alcohol syndrome in the United States ranges from 0.5 to 2.0 per 1,000 live births in the general population to 1.4 to 9.8 per 1,000 live births in high‐risk groups (May and Gossage, 2001). Based on active case ascertainment methods, the prevalence of the entire continuum of fetal alcohol spectrum disorder (FASD) has been estimated to be as high as 24 to 48 per 1,000 live births, or 2 to 5% in the United States (May et al., 2014). However, the identification of children adversely affected by prenatal alcohol exposure (PAE) can be difficult because many of them do not exhibit any of the physical features associated with PAE and mainly manifest with neurocognitive deficits later in life. In the absence of the physical features and without accurate information on PAE, it is difficult to make a diagnosis of FASD. While ethanol (EtOH) biomarkers, as an objective measure, can help to overcome the limitations of self‐report on alcohol use during pregnancy, none of the existing biomarkers of alcohol consumption are 100% sensitive or specific (Bakhireva and Savage, 2011). Given these limitations, the identification of more sensitive and specific biomarkers that either alone or in combination with other clinical and self‐report measures can provide more accurate information on PAE would result in better identification of children with FASD and increase the opportunities for earlier diagnosis and interventions.
MicroRNAs (miRNAs) are approximately 22‐nucleotide single‐stranded non‐protein‐coding RNA molecules that serve primarily to silence the expression of mRNA transcripts to which they hybridize by base pairing. miRNA biogenesis involves the processing of primary miRNA transcripts (pri‐miRNA) to hairpin precursor structures (pre‐miRNA), which are then exported to the cytoplasm where either strand may be loaded into the RNA‐induced silencing complex (RISC). The strand previously called the miRNA star (miRNA*) was generally thought to be rapidly degraded. However, miRNA* sequences have been found in RISC and shown to impact disease states such as cancer (Jazdzewski et al., 2009). The nomenclature is now changing to describe from which arm of the precursor the miRNA originates, 5′ (5p) or 3′ (3p). Mature miRNAs can bind thousands of transcripts in a cell, making them “master regulators” of gene expression (Bartel, 2004). Moreover, miRNAs have been shown to control many complex biological processes, from embryonic development to the promotion (or inhibition) of various pathological conditions, including cancers and neurodevelopmental disorders (Sayed and Abdellatif, 2011).
Recent animal model and cell culture studies suggest that miRNAs play an important role in the mechanisms underlying the deleterious effects of PAE (Balaraman et al., 2013). EtOH suppresses miRs‐9, ‐21, ‐153, and ‐335 in cultured fetal mouse neural stem cells, coordinately regulating genes that make cells resistant to apoptosis, with increased proliferation and aberrant differentiation (Sathyan et al., 2007). miRs‐9(‐5p), 9*(‐3p), and ‐153 are repressed by EtOH exposure in the developing zebrafish brain (Tal et al., 2012). EtOH exposure in the developing mouse brain increases the expression of miRs‐10a and ‐10b, with a concomitant decrease in the target gene homeobox A1 (HOXA1; Wang et al., 2009), which is essential for normal embryonic development. Alterations in miRNA expression were also found in primary neuronal cultures from the cortex of mice at embryonic day 15 following chronic intermittent EtOH exposure and withdrawal (Guo et al., 2012). Finally, differences in miRNA expression were seen in the amygdala and striatum of rats at postnatal day 42 that were exposed to EtOH prenatally and subjected to social enrichment training postnatally (Ignacio et al., 2014).
In addition to working inside the cell, miRNAs can be secreted and transferred between cells, indicating that they participate in cell‐to‐cell communication (Valadi et al., 2007). miRNAs are present not only in blood plasma and serum but also in a variety of cell‐free body fluids including urine and saliva, making them attractive as potential biomarkers. In the circulation, they are found in vesicles such as exosomes, microvesicles, and apoptotic bodies (Valadi et al., 2007; Zernecke et al., 2009) as well as in association with proteins (Vickers et al., 2011). Because of their association with vesicles and proteins, circulating miRNAs are resistant to degradation and can be accurately measured even when present at very low levels (Mitchell et al., 2008). Thus, circulating miRNAs have great promise as biomarkers of a variety of disease conditions from cancer to myocardial infarction (Etheridge et al., 2011). Although extracellular miRNAs have been investigated in animal models of PAE (Balaraman et al., 2014), the utility of these biomarkers in humans has not been reported thus far. The goal of the present study was to identify alterations in miRNA expression in maternal serum that could predict maternal alcohol consumption in pregnant women and further elucidate the mechanisms of fetal alcohol effects.
Materials and Methods
Patient Recruitment and Assessment of Substance Use
Study participants for this project were selected from a prospective cohort study at the University of New Mexico (UNM), which was described in detail elsewhere (Bakhireva et al., 2012, 2014; Gutierrez et al., 2015). Briefly, patients were recruited from a UNM clinic, which provides comprehensive care to pregnant women with current or past history of substance abuse, and followed until early postpartum. Use of alcohol and other substances was ascertained by timeline follow back interviews at enrollment and during the hospital stay after delivery. The composite index, which was based on self‐reported measures of alcohol use, was used to classify subjects into alcohol and control groups. Alcohol consumers had to report ≥0.21 ounces of absolute alcohol per day (AAD), equivalent to ≥3 drinks/wk, or ≥2.0 ounces of absolute alcohol per drinking day (AADD), equivalent to at least 1 binge‐drinking episode, at enrollment. Nonalcohol consumers reported no binge‐drinking episodes in the periconceptional period, ≤0.14 ounces AAD (equivalent to ≤2 drinks/wk) in the periconceptional period, and zero ounces AAD and AADD at enrollment and at the follow‐up. Patients were also administered a standard Alcohol Use Disorders Identification Test (AUDIT) questionnaire to assess alcohol use over the past 12 months.
Self‐reported use of other substances, ascertained at each visit, was supplemented by urine drug screens for amphetamines, barbiturates, benzodiazepines, buprenorphine, cannabinoids, cocaine, methadone, and opiates (analyzed at Tricore Reference Laboratories, Albuquerque, NM). Information on hepatitis C status and other hepatobiliary conditions was extracted from electronic medical records.
Sample Preparation and Analysis of EtOH Biomarkers
Blood was collected at enrollment (Visit 1) and again upon admission for labor and delivery (Visit 2). Serum was isolated from the whole blood. A portion of the serum was tested for serum gamma‐glutamyltranspeptidase (GGT) and % disialotransferrin (carbohydrate deficient) from total transferrin (%dCDT) analysis. Urine was also collected and tested for urine ethyl glucuronide (UEtG) and urine ethyl sulfate (UEtS). Phosphatidylethanol in dried blood spot cards (PEth‐DBS), collected from newborns by heel stick, was also analyzed. The remaining serum was stored at −80°C until further processing.
RNA Extraction and Microarray Analysis
Total RNA, including miRNA, was extracted from 200 μl serum collected at Visit 2 using the miRNeasy Serum/Plasma Kit (Qiagen, Valencia, CA). Bacteriophage MS2 RNA (1 μg) was included as a carrier, and Caenorhabditis elegans miR‐39 (5.6 × 108 copies) was added as a spike‐in control for normalization purposes. miRNA was prepared for microarray analysis using the FlashTag™ Biotin HSR RNA Labeling Kit (Affymetrix, Santa Clara, CA). Biotin‐labeled RNA targets were then hybridized to GeneChip® miRNA 3.0 Arrays (Affymetrix). The probe signal intensities were log2‐transformed and normalized to the total intensity of the array as described in Supplementary methods (Appendix S1). All miRNA nomenclature in the body of this article has been corrected to reflect that of miRBase Release 21 (Kozomara and Griffiths‐Jones, 2014), the latest version at the time of submission.
Quantitative Reverse Transcription Polymerase Chain Reaction
Total RNA was used for reverse transcription followed by quantitative polymerase chain reaction (qPCR) using the TaqMan® MicroRNA Reverse Transcription Kit and TaqMan® MicroRNA Assays (Life Technologies, Carlsbad, CA) as described in Supplementary methods.
Statistical and Other Data Analyses
Batch Effect Corrections
The 30 samples analyzed by microarray were completed in 2 batches with each batch including samples from both alcohol and nonalcohol groups. Principal component analysis and hierarchical clustering of the normalized expression data revealed batch effects in the data reflecting the sample processing batches. The batch effects were corrected using the ComBat method implemented in the R “sva” package as described in Supplementary methods.
Analyses of Covariance
Although there were no significant differences in the presence of hepatitis C or drugs of abuse between alcohol‐consuming and nonconsuming subjects (Tables 1 and 2), women on opioid maintenance therapy (OMT) had increased prevalence of smoking, marijuana use, and hepatitis C (Tables S1–S3). Thus, these factors were included as covariates in the analysis of covariance (ANCOVA) that was performed for each miRNA to evaluate the effects of alcohol and OMT (Table S4). Statistical significance was evaluated using F‐tests, which were corrected for multiple testing using the false discovery rate (FDR) method (Benjamini and Hochberg, 1995) with a cutoff p‐value of <0.05.
Table 1.
Demographic Characteristics of the Study Sample (N = 30)
| Characteristics | Alcohol (N = 14) | Control (N = 16) | p‐Value |
|---|---|---|---|
| mean ± SD | mean ± SD | ||
| Maternal age, years | 29.1 ± 6.7 | 25.7 ± 3.7 | 0.09 |
| Gestational age at enrollment, mo | 25.1 ± 8.7 | 26.8 ± 9.0 | 0.63 |
| Gestational age at delivery, mo | 38.2 ± 2.2 | 37.7 ± 3.7 | 0.65 |
| Ethnicity: Hispanic/Latina | 50% | 63% | 0.71 |
| Race | |||
| White | 64% | 62% | 1.0 |
| American Indian | 21% | 6% | 0.32 |
| Black/African American | 14% | 6% | 0.59 |
| Marital status | |||
| Single/separated/divorced | 50% | 50% | 1.0 |
| Married/co‐habitating | 50% | 50% | 1.0 |
| Health insurance | |||
| No insurance | 0% | 0% | — |
| Any other insurance | 100% | 100% | 1.0 |
| Education | |||
| Less than high school | 43% | 19% | 0.46 |
| High school or equivalent | 21% | 31% | 0.69 |
| Some college or higher | 36% | 50% | 0.48 |
| Primigravida | 43% | 25% | 0.44 |
| Maternal chronic conditions | |||
| Hepatitis C | 14% | 38% | 0.22 |
| Other hepatobiliary disorders | 0% | 6% | 1.0 |
| Pre‐eclampsia/PIH | 21% | 0% | 0.09 |
PIH, pregnancy‐induced hypertension.
p‐Values for age were calculated using t‐tests, and those for % in each category were calculated using 2‐tailed Fisher's exact tests.
Table 2.
Use of Alcohol and Other Substances by Study Group (N = 30)
| Substances of abuse | Alcohol (N = 14) | Control (N = 16) | p‐Value |
|---|---|---|---|
| Alcohol use in past 12 months | |||
| AUDIT (mean ± SD) | 10.1 ± 8.5 | 1.2 ± 1.0 | <0.01 |
| AUDIT ≥ 8 | 43% | 0% | <0.01 |
| Alcohol use in periconceptional period | |||
| Frequency of binge (≥4 drinks) | |||
| No binge episodes | 14% | 100% | <0.01 |
| Once/wk or less | 64% | 0% | <0.01 |
| 2 to 4 times a week | 14% | 0% | 0.21 |
| ≥5 times/wk | 7% | 0% | 0.47 |
| Alcohol use at Visit 1 | |||
| At least 1 episode of binge since LMP | 85% | 0% | <0.01 |
| At least 3 drinks in 1 week since last LMP | 15% | 0% | <0.01 |
| Alcohol use at Visit 2 | |||
| Admitted any alcohol use during the past 2 weeks | 0% | 0% | — |
| Ethanol biomarkers | |||
| GGT > 40 U/l | 7% | 0% | 0.47 |
| %dCDT > 2.0% | 29% | 0% | 0.04 |
| UEtG ≥ 25 ng/ml | 7% | 0% | 0.47 |
| UEtS ≥ 7 ng/ml | 14% | 0% | 0.21 |
| PEth‐DBS on a newborn > 8 ng/ml | 43% | 0% | 0.02 |
| Illicit drug and tobacco use | |||
| Marijuana | 7% | 19% | 0.60 |
| Cocaine/crack‐cocaine | 0% | 6% | 1.0 |
| Opioid maintenance therapy | 36% | 44% | 0.72 |
| Opioids | 14% | 12% | 1.0 |
| Benzodiazepines | 0% | 0% | — |
| Amphetamines | 0% | 0% | — |
| Tobacco | 29% | 44% | 0.47 |
AUDIT, Alcohol Use Disorders Identification Test; LMP, last menstrual period; GGT, gamma‐glutamyltranspeptidase; %dCDT, % disialotransferrin (carbohydrate deficient) from total transferrin; UEtG, urine ethyl glucuronide; UEtS, urine ethyl sulfate; PEth‐DBS, phosphatidylethanol in dried blood spot cards.
Opioid maintenance therapy: methadone and/or buprenorphine; opioids: heroin and/or prescription opioids. Visit 1: enrollment. Visit 2: labor and delivery.
p‐Values for AUDIT values were calculated using t‐tests, and those for % in each category were calculated using 2‐tailed Fisher's exact tests.
Cluster Analyses
Hierarchical clustering of normalized batch‐corrected data was performed for both the samples and the miRNAs that passed FDR‐corrected ANCOVAs (α = 0.05), as described in Supplementary methods.
Discriminant Analyses and Leave‐One‐Out Cross‐Validation Analyses
Orthogonal partial least‐squares discriminant analysis (O‐PLS‐DA) was performed on the miRNAs that passed FDR‐corrected ANCOVAs (α = 0.05) to identify miRNAs that best discriminated between groups. Using the Multibase add‐in for Excel, a 2‐component model was calculated (Table S5). Leave‐one‐out cross‐validation (LOOCV) analyses (Stone, 1974) were performed repeating ANCOVAs leaving each of the 30 samples out (Table S6).
Pathway Analyses
TargetScanHuman Release 6.2 (http://www.targetscan.org/vert_61/) or MicroCosm Targets version 5 (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/) was used to identify transcripts bioinformatically predicted to be targets of miRNAs of interest based on complementarity to the seed region as described in Supplementary methods. We then used Ingenuity Pathway Analysis (IPA; Qiagen) to search the putative targets for common molecular pathways.
Results
Patient Characteristics and Substance Use
As shown in Table 1, there were no significant differences between the study groups for maternal age, gravidity, gestational age at enrollment or delivery, and the presence of hepatobiliary disorders, such as hepatitis C (all p > 0.05). The 2 groups were also similar in all other sociodemographic characteristics.
The use of alcohol and other substances among study participants is presented in Table 2. Almost half (43%) of subjects in the alcohol group had AUDIT ≥8, almost a quarter (21%) reported ≥2 binge‐drinking episodes per week in the periconceptional period, and the vast majority (85%) admitted to at least 1 binge episode since the last menstrual period (LMP). The most prevalent positive biomarkers in the alcohol group were PEth‐DBS (43%) and %dCDT (29%). None of the controls reported any risky drinking in the periconceptional period. All of the controls reported abstinence from alcohol after the LMP and tested negative on all 5 biomarkers (i.e., GGT, %dCDT, UEtG, UEtS, and PEth‐DBS). As might be expected of patients recruited from substance‐abuse clinics, the use of drugs other than alcohol, that is, marijuana, cocaine, OMT or other opioids, and tobacco, was prevalent. However, there were no significant differences between the groups for any of these substances.
Microarray Analysis Reveals Serum miRNAs Altered by Alcohol Use
We extracted total RNA from the serum samples and measured miRNA levels using Affymetrix GeneChip® miRNA Arrays. The advantage of this system is that its robust signal amplification is very sensitive without the need for nucleic acid amplification. This is important because PCR inhibitors present in serum and plasma may carry over into RNA samples. By our analysis, the most abundant miRNAs were miRs‐3613‐3p, 4668‐5p, ‐16, ‐92a, and let‐7b. We detected miRNAs previously identified in the serum of pregnant women, such as the placenta‐specific miR‐498 cluster members (Gilad et al., 2008; Williams et al., 2013), validating this detection method (Table S7).
Several studies have reported that high levels of hemolysis can affect accurate identification and quantitation of miRNAs in circulation (Pritchard et al., 2012). We measured free hemoglobin in all of our serum samples and found low levels across all samples, with no difference between the alcohol and control groups (data not shown). We also evaluated the levels of miRNAs specific to red blood cells (RBCs), platelets, and white blood cells (WBCs). Although miRNAs that are specific to RBCs, such as miR‐451, were detected in our samples, none were significantly altered between groups (Fig. 1 A,B), indicating that hemolysis was not a significant confounder in the study. The levels of WBC miRNAs, such as the lymphoid line marker miR‐125b (Malumbres et al., 2009) and the WBC and platelet marker miR‐223 (Pan et al., 2014), also were not different between the groups (Fig. 1 C,D).
Figure 1.
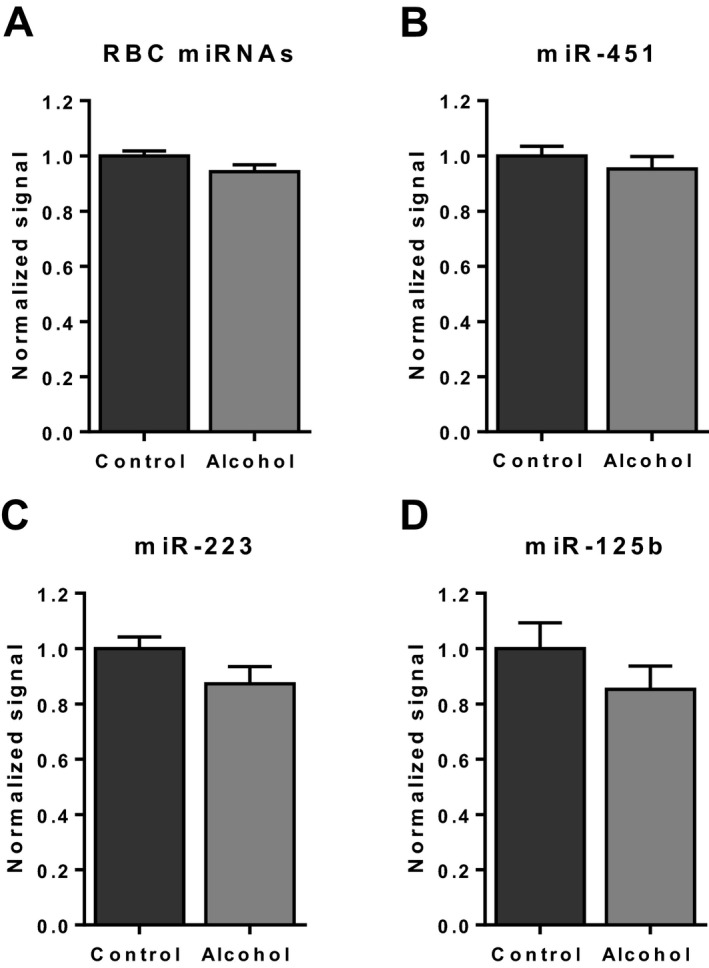
Red blood cell (RBC) and white blood cell (WBC) microRNA (miRNA) levels in serum. Relative levels of RBC miRNAs (miR‐451, miR‐486‐5p, miR‐92a, let‐7a, and miR‐16) (A), and in particular miR‐451 (B), showed no difference between groups, indicating that hemolysis did not affect the results. There was also no difference between groups for WBC miRNAs, miR‐223 (C) and miR‐125b (D). Data represent microarray signal intensities normalized to the average values of the control group (mean ± SEM).
ANCOVAs adjusting by hepatitis C, tobacco, and marijuana use identified 55 miRNAs that were significantly altered between the alcohol‐consuming and control groups (FDR‐corrected p < 0.05; Table 3). Forty‐six miRNAs were elevated and 9 were reduced in the alcohol‐consuming group. The top 10 miRNAs with increased fold changes in the alcohol group were miRs‐509‐5p, ‐3119, ‐26a‐2‐3p, ‐1279, ‐4743, ‐4799‐3p, ‐4657, ‐3942‐3p, ‐3126‐3p, and ‐514b‐5p. The top 4 miRNAs whose levels decreased in the alcohol group were miRs‐125a‐5p, ‐602, ‐126, and ‐3180‐3p. Although there was a significant increase in the frequency of tobacco use in subjects receiving OMT (Fisher's exact test p = 0.0004; Table S3), we found no effect of OMT on the levels of the 55 miRNAs (Table S4).
Table 3.
Significantly Altered Serum MicroRNAs (miRNAs) Between Alcohol and Control Groups
| miRNA | Mean Alcohol | Mean Control | Fold change | FDR (α = 0.05) |
|---|---|---|---|---|
| hsa‐miR‐509‐5p | 2.4966 | 1.3980 | 1.79 | 0.000901 |
| hsa‐miR‐3119 | 2.0249 | 1.1922 | 1.70 | 0.021064 |
| hsa‐miR‐26a‐2 star | 2.6341 | 1.5560 | 1.69 | 0.018766 |
| hsa‐miR‐1279 | 2.9474 | 1.7543 | 1.68 | 0.041649 |
| hsa‐miR‐4743 | 2.6060 | 1.5843 | 1.64 | 0.007583 |
| hsa‐miR‐4799‐3p | 2.6154 | 1.5950 | 1.64 | 0.013677 |
| hsa‐miR‐4657 | 3.0696 | 1.8938 | 1.62 | 0.004073 |
| hsa‐miR‐3942‐3p | 2.9562 | 1.8385 | 1.61 | 0.007583 |
| hsa‐miR‐3126‐3p | 2.7875 | 1.7392 | 1.60 | 0.040425 |
| hsa‐miR‐514b‐5p | 3.1947 | 2.0040 | 1.59 | 0.007583 |
| hsa‐miR‐489 | 2.4800 | 1.5605 | 1.59 | 0.047477 |
| hsa‐miR‐890 | 3.3764 | 2.1307 | 1.58 | 0.013677 |
| hsa‐miR‐122 star | 3.8076 | 2.4442 | 1.56 | 0.014252 |
| hsa‐miR‐542‐3p | 2.6132 | 1.6777 | 1.56 | 0.015460 |
| hsa‐miR‐556‐3p | 3.0859 | 1.9935 | 1.55 | 0.019162 |
| hsa‐miR‐541 | 1.7869 | 1.1610 | 1.54 | 0.013677 |
| hsa‐miR‐202 | 2.6232 | 1.7098 | 1.53 | 0.013677 |
| hsa‐miR‐3606 | 2.9914 | 1.9513 | 1.53 | 0.021064 |
| hsa‐miR‐216b | 2.7549 | 1.8007 | 1.53 | 0.046808 |
| hsa‐miR‐635 | 3.3317 | 2.1867 | 1.52 | 0.013677 |
| hsa‐miR‐1911 star | 3.2525 | 2.1360 | 1.52 | 0.015594 |
| hsa‐miR‐550a star | 3.1434 | 2.0700 | 1.52 | 0.013677 |
| hsa‐miR‐4791 | 4.2234 | 2.8288 | 1.49 | 0.022087 |
| hsa‐miR‐3942‐5p | 4.6259 | 3.1228 | 1.48 | 0.029362 |
| hsa‐miR‐4439 | 1.6874 | 1.1418 | 1.48 | 0.046808 |
| hsa‐miR‐3927 | 3.7734 | 2.5616 | 1.47 | 0.015594 |
| hsa‐miR‐31 star | 1.4057 | 0.9707 | 1.45 | 0.007583 |
| hsa‐miR‐221 star | 3.4809 | 2.4129 | 1.44 | 0.047477 |
| hsa‐miR‐3152‐3p | 3.2677 | 2.2680 | 1.44 | 0.007583 |
| hsa‐miR‐4742‐5p | 4.4185 | 3.0798 | 1.43 | 0.005426 |
| hsa‐miR‐30c‐2 star | 2.1195 | 1.4783 | 1.43 | 0.032213 |
| hsa‐miR‐509‐3‐5p | 3.0559 | 2.1455 | 1.42 | 0.041649 |
| hsa‐miR‐25 star | 2.6124 | 1.8363 | 1.42 | 0.005551 |
| hsa‐miR‐4772‐5p | 3.4997 | 2.4684 | 1.42 | 0.047864 |
| hsa‐miR‐3121‐3p | 3.0179 | 2.1405 | 1.41 | 0.040425 |
| hsa‐miR‐183 star | 2.2611 | 1.6059 | 1.41 | 0.016625 |
| hsa‐miR‐488 | 2.5163 | 1.8040 | 1.39 | 0.023965 |
| hsa‐miR‐496 | 1.6834 | 1.2162 | 1.38 | 0.041649 |
| hsa‐miR‐31 | 2.2425 | 1.6210 | 1.38 | 0.032213 |
| hsa‐miR‐30b star | 1.6035 | 1.1607 | 1.38 | 0.016625 |
| hsa‐miR‐510 | 3.4185 | 2.4785 | 1.38 | 0.047864 |
| hsa‐miR‐4704‐3p | 1.5545 | 1.1368 | 1.37 | 0.047864 |
| hsa‐miR‐4736 | 1.9879 | 1.4964 | 1.33 | 0.007583 |
| hsa‐miR‐4670‐5p | 1.5577 | 1.1736 | 1.33 | 0.045832 |
| hsa‐miR‐1269b | 1.2163 | 0.9372 | 1.30 | 0.040425 |
| hsa‐miR‐1323 | 5.6209 | 4.4762 | 1.26 | 0.026218 |
| hsa‐miR‐4507 | 4.7098 | 5.4010 | −1.15 | 0.047477 |
| hsa‐miR‐567 | 0.9540 | 1.1065 | −1.16 | 0.021064 |
| hsa‐miR‐523 | 0.8503 | 0.9865 | −1.16 | 0.023250 |
| hsa‐miR‐1184 | 4.3004 | 5.2474 | −1.22 | 0.035926 |
| hsa‐miR‐2277‐5p | 3.8111 | 4.6653 | −1.22 | 0.021064 |
| hsa‐miR‐3180‐3p | 2.8494 | 3.5929 | −1.26 | 0.007583 |
| hsa‐miR‐126 | 4.4745 | 5.8536 | −1.31 | 0.041649 |
| hsa‐miR‐602 | 2.3604 | 3.2803 | −1.39 | 0.005551 |
| hsa‐miR‐125a‐5p | 2.0362 | 2.8528 | −1.40 | 0.013677 |
Table shows the mean serum miRNA levels (expressed as array‐normalized and batch‐corrected log2 of probe signal intensity), fold changes, and false discovery rate (FDR)‐corrected p‐values.
To verify our microarray results, we performed quantitative RT‐PCR for 4 miRNAs (miRs‐509‐5p, ‐4657, ‐542‐3p, and ‐602) with high fold changes and low FDR values. Our microarray and reverse transcription (RT)‐qPCR data correlated well as miRs‐509‐5p, ‐4657, and 542‐3p, which were elevated in the microarray data, were also increased in our RT‐qPCR data, and miR‐602 was decreased in both data sets (Fig. 2 A,B).
Figure 2.
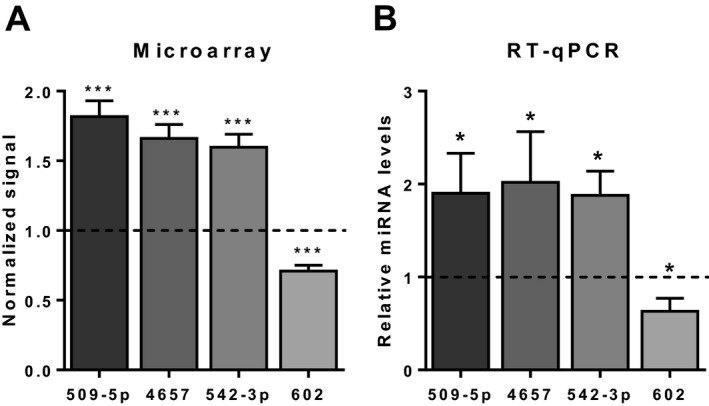
Reverse transcription quantitative polymerase chain reaction (RT‐qPCR) confirms results of microarray analysis. (A) Relative levels by microarray of 4 microRNAs (miRNAs) that passed false discovery rate‐corrected ANCOVAs (α = 0.05), compared to control (dotted line). (B) Relative levels by RT‐qPCR of the same miRNAs. Spike‐in synthetic Caenorhabditis elegans miR‐39 was used for normalization for RT‐qPCR. Data represent mean ± SEM. *p < 0.05, ***p < 0.001.
Clustering Analysis Shows that Serum miRNA Levels Can Classify Subjects According to Alcohol Status
To determine whether serum miRNA levels can be used to categorize patients, we performed hierarchical clustering applying only the significantly altered miRNAs. This analysis grouped the subjects into 2 clusters with each cluster consisting mostly of either alcohol‐consuming or non‐alcohol‐consuming patients (Fig. 3). One alcohol‐consuming subject (A08) who was positive for 1 of the EtOH biomarkers (2.2% dCDT) was grouped with the controls. In addition, 3 control subjects (C1, C9, and C15) were grouped with the alcohol subjects. One explanation for this result could be that alcohol consumption was self‐reported and may not reflect the true/complete nature of alcohol use for some women. Overall, based on the levels of serum miRNAs, most alcohol‐consuming subjects were clustered together separately from non‐alcohol‐consuming subjects.
Figure 3.
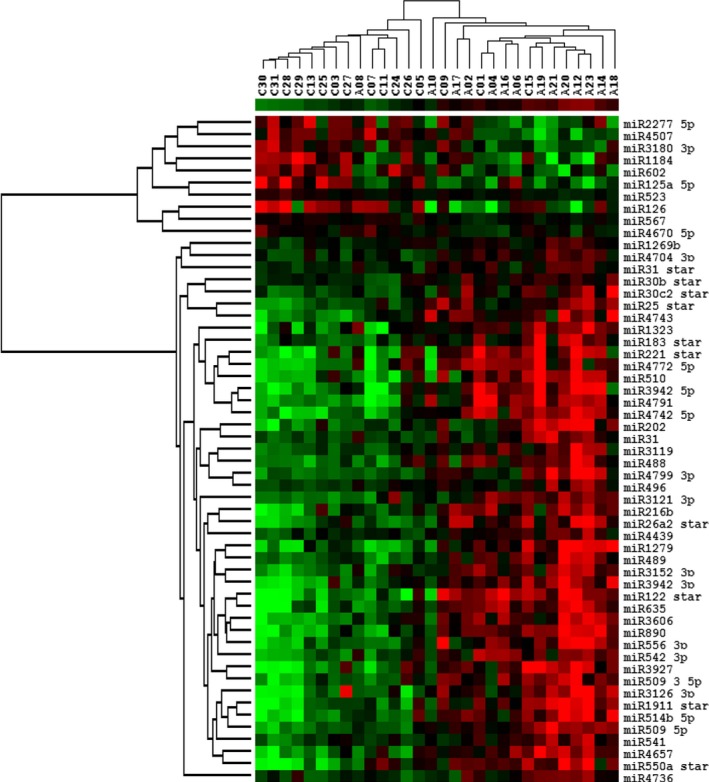
Hierarchical clustering of subjects and microRNAs (miRNAs). The levels of 55 serum miRNAs that passed false discovery rate‐corrected ANCOVAs (α = 0.05) were used for analyses of the 30 subjects. An “A” followed by a number designates all the alcohol‐consuming subjects, while a “C” designates control subjects. Subjects were clustered into 2 groups with most alcohol‐consuming and control individuals present in separate clusters. miRNAs were clustered into 2 groups, those that were increased in the alcohol group and those that were decreased in this group.
miRNAs were grouped into 2 clusters consisting of miRNAs that were either increased or decreased in the alcohol‐consuming patients (Fig. 3). miR‐509‐5p, which displayed the highest increase in fold change in the alcohol group and the most significant p‐value (1.8‐fold, p < 0.001; Table 3), was clustered with miRs‐514b‐5p, ‐1911‐3p, ‐3136‐3p, ‐541, ‐4657, and ‐550a‐3p. miR‐3119, which had the second highest increase in fold change in the alcohol group (1.7‐fold; Table 3), was clustered with miRs‐31, ‐488, and ‐4799‐3p. miR‐125a‐5p, which displayed the largest decrease in fold change (−1.4‐fold; Table 3), was clustered with miR‐523. miR‐602, which had the most significant p‐value among miRNAs exhibiting decreased levels compared to controls (p = 0.006; Table 3), was clustered with miRs‐1184, ‐3180‐3p, ‐4507, and ‐2277‐5p.
Discriminant Analysis Identifies the Most Discerning miRNAs
We next performed O‐PLS‐DA to determine which miRNAs contributed most to the segregation of the alcohol and control groups. We found that miRs‐122*(‐3p), ‐126, ‐216b, ‐221*(‐5p), ‐3119, ‐3942‐5p, ‐4704‐3p, ‐4743, ‐514b‐5p, and ‐602 were the top 10 discriminators between groups (Fig. 4). As described in Table S7 and discussed below, some of these miRNAs are expressed in the liver (miRs‐122*, ‐216b, and ‐602), endothelial cells (miR‐126), or other tissues, where they participate in specific gene regulation and/or are associated with several disease processes. To better visualize the difference between groups for individual miRNAs, we generated bar graphs for the 10 discriminating miRNAs depicting that the levels of all 10 miRNAs were significantly altered in the alcohol‐consuming group (Fig. 5).
Figure 4.
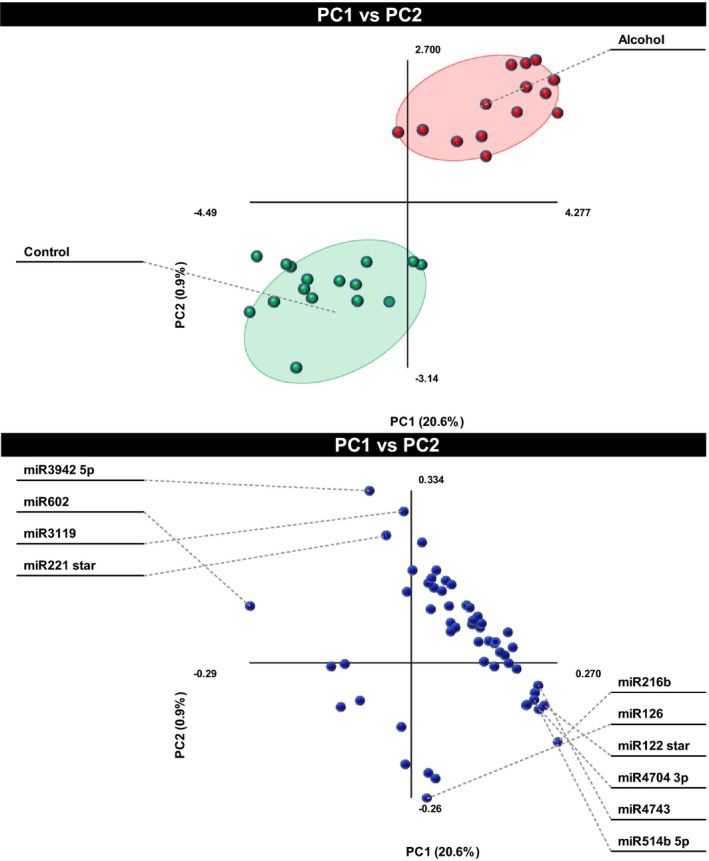
Identification of the top 10 most discriminant microRNAs (miRNAs). The 55 significantly altered miRNAs were subjected to orthogonal partial least‐squares discriminant analysis to determine which miRNAs contributed most to the segregation of the 2 groups. The top panel shows the separation of subjects according to the alcohol consumption. The top 10 contributors are labeled in the bottom panel.
Figure 5.
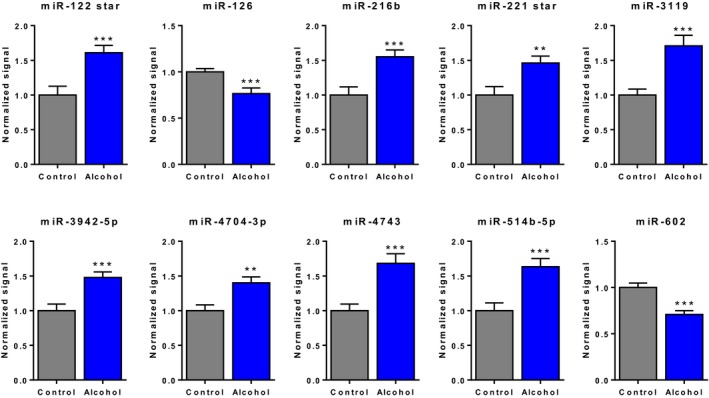
Bar graphs illustrating the difference between alcohols and controls for the top 10 discriminating microRNAs (miRNAs) identified by discriminant analysis. Data represent batch‐corrected and log2‐normalized microarray signal intensities (mean ± SEM). **p < 0.01, ***p < 0.001.
Finally, to assess how the results of our analyses could be generalized to a larger independent dataset, we used LOOCV tests. All leave‐one‐out analyses yielded effect predictions in the predicted direction, with average cross‐validation prediction errors ranging from 0.49 to 4.02% depending on the microRNA (Table S6). The analyses generated significant results even when each of the 30 samples was excluded from the analysis (Table S6).
Pathway Analysis Reveals Putative Targets of Alcohol‐Associated miRNAs
miRNAs largely function by binding to the 3′UTR of target mRNAs, thereby inhibiting translation or triggering degradation of the target. To gain a better understanding of the functional consequences of the presence of particular miRNAs in serum, we analyzed their putative targets, which we obtained from TargetScan or MicroCosm Targets, using IPA. The 10 miRNAs identified by O‐PLS‐DA and the miRNAs confirmed by RT‐qPCR were selected for the analysis. In Figs 6 and 7, we present the top 10 canonical pathways and 1 molecular network for each of 2 miRNAs identified by O‐PLS‐DA, miRs‐126 and ‐216b, and 2 miRNAs confirmed by RT‐qPCR, miRs‐509‐5p, and ‐542‐3p, respectively. Canonical pathways for other miRNAs are shown in Figs S1–S3. miR‐126 targets were included in canonical pathways such as insulin signaling, cardiac hypertrophy signaling, glial cell‐derived neurotrophic factor (GDNF) signaling, and phosphoinositide 3‐kinase (PI3K) signaling (Fig. 6 A). The top network for miR‐126 contained validated targets such as insulin receptor substrate 1 (IRS1) and phosphatidylinositol 3‐kinase regulatory subunit 2 (PIK3R2) and putative targets such as nitric oxide synthase 1 (NOS1) and protein phosphatase 3, catalytic subunit beta (PPP3CB), as well as cAMP‐response element–binding protein (CREB), extracellular signal‐regulated kinases 1/2 (ERK1/2), mitogen‐activated protein kinase kinase 1/2 (MAP2K1/2), mammalian target of rapamycin complex 1 (MTORC1), and protein kinase A (PKA), all of which regulate the effects of alcohol in various systems (Ron and Messing, 2013; Fig. 6 A). miR‐216b targets are involved in prolactin signaling and insulin signaling, as might be expected because it regulates insulin‐like growth factor‐binding protein 2 (IGFBP2; Liu et al., 2015). They also promote cardiogenesis, adipogenesis, and growth hormone signaling (Fig. 6 B). Putative targets of miR‐216b such as neuron‐derived neurotrophic factor (NDNF) and prolylcarboxypeptidase (PRCP) converge on Ak strain transforming (AKT; also known as protein kinase B) in the functional network termed “cell cycle, cellular movement, and cellular function and maintenance” (Fig. 6 B).
Figure 6.
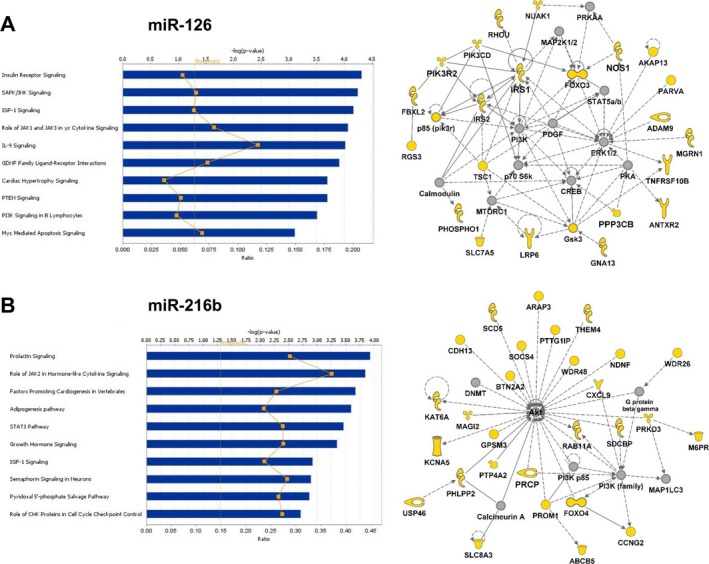
Ingenuity Pathway Analysis canonical pathways and functional networks for putative targets of miR‐126 (A) and miR‐216b (B). The top 10 canonical pathways are shown. Threshold was set at p < 0.05. Molecules in yellow are predicted targets. Solid and dashed lines denote direct and indirect interactions, respectively.
Figure 7.
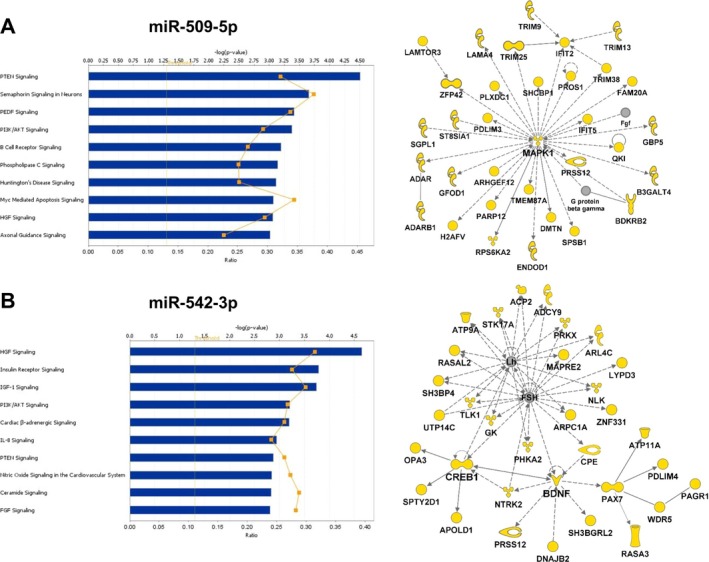
Ingenuity Pathway Analysis canonical pathways and functional networks for putative targets of miR‐509‐5p (A) and miR‐542‐3p (B). The top 10 canonical pathways are shown. Threshold was set at p < 0.05. Molecules in yellow are predicted targets. Solid and dashed lines denote direct and indirect interactions, respectively.
miR‐509‐5p targets genes involved in canonical pathways such as phosphate and tensin homolog (PTEN) signaling, pigment epithelium‐derived factor (PEDF) signaling, PI3K/AKT signaling, and hepatocyte growth factor (HGF) signaling (Fig. 7 A). These pathways are important for cell growth, angiogenesis, and signal transduction. One network for miR‐509‐5p revealed MAPK1 as a putative target as well as other MAPK1‐interacting molecules (Fig. 7 A). Other networks included serotonin receptors HTR2A and HTR2C and the glutamate receptor GRM2 (data not shown). miR‐542‐3p also affects PI3K/AKT signaling and HGF signaling, as well as insulin signaling, cardiac β‐adrenergic signaling, and nitric oxide signaling in the cardiovascular system (Fig. 7 B). Importantly, it is predicted to target brain‐derived neurotrophic factor (BDNF) and CREB1 within a functional network that includes luteinizing hormone (LH) and follicle‐stimulating hormone (FSH) (Fig. 7 B). Among all miRNAs, the most common pathways involved cardiovascular system development and maintenance (Figs 6 and 7, and Figs S1–S3).
Additional pathways of interest included several involved in stress responses such as the production of nitric oxide and reactive oxygen species, iNOS signaling, nNOS signaling in neurons, eNOS signaling in the cardiovascular system, hypoxia signaling, and the endoplasmic reticulum (ER) stress pathway. Specific molecules implicated in stress responsive pathways, such as NOS1, hypoxia‐inducible factors 1A and 3A (HIF1A and HIF3A), ras‐related C3 botulinum toxin substrate 1 (RAC1), heat shock transcription factor 1 (HSF1), and other heat shock proteins, were identified as putative targets of miRs‐4657, ‐509‐5p, ‐542‐3p, ‐602, and ‐216b (Figs S4 and S5). Pathways associated with cognition, such as synaptic long‐term potentiation and synaptic long‐term depression, were also observed. Cognitive deficit‐related factors, such as fragile × mental retardation syndrome‐related protein 1 (FXR1), autism susceptibility candidate 2 (AUTS2), alpha thalassemia/mental retardation syndrome X‐linked (ATRX), methyl‐CpG binding protein 2 (MECP2), and enhancer of zeste 2 (EZH2), were identified as putative targets of miRs‐4657, ‐602, ‐4704‐3p, ‐3119, and ‐126 (Figs S4 and S5).
Discussion
This study evaluated the effect of drinking during pregnancy on the levels of serum miRNAs in order to identify those that might be of use as biomarkers of alcohol consumption. By microarray analysis, 55 miRNAs were identified that were significantly altered by drinking. Discriminant analyses underscored the importance of miRs‐122*, ‐126, ‐216b, ‐221*, ‐3119, ‐3942‐5p, ‐4704‐3p, ‐4743, ‐514b‐5p, and ‐602 for discriminating between the alcohol‐consuming and nonconsuming groups. IPA revealed common pathways affected by all identified miRNAs, such as cell growth and signaling, especially in the cardiovascular system. These serum miRNAs are distinct from those identified in a recent study of patients with alcohol use disorder (AUD; Ignacio et al., 2015), suggesting that the effects of alcohol on pregnant women may be distinct from those in nonpregnant adults. A comparison of alcohol‐sensitive miRNAs identified here with those identified in pregnant ewes exposed to EtOH (Balaraman et al., 2014) revealed only 1 miRNA, miR‐26a‐2*, in common. These findings attest to the complexity of alcohol use in human populations and the importance of understanding the interaction of alcohol with other drugs of abuse.
While many studies have shown that the transfer of exosomes between cells in close proximity is a common form of intercellular communication (Mittelbrunn et al., 2011; Valadi et al., 2007), less is known about the remote function of extracellular miRNAs in circulation. Do they reflect cellular debris, are they merely a reflection of cellular insult by toxins, such as EtOH, or inflammation and other pathogenic processes? Recent studies demonstrate that circulating miRNAs are not just a byproduct of tissue injury, but rather mediate important biological functions. For example, Zhang and colleagues (2010) showed that plasma from patients with atherosclerosis contain higher levels of miR‐150‐containing microvesicles than from healthy controls. These microvesicles can be taken up by endothelial cells, where miR‐150 represses v‐Myb avian myeloblastosis viral oncogene homolog (c‐Myb), thereby regulating cell migration (Zhang et al., 2010). Also, miR‐143/145‐containing extracellular vesicles can be taken up by smooth muscle cells in the aorta of apolipoprotein E −/− (ApoE−/−) mice, reducing atherosclerotic lesions (Hergenreider et al., 2012).
Additional evidence for the functional role of extracellular miRNAs is the finding that the repertoire of miRNAs found in exosomes is distinct from that found in the cells from which they are derived. Two recent publications provide compelling evidence that certain miRNAs are actively selected for secretion. One study identified sequence motifs in a set of miRNAs that were specific to the exosomal (EXOmotif) or cell‐retained (CLmotif) subset (Villarroya‐Beltri et al., 2013). This group also found that the RNA‐binding protein heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNPA2B1) recognizes the EXOmotifs and loads EXOmotif‐containing miRNAs into exosomes accordingly (Villarroya‐Beltri et al., 2013). Intriguingly, 2 miRNAs identified by discriminant analysis, miRs‐514b‐5p and ‐4743, contain the EXOmotif GGAG, suggesting that miRs‐514b‐5p and ‐4743 are targeted for secretion in exosomes. In another study, specific nontemplate terminal nucleotide additions (NTAs) were found to be associated with either exosomal (i.e., uridylation) or cell‐retained miRNAs (i.e., adenylation; Koppers‐Lalic et al., 2014).
The above finding that certain NTAs are associated with miRNA secretion may have implications for the miRNAs identified in our study, especially miR‐122*(‐3p), which was elevated in the serum of pregnant women who consumed alcohol. miR‐122*(‐3p) is a liver‐specific miRNA produced from the same hairpin precursor structure as miR‐122(‐5p), which interacts with the hepatitis B and C viruses (HBV and HCV) and whose expression is highly correlated with liver disease and cancer (Bandiera et al., 2015). Consistent with its abundant expression in the liver, the majority of miR‐122‐5p was found to be adenylated, with a small proportion being uridylated (Li et al., 2012). Both miRs‐122(‐5p) and ‐122*(‐3p) were shown to be elevated in the plasma of children with chronic HBV, although NTA modification was not evaluated in the study (Winther et al., 2013). We found that only miR‐122*(‐3p) was significantly increased in the serum of the alcohol‐consuming group, even though miR‐122(‐5p) was more abundant overall (Table S7).
Whether the miRNAs identified in this study have functional consequences is unknown. miR‐126 is an endothelial‐specific miRNA, processed from intron 7 of the epidermal growth factor‐like 7 gene (EGFL7; a.k.a. vascular endothelial statin), that regulates angiogenesis by targeting several genes including sprout‐related, EVH1 domain containing 1 (Spred‐1) and vascular endothelial growth factor (VEGF; Liu et al., 2009; Wang et al., 2008) as well as EGFL7 itself (Sun et al., 2010). This miRNA is also secreted and present at high levels in exosomes (Koppers‐Lalic et al., 2014). miR‐126 was reduced in the serum of our alcohol group (Table 3, Fig. 5). It is also involved in insulin signaling, and its targets affect PI3K, ERK, and MAPK signaling pathways (Fig. 6 A). Intriguingly, miR‐126 delivered in apoptotic bodies can induce vascular protection in a VEGF‐dependent manner (Zernecke et al., 2009). miR‐126 is also expressed in the brain where it has been associated with both normal neuronal function and neurological disorders (Sonntag et al., 2012). miR‐216b, which was elevated in our alcohol group (Table 3, Fig. 5), is expressed in many tissues and its predicted targets are involved in AKT and p38 MAPK signaling (Fig. 6 B and Fig. S4). miR‐602 is expressed in the liver and associated with HBV‐induced hepatocellular carcinoma (HCC; Yang et al., 2010). Interestingly, 1 putative target, HOXA1 (Fig. S4), is also a target of miRs‐10a and ‐10b, which are altered in the brains of mice prenatally exposed to EtOH (Wang et al., 2009). In human chondrocytes, miR‐602 regulates sonic hedgehog (SHH), important for skeletal formation during development and following birth (Akhtar et al., 2015).
miR‐509‐5p was the most significantly elevated miRNA in the alcohol group, and the related miR‐509‐3‐5p was also increased (Table 3). These miRNAs derive from a cluster of 3 MIR509 genes located in the X chromosome and are highly expressed during development (Zhang et al., 2007). Pathway analysis showed that miR‐509‐5p affects PI3K/AKT signaling and may target MAPK1 and HIF1A (Fig. 7 A and Fig. S4). Importantly, miR‐509‐5p is abundant in many extracellular niches, such as amniotic fluid, saliva, urine, breast milk, cerebrospinal fluid, and colostrum (Weber et al., 2010). miR‐542‐3p was not only elevated in our alcohol group (Table 3) but also in the serum of pregnant rats voluntarily consuming moderate quantities of EtOH as a model of social drinking (Davies S, Gardiner A, Perrone‐Bizzozero N, Savage D, in preparation). miR‐542‐3p is involved in PI3K/AKT signaling, insulin signaling, and cardiovascular maintenance (Fig. 7 B). A recent study identified Syk as a direct target, implicating this miRNA in the proliferation of vascular smooth muscle cells (Qian et al., 2015). Other miRNAs that were altered in the alcohol group include 2 expressed in placenta (miR‐1323 and ‐202), 2 expressed in brain (miR‐488 and ‐541), and 1 that was shown to be down‐regulated in fetal brains of a PAE mouse model (Wang et al., 2009). Additional information and references from miRBase Release 21 for the 55 altered miRNAs between the alcohol and control groups are provided in Table S8. In future studies, it will be interesting to see how the miRNAs identified here are delivered to various tissues and participate in gene regulation. One intriguing possibility is that circulating miRNAs may cross the placental barrier to the developing fetus. A recent study reported that plasma miRNA profiles of newborns (collected from umbilical cord blood) are more similar to their pregnant mothers than unrelated pregnant women or their fathers (Williams et al., 2013). Similar results were observed in control lambs and pregnant ewes; however, the levels of several alcohol‐sensitive plasma miRNAs in PAE lambs were different from their mothers (Balaraman et al., 2014). Thus, circulating miRNA activity may have important implications for both the pregnant woman and the developing fetus.
As in the case of other human tissue studies, some limitations need to be considered. First, similar to another recent serum miRNA microarray study of adults with AUD (Ignacio et al., 2015), our study has a relatively small number of subjects, which may increase the risk that 1 sample may be driving the observed effect. However, the reproducibility of the results using the LOOCV tests indicates that this is not likely the case. Second, it is hard to assess the specific impact of each of the covariates identified in this study, such as tobacco use, on the levels of miRNAs. This is particularly important as nicotine was found to have the opposite effect as EtOH on a panel of EtOH‐sensitive miRNAs identified in neural progenitor cells (Balaraman et al., 2012). Future studies in animal models could help elucidate the effect of different exposures to alcohol alone and in combination with other drugs of abuse on the levels of the miRNAs identified in this study. Third, other important variables such as the window of detectability for miRNA changes at the time of delivery and the effect of different doses of alcohol will need to be assessed in future studies.
In summary, this study identified a panel of miRNAs that may serve as markers of EtOH consumption in pregnant women. The future validation of these miRNA candidate biomarkers along with studies in newborns could facilitate the identification of children at risk for neurodevelopmental disorders, creating opportunities for earlier interventions.
Conflicts of Interest
The authors declared no conflict of interest.
Supporting information
Fig. S1. Top 10 IPA canonical pathways for putative targets of miRs ‐122*(‐3p), ‐221*(‐5p), and ‐514b‐5p.
Fig. S2. Top 10 IPA canonical pathways for putative targets of miRs ‐602, ‐3119, and ‐3942‐5p.
Fig. S3. Top 10 IPA canonical pathways for putative targets of miRs ‐4704‐3p, ‐4743, and ‐4657.
Fig. S4. IPA networks for putative targets of miRs ‐509‐5p, ‐542‐3p, ‐602, and ‐216b.
Fig. S5. IPA networks for putative targets of miRs ‐4657, ‐4704‐3p, ‐3119, and ‐126.
Table S1. Demographic characteristics of the study sample considering OMT status.
Table S2. Subject use of alcohol and other substances considering OMT status.
Table S3. Frequency of smoking and HepC in subjects with and without OMT.
Table S4. ANCOVA analyses of batched corrected and log2 normalized signal intensity adjusted for HepC, smoking, and marijuana.
Table S5. The results of the O‐PLS‐DA.
Table S6. LOOCV analyses.
Table S7. Microarray data for all miRNAs after batch correction and log2 transformation.
Table S8. Additional information and references from miRBase Release 21 for the 55 significant microRNAs.
Appendix S1. Supplementary methods and references.
Acknowledgments
We wish to thank Jamie Padilla and Dr. Gavin Pickett at the Keck‐UNM Genomic Resource facility for performing the microarray assays and initial analyses of the data. This work was supported by a component of the New Mexico Alcohol Research Center (1 P20 AA017068, 1 P50 AA0222534, DDS, PI) to NIP‐B and LNB.
References
- Akhtar N, Makki MS, Haqqi TM (2015) MicroRNA‐602 and microRNA‐608 regulate sonic hedgehog expression via target sites in the coding region in human chondrocytes. Arthritis Rheumatol 67:423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhireva LN, Cano S, Rayburn WF, Savich RD, Leeman L, Anton RF, Savage DD (2012) Advanced gestational age increases serum carbohydrate‐deficient transferrin levels in abstinent pregnant women. Alcohol Alcohol 47:683–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhireva LN, Leeman L, Savich RD, Cano S, Gutierrez H, Savage DD, Rayburn WF (2014) The validity of phosphatidylethanol in dried blood spots of newborns for the identification of prenatal alcohol exposure. Alcohol Clin Exp Res 38:1078–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhireva LN, Savage DD (2011) Focus on: biomarkers of fetal alcohol exposure and fetal alcohol effects. Alcohol Res Health 34:56–63. [PMC free article] [PubMed] [Google Scholar]
- Balaraman S, Lunde ER, Sawant O, Cudd TA, Washburn SE, Miranda RC (2014) Maternal and neonatal plasma microRNA biomarkers for fetal alcohol exposure in an ovine model. Alcohol Clin Exp Res 38:1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaraman S, Tingling JD, Tsai PC, Miranda RC (2013) Dysregulation of microRNA expression and function contributes to the etiology of fetal alcohol spectrum disorders. Alcohol Res 35:18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaraman S, Winzer‐Serhan UH, Miranda RC (2012) Opposing actions of ethanol and nicotine on microRNAs are mediated by nicotinic acetylcholine receptors in fetal cerebral cortical‐derived neural progenitor cells. Alcohol Clin Exp Res 36:1669–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandiera S, Pfeffer S, Baumert TF, Zeisel MB (2015) miR‐122—a key factor and therapeutic target in liver disease. J Hepatol 62:448–457. [DOI] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B (Methodol) 57:289–300. [Google Scholar]
- Etheridge A, Lee I, Hood L, Galas D, Wang K (2011) Extracellular microRNA: a new source of biomarkers. Mutat Res 717:85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, Bentwich Z, Hod M, Goren Y, Chajut A (2008) Serum microRNAs are promising novel biomarkers. PLoS One 3:e3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Chen Y, Carreon S, Qiang M (2012) Chronic intermittent ethanol exposure and its removal induce a different miRNA expression pattern in primary cortical neuronal cultures. Alcohol Clin Exp Res 36:1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez HL, Hund L, Shrestha S, Rayburn WF, Leeman L, Savage DD, Bakhireva LN (2015) Ethylglucuronide in maternal hair as a biomarker of prenatal alcohol exposure. Alcohol 49:617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergenreider E, Heydt S, Treguer K, Boettger T, Horrevoets AJ, Zeiher AM, Scheffer MP, Frangakis AS, Yin X, Mayr M, Braun T, Urbich C, Boon RA, Dimmeler S (2012) Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol 14:249–256. [DOI] [PubMed] [Google Scholar]
- Ignacio C, Hicks SD, Burke P, Lewis L, Szombathyne‐Meszaros Z, Middleton FA (2015) Alterations in serum microRNA in humans with alcohol use disorders impact cell proliferation and cell death pathways and predict structural and functional changes in brain. BMC Neurosci 16:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignacio C, Mooney SM, Middleton FA (2014) Effects of acute prenatal exposure to ethanol on microRNA expression are ameliorated by social enrichment. Front Pediatr 2:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazdzewski K, Liyanarachchi S, Swierniak M, Pachucki J, Ringel MD, Jarzab B, de la Chapelle A (2009) Polymorphic mature microRNAs from passenger strand of pre‐miR‐146a contribute to thyroid cancer. Proc Natl Acad Sci USA 106:1502–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppers‐Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MA, Sadek P, Sie D, Zini N, Middeldorp JM, Ylstra B, de Menezes RX, Wurdinger T, Meijer GA, Pegtel DM (2014) Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep 8:1649–1658. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths‐Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SC, Tsai KW, Pan HW, Jeng YM, Ho MR, Li WH (2012) MicroRNA 3′ end nucleotide modification patterns and arm selection preference in liver tissues. BMC Syst Biol 6(Suppl 2):S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Peng XC, Zheng XL, Wang J, Qin YW (2009) MiR‐126 restoration down‐regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer 66:169–175. [DOI] [PubMed] [Google Scholar]
- Liu FY, Zhou SJ, Deng YL, Zhang ZY, Zhang EL, Wu ZB, Huang ZY, Chen XP (2015) MiR‐216b is involved in pathogenesis and progression of hepatocellular carcinoma through HBx‐miR‐216b‐IGF2BP2 signaling pathway. Cell Death Dis 6:e1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres R, Sarosiek KA, Cubedo E, Ruiz JW, Jiang X, Gascoyne RD, Tibshirani R, Lossos IS (2009) Differentiation stage‐specific expression of microRNAs in B lymphocytes and diffuse large B‐cell lymphomas. Blood 113:3754–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May PA, Baete A, Russo J, Elliott AJ, Blankenship J, Kalberg WO, Buckley D, Brooks M, Hasken J, Abdul‐Rahman O, Adam MP, Robinson LK, Manning M, Hoyme HE (2014) Prevalence and characteristics of fetal alcohol spectrum disorders. Pediatrics 134:855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May PA, Gossage JP (2001) Estimating the prevalence of fetal alcohol syndrome. A summary. Alcohol Res Health 25:159–167. [PMC free article] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova‐Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M (2008) Circulating microRNAs as stable blood‐based markers for cancer detection. Proc Natl Acad Sci USA 105:10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelbrunn M, Gutierrez‐Vazquez C, Villarroya‐Beltri C, Gonzalez S, Sanchez‐Cabo F, Gonzalez MA, Bernad A, Sanchez‐Madrid F (2011) Unidirectional transfer of microRNA‐loaded exosomes from T cells to antigen‐presenting cells. Nat Commun 2:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Liang H, Liu H, Li D, Chen X, Li L, Zhang CY, Zen K (2014) Platelet‐secreted microRNA‐223 promotes endothelial cell apoptosis induced by advanced glycation end products via targeting the insulin‐like growth factor 1 receptor. J Immunol 192:437–446. [DOI] [PubMed] [Google Scholar]
- Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, Tait JF, Tewari M (2012) Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 5:492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian DH, Gao P, Feng H, Qin ZX, Li JB, Huang L (2015) Down‐regulation of mir‐542‐3p promotes neointimal formation in the aging rat. Vascul Pharmacol 72:118–129. [DOI] [PubMed] [Google Scholar]
- Ron D, Messing RO (2013) Signaling pathways mediating alcohol effects. Curr Top Behav Neurosci 13:87–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyan P, Golden HB, Miranda RC (2007) Competing interactions between micro‐RNAs determine neural progenitor survival and proliferation after ethanol exposure: evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J Neurosci 27:8546–8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed D, Abdellatif M (2011) MicroRNAs in development and disease. Physiol Rev 91:827–887. [DOI] [PubMed] [Google Scholar]
- Sonntag KC, Woo TU, Krichevsky AM (2012) Converging miRNA functions in diverse brain disorders: a case for miR‐124 and miR‐126. Exp Neurol 235:427–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone M (1974) Cross‐validatory choice and assessment of statistical predictions. J R Stat Soc B (Methodol) 36:111–147. [Google Scholar]
- Sun Y, Bai Y, Zhang F, Wang Y, Guo Y, Guo L (2010) miR‐126 inhibits non‐small cell lung cancer cells proliferation by targeting EGFL7. Biochem Biophys Res Commun 391:1483–1489. [DOI] [PubMed] [Google Scholar]
- Tal TL, Franzosa JA, Tilton SC, Philbrick KA, Iwaniec UT, Turner RT, Waters KM, Tanguay RL (2012) MicroRNAs control neurobehavioral development and function in zebrafish. FASEB J 26:1452–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CH, Denny CH, Cheal NE, Sniezek JE, Kanny D (2015) Alcohol use and binge drinking among women of childbearing age—United States, 2011–2013. MMWR Morb Mortal Wkly Rep 64:1042–1046. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO (2007) Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659. [DOI] [PubMed] [Google Scholar]
- Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT (2011) MicroRNAs are transported in plasma and delivered to recipient cells by high‐density lipoproteins. Nat Cell Biol 13:423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya‐Beltri C, Gutierrez‐Vazquez C, Sanchez‐Cabo F, Perez‐Hernandez D, Vazquez J, Martin‐Cofreces N, Martinez‐Herrera DJ, Pascual‐Montano A, Mittelbrunn M, Sanchez‐Madrid F (2013) Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun 4:2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LL, Zhang Z, Li Q, Yang R, Pei X, Xu Y, Wang J, Zhou SF, Li Y (2009) Ethanol exposure induces differential microRNA and target gene expression and teratogenic effects which can be suppressed by folic acid supplementation. Hum Reprod 24:562–579. [DOI] [PubMed] [Google Scholar]
- Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel‐Duby R, Olson EN (2008) The endothelial‐specific microRNA miR‐126 governs vascular integrity and angiogenesis. Dev Cell 15:261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K (2010) The microRNA spectrum in 12 body fluids. Clin Chem 56:1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams Z, Ben‐Dov IZ, Elias R, Mihailovic A, Brown M, Rosenwaks Z, Tuschl T (2013) Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc Natl Acad Sci USA 110:4255–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winther TN, Bang‐Berthelsen CH, Heiberg IL, Pociot F, Hogh B (2013) Differential plasma microRNA profiles in HBeAg positive and HBeAg negative children with chronic hepatitis B. PLoS One 8:e58236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Ma Z, Wang D, Zhao W, Chen L, Wang G (2010) MicroRNA‐602 regulating tumor suppressive gene RASSF1A is overexpressed in hepatitis B virus‐infected liver and hepatocellular carcinoma. Cancer Biol Ther 9:803–808. [DOI] [PubMed] [Google Scholar]
- Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Koppel T, Jahantigh MN, Lutgens E, Wang S, Olson EN, Schober A, Weber C (2009) Delivery of microRNA‐126 by apoptotic bodies induces CXCL12‐dependent vascular protection. Sci Signal 2:ra81. [DOI] [PubMed] [Google Scholar]
- Zhang R, Peng Y, Wang W, Su B (2007) Rapid evolution of an X‐linked microRNA cluster in primates. Genome Res 17:612–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X, Sun Q, Wang K, Ba Y, Wang Q, Wang D, Yang J, Liu P, Xu T, Yan Q, Zhang J, Zen K, Zhang CY (2010) Secreted monocytic miR‐150 enhances targeted endothelial cell migration. Mol Cell 39:133–144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Top 10 IPA canonical pathways for putative targets of miRs ‐122*(‐3p), ‐221*(‐5p), and ‐514b‐5p.
Fig. S2. Top 10 IPA canonical pathways for putative targets of miRs ‐602, ‐3119, and ‐3942‐5p.
Fig. S3. Top 10 IPA canonical pathways for putative targets of miRs ‐4704‐3p, ‐4743, and ‐4657.
Fig. S4. IPA networks for putative targets of miRs ‐509‐5p, ‐542‐3p, ‐602, and ‐216b.
Fig. S5. IPA networks for putative targets of miRs ‐4657, ‐4704‐3p, ‐3119, and ‐126.
Table S1. Demographic characteristics of the study sample considering OMT status.
Table S2. Subject use of alcohol and other substances considering OMT status.
Table S3. Frequency of smoking and HepC in subjects with and without OMT.
Table S4. ANCOVA analyses of batched corrected and log2 normalized signal intensity adjusted for HepC, smoking, and marijuana.
Table S5. The results of the O‐PLS‐DA.
Table S6. LOOCV analyses.
Table S7. Microarray data for all miRNAs after batch correction and log2 transformation.
Table S8. Additional information and references from miRBase Release 21 for the 55 significant microRNAs.
Appendix S1. Supplementary methods and references.


