Abstract
Shifts in the maternal gut microbiome have been implicated in metabolic adaptations to pregnancy. We investigated how pregnancy and diet interact to influence the composition of the maternal gut microbiota. Female C57BL/6 mice were fed either a control or a high fat diet for 8 weeks prior to mating. After confirmation of pregnancy, maternal weight gain and food intake were recorded. Fecal pellets were collected at 2 timepoints prior to mating (at the beginning of the experiment, and after 6 weeks of the specified diet) and at 4 timepoints during pregnancy (gestation day 0.5, 5.5, 10.5, and 15.5). The microbial composition and predicted metabolic functionality of the non-pregnant and pregnant gut was determined via sequencing of the variable 3 region of the 16S rRNA gene. Upon conception, differences in gut microbial communities were observed in both control and high fat-fed mice, including an increase in mucin-degrading bacteria. Control versus high fat-fed pregnant mice possessed the most profound changes to their maternal gut microbiota as indicated by statistically significant taxonomic differences. High fat-fed pregnant mice, when compared to control-fed animals, were found to be significantly enriched in microbes involved in metabolic pathways favoring fatty acid, ketone, vitamin, and bile synthesis. We show that pregnancy-induced changes in the female gut microbiota occur immediately at the onset of pregnancy, are vulnerable to modulation by diet, but are not dependent upon increases in maternal weight gain during pregnancy. High fat diet intake before and during pregnancy results in distinctive shifts in the pregnant gut microbiota in a gestational-age dependent manner and these shifts predict significant differences in the abundance of genes that favor lipid metabolism, glycolysis and gluconeogenic metabolic pathways over the course of pregnancy.
Keywords: gut, high fat diet, intestine, microbiome, obesity, pregnancy
Background
Obesity affects more than 500 million people worldwide.1 The steady rise in the prevalence of obesity over the past 3 decades cannot be solely attributed to genetic factors and has led researchers to examine other causes that may contribute to disease risk. Epidemiological and experimental data has shown a relationship between the in utero environment and the risk of developing chronic diseases, such as obesity, later in life.2 Prenatal signals, such as nutrition, modulate disease risk by inducing gene-environment interactions in fetal homeostasis leading to persistent changes in key signaling pathways. Maternal obesity, a key predictor of childhood obesity, is associated with abnormal feto-placental function,3 offspring obesity risk,4 and increased disease risk in general.5
The gut microbiota is emerging as a disease risk factor due to its role in regulating energy extraction and whole body metabolism. These microbes are essential for metabolizing indigestible polysaccharides, producing essential nutrients, and regulating fat storage.6 The presence and abundance of each bacterial taxa is crucial since different populations differentially metabolize nutrients and have varying capacities of energy harvest.7 The mammalian gut is generally dominated by 2 phyla, Firmicutes and Bacteroidetes,8 and shifts in the relative abundances of these groups have been observed in obesogenic states when compared to the gut microbiota of healthy individuals.9 Both dietary factors 10 and genetics 9 influence gut microbial composition.11 High fat (HF) intake is associated with an increased Firmicutes to Bacteroidetes ratio, increased gut inflammation and intestinal permeability.12,13 Comparisons of the gut microbiota of obese and lean twins has shown that obesity is associated with a decrease in abundance of Bacteroidetes and altered expression of genes involved in metabolic pathways that favor increased energy uptake and adipogenesis.7
There is evidence, although limited, to suggest that pregnancy is a physiological state that is also associated with shifts in gut microbiota.14 The biological purpose of these shifts may contribute to the ability of the mother to adapt to pregnancy and facilitates optimal fetal growth and development, but the mechanisms regulating these adaptations are unclear. Over the course of normal pregnancy, key metabolic changes occur in the mother that resemble metabolic dysfunction, including insulin resistance, dyslipidemia and hypertension.15 Late pregnancy is characterized by increased insulin and leptin resistance and depletion of maternal adipose tissue deposits, facilitating the growth of the placenta and the fetus. Recent studies have suggested that changes to the microbiota may influence these pregnancy-associated metabolic changes, but whether this occurs immediately at the onset of pregnancy or occurs slowly over the course of gestation is unknown. The biomass present in the maternal gut microbiota is known to increase from early to late pregnancy and is accompanied by an increase in diversity.14,16 Of particular interest, when germ-free mice are colonized with third trimester maternal microbiota, these mice demonstrate heightened inflammatory responses and adipose tissue accumulation.14 This observation is suggestive of pregnancy-induced changes to the gut microbiota, which appear similar to that observed in obese individuals.
The factors that modulate maternal gut microbial changes are currently unknown; however, body weight may be an associated factor. It has been identified that the maternal gut microbiota may be influenced by pre-pregnancy weight and weight gain over the course of pregnancy.16 In human studies, overweight pregnant women were found to have significantly higher levels of Bacteroides and Staphylococcus, where pregnancy weight gain was correlated with Bacteroides abundance.16 However, the relationships between advancing gestation, maternal weight gain, and maternal diet-induced obesity remain unclear. Critically, it is also unknown whether the maternal pre-pregnancy condition modulates pregnancy-associated changes in maternal gut microbial population. Thus, using a mouse model of high fat (HF) diet intake, we investigated pregnancy-associated shifts as well as diet-induced shifts in the maternal gut microbiota and predicted the impacts of these variables on metabolic pathways. This model is novel in that it allows investigations of the interactions between diet and pregnancy and will determine the extent to which microbial shifts in HF pregnancies are due to diet, pregnancy, or an interaction between the 2 conditions. We suggest that since microbiota are responsive to changes in the nutritional environment, changes in the mother's gut niche may impact maternal adaptation to pregnancy, the function of the placenta, and/or the growth of the fetus, and impart an increased risk of obesity and metabolic dysfunction in the offspring.
Results
Body weight and caloric intake
Fecal samples were collected from all mice at all timepoints (Fig. 1). Mating of control (n = 5) and high fat (n = 5) fed female mice resulted in samples collected from n = 5 control and n = 3 HF pregnant mice. Females fed a high fat (HF n = 5) diet for 8 weeks were not statistically heavier than low fat control diet-fed females (Con n = 5) at the time of mating. Consistent with previous reports indicating that nutritional excess and obesity reduces fertility in females,17 3 out of the 5 HF fed mice became pregnant. Gestational weight gain increased steadily throughout gestation and was not different between treatment groups (Fig. 2). Caloric intake was not statistically different between groups (Fig. 2).
Figure 1.
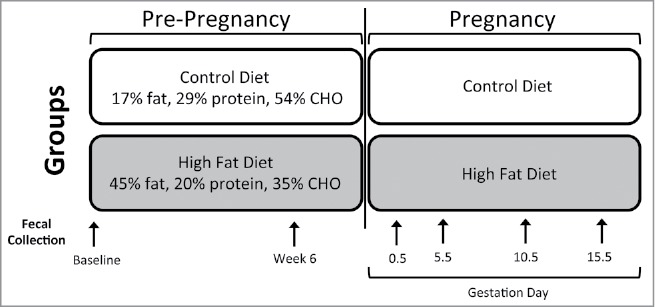
Schematic of mouse model. Schematic representation of the experimental design. Six-week-old female C57BL/6J mice (n = 10) were randomly assigned to one of 2 nutritional groups: 1) Control diet (Con; n = 5 17% fat, 29% protein, 54% CHO, 3 kcal/g) or 2) high fat diet (HF; n = 5; 45% fat, 20% protein, 35% CHO, 4.73 kcal/g), fed for 8 weeks and then mated with viable males. Pregnancy was identified when a vaginal plug was visible and designated as gestational day (E) 0.5 in both control (n = 5) and high fat (n = 3) groups. Fecal samples were collected from control mice prior to nutritional randomization (Baseline), after 6 weeks of either control or high fat diet feeding (Week 6) and then at timepoints during pregnancy (0.5, 5.5, 10.5 and 15.5; n = 5 control, n = 3 high fat fed).
Figure 2.
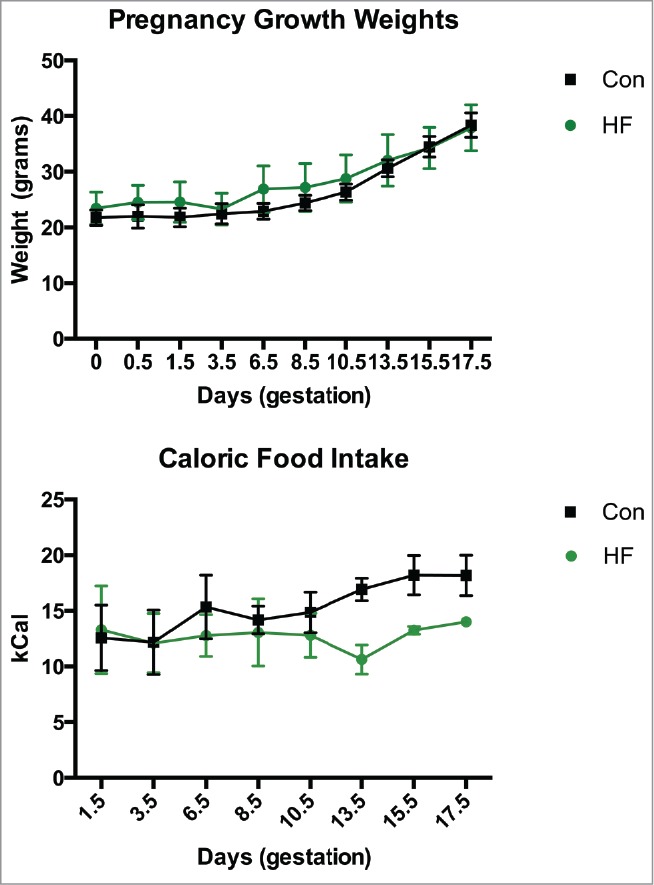
Maternal weight gain and caloric intake. Maternal weight gain over the course of pregnancy and caloric intake (expressed as calories consumed / gram body weight) in mothers during pregnancy. Data are mean ± SEM; n = 5 in control group, n = 3 in the HF group. Con = control diet, HF = high fat diet.
Pregnancy induces a shift in the gut microbiota of control-fed females
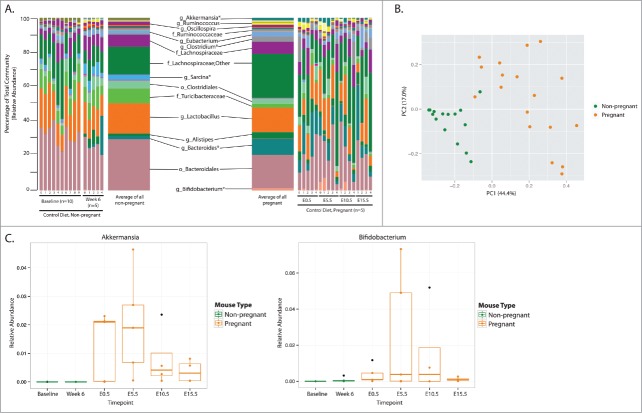
In order to study the impact of pregnancy on the maternal gut microbiota, mice were fed a control diet leading up to and during pregnancy. Fecal samples were collected pre- and post-conception, and the microbial communities were examined at 2 pre-conception and 4 post-conception timepoints (Figs. 1 and 3). Significant differences in the relative abundance of 21 genera were identified between non-pregnant and pregnant animals fed a control diet (Fig. S1 and Table S1). This includes 4 abundant genera (present at greater than 1%), namely Akkermansia, Clostridium, Bacteroides and Bifidobacterium, that were significantly increased during pregnancy compared to non-pregnant females (Fig. 3a, c, Table S1). Five taxa were reduced during pregnancy but these were present at low relative abundance (<0.5%), with the exception of Sarcina which decreased from 2.94% mean relative abundance to 0.14% with pregnancy (Fig. S1 and Table S1). These taxonomic differences indicate some variation within the non-pregnant and pregnant mice, which were further visualized using a Principal Coordinates Analysis (PCoA) of the Bray Curtis distance between microbial communities (Fig. 3b). Here, we see a distinct separation of the non-pregnant and pregnant animals despite some variation within both groups.
Figure 3.
Pregnancy alters the maternal gut microbial community of Control-fed mice. Mice fed a control diet were sampled twice during an 8-week period prior to impregnation (Baseline (BL), Week 6 (Wk6)), at 0.5 days of pregnancy (E0.5) and then every 5 days during pregnancy (E5.5, E10.5, E15.5). (A) Taxonomic summaries of microbial relative abundance for each mouse sampled at each timepoint are displayed; the microbial communities of non-pregnant and pregnant mice are summarized and taxonomic classifications, resolved to the order (o), family (f), or genus (g) level, are displayed and those with relative abundance > 1.0% are labeled. Asterisks identify genera that were significantly different between non-pregnant and pregnant mice; additional low abundance genera identified as significantly different are listed in Additional File 2. Further, an average taxonomic summary of each group displays the mean relative abundance of each genus. (B) A Principal Coordinates Analysis (PcoA) using the Bray Curtis distance metric displays separation of non-pregnant and pregnant animals (PERMANOVA, P = 0.001). (C) The relative abundance of the genera with the largest change in relative abundance between non-pregnant and pregnant animals are displayed. The x-axis indicates the sampling timepoints and samples are colored by pregnancy information. All other significant genera are displayed in Figure S1.
A more detailed investigation of the relative abundance of the most significantly different taxa between non- and pregnant Con diet animals shows that 3 taxa are significantly increased in the relative amounts of each bacteria over the course of pregnancy. Akkermansia is most abundant during early gestation (E0.5-E5.5) after which it drops to less than half its abundance during mid to late gestation (E10.5–15.5) (Fig. 3c). The relative abundance of Bifidobacterium also increases modestly at the onset of pregnancy, peaks dramatically in early gestation (E5.5) after which levels gradually decrease similar to those seen in the non-pregnant mice (Fig. 3c). Bacteroides increases with the onset of pregnancy at E0.5 and remains relatively elevated throughout pregnancy compared to non-pregnant females (Fig. S1). Examining these taxa, as well as others, which differ significantly (Fig. S1 and Table S1), we found changes to the gut microbial communities that occur not just at the onset of pregnancy, but also over the course of gestation (Fig. 3c, Fig. S1). In the 5 taxa that were suppressed with pregnancy, and did not display any changes in abundance over the course of gestation, all fell into the Firmicutes and Tenericute phyla, of which the genus Coprobacillus and Clostridium (phylum Tenericutes, family Erysipelotrichaceae) and Sarcina (phylum Firmicutes, family Clostridia) decreased the most upon conception (Fig. S1).
Pre-conception high fat diet intake modulates pregnancy-induced changes in female gut microbiota: a diet and pregnancy interaction
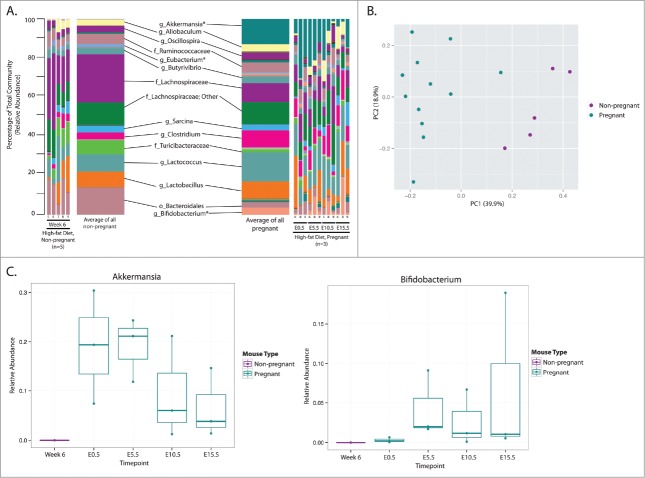
In parallel, we imposed a HF diet challenge in female mice to investigate the interactive effects between diet and pregnancy on the maternal gut microbiota. Eleven genera were identified as being significantly different between the non- and pregnant HF-fed groups most of which were Firmicutes and Bacteroidetes (increased with pregnancy) (Fig. S2 and Table S2) as well as a dramatic increase in Akkermansia (16.8 fold increase, phylum Verrucomicrobia) and Bifidobacterium (13.9 fold increase, phylum Actinobacteria) with gestation (Fig. 4a). A distinct set of pregnancy associated changes in microbial relative abundance occurs in mice fed a high fat diet compared to those fed a control diet where approximately half of the 11 genera that changed in high fat fed mice did not change in control mice (Table S1 and Table S2). Where pregnancy, independent of diet, induced an increased abundance of Allobaculum and Clostridium (family Lachnospiracea), and decreased 5 genera in control mice, HF-exposed mice did not (Table S1 and Table S2). Rather, HF exposed mice display no significant decrease in any genera with the onset of pregnancy.
Figure 4.
The maternal gut microbiota of animals fed a High fat diet is altered upon pregnancy. High fat-fed mice were sampled following 6 weeks of high fat feeding (Wk 6) prior to impregnation at 0.5 days of pregnancy (E0.5) and then every 5 days during pregnancy (E5.5, E10.5, E15.5). (A) Taxonomic summaries of microbial relative abundance for each mouse sampled at each timepoint are displayed; the microbial communities of non-pregnant and pregnant mice are summarized and taxonomic classifications, resolved to the order (o), family (f), or genus (g) level, are displayed and all genera present at a relative abundance of >1.0% are labeled. Asterisks identify genera that were significantly different between non-pregnant and pregnant mice on this diet; significantly different low abundance genera are listed in Additional File 4. Also, the mean relative abundance of each genus per group is displayed as an average taxonomic summary. (B) A Principal Coordinates Analysis (PcoA) using the Bray Curtis distance metric displays separation of non-pregnant and pregnant animals (PERMANOVA, P = 0.001). (C) The relative abundances of 2 genera identified as significantly different between non-pregnant and pregnant HF-fed animals are visualized. The x-axis indicates the sampling timepoints and samples are colored by pregnancy information. All other significant genera are displayed in Figure S2.
Consistent with Con fed mice, examination via PCoA indicates a distinct separation between non-pregnant and pregnant HF fed animals along the first axis despite some variation between the mice with each group (Fig. 4b).
When the relative abundances of the 2 genera with the greatest increase in relative abundance are visualized, similar patterns of abundance in Akkermansia over the course of gestation were observed in HF-fed pregnancies as in the Con-fed pregnancies (Figs. 3c and 4c). This indicates that the presence and abundance trends of this microbe may be universal in pregnancy. However, it appears that maternal diet may impact the fold increase of this microbe at the onset of pregnancy (Fig. 4b), as the relative abundance of Akkermansia is ∼10 fold greater in HF-fed pregnancies (Table S1 and Table S2). Similarly, over the course of pregnancy in HF-fed mice, the relative abundance of Bifidobacterium generally increases from E0.5–5.5, after which it remains elevated; this trend is unique to HF-fed mice and this genera (Figs. S1, S2, and 4c). Detailed abundances of other significantly different taxa are displayed in Figure S2.
Pregnancy-induced shifts in maternal gut microbiota: control vs. high fat nutrition
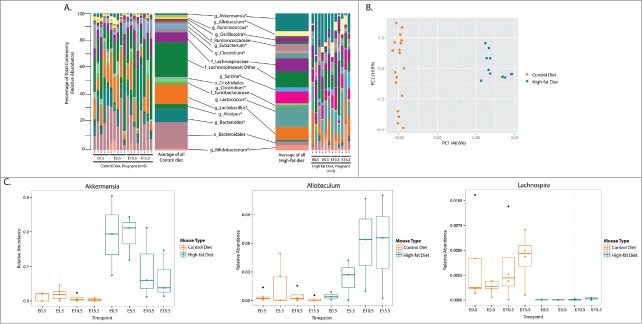
We have previously shown that imbalances in maternal macronutrient intake modifies both maternal and offspring metabolic function.18-20 Given the established relationship between diet-induced shifts in gut microbiota and metabolic function in non-pregnant animals,10 we set out to investigate whether there exists an interaction between diet and pregnancy on the gut microbiota. We observed the greatest differences in the gut microbiota, in terms of significantly altered taxa, between Con pregnant and HF pregnant mice (Fig. 5, Fig. S3 and Table S3), where 26 genera were identified as significantly different between Con and HF pregnancies. Taxonomic summaries of these mice over the course of pregnancy (Fig. 5a) and tests of significance across taxonomic groups (Table S3), demonstrate that pregnancy-induced shifts in the maternal gut microbiota are dependent upon the mother's diet before and during pregnancy. Indeed, when all 4 groups are visualized together, the first axis separates the mice groups based on diet, and the second axis by pregnancy (Fig. S4). Similarly, these differences are reflected when the 2 pregnant mouse groups are examined; pregnant mice fed a HF diet show distinctive changes in gut bacterial populations compared to pregnant controls (Fig. 5b). These data are robust, given that we observe distinct and significant differences despite relatively small numbers of mice that were successfully fertilized under the high fat diet regimen.
Figure 5.
The maternal gut microbiota during pregnancy is dependent on maternal diet. Pregnant mice in the Con and HF groups are compared to each other. (A) Taxonomic summaries of microbial relative abundance reveal more significant differences in taxa then in any other comparisons made. Each mouse sampled at each timepoint are displayed; the microbial communities of Con and HF pregnant mice are summarized and taxonomic classifications, resolved to the order (o), family (f), or genus (g) level, are displayed and all genera present at a relative abundance of >1.0% are labeled. Asterisks identify genera that were significantly different between Con and HF pregnant mice on this diet; significantly different low abundance genera are listed in Additional File 6. Further, an average taxonomic summary displays the mean relative abundance of each genus per grouping. (B) A Principal Coordinates Analysis (PcoA) using the Bray Curtis distance metric displays distinct separation of pregnant animals on differing diets, and distinct clustering of mice from each diet type (PERMANOVA, P = 0.001). (C) The relative abundances of 3 genera identified as significantly different between Con-fed and HF-fed pregnant animals are visualized. The x-axis indicates the sampling timepoints and samples are colored by pregnancy information. All other significant genera are displayed in Figure S3.
In total, 26 genera were found to be significantly different between Con and HF fed pregnant mice. This is greater than the 21 and 11 taxonomic groups that were altered with pregnancy alone in either Con or HF fed females respectively (Tables S1, S2 and S3). In order to further elucidate whether some of these changes were due more to diet then to pregnancy, we compared the non-pregnant Con to non-pregnant HF gut microbial composition in which we found 11 significantly different genera, all of which were also different during pregnancy. Thus, 15 genera are specifically related to pregnancy effects and changes in these genera are not due to an underlying diet-associated difference in their gut microbiota before pregnancy (Table S3). In contrast to the diet x pregnancy-induced changes in Akkermansia, other bacterial taxa are elevated during pregnancy only when mothers are fed a specific diet. For example, Allobaculum steadily increases in relative abundance during pregnancy in HF fed mothers, but has a significantly lower and unchanged abundance in Con mice (Fig 5c). Overall, of the 26 taxa that are different between Con- and HF-fed pregnant mice, the majority of them fall into the phylum Firmicutes, many of the order Clostridiales. Of those genera identified as significant, 14 are increased with the HF diet and 12 are increased with the Con diet (Fig. S3 and Table S3). Together, these observations suggest that there is an interactive effect of the mother's periconceptional diet on pregnancy-induced changes in the maternal gut microbiota.
Since previous reports have demonstrated that high fat intake is associated with an increased Firmicutes to Bacteroidetes ratio which is suggested to contribute to increased gut inflammation and intestinal permeability,12,13 the ratio of these 2 phyla was evaluated in non-pregnant and pregnant states with the addition of a dietary challenge. At the onset of pregnancy, control mice show little change in the Firmicutes to Bacteroidetes ratio, but this ratio increases with advancing gestation (Fig. S5). In high fat fed mice, this ratio is significantly increased in non-pregnant mice compared to controls and is not further altered with the onset of pregnancy. Our data support previous reports that high fat dietary intake increases the Firmicutes to Bacteroidetes ratio, but pregnancy had no further impact, and the ratio remained relatively constant in HF-fed pregnant mice. This suggested that changes in the ratio of these 2 phyla in pregnant obesogenic states may be primarily due to diet, not due to pregnancy.
Maternal high fat diet alters the abundance of microbes containing genes predicted to regulate metabolic pathways related to fuel and vitamin metabolism
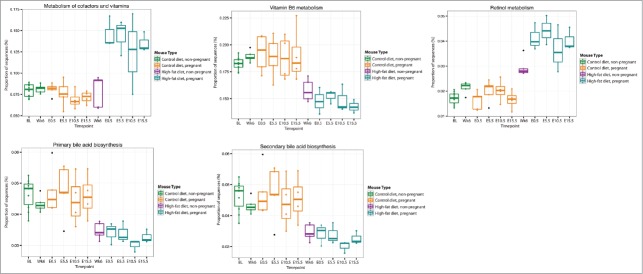
It has been shown that the gut microbiota of obese male mice is enriched in genes encoding for enzymes involved in energy extraction, where these bacterial populations facilitate obese mice to harvest more energy from their diet.4 We used PICRUSt 21 in combination with our 16S rRNA gene sequencing to estimate changes in microbial metabolic pathways involved in fuel and vitamin metabolism. We demonstrate that both maternal diet and gestational age significantly affect pathways involved in fuel metabolism, (Fig. 6, Table S4). While numerous pathways were identified as significantly different between the dietary groups, pathways particularly relevant to maternal metabolism during pregnancy are highlighted in Figure 6. The gut microbiota of HF pregnant mice is predicted by PICRUSt to be significantly enriched in genes involved in fatty acid and sulfur-containing amino acid metabolism, and glycolysis and gluconeogenesis compared to Con pregnant mice, and these increases are maintained throughout gestation. Interestingly, although there is little effect of a HF diet on unsaturated fatty acid synthesis in non-pregnant animals (Week 6 (Wk6), Fig. 6) pregnancy onset in HF-fed mice resulted in a significant rise in the abundance of these genes. Genes involved in the biosynthesis of unsaturated fatty acids are twice as high in HF mothers during early gestation, but these levels decline by mid-gestation (E10.5) (Fig. 6). Similarly, there appears to be a diet x gestational age interaction in genes associated with cysteine and methionine metabolism, and in the synthesis and degradation of ketone bodies in HF-fed mice compared to Con mice; where abundance of these sequences is elevated in HF mice and increase further with advancing gestation (Fig. 6).
Figure 7.
Maternal diet- and pregnancy-induced changes in the abundance of microbe containing genes predicted to regulate metabolic pathways related to vitamin and bile acid metabolism. Mice fed a control diet were sampled twice during an 8 week period prior to impregnation (BL, Wk6), at 0.5 days of pregnancy (E0.5) and then every 5 days during pregnancy (E5.5, E10.5, E15.5). HF fed mice were sampled following 6 weeks of high fat feeding (Wk 6) prior to impregnation, and at 0.5 days of pregnancy (E0.5) and then every 5 days during pregnancy (E5.5, E10.5, E15.5). Here, estimations of changes in metabolic pathways between Con pregnant and HF pregnant groups were calculated using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt)21 and the proportion of genes predicted to be present for significantly different pathways of interest are visualized. The corresponding abundances for non-pregnant groups are show for the sake of comparison. The x-axis indicates the sampling timepoints and samples are colored by diet and pregnancy information. All other significant pathways are listed in Table S4.
Figure 6.
Maternal diet- and pregnancy-induced changes in the abundance of microbe containing genes predicted to regulate metabolic pathways related to fuel metabolism. Mice fed a control diet were sampled twice during an 8 week period prior to impregnation (BL, Wk6), at 0.5 days of pregnancy (E0.5) and then every 5 days during pregnancy (E5.5, E10.5, E15.5). HF fed mice were sampled following 6 weeks of high fat feeding (Wk 6) prior to impregnation, and at 0.5 days of pregnancy (E0.5) and then every 5 days during pregnancy (E5.5, E10.5, E15.5). Here, estimations of changes in metabolic pathways between Con pregnant and HF pregnant groups were calculated using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt)21 and the proportion of genes predicted to be present for significantly different pathways of interest are visualized. The corresponding abundances for non-pregnant groups are show for the sake of comparison. The x-axis indicates the sampling timepoints and samples are colored by diet and pregnancy information. All other significant pathways are listed in Table S4.
Microbial genes involved in vitamin metabolism and bile metabolism are also predicted to differ between Con- and HF-fed pregnant mice. The microbial communities of Con-fed pregnant mice are enriched in genes involved in vitamin B6 metabolism compared to the microbiome of HF mothers and in contrast, the microbiome of mice fed a HF diet during pregnancy is enriched in genes involved in cofactor and retinol metabolism (Fig. 7). Genes involved in primary and secondary bile acid biosynthesis are predicted to be significantly decreased in the microbiome of HF mothers. All other significant metabolic pathways are listed in Table S4.
Similar to our investigations into the microbial composition, we performed similar analyses to predict metabolic pathways between non-pregnant Con and HF fed mice (Table S5). In parallel to changes in taxonomic composition, we found a subset of the pathways increased in pregnancy x diet to also be increase with diet, indicating that these effects are multi-factorial.
Discussion
We present novel data in mice to suggest that diet and pregnancy have interactive effects on the composition of the female gut microbiota. We show that pregnancy-induced changes in the gut microbiota occur immediately at the onset of pregnancy, and are vulnerable to modulation by diet. This is entirely diet x pregnancy related and does not appear to be dependent upon increases in maternal weight gain during pregnancy, consistent with our previous study where a HF diet containing 45% kcal from fat did not increase weight gain during pregnancy but did increase adipose depots.22 HF diet intake before and during pregnancy resulted in distinctive shifts in the maternal gut microbiota in a gestational-age dependent manner and these shifts resulted in significant differences in the abundance of genes that favor lipid metabolism, glycolysis and gluconeogenesis metabolic pathways over the course of pregnancy.
Pregnancy is a state that induces shifts in the gut microbiota of control females: shifts that change over the course of gestation
Previous work demonstrates that the pregnant gut microbiota changes from the first to the third trimester of pregnancy in humans.14,16 We now show in mice that these shifts occur as early at 0.5 days of pregnancy. Most notably, Akkermansia abundance is significantly elevated at the onset of pregnancy (at 0.5 days) and, consistent with other reports,14 remains elevated throughout pregnancy. It is possible that this immediate shift is linked to reproductive cycle changes in the non-pregnant female, although we did not test this. Previous studies have shown shifts in the vaginal microbiome with reproductive cycles 23 but to our knowledge similar changes in gut microbiota have not been tested.
We hypothesize that gut microbes could participate in or be facilitated by pregnancy-induced changes in maternal endocrine and metabolic function. Early pregnancy is characterized by a maternal anabolic state; where maternal metabolic adaptations, mediated by pregnancy hormones, facilitate lipogenesis, glycogenesis, and adipocyte hypertrophy.24 By late pregnancy, the mother is catabolic, where increasing production of progesterone, estradiol and placental lactogen participate in mediating insulin and leptin resistance thus ensuring high levels of maternal-fetal glucose transfer for normal growth and development.24 Gut microbiota can affect host metabolism by modulating energy extraction,7 immunity,12,13 and lipid metabolism.25 Whether maternal endocrine related changes in metabolic function may be mediated by gut microbiota is unknown.
The intestinal epithelium is the interface between the host and the gut microbiota. The gut mucosal layer serves as both a barrier to pathogenic bacteria and a source of oligosaccharides for mucin-degrading bacteria including Ruminococcus, Bacteroides, Bifidobacterium, Clostridium 26 and Akkermansia.27 The relative abundance of Akkermansia has recently been shown to be inversely proportional to body weight and is decreased in genetically and diet induced obese mice.28 Our observed increase in the abundance of Akkermansia during pregnancy does not correlate with increased maternal body weight. It may be that increased mucin-degrading bacteria serve to provide substrates for other commensal bacteria (Allobaculum) and facilitate metabolic pathways that serve as barriers to pathogens and permeability. Estrogen receptors (ERs) have been localized in all segments of the gut epithelium 29 and are known to mediate epithelial function.30 Whether estrogen participates in changing the composition of the mucus layer during pregnancy is unknown, although it seems likely that an interactive effect between pregnancy-related hormonal changes and microbial population shifts would impact the gut microbial niche environment. Indeed, data in hamsters has shown a positive correlation between circulating leptin levels (an adipokine) and the presence of Akkermansia and Allobaculum31 and it is known that leptin levels rise with pregnancy.32
Generally, high caloric intake has been shown to favor Firmicutes over Bacteroidetes where in cases of obesity, the ratio of Firmicutes to Bacteroidetes increases,33 although data are conflicting.34 Despite similarities in the metabolic constitution of pregnancy and obesity, including insulin and leptin resistance, our data show no significant shift in the Firmicutes to Bacteroidetes phyla ratio either with the onset of pregnancy or throughout advancing gestation in control fed mice, whereas Akkermansia increased with advancing gestation. This is consistent with human studies investigating early versus late pregnancy states 14 although maternal gut bacterial counts increase,16 and an expansion of β diversity occurs with advancing gestational age.14
Pregnancy induced microbial shifts are dependent upon maternal dietary intake before and during pregnancy
We show that pregnancy-associated shifts in the maternal gut microbiota are dependent upon the mother's diet prior to and during pregnancy. Akkermansia (phylum Firmicutes) and Bifidobacterium abundance are increased in mothers fed a HF diet before and during pregnancy. Notably, only half of the genera that increased with pregnancy in control animals were also increased in high fat pregnancies; where in high fat pregnancies distinct genera were increased. We recognize the fact that our data are based upon 3 HF pregnancies, but despite this relatively low sample, our data are robust enough to show distinct differences due to pregnancy and to diet. Taken together, these data suggest that a high fat diet before pregnancy significantly alters the type of microbial changes that occur at the onset of pregnancy.
Despite our lack of knowledge regarding the exact role of these pregnancy-associated microbial shifts on maternal metabolism, the predicted metabolic pathways suggest that high fat diet before and during pregnancy increases numerous metabolic signaling pathways. In particular, those of fatty acid, ketone, cysteine, methionine, and vitamin metabolism are significantly elevated in HF pregnancies compared to control pregnancies. Of note, there appears to be a diet x pregnancy interaction in these predicated pathways, where the pregnancy-induced changes in these pathways are dependent upon the preconception diet. It is presently unknown whether these pathways provide substrates for the bacterial niche environment or whether these are substrates that could be utilized by the mother. The uptake of fatty acid, ketone, and sulfur containing amino acids by the mother would be consistent with the metabolic characteristics of late pregnancy where lipolysis results in increased maternal circulating free fatty acids, triglycerides and ketones.24
It is interesting to note that predicted primary and secondary bile acid metabolism pathways are decreased in HF pregnancies. Primary bile acid synthesis occurs in the liver where products are transported to the gut within which bacterial dehydroxylation occurs to form secondary bile acids that are integral for cholesterol, glucose and lipid metabolism, as well as fat soluble vitamin absorption 35 and more recently bile acids are thought to play a role as signaling molecules.36 Bile biosynthesis during pregnancy is thought to remain similar to control values.37 In cases of obesity, taurine-conjugated bile acids have been shown to be increased in the cecum of high fat-fed mice and may act as a marker of energy utilization behavior between the bacterial community and the host.38 Whether this occurs during pregnancies complicated by obesity, however, is unknown, but would be consistent with our previously observed changes in maternal metabolic function.19
We have predicted changes in metabolic capabilities due to the shift in microbial populations due to periconceptional HF diet using PiCRUST.21 Further metabolomics studies will be required in order to establish these inferred microbial relationships to metabolic functionality. Nevertheless, these predicated pathways can be used to probe further into the impact of maternal HF diet on adaptation to pregnancy.
Human data have suggested that pregnancy imposes changes in the maternal gut microbiome14,16 and our data in mice are consistent with these observations, although the functional consequences are still unclear. No doubt there are significant species differences in gut microbial populations and their influences on host metabolic function both in the pregnant and non pregnant state, but the use of animal models are an essential starting point for studying the gut microbiota especially in circumstances that do not permit invasive investigations in humans–particularly during pregnancy.
In conclusion, we show in mice that diet and pregnancy have interactive effects on the composition of the female gut microbiota. We show that pregnancy-induced changes in the female microbiota occur immediately at the onset of pregnancy, and are vulnerable to modulation by maternal diet. HF diet intake before and during pregnancy results in distinctive shifts in the maternal gut microbial community in a gestational-age dependent manner and that these shifts result in significant differences in the abundance of genes that favor lipid metabolism, glycolysis and gluconeogenic metabolic pathways over the course of pregnancy. Future studies are set to determine whether these shifts result in changes in maternal nutrient absorption and how maternal dietary induced microbiome shifts impact on fetal growth and placental function.
Methods
Animal model
All experiments were approved by the Animal Ethics Committee at McMaster University (AUP 12-38-10). Ten six-week-old female C57BL/6J mice were obtained from Jackson Laboratories. All mice were fed a control diet and after one week in our facility, baseline fecal samples were collected (n = 10, identified as Baseline), after which, mice were randomly assigned to one of 2 nutritional groups: 1) Control diet (Con; n = 5 17% fat, 29% protein, 54% CHO, 3 kcal/g; Harlan 8640 Teklad 22/5 Rodent Diet) or 2) high fat diet (HF; n = 5; 45% fat, 20% protein, 35% CHO, 4.73 kcal/g, Research Diets Inc.. D12451) (Fig. 1). All mice were housed individually in standard mouse cages with free access to water and 150 g of food, in the same room with a constant temperature of 25°C, a 12-h light, 12-h dark cycle and maintained on their respective diets for 8 weeks. Body weight and food intake were recorded. Following 6 weeks of nutritional intervention, fecal samples were collected (identified as Week 6) to examine the effect of diet on non-pregnant female microbiota.
After 8 weeks of dietary intervention all mice were housed with a C57BL/6J male and mating was confirmed by the appearance of a vaginal plug. If no plug was sighted after 7 days of consecutive mating, the mice were excluded from the study. Of the n = 5 Con and n = 5 HF females, we were able to confirm pregnancy in n = 5 and n = 3, respectively. This is not inconsistent with high fat diet-induced difficulties in fertility in mice.17 The morning of vaginal plug identification is described as day (embryonic day) E0.5 of gestation. Pregnant females were housed individually and provided with free access to water. Throughout pregnancy maternal gestational weight and food intake were recorded, and maternal fecal samples were collected (in the same manner described above) at embryonic days (E) 0.5, E5.5, E10.5, and E15.5 and stored at −80 °C for future analysis. Fecal samples were collected from all mice throughout pregnancy.
Genomic DNA extraction and 16S rRNA gene sequencing
Fecal pellets collected from these mice were used to infer gut microbial communities. Genomic DNA was extracted from fecal pellets as previously described39 with a few modifications. Briefly, 0.2 g of fecal material was mechanically lysed using 800μl 200 mM NaPO4, 100 μl guanidine thiocyanate-ethylenediaminetetraacetic-Sarkosyl, and 0.2 g of 2.8 mm ceramic beads (Mo Bio). A further mechanical and enzymatic lysis was performed using 0.2 g of 0.1-mm glass beads (Mo Bio), 50 μl lysozyme (100 mg/mL), 50 μl mutanolysin (10 U/μl), and 10 μl of RNase A (10 mg/mL) and incubated for 1 hour at 37°C. A second enzymatic lysis using 25 μl 25% sodium dodecyl sulfate, 25 μl proteinase K, and 62.5 μl 5 M NaCl was further incubated for 1 hour at 65°C. The resulting solution was centrifuged (12,000 x g), supernatant removed, and combined with an equal volume of phenol-chloroform-isoamyl alcohol in a new microcentrifuge tube. This was centrifuged (12,000 x g) and the solution of the lowest density removed and combined with 200 μl DNA binding buffer (Zymo), which was transferred to a DNA column (Zymo), washed, and eluted into sterile H2O.
PCR amplification of the variable 3 (V3) region of the 16S rRNA gene was subsequently performed on the extracted DNA from each sample independently using methods described previously.39,40 341F and 518R rRNA primers were used, modified to include adapter sequences specific to the Illumina technology, and 6-base pair barcodes to allow multiplexing of samples. DNA products of this PCR amplification were subsequently sequenced using the Illumina MiSeq platform, resulting in 150bp paired-end reads.
Processing and analysis of 16S rRNA gene sequencing data
Resultant FASTQ files were processed using a custom in-house developed pipeline for the processing of 16S rRNA gene sequencing data used previously.39 In this pipeline, Cutadapt,41 is used to trim any reads exceeding the length of the 16S rRNA V3 region, PANDASeq42 to align paired-end reads, AbundantOTU+43 to group reads into Operational Taxonomic Units (OTUs) based on 97% similarity, and the RDP Classifier44 as implemented in Quantitative Insights Into Microbial Ecology (QIIME)45 against the Feb 4 2011 release of the Greengenes reference database 47 to assign a taxonomy to each OTU. Any OTU not assigned to the Bacterial domain were culled, as was any OTU to which only 1 sequence was assigned. This processing resulted in a total of 7985925 reads (mean 120999 reads per sample; range: 16524–237026) and 4982 OTUs (mean 631 OTUs per sample; range: 200–1242).
Analyses of these data were completed using various open source software. Taxonomic summaries were computed using QIIME.45 Measures of β diversity were computed using Phyloseq's implementation of the Bray Curtis distance metric, and tested for whole community differences across groups using vegan's implementation of permutational multivariate analysis of variance (PERMANOVA) in the adonis command.48 These results were visualized via Principal Coordinates Analysis (PCoA) ordination.46 Calculations of genera which differed significantly between mouse groups were computed using DESeq250 (considered significant if the p-value was <0.01 after adjustment for multiple testing via DESeq2's implementation of the Benjamini-Hochberg multiple testing adjustment procedure) and visualized using R's ggplot2 package.49 Estimations of changes in metabolic pathways were calculated using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt)21 and significantly different pathways of interest (considered significant if the p-value was <0.01 after adjustment for multiple testing) were visualized using R's ggplot2.49
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
The authors would like to thank Jay Patel with his assistance in the animal model.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Authors' Contributions
WG assisted in the animal study, performed genomic DNA extraction, prepared samples for sequencing, interpreted data, and drafted the manuscript. FJW processed and analyzed sequencing data, performed statistical analysis, and drafted the manuscript. WG and FJW contributed equally to the preparation of this manuscript. MGS interpreted data and assisted in drafting the manuscript. CM carried out the animal work and collected samples. JDS interpreted data and helped to draft the manuscript. DSM designed the study, participated in animal work and data interpretation, and drafted the manuscript.
Funding
DMS and MGS are Canada Research Chairs, WG is supported by the Natural Sciences and Engineering Research Council of Canada, FJW is supported by a studentship from the Canadian Institutes of Health Research. JDS is a Canadian Diabetes Association Scholar. This study was partially funded by an operating grant from the Natural Sciences and Engineering Research Council of Canada and a Catalyst grant from the Canadian Institutes for Health Research.
References
- 1.Organization WH Obesity 2013. [Google Scholar]
- 2.Gluckman PD, Hanson MA. Developmental plasticity and human disease: research directions. J Intern Med 2007; 261:461-71; PMID:17444885; http://dx.doi.org/ 10.1111/j.1365-2796.2007.01802.x [DOI] [PubMed] [Google Scholar]
- 3.Kristensen J, Vestergaard M, Wisborg K, Kesmodel U, Secher NJ. Pre-pregnancy weight and the risk of stillbirth and neonatal death. BJOG 2005; 112:403-8; PMID:15777435; http://dx.doi.org/ 10.1111/j.1471-0528.2005.00437.x [DOI] [PubMed] [Google Scholar]
- 4.Patro B, Liber A, Zalewski B, Poston L, Szajewska H, Koletzko B. Maternal and paternal body mass index and offspring obesity: a systematic review. Ann Nutr Metab 2013; 63:32-41; PMID:23887153; http://dx.doi.org/ 10.1159/000350313 [DOI] [PubMed] [Google Scholar]
- 5.Poston L. Maternal obesity, gestational weight gain and diet as determinants of offspring long term health. Best Pract Res Clin Endocrinol Metab 2012; 26:627-39; PMID:22980045; http://dx.doi.org/ 10.1016/j.beem.2012.03.010 [DOI] [PubMed] [Google Scholar]
- 6.Chow J, Lee SM, Shen Y, Khosravi A, Mazmanian SK. Host-bacterial symbiosis in health and disease. Adv Immunol 2010; 107:243-74; PMID:21034976; http://dx.doi.org/ 10.1016/B978-0-12-381300-8.00008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbaugh P, Ley RE, Mahowald MA, Magrini V, Mardis E, Gordon JI. An obesity associated gut microbiome with increased capacity for energy harvest. Nature 2006; 444:1027-31; PMID:17183312; http://dx.doi.org/ 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 8.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, et al.. Evolution of mammals and their gut microbes. Science 2008; 320:1647-51; PMID:18497261; http://dx.doi.org/ 10.1126/science.1155725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 2005; 102:11070-5; PMID:16033867; http://dx.doi.org/ 10.1073/pnas.0504978102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-Induced Obesity Is Linked to Marked but Reversible Alterations in the Mouse Distal Gut Microbiome. Cell Host Microbe 2008; 3:213-23; PMID:18407065; http://dx.doi.org/ 10.1016/j.chom.2008.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W, Mao Y, Zhang X, Pang X, Wei C, et al.. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J 2010; 4:232; PMID:19865183; http://dx.doi.org/ 10.1038/ismej.2009.112 [DOI] [PubMed] [Google Scholar]
- 12.de La Serre CB, Ellis CL, Lee J, Hartman AL, Rutledge JC, Raybould HE. Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am J Physiol Gastrointest Liver Physiol 2010; 299:G440-G8; PMID:20508158; http://dx.doi.org/ 10.1152/ajpgi.00098.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim K-A, Gu W, Lee I-A, Joh E-H, Kim D-H. High Fat Diet-Induced Gut Microbiota Exacerbates Inflammation and Obesity in Mice via the TLR4 Signaling Pathway. PLoS ONE 2012; 7:e47713; PMID:23091640; http://dx.doi.org/ 10.1371/journal.pone.0047713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Backhed HK, Gonzalez A, Werner JJ, Angenent LT, Knight R, et al.. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 2012; 150:470-80; PMID:22863002; http://dx.doi.org/ 10.1016/j.cell.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmet P, Alberti KG, Shaw J. Global and societal implications of the diabetes epidemic. Nature 2001; 414:782-7; PMID:11742409; http://dx.doi.org/ 10.1038/414782a [DOI] [PubMed] [Google Scholar]
- 16.Collado MC, Isolauri E, Laitinen K, Salminen S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Ame J Clin Nutr 2008; 88:894-9; PMID:18842773 [DOI] [PubMed] [Google Scholar]
- 17.Sharma S, Morinaga H, Hwang V, Fan W, Fernandez MO, Varki N, Olefsky JM, Webster NJ. Free fatty acids induce Lhb mRNA but suppress Fshb mRNA in pituitary LbetaT2 gonadotropes and diet-induced obesity reduces FSH levels in male mice and disrupts the proestrous LH/FSH surge in female mice. Endocrinology 2013; 154:2188-99; PMID:23525221; http://dx.doi.org/ 10.1210/en.2012-2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vickers MH, Clayton ZE, Yap C, Sloboda DM. Maternal Fructose Intake during Pregnancy and Lactation Alters Placental Growth and Leads to Sex-Specific Changes in Fetal and Neonatal Endocrine Function. Endocrinology 2011; 152(4):1378-87:en.2010–1093 [DOI] [PubMed] [Google Scholar]
- 19.Howie G, Sloboda D, Kamal T, Vickers M. Maternal nutritional history predicts obesity in adult offspring independent of postnatal diet. The J Physiol 2009; 587:905-15; PMID:19103681; http://dx.doi.org/ 10.1113/jphysiol.2008.163477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mark PJ, Sisala C, Connor K, Patel R, Lewis JL, Vickers MH, Waddell BJ, Sloboda DM. A maternal high-fat diet in rat pregnancy reduces growth of the fetus and the placental junctional zone, but not placental labyrinth zone growth. J Dev Origins Health Dis 2011; 2:63-70; http://dx.doi.org/ 10.1017/S2040174410000681 [DOI] [Google Scholar]
- 21.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, et al.. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013; 31:814-21; PMID:23975157; http://dx.doi.org/ 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howie GJ, Sloboda DM, Kamal T, Vickers MH. Maternal nutritional history predicts obesity in adult offspring independent of postnatal diet. J Physiol 2009; 587:905-15; PMID:19103681; http://dx.doi.org/ 10.1113/jphysiol.2008.163477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uchihashi M, Bergin IL, Bassis CM, Hashway SA, Chai D, Bell JD. Influence of age, reproductive cycling status, and menstruation on the vaginal microbiome in baboons (Papio anubis). Am J Primatol 2015; 77:563-78; PMID:25676781; http://dx.doi.org/ 10.1002/ajp.22378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butte N. Carbohydrate and lipid metabolism in pregnancy: normal compared with gestational diabetes mellitus. Am J Clin Nutr 2000; 71:1256S; PMID:10799399 [DOI] [PubMed] [Google Scholar]
- 25.Harris K, Kassis A, Major G, Chou CJ. Is the gut microbiota a new factor contributing to obesity and its metabolic disorders? J Obes 2012; 2012:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salyers AA, West SE, Vercellotti JR, Wilkins TD. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl Environ Microbiol 1977; 34:529-33; PMID:563214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 2004; 54:1469-76; PMID:15388697; http://dx.doi.org/ 10.1099/ijs.0.02873-0 [DOI] [PubMed] [Google Scholar]
- 28.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al.. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci 2013; 110:9066-71; http://dx.doi.org/ 10.1073/pnas.1219451110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas ML, Xu X, Norfleet AM, Watson CS. The presence of functional estrogen receptors in intestinal epithelial cells. Endocrinology 1993; 132:426-30; PMID:8419141 [DOI] [PubMed] [Google Scholar]
- 30.Looijer-van Langen M, Hotte N, Dieleman LA, Albert E, Mulder C, Madsen KL. Estrogen receptor-β signaling modulates epithelial barrier function. Am J Physiol Gastrointest Liver Physiol 2011; 300(4):G621-6; PMID:21252046 [DOI] [PubMed] [Google Scholar]
- 31.Ravussin Y, Koren O, Spor A, LeDuc C, Gutman R, Stombaugh J, Knight R, Ley RE, Leibel RL. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity 2012; 20:738-47; PMID:21593810; http://dx.doi.org/ 10.1038/oby.2011.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukherjea R, Castonguay TW, Douglass LW, Moser-Veillon P. Elevated leptin concentrations in pregnancy and lactation: possible role as a modulator of substrate utilization. Life sciences 1999; 65:1183-93; PMID:10503934; http://dx.doi.org/ 10.1016/S0024-3205(99)00352-5 [DOI] [PubMed] [Google Scholar]
- 33.Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, Gordon JI, Krakoff J. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr 2011; 94:58-65; PMID:21543530; http://dx.doi.org/ 10.3945/ajcn.110.010132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walters WA, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett 2014; 588(22):4223-33; PMID:25307765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li T, Chiang JYL. Bile acid signaling in liver metabolism and diseases. J Lipids 2012; 2012:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma H, Patti ME. Bile acids, obesity, and the metabolic syndrome. Best Pract Res Clin Gastroenterol 2014; 28:573-83; PMID:25194176; http://dx.doi.org/ 10.1016/j.bpg.2014.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu QN, Xie HM, Zhang D, Liu J, Lu YF. Hepatic bile acids and bile acid-related gene expression in pregnant and lactating rats. PeerJ 2013; 1:e143; PMID:24010021; http://dx.doi.org/ 10.7717/peerj.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker A, Pfitzner B, Neschen S, Kahle M, Harir M, Lucio M, Moritz F, Tziotis D, Witting M, Rothballer M, et al.. Distinct signatures of host-microbial meta-metabolome and gut microbiome in two C57BL/6 strains under high-fat diet. ISME J 2014; 8(12):2380-96; PMID:24906017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whelan FJ, Verschoor CP, Stearns JC, Rossi L, Luinstra K, Loeb M, Smieja M, Johnstone J, Surette MG, Bowdish DM. The loss of topography in the microbial communities of the upper respiratory tract in the elderly. Ann Am Thoracic Soci 2014; 11:513-21; PMID:24601676; http://dx.doi.org/ 10.1513/AnnalsATS.201310-351OC [DOI] [PubMed] [Google Scholar]
- 40.Bartram AK, Lynch MD, Stearns JC, Moreno-Hagelsieb G, Neufeld JD. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end illumina reads. Appl Environ Microbiol 2011; 77:3846-52; PMID:21460107; http://dx.doi.org/ 10.1128/AEM.02772-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011. 2011; 17 [Google Scholar]
- 42.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics 2012; 13:31; PMID:22333067; http://dx.doi.org/ 10.1186/1471-2105-13-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye Y. Identification and Quantification of Abundant Species from Pyrosequences of 16S rRNA by Consensus Alignment. Proc IEEE Int Conf Bioinformatics Biomed 2011; 2010:153-7; PMID:22102981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007; 73:5261-7; PMID:17586664; http://dx.doi.org/ 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al.. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7:335-6; PMID:20383131; http://dx.doi.org/ 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013; 8:e61217; PMID:23630581; http://dx.doi.org/ 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069-72; PMID:16820507; http://dx.doi.org/ 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jari Oksanen FGB, Roeland Kindt, Pierre Legendre, Peter R, Minchin RB O'Hara Gavin L, Simpson Peter, Solymos M, Henry H. Stevens and Helene Wagner vegan: Community Ecology Package. 2013. [Google Scholar]
- 49.H. Wickham ggplot2: elegant graphics for data analysis. Springer New York, 2009. [Google Scholar]
- 50.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol 2014; 15(12):550; PMID:25516281 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.