Abstract
The DHR3 and Hr4 early-late genes of the ecdysone cascade are described as models for transcriptional studies in Drosophila cells. In a set of experiments, it became clear that these genes are a convenient and versatile system for research into the physiological conditions upon 20-hydroxyecdysone induction. DHR3 and Hr4 gene transcription is characterized by fast activation kinetics, which enables transcriptional studies without the influence of indirect effects. A limited number of activated genes (only 73 genes are induced one hour after treatment) promote the selectivity of transcriptional studies via 20-hydroxyecdysone induction. DHR3 and Hr4 gene expression is dose dependent, is completely controlled by the hormone titer and decreases within hours of 20-hydroxyecdysone withdrawal. The DHR3 and Hr4 gene promoters become functional within 20 minutes after induction, which makes them useful tools for investigation if the early activation process. Their transcription is controlled by the RNA polymerase II pausing mechanism, which is widespread in the genome of Drosophila melanogaster but is still underinvestigated. Uniform expression activation of the DHR3 and Hr4 genes in a cell population was confirmed at both the RNA and protein levels. Homogeneity of the transcription response makes DHR3/Hr4 system valuable for investigation of the protein dynamics during transcription induction.
Keywords: coactivator, early-late genes, RNA polymerase II, transcription, 20-hydroxyecdysone
Introduction
In the whole-genome sequencing era, it may appear that there is no need to develop novel models for transcriptional investigation. Now, we have the ability to investigate every gene in the genome rather than each gene individually. The distribution of many transcriptional factors and coactivators was previously determined in Drosophila and mammalian genomes.1,2 However, we unfortunately remain far from understanding the mechanisms of their functions. Most of the investigated proteins that are involved in transcriptional regulation are localized to the same genomic regions or demonstrate mostly overlapping recruitment profiles. This fact significantly complicates investigations of the interactions between different transcriptional factors and coactivators. Finding an exact mechanism of interaction between these factors at the same site in the genome is quite a challenging goal. It is equally important to demonstrate which of these proteins are indeed bound to the same genomic region at the same time. We believe that inducible transcription systems could be a priceless tool for investigating the answers to these questions. Investigation of the dynamics of protein recruitment during transcriptional activation would reveal real partners that are functionally connected and dismiss unrelated. Whole-genome determination of protein recruitment profiles at different time points after activation of transcription remains quite expensive and labor-intensive. Thus, verification experiments could be performed with model genes.
Drosophila melanogaster is a well-described model organism that has been widely used in molecular biology for several decades. Detailed descriptions of heat-shock activation made Drosophila a popular model organism in transcriptional studies.3 Numerous Drosophila transcriptional complexes have been described to date. The number of subunits with redundant functions in these complexes is relatively small when compared with human orthologues, thus making the mechanisms of transcriptional complex functioning simpler and easier to investigate. Drosophila transcripts rarely undergo alternative splicing, which is why transcriptional factors are often expressed in a singular form that significantly simplifies investigation of their functions. The main drawback of Drosophila as a model organism is a restricted number of characterized model inducible genes. We hypothesize that this limitation arises from the excessive commitment of investigators to perform their studies on previously characterized models. The most popular model systems are heat shock-inducible genes. 4 Investigators continue to use heat shock genes to investigate new factors in transcription activation. 5,6 These investigations demonstrated that this system is a useful model and has led to a detailed description of its transcriptional processes. A significant disadvantage of the heat shock model is the non-physiological, stress conditions of its activation, which involves the activation of a cell protective system against misfolding and total repression of most genes. Extremely fast activation of the heat shock genes complicates investigation of the early transcription activation process and requires considerable modifications of routine methods to achieve the goal.7
The hormone-dependent transcription activation process is widely investigated both in Drosophila and mammals.8-10 The ecdysone cascade is the most described hormone-inducible system in Drosophila.11 Currently, transcription induction with ecdysone is often used to investigate the dynamics of factors influencing transcription.12,13 Recently, our group characterized the role of the SWI/SNF chromatin remodeling complex in the transcriptional activation of ecdysone-inducible genes.14 Transcription of ftz-f1, an early-late gene, was described in detail.15 We mimicked a portion of the ecdysone cascade in Drosophila S2 Schneider cells and investigated the different stages of ftz-f1 gene transcription together with the SWI/SNF complex functions. However, an important limitation of the described system for ftz-f1 activation involves its quite complex, multi-step mode of induction. ftz-f1 gene transcription starts only after complete removal of the ecdysone from the culture medium, which makes the system very sensitive to the performance of this procedure. Thus, we did not consider ftz-f1 gene activation as a robust inducible system for the comprehensive study of transcription.
Here, we describe the DHR3 (also referred as Hr46) and Hr4 early-late genes of the ecdysone cascade as promising systems for transcription investigation. Fast and robust induction of these genes in Drosophila S2 Schneider cells allows them to be available for multiple types of analysis. Here, we review a set of transcriptional characteristics of these genes. We consider this primary description to be useful for further investigation of these genes and in particular the study of the transcriptional processes in their dynamic action.
Results
Early-late genes of the ecdysone cascade demonstrate fast activation kinetics in S2 Schneider cells
Previously, we demonstrated that DHR3 gene (FBgn0000448) induction occurs by switching on the ecdysone cascade in S2 Schneider cells.14 In the current work, we describe the precise kinetics of DHR3 transcription activation. Transcription of the DHR3 gene was measured in S2 Schneider cells within several hours after the addition of 20-hydroxyecdysone (hereinafter referred as 20H-ecdysone) to the culture medium (Fig. 1A). DHR3 gene transcription is characterized by fast activation kinetics within the first 2 hours after induction, reaching a one hundredfold greater level at the end of the 2 hour period. Subsequently, the speed of DHR3 gene activation decreases manyfold, demonstrating an approximately fivefold activation within the following 18 hours. We interpret the fast activation of the DHR3 gene within the first 2 hours after induction as a result of the direct action of primary transcription activation mechanisms. Subsequent activation with slower kinetics is likely connected to the process of transcription re-initiation, i.e., recycling of transcriptional factors at actively transcribed loci at the end of a single transcriptional act.16
Figure 1.
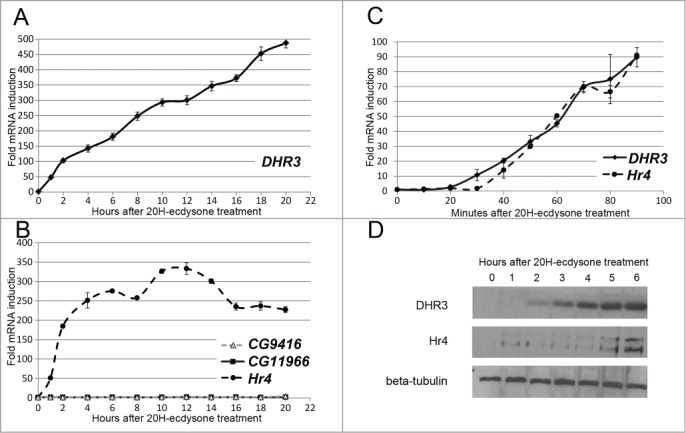
DHR3 and Hr4 gene expression is rapidly induced by ecdysone in S2 Schneider cells. Levels of DHR3 (A), Hr4, CG11966, and CG9416 (B) gene transcription in S2 Schneider cells after treatment with 20-hydroxyecdysone as evidenced by qPCR. The x-axis represents the time (hours) after 20-hydroxyecdysone addition, and the y-axis represents the fold induction relative to the corresponding gene level in untreated cells. Experiments were performed at least thrice. Error bars represent the standard error of the mean (SEM). (C) Levels of DHR3 and Hr4 gene transcription in S2 Schneider cells after the addition of 20-hydroxyecdysone as evidenced by qPCR. The x-axis represents the time (minutes) after 20-hydroxyecdysone addition, and the y-axis represents the fold induction relative to the corresponding gene level in untreated cells. Experiments were performed at least thrice. Error bars represent the standard error of the mean (SEM). (D) DHR3 and Hr4 protein levels in S2 Schneider cells after the addition of 20-hydroxyecdysone as evidenced by western blot analysis. The membranes were treated with an anti-β-tubulin antibody for normalization of protein extract amount.
To reinforce the described inducible system, we decided to determine at least one additional model gene with comparable activation kinetics. Transcriptional research performed on 2 model genes instead of one will be less sensitive to accusations of gene-specificity of the results. Ecdysone hormone, a major controller of Drosophila development, stimulates induction of ecdysone-dependent genes at different stages. At the beginning of our study, we did not have any information regarding the subset of genes that are activated in S2 cells upon 20H-ecdysone treatment. Therefore, we based our search of genes with transcriptional properties similar to DHR3 on information about their expression at different stages of Drosophila development. A list of genes with developmental expression profile similar to the DHR3 gene was created with www.Flybase.org on the basis of modENCODE data.1 The Hr4 (FBgn0264562), CG11966 (FBgn0037645) and CG9416 (FBgn0034438) genes represent the top 3 hits from the compiled list. The Hr4 gene, one of the candidates, indeed demonstrated fast activation in S2 Schneider cells with no further decrease in transcription level (Fig. 1B). CG11966 and CG9416 did not demonstrate any significant activation in S2 cells upon 20H-ecdysone treatment. We suggest that this finding is due to selective activation with substantial differences in the protein repertoire between S2 cultured cells and organismal cells at the particular developmental stage. Cultured embryonic cells could express undescribed repressor proteins whose action is not inhibited by the addition of 20H-ecdysone in the culture medium. Another possibility is that undescribed transcriptional activators, in addition to the 20H-ecdysone receptor, may determine the activation of CG11966 and CG9416 during appropriate developmental stages. Hr4 and DHR3 genes were selected for further assessment as possible models of an ecdysone-inducible system.
The primary stage of activation is of particular interest for transcriptional research. The induction kinetics of DHR3 and Hr4 genes in S2 Schneider cells during the initial 90 minutes were defined (Fig. 1C). Both genes demonstrated a significant level of activation 30 to 40 minutes after ecdysone treatment. On the one hand, the fast course of DHR3 and Hr4 induction protects transcriptional research performed in this model system from indirect effects caused by serial activations events. On the other hand, the 30-minute delay in DHR3 and Hr4 gene induction still allows investigation of this interesting preparatory stage of transcription via conventional methods.
To support our results of the DHR3 and Hr4 transcription induction, we analyzed their expression levels by western blotting (Fig. 1D). Both proteins exhibited increased expression in S2 Schneider cells within hours after ecdysone treatment. This finding demonstrates that DHR3 and Hr4 gene expression is regulated predominantly at the transcriptional level in S2 Schneider cells.
DHR3 and Hr4 gene transcription in S2 Schneider cells depends on a 20H-ecdysone titer
Transcription of hormone-inducible genes can be sensitive to a variety of different external stimuli and exhibit complex regulation. For example, transcription of ecdysone cascade genes was recently demonstrated to be regulated partially by nitric oxide (NO) signaling.17 We have determined whether transcription of our model genes is primarily controlled by 20H-ecdysone. The DHR3 and Hr4 gene transcription induction levels were determined 2 hours after treatment with different 20H-ecdysone concentrations (Fig. 2A). Both genes demonstrated a similar response to the hormone titer. Their induction levels increased with the increasing hormone concentration, reaching a plateau at 0.3 µM (this concentration was used in all further experiments).
Figure 2.
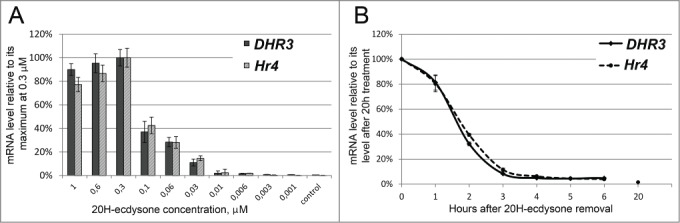
DHR3 and Hr4 gene induction level in S2 Schneider cells depends on the 20H-ecdysone titer and decreases after hormone withdrawal. (A) DHR3 and Hr4 gene transcription levels in S2 Schneider cells 2 hours after treatment with different concentrations of 20-hydroxyecdysone as evidenced by qPCR. The x-axis demonstrates the concentration of 20-hydroxyecdysone used for treatment, and the y-axis represents the level of DHR3 or Hr4 transcription activation relative to its maximum, which was estimated at 0.3 µM 20-hydroxyecdysone treatment. (B) DHR3 and Hr4 gene transcription levels at different time points after 20-hydroxyecdysone removal from the S2 Schneider cells after 20 hours of 20-hydroxyecdysone treatment as evidenced by qPCR. The x-axis represents the time (hours) after 20-hydroxyecdysone removal, and the y-axis represents the level of DHR3 and Hr4 transcription activation relative to the maximum of the corresponding gene transcription, which was estimated after 20 hours of hormone treatment. Both experiments were performed at least thrice. Error bars represent the standard error of the mean (SEM).
As mentioned above, DHR3 and Hr4 gene transcription remains activated for many hours after induction (Figs. 1A and B). To study the repression kinetics of our model genes, we removed the 20H-ecdysone hormone from the culture medium after 20 hours of induction and analyzed the subsequent decline in DHR3 and Hr4 transcription levels (Fig. 2B). Both genes demonstrated similar profiles of transcription repression. A 20% decrease in the transcription of DHR3 and Hr4 genes was detected an hour after ecdysone withdrawal. A ten-fold repression in transcription was achieved 3 hours after ecdysone withdrawal for each gene. The fast activation and repression kinetics of the studied genes confirms the exclusive dependence of their transcription on the 20H-ecdysone hormone in the culture medium.
20H-ecdysone induction affects a limited number of genes in S2 Schneider cells
To determine the genes that exhibit altered transcription upon 20H-ecdysone treatment, whole-transcriptome analysis of S2 Schneider cells was performed. For this experiment, 20H-ecdysone was added to the culture medium for one hour, and DMSO (a hormone solvent) was used as a negative control. Two biological replicates were performed to enable statistical verification of the results. The RNA fraction containing PolyA was isolated with the NEBNext® Ultra™ Directional RNA Library Prep Kit and prepared for next-generation sequencing (NGS) by Illumina. Obtained sequences were aligned against the Drosophila melanogaster genome (BDGP5.78 version) using TopHat2 software.18 The DHR3 and Hr4 genes demonstrated significant activation in the RNA-Seq experiment, while constitutive actin5C gene transcription remained unchanged (Supplementary fig 1). The transcription levels in experimental samples were compared with the Cuffdiff program and expressed as FPKM (fragments per kilobase of exon per million fragments mapped) (File_1).19 Treatment with 20H-ecdysone affected the transcription level of 86 genes (File_2). Genes with insignificant changes in transcription were excluded.
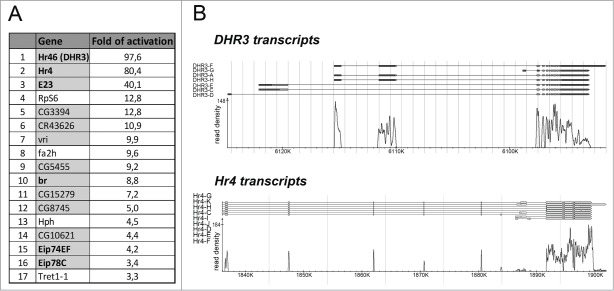
A RNA-Seq experiment demonstrated that the cellular response to 20H-ecdysone is characterized by preferential activation of transcription. Thus, only 13 of the 86 genes with differential transcription were repressed after hormone treatment, and a decrease of no greater than 2-fold was observed. The remainder of the altered genes was activated. Among the 73 genes, 11 (15.1%) were previously described as ecdysone-inducible genes, and 52 (71.2%) had expression profiles in development with a peak during pupariation (which is controlled by an ecdysone pulse) (FileS_2 and Fig. 3A). These data demonstrate that 20H-ecdysone treatment has a highly selective influence on transcription in S2 Schneider cells. We believe that the observed induction of a small set of developmental genes upon 20H-ecdysone treatment causes rather small indirect effects on the experiments performed in the DHR3/Hr4 model system.
Figure 3.
Ecdysone treatment activates transcription of a limited number of genes in S2 Schneider cells. (A) A set of genes exhibiting activated transcription (more than threefold) in S2 Schneider cells one hour after 20-hydroxyecdysone treatment as revealed by RNA-seq analysis. The lines with genes that have a peak in their expression profile during puparium formation are indicated with a gray color. The names of the genes previously described as participants of ecdysone activation are noted in bold. (B) Transcription activation at DHR3 and Hr4 loci in S2 Schneider cells after one hour of 20-hydroxyecdysone treatment determined by RNA-seq analysis. The coordinates of the x-axis correspond to the BDGP5.78 version of the Drosophila genome. The y-axis represents the read density at the corresponding genome regions.
Twenty minutes is required to prepare promoters of the DHR3 and Hr4 genes for an active transcription
To make appropriate models of the DHR3 and Hr4 gene activation, we characterized the mode of their transcription induction. The most significant difference in the transcription induction of eukaryotic genes is determined by the time required for RNA polymerase II to engage the promoters. Some genes do not contain a RNA polymerase II complex on their promoters in the resting or repressed state. Instead, these genes utilize the long type of transcription activation, which starts from the preinitiation complex formation. The promoter-proximal pausing path is another, short type of transcription induction. This mechanism includes pre-engagement of the RNA polymerase II complex to the promoters of genes in their inactive state. Promoter-proximal pausing of RNA polymerase II was recently identified as a widespread mechanism of transcriptional regulation. 20
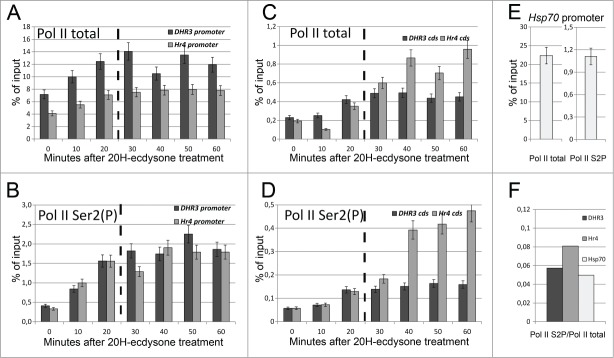
DHR3 and Hr4 genes have a complex structure and contain multiple exon-intron junctions and several alternative promoters. To identify the exact promoters that are activated in S2 Schneider cells upon 20H-ecdysone treatment, we analyzed the RNA-Seq results. As illustrated in Figure 3B and in Supplementary fig 1, promoters of the A, F, and H transcripts of DHR3 and the C, D, G, H, I, J, and K transcripts of Hr4 are functional in the 20H-ecdysone-treated cells. To determine the precise start of transcription, we performed 5′ rapid amplification of cDNA ends (RACE) analysis for each gene after activation (the results are provided in the Materials and Methods section). Determination of the promoter positions enables us to describe the properties of RNA polymerase II recruitment during DHR3 and Hr4 gene activation. Chromatin immunoprecipitation (ChIP) analysis with specific antibodies against total and elongation-specific (CTD Ser2 phosphorylated) forms of RNA polymerase II helped us to determine the modes of gene activation. RNA polymerase II status on the activated promoters of DHR3 and Hr4 was determined every 10 minutes for one hour after 20H-ecdysone treatment (Figs. 4A and B). We demonstrated that both DHR3 and Hr4 gene promoters contain a significant amount of RNA polymerase II before transcription induction. Transcriptional activation resulted in a doubling in the total amount of RNA polymerase II (Pol II total) at the promoters of the investigated genes. However, the amount of elongating form of the RNA polymerase II (Pol II Ser2(P)) at the promoters was increased by fivefold and sixfold for the DHR3 and Hr4 genes, respectively. We assume that the pre-recruited RNA polymerase II that binds the promoters of the studied genes in their inactive states exists in the hypophosphorylated form. Phosphorylation of the CTD Ser2 of RNA polymerase II significantly increases after DHR3 and Hr4 activation and reaches a plateau in 20 minutes. We consider this time point as the termination of the initiation stage of transcription, which leads to the formation of the RNA polymerase II complex that is capable of elongation. This conclusion is supported by the observed increase in the RNA polymerase II level at the coding regions of the DHR3 and Hr4 genes 20 minutes after transcription activation (Figs. 4C and D). Altogether, the DHR3 and Hr4 promoters are prepared for forthcoming transcription and become functional 20 minutes after ecdysone treatment.
Figure 4.
DHR3 and Hr4 promoters are regulated via a Pol II pausing mechanism and activated 20 minutes after ecdysone treatment. ChIP analysis of the total (A and C) and the CTD Ser2 phosphorylated (B and D) RNA polymerase II recruitment on DHR3 (dark gray) and Hr4 (light gray) promoters and coding regions after transcription induction of corresponding genes with 20-hydroxyecdysone treatment. The x-axis represents the time (minutes) after 20-hydroxyecdysone addition, and the y-axis represents the amount of precipitated DNA, which is presented as a percentage of the input. ChIP analysis of the total and the CTD Ser2 phosphorylated RNA polymerase II (E) recruitment on the Hsp70 promoter in untreated (corresponding to 0 min point) S2 Schneider cells. All experiments were performed at least thrice. Error bars represent the accuracy of measurement. Panel (F) graphs demonstrate the ratio between phosphorylated and total RNA polymerase II in the untreated (corresponding to 0 min point) S2 Schneider cells, which have been calculated for the DHR3, Hr4 and Hsp70 promoters.
To further clarify the RNA polymerase II status at the DHR3 and Hr4 promoters, we analyzed the CTD Ser2 phosphorylated and total Pol II at the Hsp70 promoter in untreated S2 Schneider cells (corresponding to the 0 min time point) (Fig. 4E). The ratio between the CTD Ser2 phosphorylated and total forms of RNA polymerase II at the DHR3 and Hr4 inactive promoters were quite close to the value obtained for the inactive Hsp70 gene promoter (Fig. 4F). As transcription of the Hsp70 gene was previously shown to be regulated through RNA polymerase II promoter-proximal pausing, which is triggered by CTD Ser2 phosphorylation, we assume that transcription of our model genes are governed by the same mechanism.
DHR3 and Hr4 gene expressions is activated uniformly in cell population
The validity of standard techniques, such as reverse transcription polymerase chain reaction (RT-PCR), ChIP, and chromatin conformation capture (3C) for investigation of the transcription activation could be questioned given the heterogeneity of these processes in a cell population.21 Indeed, changes in the ratio of induced/uninduced cells could significantly impair results and demonstrate an incorrect pattern of factor recruitment or chromatin restructuration.
To describe the uniformity of gene activation in our inducible model system in a cell population, we analyzed transcription of DHR3 and Hr4 genes using the RNA fluorescence in situ hybridization (FISH) method (Figs. 5A and B). As expected, we detected no transcription of our model genes prior to ecdysone treatment. Two hours after transcription induction, we observed an accumulation of DHR3 and Hr4 transcripts in all cells of the population. This homogeneity of the gene expression response was also confirmed at the protein level (Fig. 6A). Indeed, DHR3 and Hr4 proteins appeared in S2 Schneider cells soon after ecdysone treatment and were present in all cells of the population.
Figure 5.
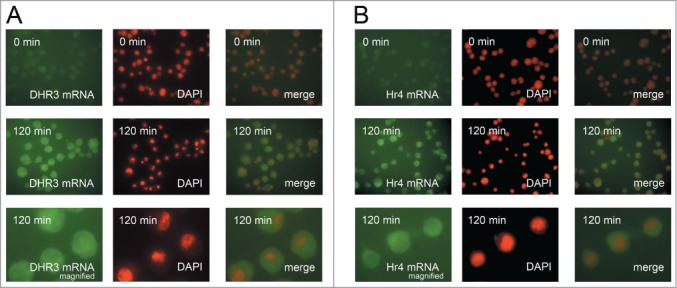
DHR3 and Hr4 genes transcription is induced uniformly in the cell population. FISH analysis of DHR3 (A) and Hr4 (B) genes transcription in S2 Schneider cells before and after 2 hours of 20-hydroxyecdysone treatment. The lower panel shows an enlarged image of the corresponding FISH analysis. Images before and after activation were made with the same exposure settings.
Figure 6.
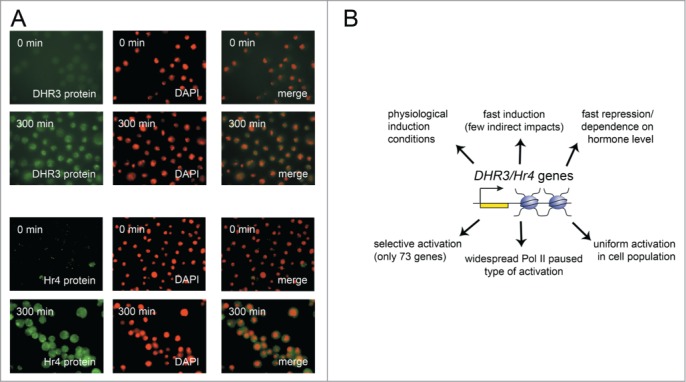
(A) DHR3 and Hr4 genes are expressed uniformly in a cell population. DHR3 and Hr4 proteins accumulation in S2 Schneider cells after 5 hours of 20-hydroxyecdysone treatment as evidenced by immunostaining with corresponding specific antibodies. (B) The summary of benefits of the DHR3 and Hr4 genes as a system for transcriptional investigation.
A uniform course of activation in a cell population is an important factor for the research of transcriptional kinetics. Thus, the demonstrated synchronous activation of the DHR3 and Hr4 genes is a considerable advantage of the described model system.
Discussion
We described a system enabling the induction of the DHR3 and Hr4 developmental genes in S2 Schneider cells by the hormone 20H-ecdysone. Due to the fast activation kinetics of the genes and the simplicity of the protocol execution, we propose these genes as a valuable model system for transcriptional research (Fig. 6B). In fact, 20H-ecdysone induction has quite a long history of use in molecular biology studies in both cultured cells and tissues.22-24 However, different research groups use different timings of induction with this hormone. We suggest that a rather short induction time, such as that utilized in this DHR3/Hr4 model system, offers benefits compared with other systems. Only two hours are required for the DHR3 and Hr4 genes to achieve a high level of activation, which is an insufficient amount of time to complete an expression cycle for any other gene. Thus, in the proposed system, indirect effects are significantly minimized. Fast induction in the DHR3/Hr4 system, unlike the heat shock model, does not require an adaptation of any common laboratory techniques that are used for transcription studies. The timing of DHR3 and Hr4 transcription activation lies within the limits required for usage of a wide spectrum of methods and corresponds to the time periods that are commonly described in mammalian models. 8,9
A high 20H-ecdysone titer was shown to maintain DHR3 and Hr4 activated transcription in S2 Schneider cells for a long time. This fact demonstrates that the DHR3 and Hr4 proteins, which are transcriptional factors per se, do not effect expression of their own genes. In other words, this model system has no negative-feedback loop, which is very common for transcriptional regulation of genes controlling development. We demonstrated that transcription of the DHR3 and Hr4 model genes could be rapidly repressed by removal of 20H-ecdysone from the culture medium. DHR3 and Hr4 gene transcription is induced directly by 20H-ecdysone treatment in a dose-dependent manner. These findings demonstrate a straightforward and relatively simple mechanism of hormone-dependent transcriptional regulation of the DHR3 and Hr4 genes, which facilitates its study. The rapid course of transcription reduction upon hormone withdrawal makes the DHR3/Hr4 system potentially valuable for transcription repression research.
In the current manuscript, we provided a list of genes exhibiting transcription that is induced one hour after 20H-ecdysone treatment. We are enabling researchers to extend the existing model and investigate the transcription activation/repression of other ecdysone-dependent genes. The selectivity of the impact of 20H-ecdysone on S2 Schneider cells makes it possible to assume that a subset of these activated genes would demonstrate features comparable to the DHR3 and Hr4 genes. This information makes it possible to expand our model system in a short period of time, which will make it even more reliable.
For a long time, a complete mechanism of transcription activation, including RNA polymerase II recruitment to promoters and transcription initiation, was considered as a preferable method of induction for most genes. Recently, it has been demonstrated that RNA polymerase II promoter-proximal pausing is a widespread and even dominant type of transcription regulation.25 Although this phenomenon was discovered 2 decades ago, the detailed molecular mechanism of this process remains unresolved.26,27 RNA polymerase II pausing is certainly a composite mechanism, which is considered to be regulated by distinct protein complexes, but only 3 participants of the process have been described to date.28 Most of the information regarding the exact mechanism of RNA polymerase II promoter-proximal pausing in Drosophila was obtained in the heat-shock gene system. We believe that our finding of DHR3 and Hr4 gene regulation by the RNA polymerase II pausing mechanism will be useful for further investigation of the precise mechanism of this phenomenon. The slow kinetics of DHR3 and Hr4 gene transcription activation (compared with the heat shock system) provides researchers with an opportunity to determine additional protein candidates that are responsible for the release of transcription from the paused state.
In conclusion, we would like to highlight a few prospective applications of the described DHR3/Hr4 model system. We propose the study of transcriptional factors recruitment as a primary application for the characterized system. Indeed, not all the molecular participants of the transcription cycle are defined. Moreover, the particular order of transcription factors involvement in this process is currently obscure. Despite recent breakthroughs in single-cell investigation techniques, transcription activation kinetic studies on cell populations are still considered relevant. Investigation of the protein dynamics on chromatin during transcription induction is still an actively developing area of research with a great potential for future breakthrough.21
Transcriptional studies in the DHR3/Hr4 system would also be of use for broadening our understanding of the ecdysone cascade molecular mechanism. Although this developmental cascade was described a long time ago, it still remains underinvestigated.29 Only the transcriptional activators, the key components of the cascade, are relatively well described to date.11 The presence of specific transcriptional co-activators in the cascade is possible but mainly remains unknown. Some difficulties in investigation in this area are associated with the short period of time during Drosophila development when the ecdysone cascade is active. Transcriptional studies in living tissues are complicated and limited in the number of applicable research techniques. Using the described DHR3/Hr4 gene system allows the investigation of the ecdysone cascade activation in cultural cells. Thus, the DHR3/Hr4 model system provides numerous opportunities for investigation of ecdysone-dependent transcription by the wide range of methods.
In addition to its fundamental value, the DHR3/Hr4 system could be useful for some practical applications. Various systems used for recombinant protein expression both in insects and mammals utilize ecdysone-dependent inducible transcription as a part of their processes.30 A detailed characterization of the previously undescribed DHR3 and Hr4 promoters could be used in the construction of expression vectors. A possible spectrum of applications for such constructs could be quite extensive, ranging from the production of transgenic organisms to the quantitative expression of potential drug proteins.31-33
Materials and methods
Protocols for Immunostaining,34 RT-PCR 15 and ChIP 35 analysis were previously described.
Experiments with S2 cell culture (ecdysone-dependent genes induction)
Drosophila Schneider line 2 (S2) cells were maintained at 25°C in ecdysone-free Schneider's insect medium (Sigma) containing 10% FBS (HyClone). The cells were treated with 0.3 µM 20-hydroxyecdysone (Sigma, http://www.sigmaaldrich.com/catalog/product/sigma/h5142) for ecdysone cascade initiation. The influence of 20-hydroxyecdysone in the cell medium on DHR3 and Hr4 genes activation was determined 2 hours after hormone addition. For analysis of the repression dynamics, cells were treated with 20-hydroxyecdysone overnight, subsequently washed 3 times with 100-fold amount of fresh medium, and then cultivated for the required periods of time.
Antibodies and western blot analysis
We used antibodies against DHR3,14 total Pol II 36 and phosphoS2 Pol II Ab5095 (Abcam, http://www.abcam.com/rna-polymerase-ii-ctd-repeat-ysptsps-phospho-s2-antibody-chip-grade-ab5095.html). Antibodies against Hr4 (1211-1518 aa. fragment of C isoform NP_001033823.1) were raised in rabbits immunized with the corresponding His6-tagged fragments in our laboratory. An antibody against β-tubulin, obtained from M. Klymkowsky (University of Colorado, Boulder), was from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained at the University of Iowa, Department of Biological Sciences.
RNA-FISH
DHR3 and Hr4 probes for RNA-FISH were synthesized with a DIG RNA Labeling Kit (Roche) according to manufacturer's instructions. S2 Schneider cells were fixed on glass slides with 3.7% FA for 10 min, permeabilized with 0.5% Triton X-100, washed with PBT buffer and treated with Proteinase K (50 µg/ml) for 4 minutes at 37°C. Then, the reaction was stopped with a 2-minute incubation in glycine (2 mg/ml), and the cells were postfixed in 3.7% FA for 20 minutes. After 1.5 hours of pre-hybridization at 37°C, the cells were hybridized with the corresponding pre-denatured RNA probe for 16 hours at 37°C in buffer containing 5xSSC (saline sodium citrate), 50% formamide, 0.1% Tween 20, 100 mkg/ml salmon sperm DNA, 50 mkg/ml heparin, and 1U/mkl Ribolock. The unbound probe was washed from the cells by washing with 5xSSC and 2xSSC at 37°C, 2xSSC and 0.5xSSC at 55°C, and 0.5xSSC at RT. Then, glass slides were treated with a blocking solution (1xPBS, 0.1% Tween 20, 0.1% Triton X-100, and 1% bovine serum albumin (BSA)) for one hour at room temperature (RT) and stained with anti-DIG-rhodamine (Roche) for an hour at RT.
RNA-Seq
Drosophila Schneider line 2 (S2) cells were treated with 0.3 µM 20-hydroxyecdysone (Sigma) for one hour. Two biological replicas were performed for both treated and control cells. Total RNA was extracted with the TRI reagent (Ambion). The PolyA comprising RNA fraction was isolated with the NEBNext® Ultra™ Directional RNA Library Prep Kit and subjected to NGS by Illumina. Library preparation and NGS were performed by the Evrogen JSC Company.
5′-RACE
cDNA was synthesized and amplified with the Mint RACE kit (Evrogen JSC) using RNA isolated from S2 Schneider cells treated with 20-hydroxyecdysone for 2 hours as a matrix. Primers located in first exon of Hr4 (5′-TATCGATAATGGTGGTGGCG) and DHR3 (5′-ACACACTGGCACACACTCGC) induced transcripts were used for amplification of 5′ fragments. The start site of the DHR3 gene transcripts, induced in S2 Schneider cells by 20-hydroxyecdysone hormone, is located 6 base pairs (b.p.) downstream of the previously described starts of the A, F, and H transcripts. The start site of the Hr4 gene 20-hydroxyecdysone-induced transcripts is located 20 b.p. upstream of the previously determined starts of the C, D, G, H, I, J, and K transcripts.
Primers
The following primers were used for quantitative PCR (qPCR) detection in RT-PCR analysis: 5′-CGGTTGCGATTAACACGGTCC and 5′- CTGCAAGGGATTCTTTCGAAGATC (DHR3 gene); 5′-GTGCGTCTGCACAATGTTGG and 5′-GGAACAGTCCATCAGCTCCTCG (Hr4 gene); 5′-TCGGGATCTCTTGGCCTTGG and 5′-GTGGCTATTCCATGCAAGCATCG (CG11966 gene); 5′-GAATATGCTCGTGTGATCCG and 5′-ACAGCTATTTTGTTCCTGGTCC (CG9416 gene).
The following primers were used for qPCR detection in ChIP analysis: 5′-CACGACGACAACGAACAGTC and 5′-CTCAATACGAGGTGTAGCTGAAG (DHR3 ecdysone-inducible promoter); 5′-AACAATCCACCACCGACTGG and 5′-AGAGAACCCTCTCTTGAGCGC (Hr4 ecdysone-inducible promoter); 5-ATTCGGTTTTCGCTTAGATCTTAG and 5-CGATTGGCTAATACAATTGGAACC (coding region of the DHR3 gene); 5-TTTATTTCGGCCTGGTTTCG and 5-TAGACTTGAGTGCTTGCATACTATGTGG (coding region of the Hr4 gene); 5-ACATACTGCTCTCGTTGGTTCG and 5-TTGAATTGAATTGTCGCTCCGTAG (Hsp70 gene promoter).
The following primers were used as probes for RNA-FISH: 5′-CACATCGTTGATATCGCCATCC and 5′-TCTTTCGAAGATCGCAGAGCTCC (DHR3 gene); 5′-GTGAACGGAACGATACTGAAG and 5′-AGCTGCTCCATCGCTATCGG (Hr4 gene).
Acknowledgments
We are grateful to L. Tora for the gift of antibodies against total RNA polymerase II (Institute of Genetics and Molecular and Cellular Biology, Strasburg).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was supported by the Russian Science Foundation (Project №14-14-01032).
References
- 1.Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al.. Unlocking the secrets of the genome. Nature 2009; 459:927-30; PMID:19536255; http://dx.doi.org/ 10.1038/459927a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siggens L, Ekwall K. Epigenetics, chromatin and genome organization: recent advances from the ENCODE project. J Intern Med 2014; 276:201-14; PMID:24605849; http://dx.doi.org/ 10.1111/joim.12231 [DOI] [PubMed] [Google Scholar]
- 3.Ashburner M, Bonner JJ. The induction of gene activity in drosophilia by heat shock. Cell 1979; 17:241-54; PMID:110462; http://dx.doi.org/ 10.1016/0092-8674(79)90150-8 [DOI] [PubMed] [Google Scholar]
- 4.Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 1988; 54:795-804; PMID:3136931; http://dx.doi.org/ 10.1016/S0092-8674(88)91087-2 [DOI] [PubMed] [Google Scholar]
- 5.Murawska M, Hassler M, Renkawitz-Pohl R, Ladurner A, Brehm A. Stress-induced PARP activation mediates recruitment of Drosophila Mi-2 to promote heat shock gene expression. PLoS Genet 2011; 7:e1002206; PMID:21829383; http://dx.doi.org/ 10.1371/journal.pgen.1002206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kopytova DV, Orlova AV, Krasnov AN, Gurskiy DY, Nikolenko JV, Nabirochkina EN, Shidlovskii YV, Georgieva SG. Multifunctional factor ENY2 is associated with the THO complex and promotes its recruitment onto nascent mRNA. Genes Dev 2010; 24:86-96; PMID:20048002; http://dx.doi.org/ 10.1101/gad.550010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 2008; 134:74-84; PMID:18614012; http://dx.doi.org/ 10.1016/j.cell.2008.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beato M, Vicent GP. Impact of chromatin structure and dynamics on PR signaling. The initial steps in hormonal gene regulation. Mol Cell Endocrinol 2012; 357:37-42; PMID:21945605; http://dx.doi.org/ 10.1016/j.mce.2011.09.004 [DOI] [PubMed] [Google Scholar]
- 9.Metivier R, Penot G, Carmouche RP, Hubner MR, Reid G, Denger S, Manu D, Brand H, Kos M, Benes V, et al.. Transcriptional complexes engaged by apo-estrogen receptor-alpha isoforms have divergent outcomes. EMBO J 2004; 23:3653-66; PMID:15343269; http://dx.doi.org/ 10.1038/sj.emboj.7600377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernardo TJ, Dubrovskaya VA, Xie X, Dubrovsky EB. A view through a chromatin loop: insights into the ecdysone activation of early genes in Drosophila. Nucleic Acids Res 2014; 42:10409-24; PMID:25143532; http://dx.doi.org/ 10.1093/nar/gku754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ou Q, King-Jones K. What goes up must come down: transcription factors have their say in making ecdysone pulses. Curr Top Dev Biol 2013; 103:35-71; PMID:23347515; http://dx.doi.org/ 10.1016/B978-0-12-385979-2.00002-2 [DOI] [PubMed] [Google Scholar]
- 12.Shlyueva D, Stelzer C, Gerlach D, Yanez-Cuna JO, Rath M, Boryn LM, Arnold CD, Stark A. Hormone-responsive enhancer-activity maps reveal predictive motifs, indirect repression, and targeting of closed chromatin. Mol Cell 2014; 54:180-92; PMID:24685159; http://dx.doi.org/ 10.1016/j.molcel.2014.02.026 [DOI] [PubMed] [Google Scholar]
- 13.Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 2010; 143:540-51; PMID:21074046; http://dx.doi.org/ 10.1016/j.cell.2010.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vorobyeva NE, Nikolenko JV, Krasnov AN, Kuzmina JL, Panov VV, Nabirochkina EN, Georgieva SG, Shidlovskii YV. SAYP interacts with DHR3 nuclear receptor and participates in ecdysone-dependent transcription regulation. Cell Cycle 2011; 10:1821-7; PMID:21519192; http://dx.doi.org/ 10.4161/cc.10.11.15727 [DOI] [PubMed] [Google Scholar]
- 15.Vorobyeva NE, Nikolenko JV, Nabirochkina EN, Krasnov AN, Shidlovskii YV, Georgieva SG. SAYP and Brahma are important for ‘repressive’ and ‘transient’ Pol II pausing. Nucleic Acids Res 2012; 40:7319-31; PMID:22638575; http://dx.doi.org/ 10.1093/nar/gks472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH. Crosstalk between mRNA 3′ end processing and transcription initiation. Mol Cell 2010; 40:410-22; PMID:21070967; http://dx.doi.org/ 10.1016/j.molcel.2010.10.012 [DOI] [PubMed] [Google Scholar]
- 17.Johnston DM, Sedkov Y, Petruk S, Riley KM, Fujioka M, Jaynes JB, Mazo A. Ecdysone- and NO-mediated gene regulation by competing EcR/Usp and E75A nuclear receptors during Drosophila development. Mol Cell 2011; 44:51-61; PMID:21981918; http://dx.doi.org/ 10.1016/j.molcel.2011.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14:R36; PMID:23618408; http://dx.doi.org/ 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 2013; 31:46-53; PMID:23222703; http://dx.doi.org/ 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Core LJ, Waterfall JJ, Gilchrist DA, Fargo DC, Kwak H, Adelman K, Lis JT. Defining the status of RNA polymerase at promoters. Cell Rep 2012; 2:1025-35; PMID:23062713; http://dx.doi.org/ 10.1016/j.celrep.2012.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coulon A, Chow CC, Singer RH, Larson DR. Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nat Rev Genet 2013; 14:572-84; PMID:23835438; http://dx.doi.org/ 10.1038/nrg3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horner MA, Chen T, Thummel CS. Ecdysteroid regulation and DNA binding properties of Drosophila nuclear hormone receptor superfamily members. Dev Biol 1995; 168:490-502; PMID:7729584; http://dx.doi.org/ 10.1006/dbio.1995.1097 [DOI] [PubMed] [Google Scholar]
- 23.Sempere LF, Dubrovsky EB, Dubrovskaya VA, Berger EM, Ambros V. The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Dev Biol 2002; 244:170-9; PMID:11900466; http://dx.doi.org/ 10.1006/dbio.2002.0594 [DOI] [PubMed] [Google Scholar]
- 24.Gvozdev VA, Kakpakov VT, Mukhovatova LM, Polukarova LY, Tarantul VZ. Influence of ecdysterone on the growth of cells and synthesis of macromolecules in established cell lines of Drosophila melanogaster. Sov J Dev Biol 1975; 5:29-36; PMID:803716 [PubMed] [Google Scholar]
- 25.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 2012; 13:720-31; PMID:22986266; http://dx.doi.org/ 10.1038/nrg3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev 1992; 6:2201-13; PMID:1427080; http://dx.doi.org/ 10.1101/gad.6.11.2201 [DOI] [PubMed] [Google Scholar]
- 27.Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci U S A 1993; 90:7923-7; PMID:8367444; http://dx.doi.org/ 10.1073/pnas.90.17.7923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Gilmour DS. Distinct mechanisms of transcriptional pausing orchestrated by GAGA factor and M1BP, a novel transcription factor. EMBO J 2013; 32:1829-41; PMID:23708796; http://dx.doi.org/ 10.1038/emboj.2013.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamanaka N, Rewitz KF, O'Connor MB. Ecdysone control of developmental transitions: lessons from Drosophila research. Ann Rev Entomol 2013; 58:497-516; PMID:23072462; http://dx.doi.org/ 10.1146/annurev-ento-120811-153608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karzenowski D, Potter DW, Padidam M. Inducible control of transgene expression with ecdysone receptor: gene switches with high sensitivity, robust expression, and reduced size. BioTechniques 2005; 39:191-2, 4, 6 passim; PMID:16116792; http://dx.doi.org/ 10.2144/05392ST01 [DOI] [PubMed] [Google Scholar]
- 31.Moraes AM, Jorge SA, Astray RM, Suazo CA, Calderon Riquelme CE, Augusto EF, Tonso A, Pamboukian MM, Piccoli RA, Barral MF, et al.. Drosophila melanogaster S2 cells for expression of heterologous genes: From gene cloning to bioprocess development. Biotechnol Adv 2012; 30:613-28; PMID:22079894; http://dx.doi.org/ 10.1016/j.biotechadv.2011.10.009 [DOI] [PubMed] [Google Scholar]
- 32.Albanese C, Hulit J, Sakamaki T, Pestell RG. Recent advances in inducible expression in transgenic mice. Semin Cell Dev Biol 2002; 13:129-41; PMID:12240598; http://dx.doi.org/ 10.1016/S1084-9521(02)00021-6 [DOI] [PubMed] [Google Scholar]
- 33.Kumpun S, Girault JP, Dinan L, Blais C, Maria A, Dauphin-Villemant C, Yingyongnarongkul B, Suksamrarn A, Lafont R. The metabolism of 20-hydroxyecdysone in mice: relevance to pharmacological effects and gene switch applications of ecdysteroids. J Steroid Biochem Mol Biol 2011; 126:1-9; PMID:21439380; http://dx.doi.org/ 10.1016/j.jsbmb.2011.03.016 [DOI] [PubMed] [Google Scholar]
- 34.Vorobyeva NE, Soshnikova NV, Nikolenko JV, Kuzmina JL, Nabirochkina EN, Georgieva SG, Shidlovskii YV. Transcription coactivator SAYP combines chromatin remodeler Brahma and transcription initiation factor TFIID into a single supercomplex. Proc Natl Acad Sci U S A 2009; 106:11049-54; PMID:19541607; http://dx.doi.org/ 10.1073/pnas.0901801106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vorobyeva NE, Mazina MU, Golovnin AK, Kopytova DV, Gurskiy DY, Nabirochkina EN, Georgieva SG, Georgiev PG, Krasnov AN. Insulator protein Su(Hw) recruits SAGA and Brahma complexes and constitutes part of Origin Recognition Complex-binding sites in the Drosophila genome. Nucleic acids research 2013; 41:5717-30; PMID:23609538; http://dx.doi.org/ 10.1093/nar/gkt297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Georgieva S, Kirschner DB, Jagla T, Nabirochkina E, Hanke S, Schenkel H, de Lorenzo C, Sinha P, Jagla K, Mechler B, et al.. Two novel Drosophila TAF(II)s have homology with human TAF(II)30 and are differentially regulated during development. Mol Cell Biol 2000; 20:1639-48; PMID:10669741; http://dx.doi.org/ 10.1128/MCB.20.5.1639-1648.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.