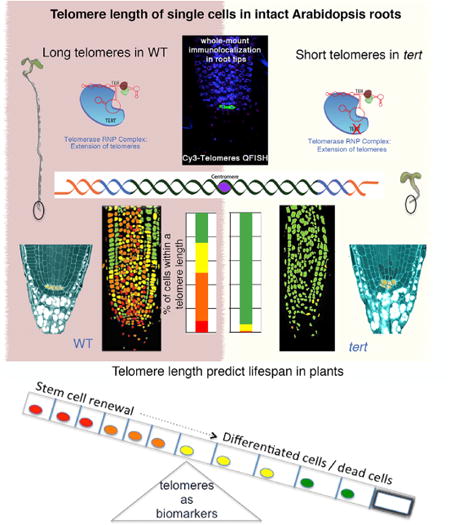
Summary
Telomeres are specialized nucleoprotein caps that protect chromosome ends assuring cell division. Single-cell telomere quantification in animals established a critical role for telomerase in stem cells, yet, in plants, telomere-length quantification has been reported only at the organ level. Here, a quantitative analysis of telomere length of single cells in Arabidopsis root apex uncovered a heterogeneous telomere-length distribution of different cell lineages showing the longest telomeres at the stem cells. The defects in meristem and stem cell renewal observed in tert mutants demonstrate that telomere lengthening by TERT sets a replicative limit in the root meristem. Conversely, the long telomeres of the columella cells and the premature stem cell differentiation plt1,2 mutants suggest that differentiation can prevent telomere erosion. Overall, our results indicate that telomere dynamics are coupled to meristem activity and continuous growth, disclosing a critical association between telomere length, stem cell function, and the extended lifespan of plants.
Graphical abstract
Introduction
Telomeres are nucleoprotein structures at chromosome ends that allow proper chromosome segregation and are essential to maintain genomic stability. Since their original discovery in maize (Zea mays) (Sax and Enzmann, 1939), telomeres have become a central focus in the study of cancer, aging, and stem cell biology in mammals. Telomeres are maintained by telomerase, a ribonucleoprotein reverse transcriptase that employs a catalytic subunit TERT to reiteratively synthesize telomeric DNA on chromosome ends. Telomerase compensates for the inability of conventional DNA polymerases to replicate the ends of linear chromosomes and thereby circumvents the erosion of telomere tracts that naturally occurs with successive rounds of cell division (Lingner et al., 1995). The study of telomerase in mice demonstrated a rate-limiting role for telomerase in longevity and regeneration capacity due to stem cell dysfunction (Blasco et al., 1997b; Flores et al., 2005), and increasing telomerase levels can enhance the regenerative capacity of cells and increase both lifespan and health span (Bernardes de Jesus et al., 2012; Tomás-Loba et al., 2008).
Telomeric chromatin for most angiosperms consist of tandem repeats of the TTTAGGG DNA sequence bound by a set of telomere binding proteins that protect the chromosome ends and that have a high structural and functional similarity with their homologs in animals (Shakirov et al., 2010; Song et al., 2008; Surovtseva et al., 2009). As in animals, Arabidopsis tightly regulates telomerase expression and enzyme activity is confined to dividing tissues/organs (Watson and Riha, 2010). The absence of telomerase activity in mutant slacking TERT causes progressive telomere shortening and aberrant shoot development (Riha et al., 2001) arguing that telomere maintenance is essential for plant viability. However, the contributions of telomerase to most fundamental aspects of plant growth and development are largely unexplored.
Conventional molecular methods are available in Arabidopsis to assess bulk telomere length and the length of telomeres on individual chromosome arms using whole plants/organs (Heacock et al., 2004), yet the precise quantification of individual telomeres within a tissue or specific organ has not been examined. These techniques established that the average telomere length ranges between 2 and 5 kb in the Columbia ecotype (Richards and Ausubel, 1988; Shakirov and Shippen, 2004), and further that telomeres must exceed a critical length threshold of approximately 1 kb for genome stability (Heacock et al., 2004).
Based on the idea that telomeres progressively shorten with successive divisions in cells lacking telomerase, confocal telomere quantitative-fluorescence in situ hybridization (Q-FISH) has been employed in animal models to trace the proliferative history of tissues and thus define the position of stem cell compartments (Flores et al., 2008; Jung et al., 2011; Martens et al., 1998). Although confocal telomere Q-FISH has provided a means of measuring telomere-length distribution along a given tissue section in animals, the Arabidopsis primary root is a superior system for imaging development in an intact organ. Its thin roots (∼150 μm) can be captured within a single confocal stack of images, with low autofluorescence. Both characteristics allow in vivo nuclear imaging of an intact organ. In the roots, the meristem divisions of the different root lineages can be traced back to the position of the stem cells, thus offering an excellent system to trace cell division history in plant organs. The stem cell niche is formed by a small group (3–7) of slowly dividing cells that form quiescent center (QC) cells surrounded by the stem cell initials (Petricka et al., 2012; Scheres et al., 2002).
For these reasons, the primary root of Arabidopsis was chosen in this study to establish a high-throughput methodology able to assess the length of individual telomeres. Our analysis in the cells of the intact Arabidopsis root apex defines a telomere distribution map uncovering the existence of telomere gradients within plant cell types and demonstrates that telomere length is tightly coupled to meristem activity. Interestingly, these results explain the dramatically reduced stem cell renewal of tert roots, further substantiating the importance of telomere length in preserving the potential for cell division of plant stem cells. Collectively, our data demonstrated that telomere length assures the continuous stem cell renewal during root growth in plants.
Results
Telomere Q-FISH Analysis in Intact Roots Enables the Quantification of Telomere Length with Tissue Resolution
Quantification of telomere length in plants has been reported using bulk tissue and organs by conventional molecular biology techniques (Fajkus et al., 1998; Riha et al., 1998), yet telomere length distribution within a plant organ has not been previously reported. In this study, we set up a whole-mount telomere Q-FISH-based (quantitative fluorescence in situ hybridization) method to quantify telomere fluorescence intensity in an intact organ with tissue resolution based on Flores et al. (2008). We used Arabidopsis root to capture confocal z stack of images within an intact organ and to quantify the telomere length of different cell layers along the longitudinal root apex (Figures 1A and 1B). This approach enables the analysis of single cells and preserves the structure of the cells (Figure S1; Movie S1).
Figure 1. A Q-FISH-Based Telomere Distribution Map in the Arabidopsis Root Apex.
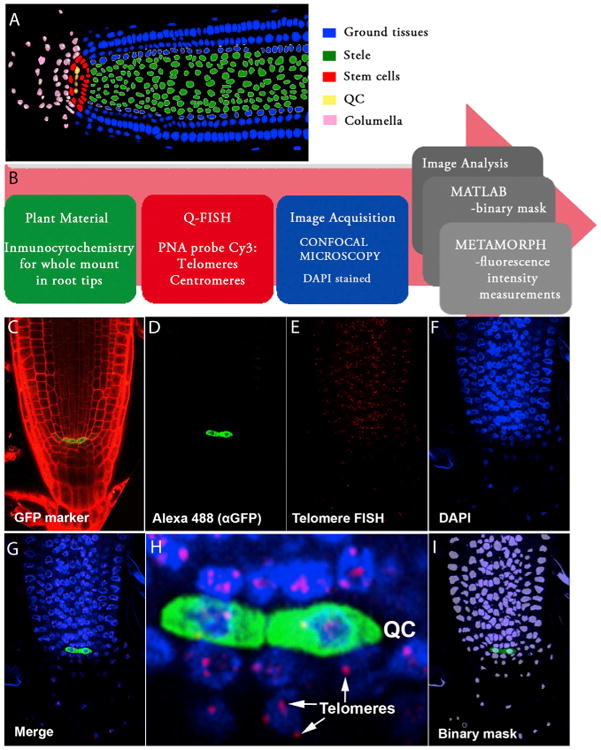
(A) Schematic representation of meristem organization in a 6-day-old Arabidopsis root. The color code identifies the different cell types: ground tissues (epidermis, cortex, endodermis, and lateral root cap) in blue, stele in green, stem cell niche in red, quiescent center (QC) in yellow, and columella cells in pink.
(B) Pipeline describing the experimental setup for whole-mount root telomere Q-FISH. Six-day-old Arabidopsis roots were subjected to a whole immunocytochemistry process using GFP markers (green box). GFP immunofluorescence was combined with confocal Q-FISH directly using a PNA Cy3-labeled telomeric/centromeric probe (red box). DAPI, Cy3, and Alexa 488 signals were acquired simultaneously into separate channels using a confocal microscope. The DAPI image was used to define the nuclear area to create a binary mask that allows isolation of each nucleus (blue box). The segmentation method was implemented using MATLAB (gray box). In the final step of the pipeline, Metamorph software was used to combine the binary DAPI mask and the matching Cy3 image. Cy3 fluorescence intensity (telomere fluorescence) was measured as “average gray values” units (a.u. of fluorescence). A code of four colors was used to classify the nuclei according to their average telomere fluorescence (gray box). A large pink arrow indicates the flow direction of the whole mount root Q-FISH procedure.
(C) Six-day-old roots stained with PI. The GFP marker (green) stains the pWOX5:GFP expression domain, coinciding with the quiescent cells.
(D–G) Confocal images showing the QC labeled by pWOX5:GFP (D), Q-FISH using a telomeric PNA probe stained with Cy3 (E), cell nuclei stained with DAPI (F), and a merged image for all three channels (G).
(H) Inset showing, with cellular resolution, the QC and surrounding cells, together with their corresponding telomeres indicated by arrows.
(I) Binary mask generated in MATLAB and used for individual nuclei fluorescence intensity quantification (see the Supplemental Information).
As individual z-planes do not enable the visualization of all centromeres/telomeres present in the nuclei, the fluorescence intensity values were normalized with the number of fluorescence spots by dividing the sum of the intensities of all the individual centromeres/telomeres observable in a given cell, by their number. The averaged spots intensity value per cell was shown to avoid the detection of changes in fluorescence caused by ploidy, and/or nuclear size (see Supplemental Information). Furthermore, a 3D model for individual cells at the root apex was built from the stack of confocal images. A semi-supervised 3D segmentation process was conducted to create a three-dimensional model of the cell in which the centromeres/telomeres detected in the layer-wise quantization process were represented by red spheres. The diameter of these spheres is proportional to the measured size of the fluorescence spots. Moreover, the cell nucleus boundaries are used to build a 3D mesh that constitutes a faithful virtual reconstruction of the cell nucleus (Figure S1; Movie S2).
Initially, whole-mounted immunofluorescence using cell-specific GFP markers was used to visualize the position of specific cell types in the root under a confocal microscope. To mark the quiescence center (QC) or the bona fide stem cells, which are located at the median longitudinal plane of the root apex, we used the WUSCHEL-related homeobox 5 pWOX5:GFP (Figures 1C and 1D, rendered in green) (Sarkar et al., 2007). Subsequently, we performed quantitative FISH with a plant-specific telomere fluorescent peptide nucleic acid (PNA) probe (Cy3-[CCCAGGG]) to visualize and quantify individual telomere fluorescence signals at a cell level in the Arabidopsis root (Figure 1E). A merged image of GFP, Cy3, and DAPI channels enabled the visualization of telomeres inside individual nuclei of the root apex (Figures 1D–1G). The GFP labeling of QC allowed the precise identification of the stem cell compartment (Figure 1H; Movie S1). In the confocal Z-scan at the median longitudinal plane, DAPI-staining of the nuclei was used for nuclear area segmentation and binary mask generation (Figure 1I; Supplemental Information). Finally, the fluorescence quantification of individual telomere spots inside each nucleus in the confocal Z-scan was achieved by merging the binary mask with the Cy-3-labeled confocal image and using the Granularity module of the Metamorph platform (Supplemental Information). Collectively, this method allows the precise quantification of telomere length in an intact plant organ with cellular resolution.
A Telomere-Length Distribution Map for the Arabidopsis Primary Root Apex
The combination of immunofluorescence and telomere Q-FISH with quantitative imaging technology revealed a telomere-length distribution map for the Arabidopsis root apex (n = 2,541 nuclei) (Figure 2A). We found telomere-length heterogeneity between the different cells in the root meristem, suggesting that telomere length may be coupled to specific cells or cellular activities. The same pattern was observed among all individuals tested in our study (see Experimental Procedures; Figure S2). The fluorescence values representing individual telomere length per cell were quantified and plotted into frequency histograms and percentiles were calculated (Figures 2B and 2I; Supplemental Information). The value for the average telomere fluorescent intensity calculated was 636 ± 7 arbitrary units of fluorescence (a.u.f.), with 8% of the cells being significantly longer than their counterparts in the wild-type (WT) root (Figures 2A, 2B, and 2I). Next, we used Arabidopsis tert mutants to validate the ability of this technology to detect changes in telomere length. To this end, successive generations of Arabidopsis tert mutants (generations 3–5 [G3–G5] [Riha et al. 2001]) were subjected to the telomere Q-FISH analysis. The significant telomere shortening observed in successive generation of tert mutants (Figures 2C–2I; p < 0.005) further validates the sensitivity of the telomere Q-FISH method in the root. The reduction in the average telomere length in the root apex of G3 tert (524 ± 7 a.u.f.; Figures 2C and 2D) and G4 tert (475 ± 9 a.u.f.; Figures 2E and 2F) was even more dramatic in the roots of G5 tert (250 ± 8 a.u.f.; Figures 2G and 2H) compared to WT plants (636 ± 7 a.u.f.; Figures 2A, 2B, and 2I; Figure S2).
Figure 2. Telomere-Length Analysis by Whole-Mount Root Q-FISH of Successive tert Mutant Generations.
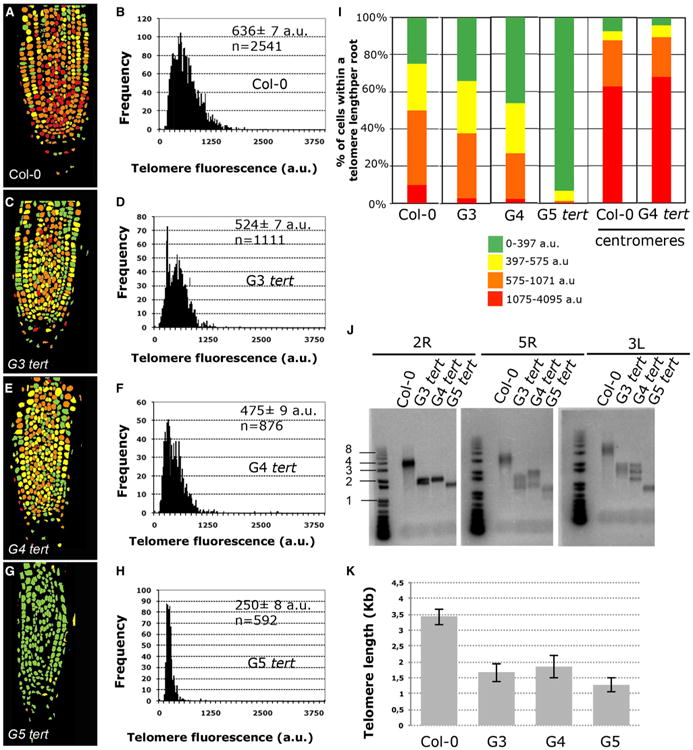
(A, C, E, and G) Representative telomere-length pseudo-color images in 6-day-old roots of WT (A), G3 tert (C), G4 tert (E), and G5 tert (G).
(B, D, F, and H) Telomere Q-FISH histograms showing telomere fluorescence frequencies in WT (B), G3 tert (D), G4 tert (F), and G5 tert plants (H). Average telomere fluorescence and SEM are indicated. See also Figure S2.
(I) Percentage of cells showing a given telomere fluorescence in WT, G3 tert, G4 tert, and G5 tert and centromere fluorescence in WT and G4 tert plants. Note the significant enrichment in cells with short telomeres in tert mutants (green color) and also the lack of changes in centromere fluorescence (see also Figures S1 and S2).
(J) PETRA analysis of telomeres on 2R, 5R and 3L chromosome arms in WT and (G3–G5) tert mutant generations. A telomeric probe was used to detect PETRA products (see Experimental Procedures). See also Figure S2.
(K) Quantification of the average telomere length in Kb of PETRA bands of different chromosome arms of DNA extracted from roots of WT and (G3–G5) tert plants. The data shown in (B), (D), (F), (H), and (I) were collected by pooling seven individuals plants of WT Col-0, three G3 tert, three G4 tert, and three G5 tert.
Notably, telomere-length heterogeneity was lost in later-generation tert mutants, pointing to eventual exhaustion of telomere signals owing to the prolonged absence of telomerase activity (Figure 2I). Notably, in G5 tert roots a very small fraction of cells (1%) had telomeres longer than the rest of the cells in the root apex (Figure 2I; represented in red). This phenotype was even more dramatic in G6 tert roots (Figure S3).
Next, we validated the differences in telomere length observed using telomere Q-FISH in tert mutants by performing an independent technique based on qPCR. To this end, we used primer extension telomere repeat amplification (PETRA), which allows for the quantification of telomere length of individual chromosome arms (Heacock et al., 2004) in bulk tissues. PETRA was performed on DNA extracted from root tips to measure the telomere length for the right arm of chromosomes 2 (2R) and 5 (5R) and the left arm of chromosome 3 (3L). PETRA products confirmed telomere shortening in roots from successive generations of plants lacking TERT (Figures 2J and 2K). As the fluorescence intensity resulting from telomere Q-FISH is directly proportional to the telomere length (Vera and Blasco, 2012), we used PETRA values to convert a.u.f. values into kilobases. To this end, we plotted average telomere fluorescence values against the sizes of PETRA fragments in WT and G3–G5 tert mutants. Linear regression analysis estimated that 1 kb corresponds to 215 a.u.f. (Figure S2). We observed that the telomeric distributions were not normal, with positive skewness and with minimal values above 75 a.u.f. This cutoff might be due to an optimal telomere length value required for the normal functioning of the cell and not by our method, since our technique is sensitive enough to detect telomeres in G6 tert mutants that were shown to be extremely short by PETRA experiments (Figure S3).
Finally, to verify that the observed differences in telomere length did not reflect changes in probe accessibility,a PNA probe specific for the Arabidopsis centromere repeat sequence (Cy3-GACTCCAAAACACTAACC) was applied following the same methodology (Experimental Procedures; Supplemental Information). Centromere fluorescence intensity of WT and G4 tert roots was quantified (Figure 2I). The average fluorescence intensity of WT plants (1,274 ± 17 a.u.f.; n = 1,088 nuclei) was compared to G3 tert (1,601 ± 26 a.u.f.; n = 693 nuclei) and to G4 tert (1,305 ± 19 a.u.f.; n = 753 nuclei; Figure S1). The absence of significant changes in centromere intensity between the different generations of root cells confirms that the observed changes in telomere intensity are not caused by differences in the hybridization procedure but instead reflects progressive telomere shortening owing to telomerase deficiency. Furthermore, our data reveal that TERT maintains heterogeneous telomere length distribution within the root apex.
Cells within the Stem Cell Compartment Display the Longest Telomeres
To further investigate the origin of the telomere length variability observed in WT primary root and to check whether it can be attributed to specific cellular activities, we measured the telomere length in different root cell types.
The Arabidopsis root system offers a set of specific cell-type markers based on promoter-GFP fusions that can be used to trace the location of different root cell types (Figure S4). Using these markers, we analyzed the telomere length in the outer cell layers (ground tissues), the stele (inner cells layers), the stem cell niche (SCN), and the columella cells (Figure 1A). Interestingly, the cells with the longest telomeres were enriched at the position of the known SCN (791 ± 36 a.u.f.; p < 0.05) (Figure 3A), although no significant differences in telomere length could be detected between the QC and the surrounding stem cells (Figure S2). The telomere length of the SCN was undistinguishable from that of the columella cells (771 ± 32 a.u.f.), which differentiated after a single columella stem cell division (Scheres et al., 2002; Figure 3B). Telomere length was significantly shorter in the stele (674 ± 9 a.u.f., p < 0.05) (Figure 3C) and the ground tissues (578 ± 9 a.u.f., p < 0.05) (Figure 3D). Next, we analyzed the stem cell niche telomere-length distributions for G3–G5 tert mutants, which appeared increasingly shorter than those of the corresponding WT controls (Figure 3E; p < 0.001). Remarkably, no differences in average telomere length were observed for the different cell types of G5 and G6 tert mutants, consistent with the loss of heterogeneity shown in the tert mutant heatmap (Figure 3F; Figure S3).
Figure 3. The Longest Root Telomeres Are Mapped in the Stem Cell Niche.
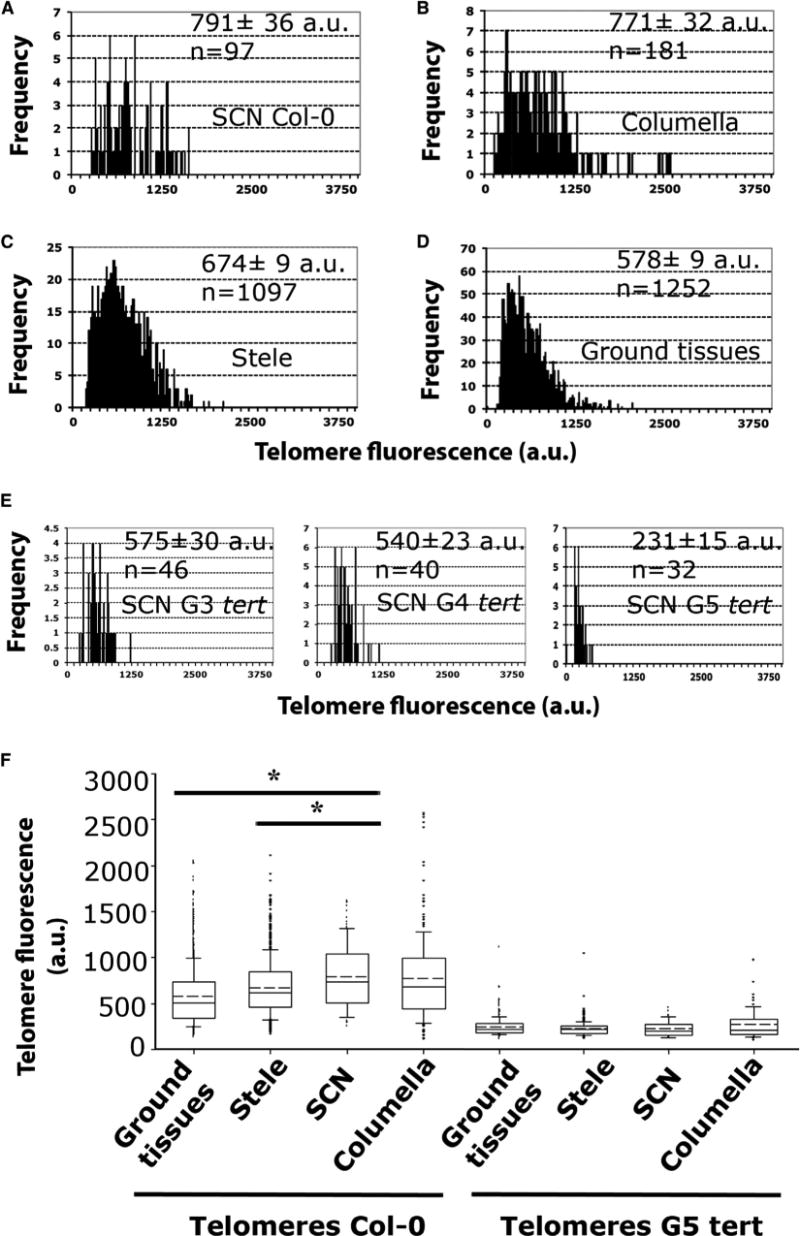
(A–D) Q-FISH histogram showing telomere fluorescence frequencies in the different cell types of Col-0 root: stem cell niche (SCN) (A), columella (B), stele (C), and ground tissues (D). Average telomere fluorescence and SEM are indicated. See also Figure S2.
(E) Q-FISH histograms of G3, G4, G5 tert stem cell niche. Note the high difference in telomere length compared to Col-0 stem cell niche.
(F) Boxplots showing the heterogeneity between cell types in telomere-length distributions of Col-0 compared to G5 tert. Boxes represent the interquartile range (25th–75th percentiles, median indicated by the black horizontal line, average indicated by the dashed line) of the distribution, whiskers extend to the 10th and 90th percentiles and outliers are represented by black dots. n represents the number of nuclei pooled for the histograms from seven individual Col-0 plants and from three plants for each G3, G4, and G5 tert mutants. For Col-0 and G5 tert roots, SCN telomere-length values were compared to ground tissues, stele, and columella values. Statistical significant differences were observed in Col-0 only between SCN and stele and between SCN and ground tissues. In the G5 tert mutants, no differences were observed between cell types. *p < 0.05.
These differences in telomere length between different cell populations within the Arabidopsis root suggest the use of whole-mount telomere Q-FISH as a powerful method to visualize telomere length distribution in the Arabidopsis roots that can be associated with specific cells and/or cellular activities, such as telomerase activity. The lack of differences in telomere length between SCN and columella cells suggests that telomere length correlates to the number of cell division prior differentiation.
Telomerase Sustain Cell Division at the Root Meristem
Previous studies showed that telomerase activity is present in rapidly dividing plant cells but undetectable in differentiated tissues (Fitzgerald et al., 1996, 1999). Here, we sought to investigate the functional consequences of critical telomere shortening owing to telomerase deficiency in the potency of meristematic cells in Arabidopsis. We first confirmed abrogation of telomerase activity in the tert-deficient Arabidopsis roots by using the TRAP (telomere repeat amplification protocol) assay. As expected, the root tips of 6-day-old WT seedlings exhibited telomerase activity, which was undetectable in G4 tert mutants (Figure S5). To test whether TERT is required post-embryonically to restore telomere shortening associated with divisions during the primary root growth, we analyzed meristem development in roots from tert mutants (Figures 4A–4D). Concomitant with the loss of telomerase activity, successive generations of tert (G4–G6) exhibited a progressive reduction of root growth and meristem size compared to WT (Figures 4B and 4D). Next, we studied the expression of D-box pCYCB1;1:GFP reporter (González-Garcia et al., 2011; Ubeda-Tomás et al., 2009), which marks proliferating cells, and performed immunostaining using the cytokinesis-specific syntaxin KNOLLE (Völker et al., 2001) in WT and increasing generation of tert mutant roots. We observed that tert-deficient roots showed a reduction in the number of mitotically active cells, as marked by pCYCB1;1:GFP (Figures 4J and 4L) as well as in the number of cell plates labeled by anti-KNOLLE antibodies (Figures 4H and 4K) with increasing plant generations in contrast to WT (Figures 4G and 4I). Furthermore, late-generation tert mutants displayed increased levels of the plant-specific cell-cycle inhibitor pICK2/KRP2:GUS (De Veylder et al., 2001) as compared to the WT (Figures 4E and 4F). To further confirm a relationship between telomere length and meristem activity, we studied roots with null mutation in KU70, a negative regulator of telomere length, and thus presenting longer telomeres than WT plants (Riha et al., 2002). Interestingly, we found that KU70 deficiency leads to both longer telomeres and increased meristem size relative to WT roots (Figure S3, p < 0.005). Together, these results indicate that telomere length is linked to meristem potency in plants.
Figure 4. Active Telomerase Is a Requirement to Preserve Cell Division during Root Growth.
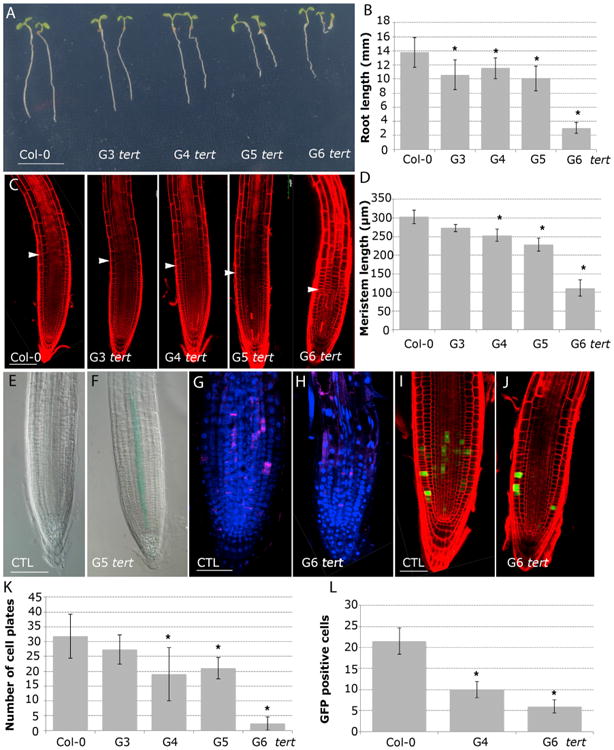
(A) Morphology of 6-day-old seedlings of WT and (G3–G6) tert plants. Scale bar, 10 mm. (See also Figure S3.)
(B) Root-length measurements of (G3–G6) tert seedlings compared to the WT. Asterisks denote a statistically significant difference with WT (p < 0.005).
(C) Confocal images of 6-day-old WT and (G3–G6) tert roots stained with PI. Arrows mark the boundary between the proximal meristem and the elongation zone of the root. Scale bars, 100 μm.
(D) The meristem length of 6-day-old roots of (G3–G6) tert mutant generations compared to WT. Asterisks indicate significant differences relative to WT for each day (p < 0.005).
(E and F) GUS staining of 6-day-old roots expressing pICK2/KRP2:GUS (E) and G5 tert; pICK2/KRP2:GUS (F). Scale bar, 50 μm.
(G and H) Whole-mount immunofluorescence using anti-KNOLLE antibodies in WT (G) and G6 tert (H). Scale bar, 50 μm.
(I and J) pCYCB1;1:GFP expression in WT roots (I) and G6 tert (J). Scale bar, 50 μm.
(K) Number of cell plates calculated in whole-mount immunofluorescence with anti-KNOLLE antibodies present in (G3–G6) tert seedlings compared to the WT.
(L) Number of GFP cells in the meristem of pCYCB1;1:GFP (p < 0.001) and G4 tert and G6 tert seedlings (p < 0.001). Asterisks denote a statistically significant difference with the WT (p < 0.001).
Telomere Length Sets a Replicative Limit in the Stem Cells
Our observations showing that cells with the longest telomeres are enriched at the root stem cell compartment (Figure 3) together with the loss of meristem activity of tert mutants (Figure 4) prompted us to investigate the impact of telomere length on plant stem cell function. Microscopic analysis of roots revealed that, relative to WT, tert mutants displayed striking differences in the anatomy of the stem cell niche. We observed an increased cell division rates in the QC of tert mutants (Figures 5A–5G). In particular, the majority of G6 tert plants (86%) had additional QC divisions while only 7% of WT plants showed this phenotype (Figure 5J). Concomitantly, confocal images of modified pseudo-Schiff (mPS)-PI-stained roots revealed the presence of starch granules in former columella stem cells, indicative of increased stem cell differentiation dynamics in tert mutants (Figures 5B–5E and 5K), whereas in the WT starch granules were normally absent from columella stem cells (Figures 5A and 5K). Consistent with these phenotypic defects at the stem cell niche, tert mutants exhibited an altered expression of QC-specific marker pWOX5:GFP (Sarkar et al., 2007) (Figures 5F and 5G) and the death of stem cells (Figures 5H and 5I). The cell death phenotype worsened in late tert generations so that propidium iodide (PI) staining was present in 100% of G6 tert mutants analyzed (Figure 5L).
Figure 5. Telomere-Length Gradients Control Stem Cell Function in the Root Apex.
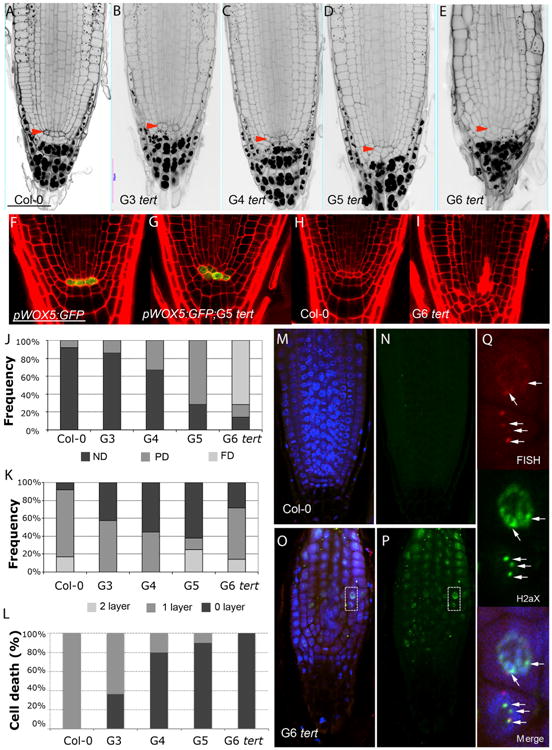
(A–E) mPS-PI-stained root tips of Col-0 (A) and G3 tert (B), G4 tert (C), G5 tert (D), and G6 tert (E) roots. Col-0 roots show normally one tier of columella stem cells (CSC). Red-arrow indicated QC cells.
(F and G) Confocal images of primary roots expressing pWOX5:GFP in the QC cells in WT (F) and G5 tert mutants (G).
(H and I) Cell death in the stem cells and adjacent daughter cells detected in WT (H) and G6 tert roots (I).
(J) Frequency distribution of QC division in (G3–G6) tert seedlings compared to the WT. PD, partially divided; ND, non-divided; FD, fully divided.
(K) Quantitative analysis of the effects of telomere length on CSC differentiation. Frequency distribution of the number of cell layers is given between the QC and the first differentiated columella cells that contain starch granules. Gradual differentiation of CSC in tert mutants is revealed by the frequency of roots without CSC layer.
(L) Cellular damage at the stem cells and vascular descendants was scored by counting intense PI-stained roots of WT and (G3–G6) tert plants.
(M and N) Immunostaining with γ-H2AX antibodies and telomere Q-FISH, cell nuclei stained with DAPI, telomeric PNA probe stained with Cy3, γ-H2AX foci colored in green, and a merged image for all the three channels of the WT.
(O and P) Immunostaining with γ-H2AX antibodies and telomere Q-FISH, cell nuclei stained with DAPI (blue), telomeric PNA probe stained with Cy3, γ-H2AX foci colored in green, and a merged image for all the three channels of the G6 tert. Note: for better visualization the laser intensity of the Cy3, channel was increased.
(Q) Inset shows magnification (dotted box) revealing the colocalization of γ-H2AX foci with the Cy3-telomere probe (white arrows). Due to the extremely short telomeres of G6 tert mutants, these images were captured by increasing the confocal laser power.
Similar to what has been described for mammals (d'Adda di Fagagna et al., 2003; Herbig et al., 2004), plant telomere dysfunction generates a DNA-damage response (DDR) that activates ATM/ATR kinase pathways and results in programmed cell death (PCD) (Boltz et al., 2012). To assess early DDR responses dependent on ATM/ATR kinases, we analyzed the phosphorylation of g-H2AX (Amiard et al., 2011). Confocal immunofluorescence using γ-H2AX antibodies in G6 tert roots revealed the presence of γ-H2AX-labeled foci co-localizing with telomeres (the so-called TIFs or telomere-damage-induced foci) in the majority of living cells at the G6 tert mutants root meristem (Figures 5O and 5P and inset in Figure 5Q) compared to the WT controls where the labeling with γ-H2AX was undetectable (Figures 5M and 5N). These results show that telomerase preserves genomic stability by preventing critical telomere loss and the activation of DDR downstream signaling events that cause stem cell loss and meristem exhaustion.
Telomere Q-FISH Reveals Longer Telomeres in plt1 plt2 Mutants
To further investigate whether cell differentiation can prevent telomere erosion and how telomere attrition affects the behavior of different stem cells in the root, we analyzed telomere length in plt1 plt2 mutants (Aida et al., 2004). PLETHORA (PLT) transcription factors are central regulators of stem cell differentiation and meristem maintenance in the Arabidopsis root apex. Mutations in PLT cause premature stem cell differentiation, leading to the formation of dramatically shortened, aberrant roots (Figures 6A, 6B, and S6) in agreement with Aida et al. (2004) and Galinha et al. (2007). Strikingly, telomere Q-FISH analysis in whole-mounted roots of plt1 plt2 revealed a significant increase (p < 0.001) in average telomere fluorescence (1,214 ± 32 a.u.f.; n = 324 nuclei; n = 3 roots; Figures 6G and 6H) compared to WT (Ws-2) plants (934 ± 14 a.u.f.; n = 1,152 nuclei; n = 3 roots; Figures 6E and 6F). These results were confirmed molecularly by TRF (Figure 6C) and PETRA assays (Figure 6D). The increase in telomere length in plt1 plt2 plants relative to WT can be explained by the reduced replicative history of plt1 plt2 cells before they undergo differentiation (Aida et al., 2004).
Figure 6. Early Differentiation in the Meristems of plt1 plt2 Mutants Prevents Telomere Erosion in the Primary Root.
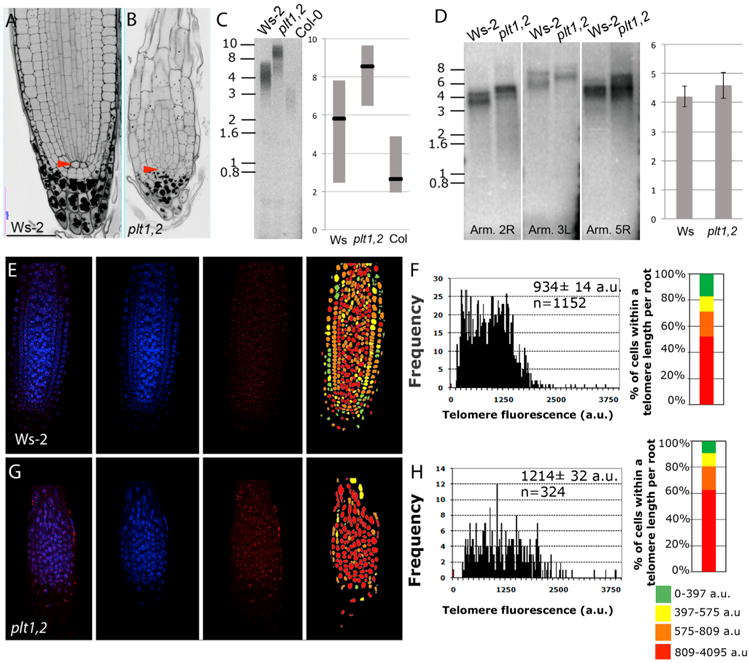
(A and B) mPS-PI-stained root tips of 6-day-old WT (Ws-2) (A) and plt1 plt2 (B) roots. Scale bars, 100 μm. See also Figure S6.
(C) TRF analysis in shoot of WT (Ws-2) and plt1 plt2 mutants. The blot was hybridized with a radiolabeled telomeric (T3AG3)4 probe. Molecular weight markers and a schematic representation of the chromosome length are indicated.
(D) PETRA analysis of the 2R, 5R, and 3L telomeres in WT (Ws-2) and plt1 plt2. A telomeric probe was used to detect PETRA products. Molecular weight markers are indicated. Quantification of the average telomere length in kilobases of PETRA bands of different chromosome arms in WT (Ws-2) and plt1 plt2.
(E) Q-FISH in Ws-2 root telomeric PNA probe stained with Cy3, cell nuclei stained with DAPI, and a merged image for both channels. Representative telomere-length pseudo-color images of WT. Nuclei are colored according to their average telomere fluorescence in a.u.
(F) Q-FISH histogram showing telomere fluorescence frequencies in Ws-2. Average telomere fluorescence, SEM and the number of nuclei n, pooled from three individual Ws-2 plants, are indicated.
(G) Q-FISH in plt1 plt2 root telomeric PNA probe stained with Cy3 cell nuclei stained with DAPI and a merged image for all the two channels. Representative telomere-length pseudocolor images of WT. Nuclei are colored according to their average telomere fluorescence in a.u. See also Figure S6.
(H) Q-FISH histogram showing telomere fluorescence frequencies in plt1 plt2. Average telomere fluorescence and SEM are indicated. Average telomere fluorescence, SEM, and the number of nuclei n, pooled from three individual plt1 plt2 plants, are indicated.
Discussion
The plant meristem sustains the production of cells through an organismal lifespan that reaches thousands of years in some plant species. Whether telomeres contribute to the replicative senescence in plants has been subject of a long-standing controversy (Gan, 2003; Watson and Riha, 2011). In this study, we integrated genetic, cellular, and molecular tools to dissect the contribution of telomere maintenance to plant stem cell renewal. We first describe here that, similar to that found within the normal architecture of mammalian tissues (Flores et al., 2008; Vera and Blasco, 2012), telomere length is not uniformly distributed among root cell types in the meristem of Arabidopsis. Instead, cells with the longest telomeres are enriched at the known stem cell compartments, and proper telomere maintenance in these compartments is essential for their ability to sustain meristem growth.
In animals, gradual telomere shortening in the absence of telomerase is proposed to act as mitotic clock that limits cell proliferation capacity (Harley et al., 1990). While such a mechanism is useful to inhibit rapid growth of tumors, telomere attrition in stem cells exhausts their proliferation capacity and contributes to organismal aging (Blasco et al., 1997a). In plants, telomerase activity is associated with all proliferative organs and is absent in fully differentiated tissues such as stem and leaves (Riha et al., 1998) suggesting that differential expression of telomerase influences plant development (Gan, 2003; Thomas, 2013). Our analysis documents that meristem activity and stem cell function are intimately connected to the ability of telomeres to maintain genomic stability in plants.
Here, we exploited the amenability of experimenting with Arabidopsis primary roots to address how telomere-length dynamics is coupled to plant meristem development. We found that telomerase activity is maintained post-embryonically in specific root cells in the root apex together with the preferential transcription of TERT mRNA in specific cells at the meristem (Brady et al., 2007) supporting a role for telomerase in maintenance of meristem growth. In the root, telomere length defines two important features of root growth and development: meristem division and stem cell replenishment. Both are essential to ensure the reproductive phase of development and lifespan in plants (Mencuccini et al., 2005). We found that telomere shortening in Arabidopsis roots causes cell-cycle arrest that is associated with phenotypic changes resulting in reduced root growth as shown by a marked reduction of meristem cell number, reduced mitotic activity, and the accumulation of the cell-cycle inhibitors ICK2/KRP2. These changes, together with the presence of telomere-localized γ-H2AX in the meristematic cells, support a model in which telomere length sets a replicative limit for a functional and living meristem in plants.
Additional evidence supporting the hypothesis that cell division can be recorded by telomere length in plants is provided by the longer telomeres in the roots of stem cell mutants plt1 plt2, which have undergone premature differentiation (Aida et al., 2004). While short telomeres limit cell divisions in the meristem, long telomeres cannot prevent cell differentiation as indicated by the increased telomere length in differentiated columella cells in WT and plt1 plt2. Rather plt1 plt2 mutants maintain their regenerative potential when their root tips are excised (Galinha et al., 2007). Thus, preserving telomere length during the division of stele cells appears to be a critical mechanism controlling root growth and development.
Intriguingly, we found that short telomeres in late-generation tert mutant plants show an increased frequency of QC division. These unscheduled divisions resembled those of plants subjected to genotoxic stress consistent with known responses of the QC to DNA damage (Cruz-Ramírez et al., 2013; Vilarrasa-Blasi et al., 2014) and reflecting the need of telomerase to ensure stem cell renewal in plants. Moreover, in tert stem cells, the absence of telomeric γ-H2AX foci and the increase of cell-cycle inhibitors pICK2/KRP2 argue that critically short telomeres activate an irreversible DNA damage signal that not only promotes death but also increase susceptibility to DNA damage for stem cells relative to their descendants in the meristem (Fulcher and Sablowski, 2009). This proposition is consistent with current models for premature senescence of mammalian stem cells (López-Otín et al., 2013; Rossi et al., 2008), uncovering the potential of plants as a model for studying telomere length control of stem cell function in eukaryotes. Further analysis of telomere-length gradients can reveal new insights into developmental circuitry and functional stem cell pools, and also serve as a basis for mechanistic comparison of plant and animal regeneration and lifespan.
Finally, while there are no technical constrains that limit the quantification of telomeres by Q-FISH in additional plant species, this technology opens new possibilities for the study of meristem and stem cell function in other plant systems, especially in perennial species.
Experimental Procedures
Plant Material and Microscopy
Arabidopsis thaliana (L.) Heyhn. seed accessions Columbia (Col-0), Wassilewskija (Ws-2), plt1 plt2 mutants (Aida et al., 2004; Galinha et al., 2007), tert mutants (Riha et al., 2001), ku70 (Riha and Shippen, 2003), and all marker lines pWOX5:GFP (Sarkar et al., 2007), pGL2:GFP (Lin and Schiefelbein, 2001), pSCR:GFP (Sabatini et al., 1999), pWOLGFP (Mähönen et al., 2000), QC12 (Lee et al., 2006), DR5:GFP (Ulmasov et al., 1997), pARF7:ARF7-GFP (Rademacher et al., 2011) were used in this study. Seeds were surface-sterilized in 35% sodium hypochlorite, vernalized 72 hr at 4°C in darkness, and grown on vertically oriented plates containing 1 × Murashige and Skoog (MS) salt mixture, 1% sucrose, and 0.8% agar. Plates were incubated at 22°C and 70% humidity under long-day conditions (16 hr light/8 hr dark).
A FV 1000 confocal microscope (Olympus) was used throughout the study. Roots were stained in 10 μg/ml propidium iodide (PI) for 2–5 min, rinsed, mounted in dH20, and visualized after excitation by a Kr/Ar 488-nm laser line. PI and GFP were detected with a band-pass 570- to 670-nm filter and 500- to 545-nm filter, respectively. For the yellow and cyan fluorescent protein GFP-tag reporter, the excitation wavelengths were 488 nm fluorescence collected in the ranges of 493-536 nm (rendered in green). For every fluorochrome, image z stacks were taken with a sampling distance of 1 μm along the z axis and 200 μm in the x, y direction. To standardize the fluorescence between samples, the same exposure time was used for all Cy3 images. Starch granules in columella cells were visualized by a modified pseudo-Schiff (mPS)-PI staining method (Truernit et al., 2008).
Immunofluorescence and Q-FISH
Whole-mount immunolocalization in 6-day-old roots was performed as described (González-García et al., 2011) with minor modifications. Immunolocalization using anti-GFP and anti-KNOLLE antibodies was done as reported by González-García et al. (2011) and for γ-H2AX immunostaining antiserum diluted 1:600 using Alexa 488 as a secondary antibody in all cases (Amiard et al., 2011). DAPI and Alexa 488 signals were acquired sequentially in two separated channels. Combined immunostaining with telomere Q-FISH was done to precisely mark by GFP the position of the stem cell niche in the whole-mounted roots. For Q-FISH, microscope slides were rinsed in 4% formaldehyde for 2′ and then washed with 1 × PBS. FISH was hybridized with a Cy3-labeled plant-telomere PNA specific probe (TTAGGG)3 and Cy3-labeled Arabidopsis centromeres PNA probe (5-GACTCCAAAACACTA ACC-3; see the Supplemental Information). Nuclei were counterstained with DAPI Vectashield and analyzed with a FV 1000 confocal microscope (Olympus). The DAPI image was used to define a nuclear area or ROI of each cell types to measure centromere and fluorescence intensities of the Cy3-labeled probes were measured as detailed in the Supplemental Information. Acquired images were quantified and processed using a Metamorph software package (v.6.3r6, Molecular Devices).
TRF, PETRA, Telomere Fusions, and Telomerase Activity Assays
DNA from root tips and shoots of 6-day-old was extracted by the CTAB method. TRF analysis was performed as described (Shakirov and Shippen, 2004). PETRA analysis and fusion PCR on tert mutants and WT Col-0 was done using 2 μg of root tip DNA as described (Heacock et al., 2004). The range of telomere length was determined using ImageQuant software. The average length of bulk telomeres was determined by ImageJ software (http://rsb.info.nih.gov/ij/). TRAP in root tips were performed as described (Kannan et al., 2008; Shakirov and Shippen, 2004).
For telomere Q-FISH quantification and statistical analysis of the data, see the Supplemental Information.
Supplementary Material
Highlights.
Heterogeneous telomere-length distribution in different root cell lineages
Postembryonic telomerase activity is required for stem cell function in roots
Telomerase sustains cell division at the meristem
Premature cell differentiation can prevent telomere erosion in roots
In Brief.
González-García et al. establish a Q-FISH methodology able to assess telomere length in individual cells at the Arabidopsis root apex, providing the first telomere-length distribution map in plants. The study reveals that telomere length is critical for meristem activity and stem cell renewal, thus coupling telomere dynamics to plant development.
Acknowledgments
We thank M. Gallego for providing anti-H2AX antibodies, I.Flores and C.Vilella for help with data analysis and comments on the manuscript. This work was supported by NIH R01-GM065383 to D.E.S. Research in the M.A.B. lab is funded by European Research Council (ERC) Project TEL STEM CELL (GA#232854), European Union FP7 Projects 2007-A-20088 (MARK-AGE) and 2010-259749 (EuroBATS), Spanish Ministry of Economy and Competitiveness Projects SAF2008-05384 and CSD2007-00017, Regional of Government of Madrid Project S2010/BMD-2303 (ReCaRe), AXA Research Fund (Life Risks Project), and Lilly 2010 Preclinical Biomedicine Research Award and Fundación Botín (Spain). M.I. acknowledges support from the Spanish Ministry of Science and Innovation through grant FIS2012-37655-C02-02 and to the Generalitat de Catalunya through grant 2014 SGR 878. A.I.C.-D. is funded by the Spanish Ministry of Economy and Competitiveness (BIO2010-16673 and BIO2013-43873) and a Marie-Curie Initial Training Network (grant no. PITN-GA-2008-215118). M.-P.G.-G. was the recipient of a postdoctoral contract from BIO2010-16673 and an EMBO short-term fellowship and I.P. is funded by a JAE-CSIC PhD fellowship in the A.I.C.-D. laboratory.
Footnotes
Supplemental Information: Supplemental Information includes Supplemental Experimental Procedures, six figures, and two movies and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2015.04.013.
References
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119:109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Amiard S, Depeiges A, Allain E, White CI, Gallego ME. Arabidopsis ATM and ATR kinases prevent propagation of genome damage caused by telomere dysfunction. Plant Cell. 2011;23:4254–4265. doi: 10.1105/tpc.111.092387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardes de Jesus B, Vera E, Schneeberger K, Tejera AM, Ayuso E, Bosch F, Blasco MA. Telomerase gene therapy in adult and old mice delays aging and increases longevity without increasing cancer. EMBO Mol Med. 2012;4:691–704. doi: 10.1002/emmm.201200245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997a;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Rizen M, Hanahan D, DePinho R, Greider CW. Mouse models for the study of telomerase. Ciba Found Symp. 1997b;211:160–170. doi: 10.1002/9780470515433.ch11. discussion 170–176. [DOI] [PubMed] [Google Scholar]
- Boltz KA, Leehy K, Song X, Nelson AD, Shippen DE. ATR cooperates with CTC1 and STN1 to maintain telomeres and genome integrity in Arabidopsis. Mol Biol Cell. 2012;23:1558–1568. doi: 10.1091/mbc.E11-12-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Díaz-Triviño S, Wachsman G, Du Y, Arteága-Vázquez M, Zhang H, Benjamins R, Blilou I, Neef AB, Chandler V, Scheres B. A SCARECROW-RETINOBLASTOMA protein network controls protective quiescence in the Arabidopsis root stem cell organizer. PLoS Biol. 2013;11:e1001724. doi: 10.1371/journal.pbio.1001724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inzé D. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell. 2001;13:1653–1668. doi: 10.1105/TPC.010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajkus J, Fulnecková J, Hulánová M, Berková K, Ríha K, Matyásek R. Plant cells express telomerase activity upon transfer to callus culture, without extensively changing telomere lengths. Mol Gen Genet. 1998;260:470–474. doi: 10.1007/s004380050918. [DOI] [PubMed] [Google Scholar]
- Fitzgerald MS, McKnight TD, Shippen DE. Characterization and developmental patterns of telomerase expression in plants. Proc Natl Acad Sci USA. 1996;93:14422–14427. doi: 10.1073/pnas.93.25.14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald MS, Riha K, Gao F, Ren S, McKnight TD, Shippen DE. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc Natl Acad Sci USA. 1999;96:14813–14818. doi: 10.1073/pnas.96.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores I, Cayuela ML, Blasco MA. Effects of telomerase and telomere length on epidermal stem cell behavior. Science. 2005;309:1253–1256. doi: 10.1126/science.1115025. [DOI] [PubMed] [Google Scholar]
- Flores I, Canela A, Vera E, Tejera A, Cotsarelis G, Blasco MA. The longest telomeres: a general signature of adult stem cell compartments. Genes Dev. 2008;22:654–667. doi: 10.1101/gad.451008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher N, Sablowski R. Hypersensitivity to DNA damage in plant stem cell niches. Proc Natl Acad Sci USA. 2009;106:20984–20988. doi: 10.1073/pnas.0909218106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449:1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- Gan S. Sci. Vol. 2003. SAGE KE; 2003. Mitotic and postmitotic senescence in plants. RE7. [DOI] [PubMed] [Google Scholar]
- González-García MP, Vilarrasa-Blasi J, Zhiponova M, Divol F, Mora-Gar-cía S, Russinova E, Caño-Delgado AI. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development. 2011;138:849–859. doi: 10.1242/dev.057331. [DOI] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Heacock M, Spangler E, Riha K, Puizina J, Shippen DE. Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J. 2004;23:2304–2313. doi: 10.1038/sj.emboj.7600236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Mol Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- Jung P, Sato T, Merlos-Suárez A, Barriga FM, Iglesias M, Rossell D, Auer H, Gallardo M, Blasco MA, Sancho E, et al. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- Kannan K, Nelson AD, Shippen DE. Dyskerin is a component of the Arabidopsis telomerase RNP required for telomere maintenance. Mol Cell Biol. 2008;28:2332–2341. doi: 10.1128/MCB.01490-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN. Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci USA. 2006;103:6055–6060. doi: 10.1073/pnas.0510607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Schiefelbein J. Embryonic control of epidermal cell patterning in the root and hypocotyl of Arabidopsis. Development. 2001;128:3697–3705. doi: 10.1242/dev.128.19.3697. [DOI] [PubMed] [Google Scholar]
- Lingner J, Cooper JP, Cech TR. Telomerase and DNA end replication: no longer a lagging strand problem? Science. 1995;269:1533–1534. doi: 10.1126/science.7545310. [DOI] [PubMed] [Google Scholar]
- López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 2000;14:2938–2943. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens UM, Zijlmans JM, Poon SS, Dragowska W, Yui J, Chavez EA, Ward RK, Lansdorp PM. Short telomeres on human chromosome 17p. Nat Genet. 1998;18:76–80. doi: 10.1038/ng0198-018. [DOI] [PubMed] [Google Scholar]
- Mencuccini M, Martínez-Vilalta J, Vanderklein D, Hamid HA, Korakaki E, Lee S, Michiels B. Size-mediated ageing reduces vigour in trees. Ecol Lett. 2005;8:1183–1190. doi: 10.1111/j.1461-0248.2005.00819.x. [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN. Control of Arabidopsis root development. Annu Rev Plant Biol. 2012;63:563–590. doi: 10.1146/annurev-arplant-042811-105501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher EH, Möller B, Lokerse AS, Llavata-Peris CI, van den Berg W, Weijers D. A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J. 2011;68:597–606. doi: 10.1111/j.1365-313X.2011.04710.x. [DOI] [PubMed] [Google Scholar]
- Richards EJ, Ausubel FM. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell. 1988;53:127–136. doi: 10.1016/0092-8674(88)90494-1. [DOI] [PubMed] [Google Scholar]
- Riha K, Shippen DE. Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proc Natl Acad Sci USA. 2003;100:611–615. doi: 10.1073/pnas.0236128100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K, Fajkus J, Siroky J, Vyskot B. Developmental control of telomere lengths and telomerase activity in plants. Plant Cell. 1998;10:1691–1698. doi: 10.1105/tpc.10.10.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K, McKnight TD, Griffing LR, Shippen DE. Living with genome instability: plant responses to telomere dysfunction. Science. 2001;291:1797–1800. doi: 10.1126/science.1057110. [DOI] [PubMed] [Google Scholar]
- Riha K, Watson JM, Parkey J, Shippen DE. Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 2002;21:2819–2826. doi: 10.1093/emboj/21.11.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi DJ, Jamieson CH, Weissman IL. Stems cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Ben-fey P, Leyser O, Bechtold N, Weisbeek P, Scheres B. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell. 1999;99:463–472. doi: 10.1016/s0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- Sax K, Enzmann EV. The effect of temperature on X-ray induced chromosome aberrations. Proc Natl Acad Sci USA. 1939;25:397–405. doi: 10.1073/pnas.25.8.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Benfey P, Dolan L. Root development. Arabidopsis Book. 2002;1:e0101. doi: 10.1199/tab.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov EV, Shippen DE. Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell. 2004;16:1959–1967. doi: 10.1105/tpc.104.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov EV, Perroud PF, Nelson AD, Cannell ME, Quatrano RS, Shippen DE. Protection of Telomeres 1 is required for telomere integrity in the moss Physcomitrella patens. Plant Cell. 2010;22:1838–1848. doi: 10.1105/tpc.110.075846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Leehy K, Warrington RT, Lamb JC, Surovtseva YV, Shippen DE. STN1 protects chromosome ends in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:19815–19820. doi: 10.1073/pnas.0807867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell. 2009;36:207–218. doi: 10.1016/j.molcel.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H. Senescence, ageing and death of the whole plant. New Phytol. 2013;197:696–711. doi: 10.1111/nph.12047. [DOI] [PubMed] [Google Scholar]
- Tomás-Loba A, Flores I, Fernández-Marcos PJ, Cayuela ML, Maraver A, Tejera A, Borrás C, Matheu A, Klatt P, Flores JM, et al. Telomerase reverse transcriptase delays aging in cancer-resistant mice. Cell. 2008;135:609–622. doi: 10.1016/j.cell.2008.09.034. [DOI] [PubMed] [Google Scholar]
- Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthélémy J, Palauqui JC. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell. 2008;20:1494–1503. doi: 10.1105/tpc.107.056069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomás S, Federici F, Casimiro I, Beemster GT, Bhalerao R, Swarup R, Doerner P, Haseloff J, Bennett MJ. Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr Biol. 2009;19:1194–1199. doi: 10.1016/j.cub.2009.06.023. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera E, Blasco MA. Beyond average: potential for measurement of short telomeres. Aging. 2012;4:379–392. doi: 10.18632/aging.100462. Albany, N.Y Online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilarrasa-Blasi J, González-García MP, Frigola D, Fábregas N, Alexiou KG, López-Bigas N, Rivas S, Jauneau A, Lohmann JU, Benfey PN, et al. Regulation of plant stem cell quiescence by a brassinosteroid signaling module. Dev Cell. 2014;30:36–47. doi: 10.1016/j.devcel.2014.05.020. [DOI] [PubMed] [Google Scholar]
- Völker A, Stierhof YD, Jürgens G. Cell cycle-independent expression of the Arabidopsis cytokinesis-specific syntaxin KNOLLE results in mistargeting to the plasma membrane and is not sufficient for cytokinesis. J Cell Sci. 2001;114:3001–3012. doi: 10.1242/jcs.114.16.3001. [DOI] [PubMed] [Google Scholar]
- Watson JM, Riha K. Comparative biology of telomeres: where plants stand. FEBS Lett. 2010;584:3752–3759. doi: 10.1016/j.febslet.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JM, Riha K. Telomeres, aging, and plants: from weeds to Methuselah - a mini-review. Gerontology. 2011;57:129–136. doi: 10.1159/000310174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


