Maize histone deacetylase HDA101 mainly targets active genes regulating H4K5 acetylation and represses a small subset of targets whose expression must be kept at a low level during seed development.
Abstract
Histone deacetylases (HDACs) regulate histone acetylation levels by removing the acetyl group from lysine residues. The maize (Zea mays) HDAC HDA101 influences several aspects of development, including kernel size; however, the molecular mechanism by which HDA101 affects kernel development remains unknown. In this study, we find that HDA101 regulates the expression of transfer cell-specific genes, suggesting that their misregulation may be associated with the defects in differentiation of endosperm transfer cells and smaller kernels observed in hda101 mutants. To investigate HDA101 function during the early stages of seed development, we performed genome-wide mapping of HDA101 binding sites. We observed that, like mammalian HDACs, HDA101 mainly targets highly and intermediately expressed genes. Although loss of HDA101 can induce histone hyperacetylation of its direct targets, this often does not involve variation in transcript levels. A small subset of inactive genes that must be negatively regulated during kernel development is also targeted by HDA101 and its loss leads to hyperacetylation and increased expression of these inactive genes. Finally, we report that HDA101 interacts with members of different chromatin remodeling complexes, such as NFC103/MSI1 and SNL1/SIN3-like protein corepressors. Taken together, our results reveal a complex genetic network regulated by HDA101 during seed development and provide insight into the different mechanisms of HDA101-mediated regulation of transcriptionally active and inactive genes.
INTRODUCTION
Histone acetylation plays a key role in modulating chromatin structure and function (Shahbazian and Grunstein, 2007). The level of histone acetylation is regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs), which interact with various coactivators and corepressors in different chromatin modifying complexes (Peserico and Simone, 2011). HATs acetylate lysines of histone proteins, usually resulting in the relaxation of chromatin structure and thus facilitating gene activation. Conversely, HDACs remove acetyl groups from acetylated histones, usually leading to a tight chromatin structure. Although HDACs were originally associated with gene repression, recent evidence indicates that, in combination with HATs, HDACs also bind highly transcribed genes to regulate the turnover of acetylated histones and to reset chromatin after transcription (Shahbazian and Grunstein, 2007; Wang et al., 2009). Current models of how histone acetylation modulates chromatin structure and gene transcription hold that acetylation affects the electrostatic histone-DNA interaction and higher order folding of chromatin (Bannister and Kouzarides, 2011). However, acetylation of specific lysine residues can also act as a signal to modulate the recruitment of chromatin remodeling complexes and transcription factors that, in turn, affect the transcriptional status of chromatin.
Studies performed in diverse plant species have shown that histone acetylation is associated with several aspects of development (Wang et al., 2014). Different plant HDACs have been characterized in detail (Liu et al., 2014). Based on sequence similarity and cofactor dependency, HDACs in all eukaryotes are grouped into three families: Rpd3/HDA1, SIR2, and the plant-specific HD2-related protein families (Pandey et al., 2002). Studies in Arabidopsis thaliana showed that many of these HDACs have critical functions, including maintenance of genome stability (Probst et al., 2004; To et al., 2011; Liu et al., 2012), determination of cell-type specificity (Xu et al., 2005; Hollender and Liu, 2008), transition between developmental stages (Tanaka et al., 2008; Yu et al., 2011), and responses to biotic or abiotic stress (Zhou et al., 2005; Chen et al., 2010; Perrella et al., 2013). In rice (Oryza sativa), HDACs regulate plant innate immunity, root growth, peduncle development, and fertility (Chung et al., 2009; Hu et al., 2009; Ding et al., 2012).
In maize (Zea mays), the Rpd3-type HDAC protein HDA101 affects transcription and sequence-specific modulation of histone modification of genes and repeats (Rossi et al., 2007). Transgenic maize plants with up- and downregulation of hda101 expression exhibit pleiotropic effects on plant development; for example, hda101 overexpression causes smaller grains (Rossi et al., 2007). However, the molecular mechanism of HDA101 function in kernel development is unknown. Although the involvement of HDACs in regulating plant seed dormancy, maturation, and germination has been extensively documented (Wang et al., 2014), only limited information is available regarding HDAC function during seed formation and endosperm development. For example, it has been reported that three maize Rpd3-type HDAC genes (hda101, hda102, and hda108) have similar expression profiles, with higher transcript accumulation in the endosperm (Varotto et al., 2003). Wu et al. (2000) reported that silencing of HD2A expression results in aborted seed development in transgenic Arabidopsis plants. Also, plants with artificial microRNA-mediated downregulation of the rice Rpd3-like gene HDA703 display partial or complete sterility and generate seeds with awns, related to increased levels of histone H4 acetylation (Hu et al., 2009). Although these findings suggest that HDACs play important roles in regulating seed development and morphology, further efforts are required to identify the specific targets and mechanisms of HDACs in regulating seed and endosperm formation and size.
The maize endosperm plays an essential role in supporting embryo development and seed germination and also provides humanity with food, livestock feed, and renewable resources (Li et al., 2014). Thus, the development of endosperm largely determines the value of maize both in quantitative and qualitative terms. During the early stages of seed development, key developmental processes in endosperm, including coenocytic development, cellularization, cellular differentiation, and the early mitotic phase, can affect seed size (Mizutani et al., 2010). At 0 to 4 d after pollination (DAP), seed development involves double fertilization, syncytium formation, and cellularization (Lopes and Larkins, 1993; Olsen, 2001, 2004; Li et al., 2014). The differentiation stage occurs at 4 to 6 DAP; at this stage, the endosperm differentiates into four major cell types: the starchy endosperm, the basal endosperm transfer layer (BETL), the aleurone, and the embryo-surrounding region (Olsen, 2001, 2004). Starch and proteins accumulate in the starchy endosperm cells. The BETL tissue functions in nutrient uptake from maternal tissue to the endosperm. During seed germination, aleurone cells are activated to produce hydrolases for starch and protein degradation. The embryo-surrounding region, located around and below the developing embryo, appears to be involved in signaling and/or in defense of the developing embryo against pathogens (Olsen, 2004; Li et al., 2014). Only a little research has been devoted to understanding the mechanisms regulating early endosperm development. Recent studies have provided a comprehensive profile of gene expression during seed development, especially at the early stages (Chen et al., 2014; Li et al., 2014; Wang et al., 2014), thus improving our understanding of the regulatory genetic networks controlling these processes. Seed development also involves epigenetic-related mechanisms. For example, genome-wide chromatin modification of maternal and paternal genomes, associated with gene imprinting, regulates early endosperm development (Berger and Chaudhury, 2009; Zhang et al., 2014).
In this study, we investigated HDA101 function in early seed development in maize. First, our results show that the smaller grain size and defects of transfer cell development observed in hda101 mutant lines are associated with altered expression of transfer cell-specific marker genes. Second, we used chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) to conduct a genome-wide mapping of HDA101 binding sites in maize seed at an early developmental stage. Although genome-wide mapping has been employed to investigate HDAC function in yeast and mammals (Kurdistani et al., 2002; Wang et al., 2002; Robert et al., 2004; Wang et al., 2009; Mundade et al., 2014), it has not previously been used plants. Our results indicate that HDA101 is mainly targeted to genes with high and intermediate expression levels. Loss of HDA101 induces hyperacetylation of histone H4 lysine 5 (H4K5ac) of direct target genes; however, this does not affect the transcript level for most of the target genes. Conversely, HDA101 represses the expression of a small subset of its direct targets, which must be kept at low expression levels during seed development. Finally, we found that HDA101 interacts with two corepressors, NFC103/MSI1 and SNL1/SIN3-like proteins. Taken together, our results identify a complex genetic network associated with HDA101 function in maize seed, thus significantly improving our understanding about the mechanisms of HDA101-mediated gene regulation and the role of histone deacetylation during seed development.
RESULTS
Characterization of HDA101 Antibody and Analysis of HDA101 Protein Levels during Seed Development of hda101 Mutant Lines
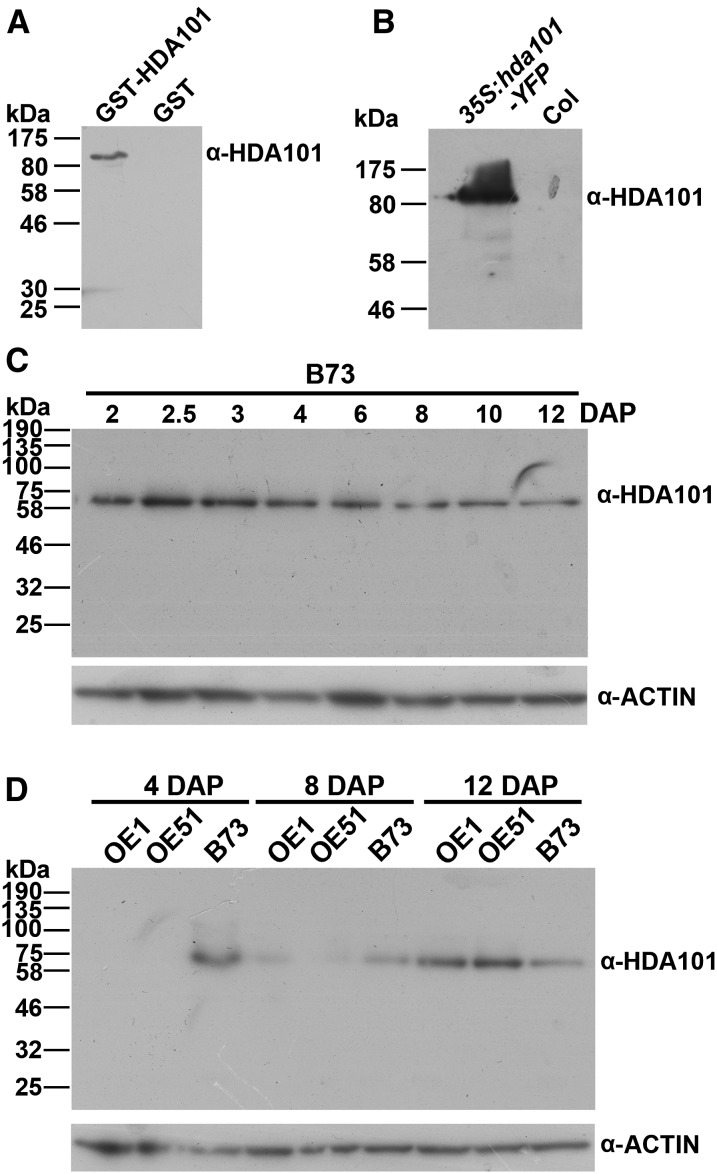
The phenotypes observed in hda101 overexpression (OE) maize lines include smaller kernels (Rossi et al., 2007). To analyze changes in HDA101 protein levels in these lines and to produce tools for analyzing HDA101 function, we generated an HDA101-specific antibody. We used a peptide epitope located outside the highly conserved domains present in all plant HDACs to produce a specific mouse monoclonal antibody (see Methods and Supplemental Figure 1). Immunoblot experiments showed that the antibody recognizes the recombinant GST-HDA101 protein in Escherichia coli extracts (Figure 1A). The antibody specificity was further corroborated by showing that it specifically detects an 80-kD polypeptide in protein extracts from seedlings of transgenic Arabidopsis lines overexpressing a maize HDA101:YFP fusion protein, while this polypeptide is not observed in wild-type Arabidopsis seedlings (Figure 1B). Analysis of HDA101 protein level in seeds of the wild-type maize B73 inbred specifically detected a 60-kD polypeptide, corresponding to the predicted HDA101 molecular mass, and this polypeptide was present throughout seed development (Figure 1C). Taken together, these findings support the specificity of the antibody for the HDA101 protein.
Figure 1.
Characterization of Anti-HDA101 Antibody and Analysis of HDA101 Protein Level.
(A) Immunoblot for detection of GST-HDA101 fusion protein in E. coli extracts.
(B) Immunoblot for detection of GST-HDA101:YFP fusion protein in extracts of wild-type Columbia (Col) and transgenic Arabidopsis seedlings overexpressing maize hda101(35S:hda101-YFP).
(C) Immunoblot for detection of HDA101 protein level in seeds of the wild-type B73 inbred line, harvested at different developmental stages. Immunoblot with anti-ACTIN antibody was used as a loading control. The seeds were harvested at the indicated developmental stages.
(D) Immunoblot for detection of HDA101 level in seeds of the wild-type B73 inbred line and in maize transgenic lines OE1 and OE51 (Rossi et al., 2007) overexpressing the hda101 gene. Immunoblot with anti-ACTIN antibody was used as a loading control. The seeds were harvested at the indicated developmental stages.
Subsequently, we measured HDA101 protein levels in seeds of hda101-overexpressing maize lines (OE1 and OE51 lines; Rossi et al., 2007). Surprisingly, we found that the HDA101 protein level was drastically reduced in early seed development of OE1 and OE51 lines (Figure 1D) compared with the wild-type B73 inbred. In the OE1 and OE51 lines, no HDA101 protein was detected in 4-DAP seeds. HDA101 protein was detected in 8-DAP seeds of the OE lines, although at a lower level with respect to the wild type. In 12-DAP seeds, the HDA101 protein level was higher in OE lines than in wild-type plants (Figure 1D).
The level of the endogenous and of the total (i.e., endogenous plus transgenic) hda101 transcript was measured by RT-qPCR in seeds harvested at different developmental stages of OE1 and OE51 transgenic lines, as well as of B73 wild-type plants. The results showed that the endogenous hda101 RNA was almost undetectable in 4-DAP seeds and strongly downregulated in 8-DAP seeds of the OE transgenic lines but was restored to wild-type levels at 12 DAP (Supplemental Figure 2). Conversely, the level of endogenous plus transgenic hda101 transcript was upregulated in the transgenic lines. These data, supported also by observations from RNA-seq experiments (Supplemental Figure 2), suggest that loss of HDA101 protein in 4-DAP seeds of OE lines could be due to a cosuppression mechanism (De Paoli et al., 2009). Nevertheless, the precise mechanisms leading to overexpression of hda101 in the subsequent seed developmental stages remain unknown. In any case, these results allow us to employ the transgenic lines as an important tool for studying hda101 function during the early stages of seed development. In this context, we conducted molecular and cytological analysis of the 4-DAP seeds of the OE1 and OE51 transgenic lines, as well as of the wild-type B73 inbred. Hereafter, we refer to the two independent transgenic lines, OE1 and OE51, as the hda101-1 and hda101-2 mutant lines, respectively.
hda101 Mutants Produce Smaller Kernels with Defects in Transfer Cells
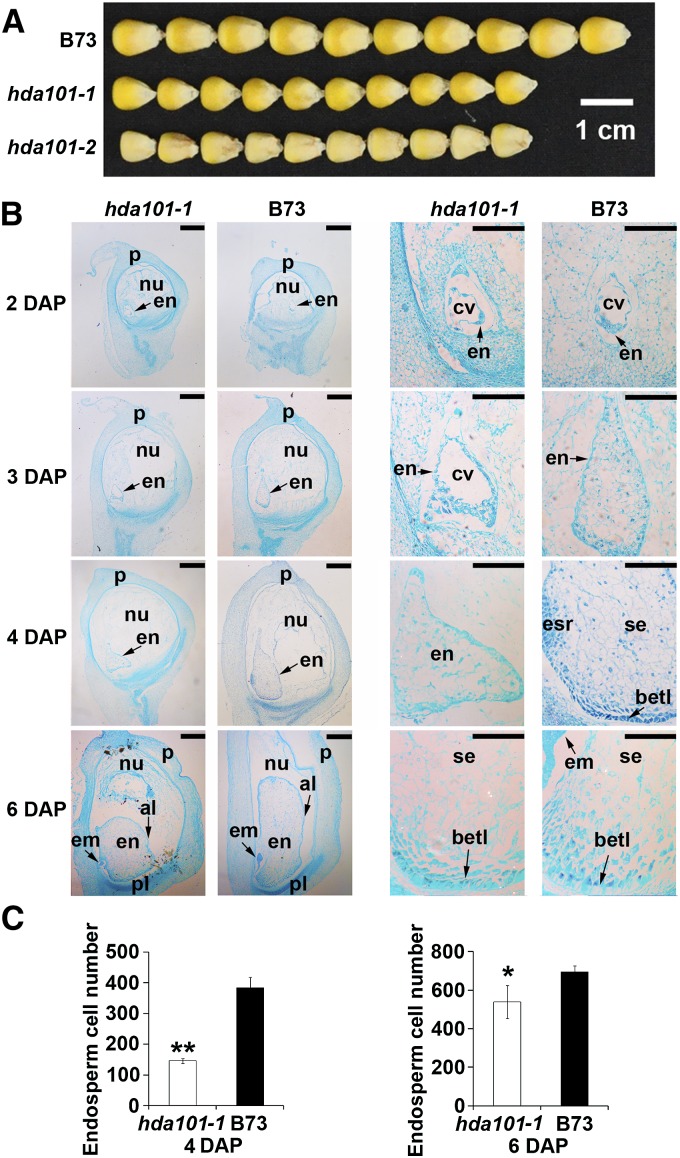
In agreement with a previous report (Rossi et al., 2007), we found that both hda101-1 and hda101-2 mutant lines produce smaller kernels compared with wild-type B73 (Figure 2A). We used microscopy of histological sections stained with safranin-fast green to examine the morphology of developing endosperms of B73 and hda101-1 mutant seeds collected at 2, 3, 4, and 6 DAP (Figure 2B). At 2 DAP, when the endosperms are at the coenocytic stage, seeds of B73 and the hda101-1 mutant showed similar progress, with the primary endosperm nucleus undergoing a series of divisions without cytokinesis and the nuclei distributing to the wall of the endosperm. At 3 DAP, the endosperm of B73 was almost fully cellularized, while the endosperm of the hda101-1 mutant remained partially cellularized containing the central vacuole. At 4 to 6 DAP, we found that the hda101-1 mutant generated a smaller endosperm, with fewer cells compared with B73 (Figure 2C). Notably, the BETL of hda101-1 mutants showed irregularities in the organization and differentiation of the transfer cells. In particular, the length and depth of the BETL were restricted in the hda101-1 mutant, with discrete clusters of transfer cells, while in B73 seeds, the transfer cells were organized regularly and showed deeper cell wall ingrowth (Figure 2B). These results indicate that the hda101-1 mutant line exhibits defects of the BETL cells, which might lead to smaller grain size.
Figure 2.
The hda101 Mutant Lines Exhibit Smaller Endosperm Size with Defects in Transfer Cells.
(A) The hda101-1 and hda101-2 mutant lines generate smaller kernels compared with wild-type B73.
(B) The morphology of developing maize endosperms of hda101-1 and wild-type B73 plants collected at 2, 3, 4, and 6 DAP were determined by examining histological sections stained with safranin-fast green to identify endosperm development defects. em, embryo; en, endosperm; nu, nucellus; p, pericarp; cv, central vacuole; betl, basal endosperm transfer layer; esr, embryo-surrounding region; se, starch endosperm; pl, placentochalaza; al, aleurone. Short bars = 500 µm; long bars = 200 µm.
(C) Endosperm cell size in B73 and hda101-1 mutant line at 4 and 6 DAP. The single and double asterisks represent significant differences between B73 and the hda101-1 mutant determined by Student’s t test at P < 0.05 and P < 0.01, respectively.
Expression of Transfer Cell-Specific Marker Genes Is Altered in the hda101 Mutant
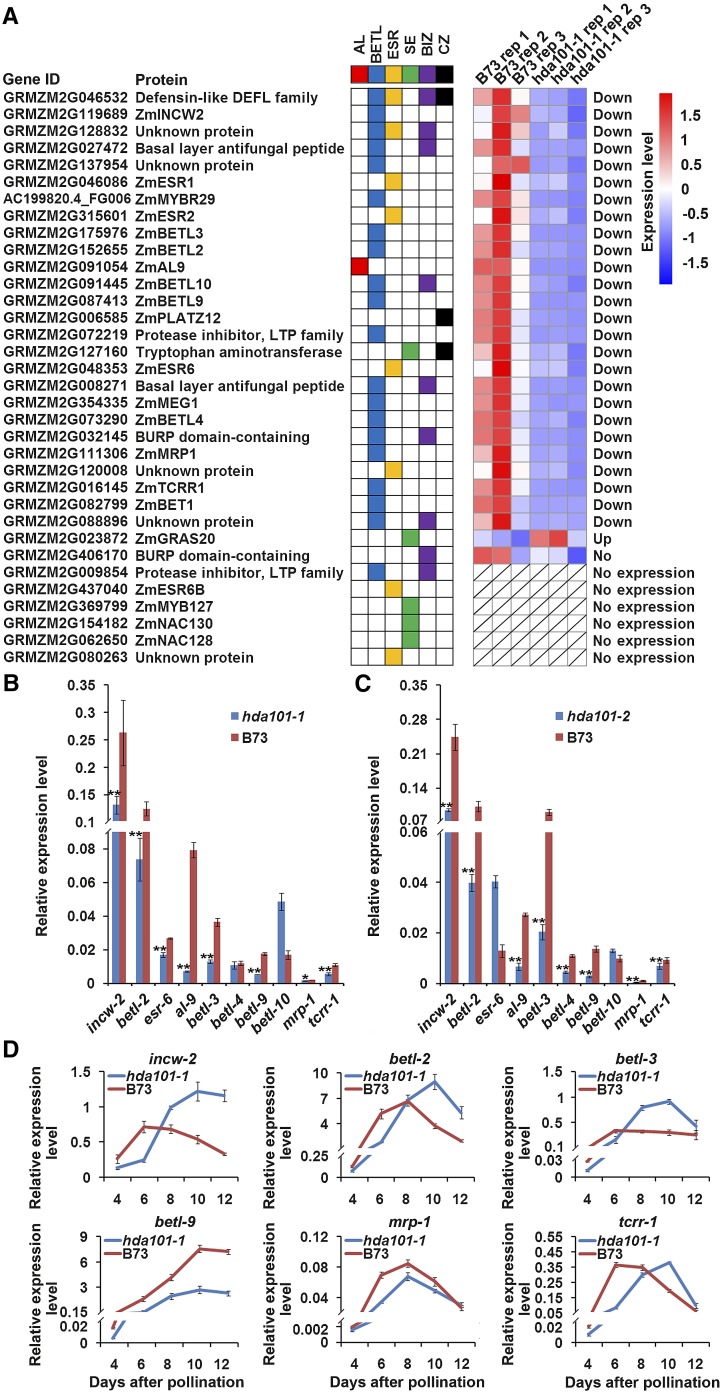
Previous studies identified 34 genes that show cell type-specific expression during endosperm differentiation (Gómez et al., 2009; Li et al., 2014; Muñiz et al., 2014) (Figure 3A). Consistent with the morphological abnormalities observed in seeds of the hda101-1 mutant, our results from the RNA-seq analysis, performed using three biological replications, showed that 26 out of these 34 genes exhibited a statistically significant decrease of their transcript level in 4-DAP seeds of the hda101-1 mutant compared with B73 (Figure 3A). Conversely, the gras20 gene, encoding a DELLA-like protein that functions as a negative regulator in the gibberellic acid pathway, was upregulated in the hda101-1 mutant. The GRMZM2G416170 gene did not show statistically significant differences between the hda101-1 mutant and B73 and the remaining six genes were expressed at very low levels, if at all, in both lines (see below and Methods for details of RNA-seq analysis). The expression of 10 randomly selected genes exhibiting RNA level variation in the hda101-1 mutant was validated by performing RT-qPCR analysis in both hda101-1 and hda101-2 mutants. The results (Figures 3B and 3C) indicated that seven of the genes (i.e., cell wall invertase-2 [incw-2], betl-2, aleurone9, betl-3, betl-9, Myb related protein1 (mrp-1), and transfer cell response regulator1 [tcrr1]) are downregulated in 4-DAP seeds of both mutant lines, while three genes are downregulated only in one of the two mutant lines (i.e., esr-6 in the hda101-1 line and betl-4 in the hda101-2 line). Therefore, these results indicate that many of the genes exhibiting specific expression in the endosperm are regulated, directly or indirectly, by HDA101. Since a total of 20 genes, among the 34 analyzed, were specifically or mainly expressed in the BETL region and because our RNA-seq results showed that 19 of them were downregulated in the hda101-1 mutant line (Figure 3A), we envisage that HDA101 regulates the majority of transfer cell-specific marker genes.
Figure 3.
HDA101 Is Required for the Normal Expression of Genes Involved in Endosperm Cell Differentiation.
(A) Tissue-specific expression patterns of genes associated with endosperm differentiation during early endosperm development are reported on the left column and were calculated on the basis of results from Li et al. (2014). Colored squares represent a specific tissue in kernel: AL (red), aleurone; BETL (blue); ESR (yellow), embryo-surrounding region; SE (green), starchy endosperm; BIZ (purple), basal intermediate zone; CZ (black), conducting zone. The differential expression of these genes in 4-DAP kernels of hda101-1 mutant and wild-type B73 plants were identified based on RNA-seq analysis performed in this study and is illustrated in the heat map at right. Individual gene IDs are shown.
(B) and (C) Real-time RT-qPCR was used to assess the RNA level of 10 selected genes in 4-DAP kernel of wild-type B73 and the hda101-1 (B) or hda101-2 (C) mutant plants. The RT-qPCR data were normalized to the GRMZM2G027378 gene and represented as means ±se (n = 3). GRMZM2G027378 was constitutively expressed during grain development (Li et al., 2014). The single and double asterisks represent significant differences determined by the Student’s t test at P < 0.05 and P < 0.01, respectively.
(D) RT-qPCR quantification of the RNA level of six BETL-specific genes in seeds of wild-type B73 and hda101-1 mutant plants. Seeds were harvested at the indicated developmental stages. The data were normalized to GRMZM2G027378.
BETL-specific genes are often expressed in a dynamic pattern during endosperm development, characterized by significant upregulation at 4 to 8 DAP, when rapid cell differentiation and dramatic reprogramming of the kernel transcriptome take place (Gómez et al., 2009; Li et al., 2014; Muñiz et al., 2014). We therefore used RT-qPCR to assess whether loss of HDA101 protein in early seed development affects the temporal expression pattern of six BETL-specific genes (Figure 3D). All these genes showed decreased expression in 4- and 6-DAP seeds of the hda101-1 mutant compared with B73. In addition, we found that expression of tcrr-1, betl-2, berl-3, and incw-2 peaked at 6 to 8 DAP in B73, while it peaked at 8 to 10 DAP in the hda101-1 mutant. Furthermore, the expression of mrp-1 and betl-9 remained low in the hda101-1 mutant compared with B73. Hence, our results indicate that HDA101 is required to maintain the normal expression pattern of these BETL-specific genes during the different stages of endosperm development.
Together, these findings, along with previous histological observations, suggest that an alteration of transfer cell development occurs in the hda101 mutants and that this can be related to specific involvement of HDA101 in regulating the expression of BETL marker genes.
Genome-Wide Mapping of HDA101 Protein Binding Sites in Maize Seed Indicates That It Mainly Associates with Transcriptionally Active Genes
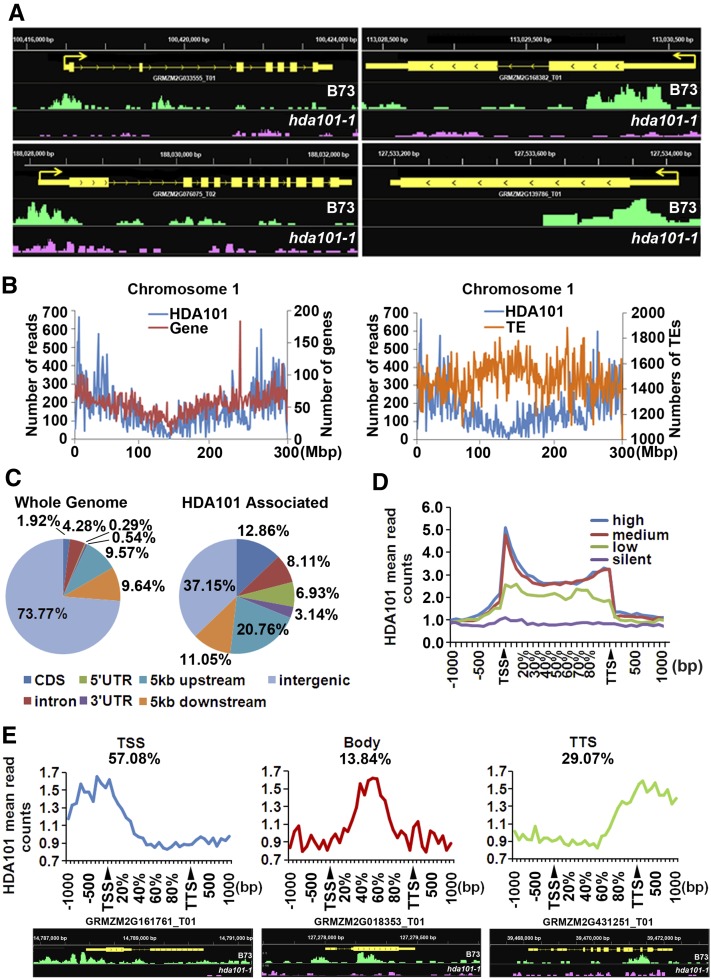
To study the molecular function of HDA101 during seed development in detail, we identified HDA101 binding sites at the genome-wide level. To this end, we employed the HDA101-specific antibody (Figure 1) to carry out ChIP-seq. The experiments were performed using chromatin extracted from 4-DAP seeds of wild-type B73 plants. The hda101-1 mutant line, which has no detectable HDA101 protein in 4-DAP seeds (Figure 1D), was used as a negative control. Two biological replicates were performed for both maize lines. The resulting sequencing reads were mapped to the B73 reference genome sequence (release AGPv3). Input B73 and hda101-1 mutant chromatin fragments were also sequenced and used as a negative control in enrichment analysis. The abundance of the ChIP-seq reads was normalized to the corresponding input, the relative enrichment of HDA101 was calculated as a ratio of the values for B73 over those of the hda101-1 mutant, and HDA101 binding sites were defined as regions with 5x or higher HDA101 signal in B73 with respect to the hda101-1 mutant line. A total of 10,381 peaks, corresponding to 7578 protein-coding genes, were identified as candidate HDA101 binding sites (Figure 4A; Supplemental Data Set 1). Interestingly, these genes did not include the BETL-specific genes downregulated in hda101 mutant lines (see above), indicating that their expression is indirectly controlled by HDA101. ChIP-seq data were validated by ChIP assays followed by qPCR quantification (ChIP-qPCR) of 12 loci, randomly selected among those that showed HDA101 binding. Results for nine of the loci are consistent with ChIP-seq analysis (Supplemental Figure 3).
Figure 4.
Genome-Wide Mapping of HDA101 Binding Sites in Maize 4-DAP Seeds.
(A) Representative HDA101 binding peaks shown by the Integrated Genome Browser. These peaks are uniquely or statistically significantly enriched in wild-type B73 compared with the hda101-1 mutant. The gene structure is shown (in yellow), and peaks in B73 and the hda101-1 mutant are indicated in green and pink, respectively.
(B) Distribution of HDA101 protein binding sites with respect to genes (left panel) and to transposable elements (TEs) (right panel) along maize chromosome 1. The x axis represents different positions in chromosome 1; y axis of left-side scale represents number of HDA101 binding reads per 1 Mb; y axis of right-side scale represents number of genes per 1 Mb (left panel) and number of repeat regions per 1 Mb (right panel).
(C) Distribution of HDA101 binding regions (right) relative to the overall maize genome gene structure (left).
(D) Profiles of HDA101 binding across 5′ gene ends, 3′ gene ends, and gene body regions of genes with high, medium, low, and silent expression. TSS and TTS represent the transcription start site and transcription termination site, respectively.
(E) The target genes bound by HDA101 are divided into three types, based on HDA101 distribution in the TSS, gene body, and TTS region. Representative HDA101 binding peaks by the Integrated Genome Browser are shown at the bottom. The gene structure is shown (in yellow), and peaks in B73 and the hda101-1 mutant are indicated in green and pink, respectively.
Analysis of HDA101 protein distribution on the chromosomes indicated that HDA101 is highly enriched in the gene-rich euchromatin and depleted from regions containing transposable elements and other repetitive sequences (Figure 4B; Supplemental Figure 4). Next, we analyzed the HDA101 distribution relative to gene structure in the overall maize genome. We defined genomic segments as comprising the gene coding sequence, the 5 kb upstream of the transcription start site (TSS; or ATG if the TSS is not yet determined), and 5 kb downstream of the poly(A) site (Morohashi et al., 2012). In maize, genes represent only 26.2% of the genome, and among the HDA101-enriched peaks, ∼63% of them occurred in gene regions, suggesting that HDA101 binding occurs mainly in genes (Figure 4C). In particular, HDA101 binding sites were significantly enriched in the 5-kb upstream regions (21%) and in the coding sequence (13%), although these two regions represent only 9.6 and 1.9%, respectively, of the maize genome (Figure 4C).
To investigate in more detail the HDA101 binding profile within the gene region, we determined HDA101 binding distribution in the region starting from 1 kb upstream of the TSS to 1 kb downstream of the transcription termination sites (TTSs) (Figure 4D). Within the gene body, reads were summed in windows equal to 5% of the gene length. We also investigated the correlation between HDA101 binding and gene expression by dividing genes into quartiles, based on their expression levels including high, medium, low, and silent expression (Figure 4D). Our data revealed that HDA101 is mainly enriched in genes with high and intermediate transcription and that, in these classes of genes, HDA101 binding occurs mainly around the TSS and, at a lower level, in the gene body and in the TTS regions. We observed only a minor enrichment in genes with low expression. Because HDACs are generally considered to be transcriptional repressors, it may be considered surprising to find that the enrichment of HDA101 was much higher in highly expressed genes. Nevertheless, studies of mammalian HDACs reported similar results (Wang et al., 2009).
We subsequently analyzed whether HDA101 is enriched along the TSS to gene body region in the same gene or whether HDA101 binds different target genes in either the TSS or gene body regions. The results indicated that 92% of target genes contain a single peak and only 8% of genes contain two peaks. Among the target genes with one HDA101 peak, 57% of genes are bound by HDA101 in the TSS, 29% in the TTS, and 14% in the gene body region (Figure 4E). We further tested the relationship between HDA101 binding location in the TSS, gene body, and TTS region with gene expression, gene length, and tissue specificity of the target genes; however, we found that the HDA101 location within a gene does not relate to any of these features (Supplemental Figure 5).
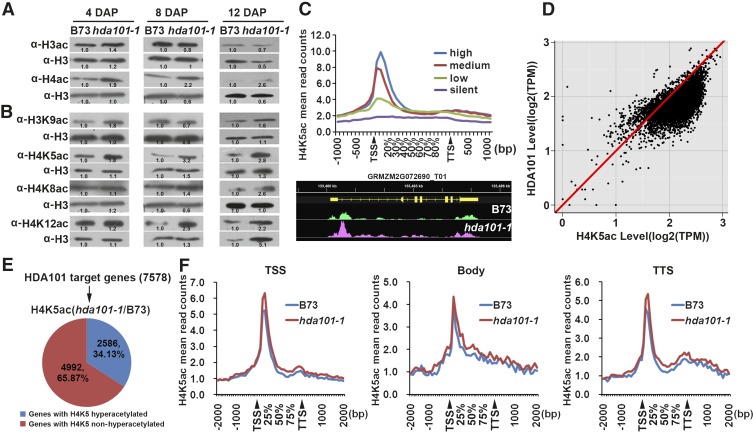
Loss of HDA101 Binding Correlates with Increased Histone Acetylation
Previous reports indicated that HDA101 has HDAC activity and its overexpression in maize seedlings correlates with a decrease of acetylation in different lysine residues (Lechner et al., 2000; Rossi et al., 2003, 2007). Accordingly, we assessed the correlation between HDA101 protein levels and histone acetylation in 4-, 8-, and 12-DAP seeds of the hda101-1 mutant and B73 (Figures 5A and 5B). The level of histone H3 acetylated at lysine 9 and 14 (H3ac) and of histone H4 acetylated at lysine 5, 8, 12, and 16 (H4ac) was detected by immunoblot with specific antibodies. We found that at 4 DAP, when HDA101 protein is not detectable in the hda101-1 mutant (Figure 1D), the H3ac and H4ac levels were higher in hda101-1 mutants compared with B73, with the major difference detectable for H4ac (Figure 5A). In the later seed developmental stages, when the amount of HDA101 protein in the hda101-1 mutant line returned to the wild-type level or is more abundant than B73 (Figure 1D), the hda101-1 mutant and B73 had similar levels of H3ac and H4ac (Figure 5A). Analysis of the histone acetylation level at single lysine residues, including H4K5ac, H4K8ac, H4K12ac, and histone H3 lysine 9 (H3K9ac), showed similar results, although we found that the opposite relationship between HDA101 protein level and acetylation differs for different lysine residues (e.g., HDA101 affects H4K5ac level more than other lysine residues; Figure 5B). In conclusion, these experiments indicate that loss of HDA101 in 4-DAP seeds is associated with a global increase of histone acetylation.
Figure 5.
Correlation between HDA101 Loss and Histone Acetylation.
(A) Immunoblot to detect the global H3ac and H4ac levels in 4-, 8-, and 12-DAP seeds of hda101-1 mutant and wild-type B73 plants. Protein loading was assessed using an antibody that recognizes histone H3, independently by its posttranslational modifications (indicated in the figure as H3). Three replications were performed and the signals for the bands were measured using Quantity One software (Bio-Rad). The value of the optical density is labeled in the figure and used to quantify the abundance of the hybridizing polypeptide.
(B) As in (A), but antibodies specific for distinct lysine residues were used (i.e., H3K9ac, H4K5ac, H4K8ac, and H4K12ac).
(C) Profiles of H4K5ac level across 5′ gene ends, 3′ gene ends, and gene body regions of genes with high, medium, low, and silent expression. TSS and TTS represent the transcription start site and transcription termination site, respectively. Representative H4K5ac level peaks by the Integrated Genome Browser are shown, the gene structure is reported in yellow, and peaks of H4K5ac in B73 and the hda101-1 mutant are indicated in green and pink, respectively.
(D) The correlation between HDA101 binding and H4K5ac level among the HDA101 target genes.
(E) Variation of H4K5ac level in the HDA101 targets by comparison of hda101-1 mutant and B73 as determined by ChIP-seq analysis.
(F) Variation of H4K5ac level in 4-DAP seeds of wild-type B73 and the hda101-1 mutant for three types of HDA101 direct targets, subdivided on the basis of the region bound by HDA101 (i.e., TSS, gene body, and TTS region).
Since HDACs remove the acetyl group from histones of direct target genes, we examined the correlation between HDA101 binding and histone acetylation in its direct targets. To this end, we performed ChIP-seq experiments with three biological replications in 4-DAP seeds of B73 wild-type plants to map the distribution of H4K5ac, which is a favored substrate of HDA101 in these maize tissues (Figure 5B). As expected (Zhou et al., 2010; Lauria and Rossi, 2011), we observed that H4K5ac was enriched around the TSS of highly expressed genes, but not in silent genes (Figure 5C; Supplemental Data Set 2). Interestingly, the level of HDA101 binding also appeared to be positively correlated with the amount of H4K5ac (Figure 5D).
To better investigate the relationship between HDA101 binding and the level of histone acetylation at specific HDA101 direct targets, we performed ChIP-seq experiments with three replications in the hda101-1 mutant line (Supplemental Data Set 3). Among the HDA101-bound genes, 34.13% exhibited increased H4K5ac levels in the hda101-1 mutant compared with B73 (Figure 5E). The increase of H4K5ac in other direct HDA101 targets was not detected, likely due to different effects associated with HDA101 activity (e.g., other histone lysine residues may be targeted by HDA101). These results were validated by ChIP-qPCR on six randomly selected HDA101 direct targets that exhibited increased H4K5ac in the hda101-1 mutant. The experiment was performed using chromatin extracted from both hda101-1 and hda101-2 mutant lines. The results showed that, in five of these genes, the level of H4K5ac increased in 4-DAP seeds of hda101-1 and hda101-2 mutants compared with wild-type B73 (Supplemental Figure 6).
Subsequently, we identified the gene regions where H4K5ac level increases following the loss of HDA101 binding. To this end, the genes where loss of HDA101 binding in the hda101-1 mutant corresponds with an increase of H4K5ac level were subdivided into three categories, depending on the localization of HDA101 binding in the TSS, gene body, and TTS regions. The results (Figure 5F) show that loss of HDA101 binding in the TSS and TTS regions induces an increase of H4K5ac level around the TSS, where most of this histone mark localizes (Figure 5C), although removal of HDA101 from the TTS also led to a slight increase in other gene regions. Conversely, when loss of HDA101 binding occurred in the gene body region, increased H4K5ac was detected throughout the entire gene.
Collectively, these results suggest that one major function of HDACs is to remove the acetyl group from its direct targets.
Loss of HDA101 Differentially Affects Expression of Direct Targets
To analyze the relationship between loss of HDA101 binding, histone acetylation, and gene expression, we performed RNA-seq experiments with three biological replications to examine the differences of transcript levels between 4-DAP seeds of B73 and the hda101-1 mutant. We found that 2030 differentially expressed transcripts were upregulated and 1784 were downregulated in the hda101-1 mutant (P value < 0.05; Supplemental Data Set 4). We validated the results by performing RT-qPCR of 17 randomly selected sequences, showing that transcript variation of 13 of these sequences was consistent with the RNA-seq analysis (Supplemental Figure 7). The Gene Ontology (GO) functional annotation of differentially expressed genes revealed that upregulated genes were highly enriched in pathways related to regulation of gene expression, transcription, regulation of macromolecule metabolic process, response to abiotic stimulus, and regulation of metabolic process. Conversely, the downregulated genes were enriched in pathways related to carbohydrate metabolic process, response to stimulus, and stress (Supplemental Figure 8). If the molecular function pathway is considered, the GO analysis showed that upregulated genes were enriched in transcription regulator, transcription factor and transferase activity, and kinase activity, while downregulated genes were enriched in transporter, chromatin binding, catalytic, and hydrolase activity (Supplemental Figure 8). This observation corroborated the previous findings that HDA101 affects several biological processes (Rossi et al., 2007).
Subsequently, we determined the expression alteration of the 2586 HDA101 direct targets that exhibited an increase of H4K5ac levels in the hda101-1 mutant. The results indicated that among the 2586 HDA101 targets, 1773 (68.56%) genes are highly expressed or intermediately active, and 92.4% of these do not show changes in expression (Supplemental Figure 9). These findings indicate that HDA101 preferentially binds transcriptionally active genes to decrease their histone acetylation levels; however, this does not affect transcription of the majority of target genes. We also found that only 127 out of the 2586 genes were upregulated in the hda101-1 mutant compared with B73 (Supplemental Data Set 5).
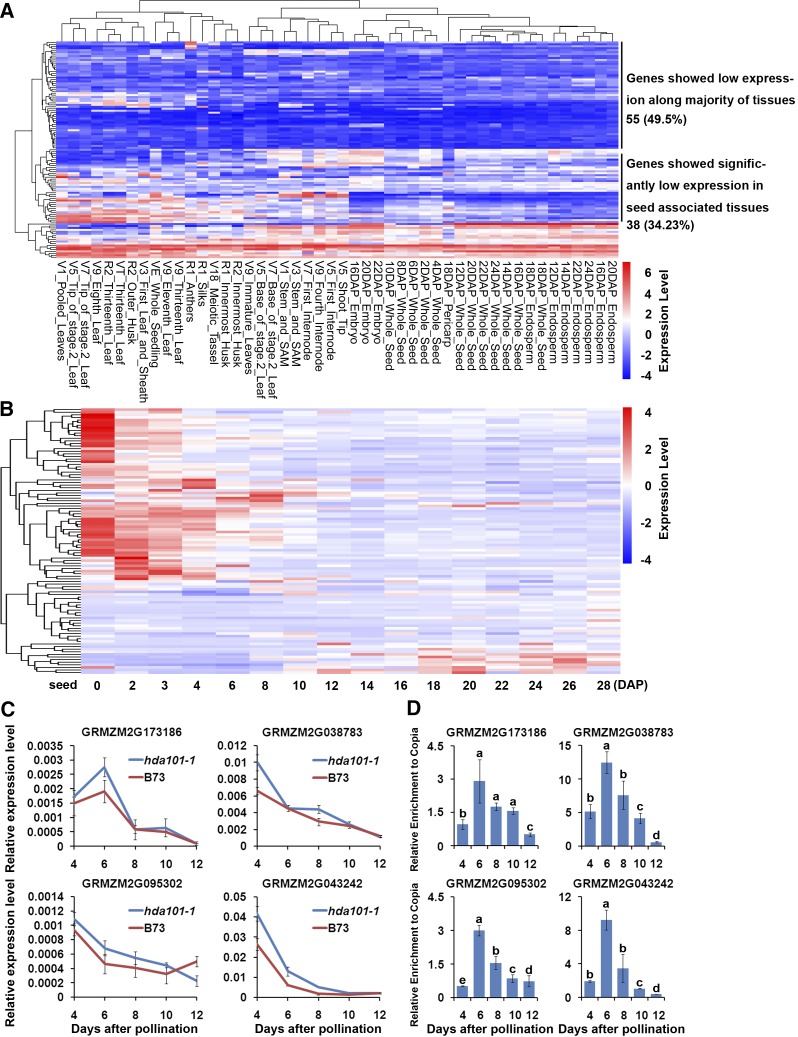
We next conducted a more detailed analysis of these genes, where loss of HDA101 binding occurs along with histone hyperacetylation and upregulation. The tissue-specific expression of these 127 genes was assessed in 48 different maize tissues, using publicly available data from the PLEXdb database (http://www.plexdb.org/; Sekhon et al., 2011). The heat map of 111 genes (the database lacked expression data for the remaining genes) indicated that 93 (83.78%) genes show lower expression in maize seed tissues, among which 55 genes show a low transcript level in the majority of tissues (Figure 6A). This result suggests that these genes are silenced or expressed at a low level in seed tissues of wild-type plants. We further examined the expression of the 127 genes during seed development from 0 to 28 DAP, using the data produced by Chen et al. (2014). The results indicated that 66 (64.71%) genes show a transition to downregulation from early stage of seed development (0 to 4 DAP) to later stage (10 to 28 DAP), and 24 (23.53%) genes have low expression levels from 0 to 28 DAP (Figure 6B).
Figure 6.
HDA101 Is Associated with Negative Regulation of Gene Expression of a Subset of Direct Targets.
(A) The heat map shows transcript variation in 48 maize tissues for a subset of HDA101 direct targets that, in 4-DAP endosperm of hda101-1 mutant versus wild-type B73, exhibit an increase of both H4K5ac and transcription. These data are publicly available from the PLEXdb database (http://www.plexdb.org/; Sekhon et al., 2011).
(B) The heat map shows transcript variation of these genes (same as in [A]) during maize seed development from 0 to 28 DAP. The expression data were reported by Chen et al. (2014).
(C) The RNA level of four HDA101 target genes in seeds at 4, 6, 8, 10, and 12 DAP of the hda101-1 mutant. These genes are negatively regulated during seed development and upregulated in transcription with H4K5ac increase in the hda101 mutant. The relative expression of each gene (y axis) was normalized to GRMZM2G027378 and is represented as means ± se (n = 3).
(D) ChIP assays followed by qPCR quantification of the HDA101 binding profile during seed development of B73 for four of HDA101 direct targets, which exhibit an increase of both H4K5ac and mRNA level in 4-DAP endosperms of hda101-1 and hda101-2 mutants compared with wild-type B73 plants. The HDA101 binding enrichment (y axis) was normalized to input and the non-target transposon Copia (AF398212) and represented as means ± se (n = 3).
These results suggest that expression of these genes is negatively regulated during seed development. Hence, we asked whether this feature is associated with HDA101 binding. To test this hypothesis, we measured the expression of six randomly selected genes from the set of 127 genes at 4, 6, 8, 10, and 12 DAP in the hda101-1 seeds and at 4 and 8 DAP in the hda101-2 seeds, compared with B73. We found that four genes showed higher expression levels in both hda101-1 and hda101-2 mutants during early seed development (Figure 6C; Supplemental Figure 10). Subsequently, ChIP assays with anti-HDA101 antibody using chromatin extracted from B73 wild-type seeds harvested at different developmental stages were performed and indicated that all four genes are bound by HDA101 and that the binding is maintained throughout the early development of the seed (Figure 6D). These findings suggest that the silencing or the low expression of a small subset of direct HDA101 targets is, in fact, associated with HDA101 binding and that it may be required for the maintenance of lower expression during seed development.
Identification of HDA101-Interacting Proteins
HDACs can be recruited by different transcription factors and form multiple protein complexes, involved in various developmental processes (Liu et al., 2014). To identify the proteins interacting with HDA101, we used the HDA101-specific antibody for coimmunoprecipitation assays with extracts from 4-DAP kernels of B73. A protein extract from 4-DAP kernels of the hda101-1 mutant was used as a negative control. The immunoprecipitated proteins were separated on SDS gels and visualized by silver staining. This experiment detected the presence of a polypeptide with the predicted molecular mass of HDA101 in extracts from B73, but not from the hda101-1 mutant (Supplemental Figure 11A). However, additional polypeptides were detected only in B73 extracts and they were recovered from the SDS-PAGE gel for analysis by liquid chromatography-tandem mass spectrometry. Seventeen proteins were identified as potential HDA101 interactors (Supplemental Data Set 6). Among them, the protein with highest coverage was HDA101, which is recognized by HDA101 antibody. One of the putative HDA101-interacting proteins was the MSI/RbAp-like protein NFC103/MSI1, belonging to the WD40 repeat-containing family (Hennig et al., 2005). Another interesting polypeptide corresponded to the maize homolog of the SIN3-like protein, the SNL1 corepressor, which is a subunit of different HDAC-containing complexes in Arabidopsis, mammals, and yeast (Song et al., 2005).
The interaction of HDA101 with NFC103 and SNL1 was verified using yeast two-hybrid assays (Supplemental Figures 11B and 11C). Results indicated that full-length HDA101 interacts with both NFC103 and SNL1 full-length proteins. To determine the specific regions of HDA101, NFC103, and SNL1 that are required for interaction, we generated constructs expressing deleted versions of the proteins. Similar to results reported in yeast (Laherty et al., 1997), we found that the SNL1 HID, but not the PAH domain, is required for interaction with HDA101, whereas the HDAC domain of HDA101 is essential for the interaction between the two proteins (Supplemental Figure 11C). The α-helix and WD40 domain of NFC103 and the N-terminal part of the HDAC domain of HDA101 are required for interaction between NFC103 and HDA101 (Supplemental Figure 11C). Overall, our findings indicate that HDA101 interacts with the two maize putative corepressors NFC103 and SNL1, thus suggesting that, similar to other eukaryotic systems, maize HDA101 is a subunit of distinct transcriptional repressor complexes.
DISCUSSION
Distinct Functions of Maize HDA101 in Active and Inactive Genes
A previous study showed that HDA101 is involved in the regulation of gene transcription and in sequence-specific modulation of histone modification (Rossi et al., 2007). However, the molecular mechanisms of HDA101 in transcriptional regulation remain unclear. HDACs reset chromatin by removing acetylation; nevertheless, the mode of association between HDACs and gene expression in yeast and human is a topic of debate (Wang et al., 2009). In yeast, some reports suggest that HDACs act as transcriptional corepressors, as they bind repressed genes and are replaced by HATs only upon gene activation (Robert et al., 2004; Berger, 2007). However, other reports from yeast and studies performed in human cells indicate that HDACs are mainly targeted to transcriptionally active genes by phosphorylated RNA Pol II, while only a small subset of transcriptionally inactive genes are directly bound by HDACs to repress their transcription (Kurdistani et al., 2002; Wang et al., 2002; Wang et al., 2009). Similar to the latter reports from yeast and mammals, our results provide evidence that the maize Rpd3-type HDAC HDA101 employs two different regulatory strategies for regulating transcriptionally active and inactive genes in seeds at early developmental stages.
We found that transcriptionally active genes are the preferred targets of HDA101. The binding occurs mainly in the TSS and to a lesser extent in gene body regions. Moreover, loss of HDA101 binding is associated with an increase of H4K5ac in many of the HDA101 direct targets. As suggested for mammalian HDACs, HDA101 may be recruited to active genes to reset their chromatin modification state after initiation of transcription, thus maintaining an adequate level of histone acetylation (Wang et al., 2002; Wang et al., 2009). In mammals and yeast, this activity is required to prevent promiscuous transcription initiation from cryptic start sites (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005; Wang et al., 2009). Our observation that loss of HDA101 binding in the gene body region produces an increase of H4K5ac throughout the entire gene region supports this scenario. Moreover, an additional function of HDA101 binding at transcriptionally active genes may be related to its requirement for regulation of the turnover of acetylated histones, which is faster in genes with a high transcription rate (Waterborg, 2002).
Interestingly, our RNA-seq data indicate that loss of HDA101 does not affect the transcript level of the majority of active genes bound by HDA101. Wang et al. (2009) reported similar results in mammalian cells, where treatment with HDAC inhibitors increased the expression of only a minor fraction of hyperacetylated and highly expressed genes (Keji Zhao, personal communication). This finding can be explained by considering that, as mentioned above, the major function of HDACs in active genes is not to regulate their transcriptional rate, but to prevent the formation of aberrant transcripts due to initiation of transcription from cryptic start sites. Moreover, in our study, we employed transgenic lines where the function of only one member of the large HDAC family was impaired (i.e., only HDA101 protein is altered). This means that other Rpd3-HDAC-like enzymes are still active and may, at least partially, compensate for the loss of HDA101. Different HDAC types have some functional differences, associated with preference toward a specific lysine residue (Kölle et al., 1999). Therefore, the loss of HDA101 should mainly determine the increase of H4K5ac, which is its preferred substrate, and other HDACs may not be able to compensate for this activity. However, it is also unlikely that the increase of H4K5ac is sufficient to account for major changes in gene expression of active genes. Indeed, it was reported that gene expression is principally regulated by the cumulative effect of acetylation at various histone lysine residues, leading to alterations of chromatin accessibility (Dion et al., 2005; Henikoff, 2005; Bannister and Kouzarides, 2011). That only a global alteration of histone acetylation can affect expression should be especially true for highly transcribed genes, where the chromatin is easily accessible to RNA Pol II and only a simultaneous alteration of various factors should significantly affect the transcriptional rate. In any case, further experiments will be required to reveal the precise roles and the mechanisms of HDA101 function in regulating transcriptionally active genes. Furthermore, additional efforts are required to establish whether binding of active genes is specific for maize HDA101 and for its function in seed development, or if this is a widespread feature of different plant HDACs, in regulating histone acetylation in various organs.
For inactive genes, we also observed results analogous to those reported for mammalian HDACs (Wang et al., 2002; Wang et al., 2009). Indeed, our results show that HDA101 binding is required for initiating and/or maintaining the repression and histone deacetylation of a small subset of genes that have low expression or are silenced during seed development. Two distinct, although not mutually exclusive, mechanisms can explain the HDA101-mediated repression of these transcriptionally inactive genes. The first mechanism involves the transient binding of HATs and HDA101 at inactive genes, leading to the establishment of a chromatin state that is potentiated for activation or silencing upon receiving external signals (Wang et al., 2009). The second mechanism involves HDA101 binding to inactive genes along with corepressors and in response to signals that induce repression (Shahbazian and Grunstein, 2007). Our finding that HDA101 interacts with NFC103 and SNL1 supports this mechanism. In addition, other members of the maize NFC/MSI family, namely, the NFC101/NFC102 proteins, recruit Rpd3-type HDACs at genes involved in maize flowering, such as indeterminate1 and the florigen gene zea centroradialis8, as well as at transposable elements, to promote their repression (Mascheretti et al., 2013). Furthermore, Rpd3-type HDACs are present in multiprotein complexes along with distinct corepressors in a wide range of eukaryotes (Shahbazian and Grunstein, 2007). Similarly, in plants, these complexes act to repress transcription during various physiological processes, including flowering, seed maturation, and leaf development (Ma et al., 2013; Liu et al., 2014).
Relationship between HDA101 and Histone Acetylation
Maize was used as a model system to examine HDAC activity and pioneering studies employed purified extracts from germinating embryos to show that different proteins possess HDAC activity (Lechner et al., 1996; Lusser et al., 2001). Subsequent studies demonstrated that these proteins are encoded by hda genes, including hda101 (originally named HD1B-I and ZmRpd3I) (Rossi et al., 1998; Lechner et al., 2000). A previous study (Rossi et al., 2007) and our results provide further evidence that HDA101 is associated with the removal of acetyl groups from histone tail lysine residues. Indeed, we show that loss of HDA101 affects the global levels of different acetylated histones and induces an increase of H4K5ac in 34% of the HDA101-bound genes. Nevertheless, 63% of its direct targets do not show alterations of H4K5ac levels. This observation can be explained in several ways. First, HDA101 may affect different lysine residues in different targets. Indeed, posttranslational modification of HDACs, possibly affecting the presence of these enzymes in distinct complexes, can modulate the substrate specificity of HDACs (Kölle et al., 1999). In addition, HDAC substrate specificity can be influenced by distinct patterns of preexisting histone modifications, which can specify different targets (Rossi et al., 2007). A second possibility is that loss of HDA101 is not sufficient to induce hyperacetylation of all direct targets because these genes can also be targeted by different HDACs that act redundantly with HDA101. In this context, it is worth noting that Rossi et al. (2007) found that the seedlings of an hda101 mutant line showed no changes in the transcript and protein levels of other Rpd3-type HDACs. Finally, HDA101 binding may not necessarily lead to histone deacetylation in all direct targets. Indeed, HDAC activity may be induced only upon receiving specific signals and/or along with the binding of additional components (e.g., cofactors and corepressors) of the histone-modifying complexes.
HDA101-Mediated Regulation of Specific Genes during Seed Development
In this study, we used the ChIP-seq approach to identify genes directly bound by HDA101 (i.e., HDA101 direct targets) in 4-DAP seeds of the B73 inbred line. In addition, we employed RNA-seq to analyze transcriptome changes in 4-DAP seeds of hda101 mutants compared with B73, thus revealing the totality of genes (i.e., direct plus indirect HDA101 targets) whose expression is affected by HDA101. The results provide several insights into the HDA101-mediated regulation of maize seed development. For example, an important observation arising from our study is that most of the genes associated with HDA101 binding, which concomitantly exhibit an alteration of their transcript levels, have a tissue-specific expression, with lower expression in the seed. In B73, this group of genes is downregulated or silenced by HDA101. Although the exact biological functions of these genes during seed development are currently unknown, our findings suggest that their expression must be negatively regulated to allow correct seed development. We also report that most of the markers for seed cell differentiation and especially for BETL formation are downregulated in the hda101 mutants. These genes are not bound by HDA101; hence, they represent indirect targets. These targets include mrp-1, which encodes a MYB-like transcription factor, identified as a key regulator of transfer cell differentiation (Gómez et al., 2009). MRP-1 protein transcriptionally activates a number of transfer cell-specific genes (Gómez et al., 2002), including maternally expressed gene1 (meg1) (Gutiérrez-Marcos et al., 2004), betl-1 and betl-2 (Gómez et al., 2002), and tcrr-1 (Muñiz et al., 2006). Our results showed that, in addition to mrp-1, all of the targets of MRP1 are also downregulated in the hda101 mutants. These findings suggest that HDA101 acts upstream of mrp-1 and possibly of other transcription factor-encoding loci to regulate transfer cell differentiation. Since we found that most of the HDA101 direct targets do not exhibit variation in their transcript level following loss of HDA101 in 4-DAP seeds, it can be predicted that, similarly to mrp-1, the major effect of HDA101 in regulating gene expression occurs indirectly, as HDA101 may bind and modulate expression of only a few key regulators.
HDA101-Mediated Regulation Is a Key Step in Transfer Cell Development and May Determine Seed Size
The BETL is one of the first tissue types to be recognizable in the early developing endosperm (Olsen, 2004). It forms in the peripheral cell layer of the endosperm at the base of the kernel, adjacent to the pedicel tissues of the maternal plant and facilitates the flow of assimilates into the starchy cells. Several lines of evidence indicate that defects of transfer cell development affect solute uptake by the developing kernel, leading to a reduction of grain size. For example, meg1 positively regulates transfer tissue development and function, thereby promoting the nutrient uptake capacity of the seed and ultimately increasing biomass yield (Costa et al., 2012). The empty pericarp6 mutant also affects the differentiation of BETL cells and the expression of BETL marker genes, leading to early arrest in grain development (Chettoor et al., 2015). Similarly, the reduced grain filling1 mutation is associated with reduced expression of BETL markers and induces a 70% decrease in seed weight at maturity (Maitz et al., 2000). Finally, the globby1, empty pericarp4, and baseless1 mutants exhibit abnormal BETL in early seed development and ultimately show aborted seed phenotypes (Costa et al., 2003; Gutiérrez-Marcos et al., 2006, 2007). In agreement with the previously mentioned reports, our results indicate that HDA101 loss is associated with defects in the BETL region and with indirect downregulation of BETL-specific marker genes, suggesting that these alterations lead to the phenotype of smaller seeds observed in hda101 mutant lines. This observation increases our knowledge of the involvement of chromatin-related mechanisms in the regulation of maize grain size, thus opening up new strategies to modulate this key agronomic trait that is associated with improved yield and seed quality.
METHODS
Plant Materials
Production of hda101-overexpressing OE1 and OE51 transgenic lines (renamed hda101-1 and hda101-2 mutant lines in this article) and their introgression into the B73 genetic background was described by Rossi et al. (2007). These plants, along with the wild-type B73 control, were grown in the experimental field in Shangzhuang (China Agricultural University, Beijing). The kernels of hda101-1 were harvested at 2, 3, 4, 6, 8, 10, and 12 DAP, and the kernels of hda101-2 were harvested at 4 and 8 DAP and stored at −80°C for experiments during the years 2012 to 2014.
Generation of Transgenic Arabidopsis thaliana
The cDNA of hda101 containing the full-length open reading frame was amplified by PCR with the primer pair hda101-ORF-L and hda101-ORF-R (Supplemental Data Set 7). The amplified fragment was cloned into the pENTR/D-TOPO entry vector (Life Technologies) and was then recombined into the pEarleyGate 101 (C-YFP-HA) destination vector, following the manufacturer’s instructions. This construct was used to transform Arabidopsis (Col-0) via the floral dip method using Agrobacterium tumefaciens strain GV3101 (Clough and Bent, 1998). Transgenic plants were selected on 0.5× Murashige and Skoog medium containing 0.1% herbicide (BASTA). The herbicide-resistant seedlings were transferred to a mixture of soil and vermiculite (2:1), and homozygous lines were generated by self-fertilization.
Immunoblots
The harvested kernels were ground in liquid nitrogen and the powder was extracted with 4× loading buffer (200 mM Tris-HCl, pH 6.8, 40% glycerol, 8% SDS, and 20% β-mercaptoethanol) at 65°C for 5 min. The solution was placed on ice for 5 min and centrifuged for 10 min at 12,000g at room temperature. The protein extracts were then separated on 10% SDS-PAGE and transferred to a PVDF membrane (Bio-Rad 162-0177) for 60 min, 300 mA in temperature-controlled conditions. The membrane was blocked using incubation with 5% defatted milk for 1 h. The primary antibody (1:5000), dissolved in 5% defatted milk in 1× TBST (1× TBS with 0.1% Tween 20), was incubated at 4°C overnight. After washing three times for 10 min with 1× TBST, the secondary antibody (1:3000) was added in 5% defatted milk-1× TBST solution and incubated for 1 h, followed by three additional washing steps of 10 min with 1× TBST. For detection, the two-component reagent Clarity Western ECL Substrate (Bio-Rad 170-5060) was used. The signal was exposed and detected with x-ray film. Three replications were performed and the band signals were measured using Quantity One software (Bio-Rad). The value of the optical density was used to quantify the abundance of the hybridizing polypeptide. The difference was analyzed by Fisher's LSD test, and different letters indicate means that differ significantly (P < 0.05).
Histological Analysis
Immature B73 and hda101-1 mutant seeds were sampled from the cobs of ears from 2 to 6 DAP. Kernels were fixed in FAA solution (63% ethanol, 5% acetic acid, and 2% formaldehyde) for 10 min at room temperature. After dehydration in a graded ethanol series, sections were slowly infiltrated with Steedman’s wax, sectioned at 10 μm with a microtome, adhered to cover slips coated with Mayer’s egg albumin adhesive, and dewaxed with absolute ethanol. Sections (10 μm) were stained with safranin-fast green. Images were collected using a Nikon Eclipse Ti inverted research microscope. The endosperm cell numbers were manually calculated for three kernels and the differences between B73 and the hda101-1 line were determined by Student’s t test.
RT-qPCR Analysis
Reverse transcription was performed using the Reverse Transcriptase M-MLV (RNase H−) following the manufacturer’s instructions. Real-time PCR was performed using the SYBR Green I Master Mix on a CFX96 real-time system (Bio-Rad). PCR conditions were as follows: 94°C for 5 min, followed by 40 cycles of 94°C for 15 s, 60°C for 15 s, 72°C for 20 s, and then 72°C for 5 min. Relative transcript amount differences were calculated using the 2−ΔCT method (Livak and Schmittgen, 2001). For each sample, the PCR amplification was repeated three times, and the average values of 2−ΔCT were used to determine the differences of gene expression by Student’s t test. Three biological replications were performed and the results showed similar trends; thus, one replication was shown in the figures.
ChIP Assay and Analysis of ChIP-Seq Data
ChIP-seq using antibody against HDA101 in B73 and the hda101-1 mutant was performed with two replications. ChIP-seq using antibody against H4K5ac in B73 and the hda101-1 mutant was performed with three replications. Maize (Zea mays) kernels were cross-linked with 1% formaldehyde in GB buffer (0.4 M sucrose, 10 mM Tris-HCl, pH 8.0, and 1 mM EDTA). Fixation was done with 20 min of vacuum infiltration at room temperature and was stopped by adding glycine at 0.17 M final concentration followed by 5 min of vacuum infiltration at room temperature. Fixed samples were washed three times with water at 4°C, dried with towels, frozen, and ground to powder in liquid N2. Nuclei pellets were suspended in a buffer containing 0.25 M sucrose, 10 mM Tris-HCl, pH 8, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF (Sigma-Aldrich; P7626), and protease inhibitors (one mini tablet per milliliter; Roche). The suspensions were transferred to microfuge tubes and centrifuged at 12,000g for 10 min. The pellets were suspended in 1.7 M sucrose, 10 mM Tris-HCl, pH 8, 2 mM MgCl2, 0.15% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitors (one tablet in 30 mL solution; Roche 11873580001), and centrifuged through a layer of the same buffer in microfuge tubes. The nuclear pellets were lysed in a buffer containing 50 mM Tris-HCl, pH 8, 10 mM EDTA, 1% SDS, and protease inhibitors (one tablet in 30 mL solution; Roche 11873580001). The lysed nuclei were sonicated with a Bioruptor (UCD-200) in a water bath at 4°C for 30 cycles with 30 s on and 30 s off. Immunoprecipitation was performed using 10 µg of chromatin, following a previously described protocol (Locatelli et al., 2009). Typically, 10 μL of affinity-purified anti-HDA101 and 5 μg of H4K5ac (Millipore 07-327) were used for immunoprecipitation. The precipitated DNA was dissolved in 30 μL 10 mM Tris-HCl, pH 7.5, and treated with RNase (DNase-free).
The ChIP-Seq DNA sample prep kit (Illumina) was used to prepare ChIP-seq libraries following the manufacturer’s instructions. The HiSeq 2500 sequencing system was used to sequence the libraries. Finally, ∼2 GB high-quality 100- or 125-bp paired-end reads were generated from each library. ChIP-seq reads were mapped to the B73 genome (release AGPv3) using Bowtie2 (version 2.2.4; parameters:–no-unal–sensitive -I 0 -X 600–fr–end-to-end -3 5–non-deterministic -p 15–phred33–no-discordant) (Langmead and Salzberg, 2012) with default parameters, and only unique alignments were kept by a customized PERL script. The SAM-formatted output files generated by Bowtie2 were transformed to BAM format, followed by sorting and index using Samtools 0.1.19 (Li et al., 2009). Peaks were called by MACS2 (-g 2.06e+9 -B–SPMR) (Zhang et al., 2008), and all duplicate reads were included in HDA101 and H4K5ac ChIP-seq and the q-value cutoff applied to calculate statistical significance was < 0.05. Other parameters were set to default values. The abundance of the HDA101 ChIP-seq reads was normalized to the corresponding input, the relative enrichment was calculated as a ratio of the values for B73 over those of the hda101-1 mutant, and HDA101 binding sites were defined as regions with 5× or higher HDA101 signal in B73 with respect to the hda101-1 mutant line. The genomic regions enriched with H4K5ac were determined by comparing the ChIP library with the input DNA library. The differential analysis between hda101-1 mutant and B73 were determined by running MACS2 on hda101-1 against B73 and vice versa. The peak summits were employed to define the location types in the genomes, using the subcommand intersectBed included in BEDTools (Quinlan and Hall, 2010) and custom-made PERL scripts. The WindowBed subcommand, contained in BEDTools, was used to identify the peaks within the genic regions (including 5 kb up- and downstream of the gene). Peak annotation was performed using PeakAnalyzer (Salmon-Divon et al., 2010).
ChIP-qPCR
The ChIP experiments were performed with three biological replications with similar results. The immunoprecipitated and purified DNA was used for quantitative real-time PCR in three replications to amplify the examined target sequences. The fold enrichment was normalized to the input chromatin and then the control gene, which is indicated in the figure legend. The fold enrichment is represented as mean ± se.
RNA Sequencing
Total RNA was isolated from 4-DAP seeds of B73 and the hda101-1 mutant line with three biological replications using a TRIzol kit (Invitrogen), according to the manual. Paired-end sequencing libraries with an average insert size of 400 bp were prepared with a TruSeq RNA Sample Preparation Kit v2 (Illumina) and sequenced on the HiSeq2000 (Illumina) according to the manufacturer’s instructions. Raw data obtained from Illumina sequencing were processed and filtered using the Illumina pipeline (http://www.Illumina.com) to generate FastQ files. Finally, ∼4 Gb high-quality 100- or 125-bp paired-end reads were generated from each library. RNA-seq reads were aligned to the maize B73 reference genome AGPv3 (Schnable et al., 2009) using Splice Junction Mapper TopHat2 version 2.0.9 with parameters–segment-mismatches 1–segment-length 50–no-coverage-search–microexon-search–mate-std-dev 120 -p 10 -i 30 -I 8000 -g 20 -a 4 -N 5–read-edit-dist 5 –r 40 or 100 (Kim et al., 2013) with build-in Bowtie2 mapping program. HTseq (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html) was used to count the read counts mapped to each of the hgenes. The read counts were then normalized to the reads per kilobases per million reads to show the relative level of expression. Bioconductor package “edgeR” (ver. 3.2.3) was used for differential expression analysis (Robinson et al., 2010). The genes showing an absolute value of log2 (fold change; hda101-1 mutant/B73) ≥1 and adjusted P value (false discovery rate) of < 0.05 were considered as differentially expressed genes. Identification of GO categories significantly enriched within up- and downregulated genes (P value ≤ 0.05) was done using AgriGO (Du et al., 2010).
Coimmunoprecipitation Assays
B73 kernels at 4 DAP were collected and ground in liquid nitrogen. Protein extracts were prepared in immunoprecipitation buffer (2.7 mM KCl, 137 mM NaCl, 10 mM Na2HPO4, 2 mM KH2PO4, 0.5 mM β-mercaptoethanol, and 1 mM PMSF) and 1% protease cocktail (Roche 04693132001). The suspension was homogenized with a homogenizer for 2 min on ice and filtered through two layers of Miracloth. The soluble protein fraction was collected by two rounds of centrifugation at 13,000g for 20 min at 4°C. The supernatant was precleared by adding 40 μL of protein A-decorated Dynabeads (Invitrogen 10002D) at 4°C for 1 h. The tube was placed on the magnet to remove the beads. The solution was incubated with 8 μL of HDA101 antibody at 4°C overnight and then 60 μL of Dynabeads was added to the solution and incubated at 4°C for 3 h. The beads were collected and washed three times for 10 min with 1 mL immunoprecipitation buffer. Beads were heated for 10 min at 70°C with 40 μL 1× loading buffer (50 mM Tris HCl, pH 6.8, 10% glycerol, 2% SDS, and 5% β-mercaptoethanol). Purified proteins were separated by 10% SDS-PAGE. The gels were stained by silver staining, and all visualized bands were digested with trypsin (final concentration 0.067 µg/µL) and identified using liquid chromatography-tandem mass spectrometry analysis as previously described (Nallamilli et al., 2013).
Yeast Two-Hybrid Assays
The coding sequences of hda101 were amplified by primer sets listed in Supplemental Data Set 7 and cloned into the pGADT7 vector. The snl1 and nfc103 sequences were introduced into the pGBKT7 vector. The plasmids were transformed into the yeast strain AH109 and yeast cells were grown on minimal medium (-Leu/-Trp) at 30°C for 2 d according to the manufacturer’s instructions (Clontech). The positive colonies were plated onto minimal medium (-Leu/-Trp/-His/-Ade) containing 20 mg/mL 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (X-alpha-gal; Clontech 630462) to test for possible interactions.
Antibodies
The peptide epitope located between amino acid 466 and 475 of the HDA101 sequence was used to produce a specific mouse monoclonal antibody, which was performed by Abmart. The antibodies used in immunoblots and ChIP were purchased from Sigma-Aldrich (actin, A0480), Millipore (H3ac [06-599], H4ac [06-598], H3K9ac [07-352], H4K5ac [07-327], H4K8ac [06-760], H4K12ac [07-595]), and Abcam (H3, ab1791).
Primers for RT-qPCR and ChIP-qPCR
The PCR primer sequences used in this study are listed in Supplemental Data Set 7.
Accession Numbers
Gene ID numbers for all sequences described in this research can be found in Supplemental Data Set 7.All high-throughput sequencing data are available at the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP067369.
Supplemental Data
Supplemental Figure 1. Alignment of amino acid sequences of different maize Rpd3-type HDACs.
Supplemental Figure 2. Analysis of the hda101 transcript level in transgenic lines.
Supplemental Figure 3. ChIP-quantitative PCR assay for 12 loci bound by HDA101.
Supplemental Figure 4. Distribution of HDA101 protein binding sites with respect to genes and to transposable elements along maize chromosomes 2-10.
Supplemental Figure 5. Relationship between HDA101 binding location in TSS, gene body, and TTS region with gene expression, gene length, and tissue specificity of the target genes.
Supplemental Figure 6. H4K5ac level in a selected number of HDA101 target genes.
Supplemental Figure 7. RT-qPCR analysis of 17 sequences differentially expressed in 4-DAP seeds of hda101-1 mutant and B73 plants.
Supplemental Figure 8. GO functional analysis of genes differentially expressed in 4-DAP seeds of hda101-1 mutant and B73 plants.
Supplemental Figure 9. Expression alteration of HDA101 target genes in the hda101-1 mutant compared to B73.
Supplemental Figure 10. The RNA level of four HDA101 target genes in seeds at 4 and 8 DAP of the hda101-2 mutant and wild-type B73.
Supplemental Figure 11. Identification of HDA10-interacting proteins.
Supplemental Data Set 1. Peaks corresponding to HDA101 binding sites, which are uniquely or significantly enriched in 4-DAP seeds of wild-type B73 compared to the hda101-1 mutant plants.
Supplemental Data Set 2. H4K5ac distribution in wild-type B73 and hda101-1 mutant.
Supplemental Data Set 3. HDA101 target genes with altered H4K5ac levels in the 4-DAP seeds of hda101-1 mutant compared to wild-type B73 plants.
Supplemental Data Set 4. List of transcripts differentially expressed in 4-DAP seeds of hda101-1 mutant and wild-type B73 plants.
Supplemental Data Set 5. List of HDA101 direct targets exhibiting an increase of H4K5ac level in the hda101-1 mutant.
Supplemental Data Set 6. Putative HDA101-interacting proteins identified by co-IP assay.
Supplemental Data Set 7. Primers and gene IDs for sequences used in this study.
Supplementary Material
Acknowledgments
We thank Xiaoyu Zhang (University of Georgia) for suggestions about experimental design, Xiangfeng Wang (China Agricultural University, China) for help with data analysis, and Keji Zhao (National Institutes of Health), Levi Yant (Harvard University), Alexandra Lusser (Innsbruck Medical University, Austria), and Joe Colasanti (University of Guelph, Canada) for critical reading of this manuscript. This work was supported by the National Basic Research Program of China (973 Program) (2012CB910900), the 863 Project Grant (2012AA10A309), and the National Natural Science Foundation of China (31471478). Work in the V.R. lab was supported by special grants from the Italian Ministry of Education, University, and Research (MIUR) and the National Research Council of Italy (CNR) for the Epigenomics Flagship Project (EPIGEN).
AUTHOR CONTRIBUTIONS
H.Y., Q.S., Z.N., and Y.Y. conceived this project and designed all experiments. H.Y., X.L., M.X., J.D., Z.H., and H.P. performed experiments. V.R. provided seeds of transgenic lines and contributed to manuscript revision. X.L. analyzed data. Y.Y. wrote the article.
Glossary
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- DAP
days after pollination
- BETL
basal endosperm transfer layer
- ChIP-seq
chromatin immunoprecipitation followed by high-throughput sequencing
- OE
overexpression
- TSS
transcription start site
- GO
Gene Ontology
- TTS
transcription termination sites
Footnotes
Articles can be viewed online without a subscription.
References
- Bannister A.J., Kouzarides T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F., Chaudhury A. (2009). Parental memories shape seeds. Trends Plant Sci. 14: 550–556. [DOI] [PubMed] [Google Scholar]
- Berger S.L. (2007). The complex language of chromatin regulation during transcription. Nature 447: 407–412. [DOI] [PubMed] [Google Scholar]
- Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., Workman J.L. (2005). Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592. [DOI] [PubMed] [Google Scholar]
- Chen J., Zeng B., Zhang M., Xie S., Wang G., Hauck A., Lai J. (2014). Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol. 166: 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.T., Luo M., Wang Y.Y., Wu K. (2010). Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61: 3345–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chettoor A.M., Yi G., Gomez E., Hueros G., Meeley R.B., Becraft P.W. (2015). A putative plant organelle RNA recognition protein gene is essential for maize kernel development. J. Integr. Plant Biol. 57: 236–246. [DOI] [PubMed] [Google Scholar]
- Chung P.J., Kim Y.S., Jeong J.S., Park S.H., Nahm B.H., Kim J.K. (2009). The histone deacetylase OsHDAC1 epigenetically regulates the OsNAC6 gene that controls seedling root growth in rice. Plant J. 59: 764–776. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Costa L.M., Gutierrez-Marcos J.F., Brutnell T.P., Greenland A.J., Dickinson H.G. (2003). The globby1-1 (glo1-1) mutation disrupts nuclear and cell division in the developing maize seed causing alterations in endosperm cell fate and tissue differentiation. Development 130: 5009–5017. [DOI] [PubMed] [Google Scholar]
- Costa L.M., Yuan J., Rouster J., Paul W., Dickinson H., Gutierrez-Marcos J.F. (2012). Maternal control of nutrient allocation in plant seeds by genomic imprinting. Curr. Biol. 22: 160–165. [DOI] [PubMed] [Google Scholar]
- De Paoli E., Dorantes-Acosta A., Zhai J., Accerbi M., Jeong D.H., Park S., Meyers B.C., Jorgensen R.A., Green P.J. (2009). Distinct extremely abundant siRNAs associated with cosuppression in petunia. RNA 15: 1965–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Bellizzi Mdel.R., Ning Y., Meyers B.C., Wang G.L. (2012). HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell 24: 3783–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion M.F., Altschuler S.J., Wu L.F., Rando O.J. (2005). Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl. Acad. Sci. USA 102: 5501–5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez E., Royo J., Guo Y., Thompson R., Hueros G. (2002). Establishment of cereal endosperm expression domains: identification and properties of a maize transfer cell-specific transcription factor, ZmMRP-1. Plant Cell 14: 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez E., Royo J., Muñiz L.M., Sellam O., Paul W., Gerentes D., Barrero C., López M., Perez P., Hueros G. (2009). The maize transcription factor myb-related protein-1 is a key regulator of the differentiation of transfer cells. Plant Cell 21: 2022–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Marcos J.F., Costa L.M., Biderre-Petit C., Khbaya B., O’Sullivan D.M., Wormald M., Perez P., Dickinson H.G. (2004). maternally expressed gene1 Is a novel maize endosperm transfer cell-specific gene with a maternal parent-of-origin pattern of expression. Plant Cell 16: 1288–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Marcos J.F., Costa L.M., Evans M.M. (2006). Maternal gametophytic baseless1 is required for development of the central cell and early endosperm patterning in maize (Zea mays). Genetics 174: 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Marcos J.F., Dal Prà M., Giulini A., Costa L.M., Gavazzi G., Cordelier S., Sellam O., Tatout C., Paul W., Perez P., Dickinson H.G., Consonni G. (2007). empty pericarp4 encodes a mitochondrion-targeted pentatricopeptide repeat protein necessary for seed development and plant growth in maize. Plant Cell 19: 196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. (2005). Histone modifications: combinatorial complexity or cumulative simplicity? Proc. Natl. Acad. Sci. USA 102: 5308–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L., Bouveret R., Gruissem W. (2005). MSI1-like proteins: an escort service for chromatin assembly and remodeling complexes. Trends Cell Biol. 15: 295–302. [DOI] [PubMed] [Google Scholar]
- Hollender C., Liu Z. (2008). Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 50: 875–885. [DOI] [PubMed] [Google Scholar]
- Hu Y., Qin F., Huang L., Sun Q., Li C., Zhao Y., Zhou D.X. (2009). Rice histone deacetylase genes display specific expression patterns and developmental functions. Biochem. Biophys. Res. Commun. 388: 266–271. [DOI] [PubMed] [Google Scholar]
- Joshi A.A., Struhl K. (2005). Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell 20: 971–978. [DOI] [PubMed] [Google Scholar]
- Keogh M.C., et al. (2005). Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölle D., Brosch G., Lechner T., Pipal A., Helliger W., Taplick J., Loidl P. (1999). Different types of maize histone deacetylases are distinguished by a highly complex substrate and site specificity. Biochemistry 38: 6769–6773. [DOI] [PubMed] [Google Scholar]
- Kurdistani S.K., Robyr D., Tavazoie S., Grunstein M. (2002). Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 31: 248–254. [DOI] [PubMed] [Google Scholar]
- Laherty C.D., Yang W.M., Sun J.M., Davie J.R., Seto E., Eisenman R.N. (1997). Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89: 349–356. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauria M., Rossi V. (2011). Epigenetic control of gene regulation in plants. Biochim. Biophys. Acta 1809: 369–378. [DOI] [PubMed] [Google Scholar]
- Lechner T., Lusser A., Brosch G., Eberharter A., Goralik-Schramel M., Loidl P. (1996). A comparative study of histone deacetylases of plant, fungal and vertebrate cells. Biochim. Biophys. Acta 1296: 181–188. [DOI] [PubMed] [Google Scholar]
- Lechner T., Lusser A., Pipal A., Brosch G., Loidl A., Goralik-Schramel M., Sendra R., Wegener S., Walton J.D., Loidl P. (2000). RPD3-type histone deacetylases in maize embryos. Biochemistry 39: 1683–1692. [DOI] [PubMed] [Google Scholar]
- Li G., et al. (2014). Temporal patterns of gene expression in developing maize endosperm identified through transcriptome sequencing. Proc. Natl. Acad. Sci. USA 111: 7582–7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yang S., Zhao M., Luo M., Yu C.W., Chen C.Y., Tai R., Wu K. (2014). Transcriptional repression by histone deacetylases in plants. Mol. Plant 7: 764–772. [DOI] [PubMed] [Google Scholar]
- Liu X., Yu C.W., Duan J., Luo M., Wang K., Tian G., Cui Y., Wu K. (2012). HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol. 158: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Locatelli S., Piatti P., Motto M., Rossi V. (2009). Chromatin and DNA modifications in the Opaque2-mediated regulation of gene transcription during maize endosperm development. Plant Cell 21: 1410–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M.A., Larkins B.A. (1993). Endosperm origin, development, and function. Plant Cell 5: 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusser A., Kölle D., Loidl P. (2001). Histone acetylation: lessons from the plant kingdom. Trends Plant Sci. 6: 59–65. [DOI] [PubMed] [Google Scholar]
- Ma X., Lv S., Zhang C., Yang C. (2013). Histone deacetylases and their functions in plants. Plant Cell Rep. 32: 465–478. [DOI] [PubMed] [Google Scholar]
- Maitz M., Santandrea G., Zhang Z., Lal S., Hannah L.C., Salamini F., Thompson R.D. (2000). rgf1, a mutation reducing grain filling in maize through effects on basal endosperm and pedicel development. Plant J. 23: 29–42. [DOI] [PubMed] [Google Scholar]
- Mascheretti I., Battaglia R., Mainieri D., Altana A., Lauria M., Rossi V. (2013). The WD40-repeat proteins NFC101 and NFC102 regulate different aspects of maize development through chromatin modification. Plant Cell 25: 404–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M., Naganuma T., Tsutsumi K., Saitoh Y. (2010). The syncytium-specific expression of the Orysa;KRP3 CDK inhibitor: implication of its involvement in the cell cycle control in the rice (Oryza sativa L.) syncytial endosperm. J. Exp. Bot. 61: 791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K., et al. (2012). A genome-wide regulatory framework identifies maize pericarp color1 controlled genes. Plant Cell 24: 2745–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundade R., Ozer H.G., Wei H., Prabhu L., Lu T. (2014). Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond. Cell Cycle 13: 2847–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñiz L.M., Gómez E., Guyon V., López M., Khbaya B., Sellam O., Peréz P., Hueros G. (2014). A PCR-based forward genetics screening, using expression domain-specific markers, identifies mutants in endosperm transfer cell development. Front. Plant Sci. 5: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñiz L.M., Royo J., Gómez E., Barrero C., Bergareche D., Hueros G. (2006). The maize transfer cell-specific type-A response regulator ZmTCRR-1 appears to be involved in intercellular signalling. Plant J. 48: 17–27. [DOI] [PubMed] [Google Scholar]
- Nallamilli B.R., Zhang J., Mujahid H., Malone B.M., Bridges S.M., Peng Z. (2013). Polycomb group gene OsFIE2 regulates rice (Oryza sativa) seed development and grain filling via a mechanism distinct from Arabidopsis. PLoS Genet. 9: e1003322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen O.A. (2001). Endosperm development: Cellularization and cell fate specification. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 233–267. [DOI] [PubMed] [Google Scholar]
- Olsen O.A. (2004). Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 16 (Suppl): S214–S227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R., Müller A., Napoli C.A., Selinger D.A., Pikaard C.S., Richards E.J., Bender J., Mount D.W., Jorgensen R.A. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30: 5036–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrella G., Lopez-Vernaza M.A., Carr C., Sani E., Gosselé V., Verduyn C., Kellermeier F., Hannah M.A., Amtmann A. (2013). Histone deacetylase complex1 expression level titrates plant growth and abscisic acid sensitivity in Arabidopsis. Plant Cell 25: 3491–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peserico A., Simone C. (2011). Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J. Biomed. Biotechnol. 2011: 371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst A.V., Fagard M., Proux F., Mourrain P., Boutet S., Earley K., Lawrence R.J., Pikaard C.S., Murfett J., Furner I., Vaucheret H., Mittelsten Scheid O. (2004). Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16: 1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F., Pokholok D.K., Hannett N.M., Rinaldi N.J., Chandy M., Rolfe A., Workman J.L., Gifford D.K., Young R.A. (2004). Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 16: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi V., Hartings H., Motto M. (1998). Identification and characterisation of an RPD3 homologue from maize (Zea mays L.) that is able to complement an rpd3 null mutant of Saccharomyces cerevisiae. Mol. Gen. Genet. 258: 288–296. [DOI] [PubMed] [Google Scholar]
- Rossi V., Locatelli S., Lanzanova C., Boniotti M.B., Varotto S., Pipal A., Goralik-Schramel M., Lusser A., Gatz C., Gutierrez C., Motto M. (2003). A maize histone deacetylase and retinoblastoma-related protein physically interact and cooperate in repressing gene transcription. Plant Mol. Biol. 51: 401–413. [DOI] [PubMed] [Google Scholar]
- Rossi V., Locatelli S., Varotto S., Donn G., Pirona R., Henderson D.A., Hartings H., Motto M. (2007). Maize histone deacetylase hda101 is involved in plant development, gene transcription, and sequence-specific modulation of histone modification of genes and repeats. Plant Cell 19: 1145–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon-Divon M., Dvinge H., Tammoja K., Bertone P. (2010). PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics 11: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Sekhon R.S., Lin H., Childs K.L., Hansey C.N., Buell C.R., de Leon N., Kaeppler S.M. (2011). Genome-wide atlas of transcription during maize development. Plant J. 66: 553–563. [DOI] [PubMed] [Google Scholar]
- Shahbazian M.D., Grunstein M. (2007). Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76: 75–100. [DOI] [PubMed] [Google Scholar]
- Song C.P., Agarwal M., Ohta M., Guo Y., Halfter U., Wang P., Zhu J.K. (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17: 2384–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Kikuchi A., Kamada H. (2008). The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 146: 149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To T.K., et al. (2011). Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet. 7: e1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto S., Locatelli S., Canova S., Pipal A., Motto M., Rossi V. (2003). Expression profile and cellular localization of maize Rpd3-type histone deacetylases during plant development. Plant Physiol. 133: 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Kurdistani S.K., Grunstein M. (2002). Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 298: 1412–1414. [DOI] [PubMed] [Google Scholar]
- Wang Z., Cao H., Chen F., Liu Y. (2014). The roles of histone acetylation in seed performance and plant development. Plant Physiol. Biochem. 84: 125–133. [DOI] [PubMed] [Google Scholar]
- Wang Z., Zang C., Cui K., Schones D.E., Barski A., Peng W., Zhao K. (2009). Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138: 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterborg J.H. (2002). Dynamics of histone acetylation in vivo. A function for acetylation turnover? Biochem. Cell Biol. 80: 363–378. [DOI] [PubMed] [Google Scholar]
- Wu K., Tian L., Malik K., Brown D., Miki B. (2000). Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J. 22: 19–27. [DOI] [PubMed] [Google Scholar]
- Xu C.R., Liu C., Wang Y.L., Li L.C., Chen W.Q., Xu Z.H., Bai S.N. (2005). Histone acetylation affects expression of cellular patterning genes in the Arabidopsis root epidermis. Proc. Natl. Acad. Sci. USA 102: 14469–14474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.W., Liu X., Luo M., Chen C., Lin X., Tian G., Lu Q., Cui Y., Wu K. (2011). HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol. 156: 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., et al. (2014). Genome-wide high resolution parental-specific DNA and histone methylation maps uncover patterns of imprinting regulation in maize. Genome Res. 24: 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Zhang L., Duan J., Miki B., Wu K. (2005). HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17: 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang X., He K., Charron J.B., Elling A.A., Deng X.W. (2010). Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol. Biol. 72: 585–595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.