Abstract
Throughout cell cycle progression, the expression of multiple transcripts oscillate, and whether these are under the centralized control of the CDK-APC/C proteins or can be driven by a de-centralized transcription factor (TF) cascade is a fundamental question for understanding cell cycle regulation. In budding yeast, we find that the transcription of nearly all genes, as assessed by RNA-seq or fluorescence microscopy in single cells, is dictated by CDK-APC/C. Three exceptional genes are transcribed in a pulsatile pattern in a variety of CDK-APC/C arrests. Pursuing one of these transcripts, the SIC1 inhibitor of B-type cyclins, we use a combination of mathematical modeling and experimentation to provide evidence that, counter-intuitively, Sic1 provides a failsafe mechanism promoting nuclear division when levels of mitotic cyclins are low.
Introduction
At the center of the canonical model of eukaryotic cell cycle regulation is the CDK-APC/C oscillator. Most or all cell cycle events such as periodic DNA replication, nuclear division, and cytokinesis can be directly driven from the CDK-APC/C 'clock' [Morgan, 2007]. Hundreds of genes are expressed in synchrony with cell cycle progression, and numerous mechanisms exist for direct transcription factor (TF) regulation by CDK-APC/C. Start cyclins (Cln1-3), in complex with Cdk1, turn on Start (SBF) genes by phosphorylating Whi5; mitotic cyclin Clb2-CDK inhibits the transcription factor SBF and activates Fkh2-Ndd1 for CLB2 cluster expression; Clb2-CDK further keeps Swi5 inactive by phosphorylating it until it is released by Cdc14 [Morgan, 2007].
Genome-wide TF binding data have led to another model. TFs transcribed during one cell cycle stage can bind the promoters of the next set [Simon et al., 2001, Lee et al., 2002]. In principle, this cyclical TF chain (sketched in Fig. 2 D right) could drive the global cell cycle transcription program without CDK-APC/C regulation. Evidence for such a CDK-APC/C-independent global transcriptional oscillator (GTO) has been reported in yeast cells; hundreds of genes oscillate in mutant strains with crippled CDK-APC/C oscillations [Orlando et al., 2008, Simmons Kovacs et al., 2012, Bristow et al., 2014].
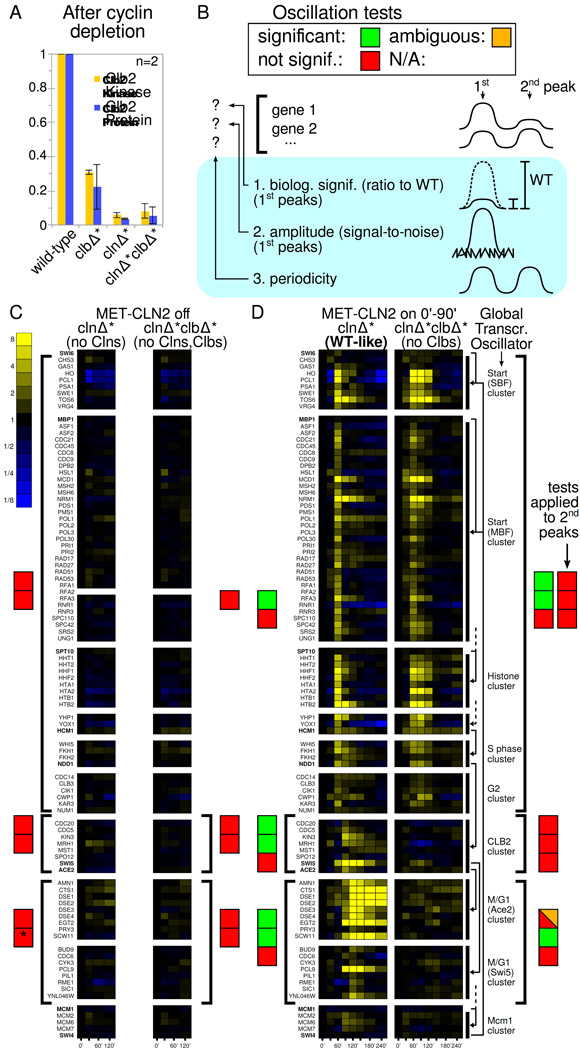
Figure 2.
Transcriptome-wide time course measurements in Cln- or Cln,Clb-depleted cells fail to show pulses predicted by the GTO model. A: Clb2 levels after cyclin-depletion protocol (time 0' in all experiments involving clnΔ*clbΔ*). B: Scheme for displaying the results of the oscillation tests for the pools of genes indicated by brackets in C and D. For thresholds, see main text. By default, the biological significance and amplitude tests are performed on the first (main) pulse; results for the second oscillation are only shown for MET-CLN2-induced Clb-depleted cells. The periodicity test evaluates the whole time course for any gene, so peaks are not specified. See ‘Detecting oscillations’ for details. C, D: RNA-seq time course measurements of cell-cycle regulated genes and their TFs. Fold induction over each gene's level at 0' is shown, i.e., each gene's mRNA level is divided by the level at 0'. The clb3Δ deletion in clnΔ*clbΔ* cells allows transcriptional dynamics to be picked up from the remaining 5' terminus. '*' indicates that the end-of-cell-cycle clusters in Cln-blocked cells are significantly upregulated (p=0.002) below our p value threshold. However, the p value is three orders of magnitude larger than the next lower p value, and the upregulation does not support the GTO model since preceding clusters are not activated. D right: Simplified wiring diagram of the proposed GTO [Orlando et al., 2008, Haase and Wittenberg, 2014]. Arrows from TFs (bold font) to clusters, which are delineated by black bars. Dashed lines indicate that important TFs have been omitted to simplify the drawing.
It is important to understand the extent to which CDK-APC/C or the GTO control cell cycle transcription. Control by multiple oscillators requires coordination. In cycling cells, without coupling mechanisms, the oscillators inevitably slip out of phase. In arrested cells, checkpoints must feed into all of the oscillators to halt them independently. However, transcriptional oscillations have not been reported at the spindle assembly checkpoint, the cell size checkpoint, or pheromone arrest, which are thought to mainly inhibit APC-Cdc20, shift the Cln3-CDK/Whi5 balance, or inhibit Cln-CDK, respectively [Morgan, 2007]. Thus, our understanding of synchrony and arrest is currently incomplete.
Using engineered strains with complete extrinsic control of all mitotic and G1 cyclins, we test the relationship between CDK-APC/C and transcription. These results support the CDK-APC/C model over the GTO model. However, a few genes (SIC1, CYK3, CDC6) do oscillate in different CDK-APC/C arrests. The oscillations of SIC1, an inhibitor of Clbs, in no-Clb arrests are surprising. Mathematical models show that robust SIC1 pulsing, counter-intuitively, can rescue cells with low Clb levels. We validate these predictions by showing that SIC1 does indeed rescue low-Clb cells in a physiological range of Clb levels.
Results
Oscillations under constitutive cyclin transcription
We constructed strains with all endogenous Cdk1 cyclins deleted, while CLN2 promoting Start and CLB2 promoting S-phase and mitotic entry can be turned on or off exogenously (cln1-3Δ MET-CLN2 clb1-6Δ GALL-CLB2, for brevity: cln1-3Δ MET-CLN2=clnΔ*, clb1-6Δ GALL-CLB2=clbΔ*). This simplifies the classical 'CDK-APC/C model' because wild-type (WT) transcriptional control of cyclins has been removed (Fig. S1 A). This strain is viable when MET-CLN2 and GALL-CLB2 are induced continuously in galactose (‘G’) and absence of methionine (‘−Met’) and shows transcriptional oscillations from the CLN2, CLB2, and SIC1 promoters, which are members of the Start (early), CLB2 (middle), and Swi5 (late) cell cycle clusters, respectively (Fig. 1). The observation that these promoters remain periodic despite constitutive expression of the sole remaining cyclins is consistent with either strong post-transcriptional regulation of cyclins or a GTO forcing oscillations. It is inconsistent with the proposed GTO being a crucial driver of CDK-APC/C oscillations by periodically transcribing cyclins; periodic transcription of the G1 cyclin Cln3 is required to restart the cycle in newborn cells in published GTO models [Simon et al., 2001, Orlando et al., 2008]. Yet, because of strong post-transcriptional regulation of cyclins, these observations alone do not fully test all GTO models. We must clamp cyclin-CDK-APC/C activity, not just cyclin transcription, and see whether transcriptional oscillations cease or continue.
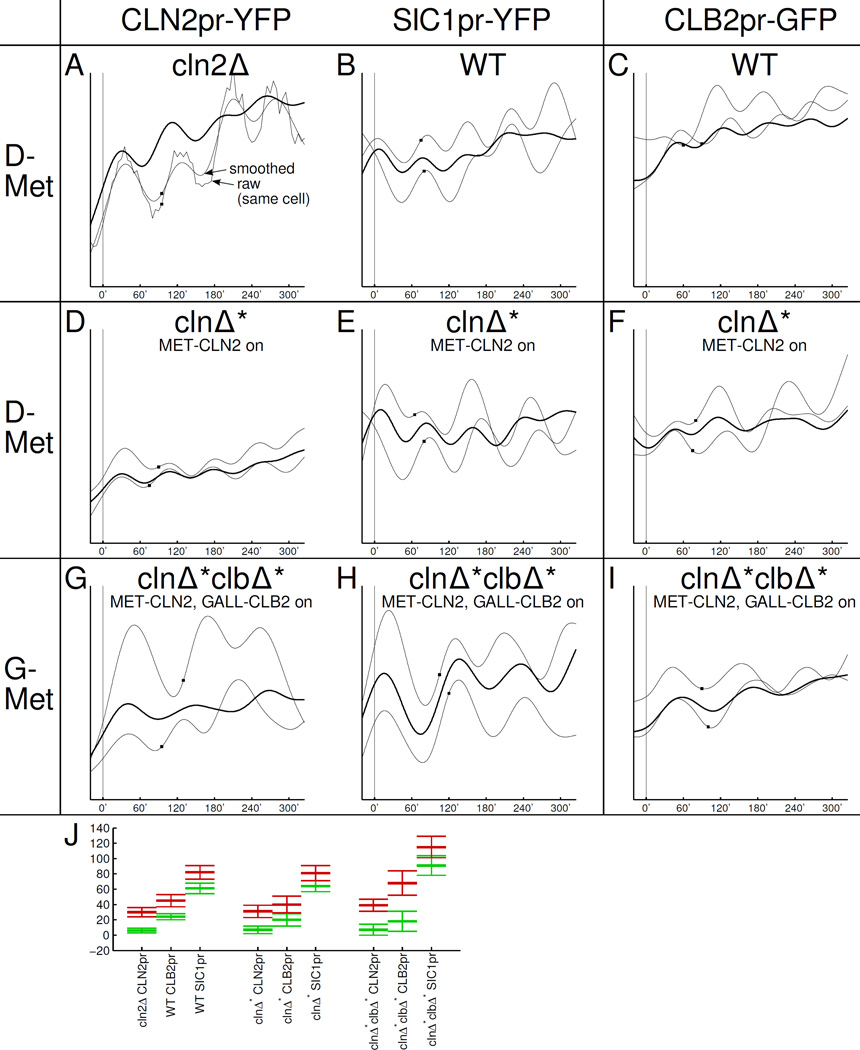
Figure 1.
Constitutive transcription of CLN2 and CLB2 in otherwise clnΔclbΔ cells suffices for viability and transcriptional oscillations. Vertical scales in A–I identical in each column (AU). Abbrev.: G: galactose (GALL-CLB2 on); −Met: absence of methionine (MET-CLN2 on). Traces are aligned so that budding occurs at 0'; the next budding event is indicated by a small rectangle on same trace. Thin traces: sample traces of individual cells; thick traces: averages over 10 cells. A: We recorded time courses for the cln2Δ:CLN2pr-YFP strain that is the parent strain of all of our clnΔ* CLN2pr-YFP strains. (The cln2Δ deletion has hardly any influence on cell cycle timing and CLN2 activation [Skotheim et al., 2008].) J: Mean ± SD for the 'on' time (=inflection point, green) and 'off' time (=maximum, red) with respect to budding at 0'. The relatively long time between activation of CLN2pr and SIC1pr in cycling clnΔ*clbΔ* cells is likely due to specific changes in cell cycle kinetics and not the carbon source (G vs. D) since the time between CLN2pr and SIC1pr is hardly affected by G-Met (not shown) vs. D-Met in clnΔ* cells.
Detecting oscillations
We apply three independent tests to distinguish random fluctuations from transcriptional oscillations: i) Biological significance test: We compare the size and timing of the fluctuations to true cell-cycle oscillations. If during the CDK-APC/C block, gene activity is very low (hardly rises), very high (hardly drops), or changes too slowly compared to normal cell cycle oscillations, we see this as incompatible with a GTO, which should exhibit roughly normal strength and timing if it is to have a biological impact. We call such fluctuations 'biologically insignificant'. We use the following thresholds: insignificant: size of rise or drop <10% compared to WT; ambiguous: rise or drop 11%–30%; significant otherwise. In some cases, the CDK-APC/C block suppresses transcription or keeps it at very high levels, yet, transcription can eventually return to near-normal levels. For such cases, we set a limit of 3*cell cycle periods, ≈200 min, above which the event is too slow to be considered a biologically significant oscillation. Such thresholds are somewhat arbitrary, but these are reasonably permissive. ii) Amplitude test: The height of an oscillation can also be compared directly to noise in the data, using a variation of the procedure of [de Lichtenberg et al., 2005]. We compare the amplitude of a given gene to fluctuations in non-cell-cycle regulated housekeeping genes representing the noise background. This test requires a reference set of housekeeping genes, so it is applicable to transcriptome data but not to single-cell/single-gene data. iii) Periodicity test: Fluctuations can be small but highly regular. So, we use a second tool from [de Lichtenberg et al., 2005] to detect periodicity. The test requires a fixed period as input; we scan periods from 50' to 130', and accept periodicity at any period (again, a permissive criterion). We add two more periodicity tests of our own for ambiguous cases (see Extended Experimental Procedures). All three tests are illustrated for one time course in Fig. S2 G.
For statistical power, we pool results for similarly regulated genes in transcriptome measurements (brackets in Fig. 2 C,D) as well as the results of individual time traces for single-cell/single-gene measurements (see Extended Experimental Procedures for details). The aggregate results are almost always unambiguous; we report p<0.01 as significant for the statistical tests ii) and iii).
Transcriptome-wide analysis of no-Clb arrests
In the first set of CDK-APC/C states we studied, we turned the mitotic cyclin CLB2 off and switched Start cyclin MET-CLN2 on or off in clnΔ*clbΔ* cells. Cln2 protein disappears quickly after turning MET-CLN2 off [Tyers et al., 1992] in +Met. The MET-CLN2 construct [Amon et al., 1994] is tight: MET-CLN2-blocked cells do not rebud for the duration of our experiments (>14 h) and show no activation of the Start gene cluster.
To clear Clb2, which is produced copiously by GALL-CLB2, we needed to devise a 'cyclin-depletion protocol' (Experimental Procedures). Simple shut-off of GALL-CLB2 was not sufficient to clear Clb2 even after 2.5 h in glucose (‘D’); using the cyclindepletion protocol we achieve negligible Clb2 levels and kinase activity, comparable to 1–2% of peak total Clb levels and activity in WT (Figs. 2 A, S2 A–E). (This is a potentially important difference from past experiments (see 'Comparison to past experiments').)
Stringent depletion of Clb2 is important to allow rigorous statements about transcriptional oscillations in the 'absence of cyclins'. On the other hand, the cyclin-depletion protocol involves extended blocks and could introduce artifacts on its own. The following observations suggest that the cyclin-depletion protocol is reasonably innocuous: (1) After the cyclin-depletion protocol is applied to clnΔ* cells, a brief activation of MET-CLN2 leads to a prompt and complete cell division cycle. A similar pulse applied to clnΔ*clbΔ* cells, which were arrested for an identical time by identical medium shifts, results in prompt bud emergence at the same time as in clnΔ* cells, though no cell division results. (2) clnΔ* and clnΔ*clbΔ* turn on early cell cycle genes almost exactly comparably in response to the MET-CLN2 pulse; thus these cells are not transcriptionally inert. (3) If MET-CLN2 is induced constitutively, some of the genes we studied remain on or oscillate in clnΔ*clbΔ* cells for hours and cells bud repeatedly during that time. Thus, cells maintain comparable functionality in Clb-depleted conditions for long periods. (4) Finally, the block in clnΔ*clbΔ* cells is reversible: if cyclin-depleted clnΔ*clbΔ* cells are reinduced in G+Met and arrest is continued for another 10 hrs before a final pulse of −Met, 75% of cells bud and 67% of these cells execute a complete cell cycle. Note that the total block time in this last experiment is far longer than in any of our experiments.
After cyclin depletion (time point 0' in all experiments with clnΔ*clbΔ* cells), we look for oscillations at the transcriptome level. We either leave clnΔ*clbΔ* and clnΔ* cells in the Cln,Clb-blocked or Cln-blocked state, respectively, (Fig. 2 C) or we induce MET-CLN2 temporarily (Fig. 2 D), which initiates one normal cell cycle in clnΔ* cells. (The ability to turn Clns on or off is another key difference with past experiments (see 'Comparison to past experiments’).) We compiled 130 genes from [Spellman et al., 1998, Colman-Lerner et al., 2001, Haase and Wittenberg, 2014] previously assigned to cellcycle specific regulons, and the known TFs for these genes. Among these, we focus on the genes that are significantly upregulated (amplitude test: p<0.01) in MET-CLN2-induced clnΔ* cells, which perform one normal cell cycle. We are left with 91 genes (see SI), which are displayed in Fig. 2 C,D together with their TFs. (We keep the non-oscillating TFs solely for illustration.)
In the normal cell cycle, we see transcriptional peaks at 60', 90', and 120' (Fig. 2 D left). Given that one cell cycle produces transcriptional peaks over a span of more than 60', we should see at least one transcriptional oscillation in Cln, Clb-depleted cells if there is an autonomous GTO. Yet, we do not see any gene pulses (Fig. 2 C right), confirmed by two oscillation tests (the periodicity test cannot be applied here). Reflecting the effectiveness of the MET-CLN2 shutoff, no budding is observed.
It is possible that we did not observe transcriptional oscillations simply because they gradually dissipate over the duration of the cyclin-depletion protocol before 0'. So, we induce MET-CLN2 briefly, which kick-starts gene transcription through the Cln1,2-Whi5-SBF positive feedback loop [Skotheim et al., 2008]. In clnΔ* cells this leads to one complete cell cycle (Fig. 2 D left).
In Clb-depleted clnΔ*clbΔ* cells, MET-CLN2 induction leads to budding (90% = 201/223 cells) but no cell or nuclear division (determined by bud counts and by timelapse microscopy with histone Htb2-mCherry). In these cells, early-to-mid cell cycle clusters activate, but only once: the oscillation tests show no periodicity and no significant second peaks. This conflicts with any closed GTO model, which requires a second cycle (Fig. 2 D right). The clusters following Start (S phase, G2 cluster) do fire, which could imply the existence of a short non-oscillatory chain of TFs. Alternatively, their firing may be due to post-transcriptional regulation of Hcm1 [Pramila et al., 2006] with Clns as key activators. The CLB2 cluster is not activated according to any of our three standards. For example, CDC20 decreases slightly with time and SWI5 and ACE2 have peaks at 8% and 10% of WT; overall, CLB2 cluster activity is consistent with noise (p=0.24).
Interestingly, a few genes in the Ace2/Swi5 cluster (CDC6, CYK3, SIC1) pulse at biologically significant levels and above-noise levels (see Fig. S2 H for 2D plots). This observation is pursued below.
Single-cell analysis of no-Clb arrests
Because bulk population measurements may miss brief or unsynchronized gene expression bursts, we also measure fluorescence from CLN2pr-YFP and CLB2pr-GFP by single-cell timelapse microscopy (Fig. 3 B–I).
Figure 3.
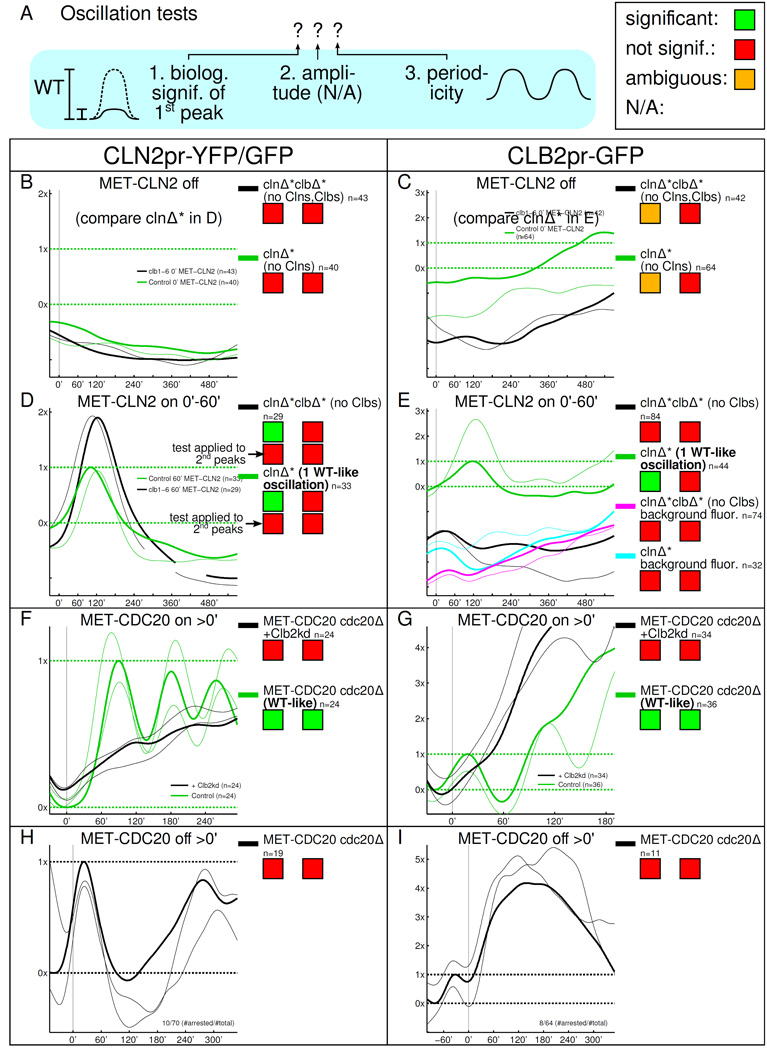
Single-cell/single-gene time course measurements fail to show pulses predicted by the GTO model. A: Same scheme as in Fig. 2. The statistical amplitude test is not available for single-cell/single-gene recordings. B–I: Smoothed, thin lines: single cell recordings; thick lines: population mean. B–G: Units of fluorescence chosen so that 0× is the mean trough and 1× is the mean peak of the first normal control cell cycle oscillation. The WT-like oscillations for B and C are plotted in D and E, respectively. H, I: 0× and 1× are defined such that the last, pre-arrest mean cell cycle response runs though 0× and 1× (trough and peak). Time courses are aligned so that at 0', cells bud for the last time before arresting for at least 350'.
For the biological relevance test, we measure the rise (Λ↑) and drop (Λ↓) of each oscillation, as illustrated in Fig. S3 A. The scatter plots in Fig. S3 B–I show the Λs for each cell as well as the population averages, which determine biological significance.
In single cell recordings, the periodicity test is applied to raw, unsmoothed data for individual cells. We then aggregate the p values, allowing detection of periodicity even if oscillations are completely out-of-phase between cells.
As in the transcriptome experiments, upon completion of the cyclin-depletion protocol, we either leave clnΔ* and clnΔ*clbΔ* cells in a Cln-blocked state (MET-CLN2 off, Fig. 3 B,C) or we induce MET-CLN2 briefly, which leads to budding (Fig. 3 D,E). Again, MET-CLN2-induced clnΔ* cells perform exactly one normal cell cycle (green plots in Fig. 3 D,E), which we use as the standard for the biological significance test (controlling for effects of the cyclin-depletion protocol, if any).
In Cln,Clb-depleted cells, neither CLN2pr-YFP (representing the Start cluster) nor CLB2pr-GFP (CLB2 cluster) show significant oscillations (Fig. 3 B,C). Cells do not bud. We observe CLB2pr-GFP fluorescence fluctuations that are between our insignificance and significance thresholds (11%-30%) but periodicity is not detected.
CLN2pr-YFP fires just once in Clb-depleted cells after short MET-CLN2 induction (Fig. 3 D). Cells bud but do not complete nuclear division. Similar to other CLB2 cluster genes, CLB2pr-GFP fluorescence fluctuates insignificantly, similar to background fluorescence (Fig. 3 E). It is interesting that the CLB2pr-GFP baseline is lower in Clbdepleted cells (explored further in Fig. S3 K).
These single-cell measurements are consistent with the transcriptome results described above.
Summary for no-Clb arrests
The lack of activation of the CLB2 cluster contradicts all previous GTO models. This is crucial because the CLB2 cluster contains key genes for entry and exit from mitosis, e.g., CLB1, CLB2, CDC5, and CDC20, and it is the key link in GTO models which activates end-of-cell-cycle regulons via SWI5 and ACE2. Indeed, most end-of-cell cycle genes are not activated in our experiments; we discuss the exceptions in 'Pulsing of end-of-cell-cycle genes in CDK-APC/C arrests'.
The absence of a second Start gene pulse argues against GTO models requiring no cyclin-Cdk activity at all [Lee et al., 2002, Simmons Kovacs et al., 2012]. However, some GTO models are hybrid models, which incorporate the Start cyclin CLN3 [Simon et al., 2001, Orlando et al., 2008], and CLN3 is not present in clnΔ* and clnΔ*clbΔ* cells. So, we analyze cln1,2Δ CLN3 MET-CLN2 clbΔ* cells. In these, CLN2pr-YFP levels reach superphysiological (8×–12×) levels and do not return back to even 1× (mean wt peak) for at least 8 h. Thus, CLN2 transcription is constantly 'on'. The slow CLN2pr-YFP fluctuations on top of these high levels are ‘biologically insignificant’ according to our criteria and are similar to fluctuations on top of high overall levels in clnΔ*clbΔ* cells when MET-CLN2 is turned on constitutively (Fig. S3 J).
Single-cell analysis of high-Clb arrests
In the second set of strains we constructed, mitotic cyclin levels can be locked at high instead of near-zero levels. We achieved this in two ways, (1) by just turning off MET-CDC20 in cdc20Δ cells (Fig. 3 H,I) or (2) by turning off MET-CDC20, expressing limited amounts of undegradable Clb2kd and turning MET-CDC20 back on (Fig. 3 F,G) [Drapkin et al., 2009, Lu and Cross, 2010]. (Clb2kd lacks Clb2's APC degradation sites and blocks mitotic exit even under control of the endogenous CLB2 promoter [Wäsch and Cross, 2002].) The CDK-APC/C model simplifies substantially under these conditions (Fig. S1 B,C). Slippage occurs in both blocks [Drapkin et al., 2009]; a single-cell analysis, in which escaping cells can be taken out, is indispensable.
Both cdc20Δ and Clb2kd arrests yield the same results. CLN2pr-GFP is noticeably suppressed but induction creeps up slowly (Fig. 3 F,H), e.g., CLN2pr-GFP reaches half-peak in about 200' in cdc20Δ cells. Suppression of CLN2 is consistent with well-known SBF inhibition by mitotic cyclins [Amon et al., 1993], whose levels are high in these arrests. We attribute the slow rise of CLN2pr-GFP to MET-CDC20 leakage or inactivation or dilution of Clb2kd, since the rise is invariably followed by cell division. Positive feedback from Clb1/2 onto the CLB2 cluster is well-known [Amon et al., 1993]; consistently, fluorescence from CLB2pr-GFP shoots up to multiple WT peak levels, only coming back down after a time equivalent to multiple cell cycles (Fig. 3 G,I). No oscillatory behavior is observed during the block.
Pulsing of end-of-cell-cycle genes in CDK-APC/C arrests
Our results so far support the CDK-APC/C model (Fig. S1) over the GTO model. However, there were clear exceptions in the RNA-seq data (Fig. 2 D). When MET-CLN2 is induced, a small subset of Swi5 cluster genes (CDC6, CYK3, SIC1) turn on in Clb-depleted cells above the biological significance threshold (30%) or nearly so (CDC6). These genes, together with others near the biological insignificance threshold (10%), pull the activity level of the whole end-of-cell-cycle pool of genes above the noise threshold (amplitude test).
The discrepancy between lack of CLB2 cluster activation, including SWI5, and activation of Swi5 cluster genes, was surprising. To ascertain whether the tiny SWI5 mRNA peak (8% of WT) could be responsible for activation of these genes, we use a Swi5-YFP construct [Sbia et al., 2008] to monitor protein level and localization. In single Clb-depleted clnΔ*clbΔ* cells, following 60' MET-CLN2 induction, we detected no increase in Swi5-YFP fluorescence (Fig. S4 M). Also, we did not detect any increase in the spatial variance of fluorescence, an unbiased measure of Swi5 nuclear entry (Fig. S4 N). Both signals, especially nuclear entry, are clearly present in WT cell cycles.
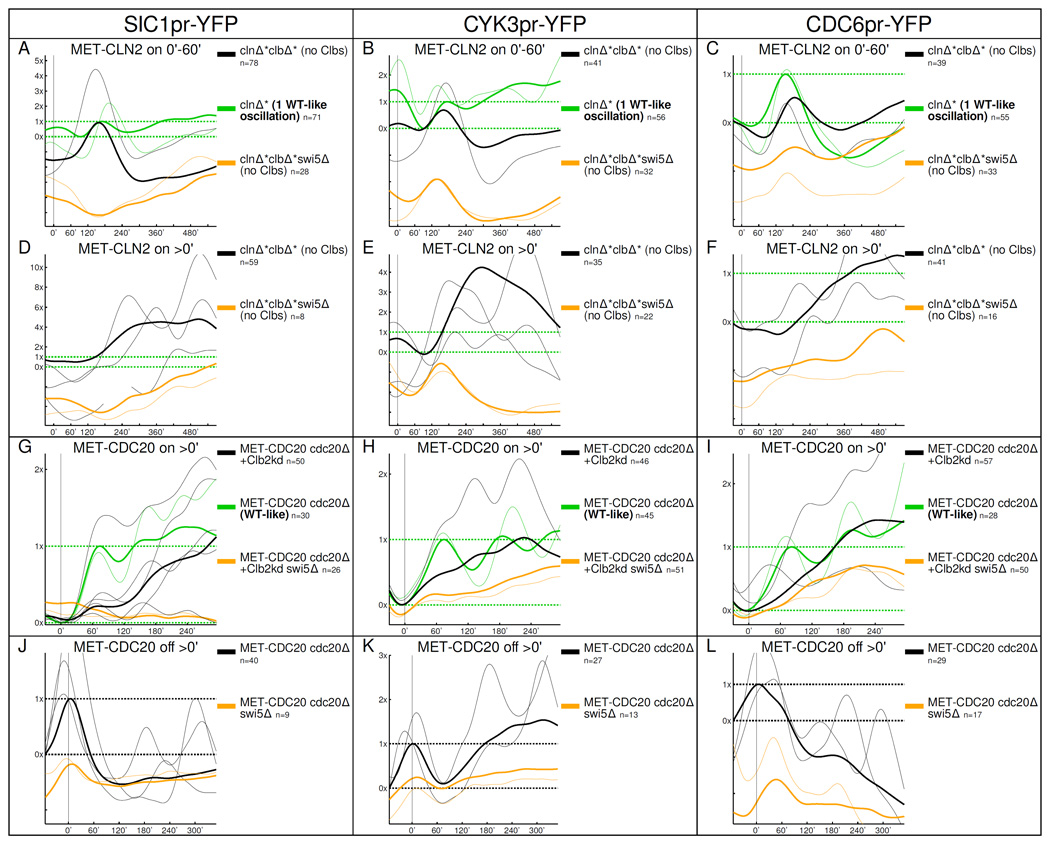
Next, we looked at CDC6, CYK3, SIC1 in single cells. All three genes respond at least as strongly in Clb-depleted clnΔ*clbΔ* cells as in WT-like cell cycles in response to a 60’ MET-CLN2 pulse (Fig. 4 A–C). The median time when the pulses come on is comparable in Clb-depleted cells and normal cell cycles (SIC1, CYK3: 120'/130', CDC6: 140'/110', respectively). The response in Clb-depleted cells is either entirely (SIC1) or largely (CYK3,CDC6) eliminated by swi5Δ deletion (CDC6 is co-regulated by Swi5 and Start TFs [Piatti et al., 1995]). If MET-CLN2 is induced continuously, transcription of these three genes oscillates (Fig. 4 D–F); these oscillations are genetically dependent on SWI5. Multiple pulses in Clb-depleted cells are roughly of the same size as in control cell cycles (Fig. 4 A–C; mean Λs are around 1×–2× for both controls and Clb-depleted cells) but the median period between oscillations is large, 190'–200' for all three genes. Thus, Swi5 does not accumulate detectably in the nucleus and its concentration does not increase, inconsistent with a transcriptional chain model. Nevertheless, it activates a subset of its targets at least as strongly as in WT cells, suggesting the possibility that low-level Swi5, either from constitutive transcription or left over from previous cell cycles, can be activated post-transcriptionally following MET-CLN2 induction.
Figure 4.
Swi5-regulated genes oscillate in different locked CDK-APC/C states. The multiple oscillations in D–F should be compared with the normal cell cycle oscillations in A–C. The results of the oscillation tests are in Fig. S4; the oscillation tests show that the sizes of the oscillations in CDK-APC/C arrested cells are comparable to WT.
In high-Clb-arrested cells, SWI5 is transcribed, and Swi5-YFP cycles in and out of the nucleus multiple times (Fig. S4 O,P; A. Procko PhD thesis). The Swi5 cluster transcripts SIC1, CDC6, and CYK3 oscillate as well (Fig. 4 G–L); SIC1 activation, and the majority of CYK3 and CDC6 activation, is dependent on SWI5. In the presence of Clbs, Swi5 is activated by Cdc14, which itself cycles in and out of the nucleolus at high-Clb arrests [Lu and Cross, 2010]. Both Cdc14 oscillations [Lu and Cross, 2010] and the interaction of Swi5 with Cdc14 are well-understood [Nasmyth et al., 1990, Visintin et al., 1998]. So, transcriptional oscillations would link up to another oscillatory mitotic exit module via Swi5.
In summary, we find a subset of Swi5 targets to be transcribed at different CDK-APC/C arrests. This transcription occurs in a pulsatile fashion and at both no-Clb (clbΔ) and high-Clb (+Clb2kd and cdc20Δ) blocks.
Biological function of SIC1 oscillations
The activation of a small number of genes (CDC6, CYK3, SIC1) at distinct CDK-APC/C blocks intrigued us. While the known mechanisms for Swi5 activation are specifically Clb-dependent [Moll et al., 1991, Visintin et al., 1998], the activation of CDC6, CYK3, and SIC1, at roughly WT amplitude, is evidently robust to large variations in Clb levels. We considered the possibility that ‘out-of-order’ transcriptional activity of these genes could make the system more robust to CDK-APC/C fluctuations.
Sic1 inhibits mitotic cyclins, and thus regulates the CDK-APC/C clock itself. Sic1 is not essential; its main biological role may be keeping B-type cyclin activity from rising too early in G1 [Lengronne and Schwob, 2002]. But in WT cell cycles, Clb levels may deviate from their 'ideal' cell-cycle trajectory, e.g., due to random noise. SIC1 turning on precociously in the cell cycle sequence at a high-Clb block could help to bring down Clb levels when they fluctuate too high. This idea is consistent with hypersensitivity of sic1Δ cells to Clb2 overexpression [Toyn et al., 1997] or stabilization [Schwab et al., 1997]. Following this line of reasoning, it is strange to see SIC1 pulses in Clb-depleted cells as well.
Clbs activate APC/C by phosphorylating it and independently by initiating transcription of CDC20 and CDC5; Cdc20 is an APC/C partner; Cdc5 activates Cdc14, which in turn activates the APC/C partner Cdh1. A negative feedback loop is formed because the APC/C-Cdc20/Cdh1 complexes destroy Clbs. Recent work has shown that negative feedback loops are uniquely fragile architectures. If low Clb levels fail to activate sufficiently strongly, the negative feedback agent (APC/C) can be slow or fail to sufficiently reduce the Clb target [Muzzey et al., 2009], leading to a trap with intermediate amounts of Clbs. Both simple models (Fig. S5 A, B) and the most realistic model to date, which accounts for all the known elements of mitotic exit, Cdc14, APC/CCdc20, APC/C-Cdh1, Sic1, exhibit these design problems (Fig. 5).
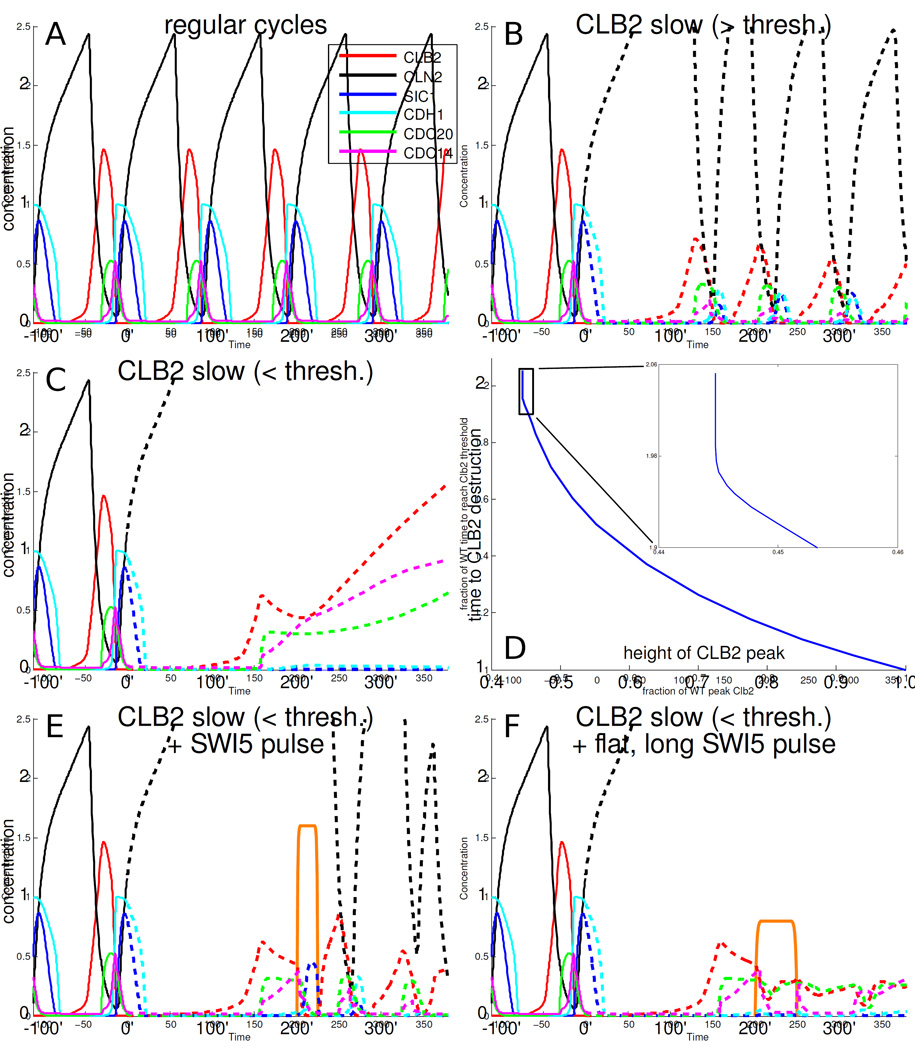
Figure 5.
The detailed model of the cell cycle from [Chen et al., 2004] shows that with low amounts of Clbs the cell cycle becomes trapped, and can be rescued by a (forced) pulse of Sic1. A: Normal cell cycle dynamics according to the model. B: When the rate of accumulation of Clb2 (red) is slowed down (dashed lines after 0'), the Clb2 peak occurs later and is smaller. C: With even slower Clb2 accumulation rates, Clb2 destruction becomes ineffective (also, the first Clb2 peak after 0' is smaller than in B). D: Above a certain threshold Clb2 accumulation rate (the height of the first Clb2 peak after 0' is larger), Clb2 destruction is successful (as in A and B), and below the threshold (small Clb2 accumulation rate, smaller Clb2 peak), Clb2 destruction is ineffective (as in C). The threshold is at about 45% of WT peak Clb2 levels. Time to Clb2 destruction and Clb2 peak height are normalized to the WT time and height in the model. Clb2 destruction time is defined as the time to return to a fixed, low Clb2 level (0.1). The time to Clb2 destruction is measured from 0'; the result is similar if the Clb2 destruction time is measured from the time of the Clb2 peak (Fig. S5 C). E: A sharp 25 min pulse of Swi5 (orange plot, equal in area to that of Swi5 in WT cycles) effectively turns off Clb2 and resets the cell cycle. F: Pulses of lower height but equal area are less effective at rescue; here, the height is reduced by half.
Based on these results, we hypothesized a role for SIC1 expression at low Clb levels (Fig. 6). This is counter-intuitive because the usual rules of genetics predict the opposite: reductions in the target (Clbs) and its inhibitor (Sic1) should compensate for one another, not exacerbate each other.
Figure 6.
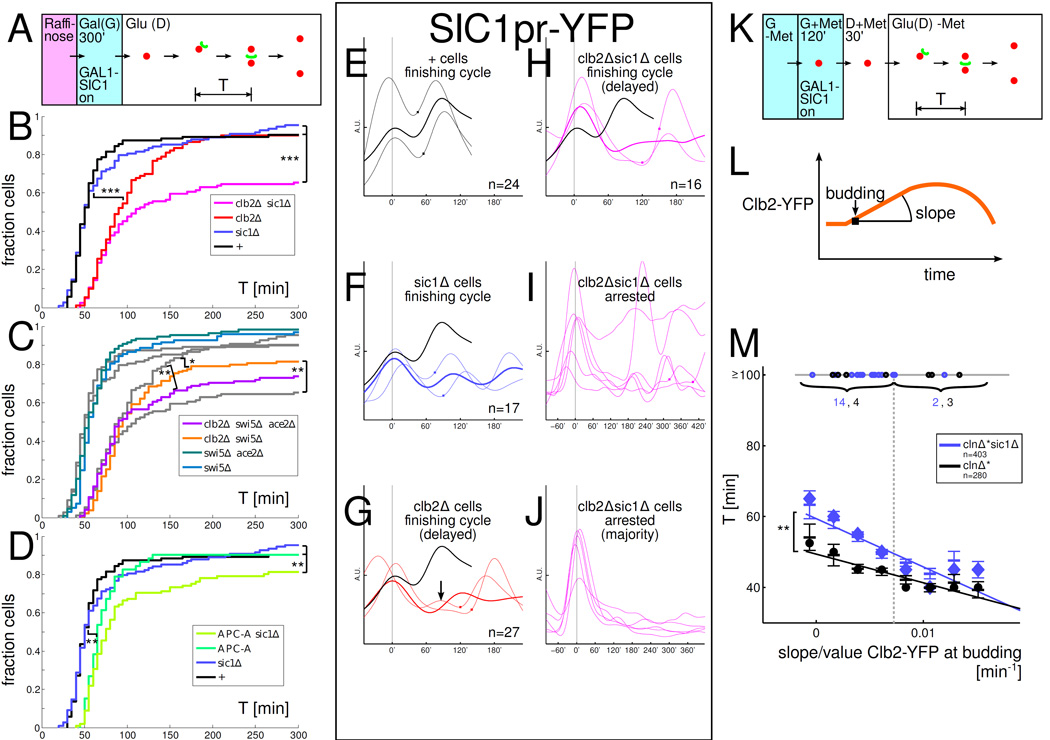
SIC1 rescues low-Clb cell cycles and some low-Clb cells show multiple SIC1pr-YFP oscillations. All strains in A-M have one copy of a GAL1-SIC1 construct. '+': WT except for GAL1-SIC1. A: Experimental set-up for panels A–J. B–D: Distribution of times T between the first budding to nuclear division in glucose (budneck marker: Myo1-GFP, nuclear marker: Htb2-mCherry). Plots from B underlaid in C in grey for comparison. E–J: A subset of low-Clb cells reveal extra peaks and oscillations in SIC1pr-YFP. Time courses aligned so that first budding after GAL1-SIC1 shut-off is at time 0'; Square symbol: time of nuclear division. Same vertical scale is used in all time course plots. EH: SIC1pr-YFP levels in cells that complete a cell cycle (note extra bump in low-Clb cell in panel G). Thin lines: sample time courses; thick lines: mean. Mean time course for '+' cells plotted in black for reference. I: Subset of arrested clb2Δsic1Δ cells 14%(6/43) show SIC1pr-YFP oscillations. J: In the majority of arrested clb2Δsic1Δ cells, strong SIC1pr-YFP activity cannot be easily detected during the arrest. K: Experimental set-up for experiment in panel M. L: Clb2-YFP was recorded in each cell using time lapse fluorescence microscopy. M: sic1Δ cells which fire Clb2-YFP more slowly take longer to finish nuclear division (and fail significantly more often) than SIC1 cells. Plot of time from budding to nuclear division T versus slope of Clb2-YFP at budding (normalized by Clb2-YFP at budding). The median Clb2-YFP slope at budding is indicated by a dashed vertical line. The circles on the horizontal line at T=100' indicate cells that took T≥100' to complete budding to nuclear division (numbers indicated). For cells with T<100', slopes were collected in 8 equal-width bins covering the range of slopes observed, the mean ± SEM is indicated by horizontal lines for each bin, and a large circle (clnΔ*) or diamond (clnΔ*sic1Δ) indicates the median in each bin; the linear regression lines for T<100' populations are also indicated. Statistical significance of differences is indicated as : p<0.1, : p<0.05, : p<0.001 (log-rank test). In B-D, the difference between strains is only statistically significant if indicated. In M, the statistical significance refers to the difference of the slopes of the linear regression lines.
To test this prediction, we lowered Clb levels by deleting the major mitotic B-cyclin Clb2, which reduces total Clb levels by about half [Cross et al., 2002]. In order for SIC1 and sic1Δ cells to start out similarly, we induce SIC1 from a Gal promoter in all strains; then, we switch off GAL1-SIC1 and measure the time from budding to nuclear division into the bud (Fig. 6 A). clb2Δ but not the sic1Δ deletion significantly lengthens cell cycle timing (Fig. 6 B), yet, similar proportions of WT, clb2Δ, and sic1Δ cells do finish (90–95%). In the clb2Δsic1Δ strain, in contrast, cells take significantly longer, and a substantial fraction (35% = 43/124) fails to finish at all. This cannot be explained by an additive influence of sic1Δ and clb2Δ since sic1Δ has no noticeable effect.
Is SIC1 uniquely important to low-Clb cells among the Swi5 cluster genes? A swi5Δ deletion partially replicates the effect of sic1Δ on clb2Δ cells while swi5Δ alone (like sic1Δ alone) has no detectable effect (Fig. 6 C). To mimic the effect of the sic1Δ deletion in the clb2Δ background by removing TFs, we have to remove both known SIC1 transcription factors, ACE2 and SWI5 (Fig. 6 C). clb2Δsic1Δ cells are statistically indistinguishable from clb2Δswi5Δace2Δ cells. So, SIC1 suffices to explain the importance of the Swi5/Ace2 cluster for cell cycle completion in low-Clb cells.
If lowering Clb levels makes the APC/C-Cdc20/Cdh1 loop less efficient, resulting in a requirement for SIC1 for efficient mitotic entry, then weakening APC/C directly should have the same effect. Low Clb levels (clb2Δ) result in incomplete APC/C phosphorylation, diminishing APC/C-Cdc20 activity [Rudner and Murray, 2000]. We tested the APC-A background, with mutated Clb2-Cdk-dependent APC/C phosphorylation sites in Cdc16, Cdc23, and Cdc27 [Rudner and Murray, 2000]. Similar to our results with clb2Δ, we observe that combination of APC-A with sic1Δ caused a significant decrease in the fraction of cells completing nuclear division, while sic1Δ and APC-A alone had no effect (Fig. 6 D). So, low Clb levels make APC/C less efficient [Rudner and Murray, 2000] and a less efficient APC/C is rescued by SIC1 to promote nuclear division.
We studied the dynamics of SIC1 firing in low-Clb cells using a SIC1pr-YFP construct. When cells do complete cell division, SIC1 firing dynamics are similar in the different strains (Fig. 6 E–H): a large rise in SIC1pr-YFP roughly coincides with nuclear division. However, a subset of clb2Δsic1Δ cells exhibits multiple SIC1pr-YFP pulses without associated nuclear division (Fig. 6 I), resembling our original observations in Clb-depleted cells (Fig. 4). This suggests that if these cells had functional SIC1, these transcriptional pulses would have released them into division. The majority of clb2Δsic1Δ cells, however, do not show clear SIC1pr-YFP pulses even if failing nuclear division (Fig. 6 J). Low (but non-zero) amounts of mitotic cyclin may be preventing SIC1pr-YFP pulsing in these cells because there is no SIC1 to complete the positive feedback loop onto itself to cause a runaway SIC1 pulse. Pinpointing the exact SIC1 dynamics which release delayed clb2Δ cells is difficult. The initial rise of SIC1 could be mainly responsible for the rescue. Interestingly, in a few clb2Δ cells, we observe an early pulse before the main SIC1 pulse (one such cell is shown in Fig. 6 G). We do not know if such a small, early SIC1 pulse matters for ultimate cell cycle completion. However, Ace2, a minor TF for SIC1 [Knapp et al., 1996], is sufficient for significant rescue in clb2Δswi5Δ cells (see above), suggesting that low Sic1 levels may be sufficient.
The experiments above involved genetic perturbations to interfere with Clb-APC/C negative feedback. In WT are fluctuations in Clb2 levels frequently sufficiently extreme to result in a SIC1 requirement at either low or high Clb2 levels? This is important to evaluate whether the effects we observe are relevant to WT physiology. We used a functional CLB2-YFP fusion protein, replacing endogenous CLB2, and correlate bud-to-nuclear division times with Clb2 expression dynamics (Fig. 6 K–M). We use Clb2 as an approximate indicator of total Clb levels because Clb2 makes up about half of total Clbs [Cross et al., 2002]. Following the mathematical modeling in Fig. 5 closely, we expect cells with slowly rising Clb2-YFP to be particularly negatively affected by the absence of SIC1. To quantify Clb2 rise, we measure the slope of Clb2-YFP at budding (Fig. 6 L). The strain backgrounds included GAL1-SIC1 (so that we could normalize SIC1 levels between sic1Δ and SIC1 strains prior to the experiment) and clnΔ* to allow synchronization. Cells are blocked in G+Met; after switching GAL1-SIC1 off in glucose, cells are released by re-inducing MET-CLN2 (Fig. 6 K).
Some outlier cells are substantially delayed in nuclear division: budding to nuclear division time T>100' (grey horizontal line at T=100') (Fig. 6 M). Among such cells in which Clb2-YFP rises slowly (to the left of the population median indicated by the dashed, grey vertical line), sic1Δ cells are significantly more likely to be substantially delayed compared to wt-SIC1 cells (7% vs. 3%, p=0.04). In contrast, in cells that fire Clb2-YFP quickly (above-median Clb2-YFP slopes), sic1Δ does not slow nuclear division (1% vs. 2%, p=0.20). The low background rate of cell cycle failure (1%–3%, except in sic1Δ cells with slow Clb2) is similar to what we have observed with other genotypes under exogenous Start control (clnΔ*, data not shown).
For the bulk of cells that divided nuclei within the movie duration, we noted first that the faster Clb2-YFP rises at budding, the faster nuclear division occurs (p<10^−6 for negative slope of linear regression lines for both sic1Δ and SIC1). This agrees with previous experiments showing a speed-up of cell cycle events with increasing Clb2 [Oikonomou and Cross, 2011].
Superimposed on this effect, sic1Δ deletion increases the time to complete nuclear division specifically in cells with slowly rising Clb2. The delay for sic1Δ cells compared to SIC1 cells is 10'–15' for slowly rising Clb2-YFP and decreases to 0'–5' for fast rising Clb2-YFP (p<0.01 for the difference in slopes of the linear regression lines). These results indicate that in typical cell cycles, SIC1 is most important for nuclear division in cells firing Clb2 slowly at Start (below median), as opposed to cells firing Clb2 strongly (above median). We conclude that SIC1 helps WT cells complete nuclear division particularly when Clb2 firing is slow. It is possible that SIC1 is important in WT cells with rapid Clb2 firing, but only for mitotic exit after nuclear division is complete; our measurements do not address this issue.
Comparison to previous experiments
It is important to compare our results with experimental results supporting the GTO model [Orlando et al., 2008, Simmons Kovacs et al., 2012, Bristow et al., 2014].
Transcriptome results for our control strain (MET-CLN2-induced clnΔ* cells), undergoing a single cell cycle, gave results highly comparable to the first cycle of the WT control in [Orlando et al., 2008] (Fig. S6 A,B), indicating good overall concordance despite numerous technical differences (RNA-seq vs. microarray, Cln-block vs. elutriation for synchronization, spacing of time points, strain backgrounds).
To test if our data analysis methods are capable of detecting oscillationswe applied our three oscillation test to the data setsfrom [Orlando et al., 2008]. They robustly detect the oscillations in agreement with the interpretations of Orlando et al., despite many analytical differences (pooling of genes for oscillation tests, statistical tests, specific oscillation test thresholds) (Fig. S6 D).
In cdc28-ts cells, transcriptional oscillations were interpreted to be completely cyclin-CDK-independent [Simmons Kovacs et al., 2012]. Our most comparable experiments (Cln,Clb depletion) showed no suggestion of any oscillatory behavior at the transcriptome (Fig. 2 C) or at the single-cell/single-gene level (Fig. 3 B,C). The discrepancy may be due to leakage of the ts mutant or to hitherto unknown Cdk1-independent but cyclin-dependent transcriptional activity. However, at least for SBF genes the latter explanation would contradict previous results [Cross and Tinkelenberg, 1991, Dirick and Nasmyth, 1991].
The report of oscillations in wt-CLN clb1-6Δ GAL1-CLB1 cells [Orlando et al., 2008] is best compared to our experiments with Cln,Clb-depleted cells with controlled pulses of MET-CLN2. Here, we see a single firing of early-expressed genes and no second firing, both at the transcriptome (Fig. 2 D) and single-cell/single-gene levels (Fig. 3 D). The ability to turn MET-CLN2 back off after kick-starting SBF/MBF transcription in Clb-depleted cells is crucial here. With endogenous CLNs as in [Orlando et al., 2008], we expect continuous transcription from SBF/MBF Start genes in the absence of CLB function [Charvin et al., 2010]; the Cln1,2-Whi5-SBF positive feedback loop should be switched on continuously, which is what we observed. Neither reintroduction of wt-CLN3 into clnΔ*clbΔ* nor expressing MET-CLN2 continuously recreated a cell-cycle-like oscillation, i.e., a rise and dip in CLN2pr-YFP below peak levels (Fig. S3 J).
In order to attain robust and reliable clearing of mitotic cyclin, we applied prolonged cell cycle arrests (‘cyclin-depletion protocol’); simple turnoff of GALL-CLB2 was not effective in depleting Clb2 since remaining Clb2 levels were comparable to ≈20–25% WT peak total Clb levels even after 2.5 h (Fig. S2 E).
In [Orlando et al., 2008], GAL1-CLB1 was shut off 45 min before elutriation and Clb1 levels were not assessed, presumably due to absence of an appropriate antibody. Based on our Fig S2E results and perhaps exacerbated by the fact that GAL1, used in [Orlando et al., 2008], is expressed more than 10× as strongly as GALL [Mumberg et al., 1994] it is reasonable to hypothesize that some significant level of Clb1 persisted in the prior experiments.
In [Orlando et al., 2008], effective Clb1 depletion was inferred from cytological failure of nuclear and cell division. Different cell cycle events are promoted at distinct Clb2 thresholds [Oikonomou and Cross, 2011]. Therefore, we considered the possibility that there could be Clb levels sufficient to regulate transcription, but insufficient for overt nuclear division.
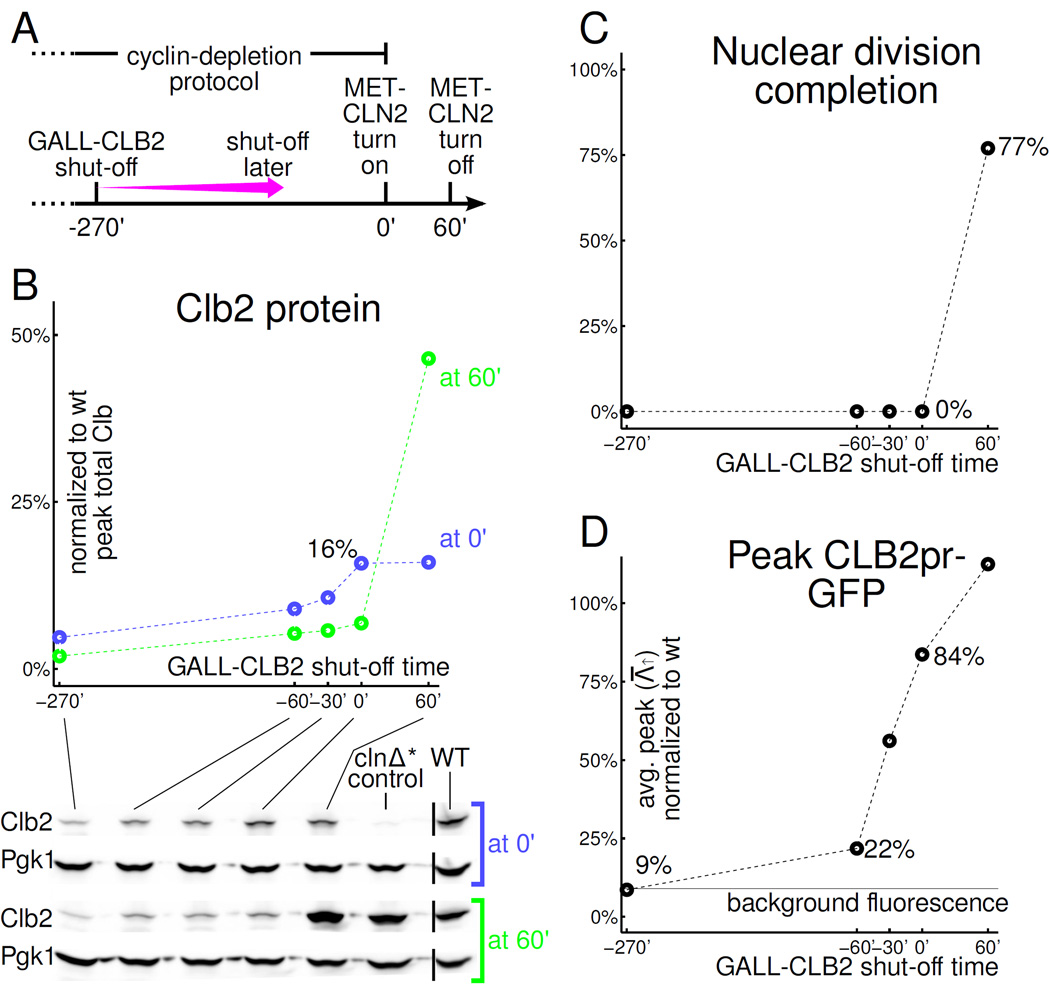
To test possible transcriptional effects of residual mitotic cyclins on transcriptional oscillations, we focus on the CLB2 cluster. With our standard protocol, (GALL-CLB2 turned off at -4.5 hrs), CLB2 cluster induction in Clb-depleted cells was near-background (Fig. 2 D, 10%, 8%, 0% of WT for ACE2, SWI5, and CDC20). By turning GALL-CLB2 off later in the protocol, we can shorten the Clb2 depletion time, keeping total arrest time constant (Fig. 7 A). This procedure will leave progressively more residual Clb2 in clnΔ*clbΔ* cells at 0' when MET-CLN2 is induced (Fig. 7 B). We then can assess both nuclear division with Htb-mCherry and CLB2pr-GFP expression, correlated with residual Clb2 levels detected by Western blot in parallel experiments.
Figure 7.
CLB2 cluster activation (CLB2pr-GFP) and cell cycle completion require different GALL-CLB2 shut-off times suggesting different thresholds. All cells are clnΔ*clbΔ*. A: In B–D, the cyclin-depletion protocol is modified by postponing GALL-CLB2 shut-off (standard protocol: −270'). MET-CLN2 is reinduced at 0'. B: Clb2 Western. Black bars indicate where the image has been spliced to remove one lane. C: 85%-90% of cells (out of 200) bud but we observe no nuclear divisions (Htb2-mCherry-marked nuclei) unless GALL-CLB2 is left on past 0' while MET-CLN2 is induced. D: CLB2pr-GFP pulses are above background levels for any GALL-CLB2 shut-off time tested except −270' (standard cyclin-depletion protocol).
We observed a dissociation between nuclear division and CLB2pr-GFP levels in this analysis: While in our standard protocol, nuclear division and CLB2pr-GFP expression were efficiently blocked, at shorter shutoff times, CLB2pr-GFP expression steadily increased, up to nearly WT peak levels, but nuclear division (Fig. 7 C,D) and DNA replication (Fig. S6 C) remained effectively blocked.
For a rough comparison to [Orlando et al., 2008]: they depleted GAL1-CLB1 for 45’ before elutriation, and observed a CLB2 cluster transcriptional pulse at 15–26% of WT, without nuclear or cell division; in our experiments, a 60’ Clb2 depletion time resulted in a CLB2pr-GFP peak 22% of WT, also without nuclear or cell division.
These results are most simply interpreted as implying much lower Clb2 thresholds for transcriptional regulation than for nuclear division, indicating that cytologically detectable failure of cell cycle events is an imperfect indicator of underlying Clb levels. We have no data to suggest actual Clb1 levels in the protocol of [Orlando et al., 2008], however.
Some (but not all) SBF-regulated genes were reported to fire in cdc20Δ-blocked cells in [Bristow et al., 2014]. We observe strong, initial repression of the SBF-regulated CLN2 promoter, consistent with the well-established ability of Clb-Cdk (which accumulates to high levels in cdc20Δ-blocked cells) to repress SBF [Amon et al., 1993] (Fig. 3 H). After long incubations, CLN2pr-GFP transcription does gradually rise; in every case, this rise is followed by overt leakage of the cdc20Δ block with cell division and arrest in the next cell cycle. Therefore, the slow rise in SBF transcription could reflect slow removal of Clbs by leaky APC/C activity or SIC1 activation. In our hands, the MET-CDC20 block is rather leaky; single-cell analysis was crucial for identifying and removing escaping cells from analysis. We have no information to suggest leakiness of the cdc20Δ block in [Bristow et al., 2014]. However, any leakage that does occur will contribute to an average increase in SBF gene expression and in our experience, leakage in cdc20Δ-blocked cells is very hard to catch in bulk culture, since the leaked cells promptly re-arrest in the next cell cycle.
Swi5 nuclear oscillations (Fig. S4 P) and SIC1 pulses (Fig. 4 J) were seen in our experiments in arrested cells but not in [Bristow et al., 2014]. These events occurred with great inter-cell timing variability, thus are nearly undetectable in a population experiment.
Discussion
Because of the broad implications of the GTO model [Simon et al., 2001, Lee et al., 2002, Orlando et al., 2008, Simmons Kovacs et al., 2012, Bristow et al., 2014] for normal and aberrant cell cycles, we sought to understand the coupling and decoupling of transcription to the CDK-APC/C oscillator. We provide a system-level overview of CDK-APC/C control of transcription, which suggests that CDK-APC/C control is strongly dominant, despite the potential for a cyclical chain of TFs to contribute to control [Orlando et al., 2008, Simmons Kovacs et al., 2012, Bristow et al., 2014].
If the CDK-APC/C state controls almost all cell-cycle-regulated transcription, global decoupling of transcription from the CDK-APC/C oscillator would be prohibited: transcription and CDK-APC/C oscillations cannot run out-of-phase in normal cycling cells. For complete cell cycle arrest, including transcriptional oscillations, checkpoints need only arrest the CDK-APC/C oscillator and do not need to target TFs directly. The GTO model could impose the need for independent checkpoint regulation of CDK-APC/C and TFs.
Lastly, we sought to elucidate potential benefits of transcriptional pulsing robustness for the few genes that escape normal CDK-APC/C order. A mathematical analysis revealed that SIC1 pulsing could, counter-intuitively, bring a benefit to low-Clb cells because the main mechanism controlling mitotic entry and exit, Clb-APC/C negative feedback, becomes inefficient. We found that SIC1 does indeed ensure cell cycle completion in cells with lowered Clb levels (either due to Clb2 deletion, or to stochastic fluctuations in Clb2 expression) or with weakened APC/C (APC-A). A contribution of Sic1 to nuclear division had not been recognized before. Like SIC1 pulsing at no-Clb arrests (Fig. 4 A,D), SIC1 rescue in clb2Δ cells is controlled by Swi5, and a number of arrested clb2Δsic1Δ cells repeatedly pulse SIC1pr-YFP (and a small number of clb2ΔSIC1 cells show two peaks per cell cycle). These cells may be attempting breakout from a low-Clb2 block by repeatedly pulsing SIC1 transcription. Beyond the requirement for Swi5, we know nothing of the mechanism for this pulsatile SIC1 expression, so at present we lack a way to address its biological significance more directly.
Experimental Procedures
Standard procedures and established disruption alleles, transgenes, and fluorescent fusions were used for strain constructions by tetrad analysis and transformation. All strains were W303 background.
Cyclin-depletion protocol (effect on clnΔ*clbΔ* cells is indicated): G-Met (cells cycling) → 180 min G+Met (cells blocked in G1) → 50 min G-Met (induce MET-CLN2, start new cell cycle) → 30 min G+Met (turn off MET-CLN2) → 270 min D+Met (turn off GALL-CLB2 near the end of the cell cycle and arrest). For +Clb2kd experiments, the protocol is as follows: R-Met (cells cycling) → 180 min R+Met (cdc20Δ arrest) → 60 min G+Met (GAL1-CLB2kd on, if exists) → 30 min D+Met (GAL1-CLB2kd off) → 300 min D-Met (MET-CDC20 on). The protocol for cdc20Δ experiments is as follows: D-Met (cells cycling) → 180 min D+Met (cdc20Δ arrest). G=galactose, R=raffinose, D=glucose, +/− Met=presence/absence of methionine.
For whole-transcriptome measurements, RNA was extracted using standard methods and cDNA libraries were prepared using the Illumina TruSeq RNA Sample Preparation Kit v2. Sequencing was carried out by Genewiz on an Illumina HiSeq2500 platform in 1×50 bp single-read configuration in Rapid Run mode.
Flowcell microscopy was performed using CellASIC technology following the manufacturer's instructions.
Image analysis, quantification, and data analysis employed custom software ([Charvin et al., 2010], this work) available upon request.
See Results section 'Detecting oscillations' for a description of the oscillation tests.
See Extended Experimental Procedures for details.
Supplementary Material
Highlights.
The CDK-APC/C oscillator controls most cell-cycle-regulated gene transcription.
This contradicts proposed global transcriptional oscillator models.
We only find three genes that oscillate during CDK-APC/C arrests.
One of them (SIC1), counter-intuitively, rescues low Clb-CDK cell cycles.
Acknowledgments
SJR thanks Nahal Mansouri for insightful discussions and Aleeza Kazmi for help with image processing. The work was supported by NIH grant 5RO1-GM078153-07 to FRC, NRSA Training Grant CA009673-36A1, and Merck and Simons Foundation Postdoctoral Fellowships to SJR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization: SJR, FRC; Methodology, writing, analysis: SJR, KP, FRC; Investigation: SJR, KP, AO, CO.
References
- Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- Bristow S, Leman A, Simmons Kovacs L, Deckard A, Harer J, Haase S. Checkpoints couple transcription network oscillator dynamics to cell-cycle progression. Genome Biol. 2014;15:446. doi: 10.1186/s13059-014-0446-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvin G, Oikonomou C, Siggia ED, Cross FR. Origin of Irreversibility of Cell Cycle Start in Budding Yeast. PLoS Biol. 2010;8:e1000284. doi: 10.1371/journal.pbio.1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KC, Calzone L, Csikasz-Nagy A, Cross FR, Novak B, Tyson JJ. Integrative analysis of cell cycle control in budding yeast. Mol. Biol. Cell. 2004;15:3841–3862. doi: 10.1091/mbc.E03-11-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman-Lerner A, Chin TE, Brent R. Yeast Cbk1 and Mob2 Activate Daughter-Specific Genetic Programs to Induce Asymmetric Cell Fates. Cell. 2001;107:739–750. doi: 10.1016/s0092-8674(01)00596-7. [DOI] [PubMed] [Google Scholar]
- Cross FR, Archambault V, Miller M, Klovstad M. Testing a mathematical model of the yeast cell cycle. Mol. Biol. Cell. 2002;13:52–70. doi: 10.1091/mbc.01-05-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR, Tinkelenberg AH. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- de Lichtenberg U, Jensen LJ, Fausbøll A, Jensen TS, Bork P, Brunak S. Comparison of computational methods for the identification of cell cycle-regulated genes. Bioinformatics. 2005;21:1164–1171. doi: 10.1093/bioinformatics/bti093. [DOI] [PubMed] [Google Scholar]
- Dirick L, Nasmyth K. Positive feedback in the activation of G1 cyclins in yeast. Nature. 1991;351:754–757. doi: 10.1038/351754a0. [DOI] [PubMed] [Google Scholar]
- Drapkin BJ, Lu Y, Procko AL, Timney BL, Cross FR. Analysis of the mitotic exit control system using locked levels of stable mitotic cyclin. Mol. Syst. Biol. 2009;5:328. doi: 10.1038/msb.2009.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase SB, Wittenberg C. Topology and control of the cell-cycle-regulated transcriptional circuitry. Genetics. 2014;196:65–90. doi: 10.1534/genetics.113.152595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp D, Bhoite L, Stillman DJ, Nasmyth K. The transcription factor Swi5 regulates expression of the cyclin kinase inhibitor p40SIC1. Mol. Cell. Biol. 1996;16:5701–5707. doi: 10.1128/mcb.16.10.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne J-B, Volkert TL, Fraenkel E, Gifford DK, Young RA. Transcriptional Regulatory Networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Lengronne A, Schwob E. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1) Mol. Cell. 2002;9:1067–1078. doi: 10.1016/s1097-2765(02)00513-0. [DOI] [PubMed] [Google Scholar]
- Lu Y, Cross FR. Periodic cyclin-Cdk Activity Entrains an Autonomous Cdc14 Release Oscillator. Cell. 2010;141:268–279. doi: 10.1016/j.cell.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll T, Tebb G, Surana U, Robitsch H, Nasmyth K. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell. 1991;66:743–758. doi: 10.1016/0092-8674(91)90118-i. [DOI] [PubMed] [Google Scholar]
- Morgan DO. The cell cycle: Principles of control. Boca Raton: New Science Press; 2007. [Google Scholar]
- Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzzey D, Gómez-Uribe CA, Mettetal JT, van Oudenaarden A. A systems-level analysis of perfect adaptation in yeast osmoregulation. Cell. 2009;138:160–171. doi: 10.1016/j.cell.2009.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Adolf G, Lydall D, Seddon A. The identification of a second cell cycle control on the HO promoter in yeast: cell cycle regulation of SWI5 nuclear entry. Cell. 1990;62:631–647. doi: 10.1016/0092-8674(90)90110-z. [DOI] [PubMed] [Google Scholar]
- Oikonomou C, Cross FR. Rising Cyclin-CDK Levels Order Cell Cycle Events. PLoS ONE. 2011;6:e20788. doi: 10.1371/journal.pone.0020788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Lin CY, Bernard A, Wang JY, Socolar JES, Iversen ES, Hartemink AJ, Haase SB. Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature. 2008;453:944–947. doi: 10.1038/nature06955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a 'reductional' anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramila T, Wu W, Miles S, Noble WS, Breeden LL. The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev. 2006;20:2266–2278. doi: 10.1101/gad.1450606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner AD, Murray AW. Phosphorylation by Cdc28 Activates the Cdc20-Dependent Activity of the Anaphase-Promoting Complex. J. Cell Biol. 2000;149:1377–1390. doi: 10.1083/jcb.149.7.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbia M, Parnell EJ, Yu Y, Olsen AE, Kretschmann KL, Voth WP, Stillman DJ. Regulation of the Yeast Ace2 Transcription Factor during the Cell Cycle. J. Biol. Chem. 2008;283:11135–11145. doi: 10.1074/jbc.M800196200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Simmons Kovacs LA, Mayhew MB, Orlando DA, Jin Y, Li Q, Huang C, Reed SI, Mukherjee S, Haase SB. Cyclin-dependent kinases are regulators and effectors of oscillations driven by a transcription factor network. Mol. Cell. 2012;45:669–679. doi: 10.1016/j.molcel.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Zeitlinger J, Gifford DK, Jaakkola TS, Young RA. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/s0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- Skotheim JM, Di Talia S, Siggia ED, Cross FR. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454:291–296. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyn JH, Johnson AL, Donovan JD, Toone WM, Johnston LH. The Swi5 transcription factor of Saccharomyces cerevisiae has a role in exit from mitosis through induction of the cdk-inhibitor Sic1 in telophase. Genetics. 1997;145:85–96. doi: 10.1093/genetics/145.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S0 cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The Phosphatase Cdc14 Triggers Mitotic Exit by Reversal of Cdk-Dependent Phosphorylation. Mol. Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Wäsch R, Cross FR. APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature. 2002;418:556–562. doi: 10.1038/nature00856. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.