Abstract
Cyclin-dependent kinase 6 (CDK6) plays a vital role in regulating the progression of the cell cycle. More recently, CDK6 has also been shown to have a transcriptional role in tumor angiogenesis. Up-regulated CDK6 activity is associated with the development of several types of cancers. While CDK6 is over-expressed in cancer cells, it has a low detectable level in non-cancerous cells and CDK6-null mice develop normally, suggesting a specific oncogenic role of CDK6, and that its inhibition may represent an ideal mechanism-based and low toxic therapeutic strategy in cancer treatment. Identification of selective small molecule inhibitors of CDK6 is thus needed for drug development. Herein, we review the latest understandings of the biological regulation and oncogenic roles of CDK6. The potential clinical relevance of CDK6 inhibition, the progress in the development of small-molecule CDK6 inhibitors and the rational design of potential selective CDK6 inhibitors are also discussed.
Keywords: cyclin-dependent kinase, cell cycle, drug discovery, inhibitor design, oncogene and tumor suppressor, targeted cancer therapy
Abbreviations
- 3T3
3-day Transfer, Inoculum 3 × 105 cells
- ACTR
Activator of Thyroid and Retinoid Receptor
- AIB1
Amplified in Breast 1
- ALL
Acute Lymphoblastic Leukemia
- AML
Acute Myeloid Leukemia
- CAK
Cyclin Dependent Kinase Activating Kinase
- cAMP
Cyclic Adenosine Monophosphate
- CBP
CREB-binding Protein
- CDK
Cyclin Dependent Kinase
- CDKI
CDK Inhibitor
- Cip
CDK Interacting Protein
- CREB
cAMP-response Element-binding Protein
- DNA
Deoxyribonucleic Acid
- DP
Dimerization Partner (Protein)
- E2F
E2 Promoter Binding Factor
- EYA2
Eyes Absent Homolog 2
- Fbxo7
F-box Protein 7
- FOXM1
Forkhead Box M1
- GCN5
General Control of Amino Acid Synthesis Protein 5
- HER2
Human Epidermal Growth Factor Receptor 2
- HR+
Hormone Receptor Positive
- IC50
Half Maximal Inhibitory Concentration
- INK4
Inhibitor of CDK4
- Kip
Kinase Inhibitory Protein
- MEK
Mitogen-activated Protein Kinase Kinase
- MEP50
Methylosome Protein 50
- miRNA
MicroRNA
- MLL
Mixed Lineage Leukemia
- mRNA
Messenger RNA
- mTOR
Mammalian Target of Rapamycin
- NF-kB
Nuclear Factor kappa-B
- NOTCH1
Neurogenic Locus Notch Homolog Protein 1
- PI3K
Phosphoinositide 3-Kinase
- PLK1
Polo-like Kinase 1
- Rb
Retinoblastoma
- RNA
Ribonucleic Acid
- Runx1
Runt-related Transcription Factor 1
- shRNA
Short Hairpin RNA
- siRNA
Small Interfering RNA
- SMAD2/3
Mothers against Decapentaplegic Homolog 2/3
- STAT3
Signal Transducer and Activator of Transcription 3
- T-ALL
T-Cell Acute Lymphoblastic Leukemia
- TFIIH
Transcription Factor II Human
- Tip60
TAT-interactive Protein
- TRAPP
Transactivation Transformation Domain Associated Protein
- VEGF-A
Vascular Endothelial Growth Factor A
Introduction
Cyclin-dependent kinases (CDKs) are a family of serine/threonine protein kinases that are involved in the cell cycle, transcription and other biological processes such as translation, neurogenesis and apoptosis.1 Deregulation of CDKs is directly linked to oncogenesis. CDKs are reliant on binding a cyclin for their activation. To date, at least 20 CDKs and 30 cyclins have been reported.1,2 Among them, CDK1, CDK2, CDK4 and CDK6 regulate the transition of phases in the cell cycle while CDKs 7–11 are involved in transcription.3
CDK6 gene is located in human chromosome 7 and is translated into a kinase with 326 amino acids. Expression of this gene is upregulated in several types of cancers. CDK6 is the catalytic subunit of the CDK6-cyclin D complex involved in the G1 to S cell cycle progression and negatively regulates cell differentiation. Its activity first appears in mid-G1 phase to phosphorylate, and thus regulate the activity of tumor suppressor protein retinoblastoma (Rb).3,4 Emerging evidence suggests that certain tumor cells require CDK6 for proliferation.5 Consequently, CDK6 represents a promising target for anti-cancer therapy.
This review summarizes the latest knowledge on the function, regulation and structure of CDK6 and the recent progress in the development of pharmacological CDK6 inhibitors. In addition, the potential clinical relevance of specific CDK6 inhibition and the rational design of selective inhibitors are discussed.
Biological functions of CDK6
Phosphorylation of the retinoblastoma proteins
In 1994, Meyerson and Harlow first reported the discovery of CDK6 which is structurally and functionally similar to CDK4.4 Since then, it has been demonstrated that CDK6 and CDK4 are cyclin D activated kinases that phosphorylate Rb and its related proteins p107 and p130 in the G1 phase of the cell cycle (Fig. 1). Both Rb and its related proteins are tumor suppressors that interact with a family of transcription factors known as E2 promoter binding factors (E2F1-E2F8) and repress transcription of genes that are essential for cell cycle progression.6,7 This event involves either direct binding to the E2F transcription factors or modification of chromatin by interacting with histone deacetylases, histone methyltransferases and DNA methyltransferases.8-10
Figure 1.
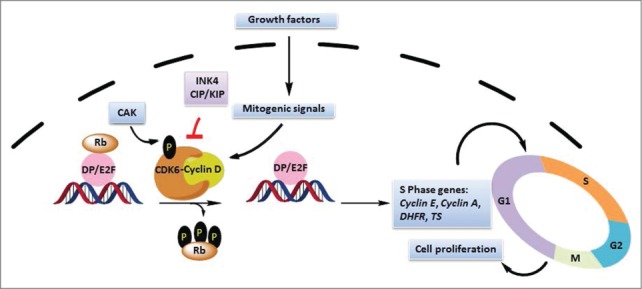
Schematic representation of the function and regulation of CDK6. CDK6 phosphorylates the retinoblastoma (Rb) and its related proteins (Rb) in the G1 phase of the cell cycle, derepressing E2F. E2F then activates the transcription of genes that encode proteins necessary for DNA replication (S-phase entry). Activation of CDK6 requires binding to D-type cyclins and phosphorylation by CAK (CDK7/cyclin H/MAT1). INK4s deactivate CDK6 and Cip/Kip proteins, acting as negative modulators of the CDK6-cyclin D complex.
The first 3 members of the E2F transcription factors, namely E2F1-E2F3, bind to Rb whereas E2F4 and E2F5 bind to any of the 3 proteins. This binding occurs at the C terminus transactivation domain of E2F1-E2F3 which is needed for the activation of gene expression and consequently prevents this site from recruiting transcription factor II D (TFIID) and transcription cofactors such as cyclic adenosine monophosphate (cAMP) response element-binding protein (CREB)-binding protein (p300/CBP), general control of amino acid synthesis protein 5 (GCN5), transactivation transformation domain associated protein (TRAPP), Tat-interactive protein (Tip60) and activator of thyroid and retinoid receptor/amplified in breast 1 (ACTR/AIB1). Rb is also capable of preventing the DNA binding activity of E2F1. Indeed, the E2F transcriptional factors E2F1-E2F6 require dimerization partner proteins (DP1-DP4) for their binding to DNA.11
An initial partial phosphorylation of the Rb proteins by CDK4/6 followed by a complete phosphorylation by CDK2-cyclin E complex leads to structural changes in the pocket domain of Rb and its related proteins, thus releasing and activating E2Fs.6,7 E2Fs subsequently activate transcription of genes necessary for DNA replication (S-phase entry) and cell cycle progression.6-11 Nevertheless, this sequential phosphorylation model has been challenged as Kozar et al. demonstrated that CDK2-cyclin E complex is capable of phosphorylating Rb in the absence of D-type cyclins to induce E2F transcription factors.12
Interestingly, genetic analysis has also revealed that many cell types can proliferate in the absence of CDK4/6 or D cyclins. Yet, these studies have also pinpointed specific CDK requirements by specialized type of cells. For example, CDK6 has been shown to play specific roles in the hematopoietic system; erythropoiesis and T-cell functions were altered in CDK6 knockout mice and this was not compensated by another CDK. CDK6 has also been found to be essential for β cell proliferation in pancreas.12-14 However, CDK6 overexpression has also been shown to reduce skin tumorigenesis and cell growth in fibroblasts.15,16
Phosphorylation of transcription factors
CDK4/6 cyclin D complexes phosphorylate transcription factors, such as forkhead box M1 (FOXM1), mothers against decapentaplegic homolog 2/3 (SMAD2/3), eyes absent homolog 2 (EYA2) and methylosome protein 50 (MEP50) (cofactor) to alter their functions. FOXM1 is needed for both S and M phases of the cell cycle. Multisite phosphorylation by CDK4/6-cyclin D complexes has been shown to both stabilize and activate FOXM1.17 Similarly, phosphorylation of SMAD2 and SMAD3 results in the elimination of the activation function of a SMAD2/3/4 trimer such as the expression of p15 and p21.18 CDK6, but not CDK4, binds to EYA2 protein to reduce its half-life. EYA2 is important in transcriptional regulation during organogenesis.19 CDK6 has also been shown to phosphorylate nuclear factor kappa-B (NF-kB) linking cancers to inflammation. NF-κB induces the expression of pro-inflammatory genes.20,21
Modulation of cell differentiation
Despite the high level of homology between CDK4 and CDK6, there is an increasing list of distinct functions reported for CDK6. Among them is the ability of CDK6 to modulate differentiation in specific cell types including murine erythroid leukemia, primary mouse astrocytes, osteoblasts and osteoclasts, thymocytes, neurons, cardiomyocytes and hematopoietic cells.22-31 In hematopoietic cells, for example, down-regulation of CDK6 allows terminal differentiation due to the release of runt-related transcription factor 1 (Runx1), a transcriptional factor required for the opening of chromatin of important hematopoietic regulator genes.27
Location of CDK6
CDK6 has been shown to reside in the cytoplasm of many cell types. However, in T cells despite the presence of CDK6/cyclin D complex in both the cytoplasm and nucleus, only the nuclear fraction exhibits kinase activity. Kohrt et al. argue that the simultaneous distribution of CDK6 in both the nucleus and cytoplasm is important for this protein to coordinate its roles in cell division and differentiation.32 It has also been located on ruffling edge of fibroblasts, promoting their spreading.33
Clinical relevance
Aberrance of the CDK4/6 cyclin D-INK4-pRb-E2F pathway is common in >80% of human cancers.34 Cyclins D1 and D3 have been shown to be amplified in breast cancer and lymphoid malignancies, respectively.35-37 CDK6 is overexpressed in lymphoma, leukemia, glioma, glioblastoma, medulloblastoma, and cancers of squamous cells, salivary gland, bladder, pancreas and prostate.38-50 In human prostate cancer cells, CDK6 has also been shown to bind androgen receptor and stimulate its activity in a kinase activity independent manner.46 Blockage of CDK6 expression by microRNAs (miRNAs) has been shown to inhibit the proliferation of gliomas, medulloblastoma, prostate, bladder, gastric, hepatocellular, and lung cancer cells, indicating the significant role of CDK6 in the initiation and progression of these cancers.51-58 These observations, coupled with the fact that mice lacking D-type cyclins and/or CDK4/6 are viable, make targeting components of the CDK4/6 cyclin D-INK4-pRb-E2F pathway a highly attractive anti-cancer strategy.12,14
CDK6 and hematological malignancies
Acute myeloid leukemia (AML) is a malignant transformation of hematopoietic cells characterized by an increase in the number of myeloid cells in the marrow and an arrest in their maturation. Chromosomal translocations of band q23 of chromosome 11 of the mixed lineage leukemia (MLL) gene are common in AML and are associated with poor prognosis.59,60 No effective treatment is currently available for patients affected by the disease. The discovery of new therapeutic agents against the disease is vital. Recently, Placke et al. proposed CDK6 as a novel therapeutic target in MLL-rearranged AML. Using a functional genetic approach based on RNA interference (RNAi), it was shown that MLL-AML cells are exceptionally reliant on CDK6, but not CDK4, and that the growth inhibition induced by CDK6 depletion is mediated through enhanced myeloid differentiation. Given that expression of a CDK6K43M mutant with disrupted kinase function or CDK4 failed to show similar effect, it was suggested that the observed induction of differentiation is dependent on the reduced kinase activity of CDK6, but not on the activity of CDK4.61 Similarly, another study has provided evidence for the validity of targeting CDK6 in an aggressive MLL fusion-driven acute lymphoblastic leukemia (MLL-ALL). It was found that CDK6 messenger RNA (mRNA) was over-activated in infant patients with MLL rearranged ALL, and that targeting CDK6 specifically using small interfering RNAs (siRNAs) reduced MLL fusion mRNA expression. A separate knock-down experiment confirmed that CDK6, but not CDK4, was required for the proliferation of MLL-ALL cells.62 Taken together, these findings suggest that CDK6 is a major oncogenic target of MLL-AML and MLL-ALL, and that CDK6 inhibitors are highly likely to find an application in treating patients with the disease.
Another study highlighted a function of CDK6 as a transcriptional regulator that is unrelated to its kinase activity.63 Forced CDK6 expression in p185BCR-ABL transformed pro-B cells decreased cell proliferation accompanied by enhanced levels of p16INK4a, the cell-cycle inhibitor and tumor suppressor, and pro-angiogenic factor VEGF-A. CDK6 has 2 opposing functions, i.e., the ability to inhibit or accelerate cell proliferation depending on whether p16INK4a is present or not. Malignancies of the B- or T-lymphoid lineage frequently display loss of p16INK4a, CDK6 thus confers a proliferative advantage to the transformed cells in the absence of p16INK4a. Furthermore, the ability of CDK6 to promote angiogenesis provides an additional advantage for the growth of cells that express high levels of CDK6. As such, inhibition of CDK6 blocks not only the cancer cell proliferation, but also the blood vessel growth that is required to meet the enhanced demand of tumors for blood supply. The new insight into the transcriptional role of CDK6 provides a rationale to develop CDK6 inhibitors, allowing the simultaneous inhibition of cell-cycle progression and kinase-independent functions.64
In another study, CDK6-deficient mice were shown to be resistant to the development of lymphoma.30 Similarly, CDK6 knock-out mice were also resistant to neurogenic locus notch homolog protein 1 (NOTCH1) driven T-cell acute lymphoblastic leukemia (T-ALL). Inhibition of CDK6-cyclin D3 by PD0332991 in human leukemic cells causes apoptosis in addition to cell cycle arrest. Due to this synthetic-lethal interaction between NOTCH1 overexpression and CDK6 inhibition, it was proposed that inhibiting the kinase activity of CDK6 in patients with NOTCH1-postive T-ALL is an attractive therapeutic strategy.65-67 Furthermore, several studies on patients suffering from lymphomas have revealed chromosomal translocations and the consequent over-expression of CDK6 is a driving force for the disease.68,69 Taken together, these findings strongly support CDK6 as a specific therapeutic target in human lymphoid malignancies.
CDK6 and colorectal carcinoma
A recent study on colorectal carcinoma, one of the leading causes of cancer-related death in the world, has shown CDK6 to be a key therapeutic target.70 Li et al. eliminated the expression of CDK4 and/or CDK6 protein(s) in COLO320 cells by shRNA and showed that CDK6 rather than CDK4 was important for the phosphorylation of Rb protein. Knock-down of CDK6 significantly repressed the growth of COLO320 cells suggesting the potential therapeutic benefit of CDK6 inhibitors against colorectal carcinoma.
CDK6 and medulloblastoma
CDK6 has also been implicated in development of medulloblastoma, the most common malignant brain tumor in children.48,54,71 Medulloblastoma comprises multiple and distinct molecular entities whose clinical and genetic differences likely require separate therapeutic strategies. Four principal sub-groups of medulloblastoma have been identified: those with Wnt pathway mutations, Sonic Hedgehog pathway mutations, and those termed Groups 3 and 4. Group 4 accounts for about 40% of all medulloblastoma cases, and metastases are common in this group. A major feature of Group 4 tumors is significant overexpression of CDK6, and this kinase is an independent prognostic factor for poor overall survival.48,71 Interfering with the production of CDK6 by shRNA or by inducing specific miRNA (i.e., miR-124) reduced proliferation and colony formation in vitro and inhibited the growth of medulloblastoma xenograft tumors in animals.47,54,72 More recently, PD0332991 has been shown to arrest cells in the G1 phase, decrease proliferation and sensitize medulloblastoma cells to ionizing radiation; the effects being modulated by its CDK6 inhibitory mechanism. As alterations in CDK4, cyclin D1, p15 and p16 genes are not commonly seen in medulloblastoma,73 targeting CDK6 specifically may prove beneficial in treating medulloblastoma with minimal toxicity.
Structural features and regulation
Currently, 13 X-ray crystallographic structures of CDK6 (Table 1) are available, 5 of which are complexed with viral cyclin (Vcyclin) and are deemed to depict the active conformation. Four of these structures illustrate inhibitors bound at the ATP active site. The crystal structures of CDK6-inhibitor complexes provide insights on the binding modes and clues on reducing promiscuity. The overall structure of CDK6 demonstrates the bilobal fold that is common to other kinases. The N-terminal lobe contains residues 1-100 and is made of 5 strands of antiparallel β-sheets and the αC-helix (also known as the α1 or PLSTIRE helix). The larger C-terminal lobe is mostly α-helical and comprises residues 101–326. The two lobes are connected by a region known as the hinge. The ATP binding site is located at the lobal interface with the hinge forming one of the edges. The C-terminal lobe portion of the ATP-binding site consists of the highly conserved DFG-motif and the activation loop (residues 163–189). The activation loop, which is also known as T-loop, spans from the DFG motif to the APE motif and includes the phosphorylation site Thr177 (Fig. 2).74-76
Table 1.
Available X-ray crystallographic structures of CDK6
| Description | Resolution (Å) | PDB Code |
|---|---|---|
| CDK6 - Vcyclin76 | 3.10 | 1JOW |
| CDK6 - Vcyclin - 9-cyclopentyl-N-(5-piperazin-1-ylpyridin-2-yl)pyrido[4,5]pyrrolo[1,2-d]pyrimidin-2-amine109 | 2.90 | 4TTH |
| CDK6 - Vcyclin - Aminopurvalanol114 | 2.80 | 2F2C |
| CDK6 - Vcyclin - Fisetin117 | 2.90 | 1XO2 |
| CDK6 - Vcyclin - PD0332991114 | 3.00 | 2EUF |
| CDK6 - Kcyclin - p18INK4c 118 | 2.90 | 1G3N |
| CDK6 - {5-[4-(dimethylamino)piperidin-1-yl]-1H-imidazo[4,5-b]pyridin-2-yl}[2-(isoquinolin-4-yl)pyridin- 4-yl]methanone111 | 2.70 | 4EZ5 |
| CDK6 - 1H-benzimidazol-2-yl(1H-pyrrol-2-yl)methanone111 | 2.31 | 4AUA |
| CDK6 - 4-[3-(1-methylethyl)-1H-pyrazol-4-yl]-N-(1-methylpiperidin-4-yl)pyrimidin-2-amine119 | 2.60 | 3NUP |
| CDK6 - 4-[5-chloro-3-(1-methylethyl)-1H-pyrazol- 4-yl]-N-(5-piperazin-1-ylpyridin-2-yl)pyrimidin-2-amine119 | 2.70 | 3NUX |
| CDK6 - p16INK4a 83 | 3.40 | 1BI7 |
| CDK6 - p19INK4d74,83 | 1.90 | 1BLX |
| 2.80 | 1BI8 |
Figure 2.
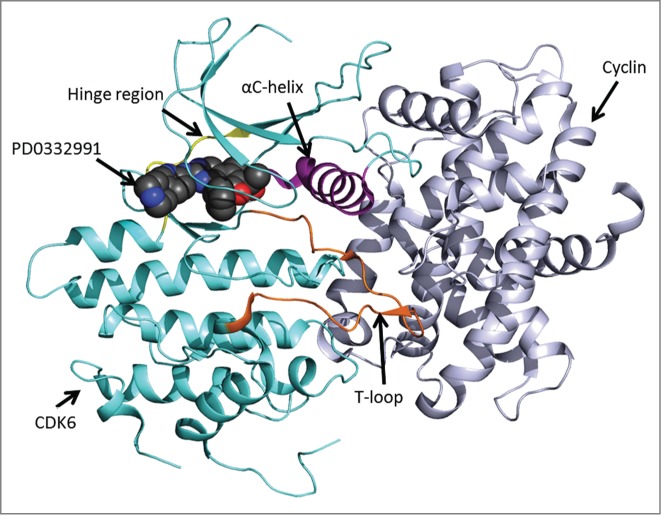
Schematic drawing of the CDK6-Vcyclin complex with inhibitor PD0332991 (PDB ID: 2EUF). CDK6 is shown in turquoise with the αC helix (PLSTIRE) in purple, the T-loop in orange, and the hinge region in yellow. Cyclin is shown in gray. PD0332991 is shown bound in the ATP binding pocket of the kinase. The figure was prepared using PyMOL1.3 (Schrödinger Inc., 2013).
In the monomeric CDK6, the T-loop obstructs the get to the catalytic site and prevents the protein substrate from accessing ATP. Furthermore, key residues involved in the catalysis, such as Glu61, Lys43 and Asp163 are not properly positioned, thus rendering CDK6 catalytically inactive.76
The activation of CDK6 requires the binding to cyclin D1, D2 or D3.4 Due to the lack of a co-crystal structure of CDK6 bound to any of the D-type cyclins, the deduction of the mechanism of partial activation of CDK6 upon binding to D-type cyclins stems from its activation by viral cyclins which are homologous to human D cyclins. As a consequence of binding to Vcyclin, the αC-helix changes its positioning so that Glu61 moves near Lys43 and Asp163 in to the binding cleft. These three residues orient the phosphate group of the ATP for nucleophilic attack by the hydroxyl group of Ser/Thr of the substrate. In addition, the conformational change due to Vcyclin binding exposes Thr177 for phosphorylation by CDK activating kinase (CAK, CDK7/cyclin H/MAT1). This phosphorylation results in the complete activation of CDK6 and stabilizes its active conformation.76 It is noteworthy that the activation of CDK4 by cyclin does not follow this model. Strikingly, unlike CDK6, the binding of cyclin D1 or D3 to the αC-helix (PISTVRE) of CDK4 does not give rise to an active conformation. Moreover, phosphorylation of Thr172 in the T-loop does not result in the activation of the enzyme bound to cyclin D1. Furthermore, there are several lines of evidence showing that CDK4 might not even be phosphorylated by CAK.77,78
The INK4 family of CDK inhibitors (CDKIs) which include p16INK4a, p15INK4b, p18INK4c, and p19INK4d are specific to CDK4 and CDK6. They bind to either the monomeric CDK or the CDK-cyclin D complex resulting in inactivation of the enzyme.79-82 Information obtained from the crystal structures of the complexes between CDK6-cyclin D and p16INK4a, p18INK4c, and p19INK4d reveal that the binding of the INK4 family members to CDK6 causes an extensive movement of the N-domain of the kinase culminating in misalignment of the catalytic residues and distortion of the ATP and cyclin binding sites. In addition, the binding of INK4 proteins to CDK6 has been shown to prevent the binding to the p27Kip1 inhibitor allowing its redistribution to other CDKs.74,83,84 Unlike INK4s, Cip/Kip (CDK interacting protein/kinase inhibitory protein) family of inhibitors (p21Cip1, p27Kip1 and p57Kip2) interact with both the cyclin and CDK parts to deactivate the catalytic CDK6 by causing conformational changes. Both sets of endogenous CDK inhibitors also prevent the binding of CAK to CDK6. Paradoxically, the Cip/Kip family of proteins have also been shown to be important for the activation of CDK4 and CDK6. In addition, phosphorylation of Tyr24 residue in the glycine rich loop by inhibitory kinases Wee1 and Myt1, which can be reversed by Cdc25 family of phosphatases, interferes with proper ATP binding to negatively regulate CDK6.85-87
Inhibiting CDK6
Discovery of highly selective pharmacological inhibitors is a challenging task. No inhibitor targeting an individual CDK has been reported to date. All CDK inhibitors identified so far are either of the pan-CDK (i.e., CDK1/2/4/6/7/9), or CDK4/6 class. Given the complexity of cancer, a combined inhibition of the multiple CDKs can be considered as an effective therapeutic approach. However, a promiscuous multi-target activity might pose risks due to unforeseeable side effects and toxicity. In addition, selective inhibitors are in need for targeting tumors where a single kinase is aberrantly regulated.88,89
Currently, a few ATP-competitive CDK4/6 dual inhibitors, i.e., Palbociclib (PD0332991), Ribociclib (LEE011) and Abemaciclib (LY2835219) are undergoing clinical trials. Mechanistically, these drug candidates inhibit CDK4/6, halt Rb phosphorylation and arrest G1 cell cycle progression of cancer cells.51,90-93 PD0332991 has also demonstrated to cause almost identical senescence phenotypes as the endogenous CDK inhibitors p16INK4a and p21Cip1 do.94
PD0332991 is a pyridopyrimidine derivative (Table 2) developed by Pfizer with relative high level of selectivity toward CDK4 and CDK6. It has been shown to inhibit various types of Rb proficient human cancer cell lines such as luminal estrogenic receptor positive subtypes of breast cancer, ovarian cancer, renal cancer, glioblastoma multiforme, maliginant rhabdoid tumor and colorectal cancer. This cellular effectiveness has also been reflected in various tumor xenograft models.95-102
Table 2.
Selective CDK4/6 inhibitors in clinical development
| Compound | Structure | CDK inhibition (IC50, nM) | Stage of Development* |
|---|---|---|---|
| PD0332991 (Pfizer)91 | 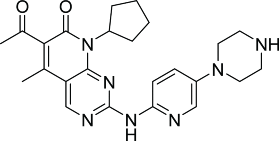 |
CDK1/B > 10,000 CDK2/A > 10,000 CDK2/E2 > 10,000 CDK4/D1 = 11 CDK4/D3 = 9 CDK5/P25 > 10,000 CDK6/D2 = 15 |
|
| LEE011 (Novartis) 90 | 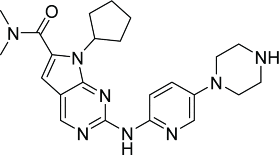 |
CDK1/B > 10,000 CDK2/A > 10,000 CDK4/D1 = 10 CDK5/P25 > 10,000 CDK6/D3 = 39 CDK9/T1 = 1,500 |
|
| LY2835219 (Eli Lilly)92 | 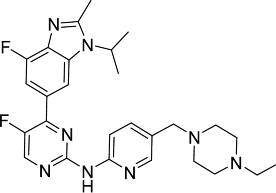 |
CDK1/B = 1,627 CDK2/E = 504 CDK4/D1 = 2 CDK6/D1 = 10 CDK7/H = 3,910 CDK9/T1 = 57 |
|
data from http://clinicaltrials.gov, accessed on 1st April 2015.
Currently, in its phase III clinical trial, PD0332991 is being tested in patients with squamous cell lung cancer and has been granted accelerated approval for use in combination with letrozole for the treatment of hormone receptor positive/human epidermal growth factor receptor 2 negative (HR+/HER2−) breast cancer in postmenopausal women for their metastatic disease (http://www.fda.gov/). Despite the exciting therapeutic outcomes obtained from this combination therapy and its advantage in combating drug resistance, there should be a certain degree of caution when considering combination regimens. Preclinical studies have shown that the resultant G1 arrest by CDK4/6 inhibitors may antagonize cytotoxic therapies that kill cancer cells in S-phase or mitotic phase. Franco et al. have demonstrated that while phosphoinositide 3-kinase/mammalian target of rapamycin (PI3K/mTOR) or mitogen-activated protein kinase kinase (MEK) inhibitors potently cooperated with CDK4/6 inhibition by PD0332991 in pancreatic ductal adenocarcinoma models, PD0332991 antagonized the cytotoxic effects of polo-like kinase 1 (PLK1) inhibitors and the antimetabolite gemcitabine. In addition, CDK4/6 inhibition has been shown to protect triple-negative breast cancer cells from doxorubicin mediated cytotoxicity.103,104
LEE011, a pyrolopyrimidine with structural features similar to PD0332991 (Table 2), was shown to be active against liposarcoma and neuroblastoma in vitro and in xenograft animal models. Twelve out of 17 human neuroblastoma cell lines were highly sensitive to LEE011 with IC50 values ranging between 126–801 nM. LEE011 is in phase III clinical trials against postmenopausal advanced breast cancer. It is also in phase I and II clinical studies as standalone or in combination with other agents against various cancers.105,106
A benzoimidazolpyrimidine derivative LY2835219 (Table 2) seems the most potent CDK4 and CDK 6 inhibitor with an IC50 value of 2 nM and 10 nM, respectively. However, it also inhibits CDK2 and CDK9 potently. The compound was shown to be efficacious against pre-clinical models of colon cancer, glioblastoma, acute myeloid leukemia, mantle cell lymphoma and lung cancer. It is currently in clinical trials for the treatment of stage IV non-small cell lung cancer and HR+/HER2− breast cancer.92,107,108
In addition to the above clinical experimental drug compounds, a few pre-clinical CDK4/6 inhibitors, i.e., 7X, AMG925, Compound 6, Compound A and PD0183812, have been reported.109-113 The chemical structures and CDK inhibitory activity of these compounds are summarized in Table 3. A close examination of the chemical structures of PD0332991, LEE011, LY2835219, AMG925 and Compound A reveals a general N-NH-N sequence of the pyrimidine-amine-pyridine or pyrazine-amine-thiazole system. This sequence is likely to play an important role in achieving CDK4/6 inhibition. Notably, substituents on the heterocyclic rings significantly differ among the 5 molcules, suggesting room for further modification to optimise potency and selectivity. Potency has been shown to be tuned by the interactions with the residues at the gate of the ATP binding pocket.114
Table 3.
Selective CDK4/6 inhibitors in pre-clinical development
| Compound | Structure | CDK inhibition (IC50, nM) |
|---|---|---|
| 7X110 | 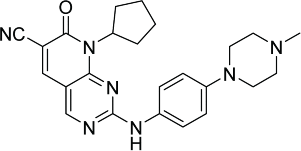 |
CDK1/A = 7,413 CDK1/B = 1,359 CDK2/A = 159 CDK2/E = 1,353 CDK3/E = 1,776 CDK4/D1 = 4 CDK4/D3 = 34 CDK5/P25 = 259 CDK5/P35 = 279 CDK6/D1 = 10 CDK6/D3 = 35 CDK7/H > 10,000 CDK9/K = 25 CDK9/T1 = 191 |
| Compound A120 | 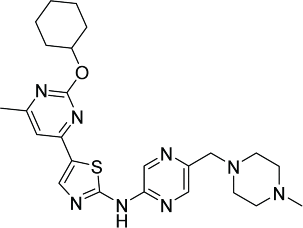 |
CDK1/B = 600 CDK2/A = 1,700 CDK4/D2 = 9.2 CDK5/P35 = 3,000 CDK6/D2 = 7.8 CDK7/H = 530 CDK9/T1 = 2,500 |
| AMG925109 |  |
CDK1/B = 1,900 CDK2/A = 375 CDK4/D1 = 3 CDK6/D1 = 8 |
| PD0183812113 |  |
CDK2/A = 210 CDK2/E = 165 CDK4/D1 = 8 CDK6/D2 = 7 CDK6/D3 = 13 |
| Compound 6111 |  |
CDK1/B > 10,000 CDK2/A > 10,000 CDK4/D1 = 15 CDK6/D3 = 120 |
Inhibitor design strategies
The majority of small-molecule kinase inhibitors developed so far target the ATP binding site (i.e., acting as ATP competitors), although alternative approaches for inhibitor design in targeting sites other than the ATP cleft are being proposed.115 Chemical scaffolds of the ATP competitive inhibitors usually consist of a planar heterocyclic system that acts as an ATP adenine mimetic. Due to highly conserved structure of the ATP binding domain of most kinases, these inhibitors may suffer from cross-reactivity with other kinases, resulting in poor safety and sometimes severe side effects.86,116 Nevertheless, many ATP competitive inhibitors, including PD0332991, have achieved high specificity and are successfully developed as therapeutics.
In PD0183812 (Table 3) the piperidine nitrogen was proposed to be protonated at the physiological pH. As a consequence, the positively charged nitrogen results in an unfavourable electrostatic repulsion with Lys89 of CDK1/2. On the other hand, due to the replacement of Lys89 by the less sterically demanding and less basic Thr102 and Thr107 in CDK4 and CDK6, respectively, the compound adopts a more elongated conformation which allows the favorable ion-pair interaction of the positively charged piperidinyl nitrogen with Glu144.116 In CDK7 and CDK9 the position of Lys89 is occupied by Val100 and Gly112, respectively, and hence specificity for these kinases might be achieved with bulky and hydrophobic substituents.85,86
Similarly, by virtue of its C5-methyl and C6-acetyl groups, PD0332991 (Fig. 3a) has been reported to have several steric clashes with the gatekeeper residue Phe80 of CDK2. The C6-acetyl group in PD0332991 forms hydrogen bond with the backbone NH of Asp163 of the DFG motif in CDK6.110 Investigation on the crystal structure of CDK6-Vcyclin in complex with PD0332991 has revealed that selectivity for CDK6 may be achieved by targeting the relatively less conserved hinge region and the pocket near the Phe98 gate keeper in the back of the ATP catalytic site. For instance, this is possible by aromatic sp2 nitrogen near the rarely conserved His100 (Phe82 in CDK2). It was also suggested that engaging in an interaction with the Phe98 gatekeeper enhances selectivity. For example, pyridine in AMG 925 has been shown to form an edge to face aromatic-aromatic contact with the gatekeeper residue Phe98.109,111 Docking and scoring of 7X (Table 3), which is highly structurally similar to PD0332991, have identified a binding orientation different from that of PD0332991. This has been attributed to the rigidity and higher electron withdrawing effect of the cyano group when compared with the acetyl group. The nitrogen of the cyano group is believed to interact with Lys43 and Ala23 of CDK6.110
Figure 3.
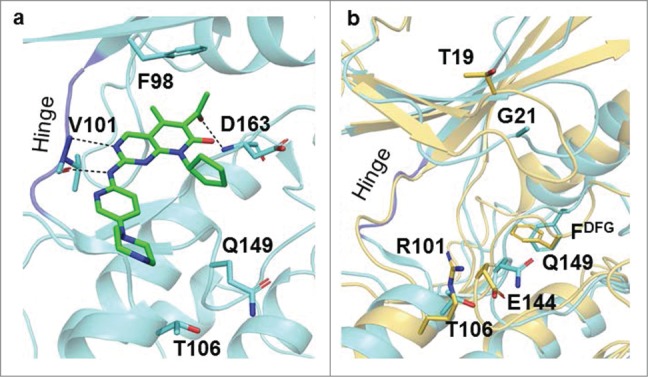
(a) PD0332991 is shown in green and bound in the ATP binding pocket of CDK6. It forms hydrogen bonds (shown in black dashed lines) with the conserved amino acids Val101 and Asp163. (PDB ID: 2EUF). (b) The ATP binding site of CDK6 (PDB ID: 2EUF) aligned with CDK4 (PDB ID: 2W96). CDK6 is shown in turquoise and CDK4 is in yellow. The differences within the ATP binding site between CDK6 and CDK4 are labeled in black. The figure was prepared using PyMOL1.3 (Schrödinger Inc., 2013).
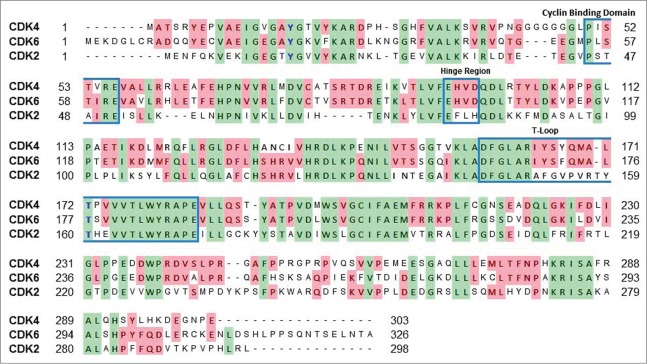
Even though, the above findings point out some of the structural requirements for the design and synthesis of potent and selective potential CDK6 inhibitors, achieving selectivity for CDK6 over CDK4 is highly challenging. Firstly, the 2 kinases are highly structurally similar. CDK6 shares 70% amino acid sequence identity with CDK4, while only 40% with CDK2 (Fig. 4). Secondly, due to the lack of a crystal structure of CDK4 bound to a ligand, structural elements contributing to CDK4 selectivity and potency remain to be fully understood. The available crystal structures of monomeric CDK4 are engineered and hence there is a possibility that the wild-type CDK4 might be different. In fact, the aforementioned molecules are relatively more selective toward CDK4 than CDK6. Hence, the interactions of these molecules with CDK6, as described above, might also occur with CDK4. In order to get better understanding of the differences in inhibitor binding (selectivity) between CDK4 and CDK6, it is crucial to determine the structures of CDK4-ligand complexes.
Figure 4.
Sequence comparison of CDK2, CDK4 and CDK6. Green and pink indicate residues conserved in all the 3 CDKs and 2 CDKs, respectively. Cyclin binding domain, hinge region and the T-loop are boxed in a blue frame. Amino acids for inhibitory phosphorylation by Wee1 and Myt1 are shown in blue. In the T-loop, the threonine labeled in blue is essential for phosphorylation by CAK for activation. The sequence alignment was generated using UniProt (http://www.uniprot.org/align/).
The overlay of the ATP binding site of CDK4 on that of CDK6 (Fig. 3b) reveals the presence of structural features in the binding pocket which are not involved with ATP binding. Exploiting these structural features would offer a strategy for the development of ATP competitive and selective CDK6 inhibitors. For instance, on the edge of the ATP binding site, Arg101 and Glu144 of CDK4 are replaced by Thr106 and Gln149 in CDK6. Therefore, this area in CDK6 provides a more spacious pocket with non-charged amino acid side chains. Furthermore, the crystal structures indicate that Lys106 in CDK4 is ∼0.9 Å further from the Val101 of the hinge – which participates in crucial hinge-ligand interactions – as measured from respective Cα atoms. This is presumably due to a few sequence changes surrounding Lys106. These structural features can be exploited by bulkier groups with different properties. For example, placing a carboxylate group on the piperazine moiety of PD0332991 or LEE011 might improve selectivity for CDK6 over CDK4. Moreover, the dynamic behavior of CDK4 and CDK6 remains barely investigated, experimentally or computationally. Their dynamics may illustrate further conformational differences that could be exploited for improving selectivity.
Though inhibition of the catalytic activity of the CDKs with small molecules that compete with ATP has proved to be the most successful strategy to date, discovery of non-ATP competitive compounds might provide a means to overcome the selectivity issue with fewer off-target side effects.
Conclusions
Although both CDK4 and CDK6 regulate the G1 to S phase of the cell cycle, CDK6 has unique functions that are cell-type specific and developmentally distinct. There are compelling reasons to develop mono-specific CDK6 inhibitors which will not only be effective against several types of cancers, including MLL-AML, MLL-ALL, glioblastomas and medulloblastoma where CDK6 is a major oncogenic factor, but also have a less risk of off-targeting toxicity. To further validate CDK6 as a pharmacological target in a diversity of cancer types and to develop effective and safe anti-cancer agents, CDK6 inhibitors with high selectivity have to be discovered.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Malumbres M. Cyclin-dependent kinases. Genome Biol 2014; 15:122; PMID:25180339; http://dx.doi.org/ 10.1186/gb4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao L, Chen F, Yang X, Xu W, Xie J, Yu L. Phylogenetic analysis of CDK and cyclin proteins in premetazoan lineages. BMC Evol Biol 2014; 14:10; PMID:24433236; http://dx.doi.org/ 10.1186/1471-2148-14-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci 2005; 30:630-41; PMID:16236519; http://dx.doi.org/ 10.1016/j.tibs.2005.09.005 [DOI] [PubMed] [Google Scholar]
- 4.Meyerson M, Harlow E. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell Biol 1994; 14:2077-86; PMID:8114739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 2009; 9:153-66; PMID:19238148; http://dx.doi.org/ 10.1038/nrc2602 [DOI] [PubMed] [Google Scholar]
- 6.Lundberg AS, Weinberg RA. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol 1998; 18:753-61; PMID:9447971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harbour JW, Luo RX, Santi AD, Postigo AA, Dean DC. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell 1999; 98:859-69; PMID:10499802; http://dx.doi.org/ 10.1016/S0092-8674(00)81519-6 [DOI] [PubMed] [Google Scholar]
- 8.Polager S, Ginsberg D. E2F–at the crossroads of life and death. Trends Cell Biol 2008; 18:528-35; PMID:18805009; http://dx.doi.org/ 10.1016/j.tcb.2008.08.003 [DOI] [PubMed] [Google Scholar]
- 9.Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene 2005; 24:2810-26; PMID:15838517; http://dx.doi.org/ 10.1038/sj.onc.1208612 [DOI] [PubMed] [Google Scholar]
- 10.Harbour JW, Dean DC. Rb function in cell-cycle regulation and apoptosis. Nat Cell Biol 2000; 2:E65-E7; PMID:10783254; http://dx.doi.org/ 10.1038/35008695 [DOI] [PubMed] [Google Scholar]
- 11.DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci U S A 1997; 94:7245-50; PMID:9207076; http://dx.doi.org/ 10.1073/pnas.94.14.7245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozar K, Sicinski P. Cell cycle progression without cyclin D-CDK4 and cyclin D-CDK6 complexes. Cell Cycle 2005; 4:388-91; PMID:15738651; http://dx.doi.org/ 10.4161/cc.4.3.1551 [DOI] [PubMed] [Google Scholar]
- 13.Fiaschi-Taesch NM, Salim F, Kleinberger J, Troxell R, Cozar-Castellano I, Selk K, Cherok E, Takane KK, Scott DK, Stewart AF. Induction of human β-cell proliferation and engraftment using a single G1/S regulatory molecule, cdk6. Diabetes 2010; 59:1926-36; PMID:20668294; http://dx.doi.org/ 10.2337/db09-1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malumbres M, Sotillo R, Santamaría D, Galán J, Cerezo A, Ortega S, Dubus P, Barbacid M. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell 2004; 118:493-504; PMID:15315761; http://dx.doi.org/ 10.1016/j.cell.2004.08.002 [DOI] [PubMed] [Google Scholar]
- 15.Nagasawa M, Gelfand EW, Lucas JJ. Accumulation of high levels of the p53 and p130 growth-suppressing proteins in cell lines stably over-expressing cyclin-dependent kinase 6 (cdk6). Oncogene 2001; 20:2889-99; PMID:11420701; http://dx.doi.org/ 10.1038/sj.onc.1204396 [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Sistrunk C, Rodriguez-Puebla ML. Unexpected reduction of skin tumorigenesis on expression of cyclin-dependent kinase 6 in mouse epidermis. Am J Pathol 2011; 178:345-54; PMID:21224071; http://dx.doi.org/ 10.1016/j.ajpath.2010.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anders L, Ke N, Hydbring P, Choi YJ, Widlund HR, Chick JM, Zhai H, Vidal M, Gygi SP, Braun P. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell 2011; 20:620-34; PMID:22094256; http://dx.doi.org/ 10.1016/j.ccr.2011.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi YJ, Anders L. Signaling through cyclin D-dependent kinases. Oncogene 2014; 33:1890-903; PMID:23644662; http://dx.doi.org/ 10.1038/onc.2013.137 [DOI] [PubMed] [Google Scholar]
- 19.Kohrt D, Crary J, Zimmer M, Patrick AN, Ford HL, Hinds PW, Grossel MJ. CDK6 binds and promotes the degradation of the EYA2 protein. Cell Cycle 2014; 13:62-71; PMID:24196439; http://dx.doi.org/ 10.4161/cc.26755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buss H, Handschick K, Jurrmann N, Pekkonen P, Beuerlein K, Müller H, Wait R, Saklatvala J, Ojala PM, Lienhard Schmitz M, et al.. Cyclin-dependent kinase 6 phosphorylates NF-κB P65 at serine 536 and contributes to the regulation of inflammatory gene expression. PloS One 2012; 7:1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Handschick K, Beuerlein K, Jurida L, Bartkuhn M, Müller H, Soelch J, Weber A, Dittrich-Breiholz O, Schneider H, Scharfe M, et al.. Cyclin-dependent kinase 6 is a chromatin-bound cofactor for NF-κB-dependent gene expression. Mol Cell 2014; 53:193-208; PMID:24389100; http://dx.doi.org/ 10.1016/j.molcel.2013.12.002 [DOI] [PubMed] [Google Scholar]
- 22.Ogasawara T, Kawaguchi H, Jinno S, Hoshi K, Itaka K, Takato T, Nakamura K, Okayama H. Bone morphogenetic protein 2-induced osteoblast differentiation requires Smad-mediated down-regulation of Cdk6. Mol Cell Biol 2004; 24:6560-8; PMID:15254224; http://dx.doi.org/ 10.1128/MCB.24.15.6560-6568.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogasawara T, Katagiri M, Yamamoto A, Hoshi K, Takato T, Nakamura K, Tanaka S, Okayama H, Kawaguchi H. Osteoclast differentiation by RANKL requires NF-kappaB-mediated downregulation of cyclin-dependent kinase 6 (Cdk6). J Bone Miner Res 2004; 19:1128-36; PMID:15176996; http://dx.doi.org/ 10.1359/jbmr.2004.19.7.1128 [DOI] [PubMed] [Google Scholar]
- 24.Ogasawara T, Mori Y, Abe M, Suenaga H, Kawase-Koga Y, Saijo H, Takato T. Role of cyclin-dependent kinase (Cdk) 6 in osteoblast, osteoclast, and chondrocyte differentiation and its potential as a target of bone regenerative medicine. Oral Sci Int 2011; 8:2-6; http://dx.doi.org/ 10.1016/S1348-8643(11)00007-3 [DOI] [Google Scholar]
- 25.Ericson KK, Krull D, Slomiany P, Grossel MJ. Expression of cyclin-dependent kinase 6, but not cyclin-dependent kinase 4, alters morphology of cultured mouse astrocytes. Mol Cancer Res 2003; 1:654-64; PMID:12861051 [PubMed] [Google Scholar]
- 26.Matushansky I, Radparvar F, Skoultchi AI. CDK6 blocks differentiation: coupling cell proliferation to the block to differentiation in leukemic cells. Oncogene 2003; 22:4143-9; PMID:12833137; http://dx.doi.org/ 10.1038/sj.onc.1206484 [DOI] [PubMed] [Google Scholar]
- 27.Fujimoto T, Anderson K, Jacobsen SEW, Nishikawa SI, Nerlov C. Cdk6 blocks myeloid differentiation by interfering with Runx1 DNA binding and Runx1-C/EBPα interaction. EMBO J 2007; 26:2361-70; PMID:17431401; http://dx.doi.org/ 10.1038/sj.emboj.7601675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beukelaers P, Vandenbosch R, Caron N, Nguyen L, Belachew S, Moonen G, Kiyokawa H, Barbacid M, Santamaria D, Malgrange B. Cdk6-dependent regulation of G1 length controls adult neurogenesis. Stem Cells 2011; 29:713-24; PMID:21319271; http://dx.doi.org/ 10.1002/stem.616 [DOI] [PubMed] [Google Scholar]
- 29.Bryan C, Blanton R, Aronovitz M, Karas R, Hu M, Hinds PW. The role of cyclin-dependent kinase 6 in cardiac development and hypertrophy. FASEB J 2013; 27:lb35 [Google Scholar]
- 30.Hu MG, Deshpande A, Enos M, Mao D, Hinds EA, Hu G-F, Chang R, Guo Z, Dose M, Mao C. A requirement for cyclin-dependent kinase 6 in thymocyte development and tumorigenesis. Cancer Res 2009; 69:810-8; PMID:19155308; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grossel MJ, Hinds PW. Beyond the cell cycle: a new role for cdk6 in differentiation. J Cell Biochem 2006; 97:485-93; PMID:16294322; http://dx.doi.org/ 10.1002/jcb.20712 [DOI] [PubMed] [Google Scholar]
- 32.Kohrt DM, Crary JI, Gocheva V, Hinds PW, Grossel MJ. Distinct subcellular distribution of cyclin dependent kinase 6. Cell Cycle 2009; 8:2837-43; PMID:19667758; http://dx.doi.org/ 10.4161/cc.8.17.9521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fåhraeus R, Lane DP. The p16INK4a tumour suppressor protein inhibits αvβ3 integrin-mediated cell spreading on vitronectin by blocking PKC-dependent localization of αvβ3 to focal contacts. EMBO J 1999; 18:2106-18; http://dx.doi.org/ 10.1093/emboj/18.8.2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta 2002; 1602:73-87; PMID:11960696 [DOI] [PubMed] [Google Scholar]
- 35.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer 2011; 11:558-72; PMID:21734724; http://dx.doi.org/ 10.1038/nrc3090 [DOI] [PubMed] [Google Scholar]
- 36.Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: cyclin D1: normal and abnormal functions. Endocrinology 2004; 145:5439-47; PMID:15331580; http://dx.doi.org/ 10.1210/en.2004-0959 [DOI] [PubMed] [Google Scholar]
- 39.Schwartz R, Engel I, Fallahi-Sichani M, Petrie HT, Murre C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc Natl Acad Sci U S A 2006; 103:9976-81; PMID:16782810; http://dx.doi.org/ 10.1073/pnas.0603728103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yagi T, Morimoto A, Eguchi M, Hibi S, Sako M, Ishii E, Mizutani S, Imashuku S, Ohki M, Ichikawa H. Identification of a gene expression signature associated with pediatric AML prognosis. Blood 2003; 102:1849-56; PMID:12738660; http://dx.doi.org/ 10.1182/blood-2003-02-0578 [DOI] [PubMed] [Google Scholar]
- 41.Bax DA, Mackay A, Little SE, Carvalho D, Viana-Pereira M, Tamber N, Grigoriadis AE, Ashworth A, Reis RM, Ellison DW. A distinct spectrum of copy number aberrations in pediatric high-grade gliomas. Clin Cancer Res 2010; 16:3368-77; PMID:20570930; http://dx.doi.org/ 10.1158/1078-0432.CCR-10-0438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Timmermann S, Hinds PW, Münger K. Elevated activity of cyclin-dependent kinase 6 in human squamous cell carcinoma lines. Cell Growth Differ 1997; 8:361-70; PMID:9101082 [PubMed] [Google Scholar]
- 43.Lee K-H, Lotterman C, Karikari C, Omura N, Feldmann G, Habbe N, Goggins MG, Mendell JT, Maitra A. Epigenetic silencing of microRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology 2009; 9:293-301; PMID:19407485; http://dx.doi.org/ 10.1159/000186051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G, Zheng L, Yu Z, Liao G, Lu L, Xu R, Zhao Z, Chen G. Increased cyclin-dependent kinase 6 expression in bladder cancer. Oncol Lett 2012; 4:43-6; PMID:22807957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pardis S, Tadbir AA, Ashkavandi ZJ, Najvani AD, Ashraf MJ, Ranjbaran H. Expression of CDK6 in salivary gland tumors. J Med Sci 2012; 12:193-7; PMID:23244127; http://dx.doi.org/ 10.3923/jms.2012.193.19723244127 [DOI] [Google Scholar]
- 37.Deshpande A, Sicinski P, Hinds PW. Cyclins and cdks in development and cancer: a perspective. Oncogene 2005; 24:2909-15; PMID:15838524; http://dx.doi.org/ 10.1038/sj.onc.1208618 [DOI] [PubMed] [Google Scholar]
- 38.Nagel S, Leich E, Quentmeier H, Meyer C, Kaufmann M, Drexler H, Zettl A, Rosenwald A, MacLeod R. Amplification at 7q22 targets cyclin-dependent kinase 6 in T-cell lymphoma. Leukemia 2007; 22:387-92; PMID:17989712; http://dx.doi.org/ 10.1038/sj.leu.2405028 [DOI] [PubMed] [Google Scholar]
- 46.Lim JT, Mansukhani M, Weinstein IB. Cyclin-dependent kinase 6 associates with the androgen receptor and enhances its transcriptional activity in prostate cancer cells. Proc Natl Acad Sci U S A 2005; 102:5156-61; PMID:15790678; http://dx.doi.org/ 10.1073/pnas.0501203102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whiteway SL, Harris PS, Venkataraman S, Alimova I, Birks DK, Donson AM, Foreman NK, Vibhakar R. Inhibition of cyclin-dependent kinase 6 suppresses cell proliferation and enhances radiation sensitivity in medulloblastoma cells. J Neurooncol 2013; 111:113-21; PMID:23138228; http://dx.doi.org/ 10.1007/s11060-012-1000-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mendrzyk F, Radlwimmer B, Joos S, Kokocinski F, Benner A, Stange DE, Neben K, Fiegler H, Carter NP, Reifenberger G. Genomic and protein expression profiling identifies CDK6 as novel independent prognostic marker in medulloblastoma. J Clin Oncol 2005; 23:8853-62; PMID:16314645; http://dx.doi.org/ 10.1200/JCO.2005.02.8589 [DOI] [PubMed] [Google Scholar]
- 49.Costello JF, Plass C, Arap W, Chapman VM, Held WA, Berger MS, Su Huang HJ, Cavenee WK. Cyclin-dependent kinase 6 (CDK6) amplification in human gliomas identified using two-dimensional separation of genomic DNA. Cancer Res 1997; 57:1250-4; PMID:9102208 [PubMed] [Google Scholar]
- 50.Chilosi M, Doglioni C, Yan Z, Lestani M, Menestrina F, Sorio C, Benedetti A, Vinante F, Pizzolo G, Inghirami G. Differential expression of cyclin-dependent kinase 6 in cortical thymocytes and T-cell lymphoblastic lymphoma/leukemia. Am J Pathol 1998; 152:209-17; PMID:9422538 [PMC free article] [PubMed] [Google Scholar]
- 51.Anderlind C, Spira A, Cardoso WV, Lü J. miR-129 regulates cell proliferation by downregulating Cdk6 expression. Cell Cycle 2010; 9:1809-18; PMID:20404570; http://dx.doi.org/ 10.4161/cc.9.9.11535 [DOI] [PubMed] [Google Scholar]
- 52.Wang X, Wu J, Lin Y, Zhu Y, Xu X, Xu X, Liang Z, Li S, Hu Z, Zheng X. MicroRNA-320c inhibits tumorous behaviors of bladder cancer by targeting cyclin-dependent kinase 6. J Exp Clin Cancer Res 2014; 33:1-12; PMID:24383517; http://dx.doi.org/ 10.1186/1756-9966-33-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu X, Li Y, Shen H, Li H, Long L, Hui L, Xu W. miR-137 inhibits the proliferation of lung cancer cells by targeting Cdc42 and Cdk6. FEBS Lett 2013; 587:73-81; PMID:23178712; http://dx.doi.org/ 10.1016/j.febslet.2012.11.004 [DOI] [PubMed] [Google Scholar]
- 54.Pierson J, Hostager B, Fan R, Vibhakar R. Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J Neurooncol 2008; 90:1-7; PMID:18607543; http://dx.doi.org/ 10.1007/s11060-008-9624-3 [DOI] [PubMed] [Google Scholar]
- 55.Xiao F, Zhang W, Zhou L, Xie H, Xing C, Ding S, Chen K, Zheng S. microRNA-200a is an independent prognostic factor of hepatocellular carcinoma and induces cell cycle arrest by targeting CDK6. Oncol Rep 2013; 30:2203-10; PMID:24009066 [DOI] [PubMed] [Google Scholar]
- 56.Feng L, Xie Y, Zhang H, Wu Y. miR-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cells. Med Oncol 2012; 29:856-63; PMID:21264532; http://dx.doi.org/ 10.1007/s12032-011-9823-1 [DOI] [PubMed] [Google Scholar]
- 57.Honeywell DR, Cabrita MA, Zhao H, Dimitroulakos J, Addison CL. miR-105 inhibits prostate tumour growth by suppressing CDK6 levels. PloS One 2013; 8:1-12; PMID:23950948; http://dx.doi.org/ 10.1371/journal.pone.0070515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen L, Zhang R, Li P, Liu Y, Qin K, Fa ZQ, Liu YJ, Ke YQ, Jiang XD. P53-induced microRNA-107 inhibits proliferation of glioma cells and down-regulates the expression of CDK6 and Notch-2. Neurosci Lett 2013; 534:327-32; PMID:23220650; http://dx.doi.org/ 10.1016/j.neulet.2012.11.047 [DOI] [PubMed] [Google Scholar]
- 59.Lowenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med 1999; 341:1051-62; PMID:10502596; http://dx.doi.org/ 10.1056/NEJM199909303411407 [DOI] [PubMed] [Google Scholar]
- 60.Ohlsson E, Hasemann MS, Willer A, Lauridsen FKB, Rapin N, Jendholm J, Porse BT. Initiation of MLL-rearranged AML is dependent on C/EBPα. J Exp Med 2014; 211:5-13; PMID:24367003; http://dx.doi.org/ 10.1084/jem.20130932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Placke T, Faber K, Nonami A, Putwain SL, Salih HR, Heidel FH, Krämer A, Root DE, Barbie DA, Krivtsov AV. Requirement for CDK6 in MLL-rearranged acute myeloid leukemia. Blood 2014; 124:13-23; PMID:24764564; http://dx.doi.org/ 10.1182/blood-2014-02-558114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van der Linden MH, Willekes M, van Roon E, Seslija L, Schneider P, Pieters R, Stam RW. MLL fusion-driven activation of CDK6 potentiates proliferation in MLL-rearranged infant ALL. Cell Cycle 2014; 13:834-44; PMID:24736461; http://dx.doi.org/ 10.4161/cc.27757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kollmann K, Heller G, Schneckenleithner C, Warsch W, Scheicher R, Ott RG, Schäfer M, Fajmann S, Schlederer M, Schiefer A-I, et al.. A kinase-independent function of CDK6 links the cell cycle to tumor angiogenesis. Cancer Cell 2013; 24:167-81; PMID:23948297; http://dx.doi.org/ 10.1016/j.ccr.2013.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kollmann K, Sexl V. CDK6 and p16INK4A in lymphoid malignancies. Oncotarget 2013; 4:1858-9; PMID:24161991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choi YJ, Li X, Hydbring P, Sanda T, Stefano J, Christie AL, Signoretti S, Look AT, Kung AL, von Boehmer H. The requirement for cyclin D function in tumor maintenance. Cancer Cell 2012; 22:438-51; PMID:23079655; http://dx.doi.org/ 10.1016/j.ccr.2012.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sawai CM, Freund J, Oh P, Ndiaye-Lobry D, Bretz JC, Strikoudis A, Genesca L, Trimarchi T, Kelliher MA, Clark M, et al.. Therapeutic targeting of the cyclin D3:CDK4/6 complex in T cell leukemia. Cancer Cell 2012; 22:452-65; PMID:23079656; http://dx.doi.org/ 10.1016/j.ccr.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choi YJ, Sicinski P. Unexpected outcomes of CDK4/6 inhibition. Oncotarget 2013; 4:176; PMID:23563596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brito-Babapulle V, Gruszka-Westwood AM, Platt G, Andersen CL, Elnenaei MO, Matutes E, Wotherspoon AC, Weston-Smith SG, Catovsky D. Translocation t(2;7)(p12; q21-22) with dysregulation of the CDK6 gene mapping to 7q21-22 in a non-Hodgkin's lymphoma with leukemia. Haematologica 2002; 87:357-62; PMID:11940479 [PubMed] [Google Scholar]
- 69.Parker EP, Siebert R, Oo TH, Schneider D, Hayette S, Wang C. Sequencing of t(2;7) translocations reveals a consistent breakpoint linking CDK6 to the IGK locus in indolent B-cell neoplasia. J Mol Diagn 2013; 15:101-9; PMID:23127611; http://dx.doi.org/ 10.1016/j.jmoldx.2012.07.006 [DOI] [PubMed] [Google Scholar]
- 70.Li C, Qi L, Bellail AC, Hao C, Liu T. PD-0332991 induces G1 arrest of colorectal carcinoma cells through inhibition of the cyclin-dependent kinase-6 and retinoblastoma protein axis. Oncol Lett 2014; 7:1673-8; PMID:24765199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho Y-J, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S, Gajjar A. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol 2012; 123:465-72; PMID:22134537; http://dx.doi.org/ 10.1007/s00401-011-0922-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu J, Qian J, Li C, Kwok L, Cheng F, Liu P, Perdomo C, Kotton D, Vaziri C, Anderlind C, et al.. miR-129 regulates cell proliferation by downregulating Cdk6 expression. Cell Cycle 2010; 9:1809-18; PMID:20404570; http://dx.doi.org/ 10.4161/cc.9.9.11535 [DOI] [PubMed] [Google Scholar]
- 73.Sato K, Schäuble B, Kleihues P, Ohgaki H. Infrequent alterations of the p15, p16, CDK4 and Cyclin D1 genes in non-astrocytic human brain tumors. Int J Cancer 1996; 66:305-8; PMID:8621248; http://dx.doi.org/ 10.1002/(SICI)1097-0215(19960503)66:3%3c305::AID-IJC6%3e3.0.CO;2-2 [DOI] [PubMed] [Google Scholar]
- 74.Brotherton DH, Dhanaraj V, Wick S, Brizuela L, Domaille PJ, Volyanik E, Xu X, Parisini E, Smith BO, Archer SJ, et al.. Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19INK4d. Nature 1998; 395:244-50; PMID:9751051; http://dx.doi.org/ 10.1038/26164 [DOI] [PubMed] [Google Scholar]
- 75.Pavletich NP. Mechanisms of cyclin-dependent kinase regulation: structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J Mol Biol 1999; 287:821-8; PMID:10222191; http://dx.doi.org/ 10.1006/jmbi.1999.2640 [DOI] [PubMed] [Google Scholar]
- 76.Schulze-Gahmen U, Sung-Hou K. Structural basis for CDK6 activation by a virus-encoded cyclin. Nat Struct Biol 2002; 9:177-81; PMID:11828325 [DOI] [PubMed] [Google Scholar]
- 77.Day PJ, Cleasby A, Tickle IJ, O'Reilly M, Coyle JE, Holding FP, McMenamin RL, Yon J, Chopra R, Lengauer C. Crystal structure of human CDK4 in complex with a D-type cyclin. Proc Natl Acad Sci U S A 2009; 106:4166-70; PMID:19237565; http://dx.doi.org/ 10.1073/pnas.0809645106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takaki T, Echalier A, Brown N, Hunt T, Endicott J, Noble M. The structure of CDK4/cyclin D3 has implications for models of CDK activation. Proc Natl Acad Sci U S A 2009; 106:4171-6; PMID:19237555; http://dx.doi.org/ 10.1073/pnas.0809674106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 1999; 13:1501-12; PMID:10385618; http://dx.doi.org/ 10.1101/gad.13.12.1501 [DOI] [PubMed] [Google Scholar]
- 80.Hannon GJ, Beach D. pl5INK4B is a potentia| effector of TGF-β-induced cell cycle arrest. Nature 1994; 371:257-61; PMID:8078588; http://dx.doi.org/ 10.1038/371257a0 [DOI] [PubMed] [Google Scholar]
- 81.Hirai H, Roussel MF, Kato JY, Ashmun RA, Sherr CJ. Novel INK4 proteins, p19 and p18, are specific inhibitors of the cyclin D-dependent kinases CDK4 and CDK6. Mol Cell Biol 1995; 15:2672-81; PMID:7739547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guan KL, Jenkins CW, Li Y, O'Keefe CL, Noh S, Wu X, Zariwala M, Matera AG, Xiong Y. Isolation and characterization of p19INK4d, a p16-related inhibitor specific to CDK6 and CDK4. Mol Biol Cell 1996; 7:57-70; PMID:8741839; http://dx.doi.org/ 10.1091/mbc.7.1.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Russo AA, Tong L, Lee J-O, Jeffrey PD, Pavletich NP. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature 1998; 395:237-43; PMID:9751050; http://dx.doi.org/ 10.1038/26155 [DOI] [PubMed] [Google Scholar]
- 84.Noh SJ, Li Y, Xiong Y, Guan KL. Identification of functional elements of p18INK4C essential for binding and inhibition of cyclin-dependent kinase (CDK) 4 and CDK6. Cancer Res 1999; 59:558-64; PMID:9973200 [PubMed] [Google Scholar]
- 85.Lolli G, Johnson LN. CAK-cyclin-dependent activating kinase: a key kinase in cell cycle control and a target for drugs? Cell Cycle 2005; 4:565-70; PMID:15876871; http://dx.doi.org/ 10.4161/cc.4.4.1607 [DOI] [PubMed] [Google Scholar]
- 86.Lolli G. Structural dissection of cyclin dependent kinases regulation and protein recognition properties. Cell Cycle 2010; 9:1551-61; PMID:20372077; http://dx.doi.org/ 10.4161/cc.9.8.11195 [DOI] [PubMed] [Google Scholar]
- 87.Cheng M, Olivier P, Diehl JA, Fero M, Roussel MF, Roberts JM, Sherr CJ. The p21Cip1 and p27Kip1 CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J 1999; 18:1571-83; PMID:10075928; http://dx.doi.org/ 10.1093/emboj/18.6.1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wesierska-Gadek J, Maurer M, Zulehner N, Komina O. Whether to target single or multiple CDKs for therapy? That is the question. J Cell Physiol 2011; 226:341-9; PMID:20836132; http://dx.doi.org/ 10.1002/jcp.22426 [DOI] [PubMed] [Google Scholar]
- 89.Shapiro GI. Cyclin-dependent kinase pathways as targets for cancer treatment. J Clin Oncol 2006; 24:1770-83; PMID:16603719; http://dx.doi.org/ 10.1200/JCO.2005.03.7689 [DOI] [PubMed] [Google Scholar]
- 90.Kim S, Loo A, Chopra R, Caponigro G, Huang A, Vora S, Parasuraman S, Howard S, Keen N, Sellers W. LEE011: an orally bioavailable, selective small molecule inhibitor of CDK4/6–reactivating Rb in cancer. Mol Cancer Ther 2013; 12:PR02-PR; http://dx.doi.org/ 10.1158/1535-7163.MCT-12-1188 [DOI] [Google Scholar]
- 91.Toogood PL, Harvey PJ, Repine JT, Sheehan DJ, VanderWel SN, Zhou H, Keller PR, McNamara DJ, Sherry D, Zhu T. Discovery of a potent and selective inhibitor of cyclin-dependent kinase 4/6. J Med Chem 2005; 48:2388-406; PMID:15801831; http://dx.doi.org/ 10.1021/jm049354h [DOI] [PubMed] [Google Scholar]
- 92.Gelbert LM, Cai S, Lin X, Sanchez-Martinez C, Del Prado M, Lallena MJ, Torres R, Ajamie RT, Wishart GN, Flack RS, et al.. Preclinical characterization of the CDK4/6 inhibitor LY2835219: in-vivo cell cycle-dependent/independent anti-tumor activities alone/in combination with gemcitabine. Invest New Drugs 2014; 32:825-37; PMID:24919854; http://dx.doi.org/ 10.1007/s10637-014-0120-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tate SC, Cai S, Ajamie RT, Burke T, Beckmann RP, Chan EM, De Dios A, Wishart GN, Gelbert LM, Cronier DM. Semi-mechanistic pharmacokinetic/pharmacodynamic modeling of the antitumor activity of LY2835219, a new cyclin-dependent kinase 4/6 inhibitor, in mice bearing human tumor xenografts. Clin Cancer Res 2014; 20:3763-74; PMID:24850847; http://dx.doi.org/ 10.1158/1078-0432.CCR-13-2846 [DOI] [PubMed] [Google Scholar]
- 94.Leontieva OV, Blagosklonny MV. CDK4/6-inhibiting drug substitutes for p21 and p16 in senescence: duration of cell cycle arrest and MTOR activity determine geroconversion. Cell Cycle 2013; 12:3063-9; PMID:23974099; http://dx.doi.org/ 10.4161/cc.26130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cen L, Carlson BL, Schroeder MA, Ostrem JL, Kitange GJ, Mladek AC, Fink SR, Decker PA, Wu W, Kim J-S. p16-Cdk4-Rb axis controls sensitivity to a cyclin-dependent kinase inhibitor PD0332991 in glioblastoma xenograft cells. Neuro Oncol 2012; 14:870-81; PMID:22711607; http://dx.doi.org/ 10.1093/neuonc/nos114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet E, Albassam M, Zheng X, Leopold WR, Pryer NK, et al.. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther 2004; 3:1427-38; PMID:15542782 [PubMed] [Google Scholar]
- 97.Li C, Qi L, Bellail AC, Hao C, Liu T. PD-0332991 induces G1 arrest of colorectal carcinoma cells through inhibition of the cyclin-dependent kinase-6 and retinoblastoma protein axis. Oncol Lett 2014; 7:1673-8; PMID:24765199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Katsumi Y, Iehara T, Miyachi M, Yagyu S, Tsubai-Shimizu S, Kikuchi K, Tamura S, Kuwahara Y, Tsuchiya K, Kuroda H. Sensitivity of malignant rhabdoid tumor cell lines to PD 0332991 is inversely correlated with p16 expression. Biochem Biophys Res Commun 2011; 413:62-8; PMID:21871868; http://dx.doi.org/ 10.1016/j.bbrc.2011.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Michaud K, Solomon DA, Oermann E, Kim J-S, Zhong W-Z, Prados MD, Ozawa T, James CD, Waldman T. Pharmacologic inhibition of cyclin-dependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res 2010; 70:3228-38; PMID:20354191; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Logan JE, Mostofizadeh N, Desai AJ, VON Euw E, Conklin D, Konkankit V, Hamidi H, Eckardt M, Anderson L, Chen HW, et al.. PD-0332991, a potent and selective inhibitor of cyclin-dependent kinase 4/6, demonstrates inhibition of proliferation in renal cell carcinoma at nanomolar concentrations and molecular markers predict for sensitivity. Anticancer Res 2013; 33:2997-3004; PMID:23898052 [PubMed] [Google Scholar]
- 101.Konecny GE, Winterhoff B, Kolarova T, Qi J, Manivong K, Dering J, Yang G, Chalukya M, Wang H-J, Anderson L. Expression of p16 and retinoblastoma determines response to CDK4/6 inhibition in ovarian cancer. Clin Cancer Res 2011; 17:1591-602; PMID:21278246; http://dx.doi.org/ 10.1158/1078-0432.CCR-10-2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res 2009; 11:R77; PMID:19874578; http://dx.doi.org/ 10.1186/bcr2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Franco J, Witkiewicz AK, Knudsen ES. CDK4/6 inhibitors have potent activity in combination with pathway selective therapeutic agents in models of pancreatic cancer. Oncotarget 2014; 5:6512; PMID:25156567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McClendon AK, Dean JL, Rivadeneira DB, Yu JE, Reed CA, Gao E, Farber JL, Force T, Koch WJ, Knudsen ES. CDK4/6 inhibition antagonizes the cytotoxic response to anthracycline therapy. Cell Cycle 2012; 11:2747; PMID:22751436; http://dx.doi.org/ 10.4161/cc.21127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rader J, Hart L, Russell M, Nakazawa M, Belcastro L, Martinez D, Carpenter E, Kim S, Parasuraman S, Caponigro G. CDK4/CDK6 inhibition is potently active in a definable subset of human neuroblastomas. Cancer Res 2013; 73:2744; http://dx.doi.org/ 10.1158/1538-7445.AM2013-2744 [DOI] [Google Scholar]
- 106.Zhang Y-X, Sicinska E, Czaplinski JT, Remillard SP, Moss S, Wang Y, Brain C, Loo A, Snyder EL, Demetri GD. Antiproliferative effects of CDK4/6 inhibition in CDK4-amplified human liposarcoma in vitro and in vivo. Mol Cancer Ther 2014; 13:2184-93; PMID:25028469; http://dx.doi.org/ 10.1158/1535-7163.MCT-14-0387 [DOI] [PubMed] [Google Scholar]
- 107.Dempsey JA, Chan EM, Burke TF, Beckmann RP. LY2835219, a selective inhibitor of CDK4 and CDK6, inhibits growth in preclinical models of human cancer. [abstract]. In: Proceedings of the 104th Annual Meeting of the American Association for Cancer Research; 2013. April 6–10; Washington, DC. Philadelphia (PA): AACR; Cancer Res 2013; 73(8 Suppl):Abstract nr LB-122. [Google Scholar]
- 108.Patnaik A, Rosen L, Tolaney S, Tolcher A, Goldman J, Gandhi L. Clinical activity of LY2835219, a novel cell cycle inhibitor selective for CDK4 and CDK6, in patients with metastatic breast cancer. Proceedings of the 105th Annual Meeting of the American Association for Cancer Research, Abstract CT232 Presented April 7, 2014. [Google Scholar]
- 109.Li Z, Wang X, Eksterowicz J, Gribble MW Jr, Alba GQ, Ayres M, Carlson TJ, Chen A, Chen X, Cho R. Discovery of AMG 925, a FLT3 and CDK4 dual kinase inhibitor with preferential affinity for the activated state of FLT3. J Med Chem 2014; 57:3430-49; PMID:24641103; http://dx.doi.org/ 10.1021/jm500118j [DOI] [PubMed] [Google Scholar]
- 110.Reddy MVR, Akula B, Cosenza SC, Athuluridivakar S, Mallireddigari MR, Pallela VR, Billa VK, Subbaiah DRCV, Bharathi EV, Vasquez-Del Carpio R, et al.. Discovery of 8-cyclopentyl-2-[4-(4-methyl-piperazin-1-yl)-phenylamino]-7-oxo-7,8-dihydro-pyrido[2,3-d]pyrimidine-6-carbonitrile (7x) as a potent inhibitor of cyclin-dependent kinase 4 (CDK4) and AMPK-related kinase 5 (ARK5). J Med Chem 2014; 57:578-99; PMID:24417566; http://dx.doi.org/ 10.1021/jm401073p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cho YS, Angove H, Brain C, Chen CHT, Cheng H, Cheng R, Chopra R, Chung K, Congreve M, Dagostin C, et al.. Fragment-based discovery of 7-azabenzimidazoles as potent, highly selective, and orally active CDK4/6 inhibitors. ACS Med Chem Lett 2012; 3:445-9; PMID:24900493; http://dx.doi.org/ 10.1021/ml200241a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shimamura T, Shibata J, Kurihara H, Mita T, Otsuki S, Sagara T, Hirai H, Iwasawa Y. Identification of potent 5-pyrimidinyl-2-aminothiazole CDK4, 6 inhibitors with significant selectivity over CDK1, 2, 5, 7, and 9. Bioorg Med Chem Lett 2006; 16:3751-4; PMID:16682184; http://dx.doi.org/ 10.1016/j.bmcl.2006.04.048 [DOI] [PubMed] [Google Scholar]
- 113.Fry DW, Bedford DC, Harvey PH, Fritsch A, Keller PR, Wu Z, Dobrusin E, Leopold WR, Fattaey A, Garrett MD. Cell cycle and biochemical effects of PD 0183812 a potent inhibitor of the cyclin D-dependent kinases CDK4 and CDK6. J Biol Chem 200; 276:16617-23; PMID:11278443; http://dx.doi.org/ 10.1074/jbc.M008867200 [DOI] [PubMed] [Google Scholar]
- 114.Lu H, Schulze-Gahmen U. Toward understanding the structural basis of cyclin-dependent kinase 6 specific inhibition. J Med Chem 2006; 49:3826-31; PMID:16789739; http://dx.doi.org/ 10.1021/jm0600388 [DOI] [PubMed] [Google Scholar]
- 115.Kirkland LO, McInnes C. Non-ATP competitive protein kinase inhibitors as anti-tumor therapeutics. Biochem Pharmacol 2009; 77:1561-71; PMID:19167366; http://dx.doi.org/ 10.1016/j.bcp.2008.12.022 [DOI] [PubMed] [Google Scholar]
- 116.McInnes C, Wang S, Anderson S, O'Boyle J, Jackson W, Kontopidis G, Meades C, Mezna M, Thomas M, Wood G. Structural determinants of CDK4 inhibition and design of selective ATP competitive inhibitors. Chem Biol 2004; 11:525-34; PMID:15123247; http://dx.doi.org/ 10.1016/j.chembiol.2004.03.022 [DOI] [PubMed] [Google Scholar]
- 117.Lu H, Chang DJ, Baratte B, Meijer L, Schulze-Gahmen U. Crystal structure of a human cyclin-dependent kinase 6 Complex with a flavonol inhibitor, fisetin. J Med Chem 2005; 48:737-47; PMID:15689157; http://dx.doi.org/ 10.1021/jm049353p [DOI] [PubMed] [Google Scholar]
- 118.Jeffrey PD, Tong L, Pavletich NP. Structural basis of inhibition of CDK-cyclin complexes by INK4 inhibitors. Genes Dev 2000; 14:3115-25; PMID:11124804; http://dx.doi.org/ 10.1101/gad.851100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cho YS, Borland M, Brain C, Chen CH, Cheng H, Chopra R, Chung K, Groarke J, He G, Hou Y, et al.. 4-(Pyrazol-4-yl)-pyrimidines as selective inhibitors of cyclin-dependent kinase 4/6. J Med Chem 2010; 53:7938-57; PMID:21038853; http://dx.doi.org/ 10.1021/jm100571n [DOI] [PubMed] [Google Scholar]
- 120.Hirai H, Shimomura T, Kobayashi M, Eguchi T, Taniguchi E, Fukasawa K, Machida T, Oki H, Arai T, Ichikawa K, et al.. Biological characterization of 2-aminothiazole-derived Cdk4/6 selective inhibitor in vitro and in vivo. Cell Cycle 2010; 9:1590-600; PMID:20372067; http://dx.doi.org/ 10.4161/cc.9.8.11306 [DOI] [PubMed] [Google Scholar]