Abstract
Aims/hypothesis
Tobacco smoking, a risk factor for diabetes, is an established modifier of DNA methylation. We hypothesised that tobacco smoking modifies DNA methylation of genes previously identified for diabetes.
Methods
We annotated CpG sites available on the Illumina Human Methylation 450K array to diabetes genes previously identified by genome-wide association studies (GWAS), and investigated them for an association with smoking by comparing current to never smokers. The discovery study consisted of 630 individuals (Bonferroni-corrected p = 1.4 × 10−5), and we sought replication in an independent sample of 674 individuals. The replicated sites were tested for association with nearby genetic variants and gene expression and fasting glucose and insulin levels.
Results
We annotated 3,620 CpG sites to the genes identified in the GWAS on type 2 diabetes. Comparing current smokers to never smokers, we found 12 differentially methylated CpG sites, of which five replicated: cg23161492 within ANPEP (p = 1.3 × 10−12); cg26963277 (p = 1.2 × 10−9), cg01744331 (p = 8.0 × 10−6) and cg16556677 (p = 1.2 × 10−5) within KCNQ1 and cg03450842 (p = 3.1 × 10−8) within ZMIZ1. The effect of smoking on DNA methylation at the replicated CpG sites attenuated after smoking cessation. Increased DNA methylation at cg23161492 was associated with decreased gene expression levels of ANPEP (p = 8.9 × 10−5). rs231356-T, which was associated with hypomethylation of cg26963277 (KCNQ1), was associated with a higher odds of diabetes (OR 1.06, p = 1.3 × 10−5). Additionally, hypomethylation of cg26963277 was associated with lower fasting insulin levels (p = 0.04).
Conclusions/interpretation
Tobacco smoking is associated with differential DNA methylation of the diabetes risk genes ANPEP, KCNQ1 and ZMIZ1. Our study highlights potential biological mechanisms connecting tobacco smoking to excess risk of type 2 diabetes.
Electronic supplementary material
The online version of this article (doi:10.1007/s00125-016-3872-0) contains peer-reviewed but unedited supplementary material, which is available to authorised users.
Keywords: DNA methylation, Gene expression, Tobacco smoking, Type 2 diabetes
Introduction
In the last decade, genome-wide association studies (GWAS) have been conducted in order to identify DNA sequence variants for a wide range of diseases including type 2 diabetes [1–3]. These GWAS have successfully identified numerous single-nucleotide polymorphisms (SNPs) located in and near genes that may be key in the development of type 2 diabetes. Up to now, a total number of 88 genetic loci have been identified for type 2 diabetes [4].
Tobacco smoking is associated with an increased risk of type 2 diabetes [5]. Several biological mechanisms have been proposed through which smoking may have an effect on the development of diabetes, including inflammation and the effect of nicotine on insulin resistance [6]. However, the exact molecular mechanisms connecting smoking to an increased risk of diabetes remain largely unknown. Previous research has established that tobacco smoking has an important role in DNA methylation, the epigenetic mechanism of attachment of a methyl group to a nucleotide [7–9]. DNA methylation has several functions in the human genome including the regulation of gene expression and maintenance of genome stability [10]. In line with this, previous studies have suggested DNA methylation as a potential pathway in the association between tobacco smoking and an increased risk of diabetes [11].
We hypothesised that tobacco smoking changes DNA methylation of susceptibility loci identified in GWAS for type 2 diabetes. We therefore investigated the association between DNA methylation in whole blood at loci identified for type 2 diabetes through GWAS and current tobacco smoking in a Dutch population-based cohort study. Furthermore, we investigated the potential effect of DNA methylation on the expression of genes near to the identified methylation sites.
Methods
Study population
The study was conducted using data from the Rotterdam Study; the design of the Rotterdam Study has been described elsewhere [12]. In brief, in 1990 all inhabitants living in the neighbourhood of Ommoord in Rotterdam, the Netherlands, aged 55 years and over, were invited to participate (RS-1). In 2000, the cohort was extended with 3,011 participants who had reached the age of 55 years or who were aged 55 years and over and had moved into the research area (RS-2). In 2006, a third cohort of 3,934 participants aged 45 years and older was initiated (RS-3). The discovery panel consisted of 630 non-diabetic participants in the first visit of RS-3 (diabetes was defined as a serum glucose level ≥ 7.0 mmol/l or the use of glucose-lowering medication) of a random subset of 747 individuals of European descent with DNA methylation data available. We sought replication of the identified CpG sites in a set of 674 non-diabetic participants from the third visit of RS-2 and the second visit of RS-3. The individuals in the replication study did not participate in the discovery study. The Rotterdam Study has been approved by the medical ethics committee according to the Population Screening Act: Rotterdam Study, executed by the Ministry of Health, Welfare and Sports of the Netherlands. All participants in the present analysis provided written informed consent to participate and to obtain information from their treating physicians.
Data collection
Data on tobacco smoking was collected during home interviews. Participants were asked about past and present cigarette, cigar and pipe smoking behaviour and were then categorised into current, former and never tobacco smokers. We asked current smokers about the age at which they started smoking and the number of cigarettes that they smoked per day. Former smokers were asked at what age they ceased smoking. Five of the participants had missing smoking status and were therefore excluded from any analysis. During the visit to the centre, weight and height were measured with the participant in standing position and wearing normal clothes. BMI was calculated as height in metres by weight in kilograms squared. All participants had blood samples taken during the visit to quantify DNA methylation, messenger RNA (mRNA) expression levels, DNA sequence variants and other blood measurements.
DNA methylation data
DNA was extracted from whole peripheral blood (stored in EDTA tubes) by standardised salting out methods. Genome-wide DNA methylation levels were measured using the Illumina Human Methylation 450K array [13] (Illumina, San Diego, CA, USA). In short, samples (500 ng of DNA per sample) were first treated with bisulfite using the Zymo EZ-96 DNA-methylation kit (Zymo Research, Irvine, CA, USA). Next, samples were hybridised to the arrays according to the manufacturers’ protocols. The methylation percentage of a CpG site was reported as a β value ranging between 0 (no methylation) and 1 (full methylation). Processing of the Rotterdam Study DNA methylation samples was performed at the Genetic Laboratory of Internal Medicine, Erasmus University Medical Centre, Rotterdam.
Quality control of the samples was carried out using Genome Studio (v2011.1, methylation module version 1.9.0; Illumina). In the discovery panel, a total number of 16 samples were removed: seven had a sample call rate below 99%; five had incomplete bisulfite conversion and four had sex changes. In the replication set, all samples passed the quality control based on the first two principal components obtained using principal component analysis (PCA), and no sex swaps were detected. Further quality control of the probes was done based on the detection p value calculated with Genome Studio. Probes with a detection p value of more than 0.01 in more than 1% of the samples were excluded. Additionally, sample-level quality control was performed using MethylAid (https://bioconductor.org/packages/release/bioc/html/MethylAid.html) [14]. This resulted in a total set of 474,528 probes that were normalised using the Dasen option of the WateRmelon R-package (https://www.bioconductor.org/packages/release/bioc/html/wateRmelon.html) [15].
mRNA expression data
Whole blood was collected (PAXGene Tubes; Becton Dickinson, Erembodegem, Belgium) and total RNA was isolated (PAXGene Blood RNA kits; Qiagen, Venlo, the Netherlands). To ensure the constant high quality of the RNA preparations, all RNA samples were analysed using the Labchip GX (Caliper, Hopkinton, MA, USA) according to the manufacturer’s instructions. Samples with an RNA quality score of more than 7 were amplified and labelled (TotalPrep RNA; Ambion, Austin, TX, USA) and hybridised to the Illumina HumanHT12v4 Expression Beadchips (Illumina) as described by the manufacturer’s protocol. Processing of the Rotterdam Study RNA samples was performed at the Genetic Laboratory of Internal Medicine, Erasmus University Medical Centre, Rotterdam. The RS-3 expression dataset is available at GEO (Gene Expression Omnibus) public repository under the accession GSE33828: 881 samples are available for analysis.
Illumina gene expression data was quantile-normalised to the median distribution and subsequently log2-transformed. The probe and sample means were centred to zero. Genes were declared significantly expressed when the detection p values calculated by GenomeStudio were less than 0.05 in more than 10% of all discovery samples, which added to a total number of 21,238 probes. Quality control was carried out using the eQTL-mapping pipeline (https://github.com/molgenis/systemsgenetics/tree/master/eqtl-mapping-pipeline) [16]. We only analysed probes that uniquely mapped to the human genome build 37 and represented gene mRNA expression [17].
Selection of methylation sites
A recent review summarising findings from all diabetes GWAS was used to compile a list of variants significantly associated with diabetes (88 variants) [4]. Next, the list of 88 variants was extended with polymorphisms in linkage disequilibrium (R2 > 0.8) in the HapMap panel and within 500 kb using the SNAP Proxy Search tool (https://broadinstitute.org/mpg/snap/ldsearch.php; accessed 1 October 2015). The final list included 890 SNPs, which were tested for in-gene variants and effects on expression of a gene within 1 Mb as found in a large publically available blood cis-expression-quantitative trait loci (cis-eQTL) database (false discovery rate [FDR] <0.05) [16]. We identified 525 SNPs that were in-gene (mapping to 72 unique genes) and 316 SNPs with an eQTL effect (mapping to 50 unique genes). The final number of unique genes was 111. The methylation probes within and near these diabetes-related genes as provided by Illumina were included in the analysis. We excluded probes from the Infinium HD methylation SNP list with a minor allele frequency above 1% as provided by Illumina, since variations in these SNPs can cause bias in the methylation measurement [18]. We further excluded known cross-reactive probes, since they can introduce bias in the results [19]. In total, we included 3,620 CpG sites in the analyses.
Statistical analysis
The characteristics of the discovery and replication populations were compared between current and never smokers using IBM SPSS Statistics version 21.0.0.1 (IBM, Armonk, NY, USA). The p values were calculated using independent sample t tests for continuous variables and χ2-square tests for dichotomous variables.
The 3,620 methylation probes were tested for association with tobacco smoking using a linear mixed model with the LME4 package in R version 3.1.0 with Dasen-normalised β values of the CpG sites as outcome measure (https://cran.r-project.org/web/packages/lme4/index.html) [20]. Extreme outliers (>4 SD from the mean and >4SD from the before last) in the DNA methylation values were excluded. We first compared current smokers with never smokers and then performed a sensitivity analysis on the identified CpG sites comparing former smokers with never smokers. Covariates were selected based on known association with DNA methylation. The selected covariates with fixed effects were age, sex and BMI [21–24]. Houseman-estimated white blood cell proportions were used as fixed effects to correct for cell mixture distribution [25]. Array number and position on array were added in the model as covariates with random effects to correct for batch effects. We corrected for multiple testing using a robust Bonferroni-corrected p value of 1.4 × 10−5 as the threshold for significance (0.05 / 3,620 probes).
The probes identified in the discovery analysis were tested for replication in the independent samples from the Rotterdam Study. We used identical models with the addition of cohort (RS-2 or RS-3) as a variable in the model to adjust for a potential cohort effect. A Bonferroni-corrected p value of 0.05 divided by the number of significant findings in the discovery study was used as a threshold of significant replication.
The replicated probes were further tested with total pack-years in the current smokers to test the association between tobacco smoking and cumulative exposure to smoking. We further investigated the association between the replicated probes and time since cessation in former smokers to study the change in methylation after smoking cessation. To decrease the possibility of confounding in our association, we further adjusted the model in a second analysis for other possible confounders and mediators. This analysis included total cholesterol, HDL-cholesterol, triacylglycerol levels (natural log-transformed), systolic blood pressure, daily alcohol intake and C-reactive protein levels (natural log-transformed).
Functional analysis
Since DNA methylation may have an effect on gene expression, we tested the association between DNA methylation and mRNA expression levels of nearby genes (cis) within 500 kb of the replicated CpG sites (250 kb upstream and downstream of the CpG location). First, residuals for mRNA expression were created after regressing out the measured cell counts (granulocytes, lymphocytes, monocytes, platelets and erythrocytes), fasting state, RNA quality score, plate number, age and sex on the mRNA expression levels using a linear mixed model. We then created residuals for DNA methylation regressing out the measured white blood cells, age, sex, array number and position on array on the Dasen-normalised β values of the CpG sites using a linear mixed model. The residuals of the mRNA expression levels and the residuals of the Dasen-normalised β values of the CpG sites were tested for association using a linear regression model.
We also studied the association between the replicated CpG sites and serum measures of fasting glucose and insulin combining both the discovery and replication samples. Serum glucose and insulin were measured using standard laboratory techniques. The models were adjusted for the same covariates as in the main analyses, with the addition of smoking category. Serum insulin was natural log-transformed. A Bonferroni-corrected p value for five tests was used. Furthermore, we searched for genetic variants (methylation quantitative trait loci [met-QTLs]) associated with the replicated methylation sites in the publicly available data from the paper by Grundberg et al [26]. Significant met-QTLs were then tested for an association with type 2 diabetes in the publicly available data from the DIAGRAM consortium, using a Bonferroni-corrected p value of 0.01 (0.05 / 5 met-QTLs) [3].
Results
A total of 630 participants were included in the discovery study. Clinical characteristics of the study population by smoking category are listed in Table 1. The participants were on average 59.5 ± 8.0 years old and 45% were men. The samples consisted of 175 current smokers, 184 never smokers and 271 former smokers. On average, current smokers had lower HDL-cholesterol, higher triacylglycerol and serum C-reactive protein than never smokers. Also alcohol consumption was higher in current smokers than in former smokers or never smokers. In the replication population, 68 individuals were current smokers, 238 were never smokers and 368 were former smokers. Clinical characteristics of the replication population can be found in electronic supplementary material (ESM) Table 1.
Table 1.
Baseline characteristics of the study population according to smoking status
| Characteristic | Total | Current | Former | Never | p valuea |
|---|---|---|---|---|---|
| N | 630 | 175 | 271 | 184 | |
| Age, years | 59.5 ± 8.0 | 57.9 ± 6.6 | 60.9 ± 8.5 | 59.0 ± 8.1 | 0.16 |
| Male sex (%) | 283 (45) | 85 (49) | 126 (47) | 72 (39) | 0.07 |
| BMI, kg/m2 | 27.4 ± 4.5 | 26.7 ± 4.4 | 27.6 ± 4.3 | 27.6 ± 4.8 | 0.07 |
| Fasting glucose, mmol/l | 5.35 ± 0.55 | 5.33 ± 0.58 | 5.40 ± 0.55 | 5.30 ± 0.52 | 0.65 |
| Systolic blood pressure, mmHg | 138.5 ± 63.0 | 136.4 ± 60.4 | 139.7 ± 67.4 | 138.7 ± 58.8 | 0.71 |
| Diastolic blood pressure, mmHg | 88.0 ± 65.0 | 86.0 ± 62.1 | 89.0 ± 9.8 | 88.4 ± 60.3 | 0.71 |
| Total cholesterol, mmol/l | 5.60 ± 1.03 | 5.60 ± 1.07 | 5.62 ± 1.01 | 5.56 ± 1.02 | 0.72 |
| HDL-cholesterol, mmol/l | 1.41 ± 0.40 | 1.34 ± 0.39 | 1.44 ± 0.41 | 1.44 ± 0.37 | 0.01 |
| Triacylglycerol, mmol/l | 1.45 ± 0.81 | 1.62 ± 1.02 | 1.39 ± 0.62 | 1.40 ± 0.81 | 0.02 |
| C-reactive protein, mg/l | 2.55 ± 4.74 | 3.17 ± 7.03 | 2.52 ± 3.54 | 2.03 ± 3.31 | 0.05 |
| Alcohol consumption, g/day | 18.3 ± 11.0 | 19.4 ± 12.7 | 19.0 ± 10.9 | 16.1 ± 9.3 | 0.006 |
| Fastingb, yes (%) | 628 (100) | 173 (99) | 271 (100) | 184 (100) | 0.15 |
Data are mean ± SD or n (%)
aCurrent vs never smokers
bThe participants who provided blood after an overnight fast
After correction for multiple testing (p = 1.4 × 10−5), we identified 12 differentially methylated CpG sites when comparing current smokers to never smokers in the discovery study (Table 2; results for all probes are presented in ESM Table 2). The 12 differentially methylated CpG sites were located within eight genes. The most significant finding was cg23161492 located within the gene ANPEP on chromosome 15 (p = 1.3 × 10−12). On chromosome 11, four CpG sites located within the gene KCNQ1 were significantly associated with current tobacco smoking (cg26963277, p = 1.2 × 10−9; cg13428066, p = 5.8 × 10−6; cg01744331, p = 8.0 × 10−6; cg16556677, p = 1.2 × 10−5). Within the gene ZMIZ1 on chromosome 10, two CpG sites were significant differentially methylated between current and never smokers (cg03450842, p = 3.1 × 10−8; cg21344746, p = 6.6 × 10−6). In addition, we identified CpG sites in and near INPP5E, NDUFS5, FCHSD2, PBX4 and TCF19 that were differentially methylated in current smokers compared with never smokers.
Table 2.
Significant associations between current vs never tobacco smoking and methylation of diabetes genes
| CpG site | Chromosome | Position Hg19 | Discovery | Replication | Gene | ||||
|---|---|---|---|---|---|---|---|---|---|
| β | SE | p value | β | SE | p value | ||||
| cg23161492 | 15 | 90357202 | −0.044 | 0.006 | 1.3 × 10−12 | −0.045 | 0.006 | 3.4 × 10−11 | ANPEP |
| cg26963277 | 11 | 2722407 | −0.026 | 0.004 | 1.2 × 10−9 | −0.034 | 0.004 | 3.3 × 10−14 | KCNQ1 |
| cg03450842 | 10 | 80834947 | −0.017 | 0.003 | 3.1 × 10−8 | −0.030 | 0.004 | 2.2 × 10−12 | ZMIZ1 |
| cg14024579 | 9 | 139332845 | −0.022 | 0.004 | 1.1 × 10−7 | −0.015 | 0.006 | 0.01 | INPP5E |
| cg14656441 | 1 | 39500070 | 0.026 | 0.005 | 1.5 × 10−6 | 0.016 | 0.008 | 0.05 | NDUFS5 |
| cg13912027 | 11 | 72759293 | 0.022 | 0.005 | 2.1 × 10−6 | −0.001 | 0.006 | 0.89 | FCHSD2 |
| cg00591868 | 19 | 19729048 | −0.015 | 0.003 | 4.6 × 10−6 | −0.003 | 0.005 | 0.51 | PBX4 |
| cg13428066 | 11 | 2677768 | 0.015 | 0.003 | 5.8 × 10−6 | 0.007 | 0.006 | 0.28 | KCNQ1 |
| cg21344746 | 10 | 80831230 | 0.016 | 0.004 | 6.6 × 10−6 | 0.001 | 0.005 | 0.82 | ZMIZ1 |
| cg16095155 | 6 | 31127863 | −0.013 | 0.003 | 7.2 × 10−6 | −0.007 | 0.004 | 0.12 | TCF19 |
| cg01744331 | 11 | 2722358 | −0.013 | 0.003 | 8.0 × 10−6 | −0.025 | 0.003 | 7.4 × 10−12 | KCNQ1 |
| cg16556677 | 11 | 2722401 | −0.015 | 0.003 | 1.2 × 10−5 | −0.027 | 0.004 | 3.9 × 10−10 | KCNQ1 |
Adjusted for age, sex, BMI, Houseman-estimated white blood cell proportions and batch effects
Bonferroni-corrected threshold for significance: 0.05/3,620 = 1.4 × 10−5
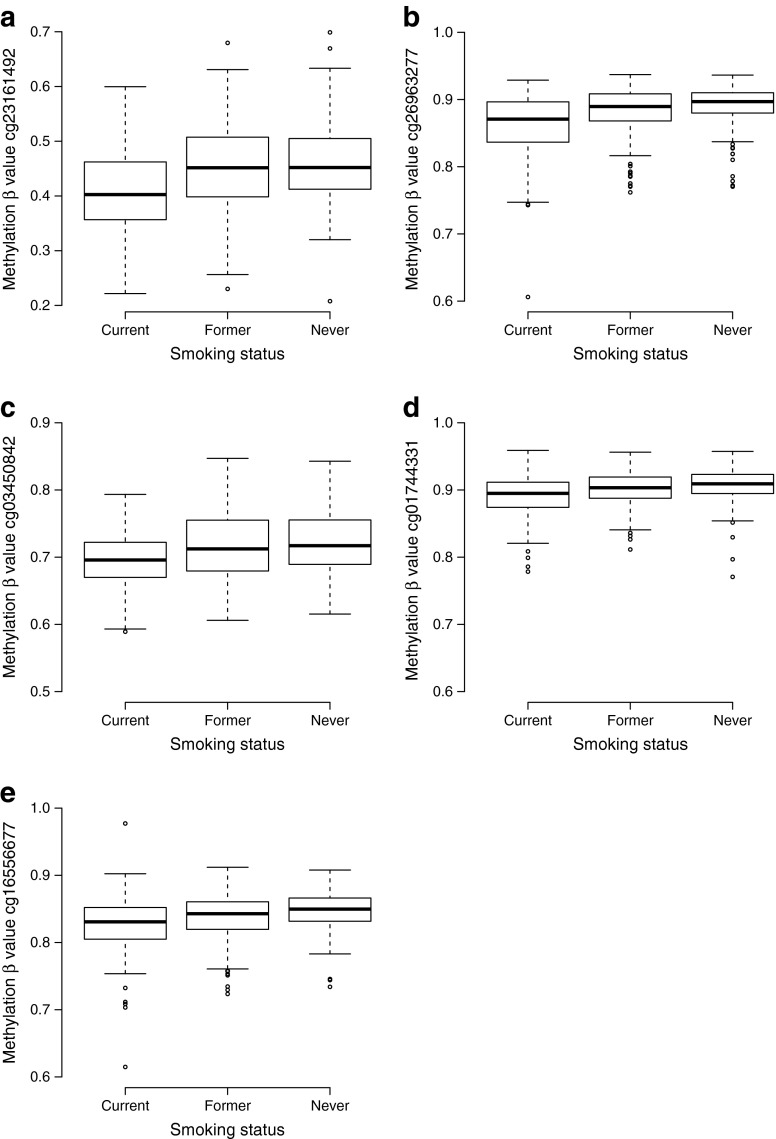
We attempted replication of the 12 differentially methylated CpG sites from the discovery study in 674 independent participants of the second and third cohort of the Rotterdam Study. We used a p value of 4.2 × 10−3 (0.05 / 12) as a threshold of significant replication. We significantly replicated the five CpG sites cg23161492 (ANPEP), cg26963277 (KCNQ1), cg03450842 (ZMIZ1), cg01744331 (KCNQ1) and cg16556677 (KCNQ1) (Table 2). Furthermore, the replicated associations were robust to further adjustment for possible confounders including systolic blood pressure, total cholesterol, HDL-cholesterol, triacylglycerol, alcohol consumption and C-reactive protein (ESM Table 3). Boxplots of replicated probe β values per smoking category are presented in Fig. 1. When we adjusted the effect of the top signal within the KCNQ1 gene (cg26963277) for the second (cg01744331) or third (cg16556677) signal within KCNQ1, cg26963277 was associated with current smoking, whereas cg01744331 and cg16556677 did not show an association (p = 0.84 and 0.35, respectively).
Fig. 1.
Boxplots depicting the methylation values in the replicated CpG sites (cg23161492 [a], cg26063277 [b], cg03450842 [c], cg01744331 [d], cg16556677 [e]) in current, former and never smokers. The bold horizontal lines represent the median methylation values, the box represents the interquartile range, the whiskers extend to 1.5 times the interquartile range or the most extreme value, and the circles represent a participant’s unique methylation value
To study the effect of smoking cessation on the replicated CpG sites, we compared former smokers with never smokers and tested the association between time since smoking cessation and DNA methylation. DNA methylation at the five CpG sites were not differentially methylated when former smokers were compared with never smokers (Table 3). Methylation at cg23161492 (p = 2.6 × 10−6), cg26963277 (p = 2.1 × 10−4), cg01744331 (p = 5.1 × 10−5) and cg16556677 (p = 1.2 × 10−3) was associated with time since smoking cessation. Additionally, methylation at the CpG sites cg23161492, cg26963277, cg03450842 and cg01744331 was associated with cumulative exposure to tobacco smoking.
Table 3.
Association between CpG sites and former smokers compared with never smokers, time since smoking cessation and cumulative smoking exposure in pack-years
| CpG site | Gene | Former vs never smokers | Cessation time | Pack-years | |||
|---|---|---|---|---|---|---|---|
| β (SE) | p value | βa (SE) | p value | βa (SE) | p value | ||
| cg23161492 | ANPEP | −0.007 (0.006) | 0.24 | 0.014 (0.003) | 2.6 × 10−6 | −0.007 (0.002) | 2.8 × 10−3 |
| cg26963277 | KCNQ1 | −0.006 (0.003) | 0.05 | 0.006 (0.002) | 2.1 × 10−4 | −0.006 (0.002) | 9.0 × 10−4 |
| cg03450842 | ZMIZ1 | −0.005 (0.002) | 0.06 | 0.002 (0.001) | 0.20 | −0.003 (0.001) | 1.6 × 10−3 |
| cg01744331 | KCNQ1 | −0.003 (0.002) | 0.21 | 0.005 (0.001) | 5.1 × 10−5 | −0.004 (0.001) | 1.1 × 10−4 |
| cg16556677 | KCNQ1 | −0.007 (0.003) | 7.1 × 10−3 | 0.005 (0.001) | 1.2 × 10−3 | −0.003 (0.001) | 0.05 |
Adjusted for age, sex, BMI, white blood cell counts and batch effects. Bonferroni corrected
aβ represents change in methylation per 10 years since smoking cessation and per 10 pack-years.
In the 630 individuals from the discovery panel, six genes out of 20 candidates were significantly expressed in the analysed whole-blood samples. The 12 methylation expression combinations are shown in ESM Table 4. The p value threshold for association was 4.2 × 10−3 (0.05/12 tests). Increased methylation at cg23161492 was negatively associated with gene expression levels of ANPEP (p = 8.9 × 10−5) (ESM Fig. 1).
We observed a putative effect of the CpG site cg26963277 with fasting serum insulin (effect: 0.004, p = 0.04). Results for the associations between all replicated CpG sites and serum fasting glucose and insulin are presented in ESM Table 5.
We identified a significant met-QTL for all replicated CpG sites, except cg0345084 (ESM Table 6). The T allele of the SNP rs231356 was associated with lower methylation of both cg26963277 and cg01744331 (KCNQ1). Also, the T allele of the SNP rs231356 was associated with an increased odds of type 2 diabetes (OR 1.06, p = 1.3 × 10−5).
Discussion
Our findings suggest that tobacco smoking is associated with differential methylation of CpG sites within the type 2 diabetes risk genes ANPEP, KCNQ1 and ZMIZ1. The associations were robust to adjustment for potential confounders and the effect of tobacco smoking appeared to be reversible after smoking cessation. In addition, methylation within ANPEP was significantly associated with gene expression levels of ANPEP. Methylation at KCNQ1 was associated with fasting insulin levels and genetic data supported a role for methylation at KCNQ1 in the development of diabetes. This study provides further insight into potential biological mechanisms underlying the association between tobacco smoking and an excess risk of type 2 diabetes.
In contrast to the findings for current vs never smokers, we found similar DNA methylation levels at the replicated CpG sites when comparing former smokers to never smokers. Furthermore, four significant CpG sites were associated with time since smoking cessation, suggesting a return after smoking cessation to DNA methylation levels similar to never smokers. This is in agreement with previous studies investigating the role of smoking cessation in DNA methylation [7, 27, 28]. DNA methylation may return to levels similar to never smokers at some sites, while other sites stay differentially methylated. Our results are in agreement with a potential beneficial effect of smoking cessation on DNA methylation at risk loci for diabetes. Furthermore, at four CpG sites we observed a dose-dependent effect of smoking underscoring the importance of cumulative tobacco exposure over time.
We identified three CpG sites within intron 11 of KCNQ1 (potassium channel, voltage gated KQT-like subfamily Q, member 1) that were differentially methylated in current smokers compared with never smokers. Previous studies have reported differential DNA methylation at the KCNQ1 locus in pancreatic islets and adipose tissue of diabetes cases and non-diabetes controls [29, 30]. Adjustment analyses suggested that cg26963277 is the driving CpG site associated with current smoking at this locus. Furthermore, we found the met-QTL (rs231356) for cg26963277 to be associated with the risk of diabetes. More specifically, the T allele of rs231356 is associated with lower methylation of cg26963277 and an increased odds of type 2 diabetes. In agreement with this observation, tobacco smoking lowers methylation at cg26963277 and is associated with an increased risk of diabetes. Additionally, our data suggest an association between cg26963277 and fasting insulin levels: increased methylation was putatively associated with increased fasting insulin levels. Although we did not observe an association between DNA methylation at cg26963277 and expression of KCNQ1, our results provide evidence that smoking may increase the risk of diabetes through decreased methylation at KCNQ1 and a subsequent decrease in fasting insulin levels.
Further, current tobacco smoking was associated with a 4.4% decrease in methylation at cg23161492 located near the 5′ untranslated region (UTR) of ANPEP and this decreased methylation was correlated with increased gene expression levels of ANPEP. ANPEP encodes the protein alanine aminopeptidase, a widely expressed enzyme involved in various cellular processes including cell proliferation, differentiation and apoptosis [31]. The observation that current smoking, which increases the risk of type 2 diabetes, may lead to higher gene expression levels of ANPEP is in line with the observation of Locke and colleagues [32]. The risk allele of the SNP rs2007084, identified by the DIAGRAM consortium, is also associated with increased gene expression of ANPEP in islet cells [32]. This suggests that increased expression of ANPEP leads to an increased risk of type 2 diabetes. The observation that DNA sequence variation and DNA methylation at this locus is associated with increased expression levels of ANPEP suggests a role for ANPEP in the pathogenesis of type 2 diabetes, rather than the gene AP3S2 proposed by prior GWAS [3].
We further identified the CpG cg03450842, near the 5′ UTR of ZMIZ1, to be differentially methylated in smokers compared with never smokers. The CpG cg03450842 has been identified previously to be associated with smoking [11]. Unfortunately, we had no expression data available in our samples for this gene and could therefore not study the effect of methylation at cg03450842 on gene expression of ZMIZ1.
The strength of the current study is the large sample size with available data on DNA methylation, gene expression and genetic variants, which allowed detailed investigation of the interrelationship between tobacco smoking, DNA methylation and gene expression. A limitation of the current work is the use of whole-blood samples for the quantification of DNA methylation and gene expression. As both methylation and expression may be tissue specific, we might have overlooked potential associations between tobacco smoking and differential methylation of diabetes-related genes in other tissues (e.g. liver, fat, pancreas or muscle tissue). Furthermore, observed associations may not be generalisable to other tissues. Another limitation is the challenge of gene annotation in GWAS. GWAS locate DNA sequence variants for phenotypes, but the underlying causal gene might be difficult to designate. To minimise this problem we limited our analysis to genes annotated to in-gene variants and known cis-eQTL effects. Therefore the diabetes risk genes selected in our study are more plausible as being the causal gene for diabetes.
In summary, our study suggests an effect of tobacco smoking on DNA methylation of the diabetes-related genes ANPEP, KCNQ1 and ZMIZ1. Our study provides further insight into potential mechanisms linking tobacco smoking to an excess risk of type 2 diabetes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 94 kb)
(PDF 21 kb)
(XLSX 160 kb)
(PDF 7 kb)
(PDF 21 kb)
(PDF 7 kb)
(PDF 20 kb)
Acknowledgements
The authors are grateful to the Rotterdam Study participants, the staff involved with the Rotterdam Study and the participating general practitioners and pharmacists. We thank M. Verbiest, M. Jhamai, S. Higgins, J. van Rooij and M. Verkerk from the Department of Internal Medicine at the Erasmus University Medical Center, Rotterdam, the Netherlands, for their help in creating the methylation and RNA array expression database.
Abbreviations
- GWAS
Genome-wide association studies
- met-QTLs
Methylation quantitative trait loci
- SNP
Single-nucleotide polymorphism
- UTR
Untranslated region
BIOS consortium members
Bastiaan T. Heijmans, Peter A. C. ’t Hoen, Joyce van Meurs, Aaron Isaacs, Rick Jansen, Lude Franke, Dorret I. Boomsma, René Pool, Jenny van Dongen, Jouke J. Hottenga, Marleen M. J. van Greevenbroek, Coen D. A. Stehouwer, Carla J. H. van der Kallen, Casper G. Schalkwijk, Cisca Wijmenga, Alexandra Zhernakova, Ettje F. Tigchelaar, P. Eline Slagboom, Marian Beekman, Joris Deelen, Diana van Heemst, Jan H. Veldink, Leonard H. van den Berg, Cornelia M. van Duijn, Albert Hofman, André G. Uitterlinden, P. Mila Jhamai, Michael Verbiest, H. Eka D. Suchiman, Marijn Verkerk, Ruud van der Breggen, Jeroen van Rooij, Nico Lakenberg, Hailiang Mei, Maarten van Iterson, Michiel van Galen, Jan Bot, Peter van ’t Hof, Patrick Deelen, Irene Nooren, Matthijs Moed, Martijn Vermaat, Dasha V. Zhernakova, René Luijk, Marc Jan Bonder, Freerk van Dijk, Wibowo Arindrarto, Szymon M. Kielbasa, Morris A. Swertz and Erik W. van Zwet
Funding
AD is supported by Netherlands Organization for Scientific Research (NWO) grant (veni, 916.12.154) and the EUR Fellowship. JBJvM is funded by the Netherlands Society for Scientific Research (NWO) VIDI Grant 917103521. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII) and the Municipality of Rotterdam. The generation and management of the Illumina 450 K methylation array data (EWAS data) for the Rotterdam Study was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Netherlands. The EWAS data was funded by the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, and by the Netherlands Organization for Scientific Research (NWO; project number 184021007) and made available as a Rainbow Project (RP3; BIOS) of the Biobanking and Biomolecular Research Infrastructure Netherlands (BBMRI-NL). The generation and management of RNA expression array data for the Rotterdam Study was executed and funded by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Netherlands.
Duality of interest
OHF works in ErasmusAGE, a centre for ageing research across the life course funded by Nestlé Nutrition (Nestec Ltd), Metagenics Inc. and AXA. Nestlé Nutrition (Nestec Ltd), Metagenics Inc. and AXA had no role in the design and conduct of the study, in data collection, management, analysis and interpretation or in the preparation, review or approval of the manuscript. All other authors declare that there is no duality of interest associated with their contribution to this manuscript.
Contribution statement
SL and RVS contributed to the design and analysis of the study and wrote the draft of the manuscript. MJP, JBJvM, EJGS, AGU, MJB, AH and OHF interpreted the results and reviewed and edited the manuscript. AD contributed to the design of the study and reviewed and edited the manuscript, and is the guarantor of this work. The BIOS consortium contributed to the design of the study. All authors have approved the final version of the manuscript.
Footnotes
Symen Ligthart and Rebecca V. Steenaard contributed equally to this manuscript.
A full list of BIOS Consortium members can be found at the end of the article.
References
- 1.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marullo L, Moustafa JSE-S, Prokopenko I. Insights into the genetic susceptibility to type 2 diabetes from genome-wide association studies of glycaemic traits. Cur Diab Rep. 2014;14:1–17. doi: 10.1007/s11892-014-0551-8. [DOI] [PubMed] [Google Scholar]
- 5.Willi C, Bodenmann P, Ghali WA, Faris PD, Cornuz J. Active smoking and the risk of type 2 diabetes: a systematic review and meta-analysis. Jama. 2007;298:2654–2664. doi: 10.1001/jama.298.22.2654. [DOI] [PubMed] [Google Scholar]
- 6.Xie X-t, Liu Q, Wu J, Wakui M. Impact of cigarette smoking in type 2 diabetes development. Acta Pharmacol Sin. 2009;30:784–787. doi: 10.1038/aps.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeilinger S, Kühnel B, Klopp N, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One. 2013;8 doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shenker NS, Polidoro S, van Veldhoven K, et al (2012) Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet dds488 [DOI] [PubMed]
- 9.Steenaard RV, Ligthart S, Stolk L, et al. Tobacco smoking is associated with methylation of genes related to coronary artery disease. Clin Epigenetics. 2015;7:54. doi: 10.1186/s13148-015-0088-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 11.Besingi W, Johansson Å (2013) Smoke related DNA methylation changes in the etiology of human disease. Hum Mol Genet ddt621 [DOI] [PubMed]
- 12.Hofman A, Brusselle GGO, Murad SD, et al. The Rotterdam study: 2016 objectives and design update. Eur J Epidemiol. 2015;30:661–708. doi: 10.1007/s10654-015-0082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandoval J, Heyn H, Moran S, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 14.van Iterson M, Tobi EW, Slieker RC, et al. MethylAid: visual and interactive quality control of large Illumina 450k datasets. Bioinformatics. 2014;30:3435–3437. doi: 10.1093/bioinformatics/btu566. [DOI] [PubMed] [Google Scholar]
- 15.Pidsley R, Wong CCY, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westra H-J, Peters MJ, Esko T, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schurmann C, Heim K, Schillert A, et al. Analyzing illumina gene expression microarray data from different tissues: methodological aspects of data analysis in the metaxpress consortium. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhi D, Aslibekyan S, Irvin MR, et al. SNPs located at CpG sites modulate genome-epigenome interaction. Epigenetics. 2013;8:802–806. doi: 10.4161/epi.25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Y-a C, Lemire M, Choufani S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Team RC . R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2012. [Google Scholar]
- 21.Koestler DC, Christensen BC, Karagas MR, et al. Blood-based profiles of DNA methylation predict the underlying distribution of cell types: a validation analysis. Epigenetics. 2013;8:816–826. doi: 10.4161/epi.25430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dick KJ, Nelson CP, Tsaprouni L, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;383:1990–1998. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 23.Florath I, Butterbach K, Müller H, Bewerunge-Hudler M, Brenner H. Cross-sectional and longitudinal changes in DNA methylation with age: an epigenome-wide analysis revealing over 60 novel age-associated CpG sites. Hum Mol Genet. 2014;23:1186–1201. doi: 10.1093/hmg/ddt531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang FF, Cardarelli R, Carroll J, et al. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics. 2011;6:623–629. doi: 10.4161/epi.6.5.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grundberg E, Meduri E, Sandling JK, et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am J Hum Genet. 2013;93:876–890. doi: 10.1016/j.ajhg.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsaprouni LG, Yang T-P, Bell J, et al. Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics. 2014;9:1382–1396. doi: 10.4161/15592294.2014.969637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nilsson E, Jansson PA, Perfilyev A, et al. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes. 2014;63:2962–2976. doi: 10.2337/db13-1459. [DOI] [PubMed] [Google Scholar]
- 30.Dayeh T, Volkov P, Salö S, et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mina-Osorio P. The moonlighting enzyme CD13: old and new functions to target. Trends Mol Med. 2008;14:361–371. doi: 10.1016/j.molmed.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Locke JM, Hysenaj G, Wood AR, Weedon MN, Harries LW (2014) Targeted allelic expression profiling in human islets identifies cis-regulatory effects for multiple variants identified by type 2 diabetes genome-wide association studies. Diabetes DB_140957 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 94 kb)
(PDF 21 kb)
(XLSX 160 kb)
(PDF 7 kb)
(PDF 21 kb)
(PDF 7 kb)
(PDF 20 kb)