Abstract
Nematode dauer formation represents an essential survival and dispersal strategy and is one of a few ecologically relevant traits that can be studied in laboratory approaches. Under harsh environmental conditions, the nematode model organisms Caenorhabditis elegans and Pristionchus pacificus arrest their development and induce the formation of stress-resistant dauer larvae in response to dauer pheromones, representing a key example of phenotypic plasticity. Previous studies have indicated that in P. pacificus, many wild isolates show cross-preference of dauer pheromones and compete for access to a limited food source. When investigating the genetic mechanisms underlying this intraspecific competition, we recently discovered that the orphan gene dauerless (dau-1) controls dauer formation by copy number variation. Our results show that dau-1 acts in parallel to or downstream of steroid hormone signaling but upstream of the nuclear hormone receptor daf-12, suggesting that DAU-1 represents a novel inhibitor of DAF-12. Phylogenetic analysis reveals that the observed copy number variation is part of a complex series of gene duplication events that occurred over short evolutionary time scales. Here, we comment on the incorporation of novel or fast-evolving genes into conserved genetic networks as a common principle for the evolution of phenotypic plasticity and intraspecific competition. We discuss the possibility that orphan genes might often function in the regulation and execution of ecologically relevant traits. Given that only few ecological processes can be studied in model organisms, the function of such genes might often go unnoticed, explaining the large number of uncharacterized genes in model system genomes.
Keywords: daf-12, dauer formation, dau-1, evo-devo, intraspecific competition, nuclear hormone receptors, orphan genes, phenotypic plasticity, Pristionchus pacificus
Introduction
Evolutionary developmental biology (evo-devo) aims for a comprehensive understanding of the evolutionary forces and the developmental processes that generate biological diversity.1-3 The fact that diverse organisms have highly conserved developmental control genes but fundamentally different regulatory mechanisms for the specification of homologous structures resulting in developmental systems drift is a central finding of evo-devo research and by now, a truism of modern biology.4,5 However, explaining the link between genetic variation and phenotypic diversity requires a more integrative approach combining evo-devo with population genetics and evolutionary ecology.6,7 Over the last 2 decades, evoluti-onary biologists have attributed increasing importance to the influence of ecology on evolution and development, demonstrated by the growing number of studies on phenotypically plastic traits.8,9
Phenotypic plasticity describes the ability of an individual organism to develop alternative forms of a trait in response to changing environmental conditions. In the last decade, phenotypic plasticity has been suggested to not only be of ecological but also evolutionary importance because it can facilitate morphological innovations that result in the origin and evolution of novel traits.8 Nematode dauer formation is an example of phenotypic plasticity in an ecologically relevant trait, which exists as a choice that many free-living nematodes have between direct development into reproductive adults and indirect development into stress-resistant dauer larvae (Fig. 1A).10,11 The dauer stage is an alternative third larval stage, induced by a dauer pheromone, and formed under harsh environmental conditions, such as food shortage or high population density.12,13 Dauer formation is also considered to be of relevance for the evolution of parasitism. Many theoretical studies have discussed dauer larvae of free-living nematodes and their morphological similarities to infective juveniles of parasitic nematodes as a preadaptation toward the evolution of parasitism.14-17 In this context, it is important to note that in the wild P. pacificus predominantly exists as dauer larvae, which live in a necromenic association with scarab beetles.18,19 Only after the natural death of its host, does P. pacificus exit the dauer stage and resume its development by feeding on bacteria, fungi, and other organisms growing on the dead beetle. Therefore, dauer formation represents a survival and dispersal strategy, enabling individuals of a population to endure and escape unfavorable conditions. In contrast to many other nematode associations with invertebrates that are often unspecific, such as C. elegans with slugs and isopods,20 the association of Pristionchus nematodes with scarab beetles is mostly species-specific, an important prerequisite for the evolution of parasitism.15,16
Figure 1.
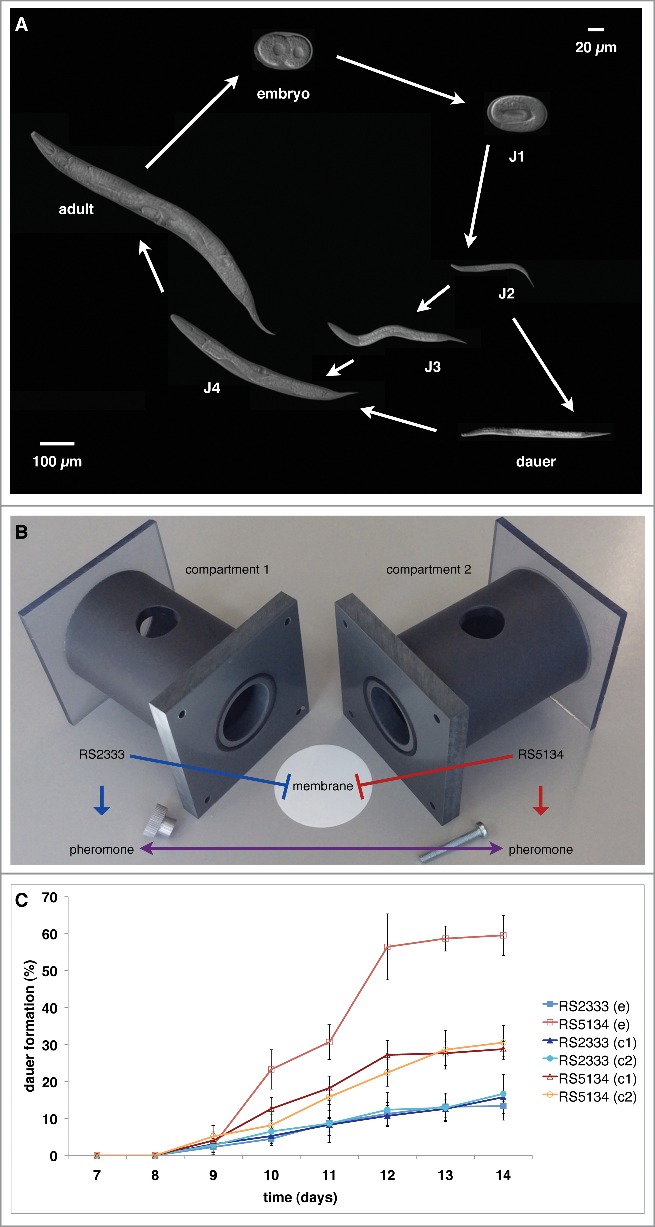
P. pacificus dauer formation and intraspecific competition. (A) P. pacificus life cycle. (B) Experimental design of Ussing chamber competition assay. (C) Dauer formation observed over time, showing intraspecific competition between RS2333 and RS5134. In the competition experiment (e) one compartment of the Ussing chamber contains RS2333 and the other RS5134, whereas in control experiments (c) the same strain is grown in both compartments.24
Cross-preference of dauer pheromones and intraspecific competition
The huge advantage of working with P. pacificus is our extensive understanding of its natural ecology and population genetics. Since P. pacificus is a cosmopolitan species, many wild isolates have been collected and are available for ecological studies.7,21 We previously used a natural variation approach to compare various aspects of dauer formation among diverse P. pacificus strains.22,23 Our results indicate high levels of natural variation in dauer longevity, as well as in dauer formation in response to individual small molecules and dauer pheromone, the complex blend of small molecules extracted from each strain. Furthermore, we observed significant qualitative and quantitative differences in production of and response to individual small molecules. Specifically, some strains synthesize considerable amounts of a certain compound, although they do not respond to it, whereas other strains form many dauers in response to compounds they do not produce themselves.23 Indeed, most strains show cross-preference of dauer pheromones, whereby a strain forms more dauers in response to another strain's pheromone extract than in response to its own pheromone, suggesting intraspecific competition as a previously unconsidered aspect of dauer formation.
Following these initial experiments, we developed a novel assay to directly test for competition.23 In contrast to our previous dauer formation assays, involving the response of an individual strain to a specific small molecule or pheromone extract, the competition assay was designed to enable the observation of the dauer formation of 2 strains in a common environment in response to their naturally secreted pheromones. Using the Ussing chamber system (Fig. 1B, C), our results confirm the presence of cross-preference of dauer pheromones and reveal intraspecific competition among both allopatric and sympatric strains.23,24 Competitive interactions among organisms have been suggested to be associated with evolutionary arms races.25 Intraspecific competition for survival and access to limited resources under changing environmental conditions represents a strong selective force driving the divergence among different populations of the same species.8 Strategies for surviving and competing in changing environments are well known in bacteria and involve the regulation of the expression of specific genes in response to different environmental conditions.26 However, little is known about the molecular mechanisms of competitive interactions in eukaryotes.
Intraspecific competition in P. pacificus dauer formation probably occurs at multiple levels of dauer entry but also after dauer exit when the dauer larvae start to feed on a limited food source, such as bacteria and other organisms growing on a beetle carcass. Such competition for a limited food source may result in an evolutionary arms race regarding the dauer formation phenotypes of different P. pacificus strains and lead to changes in dauer pheromone production and response. Two different survival strategies may have evolved due to intraspecific competition in P. pacificus dauer formation. Strains with low dauer formation phenotypes, such as the P. pacificus reference strain RS2333 from California, can drive other strains into early dauer formation, thereby gaining the advantage of reproducing for a longer time. In contrast, strains with high dauer formation phenotypes, such as RS5134 from Ohio, can avoid competition, overpopulation, and potential starvation by optimizing their dispersal as dauer larvae. Indeed, RS5134/Ohio is a champion of survival in the dauer stage. Under stable laboratory conditions, this strain survives for almost one year in distilled water at 8°C, in contrast to P. pacificus RS2333 and C. elegans N2, the latter of which only survives for 18 weeks.22
A new element in the dauer regulatory network
Since RS2333/California and RS5134/Ohio show a large difference in their dauer formation phenotypes (Fig. 1C), we selected these 2 strains to investigate the molecular mechanisms that determine intraspecific competition by generating recombinant inbred lines and performing quantitative-trait-loci mapping.24 We discovered that the orphan gene dauerless (dau-1) regulates dauer formation by copy number variation (CNV) (Fig. 2). A single dau-1 copy is responsible for the high dauer formation phenotype of RS5134/Ohio, whereas 2 dau-1 copies, resulting from a very recent gene duplication event, lead to reduced dauer formation in R2333/California. These findings are supported by experimental studies using transgenic animals. Specifically, transgenic animals expressing multiple dau-1 copies in either strain do not form dauer larvae at all. We detected dau-1 expression in the CAN neurons, indicating a previously unknown role of these neurons in dauer formation. Ablation of the CAN neurons, as well as mutations in the dau-1 locus generated by the CRISPR/Cas9 system, causes increased dauer formation. Finally, we performed epistasis experiments to determine where dau-1 acts in the dauer regulatory network. The key factor in the dauer regulatory network is the nuclear hormone receptor DAF-12 that acts as a developmental switch, induces dauer formation in its ligand-free form, and is conserved among many free-living and parasitic nematodes.27,28 Our results show that dau-1 acts in parallel to or downstream of steroid hormone signaling but upstream of daf-12, suggesting that DAU-1 represents a novel inhibitor of DAF-12.
Figure 2.
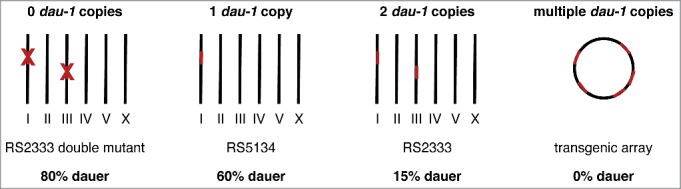
dau-1 regulates dauer formation by CNV. One copy leads to high dauer formation in RS5134, 2 copies cause low dauer formation in RS2333, and the expression of multiple copies completely inhibits dauer formation in transgenic animals. In contrast, the absence of dau-1 results in an extremely high dauer formation phenotype.24
CNV, gene duplication, and orphan genes
Phylogenetic analysis reveals that the observed CNV of dau-1 is part of a complex series of gene duplication events that occurred over short evolutionary time scales.24 Gene duplication is a form of CNV and represents an important mechanism for adapting to changing environmental conditions, which has been studied extensively in bacteria.29 Prevalent in a variety of organisms, gene duplication has been suggested to facilitate the evolution of novel phenotypic traits.30 After a duplication event, one gene copy may retain the ancestral function, while the other copy may acquire a new function.30,31 Therefore, gene duplication may emerge as a common principle for the evolution of novel traits and may drive the evolution of genetic networks and the divergence among populations.30,32 An interesting aspect of gene duplication events is their ability to promote the rapid evolution of certain gene families and thus induce the formation of orphan genes.33
Orphan genes lack significant sequence similarity to genes in other organisms and thus, they are defined as not being conserved above the genus level.34 They may evolve de novo from previously non-coding sequences or originate from gene duplication events followed by rapid evolution.33 Comparative genome sequencing projects have revealed a large number (10–20%) of orphan genes in all taxonomic groups, including nematodes.34,35 For example, in P. pacificus, orphan genes constitute approximately one third of the predicted 27,000 protein-coding genes.36,37 Interestingly, follow-up experiments have indicated that more than half of these orphan genes are transcribed.37 Approximately 30% of the P. pacificus orphan genes can be grouped into distinct protein families that have homologues in very closely related species of the same genus but not in C. elegans. This suggests that these orphan genes belong to fast-evolving gene families and diverge so rapidly that no homologues can be identified at greater phylogenetic distances,37 an assumption that has now been confirmed in the case of dau-1.
Since orphan genes are taxonomically restricted, they may have evolved to enable the adaptation to the ecology of the organism and specific environmental conditions.35 For example, it has been shown that the expression of many P. pacificus orphan genes is upregulated in the dauer stage, suggesting a crucial role in the evolution of P. pacificus dauer formation, which differs in its specific functions from that of C. elegans.38 Furthermore, proteomic and transcriptomic studies have provided strong evidence that orphan genes are expressed and of functional importance,37,38 yet they are not conserved. Thus, although developmental control genes and signaling pathways are often conserved throughout the animal kingdom – the truism of modern biology discussed above, a relatively large part of genes in an animal shows limited or no sequence similarity to genes in other organisms. Our work on the orphan gene dau-1 and its role as a novel inhibitor of dauer formation therefore shows that rapidly evolving genes are of developmental and ecological relevance. In addition, our studies suggest that the incorporation of orphan genes into conserved regulatory networks may represent a general evolutionary mechanism.
Conclusions
Ever since the C. elegans genome was sequenced as the first metazoan representative,39 researchers have been puzzled by the following overarching pattern. While a small number of conserved genes was identified as developmental control genes that are shared between C. elegans, Drosophila, and several vertebrate models, the vast majority of genes has no associated function and limited or no sequence similarity to genes in other taxonomic groups. This observation, when combined with the results of developmental genetics at large, resulted in an ascertainment bias. While developmental geneticists identified the same (homologous) genes over and over again, most genes found in genome sequencing projects are not conserved and often novel.31 As a result, we are clueless about the function of the majority of genes, even in some of the best-studied model organisms of the life sciences, such as C. elegans.40 Do all of these genes have any function at all or are they just rapidly evolving units of the genome that come and go? To answer these questions, we have to appreciate that the highly successful model system approach has one important downside: the ecology and the environment, in which organisms normally live and evolve, is not considered. Rather, we study them in (sterile) laboratory environments. Could it be that many orphan genes have functions in the environment? These questions will only begin to be addressed in future studies. However, the story of P. pacificus dau-1 tells us that orphan genes can find a home in ecology and are incorporated into preexisting regulatory networks.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Raff RA. The shape of life: genes, development, and the evolution of animal form. Chicago: The University of Chicago Press 1996. 544. [Google Scholar]
- 2.Gerhart J, Kirschner M. The theory of facilitated variation. PNAS 2007; 104:8582-89; PMID:17494755; http://dx.doi.org/ 10.1073/pnas.0701035104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 2008; 134:25-36; PMID:18614008; http://dx.doi.org/ 10.1016/j.cell.2008.06.030 [DOI] [PubMed] [Google Scholar]
- 4.True JR, Haag ES. Developmental system drift and flexibility in evolutionary trajectories. Evol Dev 2001; 3:109-19; PMID:11341673; http://dx.doi.org/ 10.1046/j.1525-142x.2001.003002109.x [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Sommer RJ. Antagonism of LIN-17/Frizzled and LIN-18/Ryk in nematode vulva induction reveals evolutionary alterations in core developmental pathways. PLOS Biol 2011; 9:e1001110; PMID:21814488; http://dx.doi.org/ 10.1371/journal.pbio.1001110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sommer RJ. The future of evo-devo: model systems and evolutionary theory. Nat Rev Genet 2009; 10:416-22; PMID:19369972 [DOI] [PubMed] [Google Scholar]
- 7.Sommer RJ, McGaughran A. The nematode Pristionchus pacificus as a model system for integrative studies in evolutionary biology. Mol Ecol 2013; 22:2380-93; PMID:23530614; http://dx.doi.org/ 10.1111/mec.12286 [DOI] [PubMed] [Google Scholar]
- 8.West-Eberhard MJ. Developmental Plasticity and Evolution. Oxford: Oxford University Press; 2003. 816. [Google Scholar]
- 9.Gilbert SF, Epel D. Ecological developmental biology: integrating epigenetics, medicine and evolution. Sunderland (MA): Sinauer Associates Inc 2009. 480. [Google Scholar]
- 10.Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev 2008; 22:2149-65; PMID:18708575; http://dx.doi.org/ 10.1101/gad.1701508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sommer RJ, Ogawa A. Hormone signaling and phenotypic plasticity in nematode development and evolution. Curr Biol 2011; 21:R758-66; PMID:21959166; http://dx.doi.org/ 10.1016/j.cub.2011.06.034 [DOI] [PubMed] [Google Scholar]
- 12.Cassada RC, Russell RL. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev Biol 1975; 46:326-42; PMID:1183723; http://dx.doi.org/ 10.1016/0012-1606(75)90109-8 [DOI] [PubMed] [Google Scholar]
- 13.Golden JW, Riddle DL. A pheromone influences larval development in the nematode Caenorhabditis elegans. Science 1982; 218:578-80; PMID:6896933; http://dx.doi.org/ 10.1126/science.6896933 [DOI] [PubMed] [Google Scholar]
- 14.Osche G. Die Präadaptation freilebender Nematoden an den Parasitismus. Zool Anz 1956; 19:391-96. [Google Scholar]
- 15.Poulin R. Evolutionary ecology of parasites. 2nd ed. Princeton. (NJ): Princeton University Press; 2007. 332. [Google Scholar]
- 16.Dieterich C, Sommer RJ. How to become a parasite - lessons from the genomes of nematodes. Trends Genet 2009; 25:203-9; PMID:19361881; http://dx.doi.org/ 10.1016/j.tig.2009.03.006 [DOI] [PubMed] [Google Scholar]
- 17.Sudhaus W. Preadaptive plateau in Rhabditida (Nematoda) allowed the repeated evolution of zooparasites, with an outlook on evolution of life cycles within Spiroascarida. Palaeodiversity 2010; 3:117-30. [Google Scholar]
- 18.Herrmann M, Mayer WE, Sommer RJ. Nematodes of the genus Pristionchus are closely associated with scarab beetles and the Colorado potato beetle in Western Europe. Zoology 2006; 109:96-108; PMID:16616467; http://dx.doi.org/ 10.1016/j.zool.2006.03.001 [DOI] [PubMed] [Google Scholar]
- 19.Ragsdale EJ. Pristionchus pacificus: a nematode model for comparative and evolutionary biology Leiden, Netherlands: Brill 2015. Chapter 11, Mouth dimorphism and the evolution of novelty and diversity. 301–329. [Google Scholar]
- 20.Petersen C, Hermann RJ, Barg MC, Schalkowski R, Dirksen P, Barbosa C, Schulenburg H. Travelling at a slug's pace: possible invertebrate vectors of Caenorhabditis nematodes. BMC Ecol 2015; 15:19; PMID:26170141; http://dx.doi.org/ 10.1186/s12898-015-0050-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgan K, McGaughran A, Villate L, Herrmann M, Witte H, Bartelmes G, Rochat J, Sommer RJ. Multi locus analysis of Pristionchus pacificus on La Réunion Island reveals an evolutionary history shaped by multiple introductions, constrained dispersal events and rare out-crossing. Mol Ecol 2012; 21:250-66; PMID:22126624; http://dx.doi.org/ 10.1111/j.1365-294X.2011.05382.x [DOI] [PubMed] [Google Scholar]
- 22.Mayer MG, Sommer RJ. Natural variation in Pristionchus pacificus dauer formation reveals cross-preference rather than self-preference of nematode dauer pheromones. Proc R Soc B 2011; 278:2784-90; PMID:21307052; http://dx.doi.org/ 10.1098/rspb.2010.2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bose N, Meyer JM, Yim JJ, Mayer MG, Markov GV, Ogawa A, Schroeder FC, Sommer RJ. Natural variation in dauer pheromone production and sensing supports intraspecific competition in nematodes. Curr Biol 2014; 24:1536-41; PMID:24980503; http://dx.doi.org/ 10.1016/j.cub.2014.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayer MG, Rödelsperger C, Witte H, Riebesell M, Sommer RJ. The orphan gene dauerless regulates dauer development and intraspecific competition in nematodes by copy number variation. PLOS Genet 2015; 11:e100514624.; http://dx.doi.org/ 10.1371/journal.pgen.1005146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dawkins R, Krebs JR. Arms races between and within species. Proc R Soc B 1979; 205:489-511; http://dx.doi.org/ 10.1098/rspb.1979.0081 [DOI] [PubMed] [Google Scholar]
- 26.Dubnau D, Losick R. Bistability in bacteria. Mol Microbiol 2006; 61:564-72; PMID:16879639; http://dx.doi.org/ 10.1111/j.1365-2958.2006.05249.x [DOI] [PubMed] [Google Scholar]
- 27.Ogawa A, Streit A, Antebi A, Sommer RJ. A conserved endocrine mechanism controls the formation of dauer and infective larvae in nematodes. Curr Biol 2009; 19:67-71; PMID:19110431; http://dx.doi.org/ 10.1016/j.cub.2008.11.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antebi A. Nuclear receptor signal transduction. WormBook. The C. elegans Research Community 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersson DI, Hughes D. Gene amplification and adaptive evolution in bacteria. Ann Rev Genet 2009; 43:167-95; PMID:19686082; http://dx.doi.org/ 10.1146/annurev-genet-102108-134805 [DOI] [PubMed] [Google Scholar]
- 30.Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol 2003; 18:292-98; http://dx.doi.org/ 10.1016/S0169-5347(03)00033-8 [DOI] [Google Scholar]
- 31.Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution 2007; 61:995-1016; PMID:17492956; http://dx.doi.org/ 10.1111/j.1558-5646.2007.00105.x [DOI] [PubMed] [Google Scholar]
- 32.Ohno S. Evolution by gene duplication. New York: Springer-Verlag; 1970. 160. [Google Scholar]
- 33.Tautz D, Domazet-Lošo T. The evolutionary origin of orphan genes. Nat Rev Genet 2011; 12:692-702; PMID:21878963; http://dx.doi.org/ 10.1038/nrg3053 [DOI] [PubMed] [Google Scholar]
- 34.Rödelsperger C, Streit A, Sommer RJ. Structure, function and evolution of the nematode genome. In: eLS. Chichester: John Wiley & Sons Ltd; 2013. http://dx.doi.org/ 10.1002/9780470015902.a0024603 [DOI] [Google Scholar]
- 35.Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TCG. More than just orphans: are taxonomically restricted genes important in evolution? Trends Genet 2009; 25:404-13; PMID:19716618; http://dx.doi.org/ 10.1016/j.tig.2009.07.006 [DOI] [PubMed] [Google Scholar]
- 36.Dieterich C, Clifton SW, Schuster LN, Chinwalla A, Delehaunty K, Dinkelacker I, Fulton L, Fulton R, Godfrey J, Minx P et al.. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat Genet 2008; 40:1193-98; PMID:18806794; http://dx.doi.org/ 10.1038/ng.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borchert N, Dieterich C, Krug K, Schütz W, Jung S, Nordheim A, Sommer RJ, Macek B. Proteogenomics of Pristionchus pacificus reveals distinct proteome structure of nematode models. Genome Res 2010; 20:837-46; PMID:20237107; http://dx.doi.org/ 10.1101/gr.103119.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sinha A, Sommer RJ, Dieterich C. Divergent gene expression in the conserved dauer stage of the nematodes Pristionchus pacificus and Caenorhabditis elegans. BMC Genomics 2012; 13:254-70; PMID:22712530; http://dx.doi.org/ 10.1186/1471-2164-13-254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.C. elegans Sequencing Consortium . Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 1998; 282:2012-18; PMID:9851916; http://dx.doi.org/ 10.1126/science.282.5396.2012 [DOI] [PubMed] [Google Scholar]
- 40.Sternberg PW. Pristionchus pacificus: a nematode model for comparative and evolutionary biology Leiden, Netherlands: Brill 2015. Chapter 1, Why Caenorhabditis elegans is great and Pristionchus pacificus might be better. 1–17. [Google Scholar]


