Abstract
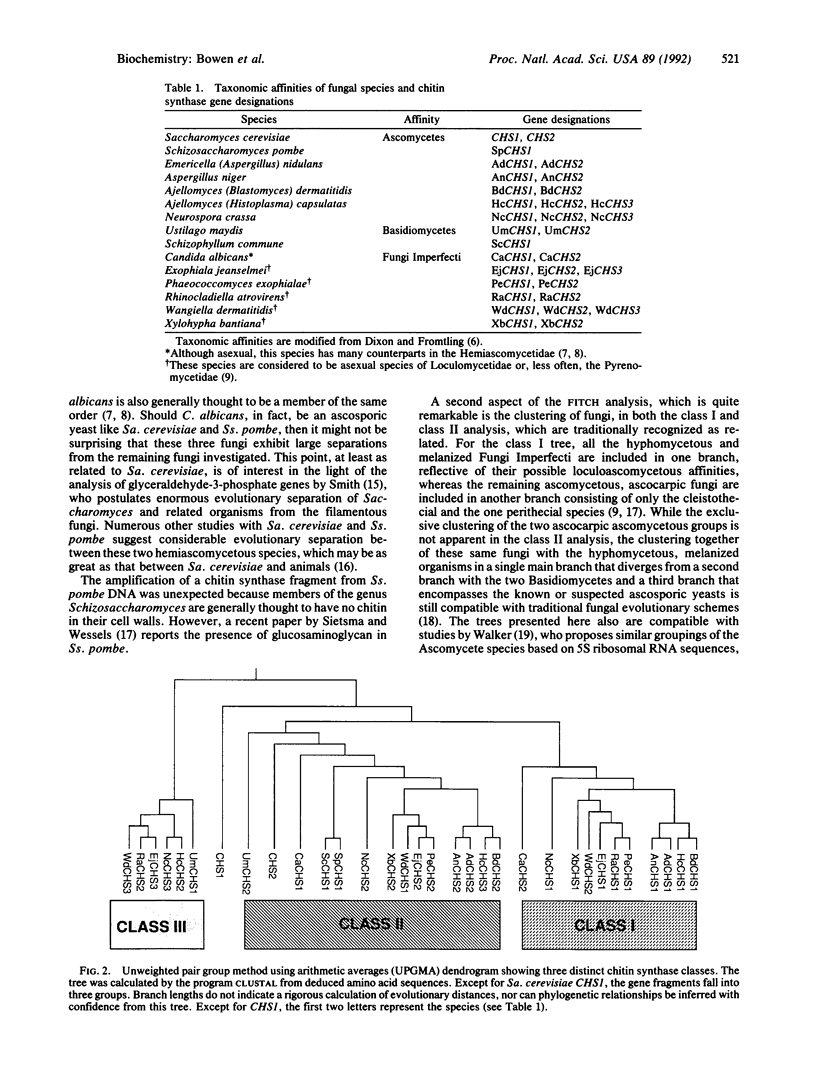
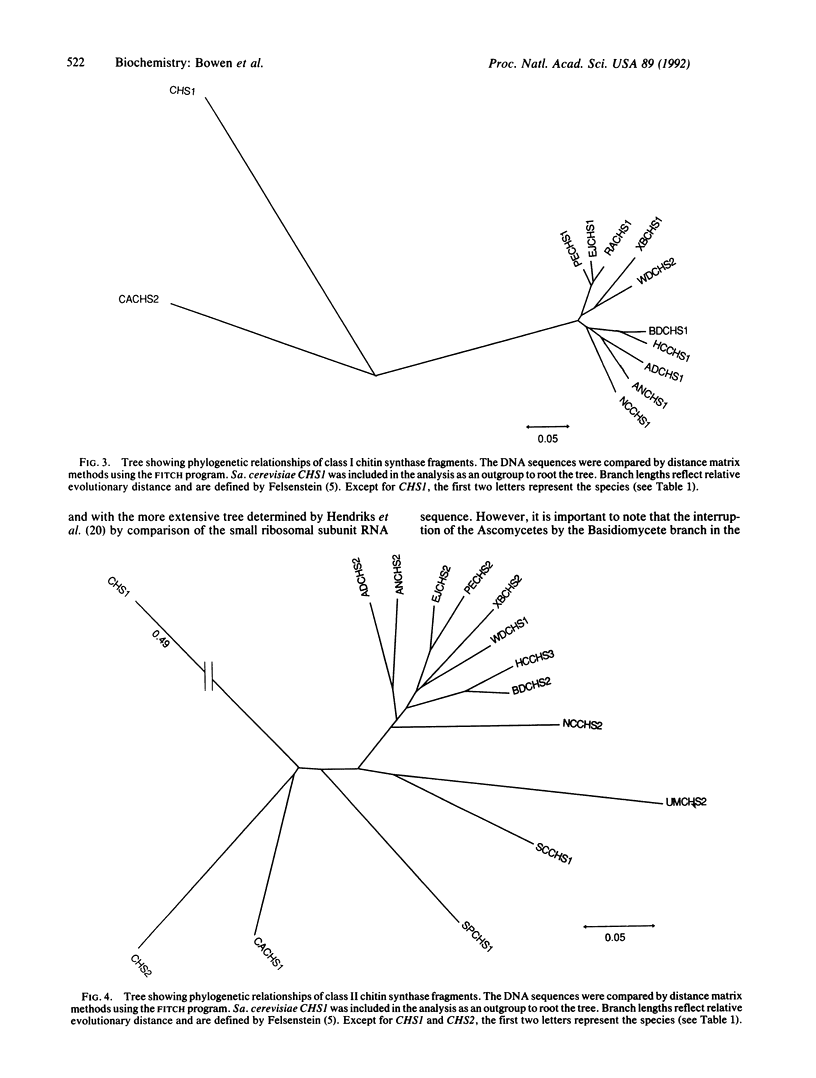
Comparison of the chitin synthase genes of Saccharomyces cerevisiae CHS1 and CHS2 with the Candida albicans CHS1 gene (UDP-N-acetyl-D-glucosamine:chitin 4-beta-N-acetylglucosaminyltransferase, EC 2.4.1.16) revealed two small regions of complete amino acid sequence conservation that were used to design PCR primers. Fragments homologous to chitin synthase (approximately 600 base pairs) were amplified from the genomic DNA of 14 fungal species. These fragments were sequenced, and their deduced amino acid sequences were aligned. With the exception of S. cerevisiae CHS1, the sequences fell into three distinct classes, which could represent separate functional groups. Within each class phylogenetic analysis was performed. Although not the major purpose of the investigation, this analysis tends to confirm some relationships consistent with current taxonomic groupings.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barns S. M., Lane D. J., Sogin M. L., Bibeau C., Weisburg W. G. Evolutionary relationships among pathogenic Candida species and relatives. J Bacteriol. 1991 Apr;173(7):2250–2255. doi: 10.1128/jb.173.7.2250-2255.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulawa C. E., Osmond B. C. Chitin synthase I and chitin synthase II are not required for chitin synthesis in vivo in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7424–7428. doi: 10.1073/pnas.87.19.7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Kang M. S., Elango N., Mattia E., Au-Young J., Robbins P. W., Cabib E. Isolation of chitin synthetase from Saccharomyces cerevisiae. Purification of an enzyme by entrapment in the reaction product. J Biol Chem. 1984 Dec 10;259(23):14966–14972. [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980 Dec;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Orlean P. Two chitin synthases in Saccharomyces cerevisiae. J Biol Chem. 1987 Apr 25;262(12):5732–5739. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sietsma J. H., Wessels J. G. The occurrence of glucosaminoglycan in the wall of Schizosaccharomyces pombe. J Gen Microbiol. 1990 Nov;136(11):2261–2265. doi: 10.1099/00221287-136-11-2261. [DOI] [PubMed] [Google Scholar]
- Smith T. L. Disparate evolution of yeasts and filamentous fungi indicated by phylogenetic analysis of glyceraldehyde-3-phosphate dehydrogenase genes. Proc Natl Acad Sci U S A. 1989 Sep;86(18):7063–7066. doi: 10.1073/pnas.86.18.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia A., Chardin P., Wittinghofer A., Sander C. The ras protein family: evolutionary tree and role of conserved amino acids. Biochemistry. 1991 May 14;30(19):4637–4648. doi: 10.1021/bi00233a001. [DOI] [PubMed] [Google Scholar]



