Light-dependent degradation of LIR1 regulates the distribution of LFNR, which may play an important role in the allocation of reducing power depending upon environmental conditions.
Abstract
LIR1 (LIGHT-INDUCED RICE1) encodes a 13-kD, chloroplast-targeted protein containing two nearly identical motifs of unknown function. LIR1 is present in the genomes of vascular plants, mosses, liverworts, and algae, but not in cyanobacteria. Using coimmunoprecipitation assays, pull-down assays, and yeast two-hybrid analyses, we showed that LIR1 interacts with LEAF-TYPE FERREDOXIN-NADP+ OXIDOREDUCTASE (LFNR), an essential chloroplast enzyme functioning in the last step of photosynthetic linear electron transfer. LIR1 and LFNR formed high molecular weight thylakoid protein complexes with the TIC62 and TROL proteins, previously shown to anchor LFNR to the membrane. We further showed that LIR1 increases the affinity of LFNRs for TIC62 and that the rapid light-triggered degradation of the LIR1 coincides with the release of the LFNR from the thylakoid membrane. Loss of LIR1 resulted in a marked decrease in the accumulation of LFNR-containing thylakoid protein complexes without a concomitant decrease in total LFNR content. In rice (Oryza sativa), photosynthetic capacity of lir1 plants was slightly impaired, whereas no such effect was observed in Arabidopsis thaliana knockout mutants. The consequences of LIR1 deficiency in different species are discussed.
INTRODUCTION
The orthologs of LIGHT-INDUCED RICE1 (LIR1), which encodes a 13-kD protein with two almost identical repeated motifs of 16 amino acids (Supplemental Figure 1; Reimmann and Dudler, 1993; Ciannamea et al., 2007), have been identified in various angiosperm and gymnosperm species (Reimmann and Dudler, 1993; Abied and Holland, 1994; Teramoto et al., 1994; Quigley et al., 1996; Ciannamea et al., 2007). Both LIR1 motifs contain two highly conserved cysteine residues (Reimmann and Dudler, 1993; Ciannamea et al., 2007) and show a high degree of conservation among plant species, suggesting an important but yet uncharacterized functional role for the motifs. LIR1 is expressed in a circadian manner, with transcript levels increasing during the light period, reaching a maximum at the end of the day and diminishing in the dark (Reimmann and Dudler, 1993; Hayama et al., 2002; Bläsing et al., 2005; Ciannamea et al., 2007). Moreover, LIR1 expression is induced by low temperature (Ciannamea et al., 2007) and repressed by cytokinins (Teramoto et al., 1994) and soluble sugars (Bläsing et al., 2005). Based on the diurnal rhythm of LIR1 expression and a minor delay in flowering time of Arabidopsis thaliana plants heterologously expressing ryegrass (Lolium perenne) LIR1, the LIR1 protein has been speculated to participate in the photoperiodic regulation of flowering (Hayama et al., 2002; Ciannamea et al., 2007). However, due to a lack of lir1 mutants, the physiological role of LIR1 has remained elusive. In addition, lack of knowledge concerning its accumulation and localization hinders conclusions about the functional properties of the LIR1 protein.
LEAF-TYPE FERREDOXIN-NADP+ OXIDOREDUCTASE (LFNR) catalyzes electron transfer from ferredoxin (FD) to NADP+ during photosynthesis and generates the reducing power (as NADPH) required for carbon fixation (Ceccarelli et al., 2004). NADPH is also required for numerous other reactions in the chloroplast such as fatty acid synthesis and reactive oxygen species scavenging. In addition to LFNR, numerous chloroplast enzymes, such as nitrite reductase, ferredoxin-thioredoxin reductase, and sulfite reductase, compete for electrons derived from photosystem I via FD (Hanke and Mulo, 2013). Thus, LFNR plays an important role in regulating FD-dependent electron partitioning in the chloroplast, acting at the branch point of photosynthetic electron transfer and reductive metabolism. Nevertheless, the mechanisms determining electron distribution among different reactions according to environmental cues are not yet understood.
In most plants studied to date, two distinct nuclear genes encode two LFNR isoforms with different pIs (Hanke et al., 2005; Gummadova et al., 2007; Higuchi-Takeuchi et al., 2011). In Arabidopsis, both LFNR isoforms are present in three different chloroplast compartments: the thylakoid membrane, stroma, and inner envelope membrane (Hanke et al., 2005; Benz et al., 2009). Recently, two Arabidopsis chloroplast proteins, At-TROL and At-TIC62, which possess conserved proline-rich LFNR binding motif(s) in their C termini, have been shown to mediate anchoring of hydrophilic LFNR to the thylakoid membrane (Küchler et al., 2002; Balsera et al., 2007; Benz et al., 2009; Jurić et al., 2009; Alte et al., 2010; Lintala et al., 2014). LFNR forms thylakoid protein complexes of ∼500 and 190 kD with At-TIC62 and At-TROL, respectively (Benz et al., 2009; Jurić et al., 2009). Accumulation of the complexes is dynamic and light dependent. Illumination results in alkalization of the stroma, which dissociates LFNR from the thylakoid protein complexes, while in the dark, acidification leads to reassembly or membrane attachment of the LFNR-containing complexes (Benz et al., 2009). Notably, the physiological roles of the soluble and membrane-bound pools of LFNR have not yet been identified. Although both soluble and membrane-bound LFNR have been shown to form a complex with FD with the same dissociation constant, the rate constant of NADP+ photoreduction is higher in the membrane-bound complex than in the soluble complex in vitro (Forti et al., 1983; Forti and Bracale, 1984). Nevertheless, as plant performance is not markedly affected in Arabidopsis fnr1 and tic62 trol mutant plants completely lacking LFNR in thylakoid membranes, the soluble LFNR pool also appears to be photosynthetically competent (Lintala et al., 2007, 2014).
The delicately regulated distribution of LFNR between the stroma and thylakoid membrane might provide an efficient mechanism for direct distribution of reducing power to various stromal reactions (Benz et al., 2010). In this study, we obtained strong evidence that, in rice (Oryza sativa), LIR1 (Os-LIR1) is an LFNR-interacting partner that strengthens the binding of LFNR to the membrane anchor. Based on experiments using rice and Arabidopsis, we show that the allocation of LFNR between the stroma and chloroplast membrane is fine-tuned by light through the action of LIR1. Specifically, the regulation of LFNR allocation appears to be achieved by a rapid degradation of LIR1 upon illumination, which also results in the disassembly of the LFNR-containing thylakoid protein complexes. In darkness, LIR1 is stabilized and LFNR accumulates in the thylakoid membrane. Moreover, we compared the physiological consequences of LIR1 deficiency in different species, revealing that the photosynthetic capacity of rice lir1 knockout plants was slightly impaired, whereas no such effect was observed in the analogous Arabidopsis knockout mutants.
RESULTS
LIR1 Deficiency in Rice Results in Retarded Growth
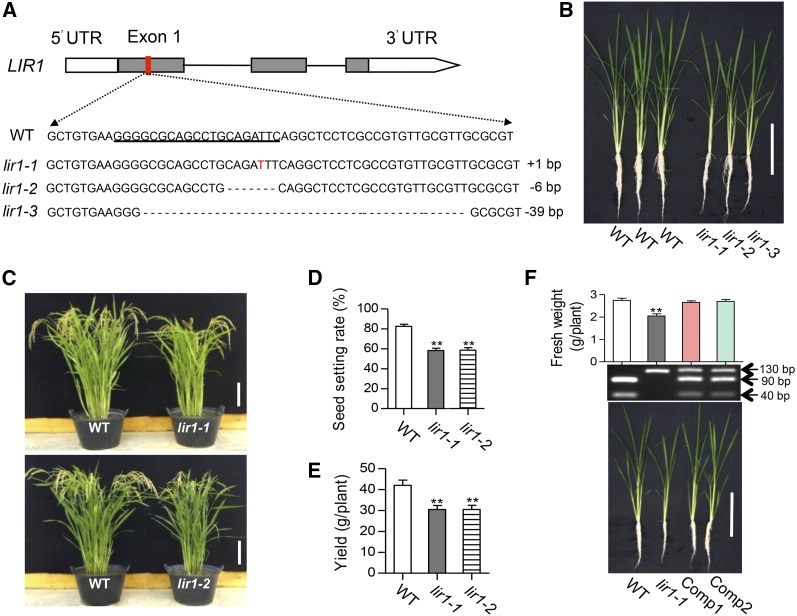
In order to assess the physiological function of LIR1 in rice, lir1 knockout plants were produced using the CRISPR-Cas9 (clustered regulatory interspaced short palindromic repeat-associated proteins) approach. Sequencing of 30 T0 plants resulted in the identification of nine transgenic lines carrying mutations in LIR1, and three unique mutants were recovered in the T1 generation. lir1-1 contained a 1-bp (T) insertion, lir1-2 contained a 6-bp deletion, and lir1-3 contained a 39-bp deletion, all in the middle of the first exon (Figure 1A). All three mutant lines exhibited similar visual phenotypes (Figure 1B), so we focused on two lines (lir1-1 and lir1-2) to study the physiological effects of LIR1 deficiency in planta. Although the mutants exhibited retarded growth when grown hydroponically (Figure 1B) and in soil (Figure 1C), they were fully viable and produced ∼75% as many seeds as wild-type plants (Figures 1D and 1E). Complementation of lir1 (lir1/PLIR1:LIR1-GFP) restored the growth of the mutant plants, indicating that the mutant phenotype in rice resulted from LIR1 deficiency (Figure 1F).
Figure 1.
Characteristics of Rice lir1 Mutants and Two Complementation Lines.
(A) Genomic structure model of rice LIR1 and sequence of mutant alleles constructed by the CRISPR-Cas system. Gray boxes show exons, black lines show introns, and white boxes show 5′- and 3′-untranslated regions (UTR). The underlined coding sequences from the region indicated by the red box in genomic structures model was designed as CRISPR-Cas target site (single guide RNA sequence). Os-lir1-1 contains a 1-bp insertion (T, red font), Os-lir1-2 has a 6-bp deletion (dashed line), and Os-lir1-3 has a 39-bp deletion (dashed line).
(B) Phenotypes of wild-type, lir1-1, lir1-2, and lir1-3 mutant plants grown in hydroponic culture for 30 d. Bar = 20 cm.
(C) Phenotypes of wild type, lir1-1, and lir1-2 mutant plants at the harvest stage. Plants were grown in soil for 100 d. Bars = 20 cm.
(D) Seed setting rate of wild-type, lir1-1, and lir1-2 mutant plants grown in soil.
(E) Yields of wild type, lir1-1, and lir1-2 mutant plants grown in soil.
(F) Phenotypes, fresh weights, and molecular characteristics of 30-d-old wild-type, lir1-1, and two independent complementation lines [lir1/PLIR1:LIR1-GFP(1) and lir1/PLIR1:LIR1-GFP(2), labeled as Comp1 and Comp2, respectively] grown in hydroponic culture. The upper panel shows the fresh weights of the plants, the middle panel shows representative results from dCAPS analysis used to verify the point mutation in lir1-1 and LIR1 transgenic DNA sequence in the complementation lines, and the lower panel shows the phenotypes. Genomic DNA from the plants was amplified using the dCAPS primers (Supplemental Table 3), which produce a 130-bp LIR1 DNA fragment. Digestion of the wild-type PCR products with EcoRI results in the production of 90- and 40-bp fragments, whereas the point mutation in lir1-1 prevents the cleavage. Bar = 20 cm.
Bars in (D) to (F) represent means ± sd (n = 3). Asterisks indicate significant difference from the wild type at P < 0.01 (Student’s t test).
Light Stimulates LIR1 Degradation in Rice
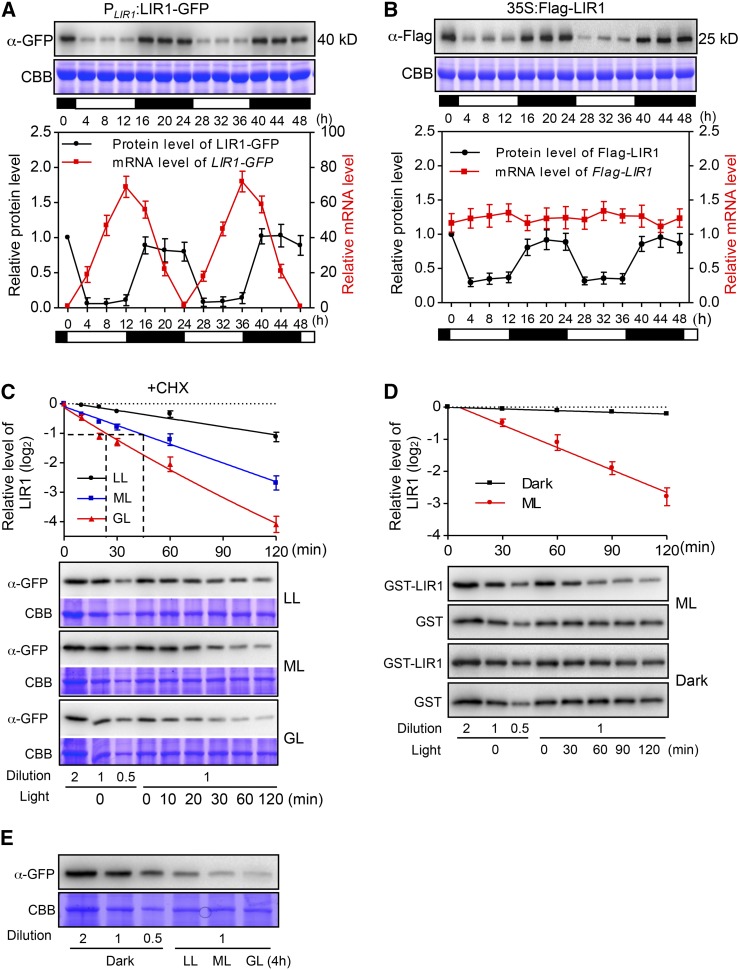
To examine the molecular function of LIR1, we studied the expression pattern of LIR1 in lir1/PLIR1:LIR1-GFP plants using quantitative RT-PCR. Levels of LIR1 transcript increased following illumination, reaching a maximum at the end of the light period and dropping to a minimum at the end of the dark period (Figure 2A), corroborating previous findings (Reimmann and Dudler, 1993; Hayama et al., 2002). Intriguingly, marked accumulation of LIR1 was observed during the dark phase, whereas the LIR1 content decreased substantially upon the onset of illumination (Figure 2A). A similar pattern of LIR1 accumulation was also detected in a transgenic rice line with constitutive expression of Flag-LIR1 (35S:Flag-LIR1) (Figure 2B). Moreover, exposure of the rice plants to low light (LL; 50 µmol photons m−2 s−1), medium light (ML; 500 µmol photons m−2 s−1), and standard growth light (GL; 1000 µmol photons m−2 s−1) levels in the presence of the translation inhibitor cycloheximide revealed that the rate of LIR1 degradation increased with increasing light intensity (Figure 2C). To verify the dependence of the LIR1 degradation on light, we further studied the in vitro degradation of recombinant GST-tagged LIR1 (expressed and purified from Escherichia coli) in wild-type rice extracts. Similar to the in vivo results, recombinant LIR1 was stable in rice extracts isolated from dark-grown plants, while rapid degradation was detected in extracts isolated from plants grown in the light (Figure 2D). Finally, we analyzed the LIR1 protein levels in the plants exposed to different light intensities and found that the LIR1 protein level corresponded with the ambient light intensity (Figure 2E).
Figure 2.
Light Promotes Degradation of LIR1 in Rice.
(A) Expression of LIR1 mRNA and LIR1 protein in lir1/PLIR1:LIR1-GFP plants grown under a 12-h-light/12-h-dark photoperiod. Leaf samples were collected at the indicated times, and total mRNA and proteins were extracted and subjected to qRT-PCR and immunoblot analysis, respectively. The white and black horizontal bars in each panel indicate day and night, respectively. The upper panel shows a representative immunoblot using an anti-GFP antibody, the middle panel shows CBB staining of the gels to verify equal loading of the gels, and the lower panel shows quantification of the results.
(B) Expression of Flag-LIR1 mRNA and Flag-LIR1 protein in the leaves of 35S:Flag-LIR1 plants grown under a 12-h-light/12-h-dark photoperiod. The leaf samples were collected at the indicated times, and total mRNA and proteins were extracted and subjected to qRT-PCR and immunoblot analysis, respectively. To detect Flag-LIR1 fusion mRNA, the forward primer on the Flag vector (Flag-qRT-F) and the reverse primer on rice LIR1 (OsLIR1-qRT-R) were used. Primer sequence information is shown in Supplemental Table 3. The white and black horizontal bars in each panel indicate day and night, respectively. The upper panel shows a representative immunoblot using the anti-Flag antibody, the middle panel shows CBB staining of the gels, and the lower panel shows quantification of the results.
(C) Degradation of LIR1-GFP in the leaves of 10-d-old lir1/PLIR1:LIR1-GFP plants. The 4-h dark-adapted plants were treated with 200 µM cycloheximide (CHX) for 60 min. The seedlings were then exposed to different light intensities (LL, 50 µmol photons m−2 s−1; ML, 500 µmol photons m−2 s−1; and GL, 1000 µmol photons m−2 s−1) for 0, 10, 20, 30, 60, or 120 min. The upper panel shows quantification of the LIR1-GFP calculated and plotted on a semilog graph, and the lower panel shows representative immunoblots using the GFP antibody. Half-lives (t1/2) for LIR1-GFP under given light conditions are indicated by a dashed line on the x axis. Dilution series (0.5× to 2×) were prepared with the corresponding negative control (0 min) in each gel to avoid possible saturation and to ensure precise quantification in immunodetection. The densitometry values of samples with 1× loading were quantified.
(D) Degradation rate of recombinant GST-LIR1 protein in vitro. GST or GST-LIR1 was expressed and purified from E. coli and incubated for 0, 30, 60, 90, or 120 min in the presence of leaf protein extracts isolated from wild-type plants treated for 4 h with ML or in the dark. Incubation was also performed either under ML or in the dark. The upper panel shows quantification of the GST-LIR1 calculated and plotted on a semilog graph, and the lower panel shows representative immunoblots using the GST antibody. GST was used as a control to confirm that there was no light-dependent degradation of the tag itself. Dilution series (0.5× to 2×) were prepared with the corresponding control (0 min) in each gel to avoid possible saturation and to ensure precise quantification in immunodetection. The densitometry values of samples with 1× loading were quantified.
(E) Expression of LIR1 in lir1/PLIR1:LIR1-GFP plants in response to light intensity. The seedlings were exposed to different light intensities (LL, ML, or GL) or darkness for 4 h before harvest for analysis. A representative immunoblot using GFP antibody is shown.
CBB staining of the gels verified equal loading of the gels. Graphs in (A) through (D) show means ± sd (n = 3).
LIR1 Interacts with LFNR1 and LFNR2
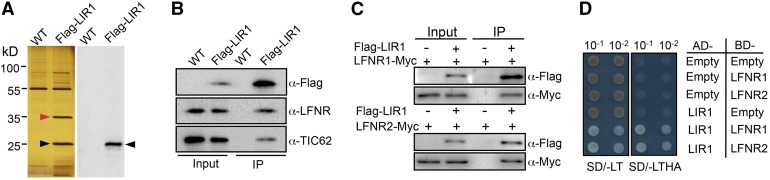
We next attempted to identify interaction partners of LIR1 in rice using in vivo coimmunoprecipitation. Protein extracts from wild-type and 35S:Flag-LIR1 transgenic rice seedlings were affinity purified using anti-Flag M2 magnetic beads. Silver staining of the resulting Flag-affinity eluent revealed two bands (35 and 25 kD) specifically enriched in the Flag-LIR1 sample (Figure 3A). Using liquid chromatography-tandem mass spectrometry (LC-MS/MS), we found that the 35-kD band contained LFNR1 and LFNR2 (Supplemental Table 1), whereas the 25-kD band was identified as LIR1 (Supplemental Table 2). Immunoblot analysis using Flag and LFNR antibodies verified the identity of the 25-kD protein as LIR1 (Figure 3A) and the 35-kD protein as LFNR (Figure 3B).
Figure 3.
Interaction of Os-LIR1 with Os-LFNR1/2.
(A) In vivo coimmunoprecipitation assay in wild-type and 35S:Flag-LIR1 rice plants. Anti-Flag M2 magnetic beads were used for purification and the components of the purified complexes were separated by SDS-PAGE. The left panel shows a silver-stained gel. The 35- and 25-kD bands were subjected to LC-MS/MS analysis and identified as LFNR1/LFNR2 (red arrowhead) and LIR1 (black arrowhead), respectively. The right panel shows immunoblot analysis of the corresponding gel using anti-Flag antiserum.
(B) In vivo coimmunoprecipitation assay using Flag-LIR1, LFNR, and TIC62. Total protein extracts prepared from 1-month-old wild-type or 35S:Flag-LIR1 transgenic rice seedlings were incubated with anti-Flag M2 magnetic beads. The precipitates (IP) and total extracts (Input) were subjected to immunoblot analysis with antibodies against Flag, LFNR, and TIC62. For each well, 0.05% of total protein extracts (10 μL from 20 mL total protein extracts) and 8.3% eluted proteins (10 μL from 120 μL eluted proteins) were loaded as input and IP, respectively.
(C) In vivo coimmunoprecipitation analysis of LIR1 with LFNR1 and LFNR2. 35S:Flag-LIR1 was transiently expressed in tobacco leaves with either 35S:LFNR1-MYC or 35S:LFNR2-MYC. Isolated total protein extracts (Input) were immunoprecipitated using anti-Flag M2 magnetic beads (IP) and immunoblotted using anti-Flag and anti-Myc antibodies.
(D) Yeast two-hybrid analysis of interactions between LIR1 and LFNR isoforms. Yeast lines harboring either the empty control plasmids (AD, activation domain; BD, bait domain) or plasmids containing the rice fusion constructs AD-LIR1, BD-LFNR1, or BD-LFNR2 were grown on synthetic medium supplied with dextrose (SD) in the absence of Trp and Leu (SD/-LT, left panel) and on SD medium in the absence of Trp, Leu, His, and Ade (SD/-LTHA, right panel). Yeast cells were incubated until OD600 = 1 and then diluted 10- or 100-fold and used for assays.
Arabidopsis LFNR (At-LFNR) was shown to interact with the LFNR binding domain of At-TIC62 and At-TROL; therefore, we investigated whether Flag-Os-LIR1, Os-LFNR, and Os-TIC62 form an analogous complex (Figure 3B). We used an antibody raised against the LFNR binding motif of pea (Pisum sativum) TIC62, which recognizes both Arabidopsis TIC62 and TROL (Benz et al., 2009; Lintala et al., 2014). To verify that this antibody also recognizes rice TIC62, we characterized an Os-tic62 mutant identified from the Genoplante T-DNA insertional line library (for details, see Supplemental Figures 2A to 2D). The TIC62 antibody identified three distinct bands in rice (Supplemental Figure 2E). The major Os-TIC62 band comigrated with the 70-kD standard, and the very faint 45-kD band produced in this analysis might be a degradation fragment of Os-TIC62 (Supplemental Figure 2E). A third band just below the 70-kD band, which was also present in the Os-tic62 mutant, might be Os-TROL (Supplemental Figure 2E). Coimmunoprecipitation analysis of rice 35S:Flag-OsLIR1 plants revealed that Os-TIC62 was indeed present in the same complex as Os-LIR1 and Os-LFNR (Figure 3B).
We further examined the interaction between LIR1 and the two LFNR isoforms. First, protein extracts from tobacco (Nicotiana tabacum) leaves transiently coexpressing Flag-LIR1 and LFNR1-Myc or LFNR2-Myc were immunoprecipitated using anti-Flag M2 beads and analyzed by immunoblotting using anti-Flag and Myc antibodies (Figure 3C). This revealed a clear interaction between LIR1 and both LFNR isoforms (LFNR1 and LFNR2). Second, we applied a yeast two-hybrid approach to examine whether LFNR1 and LFNR2 interact with LIR1 in yeast. As shown in Figure 3D, LIR1 interacted with both LFNR1 and LFNR2 in yeast. Finally, we performed bimolecular fluorescence complementation (BiFC) assays using C- and N-terminal fragments of YFP fused to LIR1, LFNR1, and LFNR2 to further demonstrate the interaction between LFNRs and LIR1 in tobacco leaves. Coexpression of LIR1-YFPC with LFNR1-YFPN or LFNR2-YFPN resulted in clear YFP signals in the chloroplast, whereas no signal was detected in the chloroplast localized negative controls (Supplemental Figure 3). Taken together, these results indicate that LIR1 is an interaction partner of LFNR.
To test whether the conserved Cys residues in the LIR1 motifs (Supplemental Figure 1) could form disulfide bridges with the conserved Cys residues in LFNR (e.g., Cys-88 and Cys-178 in At-LFNR1; Cys-90 and Cys-180 in Os-LFNR1; and Cys-94 and Cys-184 in Os-LFNR2) and provide an interaction mechanism between the proteins, we performed coimmunoprecipitation assays using rice Flag-LIR1, LFNR, and TIC62 under various redox conditions. Supplemental Figure 4 shows that addition of the oxidative agent (immunoprecipitate [IP] buffer supplemented with 5 mM diamide) to the reaction markedly increased the interaction of LIR1 with LFNR, whereas reduction of the disulfide bridges (IP buffer supplemented with 20 mM DTT) slightly decreased it. These results imply that thiol regulation may have an impact on LIR1-LFNR interaction and thereby to the membrane tethering of LFNR.
LIR1 and LFNR Are Colocalized within the Chloroplast
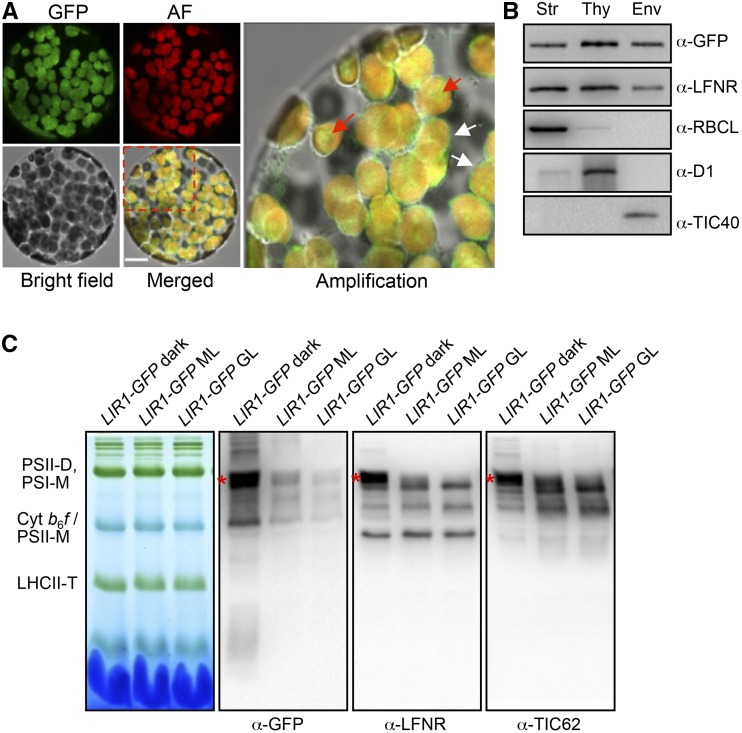
To investigate the localization of Os-LIR1, tobacco plants were used for transient expression of rice LIR1-GFP (PLIR1:LIR1-GFP). When mesophyll protoplasts isolated from the leaves of dark-adapted (4 h) PLIR1:LIR1-GFP tobacco plants were examined under a confocal microscope, the LIR-GFP signal was detected exclusively in chloroplasts (Figure 4A). In addition to the GFP signal overlapping the red autofluorescence emitted from the thylakoid membranes, the GFP signal was also detected in the stroma and envelope (Figure 4A). In accordance with these results, immunoblot analysis of the dark-adapted lir1/PLIR1:LIR1-GFP rice also demonstrated that LIR1 was present as a soluble protein in the chloroplast stroma in addition to being bound to the thylakoid and envelope membranes (Figure 4B).
Figure 4.
Colocalization of LIR1 and LFNR in the Chloroplast.
(A) Subcellular localization of rice LIR1 in tobacco mesophyll protoplasts. The PLIR1:LIR1-GFP plasmid was transformed into tobacco leaves for transient expression, and mesophyll protoplasts were scanned under a confocal microscope 2 d after transformation. The upper left panel shows the GFP signal, the upper middle panel shows chlorophyll autofluorescence (AF), the lower left panel shows the bright-field image, and the lower middle panel shows the merged image. The outlined area in the merged image is enlarged in the right panel. White arrows indicate GFP fluorescence on the envelope and red arrows indicate GFP fluorescence in the stroma. Bar = 10 µm.
(B) Subchloroplastic localization of LIR1. Chloroplast fractions comprising the stroma (Str), thylakoids (Thy), and envelopes (Env) of dark-adapted lir1/PLIR1:LIR1-GFP rice were subjected to immunoblot analysis using anti GFP, LFNR, RBCL, D1, and TIC40 antibodies. One microgram of protein was loaded in each lane.
(C) BN-PAGE analysis of the thylakoid protein complexes in lir1/PLIR1:LIR1-GFP plants. Thylakoid membranes were isolated from plants treated for 4 h in the dark, ML (500 µmol photons m−2 s−1), or GL (1000 µmol photons m−2 s−1). BN gel and immunoblots probed with anti-GFP, LFNR, and TIC62 antibodies are shown; 8 µg chlorophyll was loaded in each lane. PSII-D/M, PSII dimers/monomers; PSI-M, PSI monomers; LHCII-T, LHCII trimers.
To investigate the formation of the complex at the thylakoid membrane in more detail, the thylakoid protein complexes of lir1/PLIR1:LIR1-GFP plants were analyzed using blue native PAGE (BN-PAGE) followed by immunoblotting. The position of the LFNR-TIC62 complex on the BN-PAGE gel was first identified using the tic62 mutant (Supplemental Figure 2F). Figure 4C shows that the GFP signal at least partially overlapped with the signals obtained using LFNR or TIC62 antibodies. In accordance with previous findings in Arabidopsis (Benz et al., 2009), a strong signal originating from a LFNR-containing thylakoid protein complex was detected in plants treated for 4 h in the dark (Figure 4C, indicated by a red star). Illumination of the plants induced rapid degradation of the LIR1 protein (Figure 2) and also led to the release of LFNR from the thylakoid membrane (Figures 4C and 5A).
Figure 5.
Accumulation of LFNR-Containing Thylakoid Protein Complexes in Rice Plants.
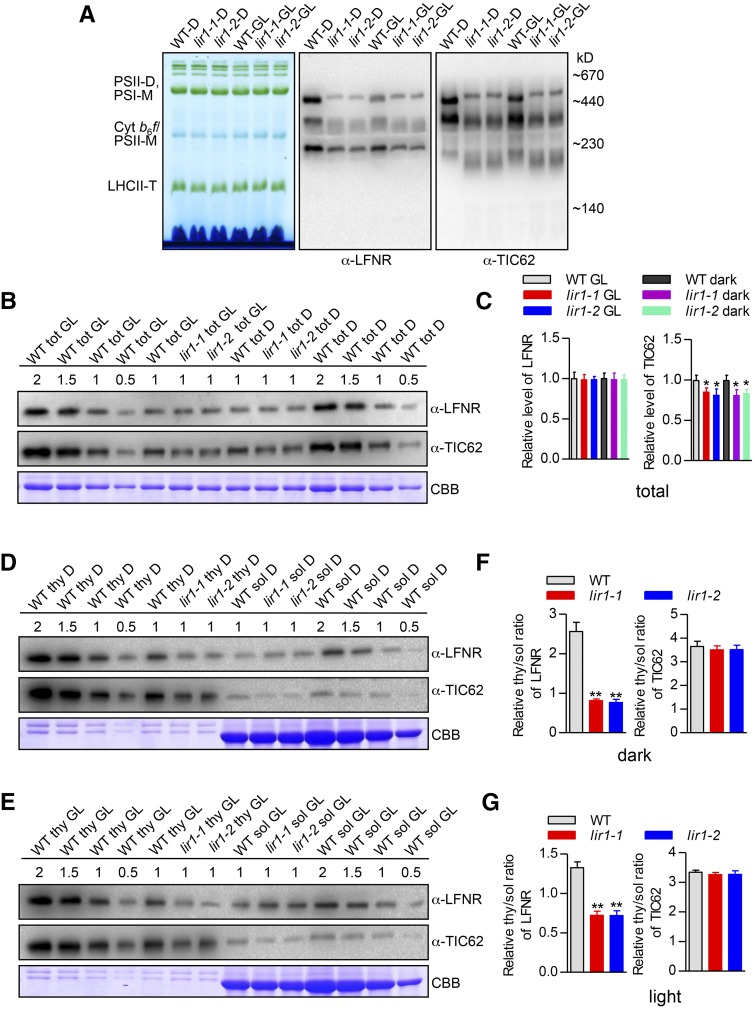
(A) BN-PAGE analysis of the thylakoid protein complexes in wild-type and lir1 plants. Thylakoid membranes were isolated from plants treated for 4 h in the dark or under standard GL (1000 µmol photons m−2 s−1). BN gel and immunoblots probed with anti-LFNR and TIC62 antibodies are shown; 5 µg chlorophyll was loaded in each lane. PSII-D/M, PSII dimers/monomers; PSI-M, PSI monomers; LHCII-T, LHCII trimers.
(B) LFNR and TIC62 total (tot) contents in wild-type and lir1 plants. Representative immunoblots of SDS-PAGE analysis of total leaf extracts isolated from plants treated for 4 h under GL or in the dark (D) are shown in upper panels. Proteins were immunodetected with anti-LFNR and TIC62 antibodies. CBB staining shows equal loading of the gel (lower panel). A dilution series (0.5× to 2×) was prepared with the corresponding control (WT tot GL and WT tot D) in each gel to avoid possible saturation and to ensure precise quantification in immunodetection. 1× loading was defined as 5 μg total proteins.
(C) Relative levels of LFNR and TIC62 in total extracts. The densitometry values of samples with 1× loading were quantified from (B). The value for the wild type tot GL was set to 1. The means ± sd from three biological replicates are shown. Asterisk indicates significant different from the wild type at P < 0.05.
(D) and (E) LFNR and TIC62 contents of thylakoid membrane (thy) and soluble (sol) leaf extracts in wild-type and lir1 plants during dark (D) and GL (E) conditions, respectively. Thylakoid membranes and soluble leaf extracts were isolated from 4 h dark (D) or 4 h GL-treated plants. Representative immunoblot results are shown in upper panels. CBB staining shows equal loading of the gels (lower panels). Dilution series (0.5× to 2×) were prepared with the corresponding control (WT thy D or GL and WT sol D or GL) in each gel to avoid possible saturation and to ensure precise quantification in immunodetection. 1× loading of thy and sol was defined as 2.5 and 10 μg proteins, respectively.
(F) and (G) The relative thy/sol ratio of LFNR (left graph) and TIC62 (right graph) in wild-type and lir1 plants according to (D) or (E). The densitometry values of samples with 1× loading were quantified and means ± sd from three biological replicates are shown. Asterisks indicate significant difference from the wild type at P < 0.01.
LIR1 Regulates the Attachment of the LFNR Complex to the Thylakoid Membrane in Rice
Next, we measured the accumulation of the LFNR-containing thylakoid protein complexes in the lir1 plants by BN-PAGE followed by immunoblotting. The analysis revealed that the bands representing all three LFNR complexes, especially the slower migrating band, were reduced in lir1 compared with the wild type under both dark and light conditions (Figure 5A; Supplemental Figure 2F). The absence of LIR1 slightly reduced the total TIC62 content but had no effect on the accumulation of LFNR (Figures 5B and 5C). However, the distribution of LFNR between the soluble and membrane fractions was markedly altered in the lir1 plants as compared with the wild type both in the light and in the dark: the membrane-bound pool of LFNR was significantly reduced with a concomitant increase in the content of soluble LFNR (Figures 5D to 5G), both LFNR isoforms responding similarly to the depletion of LIR1 (Supplemental Figure 5). Also, the ratio between the thylakoid-bound and soluble TIC62 in lir1 did not differ from that of the wild type (Figures 5F and 5G).
Loss of LIR1 Reduces Photosynthetic Electron Transfer in Rice
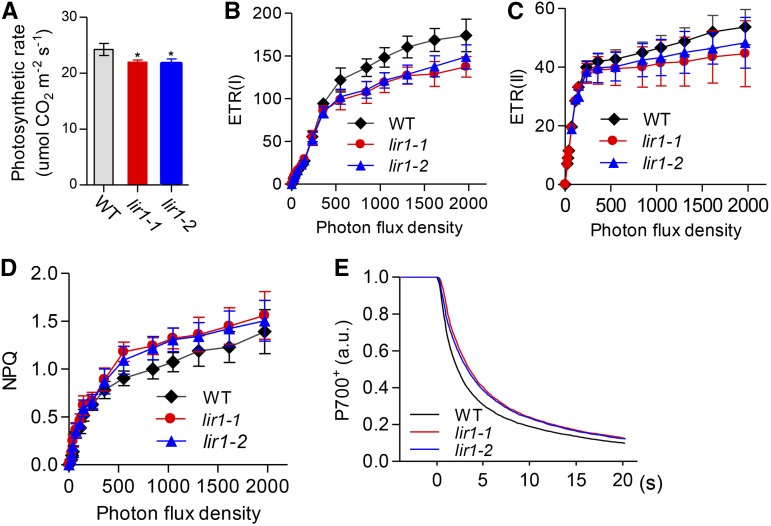
To study the effect of LIR1 deficiency on plant metabolism, we examined the photosynthetic properties of the mutant plants. The CO2 assimilation rate of the lir1 mutant was slightly but significantly reduced compared with wild type (Figure 6A). In accordance with the smaller plant size (Figure 1) and reduced CO2 fixation rate (Figure 6A) of the lir1 mutant compared with the wild type, the ETR(I) (electron transfer rate reflecting the capacity of PSI) was also reduced in the mutant, especially under higher actinic light intensities (Figure 6B). A similar trend was also observed for ETR(II), but the reduction was less prominent than that in ETR(I) (Figure 6C). Moreover, a slight increase in nonphotochemical quenching (NPQ) was detected in the lir1 mutant (Figure 6D). Next, we examined the kinetics of P700+ dark rereduction, which is used as an indicator of cyclic electron transfer (Bukhov et al., 2004; Golding et al., 2004; Fan et al., 2007; Lehtimäki et al., 2010). As shown in Figure 6E, the rereduction rate of P700+ was slower in the lir1 mutant than in the wild type.
Figure 6.
Photosynthetic Properties of Wild-Type and Os-lir1 Plants.
(A) Rate of CO2 fixation in wild-type, lir1-1, and lir1-2 plants irradiated with 1500 µmol photons m−2 s−1.
(B) and (C) Electron transfer rate (ETR) of PSI and PSII, respectively, in wild-type, lir1-1, and lir1-2 plants under different light intensities.
(D) NPQ in wild-type, lir1-1, and lir1-2 plants under different light intensities.
(E) Dark rereduction rate of P700+ in wild-type, lir1-1, and lir1-2 plants. P700 was oxidized by far-red light for 30 s, and P700+ rereduction was monitored in the dark. Curves were normalized to the maximum signal. a.u., arbitrary units.
All of the above results were obtained in at least three independent experiments. Data represent means ± sd. Asterisk indicates significant difference from the wild type at P < 0.05 (Student’s t test).
The Function of LIR1 Is Conserved in Several Dicotyledons
A BLAST search of the protein databases using the Os-LIR1 gene as a query showed that, in addition to the previously reported LIR1 orthologs in various vascular plant species (Reimmann and Dudler, 1993; Teramoto et al., 1994; Abied and Holland, 1994; Quigley et al., 1996; Ciannamea et al., 2007), LIR1 orthologs are also found in mosses (Sphagnum lescurii and Takakia lepidozioides), liverworts (Bazzania trilobata and Schistochila sp), and algae (Chara vulgaris and Chlorokybus atmophyticus), but none was identified in cyanobacteria (Supplemental Figure 6). The LIR1 domain (Reimmann and Dudler, 1993; Ciannamea et al., 2007), with two repeated motifs (SVFXXEACXXXGGEAC—TVFXXEACXXXGGEFC) separated by 35 to 53 amino acids, shows a high degree of conservation among the species (Supplemental Figure 7). Gymnosperms such as Pinus pinaster, Picea abies, and Picea sitchensis and nonflowering plants such as mosses, liverworts, and green algae exhibit only small differences in the conserved region. However, the Brassicaceae LIR1 orthologs have a notable difference at the second conserved EAC repeat (Supplemental Figure 7): Instead of the negatively charged Glu (E) or Asp (D) residue at the second EAC motif detected in all other plant LIR1 orthologs, the Brassicaceae family members contain a Tyr residue (YAC) (Supplemental Figure 7).
To investigate whether the interaction between LIR1 and LFNR is also conserved in other species, we examined this interaction by yeast two-hybrid analysis using the gene sequences from maize (Zea mays), soybean (Glycine max), and cucumber (Cucumis sativa; Supplemental Figure 8). In agreement with the results from rice, the yeast two-hybrid assays revealed a clear interaction between LIR1 and LFNR isoforms in these higher plant species (Supplemental Figure 8).
The unusual amino acid sequence (YAC) in the second EAC motif of the Brassicaceae LIR1 proteins prompted us to investigate whether the Arabidopsis LIR1 plays a similar role in the attachment of the LFNR complex to the thylakoid as does Os-LIR1. We used an Arabidopsis T-DNA insertional line (SALK_024728C), which has an insertion in LIR1 (At-lir1). The T-DNA insertion (located in the second exon of At-LIR1) leads to the absence of the full-length mRNA, causing At-lir1 to be a null mutant (Supplemental Figures 9A and 9B). Surprisingly, unlike the rice lir1 mutants, the Arabidopsis lir1-1 mutant did not display any obvious growth defect under standard growth conditions (Supplemental Figures 9C and 9D). Consistent with this, the At-lir1 mutant did not show obvious impairment in photosynthetic efficiency or electron transfer properties, except for a slight increase in NPQ (Supplemental Figures 9E to 9H).
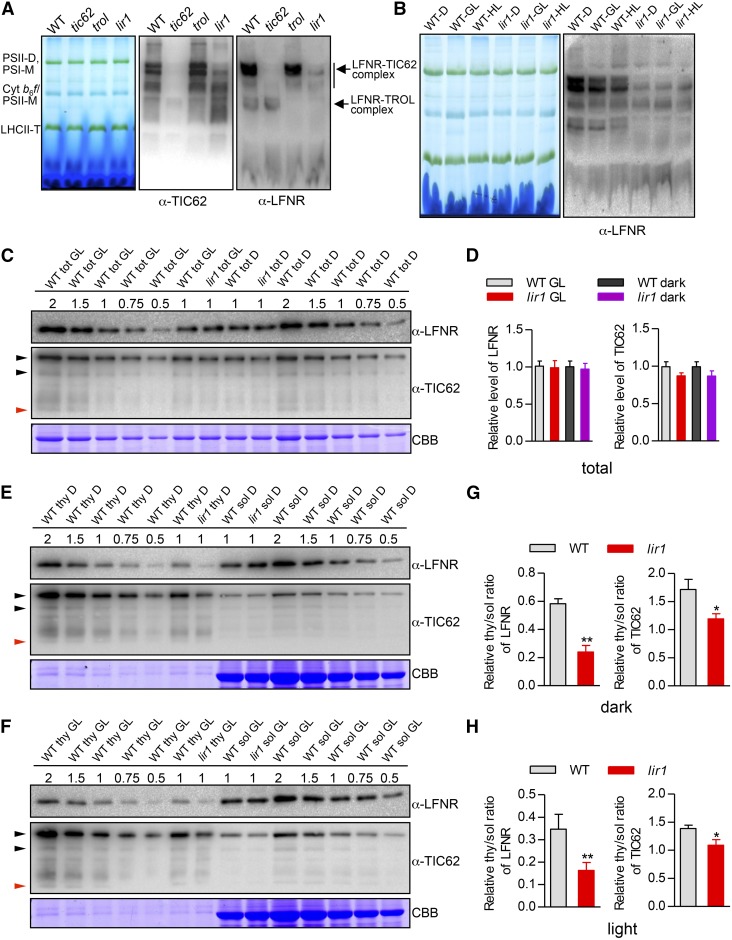
The accumulation of LFNR and TIC62/TROL in the thylakoid membranes of wild-type, tic62, trol, and lir1 Arabidopsis plants was examined by BN-PAGE. Even though the Arabidopsis LIR1 motif differs from that of rice (Supplemental Figure 7), the absence of At-LIR1 resulted in a drastic loss of binding of the At-LFNR-containing complexes to the chloroplast membrane also in Arabidopsis (Figures 7 A and 7B). Similarly to rice, the total At-LFNR content did not differ between the wild type and At-lir1, but the association of LFNR to the thylakoid was reduced in the mutant (Figures 7C and 7D). Despite its predicted molecular mass of 62 kD, TIC62 comigrated with the 110- and 85-kD markers, while TROL migrated with the 60-kD marker under our test conditions (Supplemental Figure 9I). In contrast to rice, the level of At-LFNR anchor (TIC62 or TROL) in At-lir1 did not differ significantly from that of the wild type (Figures 7C and 7D; Supplemental Figure 9I), whereas the ratio of thylakoid bound to soluble proteins (both At-LFNR and At-TIC62) was decreased in At-lir1 compared with the wild type, especially in the dark (Figures 7E to 7H). These results indicate that even if Arabidopsis LIR1 plays a similar role in the light-regulated allocation of LFNRs between the stroma and thylakoid to that of rice, the effect on the function of photosynthetic appears to differ between the species, at least under the studied conditions.
Figure 7.
Accumulation of LFNR-Containing Thylakoid Protein Complexes in Arabidopsis.
(A) BN-PAGE analysis of the thylakoid protein complexes in wild-type (Columbia), At-tic62, At-trol, and At-lir1 plants. Thylakoid membranes were isolated from plants treated for 4 h in the dark. BN gel and immunoblots probed with anti-TIC62 and LFNR antibodies are shown; 5 µg chlorophyll was loaded in each lane. PSII-D, PSII dimers; PSI-M, PSI monomers; LHCII-T, LHCII trimers.
(B) BN-PAGE analysis of the thylakoid protein complexes in wild type and At-lir plants. Thylakoid membranes were isolated from plants treated for 4 h in the dark (D), GL (150 µmol photons m−2 s−1), or high light (HL; 500 µmol photons m−2 s−1). A BN-PAGE gel and an immunoblot probed with the anti-LFNR antibody are shown; 5 µg chlorophyll was loaded in each lane.
(C) LFNR and TIC62 content in wild-type and At-lir1 plants. Total leaf extracts (tot) were isolated from plants treated for 4 h with GL (150 µmol photons m−2 s−1) or in the dark (D). Representative immunoblots using anti-LFNR and TIC62 antibodies are shown in upper panels. CBB staining shows equal loading of the gel (lower panel). Dilution series (0.5× to 2×) were prepared with the corresponding control (WT tot GL and WT tot D) in each gel to avoid possible saturation and to ensure precise quantification in immunodetection. 1× loading was defined as 5 μg total proteins. Black arrows indicate 110 and 85 kD (At-TIC62) and red arrow indicates 60 kD (At-TROL).
(D) Relative levels of At-LFNR and At-TIC62 in total extracts (C). The densitometry values of samples with 1× loading were quantified from (B). The value for wild type tot GL was set to 1. The means ± sd from three biological replicates are shown.
(E) and (F) At-LFNR and At-TIC62 contents in thylakoid membrane (thy) and soluble (sol) leaf extracts of wild-type and At-lir1 plants treated for 4 h in the dark (D) (Figure 7E) or under GL (Figure 7F). Representative immunoblots using anti-LFNR and TIC62 antibodies are shown in the upper panels. CBB staining shows equal loading of the gel (lower panel). Dilution series (0.5× to 2×) were prepared with the corresponding control (WT thy D or GL and WT sol D or GL) in each gel to avoid possible saturation and to ensure precise quantification in immunodetection. 1× loading of thy and sol was defined as 2.5 and 10 μg protein, respectively.
(G) and (H) The relative thylakoid/soluble (thy/sol) ratio of At-LFNR (left graph) and At-TIC62 (right graph) in wild-type and At-lir1 plants according to (E) or (F). The densitometry values of samples with 1× loading were quantified. Means ± sd from three biological replicates are shown. Asterisks show significant difference from the wild type at *P < 0.05 and **P < 0.01.
LIR1 Increases the Affinity between LFNR and TIC62 in Rice
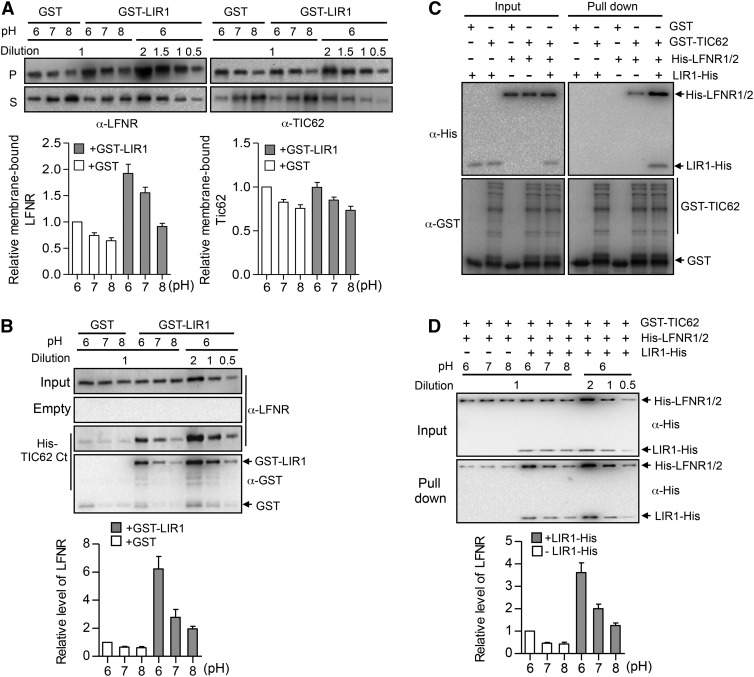
Clearly, LIR1 enhances the attachment of LFNR-containing protein complexes to the thylakoid membrane (Figures 5 and 7). To determine whether LIR1 strengthens the binding between LFNR and TIC62 or directly increases the attachment of TIC62 to the membrane, we expressed GST-LIR1 in E. coli and treated the isolated lir1 thylakoids with GST or GST-LIR1 at pH 6, 7, and 8. In agreement with a previous finding (Benz et al., 2009), more binding of LFNR to the thylakoid membrane occurred at pH 6.0 than at pH 7.0 or 8 (Figure 8A). Intriguingly, the presence of GST-LIR1 markedly shifted the distribution of LFNR toward the membrane fraction, whereas no such effect was observed for TIC62 (Figure 8A). Next, heterologously expressed and purified His(6)-TIC62 C terminus (TIC62Ct), containing the LFNR binding repeats, was bound to Ni2+ beads and used as an affinity matrix for LFNR from rice tic62 stroma (which is devoid of endogenous Tic62). GST or GST-LIR1 was added to the binding assay at pH 6, 7, and 8. As shown in Figure 8B, in samples supplemented with GST-LIR1, LFNR exhibited much higher affinity for TIC62Ct under all pH conditions compared with GST-treated samples. Notably, however, the binding affinity of LFNR for TIC62Ct was higher at pH 6.0 than at pH 7.0 or 8 (Figure 8B).
Figure 8.
LIR1 Strengthens the Binding Affinity between Os-LFNR and Its Membrane Anchor in Rice.
(A) Membrane binding affinity of LFNR and TIC62 in the presence or absence of LIR1 in isolated lir1 thylakoid membrane. Isolated thylakoids from dark-adapted lir1-1 rice plants were first incubated in 100 mM sodium phosphate buffer (pH 6.0, 7.0, and 8.0) in the presence of purified GST or GST-LIR1 proteins and then mildly washed by adding 0.1% n-decyl β-d-maltoside in corresponding sodium phosphate buffer. After centrifuging, soluble (supernatant [S]) and membrane-bound (pellet [P]) proteins were analyzed by immunoblotting using anti-LFNR and TIC62 antibodies (upper panels). Quantification of membrane-bound proteins is shown in the lower panel. Data are means ± sd (n = 3). The dilution series (0.5× to 2×) were made with GST-LIR1 under pH 6.0 to avoid possible saturation and to ensure precise quantification in the immunodetection. The densitometry value of samples with 1× loading were quantified.
(B) LFNR pull down by His-tagged TIC62 from tic62 leaf extracts in the presence or absence of LIR1. The C terminus of TIC62 (His-TIC62Ct) was purified from E. coli, bound to Ni2+ beads, and used as an affinity matrix for tic62 leaf soluble extract harvested from plants grown under GL (1000 µmol photons m−2 s−1) in the presence of GST or GST-LIR1 (pH 6, 7, or 8). Empty Ni2+ beads were used as negative control (Empty). The upper panels show immunoblots using anti-LFNR and GST antibodies, and the lower panel shows the quantification of bound LFNR. Dilution series (0.5× to 2×) were prepared with GST-LIR1 under pH 6.0 to avoid possible saturation and to ensure precise quantification in immunodetection. The densitometry values of samples with 1× loading were quantified. Data are means ± sd (n = 3).
(C) Binding affinity analysis between His-LFNR and GST-TIC62 by in vitro GST pull-down in the presence or absence of LIR1-His. GST- and GST-tagged full-length TIC62 (GST-TIC62), LIR1-His, and His-LFNR proteins were purified from E. coli, and equal amount of each protein (1 µg) was added to the pull-down buffer system (pH 7) as indicated above the panels. The molar ratio of LIR1-His:His-LFNR:GST-TIC62 is ∼2:1:0.5. His-LFNR1/2 contained a mixture of equal amounts of LFNR1 and LFNR2. Glutathione Sepharose beads were used to bind GST-TIC62 and His-LFNR1/2 in the presence or absence of LIR1-His. The amount of pulled-down LFNR and LIR1 was analyzed by immunoblotting with anti-His (upper panel) and GST (lower panel) antibodies, respectively.
(D) Binding affinity analysis of His-LFNR and GST-TIC62 by in vitro GST pull-down under different pH conditions. GST and GST-tagged full-length TIC62 (GST-TIC62), LIR1-His, and His-LFNR proteins were purified from E. coli and added to the pull-down buffer system as indicated above the panels at pH 6, 7, or 8. His-LFNR1/2 contained a mixture of equal amounts of LFNR1 and LFNR2. Glutathione Sepharose beads were used to bind GST-TIC62 in buffer with or without LIR1-His under different pH conditions. The amount of pulled-down LFNR was analyzed by immunoblotting with anti-His antibody. Dilution series (0.5× to 2×) were prepared with LIR1-His under pH 6.0 to avoid possible saturation and to ensure precise quantification in immunodetection. The densitometry values of samples with 1× loading were quantified. Data are means ± sd (n = 3).
To help confirm this result, we performed an in vitro GST pull-down assay using GST-TIC62 (full length) and His-LFNR in the presence or absence of LIR1-His. Again, the presence of LIR1 markedly increased the interaction between TIC62 and LFNR (Figure 8C). Importantly, no LIR1-His signal was detected if the binding buffer was supplemented with GST-TIC62 and LIR1-His in the absence of His-LFNR1/2 (Figure 8C, lane 2), indicating that LIR1 could not bind directly to TIC62. We verified this result by yeast two-hybrid assay, which also showed no interaction between LIR1 and TIC62 (Supplemental Figure 10). Finally, a GST pull-down assay gave further evidence that the presence of LIR1 strengthened the binding affinity of LFNR to TIC62 in a pH-dependent manner (Figure 8D). Reciprocal pull-down assays with His-tagged TIC62 ruled out the possibility that pH dependency of the interaction would result from the changes in thiol stability (Figure 8B). Taken together, these results imply LIR1 does not directly bind to TIC62, but rather, it interacts with both LFNR isoforms (Figure 3C), which increases the affinity of LFNRs for TIC62.
DISCUSSION
The quantity of light varies dramatically over the course of a day, resulting in constant adjustment of photosynthetic reactions according to environmental cues. The mechanisms underlying the modulation of the light harvesting and electron transfer processes have been extensively studied (Thornber, 1975; Horton et al., 2000; Joliot and Joliot, 2002), but the regulatory network controlling the formation of NADPH, the end product of photochemistry, is not yet well understood (Joliot and Johnson, 2011). It has been suggested that the distribution of LFNR between the membrane-bound and soluble pools might represent an elegant system that controls the allocation of reducing power not only to various assimilatory and biosynthetic reactions but also to the production and/or scavenging of reactive oxygen species that occur in the chloroplast (Benz et al., 2010; Vojta et al., 2015). Indeed, membrane tethering of LFNR via the membrane anchors TIC62 (Benz et al., 2009) and TROL (Jurić et al., 2009) is light responsive via redox regulation (Stengel et al., 2008), but the exact mechanism underlying this relocation and the functional roles of the distinct LFNR pools have remained enigmatic.
LIR1 Interacts with LFNR
We provide several lines of evidence (the results of pull-down, coimmunoprecipitation, and yeast-two hybrid assays) that LIR1 interacts with LFNR, despite the fact that LIR1 does not contain an FNR binding domain similar to that of TIC62 and TROL (Figure 3; Supplemental Figures 3 and 8). Moreover, our results indicate that the redox state of the thiol groups may be involved in the regulation of interaction between LIR1 and LFNR (Supplemental Figure 4), either directly by modulating protein-protein interactions or indirectly by inducing conformational changes in LIR1 and/or LFNR due to formation of intramolecular disulfide bridges. However, the detailed interaction mechanism remains to be elucidated.
As in Arabidopsis (Hanke et al., 2005), both rice LFNR isoforms are present in the thylakoid-bound and soluble pools in the rice chloroplast (Supplemental Figure 5). By contrast, in maize, the N termini of the three different LFNR isoforms determine their membrane associations (Twachtmann et al., 2012); Zm-LFNR1 is restricted to the thylakoid membrane, Zm-LFNR3 is exclusively a soluble protein, and Zm-LFNR2 is found in both compartments (Okutani et al., 2005). The distribution of Zm-LFNR isoforms reflects the functional requirements of C4 metabolism, as the soluble isoforms, which mainly participate in linear electron transfer, are only detected in mesophyll cells. Membrane-bound isoforms, in turn, are found in both the bundle sheath and mesophyll cells, which have high demands for cyclic electron flow (Twachtmann et al., 2012). Intriguingly, the presence of At-LFNR1 is an absolute prerequisite for the membrane attachment of At-LFNR2 in planta in Arabidopsis (Lintala et al., 2007; Hanke et al., 2008), even though both At-LFNR isoforms have equal affinity for At-TIC62 in vitro (Lintala et al., 2014). Our data reveal that Os-LIR1 interacts with both Os-LFNR isoforms (Figures 3C and 3D; Supplemental Figure 3), but it is currently not known whether the accumulation of one Os-LFNR isoform is dependent on the presence of the other isoform. It was recently shown that the distinct At-LFNR isoforms are subject to specific posttranslational modifications (Lehtimäki et al., 2014), which may affect protein-protein interactions and enzyme activity (Pejaver et al., 2014; Lehtimäki et al., 2015). It should be noted that the binding affinity of a specific LFNR isoform in vivo may be modulated by these modifications, which are not present in recombinant proteins or in vitro analyses; thus, the in vitro results may not necessarily reflect the in vivo situation.
Light-Dependent Turnover of LIR1 Regulates the Membrane Tethering of LFNR
We found that LIR1 is a component of the previously described high molecular mass thylakoid protein complexes composed of LFNR and TIC62 (Figure 4C). Because LIR1 is predicted to be a hydrophilic protein (Huang et al., 2010), it is not likely that it would serve as a membrane anchor for LFNR. Moreover, as LIR1 was not able to directly bind to TIC62 in pull-down (Figure 8C, lane 2) and yeast two-hybrid (Supplemental Figure 10) assays, and the presence of GST-LIR1 increased the accumulation of LFNR at the thylakoid membrane without having such an effect on the accumulation of TIC62 (Figure 8A), it is plausible that LIR1 does not directly regulate the attachment of the membrane anchor but rather strengthens the binding of LFNR to TIC62 (and TROL). Light-dependent degradation of LIR1 (Figure 2), which coincided with release of LFNR from the membrane, but not with that of TIC62 (Figure 5A), provides further evidence supporting this hypothesis. The possibility that LIR1-mediated conformational changes in LFNR underlie the enhancement of its binding with TIC62/TROL should be examined.
To our knowledge, the expression pattern we found for LIR1 is unique among genes for chloroplast proteins. Accumulation of LIR1 transcripts was under strict diurnal regulation, reaching its maximum at the end of the day (Figure 2), whereas the accumulation of LIR1 protein reached its peak in the dark (Figure 2). Although several chloroplast proteins have a high rate of degradation and subsequent biosynthesis upon intense illumination, their translation is usually regulated at the level of translational initiation or elongation from a stable, preexisting pool of mRNA (Mulo et al., 2012) or follows oscillations in mRNA accumulation (Chen and Chory, 2011). For example, the PSII core protein D1, exhibiting light-dependent turnover to protect PSII centers from irreversible photodamage (Järvi et al., 2015), is encoded in the plastome; thus, the regulation of its biosynthesis drastically differs from that of any nucleus-encoded protein (such as LIR1). Another example of a light-regulated chloroplast protein is Arabidopsis J8, which acts as a molecular cochaperone for HSP70 proteins (Chen et al., 2010). Transcription of the nucleus-encoded J8 gene and accumulation of the protein are induced in the dark, and its biosynthesis is negatively regulated by light (Piippo et al., 2006; Chen and Chory, 2011). Although the chloroplast proteases have been extensively studied and detailed knowledge exists, e.g., about the turnover of D1 protein (Kato and Sakamoto, 2009), the processes leading to the degradation of most chloroplast proteins have not yet been characterized.
Functional Consequences of the Subchloroplastic Location of LFNR
Although the Os-lir1 plants exhibited a visual phenotype, with reduced size and decreased grain production, the plants were fully viable under standard growth conditions (Figure 1), indicating that Os-LIR1 is not essential for plant survival. In accordance with their retarded growth, the photosynthetic capacity of the Os-lir1 plants was slightly impaired (Figure 6). However, analysis of the photosynthetic pigment-protein complexes at the thylakoid membrane of the Os-lir1 plants by BN-PAGE did not reveal any apparent changes compared with the wild type (Figure 5A), which is in agreement with earlier reports of the intact composition of photosynthetic protein complexes in Arabidopsis mutants deficient in LFNR1, LFNR2, TIC62, and TIC62 TROL (Benz et al., 2009; Lintala et al., 2009, 2014).
In contrast to rice, the Arabidopsis lir1 mutants did not exhibit obvious visual phenotypes or deficiencies in photosynthetic performance (Supplemental Figure 9). Similarly, no changes were detected in the photosynthetic properties of Arabidopsis tic62 (Benz et al., 2009), or tic62 trol double mutant plants (Lintala et al., 2014). However, the NADPH/NADP+ ratios in the tic62 trol plants were lower than that of the wild type, implying that the mutant plants had reallocated reducing power to the most important metabolic pathways required for undisturbed growth and fitness (Lintala et al., 2014). Although it is not clear what underlies the physiological differences between the Os-lir1 and At-lir1 mutants, it is worth noting that the LIR1 motif in the Brassicaceae family differs from that of other higher plants. The Brassicaceae LIR1 orthologs contain a Tyr residue (YAC) instead of negative charged residue Glu (E) or Asp (D) at the second EAC motif (Supplemental Figure 7), which might be responsible for their functional differences. Additionally, it has been reported that although photosynthetic machinery and its regulation share marked similarities between the species, there are also distinct differences. For example, the function of the NDH complex appears to differ between Arabidopsis and rice, possibly reflecting differences in the evolutionary history (i.e., temperature tolerance) of these species (Yamori et al., 2011).
Thylakoid association of LFNR mediated by TIC62 and TROL is restricted to vascular plants, which have acquired a unique proline-rich LFNR binding domain in the C termini of these proteins (Balsera et al., 2007; Jurić et al., 2009). The absence of the LFNR binding domain in TIC62 from Physcomitrella patens, green algae, and cyanobacteria suggests that the TIC62-LFNR interaction might be essential for the regulation of LFNR activity specifically in the chloroplasts of higher plants (Balsera et al., 2007). Indeed, in cyanobacteria, which lack the LFNR binding domain of TIC62, the mechanism underlying thylakoid binding of LFNR differs drastically from that of plants. The Synechocystis sp PCC6803 genome contains a single gene encoding two distinct FNR isoforms of 34 and 46 kD, which are produced by differential translational initiation (Thomas et al., 2006). The 46-kD FNR, implicated in the photoreduction of NAD(P)+, is attached to the thylakoid membrane via the phycobilisome antenna, whereas the 34-kD FNR, which likely catalyzes oxidation of NADPH, is exclusively present in the soluble stroma (Thomas et al., 2006). Thus, the evolution of LFNR isoforms together with various interactors, including TIC62 and TROL, as well as LIR1, may have enabled the establishment of regulatory circuits allowing precise coupling of light-driven electron transfer with subsequent downstream processes in the chloroplasts of higher plants.
METHODS
Plant Material and Growth Conditions
Wild-type and mutant rice plants (Oryza sativa ssp japonica cv Nipponbare) were grown in hydroponic solution using standard methods as described (Kang et al., 2013) under a 12-h-light (30°C)/12-h-dark (26°C) photoperiod with 70% humidity and a phosphorous concentration of 20 µM L−1. Wild-type (Col-0) and mutant Arabidopsis thaliana plants were grown in soil or on half-strength Murashige and Skoog medium (Sigma-Aldrich) containing 1% sucrose solidified with 0.8% agar at pH 5.7. Arabidopsis plants were grown in growth chambers under a 16-h-light (22°C; 100 μmol photons m−2 s−1)/8-h-dark (20°C) cycle.
Plasmid Construction and Plant Transformation
To produce the 35S:Flag-LIR1 construct, the sequence encoding Flag was fused with the 5′ end of the rice LIR1 coding sequence (CDS) in the pF3PZPY122 vector (Feng et al., 2003). For the PLIR1:LIR1-GFP construct, the CDS was amplified using primers OsLIR1-Kpn I-F and OsLIR1-Xba I-R, and the 2.9-kb rice LIR1 promoter was amplified using primers pOsLIR1-EcoR I-F and pOsLIR1-Kpn I-R. The CDS of rice LIR1 driven by its 2.9-kb endogenous promoter was inserted into the pCambia1300GFP vector. CRISPR-Cas9 empty vectors were obtained from the laboratory of J.K. Zhu (Feng et al., 2013). The 20-bp Os-LIR1 CDS GGGGCGCAGCCTGCAGATTC was selected as CRISPR-Cas9 target site (underlined sequence in Figure 1A) and inserted into the single guide RNA vector under the control of the rice U6-2 promoter. The LIR1 single guide RNA and 35S:Cas9 cassettes were subcloned into vector pCambia1300. These constructs were transformed into wild-type rice or lir1 mutant plants using an Agrobacterium tumefaciens-mediated transformation method (Hiei et al., 1994).
Identification of Os-lir1, Os-tic62, and At-lir1 Mutants
To screen for rice lir1 mutants, genomic DNA extracted from the 30 T0 Os-LIR1-CRISPR-Cas9 transgenic rice plants was used as a template for PCR amplification. Primers OsLIR1-CRISPR-F and OsLIR1-CRISPR-R (Supplemental Table 3) flanking the targeted region of LIR1 were used, and the purified PCR products were subjected to sequencing. Plants containing the mutated LIR1 sequence were selected. To confirm the precise mutation site, the mutated DNA fragments were cloned into T-vector (Takara), and plasmids from four to six positive clones were sequenced.
To identify the rice tic62 mutant, total DNA and RNA were extracted from wild-type and AJQE06 (hits LOC_Os10g01044; Genoplante T-DNA insertion line library) plants. Primers OsTIC62-R2 and LB2 (Supplemental Table 3) were used to verify the T-DNA insertion in AJQE06. RNA was used as a template for cDNA synthesis, and primers OsTIC62-F1 and OsTIC62-R1 (Supplemental Table 3) were used to confirm the disappearance of TIC62 mRNA in AJQE06.
To identify the Arabidopsis lir1 mutant, total DNA and RNA from wild-type and lir1 Arabidopsis plants (Salk_024728C) were isolated. Primers AtLIR1-F and Salk-LBb1 (Supplemental Table 3) were used to verify the T-DNA insertion. RNA was used as a template for cDNA synthesis, and AtLIR1-F and AtLIR1-R (Supplemental Table 3) were used to confirm the disappearance of Arabidopsis LIR1 mRNA.
Derived Cleaved Amplified Polymorphic Sequences Analysis
Identities of the lir1-1 plants and two independent complementation lines (lir1/PLIR1:LIR1-GFP) were verified by derived cleaved amplified polymorphic sequence (dCAPS) analysis. Genomic DNA used as PCR templates were extracted from 20-d-old plants. LIR1 genomic fragments (130 bp) were amplified by PCR using the LIR1 specific dCAPS primers (Supplemental Table 3), which can introduce a EcoRI restriction site. EcoRI digestion products of the PCR fragments were separated on a 3% agarose gel. The lir1-1 point mutation allele yields only one 130-bp band, whereas the complementation line shows another two bands (90 and 40 bp) besides the 130-bp band.
Cell-Free Degradation
Cell-free degradation assays were performed according to Lv et al. (2014). Shoots of 15-d-old wild-type Nipponbare rice seedlings grown under 1000 µmol photons m−2 s−1 conditions were harvested after either a 4-h light or a 4-h dark treatment and then ground into a powder in liquid nitrogen. Total proteins were subsequently extracted in degradation buffer containing 25 mM Tris-HCl, pH 7.5, 10 mM NaCl, 10 mM MgCl2, 4 mM PMSF, 5 mM DTT, and 10 mM ATP as described by Wang et al. (2009). The supernatants were collected after two 10-min centrifugations at 17,000g at 4°C. Protein concentrations were determined using the Bio-Rad protein assay. The concentrations of total protein extracts were adjusted to equal levels using degradation buffer. One hundred nanograms of recombinant GST-LIR1 was incubated in 100 μL total protein extract (containing 500 µg total protein) for each assay. The extracts were incubated in 1000 µmol photons m−2 s−1 light or in the dark. Samples were collected at the indicated intervals and used to determine GST-LIR1 levels by immunoblotting.
Coimmunoprecipitation and LC-MS/MS
Rice seedlings (wild-type and 35S:Flag-LIR1 transgenic plants) or tobacco (Nicotiana tabacum) leaves transiently expressing binary vectors following Agrobacterium infiltration were ground in liquid nitrogen and incubated in IP buffer (50 mM Tris-HCl, pH 6.8, 150 mM NaCl, Roche protease inhibitor complete, 1 tablet/50 mL IP buffer, 1 mM PMSF, and 20 μM MG132) supplemented with 0.6% Triton X-100 for 20 min at 4°C. The solution was filtered through one layer of Miracloth and centrifuged at 12,000g for 10 min. The supernatant was diluted with IP buffer to reduce the concentration of Triton X-100 to 0.2% and incubated with anti-Flag M2 magnetic beads (Sigma-Aldrich) for 1 h with gentle rotation. The beads were captured with a magnetic rack (Invitrogen) and washed six times with washing buffer (50 mM Tris-HCl, pH 7.5, and 250 mM NaCl). The protein complex was eluted with Flag elution buffer (25 mM Tris-HCl, pH 7.5. and 0.2 mg/mL 3× FLAG peptide) by rotation for 30 min. The protein complex solution was concentrated using Amicon 3KD centrifugal filters. All of the above steps were performed at 4°C. The protein complex solution was separated on SDS-PAGE gels and stained using a Silver Stain kit (Pierce) or immunoblotted with anti-Flag antibody (Sigma-Aldrich). Specific silver-stained bands were cut out of the gels and subjected to LC-MS/MS analysis (performed by Shanghai Applied Protein Technology Co.).
Yeast Two-Hybrid Assays
Yeast (Saccharomyces cerevisiae) two-hybrid assays were performed according to the Matchmaker Gold Yeast two-hybrid system manual (Clontech). Full-length LIR1 and LFNR coding sequences from different plant species and rice TIC62 coding sequences were cloned into pGADT7 and pGBKT7, respectively. The constructs for each species were cotransformed into yeast strain AH109 using the polyethylene glycol/lithium acetate method. The cotransformed yeast cells were grown on SD/-Leu/-Trp and SD/-Leu/-Trp/-His/-Ade plates to test the protein-protein interactions through the activation of two different reporter genes, His3 and Ade2. Details about the primers used for plasmid construction are shown in Supplemental Table 3.
BiFC Assays
For BiFC analysis, the coding sequences of rice LIR1, LFNR1, LFNR2, and RBCS (ribulose bisphosphate carboxylase small chain, LOC_Os12g19470) were amplified and ligated into the p35S-cYFP or p35S-nYFP vector (Yang et al., 2007). The resulting constructs were transiently expressed in tobacco leaves by Agrobacterium infiltration. YFP fluorescence of tobacco leaves was imaged 3 d after infiltration using a Zeiss LSM710NLO confocal laser scanning microscope. The excitation wavelength for YFP fluorescence was 488 nm, and fluorescence was detected at 500 to 542 nm.
RNA Extraction and qRT-PCR
Total RNA from rice was extracted using Trizol D0410 reagent according to the manufacturer’s instructions (Invitrogen). First-strand cDNA was synthesized from 2 μg DNaseI-treated total RNA with SuperScript II reverse transcriptase (Invitrogen). qRT-PCR was performed in a Roche 480 real-time PCR system using Roche SYBR Green Master Mix according to the manufacturer’s instructions. Relative expression levels were normalized to that of an internal control, ACTIN (LOC_Os03g50885). The primers used for qRT-PCR are listed in Supplemental Table 3.
Isolation and Fractionation of Chloroplasts
For chloroplast fractionation, intact chloroplasts were prepared from ∼100 g (fresh weight) of leaf tissue from 1-month-old plants according to Seigneurin-Berny et al. (2008). Stroma, thylakoid, and envelope proteins were isolated by sucrose gradient centrifugation as described by Li et al. (1991).
Thylakoid and soluble proteins were extracted according to Lintala et al. (2007). Rice leaves were frozen in liquid nitrogen and homogenized in shock buffer (10 mM HEPES-KOH, pH 7.6, 5 mM sucrose, 5 mM MgCl2, and 10 mM NaF). The homogenate was filtered through Miracloth (Millipore) and centrifuged at 2500g for 5 min at 4°C. The supernatant was defined as the soluble proteins and the pellet was defined as thylakoid proteins, which were then solubilized in shock buffer with 2% Triton X-100.
Immunoblotting Analysis
Native PAGE and BN-PAGE were performed according to Lintala et al. (2009). For native PAGE, proteins were subjected to native gel electrophoresis (12% acrylamide, 375 mM Tris-HCl, pH 8.8, and 7.6% glycerol). For BN-PAGE, fresh extracted thylakoid proteins mixed with 2× BN-PAGE loading dye (Serva; 42533.01) were separated on a 4 to 12% gradient gel. All electrophoresis were performed at 4°C. After electrophoresis, the gels were incubated in transfer buffer with 0.1% SDS for 20 min and then proteins were electroblotted onto a polyvinylidene difluoride membrane, and immunodetection was performed using an enhanced chemiluminescence immunoblot system. Anti-LFNR antibody was purchased from AntiProt, anti-Flag antibody from Sigma-Aldrich, and anti-RBCL, anti-D1, and anti-TIC40 antibodies from Agrisera. Anti-TIC62 antibody (Benz et al., 2009) was a generous gift from Bettina Bölter (LMU Munich, Germany). Immunoblots were examined using a Bio-Rad Image Analyzer and protein levels were quantified with Image Lab software (Bio-Rad).
Photosynthetic Parameters
Chlorophyll fluorescence was measured using a Dual-PAM 100 (Heinz Walz). Prior to measurements, all plants were incubated in the dark for at least 30 min. NPQ and electron transfer rate were recorded and calculated using Dual-PAM 100 software under actinic light illumination of different intensities according to He et al. (2015).
Photooxidation and P700+ rereduction were monitored by measuring changes in absorbance at 810 nm as previously described by Lintala et al. (2007). P700 was oxidized using far-red LEDs (wavelength > 705 nm; intensity, 5.2 μmol photons m−2 s−1) for 30 s, and the subsequent rereduction of P700+ was recorded in the dark.
Photosynthetic net rate was measured according to He et al. (2014). The CO2 assimilation rate was recorded as the photosynthetic efficiency using a Licor 6400 photosynthesis system at a constant CO2 concentration under standard growth conditions.
Protein Expression and Purification
To generate His-LFNR1, His-LFNR2, and HisTIC62Ct (His-322 to Pro-498), the relevant coding sequences were cloned from rice leaf cDNA and inserted into pET28a. For LIR1-His, LIR1 was cloned from rice leaf cDNA and inserted into pET22b. For GST-LIR1 and GST-TIC62, the coding sequences were cloned from rice leaf cDNA and inserted into pGEX-4T-1. For heterologous expression, the clones were transformed into Escherichia coli BL21 (DE3) cells and grown at 37°C in the presence of 50 mg/mL kanamycin sulfate (His-LFNR1, His-LFNR2, and His-TIC62Ct) or 100 mg/mL ampicillin (LIR1-His, GST-LIR1, and GST-TIC62) to an A600 of 0.5. Expression was induced by adding 0.1 mM IPTG, and the cells were grown for 4 h at 30°C (GST and GST-LIR1) or overnight at 18°C (LIR1-His, His-LFNR1, His-LFNR2, His-TIC62Ct, and GST-TIC62). LIR1-His, His-LFNR1, His-LFNR2, and His-TIC62Ct were purified via their N- or C-terminal polyhistidine tags using Ni-NTA-Sepharose (GE Healthcare) under native conditions and eluted with 250 mM imidazole. GST, GST-LIR1, and GST-TIC62 were purified via their N-terminal GST tag using a Glutathione Sepharose 4 Fast Flow kit (GE Healthcare) according to the manufacturer’s protocol. Coomassie Brilliant Blue (CBB)-stained gels of all purified proteins from E. coli used in this article are shown in Supplemental Figure 11.
Protein Binding Affinity Assays
The His-tagged C terminus of rice TIC62 (His-TIC62Ct) expressed and purified as described above was bound to Ni-NTA-Sepharose and used as an affinity matrix for leaf soluble extract, harvested from tic62 plants grown under standard GL (1000 µmol photons m−2 s−1), in the presence of GST or GST-LIR1 in different pH buffer conditions (pH 6, 7, or 8) as indicated in Figure 8B. Empty Ni2+ beads were used as a negative control. After incubation for 1 h at 4°C, the His beads were washed six times with pull-down buffer at the relevant pH plus 250 mM NaCl, and the bound proteins were eluted by boiling in an equal volume of 1× SDS protein loading buffer. The amount of LFNR pulled down was analyzed by immunoblotting with anti-LFNR antibody.
For the GST pull-down assays, 1 µg GST-tagged full-length rice TIC62 (GST-TIC62), LIR1-His, and His-LFNR1/2 proteins expressed and purified as described above were added to pull-down buffer system as indicated in Figures 8C and 8D at pH 6, 7, or 8. Equal amounts of rice LFNR1 and LFNR2 were mixed as His-LFNR1/2. Glutathione Sepharose beads (GE Healthcare) were used to bind GST-TIC62 in buffer with or without LIR1-His under different pH conditions. After incubation for 1 h at 4°C, the Glutathione Sepharose beads were washed six times with pull-down buffer at the relevant pH plus 250 mM NaCl, and then the bound proteins were eluted by boiling in an equal volume of 1× SDS protein loading buffer. The amount of pulled-down LFNR was analyzed by immunoblotting with His antibody.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis (At) TAIR or rice (Os) TIGR databases under the following accession numbers: At-LFNR1, At5g66190; At-LFNR2, At1g20020; At-LIR1, At3g26740; At-TIC62, At3g18890; At-TROL, At4g01050; Os-LFNR1, LOC_Os06g01850; Os-LFNR2, LOC_Os02g01340; Os-LIR1, LOC_Os01g01340; Os-RBCS, LOC_Os12g19470; Os-TIC62, LOC_Os10g01044; and Os-TROL, LOC_Os02g15750. Germplasm used included the Arabidopsis T-DNA insertion mutant At-lir1, SALK_024728C, and the rice tic62 insertion mutant AJQE06, Genoplante T-DNA insertion line library.
Supplemental Data
Supplemental Figure 1. Amino Acid Sequence and Structural Predictions of Rice LIR1.
Supplemental Figure 2. Characteristics of Rice tic62 Plants.
Supplemental Figure 3. Bimolecular Fluorescence Complementation Analysis of Protein Interactions in Nicotiana tabacum.
Supplemental Figure 4. Effect of DTT and Diamide on the Interaction between Rice LIR1 and LFNR.
Supplemental Figure 5. Comparison of the Two Rice LFNR Isoforms between Wild-Type and lir1 Plants.
Supplemental Figure 6. Phylogenetic Analysis of LIR1.
Supplemental Figure 7. Alignment of LIR1 Amino Acid Sequences.
Supplemental Figure 8. Interaction between LIR1 and LFNR from Maize, Soybean, and Cucumber.
Supplemental Figure 9. Characteristics of Arabidopsis lir1 Plants.
Supplemental Figure 10. Interaction Test between Rice LIR1 and TIC62 by Yeast Two-Hybrid Assay.
Supplemental Figure 11. The CBB-Stained Gels of All Purified Proteins from Escherichia coli Used in This Research.
Supplemental Table 1. List of LC-MS Identified Unique Peptide Fragments in the 35-kD Band from Coimmunoprecipitation.
Supplemental Table 2. List of LC-MS Identified Unique Peptide Fragments in 25-kD Band from Coimmunoprecipitation.
Supplemental Table 3. Primers Used in This Research.
Supplemental Methods. Phylogenetic Analysis.
Supplemental Data Set 1. Text File of the Sequences and Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 6.
Supplementary Material
Acknowledgments
We thank Shanghai Applied Protein Technology Co. for excellent technical service when performing the LC-MS/MS assay, Weihua Mao from the Center of Analysis and Measurement at Zhejiang University for sequencing, and Genoplante-Valor and CIRAD in France for supplying rice T-DNA insertional line tic62. We also thank Shelong Zhang and Fangliang Huang for their technical assistance. This work was supported by the Ministry of Agriculture of China (Grants 2016ZX08009003-005-004, 2016ZX08001003-009, 2014ZX08009003-005-005), the Ministry of Education and Bureau of Foreign Experts of China (Grant B14027), the Natural Science Foundation of Zhejiang Province (Grant LY14C020003), and the Academy of Finland (Centre of Excellence in the Molecular Biology of Primary Producers 271832). We thank Taina Tyystjärvi for critically reading the manuscript.
AUTHOR CONTRIBUTIONS
P.W. and X.M. designed the research. C.Y. studied the degradation of LIR1 and performed the blue native experiment. X.M. identified the LIR1 interacting partner. H.H. performed the pull down experiments. H.R. constructed vectors. Y.K. studied the distribution of LFNR between stroma and thylakoid. J.G. tested the interaction under various redox conditions. H.L. performed yeast two-hybrid experiments. L.W. studied the subcellular localization of LIR1. Y.H. performed photosynthetic analysis in rice. X.D. identified the T-DNA insertional line. M.G. performed photosynthetic analysis in Arabidopsis. T.C. performed BiFC. Y.L. performed rice transformation. Z.W. did the bioinformatics analysis. Y.W. monitored plant growth. P.M., X.M., and C.Y. wrote the article. C.M. revised the manuscript.
Glossary
- GL
growth light
- FD
ferredoxin
- ML
medium light
- LL
low light
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- BiFC
bimolecular fluorescence complementation
- BN-PAGE
blue native PAGE
- NPQ
nonphotochemical quenching
- CDS
coding sequence
- dCAPS
derived cleaved amplified polymorphic sequence
- CBB
Coomassie Brilliant Blue
- IP
immunoprecipitate
References
- Abied M.A., Holland D. (1994). Two newly isolated genes from citrus exhibit a different pattern of diurnal expression and light response. Plant Mol. Biol. 26: 165–173. [DOI] [PubMed] [Google Scholar]
- Alte F., Stengel A., Benz J.P., Petersen E., Soll J., Groll M., Bölter B. (2010). Ferredoxin:NADPH oxidoreductase is recruited to thylakoids by binding to a polyproline type II helix in a pH-dependent manner. Proc. Natl. Acad. Sci. USA 107: 19260–19265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsera M., Stengel A., Soll J., Bölter B. (2007). Tic62: a protein family from metabolism to protein translocation. BMC Evol. Biol. 7: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz J.P., Lintala M., Soll J., Mulo P., Bölter B. (2010). A new concept for ferredoxin-NADP(H) oxidoreductase binding to plant thylakoids. Trends Plant Sci. 15: 608–613. [DOI] [PubMed] [Google Scholar]
- Benz J.P., Stengel A., Lintala M., Lee Y.H., Weber A., Philippar K., Gügel I.L., Kaieda S., Ikegami T., Mulo P., Soll J., Bölter B. (2009). Arabidopsis Tic62 and ferredoxin-NADP(H) oxidoreductase form light-regulated complexes that are integrated into the chloroplast redox poise. Plant Cell 21: 3965–3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bläsing O.E., Gibon Y., Günther M., Höhne M., Morcuende R., Osuna D., Thimm O., Usadel B., Scheible W.R., Stitt M. (2005). Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17: 3257–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhov N.G., Govindachary S., Rajagopal S., Joly D., Carpentier R. (2004). Enhanced rates of P700(+) dark-reduction in leaves of Cucumis sativus L photoinhibited at chilling temperature. Planta 218: 852–861. [DOI] [PubMed] [Google Scholar]
- Ceccarelli E.A., Arakaki A.K., Cortez N., Carrillo N. (2004). Functional plasticity and catalytic efficiency in plant and bacterial ferredoxin-NADP(H) reductases. Biochim. Biophys. Acta 1698: 155–165. [DOI] [PubMed] [Google Scholar]
- Chen K.-M., Holmström M., Raksajit W., Suorsa M., Piippo M., Aro E.-M. (2010). Small chloroplast-targeted DnaJ proteins are involved in optimization of photosynthetic reactions in Arabidopsis thaliana. BMC Plant Biol. 10: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Chory J. (2011). Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 21: 664–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciannamea S., Jensen C.S., Agerskov H., Petersen K., Lenk I., Didion T., Immink R.G.H., Angenent G.C., Nielsen K.K. (2007). A new member of the LIR gene family from perennial ryegrass is cold-responsive, and promotes vegetative growth in Arabidopsis. Plant Sci. 172: 221–227. [Google Scholar]
- Fan D.Y., Nie Q., Hope A.B., Hillier W., Pogson B.J., Chow W.S. (2007). Quantification of cyclic electron flow around photosystem I in spinach leaves during photosynthetic induction. Photosynth. Res. 94: 347–357. [DOI] [PubMed] [Google Scholar]
- Feng S., Ma L., Wang X., Xie D., Dinesh-Kumar S.P., Wei N., Deng X.W. (2003). The COP9 signalosome interacts physically with SCF COI1 and modulates jasmonate responses. Plant Cell 15: 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Zhang B., Ding W., Liu X., Yang D.L., Wei P., Cao F., Zhu S., Zhang F., Mao Y., Zhu J.K. (2013). Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23: 1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forti G., Bracale M. (1984). Ferredoxin—ferredoxin NADP reductase interaction: Catalytic differences between the soluble and thylakoid-bound complex. FEBS Lett. 166: 81–84. [Google Scholar]
- Forti G., Cappelletti A., Nobili R.L., Garlaschi F.M., Gerola P.D., Jennings R.C. (1983). Interaction of ferredoxin and ferredoxin-NADP reductase with thylakoids. Arch. Biochem. Biophys. 221: 507–513. [DOI] [PubMed] [Google Scholar]
- Golding A.J., Finazzi G., Johnson G.N. (2004). Reduction of the thylakoid electron transport chain by stromal reductants--evidence for activation of cyclic electron transport upon dark adaptation or under drought. Planta 220: 356–363. [DOI] [PubMed] [Google Scholar]
- Gummadova J.O., Fletcher G.J., Moolna A., Hanke G.T., Hase T., Bowsher C.G. (2007). Expression of multiple forms of ferredoxin NADP+ oxidoreductase in wheat leaves. J. Exp. Bot. 58: 3971–3985. [DOI] [PubMed] [Google Scholar]
- Hanke G., Mulo P. (2013). Plant type ferredoxins and ferredoxin-dependent metabolism. Plant Cell Environ. 36: 1071–1084. [DOI] [PubMed] [Google Scholar]
- Hanke G.T., Endo T., Satoh F., Hase T. (2008). Altered photosynthetic electron channelling into cyclic electron flow and nitrite assimilation in a mutant of ferredoxin:NADP(H) reductase. Plant Cell Environ. 31: 1017–1028. [DOI] [PubMed] [Google Scholar]
- Hanke G.T., Okutani S., Satomi Y., Takao T., Suzuki A., Hase T. (2005). Multiple iso-proteins of FNR in Arabidopsis: evidence for different contributions to chloroplast function and nitrogen assimilation. Plant Cell Environ. 28: 1146–1157. [Google Scholar]
- Hayama R., Izawa T., Shimamoto K. (2002). Isolation of rice genes possibly involved in the photoperiodic control of flowering by a fluorescent differential display method. Plant Cell Physiol. 43: 494–504. [DOI] [PubMed] [Google Scholar]
- He Y., et al. (2015). Increasing cyclic electron flow is related to Na+ sequestration into vacuoles for salt tolerance in soybean. J. Exp. Bot. 66: 6877–6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Yu C., Zhou L., Chen Y., Liu A., Jin J., Hong J., Qi Y., Jiang D. (2014). Rubisco decrease is involved in chloroplast protrusion and Rubisco-containing body formation in soybean (Glycine max) under salt stress. Plant Physiol. Biochem. 74: 118–124. [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282. [DOI] [PubMed] [Google Scholar]
- Higuchi-Takeuchi M., Ichikawa T., Kondou Y., Matsui K., Hasegawa Y., Kawashima M., Sonoike K., Mori M., Hirochika H., Matsui M. (2011). Functional analysis of two isoforms of leaf-type ferredoxin-NADP(+)-oxidoreductase in rice using the heterologous expression system of Arabidopsis. Plant Physiol. 157: 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P., Ruban A.V., Wentworth M. (2000). Allosteric regulation of the light-harvesting system of photosystem II. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1361–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.-l., Yue C.-l., Qin F., Hu F., and Wang G.-x. (2010). Bioinformatics analysis of the LIR1 gene and its promoter sequence in rice. In 2010 2nd International Conference on Information Science and Engineering (IEEE Computer Society Conference Publishing Services), pp. 6482–6485. [Google Scholar]
- Järvi S., Suorsa M., Aro E.M. (2015). Photosystem II repair in plant chloroplasts--Regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim. Biophys. Acta 1847: 900–909. [DOI] [PubMed] [Google Scholar]
- Joliot P., Johnson G.N. (2011). Regulation of cyclic and linear electron flow in higher plants. Proc. Natl. Acad. Sci. USA 108: 13317–13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot P., Joliot A. (2002). Cyclic electron transfer in plant leaf. Proc. Natl. Acad. Sci. USA 99: 10209–10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurić S., Hazler-Pilepić K., Tomasić A., Lepeduš H., Jelicić B., Puthiyaveetil S., Bionda T., Vojta L., Allen J.F., Schleiff E., Fulgosi H. (2009). Tethering of ferredoxin:NADP+ oxidoreductase to thylakoid membranes is mediated by novel chloroplast protein TROL. Plant J. 60: 783–794. [DOI] [PubMed] [Google Scholar]
- Kang B., Zhang Z., Wang L., Zheng L., Mao W., Li M., Wu Y., Wu P., Mo X. (2013). OsCYP2, a chaperone involved in degradation of auxin-responsive proteins, plays crucial roles in rice lateral root initiation. Plant J. 74: 86–97. [DOI] [PubMed] [Google Scholar]
- Kato Y., Sakamoto W. (2009). Protein quality control in chloroplasts: a current model of D1 protein degradation in the photosystem II repair cycle. J. Biochem. 146: 463–469. [DOI] [PubMed] [Google Scholar]
- Küchler M., Decker S., Hörmann F., Soll J., Heins L. (2002). Protein import into chloroplasts involves redox-regulated proteins. EMBO J. 21: 6136–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtimäki N., Koskela M.M., Dahlström K.M., Pakula E., Lintala M., Scholz M., Hippler M., Hanke G.T., Rokka A., Battchikova N., Salminen T.A., Mulo P. (2014). Posttranslational modifications of FERREDOXIN-NADP+ OXIDOREDUCTASE in Arabidopsis chloroplasts. Plant Physiol. 166: 1764–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtimäki N., Koskela M.M., Mulo P. (2015). Post-translational modifications of chloroplast proteins: An emerging field. Plant Physiol. 168: 768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtimäki N., Lintala M., Allahverdiyeva Y., Aro E.M., Mulo P. (2010). Drought stress-induced upregulation of components involved in ferredoxin-dependent cyclic electron transfer. J. Plant Physiol. 167: 1018–1022. [DOI] [PubMed] [Google Scholar]
- Li H.M., Moore T., Keegstra K. (1991). Targeting of proteins to the outer envelope membrane uses a different pathway than transport into chloroplasts. Plant Cell 3: 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintala M., Allahverdiyeva Y., Kangasjärvi S., Lehtimäki N., Keränen M., Rintamäki E., Aro E.M., Mulo P. (2009). Comparative analysis of leaf-type ferredoxin-NADP oxidoreductase isoforms in Arabidopsis thaliana. Plant J. 57: 1103–1115. [DOI] [PubMed] [Google Scholar]
- Lintala M., Allahverdiyeva Y., Kidron H., Piippo M., Battchikova N., Suorsa M., Rintamäki E., Salminen T.A., Aro E.-M., Mulo P. (2007). Structural and functional characterization of ferredoxin-NADP+-oxidoreductase using knock-out mutants of Arabidopsis. Plant J. 49: 1041–1052. [DOI] [PubMed] [Google Scholar]
- Lintala M., Schuck N., Thormählen I., Jungfer A., Weber K.L., Weber A.P.M., Geigenberger P., Soll J., Bölter B., Mulo P. (2014). Arabidopsis tic62 trol mutant lacking thylakoid-bound ferredoxin-NADP+ oxidoreductase shows distinct metabolic phenotype. Mol. Plant 7: 45–57. [DOI] [PubMed] [Google Scholar]
- Lv Q., Zhong Y., Wang Y., Wang Z., Zhang L., Shi J., Wu Z., Liu Y., Mao C., Yi K., Wu P. (2014). SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 26: 1586–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulo P., Sakurai I., Aro E.M. (2012). Strategies for psbA gene expression in cyanobacteria, green algae and higher plants: from transcription to PSII repair. Biochim. Biophys. Acta 1817: 247–257. [DOI] [PubMed] [Google Scholar]
- Okutani S., Hanke G.T., Satomi Y., Takao T., Kurisu G., Suzuki A., Hase T. (2005). Three maize leaf ferredoxin:NADPH oxidoreductases vary in subchloroplast location, expression, and interaction with ferredoxin. Plant Physiol. 139: 1451–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejaver V., Hsu W.L., Xin F., Dunker A.K., Uversky V.N., Radivojac P. (2014). The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 23: 1077–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piippo M., Allahverdiyeva Y., Paakkarinen V., Suoranta U.M., Battchikova N., Aro E.M. (2006). Chloroplast-mediated regulation of nuclear genes in Arabidopsis thaliana in the absence of light stress. Physiol. Genomics 25: 142–152. [DOI] [PubMed] [Google Scholar]
- Quigley F., Dao P., Cottet A., Mache R. (1996). Sequence analysis of an 81 kb contig from Arabidopsis thaliana chromosome III. Nucleic Acids Res. 24: 4313–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimmann C., Dudler R. (1993). Circadian rhythmicity in the expression of a novel light-regulated rice gene. Plant Mol. Biol. 22: 165–170. [DOI] [PubMed] [Google Scholar]
- Seigneurin-Berny D., Salvi D., Dorne A.J., Joyard J., Rolland N. (2008). Percoll-purified and photosynthetically active chloroplasts from Arabidopsis thaliana leaves. Plant Physiol. Biochem. 46: 951–955. [DOI] [PubMed] [Google Scholar]
- Stengel A., Benz P., Balsera M., Soll J., Bölter B. (2008). TIC62 redox-regulated translocon composition and dynamics. J. Biol. Chem. 283: 6656–6667. [DOI] [PubMed] [Google Scholar]
- Teramoto H., Momotani E., Takeba G., Tsuji H. (1994). Isolation of a cDNA clone for a cytokinin-repressed gene in excised cucumber cotyledons. Planta 193: 573–579. [DOI] [PubMed] [Google Scholar]
- Thomas J.C., Ughy B., Lagoutte B., Ajlani G. (2006). A second isoform of the ferredoxin:NADP oxidoreductase generated by an in-frame initiation of translation. Proc. Natl. Acad. Sci. USA 103: 18368–18373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornber J.P. (1975). Chlorophyll proteins: Light-harvesting and reaction center components of plants. Annu. Rev. Plant Physiol. 26: 127–158. [Google Scholar]
- Twachtmann M., Altmann B., Muraki N., Voss I., Okutani S., Kurisu G., Hase T., Hanke G.T. (2012). N-terminal structure of maize ferredoxin:NADP+ reductase determines recruitment into different thylakoid membrane complexes. Plant Cell 24: 2979–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojta L., Carić D., Cesar V., Antunović Dunić J., Lepeduš H., Kveder M., Fulgosi H. (2015). TROL-FNR interaction reveals alternative pathways of electron partitioning in photosynthesis. Sci. Rep. 5: 10085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Zhu D., Huang X., Li S., Gong Y., Yao Q., Fu X., Fan L.M., Deng X.W. (2009). Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21: 2378–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamori W., Sakata N., Suzuki Y., Shikanai T., Makino A. (2011). Cyclic electron flow around photosystem I via chloroplast NAD(P)H dehydrogenase (NDH) complex performs a significant physiological role during photosynthesis and plant growth at low temperature in rice. Plant J. 68: 966–976. [DOI] [PubMed] [Google Scholar]
- Yang X., Baliji S., Buchmann R.C., Wang H., Lindbo J.A., Sunter G., Bisaro D.M. (2007). Functional modulation of the geminivirus AL2 transcription factor and silencing suppressor by self-interaction. J. Virol. 81: 11972–11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.