PH3 of petunia is a WRKY protein that is homologous to TRANSPARENT TESTA GLABRA2 from Arabidopsis.
Abstract
The WD40 proteins ANTHOCYANIN11 (AN11) from petunia (Petunia hybrida) and TRANSPARENT TESTA GLABRA1 (TTG1) from Arabidopsis thaliana and associated basic helix-loop-helix (bHLH) and MYB transcription factors activate a variety of differentiation processes. In petunia petals, AN11 and the bHLH protein AN1 activate, together with the MYB protein AN2, anthocyanin biosynthesis and, together with the MYB protein PH4, distinct genes, such as PH1 and PH5, that acidify the vacuole. To understand how AN1 and AN11 activate anthocyanin biosynthetic and PH genes independently, we isolated PH3. We found that PH3 is a target gene of the AN11-AN1-PH4 complex and encodes a WRKY protein that can bind to AN11 and is required, in a feed-forward loop, together with AN11-AN1-PH4 for transcription of PH5. PH3 is highly similar to TTG2, which regulates hair development, tannin accumulation, and mucilage production in Arabidopsis. Like PH3, TTG2 can bind to petunia AN11 and the Arabidopsis homolog TTG1, complement ph3 in petunia, and reactivate the PH3 target gene PH5. Our findings show that the specificity of WD40-bHLH-MYB complexes is in part determined by interacting proteins, such as PH3 and TTG2, and reveal an unanticipated similarity in the regulatory circuitry that controls petunia vacuolar acidification and Arabidopsis hair development.
INTRODUCTION
Regulatory complexes consisting of a MYB, basic helix-loop-helix (bHLH), and WD40 protein (MBW complexes) control multiple pathways involved in the differentiation of epidermal cells in angiosperms (reviewed in Broun, 2005; Koes et al., 2005; Ramsay and Glover, 2005; Ishida et al., 2008). The best understood function of this complex involves the biosynthesis of anthocyanin pigments that color many flowers and fruits. Closely related, and in most cases functionally exchangeable, MYB, bHLH, and WD40 proteins collaboratively activate the transcription of genes that encode enzymes of the anthocyanin pathway in a broad range of species, including distantly related dicots like Arabidopsis thaliana (a Rosid) and petunia (Petunia hybrida; an Asterid) and monocots like maize (Zea mays; Koes et al., 2005; Hichri et al., 2011; Petroni and Tonelli, 2011). In addition, related MBW complexes activate numerous other pathways and processes, such as vacuolar acidification, formation of seed mucilage, and the development of hair cells, a process that seems restricted to smaller sets of species (Koes et al., 2005; Ramsay and Glover, 2005; Serna and Martin, 2006).
In petunia flowers, the WD40 protein ANTHOCYANIN11 (AN11) and the bHLH factor AN1 activate structural anthocyanin genes in floral tissues, such as petals and anthers, and a few vegetative tissues (Quattrocchio et al., 1993; de Vetten et al., 1997; Spelt et al., 2000). To this end, AN11 and AN1 interact with paralogous and exchangeable MYB proteins, which are encoded by AN2, AN4, and DEEP PURPLE and are expressed in different tissues (Quattrocchio et al., 1999, 2006; Albert et al., 2011). Furthermore, AN1 and AN11 activate, in concert with the MYB protein PH4, a distinct pathway that alters the color of the flower by acidifying the central vacuole in epidermal petal cells, where the anthocyanins are stored (Spelt et al., 2002; Quattrocchio et al., 2006). To date, seven such PH genes have been identified in petunia via mutants that all show a similar phenotype: petals with a bluish color and an increased pH of petal homogenates (de Vlaming et al., 1983; van Houwelingen et al., 1998). Molecular analysis revealed that ph6 mutants represent specific an1 alleles that lost the capacity to acidify the vacuole but can still drive anthocyanin biosynthesis (Spelt et al., 2002) and that PH5 and PH1 are direct target genes of the AN11-AN1-PH4 complex that encodes two interacting P-ATPase transmembrane transporters that reside in the tonoplast (Verweij et al., 2008; Faraco et al., 2014).
The AN1-AN11-PH4 triumvirate is also required for the stability of anthocyanins in the vacuole, as in certain genetic backgrounds ph4 and specific an1 alleles (formerly known as ph6 alleles) trigger the complete disappearance of anthocyanins and “fading” of the flower color after opening of the bud (de Vlaming et al., 1982, 1983; Quattrocchio et al., 2006; Passeri et al., 2016). Fading is not triggered by the pH shift alone because it is not seen in ph5 and ph2 mutants but seems due to a different vacuolar defect caused by the misregulation of distinct AN1-AN11-PH4 target genes (Quattrocchio et al., 2006). In seeds, AN1 and AN11, presumably together with a yet unidentified MYB protein, influence the expression of a broad set of genes and are required for the biosynthesis of proanthocyanidins and to prevent cell divisions in the seed coat epidermis (Spelt et al., 2002; Zenoni et al., 2011).
In Arabidopsis, homologous MBW complexes, consisting of the AN11 homolog TRANSPARENTA TESTA GLABRA1 (TTG1; Walker et al., 1999) and a few selected bHLH and MYB proteins, regulate the biosynthesis of (pro)anthocyanidins (tannins) and other processes such as mucilage production in seeds and the development of hairs on aerial tissues (trichomes) and non-hair cells in the root epidermis (reviewed in Broun, 2005; Lepiniec et al., 2006; Ishida et al., 2008). In the seed coat, a complex of TTG1, the bHLH protein TRANSPARENT TESTA8 (TT8), and the MYB protein TT2 is required to induce proanthocyanidin biosynthesis (Nesi et al., 2000, 2001; Baudry et al., 2004). In various aerial tissues, TTG1 activates the biosynthesis of anthocyanins together with the partially redundant bHLH proteins GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) and a MYB protein encoded by either PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1), PAP2, MYB113, or MYB114 (Borevitz et al., 2000; Zhang et al., 2003; Gonzalez et al., 2008). In epidermal leaf and stem cells, TTG1 and GL3 or EGL3 interact with the MYB protein GL1 to promote trichome formation (Payne et al., 2000; Balkunde et al., 2011), whereas interaction of the same factors with the GL1 paralog WEREWOLF specifies the non-hair fate (atrichoblast) of certain cells in the root epidermis (Lee and Schiefelbein, 1999; Bernhardt et al., 2003). Interestingly, gain- and loss-of-function mutations in homologous WD40 and bHLH proteins do not affect the formation of hairs in either petunia or maize, indicating that this role of the MBW complexes is restricted to few species, including Arabidopsis. This contrasts with the role of MBW complexes in proanthocyanidin and anthocyanin biosynthesis, which is widely conserved among angiosperms and apparently more ancient.
It is not well understood how similar MBW complexes can activate distinct downstream pathways in different tissues and how they acquired distinct roles in different species during evolution. Because in Arabidopsis and petunia the effects of mutations of the WD40 and bHLH partners are highly pleiotropic and those of the MYB partners much less so, the specificity of distinct MBW complexes seems determined at least in part by the MYB partner (Koes et al., 2005; Ramsay and Glover, 2005). Consistent with this idea, the WD40 and bHLH genes are expressed in a much wider domain than their MYB partners. Moreover, the TTG1 homolog PALE ALEURONE COLOR1 and the bHLH protein RED, both required for anthocyanin synthesis but not trichome development in maize, are able to restore trichome differentiation, anthocyanin biosynthesis, tannin accumulation, and mucilage production defects in Arabidopsis ttg1 mutants (Lloyd et al., 1992; Carey et al., 2004). This makes it unlikely that distinct functions of MBW complexes evolved by alterations in the WD40 and bHLH proteins but rather may have resulted from alterations in partner proteins, like the MYB partner or other unknown proteins or in (cis-elements of) downstream genes.
Downstream genes of the Arabidopsis TTG1-GL3/EGL3-GL1 complex encode, among other things, a secondary tier of four transcription factors that includes the homeodomain protein GLABROUS2 (GL2) and the WRKY protein TTG2 (Ishida et al., 2007; Zhao et al., 2008; Morohashi and Grotewold, 2009). Inactivation of GL2 disrupts the development of trichomes on aerial tissues and converts non-hair cells (atrichoblasts) in the root epidermis into hair cells (trichoblasts) (Rerie et al., 1994; Di Cristina et al., 1996). Arabidopsis ttg2 mutants have defects in proanthocyanidin and mucilage production in seed coats and the development of trichomes (Johnson et al., 2002). In the root, TTG2 is specifically expressed in atrichoblasts and expression of a dominant-negative version of TTG2 causes ectopic root hair formation, indicating that TTG2 has a partially redundant role in the specification of atrichoblasts (Ishida et al., 2007).
In this study, we describe the cloning of petunia PH3, which is required for expression of PH5 and PH1 and vacuolar acidification in petals (Verweij et al., 2008; Faraco et al., 2014). We show that PH3 acts in part downstream of AN1, AN11, and PH4 and encodes a WRKY-type transcription factor that can interact with AN11. PH3 and Arabidopsis TTG2 appear to be orthologs and the encoded proteins are functionally exchangeable in protein interaction assays. Transgenic expression of TTG2 can rescue all defects in ph3 mutant. These new findings reveal an unexpected similarity between the regulatory circuitry that controls hair formation, proanthocyanidin deposition, and vacuolar acidification in different species.
RESULTS
Isolation of Unstable ph3 Alleles
The petunia line W138 contains an unstable an1 allele (an1W138) with a dTPH1 transposon insertion and consequently bears white flowers with red (AN1REV) revertant spots resulting from somatic excisions (Spelt et al., 2000). Because of the high number and activity of dTPH1 elements in W138, new mutations occur frequently among W138 progeny. When screening W138 progeny, we identified a family (V2068) that segregated for a revertant AN1REV allele and a novel flower color mutation, because of which the colored (AN1REV) spots in unstable an1W138 petals had a purplish rather than a red color, while in the revertant AN1REV background, the evenly colored corolla was purplish instead of red (Supplemental Figure 1A). Test crosses with the original ph3R49/R49 mutant (line R143) showed that the new mutation represented a new ph3 allele (ph3V2068) that was further maintained in line R144. Because neither ph3R49 nor ph3V2068 displayed any sign of somatic or germinal instability, it was unclear whether these alleles were tagged by transposon insertions.
To isolate unstable ph3 alleles, we performed a targeted mutagenesis experiment for which the petunia line R144 (ph3V2068/V2068) was crossed with PH3+ lines containing highly active dTPH1 elements (W138 and derivatives; see Methods). As expected, most progeny of these crosses (∼5400 plants in total) had a PH3+ phenotype, i.e., white flowers with red spots if homozygous for an1W138, or evenly red colored petals, if containing a revertant AN1REV allele. Three plants had the phenotype expected for a ph3 mutant (plants B2267-1, B2299-1, and B2219-1). Plant B2219-1 was homozygous for an1W138 and had white flowers with colored spots, but the spots were purplish rather than red, and within the purplish sectors occasional red spots could be seen, suggesting that B2219-1 contained a new ph3 allele that was somatically unstable. Plants B2267-1 and B2299-1 had evenly colored flowers (AN1REV) with a purplish color and occasional red (revertant) spots, suggesting they contained independent unstable ph3 alleles (Figure 1A; Supplemental Figure 1B). Test crosses of these three plants to homozygous ph3R49 and ph3V2068 mutants yielded mostly plants with ph3 mutant phenotype, and at low frequency germinal PH3 revertants, confirming that they harbored a new genetically unstable ph3 allele (Supplemental Figure 1).
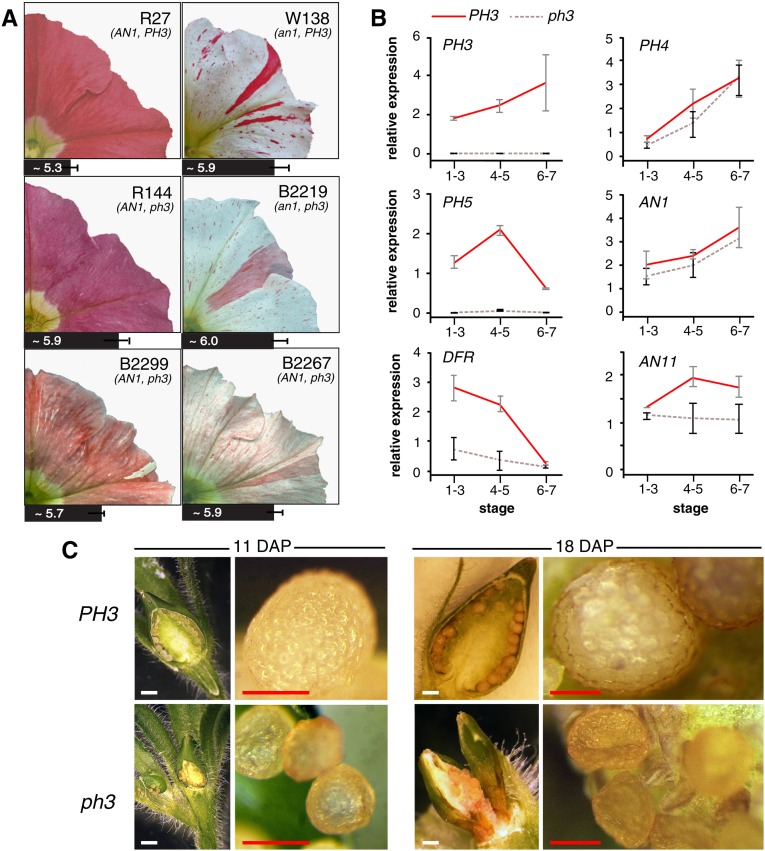
Figure 1.
Phenotype of ph3 Mutants.
(A) Petal limbs of the parental lines R27 (AN1+/+ PH3+/+) and W138 (an1m/m, PH3+/+), the derived stable recessive ph3 mutant R144 (AN1+/+ ph3V2068/V2068), and three transposon-tagged ph3 mutants B2267-1 (AN1+/m ph3V2068/B2267), B2219-1 (an1m/m ph3V2068/B2219), and B2299-1 (AN1m/Rev ph3V2068/B2299). The bars under the images represent the pH of the crude petal extract (n = 3, mean ± sd).
(B) Real-time RT-PCR analysis of mRNA from PH3, the regulatory genes, PH4, AN1, and AN11, and the structural genes PH5 and DFR in petals of lines R27 (PH3+) and R144 (ph3) from flowers of developmental stages 1 to 3 (small bud and uncolored petals), 4 to 5 (fully expanded bud and colored petals), and 6 to 7 (fully opened flower with still closed anthers and flower with dehiscent anthers). Relative expression is indicated as the mean ± sd of three biological replicates.
(C) Seedpods of a PH3 and a ph3 mutant at 11 and 18 d after pollination (DAP). White bars = 1 mm, and red bars = 0.2 mm.
Effect of ph3 Mutations on the Expression of Flower Pigmentation Genes
A characteristic of ph mutants is the reduced acidity of crude petal homogenates (de Vlaming et al., 1983). Because mutation of AN1 reduces the acidity of petal extracts in a similar way, we used progeny of the primary ph3 mutants harboring the new ph3 alleles in an AN1+ or AN1REV background to determine whether these mutations alter the pH of petal homogenates. We found that crude extracts of ph3V2068/B2267 and ph3V2068/B2219 corollas had a higher pH than those of isogenic wild types or germinal PH3 revertants (PH3V2068/B2219-Rev) (Figure 1). Petals from the ph3V2068/B2299 heterozygote (plant B2299-1) displayed a smaller pH shift of the petal extract than ph3B2267/V2068 or ph3B2219/V2068. This indicates that ph3B2299 is a weak allele and may explain why these petals are a mosaic of red and purplish cells and have a “marbled” appearance (Figure 1A).
The genetic backgrounds of the petunia lines R27 and R143, in which the ph3 alleles are maintained, contain mutations that block 5′ hydroxylation (hf1 hf2) and rhamnosylation (rt) of the anthocyanins and all subsequent modification steps (5-glucosylation, acylation, and methylation) (Supplemental Table 1). In these R27 and R143 backgrounds, ph3 had only a small effect on the amount and chemical composition of anthocyanins (Supplemental Figure 2). Thin-layer chromatography of hydrolyzed anthocyanins showed that PH3 and ph3 petals contain mostly cyanidins, consistent with the hf1 hf2 rt genotype. PH3 petals yielded in addition a small (trace) amount of peonidins, while these were reduced in ph3 siblings. HPLC analysis identified four major anthocyanin peaks (P1-P4) in PH3 petals and a minor compound (P5). In ph3 petals, we observed similar amounts of the anthocyanins in P1 to P4, whereas the minor compound in P5 was clearly reduced (Supplemental Figure 2).
Real-time RT-PCR analysis of ph3V2068 and PH3+ petals (Figure 1B) revealed that ph3 had little or no effect on the expression of regulatory anthocyanin genes (AN1 and AN11) or the regulatory PH4 gene. However, ph3V2068 almost completely abolished expression of PH5, which is consistent with previous results (Verweij et al., 2008) and can account for the reduced acidity of ph3 petal homogenates. Unexpectedly, expression of DFR, a structural anthocyanin gene encoding DIHYDROFLAVONOL REDUCTASE, was reduced 3- to 4-fold, but not abolished, in ph3 petals. DFR expression is apparently not rate-limiting for anthocyanin biosynthesis, since mutations (an6) that abolish DFR expression are fully recessive (Huits et al., 1994), which may explain why the reduced DFR expression in ph3 has little or no effect on the anthocyanin biosynthesis.
Taken together, these data show that PH3 is, together with AN1, AN11, and PH4, essential for expression of genes involved in vacuolar acidification. Although PH3 enhances DFR expression ∼3- to 4-fold, it is, unlike AN1 and AN11, not essential for DFR expression or anthocyanin biosynthesis.
Effect of ph3 on Seed Development
Petunia lines with mutations in PH3 or PH5 were reported to be female sterile (Wiering, 1974; de Vlaming et al., 1984). Although the inbred ph3 and ph5 lines that were used by these authors had been discontinued in the 1980s, we were able to recover and germinate seeds containing the same ph3 and ph5 alleles from F2 crosses with unrelated lines. With the same ph5 allele in a new genetic background, we did not observe sterility of ph5 mutants. In ph5 mutants that arose by dTPH1 insertions in the W138 background, we also observed no infertility (Verweij et al., 2008). However, the original ph3R49 allele in the new background (line R143), as well as the ph3V2068, ph3B2219, and ph3B2267 alleles in the W138 background, all caused complete female sterility. The ph3 mutation did not block fertilization or early stages of seed development, as after pollination of a ph3 flower with PH3 pollen the seedpod enlarged and contained immature seeds at 11 and 18 d after pollination (Figure 1C). However, compared with fruits on PH3 plants, those on ph3 plants remained smaller and individual seeds were also much smaller than seeds on a PH3 plant. Around 18 d after pollination, when seeds on PH3 plants were firm spheres, seeds on ph3 plants had a collapsed appearance. About 4 weeks after pollination, fruits and seeds on PH3 plants were mature, whereas seeds on ph3 plants had completely disintegrated leaving no other trace than some brownish “dust.” Strikingly, this effect of ph3 was suppressed in an an1 background. In the unstable an1W138 background in which ph3V2068 was isolated, an1W138 ph3V2068 female parents were fully fertile, while the numerous (>10) independent AN1REV ph3V2068 revertants that we tested over the years were without exception fully female sterile and displayed the seed abortion phenotype described above.
Molecular Isolation of the PH3 Locus
Because most petunia mutants isolated in W138 resulted from insertions of the high copy number transposon dTPH1 (van Houwelingen et al., 1998), we assumed that the three new ph3 alleles were tagged by dTPH1 insertions and could be used to identify the locus by transposon display.
The test cross of plant B2299-1 (ph3V2068/B2299) to line R144 (ph3V2068/V2068) yielded a family (C2124) that segregated five plants with purplish flowers (ph3V2068/V2068), 14 with purplish flowers containing a few red revertant spots (ph3V2068/B2299), and two with red flowers (PH3V2068/B2299-REV) that originated from germinal PH3 reversions (Supplemental Figure 1C). We visualized dTPH1 flanking sequences in a subset of these plants by transposon display and identified two fragments that were amplified from all seven unstable mutants (ph3V2068/B2299), but not from any of the three stable mutants or the two revertants (Supplemental Figure 3A). Sequencing revealed that these two fragments had the same 8-bp target site duplication and therefore represented the left and right flanking sequence of the same dTPH1 element.
To assess whether this fragment originated from PH3, we used fragment-specific primers to amplify this dTPH1 insertion in family C2124. With these primers we were not able to amplify the stable ph3V2068 allele, whereas all plants harboring the unstable ph3B2299 allele yielded a fragment that was ∼300 bp larger than the fragment amplified from wild-type progenitors (PH3+) or a derived revertant (PH3B2299REV) (Supplemental Figure 3B). Moreover, ph3V2068/B2219 and ph3V2068/B2267 plants also contained an ∼300-bp dTPH1 insertion in the same genomic region. Analysis of genomic and cDNA clones revealed that these three insertions disrupted a single gene, consisting of six exons, and that the alleles ph3B2219 and ph3B2267 harbor dTPH1 insertions in exon three, 9 bp apart, while ph3B2299 contains a dTPH1 insertion in the fourth exon (Figure 2A).
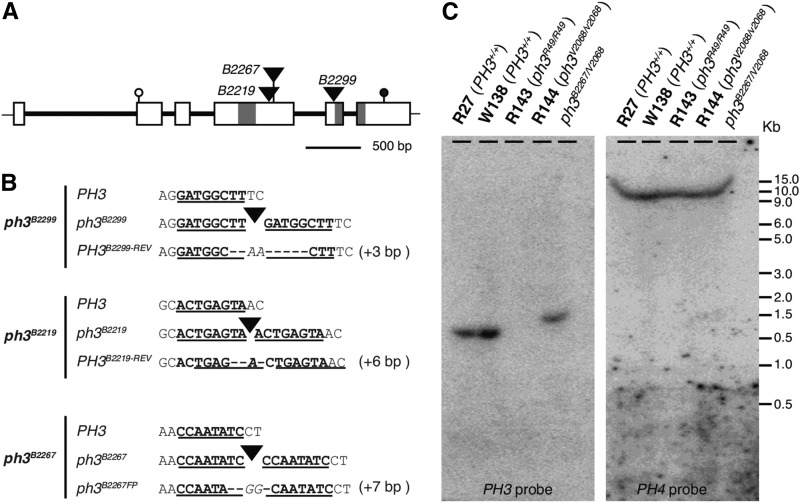
Figure 2.
Analysis of PH3 and Mutant Alleles.
(A) Structure of PH3 and mutant alleles. Exons and introns are indicated by rectangles and thick lines, start and stop codons by open and close circles, and dTPH1 insertions in three distinct ph3 alleles by triangles. Regions encoding two WRKY domains are indicated by gray shading
(B) Sequence alterations in three distinct transposon-tagged ph3 alleles and derived excision alleles. For each insertion allele, the sequence of the progenitor wild-type allele is shown above the sequence of the excision alleles. Target site duplication is indicated by boldface font and underlining. Nucleotides that were deleted during excisions are indicated by dashes and extra nucleotides that were inserted in italics.
(C) DNA gel blot analysis of the PH3 and PH4 loci in different genotypes. The blot was first hybridized with a cDNA PH3 probe spanning nucleotide 404 to 1407 and subsequently stripped and rehybridized with the full-length PH4 cDNA as a control for DNA loading.
Most progeny from backcrosses of plants B2219-1 (ph3V2068/B2219) and B2299-1 (ph3V2068/B2299) to stable ph3R49 (line R143) or ph3V2068 mutants (line R144) had purplish (ph3) flowers, whereas a few rare individuals had red flowers indicating that a (germinal) reversion of the ph3 mutation occurred (Supplemental Figure 1). The appearance of these revertant PH3B2219Rev and PH3B2299Rev alleles coincided with the excision of dTPH1 from the identified gene and the formation of a 6- and 3-bp footprint, respectively, that restored the reading frame (Figure 2B). We did not find germinal revertants among the few progeny of ph3B2267 that we grew, although we found that in one individual the dTPH1 element had excised from ph3B2267. However, this excision allele (ph3B2267FP) contained a 7-bp footprint that disrupted the reading frame of the gene, explaining why excision in this case did not cause phenotypic reversion (Figure 2C). Given that the occurrence of three ph3 alleles and subsequent reversions fully coincided with the insertion or excision of dTPH1 elements, we concluded that the identified gene is PH3.
We used various combinations of primers complementary to distinct parts of PH3 but none of them could amplify fragments from ph3R49 and ph3V2068 alleles. DNA gel blot analysis showed that a PH3 cDNA probe, spanning nucleotides 404 to 1407, detected a 2.1-kb HindIII fragment in R27 (PH3+/+) and W138 (PH3+/+). In ph3V2068/B2267 heterozygote, the probe recognized a single fragment that was slightly larger (∼2.4 kb) due to the dTPH1 insertion, but in ph3R49 and ph3V2068 homozygotes no hybridizing PH3 fragments were detected (Figure 2B). This indicates that ph3R49 and ph3V2068 are null alleles in which two independent deletions eliminated most, if not all, of the coding sequence and an unknown amount of flanking sequence.
PH3 Encodes a WRKY Protein
The cDNA of PH3 (1848 bp) contains an open reading frame of 1407 bp, encoding a predicted protein of 469 amino acids. PH3 has two WRKY domains, each consisting of a conserved WRKYGQK motif followed by a 16-amino acid zinc finger motif. These features are characteristic for the plant-specific family of WRKY transcription factors (Eulgem et al., 2000).
The Arabidopsis genome encodes some 74 WRKY proteins that can be divided in three subgroups based on the number of WRKY domains and the type of zinc finger motifs. WRKY proteins of groups II and III contain one WRKY domain with a C2-H2 or C2-HC zinc finger motif, respectively. Group I proteins have two WRKY domains containing either a C2-H2 zinc finger motif (subgroup Ia) or C2-HC motifs (subgroup Ib) (Xie et al., 2005). According to this classification, PH3 belongs to subgroup Ia (Figure 3A). Phylogenetic analysis of WRKY proteins, based on an alignment of the C-terminal WRKY domain, shows that PH3 clusters with group I WRKY proteins such as WRKY44/TTG2 from Arabidopsis, which is encoded by the TTG2 locus (Johnson et al., 2002) (Supplemental Figure 4).
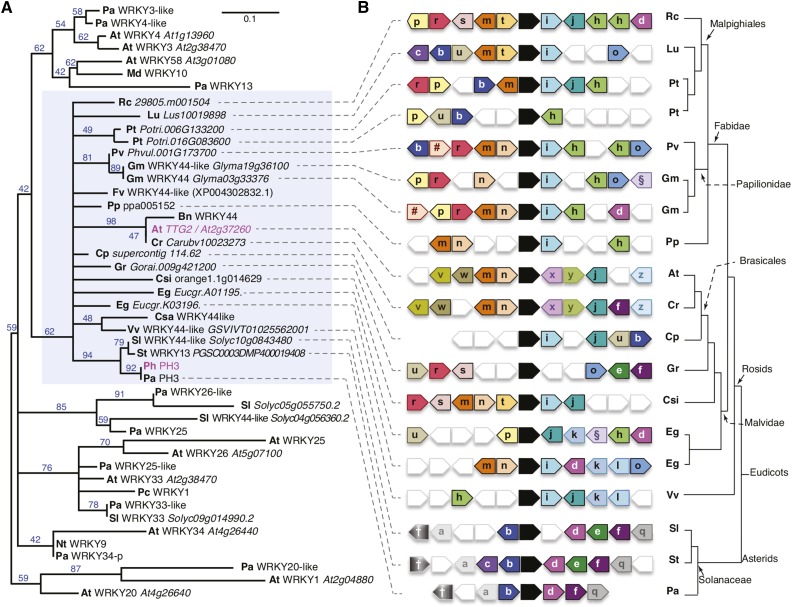
Figure 3.
PH3 Is a WRKY Protein and Homologous to Arabidopsis TTG2.
(A) Phylogenetic tree of a selection of type I WRKY proteins. Blue numbers on tree branches indicate percentage of bootstrap support (1500 replicates). The blue shaded rectangle indicates the clade containing TTG2/PH3 homologs. Prefixes denote the species of origin: Arabidopsis thaliana (At), Brassica napus (Bn), Capsella rubella (Cr), Carica papaya (Cp), Citrus sinensis (Csi), Cucumis sativum (Csa), Eucalyptus grandis (Eg), Fragaria vesca (Fv), Glycine max (Gc), Gossypium raimondii (Gr), Hordeum vulgare (Hv), Ipomea batatas (Ib), Linum usitatissimum (Lu), Malus domestica (Md), Nicotiana tabacum (Nt), Petroselinum crispum (Pc), Petunia axillaris (Pa), Petunia hybrida (Ph), Phaseolus vulgaris (Pv), Populus trichocarpa (Pt), Prunus persica (Pp), Ricinus communis (Rc), Solanum lycopersicum (Sl), Solanum tuberosum (St), and Vitis vinifera (Vv).
(B) Diagram showing relationships between genes immediately surrounding putative TTG2/PH3 homologs from distinct species inferred by Phytozome (Goodstein et al., 2012). Analysis of the P. axillaris region was done manually using unpublished sequence data from the petunia platform. The arrows denote genes and direction of transcription. Black arrows indicate PH3/TTG2 homologs. Similarity among flanking genes is indicated by similar colors and symbols/letters. Relationships of the different (groups of) species of origin are indicated on the right.
To examine the relationship of petunia PH3 with Arabidopsis TTG2 and other WRKY proteins in more detail, we performed a detailed phylogenetic analysis of type I WRKY proteins in which we included the 10 predicted petunia proteins most similar to TTG2 and the most similar predicted proteins from additional species. This analysis showed that PH3, TTG2, and related proteins from other species formed a single well defined clade, whereas the other petunia and Arabidopsis WRKY proteins grouped in distinct clades (Figure 3A). The similarity of WRKY proteins from distinct clades was confined to the two WRKY domains. However, in proteins of the PH3/TTG2 clade, similarity extended into the N-terminal domain. The same was seen for At-WRKY4, Pa-WRKY4-like, At-WRKY58, and Pa-WRKY13, strengthening the idea that these proteins are orthologous (Supplemental Figure 5).
Next, we used the Phytozome database (Goodstein et al., 2012) to compare a window of 10 genes surrounding the putative PH3/TTG2 homologs in species with known genome sequences (Figure 3B). The immediate surroundings of the PH3/TTG2 homologs from related species had several genes in common, although complete similarity was never found, even between regions containing (duplicated) PH3/TTG2 paralogs in the same species. In comparisons of more distantly related species, the number of similar genes in the examined region decreased, as might be expected. All Rosids had, besides the PH3/TTG2 homolog, at least two genes in common, the Brassicaceae Arabidopsis and Capsella rubella being the most different. The Asterids tomato (Solanum lycopersicum) and potato (Solanum tuberosum) are closely related to petunia, all belonging to the Solanaceae, and distantly related to Rosids. Despite the large evolutionary distance, tomato and potato had multiple genes in common with several Rosid species, although their similarity with Brassicaceae in this region was low (one gene in common with C. rubella and none with Arabidopsis).
Taken together, these findings indicate that PH3 and TTG2 might originate from a common ancestor and are likely to be orthologous.
Expression Pattern and Genetic Regulation of PH3
We used real-time RT-PCR to analyze the pattern and genetic regulation of PH3 mRNA expression in various tissues of petunia line R27. Although R27 is an an4 mutant (mutated in a regulator of anthocyanin biosynthesis in anthers), it contains functional alleles for all other regulatory AN loci and PH genes that govern the pigmentation of petals. In R27, PH3 was expressed in petals, mostly in the petal limb and to a lesser extent in the petal tube, and ovaries, but not in anthers, leaves stems, and sepals (Figure 4A). After pollination, PH3 mRNA expression persisted in the developing seedpod. In situ hybridization on petal sections showed that PH3 was primarily expressed in the upper epidermis of the corolla (Figure 4B), similar to AN1, PH4, and the structural genes PH5 and DFR (Quattrocchio et al., 2006; Verweij et al., 2008). However, because the PH3 mRNA signal in the epidermis was just above the background, we could not exclude that PH3 was also expressed, albeit at somewhat lower levels, in the mesophyll.
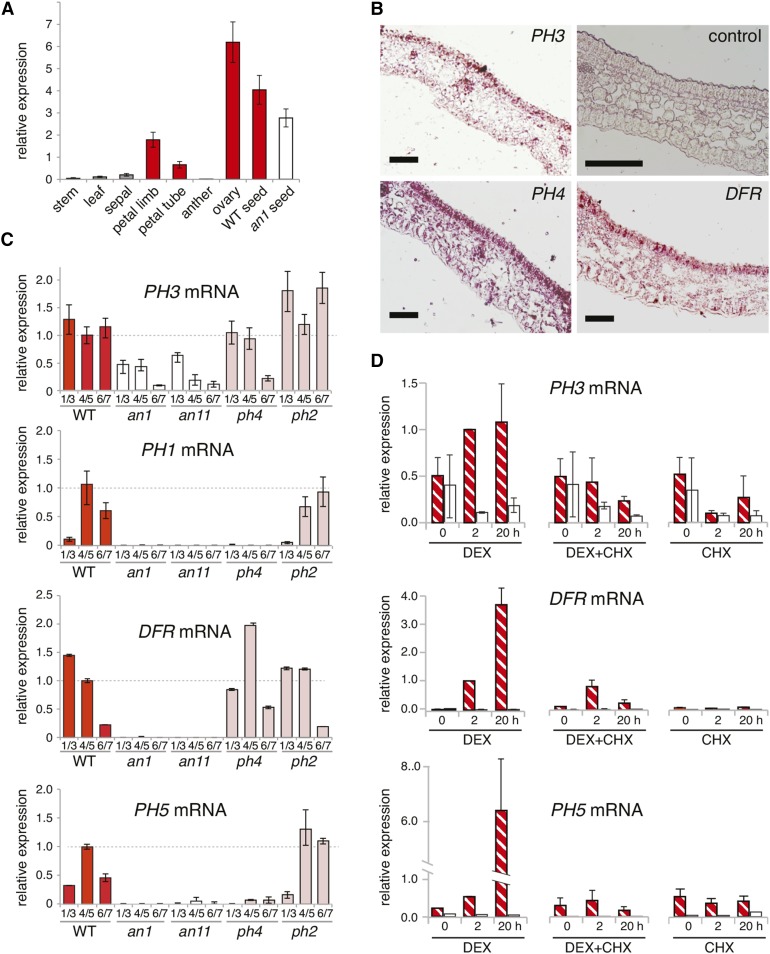
Figure 4.
Expression Pattern and Genetic Regulation of PH3.
(A) Real-time RT-PCR analysis of PH3 mRNA in organs from stage 5-7 flowers (stage 5, fully elongated bud still closed; stage 6, almost fully open flower; stage 7, fully open flower with dehiscent anthers) of the wild-type line R27 and seeds from R27 and the isogenic an1 line W225. GAPDH mRNA served as a constitutive control.
(B) In situ hybridization of PH3, DFR, and AN1 in petals. The negative control was the DFR sense probe.
(C) Real-time RT-PCR of DFR, PH3, and PH5 mRNAs at different flower developmental stages in petals of the wild-type line R27 (red bar) and isogenic lines with an1, an11 (white bars), ph4, or ph2 mutations (purplish bars).
(D) Real-time RT-PCR analysis of PH3 mRNA in the petal limbs of detached stage 4 flowers of an an1 mutant (white bars) and an1 p35S:AN1-GR transgenic line (hatched bars) that were treated for 0, 2, or 24 h with DEX and/or cycloheximide (CHX).
To determine how PH3 expression is regulated, we analyzed PH3 mRNA in petals of distinct mutants in the W138 background. We found that PH3 mRNA was expressed at comparable levels in wild-type and ph2 petals but was clearly reduced in an11, an1, and ph4 petals especially in later flower stages (Figure 4C). To examine the regulation of PH3 by AN1 in more detail, we used an1 plants that contained a transgene in which the p35S promoter drove the expression of a fusion of AN1 and the ligand binding domain of the rat glucocorticoid receptor (p35S:AN1-GR). p35S:AN1-GR completely restores DFR, PH1, and PH5 mRNA expression and pigmentation in an1 petals, but only in the presence of dexamethasone (DEX; Figure 4D), similar to previous results (Spelt et al., 2000; Verweij et al., 2008; Faraco et al., 2014). In p35S:AN1-GR petals, PH3 mRNA was expressed at a low level at stage 4-5, comparable to isogenic an1 control petals. Application of DEX to an1 p35S:AN1-GR buds increased PH3 mRNA expression within 2 h, which was due to posttranslational activation of AN1-GR, as it was not seen in an1 petals lacking the transgene (Figure 4D). Because an1 reduced the expression of PH3 much less than it did DFR or PH5, the fold induction for PH3 upon DEX treatment was lower than for DFR and PH5. In the presence of 50 µM cycloheximide, protein biosynthesis in p35S:AN1-GR petals is fully blocked (Spelt et al., 2000). Under these conditions, DFR expression remained inducible by DEX for a short period (∼2 h), suggesting that AN1 activates DFR directly. However, after a prolonged exposure to cycloheximide and DEX (20 h), DFR expression declined again to background levels (Figure 4D). This is apparently due to the decay of AN1-GR, which appears less stable than AN1, and complete turnover of DFR mRNA in 20 h (Spelt et al., 2000). In the presence of DEX and cycloheximide, PH3 mRNA remained approximately constant within the first 2 h, whereas PH3 mRNA quickly decayed in the absence of AN1-GR (Figure 4D). These results suggest that the induction of PH3 by AN1-GR was direct and did not require biosynthesis of intermediate regulators.
PH3 and TTG2 Are Functionally Homologous
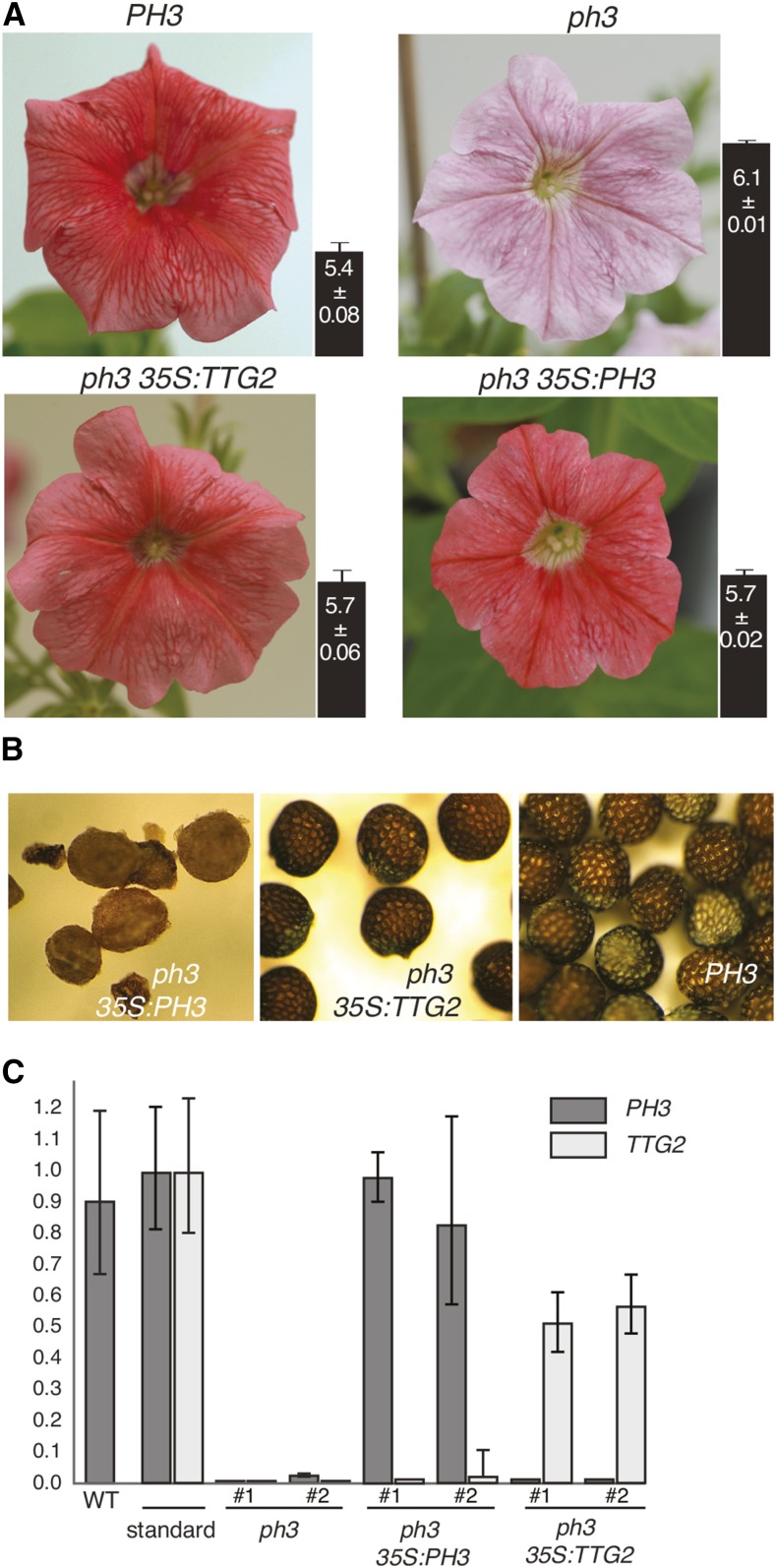
The finding that PH3 and TTG2 are homologous was unexpected, given their very different mutant phenotypes. Arabidopsis ttg2 mutants have aberrant trichomes on the leaf surface and reduced tannin accumulation in the seed coat and lack mucilage production in germinating seeds (Johnson et al., 2002). Petunia ph3 mutants do not show any effect in trichome development, but it is unclear whether PH3 is involved in tannin accumulation since ph3 homozygous seeds abort 2 to 3 weeks after pollination (Figure 1). To examine whether the function of PH3 in petunia and TTG2 in Arabidopsis diverged through changes in the encoded proteins or the downstream network, we performed a mutant complementation study. We cloned the PH3 cDNA or a genomic DNA fragment containing the TTG2 coding sequence behind the p35S promoter and transformed both constructs (p35S:PH3 and p35S:TTG2) into a transformable petunia ph3 mutant that was transheterozygous for the deletion allele ph3R49 (deletion allele) and ph3B2267FP (frame shift allele) (cf. Figure 2). The petals of this transformation host contained, compared with the wild type, a slightly reduced amount of (nonfunctional) ph3B2267FP transcripts and strongly reduced amount of PH5 mRNA (Supplemental Figure 6) and had the typical purplish color (Figure 5A). In 20 out of 21 p35S:PH3 transformants and two out of four p35S:TTG2 transformants, the transgene rescued the ph3 flower color defect and lowered the pH of the crude petal extracts, although it did not reach the same level as in PH3 flowers (Figure 5A). The expression of PH5 mRNA was also rescued, consistent with the flower phenotype, whereas no clear effect was seen on the amount of RNA from the ph3B2267 endogene (Supplemental Figure 6). The latter result suggests that the reduced level of ph3B2267FP mRNA compared with wild-type PH3 transcripts results from increased instability of the mutant mRNA, rather than from reduced transcription and activity of the PH3 promoter.
Figure 5.
Expression of p35S:PH3 and p35S:TTG2 in Petunia.
(A) Flowers of PH3 wild type, ph3 mutant, and ph3 mutants complemented by p35S:PH3 or p35S:TTG2. The pH values of the crude petal extract are indicated by the bars next to the pictures (mean ± sd, n = 4).
(B) Seeds of ph3 p35S:PH3 and ph3 p35S:TTG2 transformants with red petals and a PH3+ line.
(C) Real-time RT-PCR analysis of the expression of the PH3 endogene, p35S:PH3, and p35S:TTG2 in four transgenic lines with red flowers (shown in [A]). The standards for PH3 and TTG2 were obtained by spiking leaf cDNA with 1 fg (10−15 g) of a PH3 or TTG2 cDNA fragment, respectively.
In addition, p35S:PH3 and p35S:TTG2 also restored female fertility. Whereas all seeds on ph3 plants die and pulverize during late stages of seed development, we could recover viable seeds from ph3 p35S:PH3 and p35S:TTG2 female parent plants. Self-fertilization of ph3 p35S:PH3 plants resulted in seed capsules containing a low number of seeds, which had an abnormal seed coat that consisted of smaller cells with a lighter color than wild-type seeds (Figure 5B). However, self-fertilization of ph3 p35S:TTG2 plants resulted in seedpods filled with an almost wild-type number of seeds, having a normal seed coat.
Real-time RT-PCR analyses showed that in petal limbs of the two p35S:PH3 petunia lines with the highest transgene expression, the amount PH3 mRNA originating from the transgene was comparable to that produced by the endogene in wild-type petals (Figure 5C). Because mRNAs from PH3 originated mostly from epidermal cells, whereas the PH3 RNAs expressed from the 35S:PH3 transgene originated from both epidermal and mesophyll cells, we infer that within epidermal cells the p35S:PH3 transgene produces less RNA than the endogenous PH3 gene, which explains the partial complementation of the petal homogenate pH. To compare the amount of TTG2 mRNA expressed in the petunia transformants with PH3 mRNA, we used a standard consisting of leaf cDNA spiked with an equal amount of PH3 and TTG2 cDNA. This revealed that in petal limbs of the two complemented lines p35S:TTG2 was expressed at a similar level as p35S:PH3 in complemented mutants.
Together, these data show that Arabidopsis TTG2 can fully replace its petunia homolog PH3 in petunia, indicating that both proteins are functionally very similar.
PH3 and TTG2 Interact with the WD40 Proteins AN11 and TTG1
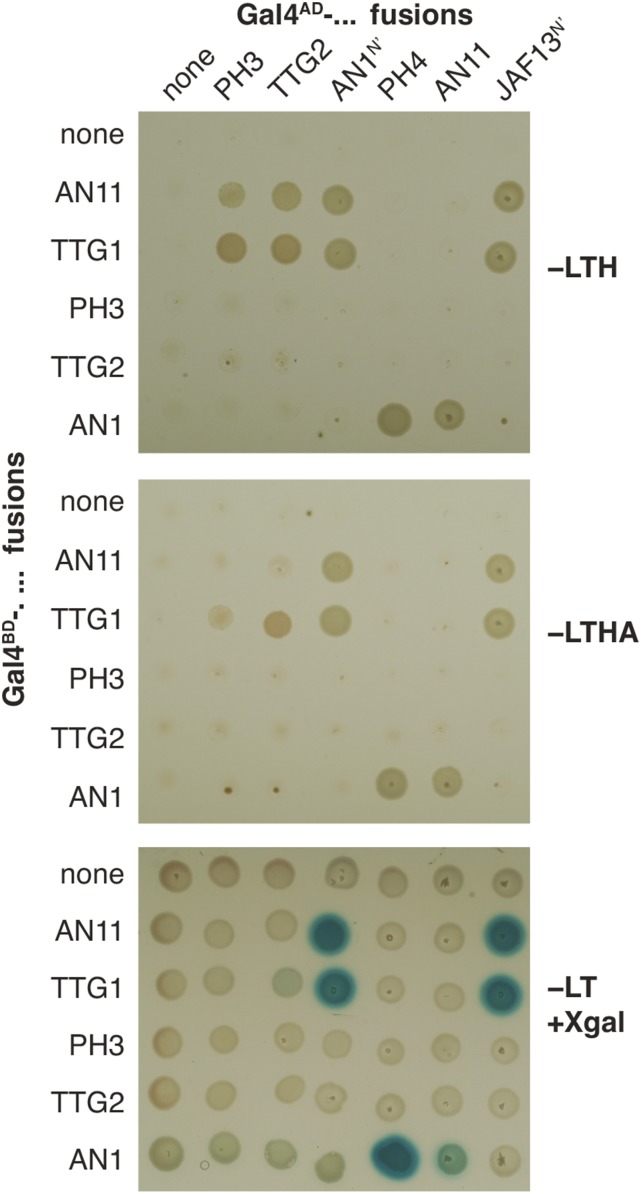
To find out whether PH3 or TTG2 can interact with known components of the petunia and Arabidopsis MBW complexes, we performed yeast two-hybrid assays. Figure 6 shows that the petunia bHLH protein AN1 interacted via its N-terminal domain with PH4, consistent with previous results (Quattrocchio et al., 2006). In addition, we observed that AN1 and the functionally related petunia bHLH protein JAF13 could interact, again via their N-terminal domain, with the petunia WD40 protein AN11 and the Arabidopsis homolog TTG1. The bHLH-MYB and WD40-bHLH interactions of these petunia factors are analogous to those reported previously for related Arabidopsis WD40, bHLH, and MYB proteins (Payne et al., 2000; Bernhardt et al., 2003; Baudry et al., 2004; Zimmermann et al., 2004).
Figure 6.
Interaction of PH3 and TTG2 with WD40 Proteins.
Yeast strains expressing the indicated GAL4AD and GAL4BD fusions were grown on dropout media lacking leucine (−L), tryptophan (−T), histidine (−H), and/or adenine (−A). Activation of the GAL4-regulated HIS and ADE reporter genes is seen as growth on –LTH and –LTHA media and of the lacZ gene as a blue staining after overlaying the cells with X-Gal.
We found that PH3 and TTG2 could interact in yeast with AN11 and TTG1 to activate the HIS3 reporter gene. Pesch et al. (2014) recently reported very similar results for TTG1 and TTG2 and confirmed direct binding of TTG1 and TTG2 in vitro. The WD40-WRKY interactions triggered a much weaker yeast two-hybrid response, being barely if at all detectable, with the more stringent and less sensitive ADE and LacZ reporter genes, than the interactions of the same WD40 proteins with AN1 or JAF13 (Figure 6). Such differences might stem from a difference in the strength of various interactions or may simply reflect differences in the stability and steady state levels of the interacting proteins in yeast. Fusion of AN11 or TTG2 to GAL4BD (or GAL4AD) strongly reduced the steady state level of GAL4BD in yeast cells to below the detection limit, whereas the GAL4BD fragment expressed from the empty vector was easily detectable on immunoblots (Supplemental Figure 7).
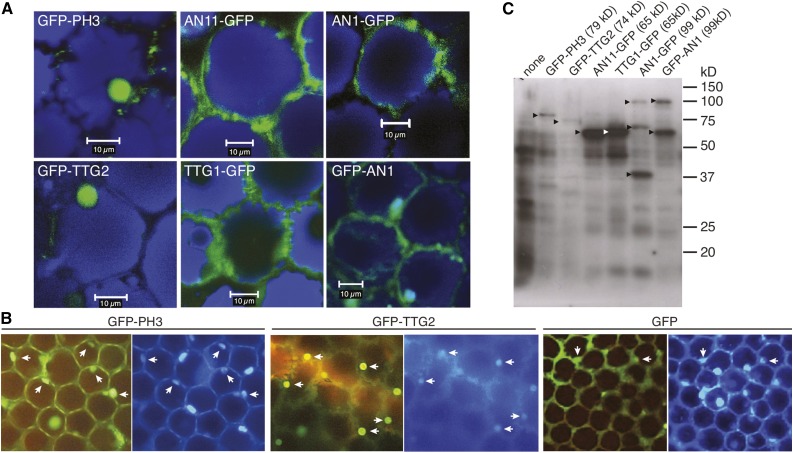
To study the significance of WD40-bHLH and WD40-WRKY interactions observed in yeast, we examined whether these interactions also take place in the epidermal petal cells where they are normally active. First, we analyzed the intracellular localization of the single proteins by agro-infecting intact wild-type petunia petals with p35S-driven transgenes encoding GFP fusions. Within cells of the adaxial epidermis, we observed fluorescence of PH3-GFP and TTG2-GFP in a ball-shaped structure, which was identified as the nucleus because it was also labeled with the DNA stain DAPI (4′,6-diamidino-2-phenylindole) (Figures 7A and 7B). Expression of AN11-GFP and TTG1-GFP labeled the cytoplasm (Figure 7A). This result is consistent with previous cell fractionation data, which showed that most AN11 protein in petunia petals is cytoplasmic (de Vetten et al., 1997). Surprisingly, expression of AN1-GFP as well GFP-AN1 also labeled primarily the cytoplasm.
Figure 7.
Cellular Localization of PH3 and TTG2 Compared with AN1 and AN11.
(A) Confocal laser scanning micrographs of agroinfected wild-type petunia petals expressing the indicated GFP fusions. GFP fluorescence is shown in green, and fluorescence of anthocyanins in blue. Bars = 10 μm.
(B) Fluorescence micrographs of agroinfected petunia petals expressing the distinct GFP fusions, after staining with DAPI. White arrowheads show that the DAPI fluorescence corresponds to the dots of GFP fluorescence from the PH3 and TTG2 constructs and confirms that these WRKY proteins localize in the nucleus.
(C) Immunoblot analysis of GFP fusion proteins expressed in agroinfected petunia petals detected with anti-GFP. Arrowheads indicate the expected molecular weight for the different fusion products.
We noticed previously that many GFP fusion proteins are cleaved, at least in petunia, because of which the separated fragments (GFP and the tagged protein) may accumulate in different cellular compartments (Quattrocchio et al., 2013). Because mutant complementation proved an unreliable means to exclude such artifacts (Quattrocchio et al., 2013; Faraco et al., 2014), we used immunoblot analysis to verify the integrity and reliability of GFP fusion proteins. We found that the GFP-PH3, GFP-TTG2, AN11-GFP, and TTG1-GFP fusions that accumulated in agroinfected petals had the expected size, indicating that little or no cleavage occurred (Figure 7C). However, petals expressing AN1-GFP accumulated GFP fusion proteins of ∼100 kD, as expected for the full size fusion, as well as two smaller proteins of ∼70 and ∼40 kD, and petals expressing GFP-AN1 accumulated the full size ∼100-kD protein along with a fragment of ∼60 kD. These findings indicate that some of the AN1 proteins with a GFP tag on the N terminus (GFP-AN1) or on the C terminus (AN1-GFP) are cleaved within the AN1 sequence.
To study interactions of the WD40 proteins with AN1, PH3, and TTG2 within petal cells, we used bimolecular fluorescence complementation (BiFC). For these experiments, we used petal protoplasts, because cotransformation of these cells with multiple constructs is nearly 100% efficient (i.e., both constructs are found in nearly all cells that are transformed) and because these retain the gene expression and protein sorting characteristics (Faraco et al., 2011). We cotransformed petal protoplasts with a transgene encoding a fusion of RFP and the Arabidopsis plasma membrane SNARE protein SYP122 (Assaad et al., 2004), to mark transformed cells including those in which no BiFC occurred, and different combinations of transgenes encoding fusions of PH3, TTG2, or AN1 to the N-terminal half of YFP (nYFP) and fusions of AN11 or TTG1 to the C-terminal half of YFP (cYFP). Since the position and relative orientation of nYFP and cYFP tags in protein pairs can profoundly affect BiFC efficiency (Bracha-Drori et al., 2004), we coexpressed in the first experiments multiple combinations of the petunia protein fusions with different orientations of the cYFP and nYFP tags.
Cotransformation of protoplasts with constructs expressing nYFP-AN1, AN11-cYFP, and cYFP-AN11 or fusions with a different arrangement of tags (AN1-cYFP, AN11-nYFP, and nYFP-AN11) caused YFP fluorescence in >90% of the cells expressing the transformation marker RFP-SYP122 (Supplemental Figure 8). Subsequent experiments showed that YFP fluorescence resulted from coexpression of nYFP-AN1 and AN11-cYFP (Figure 8), whereas coexpression of nYFP-AN1 and cYFP-AN11 yielded no detectable YFP fluorescence (Supplemental Figure 8). No interaction was detectable when we replaced nYFP-AN1 with free nYFP or cYFP-AN11 with free cYFP, even though >50% of the protoplasts expressed the transformation marker RFP-SYP122 (Figure 8). This indicates that even though nYFP and cYFP are likely to localize in the cytoplasm and nucleus of petal cells, like free GFP (Verweij et al., 2008), they do not cause any noticeable background fluorescence due to nYFP-cYFP self-assembly under the conditions used. The finding that the interaction of AN1 and AN11 fusions was orientation dependent, since coexpression cYFP-AN1 with AN11-nYFP or nYFP-AN11 did not cause YFP fluorescence (Supplemental Figure 8), also indicates that nYFP-cYFP self-assembly did not result in noticeable background fluorescence.
Figure 8.
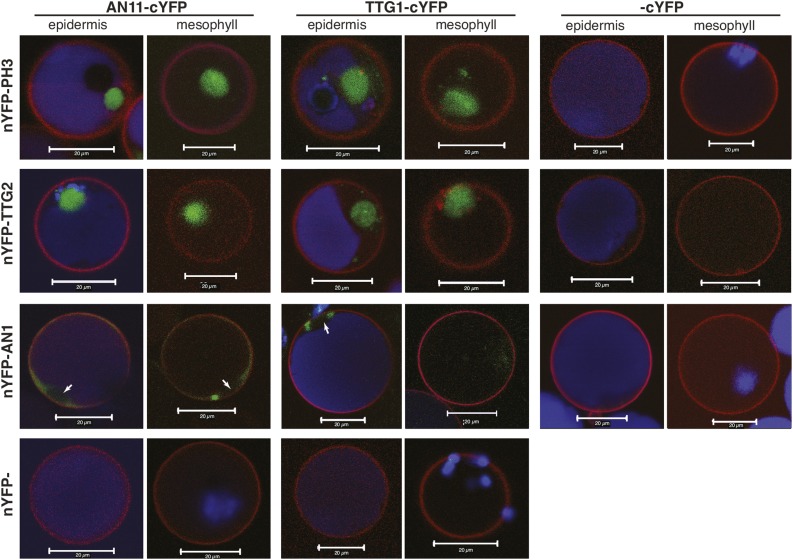
BiFC Showing Interaction of WD Proteins with bHLH and WRKY Partners.
Images are confocal laser scanning micrographs of petunia petal protoplasts derived from the epidermis, which contains anthocyanins (blue signal), or from the mesophyll with different combinations of the cYFP and nYFP fusions. Transformed cells were marked with a third construct, expressing the plasma membrane marker RFP-AtSYP122 (red signal). Bars = 20 μm.
Detailed confocal microscopy analysis of some 20 cells expressing different combinations of AN11 and AN1 fusions showed that in all cases the YFP fluorescence localized in the cytoplasm close to the nucleus but was not detectable within the nucleus (Figure 8; Supplemental Figure 8). Moreover, the observed interactions were similar, in terms of intensity and intracellular localization, in purple protoplasts originating from the epidermis and in colorless protoplasts derived from the mesophyll, indicating that it occurs independent from other epidermis-specific factors (e.g., other regulators of the anthocyanin and pH pathways). Replacement of AN11-cYFP with the homologous TTG1-cYFP fusion did not obviously alter the frequency, intensity, or intracellular localization of the resulting YFP fluorescence, indicating that Arabidopsis TTG1 and petunia AN11 are interchangeable regarding their capacity to interact with AN1 (Figure 8).
Transformation of petal protoplasts with 35S:nYFP-TTG2 or 35S:nYFP-PH3 in combination with 35S:AN11-cYFP or 35S:TTG1-cYFP caused a strong BiFC signal in all transformed cells marked by RFP-SYP122 (Figure 8). Even though untagged cYFP is thought to be present in both cytoplasm and nuclei of petal cells, like free GFP, no BiFC was seen upon coexpression with nYFP-PH3 or nYFP-TTG2 (Figure 8), and coexpression of PH3 and AN11 fusions with different orientations of cYFP and nYFP tags did not cause BiFC fluorescence either. This result indicates that the BiFC seen in cells coexpressing nYFP-PH3 or nYFP-TTG2 with AN11-cYFP or TTG1-cYFP results from the interaction of the WRKY and WD40 moieties rather than self-assembly of the cYFP and nYFP tags. Interestingly, the complexes of AN11 with PH3 or TTG2 were confined to the nuclei, both in colored (epidermal) and white (mesophyll) protoplasts (Figure 8), even though the bulk of AN11-GFP localized in the cytoplasm (Figure 7). This suggests that a small proportion of AN11 protein is nuclear, where it binds to PH3 or TTG2, or that AN11 protein reaches the nucleus only when in a complex with the WRKY factor.
These results show that PH3 and TTG2 can interact with the WD40 proteins AN11 and TTG1, indicating that that petunia PH3 and Arabidopsis TTG2 are also in this respect interchangeable and that both act as coregulators of homologous MBW complexes.
DISCUSSION
We report that PH3 of petunia encodes a WRKY transcription factor that together with the PH4-AN1-AN11 MBW complex activates downstream genes in multiple distinct pathways involved in flower pigmentation and seed development. These findings shed light on the mechanism by which MBW complexes can independently activate distinct downstream pathways and how these complexes acquired different functions during evolution.
The ph3 phenotype indicates that PH3 activates multiple processes in petunia that depend on at least partially different target gene sets. PH3 promotes the acidification of vacuoles in the petal epidermis primarily via the activation of PH5 and PH1, which encode two interacting P-ATPases in the tonoplast, since expression of PH1 and PH5 from a transgene is sufficient to rescue the pH defect of ph3 (Verweij et al., 2008; Faraco et al., 2014). In addition, PH3 is required for the stable storage of anthocyanins in the vacuole, as in genetic backgrounds containing a dominant FADING (FA) allele, ph3 triggers the disappearance of anthocyanins and consequent fading of the petal color after bud opening (de Vlaming et al., 1982). Fading is most pronounced in genotypes that accumulate peonidins, petunidins, and malvidins, which in petunia bear a 5-glucose and acylated 3-rutinoside group, while the simple cyanidin 3-glucosides that accumulate in the R27 and R143 backgrounds used here are much less sensitive to fading (de Vlaming et al., 1982). Given that R27 and R143 are FA+, the small effect of ph3 on petal anthocyanin content, i.e., the reduction of the small amount of peonidin and one anthocyanin (peak 5, Supplemental Figure 2), is most likely due to the reduced stability of these compounds (fading), rather than decreased biosynthesis. Because ph2 and ph5 mutations do not trigger fading in an FA background, fading of ph3 FA petals is not attributable to the altered vacuolar pH alone and apparently depends on the misregulation of distinct PH3 target genes that remain to be identified (Quattrocchio et al., 2006).
Furthermore, PH3 has a role in seed development that is at present poorly understood. Mutation of ph5 leads to a loss of tannins in the seed coat, most likely because ph5 decreases the proton gradient across the tonoplast that drives the vacuolar sequestration of tannin precursors via the H+-antiporter TT12 (Marinova et al., 2007; Verweij et al., 2008) but does not cause sterility. Hence, the female sterility of petunia ph3 mutants does not result from reduced expression of PH5 and vacuolar acidification, but apparently results from misregulation of a distinct pathway.
Curiously, mutations in AN1 fully suppress the female sterility of ph3. AN1 and AN11 are required in maternal tissues to suppress cell divisions in the seed coat epidermis and to activate DFR expression and proanthocyanidin biosynthesis (Huits et al., 1994; Spelt et al., 2000; Zenoni et al., 2011), but whether the downregulation of these processes or yet another unknown pathway is responsible for the rescued fertility of ph3 an1 mutants remains unclear. Genetic data indicate that both PH3 and AN1 are required in maternal rather than embryonic tissues, as pollination of a ph3 female parent with PH3 pollen does not restore fertility, while seeds on a heterozygous PH3/ph3 female parent that contain a ph3 embryo develop normally, and seeds on an an1 female parent exhibit a mutant (seed coat) phenotype, irrespective of the genotype of the embryo. Because of the low PH3 and AN1 expression levels and the characteristics of the tissue, it is difficult to assess AN1 and PH3 expression patterns in developing seeds in detail by in situ hybridization. Unraveling the molecular basis of the sterility caused by ph3 will require the identification of the downstream pathways that are affected by PH3 and AN1 in this tissue.
Position and Function of PH3 in the Regulatory MBW Network
The finding that PH3 mRNA expression is reduced in an1, an11, and ph4 mutants and can be induced by DEX in an1 35S:AN1-GR petals indicates that PH3 is yet another target gene of the AN11-AN1-PH4 complex. Importantly, an1 and an11 mutations abolish the expression of downstream genes involved in anthocyanin biosynthesis (e.g., DFR) and vacuolar acidification (PH1 and PH5) almost completely, whereas PH3 mRNA expression is reduced only ∼3-fold in mature buds and open flowers, with the strongest effect in later stages. This suggests that PH3 is activated in parallel by a second pathway that is AN1- and AN11-independent and predominantly active in younger buds. PH3 expression in seeds is also partially regulated by AN11, as a PH3-derived EST (PETIN034019) was some 3- to 5-fold downregulated in an11 seeds compared with the wild type (Zenoni et al., 2011), and by AN1 (Figure 4), whereas DFR and other anthocyanin genes are more strongly reduced (Huits et al., 1994; Zenoni et al., 2011).
Experiments with an1 p35S:AN1-GR flowers showed that AN1 acts as direct activator of the PH3 target genes PH1 and PH5 (Verweij et al., 2008; Faraco et al., 2014). This implies that PH3 does not act as an intermediate regulator in a simple linear pathway in which AN1 and AN11 activate PH3, which in turn activates (possibly together with PH4) PH5. Moreover, such a linear pathway does not account for the observed interaction of AN11 with both PH3 and AN1. Therefore, we propose a different model in which PH3 functions both as an intermediate target gene and as a cofactor of the AN1-AN11-PH4 complex that aids in the transcriptional activation of PH5 (Figure 9). As PH3 expression continues in an1 petals, though at a reduced level, DEX treatment can rapidly (and directly) induce (in <2 h) PH5 expression in an1 35S:AN1-GR petals even when protein biosynthesis is fully blocked (Verweij et al., 2008).
Figure 9.
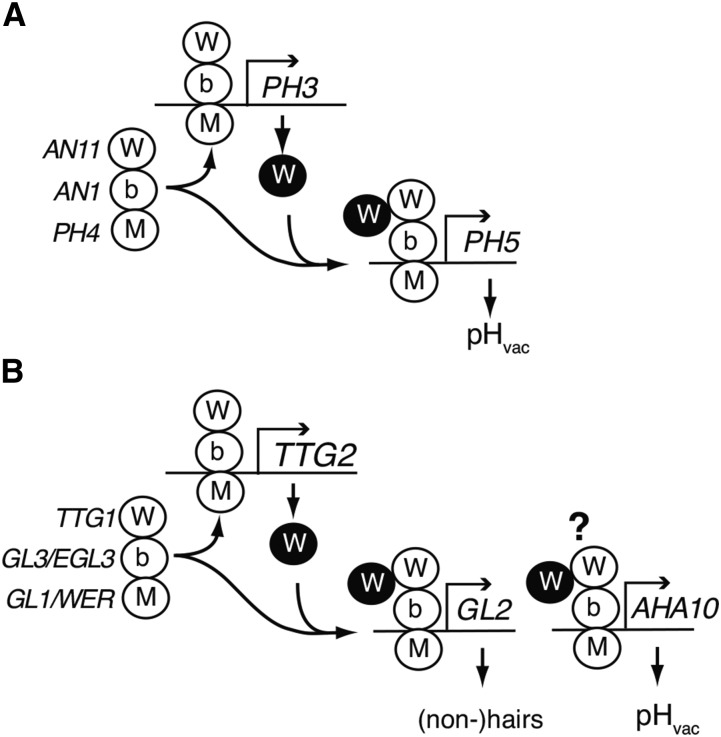
Position of PH3 and TTG2 in the Regulatory Circuitries That Control Vacuolar Acidification and Hair Development.
(A) Regulatory circuit containing PH3 in petunia.
(B) Regulatory circuit containing TTG2 in Arabidopsis.
Complexes of MYB, bHLH, and proteins are shown as white ovals, marked with M, b, and W, respectively; the encoding genes are indicated on the left in italics. WRKY proteins are indicated as a black oval, marked W. The question mark indicates that regulation of AHA10 is inferred but not experimentally proven.
We infer that the reduced amount of transcripts expressed by the frame-shift mutant ph3B2267 results from increased mRNA turnover (perhaps by nonsense-mediated decay) rather than reduced transcription due to the absence of PH3 protein because expression of active PH3 or TTG2 from a transgene does not alter the ph3B2267 mRNA level. Therefore, it is unlikely that PH3 is also required in an autoregulatory loop for the AN1-AN11-PH4-mediated activation of its own promoter.
Functional Divergence of MBW Complexes
It is thought that a single WD40 protein (AN11 or TTG1) and the largely redundant bHLH proteins (AN1 and JAF13 in petunia; GL3, EGL3, and TT8 in Arabidopsis) can independently activate different downstream pathways because they interact with distinct, less pleiotropic, MYB partners with divergent expression patterns and specificity for downstream genes (Koes et al., 2005; Ramsay and Glover, 2005). Our results show that interactions with other transcription factors, such as the WRKY proteins PH3 and TTG2, also contribute to the specificity of MBW complexes because ph3 abolishes expression of PH1, PH5, and at least eight other AN1-AN11-PH4-regulated genes of unknown function almost completely (Verweij, 2007; Verweij et al., 2008; Faraco et al., 2014) but reduces DFR expression only three fold and has no clear effect on anthocyanin accumulation. This resembles recent findings in Arabidopsis showing that ttg2 strongly reduces the expression of some MBW-regulated trichome patterning genes, such as TRYPTICHON (TRY), whereas other genes, such as CAPRICE and GL2, are reduced only moderately (Pesch et al., 2014). Given that TTG2 can bind to TTG1 and to sites in the TRY promoter, the role of TTG2, and by extension PH3, might be to specifically enhance MBW binding to a subset of target promoters that contain WRKY binding sites over promoters lacking such sites (Pesch et al., 2014).
The activation of anthocyanin biosynthesis by MBW complexes is an ancient process that is widely conserved among angiosperms and relies on highly similar MYB proteins. The finding that blue flowering soybean (Glycine max) mutants have lesions in the apparent PH4 homolog (Takahashi et al., 2013) and that AHA10 is homologous and functionally interchangeable with PH5 (Appelhagen et al., 2015) suggests that the role of MBW complexes in vacuolar acidification is also widespread. The role of the MBW complex in specification of trichome or atrichoblast fate, by contrast, was established later in evolution as it is seen in some Rosid species only (Arabidopsis) and not in dicot species belonging to the Asterids or in monocots (Serna and Martin, 2006).
The observation that PH3 and TTG2 are orthologs reveals an unexpected similarity between the circuitry that regulates hair development in Arabidopsis and vacuolar acidification in petunia. PH3 and TTG2 not only encode highly similar interchangeable proteins, but also operate in a very similar feed-forward regulatory loop (Figure 7). Expression of TTG2 is, like PH3, regulated by the MBW complex (Ishida et al., 2007; Zhao et al., 2008). Furthermore, the interaction of TTG2 with TTG1 suggests that TTG2 is, like PH3, involved in the activation of MBW target genes (Figure 7). This is consistent with results showing that GL2 and TRY are direct targets of the TTG1-GL3-GL1 complex and that GL2 and TRY expression is coregulated by TTG2 (Ishida et al., 2007; Zhao et al., 2008; Morohashi and Grotewold, 2009; Pesch et al., 2014).
Although the modulation of extracellular pH is important for the development of root hairs (Bibikova et al., 1998), there is no evidence that the regulation of vacuolar pH is required for root hair formation. Hence, the pathway(s) influenced by TTG2 in the Arabidopsis leaf or root epidermis may have little similarity with those influenced by PH3 in petunia petals. The role of TTG2 in seed pigmentation, however, is likely to share similarities with the role of PH3 in petal pigmentation. The lack of tannins in ttg2 seeds initially suggested that TTG2 is required to activate genes involved in tannin biosynthesis, such as BANYULS (BAN) or genes acting later (Johnson et al., 2002). However, even though BAN expression is dependent on a MBW complex, it does not seem to require TTG2 (Debeaujon et al., 2003). Although we cannot exclude that TTG2 is required for the activation of genes acting after BAN in the tannin biosynthetic pathway, we favor a different role for TTG2 in tannin accumulation that is more similar to the role of PH3 in petunia. PH3 promotes vacuolar acidification via activation of PH5, which encodes a tonoplast H+ pump that is also required for tannin accumulation in the seed (Verweij et al., 2008). Because a mutation in the Arabidopsis PH5 homolog AHA10 also abolishes tannin accumulation in the seed (Baxter et al., 2005; Appelhagen et al., 2015), just like ttg2, it is likely that TTG2 enables tannin accumulation, at least in part, by activating AHA10.
Given that proanthocyanidins are found in most angiosperms and more ancestral plants (gymnosperms and ferns), such a role for TTG2 and PH3 in driving vacuolar acidification is probably the ancestral function, whereas the role of TTG2 and the associated MBW complex in trichome development was established much later, most likely after the separation of Rosids and Asterids (Serna and Martin, 2006). Presumably, this involved the acquisition of TTG2-responsive elements by downstream genes involved in hair development, but direct evidence for this idea will have to await the identification of such genes.
METHODS
Plant Material
Genotypes of petunia (Petunia hybrida) lines and details of an1 and ph3 alleles used are shown in Supplemental Tables 1 and 2. They all derive from the Amsterdam petunia collection and were grown in a greenhouse at VU University (Amsterdam) with supplemental artificial lighting (cycles of 16 h light and 8 h darkness) at a (minimum) temperature of 22°C or higher.
To isolate dTPH1-tagged ph3 alleles, we crossed R144 (ph3V2068) to PH3+ line W138 or derived lines with unstable mutations in AN11 (line W137), AN10 (W237), or EXTRAPETALS (EXP) and identified three individuals with an unstable ph3 phenotype among ∼5400 progeny. Test crosses with R143 (ph3R49) confirmed these plants to be ph3 mutants and heterozygous for an11, an10, or exp, excluding that they resulted from contaminations. ph3 lines were maintained by fertilization of PH3/ph3 heterozygotes with pollen from ph3/ph3 homozygous siblings and selection of PH3/ph3 heterozygotes (red flowers) and ph3 homozygotes (purplish flowers) from the progeny.
Analysis of Anthocyanins and pH of Crude Petal Extracts
For each pH measurement, the corolla of a fresh open flower was ground in a mortar with 6 mL water, and the pH of the extract was immediately measured with a pH electrode (Checker; Hanna Instruments). Measurements were repeated on three to five different flowers and the average value and se were calculated. Thin-layer chromatography and HPLC analysis of anthocyanins was done as described previously (van Houwelingen et al., 1998; Quattrocchio et al., 2006).
DNA and RNA Methodology
Transposon display was performed as described before (Tobeña-Santamaria et al., 2002) using primers shown in Supplemental Table 3. Gels were exposed overnight and read by a phosphor imager (Molecular Dynamics).
The PH3 cDNA 3′-end was obtained by screening a R27 petal cDNA library (Quattrocchio et al., 2006), and the 5′-end was amplified by 5′ rapid amplification of cDNA ends (5′/3′-RACE KIT 2nd generation; Roche) using PH3-specific primers shown in Supplemental Table 4.
Extractions of DNA and total RNA were performed as described previously (de Vetten et al., 1997). Real-time RT-PCR analysis was performed with an iTaq Universal Syber Green kit (Bio-Rad) using primers listed in Supplemental Table 5 and an ECO P1180 real-time PCR cycler with Eco version 4.0 software (Illumina). Quantitative RT-PCR was performed as described previously (Quattrocchio et al., 1998) using gene-specific primers (Supplemental Table 6) and a reduced number of cycles, to ensure linear amplification, and detection of amplification products by hybridization of DNA gel blots.
For DNA gel blot analysis, genomic DNA was digested with HindIII, size-separated on an agarose gel, blotted to Hybond-N membrane, and hybridized overnight at 65°C with a 32P-labeled PH3 probe (nucleotides 404 to 1407). The blot was washed with 2× SSC + 0.1% SDS at 60°C for 30 min, exposed overnight, and read by a phosphor imager (Molecular Dynamics). Subsequently the blot was stripped and rehybridized with a PH4 cDNA probe.
RNA in situ hybridization was performed as described before (Souer et al., 1996). Probes were prepared by in vitro transcription using T7 polymerase (PH3 and PH4 probes) or SP6 polymerase (DFR probe) on templates that were PCR amplified from cloned cDNA fragments in pGEM-Teasy with a T7 or SP6 promoter primer and a gene-specific forward primer (Supplemental Table 7) or in case of the control DFR sense probe, a gene-specific reverse primer. The PH3 probe included nucleotides 404 to 1407 (containing both WRKY and zinc finger motifs). Because anthocyanins could mask the reddish hybridization signal, we used petals in which anthocyanin biosynthesis was blocked by a mutation in AN3, encoding the F3′H enzyme (van Houwelingen et al., 1998; Quattrocchio et al., 2006).
Phylogenetic and Synteny Analysis
Sequence alignments were generated with Multiple Sequence Comparison by Log Expectation (MUSCLE), and phylogenetic trees were constructed with maximum likelihood (PhyML) after curation with G-blocks to remove badly aligning regions, using online tools at http://phylogeny.lirmm.fr (Dereeper et al., 2008). To generate a phylogenetic tree of WRKY proteins from different subfamilies (Supplemental Figure 4), we used manually selected protein fragments spanning the C-terminal WRKY domain plus 10 upstream amino acids for alignments with MUSCLE and generation of a tree with PhyML. The alignments used to generate the threes are available as Supplemental Data Sets 1 and 2.
Synteny was analyzed using the Web-based Phytozome V9 platform (Goodstein et al., 2012). For petunia, we used unpublished sequence data from the petunia platform (www.http://flower.ens-lyon.fr/petuniaplatform/petuniaplatform.html). We compared an annotated 0.95 Mb scaffold (Peaxi162Scf00472), which contained PH3 of Petunia axillaris, and performed tBLASTn searches of flanking petunia gene sequences against the genomes of related Solanaceae (tomato [Solanum lycopersicum] and potato [Solanum tuberosum]) and vice versa by tBLASTn searches of genes flanking the tomato and potato PH3 homologs to the P. axillaris genome.
Gene Constructs and Transgenic Lines
The full-length PH3 cDNA coding sequence was amplified from R27 petal cDNA (stage 5-6) with primers #1295 and #2254 and the TTG2 coding sequence from Arabidopsis thaliana Columbia genomic DNA with primers #2224 and #1391 (see Supplemental Table 8 for primer sequences). Both fragments were digested with XbaI and BamHI and subsequently ligated into the pGreen1H vector (XbaI/BamHI) (Hellens et al., 2000).
Plasmids were introduced, together with helper plasmid pJIC, into Agrobacterium tumefaciens strain AGL0 by electroporation. A single colony was used for an overnight culture and used to infect leaf discs. Shoot regeneration was achieved in the presence of 10 μg/mL hygromycin. As lines in the R27 and R143 background are difficult to transform, we crossed a ph3V2068/B2267FP plant in the R27 background (cf. Supplemental Figure 1E) to a PH3+/R49 individual in the R143 background and used transheterozygous ph3R49/B2267FP progeny as a transformation host.
Yeast Two-Hybrid Analysis
Yeast two-hybrid analysis was done as described (Quattrocchio et al., 2006). Constructs expressing fusion of the GAL4BD or GAL4AD domain to full-size PH4, AN1, JAF13, or the N-terminal 238 amino acids were described before (Quattrocchio et al., 2006); plasmids expressing fusions of PH3, TTG2, and TTG1 were generated in the same way, using cDNAs amplified from petunia petals (PH3) or Arabidopsis leaves (TTG1 and TTG2) with primers listed in Supplemental Table 9.
Transient Expression of GFP Fusions and Split-YFP Constructs
Full-size coding sequences of PH3, TTG2, and TTG1 were amplified with primers containing attB1 and attB2 sites. These fragments were recombined with BP clonase in the entry vector pDONR P1-P2 (Invitrogen). Full-size coding sequence of AN11 was amplified with primers containing Topo sites and cloned in pENTR/D Topo (Invitrogen), and AN1 was amplified with primers containing NcoI and EcoRI restriction sites and cloned in pENTR4 (Invitrogen). All primers are shown in Supplemental Table 10. For expression of GFP fusions, coding sequences were transferred by Gateway recombination into pK7FWG2.0 or pK7WGF2.0 (Hellens et al., 2000) or, for expression of nYFP and cYFP fusions, into plasmids pGWnY (AB626693), pnYGW (AB626694), pGWcY (AB626696), and/or pcYGW (AB626696) (Hino et al., 2011).
Transient transformation of intact petals by agroinfection and transformation of petal protoplasts was described previously (Verweij et al., 2008; Faraco et al., 2014). Imaging of DAPI-stained cells was done using a Zeiss fluorescence microscope; all other fluorescence micrographs were obtained using a confocal laser scanning microscope, as described before (Verweij et al., 2008). In BiFC assays, plasmids encoding nYFP and cYFP fusions were cotransformed with a construct (35S:RFP-SYP122) that expresses the plasma membrane marker RFP-SYP122 (Assaad et al., 2004) to label transformed cells and to interpret negative BiFC results. Typically >50% of the protoplasts were transformed. For each combination of constructs, several hundred cells were examined for RFP or BiFC expression by normal fluorescence microscopy, and 5 to 10 cells were imaged by confocal microscopy.
Immunoblot Analysis
Immunoblot analysis was performed as described (de Vetten et al., 1997; Verweij et al., 2008) using anti-GAL4BD (Clontech; catalog number 630403, lot number 1107146A) or anti-GFP antibodies (Santa Cruz Biotechnology; catalog number sc-8334, lot number L1803).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers KU761265 (PH3 gene) and KU641009 (PH3 cDNA). For the protein alignments and phylogenetic analyses we used the following WRKY protein sequences: from Arabidopsis (At): AtWRKY4 (At1g13960). AtWRKY6 (At1g62300), AtWRKY9 (At1g68150), AtWRKY8 (At5g46350), AtWRKY11 (At4g31550), AtWRKY7 (At4g24240), AtWRKY14 (At1g30650), AtWRKY13 (At4g39410), AtWRKY18 (At4g31800), AtWRKY20 (At4g26640), AtWRKY23 (At2g47260), AtWRKY24 (At5g41570), AtWRKY34 (At4g26440), AtWRKY35 (At2g34830), AtWRKY39 (At3g04670), AtWRKY42 (At4g04450), AtWRKY46 (At2g46400), AtWRKY53 (At4g23810), AtWRKY44 (At2g37260), AtWRKY62 (At5g01900), AtWRKY67 (At1g66550), AtWRKY69 (At3g58710), and AtWRKY70 (At3g56400). From parsley (Pc; Petroselinum crispum) we used PcWRKY1 (AAC49527), PcWRKY3 (AAD27591.1), PcWRKY4 (AAR98818), from tobacco WIZZ (AB042973.1), from barley SUSIBA2 (AY323206.1), from Ipomoea batatas (Ib) SPF1 (D30038.1), from Solanum lycopersicum (Sl) WRKY44-like (XM_004249754.1), from Populus trichocarpa (Pt) WRKY protein (XM_002326290), from Fragaria vesca Vesca (Fv) WRKY44-like (XM_004302784.1), from Malus x domestica (Md) WRKY10 (HM122713.1), from Cucumis sativus (Cs) WRKY44-like (XM_004137750.1), and from Nicotiana tabacum (Nt) WRKY9 (AB063576).
Supplemental Data
Supplemental Figure 1. Isolation and genetic characterization of ph3 mutants.
Supplemental Figure 2. Anthocyanin content of PH3 and ph3 petals.
Supplemental Figure 3. Identification of the PH3 gene.
Supplemental Figure 4. PH3 is a type I WRKY protein with similarity to Arabidopsis TTG2.
Supplemental Figure 5. Alignment of PH3 with homologs from other species and closely related WRKY proteins.
Supplemental Figure 6. Expression p35S:PH3 and p35S:TTG2 transgenes rescues PH5 mRNA expression in ph3 mutants.
Supplemental Figure 7. Immunoblot analysis of yeast two-hybrid strains.
Supplemental Figure 8. Interaction of AN1 and AN11 in petunia petal protoplasts.
Supplemental Table 1. Petunia lines and genotypes used.
Supplemental Table 2. AN1, AN2, PH2, PH4, and PH3 alleles used.
Supplemental Table 3. Primers used for transposon display.
Supplemental Table 4. Primers used for RACE amplification of full-length PH3 cDNA.
Supplemental Table 5. Primer number and sequence used for real-time RT-PCR.
Supplemental Table 6. Primers and number of cycles used for RT-PCR analysis.
Supplemental Table 7. Primers used to generate templates for synthesis of RNA probes.
Supplemental Table 8. Primers used to generate 35S:PH3 and 35S:TTG2.
Supplemental Table 9. Primer number and sequence used for yeast two-hybrid cloning.
Supplemental Table 10. Primer number and sequence used for GFP and split-YFP constructs.
Supplemental Data Set 1. Sequence alignment used to generate the phylogenetic tree in Figure 3.
Supplemental Data Set 2. Sequence alignment used to generate the phylogenetic tree in Supplemental Figure 4.
Supplementary Material
Acknowledgments
We thank Pieter Hoogeveen, Martina Meesters, and Daisy Kloos for plant care, David Smyth for useful exchange of information, Erik Manders, Ronald Breedijk, Linda Joosen, and Dorus Gadella of the Centre for Advanced Microscopy “van Leeuwenhoek,” Section Molecular Cytology, Swammerdam Institute for Life Sciences, University of Amsterdam for the use of their facilities and technical assistance, and Tsuyoshi Nakagawa for the gift of split YFP vectors. This research was supported by the Dutch Technology Foundation STW (Grant VGC.6717), which is part of the Netherlands Organization for Scientific Research (NWO), and which is partly funded by the Ministry of Economic Affairs, and by an EMBO short-term fellowship to M.F.
AUTHOR CONTRIBUTIONS
All authors performed experimental work, discussed results, and commented on the article. W.V., F.M.Q., and R.K. wrote the article.
Glossary
- DEX
dexamethasone
- DAPI
4′,6-diamidino-2-phenylindole
- BiFC
bimolecular fluorescence complementation
References
- Albert N.W., Lewis D.H., Zhang H., Schwinn K.E., Jameson P.E., Davies K.M. (2011). Members of an R2R3-MYB transcription factor family in petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 65: 771–784. [DOI] [PubMed] [Google Scholar]
- Appelhagen I., Nordholt N., Seidel T., Spelt K., Koes R., Quattrochio F., Sagasser M., Weisshaar B. (2015). TRANSPARENT TESTA 13 is a tonoplast P3A -ATPase required for vacuolar deposition of proanthocyanidins in Arabidopsis thaliana seeds. Plant J. 82: 840–849. [DOI] [PubMed] [Google Scholar]
- Assaad F.F., Qiu J.L., Youngs H., Ehrhardt D., Zimmerli L., Kalde M., Wanner G., Peck S.C., Edwards H., Ramonell K., Somerville C.R., Thordal-Christensen H. (2004). The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15: 5118–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkunde R., Bouyer D., Hülskamp M. (2011). Nuclear trapping by GL3 controls intercellular transport and redistribution of TTG1 protein in Arabidopsis. Development 138: 5039–5048. [DOI] [PubMed] [Google Scholar]
- Baudry A., Heim M.A., Dubreucq B., Caboche M., Weisshaar B., Lepiniec L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39: 366–380. [DOI] [PubMed] [Google Scholar]
- Baxter I.R., Young J.C., Armstrong G., Foster N., Bogenschutz N., Cordova T., Peer W.A., Hazen S.P., Murphy A.S., Harper J.F. (2005). A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102: 2649–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt C., Lee M.M., Gonzalez A., Zhang F., Lloyd A., Schiefelbein J. (2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130: 6431–6439. [DOI] [PubMed] [Google Scholar]
- Bibikova T.N., Jacob T., Dahse I., Gilroy S. (1998). Localized changes in apoplastic and cytoplasmic pH are associated with root hair development in Arabidopsis thaliana. Development 125: 2925–2934. [DOI] [PubMed] [Google Scholar]
- Borevitz J.O., Xia Y., Blount J., Dixon R.A., Lamb C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha-Drori K., Shichrur K., Katz A., Oliva M., Angelovici R., Yalovsky S., Ohad N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40: 419–427. [DOI] [PubMed] [Google Scholar]
- Broun P. (2005). Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 8: 272–279. [DOI] [PubMed] [Google Scholar]
- Carey C.C., Strahle J.T., Selinger D.A., Chandler V.L. (2004). Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana. Plant Cell 16: 450–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vetten N., Quattrocchio F., Mol J., Koes R. (1997). The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 11: 1422–1434. [DOI] [PubMed] [Google Scholar]
- de Vlaming P., Cornu A., Farcy E., Gerats A.G.M., Maizonnier D., Wiering H., Wijsman H.J.W. (1984). Petunia hybrida: A short description of the action of 91 genes, their origin and their map location. Plant Mol. Biol. Rep. 2: 21–42. [Google Scholar]
- de Vlaming P., Schram A.W., Wiering H. (1983). Genes affecting flower colour and pH of flower limb homogenates in Petunia hybrida. Theor. Appl. Genet. 66: 271–278. [DOI] [PubMed] [Google Scholar]
- de Vlaming P., van Eekeres J.E.M., Wiering H. (1982). A gene for flower colour fading in Petunia hybrida. Theor. Appl. Genet. 61: 41–46. [DOI] [PubMed] [Google Scholar]
- Debeaujon I., Nesi N., Perez P., Devic M., Grandjean O., Caboche M., Lepiniec L. (2003). Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell 15: 2514–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel O. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36: W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristina M., Sessa G., Dolan L., Linstead P., Baima S., Ruberti I., Morelli G. (1996). The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 10: 393–402. [DOI] [PubMed] [Google Scholar]
- Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5: 199–206. [DOI] [PubMed] [Google Scholar]
- Faraco M., Di Sansebastiano G.P., Spelt K., Koes R.E., Quattrocchio F.M. (2011). One protoplast is not the other! Plant Physiol. 156: 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraco M., et al. (2014). Hyperacidification of vacuoles by the combined action of two different P-ATPases in the tonoplast determines flower color. Cell Reports 6: 32–43. [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Zhao M., Leavitt J.M., Lloyd A.M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53: 814–827. [DOI] [PubMed] [Google Scholar]
- Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., Rokhsar D.S. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40: D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832. [DOI] [PubMed] [Google Scholar]
- Hichri I., Barrieu F., Bogs J., Kappel C., Delrot S., Lauvergeat V. (2011). Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 62: 2465–2483. [DOI] [PubMed] [Google Scholar]
- Hino T., Tanaka Y., Kawamukai M., Nishimura K., Mano S., Nakagawa T. (2011). Two Sec13p homologs, AtSec13A and AtSec13B, redundantly contribute to the formation of COPII transport vesicles in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 75: 1848–1852. [DOI] [PubMed] [Google Scholar]
- Huits H.S.M., Gerats A.G.M., Kreike M.M., Mol J.N.M., Koes R.E. (1994). Genetic control of dihydroflavonol 4-reductase gene expression in Petunia hybrida. Plant J. 6: 295–310. [DOI] [PubMed] [Google Scholar]
- Ishida T., Hattori S., Sano R., Inoue K., Shirano Y., Hayashi H., Shibata D., Sato S., Kato T., Tabata S., Okada K., Wada T. (2007). Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 19: 2531–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Kurata T., Okada K., Wada T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59: 365–386. [DOI] [PubMed] [Google Scholar]
- Johnson C.S., Kolevski B., Smyth D.R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14: 1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes R., Verweij W., Quattrocchio F. (2005). Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10: 236–242. [DOI] [PubMed] [Google Scholar]
- Lee M.M., Schiefelbein J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483. [DOI] [PubMed] [Google Scholar]
- Lepiniec L., Debeaujon I., Routaboul J.M., Baudry A., Pourcel L., Nesi N., Caboche M. (2006). Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57: 405–430. [DOI] [PubMed] [Google Scholar]
- Lloyd A.M., Walbot V., Davis R.W. (1992). Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258: 1773–1775. [DOI] [PubMed] [Google Scholar]
- Marinova K., Pourcel L., Weder B., Schwarz M., Barron D., Routaboul J.M., Debeaujon I., Klein M. (2007). The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+ -antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell 19: 2023–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K., Grotewold E. (2009). A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet. 5: e1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N., Debeaujon I., Jond C., Pelletier G., Caboche M., Lepiniec L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12: 1863–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N., Jond C., Debeaujon I., Caboche M., Lepiniec L. (2001). The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13: 2099–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passeri V., Koes R., Quattrocchio F.M. (2016). New challenges for the design of high value plant products: stabilization of anthocyanins in plant vacuoles. Front. Plant Sci. 7: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne C.T., Zhang F., Lloyd A.M. (2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch M., Dartan B., Birkenbihl R., Somssich I.E., Hülskamp M. (2014). Arabidopsis TTG2 regulates TRY expression through enhancement of activator complex-triggered activation. Plant Cell 26: 4067–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni K., Tonelli C. (2011). Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 181: 219–229. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F., Verweij W., Kroon A., Spelt C., Mol J., Koes R. (2006). PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 18: 1274–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F., Wing J., van der Woude K., Souer E., de Vetten N., Mol J., Koes R. (1999). Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11: 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F., Wing J.F., Leppen H., Mol J., Koes R.E. (1993). Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 5: 1497–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F., Wing J.F., van der Woude K., Mol J.N.M., Koes R. (1998). Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 13: 475–488. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F.M., Spelt C., Koes R. (2013). Transgenes and protein localization: myths and legends. Trends Plant Sci. 18: 473–476. [DOI] [PubMed] [Google Scholar]
- Ramsay N.A., Glover B.J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10: 63–70. [DOI] [PubMed] [Google Scholar]
- Rerie W.G., Feldmann K.A., Marks M.D. (1994). The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8: 1388–1399. [DOI] [PubMed] [Google Scholar]
- Serna L., Martin C. (2006). Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci. 11: 274–280. [DOI] [PubMed] [Google Scholar]
- Souer E., van Houwelingen A., Kloos D., Mol J., Koes R. (1996). The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85: 159–170. [DOI] [PubMed] [Google Scholar]
- Spelt C., Quattrocchio F., Mol J.N., Koes R. (2000). anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12: 1619–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelt C., Quattrocchio F., Mol J., Koes R. (2002). ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms. Plant Cell 14: 2121–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi R., Yamagishi N., Yoshikawa N. (2013). A MYB transcription factor controls flower color in soybean. J. Hered. 104: 149–153. [DOI] [PubMed] [Google Scholar]
- Tobeña-Santamaria R., Bliek M., Ljung K., Sandberg G., Mol J.N., Souer E., Koes R. (2002). FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev. 16: 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houwelingen A., Souer E., Spelt K., Kloos D., Mol J., Koes R. (1998). Analysis of flower pigmentation mutants generated by random transposon mutagenesis in Petunia hybrida. Plant J. 13: 39–50. [DOI] [PubMed] [Google Scholar]
- Verweij C.W. (2007). Vacuolar Acidification: Mechanism, Regulation and Function in Petunia Flowers. (Amsterdam: VU-University; ). [Google Scholar]
- Verweij W., Spelt C., Di Sansebastiano G.P., Vermeer J., Reale L., Ferranti F., Koes R., Quattrocchio F. (2008). An H+ P-ATPase on the tonoplast determines vacuolar pH and flower colour. Nat. Cell Biol. 10: 1456–1462. [DOI] [PubMed] [Google Scholar]
- Walker A.R., Davison P.A., Bolognesi-Winfield A.C., James C.M., Srinivasan N., Blundell T.L., Esch J.J., Marks M.D., Gray J.C. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiering H. (1974). Genetics of flower colour in Petunia hybrida Hort. Genen. Phaenen. 17: 117–134. [Google Scholar]
- Xie Z., Zhang Z.L., Zou X., Huang J., Ruas P., Thompson D., Shen Q.J. (2005). Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 137: 176–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenoni S., D’Agostino N., Tornielli G.B., Quattrocchio F., Chiusano M.L., Koes R., Zethof J., Guzzo F., Delledonne M., Frusciante L., Gerats T., Pezzotti M. (2011). Revealing impaired pathways in the an11 mutant by high-throughput characterization of Petunia axillaris and Petunia inflata transcriptomes. Plant J. 68: 11–27. [DOI] [PubMed] [Google Scholar]
- Zhang F., Gonzalez A., Zhao M., Payne C.T., Lloyd A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869. [DOI] [PubMed] [Google Scholar]
- Zhao M., Morohashi K., Hatlestad G., Grotewold E., Lloyd A. (2008). The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135: 1991–1999. [DOI] [PubMed] [Google Scholar]
- Zimmermann I.M., Heim M.A., Weisshaar B., Uhrig J.F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40: 22–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.